ABSTRACT
Scaffold protein Receptor for Activated C Kinase 1 (RACK1) is a negative regulator of plant stress hormone – abscisic acid (ABA) mediated pathways. RACK1 has been reported to regulate global miRNA biogenesis pathway in C. elegans, humans, and in Arabidopsis. RACK1 regulates different steps of miRNA biogenesis and stability in response to different stimuli in plants. miR393s is implicated in salt stress response pathway through an antagonistic response between the stress hormone ABA-mediated salt stress and growth hormone auxin. Specifically, the known auxin receptor clade transcripts TIR1/AFB2 are the target for the miR393s. By down-regulating the auxin signaling pathways, the miR393s inhibit the regulation of salt tolerance by auxin. Here we show that genetic loss of RACK1A- the predominant member of the three genes family of RACK1 in Arabidopsis, results in the inhibition of miR393 level causing the same salt sensitivities as the individual mir393a or mir393b or the double mutant mir393ab phenotypes. We propose that down-regulation of auxin signaling through RACK1A induced miR393 biogenesis potentially regulates the Arabidopsis acclimation to salinity. Our findings fill up a molecular gap in our understanding of the role of miR393 mediated ABA and auxin-regulated salt stress responses.
KEYWORDS: Receptor for Activated C Kinase 1, abscisic acid (ABA), MicroRNAs (miRNAS), salt (NaCl)
1. Introduction
1.1. Salt as a problem in cultivation
High salt concentrations in soil or water make cultivating crops of agronomic significance difficult, especially in arid, semi-arid regions and in irrigation areas. A recent study finds that every day for more than 20 years, an average of 2,000 hectares of irrigated land in arid and semi-arid areas across 75 countries have been degraded by salt.1 The same study finds that the global annual cost of salt-induced land degradation in irrigated areas could be US$ 27.3 billion because of lost crop production.1 Even trace amounts of salt in soil not only inhibits the plants growth, but also causes changes in the plants' physiology and metabolism from germination, seedling growth, bolting, flowering and fruit yields.2,3 Salt stress affects plant physiology at both whole plant and cellular levels through osmotic and ionic stress.4,5 The accumulation of toxic levels of ions has various effects on plant physiological processes such as increased respiration rate and ion toxicity, and decreased efficiency of photosynthesis.4
1.2. The role of RACK1
Receptor for Activated C Kinase 1 (RACK1) is a 37 kDa scaffold protein that is ubiquitously expressed in every eukaryotic organism and in few prokaryotes. The protein has seven tryptophan-aspartic acid amino acid repeats (WD-40 repeat) and it influences a variety of functions from signal transduction, regulation of transcription, growth hormones, control of conditions like drought and salt stress, innate immunity and senescence.6 The structure and protein sequence of RACK1 is widely conserved in both plants and in the metazoan kingdom. The model plant, Arabidopsis thaliana (A. thaliana) genome contains three different RACK1 genes: RACK1A (predominant form of the gene used in this study), RACK1B (87% sequence identity), and RACK1C (93% sequence identity).7 Though use of genetic complementation studies in the individual loss of function mutant indicated that they are in principle functionally equivalent but based on gene activity levels, it is found that they operate on an unequal genetic redundant manner.8 It is found that RACK1A is the ancestral gene which retains most functions of RACK1 genes by inducing gene activity beyond the threshold level needed for RACK1 functions; whereas, RACK1B and RACK1C appear to be duplicated genes.8 RACK1 mediates diverse environmental stress pathways by negatively regulating the stress hormone abscisic acid (ABA) signaling.9 RACK1 has been found to interact with a plethora of proteins as a signal integrator for regulating diverse cellular and physiological process like translation, cell proliferation, hormone signaling, growth, development, photosynthesis and environmental stress.6,10 This article presents the role of RACK1 on salt stress acclimation mediated by a cross-talk between ABA and auxin signaling pathways. This integrative role is exerted by regulating the biogenesis of a small RNAs (micro RNAs- MIRNAs), more specifically miR393s.
1.3. miRNA/miR393, small RNAs and ABA
MicroRNAs (miRNAs) control many aspects of development and adaption in plants and animals by post-transcriptional control of mRNA stability and translatability.11 Previously RACK1 has been implicated in the miRNA pathway of C. elegans, humans, and Arabidopsis.11-13 It has been found that RACK1 mediates different steps of miRNA biogenesis, processing, and stability to regulate the miRNA abundance.11 A combination of functions of RACK1 has been proposed in the miRNA pathway. They include MIRNA gene transcription/stabilization, pri-miRNA processing, and interaction with ARGONAUTE (AGO) effector protein to create RNA-induced silencing complex (RISC).11 The proposed mechanism is supported by the findings of reduced miRNA level and increased accumulation of miRNA target mRNAs in rack1 mutants.11
As a result of environmental stress, specific miRNAs may be induced to perform regulation of different genes and many have been implicated in salt, drought-like stress signaling pathways.14–16 In this regard, miR393 has been found to be induced by salt and drought stresses.14 During the experiment to identify global stress-related miRNAs in Arabidopsis, it was found that miR393 is strongly upregulated by cold, dehydration, NaCl, and ABA treatments.14 The role of miRNA393 in the ABA-mediated drought and salt stresses have been elucidated.16 It is known that the target genes for the miR393 are F-box genes that encode auxin receptors TIR1, AFB2, and AFB3.17 It is now proposed that MIR393 exerts its ABA pathway function by an auxin antagonistic pathway; whereby upregulation of miR393 by ABA results in the down-regulation of auxin-based signaling pathways. The evidence for this model comes from the use of an ABA hypersensitive mutant fry1 mutants in Arabidopsis. The fry1 mutants were found to be hypersensitive to the inhibition of lateral root development by ABA while displaying auxin-resistant like phenotypes.16 The fry1 mutants exhibited enhanced cleavage of auxin receptor transcripts as well as increased expression of the ABA‐induced miR393.16 Though it is now established that MIR393 crosslinks to different hormone signaling pathways, the precise molecular mechanism of this antagonistic functions is not known.
Given the evidence that RACK1 is a negative regulator of the stress hormone ABA signaling, and a positive regulator of auxin signaling pathways,19 it is not far-fetched to suggest that RACK1 may regulate these pathways possibly through miRNA regulation. The prominent role RACK1 plays in global miRNA biogenesis, processing, and stability indicates a role for RACK1 in the miRNA mediated ABA and auxin signaling pathways. Here we report that the known pathway of ABA-induced destabilization of the TIR1/AFB transcripts via miR393‐guided cleavage potentially requires scaffold protein RACK1 function.
2. Materials and methods
2.1. Plant materials and growth conditions
A. thaliana ecotype Columbia (Col) was used in all experiments as the wild type (WT). Other A. thaliana plants used included the rack1a-1 mutants, miR393ab mutants. rack1a-1 (T-DNA is inserted into the first exon of the RACK1A gene) mutants8 and miR393 mutants seeds as reported in were obtained from Dr. Jarmolowski lab. All seeds were sterilized with 70% ethanol for 1 min, then for 10 min with bleach solution (30% bleach + 0.1% tween 20) and washed 3 times with DI H2O. Seeds were then planted on Murashige and Skoog growth media with basal salts and vitamins, Sucrose (30g/L) and Phytoblend (9g/L). MS plate grown two weeks old seedlings were transferred to the indicated plates and were allowed two additional weeks of growth. Growth chamber was set at 24–26°Celsius with 12 h of darkness and 12 h of light.
2.2. RNA isolation
All RNA was isolated using the trizol method following the manufacturer’s recommended protocol (ThermoFisher Scientific, Waltham, MA). Briefly, 25 mg of whole plant tissue was homogenized with 0.5 ml of trizol and after a brief incubation of 5 min, 0.1 ml of chloroform was added and then centrifuged. The supernatant was used to precipitate RNA with 0.25 ml of isopropyl alcohol. The precipitated RNA was washed with 75% ethanol and air-dried pellet was dissolved in RNAse free water and quantified in a nanodrop equipment.
2.3. cDNA Synthesis and qPCR
cDNA synthesis was done from 500 ng of RNA using Superscript III First-Strand Synthesis Systems following the manufacturer’s protocol. Note that the protocol calls for oligo (dT) primed cDNA synthesis that can amplify pri-miRNAs. Real-time quantitative PCR was done using standard protocols for the Bio-Rad CFX Manager 3.1 Detection System. For each reaction, 2μl of 1/25 dilution of cDNA was used with 2X Sybrgreen master mix (ThermoFisher Scientific, Waltham, MA) with 20 pmole of each primer (specific to the stem regions of miR393s) in 25 ul reaction volume. The reactions were amplified at 50° for 2 min, 95° for 10 min, then 39 cycles at 95° for 15 s and 55° for 1 min. Finally, 65° for 5 s and 95° for 5 s. All reactions had three technical replicates and two to five biological replicates. In-built BioRad software generated t-values that resulted in the pairwise significance level calculation.
2.4. PCR primers
Rack1a genomic DNA was amplified using primers: Forward: 5‘-CTGAGGCTGAAAAGGCTGACAACAG-3ʹ Reverse: 5‘-CTAGTAACGACCAATACCCCAAACTC-3‘ (~122bp). MIR393a genomic DNA was amplified using primers: miR393a-RT: Forward: AACCACATTGCTCTCAACTTTTAG and miR393a-RT: Reverse: GAGATAGCATGATCCAAAACCA (~162 bp). MIR393b genomic DNA was amplified using primers miR393b-RT: Forward: AAAACACCATTGCTCCCACCT and miR393b-RT: Reverse: CGCATGATCCGGAAAAGTAAG (~169bp). actin genomic DNA was amplified using primers: actin2-RT: Forward: GGTAACATTGTGCTCAGTGGTGG and Actin2-RT: Reverse: AACGACCTTAATCTTCATGCTGC (~108bp).
3. Results and discussion
3.1. RACK1 is required for salt-induced mir393 biogenesis
In addition, to be a negative regulator of ABA signaling pathways and a positive regulator of auxin-mediated lateral root development,19 RACK1 protein has been implicated in miRNA biogenesis for several organisms and regulation of global miRNAs biogenesis process.11 Among many other miRNAs, miR393 also specifically regulates ABA-mediated salt stress acclimatization process by targeting transcripts of auxin signaling pathways.22 It is also known that double mutants of auxin receptors tir1 afb2 is more tolerant to salt stress than wild-type plants and it has been proposed that down-regulation of auxin signaling might be part of Arabidopsis acclimation to salinity.22 This salt acclimation is mediated by the salt-induced miR393 expression that resulted in the down-regulation of auxin receptors TIR1 and AFB2.22 Several studies have also previously shown that miR393 can be induced by different stresses including salt.14,19-21,28 In order to investigate whether RACK1 has any role in the MIR393 mediated pathways, salt-induced miR393 expression level in WT and rack1a-1 mutant plants were analyzed. Two-week-old A. thaliana seedlings were exposed to 100 mM NaCl for 14 days and gene expression level of miR393a and miR393b in WT and rack1a-1 mutant plants were analyzed. As can be seen from Figure 1, NaCl induced the transcript level of both the miR393a and miR393b while in the rack1a-1 mutant plants, neither of the miR393 transcripts were expressed in the presence or in the absence of salt. This result indicates that the earlier findings of salt stress-induced upregulation of miR393 transcripts22 was based on the presence of a functional RACK1 protein. These results are consistent with the role of RACK1 in the biogenesis of the miR393 transcripts. Though the study22 found the MIR393A promoter, not MIR393B promoter, to be responsive to the salt treatment, our study shows that both of the MIR393s are induced by salt. The short treatment regimen of 2 h as opposed to our sustained 2-weeks long salt treatment may account for this discrepancy. It is possible that immediate response for salt acclimation is mediated by the MIR393A while a long sustained response may possibly be mediated by the MIR393B. It is interesting to note that salt induction of MIR393B was higher compared to that of MIR393A induction (Figure 1). A knowledge gap was present as to how the antagonistic effect of ABA and auxin during salt stress acclimation pathway operate and the findings of this result now show that scaffold protein RACK1A can potentially play the role of signal integration from these two apparently antagonistic pathways. RACK1 is known to integrate diverse signaling pathways ranging from the environmental stress to the growth and development of plants.19
Figure 1.
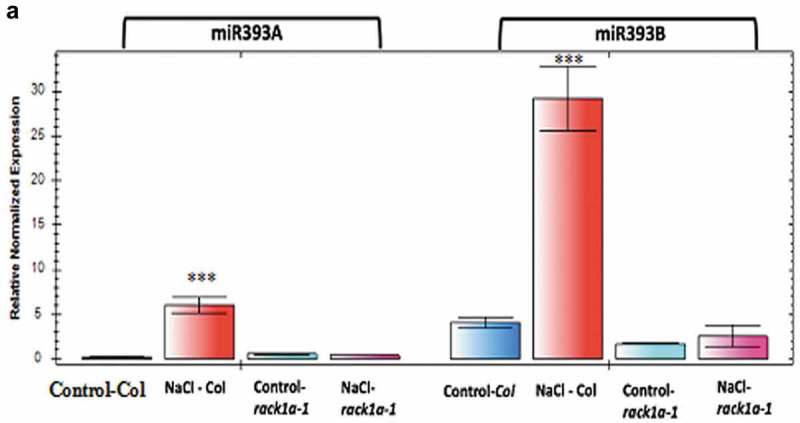
(a) RACK1A dependent miR393 transcript expression in response to exogenous salt treatment. Quantifications were normalized to the expression of house-keeping gene Actin. The error bars represent standard deviation from three PCR replicate experiments. The WT (Col) and rack1a-1 mutants were grown on MS plates with/without 100 mM NaCl for two weeks. Asterisks denote significant difference from the control plants (*** denotes p < 0.01, Student’s t-test).
3.2. Stress hormone ABA-induced mir393 expression requires RACK1A
MIR393 has been found to regulate many abiotic stresses like salt, cold, dehydration, and metal toxicity15,16,22,23 Our hypothesis of RACK1 mediated miR393 biogenesis indicate that ABA-induced miR393 transcript production16 would be altered in the rack1 mutant plants. Therefore, we investigated whether ABA-induced miR393 expression requires the active presence of RACK1A protein. As seen in Figure 2(a,b), both miR393a and miR393b show significant ABA-induced expression in the WT plants but the expression is very attenuated in the rack1a-1 mutant plant background indicating that an active RACK1A is required for the proper expression of the miR393 to regulate ABA-induced stress signaling pathways which include salinity. Note that the attenuation is greater for the miR393b transcripts which correlates with our salt-induced miR393 expression as well (Figure 1). Sustained exposure to salt resulted in the higher induction of MIR393B indicating a role in the long-term salt stress response pathway. The MIR393B dominant function has been reported before25, and it has been reported that the two MIR393 genes have distinct, but partially overlapping expression profiles.26 Our results also indicate a positive role of RACK1 in the ABA-induced miR393 generation; however, it is known that RACK1 is a negative regulator of ABA signaling and short-term (2 h) ABA treatment resulted in the down-regulation of RACK1 transcript expression.9 However, it is quite possible that in the long-term (2 weeks) exposure to ABA-induced miR393 biogenesis pathway, RACK1 has a distinct role through post-translational modification like phosphorylation of its key residues. RACK1 Y248 phosphorylation has been found to have a key role in mediating diverse stress responses.27,28 ABA-induced RACK1 phosphorylation is quite possible in the RACK1 mediated miR393 biogenesis. The results here clearly indicate that a functional RACK1 protein is required for the ABA-induced miR393 biogenesis.
Figure 2.
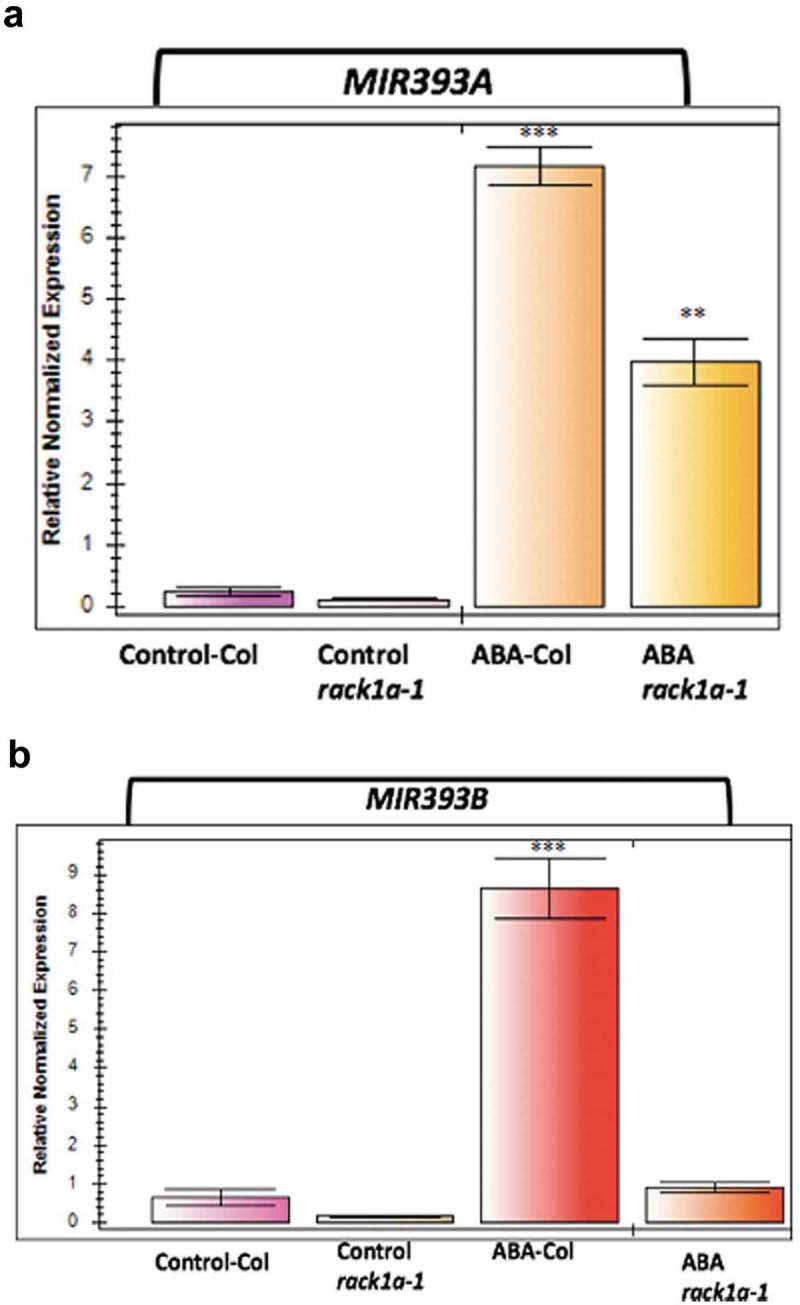
ABA treatment induced miR393 transcripts expression. qPCR analysis of miR393A (panel A) and miR393B (panel B) in response to exogenous ABA (0.5 mM). Quantifications were normalized to the expression of house-keeping gene Actin. The error bars represent standard deviation from three PCR replicate experiments. MS plate grown two-weeks old seedlings were transferred to MS plates containing ABA or methanol (vehicle control) and RNA were isolated after additional two weeks of growth. Asterisks denote significant difference from the control plants (** and *** denotes p < 0.05, p < 0.01, respectively, Student’s t-test).
3.3. Auxin-induced mir393 biogenesis
miR393-mediated posttranscriptional regulation of auxin receptors has been established as a crucial component of plant acclimation to salinity.22 It has been proposed that through a feedback mechanism, auxin-induced receptors can positively regulate the miR393 biogenesis.25 However, when we assayed auxin-induced miR393 biogenesis, we saw a modest increase (note the fold difference units on Y axis) in the MIR393A production but no significant induction was seen in the MIR393B biogenesis from auxin treatment for two weeks (Figure 3). While only MIR393A showed RACK1 dependency for its potential feedback induction, the MIR393B did not show any RACK1 regulated biogenesis (Figure 4). This indicates that possibly RACK1 regulates the MIR393 accumulation by acting upstream of the biogenesis process and only MIR393A biogenesis through a feedback mechanism is regulated by RACK1. It is known that MIR393A is responsible to generate almost all the miR393 in the root tissue26 and auxin effect on lateral root development requires a functional RACK1 protein (Figure 4). Also, we cannot rule out an epistasis relationship between the MIR393A and MIR393B. In fact, when miRNA target sequence database is used, among the MIR393A target genes, MIR393B is listed among the targets (data not shown).
Figure 3.
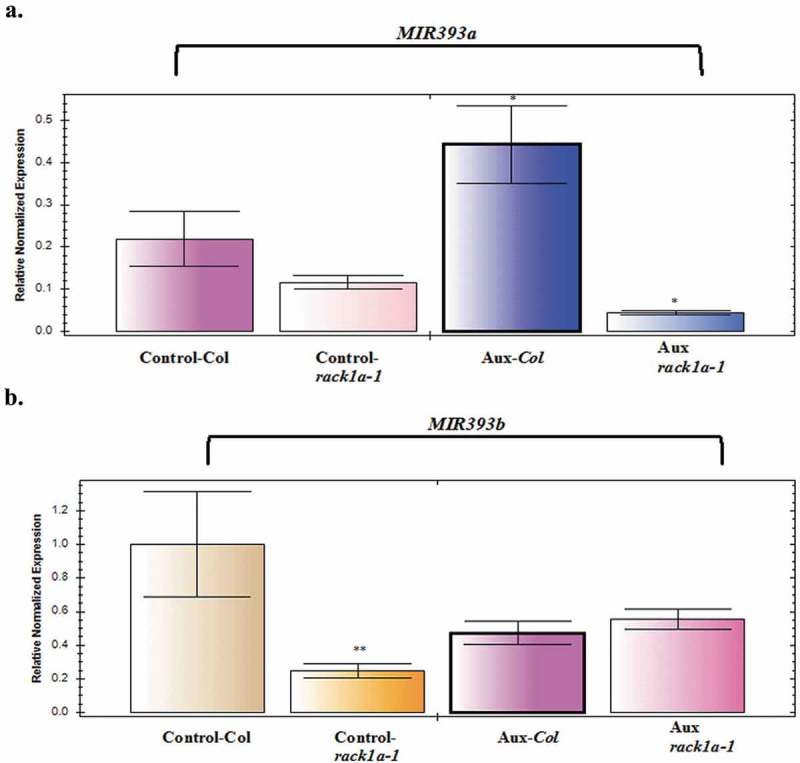
Auxin-induced miR393 transcript expression in WT and rack1a-1 mutant plants. qPCR analysis of miR393A (panel A) and miR393B (panel B) in response to exogenous Auxin (10 mM). RNA was isolated from two weeks of treatment with 10 uM of auxin (IAA). The error bars represent standard deviation from three PCR replicate experiments. Quantifications were normalized to the expression of house-keeping gene Actin. Asterisks denote significant difference from the control plants (* and ** denotes p < 0.1, p < 0.05, respectively, Student’s t-test).
Figure 4.
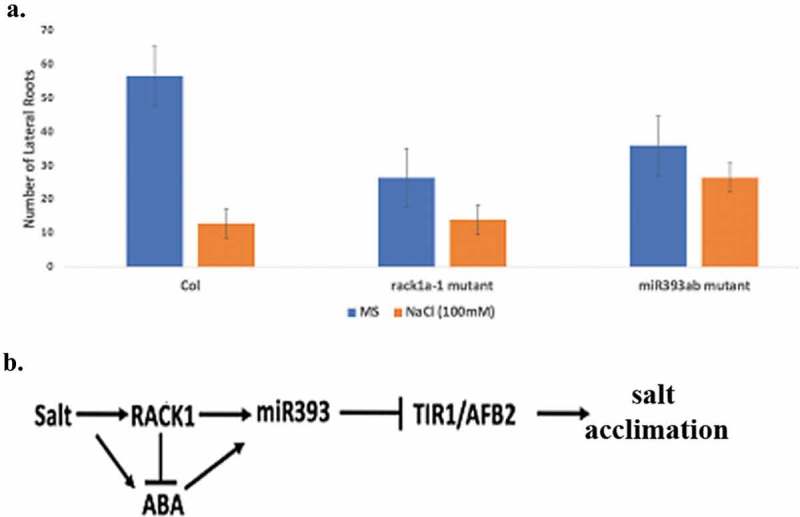
NaCl (100 mM) induced lateral root development in WT and rack1a-1 and mir393 mutant plants (a). Two weeks old seedlings were grown on NaCl plates for additional two weeks. While the WT plants showed statistically significant down-regulation in number of lateral roots the rack1a-1 and mir393ab mutants did not show any significant difference in the lateral root numbers in response to salt treatment for two weeks. (b) proposed model. Signal Integration role of RACK1 crosslinks salt and/or ABA-induced RACK1 mediated auxin receptors targeting miRNA miR393. The down-regulation of auxin signaling affects the acclimation to salinity.
RACK1 has been found to have a role in MIRNA gene transcription and/or pri-miRNA stability.11 Within the global pri-miRNA contents, some pri-miRNAs accumulated to a higher level and some pri-miRNA exhibited a reduction in the rack1 triple mutants.11 Notable among the upregulated transcripts was the pri-miR393a indicating RACK1 has a role in their transcription activation. However, those pri-miR393a transcripts were misprocessed and the mature level of miR393a was indeed lower in the rack1 triple mutants11 (Supplemental Fig.1 in reference 11) supporting the RACK1’s positive role in the biogenesis of miR393a transcripts as we have reported here.
3.4. MIR393 regulates lateral root abundance during salt stress
Chen et al.16 demonstrated that MIR393-mediated attenuation of auxin signaling is essential for inhibition of lateral root growth by ABA or osmotic stress. MIR393 was found to be involved in the repression of LR growth during salt stress.22 As RACK1 has been found to be a regulator of miR393 biogenesis, we investigated whether rack1 mutant plants show similar inhibition of lateral root abundance during the salt stress. As can be seen in Figure 4, salt-induced reduction in the number of lateral roots is prominently observed in the WT plants while mir393ab double mutants showed insensitivity to the salt-induced reduction in the lateral root development. Likewise, rack1a-1 mutant plants also showed insensitivity in the same assay indicating that active miR393 function is needed to attenuate lateral root development inhibition from salinity. Though the mutant plants started with a lower number of lateral roots, the percentage inhibition is much higher in the WT plants compared to that in both the mutant plants. This result, in conjunction with previous results, indicates that RACK1 mediated miR393 generation may regulate the lateral root development during salt stress. The importance of root architecture during the stress is of paramount importance to plants and numerous genetic and physiological evidence indicate growth hormone auxin plays a dominant role in several developmental stages of LR development.29 Our result now complements the earlier results that indicated a role of miR393 in the inhibition of LR initiation and elongation under salinity and RACK1 as a regulator of miR393 biogenesis plays an important role in this regard. In summary, it is quite possible that salt induces RACK1 that upregulates miR393 biogenesis in the pericycle cells which subsequently down-regulate auxin signal to inhibit LR organogenesis. It is not far fetched to envision a role for stress hormone ABA to regulate LR growth under salt-stress as well.
Figure 4(b) presents a model that shows the relationship of RACK1, miR393, ABA and Auxin response genes on salt tolerance. Salt upregulates RACK1 and ABA, which leads to an increase of miR393 that in turn inhibits auxin signaling and causes salt stress acclimation.
Conclusion: Survival of plants under salt stress requires rapid perception and adaptation to the stress condition. Though ABA has long been known to be a key regulator of stress responses, auxin is rapidly being implicated in regulating diverse environmental stresses including salinity stress. However, there is still many unknown as to how auxin and ABA can regulate a stress like salinity together. Here we fill a piece of the regulatory puzzle showing that scaffold protein RACK1 through the biogenesis of miR393 can mediate the crosstalk between ABA and auxin in salt stress signaling pathway. We propose that RACK1A, which has previously been implicated in both the auxin and ABA pathway, functions as an integrator of miR393 mediated ABA and auxin-regulated salinity stress responses. In this work, we showed that NaCl or ABA-mediated stress induced miR393 expression and the expression is abolished in the rack1a-1 mutant background. As the mir393ab double mutant plants show insensitivity to salt stress induced inhibition of lateral root development, we establish that it is a manifestation of miR393 mediated attenuation of auxin signaling pathways that results in salinity acclimation. By combining the results, we proposed the model (Figure 4) where RACK1A occupies a major position to integrate the miR393 mediated salt stress responses that combine opposing effects of two key plant hormones, ABA and auxin.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Qadir M, Quillérou E, Nangia V, Murtaza G, Singh M, Thomas RJ, Drechsel P, Noble AD.. Economics of salt‐induced land degradation and restoration. Nat Resour Forum. 2014. November;38(4):282–295. doi: 10.1111/1477-8947.12054. [DOI] [Google Scholar]
- 2.Bhandal IS, Malik CP. Potassium estimation, uptake, and its role in the physiology and metabolism of flowering plants. Int Rev Cytol. 1988;110:205–254. [Google Scholar]
- 3.Chaves MM, Flexas J, Pinheiro C. Photosynthesis under drought and salt stress: regulation mechanisms from whole plant to cell. Ann Bot. 2009. February 1;103(4):551–560. doi: 10.1093/aob/mcn125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sudhir P, Murthy SD. Effects of salt stress on basic processes of photosynthesis. Photosynthetica. 2004. December 1;42(2):481–486. doi: 10.1007/S11099-005-0001-6. [DOI] [Google Scholar]
- 5.Fernando VD, Schroeder DF.. Role of ABA in Arabidopsis salt, drought, and desiccation tolerance. Abiotic and biotic stress in plants – recent advances and future perspectives 2016, IntechOpen Publishers. [Google Scholar]
- 6.Kundu N, Dozier U, Deslandes L, Somssich IE, Ullah H. Arabidopsis scaffold protein RACK1A interacts with diverse environmental stress and photosynthesis related proteins. Plant Signal Behav. 2013. May 1;8(5):e24012. doi: 10.4161/psb.24012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ullah H, Scappini EL, Moon AF, Williams LV, Armstrong DL, Pedersen LC. Structure of a signal transduction regulator, RACK1, from Arabidopsis thaliana. Protein Science. 2008. October;17(10):1771–1780. doi: 10.1110/ps.035121.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guo J, Chen JG. RACK1 genes regulate plant development with unequal genetic redundancy in Arabidopsis. BMC Plant Biol. 2008. December;8(1):108. doi: 10.1186/1471-2229-8-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guo J, Wang J, Xi L, Huang WD, Liang J, Chen JG. RACK1 is a negative regulator of ABA responses in Arabidopsis. J Exp Bot. 2009. July 7;60(13):3819–3833. doi: 10.1093/jxb/erp221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Adams DR, Ron D, Kiely PA. RACK1, A multifaceted scaffolding protein: structure and function. Cell Commun Signaling. 2011. December;9(1):22. doi: 10.1186/1478-811X-9-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Speth C, Willing EM, Rausch S, Schneeberger K, Rack LS. 1 scaffold proteins influence mi RNA abundance in Arabidopsis. The Plant Journal. 2013. November;76(3):433–445. doi: 10.1111/tpj.12308. [DOI] [PubMed] [Google Scholar]
- 12.Otsuka M, Takata A, Yoshikawa T, Kojima K, Kishikawa T, Shibata C, Takekawa M, Yoshida H, Omata M, Koike K. Receptor for activated protein kinase C: requirement for efficient microRNA function and reduced expression in hepatocellular carcinoma. PLoS One. 2011. September 15;6(9):e24359. doi: 10.1371/journal.pone.0024359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jannot G, Bajan S, Giguere NJ, Bouasker S, Banville IH, Piquet S, Hutvagner G, Simard MJ. The ribosomal protein RACK1 is required for microRNA function in both C. Elegans and Humans. EMBO Reports. 2011. June 1;12(6):581–586. doi: 10.1038/embor.2011.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sunkar R, Zhu JK. Novel and stress-regulated microRNAs and other small RNAs from Arabidopsis. Plant Cell. 2004. August 1;16(8):2001–2019. doi: 10.1105/tpc.104.022830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mendoza-Soto AB, Sánchez F, Hernández G. MicroRNAs as regulators in plant metal toxicity response. Front Plant Sci. 2012. May;21(3):105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen H, Li Z, Xiong L. A plant microRNA regulates the adaptation of roots to drought stress. Chen, H., Li, Z., & Xiong, L. 2012, Febs Letters, Vol. 586(12), pp. 1742–1747. [DOI] [PubMed] [Google Scholar]
- 17.Navarro L, Dunoyer P, Jay F, Arnold B, Dharmasiri N, Estelle M, Voinnet O, Jones JD. A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science. 2006. April 21;312(5772):436–439. doi: 10.1126/science.1126088. [DOI] [PubMed] [Google Scholar]
- 18.Xia K, Wang R, Ou X, Fang Z, Tian C, Duan J, Wang Y, Zhang M. OsTIR1 and OsAFB2 downregulation via OsmiR393 overexpression leads to more tillers, early flowering and less tolerance to salt and drought in rice. PLoS One. 2012. Jan 10;7(1):e30039. doi: 10.1371/journal.pone.0030039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Islas-Flores T, Rahman A, Ullah H, Villanueva MA. The receptor for activated C kinase in plant signaling: tale of a promiscuous little molecule. Front Plant Sci. 2015;6:1090. Published 2015 Dec 8. doi: 10.3389/fpls.2015.01090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gao P, Bai X, Yang L, Lv D, Pan X, Li Y, et al. osa-MIR393: a salinity- and alkaline stress-related microRNA gene. Mol Biol Rep. 2011;38:237–242. doi: 10.1007/s11033-010-0100-8. [DOI] [PubMed] [Google Scholar]
- 21.Duan L, Dietrich D,Ng CH, Chan PM, Bhalerao R, Bennett MJ,Dinneny JR. Endodermal ABA signaling promotes lateral root quiescence during salt stress in Arabidopsis seedlings. Plant Cell. 2013. Jan 1;25(1):324–341. doi: 10.1105/tpc.112.107227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Iglesias MJ, Terrile MC, Windels D, Lombardo MC, Bartoli CG, Vazquez F, Estelle M, Casalongué CA. MiR393 regulation of auxin signaling and redox-related components during acclimation to salinity in Arabidopsis. PLoS One. 2014. Sep 15;9(9):e107678 doi: 10.137/journal.pone.0107678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kruszka K,Pieczynski M,Windels D,Bielewicz D,Jarmolowski A,Szweykowska-Kulinska Z, Vazquez F. Role of microRNAs and other sRNAs of plants in their changing environments. J Plant Physiol. 2012. Nov 1;169(16):1664–1672. doi: 10.1016/j.jplph.2012.03.009. [DOI] [PubMed] [Google Scholar]
- 24.Chen JG, Ullah H,Temple B, Liang J, Guo J, Alonso JM, Ecker JR, Jones AM. RACK1 mediates multiple hormone responsiveness and developmental processes in Arabidopsis. J Exp Bot. 2006. Jul 7;57(11):2697–2708. doi: 10.1093/jxb/erl035. [DOI] [PubMed] [Google Scholar]
- 25.Chen ZH, Bao ML, Sun YZ, Yang YJ, Xu XH, Wang JH, Han N, Bian HW, Zhu MY. Regulation of auxin response by miR393-targeted transport inhibitor response protein1 is involved in normal development in Arabidopsis. Plant Mol Biol. 2011. Dec 1;77(6):619–629. doi: 10.1007/s11103-011-9838-1. [DOI] [PubMed] [Google Scholar]
- 26.Windels D, Bielewicz D, Ebneter M, Jarmolowski A, Szweykowska-Kulinska Z, Vazquez F. miR393 is required for production of proper auxin signalling outputs. PLoS One. 2014. Apr 24;9(4):e95972. doi: 10.1371/journal.pone.0095972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sabila M, Kundu N, Smalls D, Ullah H. Tyrosine phosphorylation based homo-dimerization of Arabidopsis RACK1A proteins regulates oxidative stress signaling pathways in yeast. Front Plant Sci. 2016;7:176. doi: 10.3389/fpls.2016.00176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu HH, Tian X, Li YJ, Wu CA, Zheng CC. Microarray-based analysis of stress-regulated microRNAs in Arabidopsis thaliana. Rna. 2018:14(5), 836–843. doi: 10.1261/rna.895308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Casimiro I, Beeckman T, Graham N, Bhalerao R, Zhang H, Casero P, Sandberg G, Bennett MJ. Dissecting Arabidopsis lateral root development. Trends Plant Sci. 2003;8:165–171. doi: 10.1016/S1360-1385(03)00051-7. [DOI] [PubMed] [Google Scholar]


