Abstract
Objectives:
Phenylephrine and salbutamol are drugs which are widely used to treat diseases/disorders, such as nasal congestion, hypotension, and asthma, in individuals of different age groups. Human cytosolic sulfotransferase (SULT) SULT1A3 has been shown to be critically involved in the metabolism of these therapeutic agents. The current study was performed to investigate the effects of single nucleotide polymorphisms (SNPs) of human SULT1A3 and SULT1A4 genes on the sulfation of phenylephrine and salbutamol by SULT1A3 allozymes.
Methods:
Wild-type and SULT1A3 allozymes, previously prepared via site-directed mutagenesis in conjunction with bacterial expression and affinity purification, were analyzed for sulfating activity using an established assay procedure.
Results:
Purified SULT1A3 allozymes, in comparison to the wild-type enzyme, displayed differential sulfating activities toward phenylephrine and salbutamol. Kinetic studies showed further significant variations in their substrate-binding affinity and catalytic activity toward phenylephrine and salbutamol.
Conclusions:
The results obtained showed clearly the differential enzymatic characteristics of SULT1A3 allozymes in mediating the sulfation of phenylephrine and salbutamol. Such information may contribute to a better understanding about the pharmacokinetics of these two drugs in individuals with distinct SULT1A3 and/or SULT1A4 genotypes.
Keywords: Single nucleotide polymorphism, sulfation, SULT1A3, allozymes, phenylephrine, salbutamol
1. Introduction
Phenylephrine and salbutamol are sympathomimetic drugs widely used to treat or control a number of pathophysiological conditions. Phenylephrine acts primarily as a α1-agonist and is commonly used as a decongestant and as a vasopressor to elevate the blood pressure in patients with hypotension [1, 2]. Salbutamol (albuterol) is used as a bronchodilator for patients with asthma and chronic obstructive pulmonary disease (COPD) and as a uterine relaxant to prevent premature labor by acting as a β2-agonist [3–5]. Recent pharmacokinetic studies demonstrated that a significant fraction of these drugs is metabolized by sulfoconjugation in humans, forming inactive sulfated metabolites [4, 6]. Approximately 47% of phenylephrine and 20–60% of salbutamol administered orally have been shown to be subjected to sulfation [4, 7]. Studies have revealed that of the thirteen human cytosolic sulfotransferases (SULTs), SULT1A3 was the major SULT responsible for the sulfation of both these two drugs [8, 9]. It has been reported that orally administered salbutamol and other β2 agonists, although well absorbed from the gastrointestinal tract, may undergo extensive presystemic sulfation in the liver and intestine, leading to their inactivation and low systemic bioavailability [6, 10–12]. In support of this notion, studies have shown that inhibition of SULT1A3 by components in beverages such as tea may sustain the systemic bioavailability of salbutamol [10].
In humans and other mammals, sulfation is known to be a major Phase II metabolic pathway that is involved in the inactivation and disposal of biologically active compounds [13–15]. The SULT enzymes catalyze the transfer of a sulfonate group from the 3’-phosphoadenosine 5’-phosphosulfate (PAPS) to the hydroxyl or amino group of an acceptor substrate compound [16]. As noted above, SULT1A3 has been shown to be responsible for the sulfation of phenylephrine and salbutamol [8, 9]. Previous genomic studies have demonstrated the duplication of the gene encoding SULT1A3 during evolution process [17, 18]. Both the SULT1A3 and SULT1A4 genes were found to be located on chromosome 16 and both are considered transcriptionally active and encode identical protein products, collectively called SULT1A3 [18]. In addition to gene duplication, genetic polymorphisms of both SULT1A3 and SULT1A4 genes have been reported and distinct allele frequencies of some SULT1A3 and SULT1A4 genotypes have been found for different ethnic groups [19]. Importantly, several SULT1A3 allozymes, expressed in COS-7 cells, have been shown to display differential sulfating activities toward dopamine and ritodrine (a β2 agonist) [19, 20].
In this communication, we report functional analyses of a panel of SULT1A3 allozymes with phenylephrine and salbutamol as substrates. Moreover, kinetic studies were carried out to delineate their substrate-binding affinity and catalytic activity toward these two drug compounds.
2. Methods
2.1. Preparation of wild-type and SULT1A3 allozymes
Wild-type and SULT1A3 allozymes were generated, expressed, and purified to greater than 95% homogeneity as previously reported [21].
2.2. Enzymatic assay
A previously established assay procedure [22] was used to analyze the sulfating activity of SULT1A3 allozymes. Phenylephrine and salbutamol, at two different concentrations (1 and 10 μM for phenylephrine, and 5 and 50 μM for salbutamol), were used as substrates. The reaction mixture, with a final volume of 20 μl, contained 50 mM HEPES buffer at pH 7.4, 1 mM dithiothreitol (DTT), 14 μM PAP[35S], and a specified concentration of phenylephrine or salbutamol as substrate. A control without substrate was analyzed in parallel. The reaction was started by the addition of the enzyme (0.5 μg), allowed to proceed for 10 min at 37ºC, and terminated by heating the tubes containing the reaction mixture at 100°C for 3 min, followed by TLC separation and autoradiography to locate the spots of the [35S]sulfated phenylephrine or salbutamol. The located spots were cut out from the plate and the [35S]sulfated products therein were eluted with Milli-Q water and counted for [35S]-radioactivity using a liquid scintillation counter. Data obtained were used to compute the specific activities in nmol of sulfated product/min/mg of allozyme.
2.3. Kinetic analysis
To determine the kinetic parameters, Km, Vmax, and Vmax/Km, of SULT1A3 allozymes in mediating the sulfation of phenylephrine and salbutamol, enzymatic assays were performed using 0, 0.5, 1, 2.5, 5, 10, 20, 30, 40, 50, 60, 70 and 80 μM of phenylephrine or 0, 5, 10, 25, 50, 66.6, 100, 200, 500, 750, 1000 and 1500 μM of salbutamol as substrates.
2.4. Data analysis
Data obtained in the kinetic experiments were processed based on non-linear regression of the Michaelis-Menten equation to compute the kinetic constants. GraphPad Prism 7 software was utilized in data analysis.
2.5. Materials
Phenylephrine (chemical purity: 99.4%) and Ecolume scintillation cocktail were purchased from MP Biomedicals, LLC. (Irvine, CA, USA). Salbutamol (chemical purity: ≥98%) was from Cayman Chemical (Ann Arbor, MI, USA). Dimethyl sulfoxide (DMSO), adenosine 5’-triphosphate (ATP), N-2-hydroxylpiperazine-N’−2-ethanesulfonic acid (HEPES), and dithiothreitol (DTT) were products of Sigma Chemical Company (St. Louis, MO, USA). 3’-Phosphoadenosine-5’-phospho[35S]sulfate (PAP[35S]) was prepared using ATP and free [35S]sulfate based on a previously established protocol [23]. Cellulose TLC plates were purchased from EMD Millipore Corporation (Burlington, MA, USA). PCR kit was from G Biosciences (St. Louis, MO, USA). Prime STAR® GXL DNA Polymerase was a product of Clontech Laboratories, Inc. (Mountain View, CA, USA). QIAprep® Spin Miniprep Kit was a product of QIAGEN (Germantown, MD, USA). Protein molecular weight markers were from Bioland Scientific LLC (Paramount, CA, USA). Glutathione Sepharose™ was purchased from GE Healthcare Life Sciences (Pittsburgh, PA, USA). X-Ray films were obtained from Research Products International Corporation (Mt Prospect, IL, USA). All other chemicals were of the highest grades commercially available.
3. Results
3.1. Specific Activities of wild-type and SULT1A3 allozymes with phenylephrine and salbutamol as substrates
Specific activities of wild-type and SULT1A3 allozymes were determined using two different substrate concentrations, one approximately ten times lower than the reported Km and the other close to the reported Km of the wild-type SULT1A3 with phenylephrine or salbutamol as substrate [8, 9]. Results obtained are shown in Figures 1 and 2.
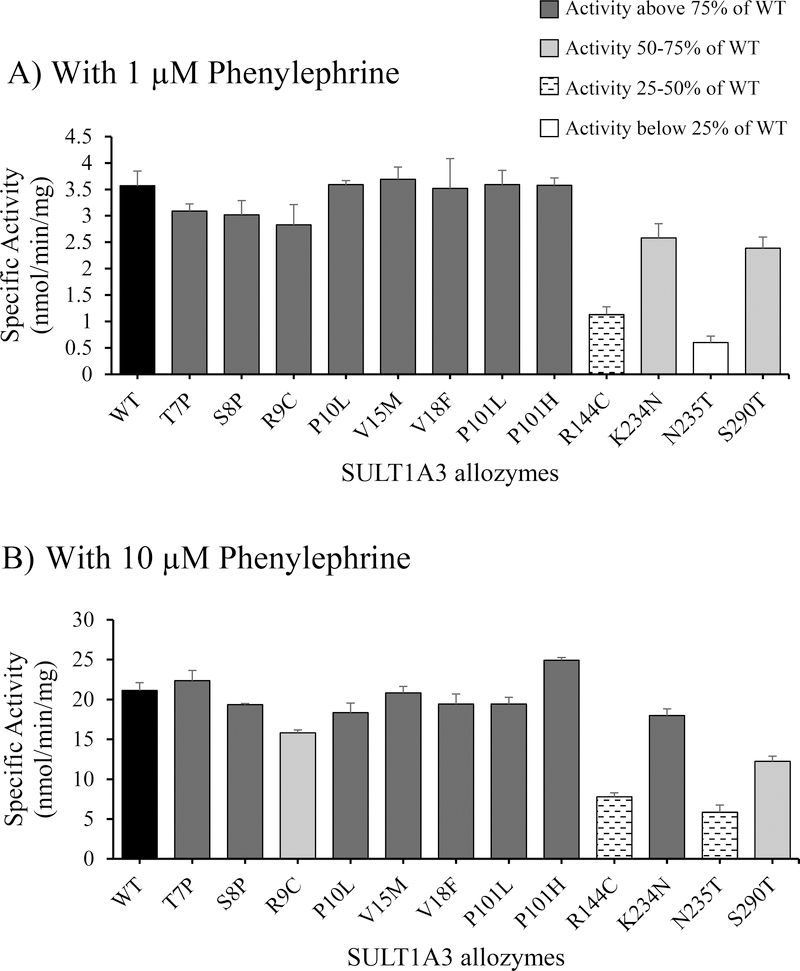
Figure 1.
Specific activities of wild-type and SULT1A3 allozymes with phenylephrine as substrate. Concentrations of phenylephrine used in the enzymatic assays were 1 μM (A) and 10 μM (B). Specific activity refers to nmol phenylephrine sulfated/min/mg of purified enzyme. Data shown represent mean ± standard deviation derived from three determinations (n=6). WT refers to wild-type SULT1A3.
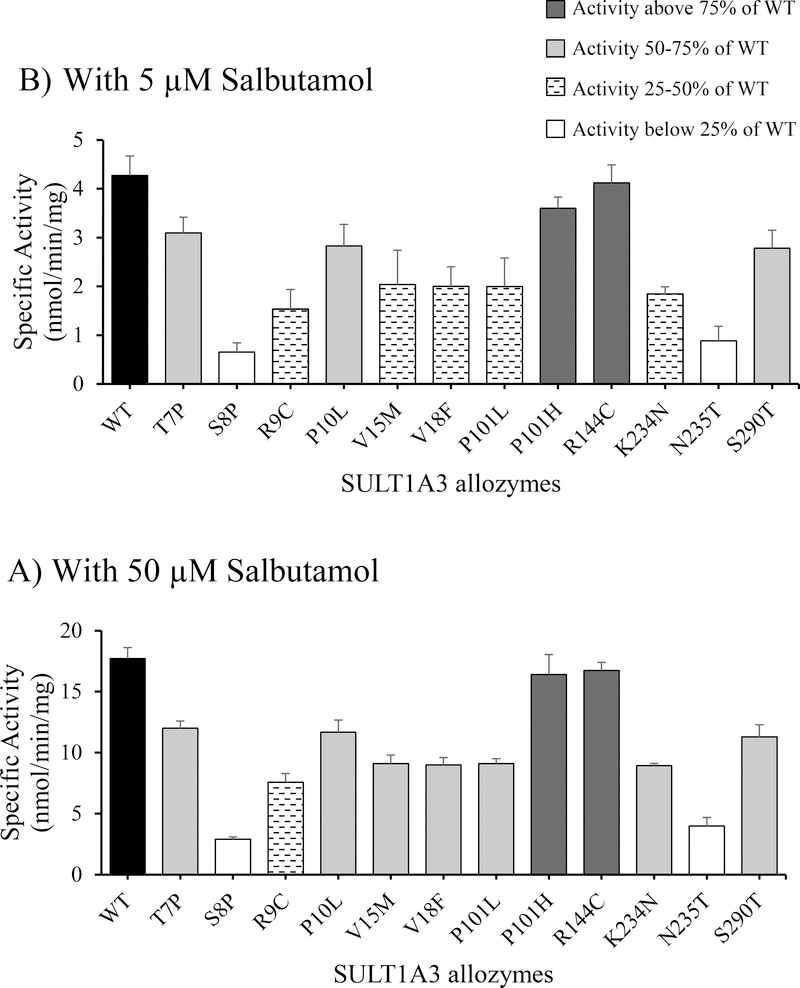
Figure 2.
Specific activities of wild-type and SULT1A3 allozymes with salbutamol as substrate. Concentrations of salbutamol used in the enzymatic assays were 5 μM (A) and 50 μM (B). Specific activity refers to nmol salbutamol sulfated/min/mg of purified enzyme. Data shown represent mean ± standard deviation derived from three determinations (n=6). WT refers to wild-type SULT1A3.
With 1 μM phenylephrine as substrate (Figure 1A), five allozymes, SULT1A3-P10L, -V15M, -V18F, -P101L, and -P101H, displayed specific activities comparable to that of the wild-type enzyme, while three allozymes, SULT1A3-T7P, -S8P, and -R9C, exhibited slightly lower (86.5%, 84.5%, and 79.2%, respectively) specific activity than that of the wild-type. The remaining four SULT1A3 allozymes exhibited much lower specific activities than the wild-type enzyme. Among these four allozymes, SULT1A3-N235T showed the lowest phenylephrine-sulfating activity, being only 16.8% of that of the wild-type. With 10 μM phenylephrine as substrate (Figure 1B), eight allozymes, SULT1A3-T7P, -S8P, -P10L, -V15M, -V18F, -P101L, -P101H, -K234N, showed specific activities that were more than 75% of that of the wild-type enzyme. Two allozymes, SULT1A3-R9C and –S290T, displayed specific activities 50–75% that of the wild-type. Specific activities determined for the remaining two allozymes, SULT1A3-R144C and -N235T, were much lower (<50%) than that of the wild-type. Notably, SULT1A3-N235T exhibited a specific activity which was only 27.7% of that of the wild-type enzyme.
With 5 μM salbutamol as substrate (Figure 2A), all SULT1A3 allozymes showed lower specific activities than the wild-type enzyme. Two, SULT1A3-P101H and -R144C, exhibited only slightly lower specific activities, while three others, SULT1A3-T7P, -P10L and -S290T, showed specific activities between 50–75% that of the wild-type. The remaining seven SULT1A3 allozymes displayed much lower specific activities (<50% that of the wild-type). Among these seven allozymes, SULT1A3-S8P and -N235T displayed the lowest specific activities, being only 15.2% and 20.6% that of the wild-type. With 50 μM salbutamol as substrate (Figure 2B), SULT1A3-P101H and -R144C showed specific activities comparable to that of the wild-type, while SULT1A3-T7P, -P10L, -V15M, -V18F, -P101L, -K234N and -S290T displayed significantly lower (50–75% that of the wild-type). In contrast, the remaining two, SULT1A3-S8P and -N235T, showed much lower activities (16.3% and 22.5% of that of the wild-type).
3.2. Kinetic parameters of wild-type and SULT1A3 allozymes in mediating the sulfation of phenylephrine and salbutamol
Kinetic constants of wild-type and SULT1A3 allozymes in mediating the sulfation of phenylephrine were determined using varying concentrations of phenylephrine as substrates. As shown in Table 1, ten allozymes displayed Km values close to that of the wild-type (10.22 ± 0.57 μM). Of the two remaining SULT1A3 allozymes, SULT1A3-R144C showed a Km value (26.99 ± 3.66 μM) 2.5 times that of the wild-type, while SULT1A3-N235T showed a Km value (207.00 ± 65.51 μM) nearly 20 times that of the wild-type. In regard to Vmax, ten of the twelve allozymes showed Vmax values comparable to, albeit slightly lower than, that of the wild-type. Of the other two allozymes, SULT1A3-N235T showed a Vmax value nearly two times that of the wild-type, while SULT1A3-S290T exhibited a Vmax value only half of that of the wild-type. Based on these Km and Vmax values, nine of the twelve SULT1A3 allozymes displayed Vmax/Km values comparable to that of the wild-type enzyme. The remaining three allozymes (SULT1A3-R144C, -N235T, and -S290T) showed Vmax/Km values that were, respectively, 27.4%, 10.1%, and 57.7% of that of the wild-type.
Table 1.
Kinetic parameters of wild-type and SULT1A3 allozymes with phenylephrine as substrate.
| SULT1A3 allozymes | Km (μM) | Vmax (nmol/min/mg) | Vmax/Km (nmol/min/mg/μM) |
|---|---|---|---|
| 1A3-WT1 | 10.22 ± 0.57 | 44.34 ± 0.58 | 4.33 |
| 1A3-T7P | 8.96 ± 1.02 | 37.17 ± 1.26 | 4.14 |
| 1A3-S8P | 9.33 ± 0.82 | 37.74 ± 1.90 | 4.04 |
| 1A3-R9C | 11.20 ± 0.83 | 33.90 ± 0.70 | 3.03 |
| 1A3-P10L | 8.91 ± 0.62 | 34.33 ± 0.61 | 3.85 |
| 1A3-V15M | 10.00 ± 0.49 | 43.48 ± 0.59 | 4.35 |
| 1A3-V18F | 8.88 ± 0.99 | 37.04 ± 1.10 | 4.17 |
| 1A3-P101L | 8.29 ± 0.53 | 35.34 ± 0.57 | 4.26 |
| 1A3-P101H | 9.15 ± 0.84 | 44.35 ± 1.01 | 4.85 |
| 1A3-R144C | 26.99 ± 3.66 | 32.19 ± 1.66 | 1.19 |
| 1A3-K234N | 8.99 ± 1.00 | 31.90 ± 0.85 | 3.55 |
| 1A3-N235T | 207.00 ± 65.51 | 90.83 ± 22.34 | 0.44 |
| 1A3-S290T | 9.55 ± 1.08 | 23.89 ± 0.71 | 2.50 |
Wild-type human SULT1A3.
Kinetic constants of wild-type and SULT1A3 allozymes in mediating the sulfation of salbutamol were also determined using varying concentrations of salbutamol as substrates. As shown in Table 2, of the twelve allozymes, two, SULT1A3-P101H and -R144C, exhibited Km values (54.26 ± 4.29 and 57.75 ± 5.51, respectively) close to that of the wild-type (54.07 ± 3.47 μM), while the other ten allozymes all showed considerably higher Km values. Among the latter, six allozymes, SULT1A3-S8P, -R9C, -V15M, -V18F, -K234N and -N235T, showed substantially higher Km values than the other four allozymes. Notably, SULT1A3-N235T exhibited a Km value which was 7.1 times that of the wild-type. In regard to Vmax, SULT1A3-R144C, displayed a slightly higher Vmax value (35.25 ± 0.66 nmol/min/mg) than that (34.27 ± 1.29 nmol/min/mg) determined for the wild-type enzyme, while the other eleven allozymes all showed lower Vmax values than the wild-type. In particular, SULT1A3-S8P exhibited a Vmax value (11.89 ± 0.31 nmol/min/mg), which was only one-third of that of the wild-type. The calculated Vmax/Km indicated that only two allozymes, SULT1A3-P101H and -R144C, showed values (0.62 and 0.61, respectively) comparable to that of wild-type (0.63), while the other ten allozymes all showed lower Vmax/Km values than the wild-type. Like with phenylephrine, six allozymes, SULT1A3-S8P, -R9C, -V15M, -V18F, -K234N and -N235T, exhibited dramatically lower catalytic efficiencies (11.1%, 20.6%, 26.9%, 43.9%, 36.5% and 11.1%, respectively) than the wild-type enzyme.
Table 2.
Kinetic parameters of wild-type and SULT1A3 allozymes with salbutamol as substrate.
| SULT1A3 allozymes | Km (μM) | Vmax (nmol/min/mg) | Vmax/Km (nmol/min/mg/μM) |
|---|---|---|---|
| 1A3-WT1 | 54.07 ± 3.47 | 34.27 ± 1.29 | 0.63 |
| 1A3-T7P | 71.83 ± 5.87 | 29.56 ± 0.50 | 0.49 |
| 1A3-S8P | 172.00 ± 19.23 | 11.89 ± 0.31 | 0.07 |
| 1A3-R9C | 243.00 ± 43.92 | 32.26 ± 1.51 | 0.13 |
| 1A3-P10L | 88.50 ± 9.05 | 30.47 ± 0.66 | 0.34 |
| 1A3-V15M | 173.20 ± 31.58 | 28.90 ± 1.26 | 0.17 |
| 1A3-V18F | 118.80 ± 18.52 | 25.74 ± 0.90 | 0.22 |
| 1A3-P101L | 90.77 ± 8.70 | 24.05 ± 0.49 | 0.26 |
| 1A3-P101H | 54.26 ± 4.29 | 33.40 ± 0.51 | 0.62 |
| 1A3-R144C | 57.75 ± 5.51 | 35.25 ± 0.66 | 0.61 |
| 1A3-K234N | 107.10 ± 13.83 | 24.30 ± 0.69 | 0.23 |
| 1A3-N235T | 386.30 ± 45.86 | 28.06 ± 1.85 | 0.07 |
| 1A3-S290T | 87.78 ± 9.04 | 30.76 ± 0.67 | 0.35 |
Wild-type human SULT1A3.
4. Discussion
Phenylephrine and salbutamol are drugs that are prescribed for treating patients suffering from a number of diseases/disorders, including nasal and sinus congestion, hypotension, asthma and COPD [1–3, 5]. Pharmacokinetic studies have demonstrated that both these two drugs are metabolized through sulfation in human body under the action of SULTs, particularly SULT1A3 [4, 7–9]. Considering the critical role SULT1A3 in their metabolism, we were interested in examining the effects of genetic polymorphism on the sulfating activity of SULT1A3 allozymes toward phenylephrine and salbutamol. Previous studies have revealed that the gene coding for SULT1A3 had undergone duplication during the evolutionary process generating two genes, designated SULT1A3 and SULT1A4, that encode identical protein products, collectively called SULT1A3 [18]. We recently employed site-directed mutagenesis in conjunction with bacterial expression and affinity purification to prepare twelve distinct SULT1A3 allozymes [21]. In this study, the twelve SULT1A3 allozymes, in comparison with the wild-type enzyme, were investigated for their sulfating activity toward phenylephrine and salbutamol. With regard to the sulfation of salbutamol, it should be pointed out that the assay procedure used in the current study made no distinction between sulfated (R)- and (S)-salbutamol. There are, nevertheless, established procedures employing enantioselective liquid chromatography-mass spectrometry [24] that are capable of measuring sulfated (R)- and (S)-salbutamol.
An initial analysis of the twelve SULT1A3 allozymes revealed that eight of them displayed phenylephrine-sulfating activities comparable to that of the wild-type enzyme (Figure 1). The other four SULT1A3 allozymes (-R144C, -K234N, -N235T, and -S290T), on the other hand, showed specific activities considerably lower than that of the wild-type. The follow-up kinetic studies indicated significant changes in Km (reflecting the substrate-binding affinity) and/or Vmax (reflecting the catalytic activity) that resulted in differential catalytic efficiency (as reflected by Vmax/Km) of these four SULT1A3 allozymes. In regard to the salbutamol-sulfating activity, a more highly variable pattern of the specific activities was detected for the twelve SULT1A3 allozymes. The follow-up kinetic studies showed more drastic changes in Km and/or Vmax, and, consequently, Vmax/Km. These results indicated clearly that the amino acid changes resulting from the cSNPs of the SULT1A3 and SULT1A4 genes indeed have a significant impact on the sulfating activity of the coded SULT1A3 allozymes.
Previous studies on the crystal structure of SULT1A3 have revealed specific amino acid residues and/or segments that play important roles in the functioning of the enzyme. Among them are the N-terminal region comprising the βA- and βB-sheets (residues Leu12-Val15 and Val18-Ile21, respectively) that has been shown to be important for the polypeptide folding [25, 26], a catalytic residue (His108), the co-substrate (PAPS) binding regions (residues 45TYPKSGTT52, Arg130, Ser138, and 257RKG259), the substrate-binding residues (residues Asp86 and Glu146) [27], and the C-terminal dimerization motif (residues Lys265 to Glu274, with KXXXTVXXXE sequence) [28]. In a recent modeling study, three loop segments, Asp66-Met77, Ser228-Gly259, and Lys85-Pro90, have been proposed to form a gate that governs the substrate selectivity [29]. In light of these critical structural elements, it is an interesting question how the amino acid substitutions in different SULT1A3 allozymes may affect their phenylephrine- and salbutamol-sulfating activity.
As noted above, two SULT1A3 allozymes, -R144C and -N235T, showed the lowest specific activities and catalytic efficiencies (approximately 27% and 10%, respectively, of that of the wild-type) toward phenylephrine. In the case of SULT1A3-R144C, Arg originally in the wild-type enzyme is positively charged and may form salt-bridges with acidic (negatively charged) amino acid residues (Asp or Glu) thereby establishing hydrogen bonds that may be critical to the functioning of the enzyme [30]. Upon replacement with Cys, however, the hydrogen bonding can no longer be formed. It should be pointed out that in the SULT1A3 molecule, Arg144 residue is located within the segment 143HRMEKA148, which has been reported to play a significant role in substrate-binding and catalysis in both SULT1A1 and SULT1A3 [31]. The Arg144Cys substitution therefore may affect the substrate-binding affinity by causing structural changes in the substrate-binding pocket. SULT1A3-N235T also exhibited low sulfating activity and catalytic efficiency toward phenylephrine. Asn235 is located within the α15 sheet secondary structure previously proposed to play an indirect role in PAPS-binding and may restrict the required conformations for substrate binding when the co-substrate, PAPS, is bound to the SULT molecule [26]. In SULT1A3-N235T, the bulky side chain of the substituting Thr residue cannot be easily accommodated within the α15 helical structure, which may consequently affect the binding of both the co-substrate and substrate. This situation may explain the very low sulfating activity of SULT1A3-N235T toward phenylephrine. It is noted that previous studies have also demonstrated that Asn235Thr substitution affected greatly the sulfating activity of SULT1A1 toward a prototype substrate, p-nitrophenol [32, 33].
With salbutamol as the substrate, seven SULT1A3 allozymes, -S8P, -R9C, -V15M, -V18F, -P101L, -K234N and -N235T, showed significantly lower sulfating activity and catalytic efficiency than the wild-type enzyme. Among them, SULT1A3-S8P exhibited the lowest sulfating activity and catalytic efficiency (~11% of that of the wild-type). It is possible that the substituting proline residue may impose an unfavorable turn in the N-terminal region, thereby affecting the conformation and thus the salbutamol-sulfating activity of this allozyme. In the case of SULT1A3-R9C, the Arg to Cys substitution may lead to the disruption of hydrogen bonding which may be critical to the functioning of the enzyme. Moreover, the potential of the consequential disulfide bond formation should not be overlooked. SULT1A3-V15M and -V18F involve the substitution of an aliphatic side chain with an S-methyl thioether (SULT1A3-V15M) or an aromatic moiety (SULT1A3-V18F). Both Met and Phe have bulkier side chains than Val, which at their respective locations may lead to more restricted conformations of the βA- and βB-sheets (spanning residues Leu12-Val15 and Val18-Ile21, respectively) in the N-terminal region. The lower sulfating activity and catalytic efficiency of the allozymes with amino acids substitutions in the N-terminal region (SULT1A3-S8P, -R9C, -V15M, -V18F) provide support for the important structural role of this region in the SULT1A3 molecule [26]. SULT1A3-P101L involves the substitution of Pro with Leu within the loop connecting α6 and βD sheets, which is located near the catalytic residue His108 and is within the segment 84–104. It is noted that the segment 84–104 has been proposed to be involved in substrate binding and, together with the segment 145MEKAHPEPGT154, plays a role in reshaping the conformation of the substrate-binding pocket [27]. Both SULT1A3-K234N and -N235T showed decreased salbutamol-sulfating activity and catalytic efficiency (36.5% and 11.1%, respectively) compared with the wild-type enzyme. As noted earlier, both N235 and K234 are located within the α15 sheet, which is involved in the binding of the co-substrate, PAPS, and the substrate [26], which may be the reason for their decreased salbutamol-sulfating activity.
It is interesting to note that while SULT1A3 is the major SULT responsible for the sulfation of phenylephrine and salbutamol [8, 9], the twelve SULT1A3 allozymes showed quite different patterns of differential sulfating activities toward these two substrates (cf. Figures 1 and 2). Moreover, kinetic analyses indicated different patterns of variations in regard to Km and Vmax, and thus Vmax/Km, of the twelve SULT1A3 allozymes (cf. Tables 1 and 2). It is possible that these differences may be due to the chemical structures of the two substrates. Compared with phenylephrine, salbutamol has a bulkier structure, which may affect its passage through the gate [29] that governs the substrate selectivity, as well as the chemical interactions of the salbutamol molecule once inside the substrate-binding pocket of SULT1A3. Salbutamol as a chiral compound raises another important issue concerning the differential sulfation of (R)- vs. (S)-salbutamol. While the assay procedure used in the current study could not distinguish sulfated (R)-salbutamol from sulfated (S)-salbutamol, previous studies have demonstrated much greater area under the curve (AUC), maximum plasma concentration (Cmax), and time from administration to time of maximum plasma concentration (tmax) for(S)-salbutamol than for (R)-salbutamol in healthy volunteers upon inhaled administration with racemic salbutamol [34]. Since salbutamol is known to be primarily metabolized by SULT1A3 [35], an intriguing question is whether genetic polymorphisms of its coding genes, SULT1A3 and SULT1A4, may influence the pharmacokinetics of (R)- vs. (S)-salbutamol. In a more recent study, the effect of a common SULT1A3 SNP 105A>G was investigated [24], and the results indicated that upon inhaled administration with racemic salbutamol, similar pharmacokinetic parameters (t1/2, Cmax, and AUC0–4h) for (S)- vs. (R)-salbutamol were detected for individuals with this SNP or wild-type SULT1A3. Since SULT1A3 SNP 105A>G is a synonymous cSNP, it is an interesting question whether non-synonymous SULT1A3/SULT1A4 cSNPs may exert any significant effects on the pharmacokinetics of (S)- vs. (R)-salbutamol.
To summarize, the current study was performed to collect data relevant to the impact of SULT1A3/SULT1A4 genetic polymorphisms on the sulfating activity of the human SULT1A3 toward phenylephrine and salbutamol. Both the specific activity and kinetic data revealed that the twelve SULT1A3 allozymes indeed displayed differential sulfating activity and catalytic efficiency toward the two substrates. These results may underscore the differential metabolism of phenylephrine and salbutamol in individuals with different SULT1A3/SULT1A4 genotypes. Such information may in the future help design personalized regimens of these two drugs in order to improve their therapeutic efficacy and minimize their side effects.
Acknowledgments:
This work was supported in part by a grant from National Institutes of Health (Grant # R03HD071146).
Abbreviations:
- PAP[35]S
3’-phosphoadenosine-5’-phosphosulfate
- SULT
cytosolic sulfotransferase
- TLC
thin-layer chromatography
- SNP
single nucleotide polymorphism
Footnotes
Conflict of interest: The authors declare no conflicts of interest.
References
- [1].Johnson DA, Hricik JG. The pharmacology of alpha-adrenergic decongestants. Pharmacotherapy. 1993;13:110S–115S. [PubMed] [Google Scholar]
- [2].Horak F, Zieglmayer P, Zieglmayer R, Lemell P, Yao R, Staudinger H, et al. A placebo-controlled study of the nasal decongestant effect of phenylephrine and pseudoephedrine in the Vienna Challenge Chamber. Ann Allergy Asthma Immunol. 2009;102:116–120. [DOI] [PubMed] [Google Scholar]
- [3].Ullah MI, Newman GB, Saunders KB. Influence of age on response to ipratropium and salbutamol in asthma. Thorax. 1981;36:523–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Boulton DW, Fawcett JP. The pharmacokinetics of levosalbutamol: what are the clinical implications? Clin Pharmacokinet. 2001;40:23–40. [DOI] [PubMed] [Google Scholar]
- [5].Starkey ES, Mulla H, Sammons HM, Pandya HC. Intravenous salbutamol for childhood asthma: evidence-based medicine? Arch Dis Child. 2014;99:873–877. [DOI] [PubMed] [Google Scholar]
- [6].Hochhaus G, Möllmann H. Pharmacokinetic/pharmacodynamic characteristics of the beta-2-agonists terbutaline, salbutamol and fenoterol. Int J Clin Pharmacol Ther Toxicol. 1992;30:342–362. [PubMed] [Google Scholar]
- [7].Ibrahim KE, Midgley JM, Crowley JR, Williams CM. The mammalian metabolism of R-(−)-m-synephrine. J Pharm Pharmacol. 1983;35:144–147. [DOI] [PubMed] [Google Scholar]
- [8].Yamamoto A, Kim J, Liu MY, Kurogi K, Sakakibara Y, Suiko M, et al. Sulfation of phenylephrine by the human cytosolic sulfotransferases. Drug Metab Lett. 2014;8:96–100. [DOI] [PubMed] [Google Scholar]
- [9].Ko K, Kurogi K, Davidson G, Liu MY, Sakakibara Y, Suiko M, et al. Sulfation of ractopamine and salbutamol by the human cytosolic sulfotransferases. J Biochem. 2012;152:275–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Nishimuta H, Ohtani H, Tsujimoto M, Ogura K, Hiratsuka A, Sawada Y. Inhibitory effects of various beverages on human recombinant sulfotransferase isoforms SULT1A1 and SULT1A3. Biopharm Drug Dispos. 2007;28:491–500. [DOI] [PubMed] [Google Scholar]
- [11].Morgan DJ. Clinical pharmacokinetics of beta-agonists. Clin Pharmacokinet. 1990;18:270–294. [DOI] [PubMed] [Google Scholar]
- [12].Morgan DJ, Paull JD, Richmond BH, Wilson-Evered E, Ziccone SP. Pharmacokinetics of intravenous and oral salbutamol and its sulphate conjugate. Br J Clin Pharmacol. 1986;22:587–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Falany CN, Roth JA. Properties of human cytosolic sulfotransferases involved in the drug metabolism Human Drug Metabolism In: Jeffery HE, ed. From Molecular Biology to Man. Boca Raton: CRC Press, 1993: 101–115. [Google Scholar]
- [14].Mulder GJ, Jakoby WB. Sulfation In: Mulder GJ, Jakoby WB, eds. Conjugation Reactions in Drug Metabolism. London: Taylor and Francis, 1990: 107–161. [Google Scholar]
- [15].Weinshilboum RM, Otterness DM. Sulfotransferase enzymes In: Kaufman FC, ed. Conjugation-deconjugation reactions in Drug Metabolism and Toxicity. Berlin: Springer-Verlag, 1994: 45–78. [Google Scholar]
- [16].Lipmann F Biological sulfate activation and transfer. Science. 1958;128:575–580. [DOI] [PubMed] [Google Scholar]
- [17].Dooley TP. Cloning of the human phenol sulfotransferase gene family: three genes implicated in the metabolism of catecholamines, thyroid hormones and drugs. Chem Biol Interact. 1998;109:29–41. [DOI] [PubMed] [Google Scholar]
- [18].Hildebrandt MA, Salavaggione OE, Martin YN, Flynn HC, Jalal S, Wieben ED, et al. Human SULT1A3 pharmacogenetics: gene duplication and functional genomic studies. Biochem Biophys Res Commun. 2004;321:870–878. [DOI] [PubMed] [Google Scholar]
- [19].Thomae BA, Rifki OF, Theobald MA, Eckloff BW, Wieben ED, Weinshilboum RM. Human catecholamine sulfotransferase (SULT1A3) pharmacogenetics: functional genetic polymorphism. J Neurochem. 2004;87:809–819. [DOI] [PubMed] [Google Scholar]
- [20].Hui Y, Liu MC. Sulfation of ritodrine by the human cytosolic sulfotransferases (SULTs): Effects of SULT1A3 genetic polymorphism. Eur J Pharmacol. 2015;761:125–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Bairam AF, Rasool MI, Alherz FA, Abunnaja MS, El Daibani AA, Gohal SA, et al. Sulfation of catecholamines and serotonin by SULT1A3 allozymes. Biochem Pharmacol. 2018;151:104–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Kurogi K, Chepak A, Hanrahan MT, Liu MY, Sakakibara Y, Suiko M, et al. Sulfation of opioid drugs by human cytosolic sulfotransferases: metabolic labeling study and enzymatic analysis. Eur J Pharm Sci. 2014;1:40–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Yanagisawa K, Sakakibara Y, Suiko M, Takami Y, Nakayama T, Nakajima H, et al. cDNA cloning, expression, and characterization of the human bifunctional ATP sulfurylase/adenosine 5’-phosphosulfate kinase enzyme. Biosci Biotechnol Biochem. 1998;62:1037–1040. [DOI] [PubMed] [Google Scholar]
- [24].Jacobson GA, Yee KC, Wood-Baker R, Walters EH. SULT 1A3 single-nucleotide polymorphism and the single dose pharmacokinetics of inhaled salbutamol enantiomers: are some athletes at risk of higher urine levels? Drug Test Anal. 2015;7:109–113. [DOI] [PubMed] [Google Scholar]
- [25].Bidwell LM, McManus ME, Gaedigk A, Kakuta Y, Negishi M, Pedersen L, et al. Crystal structure of human catecholamine sulfotransferase. J Mol Biol. 1999;293:521–530. [DOI] [PubMed] [Google Scholar]
- [26].Allali-Hassani A, Pan PW, Dombrovski L, Najmanovich R, Tempel W, Dong A, et al. Structural and chemical profiling of the human cytosolic sulfotransferases. PLoS Biol. 2007;5:e97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Lu JH, Li HT, Liu MC, Zhang JP, Li M, An XM, et al. Crystal structure of human sulfotransferase SULT1A3 in complex with dopamine and 3’-phosphoadenosine 5’-phosphate. Biochem Biophys Res Commun. 2005;335:417–423. [DOI] [PubMed] [Google Scholar]
- [28].Petrotchenko EV, Pedersen LC, Borchers CH, Tomer KB, Negishi M. The dimerization motif of cytosolic sulfotransferases. FEBS Lett. 2001;490:39–43. [DOI] [PubMed] [Google Scholar]
- [29].Cook I, Wang T, Almo SC, Kim J, Falany CN, Leyh TS. The gate that governs sulfotransferase selectivity. Biochemistry. 2013;52:415–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Betts MJ, Russell RB. Amino Acid Properties and Consequences of Substitutions In: Barnes MR, Gray IC, eds. Bioinformatics for Geneticists. Chichester: John Wiley & Sons Ltd, 2003. [Google Scholar]
- [31].Chen G, Chen X. Arginine Residues in the Active Site of Human Phenol Sulfotransferase (SULT1A1). J Biol Chem. 2003;278:36358–36364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Coughtrie MW. Sulfation through the looking glass-recent advances in sulfotransferase research for the curious. Pharmacogenomics J. 2002;2:297–308. [DOI] [PubMed] [Google Scholar]
- [33].Zhu X, Veronese ME, Iocco P, McManus ME. cDNA cloning and expression of a new form of human aryl sulfotransferase. Int J Biochem Cell Biol. 1996;28:565–571. [DOI] [PubMed] [Google Scholar]
- [34].Schmekel B, Rydberg I, Norlander B, Sjöswärd KN, Ahlner J, Andersson RG. Stereoselective pharmacokinetics of S-salbutamol after administration of the racemate in healthy volunteers. Eur Respir J. 1999;13:1230–1235. [DOI] [PubMed] [Google Scholar]
- [35].Boulton DW, Fawcett JP. The pharmacokinetics of levosalbutamol: what are the clinical implications? Clin Pharmacokinet. 2001;40:23–40. [DOI] [PubMed] [Google Scholar]