ABSTRACT
The mechanisms of efficacy for fecal microbiota transplantation (FMT) in treating recurrent Clostridioides difficile infection (rCDI) remain poorly defined, with restored gut microbiota-bile acid interactions representing one possible explanation. Furthermore, the potential implications for host physiology of these FMT-related changes in gut bile acid metabolism are also not well explored. In this study, we investigated the impact of FMT for rCDI upon signalling through the farnesoid X receptor (FXR)-fibroblast growth factor (FGF) pathway. Herein, we identify that in addition to restoration of gut microbiota and bile acid profiles, FMT for rCDI is accompanied by a significant, sustained increase in circulating levels of FGF19 and reduction in FGF21. These FGF changes were associated with weight gain post-FMT, to a level not exceeding the pre-rCDI baseline. Collectively, these data support the hypothesis that the restoration of gut microbial communities by FMT for rCDI is associated with an upregulated FXR-FGF pathway, and highlight the potential systemic effect of FMT.
Keywords: Microbiota, fecal microbiota transplantation (FMT), recurrent Clostridium difficile infection (rCDI), bile acid metabolism, fibroblast growth factor (FGF)19
Introduction
Fecal microbiota transplantation (FMT) is a highly effective therapy against recurrent Clostridioides difficile infection (rCDI). However, the mechanisms by which FMT exerts its efficacy in rCDI remain unclear. In recent years, restoration of pre-morbid gut bile acid metabolism has become one of the better known potential mechanisms supported by both human and animal studies. Secondary bile acids inhibit C. difficile vegetative growth, while certain primary bile acids (and particularly taurocholic acid) promote germination.1 It has been demonstrated that secondary bile acid concentrations are much reduced while primary bile acid levels are elevated in rCDI patients compared to healthy controls, potentially perpetuating C. difficile proliferation.2 Following FMT, which restores the diversity and composition of the intestinal microbiota, bile acid homeostasis is re-established.2 Furthermore, there is also evidence that the loss of microbiota-derived bile-metabolising enzymes may contribute to the pathogenesis of CDI both in mice and in humans.3–5
Bile acid metabolism is not only regulated by commensal bacteria, but also through farnesoid X receptors (FXR), which are abundantly expressed in the liver and ileum.6 In humans, the most potent endogenous ligand for FXR is the primary bile acid chenodeoxycholic acid (CDCA); the secondary bile acids deoxycholic acid (DCA) and lithocholic acid (LCA) are moderate FXR agonists, whilst the primary bile acid cholic acid (CA) also has modest agonist activity.7 Upon ileal FXR activation, fibroblast growth factor (FGF)19 is secreted into the portal circulation, where it binds to the FGFR4/β Klotho receptor complex on hepatocytes. This interaction acts as both a negative feedback control on hepatic bile acid synthesis through inhibition, and also as a modulator of key metabolic pathways involved in glucose, lipid, and energy metabolism.8 Although both FGF19 and FGF21 are involved in regulating multiple metabolic processes, they have an inverse relationship that collectively maintains metabolic homeostasis, since FGF19 is produced during feeding while FGF21 is secreted during fasting.9 Perturbation of FXR signalling and altered FGF levels have been found in a number of disease states, including type 2 diabetes, metabolic syndrome and Crohn’s disease.10,11 Furthermore, surgically-induced weight loss is associated with an increase in FGF19 and a decrease in FGF21 levels.10
There is growing evidence from murine studies that altered interaction between the gut microbiota and bile acids may directly affect FXR signalling. Germ-free and antibiotic-treated mice have markedly reduced ileal Fgf15 gene expression (murine orthologue of human FGF19).12 The resultant accumulation of tauro-β-muricholic acid (an FXR antagonist) is thought to be the link between alterations of the gut microbiota and FXR signalling in mice. However, given that this bile acid is only present at very modest levels in humans, coupled with the differences in FGF orthologues and microbiota between humans and mice, extrapolating these data to humans is problematic. Presently, there are no human studies to our knowledge examining the impact of antibiotic-induced dysbiosis on FXR signalling. Given the apparent key contribution of the microbiota-bile acid axis to CDI pathogenesis, we investigated the association between changes in bile acid composition and FGF19 and 21 following FMT for the treatment of rCDI in humans. To do this, we analysed samples collected from a recent randomised trial of capsulized vs colonoscopic FMT for the treatment of rCDI.13 We undertook metagenomic, metabonomic and proteomic analyses for these samples, and correlated with weight changes following rCDI eradication.
Results
Stool metagenome analysis
We have previously described that successful FMT for rCDI is associated with both marked increases in stool microbial diversity and altered microbial community composition to resemble that of healthy donors, maintained up to at least 12 weeks post-FMT.13 Further analysis here demonstrated that successful FMT was particularly associated with enrichment of a number of bacterial genera including Bacteroides, Faecalibacterium, Ruminococcus, Blautia and Eubacterium (all of which contain members with bile acid-metabolising function) and loss of Klebsiella, Escherichia and Veillonella (which generally lack these functions) (Supplementary Figure 1).7
Ultra-performance liquid chromatography-mass spectrometry (UPLC-MS) stool bile acid profiling
Successful FMT was also associated with significantly decreased stool levels of the primary bile acids CDCA and CA, and significantly increased levels of the secondary bile acids DCA and LCA (Figure 1). In all cases, these changes were observed at four weeks post-FMT and were maintained at 12 weeks post-therapy.
Figure 1.
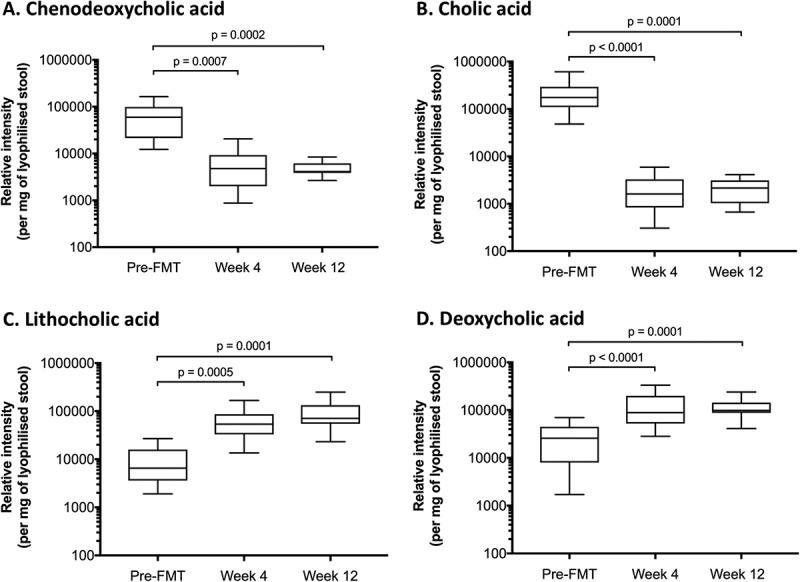
Effect of FMT for rCDI upon bile acid profiles.
A significant decrease in chenodeoxycholic acid (A) and cholic acid (B) is seen between screening and 4 weeks, and maintained up to 12 weeks post-FMT. A significant increase in lithocholic acid (C) and deoxycholic acid (D) is observed between screening and 4 weeks, and maintained up to 12 weeks post-FMT. X-axis depicts time, and y-axis depicts relative intensity of each bile acid. Pre-FMT = screening; week 4 = 4 weeks after fecal microbiota transplantation (FMT); week 12 = 12 weeks post-FMT.
Proteomic analysis
Of 73 compared proteomic markers (Supplementary Table 1), the differences were statistically significant for only two: FGF19 and FGF21 (Figure 2). FGF19 had significantly higher Normalized Protein eXpression (NPX) values at weeks 4 and 12 compared with screening, while FGF21 had significantly lower NPX values at weeks 4 and 12 compared with screening. There was no significant difference in the levels of FGF19 and FGF21 between the groups receiving FMT by either capsules or colonoscopy (data not shown).
Figure 2.
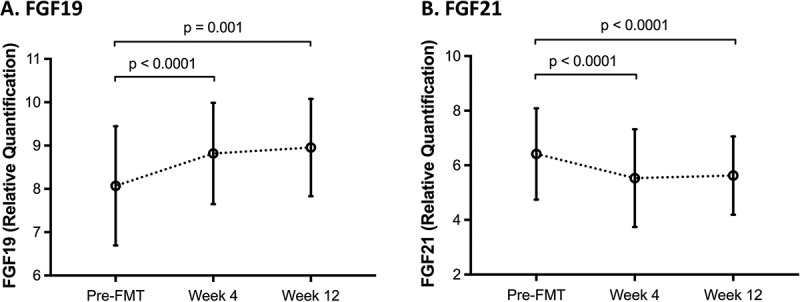
Normalized Protein eXpression (NPX) values for serum fibroblast growth factor (FGF)19 (A) and FGF21 (B) over time.
There is a statistically significant increase in FGF19 level 4 and 12 weeks after FMT compared to screening, while a statistically significant decrease in FGF21 level is observed 4 and 12 weeks following FMT. X-axis depicts time, and y-axis depicts relative quantification of respective FGF. Circles represent mean; error bars represent standard deviation.
Differences in weight before and after FMT
Following successful FMT, there was a statistically significant increase in mean BMI at 12 weeks following FMT compared to screening, but this did not exceed pre-rCDI baseline (mean BMI difference [95%CI], 0.5 [0.2, 0.8]; p = 0.003, Table 1.
Table 1.
Comparison of the mean body mass index (BMI) over time. At week 4, the mean BMI was not significantly different from the mean BMI prior to FMT. At week 12, patients had significantly higher BMI relative to pre-FMT, but did not exceed pre-rCDI baseline.
| Mean BMI Difference (95% CI) | p value | |
|---|---|---|
| Week 4 – Pre-FMT | 0.0 (−0.3, 0.3) | 0.84 |
| Week 12 – Pre-FMT | 0.5 (0.2, 0.8) | 0.003 |
| Week 12 – Week 4 | 0.4 (0.1, 0.8) | 0.006 |
Discussion
While it has already been observed that FMT in humans with rCDI restores gut microbiota and bile acid composition, we demonstrate for the first time that this procedure is also associated with activation of ileal FXR signalling, manifested by increased FGF19 and reduced FGF21 expression. CDCA is the most potent endogenous ligand for FXR, although the secondary bile acids DCA and LCA are also moderate FXR agonists. Our data suggest that the reduced level of a potent FXR agonist (CDCA) is offset by increased levels of two moderate FXR agonists (DCA and LCA),14 with a net upregulation of the ileal FXR-FGF pathway following successful FMT. Some phases of this bile acid transformation process (e.g. 7-α dehydroxylation) occur within the colon, implying that for secondary bile acids to affect ileal FXR signaling, they must be reabsorbed in the colon and re-secreted in bile into the small intestine.15 Although metabolism of bile acids in the gut is a bacterially-driven process,7 further studies are needed to examine the specific contribution of different bacteria to this process.
In addition to its well-defined roles in the regulation of metabolism and bile acid production, there is also evidence in how FXR signalling plays a role in other systemic processes relevant to CDI. For example, FXR activation has been shown to inhibit bacterial overgrowth and block mucosal injury in mouse ileum,16 and is associated with reduced expression of key cytokines (including TNF-α and IL-1β) that regulate the host innate immune response.17 Moreover, the inflammatory response or C. difficile itself could reciprocally inhibit activation of FXR and its target FGF genes, and this therefore merits further study.
Although our data are consistent with previous studies in observing restoration of the gut microbiota and bile acid composition post-FMT for rCDI, there is no direct demonstration that this pathway is a key mechanism underpinning the efficacy of FMT for rCDI. In addition, it is not clear if the observed weight gain following FMT is directly mediated through changes in FGF19/21 levels. Future mechanistic studies involving mouse models of CDI would be required to determine causality, and such studies should consider including analysis of the effect of FMT upon FXR signalling. Should these experiments validate our preliminary findings, the bile acid-FXR axis may become a novel therapeutic target for the treatment of rCDI. There are already some data that would appear to support this strategy; specifically, the potent FXR agonist obeticholic acid (INT-747) has been shown to display anti-C. difficile potential in murine models of CDI.18 However, the benefits and risks of synthetic FXR ligands require further evaluation.
In conclusion, our data suggest that FMT is associated with upregulation of the bile acid-FXR-FGF signalling pathway, and this may possibly explain the rapid improvement in energy and well-being many patients experience following FMT. Although these findings are intriguing, we acknowledge several limitations, including small sample size, short follow-up period, the observational nature of the data, lack of mucosal inflammatory protein expression data, and non-consideration of diet or host genetics. Insights gleaned from better understanding of FMT mechanisms of action using a multi-omics approach could enable development of tailored therapies that target key signaling pathways or specific constituents of those pathways that may regulate host defence to circumvent various concerns surrounding FMT.
Materials and methods
Patient clinical data, sample collection and storage
Participants (n = 116) in the capsule vs colonoscopy-delivered FMT trial were included this pilot study.13 Blood and stool samples were collected, and body mass indices (BMI) documented at screening and subsequent follow-up visits at weeks 4 and 12 after FMT. Of the 64 patients recruited from Edmonton, 43 had complete sets of archived blood samples and were subjected to proteomic analyses using the Olink inflammation panel. Of these 43 patients, 23 were chosen at random to have their stool samples undergo microbial composition analysis by shotgun metagenomics sequencing. From these 23 patients, 17 randomly-selected patients had stool bile acid profiling by ultra-performance liquid chromatography-mass spectrometry (UPLC-MS). Patient baseline characteristics are shown in Table 2. The metagenomic, metabonomic, and proteomic results were correlated with weight changes following rCDI eradication. This study was approved by the research ethics board of the University of Alberta (Pro49006).
Table 2.
Patient baseline characteristics.
| Variable | Blood proteomics analysis (n = 43) |
Stool bile acid analysis (n = 17) |
|---|---|---|
| Age, mean (SD), y | 58.8 (19.2) | 58.1 (17.1) |
| Female, No. (%) | 27 (62.8%) | 13 (76%) |
| Charlson Comorbidity Index score, median (Q1-Q3) | 3 (1–5) | 3 (1–4) |
| Immunosuppressed patients, No. (%) | 5 (11.6%) | 0 |
| BMI, mean (SD) | 25.9 (5.9) | 27.5 (6.0) |
| PPI use prior to FMT, No. (%) | 5 (11.6%) | 2 (11.7%) |
| Hemoglobin, median (Q1-Q3), g/dL | 13.8 (13.0–14.4) | 13.9 (13.1–14.5) |
| White blood cell count, median (Q1-Q3),/uL | 7100 (5850–8500) | 6750 (5620–8130) |
| Albumin, median (Q1-Q3), g/dL | 4.0 (3.8–4.3) | 4.0 (3.9–4.3) |
| C-reactive protein, median (Q1-Q3), mg/dL | 0.29 (0.085–0.10) | 0.30 (0.075–0.89) |
| Creatinine, median (Q1-Q3), mg/dL | 0.75 (0.66–0.87) | 0.74 (0.66–0.94) |
| Capsule delivered FMT, No. (%) | 25 (58.1%) | 11 (64.7%) |
Abbreviations: BMI, body mass index; FMT, fecal microbiota transplantation; PPI, proton pump inhibitor; Q1, first quartile; Q3, third quartile.
Stool metagenomics
Whole-genome shotgun sequencing was performed as previously described.13 More specifically, taxonomic classification of reads from each library was conducted with Kraken.19 The database used consisted of all bacteria, archaea, viruses, fungi, and protozoa full-length genomes from NCBI RefSeq, the human genome assembly GRCh38, and reference bacterial assemblies from the Human Microbiome Project.20 Read assignments were filtered with Kraken-filter using a threshold of 10%.
UPLC-MS profiling of fecal bile acids
Sample preparation was performed using protocols as previously-described.21 Bile acid analysis of faecal extracts was performed using ACQUITY UPLC (Waters Ltd, Elstree, UK) coupled to a Xevo G2 Q-ToF mass spectrometer equipped with an electrospray ionization source operating in negative ion mode (ESI-), using the method described by Sarafian and colleagues.22 Waters raw data files were converted to NetCDF format and data extracted using the XCMS (v1.50) package in R (v3.1.1) software. Probabilistic quotient normalisation23 was used to correct for dilution effects and chromatographic features with coefficient of variation higher than 30% in the QC samples were excluded from further analysis. The relative intensities of the features were corrected to the dry weight of the faecal samples.
Proteomics
The relative levels of serum inflammatory proteins were analyzed with Olink® Inflammation I panel (Olink Proteomics AB, Uppsala, Sweden) using Proximity Extension Assay (PEA) according to the manufacturer’s instructions.24,25 A list of the 92 inflammation-related markers is listed in Supplementary Table 2. In brief, serum samples (1µL) were incubated with 92 oligonucleotide labelled antibody probe pairs that bind to their respective target in the sample. A PCR reporter sequence was formed by a proximity dependent DNA polymerization event and was subsequently detected and amplified using a microfluidic real-time PCR instrument (Biomark HD, Fluidigm). Data was then quality controlled and normalized using an internal extension control and an inter-plate control, to adjust for intra- and inter-run variation. The final assay read-out is presented in Normalized Protein eXpression (NPX) values, which is an arbitrary unit on a log2-scale where a high value corresponds to a higher protein expression. All assay validation data (detection limits, intra- and inter-assay precision data, etc) are available on the manufacturer’s website (http://www.olink.com). Samples failing technical quality controls or that fell below lower limits of detection were excluded from analyses.
Statistical analysis
Full methodology for statistical analysis is provided in the Supplementary Material.
Biography
TM, BHM, JRM and DK contributed to study design, data analysis and interpretation, drafting of manuscript and critical revision of manuscript. HX, JP, GKSW, and TJ contributed to data analysis, drafting of manuscript and critical revision of manuscript. TM and DK contributed equally to this manuscript.
Funding Statement
This work was supported by University of Nottingham under Grant RPA22082017; Alberta Health Services under Grant 0022725; University of Alberta Hospital Foundation under Grant 3630; clinical research training fellowship by UK Medical Research Council under Grant MR/R000875/1; Division of Integrative Systems Medicine and Digestive Disease at Imperial College London receive financial support from the National Institute of Health Research (NIHR) Imperial Biomedical Research Centre (BRC) based at Imperial College Healthcare NHS Trust and Imperial College London; Medical Research Foundation [MR/R000875/1];University of Alberta Hospital Foundation [3630].
Abbreviations
- BMI
body mass index
- CA
cholic acid
- CDI
Clostridioides difficile infection
- CDCA
chenodeoxycholic acid
- DCA
deoxycholic acid
- FGF
fibroblast growth factor
- FMT
fecal microbiota transplantation
- FXR
farnesoid X receptor
- LCA
lithocholic acid
- rCDI
recurrent Clostridioides difficile infection
- NPX
Normalized Protein eXpression
Disclosure Statement
No potential conflicts of interest were disclosed.
Supplementary Material
Supplementary data for this article can be found online at www.tandfonline.com/kgmi
References
- 1.Thanissery R, Winston JA, Theriot CM.. Inhibition of spore germination, growth, and toxin activity of clinically relevant C. difficile strains by gut microbiota derived secondary bile acids. Anaerobe. 2017;45:86–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Weingarden AR, Chen C, Bobr A, Yao D, Lu Y, Nelson VM, Sadowsky MJ, Khoruts A. Microbiota transplantation restores normal fecal bile acid composition in recurrent Clostridium difficile infection. AJP Gastrointest Liver Physiol. 2014;306(4):G310–G319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allegretti JR, Kearney S, Li N, Bogart E, Bullock K, Gerber GK, Bry L, Clish CB, Alm E, Korzenik JR. Recurrent Clostridium difficile infection associates with distinct bile acid and microbiome profiles. Aliment Pharmacol Ther. 2016;43(11):1142–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buffie CG, Bucci V, Stein RR, McKenney PT, Ling L, Gobourne A, No D, Liu H, Kinnebrew M, Viale A, et al. Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature. 2014;517(7533):205–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Studer N, Desharnais L, Beutler M, Brugiroux S, Terrazos MA, Menin L, Schurch CM, McCoy KD, Kuehne SA, Minton NP, et al. Functional Intestinal Bile Acid 7α-Dehydroxylation by Clostridium scindens associated with protection from Clostridium difficile Infection in a Gnotobiotic Mouse Model. Front Cell Infect Microbiol. 2016;6:191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lefebvre P, Cariou B, Lien F, Kuipers F, Staels B. Role of Bile Acids and Bile Acid Receptors in Metabolic Regulation. Physiol Rev. 2009;89(1):147–191. [DOI] [PubMed] [Google Scholar]
- 7.Wahlströ A, Sayin SI, Marschall H-U, Bä Ckhed F. Intestinal crosstalk between bile acids and microbiota and its impact on host metabolism. Cell Metab. 2016;24:41–50. [DOI] [PubMed] [Google Scholar]
- 8.Benoit B, Meugnier E, Castelli M, Chanon S, Vieille-Marchiset A, Durand C, Bendridi N, Pesenti S, Monternier PA, Durieux AC, et al. Fibroblast growth factor 19 regulates skeletal muscle mass and ameliorates muscle wasting in mice. Nat Med. 2017;23(8):990–996. [DOI] [PubMed] [Google Scholar]
- 9.Zhang F, Yu L, Lin X, Cheng P, He L, Li X, Lu X, Tan Y, Yang H, Cai L, et al. Minireview: roles of fibroblast growth factors 19 and 21 in metabolic regulation and chronic diseases. Mol Endocrinol. 2015;29(10):1400–1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gomez-Ambrosi J, Gallego-Escuredo JM, Catalan V, Rodriguez A, Domingo P, Moncada R, Valenti V, Salvador J, Giralt M, Villarroya F, et al. FGF19 and FGF21 serum concentrations in human obesity and type 2 diabetes behave differently after diet- or surgically-induced weight loss. Clin Nutr. 2017;36(3):861–868. [DOI] [PubMed] [Google Scholar]
- 11.Nolan JD, Johnston IM, Pattni SS, Dew T, Orchard TR, Walters JR. Diarrhea in crohn’s disease: investigating the role of the ileal hormone fibroblast growth factor 19. J Crohn’s Colitis. 2015;9(2):125–131. [DOI] [PubMed] [Google Scholar]
- 12.Sayin SI, Wahlstrom A, Felin J, Jantti S, Marschall HU, Bamberg K, Angelin B, Hyotylainen T, Oresic M, Backhed F. Gut microbiota regulates bile acid metabolism by reducing the levels of tauro-beta-muricholic acid, a naturally occurring fxr antagonist. Cell Metab. 2013;17(2):225–235. [DOI] [PubMed] [Google Scholar]
- 13.Kao D, Roach B, Silva M, Beck P, Rioux K, Kaplan GG, Chang HJ, Coward S, Goodman KJ, Xu H, et al. Effect of oral capsule– vs colonoscopy-delivered fecal microbiota transplantation on recurrent Clostridium difficile Infection. JAMA. 2017;318(20):1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Parks DJ, Blanchard SG, Bledsoe RK, Chandra G, Consler TG, Kliewer SA, Stimmel JB, Willson TM, Zavacki AM, Moore DD, et al. Bile acids: natural ligands for an orphan nuclear receptor. Science. 1999;284(5418):1365–1368. [DOI] [PubMed] [Google Scholar]
- 15.Ridlon JM, Kang D-J, Hylemon PB. Bile salt biotransformations by human intestinal bacteria. J Lipid Res. 2006;47(2):241–259. [DOI] [PubMed] [Google Scholar]
- 16.Inagaki T, Moschetta A, Lee Y-K, Peng L, Zhao G, Downes M, Yu RT, Shelton JM, Richardson JA, Repa JJ, et al. Regulation of antibacterial defense in the small intestine by the nuclear bile acid receptor. Proc Natl Acad Sci. 2006;103(10):3920–3925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vavassori P, Mencarelli A, Renga B, Distrutti E, Fiorucci S. The bile acid receptor fxr is a modulator of intestinal innate immunity. J Immunol. 2009;183(10):6251–6261. [DOI] [PubMed] [Google Scholar]
- 18.Tessier MEM, Andersson H, Ross C, Peniche-Trujillo A, Dann S, Francis M, Sorg J, Thevananther S, Conner ME, Savidge T. Mo1850 obeticholic acid (INT-747) confers disease protection against Clostridium difficile infection. Gastroenterology. 2015;148(4):S–726. [Google Scholar]
- 19.Wood DE, Salzberg SL. Kraken: ultrafast metagenomic sequence classification using exact alignments. Genome Biol. 2014;15(3):R46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gevers D, Knight R, Petrosino JF, Huang K, McGuire AL, Birren BW, Nelson KE, White O, Methe BA, Huttenhower C. The human microbiome project: A community resource for the healthy human microbiome. PLoS Biol. 2012;10(8):e1001377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mullish BH, Pechlivanis A, Barker GF, Thursz MR, Marchesi JR, McDonald JAK. Functional microbiomics: evaluation of gut microbiota-bile acid metabolism interactions in health and disease. Methods. 2018. DOI: 10.1016/j.ymeth.2018.04.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sarafian MH, Lewis MR, Pechlivanis A, Ralphs S, McPhail MJ, Patel VC, Dumas ME, Holmes E, Nicholson JK. Bile acid profiling and quantification in biofluids using ultra-performance liquid chromatography tandem mass spectrometry. Anal Chem. 2015;87(19):9662–9670. [DOI] [PubMed] [Google Scholar]
- 23.Veselkov KA, Vingara LK, Masson P, Robinette SL, Want E, Li JV, Barton RH, Boursier-Neyret C, Walther B, Ebbels TM, et al. Optimized preprocessing of ultra-performance liquid chromatography/mass spectrometry urinary metabolic profiles for improved information recovery. Anal Chem. 2011;83(15):5864–5872. [DOI] [PubMed] [Google Scholar]
- 24.Assarsson E, Lundberg M, Holmquist G, Bjorkesten J, Thorsen SB, Ekman D, Eriksson A, Rennel Dickens E, Ohlsson S, Edfeldt G, et al. Homogenous 96-plex pea immunoassay exhibiting high sensitivity, specificity, and excellent scalability. PLoS One. 2014;9(4):e95192 Hoheisel JD, ed DOI: 10.1371/journal.pone.0095192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lundberg M, Eriksson A, Tran B, Assarsson E, Fredriksson S. Homogeneous antibody-based proximity extension assays provide sensitive and specific detection of low-abundant proteins in human blood. Nucleic Acids Res. 2011;39(15):e102–e102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


