ABSTRACT
Understanding how viruses and subviral agents initiate disease is central to plant pathology. Whether RNA silencing mediates the primary lesion triggered by viroids (small non-protein-coding RNAs), or just intermediate-late steps of a signaling cascade, remains unsolved. While most variants of the plastid-replicating peach latent mosaic viroid (PLMVd) are asymptomatic, some incite peach mosaics or albinism (peach calico, PC). We have previously shown that two 21-nt small RNAs (PLMVd-sRNAs) containing a 12–13-nt PC-associated insertion guide cleavage, via RNA silencing, of the mRNA encoding a heat-shock protein involved in chloroplast biogenesis. To gain evidence supporting that such event is the initial lesion, and more specifically, that different chloroses have different primary causes, here we focused on a PLMVd-induced peach yellow mosaic (PYM) expressed in leaf sectors interspersed with others green. First, sequencing PLMVd-cDNAs from both sectors and bioassays mapped the PYM determinant at one nucleotide, a notion further sustained by the phenotype incited by other natural and artificial PLMVd variants. And second, sRNA deep-sequencing and RNA ligase-mediated RACE identified one PLMVd-sRNA with the PYM-associated change that guides cleavage, as predicted by RNA silencing, of the mRNA encoding a thylakoid translocase subunit required for chloroplast development. RT-qPCR showed lower accumulation of this mRNA in PYM-expressing tissues. Remarkably, PLMVd-sRNAs triggering PYM and PC have 5ʹ-terminal Us, involving Argonaute 1 in what likely are the initial alterations eliciting distinct chloroses.
KEYWORDS: Chloroplast biogenesis, non-protein-coding RNAs, PLMVd, RNA silencing, thylakoid translocase, viroid pathogenesis
Introduction
One key question is how viruses, satellite RNAs and viroids incite plant diseases and, specifically, what is the molecular lesion initiating a signaling cascade ultimately resulting in symptoms. Due to their extreme simplicity, viroids appear very appropriate to tackle this question. They are circular, non-protein-coding RNAs of 246–434 nucleotides (nt), which infect and frequently cause disease in certain higher plants [1–6]. Those within the family Pospiviroidae, like potato spindle tuber viroid (PSTVd), have a central conserved region (CCR), lack hammerhead ribozymes and replicate in the nucleus through an asymmetric RNA-RNA rolling-circle mechanism [7,8]. In contrast, those of the family Avsunviroidae, do not have a CCR but can form hammerhead ribozymes that mediate co-transcriptional self-cleavage of the oligomeric RNAs of both polarities generated during replication in plastids (mostly chloroplasts) through a symmetric rolling-circle mechanism [9–12].
Viroids are targeted by the RNA silencing defensive response of their hosts [3]. RNA silencing, which additionally regulates gene expression and genome stability [13,14], is triggered in plants by double-stranded (ds)RNAs or structured single-stranded RNAs that are processed by Dicer-like (DCL) enzymes into small RNAs (sRNAs): micro RNAs (miRNAs, 21–22 nt) of endogenous origin, and small interfering RNAs (siRNAs, 21, 22 and 24 nt) of endogenous and foreign origin. The guide strand of miRNAs and siRNAs, incorporated into Argonaute (AGO) proteins [15,16] at the core of the RNA inducing silencing complex (RISC), inactivate their complementary RNA/DNA.
While most isolates of peach latent mosaic viroid (PLMVd, family Avsunviroidae) [17] are latent, some cause different peach mosaics (PM) or albinism (peach calico, PC). PC-inducing variants have a specific hairpin insertion of 12–13 nt and accumulate preferentially in symptomatic rather than in the surrounding green foliar sectors [18–20]. We have shown that PLMVd infection results in the accumulation of viroid-derived sRNAs (vd-sRNAs) [21], and that two 21-nt vd-sRNAs containing the PC-associated insertion (PC-sRNAs), acting like miRNAs, guide cleavage of the mRNA encoding the chloroplastic heat-shock protein 90 (cHSP90) as predicted by AGO1-mediated RNA silencing [3,22]. Such event possibly represents the primary alteration eventually resulting in symptoms. To provide further support for this concept and for the molecular basis underlying different PLMVd-induced chloroses, we have dealt in this work with one particular PM. This is a complex issue because it entails previous identification of the molecular determinant, not associated with any specific insertion.
Here we report that a characteristic peach yellow mosaic (PYM) is strictly associated with PLMVd variants with one single nucleotide change. Moreover, this symptomatology is most probably triggered by a 21-nt vd-sRNA containing the single nucleotide change, which directs cleavage via AGO1-mediated RNA silencing of the mRNA coding for a thylakoid translocase subunit required for chloroplast biogenesis. A similar mechanism may not necessarily operate in initiating pathogenesis in members of the family Pospiviroidae.
Results
Different PLMVd populations in green and PYM-expressing adjacent leaf sectors
We focused on one intense PYM expressed in leaf and young fruit sectors flanked by others asymptomatic (green) (Figure 1(a)). Such feature, which facilitated their dissection and further analysis of the corresponding PLMVd populations, turned out critical for our first purpose: identifying the PYM determinant.
Figure 1.

Peach yellow mosaic (PYM). (a) Symptoms in leaves and young fruits of peach trees cv. ‘O’Henry’. (b) Leaf phenotypes induced in GF-305 peach seedlings by the severe PLMVd variant y4. (c) Leaf phenotype induced in GF-305 peach seedlings by the mild/latent PLMVd variant y7. Notice that this variant, in contrast with the extended yellow phenotype induced by y4, causes no symptoms or (in some leaves) just small yellow sectors, in which the nucleotide substitution (U283) characteristic of y4 and strictly associated with PYM reappeared.
To gather data underpinning that the PYM observed in field trees was associated with and possibly incited by PLMVd, and not by another infectious agent or physiological disorder, nucleic acid preparations from yellow and green sectors (Figure S1) were subjected to RT-PCR with PLMVd-specific primers [23,24]. Non-denaturing polyacrylamide gel electrophoresis (PAGE) revealed products of the expected size (around 330 bp), which were cloned and sequenced. All clones corresponded to full-length PLMVd variants of 337–338 nt without the PC-associated 12–13-nt insertion [18], excluding that PYM might represent a sort of attenuated PC. A multiple alignment of seven and eight variants from the yellow (y) and green (g) sectors, respectively, revealed discriminating changes (Figure S2) mapping at different motifs in the PLMVd secondary structure (Figure S3). Thus, as in PC [19,20,22], PLMVd variants distribute unevenly in PYM-expressing leaves.
Only some PLMVd variants from the yellow sectors incite PYM
To establish a causal relationship between PYM and specific PLMVd variants, we selected four according to bioassays in GF-305 peach seedlings: y2, y4 and y7 (338 nt) from yellow sectors, and g7 (337 nt) from green sectors. Variants y4 and y7 differed in one nucleotide substitution, U283 (y4) and A283 (y7) (numbering according to y4, GenBank MK212036), while y2 (A283) and g7 (this position deleted), differed in additional changes between them and with y4 and y7 (Figure S2).
In vitro transcripts from recombinant plasmids containing dimeric head-to-tail cDNA inserts of the four variants were mechanically inoculated in blocks of eight peach seedlings. RT-PCR with PLMVd-specific primers, or dot-blot hybridization with a digoxigenin-labeled full-length riboprobe for detecting PLMVd (+) strands, showed that most plants became infected six-eight weeks post-inoculation. Variants y2 and g7 did not cause any reproducible phenotype, whereas y4 incited an extended yellow phenotype and y7 no symptoms or just small yellow sectors (Figure 1(b)), thus implicating PLMVd nucleotide at position 283 in eliciting PYM. RT-PCR, cloning and sequencing of the progenies further favored this view because: i) in the progeny of y4 the nucleotide substitution (U283) was preserved in 5/6 variants from the symptomatic sectors, but only in 1/6 and 0/6 of those from the flanking green sectors and from a fully green leaf of the same plant, respectively, ii) remarkably, in the progeny of y7 the PYM-associated nucleotide substitution reappeared in 5/5 of the variants from the small symptomatic sectors (Figure 1(c)), but not in any of the two sets of five variants from the surrounding green sectors and from a fully green leaf of the same plant, respectively, and iii) 0/7 and 0/6 variants from the progeny of y2 and g7, respectively, presented the PYM-associated nucleotide substitution. Altogether, these results supported a direct involvement of U283 in triggering PYM.
To provide additional data upholding this view, we introduced changes at this position in some PLMVd variants that were then bioassayed in blocks of eight peach seedlings for infectivity, symptom expression and progeny analysis. Dot-blot hybridization revealed that essentially all seedlings became infected. First, variant y4 (U283), serving as control, induced symptoms in all plants, with the progeny of symptomatic sectors showing a mixture of variants with U283, A283 and with this position deleted, while the progeny of green sectors only had variants with A283. Second, a mutated version of y4, in which U283 was deleted, incited no symptoms and all progeny variants presented the same deletion. Third, latent variant ls11 [23], with 10 changes with respect to y4 including U283 deletion, did not induce symptoms and its progeny contained only variants with the same deletion. And fourth, a mutated version of ls11, in which the U283 was inserted (and the U271 was mutated to A to keep the stability of the hybrid between the corresponding vd-sRNA and its mRNA target, see below), induced symptoms in all plants, with the progeny of symptomatic sectors displaying a mixture of variants with U283 and A283, while the progeny of green sectors was composed exclusively by variants with this position deleted. Collectively these data highlighted the key role of U283 in PYM. Further supporting this notion, an analysis of PLMVd variants bioassayed previously shows that those reported as latent, including ls1, ls8, ls14b, esc10 and esc 14 [23,24], and v1.1 [25], lack the U283.
Histological and cytological analyses provide clues on the PYM-associated chlorosis
Next, yellow and green sectors of the same leaves from a peach seedling infected with PYM variant y4, were examined by light and transmission electron microscopy and compared with those from a mock-inoculated seedling. Characteristic epidermal, columnar palisade and spongy mesophyll cell layers were observed in transversal sections in all instances, with those from the yellow and green sectors of the symptomatic leaves appearing slightly thinner than those from the mock-inoculated control (Figure 2(a)). Transmission electron microscopy of mesophyll cells from the yellow sectors of PYM-affected leaves showed irregularly-shaped plastids with thylakoids lacking the typical organization in grana and inter-grana, whereas such organization was maintained in plastids from the green sectors of the same leaf, although with slightly larger thylakoid interspaces than in the mock-inoculated control (Figure 2(b)). Interestingly, the ultra-structural alterations in PYM-affected leaves were different from those previously observed in PC-expressing leaves, where the columnar palisade was strongly disorganized and thylakoids almost absent [20].
Figure 2.
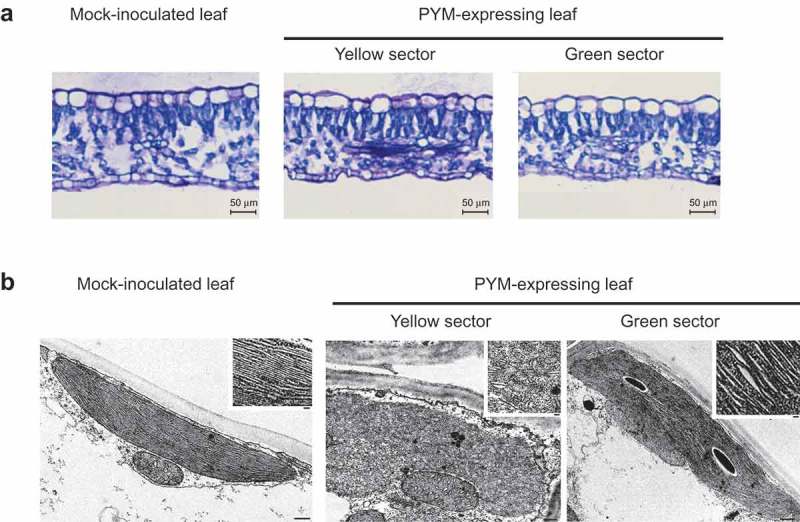
Light and transmission electron microscopy (TEM) images of leaf sections and plastids of GF-305 peach seedlings. (a) Transversal sections of a leaf from a mock-inoculated plant and from the yellow and green sectors of a leaf infected by variant PYM-y4. Layers corresponding to epidermal, columnar palisade and spongy mesophyll cells are visible in the three instances. In the symptomatic leaves the sections from the yellow and green sectors appear slightly thinner than that from the mock-inoculated control. (b) TEM of mesophyll cells of a PYM-expressing leaf shows in the yellow sectors irregularly shaped plastids with thylakoids without the characteristic organization in grana and inter-grana, while such ultra-structural organization is preserved in plastids from the green sectors of the same leaf, in which the thylakoid interspaces are moderately larger than in the mock-inoculated control (bars 500 nm). Insets show magnifications of the respective thylakoids (bars 100 nm).
PLMVd-sRNAs containing the PYM determinant accumulate preferentially in PYM-expressing sectors
Having mapped the pathogenic determinant of PYM, we addressed our second aim: how one single nucleotide change triggers this phenotypic alteration. If PYM is elicited via RNA silencing, as presumed for PC [22], a corollary is the accumulation in symptomatic tissue of (+) or (−) PLMVd-sRNAs with the PYM-associated substitution. To test this alternative, starting from an y4-infected peach seedling, we generated two PYM sRNA libraries from yellow and green leaf sectors hoping to enrich the libraries in vd-sRNAs with and without the PYM-associated substitution, respectively. We also generated another sRNAs library from a fully green leaf from the same plant, which should essentially lack vd-sRNAs with the PYM-associated substitution.
Reads for sRNAs between 18 and 26 nt (133,656,205 and 92,705,082 for the yellow and green leaf sectors, respectively, and 100,063,287 for the fully green leaf), displayed similar profiles with two 21- and 24-nt peaks (Figure S4). PLMVd-sRNA reads with zero or one mismatch with respect to (+) and (−) sequences of variant y4 (Figure S2) were 13,013,103 (9.7% of the total sRNAs) and 7,870,169 (8.5% of the total sRNAs) for the yellow and green leaf sectors, respectively, and 6,916,627 (6.9% of the total sRNAs) for the fully green leaf. Moreover, in agreement with our previous deep-sequencing data from GF-305 seedlings infected with other PLMVd variants [22]: i) vd-sRNAs of 21 nt, and to a lower extent of 22 nt, were dominant (Figure S5) and, ii) reads of vd-sRNAs of (+) and (−) polarity were similar in number (Figure S5). Mapping vd-sRNA reads of 21 nt along the PLMVd (+) and (−) RNAs revealed remarkably similar hot spot profiles in the three samples (Figure S6).
A screening for PLMVd-sRNAs containing the PYM-associated nucleotide disclosed 825, 284 and 209 non-redundant (+) and (−) 21-nt RNAs (2,825, 1,373 and 1,321 reads per million) in the yellow and green leaf sectors and in the fully green leaf, respectively. In addition, 634, 217 and 177 non-redundant (+) and (−) vd-sRNAs of 22 nt (3,255, 1,618 and 1,729 reads per million) were retrieved from the yellow and green leaf sectors and from the fully green leaf, respectively. Thus, the PLMVd region around the PYM-associated nucleotide is a source of vd-sRNAs (hereafter PYM-sRNAs), although we did not know whether one or more, acting like genuine miRNAs, might target and inactivate host mRNAs.
A search for PYM-sRNAs potentially targeting host mRNAs reveals several candidates
Drawing on the availability of the complete peach genome sequence, we applied the psRNATarget program to look for peach transcripts potentially targeted for inactivation, via RNA silencing, by (+) and (−) vd-sRNAs containing the PYM-associated U283 (or the complementary A283), and prevailing in yellow leaf sectors. We restricted our quest to 21-nt sRNAs, because this is the predominant size of PLMVd-sRNAs [22] (Figure S5) and plant miRNAs [26]. For predicting vd-sRNA targets, a position-dependent scoring matrix was applied to all potential duplexes adopting a score cut-off of 2.5, lower than the limit of 3.5 set in arabidopsis for credible specificity and sensitivity [27]. Furthermore, this cut-off appears reasonable considering that the duplexes between representative miRNAs retrieved from our libraries and conserved in other species, and their prospective targets in the complete peach genome, showed scores from 0 to 3 [22].
In total 20 PYM-sRNAs, 6 of (+) and 14 of (−) polarity, potentially targeting seven predicted peach mRNAs were found (Table 1). Two of the encoded proteins display a predicted chloroplastic transit peptide (cTP) according to ChloroP1.1 software. Therefore, PYM-sRNAs with plausible ability to target peach mRNAs are produced in PYM-expressing leaf sectors, opening up the prospect that some could function like miRNAs.
Table 1.
Predicted peach mRNAs, annotated as transcripts, targeted by PLMVd-sRNAs containing the pathogenic determinant for PYM strictly associated with U238 (or the complementary A238).
| sRNAa | sRNA 5´endb |
Viroid strand |
sRNA RPMc |
mRNA target:sRNA duplexd | mfee | Scoref | mRNA target accession numberg | Target predicted functiong | EST assembly of target transcriptg | Chloroplast localization of the encoded proteinh |
|---|---|---|---|---|---|---|---|---|---|---|
| PYM-sRNA15 | 269 | (+) | 109.7/9.0/3.7 | −34.8 | 2.5 | Prupe.1G480500 | Phospholipase D alpha1, hormonal action and stress-response | asmbl_18654.peachv21_pasa0 | No | |
| PYM-sRNA16 | 268 | (+) | 49.7/3.2/2.4 | −34.8 | 2.5 | Prupe.1G480500 | Phospholipase D alpha1, hormonal action and stress-response | asmbl_18654.peachv21_pasa0 | No | |
| PYM-sRNA16 | 268 | (+) | 49.7/3.2/2.4 | −27.7 | 2.5 | Prupe.2G151200 | MADS-box protein SOC1, RNA-polymerase II regulator | asmbl_7685.peachv21_pasa1 | No | |
| PYM-sRNA17 | 267 | (+) | 19.2/1.3/0.7 | −36.3 | 2.5 | Prupe.3G302400 | COP9 signalosome complex subunit 3 | asmbl_12131.peachv21_pasa2 | No | |
| PYM-sRNA18 | 266 | (+) | 0.6/0.1/0.0 | −36.2 | 1.5 | Prupe.3G302400 | COP9 signalosome complex subunit 3 | asmbl_12131.peachv21_pasa2 | No | |
| PYM-sRNA19 | 265 | (+) | 0.1/0.0/0.0 | −34.2 | 2.5 | Prupe.3G302400 | COP9 signalosome complex subunit 3 | asmbl_12131.peachv21_pasa2 | No | |
| PYM-sRNA41 | 284 | (−) | 0.1/0.0/0.0 | −35.4 | 2.0 | Prupe.3G002700 | Protein translocase subunit SecA1 | asmbl_124.peachv21_pasa2 | Yes | |
| PYM-sRNA40 | 285 | (−) | 14.6/0.8/0.5 | −35.6 | 1.5 | Prupe.3G002700 | Protein translocase subunit SecA1 | asmbl_124.peachv21_pasa2 | Yes | |
| PYM-sRNA39 | 286 | (−) | 64.1/3.2/1.6 | −35.7 | 2.5 | Prupe.3G002700 | Protein translocase subunit SecA1 | asmbl_124.peachv21_pasa2 | Yes | |
| PYM-sRNA38 | 287 | (−) | 31.2/2.5/1.3 | −35.0 | 2.0 | Prupe.3G002700 | Protein translocase subunit SecA1 | asmbl_124.peachv21_pasa2 | Yes | |
| PYM-sRNA37 | 288 | (−) | 16.1/1.6/0.7 | −33.1 | 2.0 | Prupe.3G002700 | Protein translocase subunit SecA1 | asmbl_124.peachv21_pasa2 | Yes | |
| PYM-sRNA32 | 293 | (−) | 12.8/0.9/0.3 | −26.2 | 2.5 | Prupe.8G251600 | RNA-binding protein BRN1, floral development, mRNA catabolism | asmbl_9675.peachv21_pasa7 | No | |
| PYM-sRNA29 | 296 | (−) | 4.8/0.7/0.2 | −38.0 | 2.0 | Prupe.1G553100 | RING domain-containing protein 2, proteins traffic across vacuole system | asmbl_21682.peachv21_pasa0 | Yes | |
| PYM-sRNA28 | 297 | (−) | 21.7/0.7/0.8 | −39.4 | 2.0 | Prupe.1G553100 | RING domain-containing protein 2, proteins traffic across vacuole system | asmbl_21682.peachv21_pasa0 | Yes | |
| PYM-sRNA27 | 298 | (−) | 65.6/2.4/1.9 | −38.3 | 1.5 | Prupe.1G553100 | RING domain-containing protein 2, proteins traffic across vacuole system | asmbl_21682.peachv21_pasa0 | Yes | |
| PYM-sRNA26 | 299 | (−) | 676.3/29.8/11.1 | −36.9 | 1.5 | Prupe.1G553100 | RING domain-containing protein 2, proteins traffic across vacuole system | asmbl_21682.peachv21_pasa0 | Yes | |
| PYM-sRNA26 | 299 | (−) | 676.3/29.8/11.1 | −33.5 | 2.0 | Prupe.8G265600 | Kinesin-like protein KIN-14I, ATPase in microtubules activity | asmbl_10074.peachv21_pasa7 | No | |
| PYM-sRNA725 | 300 | (−) | 0.6/0.1/0.0 | −36.9 | 2.0 | Prupe.1G553100 | RING domain-containing protein 2, proteins traffic across vacuole system | asmbl_21682.peachv21_pasa0 | Yes | |
| PYM-sRNA25 | 300 | (−) | 0.6/0.1/0.0 | −33.5 | 2.0 | Prupe.8G265600 | Kinesin-like protein KIN-14I, ATPase in microtubules activity | asmbl_10074.peachv21_pasa7 | No | |
| PYM-sRNA24 | 301 | (−) | 0.9/0.1 | −37.2 | 2.0 | Prupe.8G265600 | Kinesin-like protein KIN-14I, ATPase in microtubules activity | asmbl_10074.peachv21_pasa7 | No |
asRNAs containing U283/A283 in (+) and (−) polarities, respectively
bLocation of sRNA 5´-termini in the genomic RNA of variant y4
cRPM of sRNA in yellow sector/green sector/green leaf libraries
dmRNA:sRNA duplexes predicted by psRNATarget (Xinbin Dai et al., 2018); targeted host mRNAs are on top (5´-3´ orientation) and sRNAs below (3´-5´orientation); U283/A283 are in bold; |, Watson-Crick base pairs; o, G:U wobble base pairs; -, mismatches or bulges
eMinimum free energy (mfe, kcal mol−1) predicted by RNA hybrid (Rehmsmeier et al., 2004) for the mRNA target:sRNA duplex
fScore of the mRNA target:sRNA duplex, estimated by psRNATarget (Xinbin Dai et al., 2018); the lower the score, the more reliable the prediction
gAnnotations from Prunus persica Whole Genome Assembly v2.0 & Annotation v2.1 (v2.0.a1) (https://www.rosaceae.org/analysis/154)
hChloroplast targeting of the protein predicted by CHLOROP software (http://www.cbs.dtu.dk/sevices/ChloroP)
A PYM-associated sRNA targets for sequence-specific cleavage the mRNA encoding a thylakoid translocase subunit (cpSecA)
Of the potential candidates, we concentrated on peach transcript Prupe.3G002700.1 for the following reasons. First, the duplex between this transcript and PYM-sRNA40 presents one of the lowest scores (Table 1). Second, like the two PC-sRNAs [22], PYM-sRNA40 derives from the viroid (−) strand and has two 5ʹ-terminal Us, implying its preferential loading by AGO1 (see below). Third, the U283 (A283 in the minus polarity) associated with PYM is located in position 3 of PYM-sRNA40 (counting from its 5ʹ-end), a position that does not tolerate mismatches, G:U pairs or bulges in the hybrids formed by miRNAs and their target mRNAs [27]. Thus, the changes in U283 observed in certain PLMVd variants are consistent with their latent phenotype (Table S1). Moreover, while the U283 is necessary to incite PYM, additional nearby changes may weaken the hybrid between PLMVd-sRNA40 and its target mRNA and, thus, attenuate the intensity of this symptomatology. Those are the cases of variants y6 (Figure S2) (latent or mild green mosaic, data not shown), and gds3 and gds18 (green mosaics) [23], wherein a substitution at position 15 generates a mismatch that is disfavored in the hybrids formed by miRNAs and their target mRNAs [27] (Table S1). Fourth, the reads per million of PYM-sRNA40 were 14.6 in the yellow sector, 0.8 in the green sector and 0.5 in the fully green leaf of the same plant (Table 1). And fifth, peach transcript Prupe.3G002700.1 codes for a protein of 1036 amino acids annotated as homologous (86.9% amino acid identity) (Figure 3) with cpSecA, the ATPase subunit of the chloroplast Sec translocation machinery of arabidopsis (AT4G01800.1), a thylakoid protein essential for chloroplast biogenesis as revealed by the albino/glassy yellow phenotype displayed by a T-DNA insertion mutant of gene cpSecA [28]. Altogether, these data strongly suggested that PYM-sRNA40 down-regulates the expression of this gene by targeting for cleavage its mRNA.
Figure 3.

Comparison of chloroplast-targeted cpSecA from distinct species. Multiple alignment of the amino acid sequences of cpSecA from Prunus persica (ppcpSecA, Prupe.3G002700.1) and its homologues from Oryza sativa (OscpSecA, BAD44978), Pisum sativum (PscpSecA, CAA57798), Spinacia oleracea (SocpSecA, CAA88933) and the cyanobacterium Trichormus variabilis (TvSecA, ABA21744). High-affinity ATP-binding domains (a, b) and low-affinity ATP-binding domains (c, d), conserved among the prokaryotic SecA homologues, are indicated. Arrow marks the cleavage site of the chloroplast transit peptide of PpcpSecA predicted by ChloroP1.1. Sequences were aligned using Clustal Omega. Small and hydrophobic amino acids including the aromatic ones (AVFPMILW) are in red, acidic amino acids (DE) in blue, basic amino acids (RK) in magenta, and polar (STNQ) and aromatic (YH) amino acids together with cystein (C) and glycine (G) in green. (*) indicates positions with a conserved amino acid, (:) conservation between amino acid groups of strongly similar properties, and (.) conservation between amino acids groups of weakly similar properties.
To provide supporting quantitative data, we assessed by reverse transcription-quantitative polymerase chain reaction (RT-qPCR) the levels of the cpSecA and other mRNAs in yellow sectors of PYM-expressing leaves, in fully green leaves from the same plant, and in leaves of mock-inoculated peach seedlings. While no significant differences were detected between the green leaves of the mock-inoculated controls and of PYM-infected plants, significant changes were observed in the corresponding yellow sectors (Figure 4). Previously, the levels of the mRNAs coding for rbcL (the large subunit of the ribulose-1,5-bisphosphate carboxylase/oxygenase) transcribed by the plastid-encoded polymerase, and for rpoB (a subunit of the plastid-encoded polymerase) transcribed by a plastidic nuclear-encoded polymerase, were respectively found lower and higher in albino than in green tissues [20,22]. Resembling this situation, the levels of rbcL and rpoB transcripts were also lower and higher in PYM-expressing than in green tissues, respectively (Figure 4). However, the pronounced reduction of the mRNA encoding the cHSP90 (chloroplastic heat shock protein 90) in albino with respect to green tissues [22], was quite mild in PYM-expressing sectors. In contrast, a remarkable decrease of the cpSecA transcript was observed in PYM-expressing with respect to green tissues. These results are consistent with the prominent decrease of the cpSecA mRNA level being the primary alteration eliciting PYM.
Figure 4.
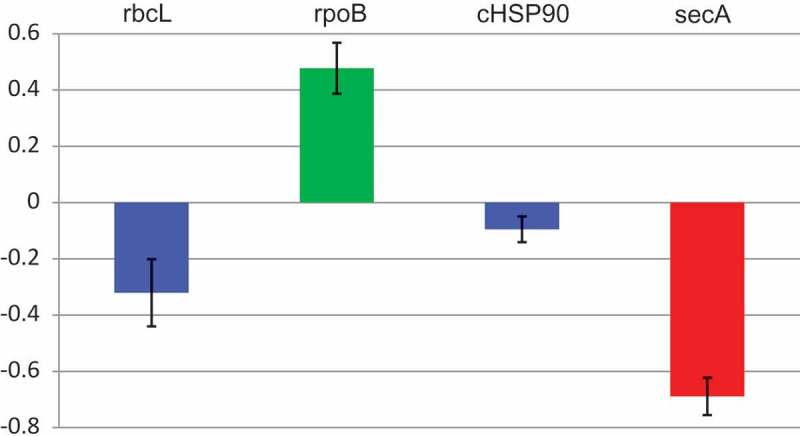
Levels of rbcL, rpoB, cHSP90 and cpSecA mRNAs in PYM-expressing and non-expressing tissues. RT-qPCR assessment of the transcript level, with respect to that of EF1α, of the four genes in yellow sectors of a PYM-inoculated plant relative to those of green leaves of the same plant. Results are presented in log10 scale. Bars indicate standard errors, with columns in blue denoting that transcript levels do not differ significantly at P ≤ 0.05 according to the Duncan’s multiple-range test, while those in green and red do.
To further validate that the lower level of the cpSecA transcript in PYM-expressing leaf sectors resulted from its targeting by PYM-sRNA40 for RISC(AGO1)-mediated cleavage, we examined by RNA ligase-mediated rapid amplification of cDNA ends (RLM-RACE) two RNA preparations from a peach seedling inoculated with PLMVd variant y4: one from fully green leaves and the other from yellow sectors of PYM-expressing leaves. While PAGE of the nested RT-PCR products failed to reveal any discernible band in the asymptomatic material, a band of the expected size was detected in the PYM-expressing material (Figure 5(a)). Moreover, cloning and sequencing of the amplified product revealed that 12/12 clones derived from the cpSecA transcript and that they had 5ʹ termini identical to the cleavage site predicted by PYM-sRNA40 (Figure 5(b)), between positions 10 and 11 from its 5ʹ terminus. These data support firmly that PYM-sRNA40 (and, importantly, no the other four PYM-sRNAs targeting the cpSecA transcript reported in Table 1) directs RISC(AGO1)-mediated cleavage of this transcript (Figure 5(c)), and that such a cleavage only occurs in PYM-expressing tissue, where PYM-sRNA40 accumulates preferentially. In summary, RNA silencing plays a major role in inciting the primary PYM lesion.
Figure 5.
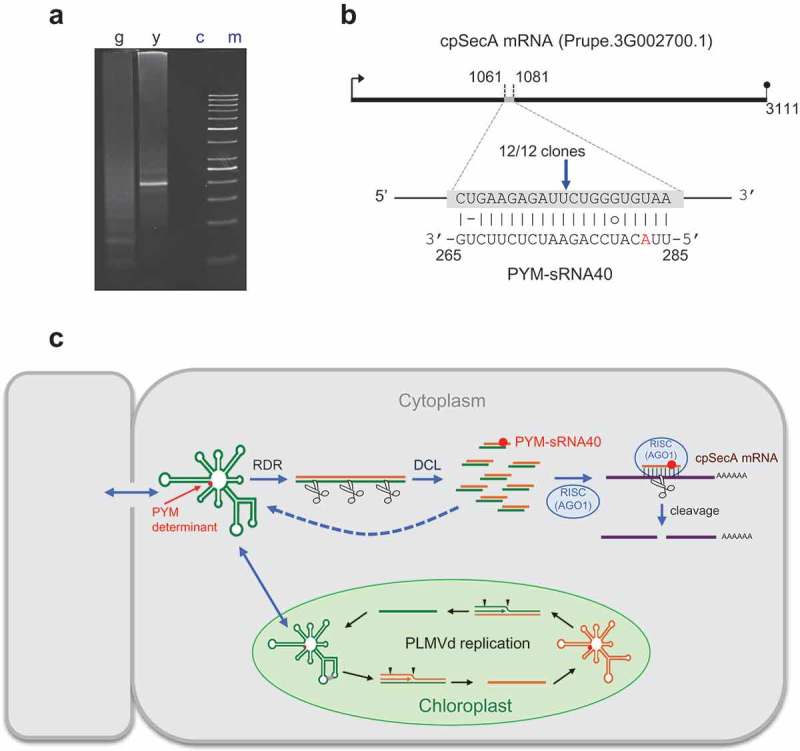
Site-specific cleavage of peach cpSecA mRNA guided by PYM-sRNA40. (a) RLM-RACE of two RNA preparations from a GF-305 peach seedling inoculated with PLMVd variant y4, one from fully green (g) leaves and the other from yellow (y) sectors of PYM-expressing leaves; (c) refers to a control without added template and (m) to the size markers. PAGE of the nested RT-PCR products reveals, only in the yellow sectors, a band of the expected size (195 bp), essentially co-migrating with the 200-bp DNA marker. (b) Duplex formed between the cpSecA mRNA and PYM-sRNA40; notice that the latter derives from the PLMVd (−) strand, with the nucleotide strictly associated with PYM (position 283 in PLMVd variant y4) highlighted in red. Arrow points to the predicted cleavage site (within the sixth exon) validated by RLM-RACE, with the fraction showing the number of independent clones producing identical results. (c) Model for the genesis of the PYM-sRNA40 (in the cytoplasm) that also includes PLMVd replication (within chloroplasts, with arrowheads marking hammerhead self-cleavage sites). RDR and DCL refer to RNA-directed RNA polymerase(s) and dicer-like ribonuclease(s), respectively. Green and yellow lines denote PLMVd (+) and (−) strands, respectively. PLMVd-sRNAs likely target the genomic viroid RNAs for their RISC(AGO)-mediated cleavage (with some fragments being transformed by RDR into additional DCL substrates); PYM-sRNA40 guides RISC(AGO1) for cleaving the cpSecA mRNA. Red dot indicates the PYM-associated nucleotide.
Discussion
Given the lack of protein-coding ability, early conjectures on how viroids initiate pathogenesis concentrated on a direct interaction of the genomic RNA (or its complementary strand) with host components, particularly proteins, which might be diverted from their normal physiological roles thus inciting disease. Later, with the discovery of RNA silencing, symptoms were proposed to result from vd-sRNAs targeting host DNA or mRNA sequences for their transcriptional or post-transcriptional gene silencing (PTGS), a view that could also explain the pathogenesis of non-protein-coding small satellite RNAs [[29,30], see for reviews [3,31]]. In the last context, the phenotype induced in tobacco by the yellow satellite RNA (Y-sat) of cucumber mosaic virus (CMV) is initially triggered by a 22-nt Y-sat sRNA comprising the pathogenic determinant dissected previously [32]. This sRNA hybridizes with a complementary sequence of the mRNA of the chlorophyll biosynthetic gene encoding the magnesium protoporphyrin chelatase subunit I (CHLI), and guides it for cleavage as predicted by RNA silencing (in the bond between positions 10 and 11 of the complementary Y-sat sRNA) [33,34]. A similar situation has been documented for two 21-nt sRNAs containing the insertion PC-associated [22]. Remarkably, these two 21-nt PC-sRNAs and the 22-nt Y-sat sRNA, besides complying with the criteria for functional sRNAs [3,35], have two or more Us at their 5ʹ termini and are most likely loaded into AGO1 that recruits specifically sRNAs with a 5ʹ-terminal U [36], including vd-sRNAs [37]. RISC(AGO1), guided by the Y-sat sRNA or PC-sRNAs, would catalyze cleavage of the CHLI and cHSP90 mRNAs, respectively [3,22,33], in line with its major role in PTGS [38–41]. Pertinent to this context is the high (79%) amino acid identity between AGO1 from arabidopsis and peach (data not shown). Moreover, arabidopsis AGO10, which also shares a high (86%) amino acid identity with its peach homolog (data not shown) and like AGO1 has a preference for endogenous and viral sRNAs with a 5ʹ-terminal U [42,43], does not bind PSTVd-sRNAs [37].
In comparing with PC [22], our present results on the PYM determinant appear different because: i) it is restricted to one nucleotide, instead of the PC-associated 12–13 nt insertion, and, ii) it maps far away from nucleotide change proposed to cause yellowing or chlorotic edge [44], although such data require further validation – PLMVd variants from symptomatic and asymptomatic tissue were not analyzed comparatively – as well as identification of the peach mRNAs hypothetically targeted by the vd-sRNAs with this polymorphism. On the other hand, because the most abundant PLMVd-sRNAs do not derive from the most structured regions predicted in the genomic (+) and (−) RNAs, it is unlikely that the latter could represent main DCL substrates. Instead, the viroid dsRNAs generated by an RNA-directed RNA polymerase(s) appear better candidates [3], but not the dsRNAs produced during replication because DCLs/AGOs have not been identified in plastids, wherein PLMVd replicates (Figure 5(c)) [20,45].
When attempting to identify the initial molecular lesion leading to PYM, we were inspired by our previous results with PC [22]. Given the differences between both chloroses, with the former being less dramatic than the latter (albinism), and with histological and ultra-structural observations showing differences between PC- and PYM-affected tissues [[20], this work], we presumed that the vd-sRNAs with the pathogenic determinants should target distinct peach mRNAs, although both encoding proteins directly related with chlorophyll metabolism. We also presumed that, like in PC, AGO1 should be involved in eliciting PYM and, consequently, that the guiding PYM-sRNA should contain one or more 5ʹ-terminal U. That is indeed the case, with the PYM-sRNA40 having two Us at its 5ʹ-terminus and, intriguingly, being of (−) polarity like the two PC-sRNAs [22]. And, as anticipated, the mRNA target of PYM-sRNA40 codes for a thylakoid translocase subunit (cpSecA) essential for photosynthetic development in Arabidopsis [28], and most likely in peach given the high amino acid identity between both proteins. Identification of the PYM determinant and, then, of the PYM-sRNA and its target mRNA, has been possible by focusing on the PLMVd variants accumulating in yellow and green sectors of the same leaf. Since the mRNAs targeted by PC- and PYM-sRNAs (and by Y-Sat-sRNA) code for proteins involved in chloroplast biogenesis, the initial molecular lesion and the expected phenotype (distinct chloroses) are connected by short signaling cascades that permit tracing a direct cause-effect relationship. We assume that other PLMVd-incited mosaics should result from vd-sRNAs (containing the specific pathogenic determinants) targeting for AGO1-mediated cleavage other mRNAs encoding chloroplastic proteins, and we already know that this is also the case for the chlorotic mottle induced by CChMVd (Serra et al., unpublished data).
The situation is less clear in members of the family Pospiviroidae, like PSTVd and a close relative, in which three groups have proposed that vd-sRNAs from their virulence modulating region (VMR) trigger symptoms by targeting three unrelated mRNAs [46–48]. In the most detailed of these studies [47], vd-sRNAs from the VMR are deemed to mediate cleavage of the tomato mRNAs encoding two callose-like synthases (CalS11-like and CalS12-like), thereby facilitating viroid movement through more permeable plasmodesmata (PD). Although this is an attractive hypothesis, there is no evidence that CalS11- and CalS12-like isozymes function at PD. Moreover, their arabidopsis homologs are not implicated in callose deposition at these intercellular bridges, wherein the responsible genes are CalS3, CalS7 and CalS10 [49]. On the other hand, most of the emphasis in this and in other cases has been put on systemic and non-specific symptoms, like stunting, which possibly result from a complex and late imbalance in growth hormones [47,50–53], making it difficult to establish a direct link between these symptoms and the down-regulation of the mRNA initially targeted by vd-sRNAs. Finally, the issue of the 5ʹ-terminal nucleotide of these vd-sRNAs and, hence, of the specific AGO(s) involved, has not been addressed. For similar reasons, even if global gene-expression and transcriptome/degradome analyses [[47,50–60], Navarro et al., unpublished data], or studies on changes in DNA methylation [61–63], offer valuable insights of genome-wide modifications caused by PSTVd and other members of the family Pospiviroidae at intermediate/late infection states, it is problematic that such investigations may provide clues on the initial pathogenic lesion, if indeed it is mediated by RNA silencing in members of this family.
Materials and methods
Plant material and inoculation
The initial leaf material was collected from field trees of peach [Prunus persica (L.) Batsch] cv. ‘O’Henry’ (Aragón, Spain) displaying intense PYM on leaves. Subsequently, blocks of eight young GF-305 peach seedlings were inoculated in Lanxade (France) and Valencia (Spain) by slashing their stems with buffer (50 mM K2HPO4) or dimeric head-to-tail transcripts generated in vitro from cDNA clones of natural and artificial PLMVd variants (GenBank accession numbers MK212033 to MK212093) resuspended in this buffer. Seedlings were kept in the glasshouse for six-eight weeks for symptom expression and sample collection.
Dot-blot hybridization, RT-PCR, cloning and sequencing
To examine infectivity of PLMVd variants by dot-blot hybridization, leaves (1 g) of the inoculated peach seedlings were processed for nucleic acid extraction and partial purification [64,65]. Nucleic acid preparations were resuspended in 50 μl of bidistilled water and aliquots of 5 μl and of a 1/5 dilution were applied onto positively-charged nylon membranes (Roche Diagnostics GmbH) that were hybridized with a full-length digoxigenin-labeled riboprobe complementary to PLMVd (+) strands [19].
For cloning the PLMVd-cDNAs, nucleic acids preparations (100 μl) were obtained from leaf portions (100 mg) of the initial material or of the inoculated peach seedlings using a silica-gel system [66]; when indicated, yellow and green sectors from symptomatic leaves were previously excised with a razor blade. RNA yield and integrity were assessed by PAGE in 5% gels. Aliquots (2.5 μl) were employed for RT (40 μl final volume) with 100 ng of the antisense primer RF43 (5ʹ-CTGGATCACACCCCCCTCGGAACCAACCGCT-3ʹ) and the reverse transcriptase Superscript II according to the supplier (Invitrogen). PCR of full-length PLMVd-cDNAs was performed using 1 μl of a 1/10 dilution of the RT reaction mixture, the same primer RF43 together with the sense primer RF44 (5ʹ-TGTGATCCAGGTACCGCCGTAGAAACT-3ʹ) (400 ng each) [23,24], and the Expand High Fidelity PCR system (Roche Diagnostics GmbH) in a final volume of 50 μl. The amplification profile consisted of 35 cycles of 30 s at 94ºC, 30 s at 60ºC, and 1 min at 72ºC, with a final extension of 10 min at 72ºC. Amplicons of the expected size were cloned into plasmid pTZ57R-T (Fisher Scientific) and sequenced by capillary electrophoresis using an ABI 3130XL apparatus (Life Technologies) and the Big Dye Terminator v3.1 cycle sequencing kit (Applied Biosystems).
Site-directed mutagenesis of PLMVd-cDNAs and transcript infectivity
Site-directed mutagenesis was performed as detailed previously [18,19] on recombinant plasmids bearing specific full-length PLMVd-cDNAs. The mutated inserts were PCR-amplified and those products of the expected monomeric length were ligated into dimers, which were confirmed to have proper head-to-tail orientation and only the desired mutations. The resulting recombinant plasmids were opened with pertinent restriction enzymes and transcribed in vitro with T7 or T3 RNA polymerases. Following examination by denaturing PAGE (in 1X TBE buffer and 8 M urea) and staining with ethidium bromide, the products were extracted with phenol, precipitated with ethanol, resuspended and inoculated by slashing young stems of peach seedlings.
Light and transmission electron microscopy
Small leaf pieces were fixed at 4°C overnight with 4% p-formaldehyde in 0.1 M sodium phosphate, pH 7.2, containing 0.1% Tween 20, dehydrated in ethanol, embedded in paraffin, and sectioned to 7 µm. Transverse leaf sections were attached to glass slides using poly-L-Lys, and stained with 1% (w/v) toluidine blue. 50 sections from each three leaves (150 in total) of mock-inoculated and PYM-expressing peach seedlings (the latter being dissected into green and yellow sectors) were examined. For sample visualization and photography a bright-field microscope (E600, Nikon) was used [20]. For thin sectioning, excised leaf pieces were processed as reported previously [20,67], and stained with lead citrate before observation with a Philip Morgagni 282D electron microscope. 30 sections from the same material indicated immediately above (90 in total) were analyzed.
RT-qPCR
RNA preparations were obtained with TRIzol (Invitrogen) of 100 mg of yellow sectors from PYM-expressing leaves and from fully green leaves of the same plant and of mock-inoculated peach seedlings, resuspended in 100 μl, and treated with 2 U of Turbo DNase (Ambion Life Technologies). Aliquots (2.5 μl) were mixed with 200 ng of random hexanucleotides (Isogen Life Sciences), heated at 95°C for 1.5 min, and reverse transcribed at 42°C for 1 h after adding 10 mM dithiothreitol (DTT), 0.5 mM dNTPs, 300 U of Superscript II and 20 U of placental ribonuclease inhibitor (PRI) (Fisher Scientific). Transcript levels were referred to those of translation elongation factor alpha (EF1α), usually taken as control in this kind of experiments [68]. The qPCR (in 20 μl with an annealing temperature of 60°C) was performed by mixing 0.6 μM of primers EF1α-F2 (5‘-GGTAACGGATATGCTCCAGTTC-3ʹ) and EF1α-R2 (5‘-TACCAGACCTCCTGTCAATCT-3ʹ), or cSecA-F3 (5‘-AACAGGGTTATGAAGACTCTGAAG-3ʹ) and cSecA-R3 (5‘-ACATAAGATGCCCACTGTTCTC-3ʹ), or rbcLF (5ʹ-AGGGAGATCACTTTAGGCTTTG-3ʹ) and rbcLR (5ʹ-CTGGCAAAGAGACCCAATCT-3ʹ), or rpoBF2 (5ʹ-GGTGGTAGAGGTCGAGTTATTG-3ʹ), and rpoBR2 (5ʹ-TTTCCATGTCTTCCAGCTACTT-3ʹ), or hsp90F: (5ʹ-GCTTCACTTCCTTCCTCATCTC-3ʹ) and hsp90R: (5ʹ-TGCTCTCAAGCTTCCACTTC-3ʹ) with 1 μl of the RT reactions and the SYBR green PCR master mix following supplier instructions (Applied Biosystems). Each qPCR was repeated at least by triplicate, on three biological replicas. Controls without template were carried out with each of the two primer pairs. Ct values were used to analyze relative gene expression in yellow sectors of PYM-expressing leaves, and in green leaves from the same plant and from mock-inoculated peach seedlings, as reported previously [69]. Relative quantification data were subjected to a logarithm transformation to resemble a normal distribution, and statistical significance was assessed by One-Way ANOVA with Duncan’s multiple-range test for comparison between groups [70]. Statistical analyses were performed using the Statgraphics Centurion software.
RLM-RACE
Aliquots (6.5 μl) of nucleic preparations obtained by the silica-gel system from yellow and green leaf sectors of PYM-expressing peach seedlings, pretreated with 2 U of Turbo-DNAse, were mixed with 0.25 μg of the RNA adaptor (5´-CGACUGGAGCACGAGGACACUGACAUGGACUGAAGGAGUAGAAA-3ʹ), heated at 65ºC for 5 min and snap-cooled on ice. The mixtures were then incubated at 20ºC for 6 h with 1 mM ATP, 10 U of PRI and 5 U of T4 RNA ligase in the buffer recommended by the supplier (Epicentre). Following phenol extraction, nucleic acids were precipitated with ethanol in the presence of 2 μg of glycogen (Roche Diagnostics GmbH) and resuspended in bidistilled water (30 μl). Aliquots (15 μl) were mixed with 10 ng of primer P2-R (5ʹ-GGCAGACCTTCTTTTGCTTC-3ʹ) (complementary to a segment of gen SecA), heated at 95ºC for 1.5 min and snap-cooled on ice. RT was at 42ºC for 1 h in a final volume (40 μl) containing 0.5 mM dNTPs, 10 mM DTT, 10 U of PRI and 300 U of Superscript II. After phenol extraction and ethanol precipitation, the products were resuspended in bidistilled water (40 μl). The first PCR was performed with an aliquot (20 μl), 400 ng each of primers P2-R-N (5ʹ-TGCTTGGTGAAGTCCATCAC-3ʹ) (complementary to a segment of gen SecA and internal with respect to P2-R) and RF-553 (5ʹ-TGGAGCACGAGGACACTGACATG-3ʹ) (homologous to the RNA adaptor), 0.4 mM dNTPs and 1 U of the Expand High Fidelity PCR system in a final volume of 50 μl with the cycling profile indicated above. Half of the reaction mixture was analyzed by PAGE and the expected product of ~230 bp was eluted and subjected to a second (nested) PCR with primers RF-1377 (5ʹ-ATCGCCTCCCCTGCATAACT-3ʹ) (complementary to a segment of gen SecA and internal with respect to P2-R-N) and RF-554 (5ʹ-GACACTGACATGGACTGAAGGAGTAG-3ʹ) (homologous to the RNA adaptor and internal with respect to RF-553). The product of ~195 bp observed by PAGE in the sample from the yellow leaf sector was eluted and cloned in the vector pTZ57R-T and the inserts were sequenced.
Sequencing and bioinformatic treatment of data
Full-length PLMVd-cDNA inserts were sequenced by capillary electrophoresis (see above), with the resulting sequences being aligned using Clustal Omega [71]. Purification of the sRNAs, ligation of bar-coded adaptors, RT-PCR, library purification and high-throughput DNA sequencing in the same channel (on the Illumina HiSeq 4000, Fasteris SA, http://www.fasteris.com) have been reported [22]. The three sRNA libraries sequenced were from PYM-expressing and green dissected leaf sectors of a peach seedling inoculated with variant y4, and from a fully green leaf from the same plant. To search for peach transcripts potentially targeted by (+) and (−) 21-nt vd-sRNAs with the U283 (or the complementary A283) associated with PYM, we applied the psRNATarget program [72] on exon sequence fragments of the complete peach genome (Peach v2.1, https://phytozome.jgi.doe.gov/pz/portal.html#!info?alias=Org_Ppersica) [73]. The quality of the hybrids between PYM-sRNAs and peach mRNAs was assessed as previously [27], with the latter being further analyzed with the ChloroP1.1 software (http://www. cbs.dtu.dk/services/ChloroP) [74] for the presence of a potential chloroplastic transit peptide (cTP) in the encoded proteins.
Abbreviations
- AGO
argonaute
- CCR
central conserved region
- PAGE
polyacrylamide gel electrophoresis
- PD
plasmodesmata
- PTGS
post-transcriptional gene silencing
- PLMVd
peach latent mosaic viroid
- PSTVd
potato spindle tuber viroid
- RISC
RNA inducing silencing complex
- RLM-RACE
RNA ligase-mediated rapid amplification of cDNA ends
- RT-qPCR
reverse transcription-quantitative polymerase chain reaction
- vd-sRNAs
viroid-derived small RNAs
Funding Statement
This work was funded by grant BFU2014-56812-P (to R.F.) from the Ministerio de Economía y Competitividad of Spain (MINECO). S.D. and P.S. were partly supported by postdoctoral contracts from MINECO. We thank the technical assistance of Amparo Ahuir and María Pedrote in handling the plants. We apologize to colleagues whose work was not quoted for space limitations.
Authors contribution
S.D., B.N., P.G. and A.D.S. performed the experiments. P.S. and M.C. analyzed part of the data. M.A.C. provided the intial material. B.N., F.D.S. and R.F. conceived the project, and supervised the experiments. R.F. wrote the article with the inputs of all authors.
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplementary material
Supplemental data for this article can be accessed here.
References
- [1].Diener TO. Discovering viroids-a personal perspective. Nat Rev Microbiol. 2003;1:75–80. [DOI] [PubMed] [Google Scholar]
- [2].Kovalskaya N, Hammond RW. Molecular biology of viroid-host interactions and disease control strategies. Plant Sci. 2014;228:48–60. [DOI] [PubMed] [Google Scholar]
- [3].Flores R, Minoia S, Carbonell A, et al. Viroids, the simplest RNA replicons: how they manipulate their hosts for being propagated and how their hosts react for containing the infection. Virus Res. 2015;209:136–145. [DOI] [PubMed] [Google Scholar]
- [4].Rao AL, Kalantidis K. Virus-associated small satellite RNAs and viroids display similarities in their replication strategies. Virology. 2015;479-480:627–636. [DOI] [PubMed] [Google Scholar]
- [5].Hadidi A, Flores R, Randles JW, et al, editors. Viroids and satellites. Boston: Academic Press; 2017. [Google Scholar]
- [6].Steger G, Riesner D. Viroid research and its significance for RNA technology and basic biochemistry. Nucleic Acids Res. 2018;46:10563–10576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Branch AD, Benenfeld BJ, Robertson HD. Evidence for a single rolling circle in the replication of potato spindle tuber viroid. Proc Natl Acad Sci USA. 1988;85:9128–9132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Feldstein PA, Hu Y, Owens RA. Precisely full length, circularizable, complementary RNA: an infectious form of potato spindle tuber viroid. Proc Natl Acad Sci USA. 1998;95:6560–6565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Daròs JA, Flores R. Arabidopsis thaliana has the enzymatic machinery for replicating representative viroid species of the family Pospiviroidae. Proc Natl Acad Sci USA. 2004;101:6792–6797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Hutchins C, Rathjen PD, Forster AC, et al. Self-cleavage of plus and minus RNA transcripts of avocado sunblotch viroid. Nucleic Acids Res. 1986;14:3627–3640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Daròs JA, Marcos JF, Hernández C, et al. Replication of avocado sunblotch viroid: evidence for a symmetric pathway with two rolling circles and hammerhead ribozyme processing. Proc Natl Acad Sci USA. 1994;91:12813–12817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Carbonell A, De la Peña M, Flores R, et al. Effects of the trinucleotide preceding the self-cleavage site on eggplant latent viroid hammerheads: differences in co-and post-transcriptional self-cleavage may explain the lack of trinucleotide AUC in most natural hammerheads. Nucleic Acids Res. 2006;34:5613–5622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Carthew RW, Sontheimer EJ. Origins and mechanisms of miRNAs and siRNAs. Cell. 2009;136:642–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Axtell MJ. Classification and comparison of small RNAs from plants. Annu Rev Plant Biol. 2013;64:137–159. [DOI] [PubMed] [Google Scholar]
- [15].Mallory A, Vaucheret H. Form, function, and regulation of ARGONAUTE proteins. Plant Cell. 2010;22:3879–3889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Fang X, Qi Y. RNAi in plants: an argonaute-centered view. Plant Cell. 2016;28:272–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Hernández C, Flores R. Plus and minus RNAs of peach latent mosaic viroid self-cleave in vitro through hammerhead structures. Proc Natl Acad Sci USA. 1992;89:3711–3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Malfitano M, Di Serio F, Covelli L, et al. Peach latent mosaic viroid variants inducing peach calico contain a characteristic insertion that is responsible for this symptomatology. Virology. 2003;313:492–501. [DOI] [PubMed] [Google Scholar]
- [19].Rodio ME, Delgado S, Flores R, et al. Variants of peach latent mosaic viroid inducing peach calico: uneven distribution in infected plants and requirements of the insertion containing the pathogenicity determinant. J Gen Virol. 2006;87:231–240. [DOI] [PubMed] [Google Scholar]
- [20].Rodio ME, Delgado S, De Stradis AE, et al. A viroid RNA with a specific structural motif inhibits chloroplast development. Plant Cell. 2007;19:3610–3626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Martínez de Alba AE, Flores R, Hernández C. Two chloroplastic viroids induce the accumulation of the small RNAs associated with post-transcriptional gene silencing. J Virol. 2002;76:13094–13096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Navarro B, Gisel A, Rodio ME, et al. Small RNAs containing the pathogenic determinant of a chloroplast-replicating viroid guide the degradation of a host mRNA as predicted by RNA silencing. Plant J. 2012;70:991–1003. [DOI] [PubMed] [Google Scholar]
- [23].Ambrós S, Hernández C, Desvignes JC, et al. Genomic structure of three phenotypically different isolates of peach latent mosaic viroid: implications of the existence of constraints limiting the heterogeneity of viroid quasi-species. J Virol. 1998;72:7397–7406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Ambrós S, Hernández C, Flores R. Rapid generation of genetic heterogeneity in progenies from individual cDNA clones of peach latent mosaic viroid in its natural host. J Gen Virol. 1999;80:2239–2252. [DOI] [PubMed] [Google Scholar]
- [25].Serra P, Bertolini E, Martínez MC, et al. Interference between variants of peach latent mosaic viroid reveals novel features of its fitness landscape: implications for detection. Sci Rep. 2017;7:42825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Rogers K, Chen X. Biogenesis, turnover, and mode of action of plant microRNAs. Plant Cell. 2013;25:2383–2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Fahlgren N, Carrington JC. miRNA target prediction in plants. Methods Mol Biol. 2010;592:51–57. [DOI] [PubMed] [Google Scholar]
- [28].Liu D, Gong Q, Ma Y, et al. cpSecA, a thylakoid protein translocase subunit, is essential for photosynthetic development in Arabidopsis. J Exp Bot. 2010;61:1655–1669. [DOI] [PubMed] [Google Scholar]
- [29].Papaefthimiou I, Hamilton AJ, Denti MA, et al. Replicating potato spindle tuber viroid RNA is accompanied by short RNA fragments that are characteristic of post-transcriptional gene silencing. Nucleic Acids Res. 2001;29:2395–2400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Wang MB, Bian XY, Wu LM, et al. On the role of RNA silencing in the pathogenicity and evolution of viroids and viral satellites. Proc Natl Acad Sci USA. 2004;101:3275–3280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Wang MB, Masuta C, Smith NA, et al. RNA silencing and plant viral diseases. Mol Plant-Microbe Interact. 2012;25:1275–1285. [DOI] [PubMed] [Google Scholar]
- [32].Masuta C, Takanami Y. Determination of sequence and structural requirements for pathogenicity of a cucumber mosaic virus satellite RNA (Y-satRNA). Plant Cell. 1989;1:1165–1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Shimura H, Pantaleo V, Ishihara T, et al. A viral satellite RNA induces yellow symptoms on tobacco by targeting a gene involved in chlorophyll biosynthesis using the RNA silencing machinery. PLoS Pathog. 2011;7:e1002021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Smith NA, Eamens AL, Wang MB. Viral small interfering RNAs target host genes to mediate disease symptoms in plants. PLoS Pathog. 2011;7:e1002022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Iwakawa HO, Tomari Y. Molecular insights into microRNA-mediated translational repression in plants. Mol Cell. 2013;52:591–601. [DOI] [PubMed] [Google Scholar]
- [36].Mi S, Cai T, Hu Y, et al. Sorting of small RNAs into Arabidopsis argonaute complexes is directed by the 5’ terminal nucleotide. Cell. 2008;133:116–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Minoia S, Carbonell A, Di Serio F, et al. Specific ARGONAUTES bind selectively small RNAs derived from potato spindle tuber viroid and attenuate viroid accumulation in vivo. J Virol. 2014;88:11933–11945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Morel JB, Godon C, Mourrain P, et al. Fertile hypomorphic ARGONAUTE (ago1) mutants impaired in post-transcriptional gene silencing and virus resistance. Plant Cell. 2002;14:629–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Baumberger N, Baulcombe DC. Arabidopsis ARGONAUTE1 is an RNA slicer that selectively recruits microRNAs and short interfering RNAs. Proc Natl Acad Sci USA. 2005;102:11928–11933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Qi Y, Denli AM, Hannon GJ. Biochemical specialization within Arabidopsis RNA silencing pathways. Mol Cell. 2005;19:421–428. [DOI] [PubMed] [Google Scholar]
- [41].Carbonell A, Carrington JC. Antiviral roles of plant ARGONAUTES. Curr Opin Plant Biol. 2015;27:111–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Zhu H, Hu F, Wang R, et al. Arabidopsis Argonaute10 specifically sequesters miR166/165 to regulate shoot apical meristem development. Cell. 2011;145:242–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Garcia-Ruiz H, Carbonell A, Hoyer JS, et al. Roles and programming of Arabidopsis ARGONAUTE proteins during turnip mosaic virus infection. PLoS Pathog. 2015;11:e1004755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Wang LP, He Y, Kang YP, et al. Virulence determination and molecular features of peach latent mosaic viroid isolates derived from phenotypically different peach leaves: a nucleotide polymorphism in L11 contributes to symptom alteration. Virus Res. 2013;177:171–178. [DOI] [PubMed] [Google Scholar]
- [45].Bussière F, Lehoux J, Thompson DA, et al. Subcellular localization and rolling circle replication of peach latent mosaic viroid: hallmarks of group A viroids. J Virol. 1999;73:6353–6360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Eamens AL, Smith NA, Dennis ES, et al. In Nicotiana species, an artificial microRNA corresponding to the virulence modulating region of potato spindle tuber viroid directs RNA silencing of a soluble inorganic pyrophosphatase gene and the development of abnormal phenotypes. Virology. 2014;450-451:266–277. [DOI] [PubMed] [Google Scholar]
- [47].Adkar-Purushothama CR, Brosseau C, Giguère T, et al. Small RNA derived from the virulence modulating region of the potato spindle tuber viroid silences callose synthase genes of tomato plants. Plant Cell. 2015;8:2178–2194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Avina-Padilla K, Martínez de la Vega O, Rivera-Bustamante R, et al. In silico prediction and validation of potential gene targets for pospiviroid-derived small RNAs during tomato infection. Gene. 2015;564:197–205. [DOI] [PubMed] [Google Scholar]
- [49].De Storme N, Geelen D. Callose homeostasis at plasmodesmata: molecular regulators and developmental relevance. Front Plant Sci. 2014;5:138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Owens RA, Tech KB, Shao JY, et al. Global analysis of tomato gene expression during Potato spindle tuber viroid infection reveals a complex array of changes affecting hormone signaling. Mol Plant Microbe Interact. 2012;25:582–598. [DOI] [PubMed] [Google Scholar]
- [51].Katsarou K, Wu Y, Zhang R, et al. Insight on genes affecting tuber development in potato upon potato spindle tuber viroid (PSTVd) infection. PLoS One. 2016;11:e0150711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Zheng Y, Wang Y, Ding B, et al. Comprehensive transcriptome analyses reveal that potato spindle tuber viroid triggers genome-wide changes in alternative splicing, inducible trans-acting activity of phased secondary small interfering RNAs, and immune responses. J Virol. 2017;91:e00247–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Aviña-Padilla K, Rivera-Bustamante R, Kovalskaya NY, et al. Pospiviroid infection of tomato regulates the expression of genes involved in flower and fruit development. Viruses. 2018;10:E516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Itaya A, Matsuda Y, Gonzales RA, et al. Potato spindle tuber viroid strains of different pathogenicity induces and suppresses expression of common and unique genes in infected tomato. Mol Plant-Microbe Interact. 2002;15:990–999. [DOI] [PubMed] [Google Scholar]
- [55].Kappagantu M, Bullock JM, Nelson ME, et al. Hop stunt viroid: effect on host (Humulus lupulus) transcriptome and its interactions with hop powdery mildew (Podospheara macularis). Mol Plant Microbe Interact. 2017;30:842–851. [DOI] [PubMed] [Google Scholar]
- [56].Pokorn T, Radišek S, Javornik B, et al. Development of hop transcriptome to support research into host-viroid interactions. PLoS One. 2017;12:e0184528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Xia C, Li S, Hou W, et al. Global transcriptomic changes induced by infection of cucumber (Cucumis sativus L.) with mild and severe variants of hop stunt viroid. Front Microbiol. 2017;8:2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Mishra AK, Kumar A, Mishra D, et al. Genome-wide transcriptomic analysis reveals insights into the response to citrus bark cracking viroid (CBCVd) in hop (Humulus lupulus L.). Viruses. 2018;10:570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Olivier T, Bragard C. Innate immunity activation and RNAi interplay in citrus exocortis viroid-tomato pathosystem. Viruses. 2018;10:587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Więsyk A, Iwanicka-Nowicka R, Fogtman A, et al. Time-course microarray analysis reveals differences between transcriptional changes in tomato leaves triggered by mild and severe variants of potato spindle tuber viroid. Viruses. 2018;10:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Martínez G, Castellano M, Tortosa M, et al. A pathogenic non-coding RNA induces changes in dynamic DNA methylation of ribosomal RNA genes in host plants. Nucleic Acids Res. 2014;42:1553–1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Lv DQ, Liu SW, Zhao JH, et al. Replication of a pathogenic non-coding RNA increases DNA methylation in plants associated with a bromodomain-containing viroid-binding protein. Sci Rep. 2016;6:35751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Torchetti EM, Pegoraro M, Navarro B, et al. A nuclear-replicating viroid antagonizes infectivity and accumulation of a geminivirus by upregulating methylation-related genes and inducing hypermethylation of viral DNA. Sci Rep. 2016;6:35101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Dellaporta SL, Wood J, Hicks JB. A plant DNA minipreparation: version II. Plant Mol Biol Rep. 1983;1:19–21. [Google Scholar]
- [65].Pallás V, Navarro A, Flores R. Isolation of a viroid-like RNA from hop different from hop stunt viroid. J Gen Virol. 1987;68:3201–3205. [Google Scholar]
- [66].Foissac X, Svanella-Dumas L, Dulucq MJ, et al. Polyvalent detection of fruit tree tricho, capillo and foveaviruses by nested RT-PCR using degenerated and inosine containing primers (PDO RT-PCR). Acta Hortic. 2001;550:37–43. [Google Scholar]
- [67].Martelli GP, Russo M. Use of thin sectioning for visualization and identification of plant viruses. Methods Virol. 1984;8:143–224. [Google Scholar]
- [68].Liu D, Shi L, Han C, et al. Validation of reference genes for gene expression studies in virus-infected Nicotiana benthamiana using quantitative real-time PCR. PLoS One. 2012;7:e46451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2–∆∆Ct method. Methods. 2001;25:402–408. [DOI] [PubMed] [Google Scholar]
- [70].Bewick V, Cheek L, Ball J. Statistics review 9: one-way analysis of variance. Crit Care. 2004;8:130–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Sievers F, Wilm A, Dineen DG, et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 2011;7:539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Dai X, Zhuang Z, Zhao PX. psRNATarget: a plant small RNA target analysis server (2017 release). Nucleic Acids Res. 2018;46(W1):W49–W54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].The International Peach Genome Initiative The high-quality draft genome of peach (Prunus persica) identifies unique patterns of genetic diversity, domestication and genome evolution. Nat Genet. 2013;45:487–494. [DOI] [PubMed] [Google Scholar]
- [74].Emanuelsson O, Nielsen H, von Heijne G. ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci. 1999;8:978–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


