ABSTRACT
Population bottlenecks often cause strong reductions in genetic diversity and alter population structure. In the context of host-parasite interactions, bottlenecks could in theory benefit either the host or the pathogen. We predicted that bottlenecking of bacterial populations that evolve CRISPR immunity against bacteriophages (phage) would benefit the pathogen, because CRISPR spacer diversity can rapidly drive phages extinct. To test this, we bottlenecked populations of bacteria and phage, tracking phage persistence and the evolution of bacterial resistance mechanisms. Contrary to our prediction, bottlenecking worked in the advantage of the host. With some possible exceptions, this effect was not caused by CRISPR immunity. This host benefit is consistent with a dilution effect disproportionately affecting phage. This study provides further insight into how bottlenecking influences bacteria-phage dynamics, the role of dilution in bacteria-phage interactions, and the evolution of host immune systems.
KEYWORDS: CRISPR-Cas, bacteriophage, bottlenecks, diversity-generating mechanisms, host-pathogen coevolution, dilution effect
Introduction
Many bacteria encode CRISPR-Cas immune systems [Clustered Regularly Interspaced Short Palindromic Repeats, CRISPR-associated], a RNA-guided mechanism used to defend against phage infection [1]. In response to phage infection, CRISPR-Cas immune systems can incorporate short DNA fragments of about 30 base pairs derived from the phage genome into CRISPR loci on the host genome, termed spacers [2,3]. CRISPR transcripts are processed into small RNAs (crRNA) that bind to CRISPR-associated (Cas) proteins, which guide the recognition and cleavage of complementary nucleic acid sequences. In the case of reinfection by the same phage genotype (or a related phage genotype carrying the cognate sequence), CRISPR-Cas systems can target and cleave the invading phage genome, preventing successful re-infections [4–6]. CRISPR loci in bacterial populations are often diverse [7–9]. High levels of CRISPR spacer diversity naturally evolve in bacterial populations with type I-F CRISPR-Cas systems due to primed spacer acquisition [10]. Priming relies on a partial match between a pre-existing spacer and the phage genome [9,11], and causes an increase in the rate of spacer acquisition. Increasing CRISPR diversity can contribute to synergistic host benefits and cause increasingly rapid phage extinction [12–14].
The levels of genetic diversity in populations of hosts and parasites are known to play a key role in determining the spread and evolution of infectious diseases. For example, host genetic diversity can limit the spread of pathogens (reviewed in [15,16]) whereas parasite genetic diversity can increase the ability of parasites to adapt to local hosts [17]. Population bottlenecks – characterised by sudden, often repeated and usually drastic reductions in population size – are common in host-pathogen populations and can strongly reduce their genetic diversity [18,19]. Bottlenecks in host-pathogen systems can be levied by clinical treatment, such as antibiotics [20]; as part of the normal infection cycle of a pathogen (e.g. Lyme Disease [21], HIV-1 [22], Hepatitis C virus [23], reviewed in [24]; or sudden changes in the abiotic environment, such as soil structure [25].
Bottlenecks can influence host-pathogen interactions in several ways. Bottlenecking can benefit the pathogen, as the reduction in host diversity increases the ability of pathogens to adapt to overcome host defenses [17,26,27]. Conversely, bottlenecks can limit the spread of parasites by a dilution effect [28,29], where low host density reduces contact rate between susceptible individuals (e.g. [30–32]), or by stochastic fixation of resistance alleles [33,34]. The only experimental test of bottlenecking in a bacteria-bacteriophage (phage) system supported a host benefit, which the authors suggest was due to highly-frequent resistant host phenotypes being more likely to pass through successive bottlenecks and fix in the population [35]. Dennehy et al. [30] also found that wildtype Pseudomonas phaselicola, which are susceptible to phage, benefited from a dilution effect when in mixed cultures with a pilus mutant strain that prevents phage attachment. However, bottlenecking would be expected to generally work in the advantage of the parasite when hosts rely on a diversity-generating immune mechanism [36] such as CRISPR.
Here we set out to explore the consequences of bottlenecking in the context of CRISPR using P. aeruginosa strain PA14, which has a type I-F CRISPR-Cas system and readily evolves CRISPR immunity against its phage DMS3vir [9,10]. Given that the benefits of CRISPR immunity can depend on the levels of population-level spacer diversity in a pre-immunized host population [14], we predicted that increased bottlenecking of an initially susceptible host population, where CRISPR diversity evolves upon phage infection, could cause a breakdown in host diversity, removing the synergistic benefits of CRISPR, and hence cause an increase in phage persistence.
Results
To understand the effect of bottlenecking on host and phage population dynamics, we infected either the WT PA14 strain or PA14 csy3::lacZ strain, which carries an inactive CRISPR-Cas system (hereafter referred to as ΔCRISPR) with phage DMS3vir in liquid media and transferred daily into fresh medium while manipulating the bottleneck size by varying the dilution factor at each daily transfer from 10–2 (weakest bottleneck) to 10–9 (strongest bottleneck). Ancestral WT PA14 carries no spacers targeting DMS3vir, so is sensitive to infection.
Surprisingly, phage went (close to) extinct in all treatments by 5 days post-infection (d.p.i.), irrespective of the bottleneck strength (Fig. 1), and importantly CRISPR background had no significant effect on phage titres (F1,334 = 2.12, p = 0.15), although there appeared to be a transient effect at the 10−4 and 10−5 bottleneck treatments (see Discussion). Phage extinction risks were also almost identical between these host genetic backgrounds for all bottlenecks (Table S1). Although there was a significant effect of CRISPR background on host titres (F1,432 = 143.2, p < 0.0001), this was likely due to the transiently higher average host densities at 2 d.p.i. in all treatments (Fig. 2) (see Discussion). Adjusted R2 comparisons of nested models showed that the effect of CRISPR background explained only 10% more variance than a model excluding the CRISPR background interaction. Otherwise, host dynamics within comparable treatments were similar in CRISPR and ΔCRISPR backgrounds. These data therefore suggest that CRISPR-Cas systems overall have a negligible and transient impact on the short-term phage population dynamics under the bottlenecking regimes that were explored here.
Figure 1.
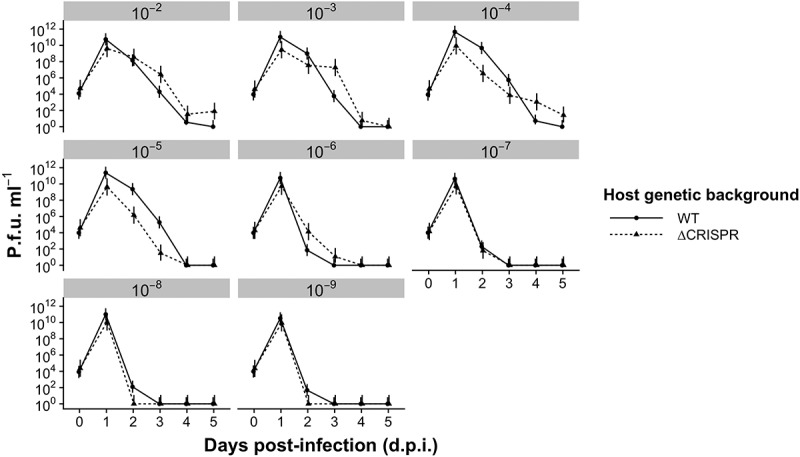
Phage population dynamics when both host and phage are bottlenecked. Mean plaque-forming units (p.f.u.) ml–1 for the WT host (i.e. encoding a functional CRISPR-Cas immune system) and the ΔCRISPR strain are shown for different bottleneck treatments (ranging from 10−2–10−9 dilutions at each transfer, as indicated above each panel). The detection limit is 200 p.f.u. ml−1. Error bars correspond to 95% confidence intervals (CIs). N = 6 for all treatments.
Figure 2.
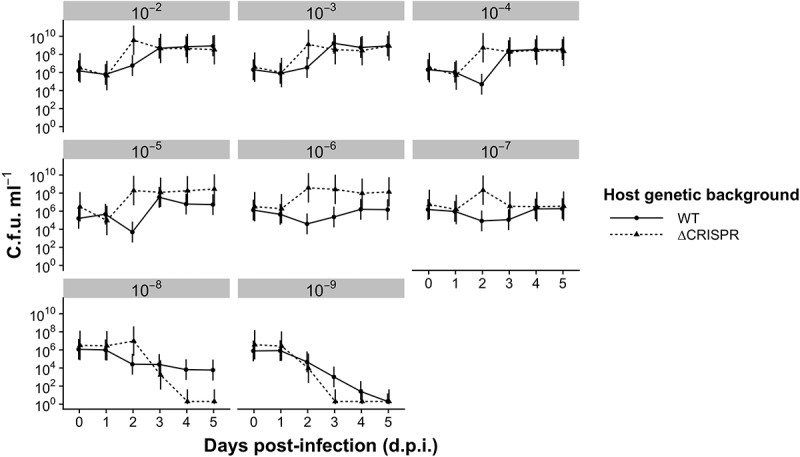
Host population dynamics when both host and phage are bottlenecked. Mean colony-forming units (c.f.u.) ml–1 for the WT host (i.e. encoding a functional CRISPR-Cas immune system) and the ΔCRISPR strain are shown for different bottleneck treatments (ranging from 10−2–10−9 dilutions at each transfer, as indicated above each panel). The detection limit is 200 c.f.u. ml−1. Error bars correspond to 95% confidence intervals (CIs). N = 6 for all treatments.
Although the presence or absence of a functional CRISPR-Cas system did not have a strong impact on the phage population dynamics, the bottlenecking regime itself did have a clear impact with stronger bottlenecks being associated with a more rapid decline in phage titers. This is supported by a control without phage, which shows that host densities remain relatively stable through all but the strongest bottleneck treatments (Fig. S1). We also ruled out any abiotic effects of fresh media that may have disproportionately impacted phage survival ([37]; Fig. S2). We hypothesized this might be caused by a dilution effect, i.e. a decrease in host densities resulting in reduced parasite spread [16,17]. To explicitly test this, we first selected two bottleneck treatments (10–4 and 10–6) that showed clear differences in phage titers over time. We then set up an experiment as described above with WT PA14 and DMS3vir, and at 1 d.p.i. diluted the culture 100-fold and transferred daily 0.6 µl into either 6 ml or 600 ml of fresh media, generating either 10–4 or 10–6 dilutions while maintaining the same degree of bottlenecking on the population. We found that the small and large dilutions of bacterial populations led to a qualitatively similar phage population dynamics as those associated with the 10–4 and 10–6 bottleneck treatments, respectively (Fig. 3). Phage persisted in most small dilution replicates until 4 d.p.i, while in the large dilution it was driven extinct in all but one replicate by 3 d.p.i. Host persisted for the duration of the experiment in the small dilution treatment, but were undetectable in 5 out of 6 large dilution replicates by 3 d.p.i., and in 3 out of 6 replicates of the 10–6 bottleneck treatment, also by 3 d.p.i. These data therefore support the conclusion that phage population dynamics observed in the bottleneck experiment were driven primarily by dilution of the phage. Interestingly, phage titre did not covary significantly with host density in the bottleneck (F68,247 = 1.06, p = 0.37) or dilution experiment (F13,98 = 0.43, p = 0.96), which suggests that phage dynamics are not correlated with those of the host, and consequently are likely more negatively affected by bottlenecking than the host.
Figure 3.
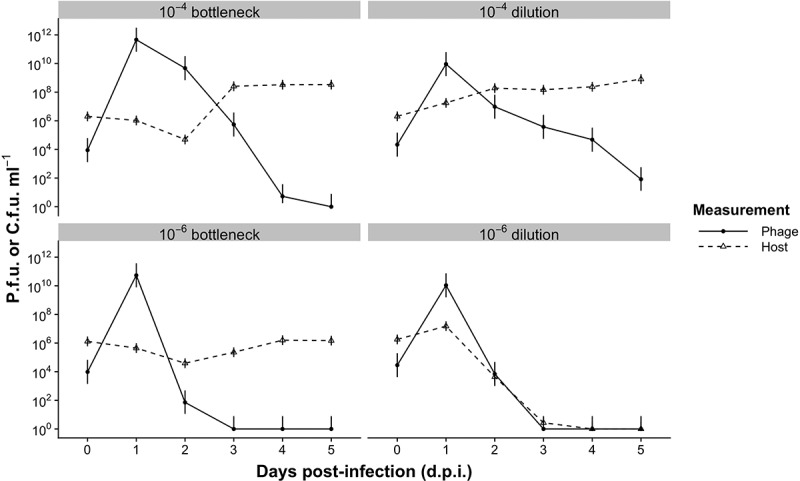
Phage and host dynamics when both WT host and phage are diluted or bottlenecked. A 10−4 or 10−6 bottleneck corresponds to either 0.6 µl or 0.006 µl culture, respectively, in 6 ml of fresh media at each transfer. A 10–4 dilution corresponds to 0.6 µl culture in 6 ml at each transfer, and a 10−6 dilution corresponds to 0.6 µl culture in 600 ml at each transfer (see Materials & methods). Mean plaque-forming units (p.f.u.) ml–1 and colony-forming units (c.f.u.) for the dilution and comparable bottleneck treatments are shown. The detection limit is 200 p.f.u. or c.f.u. ml−1. Error bars correspond to 95% confidence intervals (CIs). N = 6 for all treatments.
Given the observed effect of bottlenecking on the phage population dynamics, we investigated whether this could have knock-on effects for the evolution of host resistance to the phage, and the mechanistic basis for resistance evolution. For consistency with previous studies [9], we examined the phage resistance phenotypes at 3 d.p.i. Hosts with CRISPR immunity were most frequent in the 10−2–10−5 bottleneck treatments, which all contained detectable levels of phage at 3 d.p.i. However, bottlenecks of 10–6 – 10−9 contained mostly sensitive hosts (Fig. 4A & Table S2), which we hypothesized was due to the lower levels of phage in those treatments. A qualitatively similar pattern in the evolution of immune phenotypes was observed in the dilution experiment (Fig. S3 & Table S4). This shift from CRISPR immunity to sensitive bacteria was therefore potentially due to relaxed selection for host immunity as a result of phage dilution. To test this idea, performed a similar experiment where we bottlenecked cultures (both WT and the ΔCRISPR strain) in the same way as described above but with a fixed dilution of 10−2 for the phage. This experimental design therefore results in bottlenecking and dilution of the hosts while maintaining a similar phage pressure across the bottleneck treatments. Under these conditions, CRISPR immunity was maintained across the range of treatments and there was no invasion of sensitives (Fig. 4B & Table S3). There was again no significant covariance between phage titre and host density in this control (F65,250 = 1.23, p = 0.14), so that even when phage levels were maintained through each bottleneck, host and phage dynamics were not correlated. Although sensitive invasion did not occur in a ΔCRISPR background when phage were supplemented, intriguingly there was also no sensitive invasion when both host and phage were bottlenecked (Fig. S4).
Figure 4.
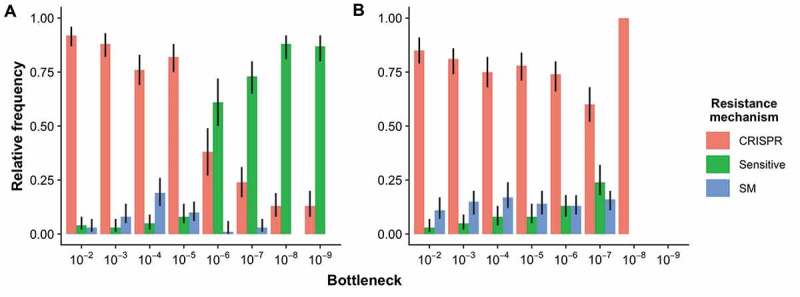
Evolution of host resistance of the WT strain. Relative frequencies of bacterial clones with CRISPR immunity, surface mutation (SM) resistance, or sensitive phenotypes, at 3 days post-infection when (A) both host and phage were bottlenecked by dilution into fresh medium as indicated on the x-axis s and (B) host was bottlenecked by dilution into fresh medium as indicated on the x-axis and phage was bottlenecked at a fixed 10−2 dilution at each transfer. Error bars correspond to 95% confidence intervals (CIs) of the mean. N = 72 for the 10−6 treatment in panel A, and N = 48 for the 10−8 treatment in panel B. No bacterial cells were recovered from any replicate in the 10−9 treatment in panel B. N = 144 for all other treatments.
Discussion
Population bottlenecks often cause strong reductions in genetic diversity [19]. In the context of host-parasite interactions, bottlenecks can in theory benefit either the host [33,34] or the pathogen [17,26,27] by affecting host-pathogen coevolution. Bottlenecks may also benefit the host through a dilution effect [28,29].While there is limited empirical support for a dilution effect in a bacteria-phage system [30], and that bottlenecking benefits bacterial hosts [35], we predicted that bottlenecking of bacterial populations that evolve CRISPR immunity against phages would benefit the pathogen [36], because CRISPR spacer diversity can rapidly drive phages extinct [14].
In contrast to our predictions, bottlenecking did not provide a clear advantage to the phage. Instead, phage were always driven extinct by 5 d.p.i. irrespective of the degree of population bottlenecking. Surprisingly, this effect was not CRISPR-specific but seemed to be driven instead by bottlenecking per se, as suggested by similar phage dynamics in the context of a CRISPR-knockout strain. Exploring the potential role of a dilution effect through bottlenecking, we found that dilution alone is sufficient to drive phage extinct in a CRISPR background. In support of this, phage density was uncoupled from host density in all experiments and phage extinction was more rapid after 10–6 bottlenecks in cultures with surviving host. Both findings are consistent with dilution disproportionately affecting phage. This biased effect on phage may be related to the different resource requirements of bacteria and phage. After passing through even very small bottlenecks, bacterial cells can still replicate if environmental resources are available. By contrast, phage cannot replicate independent of hosts – they require a sufficient number and density of host cells to recover after a bottleneck has been applied. Such an imbalance in resource dependence between bacteria and phage, coupled with a reduction in numbers of individual hosts, might explain why phage were more susceptible to dilution. While dilution of hosts can suppress phage epidemics, this will quantitatively depend on phage life history traits, such as adsorption rates, latent period and burst size of the phages. It would be interesting therefore to further explore how these factors affect phage persistence and evolution in the context of host dilution.
From the perspective of the bacterial host, a dilution effect relaxed selection for CRISPR immunity and allowed sensitive hosts to invade, particularly in the stronger bottleneck treatments of 10–6 and above. However, we did not see a similar invasion of sensitives in the ΔCRISPR background. It is unclear why sensitives did not invade, but we speculate that it might be attributable to either hosts with CRISPR immunity losing their resistance [38,39], or the CRISPR fraction of the host population ‘protecting’ sensitive hosts at the start of an infection [40], allowing them to invade after already dilute phage have been cleared. When phage were supplemented, sensitives could not invade and CRISPR immunity was most relatively frequent.
Finally, although our results are generally consistent with a dilution being the most important determinant of the phage population dynamics, we did detect some transient effects which are likely driven by CRISPR immunity. First, transiently higher average host densities at 2 d.p.i. in all treatments (Fig. 2) may at least be partially due to a difference in how quickly hosts with CRISPR immunity versus surface modification (SM) invade the culture. Second, in the experiment where only the host was bottlenecked (resulting in high levels of CRISPR resistance evolution at all treatments), phage titres were statistically similar across bottleneck treatments at all timepoints for the WT bacteria with functional CRISPR-Cas (Fig. S5A), but not for the ΔCRISPR bacteria (Fig. S5B). Third, in the experiment where the whole culture was bottlenecked, phage titres in the 10–4 and 10–5 treatments were consistently significantly higher at most timepoints compared to the ΔCRISPR background (Fig. 1), supported by a one-way ANOVA of the effect of host background on phage titre across the whole 6-point time series (F1,36 = 8.92, p < 0.01). These phage titres were maintained despite 10,000- and 100,000-fold reductions of phage immediately after the bottlenecking had been applied, suggesting that phage were still able to successfully amplify on sensitive hosts. High phage titres in these treatments occurred in the context of high relative frequency of CRISPR immunity in the host, raising the possibility of an interaction between CRISPR, host density, and bottlenecking of a certain degree that appears to temporarily favour phage. This suggests that within a certain range of bottleneck treatments CRISPR defenses may become less effective in driving phages extinct, either because of population dynamics or evolutionary effects, or both.
Altogether, this study provides further insight into how bottlenecking influences bacteria-phage dynamics and the evolution of host resistance. Bottlenecking does impact the ability of phage to coexist with the host, but this is generally independent of whether or not the host has a functional CRISPR-Cas immune system. Instead, our findings support the role of a dilution effect caused by bottlenecking that disproportionately affects phage titre and survival. However, we also find some CRISPR-specific effects under certain conditions that may be related to a complex interaction between host immune systems and host-pathogen dynamics.
Materials & methods
Bacterial strains & phage
The bacterial strains and phages used in this study have all been described previously. Evolution experiments were carried out using Pseudomonas aeruginosa UCBPP-PA14 (WT) and phage DMS3vir [41]. We used P. aeuruginosa UCBPP-PA14 csy3::lacZ [42] in CRISPR-negative control experiments and streak assays, and DMS3vir-AcrF1 [14] in streak assays.
Bottleneck experiments
All experiments were established in glass vials by inoculating 6 ml of M9 minimal media (supplemented with 0.2% glucose) with ~1 x 106 c.f.u. (colony-forming units) from an overnight culture of WT P. aeruginosa PA14. Approximately 1 × 104 p.f.u. (plaque-forming units) of DMS3vir was added to each glass vial. A phage-negative control was established similarly but without the addition of phage. A CRISPR-negative control was also established by inoculating 6 ml of M9 minimal media with ~1 x 106 c.f.u. of P. aeruginosa PA14 csy3::lacZ and ~1 x 104 of DMS3vir. At this point (0 d.p.i.), 180 μl of culture was taken from each vial and phage was extracted using chloroform. To determine phage titres, extracted phage was then serially diluted eight times in 96-well plates, and 5 μl of each phage dilution was spotted on a top lawn of P. aeruginosa PA14 csy3::lacZ. The detection limit of phage spot assays is 102 p.f.u. ml–1. Samples of culture were serially diluted in M9 minimal media, plated on LB agar and incubated overnight at 37°C. C.f.u.s were counted to determine host densities. The vials were then incubated at 37°C while shaking at 180 rpm.
After 24 hrs of growth (1 d.p.i.) the phage and host sampling protocol, described above, was repeated. To generate bottleneck treatments, samples of culture were then serially diluted in M9 minimal media from 10–1, 10–2, …10–7. 60 μl from undiluted culture and each dilution were then used to inoculate 6 ml of fresh M9 minimal media. This method of dilution and transfer gives eight bottleneck treatments of 10–2, 10–3, …10–9. To ensure only the host was bottlenecked in the phage-supplemented control, we added 60 μl of chloroform-extracted phage from the corresponding replicate into each inoculated microcosm. Each bottleneck was performed in six independent replicates (N = 6). Host and phage sampling, as well as the bottlenecking procedure in each treatment, was repeated 24 hrs after growth in fresh medium until 5 d.p.i. Samples of the culture were taken prior to bottlenecking.
Dilution experiment
We explicitly tested the effect of culture dilution on population dynamics and immunity evolution. Twelve glass vials with 6 ml M9 minimal media were established with WT P. aeruginosa PA14 and DMS3vir as described above, and the host and phage were assayed. We then established two dilution treatments that corresponded to the 10–4 and 10–6 bottleneck treatments, treatments which showed clear differences in phage titers over time. At 1 d.p.i., all twelve cultures were serially diluted. Six vials (N = 6) of 6 ml M9 minimal media were inoculated with 60 μl of the 10–2 dilution (0.0006 ml/6 ml = 1 x 10–4). Six bottles (N = 6) of 600 ml M9 minimal media were also inoculated with 60 μl of the 10–2 dilution (0.0006 ml/600 ml = 1 x 10–6). Sampling (as described above) and dilution were repeated 24 hrs after growth in fresh medium until 5 d.p.i. Samples of the culture were taken prior to dilution.
Determining host immune phenotype
Bacterial immunity against ancestral phage was determined at 3 d.p.i, using two independent assays as described in Westra et al [9]. First, bacteria were plated on LB-agar, and 24 randomly-selected individual clones per replicate were streaked through either DMS3vir or DMS3vir-AcrIF1, which encodes an anti-CRISPR gene. Clones sensitive to both phage genotypes were scored as ‘sensitive’; those resistant to the DMS3vir but sensitive to DMS3vir-AcrIF1 were scored as ‘CRISPR resistant’; and those resistant to both were scored as ‘surface mutant (SM)’. Second, each clone was also grown for 24 hrs in M9 media in the presence or absence of DMS3vir, and the OD600 was measured. Cultures that had a reduced growth rate were scored as sensitive.
Statistical analysis
All statistical analyses were conducted in R [43]. We took a nested Generalized Linear Model (GLM) approach, and model selection was performed using Aikaike’s Information Criteria (AIC) [44,45]. Models of phage titre and host densities used log-transformed residuals to fit the assumption of normality, and coefficients were back-transformed prior to presentation. Models of phenotype relative frequency used a binomial family with a logit link, and logit coefficients were back-transformed to probability values. We used a Cox proportional hazards model to assess the effect of bottleneck size on phage survivorship over the course of an experiment, with hazard ratio coefficients expressing the relative risk of phage extinction over time. ‘Bottleneck’ was treated as a discrete variable in all models. The package ggplot2 was used to generate graphics [46].
Funding Statement
JC acknowledges the Biotechnology and Biological Sciences Research Council funded South West Biosciences Doctoral Training Partnership (https://www.bbsrc.ac.uk/) (BB/J014400/1 & BB/M009122/1) for funding. ERW acknowledges the Natural Environment Research Council (http://www.nerc.ac.uk) (NE/M018350/1), the BBSRC (BB/N017412/1) and the European Research Council (https://erc.europa.eu) (ERC STG-2016-714478 - EVOIMMECH) for funding.
Acknowledgments
We would like to thank Elze Hesse for advice on survival analyses and comments on the draft of the paper.
Disclosure Potential Conflicts of Interest
No potential conflict of interest were disclosed.
Supplementary material
Supplemental data for this article can be accessed here.
References
- [1].van Houte S, Buckling A, Er W.. Evolutionary ecology of prokaryotic immune mechanisms. Microbiol Mol Biol Rev. 2016;80(3):745–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Semenova E, Jore MM, Datsenko KA, et al. Interference by clustered regularly interspaced short palindromic repeat (CRISPR) RNA is governed by a seed sequence. Proc Nat Acad Sci. 2011;108(25):10098–10103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Swarts DC, Mosterd C, Van Passel MW, et al. CRISPR interference directs strand specific spacer acquisition. PloS one. 2012;7(4):e35888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Brouns SJJ, Jore MM, Lundgren M, et al. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science. 2008;321(5891):960–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Garneau JE, Dupuis M-E, Villion M, et al. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature. 2010;468(7320):67–71. [DOI] [PubMed] [Google Scholar]
- [6].Datsenko KA, Pougach K, Tikhonov A, et al. Molecular memory of prior infections activates the CRISPR/Cas adaptive bacterial immunity system. Nat Commun. 2012;3:945. [DOI] [PubMed] [Google Scholar]
- [7].Andersson AF, Banfield JF. Virus population dynamics and acquired virus resistance in natural microbial communities. Science. 2008;320(5879):1047–1050. [DOI] [PubMed] [Google Scholar]
- [8].Paez-Espino D, Morovic W, Sun CL, et al. Strong bias in the bacterial CRISPR elements that confer immunity to phage. Nat Commun. 2013;4:1430. [DOI] [PubMed] [Google Scholar]
- [9].Westra Edze R, van Houte S, Oyesiku-Blakemore S, et al. Parasite exposure drives selective evolution of constitutive versus inducible defense. Curr Biol. 2015;25(8):1043–1049. [DOI] [PubMed] [Google Scholar]
- [10].Richter C, Dy RL, McKenzie RE, et al. Priming in the Type IF CRISPR-Cas system triggers strand-independent spacer acquisition, bi-directionally from the primed protospacer. Nucleic Acids Res. 2014;42(13):8516–8526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Cady KC, O’Toole GA. Non-identity-mediated CRISPR-bacteriophage interaction mediated via the Csy and Cas3 proteins. J Bacteriol. 2011;193(14):3433–3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Childs LM, England WE, Young MJ, et al. CRISPR-induced distributed immunity in microbial populations. PLoS One. 2014;9(7):e101710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Morley D, Broniewski JM, Westra ER, et al. Host diversity limits the evolution of parasite local adaptation. Mol Ecol. 2017;26(7):1756–1763. [DOI] [PubMed] [Google Scholar]
- [14].van Houte S, Ekroth AKE, Broniewski JM, et al. The diversity-generating benefits of a prokaryotic adaptive immune system. Nature. 2016;532(7599):385–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Ashby B, King KC. Diversity and the maintenance of sex by parasites. J Evol Biol. 2015;28(3):511–520. [DOI] [PubMed] [Google Scholar]
- [16].Jackson SA, McKenzie RE, Fagerlund RD, et al. CRISPR-Cas: adapting to change. Science. 2017;356(6333):eaal5056. [DOI] [PubMed] [Google Scholar]
- [17].Gandon S, Michalakis Y. Local adaptation, evolutionary potential and host–parasite coevolution: interactions between migration, mutation, population size and generation time. J Evol Biol. 2002;15(3):451–462. [Google Scholar]
- [18].Bohannan BJ, Lenski RE. Linking genetic change to community evolution: insights from studies of bacteria and bacteriophage. Ecol Lett. 2000;3(4):362–377. [Google Scholar]
- [19].Hudson PJ, Dobson AP, Newborn D. Prevention of population cycles by parasite removal. Science. 1998;282(5397):2256–2258. [DOI] [PubMed] [Google Scholar]
- [20].Flanagan J, Brodie E, Weng L, et al. Loss of bacterial diversity during antibiotic treatment of intubated patients colonized with Pseudomonas aeruginosa. J Clin Microbiol. 2007;45(6):1954–1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Rego RO, Bestor A, Štefka J, et al. Population bottlenecks during the infectious cycle of the lyme disease spirochete Borrelia burgdorferi. PLoS One. 2014;9(6):e101009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Gijsbers EF, Schuitemaker H, Kootstra NA. HIV-1 transmission and viral adaptation to the host. Future Virol. 2012;7(1):63–71. [Google Scholar]
- [23].Bull RA, Luciani F, McElroy K, et al. Sequential bottlenecks drive viral evolution in early acute hepatitis C virus infection. PLoS Pathog. 2011;7(9):e1002243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Gutiérrez S, Michalakis Y, Blanc S. Virus population bottlenecks during within-host progression and host-to-host transmission. Curr Opin Virol. 2012;2(5):546–555. [DOI] [PubMed] [Google Scholar]
- [25].Torsvik V, Øvreås L. Microbial diversity and function in soil: from genes to ecosystems. Curr Opin Microbiol. 2002;5(3):240–245. [DOI] [PubMed] [Google Scholar]
- [26].Morgan AD, Gandon S, Buckling A. The effect of migration on local adaptation in a coevolving host–parasite system. Nature. 2005;437(7056):253–256. [DOI] [PubMed] [Google Scholar]
- [27].Morran LT, Schmidt OG, Gelarden IA, et al. Running with the red queen: host-parasite coevolution selects for biparental sex. Science. 2011;333(6039):216–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Keesing F, Holt RD, Ostfeld RS. Effects of species diversity on disease risk. Ecol Lett. 2006;9(4):485–498. [DOI] [PubMed] [Google Scholar]
- [29].Ostfeld RS, Keesing F. Effects of host diversity on infectious disease. Annu rev ecol evol syst. 2012;43. [Google Scholar]
- [30].Dennehy JJ, Friedenberg NA, Yang YW, et al. Virus population extinction via ecological traps. Ecol Lett. 2007;10(3):230–240. [DOI] [PubMed] [Google Scholar]
- [31].Mitchell CE, Tilman D, Groth JV. Effects of grassland plant species diversity, abundance, and composition on foliar fungal disease. Ecology. 2002;83(6):1713–1726. [Google Scholar]
- [32].Roscher C, Schumacher J, Foitzik O, et al. Resistance to rust fungi in Lolium perenne depends on within-species variation and performance of the host species in grasslands of different plant diversity. Oecologia. 2007;153(1):173–183. [DOI] [PubMed] [Google Scholar]
- [33].Gokhale CS, Papkou A, Traulsen A, et al. Lotka-Volterra dynamics kills the red queen: population size fluctuations and associated stochasticity dramatically change host-parasite coevolution. BMC Evol Biol. 2013;13(1):254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Quigley BJ, López DG, Buckling A, et al. The mode of host–parasite interaction shapes coevolutionary dynamics and the fate of host cooperation. Proc R Soc B. 2012;279(1743):3742–3748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Hesse E, Buckling A. Host population bottlenecks drive parasite extinction during antagonistic coevolution. Evolution. 2016;70(1):235–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Westra ER, Sunderhauf D, Landsberger M, et al. Mechanisms and consequences of diversity-generating immune strategies. Nat Rev Immunol. 2017;17(11):719–728. [DOI] [PubMed] [Google Scholar]
- [37].Jończyk E, Kłak M, Międzybrodzki R, et al. The influence of external factors on bacteriophages. Folia Microbiol (Praha). 2011;56(3):191–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Weissman JL, Holmes R, Barrangou R, et al. Immune loss as a driver of coexistence during host-phage coevolution. ISME J. 2018;12(2):585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Chaudhry WN, Pleška M, Shah NN, et al. Leaky resistance and the conditions for the existence of lytic bacteriophage. PLoS Biol. 2018;16(8):e2005971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Payne P, Geyrhofer L, Barton NH, et al. CRISPR-based herd immunity can limit phage epidemics in bacterial populations. eLife. 2018;7:e32035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Cady KC, Bondy-Denomy J, Heussler GE, et al. The CRISPR/Cas adaptive immune system of Pseudomonas aeruginosa mediates resistance to naturally occurring and engineered phages. J Bacteriol. 2012;194(21):5728–5738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Zegans ME, Wagner JC, Cady KC, et al. Interaction between bacteriophage DMS3 and host CRISPR region inhibits group behaviors of Pseudomonas aeruginosa. J Bacteriol. 2009;191(1):210–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].R Core Team R: A language and environment for statistical computing. 3.4.1. “single candle” ed. Vienna, Austria: R Foundation for Statistical Computing; 2017. [Google Scholar]
- [44].Akaike H. Information theory and an extension of the maximum likelihood principle In: Selected papers of hirotugu akaike. New York: Springer; 1973. p. 199–213. [Google Scholar]
- [45].Burnham KP, Anderson DR. Multimodel inference: understanding AIC and BIC in model selection. Sociol Methods Res. 2004;33(2):261–304. [Google Scholar]
- [46].Wickham H. ggplot2: elegant graphics for data analysis. New York: Springer-Verlag; 2009. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


