ABSTRACT
Circular RNAs (circRNAs) represent a new type of regulatory RNA which forms a covalently closed continuous loop from back-splicing events, a process in which the downstream 5′ splice site and the 3′ splice site are covalently linked. Emerging evidence indicates that circRNAs exert a new layer of transcriptional and post-transcriptional regulation of gene expression. However, there is no standard nomenclature of circRNA, although the study of circRNAs has exploded in the past few years. Here we present circbank (www.circbank.cn), a comprehensive database for human circRNAs, where a novel naming system of circRNAs based on the host genes of circRNAs was implemented. In addition to the new naming system, circbank collected other five features of circRNAs including the miRNA binding site, conservation of circRNAs, m6A modification of circRNAs, mutation of circRNAs and protein-coding potential of circRNAs. Circbank is publicly available and allows users to query, browse and download circRNAs with all six features we provided, based on different search criteria. The database may serve as a resource to facilitate the research of function and regulation of circRNAs.
KEYWORDS: CircRNAs, database, nomenclature, circbank
Introduction
Circular RNAs (circRNAs) constitute a class of RNAs that is characterized by a covalently closed cyclic structure lacking poly-adenylated tails [1,2]. circRNAs are generally expressed at low levels and often exhibit cell type-specific and tissue-specific patterns [3–5]. Although the function of most circRNA remains largely unknown, increasing evidence showed that circRNAs can function as a microRNA sponge, RNA binding protein sponge, modulator of transcription and splicing and even coding RNA of translation potential [6–8]. More and more studies of circRNAs have shown that circRNAs are linked to physiological development and various diseases [9–11]. In addition, circRNAs have the potential to be used as biomarkers of diseases due to their stability, specific expression and relation to diseases both in cells and in extracellular fluid [12–15].
Increasing evidences have shown that circRNAs interact with miRNA and function as miRNA sponges through the ceRNA network [16–18]. The ceRNA hypothesis proposes that specific RNAs can impair miRNAs activity through sequestration of miRNAs like a sponge, thereby modulating the expression of miRNAs target genes [19–21]. CDR1as is the best example of circRNA as the sponge of miRNA, which contains more than 60 conserved binding sites for miR-7 [4,19,21]. Inhibition of CDR1as expression caused the reduced expression of miR-7 targeting mRNAs, indicating that CDR1as competes for miR-7 binding as miR-7 sponges to participate in the gene expression network. In addition to CDR1as, a few other circRNAs in mammals are known to function as potential miRNA sponges. For example, the testis-specific circRNA circSRY contains 16 target sites for miR-138 in mice [16].
Recent studies have suggested the potential of circRNAs in proteins translation [22–24]. For example, the human circZNF609 was found to be translated into protein and play a role in controlling myoblast proliferation [25]. Furthermore, it has been confirmed that circMbl3 was able to produce a protein in fly head extracts [22]. Of note, an IRES is embedded within circZNF609 and circMbl3 and the translation is in a splice-dependent/cap-independent manner. In addition to IRESs, m6A modification can also promote the initiation of protein translation from circRNA in human cells [26]. Another study of transcriptome-wide identification of m6A circRNAs defined thousands of m6A circRNAs with cell-type-specific expression, extending the concept of the RNA epitranscriptome to circRNAs [27]. These observations suggest that the cap-independent circRNA translation driven by IRES and m6A modification may provide a new direction for the functional studies of circRNAs.
CircRNAs were found to be expressed across various species including human, mouse, fly, worm, plant and yeast [3,5,28,29]. For example, circRNAs were significantly enriched in the mammalian brain, well conserved in sequence, often expressed as circRNAs in both human and mouse, and sometimes even detected in Drosophila brains [3]. However, comparison of circRNA expression from human and mouse revealed that only 10%-20% of human circRNAs are completely conserved in mouse with regard to splice site use, indicating a species-specific manner of circRNA expression [5,28]. This is likely due to the different compositions of orientation-opposite complementary sequences across species. In all, species-specific circRNAs also exist, although there is a substantial level of conservation of circRNAs.
As the studies of circRNA have exploded recently, the development of a standard nomenclature of circRNA is becoming more and more necessary for clear communication of circRNAs research. The current terminology of circRNAs is very confusing for both bioinformatic and experimental research. For example, the name ‘circZNF609’ would be ambiguous, because several circRNAs can arise from ZNF609. Circbase used some arbitrary number to name the circRNA which provide little information about the host gene or the chromosome location. Here we develop a novel naming system of circRNAs based on the circRNAs host gene and the starting/ending position of circRNAs in the host gene. Several databases are available now including CircBase [30], Circ2Traits [31], CircRNADb [24] and CicrNet [32]. However, none of them provide a meaningful nomenclature of circRNA in their database design. We incorporated the new naming system in our recently constructed database named as circbank. Circbank collects 140,790 human circRNAs named using our new naming system. All circRNAs collected in the circbank included below information: (1) miRNAs binding site, (2) conservation across species, (3) m6A modification, (4) mutation in circRNA, (5) protein-coding potential, and (6) predicted IRES sites. In addition, we have established UCSC genome trackhub which integrated all these features for data visualization.
Result and discussion
Nomenclature system of circRNAs
To facilitate the communication among circRNA researchers, we have developed a standard nomenclature of circRNA genes. Since multiple circRNAs may arise from the same host gene, the human circRNAs are named based on the HUGO symbol of host gene using the following scheme: ‘hsa-circHUGO-#’. As shown in Figure 1(a), the circRNAs from the same host gene are numbered based on the position of cirRNAs in the host gene and the number start with the most upstream one. For circRNAs with the same starting site and different ending site, the more upstream of the ending site, the less the number is (Figure 1(a)). For circRNAs with the same starting site and ending site, suggesting the alternative splicing of the circRNAs, the scheme of circRNAs nomenclature is ‘hsa-circHUGO-#_V#’. ‘V’ means variant and the number after ‘V’ is based on the length of circRNA. The shorter the circRNA length is, the less the number is. For the circRNA arising from intergenic regions, the scheme of circRNA nomenclature is ‘hsa-circChrom#_#’. For this scheme, the first number is the chromosome number and the second is the circRNA order number which follows the same rule as the circRNA form coding genes. Taken circRNAs from EGFR gene as an example, there are 15 circRNAs from EGFR gene and they are named as ‘hsa_circEGFR_001’to ‘hsa_circEGFR_020’, respectively (Figure 1(b)).
Figure 1.
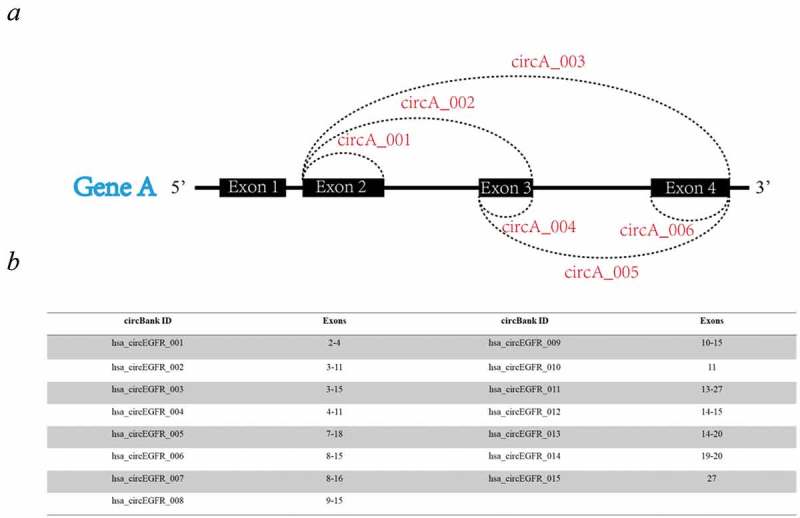
Nomenclature of circRNA at circbank. (a) schematic diagram for nomenclature of circRNAs (b) an example of circRNAs from EGFR gene (c) UCSC genome visualization of 15 circRNAs from EGFR gene.
Prediction of miRNAs binding
Although acting as miRNAs sponge is most intensively studied and well-accepted mechanism for the circRNA-mediated gene regulation, some recent studies have shown that circRNAs in mammals are expressed at low levels and they rarely contain multiple binding sites for the same miRNAs [5]. To investigate the binding relationship of circRNA and miRNA, we performed a systematic interaction prediction between 140,790 human circRNAs and 1,917 human miRNA using Miranda and TargetScan program. We found 42,917 circRNA-miRNA pairs have more than five binding sites and 3,545 circRNA-miRNA pairs have more than 10 binding sites for the same miRNA based on the prediction result from Miranda program (Figure 2). For example, we found hsa_circSH3YL1_005 (hsa_circ_0052415) harbours 39 binding sites for miR107, indicating that hsa_circSH3YL1_005 can function as the sponge of miR107.
Figure 2.
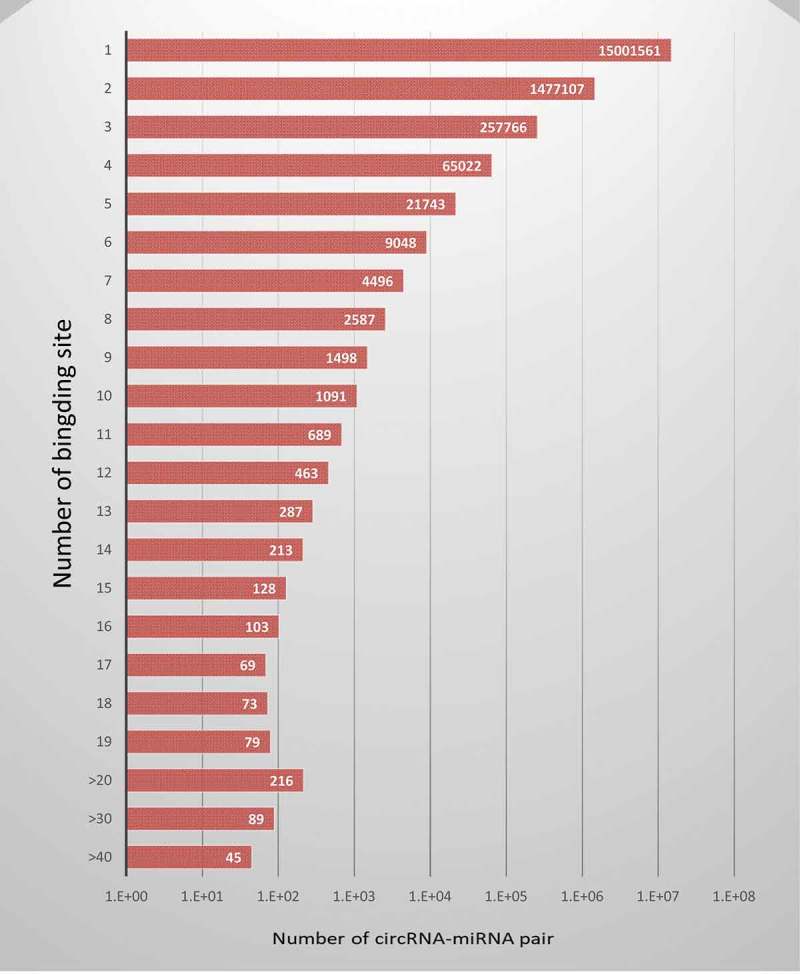
The summary of binding sites of circRNA-miRNA pairs.
CircRNAs conservation and protein coding potential
To perform the conservation analysis of human circRNA, we converted the coordinates of the backsplice site of each human circRNA to the mouse genome coordinates using the UCSC liftOver tool [33]. The converted mouse genome coordinates were compared to the backsplice sites of mouse circRNA to determine the conservation of circRNAs. If the difference is less than 2 bp, the circRNA was considered as conserved circRNA. We found 12,348 in 140,790 human circRNAs were conserved between human and mice.
To assess the protein-coding potential of the circRNAs, we used CPAT (Coding-Potential Assessment Tool) [34]to calculate the coding potential score of each human circRNA. The higher the predicted score is, the higher the coding potential of the circRNA. It has been reported engineered circRNAs can be translatable when containing IRES element [35–37]. We used the IRESfinder [38] program to predict the IRES element of each circRNA. The user can combine the coding potential data and the IRES prediction data for the translation research of the circRNAs of interest.
Query panel of circRNAs
Circbank provided two query panels for cirRNA searching: ‘circRNA’ panel and ‘miRNA’ panel. At ‘circRNA’ panel, users can perform a basic query of the database using the host gene symbol, circbank ID or circBase ID (Figure 3(a)). Users also can perform an advanced search through selecting other features of circRNAs including conservation, m6A modification and coding potential to get the circRNAs of specific interest. The output table of the query includes the circbank_Id, circbase_Id, position, strand, length, miRNA, Gene_Symbol and conserved_mouse_circRNA. Users can get more detailed information of the circRNA through clicking the circbank_Id of circRNA of interest. Users also can get the miRNA binding information of specific circRNA through clicking the ‘miRNA’ tab in the query output table. For a certain circRNA, circbank database collected the basic and functional information including (1) the circRNA sequence; (2) conservation compared with a mouse; (3) protein-coding potential; (4) point mutations circRNA based on the COSMIC database; (5) m6A modification information. Taken hsa_circHIPK3_004 as sample, basic information includes the circbank ID, host gene symbol, circBase ID, best transcript, chromosome position, circRNA annotation and circRNA length. Since hsa_circHIPK3_004 is a conserved circRNA between human and mouse, the sequence of the mouse ortholog circRNA of hsa_circHIPK3_004 was also showed, in addition to the sequence of hsa_circHIPK3_004. In the ‘coding_potential_assesement’ part, the protein-coding potential of hsa_circHIPK3_004 predicted by CPAT program was showed (Figure 3(c)). In the ‘IRES element’ part, all predicted IRES elements by IRESfinder are listed with the prediction score. The higher the score is, the higher probability the predicted IRES element is a real one. If no IRES elements were predicted, this part will be blank (Figure 3(c)). All COSMIC mutation information in the hsa_circHIPK3_004 are listed in the ‘mutation’ part, (Figure 3(d)). In the ‘RNA modification’ part, the m6A modification information of hsa_circHIPK3_004 are listed (Figure 3(d)).
Figure 3.
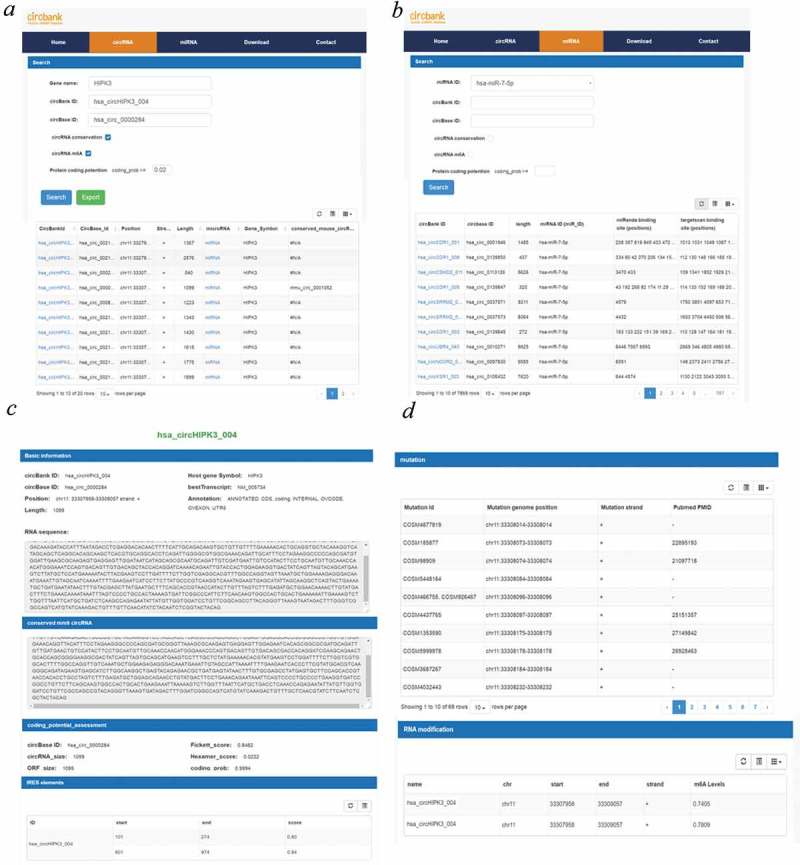
Overview of web interface in circbank. (a) ‘circRNA’ search panel web page information; (b) ‘miRNA’ search panel web page information; (c, d) detailed information page of hsa_circHIPK3_004.
At ‘circRNA’ panel, users can perform a basic query of the database using the miRNA ID, circbank ID or circBase ID (Figure 3(b)). Users also can restrict the output to certain cirRNAs of specific interest through selecting ‘circRNA conservation’ and ‘circRNA m6A’. The output table will include the miRNA-circRNA pairs predicted by two different algorithms including Miranda and Targetscan. As is known, the more binding sites for one miRNA-circRNA pair, the higher possibility the circRNA act as the sponge of the miRNA. We ranked the order of the miRNA-circRNA pairs based on the number of binding sites, and the pairs with more binding sites will appear on the top of the output table. Users will find out the potential circRNA acting as miRNA sponge easily.
UCSC genome trackhub of circRNA in circbank
For the visualization of circRNAs in the genome, we established a trackhub of the circRNAs in the UCSC genome browser which also integrated the miRNA binding site, IRES site, COSMIC mutation and m6A modified circRNA. As shown in Figure 4, the 15 circRNAs from EGFR gene were aligned to the UCSC genome. The predicted miRNA binding site and IRES site were also shown in the genome browser. The user can visualize the circRNAs of interest easily in our USCS trackhub, the link of which is in the help page of our website.
Figure 4.
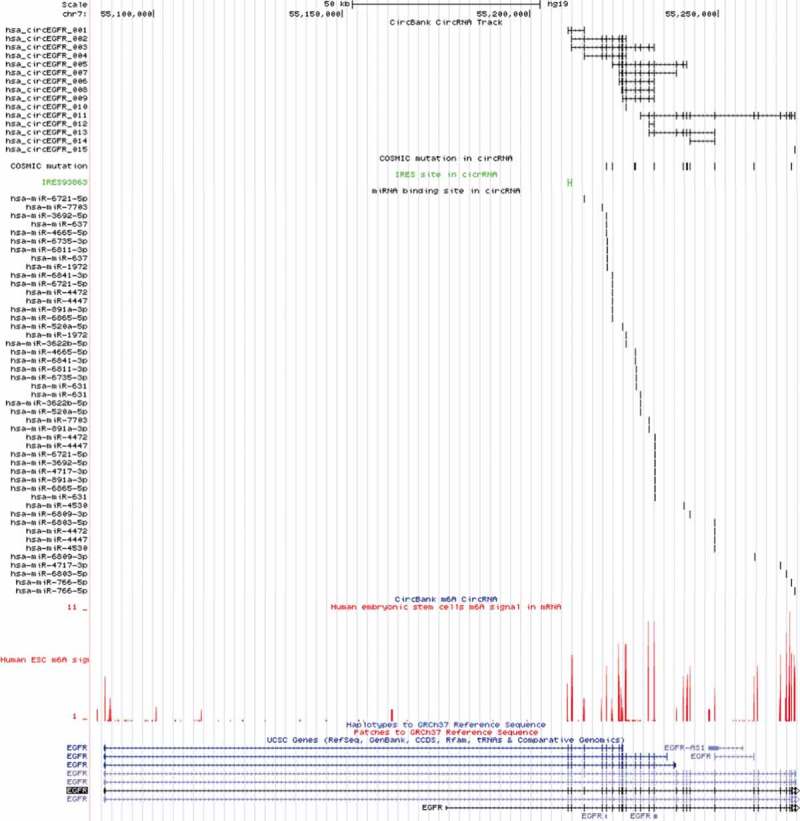
The UCSC genome visualization of 15 circRNAs from EGFR gene.
Conclusion and future direction
Circbank is a new comprehensive circRNAs database with a standard nomenclature implemented to ease the communication among the circRNAs research community. In addition to the naming system, circbank provides several other features of circRNAs such as miRNA interaction, conservation, coding potential, mutation and m6A modification. Based on these features provided by circbank, users can get more clues to advance their circRNAs study. In all, circbank is expected to be a valuable resource among circRNA research community.
We will continue to update the features of circRNAs in circbank as the new data becomes available. The current release of circbank includes 12,348 conserved circRNAs and 4,388 circRNAs with m6A modifications. Of note, we have integrated the m6A modification of both circRNAs and mRNA in our UCSC genome trackhub. The user can visualize the difference of m6A modification between circRNA and its host gene. With more sequencing data available, more conserved circRNAs and m6A modified circRNAs will be identified and we will update our database periodically. We also plan to add the circRNAs of other species such as mouse, rat and fly to our database.
Materials and methods
Data collection
The basic information including the genomic coordinates, host genes and sequences of 140,790 human circRNAs were downloaded from circbase website. The sequences of 1,917 human miRNA were downloaded from miRBase website. The data of m6A modification of circRNA was collected from related literature [27]. The human somatic mutation data were downloaded from COSMIC v85 database. The mutations located in the human circRNAs were collected in our database.
miRNA-circRNA interaction prediction
The circRNA sequences were extracted from the circBase database and miRNA sequences were extracted from the miRBase database V21. The sequence of seed region of miRNA was generated by the bioawk software. Two different algorithms including Miranda [39] and targetscan [40] were used to predict the miRNA which can target the circRNAs sequence.
CircRNAs conservation analysis
The circRNAs using the homologous backsplice sites between human and mouse were considered as the conserved circRNAs. The coordinates of backsplice sites of circRNAs were converted between human and mouse using the UCSC liftOver tool. If the difference of orthologous locus of backsplice sites between human and mouse is within 2 bp, the circRNA was identified as the conserved circRNA.
CircRNAs coding potential analysis
CPAT calculate the coding probability purely based on the sequence of the circRNAs [34]. The logistic regression model based on four features including ORF size, ORF coverage, Fickett TESTCODE and Hexamer usage bias were used to predict coding potential of the circRNAs. This method is alignment-free and is not dependent on conservation information. IRESfinder [38]was used to predict the IRES element in the circRNAs. IRESfinder predicts the IRES element based on a logit model with 19 carefully selected framed k-mer features.
Database design and web interface
All the data, including the features of circRNA, miRNA and circRNA-miRNA interaction were organized into a set of tables and stored in the MySQL database. The Java Spring framework and JavaScript library were used to implement the web interface. Circbank is freely accessible through http://www.circbank.cn
Funding Statement
This work was supported by grants from the Guangzhou Economic and Technological Development Zone.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- [1].Lasda E, Parker R.. Circular RNAs: diversity of form and function. RNA. 2014;20:1829–1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Chen LL, Yang L. Regulation of circRNA biogenesis. RNA Biol. 2015;12:381–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Rybak-Wolf A, Stottmeister C, Glazar P, et al. Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol Cell. 2015;58:870–885. [DOI] [PubMed] [Google Scholar]
- [4].Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. [DOI] [PubMed] [Google Scholar]
- [5].Guo JU, Agarwal V, Guo H, et al. Expanded identification and characterization of mammalian circular RNAs. Genome Biol. 2014;15:409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Barrett SP, Salzman J. Circular RNAs: analysis, expression and potential functions. Development. 2016;143:1838–1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Li X, Yang L, Chen LL. The biogenesis, functions, and challenges of circular RNAs. Mol Cell. 2018;71:428–442. [DOI] [PubMed] [Google Scholar]
- [8].Du WW, Yang W, Liu E, et al. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res. 2016;44:2846–2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Lu D, Xu AD. Mini review: circular RNAs as potential clinical biomarkers for disorders in the central nervous system. Front Genet. 2016;7:53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Greene J, Baird AM, Brady L, et al. Circular RNAs: biogenesis, function and role in human diseases. Front Mol Biosci. 2017;4:38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Du WW, Yang W, Li X, et al. A circular RNA circ-DNMT1 enhances breast cancer progression by activating autophagy. Oncogene. 2018;37:5829–5842. [DOI] [PubMed] [Google Scholar]
- [12].Lyu D, Huang S. The emerging role and clinical implication of human exonic circular RNA. RNA Biol. 2017;14:1000–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Chen S, Zhang L, Su Y, et al. Screening potential biomarkers for colorectal cancer based on circular RNA chips. Oncol Rep. 2018;39:2499–2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Li Y, Zeng X, He J, et al. Circular RNA as a biomarker for cancer: a systematic meta-analysis. Oncol Lett. 2018;16:4078–4084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Zhang Z, Yang T, Xiao J. Circular RNAs: promising biomarkers for human diseases. EBioMedicine. 2018;34:267–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. [DOI] [PubMed] [Google Scholar]
- [17].Caiment F, Gaj S, Claessen S, et al. High-throughput data integration of RNA-miRNA-circRNA reveals novel insights into mechanisms of benzo[a]pyrene-induced carcinogenicity. Nucleic Acids Res. 2015;43:2525–2534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Zhong Y, Du Y, Yang X, et al. Circular RNAs function as ceRNAs to regulate and control human cancer progression. Mol Cancer. 2018;17:79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Piwecka M, Glazar P, Hernandez-Miranda LR, et al. Loss of a mammalian circular RNA locus causes miRNA deregulation and affects brain function. Science. 2017;357:eaam8526. [DOI] [PubMed] [Google Scholar]
- [20].Dudekula DB, Panda AC, Grammatikakis I, et al. CircInteractome: a web tool for exploring circular RNAs and their interacting proteins and microRNAs. RNA Biol. 2016;13:34–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Kleaveland B, Shi CY, Stefano J, et al. Network of noncoding regulatory RNAs acts in the mammalian brain. Cell. 2018;174:350–62 e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Pamudurti NR, Bartok O, Jens M, et al. Translation of circRNAs. Mol Cell. 2017;66:9–21 e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Yang Y, Gao X, Zhang M, et al. Novel role of FBXW7 circular RNA in repressing glioma tumorigenesis. J Natl Cancer Inst. 2018;110:304–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Chen X, Han P, Zhou T, et al. circRNADb: a comprehensive database for human circular RNAs with protein-coding annotations. Sci Rep. 2016;6:34985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Legnini I, Di Timoteo G, Rossi F, et al. Circ-ZNF609 is a circular RNA that can be translated and functions in myogenesis. Mol Cell. 2017;66:22–37 e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Yang Y, Fan X, Mao M, et al. Extensive translation of circular RNAs driven by N(6)-methyladenosine. Cell Res. 2017;27:626–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Zhou C, Molinie B, Daneshvar K, et al. Genome-wide maps of m6A circRNAs identify widespread and cell-type-specific methylation patterns that are distinct from mRNAs. Cell Rep. 2017;20:2262–2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Dong R, Ma XK, Chen LL, et al. Increased complexity of circRNA expression during species evolution. RNA Biol. 2017;14:1064–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Veno MT, Hansen TB, Veno ST, et al. Spatio-temporal regulation of circular RNA expression during porcine embryonic brain development. Genome Biol. 2015;16:245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Glazar P, Papavasileiou P, Rajewsky N. circBase: a database for circular RNAs. Rna. 2014;20:1666–1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Ghosal S, Das S, Sen R, et al. Circ2Traits: a comprehensive database for circular RNA potentially associated with disease and traits. Front Genet. 2013;4:283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Liu YC, Li JR, Sun CH, et al. CircNet: a database of circular RNAs derived from transcriptome sequencing data. Nucleic Acids Res. 2016;44:D209–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Hinrichs AS, Karolchik D, Baertsch R, et al. The UCSC genome browser database: update 2006. Nucleic Acids Res. 2006;34:D590–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Wang L, Park HJ, Dasari S, et al. CPAT: coding-potential assessment tool using an alignment-free logistic regression model. Nucleic Acids Res. 2013;41:e74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Chen CY, Sarnow P. Initiation of protein synthesis by the eukaryotic translational apparatus on circular RNAs. Science. 1995;268:415–417. [DOI] [PubMed] [Google Scholar]
- [36].Li X, Liu CX, Xue W, et al. Coordinated circRNA biogenesis and function with NF90/NF110 in viral infection. Mol Cell. 2017;67:214–27 e7. [DOI] [PubMed] [Google Scholar]
- [37].Wang Y, Wang Z. Efficient backsplicing produces translatable circular mRNAs. Rna. 2015;21:172–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Zhao J, Wu J, Xu T, et al. IRESfinder: identifying RNA internal ribosome entry site in eukaryotic cell using framed k-mer features. J Genet Genomics. 2018;45:403–406. [DOI] [PubMed] [Google Scholar]
- [39].Enright AJ, John B, Gaul U, et al. MicroRNA targets in Drosophila. Genome Biol. 2003;5:R1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. [DOI] [PubMed] [Google Scholar]


