ABSTRACT
Novel CRISPR-Cas systems possess substantial potential for genome editing and manipulation of gene expression. The types and numbers of CRISPR-Cas systems vary substantially between different organisms. Some filamentous cyanobacteria harbor > 40 different putative CRISPR repeat-spacer cassettes, while the number of cas gene instances is much lower. Here we addressed the types and diversity of CRISPR-Cas systems and of CRISPR-like repeat-spacer arrays in 171 publicly available genomes of multicellular cyanobacteria. The number of 1328 repeat-spacer arrays exceeded the total of 391 encoded Cas1 proteins suggesting a tendency for fragmentation or the involvement of alternative adaptation factors. The model cyanobacterium Anabaena sp. PCC 7120 contains only three cas1 genes but hosts three Class 1, possibly one Class 2 and five orphan repeat-spacer arrays, all of which exhibit crRNA-typical expression patterns suggesting active transcription, maturation and incorporation into CRISPR complexes. The CRISPR-Cas system within the element interrupting the Anabaena sp. PCC 7120 fdxN gene, as well as analogous arrangements in other strains, occupy the genetic elements that become excised during the differentiation-related programmed site-specific recombination. This fact indicates the propensity of these elements for the integration of CRISPR-cas systems and points to a previously not recognized connection. The gene all3613 resembling a possible Class 2 effector protein is linked to a short repeat-spacer array and a single tRNA gene, similar to its homologs in other cyanobacteria. The diversity and presence of numerous CRISPR-Cas systems in DNA elements that are programmed for homologous recombination make filamentous cyanobacteria a prolific resource for their study.
Abbreviations: Cas: CRISPR associated sequences; CRISPR: Clustered Regularly Interspaced Short Palindromic Repeats; C2c: Class 2 candidate; SDR: small dispersed repeat; TSS: transcriptional start site; UTR: untranslated region.
KEYWORDS: CRISPR, cyanobacteria, heterocyst, nitrogen fixation, programmed DNA recombination
Introduction
Genetic tools based on CRISPR-Cas technology are currently the most popular technology for the manipulation of gene expression and genome editing. In most of these approaches, the CRISPR-Cas Type II enzyme Cas9 is used together with a guide RNA to target specific regions in chromosomal DNA [1]. In addition, large potential exists for alternative CRISPR-Cas systems and novel applications, e.g., in the markerless generation of point mutations using Type I and Type III CRISPR-Cas systems for genome editing [2] or the use of the Type V-A Cas12a (previously known as Cpf1) for the rapid engineering of markerless knock-ins, knock-outs and point mutations [3,4]. Such facts underline that the search for additional types of CRISPR-Cas modules can lead to productive innovation.
Currently, six major types of CRISPR-Cas systems are known, which belong to two major classes and can be further subdivided into multiple subtypes [5]. The functions of the diverse genes and gene products involved in these systems can be classified into three primary functions: adaptation, processing and interference [6]. During adaptation, CRISPR-associated (Cas) proteins excise the protospacer sequence from an invader DNA directly or after reverse transcription of RNA into cDNA [7] and insert it into the first repeat of the CRISPR locus. The CRISPR RNAs (crRNAs) are transcribed from the repeat-spacer array in the form of a long precursor (pre-crRNA) that is processed into the individual crRNAs each consisting of a single spacer sequence and part of the adjoining repeat sequences. During the interference stage, sequences on either invading DNA elements or their transcripts become recognized by crRNAs as guides for the Cas protein complexes that cleave the targeted nucleic acid.
CRISPR-Cas systems have been classified into two classes with regard to the complexity of the effector ribonucleoprotein complexes. Class 1 systems consist of several different subunits, whereas Class 2 systems utilize a single modularized large protein, such as Cas9, Cas12a or Cas13a [5]. Proteins implicated in adaptation are the endonuclease Cas1, Cas2 and, in some systems, Cas4, which facilitates the integration of PAM-compatible spacers [8,9]. The PAM (protospacer adjacent motif) is a short sequence motif in the target DNA that flanks the crRNA-DNA duplex and is crucial for avoiding self-cleavage [10,11]. Despite the impressive general variation in gene content and sequence diversity among different types of CRISPR-Cas systems, all systems have been assumed until very recently to possess a single Cas1 protein, which is less diverse than other Cas proteins and therefore has served as a marker for CRISPR loci. However, this notion has been challenged by recent observations of C2c (Class 2 candidate) systems lacking the cas1 gene, as they apparently only contain a CRISPR array and single gene encoding a large protein with no sequence similarity to Cas12a, Cas12b, Cas13a, or Cas9 [5]. Additionally, it was speculated that these systems might rely on an adaptation module (cas1-cas2) encoded elsewhere in the genome [12,13]. Therefore, the detection of putative novel CRISPR systems is not trivial: the numbers and types of CRISPR-Cas systems vary greatly, even between closely related strains, the similarity between Cas proteins can be very remote, and the existence of direct sequence repeats may also relate to different (non-CRISPR) genetic elements.
Cyanobacteria are the only bacteria that perform oxygenic photosynthesis. They occur in widely different environments as long as there is at least some light. Many cyanobacteria are also able to convert atmospheric nitrogen, N2, into organic biomass, hence sustaining a diazotrophic lifestyle. This process, called N2 fixation, is catalyzed by nitrogenase, an enzyme that can be irreversibly damaged by oxygen [14]. The need to protect the oxygen-sensitive nitrogenase from photosynthetically produced oxygen has probably driven the evolution of heterocysts, a type of differentiated cells providing a microoxic environment compatible with N2 fixation in some filamentous cyanobacteria. The evolution of this specialized cell type has driven the division of cellular functions and processes between heterocysts vegetative cells along the filaments [15]. Heterocysts transfer fixed nitrogen to the neighboring vegetative cells whereas vegetative cells provide heterocysts with photosynthetically fixed carbon in return. Hence, heterocyst-forming cyanobacteria are true multicellular organisms. Heterocysts are formed in a complex differentiation process that includes the programmed site-specific deletion of large genetic elements that interrupt the reading frames of critical genes by homologous recombination between direct repeats [15–17].
It has been suggested that the Cas9 effector proteins of Class 2 CRISPR-Cas systems evolved from a type of TnpB-like transposase with an HNH nuclease insert that is particularly abundant in cyanobacteria [12]. Type I and Type III (Class 1) CRISPR-Cas systems are frequent in cyanobacteria [18]; however, no proteins with sequence similarity to the hitherto characterized Class 2 effectors such as Cas12a, Cas12b, Cas13a or Cas9 have been identified thus far. Therefore, novel Class 2 CRISPR systems could exist in some cyanobacteria, a view consistent with multiple instances of CRISPR-Cas candidate systems classified as subtype V-U [5,13].
The genomes of multicellular cyanobacteria are complex (up to 12.29 Mb, > 10,000 annotated genes) and rich in the number of transposable elements and transposase genes including some encoding TnpB-like transposases. Therefore, we scanned 171 publicly available genomes of multicellular cyanobacteria for the presence of CRISPR-like repeat-spacer cassettes (Table S1). We report a high number of CRISPR-Cas candidate systems, including some with likely Class 2 effector proteins that are associated with a repeat-spacer array that is almost invariably adjacent to a tRNA gene. We then focus on Anabaena (Nostoc) sp. PCC 7120 (from here: Anabaena 7120) in greater detail and demonstrate crRNA-typical expression patterns for three Class 1, one Class 2 and five orphan repeat-spacer arrays in this well-established model for filamentous cyanobacteria.
Results
CRISPR-Cas systems are frequent in multicellular cyanobacteria
When searching for arrangements of direct repeats that match the criteria of CRISPR repeat-spacer arrays with relaxed parameters, some cyanobacteria, e.g., Tolypothrix bouteillei VB521301, host many, up to 44 widely different CRISPR-Cas systems (Table 1, see Table S1 for the full results). Together with Cas2, the Cas1 DNA-specific endonuclease makes up the core machinery of the CRISPR adaptation process. Therefore Cas1 is, with very few exceptions [5], almost universally conserved among different types of CRISPR systems. The here studied genomes of 171 filamentous cyanobacteria contain altogether 391 cas1 genes with different domain composition. Forty of the deduced Cas1 proteins are fused to a reverse transcriptase (RT) domain and 31 to a Cas4 protein domain. Such gene fusions are in line with findings that Cas4 promotes the integration of spacers from invading DNA with the correct PAM [8,9] and that the fused RT domains facilitate the direct CRISPR spacer acquisition from RNA [7]. Phylogenetic analysis of our set of Cas1 proteins yielded seven distinct clusters (Figure 1 and Figure S1). Three of these clusters consist of free-standing Cas1 sequences, whereas two clusters each contain Cas1-Cas4 or Cas1-RT fusions. The three clusters of Cas1 proteins encoded by free-standing genes each contain one of the three Cas1 proteins of Synechocystis sp. PCC 6803. We included them in this analysis because the unicellular Synechocystis sp. PCC 6803 is the best-studied model for CRISPR-Cas systems in cyanobacteria [8,19–23]. The clustering of the three Synechocystis sp. PCC 6803 Cas1 sequences suggests that they match well to the three major groups of Cas1 proteins lacking other fused domains of filamentous cyanobacteria. The set of 391 Cas1 proteins is available in Supplemental Dataset 1.
Table 1.
GenBank assembly accession numbers and Cas1 protein features of selected cyanobacterial strains. Morphological subsections were assigned according to Rippka et al. [53]. Cas1 gene types were defined as free-standing cas1, cas4_cas1 or RT_cas1 fusions. Accession numbers labelled by an asterisk refer to the JGI genome portal, all other refer to Genbank.
| Cyanobacterial strains | Subsection | Habitat | Accession | Number of Cas1 genes | Domain types of Cas1 | Number of CRISPR cassettes | Identified C2c5 homologs |
|---|---|---|---|---|---|---|---|
| Anabaena sp. PCC 7120 | IV | Freshwater | GCA_000009705.1 | 3 | RT_Cas1, 2x Cas1 | 11 | All3613, Alr2691 |
| Anabaena cylindrica sp. PCC 7122 | IV | Freshwater | GCA_000317695.1 | 5 | RT_Cas1, 4x Cas1 | 13 | Anacy_2856, Anacy_0603 |
| Calothrix sp. HK-06 | IV | Terrestrial | GCA_001904745.1 | 1 | Cas1 | 10 | 0 |
| Calothrix sp. PCC 7507 | IV | Terrestrial | GCA_000316575.1 | 9 | Cas4_Cas1, RT_Cas1, 7x Cas1 | 10 | 0 |
| Calothrix desertica sp. 7102 | IV | Terrestrial | 2509887024* | 6 | Cas4_Cas1, RT_Cas1, 4x Cas1 | 14 | 0 |
| Fischerella major NIES-592 | V | Hot spring | GCA_001904645.1 | 2 | RT_Cas1, Cas1 | 4 | 0 |
| Nostoc sp. PCC 7107 | IV | Freshwater | GCA_000316625.1 | 3 | RT_Cas1, 2x Cas1 | 14 | Nos7107_4709 |
| Nostoc calcicola FACHB-389 | IV | Terrestrial | GCA_001904715.1 | 2 | 2x Cas1 | 19 | 0 |
| Nostoc punctiforme PCC 73102 | IV | Terrestrial/symbiontic | GCA_000020025.1 | 1 | Cas1 | 6 | Npun_R5656 |
| Limnothrix rosea IAM M-220 | III | Marine | GCA_001904615.1 | 3 | RT_Cas1, 2x Cas1 | 9 | 0 |
| Phormidium am-biguum IAM M-71 | III | Freshwater | GCA_001904725.1 | 8 | RT_Cas1, 7x Cas1 | 14 | 0 |
| Rivularia sp. PCC 7116 | IV | Marine | GCA_000316665.1 | 3 | Cas1, 2x Cas1 | 14 | 0 |
| Scytonema hofmannii sp. PCC 7110 | IV | Terrestrial (limestone) | GCA_000346485.2 | 12 | 2x Cas4_Cas1, 2x RT_Cas1, 8x Cas1 | 29 | WA1_24145 |
| Tolypothrix sp. PCC 7601 | IV | Terrestrial | GCA_000300115.1 | 1 | Cas1 | 19 | FDUTEX481_03012, FDUTEX481_08898 |
| Tolypothrix bouteillei VB521301 | IV | Stone surface | GCA_000760695.2 | 5 | Cas4_Cas1, RT_Cas1, 3x Cas1 | 44 | 0 |
* JGI Taxon ID
Figure 1.
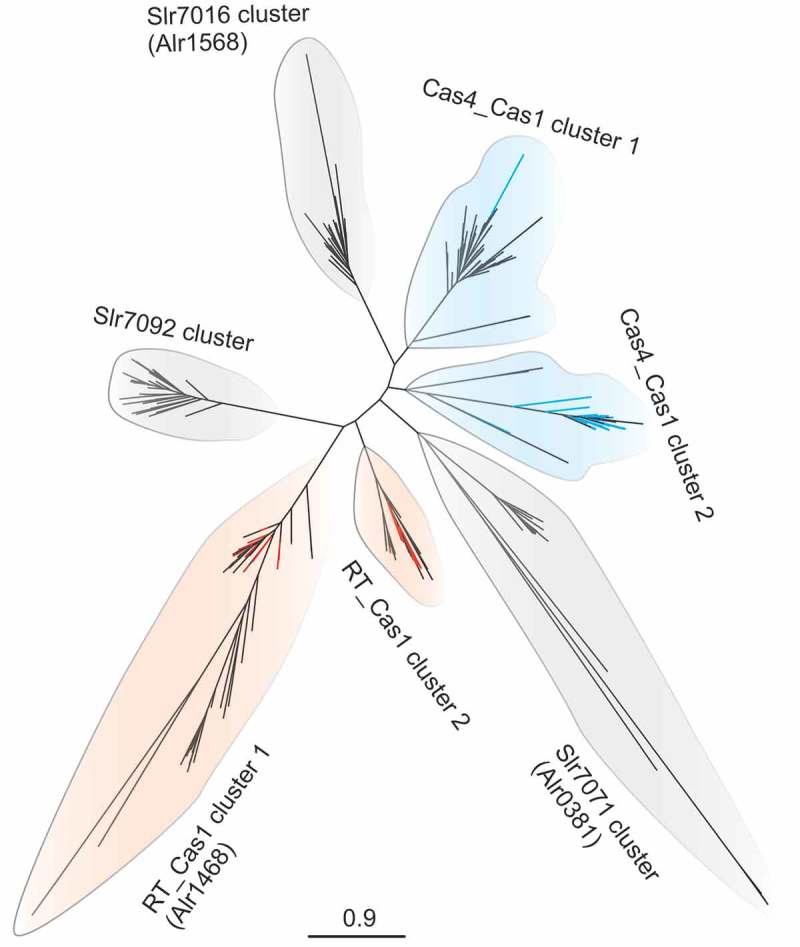
Phylogenetic analysis of 391 Cas1 sequences from 171 filamentous cyanobacteria. The three Cas1 protein sequences from Synechocystis sp. PCC 6803 were included in this unrooted phylogenetic tree, each representing a Cas1 cluster (grey shaded clusters) of 7 major clusters. Dark blue branches within light blue shaded clusters represent Cas1 proteins fused with a Cas4 domain, while red branches within light pink shaded clusters represent Cas1 proteins fused with a RV_1 domain. The positions of Cas1 proteins from Anabaena 7120 are given in brackets. The detailed Cas1 tree with sequence names and bootstrap values is presented in Figure S1, the protein sequences can be found in Supplemental Dataset 1.
We noticed a striking discrepancy in the number of cas1 genes and the number of repeat-spacer arrays. There are 5 cas1 genes and 44 repeat-spacer instances in Tolypothrix bouteillei VB521301, 12 cas1 genes and 29 repeat-spacer arrays in Scytonema hofmannii sp. PCC 7110, 5 and 15 in Aphanizomenon flos-aquae NIES-81, 3 and 14 in Rivularia sp. PCC 7116, 1 and 6 in Nostoc punctiforme PCC 73102, and 3 and 11 in Anabaena 7120 (Table 1), respectively. The genome sequences of aforementioned organisms are complete or in draft state with rigorous quality control [24], excluding assembly artefacts as a possible source of overestimation. To look further into this obvious discrepancy, we filtered the available sequences (Table S1) for completion of sequencing, yielding 36 finished genome sequences. From these, we chose three representative examples for which we performed a detailed re-annotation of the CRISPR-Cas systems (available upon request). Calothrix sp. PCC 7507 possesses 10 repeat-spacer arrays, 9 cas1 genes and four identifiable CRISPR-cas loci (Figure 2). However, three cas1 genes are fragmented and constitute pseudogenes. The remaining six include two gene copies encoding a Cas1-Cas4 and a Cas1-RT fusion (Figure 2). Rivularia sp. PCC 7116 and Nostoc sp. PCC 7107 both have 14 separate instances of repeat-spacer arrays but possess only three cas1 genes. Hence, in all these cases the numbers of repeat-spacer arrays is larger than the number of cas1 gene copies. Moreover, the two instances of genes encoding Cas1-RT fusions (in Calothrix sp. PCC 7507 and in Nostoc sp. PCC 7107) co-locate with an additional free-standing cas1 gene and are part or very close to subtype III-D systems (Figure 2). From these observations we conclude that genes encoding Cas1-RT fusions may become integrated in addition to existing cas1-cas2 adaptation modules and that the number of repeat-spacer arrays regularly exceeds the number of cas1 gene copies.
Figure 2.
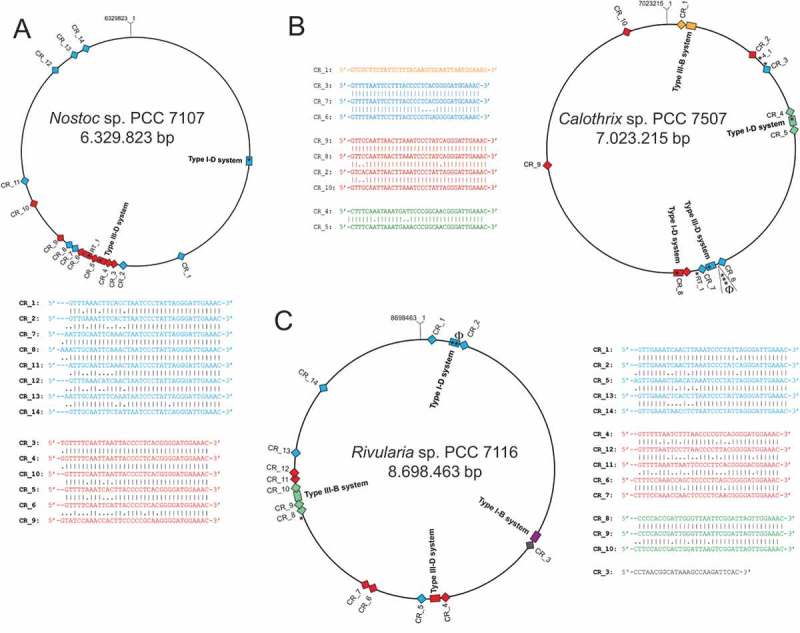
CRISPR-Cas systems and repeat-spacer arrays in three representative species, (A) Nostoc sp. PCC 7107, (B) Calothrix sp. PCC 7507 and (C) Rivularia sp. PCC 7116. The location of cas1 genes is indicated by asterisks, an added ‘RT_1’ or ‘1_4’ indicates RT or Cas4 fusions. Pseudogenes are labelled by a Φ symbol.
Unicellular cyanobacteria do not share this feature, e.g., the model cyanobacteria Synechocystis sp. PCC 6803 and PCC 6714 each harbor three different cas1 genes, matching the number of three different repeat-spacer arrays [19,20]. Therefore, the cas1-lacking systems in multicellular cyanobacteria might rely on adaptation modules encoded elsewhere in the genome [12,13] or depend on other mechanisms for recombination.
In the model Anabaena 7120, five DNA recombinase proteins are involved in the recombination in heterocyst differentiation. XisA mediates the excision of the 11 kb element from the nifD gene [25–27], the three-subunit enzyme encoded by the xisF, xisH and xisI genes excises the 55 kb element from the fdxN gene [28–30] (see Figure 3 for their location), while the XisC recombinase deletes the 10.5 kb element from the hupL gene [31,32]. The XisI recombinase has recently been identified as a candidate protein for an anti-phage defense system based on its pfam08869 domain [33]. Moreover, there are at least eight additional recombinase genes in the genome of Anabaena 7120 (all3124, alr0083, alr0084, alr2075, alr3224, alr3645, asl0560 and asl0561). The involvement of host-encoded factors such as IHF (integration host factor) in Cas1-Cas2 mediated adaptation has been reported for some types of CRISPR systems [34–36]. Therefore, it is tempting to speculate that one or several of the cyanobacterial recombinases are involved in CRISPR adaptation by functionally replacing the Cas1-Cas2 integrase complex that normally is facilitating the site-specific integration of new spacers into the CRISPR array [37,38].
Figure 3.
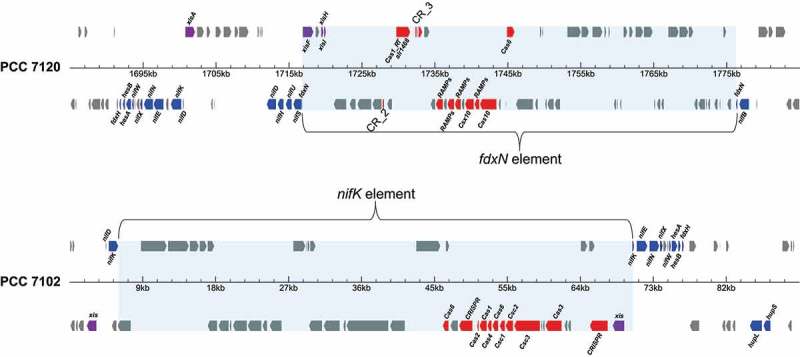
Examples of CRISPR-Cas systems that are encoded on genetic elements that are excised during cell differentiation into nitrogen-fixing heterocysts. The upper example is in the model organism Anabaena 7120, in which a CRISPR-Cas system is present within the fdxN element. The gene alr1468 encoding a reverse transcriptase-Cas1 fusion protein is annotated; details for the repeat spacer arrays CR_2 and CR_3 can be found in Table 2. The lower example presents the nifK element in Calothrix desertica sp. PCC 7102. CRISPR repeat-spacer arrays are labelled ‘CRISPR’. Note that the Anabaena 7120 element contains a cas1-RT fusion gene that is framed by split instances of the repeat-spacer arrays CR_2 and CR_3. Recombinase genes are labeled xis and xisAFHI and colored purple, cas genes are colored red, genes related to nitrogen fixation are colored blue, all other genes are in grey.
CRISPR-Cas systems are present in genetic elements that are excised during cell differentiation by homologous recombination
The appearance of fusions between the Cas1 protein and an RT domain is typical of certain types of CRISPR-Cas systems in cyanobacteria [39]. We observed that the presence of genes encoding RT_Cas1 fusions is frequently linked to the occurrence of two separate CRISPR repeat-spacer units framing the cas1-RT gene on the forward and reverse DNA strands (e.g., in Anabaena 7120, Figure 3). This suggests that an unknown DNA recombination event is involved in the evolution of some of the cassettes that contain these genes. The cas1-RT gene in Anabaena 7120 is not fused to a cas6 gene encoding the maturation endoribonuclease activity as observed in some other bacteria, e.g., Marinomonas [7,39].
Heterocyst differentiation includes the deletion of large genetic elements that interrupt the reading frames of critical genes. In different cyanobacteria, there are altogether more than ten different genes known that can be interrupted by such elements. Some of the frequently interrupted genes are the nifH, nifD and nifK (encoding nitrogenase Fe protein and subunits alpha and beta), hupL (encoding a subunit of heterocyst-specific uptake hydrogenase), fdxN (heterocyst ferredoxin) and hglE (encoding heterocyst glycolipid synthase) genes [25,28–32]. We observed that in several cases, CRISPR-cas systems are associated with these genetic elements that are precisely excised from the genome during the differentiation of heterocyst cells. In Anabaena 7120, this is the case for the fdxN element (Figure 3). Similar CRISPR arrangements can be found in the nifK elements of Calothrix sp. 7102 (Figure 3), Calothrix sp. HK-06 and Calothrix sp. 7103, nifD element of Tolypothrix PCC 7601, and nifH and hglE elements of Calothrix sp. PCC 6303. The fact that different types of CRISPR-Cas systems are present in different elements suggests that they evolved independently from each other. Thus, these elements constitute a preferred site for the integration and hosting of mobile CRISPR-Cas cassettes, as was observed previously for certain types of mobile genetic elements [5]. It should be noted that the CRISPR-Cas cassettes together with the elements in which they reside are eliminated during heterocyst differentiation. These facts point further to a previously not recognized connection between CRISPR-Cas cassettes and the genetic mechanisms involved in heterocyst differentiation.
Characteristics of CRISPR-Cas systems in the model Anabaena 7120
Based on the number of cas1 genes, there are three different Class 1 CRISPR-Cas systems in Anabaena 7120 (Table 1). However, a search for interspaced direct repeats showed that there are at least 11 CRISPR and CRISPR-like repeat-spacer cassettes (designated here CR_1 to CR_11) in Anabaena 7120, with more than 100 spacers (Table 2, Figure 4), all located in the 6,413,771-bp chromosome, whereas the six plasmids are free of such cassettes. The presence of multiple small dispersed repeat (SDR) sequences was previously reported for Nostoc punctiforme and related cyanobacteria [40]; however, these SDR sequences are different from the CRISPR repeats presented here. The repeat-spacer cassettes CR_1 to CR_11 could be fragmented versions of a lower number of functional CRISPR-Cas systems, pseudogenized versions or belong to novel types of such systems. To test their transcription as an indicator of functionality, we isolated RNA from four different cultures and hybridized specific single-stranded RNA probes after gel electrophoretic separation and blotting. The observed lengths of mature crRNAs correlated well with the theoretically expected lengths of ~ 44 nt (± 5 nt) for double processing (e.g. CR_4, 7, 8, 11 in Figure 5) or ~ 73 nt (± 5 nt) in case of single processing (e.g. CR_1, 2, 3 in Figure 5). Hence, the results showed that all of the elements are transcribed and exhibit the typical pattern of precursor accumulation, processing intermediates and accumulated crRNAs (Figure 5). Thus, they are likely part of functional CRISPR-Cas systems. We included RNA from cultures grown for a nitrogen starvation time course that would be long enough to trigger heterocyst differentiation because of the affiliation of CRISPR-Cas systems, such as CR_2 and CR_3, with elements directly affected by this process. However, remarkable nitrogen-dependent differences in crRNA accumulation were not detected over the here applied time course of 32 h (Figure 5), indicating these CRISPR cassettes were actively transcribed in the vegetative cells independent of nitrogen availability.
Table 2.
Predicted repeat-spacer arrays in Anabaena 7120. Each array has been numbered (ID), followed by the nucleotide positions of the TSS and the repeat-spacer arrays in the chromosome, the orientation (O) on the forward (+) or reverse (−) strand, the repeat sequence (DR), the number of repeats, with the total number including imperfect repeats in brackets (#), the length of spacers (L) the structural motif family (M), sequence family (F) and super family (S) as classified by the CRISPRmap algorithm [49], followed by the subtype and remarks (?, unknown). For the location within the chromosome of Anabaena 7120 see also Figure 4.
| ID | TSS | Start | End | O | transcribed leader | DR | # | L | M | F | S | Subtype, remarks |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CR_1 | 445445 | 445573 | 447786 | + | 128 | GTTACTTACCATCACTTCCCCGCAAGGGGATGGAAAC | 28 (29) | 33–48 | 4 | 8 | E | subtype I-A, no cas6 |
| CR_2 | 1728349 | 1728071 | 1727817 | − | 278 | GTTTTAATTCCTTTACCCCTCACGGGGATGGAAAC | 3 (4) | 37–40 | 4 | 9 | E | subtype III-D; CR_2 and CR_3 belong to one element; RT_cas1 fusion |
| CR_3 | 1731975 | 1732269 | 1733321 | + | 294 | GTTTTAATTCCTTTACCCCTCACGGGGATGGAAAC | 9 (15) | 34–40 | 4 | 9 | E | |
| CR_4 | 1836427 | 1836813 | 1837723 | + | 386 | GTTTCTATTAACACAAATCCCTATCAGGGATTGAAAC | 13 | 33–41 | 8 | 9 | E | subtype I-D |
| CR_5 | 2179566 | 2179167 | 2178606 | − | 399 | GTTTTTAATCTCTTACCCCTCACGGGGATGGAAAC | 8 (11) | 39–42 | 4 | 2 | E | ? |
| CR_6/7 | 3518141 | 3518084 | 3516820 | − | 57 | GTTTTAACTAACAAAAATCCCTATCAGGGATTGAAAC | 13 (16) | 31–44 | 8 | 9 | E | 134 nt MITE insertion in repeat 9 |
| CR_8 | 3836504 | 3840120 | 3840737 | + | 3616 | GTTGCAACACCATATAATCCCTATTAGGGATTGAAAC | 9 | 33–44 | 8 | 9 | E | ? |
| CR_9 | 4362990 | 4362,577 | 4362255 | − | 413 | CGTTGCAACCCTCCTTCCAGTAATGGGAGGGTTGAAAG | 3 (5) | 32–35 | ? | ? | ? | C2c5, All3613 effector |
| CR_10 | 5647342 | 5,647145 | 5646379 | − | 197 | CTTTCCGATCACATCACCCCGAAAGGGGATGGAAAC | 10 (11) | 32–45 | - | 18 | C | ? |
| CR_11 | 5654075 | 5654133 | 5654384 | + | 58 | GTTAAAACCCTCTAAAATCCCTATCAGGGATTGAAAC | 3 | 34–36 | 8 | 9 | E | ? |
Figure 4.
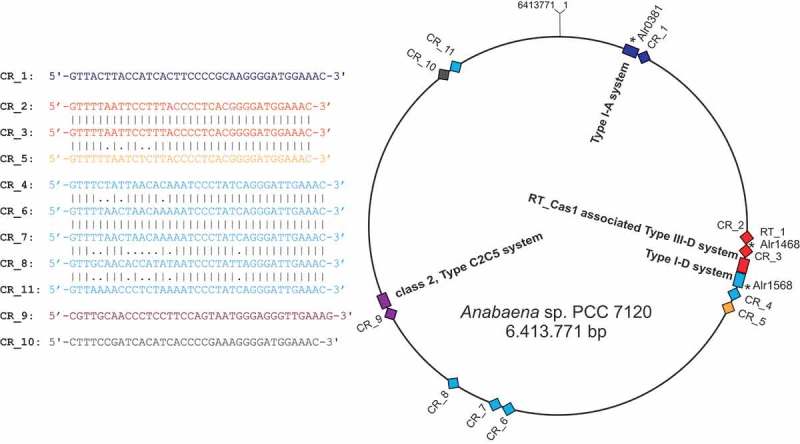
CRISPR-Cas systems in Anabaena 7120. Left: Alignments of CRISPR direct repeats. Right: Schematic distribution of CRISPR-cas systems, their subtype annotation and the location of repeat-spacer arrays in the chromosome of Anabaena 7120. The location of cas1 genes is indicated by asterisks and the respective gene IDs. For further details, see also Table 2.
Figure 5.
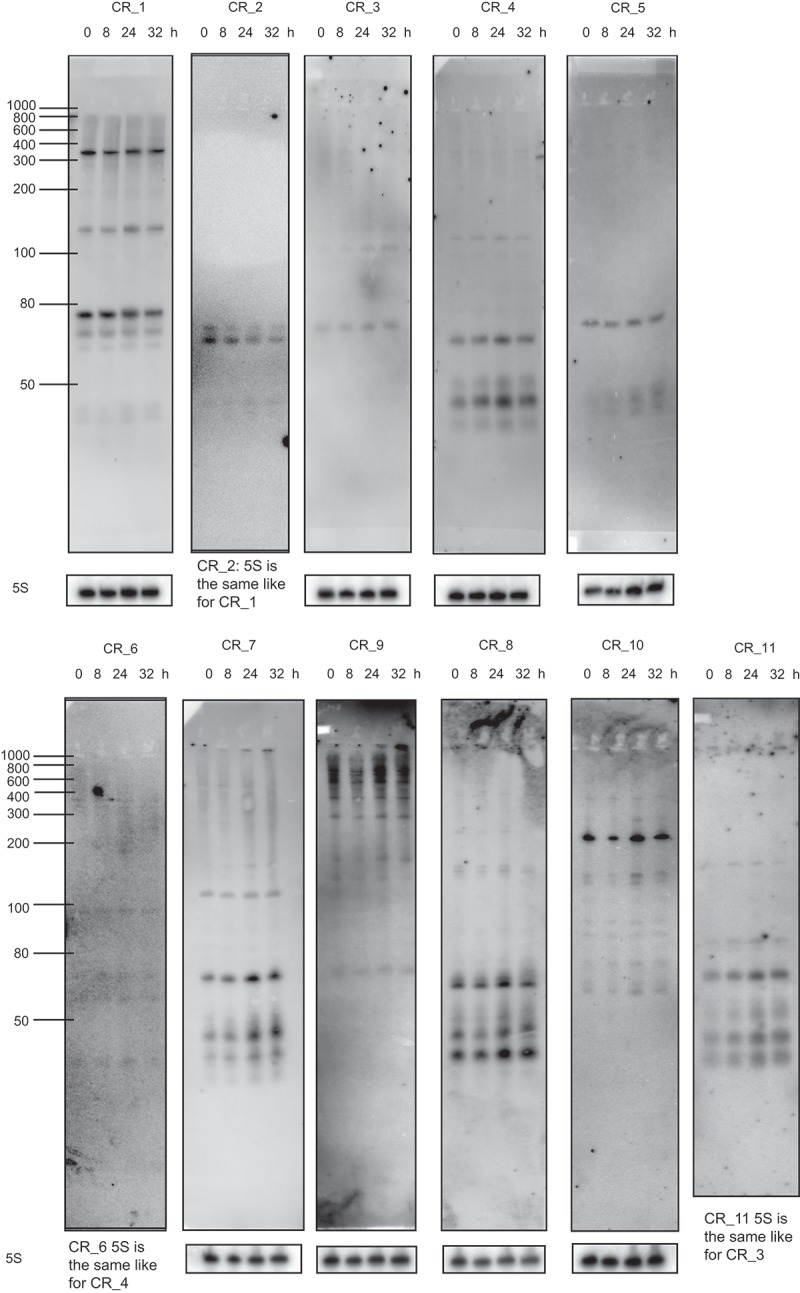
Expression of crRNAs from repeat-spacer arrays in Anabaena 7120. Total RNA was isolated from cultures grown under standard conditions for 8, 24 and 32 h after the removal of nitrogen, separated by electrophoresis on denaturing 15% PAA gels and transferred to nylon membranes. Single-stranded specific RNA probes were used for Northern hybridization. A control hybridization against 5S rRNA was performed to control for equal loading (the following membranes were used twice: for CR_3 was re-hybridized with the CR_11 probe, CR_1 with the CR_2 and CR_4 with the CR_6 probe later). The size of marker fragments is given on the left.
Organisms possessing CRISPR-Cas systems become immune to phage or other invading DNA by the insertion of DNA sequences (spacers) into the leader-repeat junction (i.e., at the 5ʹ end of the repeat-spacer array) in a site-specific process called adaptation. The leader region, especially its 3ʹ end, is indispensable for this adaptation [41–45]. Therefore, it must contain sequence determinants important for adaptation. However, the lengths of CRISPR leaders vary greatly in size, from 47 nt in some bacteria to several hundred nt in some hyperthermophilic archaea. Moreover, they possess long regions of low complexity sequence, show only limited sequence conservation [46] and therefore are difficult to predict [47]. Using differential RNA-seq, we previously experimentally defined a genome-wide map of more than 10,000 transcriptional start sites (TSS) of Anabaena 7120 at single-nucleotide resolution [48].Therefore, it is possible to precisely map the first transcribed nucleotide and to infer the length of the transcribed part of the leader when the element was expressed. This was possible for all 11 repeat-spacer cassettes. The length of the transcribed leaders varied from 57 to 3,616 nt (Table 2).
When judged by the association with known cas1 genes, the arrays CR_1 to CR_4 represent classical Class 1 CRISPR elements (the two repeat-spacer arrays CR_2 and CR_3 frame the RT_cas1 gene and belong to the same element, as depicted in Figures 3 and 4). Thus, there are at least three distinct Class 1 systems and one Class 2 (CR_9) system. The repeats CR_6 and CR_7 can be joined because they are only separated by the insertion of a 134 nt long miniature inverted repeat element (MITE) in repeat 9 of an originally contiguous array, yielding a total of five orphan repeat-spacer arrays.
Some repeats might be unified according to the similarities among their sequences, lumping CR_5 together with CR_2 and CR_3 in one group and CR_4, CR_6, CR_7, CR_8 and CR_11 in another (Figure 4), leaving CR_1, CR_9 and CR_10 separate. This is consistent with their assignment to distinct structural motif, sequence and super families as classified by the CRISPRmap algorithm [49] (Table 2). However, this unification might be an oversimplification. Hence, even when judged in a very restrictive way, there are at least five different types of arrays in total.
Novel CRISPR-Cas systems have substantial potential for genome editing and manipulation of gene expression. Therefore, it is interesting that one of the remaining systems, CR_9, is associated with a gene encoding All3613, a relatively large protein of unknown function. This protein is significantly similar in its C-terminal region to a subset of TnpB proteins encoded by transposons of the IS605 family, a feature typically associated with the Class 2 effector proteins Cas12b (C2c1) and C2c3 [12]. Among the studied 171 genomes of filamentous cyanobacteria, we found 86 All3613 homologs with a bit score ≥ 100, of which 29 were associated with a CRISPR array. This percentage is higher than expected by chance, supporting the idea that All3613 represents a novel type of CRISPR effector. This view was further supported when All3613 was analyzed by the HHpred algorithm [50], identifying a ~ 200 residues long similarity of the C terminus to the Cas12a (Cpf1) proteins of Lachnospiraceae bacterium ND2006 (probability 98.82, E-value 3.3e−10), Acidaminococcus sp. BV3L6 (probability 98.58, E-value 2.2e−9) and of Francisella tularensis subsp. novicida (probability 98.57, E-value 7.7e−9). All three proteins have been well characterized as single RNA-guided Type V effector proteins [4,51,52]. Nevertheless, proteins such as All3613 are with 648 amino acids substantially shorter than these effectors (e.g., Cas12a (Cpf1) of Lachnospiraceae bacterium ND2006 is 1231 residues long). Therefore, it is important that All3613 as well as many of its homologs are directly adjacent to a repeat-spacer cassette and that this cassette is expressed (Figure 5). A likely paralogous gene with all3613 is alr2691, encoding a protein that in a pairwise alignment exhibits 40% identical and 60% similar amino acid residues with All3613 (bit score of 447). However, alr2691 is not connected to a repeat-spacer array anywhere close in the genome.
The number of direct repeats in the CRISPR arrays associated with all3613 homologs in cyanobacteria was relatively low, with a maximum count of 13, mean count of 6, and median count of 5, pointing to a possibly inefficient insertion process of new spacers. We observed that the majority of these CRISPR arrays (24/29) were adjacent to a tRNA gene, for example, trnT(CGT) in Anabaena 7120, trnV(GAC) in Nostoc punctiforme PCC 73102, trnA(CGC) in Aphanizomenon flos-aquae NIES 81 and trnA(GGC) and trnR(CCG) in two instances in Tolypothrix sp. PCC 7601 (Figure 6). Hence, All3613 or its homologues in other cyanobacteria might constitute effector protein candidates for a novel type of CRISPR system or some kind of mobile genetic element. It cannot be excluded from consideration that the immediately adjacent tRNA genes served as integration sites of the respective cassettes. But the association with multiple different tRNA genes is puzzling in this regard (Figure 6). Although none of these putative Class 2 systems would be directly associated with a cas1 gene, the assignment of these regions as uncharacterized CRISPR system would be consistent with biocomputational analyses, which suggested that homologs of All3613 (Ava_2196 in Trichormus variabilis ATCC 29413 (previously called Anabaena variabilis sp. PCC 8801) and protein WP_027402996.1 in Aphanizomenon flos-aquae NIES 81) might constitute the core subunits of a Class 2 system called C2c5 [5,13]. Because All3613 and its homologs are shorter than the characterized Class 2 single effectors we looked for the possible synteny with other genes. Except the tRNA genes following the arrays, we only identified a gene encoding a MerR-type transcriptional regulator that is frequently located directly adjacent to the all3613/c2c5 gene (Figure 6).
Figure 6.
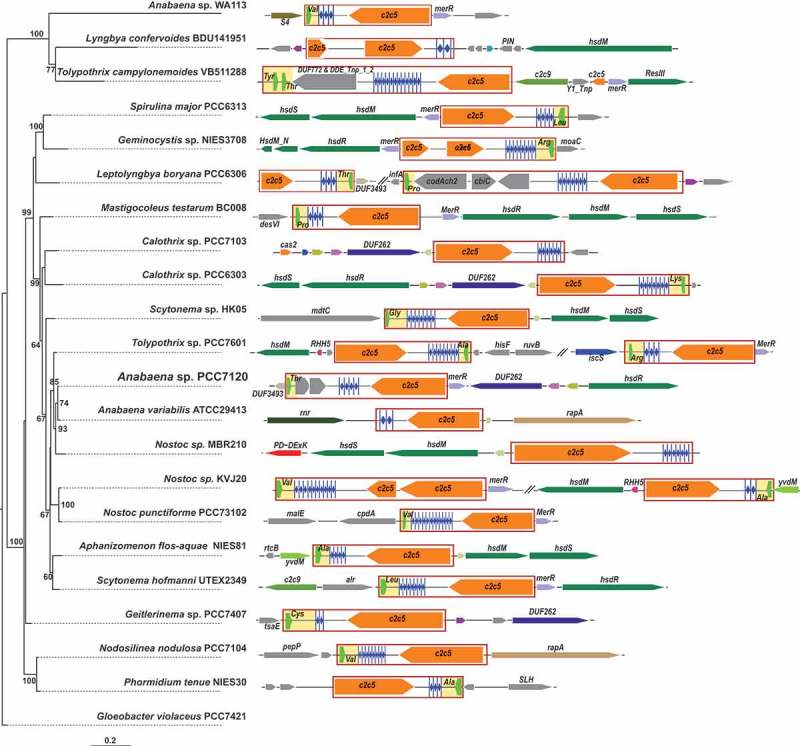
Synteny of Class 2 candidate systems. On the left the phylogenetic relationships are drawn based on 16S rRNA sequences, on the right the arrangements of putative CRISPR-cas systems are depicted that have an all3613 homolog next to the array and lack any known genes for adaptation or other known cas gene. Note the frequent presence of different tRNA genes adjacent to the repeat-spacer arrays. The units consisting of the all3613 homolog, the repeat-spacer array and tRNA gene (if present) are boxed. Known and putative cas genes are colored orange. The different tRNA genes are colored in light green and shaded in yellow for visualization, and their cognate amino acid is indicated. Numbers on the phylogenetic tree are bootstrap values and given if ≥ 60. The position of the model Anabaena 7120 is highlighted by larger fonts. 16S rDNA sequence from Gloeobacter violaceus PCC7421 was used as an outgroup to root this tree.
Summary and perspective
Certain filamentous cyanobacteria appear to be a rich source of CRISPR-Cas systems. In the case of Anabaena 7120, we show expression of the separate instances of repeat-spacer arrays by Northern hybridization with a pattern typical of the accumulation of crRNAs. We report that different types of CRISPR-Cas systems are encoded in different types of the genetic elements that are recombined during the differentiation of heterocysts, suggesting their independent evolution. All3613 might be a possible effector protein of the C2c5 type Class 2 CRISPR-Cas systems or belong to a novel genetic element.
Material and methods
The wild type strain of Anabaena 7120 was bubbled with an air/CO2 mixture (1% v/v) and grown photoautotrophically at 30°C in BG110C medium [53] lacking NaNO3 but containing 6 mM NH4Cl, 10 mM NaHCO3 and 12 mM tris(hydroxymethyl)methyl-2-aminoethanesulfonic acid-NaOH buffer (pH 7.5) until exponential phase. In order to induce nitrogen deficiency, cells grown in the presence of ammonium were collected by filtration, washed with and resuspended in nitrogen-free BG110C. Four RNA samples were isolated from cells taken at 0h, 8h, 24h and 32h after removing combined nitrogen from the media.
Total RNA was isolated using hot phenol as described [54] with some modifications. Hot phenol was added to the cells immediately after addition of lysis buffer and incubation was carried out at 65°C for 5 min. Further extractions were carried out with hot phenol, phenol:chloroform (1:1) and chloroform, followed by RNA precipitation by addition of one volume of isopropanol. CRISPR-related transcript accumulation was analyzed by Northern hybridization using single-stranded radioactively labelled RNA probes transcribed in vitro from PCR-generated templates (see Table S2 for primers) as described [55]. The oligonucleotide for the detection of 5S rRNA was 32P labelled using polynucleotide kinase (Thermo Fisher) and γ-ATP.
Phylogenetic analysis
The maximum likelihood tree was constructed based on 391 Cas1 proteins from 171 sequenced filamentous cyanobacteria and 3 Cas1 proteins from Synechocystis sp. PCC 6803. These Cas1 protein sequences were separately aligned with Clustal Omega v1.2.4 [56] and MAFFT E-INS-i v7.313 [57] with default parameters. The resulting alignments were merged using TrimAl v1.4.rev15 [58] with a minimum consistency score 0.5 (ct = 0.5) and only columns with a gap percentage < 50% were kept (gt = 0.5) for further phylogenetic analysis. ProtTest v3.4 [59] was used to find the best amino acid replacement model with best tree search operation of NNI and SPR (-s BEST) and empirical frequency estimation (-F). Based on Bayesian information criterion (BIC), the estimated best model LG+ G (-m PROTGAMMALG) was chosen to infer the maximum likelihood tree using RAxML v8.1.20 [60] with 20 best-scoring maximum likelihood searches and 1000 fast bootstrap searches (-f a -# 1000) from a random seed 12345 (-p 12345). The final phylogenetic tree was visualized using FigTree v1.4.2 (available at: http://tree.bio.ed.ac.uk/software/figtree/) and iTOL v4 online server (available at: http://itol.embl.de/[61]). The 16S rRNA gene tree was constructed based on a MAFFT E-INS-i v7.313 [57] alignment using RAxML v8.1.20 [60] with GTR nucleotide substitution model and GAMMA model of rate heterogeneity (-m GTRGAMMA). The 16S rDNA sequence of Gloeobacter violaceus PCC 7421 was used as an outgroup to root the 16S rDNA tree. 1000 fast bootstrap searches were done with the same setting as used in the Cas1 tree (-f a -# 1000 -p 12345).
Genome annotation and identification of CRISPR-Cas containing interruption elements
The genome sequences of filamentous cyanobacteria used in this study were downloaded from NCBI on March 25th, 2017 using phyloutils v1.0 (available at: https://github.com/housw/phyloutils). To keep the genome annotations consistent, all the genomes used in this study were re-annotated using Prokka v1.12-beta [62] with phyloutils wrapper. CRISPR cassettes were predicted using MinCED v0.2.0 with default parameters (MinCED is available at https://github.com/ctSkennerton/minced). Protein domains were predicted using pfam_scan.pl v1.6 (available at ftp://ftp.ebi.ac.uk/pub/databases/Pfam/Tools/) against Pfam release 30.0 [63] with an E-value cutoff of 1e−5. Interruption elements were scanned against all genomes using a xis gene-anchored algorithm with modifications [64], which required to determine the xis genes as a first step. In brief, for each genome, the xis genes were identified by searching all protein sequences against the previously identified Xis proteins [64] using blastP with an E-value cutoff of 1e−20. Then, the DNA sequences 3 kb upstream and 3 kb downstream of identified xis genes were extracted to check partial coding regions against all full-length protein sequences of Anabaena sp. PCC 7120 using blastX with default parameters. The extracted xis-containing regions were extended accordingly to cover the full length of surrounding overlapping genes. When a partial hit was identified, the target reference protein sequence was used as query to search against the whole genome sequence to find the other parts using tblastN. After that, all the partial coding regions were translated and aligned against the reference proteins to compose the full-length proteins. If successful, the excised regions were further checked for CRISPR cassettes. Motifs, families and super families of CRISPR direct repeats were identified using the CRISPRmap [49] online server (http://rna.informatik.uni-freiburg.de/CRISPRmap/Input.jsp).
Funding Statement
Financial support for this work was provided by the German Research Foundation (DFG) program FOR1680 ‘Unravelling the Prokaryotic Immune System’ (grants HE 2544/8-2 and BA 2168/5-2) to WRH and RB, by grant HE 2544/13-1, by the Ministerio de Economía y Competitividad (grant BFU2013-48282-C2-1) and the Agencia Estatal de Investigación (grant BFU2016-74943-C2-1-P) to AMP, both cofinanced by FEDER; a predoctoral contract (FPU014/05123) and a short term research stay grant (EST16/00088) by the Ministerio de Educación, Cultura y Deportes to MBA and by a China Scholarship Council grant to S.H., all of which are greatly acknowledged.
Author contributions
SH and OSA performed bioinformatics analyses, MBA and VR provided the experimental data, SH, OSA, MBA, RB, AMP and WRH analyzed data and WRH drafted the manuscript with contributions from all authors.
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplemental data
Supplemental date for this article can be accessed here.
References
- 1.Doudna JA, Charpentier E.. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014;346:1258096. [DOI] [PubMed] [Google Scholar]
- 2.Li Y, Pan S, Zhang Y, et al. Harnessing Type I and Type III CRISPR-Cas systems for genome editing. Nucleic Acids Res. 2016;44:e34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ungerer J, Pakrasi HB. Cpf1 is a versatile tool for CRISPR genome editing across diverse species of Cyanobacteria. Sci Rep. 2016;6:39681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zetsche B, Gootenberg JS, Abudayyeh OO, et al. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell. 2015;163:759–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Koonin EV, Makarova KS, Wolf YI. Evolutionary Genomics of Defense Systems in Archaea and Bacteria. Annu Rev Microbiol. 2017;71:233–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Garrett RA, Vestergaard G, Shah SA. Archaeal CRISPR-based immune systems: exchangeable functional modules. Trends Microbiol. 2011;19:549–556. [DOI] [PubMed] [Google Scholar]
- 7.Silas S, Mohr G, Sidote DJ, et al. Direct CRISPR spacer acquisition from RNA by a natural reverse transcriptase-Cas1 fusion protein. Science. 2016;351:aad4234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kieper SN, Almendros C, Behler J, et al. Cas4 facilitates PAM-compatible spacer selection during CRISPR adaptation. Cell Rep. 2018;22:3377–3384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee H, Zhou Y, Taylor DW, et al. Cas4-dependent prespacer processing ensures high-fidelity programming of CRISPR arrays. Mol Cell. 2018;70:48–59.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mojica FJM, Díez-Villaseñor C, García-Martínez J, et al. Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiol. 2009;155:733–740. [DOI] [PubMed] [Google Scholar]
- 11.Shah SA, Erdmann S, Mojica FJM, et al. Protospacer recognition motifs: mixed identities and functional diversity. RNA Biol. 2013;10:891–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shmakov S, Abudayyeh OO, Makarova KS, et al. Discovery and functional characterization of diverse Class 2 CRISPR-Cas systems. Mol Cell. 2015;60:385–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shmakov S, Smargon A, Scott D, et al. Diversity and evolution of class 2 CRISPR-Cas systems. Nat Rev Microbiol. 2017;15:169–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fay P. Oxygen relations of nitrogen fixation in cyanobacteria. Microbiol Rev. 1992;56:340–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Herrero A, Stavans J, Flores E. The multicellular nature of filamentous heterocyst-forming cyanobacteria. FEMS Microbiol Rev. 2016;40:831–854. [DOI] [PubMed] [Google Scholar]
- 16.Wolk CP. Heterocyst formation. Annu Rev Genet. 1996;30:59–78. [DOI] [PubMed] [Google Scholar]
- 17.Kumar K, Mella-Herrera RA, Golden JW. Cyanobacterial heterocysts. Cold Spring Harb Perspect Biol. 2010;2:a000315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cai F, Axen SD, Kerfeld CA. Evidence for the widespread distribution of CRISPR-Cas system in the phylum Cyanobacteria. RNA Biol. 2013;10:687–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hein S, Scholz I, Voß B, et al. Adaptation and modification of three CRISPR loci in two closely related cyanobacteria. RNA Biol. 2013;10:852–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Scholz I, Lange SJ, Hein S, et al. CRISPR-Cas systems in the cyanobacterium Synechocystis sp. PCC6803 exhibit distinct processing pathways involving at least two Cas6 and a Cmr2 protein. PloS One. 2013;8:e56470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Behler J, Sharma K, Reimann V, et al. The host-encoded RNase E endonuclease as the crRNA maturation enzyme in a CRISPR–cas subtype III-Bv system. Nat Microbiol. 2018;3:367–377. [DOI] [PubMed] [Google Scholar]
- 22.Reimann V, Alkhnbashi OS, Saunders SJ, et al. Structural constraints and enzymatic promiscuity in the Cas6-dependent generation of crRNAs. Nucleic Acids Res. 2017;45:915–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jesser R, Behler J, Benda C, et al. Biochemical analysis of the Cas6-1 RNA endonuclease associated with the subtype I-D CRISPR-Cas system in Synechocystis sp. PCC 6803. RNA Biol. 2018;[Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhu T, Hou S, Lu X, et al. Draft genome sequences of nine cyanobacterial strains from diverse habitats. Genome Announc. 2017;5:e01676-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Golden JW, Robinson SJ, Haselkorn R. Rearrangement of nitrogen fixation genes during heterocyst differentiation in the cyanobacterium Anabaena. Nature. 1985;314:419–423. [DOI] [PubMed] [Google Scholar]
- 26.Brusca JS, Chastain CJ, Golden JW. Expression of the Anabaena sp. strain PCC 7120 xisA gene from a heterologous promoter results in excision of the nifD element. J Bacteriol. 1990;172:3925–3931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Henson BJ, Pennington LE, Watson LE, et al. Excision of the nifD element in the heterocystous cyanobacteria. Arch Microbiol. 2008;189:357–366. [DOI] [PubMed] [Google Scholar]
- 28.Golden JW, Carrasco CD, Mulligan ME, et al. Deletion of a 55-kilobase-pair DNA element from the chromosome during heterocyst differentiation of Anabaena sp. strain PCC 7120. J Bacteriol. 1988;170:5034–5041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carrasco CD, Ramaswamy KS, Ramasubramanian TS, et al. Anabaena xisF gene encodes a developmentally regulated site-specific recombinase. Genes Dev. 1994;8:74–83. [DOI] [PubMed] [Google Scholar]
- 30.Ramaswamy KS, Carrasco CD, Fatma T, et al. Cell-type specificity of the Anabaena fdxN-element rearrangement requires xisH and xisI. Mol Microbiol. 1997;23:1241–1249. [DOI] [PubMed] [Google Scholar]
- 31.Carrasco CD, Buettner JA, Golden JW. Programmed DNA rearrangement of a cyanobacterial hupL gene in heterocysts. Proc Natl Acad Sci USA. 1995;92:791–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carrasco CD, Holliday SD, Hansel A, et al. Heterocyst-specific excision of the Anabaena sp. strain PCC 7120 hupL element requires xisC. J Bacteriol. 2005;187:6031–6038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Doron S, Melamed S, Ofir G, et al. Systematic discovery of antiphage defense systems in the microbial pangenome. Science. 2018; 6379:eaar4120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nuñez JK, Bai L, Harrington LB, et al. CRISPR immunological memory requires a host factor for specificity. Mol Cell. 2016;62:824–833. [DOI] [PubMed] [Google Scholar]
- 35.Wright AV, Liu -J-J, Knott GJ, et al. Structures of the CRISPR genome integration complex. Science. 2017;357:1113–1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yoganand KNR, Sivathanu R, Nimkar S, et al. Asymmetric positioning of Cas1-2 complex and Integration Host Factor induced DNA bending guide the unidirectional homing of protospacer in CRISPR-Cas type I-E system. Nucleic Acids Res. 2017;45:367–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xiao Y, Ng S, Nam KH, et al. How type II CRISPR–cas establish immunity through Cas1–cas2-mediated spacer integration. Nature. 2017;550:137–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nuñez JK, Lee ASY, Engelman A, et al. Integrase-mediated spacer acquisition during CRISPR-Cas adaptive immunity. Nature. 2015;519:193–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Silas S, Makarova KS, Shmakov S, et al. On the origin of reverse transcriptase-using CRISPR-Cas systems and their hyperdiverse, enigmatic spacer repertoires. mBio. 2017;8:e00897–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Elhai J, Kato M, Cousins S, et al. Very small mobile repeated elements in cyanobacterial genomes. Genome Res. 2008;18:1484–1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wei Y, Chesne MT, Terns RM, et al. Sequences spanning the leader-repeat junction mediate CRISPR adaptation to phage in Streptococcus thermophilus. Nucleic Acids Res. 2015;43:1749–1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Erdmann S, Garrett RA. Selective and hyperactive uptake of foreign DNA by adaptive immune systems of an archaeon via two distinct mechanisms. Mol Microbiol. 2012;85:1044–1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yosef I, Shitrit D, Goren MG, et al. DNA motifs determining the efficiency of adaptation into the Escherichia coli CRISPR array. Proc Natl Acad Sci USA. 2013;110:14396–14401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Erdmann S, Le Moine Bauer S, Garrett RA. Inter-viral conflicts that exploit host CRISPR immune systems of Sulfolobus. Mol Microbiol. 2014;91:900–917. [DOI] [PubMed] [Google Scholar]
- 45.Van Orden MJ, Klein P, Babu K, et al. Conserved DNA motifs in the type II-A CRISPR leader region. PeerJ. 2017;5:e3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shah SA, Garrett RA. CRISPR/Cas and Cmr modules, mobility and evolution of adaptive immune systems. Res Microbiol. 2011;162:27–38. [DOI] [PubMed] [Google Scholar]
- 47.Alkhnbashi OS, Shah SA, Garrett RA, et al. Characterizing leader sequences of CRISPR loci. Bioinforma. 2016;32:576–585. [DOI] [PubMed] [Google Scholar]
- 48.Mitschke J, Vioque A, Haas F, et al. Dynamics of transcriptional start site selection during nitrogen stress-induced cell differentiation in Anabaena sp. PCC7120. Proc Natl Acad Sci USA. 2011;108:20130–20135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lange SJ, Alkhnbashi OS, Rose D, et al. CRISPRmap: an automated classification of repeat conservation in prokaryotic adaptive immune systems. Nucleic Acids Res. 2013;41:8034–8044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zimmermann L, Stephens A, Nam S-Z, et al. A completely reimplemented MPI bioinformatics toolkit with a new HHpred server at its core. J Mol Biol. 2017;430:2237-2243. [DOI] [PubMed] [Google Scholar]
- 51.Dong D, Ren K, Qiu X, et al. The crystal structure of Cpf1 in complex with CRISPR RNA. Nature. 2016;532:522–526. [DOI] [PubMed] [Google Scholar]
- 52.Gao P, Yang H, Rajashankar KR, et al. Type V CRISPR-Cas Cpf1 endonuclease employs a unique mechanism for crRNA-mediated target DNA recognition. Cell Res. 2016;26:901–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rippka R, Deruelles J, Waterbury JB, et al. Generic assignments, strain histories and properties of pure cultures of cyanobacteria. Microbiology. 1979;111:1–61. [Google Scholar]
- 54.Mohamed A, Jansson C. Influence of light on accumulation of photosynthesis-specific transcripts in the cyanobacterium Synechocystis 6803. Plant Mol Biol. 1989;13:693–700. [DOI] [PubMed] [Google Scholar]
- 55.Steglich C, Futschik ME, Lindell D, et al. The Challenge of Regulation in a Minimal Photoautotroph: non-Coding RNAs in Prochlorococcus. PLOS Genet. 2008;4:e1000173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sievers F, Wilm A, Dineen D, et al. Fast, scalable generation of high‐quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 2011;7:539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Katoh K, Standley DM. MAFFT Multiple Sequence Alignment Software Version 7: improvements in performance and usability. Mol Biol Evol. 2013;30:772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 2009;25:1972–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Darriba D, Taboada GL, Doallo R, et al. ProtTest 3: fast selection of best-fit models of protein evolution. Bioinforma. 2011;27:1164–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Letunic I, Bork P. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016;44:W242–W245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinforma. 2014;30:2068–2069. [Google Scholar]
- 63.Finn RD, Coggill P, Eberhardt RY, et al. The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 2016;44:D279–D285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hilton JA, Meeks JC, Zehr JP. Surveying DNA elements within functional genes of heterocyst-forming cyanobacteria. PLOS ONE. 2016;11:e0156034. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


