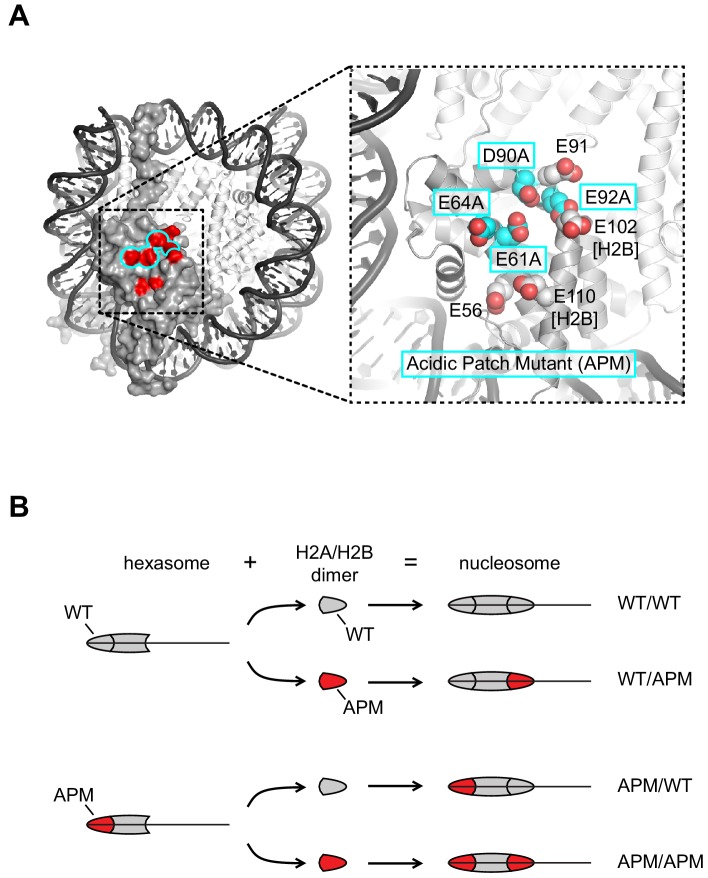
Figure 1. Design of nucleosomes with acidic patch mutations (APMs).
(A) The acidic patch of the nucleosome. In the overview, the surface is shown for one H2A/H2B dimer, with eight residues comprising the acidic patch highlighted in red. The acidic patch mutation (APM) consists of four alanine substitutions on H2A (E61A/E64A/D90A/E92A), highlighted in cyan. The structure shown is PDB code 1KX5 (Davey et al., 2002). (B) Strategy for making asymmetric nucleosomes using oriented hexasomes. As previously shown (Levendosky et al., 2016), hexasomes made with the Widom 601 (Lowary and Widom, 1998) preferentially lack an H2A/H2B dimer on the TA-poor side (here, the right side). Addition of either a wild type (gray) or APM H2A/H2B dimer (red) converts hexasomes to nucleosomes, allowing for production of all four combinations of H2A/H2B dimers with and without the APM. The locations of the H2A/H2B dimers and the H3/H4 tetramer on the Widom 601 is depicted in Figure 1—figure supplement 1.