ABSTRACT
Invading genetic elements pose a constant threat to prokaryotic survival, requiring an effective defence. Eleven years ago, the arsenal of known defence mechanisms was expanded by the discovery of the CRISPR-Cas system. Although CRISPR-Cas is present in the majority of archaea, research often focuses on bacterial models. Here, we provide a perspective based on insights gained studying CRISPR-Cas system I-B of the archaeon Haloferax volcanii. The system relies on more than 50 different crRNAs, whose stability and maintenance critically depend on the proteins Cas5 and Cas7, which bind the crRNA and form the Cascade complex. The interference machinery requires a seed sequence and can interact with multiple PAM sequences. H. volcanii stands out as the first example of an organism that can tolerate autoimmunity via the CRISPR-Cas system while maintaining a constitutively active system. In addition, the H. volcanii system was successfully developed into a tool for gene regulation.
KEYWORDS: CRISPR-Cas, Archaea, type I-B, Haloarchaea, CRISPRi, self-targeting
Introducing the key players: Cas proteins and CRISPR RNA
Repeat structures embedded in the genome of halophilic archaea were described as early as the 1990s [1, 2]. Similar alternating repeat sequences were also described in E. coli and were subsequently identified in several prokaryotic species [3–6]. Their role as key players in an adaptive and specific immune system remained elusive until the late 2000s when bioinformatics analyses confirmed that foreign genetic elements are the origin of CRISPR spacers [7–9]. CRISPR-Cas systems have since been identified in almost half of bacteria and most archaea [10]. CRISPR-Cas confers adaptive, specific and inheritable immunity through the elaborate interplay between RNA and protein components (for recent reviews see: [11, 12, 13] (Figure 1). The CRISPR loci give rise to a small RNA species, crRNA, which matches certain foreign sequences of past invaders. Both crRNA maturation and crRNA activity during the interference stage critically depend on the interaction with several Cas proteins. The latter ensure correct processing of the larger precursors into crRNAs. Subsequently, they bind each crRNA, incorporating them into a one- or multi-protein effector complex allowing the detection of the invader. Hybridization of crRNA and target triggers degradation of the targeted nucleic acid by a Cas protein. Additional Cas proteins enable the capture and integration of sequences from foreign genetic elements to expand the spacer content, and thereby the immune memory, of CRISPR loci during the adaptation process.
Figure 1.
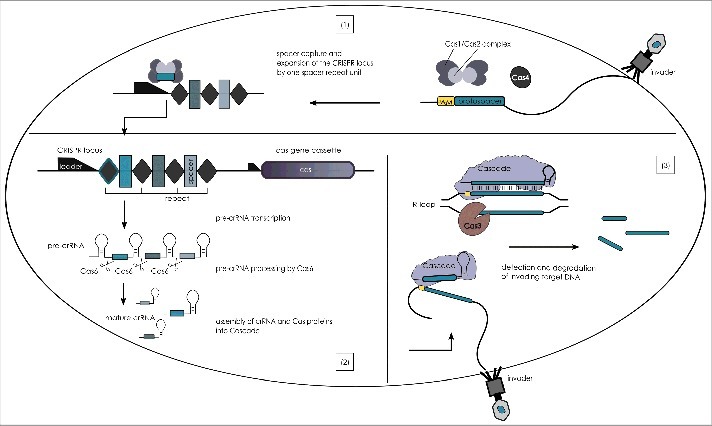
The stages of CRISPR interference in type I systems. CRISPR-Cas immunity proceeds in three stages and relies on the information stored within the unique spacers of the CRISPR loci and the Cas proteins encoded by the cas gene cassette. During the adaptation stage (1) an invading genetic element releases DNA into the cell which is recognized as such and degraded. A piece of the nucleic acid, that is flanked by a PAM sequence (yellow) is selected by the Cas1-Cas2 complex (Cas4 is also involved but its exact role has not been defined yet) and integrated as new spacer (blue-green) into the CRISPR locus (the repeat sequence is duplicated). Initiated by the promoter element within the leader sequence, the CRISPR locus is transcribed into a long precursor, the pre-crRNA, during the expression stage (2). The endonuclease Cas6 cleaves the pre-crRNA within the repeats generating a pool of crRNAs each carrying an individual spacer that are bound by Cas proteins forming the Cascade complex. Cascade complexes patrol the cell and interrogate incoming foreign DNA during the interference stage (3). If a PAM sequence is detected by Cascade the neighbouring protospacer sequence of the target is investigated by the crRNA. And if base pairing of crRNA and target ensues along the seed sequence Cascade is locked onto the targeted nucleic acid, Cas3 is recruited and activated to degrade the foreign element.
With a few exceptions, archaeal CRISPR-Cas systems are class 1 systems characterized by multi-protein effector complexes [14, 15]. Class 1 comprises a plethora of types and subtypes whose biochemical and mechanistic diversity can be substantial even within species and subtypes, e.g., as seen for the halobacterial type I-B systems [13, 16, 17]. These individual peculiarities make it difficult to draw a general conclusion from studying subtypes in only one species and illustrate how important a broad spectrum of studied organisms is for discerning common features. Although CRISPR-Cas systems are far more abundant in archaea than in bacteria, only a few archaeal CRISPR-Cas systems have been studied in detail in vivo owing to the challenging biology of most archaeal species and a general lack of genetically tractable archaeal model organisms. Haloferax volcanii is a prominent archaeal model organism, and its CRISPR-Cas system has been studied in detail with regard to its proteins and RNA components and their respective interplay [18–24]. This review will summarize current knowledge and draw connections to insights gained by studying other CRISPR-Cas systems.
The diversity of CRISPR-Cas systems within archaea is quite high, and often more than one CRISPR-Cas system is present in a genome [15]. H. volcanii is a euryarchaeon, and within this clade, almost all varieties of the type I and type III subtypes can be found [15, 16]. However, the halophilic archaea of the class Halobacteriales are quite homogenous in their CRISPR content since they contain only type I-B and I-D systems [16].
The H. volcanii CRISPR-Cas system comprises three CRISPR loci and one set of Cas proteins (Figure 2). With the exception of one CRISPR locus, all of the system´s components are encoded in a single locus on the large plasmid pHV4. The cas gene cassette encodes the genes for Cas 1–8b. The presence of a cas3 gene is indicative of a type I system and the characteristic gene synteny together with the presence of the cas8b gene marks it as a subtype I-B system [14, 25]. The cas gene complement can be subdivided into the genes of the interference module (cas6b, 8b, 7 and 5), which is separated by 190 base pairs from the genes of the adaptation module comprising cas4, cas1 and cas2. Basal transcription of the cas gene cassette in H. volcanii is rather low [18], but proteome analysis revealed constitutive production of the Cas proteins Cas5-8b in both the exponential and stationary phases, whereas adaptation proteins were not detected (Jevtic, Pfeiffer, Stoll, Harma, Urlaub, Lenz and Marchfelder, in preparation).
Figure 2.
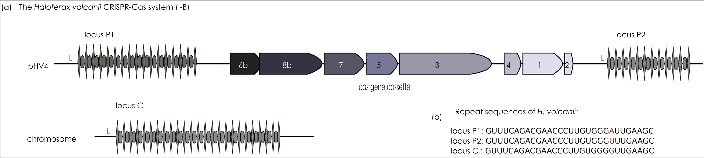
The H. volcanii CRISPR-Cas I-B system. (a) The composition and configuration of its cas gene cassette (purple) characterizes the H. volcanii CRISPR-Cas system as subtype I-B. The cas gene cassette on the chromosomal plasmid pHV4 is flanked by two of the three H. volcanii CRISPR loci (P1 and P2). The third locus (C) is encoded on the main chromosome. Each CRISPR locus encompasses unique spacer sequences (boxes) interspersed by repeat elements (diamonds). Transcription of the CRISPR loci is governed by their individual leader sequence containing the promoter elements and gives rise to the crRNAs needed for the specificity of CRISPR Cas immunity. (b) The sequence of all three H. volcanii repeat elements is identical in all but one nucleotide (red).
Transcription of all three CRISPR loci is constitutive [19]. Each possesses a leader sequence that provides the necessary promoter elements and gives rise to a crRNA precursor comprising an individual number of spacers separated by identical repeat sequences (Figure 2). The 30 nucleotide repeat sequences of the various loci only differ in one nucleotide (Figure 2B, 3A) and they all belong to the superclass A family of repeats according to the classification of repeat families by CRISPRmap [26]. Superclass A mainly comprises unstructured repeat sequences from Archaea (Euryarchaeota) and Bacteria (Firmicutes, Thermotogae and Aquificae). The amount of mature crRNA seems to be independent of stress conditions since several conditions tested, including temperature, salt or the overall growth phase, did not alter the amount of crRNAs [19].
Figure 3.
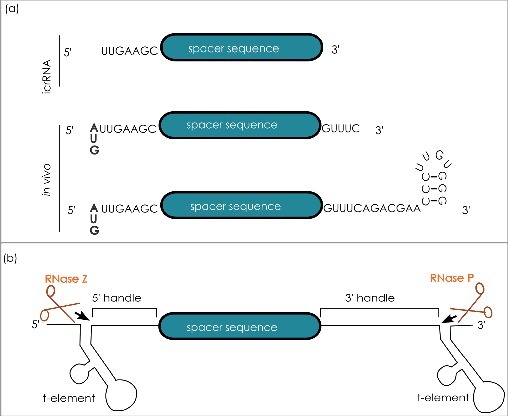
Natural and artificial crRNAs. (a) Apart from the unique spacer sequence, each crRNA present in vivo comprises an eight nucleotide long 5′ handle. In addition, the major crRNA population found in vivo contains a twenty-two-nucleotide long 3′ handle, whereas a minor crRNA population contains a five-nucleotide long 3′ handle. A systematic analysis with independently generated articificial crRNAs (icrRNAs) showed that an icrRNA with a seven nucleotide 5′ handle, the spacer sequence and without the 3′ handle is still active. (b) Cas6 independent crRNA maturation: the crRNA sequence is flanked by tRNA-like structures, so called t-elements. These are recognized by the tRNA processing enzymes RNase Z and RNase P which release the mature icrRNA.
The spacer complement of H. volcanii
The spacer content of the H. volcanii CRISPR-Cas system has interestingly changed since the publication of the genome sequence of the type strain DS2. The H. volcanii strains now used in laboratories, such as H119 and H26, are missing 23 spacers in locus P1, and currently encode a set of 51 spacers (Locus P1: 16, P2: 11 and C: 24) (Figure 2) [19]. Bioinformatics analysis of their potential targets via comparison to publicly available genome and metagenome data was performed and revealed only two matches [19]. This analysis has since been repeated with a slightly increased dataset and matches for nine spacers from all three CRISPR loci have been identified [27]. The spacer sequences matched the genomes of other haloarchaea, such as Halorubrum lacusprofundi, Haloferax elegans, Haloferax lucentense, Haloferax alexandrines, Haloterrigema jeotgali and Haloferax sp. Other sequences could be assigned to metagenomic contigs from Lake Tyrell or the Great Salt Lake, as well as their corresponding metaviriomes. The targeted regions could mainly be assigned to regions of putative or proven viral origin, either integrated proviruses or target genes that might be implicated in plasmid partitioning or the replication of foreign DNA. For one spacer a full match could be found in two other Haloferax species, all other spacer matches contained three to nine mismatches distributed over the pairing region. This rather unusual spacer origin might hint at interspecies targeting amongst Haloferax sp. (Turgeman-Grott et al., under revision) or may be reminiscent of a regulatory function towards surface proteins as seen in Francisella novicida [28]. However, the origin of the vast majority of spacers has still not been elucidated. Owing to the fast pace of viral evolution, viruses and plasmids that were captured by Haloferax CRISPR loci at the time of isolation (approximately 40 years ago) will have considerably changed or may no longer be present. In fact, the Dead Sea, the original isolation site for H. volcanii, has changed dramatically owing to evaporation since the 1970s in terms of both abiotic factors and the microbial community present, and thus H. volcanii is unlikely to have survived there. The virosphere, in general, is still under-sampled and therefore ill-represented in current publicly available databases, further hampering spacer assignment.
To gain a comprehensive view of the immunogenic potential of the CRISPR-Cas system, the cellular crRNA content was investigated with RNASeq, revealing an unequally distributed concentration of the individual crRNAs [21]. Such an uneven representation of individual spacers was also observed for other subtypes and species [29–34]. The crRNA amount in H. volcanii is not linked to its position within the array [21]. The variable presence of crRNAs in the sequencing reads might result from technical biases in RNASeq protocols or may have biological reasons, such as the differential stability of RNAs with different sequences [21]. The only variant element of the different crRNAs is the spacer sequence. The resulting differences in the ensuing steric and electrostatic interactions with associated proteins might alter Cascade loading or affinity, thus differentially exposing crRNAs to RNA degrading activities. Those ribonucleases might also be more or less active towards certain sequences within the spacers. As observed for the I-D system [35] local interactions between the different spacer sequences and the neighbouring repeat sequences could mask or facilitate access for processing or degrading enzymes. Furthermore, when tested for interference, not all spacers in H. volcanii are able to evoke interference against a matching foreign genetic element [21]. Neither abundance, spacer length, G/C content or other characteristics can account for the different efficacies. However, to date, each spacer was tested with only a single PAM, and thus, a specific activity with only one of the PAM sequences might also be possible [21]. These differences in the interference response might also reflect subtle variations in the interactions within the effector complex, hampering the target interaction or Cascade function, and have since been described in an E. coli high-through-put screen [36].
crRNA biogenesis and maintenance
The endonucleolytic activity of the Cas6 protein is responsible for processing the crRNA precursor in all type I and type III systems. Accordingly, a certain degree of coevolution between the Cas6 protein and respective repeat sequence to be processed must exist [26, 37, 38].
The entirety of known Cas6 proteins shows only limited sequence conservation, but a growing body of Cas6 structures demonstrates that they all share common structural features required for structure- and sequence-specific pre-crRNA binding and processing [39, 40]. For the Haloferax type I-B system, it has been demonstrated that Cas6 is indeed responsible for crRNA production [18]. Deletion of its coding sequence results in complete loss of crRNA maturation. In addition, Cas5 and Cas7 are required to ensure a stable steady-state level of crRNAs [18]. Individual deletions of cas5 or cas7 are detrimental to the crRNA levels in the cell. Here, the protective effect of Cas5 is more pronounced than that of Cas7 [18]. The influence of individual cas genes on the crRNA production is not strictly conserved within a given CRISPR-Cas subtype. For example, H. volcanii and H. mediterranei both possess type I-B CRISPR systems, but in contrast to H. mediterranei, the crRNA levels of H. volcanii are not influenced by deletion of cas1, cas3 or cas4 [18, 41].
The RNASeq analysis used to determine the overall crRNA complement of the H. volcanii I-B system was also used to resolve the characteristics of the mature crRNA [21]. Each Haloferax crRNA comprises a spacer sequence and an eight nucleotide 5′-handle as commonly observed in all type I systems. The 5′-handle is identical in all H. volcanii crRNAs, except for the first nucleotide, which differs between the otherwise identical repeat sequences of the three loci. However, the crRNA 3′ length is present in two different forms (Figure 3A) [21]. One fraction of the crRNAs still possesses the 22 nucleotide 3′-handle, resulting from the release of the eight-nucleotide 5′-handle of the neighbouring crRNA. Yet a second population of crRNAs possesses a 3′-handle that is reduced to only five nucleotides. The shortened crRNA form is also stably maintained within the cell as northern blot analysis shows [21]. This type of trimming appears to be characteristic of type I-B systems as it was also reported for Methanococcus maripaludis and Clostridium thermocellum [32]. Shortened crRNAs are also reported for type I-A and I-D systems, as well as type III systems [33, 34, 42–44]. In all cases the specific RNase activity responsible for crRNA 3′-handle trimming has not yet been identified. However, this additional 3´ processing may be an unspecific degradation event owing to exposure of the 3′ end protruding from Cascade rendering it accessible to non-specific RNases (see below “Composition of the H. volcanii interference complex”).
A short palindrome is present at the 3′ end of the repeat sequence, which might result in a minimal stem-loop structure with a four-nucleotide loop and a three C:G pair stem directly upstream of the endonuclease cleavage site (Figure 3). However, in vitro RNA structure analyses with RNases and 1D NMR could not confirm stable base-pairing interactions [21]. In vivo this stem-loop might be stabilized either by the high intracellular salt concentration (a characteristic feature of halophilic archaea) or protein-RNA interactions. Stabilization of the structure in the context of Cas6-mediated pre-crRNA processing is illustrated, e.g., by Cas6 of Thermus thermophilus and Sulfolobus solfataricus [45, 46]. In the I-B system of M. maripaludis, the interaction with Cas6b leads to the formation of an otherwise unfavoured alternative stem-loop for which the protein supplies a necessary base mimic [47].
The tale of Haloferax Cas6b
Owing to the rudimentary conservation of Cas6 at the sequence level and limited number of Cas6 structures, a systematic mutational approach for Haloferax Cas6b (HvoCas6b) was performed in vivo to identify the essential residues [18]. Twenty-one single-amino-acid mutants of Cas6b were investigated for crRNA production and interference. Only three mutations were shown to severely affect the crRNA level: H41A, G256A and G258A [18]. The Cas6 family of proteins typically contain two ferredoxin-like folds and Histidine 41 is located in the N-terminal ferredoxin-like fold. The tandem ferredoxin-like subdomains are angled towards each other and enclose the active center, engaging the residues of both domains [39, 40]. The Cas6 protein of Pyrococcus furiosus (PfuCas6) is the closest structural match for HvoCas6b and structural modeling of Cas6b structure according to PfuCas6 shows the HvoCas6b His41 to be part of the active site [18, 48–50] (Figure 4). Histidine residue homologous to HvoCas6b His41 are crucial for Cas6 activity in P. furiosus, Pseudomonas aeruginosa Cas6f, Thermus thermophilus Cas6a and T. thermophilus Cas6e and generally a histidine of the first α-helix is often conserved within Cas6 proteins [32, 39, 49, 51–54]. As crRNA production is not completely abolished upon mutation of HvoCas6b His41 cleavage seems not to depend on this single histidine residue in H. volcanii [18]. Similar observations have been made for the Cas6b proteins from M. maripaludis and T. thermophilus, thus this might be characteristic of type I-B Cas6b proteins [32, 51]. It seems that catalysis can be exerted by a wide variety of catalytical settings including histidine, arginine, lysine and tyrosine residues in variable combinations [39, 45].
Figure 4.
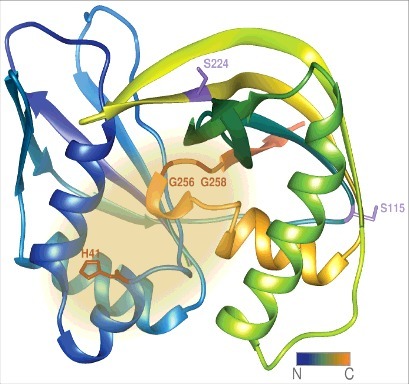
The H. volcanii Cas6b protein. The structure of the H. volcanii Cas6b protein has not been solved experimentally, yet. Depicted is a structural model created by the Phyre 2 server [50], the suspected active site is highlighted in yellow. The amino acid residues coloured in red resulted in reduced crRNA levels when mutated to alanine. Position of His41 corresponds to the conserved active site histidine residues found across Cas6 species, whereas Gly256 and Gly258 are part of the glycine-rich loop implicated in crRNA positioning. The amino acid residues coloured in lilac correspond to those resulting in elevated crRNA amounts upon mutation to alanine (S115 and S224). They are located on the averted face of the protein in the analogous Cas6 of P. furiosus responsible for substrate binding. The N-terminus is coloured in blue and colour fades to orange reaching the C-terminus.
The two glycines (HvoCas6b G256 and G258) that also affected crRNA level are part of the H. volcanii glycine-rich loop (Figure 4) [18]. The glycine-rich loop is the only feature of Cas6 proteins conserved down to the sequence level [45]. It is part of the C-terminal ferredoxin-like fold and protrudes into the protein center at the interface of both subdomains [46, 55, 56]. The G-rich loop has been found to be essential to both Cas6 folding and RNA binding [57, 58].
Interestingly two mutants (HvoCas6b S115A and S224A) have shown elevated amounts of crRNA [18]. According to the structure prediction HvoCas6b Serine115 and 224 are located on the protein surface (Figure 4) where in the analogous P. furiosus Cas6 structure the pre-crRNA is contacted [18, 48, 52]. Mutation might abrogate or weaken this interaction lowering the HvoCas6b binding affinity and increasing turnover of the crRNA substrate [18]. This has also been reported for T. thermophilus Cas6e [52]. Taken together it seems that the specific interaction of crRNA and the Cas6 protein is specific for individual protein orthologs.
The PAM sequence
In type I, II and V systems, an effective interference reaction depends on the identification of a short sequence motif located next to the protospacer called PAM (protospacer adjacent motif) [59]. PAM authentication avoids targeting the CRISPR loci themselves, protecting the cell from genomic damage. PAM sequences can be identified using bioinformatics analyses if enough spacer matches can be identified in the databases. As described earlier, this was not possible for H. volcanii since only a few matches were found. Thus, an experimental approach was used, that employed a plasmid-invader assay to identify PAM sequences for the type I-B system of H. volcanii [19]. Seven three-nucleotide long PAM sequences -TAA, TAG, TAT, TAC, TTC, ACT and CAC- that are located upstream of the protospacer were identified as determinants for prolific interference (Figure 5) [19] (Turgeman-Grott et al., under revision). The presence of multiple active PAM sequences is now a common theme and has since been described in various other species [59]. It might be a strategy to meet the fast pace of viral sequence variation and the resulting divergence of invader populations. Moreover, it renders detection of a closely related mobile genetic element more likely and decreases the likelihood of evasion through single mutations. Furthermore, the capability to recognize multiple PAMs for interference allows the successful utilization of horizontally acquired CRISPR arrays, which is advantageous for haloarchaea.
Figure 5.

Prerequisites for a successful interference – PAM and seed sequences. If a PAM sequence (yellow) located 5′ to the protospacer is detected in the target DNA by Cascade the crRNA binds to the target DNA inducing an R-loop. crRNA binding is initiated at the crRNA 5′ end and must proceed through the seed sequence (shown in red) to lock Cascade binding. The H. volcanii seed sequence is ten nucleotides long and possesses a gap at position 6. After a second gap at position 11, nucleotide 12 must be paired again. Positions 13–18 were also tested but at these positions pairing is not essential for interference.
The PAM identity is read by the Cas8 subunit of Cascade [57]. Upon deletion of cas8b in H. volcanii interference is lost. Compellingly, Cas8b variants with mutations of conserved residues respond differently when presented with different PAM sequences in an invader assay. Recent structural analysis of Cas8e (E. coli) has revealed the read-out of the PAM sequence through an intricate interplay of the minor groove of the double-stranded DNA target and residues within the Cas8e N-terminal domain [60]. A multitude of direct base contacts, base-stacking and steric fitting govern a presumably overall common mechanism for sequence- and shape-dependent PAM sensing. Comparison to the structural details of another Cas8e from Thermobifida fusca reveals species-specific differences that might very well account for the intra-subtype variability of PAM sequences, as did a biochemical analysis of the I-F system of P. aeruginosa, in which PAM identity is read via minor and major groove-interactions [60, 61, 62]. The PAM requirements during interference and adaptation often overlap, but are not identical [36, 63, 64, 65, 66]. Accordingly, PAMs have been subdivided into motifs that are essential for interference (target interference motif: TIM) and motifs that are used during adaptation (spacer acquisition motif: SAM) [64]. Since different proteins are responsible for PAM authentication during adaptation (Cas1 [67, 68]) and interference (Cas8b [57]) such a subdivision of motifs is sensible.
Determinants for crRNA-guided target recognition
The specificity of CRISPR immunity is obtained through the sequence-specific interaction of the spacer portion of crRNA and complementary site within the targeted foreign invader. Targets that are memorized in the spacer content of CRISPR loci are recognized by base-pairing to the spacer portion of crRNA. This process is governed by a seed sequence [69]. Systematic mutagenesis of the protospacer sequence in a plasmid clearance assay has been used to determine the seed sequence of the H. volcanii type I-B system (Figure 5) [21]. The 10 first nucleotides of the protospacer must perfectly match the spacer sequence except for position 6. This is in line with the seed sequences comprising the first eight nucleotides interrupted at position six, as determined for E. coli and P. aeruginosa [69–71]. Structural analysis of the E. coli type I-E complex revealed why the sixth nucleotide of the protospacer was not engaged in base pairing: the spacer runs along the backbone of Cascade, which is built by hand-shaped Cas7 subunits that have their thumb-domain flipped out each sixth base [72, 73, 74]. A recent structural analysis of the T. fusca type I-E Cascade revealed further details on the initiation of hetero-duplex formation in the seed region [62]. Upon PAM identification by the Cas8e subunit the target DNA is bent and spontaneously unwound allowing for strand-invasion by the crRNA component of Cascade. Then, a “seed-bubble” forms, engaging only the first 11 nucleotides of the target DNA. As this corresponds to the seed region, stable formation of this interaction most likely triggers full R-loop formation, engaging the target DNA in increments of six base pairs, as dictated by the “nudging” Cas7 subunits [62]. Interestingly, after the first gap at position six, mutational analysis of the Haloferax seed sequence revealed a second gap in the crRNA-target hetero-duplex at position 13 [21]. However, as detailed in the following section, the type I-B Cascade has been shown to possess more Cas7 subunits than the type I-E complex, which might account for the differences in the back-bone crRNA contact points (see below “Composition of the H. volcanii interference complex”).
Interestingly, the ability of H. volcanii to eliminate plasmid invaders is strongly dependent on the type of plasmid presented, a phenomenon that is not yet fully understood [21]. Plasmids with an origin of replication that utilize a Rep-protein dependent mechanism for replication could not be eliminated, whereas plasmids relying on an origin recognition complex for propagation were readily degraded. This appears to work against the interest of the organism since the former is more typical of smaller plasmids -which are less likely to be beneficial to their hosts- and the latter is similar to the replication of the chromosome. Further experiments are needed to show whether steric constraints owing to the placement of the protospacer in proximity to the origin of replication or functional entanglements are the cause. It is also peculiar that in the aforementioned observation certain spacers did not result in plasmid clearance when used in a plasmid-based invader assay [21]. Interestingly, only two of the seven TIMs identified for Haloferax were used in these experiments and some spacers only triggered interference when paired with one of them. An interplay of PAM and the protospacer sequence was also observed for E. coli, where sensitivity to mismatches in PAM and the seed region was differentially affected during interference [36]. As demonstrated for Streptococcus thermophilus, the Cascade spacer target interaction is “read” kinetically and not at the single-base-pair level [75]. Crosstalk for thermodynamic interactions within PAM, the spacer and the protospacer region and amongst each other and in conjunction with the contacted protein residues might differentially affect the overall stability of the Cascade R-loop conformation and its dynamic properties, resulting in an as yet unexplored interdependency of the sequences for PAM, seed and spacer.
CRISPR RNA – an in-depth analysis
The crRNA is the central player in CRISPR interference, interconnecting the interference machinery and immunological record stored within the CRISPR loci. To determine the Haloferax crRNA characteristics in vivo, a genetic system for the Cas6-independent maturation of crRNAs has been established in Haloferax to uncouple crRNA maturation and crRNA function at the interference step (Figure 3B) [22]. In this system, the sequence of mature crRNA is flanked by so called t-elements, which fold into tRNA like structures that are recognized by the tRNA processing enzymes RNase P and RNase Z. Precise cleavage of RNase Z and RNase P releases a precisely matured crRNA, (termed icrRNA for “independently generated”), which is stably maintained within the cell [22]. The only differences from the native crRNA processed by Cas6 (that generates a 5´-hydroxyl group and a 3´ or 2´-3- phosphate group), are the presence of a 5′- phosphate group and 3′-hydoxyl group. Regardless of these differences, icrRNAs elicit robust interference reactions, and thus, the divergent end groups of the icrRNA hamper neither Cascade integration nor targeting or the interference reaction in the Type I-B system [22]. This is in contrast to P. furiosus, where in the Type III system the nature of the end group determines Cmr-complex incorporation [76].
Efficient interference using an icrRNA in a ∆cas6 strain also revealed that Cas6 is not essential for the interference reaction [22]. Although Cas6 has been co-purified with other components of the Haloferax Cascade, it is not an essential part of the complex [18, 22]. Both the integration of the icrRNA into Cascade and interference reaction are possible without Cas6 [22].
The icrRNA system has also been used to analyse the importance of the 5´ and 3´ repeat-derived handles for crRNA function [22]. The 3′-handle of icrRNA has been systematically shortened and the resulting icrRNA variants were assayed for interference. Defence against a foreign genetic element by the type I-B Cascade of H. volcanii is still possible if the 3′-handle is completely absent. This is consistent with the hypothesis that Cas6 is not a crucial part of Cascade as the crRNA 3′-handle is known to mediate Cas6 binding in other systems [72–74, 77, 78]. Thus, the role of Cas6 appears to be limited to its processing activity during crRNA generation, a finding that was later also confirmed for the type I-E system of E. coli [79].
An analogous mutational analysis of the crRNA 5′-handle using the icrRNA system has included deletions and point mutations of the first and last nucleotide of the handle [22]. While the crRNA 3′-handle is very flexible in length, the crRNA 5′ end can only be shortened by one nucleotide if full activity is to be maintained (Figure 3A). The crRNA appears to be anchored in the type I-B Cascade complex by the 5′-handle binding partners.
Structural analysis of the E. coli Cascade shows that the 5′ end makes contact with a network of polar and charged residues of the Cas5 and Cas7 subunits [72, 73, 74]. However, in contrast to H. volcanii, all eight nucleotides of the crRNA 5′-handle are essential for crRNA loading into Cascade [80]. Therefore, either the crRNA binding configuration within the Cas5 binding pocket varies in the two subtypes or Cas5b can compensate for the loss of the first nucleotide explaining its minor importance.
Composition of the H. volcanii interference complex
The vital element for the interference reaction is the effector complex, termed Cascade, which is assembled by a single crRNA molecule in conjunction with a multitude of Cas proteins (Figure 6). The Cascade composition and structure have been studied for a variety of type I subtypes, and the Cascade organization and overall structure are highly similar even though the Cas building blocks share only limited sequence conservation [72, 73, 74, 77, 78, 81, 82, 83, 84]. Structural data are available for the effector complexes of subtype I-C, I-F and I-Fv, and several extensive structural analyses feature the I-E Cascade [60, 72, 73, 74, 77, 78, 81, 85, 86, 87]. The seahorse-like shape is characteristic, with the crRNA embedded in the helical backbone, comprising multiple copies of the Cas7 protein [88]. The “hand-like” shaped Cas7 subunits are interconnected via thumb-palm interactions around the crRNA. The head and tail subunits cap the crRNA at either end [88]. In the E. coli-type I-E, I-F and I-Fv systems, the head is formed by Cas6 binding to the crRNA 3′ end [72, 73, 74, 77, 78]. By contrast, no Cas6 protein is present in the I-C system of Desulfovibrio vulgaris, where the crRNA 3′ stem-loop structure halts oligomerization of Cas7 [81]. The crRNA 5′ end in both of these types is bound by the Cas5 subunit, which accommodates the 5′ end of the crRNA, forming the tail of the complex [72, 73, 74, 77, 78, 81]. The tail domain also includes the so-called large subunit Cas8, which contacts Cas5 via a short loop [77, 81, 87]. The exception is the variant I-Fv system, in which no Cas8 protein is encoded and the Cas5fv subunit accounts for the functionality of the Cas8 [78].
Figure 6.
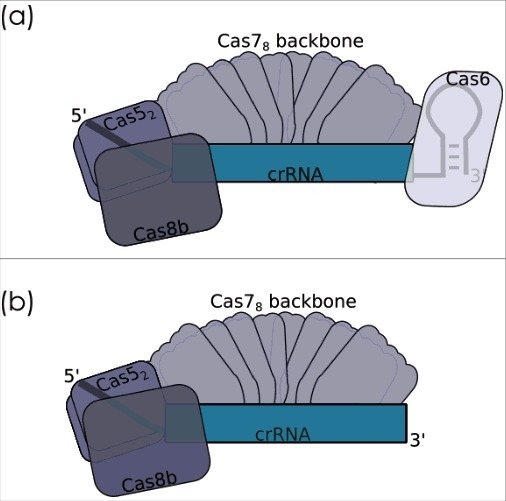
Cascade composition. Composition of the Cascade complex was experimentally determined by a co-purification/mass-spectrometry-iBAQ-quantification approach. (a) The native complex comprises an eight subunit Cas7 backbone alongside two copies of Cas5 and one copy of the Cas6b protein enclosing the crRNA. Although essential for interference, the Cas8b subunit was only weakly associated with the co-purified complex. (b) A Cas6b independent crRNA maturation approach shows that interference is still possible in the absence of Cas6b and with a crRNA missing the 3′handle.
The H. volcanii Cascade structure has not been solved, but the composition of the core complex has been determined using a co-purification assay [18]. Mass-spectrometry and intensity-based absolute quantification (iBAQ) identified Cas5, Cas6b and Cas7 as complex subunits in a stoichiometry of 1.7:1:8.5 [18]. This is in agreement with the composition of the type I-E Cascade of E. coli, which was shown to comprise Cas5, Cas6, Cas7, Cas8 and Cse2 at a 1:1:6:1:2 ratio, whereas the I-C Cascade of D. vulgaris was characterized as Cas5:Cas8:Cas7 at a ratio of 1:1:7 [81, 89, 90]. Cas6 is missing in the I-C Cascade as this subtype does not encode a cas6 gene and the Cas6 function during crRNA maturation is performed by Cas5d [83].
Although Cas6 is present in Cascade complexes of subtype I-E and I-B, in vivo experiments using Cas6-independently generated crRNAs demonstrated that Cas6 is not essential for the assembly and function of the effector complexes, as interference was still elicited in a deletion strain [22, 79]. In subtype I-A of Thermoproteus tenax, Cas6 was not identified as part of the reconstituted Cascade and the ortholog of S. solfataricus only weakly associated with the aCascade [44, 82]. Association of Cas6 with the Cascade complex might depend on its affinity towards the crRNA 3′ end, as E. coli Cas6e was shown to be a single-turnover enzyme, whereas S. solfataricus Cas6 only showed a weak binding affinity towards its cleavage product, implying multiple turnovers [40].
The Haloferax type I-B Cascade accommodates two more copies of the Cas7 backbone subunits compared to the I-E Cascade of E. coli and one more compared to the I-C Cascade [18, 81, 89, 90]. The number of additional Cas7 subunits appears to be correlated with the increase in spacer length compared to E. coli (H. volcanii spacer are 2–7 nucleotides longer). The correlation of the increase or reduction of Cas7 subunits in response to varying crRNA length was demonstrated for type I-F of Shewanella putrefaciens and type I-E of E. coli [91, 92, 93]. Those interspecies differences in spacer length and therefore Cascade composition, together with variations in the Cas7 structure, may also result in differences in the microarchitecture, especially the curvature, of the Cascade backbone. For example the I-F Cascade of P. aeruginosa with six Cas7 subunits exhibits an almost circular shape [77].
In contrast to its role in E. coli and D. vulgaris Cas8 is not an integral part of the core Haloferax Cascade [18], and only a weak association of Cas8b with the Haloferax Cascade has been demonstrated [20]. A similarly weak interaction of Cas8 with the Cascade components was also demonstrated for Cas8a2 of S. solfataricus [94], and although they stably co-purify, Cas8c and Cas8e also appear to be attached to Cascade rather weakly, too [57, 81]. Although the interaction of Cas8b and Cascade is not highly stable, the presence of Cas8b is essential for the interference reaction in subtype I-B. Deletion of cas8b abolishes interference in a plasmid-based invader assay in H. volcanii, as did certain mutations of cas8b [20]. Interestingly, two variants of Cas8b react differently to different PAM sequences presented in a plasmid-based invader assay, hinting at a role for the N-terminal part of this protein in PAM sensing. This is in agreement with structural information available for the Cas8e of E. coli and T. fusca, which revealed details for the PAM-sensing protein-DNA network of the N-terminal domain [60, 62]. However, the very low sequence conservation and extraordinary divergence of the Cas8 protein family does not allow direct extrapolation of those results to Cas8b. Even between the two Cas8e orthologs (TfusCas8e, EcoCas8e), the protein-PAM contacts are rather divergent, and a biochemical study hinted at even more profound within-subtype differences with respect to Cas8f of P. aeruginosa [60, 61, 62].
Self-targeting is tolerated in H. volcanii
The evolutionary advantages of an adaptable, heritable and specific immune system are undisputed, but CRISPR-Cas immunity might also come at a price. DNA targeting harbours the risk of self-targeting if a spacer that matches the genome is present. This turns the DNA degrading activity of the CRISPR-effector against the host genome and widely results in severe cytotoxicity [95, 96]. Acquisition of spacers that, by chance, match the host genome is a common phenomenon and often coincides with the loss of function of the corresponding CRISPR-Cas system or elimination of the targeted region, often resulting in rather large genomic deletions [95, 96, 97, 98, 99, 100]. This downside of CRISPR interference is a likely cause of the variation in the overall presence and prevalence of CRISPR-Cas systems and their patchy distribution across the phylogenetic tree [99, 101]. It could also contribute to the abundance of degenerate CRISPR-Cas systems and orphan cas genes or CRISPR loci [101].
Erroneous acquisition of genomic fragments is not the only conceivable source of self-targeting spacers. Genomic fragments may also be recruited by the continuous flux of genetic material via horizontal gene transfer [102, 103]. CRISPR-Cas systems are often encoded on mobile genetic elements; within the haloarchaea approximately 50% of the CRISPR-related DNA, either CRISPR loci, cas gene cassettes or both are encoded on mega-plasmids or mini-chromosomes [10, 24, 104]. A comprehensive bioinformatics analysis of archaeal CRISPR loci identified numerous spacers matching the genomes of other species, often from the same genus or family or integrated proviruses or plasmids [103,105]. Haloferax species are prone to the exchange of genetic material with closely related species, as mating occurs with a relatively high efficiency [102]. Via between-species mating experiments, it was also demonstrated that the exchanged genomic loci may be rather large, and explicit exchange of the CRISPR-Cas containing pHV4 was also observed [102]. Owing to strong conservation of haloarchaeal CRISPR repeat sequences, it is highly likely that a CRISPR locus acquired via horizontal gene transfer can be utilized by the host CRISPR-Cas system. Despite the danger of acquiring suicidal spacers, the overwhelming majority of haloarchaeal genomes retain a functional CRISPR-Cas system [16]. The implications of targeting an actively transcribed, yet non-essential, chromosomal gene on cellular fitness have recently been studied in H. volcanii [24]. Wild-type cells were transformed with a plasmid encoding a crRNA targeting the crtI gene responsible for carotenoid biosynthesis. Neither the transformation rate nor subsequent growth of transformants was impaired. Moreover, no mutations in the targeted region, which would lead to a loss of colony pigmentation, were observed. Under native conditions, self-targeting crRNA competes with a multitude of endogenous crRNAs. Therefore, only a small number of Cascade complexes targets the genome and the genomic damage caused can be easily repaired. This repair is relatively easy for the organism as H. volcanii, like most haloarchaeal organisms, is polyploid [106]. Intact copies of the genome that may serve as repair templates are also seen to some degree in E. coli, which has many fewer copies of its chromosome per cell [107]. Thus, naturally occurring polyploidy along with the propensity of H. volcanii to undergo homologous recombination buffer the otherwise deleterious effect of self-targeting [24]. In fact, upon growth in phosphate-limited media, resulting in a reduction of ploidy, cell-fitness and the integrity of the targeted locus was compromised [24]. To induce a higher prevalence of targeting of the competent effector complexes a strain devoid of endogenous crRNA maturation was used in follow-up experiments [24]. Even under elevated targeting-stress, only moderate toxicity occured; large deletions of up to 1.5 kilobases were observed within the targeted region. The presence of short homologous sequences flanking the deletion borders suggests that micro homology-mediated end-joining is the most likely repair pathway in this case [24].
H. volcanii can easily avoid self-targeting by recombination between the two CRISPR loci flanking the cas gene cassette, thereby eliminating the CRISPR-Cas system function (Figure 2) [19]. This genomic organization is a common theme within haloarchaea and other archaea such as P. furiosus or Sulfolobus islandicus, but still these species retained their cas operons [108, 109]. This implies that either such recombination events are extremely rare, which is unlikely, or that cas genes have an additional functions that exert a selective pressure for their retention, even in the absence of viral assaults.
CRISPRi in H. volcanii expands the archaeal tool-box
The tools for genetic manipulation of archaea are rather limited. Therefore, expansion of the archaeal toolbox is necessary for further advances in studying archaeal model organisms. CRISPR-Cas systems are a growing source of applications, one of which is CRISPR interference (CRISPRi); in this system, the specificity of CRISPR targeting is exploited in a non-interference permissive background [110]. It has been developed using the type II CRISPR-Cas system of Streptococcus pyogenes, using a variant of the effector protein Cas9 that can bind but not process target DNA and can be used for gene silencing [111]. Provided with a sgRNA that is complementary to a gene of choice, effective down-regulation of gene expression could be achieved in eukaryotes and bacteria [111, 112]. The endogenous type I-E interference complex was harnessed for CRISPRi in E. coli [113, 114]. Along these lines, a CRISPR-Cas system of type III-B has also been repurposed for targeted mRNA degradation [115, 116, 117]. Mutation of the type I complexes from interference competence to action merely as binders is relatively straightforward. DNA binding depends on a functional Cascade complex comprising a crRNA that induces DNA degradation by recruiting Cas3, and thus, cas3 deletion or inactivation strains are a good basis for CRISPRi in type I systems. Repurposing of an endogenous CRISPR-Cas system for gene regulation has become a method of choice for archaeal systems [110]. Most archaeal organisms thrive in rather harsh environments and thermophily, psychrophily, acidophily or halophily make the heterologous expression of mesophilic Cas9 proteins challenging, if not altogether impossible.
The major CRISPR-Cas type found in archaea is of type I; subtypes I-A, -B and -D are most prevalent [15]. The first archaeal type I system repurposed as a molecular biological tool was subtype I-B of H. volcanii (Figure 7) [23]. To eliminate the DNA degradation activity, cas3 was deleted and the crRNA containing Cascade complex was reprogrammed to target a sequence of interest (Figure 7). Substantial repression of a reporter gene was achieved only in the absence of endogenous crRNAs [23]. Therefore, deletion of cas6 or the genomic CRISPR loci was used to reduce competition by endogenous crRNAs. In the first approach with a cas6 deletion strain, endogenous crRNAs were not generated, while the targeting crRNA was supplied by the aforementioned icrRNA system (see above). In the other approach with a CRISPR loci deletion strain, the targeting crRNA is expressed from a synthetic targeting CRISPR locus. The versatility of the system was demonstrated by successful targeting of a plasmid-born gene, a chromosomal gene, and a gene cluster as well as an essential gene. Genes were repressed to 8% of the wild-type transcript level [23]. The targeting spacer sequence was shown to be most effective when directed against the template strand of the promoter region [23]. The high number of interference-competent PAM sequences identified for H. volcanii facilitates easy target-selection. Altogether, CRISPRi in Haloferax has been established as an efficient tool for controlled gene-regulation.
Figure 7.
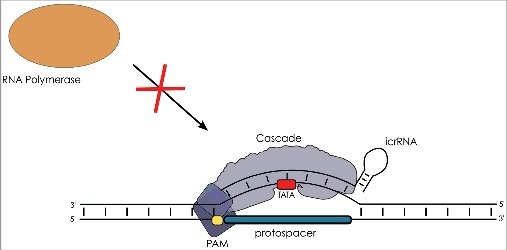
CRISPRi represses transcription initiation. The crRNA containing Cascade complex is programmed by the icrRNA to target the promoter (TATA indicated in red) of a gene of interest. Cascade binds to the promoter region blocking access of the RNA polymerase (orange) and thereby transcription initiation. The strain used for CRISPRi has both the cas3 gene and the cas6 gene deleted to avoid target DNA degradation and to optimise the repression effect.
Concluding remarks
Although their major role as defence weaponry in prokaryotes has been known for more than a decade, there is still much that is unknown regarding CRISPR-Cas systems. The details of both the interference and adaptation reactions are still not fully understood. It is becoming clearer that although different versions of the system perform the same function, they can do so using highly variable components, with crRNA perhaps being the most constant component. The diversity of versions is reflected by the more than 33 subtypes that are currently known or proposed [13], yet even within the subtypes there is great variation from organism to organism, possibly reflecting the constant evolution of this system. Therefore, studies of CRISPR-Cas systems should not focus on a few model organisms but rather expand to as many organisms as possible, including more archaeal systems that have not been explored. Indeed such explorations have resulted in several exciting discoveries such as new families of anti-CRISPR proteins that disable type I-D CRISPR-Cas immunity in crenarchaeotes [118]. Although the haloarchaea in which CRISPR-Cas systems have been studied thus far have very active systems, and probably do not encode anti-CRISPR proteins, a vast collection of related strains with sequenced genomes that encode CRISPR-Cas systems has recently become available ([119]; Turgeman-Grott et al., under revision). These strains have the potential to provide us with the first examples of euryarchaeal anti-CRISPR proteins.
A deep and thorough understanding of a variety of CRISPR-Cas types and subtypes opens the possibility of their exploitation as molecular biology tools. This is of special importance in the case of species dwelling in extreme ecological niches, where CRISPR-Cas tools used in mesophilic model organisms, such as S. pyogenes Cas9, are non-functional. Using endogenous CRISPR-Cas systems as a tool will open new possibilities, especially in studying biological processes in the archaeal domain.
An intriguing question for the H. volcanii CRISPR-Cas system is why it remains intact and constitutively active even when the strain is not exposed to invaders during laboratory cultivation for three decades, despite the fact that cas gene elimination by recombination is frequently observed under appropriate selection. This already hints at further perspectives that will focus on CRISPR-Cas functions beyond defence, which may link the CRISPR-Cas system, as a whole or individual components, to additional physiological pathways.
Funding Statement
This work was supported by the DFG under grant MA1538/16-1, 16-2, 17-1, 17-2, UR225/1-1 and 1-2, BA2168/5-1 and 5-2 (all part of FOR1680), and the Israel Science Foundation under grant 535/15.
Abbreviations
- CRISPR
Clustered Regularly Interspaced Short Palindromic Repeats
- Cas
CRISPR associated
- Cascade
CRISPR-associated complex for antiviral defense
- icrRNA
independently generated crRNA
- iBAQ
intensity-based absolute quantification
- PAM
protospacer adjacent motif
- SAM
spacer acquisition motif
- TIM
target interference motif
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
References
- [1].Mojica FJM, Ferrer C, Juez G, et al. . Long stretches of short tandem repeats are present in the largest replicons of the Archaea Haloferax mediterranei and Haloferax volcanii and could be involved in replicon partitioning. Mol. Microbiol. 1995;17:85–93. [DOI] [PubMed] [Google Scholar]
- [2].Mojica FJM, Juez G, Rodriguez-Valera F. Transcription at different salinities of Haloferax mediterranei sequences adjacent to partially modified PstI sites. Mol. Microbiol. 1993;9(3):613–621. [DOI] [PubMed] [Google Scholar]
- [3].Mojica FJ, Díez-Villaseñor C, Soria E, et al. . Biological significance of a family of regularly spaced repeats in the genomes of Archaea, Bacteria and mitochondria. Mol. Microbiol. 2000;36:244–246. [DOI] [PubMed] [Google Scholar]
- [4].Ishino Y, Shinagawa H, Makino K, et al. . Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product. J. Bacteriol. 1987;169:5429–5433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Groenen PMA, Bunschoten AE, Dv Soolingen, et al. . Nature of DNA polymorphism in the direct repeat cluster of Mycobacterium tuberculosis; application for strain differentiation by a novel typing method. Mol. Microbiol. 1993;10:1057–1065. [DOI] [PubMed] [Google Scholar]
- [6].Masepohl B, Gorlitz K, Bohme H. Long tandemly repeated repetitive (LTRR) sequences in the filamentous cyanobacterium Anabaena sp. PCC 7120. Biochim Biophys Acta. 1996;1307(1):26–30. [DOI] [PubMed] [Google Scholar]
- [7].Mojica FJ, García-Martínez J, Soria E. Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J. Mol. Evol. 2005;60:174–182. [DOI] [PubMed] [Google Scholar]
- [8].Bolotin A, Quinquis B, Sorokin A, et al. . Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology. 2005;151:2551–2561. [DOI] [PubMed] [Google Scholar]
- [9].Pourcel C, Salvignol G, Vergnaud G. CRISPR elements in Yersinia pestis acquire new repeats by preferential uptake of bacteriophage DNA, and provide additional tools for evolutionary studies. Microbiology. 2005;151:653–663. [DOI] [PubMed] [Google Scholar]
- [10].Grissa I, Vergnaud G, Pourcel C. The CRISPRdb database and tools to display CRISPRs and to generate dictionaries of spacers and repeats. BMC Bioinformatics. 2007;8:172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Amitai G, Sorek R. CRISPR-Cas adaptation: insights into the mechanism of action. Nat. Rev. Microbiol. 2016;14:67–76. [DOI] [PubMed] [Google Scholar]
- [12].Mohanraju P, Makarova KS, Zetsche B, et al. . Diverse evolutionary roots and mechanistic variations of the CRISPR-Cas systems. Science. 2016;353:aad5147. [DOI] [PubMed] [Google Scholar]
- [13].Koonin EV, Makarova KS, Zhang F. Diversity, classification and evolution of CRISPR-Cas systems. Curr. Opin. Microbiol. 2017;37:67–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Makarova KS, Wolf YI, Alkhnbashi OS, et al. . An updated evolutionary classification of CRISPR–Cas systems. Nat. Rev. Microbiol. 2015;13:722–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Vestergaard G, Garrett RA, Shah SA. CRISPR adaptive immune systems of Archaea. RNA Biology. 2014;11:156–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Maier L-K, Alkhnbashi OS, Backofen R, et al. . CRISPR and Salty: CRISPR-Cas Systems in Haloarchaea. In: Clouet-d'Orval B, editor RNA Metabolism and Gene Expression in Archaea. Vol. 32 Cham: Springer International Publishing; 2017. p. 243–269. [Google Scholar]
- [17].Makarova KS, Wolf YI, Alkhnbashi OS, et al. . An updated evolutionary classification of CRISPR-Cas systems. Nat Rev Micro. 2015;13:722–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Brendel J, Stoll B, Lange SJ, et al. . A Complex of Cas Proteins 5, 6, and 7 Is Required for the Biogenesis and Stability of Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR)-derived RNAs (crRNAs) in Haloferax volcanii. J. Biol. Chem. 2014;289:7164–7177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Fischer S, Maier L-K, Stoll B, et al. . An archaeal immune system can detect multiple protospacer adjacent motifs (PAMs) to target invader DNA. J. Biol. Chem. 2012;287:33351–33363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Cass SDB, Haas KA, Stoll B, et al. . The role of Cas8 in type I CRISPR interference. Biosci. Rep. 2015;35:e00197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Maier L-K, Lange SJ, Stoll B, et al. . Essential requirements for the detection and degradation of invaders by the Haloferax volcanii CRISPR/Cas system I-B. RNA Biology. 2013;10:865–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Maier L-K, Stachler A-E, Saunders SJ, et al. . An Active Immune Defense with a Minimal CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats) RNA and without the Cas6 Protein. J. Biol. Chem. 2015;290:4192–4201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Stachler AE, Marchfelder A. Gene Repression in Haloarchaea Using the CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats)-Cas I-B System. J Biol Chem. 2016;291(29):15226–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Stachler A-E, Turgeman-Grott I, Shtifman-Segal E, et al. . High tolerance to self-targeting of the genome by the endogenous CRISPR-Cas system in an archaeon. Nucleic Acids Res. 2017;45:5208–5216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Makarova KS, Aravind L, Wolf YI, et al. . Unification of Cas protein families and a simple scenario for the origin and evolution of CRISPR-Cas systems. Biol Direct. 2011;6:38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Lange SJ, Alkhnbashi OS, Rose D, et al. . CRISPRmap: an automated classification of repeat conservation in prokaryotic adaptive immune systems. Nucleic Acids Res. 2013;41:8034–8044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Maier L-K, Dyall-Smith M, Marchfelder A. The Adaptive Immune System of Haloferax volcanii. Life. 2015;5:521–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Sampson TR, Saroj SD, Llewellyn AC, et al. . A CRISPR/Cas system mediates bacterial innate immune evasion and virulence. Nature. 2013;497:254–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Deng L, Kenchappa CS, Peng X, et al. . Modulation of CRISPR locus transcription by the repeat-binding protein Cbp1 in Sulfolobus. Nucleic Acids Res. 2012;40:2470–2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Hale CR, Majumdar S, Elmore J, et al. . Essential features and rational design of CRISPR RNAs that function with the Cas RAMP module complex to cleave RNAs. Mol. Cell. 2012;45:292–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Nickel L, Weidenbach K, Jäger D, et al. . Two CRISPR-Cas systems in Methanosarcina mazei strain Gö1 display common processing features despite belonging to different types I and III. RNA biology. 2013;10:779–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Richter H, Zoephel J, Schermuly J, et al. . Characterization of CRISPR RNA processing in Clostridium thermocellum and Methanococcus maripaludis. Nucleic Acids Res. 2012;40:9887–9896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Scholz I, Lange SJ, Hein S, et al. . CRISPR-Cas systems in the cyanobacterium Synechocystis sp. PCC6803 exhibit distinct processing pathways involving at least two Cas6 and a Cmr2 protein. PLoS One. 2013;8:e56470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Zhang J, Rouillon C, Kerou M, et al. . Structure and mechanism of the CMR complex for CRISPR-mediated antiviral immunity. Mol. Cell. 2012;45:303–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Reimann V, Alkhnbashi OS, Saunders SJ, et al. . Structural constraints and enzymatic promiscuity in the Cas6-dependent generation of crRNAs. Nucleic Acids Res. 2017;45(2):915–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Xue C, Seetharam AS, Musharova O, et al. . CRISPR interference and priming varies with individual spacer sequences. Nucleic Acids Res. 2015;43:10831–10847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Kunin V, Sorek R, Hugenholtz P. Evolutionary conservation of sequence and secondary structures in CRISPR repeats. Genome Biol. 2007;8:R61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Wang R, Zheng H, Preamplume G, et al. . The impact of CRISPR repeat sequence on structures of a Cas6 protein-RNA complex. Protein Sci. 2012;21:405–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Hochstrasser ML, Doudna JA. Cutting it close: CRISPR-associated endoribonuclease structure and function. Trends Biochem. Sci. 2015;40:58–66. [DOI] [PubMed] [Google Scholar]
- [40].Li H. Structural Principles of CRISPR RNA Processing. Structure. 2015;23:13–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Li M, Liu H, Han J, et al. . Characterization of CRISPR RNA Biogenesis and Cas6 Cleavage-Mediated Inhibition of a Provirus in the Haloarchaeon Haloferax mediterranei. J. Bacteriol. 2013;195:867–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Hatoum-Aslan A, Maniv I, Marraffini LA. Mature clustered, regularly interspaced, short palindromic repeats RNA (crRNA) length is measured by a ruler mechanism anchored at the precursor processing site. Proc. Natl. Acad. Sci. 2011;108:21218–21222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Tamulaitis G, Venclovas C, Siksnys V. Type III CRISPR-Cas Immunity: Major Differences Brushed Aside. Trends Microbiol. 2017;25(1):49–61. [DOI] [PubMed] [Google Scholar]
- [44].Plagens A, Tripp V, Daume M, et al. . In vitro assembly and activity of an archaeal CRISPR-Cas type I-A Cascade interference complex. Nucleic Acids Res. 2014;42:5125–5138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Niewoehner O, Jinek M, Doudna JA. Evolution of CRISPR RNA recognition and processing by Cas6 endonucleases. Nucleic Acids Res. 2014;42:1341–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Shao Y, Li H. Recognition and cleavage of a nonstructured CRISPR RNA by its processing endoribonuclease Cas6. Structure. 2013;21:385–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Shao Y, Richter H, Sun S, et al. . A Non-Stem-Loop CRISPR RNA Is Processed by Dual Binding Cas6. Structure. 2016;24:547–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Carte J, Wang R, Li H, et al. . Cas6 is an endoribonuclease that generates guide RNAs for invader defense in prokaryotes. Genes & Development. 2008;22:3489–3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Carte J, Pfister NT, Compton MM, et al. . Binding and cleavage of CRISPR RNA by Cas6. RNA. 2010;16:2181–2188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Kelley LA, Mezulis S, Yates CM, et al. . The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc. 2015;10(6):845–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Niewoehner O, Jinek M, Doudna JA. Evolution of CRISPR RNA recognition and processing by Cas6 endonucleases. Nucleic Acids Res. 2014;42:1341–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Sashital DG, Jinek M, Doudna JA. An RNA-induced conformational change required for CRISPR RNA cleavage by the endoribonuclease Cse3. Nature Structural & Molecular Biology. 2011;18:680–687. [DOI] [PubMed] [Google Scholar]
- [53].Haurwitz RE, Sternberg SH, Doudna JA. Csy4 relies on an unusual catalytic dyad to position and cleave CRISPR RNA: Mechanism of CRISPR RNA cleavage. EMBO J. 2012;31:2824–2832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Sternberg SH, Haurwitz RE, Doudna JA. Mechanism of substrate selection by a highly specific CRISPR endoribonuclease. RNA. 2012;18:661–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Reeks J, Naismith James H, White Malcolm F. CRISPR interference: a structural perspective. Biochem. J. 2013;453:155–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Wang R, Preamplume G, Terns MP, et al. . Interaction of the Cas6 riboendonuclease with CRISPR RNAs: recognition and cleavage. Structure (London, England: 1993). 2011;19:257–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Sashital DG, Wiedenheft B, Doudna JA. Mechanism of foreign DNA selection in a bacterial adaptive immune system. Mol. Cell. 2012;46:606–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Wang R, Preamplume G, Terns MP, et al. . Interaction of the Cas6 Riboendonuclease with CRISPR RNAs: Recognition and Cleavage. Structure. 2011;19:257–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Leenay RT, Beisel CL. Deciphering, Communicating, and Engineering the CRISPR PAM. J Mol Biol. 2017;429(2):177–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Hayes RP, Xiao Y, Ding F, et al. . Structural basis for promiscuous PAM recognition in type I–E Cascade from E. coli. Nature. 2016;530:499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Rollins MF, Schuman JT, Paulus K, et al. . Mechanism of foreign DNA recognition by a CRISPR RNA-guided surveillance complex from Pseudomonas aeruginosa. Nucleic Acids Res. 2015;43:2216–2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Xiao Y, Luo M, Hayes RP, et al. . Structure Basis for Directional R-loop Formation and Substrate Handover Mechanisms in Type I CRISPR-Cas System. Cell. 2017;170:48–60.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Li M, Wang R, Xiang H. Haloarcula hispanica CRISPR authenticates PAM of a target sequence to prime discriminative adaptation. Nucleic Acids Res. 2014;42:7226–7235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Shah SA, Erdmann S, Mojica FJ, et al. . Protospacer recognition motifs. RNA biology. 2013;10:891–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Savitskaya E, Semenova E, Dedkov V, et al. . High-throughput analysis of type IE CRISPR/Cas spacer acquisition in E. coli . RNA biology. 2013;10:716–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Fineran PC, Gerritzen MJ, Suárez-Diez M, et al. . Degenerate target sites mediate rapid primed CRISPR adaptation. Proc. Natl. Acad. Sci. 2014;111:E1629–E1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Nuñez JK, Kranzusch PJ, Noeske J, et al. . Cas1–Cas2 complex formation mediates spacer acquisition during CRISPR–Cas adaptive immunity. Nature structural & molecular biology. 2014;21:528–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Wang J, Li J, Zhao H, et al. . Structural and Mechanistic Basis of PAM-Dependent Spacer Acquisition in CRISPR-Cas Systems. Cell. 2015;163:840–853. [DOI] [PubMed] [Google Scholar]
- [69].Semenova E, Jore MM, Datsenko KA, et al. . Interference by clustered regularly interspaced short palindromic repeat (CRISPR) RNA is governed by a seed sequence. Proc. Natl. Acad. Sci. 2011;108:10098–10103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Cady KC, Bondy-Denomy J, Heussler GE, et al. . The CRISPR/Cas Adaptive Immune System of Pseudomonas aeruginosa Mediates Resistance to Naturally Occurring and Engineered Phages. J. Bacteriol. 2012;194:5728–5738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Wiedenheft B, van Duijn E, Bultema JB, et al. . RNA-guided complex from a bacterial immune system enhances target recognition through seed sequence interactions. Proc. Natl. Acad. Sci. 2011;108:10092–10097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Jackson RN, Golden SM, van Erp PBG, et al. . Crystal structure of the CRISPR RNA-guided surveillance complex from Escherichia coli. Science. 2014;345:1473–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Mulepati S, Heroux A, Bailey S. Crystal structure of a CRISPR RNA-guided surveillance complex bound to a ssDNA target. Science. 2014;345:1479–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Zhao H, Sheng G, Wang J, et al. . Crystal structure of the RNA-guided immune surveillance Cascade complex in Escherichia coli. Nature. 2014;515:147–150. [DOI] [PubMed] [Google Scholar]
- [75].Rutkauskas M, Sinkunas T, Songailiene I, et al. . Directional R-Loop Formation by the CRISPR-Cas Surveillance Complex Cascade Provides Efficient Off-Target Site Rejection. Cell Reports. 2015;10:1534–1543. [DOI] [PubMed] [Google Scholar]
- [76].Hale CR, Cocozaki A, Li H, et al. . Target RNA capture and cleavage by the Cmr type III-B CRISPR–Cas effector complex. Genes & Development. 2014;28:2432–2443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Chowdhury S, Carter J, Rollins MF, et al. . Structure Reveals Mechanisms of Viral Suppressors that Intercept a CRISPR RNA-Guided Surveillance Complex. Cell. 2017;169:47–57.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Pausch P, Müller-Esparza H, Gleditzsch D, et al. . Structural Variation of Type I-F CRISPR RNA Guided DNA Surveillance. Mol. Cell. 2017;67:622–632.e4. [DOI] [PubMed] [Google Scholar]
- [79].Semenova E, Kuznedelov K, Datsenko KA, et al. . The Cas6e ribonuclease is not required for interference and adaptation by the E. coli type I-E CRISPR-Cas system. Nucleic Acids Res. 2015;43:6049–6061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Beloglazova N, Kuznedelov K, Flick R, et al. . CRISPR RNA binding and DNA target recognition by purified Cascade complexes from Escherichia coli. Nucleic Acids Res. 2015;43:530–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Hochstrasser ML, Taylor DW, Kornfeld JE, et al. . DNA Targeting by a Minimal CRISPR RNA-Guided Cascade. Mol. Cell. 2016;63:840–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Lintner NG, Kerou M, Brumfield SK, et al. . Structural and functional characterization of an archaeal clustered regularly interspaced short palindromic repeat (CRISPR)-associated complex for antiviral defense (CASCADE). J. Biol. Chem. 2011;286:21643–21656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Nam KH, Haitjema C, Liu X, et al. . Cas5d protein processes pre-crRNA and assembles into a cascade-like interference complex in subtype IC/Dvulg CRISPR-Cas system. Structure. 2012;20:1574–1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Majumdar S, Ligon M, Skinner WC, et al. . Target DNA recognition and cleavage by a reconstituted Type I-G CRISPR-Cas immune effector complex. Extremophiles. 2017;21:95–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].van Erp PBG Patterson A, Kant R, et al. . Conformational Dynamics of DNA Binding and Cas3 Recruitment by the CRISPR RNA-guide Cascade Complex. ACS Chem. Biol. 2017;13:481–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].van Erp Paul BG Jackson RN, Carter J, et al. . Mechanism of CRISPR-RNA guided recognition of DNA targets in Escherichia coli. Nucleic Acids Res. 2015;43:8381–8391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Jackson RN, van Erp PBG, Sternberg SH, et al. . Conformational regulation of CRISPR-associated nucleases. Curr. Opin. Microbiol. 2017;37:110–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Jackson Ryan N, Wiedenheft B. A Conserved Structural Chassis for Mounting Versatile CRISPR RNA-Guided Immune Responses. Mol. Cell. 2015;58:722–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Jore MM, Lundgren M, van Duijn E, et al. . Structural basis for CRISPR RNA-guided DNA recognition by Cascade. Nature Structural & Molecular Biology. 2011;18:529–536. [DOI] [PubMed] [Google Scholar]
- [90].Wiedenheft B, Lander GC, Zhou K, et al. . Structures of the RNA-guided surveillance complex from a bacterial immune system. Nature. 2011;477:486–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Gleditzsch D, Müller-Esparza H, Pausch P, et al. . Modulating the Cascade architecture of a minimal Type I-F CRISPR-Cas system. Nucleic Acids Res. 2016;44:5872–5882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Kuznedelov K, Mekler V, Lemak S, et al. . Altered stoichiometry Escherichia coli Cascade complexes with shortened CRISPR RNA spacers are capable of interference and primed adaptation. Nucleic Acids Res. 2016;44:10849–10861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Luo ML, Jackson RN, Denny SR, et al. . The CRISPR RNA-guided surveillance complex in Escherichia coli accommodates extended RNA spacers. Nucleic Acids Res. 2016;44(15):7385–7394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Lintner NG, Frankel KA, Tsutakawa SE, et al. . The Structure of the CRISPR-Associated Protein Csa3 Provides Insight into the Regulation of the CRISPR/Cas System. J. Mol. Biol. 2011;405:939–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Vercoe RB, Chang JT, Dy RL, et al. . Cytotoxic chromosomal targeting by CRISPR/Cas systems can reshape bacterial genomes and expel or remodel pathogenicity islands. PLos Genet. 2013;9:e1003454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Selle K, Klaenhammer TR, Barrangou R. CRISPR-based screening of genomic island excision events in bacteria. Proc Natl Acad Sci U S A. 2015;112(26):8076–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Stern A, Keren L, Wurtzel O, et al. . Self-targeting by CRISPR: gene regulation or autoimmunity? Trends Genet. 2010;26:335–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Li Y, Pan S, Zhang Y, et al. . Harnessing Type I and Type III CRISPR-Cas systems for genome editing. Nucleic Acids Res. 2016;44(4):e34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Jiang W, Bikard D, Cox D, et al. . RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat. Biotechnol. 2013;31:233–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Dy RL, Pitman AR, Fineran PC. Chromosomal targeting by CRISPR-Cas systems can contribute to genome plasticity in bacteria. Mob Genet Elements. 2013;3(5):e26831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Stern A, Keren L, Wurtzel O, et al. . Self-targeting by CRISPR: gene regulation or autoimmunity? Trends in Genetics. 2010;26:335–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Naor A, Lapierre P, Mevarech M, et al. . Low species barriers in halophilic archaea and the formation of recombinant hybrids. Curr Biol. 2012;22(15):1444–8. [DOI] [PubMed] [Google Scholar]
- [103].Brodt A, Lurie-Weinberger MN, Gophna U. CRISPR loci reveal networks of gene exchange in archaea. Biol Direct. 2011;6:65–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Lillestøl RK, Shah SA, Brügger K, et al. . CRISPR families of the crenarchaeal genus Sulfolobus: bidirectional transcription and dynamic properties. Mol. Microbiol. 2009;72:259–272. [DOI] [PubMed] [Google Scholar]
- [105].Liu G, She Q, Garrett RA. Diverse CRISPR-Cas responses and dramatic cellular DNA changes and cell death in pKEF9-conjugated Sulfolobus species. Nucleic Acids Res. 2016;44(9):4233–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Zerulla K, Chimileski S, Nather D, et al. . DNA as a phosphate storage polymer and the alternative advantages of polyploidy for growth or survival. PLoS One. 2014;9(4):e94819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Cui L, Bikard D. Consequences of Cas9 cleavage in the chromosome of Escherichia coli. Nucleic Acids Res. 2016;44(9):4243–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Terns RM, Terns MP. The RNA- and DNA-targeting CRISPR-Cas immune systems of Pyrococcus furiosus. Biochem Soc Trans. 2013;41(6):1416–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Jaubert C, Danioux C, Oberto J, et al. . Genomics and genetics of Sulfolobus islandicus LAL14/1, a model hyperthermophilic archaeon. Open Biol. 2013;3(4):130010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Gophna U, Allers T, Marchfelder A. Finally, Archaea Get Their CRISPR-Cas Toolbox. Trends in Microbiology. 2017;25:430–432. [DOI] [PubMed] [Google Scholar]
- [111].Qi LS, Larson MH, Gilbert LA, et al. . Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013;152:1173–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Bikard D, Jiang W, Samai P, et al. . Programmable repression and activation of bacterial gene expression using an engineered CRISPR-Cas system. Nucleic Acids Res. 2013;41:7429–7437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Rath D, Amlinger L, Hoekzema M, et al. . Efficient programmable gene silencing by Cascade. Nucleic Acids Res. 2015;43:237–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Luo ML, Mullis AS, Leenay RT, et al. . Repurposing endogenous type I CRISPR-Cas systems for programmable gene repression. Nucleic Acids Res. 2015;43:674–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Zebec Z, Manica A, Zhang J, et al. . CRISPR-mediated targeted mRNA degradation in the archaeon Sulfolobus solfataricus. Nucleic Acids Res. 2014;42(8):5280–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Peng W, Feng M, Feng X, et al. . An archaeal CRISPR type III-B system exhibiting distinctive RNA targeting features and mediating dual RNA and DNA interference. Nucleic Acids Res. 2015;43(1):406–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Hale Caryn R, Majumdar S, Elmore J, et al. . Essential Features and Rational Design of CRISPR RNAs that Function with the Cas RAMP Module Complex to Cleave RNAs. Mol. Cell. 2012;45:292–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].He F, Bhoobalan-Chitty Y, Van LB, et al. . Anti-CRISPR proteins encoded by archaeal lytic viruses inhibit subtype I-D immunity. Nat Microbiol. 2018;5(10):018–0120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Naor A, Altman-Price N, Soucy SM, et al. . Impact of a homing intein on recombination frequency and organismal fitness. Proc Natl Acad Sci U S 1135 A. 2016;113(32):E4654–61. [DOI] [PMC free article] [PubMed] [Google Scholar]


