ABSTRACT
MicroRNAs (miRNAs) are small, non-coding RNAs that post-transcriptionally regulate gene expression. Aberrant miRNA expression or function have close links with various human diseases. Therefore, therapeutic treatments with disease-associated miRNAs as targets are emerging. However, the intracellular miRNA networks are extremely complicated and poorly understood, which thus hinder the development of miRNA-targeted therapeutics. Small molecules that are able to regulate endogenous miRNAs hold great potential in both elucidation of miRNA networks and treatment of miRNA-related diseases. Herein, we summarize current strategies for discovery of small molecule modifiers of miRNAs, and we highlight aspects of miRNA cellular biology elucidated by using these small molecules and miRNA-targeted therapeutics realized by these small molecules. We envision that this area will expand dramatically in the near future and will ultimately contribute to a better understanding of miRNA-involved cellular processes and development of therapeutic agents for miRNA-associated diseases.
KEYWORDS: Microrna, small molecule, microRNA network, therapeutics
1. Introduction
MicroRNAs (miRNAs) are endogenous small non-coding RNAs in eukaryotic lineages, which comprise ~22 nucleotides and exert post-transcriptional gene silencing function [1,2]. The first miRNA, lin-4, that was essential for proper timing of development was discovered in C. elegans in 1993 [3,4]. However, not until let-7 was identified in C. elegans in 2000 and was found to be conserved in humans and other bilaterian animals [5,6] did the science community pay enough attention to these small RNAs. This discovery then led to a rapid expansion of miRNA library and a big revolution of the RNA world. After that, thousands of miRNAs have been identified and annotated in miRBase, which is an online miRNA database [7]. The latest miRBase release (v22, 2018) contains 38,589 miRNA loci from 271 species that can express 48,885 mature miRNAs [8]. With the development and application of small RNA deep sequencing, the number of novel miRNAs still keeps increasing.
The mode of action of miRNAs involves recognition of the 3ʹ-untranslated region (3ʹ-UTR) of target mRNAs through sequence complementarity and recruitment of nucleases to repress mRNA expression [9,10]. The two to eight nucleotides at the 5ʹ-end of miRNAs is the seed region, which is conserved among miRNA family and directs target recognition. The fate of target mRNAs depends on their complementarity with miRNAs. High complementarity causes mRNA degradation while partial complementarity prevents target mRNAs from being translated [11]. Due to their small size and seed-match property, miRNAs have a wide-spread regulation on mRNA expression. Bioinformatic analysis reveals that approximately 60% of human mRNAs are conserved targets of miRNAs [12]. Moreover, one miRNA can target multiple mRNAs and a single mRNA can also be targeted by multiple miRNAs. Thus, the miRNA-mRNA networks inside cells are extremely complicated, and it is also difficult to find a developmental or physiological process without the influence of miRNAs [13,14].
Since the wide-spread regulatory roles of miRNAs, it is no wonder that aberrant expression or function of miRNAs greatly contributes to human disorders [15,16]. The roles of miRNAs in human diseases, especially in cancers, have been extensively explored in recent years. For instance, miR-221 overexpression contributes to hepatocarcinogenesis through targeting DNA damage-inducible transcript 4 (DDIT4) [17] and miR-34 prevents the progression of lung cancer through targeting MET and BCL-2 [18]. The important discoveries of roles of miRNAs in human diseases consequently lead to an accelerating development of therapeutics with endogenous miRNAs as targets [19]. Oligonucleotides that can suppress or replace endogenous miRNAs upon proper delivery into cells are the first choice to target miRNAs and have even entered clinic trials for treatment of lung cancer or hepatitis C virus (HCV) [19–21]. Nevertheless, the poor delivery efficiency, poor tissue distribution, and severe side effects of oligonucleotides hamper their therapeutic utility [19,22]. Moreover, endogenous RNAs usually form secondary structures, which are hard to be targeted by anti-sense oligonucleotides that are most effective when targeting unstructured regions [23,24]. Small molecules that have better cellular permeability and broader tissue distribution are promising alternatives of oligonucleotides for regulating endogenous miRNAs [22,25–27].
The bioactivity and broad chemical space of small molecules also make them powerful tools in both chemical biology and biomedicine. Small molecules can be optimized via medicinal chemistry to dissect-related signalling transduction pathways and to regulate target biomolecules to achieve therapeutic purposes [28,29]. For miRNAs, the complexity of miRNA-mRNA networks and their critical roles in human diseases require proper and efficient tools for elucidation of these networks and regulation of miRNAs for therapeutic treatments. Even though RNAs have long been considered as undruggable targets for small molecules, recent work indicate that miRNAs can be targeted by small molecules obtained from high-throughput screening or appropriate design [30–36]. In this review paper, we will summarize current strategies for discovery of small molecule modifiers of miRNAs, their mechanisms of action and their use in probing miRNA-mRNA network and miRNA-targeted therapeutics.
2. Biogenesis of microRNA
As shown in Figure 1, the biogenesis of miRNA starts from the transcription of miRNA gene by RNA polymerase II (Pol II) in nucleus [37]. The long RNA transcript, dubbed primary miRNA (pri-miRNA), folds back on itself to form a hairpin structure for further processing by Drosha. Two RNase III domains of Drosha cut the two strands of stem of pri-miRNA, yielding a ~ 60–70 nt stem-loop named as precursor miRNA (pre-miRNA) [38]. After the liberation, pre-miRNA is translocated into the cytoplasm through the action of Exportin 5 and further cleaved by Dicer [39]. Dicer is an endonuclease with two RNase III domains and forms a complex with trans-activation response RNA binding protein (TRBP) for pre-miRNA processing [40]. Both strands near the loop of pre-miRNA are cut by Dicer to generate a miRNA duplex of ~22 nt in each strand. After that, the miRNA duplex, including the guide strand (miRNA) and the passenger strand (miRNA*), is loaded onto Argonaute 2 (AGO2) protein to from the RNA-induced silencing complex (RISC) [41]. After removal of the passenger strand, RISC binds to the 3ʹ-UTR of target mRNA through sequence complementarity between miRNA and target mRNA. The catalytic engine of RISC is AGO2, which carries out mRNA degradation or translational repression of mRNA [42]. More details regarding to miRNA biogenesis can be reached in several excellent review articles [1,41] and will not be introduced in depth here.
Figure 1.
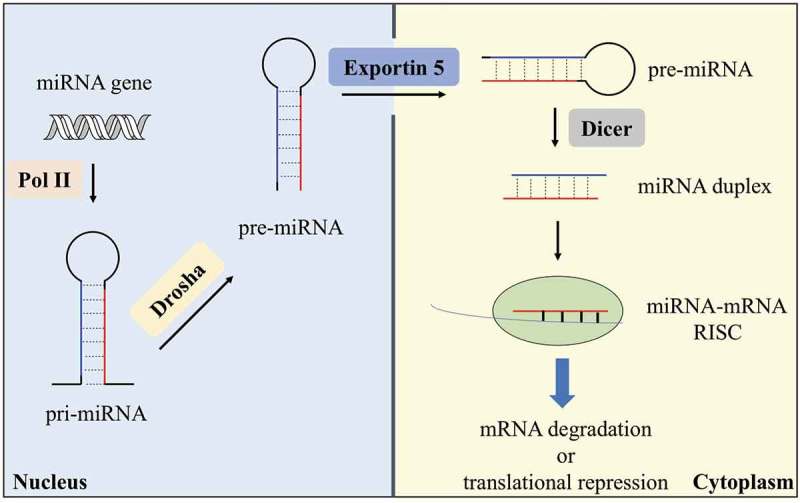
Biogenesis of a typical miRNA. MiRNA gene is transcribed by Pol II in the nucleus into pri-miRNA. The pri-miRNA is then processed by Drosha into pre-miRNA, which is transported into the cytoplasm by Exportin 5. Dicer processes pre-miRNA in the cytoplasm to give a miRNA duplex, and the mature miRNA then forms a miRNA-mRNA RISC complex to induce mRNA degradation or translational repression.
In the biogenesis pathway of miRNA, many factors are involved to control the generation of mature miRNA and the downstream mRNAs it targets. Although intense studies have been undertaken to reveal some important factors involved in miRNA biogenesis, the detailed mechanisms such as other co-factors of Drosha, Dicer or AGO2 that will determine the location and function of miRNA still wait to be explored to globally understand the biogenesis and function of miRNA. Meanwhile, the miRNA-mRNA networks inside cells are also extremely complicated, which are still far from being understood. Therefore, proper tools are needed to probe the miRNA biogenesis and miRNA-mRNA networks.
3. Roles of microRNAs in human diseases
In general, intracellular miRNA expression or function is tightly controlled. There are now considerable evidences to support that miRNAs with abnormal expression or function have close links with the development of diverse human diseases, especially cancers. These abnormal miRNAs can be generally classified into two groups, tumour-suppressive miRNAs and oncogenic miRNAs. In this section, we will discuss some prominent examples. Since roles of miRNAs are diversified, here we will just introduce their general functions. For detailed miRNA functions, we will refer to other excellent review articles [15,16,19,43,44].
MiR-34 family, one of the best-studied tumour-suppressive miRNAs, has received substantial attention. MiR-34 family has three members, miR-34a, miR-34b and miR-34c. Tumour suppressor p53 can transcriptionally regulate miR-34 during DNA damage response [45]. MiR-34 down-regulates a large pool of oncogenic mRNAs, such as cyclin-dependent kinase 4 (CDK4) and CDK6, BCL-2, MET, Notch, MYC and AXL [46]. Given its significant roles in cancer suppression, much attention has been focused on the miR-34-targeted therapeutics, which is also the first miRNA that enters clinic trials [20]. MiR-34 mimics that are used to replenish endogenous miR-34 levels showed great promise in tumour regression in phase I clinic trial. However, the trial was soon terminated due to severe immune reactions [19].
The let-7 family contains ten isoforms that also targets a wide range of oncogenic mRNAs, such as KRAS that is a proto-oncogene often activated in cancer [47]. let-7 is usually found to be down-regulated in various cancers, and the loss of let-7 results in accelerated tumour progression. Therapeutic strategies to up-regulate let-7 may be of use in cancer treatment when let-7 is down-regulated or lost [48].
MiR-200 family is another important set of tumour-suppressive miRNAs and is down-regulated in cancer. MiR-200 family contains two groups, miR-200a/b/429 and miR-200c/141, which respectively locate on chromosome 1 and chromosome 12 [49]. The miR-200 family can repress expression of proteins involved in tumour metastasis and angiogenesis, such as zinc-finger E-box-binding homeobox 1 (ZEB1) and C-X-C motif chemokine 1 (CXCL1) [50].
MiR-21, a well-known oncogenic miRNA, is usually up-regulated in cancer cells and has an anti-apoptotic function. Important factors involved in the up-regulation of miR-21 are transforming growth factor β1 (TGFβ1) and the transcription factor signal transducer and activator of transcription 3 (STAT3) [51,52]. The target genes of miR-21 include programmed cell death protein 4 (PDCD4), reversion-inducing cysteine-rich protein with Kazal motifs (RECK) and phosphatase and tensin homolog (PTEN) [53,54].
MiR-10b is remarkably up-regulated in metastatic breast cancer cells, glioblastoma and melanoma [55,56]. A transcription factor, twist family bHLH transcription factor 1 (TWIST1), can increase the expression of miR-10b in breast cancer cells through binding to the miR-10b promoter. One important target of miR-10b is homeobox D10 (HOXD10), which is a member of the homeobox DNA-binding-domain-containing transcription factors and inhibits metastasis [57].
MiR-210 is an oncogenic miRNA and is a target of hypoxia-responsive transcription factor hypoxia-inducible factor 1ɑ (HIF1ɑ)[58]. During hypoxia response, miR-210 suppresses the expression of mitochondrial electron transport chain component protein succinate dehydrogenase complex sub-unit D (SDHD), which in turn results in an increased stability of HIF1ɑ and cancer cell survival [59]. Other important targets of miR-210 include the hypoxia stress response cell death inducer mitochondrion-associated 3 (AIFM3), the hypoxia-responsive angiogenesis inhibitor ephrin A3, and cell cycle regulators E2F3 and RAD52 [60–62].
In addition to above-mentioned cancer-related miRNAs, miRNAs are also involved in other human diseases. For example, miR-122, a liver-specific miRNA, up-regulates the replication of the HCV RNA genome [63]. MiR-122 binds to the 5ʹ-end of non-coding region of HCV viral RNA, resulting in protection of HCV RNA from Xrna exoribonuclease-mediated degradation [64]. Anti-sense oligonucleotide targeting miR-122 is used to treat HCV infection and in phase II clinic trial [19,21]. MiR-1, a member of myogenic miRNAs (myomiRs) [65], is highly expressed in heart muscle cells and decreased in cardiac hypertrophy and fibrosis. MiR-1 controls heart function by targeting calcium-binding protein calmodulin, the transcription factor myocyte enhancer factor 2 (MEF2A) and insulin-like growth factor 1 (IGF1) [66,67]. Moreover, miR-19b/221/222 suppress peroxisome proliferator-activated receptor γ coactivator 1ɑ (PGC-1ɑ) to induce endothelial cell dysfunction during the progression of atherosclerosis [68] and miR-200 family controls pancreatic beta-cell survival in type 2 diabetes through targeting a series of anti-apoptotic target genes [69].
The extensive involvement of miRNAs in human diseases highlights their therapeutic potential. Many efforts have been made to target endogenous miRNAs to achieve therapeutic outcomes. Synthetic oligonucleotides that can replace or inhibit endogenous miRNAs are thus used, and some of them have entered clinic trials [19]. For instance, miRNA mimics targeting miR-16 and miR-29 are in phase I clinical trial treatments of non-small cell cancer and scleroderma, respectively. Anti-sense oligonucleotide targeting miR-155 is in phase I clinical treatment of cutaneous T cell lymphoma and mycosis fungoides. Anti-sense oligonucleotide targeting miR-103/107 is in phase II clinical trial treatment of type 2 diabetes and non-alcoholic fatty liver diseases. As promising alternatives of oligonucleotides, small molecules have also gained increasing attention for their use in regulating endogenous miRNAs to treat related diseases [22,33].
4. Assays for the discovery of small molecule modifiers of microRNAs
In order to discover small molecule modifiers of miRNAs, several assays have been developed. These assays are designed to specifically target enzymes-mediated miRNA maturation or function, or to unbiasedly target miRNA biogenesis pathway. Moreover, bioinformatic methods have also been developed to directly target miRNA secondary structure.
4.1. Enzyme-based assays
Since Dicer and AGO2 are the key enzymes that direct miRNA maturation and function, two enzyme-based assays were developed to target Dicer-mediated miRNA maturation or AGO2-mediated miRNA function. To measure the activity of Dicer, Davies and Bose et al. developed a fluorescence assay [70,71]. This assay utilized a fluorescence resonance energy transfer (FRET) probe, which was based on a pre-miRNA that carried a fluorophore at the 5ʹ-end and a quencher at the 3ʹ-end (Figure 2(a)). In light of the close proximity of the fluorophore and quencher, fluorescence was initially non-detectable. However, upon processing by Dicer to liberate the fluorophore, a significant increase in fluorescence was detected, indicating Dicer-mediated miRNA maturation. Davies and Bose et al. demonstrated this assay system in discovery of small molecule modifiers of miRNAs through screening certain peptides and small molecules, resulting in two small molecules capable of inhibiting let-7 and miR-27a maturation. As shown in Figure 2(b), to measure the loading of miRNA onto AGO2, Tan et al. developed a fluorescence polarization screening assay [72]. In this assay, TAMARA labelled miR-21 was used as the probe. In the absence of AGO2, TAMRA-labelled miR-21 was free to rotate, giving a low polarization value. In contrast, upon loading of miR-21 onto AGO2, the large miR-21-AGO2 complex rotated more slowly, resulting in a higher polarization value. Thus, the loading of miRNA onto AGO2 can be indicated by measuring the polarization values. Using this assay to screen potential small molecules, Tan et al. discovered three compounds that inhibited loading of miR-21 onto AGO2. Advantages of these two cell-free assays are their low cost and easy operation, whereas the disadvantages are that identified compounds may be miRNA-specific if they directly interact with the miRNA and intracellular activity of these compounds needs further validation.
Figure 2.
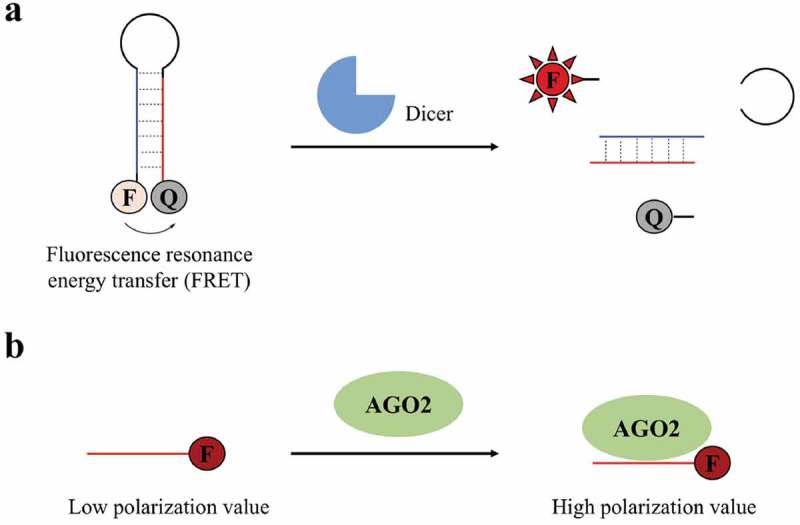
In vitro enzyme-based assays for discovery of small molecule modifiers of miRNA. (a) Fluorescence assay for monitoring Dicer-mediated miRNA maturation; (b) fluorescence polarization assay for monitoring loading of miRNA onto AGO2. F: fluorophore; Q: quencher.
4.2. Cellular assays
As miRNA biogenesis and function involve many steps and biomolecules, any perturbations in these steps will cause changes in miRNA expression or function. Therefore, to unbiasedly find small molecule modifiers of miRNA, Shan and Connelly et al. developed a cellular assay system [73–75]. As shown in Figure 3, in this assay system, green fluorescent protein (GFP) or luciferase were engineered to contain complementary sequences of miRNAs or siRNAs in their 3ʹ-UTR and used as the reporter genes. Due to miRNA and siRNA share the same RNA interference machinery system [13], small molecules identified by cellular assay for siRNA can also alter miRNA biogenesis or function. These engineered reporter genes were then introduced into cell lines through stable or transient transfection. Inside cells, miRNA or siRNA will inhibit expression of these reporter genes in sequence-specific manner. Therefore, the activity of endogenous miRNA can be read out through measuring expression of GFP or luciferase. Using these two cellular assays, Shan and Connelly et al. identified several small molecules that could regulate endogenous miRNAs. The cellular assays have the advantages that they are unbiased and identified molecules can regulate any step in miRNA biogenesis or function, giving the chance to investigate miRNA-involved cellular processes. One disadvantage of the cellular assays is that it is difficult to tell the specificity of identified molecules that is towards miRNA or other biomolecules.
Figure 3.
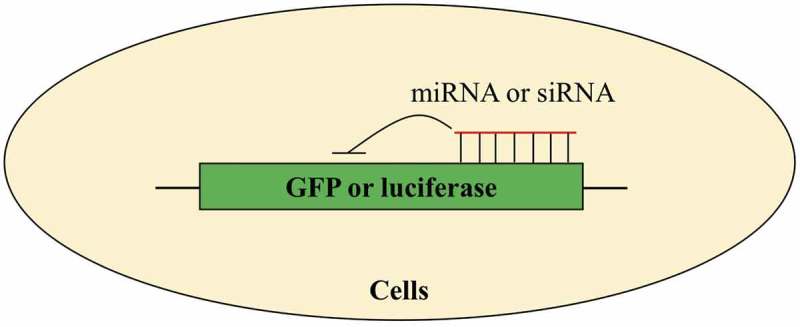
Cellular assays for discovery of small molecule modifiers of miRNA. Green fluorescent protein (GFP) or luciferase is engineered to contain complementary sequences of miRNA or siRNA and used as reporter genes.
4.3. InfoRNA
Except above-mentioned experimental assays for high-throughput screening of small molecule modifiers of miRNA, bioinformatic strategies for rational design of small molecules to directly target miRNA have also been developed [25,76]. Disney et al. devised a method to predict small molecule modifiers of miRNAs, named as infoRNA (Figure 4). This method first used a two-dimensional combinatorial screening (2DCS) to identify ligands that bound to RNA secondary structural motifs (hairpin, internal loops or bulges) [22,33]. During 2DCS, a small molecule microarray was used to incubate with a radio-labelled RNA library, followed by analysis of bound RNA. This screening enables simultaneous probing and identification of specific small molecule-RNA motif interactions. To utilize obtained information, Disney et al. then developed a statistical method called high-throughput structure-activity relationships through sequencing (HiT-StARTS) to obtain features of RNA motif-small molecule interactions. InfoRNA, a database comprising of the selected RNA motif-small molecule interactions and their associated affinity data, can thus be used to predict small molecules that directly target RNA secondary structures [77]. Since human miRNA hairpin precursors have ~3,808 unique secondary motifs in Drosha- or Dicer-processing sites [78], infoRNA can be used to find small molecules that regulate miRNA maturation in a specific manner. Using infoRNA, Disney et al. have discovered several small molecule modifiers of oncogenic miRNAs [22,25,33].
Figure 4.
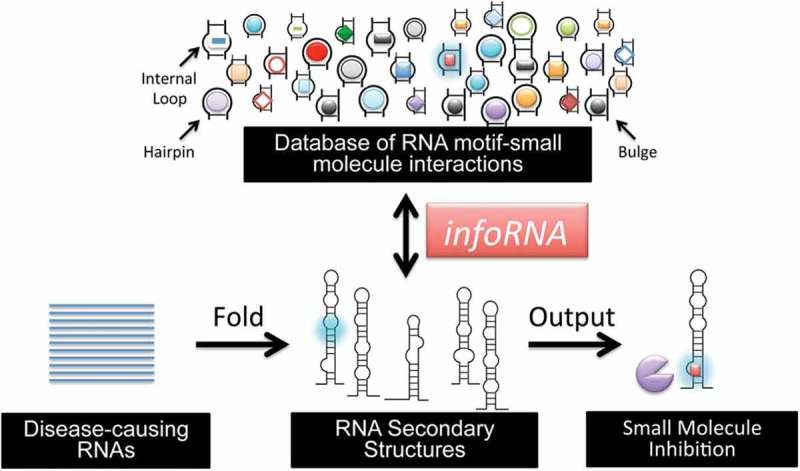
InfoRNA for design of small molecules that directly target miRNA (or other RNAs). InfoRNA extracts motifs from the secondary structure of target miRNA and compares them to the database, giving lead small molecules. Adapted from reference [77] with permission.
5. Small molecules in microRNA chemical biology and microRNA-targeted therapeutics
Using assay systems described above, diverse small molecule modifiers that can regulate miRNA biogenesis or function have been discovered, and some of them have been successfully used to probe miRNA-mRNA networks and treat related diseases. However, the exact mechanisms of action of most small molecule modifiers except those that directly targeting miRNA secondary structure still remain unclear. Therefore, we classify these small molecule modifiers into three groups and discuss some prominent examples in the following three sections.
5.1. Small molecules regulating microRNA biogenesis
Discovery of the first small molecule modifier of miRNA could be dated back to 2008, when enoxacin (1, Figure 5) was identified by Shan et al. as an activator that promoted miRNA processing [73]. Shan et al. used GFP-based cellular assays to screen a collection of 2,000 Food and Drug Administration (FDA)-approved small molecules, resulting in the discovery of compound 1 that increased siRNA-mediated GFP knockdown. The median effective concentration (EC50) of compound 1 was ~30 µM. To investigate the effect of compound 1 on endogenous miRNAs, Shan et al. profiled expression levels of 157 miRNAs. A total of 13 miRNAs had approximately twofold increase in expression of mature form, two miRNAs had decreased levels of mature forms, and the remaining miRNAs were not significantly altered. Moreover, the up-regulated miRNAs had decreased levels of pri- and pre-miRNAs, suggesting that compound 1 promotes miRNA processing. Since Dicer and TRBP play critical roles in miRNA processing, Shan et al. used Dicer and TRBP-mediated miRNA processing assay to determine the biomolecule target involved in compound 1-promoted miRNA processing, showing that the activity of compound 1 dependent on TRBP. Therefore, compound 1 is a TRBP-dependent activator of miRNA that promotes miRNA processing. Further studies carried out by Melo et al. revealed that compound 1 was a cancer-specific growth inhibitor [79]. Melo et al. tested the effect of compound 1 on the growth of 12 cancer cell lines and eight normal cells, resulting in the discovery of cancer-specific growth-suppressive activity of compound 1, with an EC50 value of 124 µM. They also validated that compound 1 enhanced miRNA production in these cancer cells, which was consistent with results reported by Shan et al. To investigate the mechanism of action of compound 1, they further used surface plasmon resonance (SPR) and isothermal titration calorimetry (ITC) to demonstrate that compound 1 directly bound to TRBP. Importantly, TRBP mutant cancer cells did not respond to compound 1, suggesting that compound 1 exerts growth-inhibitory effect in cancer cells by promoting miRNA biogenesis in a TRBP-dependent manner.
Figure 5.
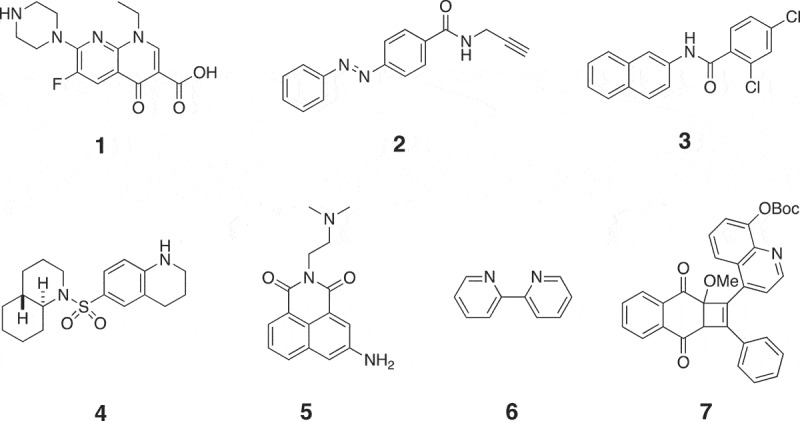
Chemical structures of small molecule modifiers of miRNAs that regulate miRNA biogenesis. 1: Activator of miRNA that promotes miRNA processing; 2: inhibitor of miR-21 transcription; 3, 4: inhibitors of miR-122 transcription; 5: activator of miR-122 transcription; 6: activator of miRNA that promotes miRNA processing; 7: inhibitor of myomiRs.
Except the discovery of compound 1 by Shan et al., Gumireddy et al. also identified a first small molecule inhibitor of miR-21 in 2008 [80]. They used a luciferase-based cellular assay to screen a library of more than 1,000 compounds from they own compound collection and the Library of Pharmacologically Active Compounds (LOPAC) towards miR-21, resulting in the discovery of a lead compound that produced a ~ 2.5-fold increase in luciferase signals at a concentration of 10 µM. Structure-activity optimization led to the discovery of a high active compound 2 (Figure 5), which increased luciferase signals by approximately fivefold at a concentration of 10 µM. To investigate the mode of action of compound 2, Gumireddy et al. checked its effect on miR-30 and miR-93. No increase in luciferase signals was detected, suggesting that it may be a specific inhibitor of miR-21. Through measuring miR-21 expression levels, Gumireddy et al. found that compound 2 significantly decreased expression levels of both pri-miR-21 and miR-21. These results suggest that compound 2 is an inhibitor of miR-21 that targets miR-21 transcription. However, the detailed mechanism for compound 2 to inhibit miR-21 still remains unclear. Young et al. also used the luciferase-based cellular assays to screen a library of 1,364 small molecules from the NCI Developmental Therapeutics Programme towards miR-122 [81]. After screening, they found three potent small molecules 3–5, which are shown in Figure 5. Similar as compound 2, compounds 3–5 only regulated miR-122 but not other miRNA, indicating that they may also be specific regulators of miR-122. Compounds 3 and 4 were inhibitors of miR-122 with EC50 values of 3 and 0.6 µM, respectively. Compound 5 was an activator of miR-122 with half maximal inhibitory concentration (IC50) value of 3 µM. Compounds 3–5 also regulated miR-122 at the transcriptional level, and the detailed mechanisms of action are still unknown. Nevertheless, since up-regulation of miR-122 was known to promote HCV replication and decrease of miR-122 could inhibit apoptosis of liver cancer cells [64,82], Young et al. further used compounds 3 and 4 to treat HCV and exploited compound 5 to induce apoptosis of liver cancer cells, all showing positive results (Figure 6).
Figure 6.
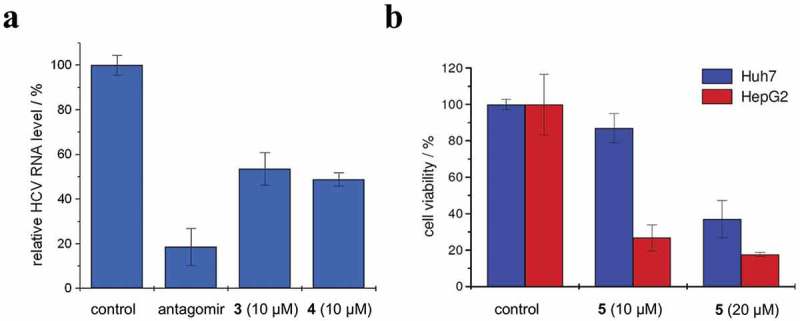
Effects of (a) compounds 3 and 4 on HCV replication in Huh7 cells and (b) compound 5 on viability of Huh7 and HepG2 cells. Adapted from reference [81] with permission.
Small molecule modifiers that regulate miRNA biogenesis also hold great promise in dissecting miRNA-involved cellular processes [34]. For example, by using the same GFP-based cellular assay systems for discovery of compound 1, Li et al. found a common iron chelator 2,2ʹ-dipyridyl (6, Figure 5) as an activator of miRNA that promoted miRNA processing [83]. However, the isomer of compound 6, 4,4ʹ-dipyridyl, did not show this activity, suggesting that iron chelating activity is required for its enhancement of miRNA processing and iron may involve in miRNA biogenesis. They then found that cytosolic iron could modulate the RNA interference activity and poly(rC)-binding protein 2 (PCBP2) that is known to facilitate the loading of cytosolic iron into ferritin was a novel biomolecule involved in miRNA processing. Collectively, with compound 6 as the chemical probe, Li et al. revealed that cytosolic iron modulates PCBP2 multimerization and PCBP2 associates with pre-miRNAs to modulate miRNA processing. Another example reported by Tan et al. used a small molecule modifier of myomiRs to unveil a novel myomiR-involved regulatory pathway in muscle cells [84]. Using the luciferase-based cellular assay system, Tan et al. screened their own compound library towards myomiRs in muscle cells, resulting in the discovery of compound 7 (Figure 5) that inhibited myomiRs including miR-1, miR-133a and miR-206. Compound 7 did not regulate cancer-related miRNAs such as miR-150 and miR-21 in other cancer cells, indicating that it may be a specific inhibitor of myomiRs. Tan et al. found that compound 7 inhibited the transcription of myomiRs and they further measured the expression levels of transcription factor of myomiRs that is myoD, showing that compound 7 decreased myoD protein level but not mRNA level. The inconsistency between myoD mRNA level and protein level led to a discovery of upstream regulator of myoD, which is miR-221/222. Therefore, using compound 7 as a chemical probe, Tan et al. revealed a novel signalling transduction pathway in muscle cells, which is miR-221/222 indirectly regulate myomiRs through targeting myoD.
5.2. Small molecules regulating microRNA function
The key step of miRNA function is the loading of miRNA onto AGO2. Small molecules that interfere with this process without affecting miRNA expression can directly modulate miRNA function. Using the in vitro fluorescence polarization assay, Tan et al. screened a total of over 2,000 compounds from LOPAC towards miR-21 function, resulting in the discovery of three potential compounds [72]. One of the three compounds, 8, is shown in Figure 7. The IC50 value of compound 8 was 0.47 µM. However, compound 8 could not regulate loading of endogenous let-7 onto AGO2 inside cultured cells, whereas compound 8 could inhibit RISC loading of exogenous siRNA. This result suggests that small molecule identifiers identified by in vitro assays need further validation in cellular assays.
Figure 7.
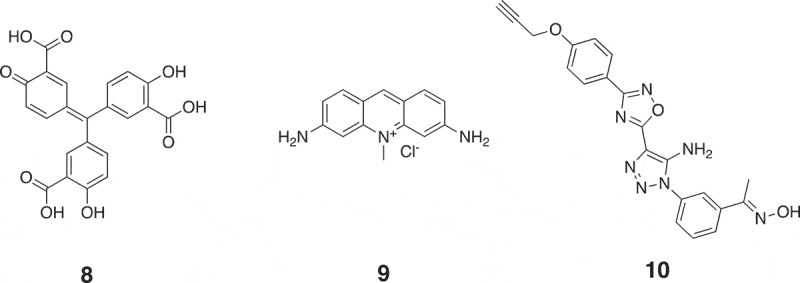
Chemical structures of small molecule modifiers of miRNAs that regulate miRNA function. 8: Inhibitor of miR-21 loading onto AGO2; 9: inhibitor of the association between miRNA with AGO2; 10: inhibitor of miR-21 function with unknown mechanism.
Watashi et al. used the luciferase-based cellular assay system to screen 530 compounds, resulting in the discovery of a small molecule modifier 9 (Figure 7) [85]. Compound 9 (10 µM) inhibited expression levels of AGO2-associated miRNA without significantly affecting total miRNA levels, suggesting that it is a modifier that inhibits miRNA loading onto AGO2. Since AGO2 overexpression is correlated with the development of cancers, Watashi et al. used compound 9 to treat miRNA-dependent tumour models, resulting in a significant reduction in tumour growth. Except using compound 9 to treat tumour models, Madsen et al. tested its effect in decreasing Venezuelan equine encephalitis virus (VEEV) replication [86]. 2.5 µM compound 9 caused an approximately 7-log10 decrease in viral titre. When using compound 8 to treat VEEV, a ~ 3-log10 decrease in viral titre was detected.
Using previously established luciferase-based cellular assay system, Naro et al. further screened a collection of 333,519 compounds towards miR-21, resulting in the discovery of a small molecule 10 (Figure 7) [87]. Compound 10 increased luciferase signals by ~12-fold at a concertation of 10 µM. To investigate the mechanism of action of compound 10, Naro et al. measured the expression levels of miR-21, showing that neither mature nor primary miR-21 levels were affected. This result indicates that compound 10 inhibits miR-21 function without affecting miR-21 expression. However, compound 10 did not modulate other miRNAs such as miR-122, miR-221 and miR-182, suggesting that it is a specific inhibitor of miR-21. Naro et al. tried to explain the mechanism of action of compound 10, whereas it is still elusive. Since overexpression of miR-21 in renal carcinoma (RCC) contributes to chemoresistance [88], Naro et al. used compound 10 to reverse the chemosensitivity of RCC cells to topotecan, an FDA-approved chemotherapeutic drug. As shown in Figure 8(a), the combination of topotecan with compound 10 decreased the IC50 value of topotecan from 1 µM to 90 nM, resulting in an over 11-fold increase in the potency of topotecan. In a 2-week clonogenic assay, 10 µM compound 10 decreased the IC50 value of topotecan from 2 µM to 74 nM, representing a 27-fold enhancement in activity of topotecan (Figure 8(b)).
Figure 8.
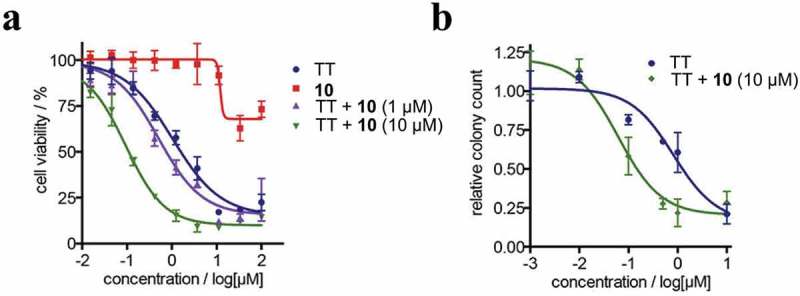
Therapeutic evaluation of compound 10 in the restoration of chemosensitivity of A498 RCC cells to topotecan (TT). (a) Combination treatment of A498 cells with TT and compound 10 in a dose-dependent cell viability assay. (b) Combination treatment of A498 cells with TT and compound 10 in a 2-week cologenic assay. Adapted from reference [87] with permission.
5.3. Small molecules directly targeting microRNA secondary structure
Many sites within miRNA precursors can be bound by small molecules, which will cause blockade of miRNA production or function. These small molecules can be given by either high-throughput screening or rational design. Unlike most of those small molecule modifiers described above, the mode of action of these small molecules can be explained since they directly bind to miRNA precursors to prevent cleavage by Drosha or Dicer.
As aminoglycosides are known to bind to secondary structures of RNAs, Bose et al. investigated the potential of aminoglycosides to inhibit oncogenic miR-21 [89]. A luciferase-based cellular assay system was used to screen 15 known aminoglycosides, resulting in the discovery of streptomycin 11 (Figure 9) that inhibited miR-21 activity at a concentration of 10 µM. Bose et al. also measured the expression levels of intracellular miR-21, showing a ~ 80% decrease in miR-21 level when using 5 µM compound 11. Meanwhile, several other endogenous miRNAs excluding miR-27a remained unchanged. To determine the mode of action of compound 11, they did several in vitro experiments including thermal melting experiment, docking study and enzyme-cleavage pattern analysis, showing that compound 11 directly bound to pre-miR-21. As miR-21 targets PDCD4 to exert its oncogenic function, Bose et al. also checked the levels of PDCD4 inside cancer cells and cell apoptosis rates, showing approximately twofold increases in both PDCD4 protein level and apoptotic rate when using 5 µM compound 11 to treat cancer cells.
Figure 9.
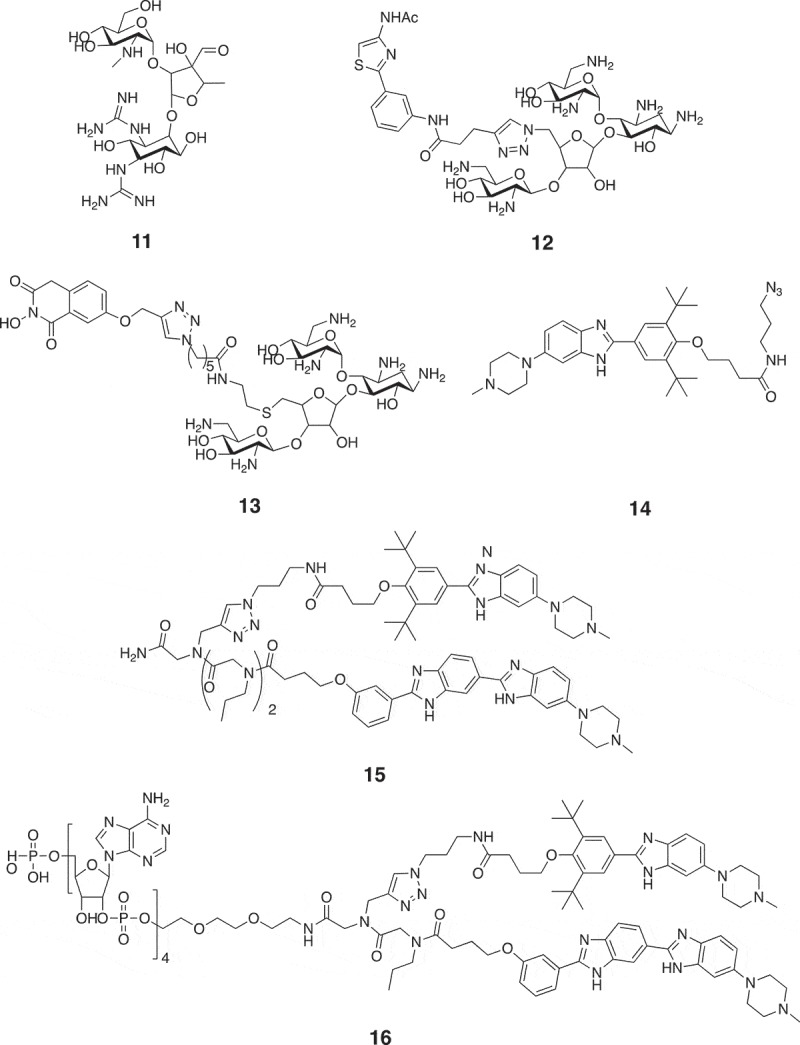
Chemical structures of small molecule modifiers of miRNAs that directly target miRNA secondary structure. 11: Inhibitor of miR-21 that directly binds to pre-miR-21; 12: inhibitor of miR-372 that blocks Dicer-mediated cleavage of pre-miR-372; 13: inhibitor of miR-21 that blocks Dicer-mediated cleavage of pre-miR-21; 14, 15: inhibitors of miR-96 that block Drosha-mediated cleavage of pri-miR-96; 16: inhibitor of miR-96 that recruits RNase L to pri-miR-96 to facilitate its destruction.
To find small molecule modifiers that directly bind to pre-miR-372 and pre-miR-373 that are implicated in gastric cancer, Vo et al. also used aminoglycosides as scaffolds to design the ligands [90]. They first used the in vitro FRET assay for Dicer-mediated cleavage of pre-miR-372 to screen seven aminoglycosides that may inhibit the cleavage by Dicer, showing that neomycin was able to completely inhibit Dicer cleavage at a concentration of 250 µM. Vo et al. then modified neomycin with artificial bases and further screened these modified small molecules, resulting in the discovery of a most potent small molecule 12 (Figure 9) that inhibited cleavage of pre-miR-372 by Dicer with a IC50 value of 2.42 µM. They then treated gastric cancer cells with compound 12 and found that ~50% decreases in both expression levels of miR-372/373 and cell viability were induced by compound 12 at a concentration of 50 µM.
Yan et al. developed a bifunctional small molecule that bound to pre-miR-21 to inhibit miR-21 maturation [91]. To find small molecules that can directly bind to pre-miR-21, they used in vitro fluorescence polarization assay that determines the binding between small molecule and pre-miR-21 to screen aminoglycosides, resulting in the discovery of neomycin. Through modifying neomycin with a Dicer inhibiting unit, the bifunctional small molecule modifier of miR-21 (13, Figure 9) was obtained. In vitro Dicer-mediated pre-miR-21 cleavage confirmed that compound 13 was able to inhibit this process. Cellular assays also showed that compound 13 down-regulated miR-21 level (~20%) and de-repressed miR-21 target PDCD4 (approximately twofold increase) at 5 µM.
Except using screening methods to find small molecules that directly bind to pre-miRNA, infoRNA also showed great promise in rational design of small molecule modifiers of miRNAs [77]. Using infoRNA, Velagapudi et al. designed a small molecule 14 (Figure 9) to target Drosha binding sites in the primary transcript of oncogenic miR-96 [92]. In cellular assays, compound 14 induced a ~ 90% decrease in miR-96 level at a concentration of 40 µM. Moreover, compound 14 did not modulate expression of other 149 disease-associated and abundant miRNAs, suggesting that it is a selective inhibitor of miR-96. Furthermore, compound 14 also de-repressed expression of oncogenic Forkhead Box O1 (FOXO1) and triggered apoptosis in breast cancer cells. However, the low micromolar activity of compound 14 did not satisfy the need of therapeutic intervention in in vivo model. Thus, Velagapudi et al. used infoRNA to design a dimeric small molecule 15 (Figure 9) that could also bind to motif that was adjacent to Drosha site [93]. Compound 15 had nanomolar affinity for miR-96. 50 nM compound 15 was sufficient to inhibit miR-96 production (~50%) and boosted expression of FOXO1 by approximately twofold and triggered apoptosis of breast cancer cells (~50%). Moreover, they appended compound 15 with a cross-linking module and a biotin tag to pull down direct targets of compound 15, confirming the direct binding between compound 15 and pri-miR-96 inside cells. Recently, Costales et al. further appended the dimeric molecule with a short 2ʹ-5ʹ poly(A) oligonucleotide that could recruit RNase L to pri-miR-96 to facilitate its destruction [94]. In breast cancer cells, compound 16 (Figure 9) could induce a ~ 40% decrease in pri-miR-96 level at 20 nM and a ~ 80% reduction at 200 nM. In comparison, compound 15 that did not induce degradation of pri-miR-96 increased pri-miR-96 levels. When using compound 16 (200 nM) to treat breast cancer cells, FOXO1 expression was increased by approximately twofold and a ~ 80% cell apoptosis was detected (Figure 10). Except discovery of small molecule modifiers of miR-96, infoRNA has also been used to identify small molecules that directly target oncogenic miRNAs including miR-210[95], miR-544[96], miR-21[97], miR-10b [76], etc.
Figure 10.
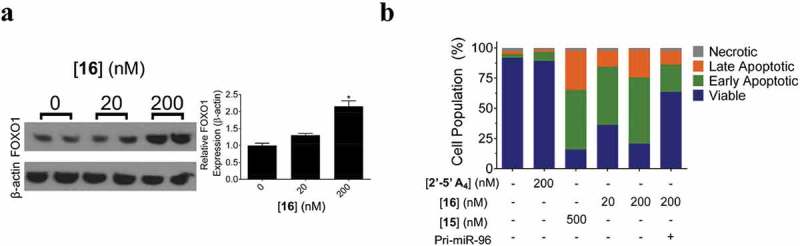
Effects of compound 16 on expression of FOXO1 and cell apoptosis. (a) Protein expression levels of FOXO1 in breast cancer MDMA-MB-231 cells after treatment with compound 16. (b) Annexin V/PI staining of apoptotic cells in MDMA-MB-231 cells after treatment with compound 16. Adapted from reference [94] with permission.
These small molecule modifiers have the advantage that they directly interact with miRNA precursors to block Drosha or Dicer cleavages. Even though these small molecules most likely to directly bind to pri- or pre-miRNAs, whether they regulate other biomolecules such as other RNA motifs or unknown proteins involved in miRNA biogenesis remains unclear. Nevertheless, exquisite selectivity may not be a perquisite for miRNA-targeting small molecules, provided that they have lower affinity to off-target sites. Since most of these reported small molecule modifiers have strong binding affinity towards miRNA precursors, the function output can be ascribed to their predominant regulation on miRNA biogenesis.
6. Conclusion and outlook
The past two decades have witnessed an accelerating development of miRNA and miRNA-targeted therapy. With improved methods of sequencing, it is becoming easier to identify new miRNAs. Their correlation with pathologies is also getting clear with the aid of genetic methods. Nevertheless, the miRNA-mRNA networks are extremely complicated, which require proper tools to dissect them. Identification of these underlying information will help to develop therapeutics designed to target these specific miRNAs. Meanwhile, in order to target miRNAs to achieve therapeutic outcome, oligonucleotides are often used to replenish or block endogenous miRNAs. Although oligonucleotides suffer from some disadvantages as mentioned above, continuous efforts have been made towards overcoming limitations, leading to clinical trials of several oligonucleotide-based drugs targeting miRNAs and one recently approved oligonucleotide-based drug targeting disease-associated gene [19,98]. Alternatively, small molecules that can regulate endogenous miRNAs are potential tools to elucidate miRNA-mRNA networks and treat miRNA-related diseases, as evidenced by the examples discussed above.
Among reported small molecule modifiers of miRNAs, small molecules can be identified either by high-throughput screening or by rational design. Small molecules discovered by cellular screening can regulate multi-steps in miRNA biogenesis or function, and their modes of action are thus complicated to be elucidated. Up to now, only a few of them have been explained in details. However, these small molecules can be used to probe unknown signaling transduction pathways that are hard to be revealed by traditional biochemical methods [83,84]. Small molecules that directly interact with miRNA secondary structures have clear mechanism of action. Since these small molecules are designed to target known miRNAs, their roles in elucidation of miRNA networks are thus limited. With regard to miRNA-targeted therapeutics, small molecule modifiers of miRNAs have also shown great promise. Introduction of other elements into these small molecules have also enhanced their therapeutic utility. It is worth noting that combination of small molecule modifiers of miRNAs with oligonucleotides could also enhance miRNA-targeted therapeutics [99,100]. Moreover, small molecules could also be hybridized with peptide to inhibit the maturation of pre-miRNA, thereby reversing miRNA function in disease development [101]. Currently reported studies mainly focused on the identification of small molecules that target cancer-related miRNAs and using them for cancer treatment. However, since miRNAs closely correlate with many human diseases other than cancers, the potential of small molecule modifiers for these diseases also waits to the explored. In the years to come, the accumulation of more small molecule modifiers of miRNAs will further revolutionize the field of miRNA chemical biology and miRNA-targeted therapeutics.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- [1].Bartel DP. Metazoan MicroRNAs. Cell. 2018;173:20–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75:855–862. [DOI] [PubMed] [Google Scholar]
- [4].Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. [DOI] [PubMed] [Google Scholar]
- [5].Reinhart BJ, Slack FJ, Basson M, et al. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–906. [DOI] [PubMed] [Google Scholar]
- [6].Pasquinelli AE, Reinhart BJ, Slack F, et al. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature. 2000;408:86–89. [DOI] [PubMed] [Google Scholar]
- [7].Kozomara A, Griffiths-Jones S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014;42:D68–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].miRBase: the microRNA database. [cited 2018 Aug]. Available from: http://www.mirbase.org.
- [9].Hausser J, Zavolan M. Identification and consequences of miRNA-target interactions–beyond repression of gene expression. Nat Rev Genet. 2014;15:599–612. [DOI] [PubMed] [Google Scholar]
- [10].Huntzinger E, Izaurralde E. Gene silencing by microRNAs: contributions of translational repression and mRNA decay. Nat Rev Genet. 2011;12:99–110. [DOI] [PubMed] [Google Scholar]
- [11].Ameres SL, Zamore PD. Diversifying microRNA sequence and function. Nat Rev Mol Cell Biol. 2013;14:475–488. [DOI] [PubMed] [Google Scholar]
- [12].Friedman RC, Farh KK, Burge CB, et al. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522–531. [DOI] [PubMed] [Google Scholar]
- [14].Xiao C, Rajewsky K. MicroRNA control in the immune system: basic principles. Cell. 2009;136:26–36. [DOI] [PubMed] [Google Scholar]
- [15].Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–269. [DOI] [PubMed] [Google Scholar]
- [16].Lujambio A, Lowe SW. The microcosmos of cancer. Nature. 2012;482:347–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Pineau P, Volinia S, McJunkin K, et al. miR-221 overexpression contributes to liver tumorigenesis. Proc Natl Acad Sci U S A. 2010;107:264–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Kasinski AL, Slack FJ. miRNA-34 prevents cancer initiation and progression in a therapeutically resistant K-ras and p53-induced mouse model of lung adenocarcinoma. Cancer Res. 2012;72:5576–5587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Rupaimoole R, Slack FJ. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discovery. 2017;16:203–222. [DOI] [PubMed] [Google Scholar]
- [20].Bouchie A. First microRNA mimic enters clinic. Nat Biotechnol. 2013;31:577. [DOI] [PubMed] [Google Scholar]
- [21].Janssen HL, Reesink HW, Lawitz EJ, et al. Treatment of HCV infection by targeting microRNA. N Engl J Med. 2013;368:1685–1694. [DOI] [PubMed] [Google Scholar]
- [22].Angelbello AJ, Chen JL, Childs-Disney JL, et al. Using genome sequence to enable the design of medicines and chemical probes. Chem Rev. 2018;118:1599–1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Vickers TA, Wyatt JR, Freier SM. Effects of RNA secondary structure on cellular antisense activity. Nucleic Acids Res. 2000;28:1340–1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Jayaraj GG, Nahar S, Maiti S. Nonconventional chemical inhibitors of microRNA: therapeutic scope. Chem Commun. 2015;51:820–831. [DOI] [PubMed] [Google Scholar]
- [25].Disney MD, Angelbello AJ. Rational design of small molecules targeting oncogenic noncoding RNAs from sequence. Acc Chem Res. 2016;49:2698–2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Di Giorgio A, Tran TP, Duca M. Small-molecule approaches toward the targeting of oncogenic miRNAs: roadmap for the discovery of RNA modulators. Future Med Chem. 2016;8:803–816. [DOI] [PubMed] [Google Scholar]
- [27].Monroig Pdel C, Chen L, Zhang S, et al. Small molecule compounds targeting miRNAs for cancer therapy. Adv Drug Deliv Rev. 2015;81:104–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Lee JS, Lee JW, Kang N, et al. Diversity-oriented approach for chemical biology. Chem Rec. 2015;15:495–510. [DOI] [PubMed] [Google Scholar]
- [29].Spiegel J, Cromm PM, Zimmermann G, et al. Small-molecule modulation of Ras signaling. Nat Chem Biol. 2014;10:613–622. [DOI] [PubMed] [Google Scholar]
- [30].Deiters A. Small molecule modifiers of the microRNA and RNA interference pathway. Aaps J. 2010;12:51–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Li J, Tan S, Kooger R, et al. MicroRNAs as novel biological targets for detection and regulation. Chem Soc Rev. 2014;43:506–517. [DOI] [PubMed] [Google Scholar]
- [32].Li J, Zhang W, Zhou M, et al. Small molecules modulating biogenesis or processing of microRNAs with therapeutic potentials. Curr Med Chem. 2013;20:3604–3612. [DOI] [PubMed] [Google Scholar]
- [33].Velagapudi SP, Vummidi BR, Disney MD. Small molecule chemical probes of microRNA function. Curr Opin Chem Biol. 2015;24:97–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Li Y, Ji P, Jin P. Probing the microRNA pathway with small molecules. Bioorg Med Chem. 2013;21:6119–6123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Lorenz DA, Garner AL. Approaches for the discovery of small molecule ligands targeting microRNAs. Top Med Chem. 2017;27:79–110. [Google Scholar]
- [36].Zhang S, Chen L, Jung EJ, et al. Targeting microRNAs with small molecules: from dream to reality. Clin Pharmacol Ther. 2010;87:754–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Lee Y, Kim M, Han J, et al. MicroRNA genes are transcribed by RNA polymerase II. Embo J. 2004;23:4051–4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Lee Y, Ahn C, Han J, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. [DOI] [PubMed] [Google Scholar]
- [39].Lund E, Guttinger S, Calado A, et al. Nuclear export of microRNA precursors. Science. 2004;303:95–98. [DOI] [PubMed] [Google Scholar]
- [40].Chendrimada TP, Gregory RI, Kumaraswamy E, et al. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature. 2005;436:740–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Ha M, Kim VN. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol. 2014;15:509–524. [DOI] [PubMed] [Google Scholar]
- [42].Liu J, Carmell MA, Rivas FV, et al. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305:1437–1441. [DOI] [PubMed] [Google Scholar]
- [43].Lin S, Gregory RI. MicroRNA biogenesis pathways in cancer. Nat Rev Cancer. 2015;15:321–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. [DOI] [PubMed] [Google Scholar]
- [45].Okada N, Lin CP, Ribeiro MC, et al. A positive feedback between p53 and miR-34 miRNAs mediates tumor suppression. Genes Dev. 2014;28:438–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Misso G, Di Martino MT, De Rosa G, et al. Mir-34: a new weapon against cancer? Mol Ther Nucleic Acids. 2014;3:e194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Johnson SM, Grosshans H, Shingara J, et al. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635–647. [DOI] [PubMed] [Google Scholar]
- [48].Johnson CD, Esquela-Kerscher A, Stefani G, et al. The let-7 microRNA represses cell proliferation pathways in human cells. Cancer Res. 2007;67:7713–7722. [DOI] [PubMed] [Google Scholar]
- [49].Gregory PA, Bert AG, Paterson EL, et al. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol. 2008;10:593–601. [DOI] [PubMed] [Google Scholar]
- [50].Pecot CV, Rupaimoole R, Yang D, et al. Tumour angiogenesis regulation by the miR-200 family. Nat Commun. 2013;4:2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Li Q, Zhang D, Wang Y, et al. MiR-21/Smad 7 signaling determines TGF-beta1-induced CAF formation. Sci Rep. 2013;3:2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Iliopoulos D, Jaeger SA, Hirsch HA, et al. STAT3 activation of miR-21 and miR-181b-1 via PTEN and CYLD are part of the epigenetic switch linking inflammation to cancer. Mol Cell. 2010;39:493–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Asangani IA, Rasheed SA, Nikolova DA, et al. MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor suppressor Pdcd4 and stimulates invasion, intravasation and metastasis in colorectal cancer. Oncogene. 2008;27:2128–2136. [DOI] [PubMed] [Google Scholar]
- [54].Krichevsky AM, Gabriely G. miR-21: a small multi-faceted RNA. J Cell Mol Med. 2009;13:39–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Ma L, Teruya-Feldstein J, Weinberg RA. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature. 2007;449:682–688. [DOI] [PubMed] [Google Scholar]
- [56].Saldanha G, Elshaw S, Sachs P, et al. microRNA-10b is a prognostic biomarker for melanoma. Mod Pathol. 2016;29:112–121. [DOI] [PubMed] [Google Scholar]
- [57].Ma L, Reinhardt F, Pan E, et al. Therapeutic silencing of miR-10b inhibits metastasis in a mouse mammary tumor model. Nat Biotechnol. 2010;28:341–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Kulshreshtha R, Ferracin M, Wojcik SE, et al. A microRNA signature of hypoxia. Mol Cell Biol. 2007;27:1859–1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Puissegur MP, Mazure NM, Bertero T, et al. miR-210 is overexpressed in late stages of lung cancer and mediates mitochondrial alterations associated with modulation of HIF-1 activity. Cell Death Differ. 2011;18:465–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Yang W, Sun T, Cao J, et al. Downregulation of miR-210 expression inhibits proliferation, induces apoptosis and enhances radiosensitivity in hypoxic human hepatoma cells in vitro. Exp Cell Res. 2012;318:944–954. [DOI] [PubMed] [Google Scholar]
- [61].Fasanaro P, D’Alessandra Y, Di Stefano V, et al. MicroRNA-210 modulates endothelial cell response to hypoxia and inhibits the receptor tyrosine kinase ligand Ephrin-A3. J Biol Chem. 2008;283:15878–15883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Crosby ME, Kulshreshtha R, Ivan M, et al. MicroRNA regulation of DNA repair gene expression in hypoxic stress. Cancer Res. 2009;69:1221–1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Jopling CL, Yi M, Lancaster AM, et al. Modulation of hepatitis C virus RNA abundance by a liver-specific microRNA. Science. 2005;309:1577–1581. [DOI] [PubMed] [Google Scholar]
- [64].Thibault PA, Huys A, Amador-Canizares Y, et al. Regulation of Hepatitis C Virus genome replication by Xrn1 and microRNA-122 binding to individual sites in the 5ʹ untranslated region. J Virol. 2015;89:6294–6311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Rao PK, Kumar RM, Farkhondeh M, et al. Myogenic factors that regulate expression of muscle-specific microRNAs. Proc Natl Acad Sci U S A. 2006;103:8721–8726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Ikeda S, He A, Kong SW, et al. MicroRNA-1 negatively regulates expression of the hypertrophy-associated calmodulin and Mef2a genes. Mol Cell Biol. 2009;29:2193–2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Shan ZX, Lin QX, Fu YH, et al. Upregulated expression of miR-1/miR-206 in a rat model of myocardial infarction. Biochem Biophys Res Commun. 2009;381:597–601. [DOI] [PubMed] [Google Scholar]
- [68].Xue Y, Wei Z, Ding H, et al. MicroRNA-19b/221/222 induces endothelial cell dysfunction via suppression of PGC-1alpha in the progression of atherosclerosis. Atherosclerosis. 2015;241:671–681. [DOI] [PubMed] [Google Scholar]
- [69].Belgardt BF, Ahmed K, Spranger M, et al. The microRNA-200 family regulates pancreatic beta cell survival in type 2 diabetes. Nat Med. 2015;21:619–627. [DOI] [PubMed] [Google Scholar]
- [70].Davies BP, Arenz C. A homogenous assay for micro RNA maturation. Angew Chem Int Ed. 2006;45:5550–5552. [DOI] [PubMed] [Google Scholar]
- [71].Bose D, Jayaraj GG, Kumar S, et al. A molecular-beacon-based screen for small molecule inhibitors of miRNA maturation. ACS Chem Biol. 2013;8:930–938. [DOI] [PubMed] [Google Scholar]
- [72].Tan GS, Chiu CH, Garchow BG, et al. Small molecule inhibition of RISC loading. ACS Chem Biol. 2012;7:403–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Shan G, Li Y, Zhang J, et al. A small molecule enhances RNA interference and promotes microRNA processing. Nat Biotechnol. 2008;26:933–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Connelly CM, Thomas M, Deiters A. High-throughput luciferase reporter assay for small-molecule inhibitors of microRNA function. J Biomol Screening. 2012;17:822–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Connelly CM, Deiters A. Identification of inhibitors of microRNA function from small molecule screens. Methods Mol Biol. 2014;1095:147–156. [DOI] [PubMed] [Google Scholar]
- [76].Velagapudi SP, Disney MD. Two-dimensional combinatorial screening enables the bottom-up design of a microRNA-10b inhibitor. Chem Commun. 2014;50:3027–3029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Disney MD, Winkelsas AM, Velagapudi SP, et al. Inforna 2.0: a platform for the sequence-based design of small molecules targeting structured RNAs. ACS Chem Biol. 2016;11::1720–1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Liu B, Childs-Disney JL, Znosko BM, et al. Analysis of secondary structural elements in human microRNA hairpin precursors. BMC Bioinf. 2016;17:112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Melo S, Villaneuva A, Moutinho C, et al. Small molecule enoxacin is a cancer-specific growth inhibitor that acts by enhancing TAR RNA-binding protein 2-mediated microRNA processing. Proc Natl Acad Sci U S A. 2011;108:4394–4399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Gumireddy K, Young DD, Xiong X, et al. Small-molecule inhibitors of microRNA miR-21 function. Angew Chem Int Ed. 2008;47:7482–7484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Young DD, Connelly CM, Grohmann C, et al. Small molecule modifiers of microRNA miR-122 function for the treatment of Hepatitis C Virus infection and hepatocellular carcinoma. J Am Chem Soc. 2010;132:7976–7981. [DOI] [PubMed] [Google Scholar]
- [82].Lin CJ, Gong HY, Tseng HC, et al. miR-122 targets an anti-apoptotic gene, Bcl-w, in human hepatocellular carcinoma cell lines. Biochem Biophys Res Commun. 2008;375:315–320. [DOI] [PubMed] [Google Scholar]
- [83].Li Y, Lin L, Li Z, et al. Iron homeostasis regulates the activity of the microRNA pathway through poly(C)-binding protein 2. Cell Metab. 2012;15:895–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Tan S-B, Li J, Chen X, et al. Small molecule inhibitor of myogenic microRNAs leads to a discovery of miR-221/222-myoD-myomiRs regulatory pathway. Chem Biol. 2014;21:1265–1270. [DOI] [PubMed] [Google Scholar]
- [85].Watashi K, Yeung ML, Starost MF, et al. Identification of small molecules that suppress microRNA function and reverse tumorigenesis. J Biol Chem. 2010;285:24707–24716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Madsen C, Hooper I, Lundberg L, et al. Small molecule inhibitors of Ago2 decrease Venezuelan equine encephalitis virus replication. Antiviral Res. 2014;112:26–37. [DOI] [PubMed] [Google Scholar]
- [87].Naro Y, Ankenbruck N, Thomas M, et al. Small molecule inhibition of microRNA miR-21 rescues chemosensitivity of Renal-Cell carcinoma to topotecan. J Med Chem. 2018;61:5900–5909. [DOI] [PubMed] [Google Scholar]
- [88].Gaudelot K, Gibier JB, Pottier N, et al. Targeting miR-21 decreases expression of multi-drug resistant genes and promotes chemosensitivity of renal carcinoma. Tumour Biol. 2017;39:1010428317707372. [DOI] [PubMed] [Google Scholar]
- [89].Bose D, Jayaraj G, Suryawanshi H, et al. The tuberculosis drug streptomycin as a potential cancer therapeutic: inhibition of miR-21 function by directly targeting its precursor. Angew Chem Int Ed. 2012;51:1019–1023. [DOI] [PubMed] [Google Scholar]
- [90].Vo DD, Staedel C, Zehnacker L, et al. Targeting the production of oncogenic microRNAs with multimodal synthetic small molecules. ACS Chem Biol. 2014;9:711–721. [DOI] [PubMed] [Google Scholar]
- [91].Yan H, Bhattarai U, Guo ZF, et al. Regulating miRNA-21 biogenesis by bifunctional small molecules. J Am Chem Soc. 2017;139:4987–4990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Velagapudi SP, Gallo SM, Disney MD. Sequence-based design of bioactive small molecules that target precursor microRNAs. Nat Chem Biol. 2014;10:291–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Velagapudi SP, Cameron MD, Haga CL, et al. Design of a small molecule against an oncogenic noncoding RNA. Proc Natl Acad Sci U S A. 2016;113:5898–5903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Costales MG, Matsumoto Y, Velagapudi SP, et al. Small molecule targeted recruitment of a nuclease to RNA. J Am Chem Soc. 2018;140:6741–6744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Costales MG, Haga CL, Velagapudi SP, et al. Small molecule inhibition of microRNA-210 reprograms an oncogenic hypoxic circuit. J Am Chem Soc. 2017;139:3446–3455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Haga CL, Velagapudi SP, Strivelli JR, et al. Small molecule inhibition of miR-544 biogenesis disrupts adaptive responses to hypoxia by modulating ATM-mTOR signaling. ACS Chem Biol. 2015;10:2267–2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Velagapudi SP, Costales MG, Vummidi BR, et al. Approved anti-cancer drugs target oncogenic non-coding RNAs. Cell Chem Biol. 2018;25:1086–1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Ottesen EW. ISS-N1 makes the first FDA-approved drug for spinal muscular atrophy. Transl Neurosci. 2017;8:1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Yu C, Qian L, Ge J, et al. Cell-penetrating poly(disulfide) assisted intracellular delivery of mesoporous silica nanoparticles for inhibition of miR-21 function and detection of subsequent therapeutic effects. Angew Chem Int Ed. 2016;55:9272–9276. [DOI] [PubMed] [Google Scholar]
- [100].Yu C, Qian L, Uttamchandani M, et al. Single-vehicular delivery of antagomir and small molecules to inhibit miR-122 function in hepatocellular carcinoma cells by using “smart” mesoporous silica nanoparticles. Angew Chem Int Ed. 2015;54:10574–10578. [DOI] [PubMed] [Google Scholar]
- [101].Ghosh A, Degyatoreva N, Kukielski C, et al. Targeting miRNA by tunable small molecule binders: peptidic aminosugar mediated interference in miR-21 biogenesis reverts epithelial to mesenchymal transition. MedChemComm. 2018;9:1147–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]


