Abstract
Cardiac tissue is characterized by being dynamic and contractile, imparting the important role of biomechanical cues in the regulation of normal physiological activity or pathological remodeling. However, the dynamic mechanical tension ability also varies due to extracellular matrix remodeling in fibrosis, accompanied with the phenotypic transition from cardiac fibroblasts (CFs) to myofibroblasts. We hypothesized that the dynamic mechanical tension ability would regulate cardiac phenotypic transition within fibrosis in a strain-mediated manner. In this study, we developed a microdevice that is able to simultaneously and accurately mimic the biomechanical properties of the cardiac physiological and pathological microenvironment. The microdevice could apply cyclic compressions with gradient magnitudes (5–20%) and tunable frequency onto gelatin methacryloyl (GelMA) hydrogels laden with CFs, and also enabled the integration of cytokine. The strain-response correlations between mechanical compression and CFs spreading, and proliferation and fibrotic phenotype remolding were investigated. Results revealed that mechanical compression played a crucial role in the CFs phenotypic transition, depending on the strain of mechanical load and myofibroblast maturity of CFs encapsulated in GelMA hydrogels. The results provided evidence regarding the strain-response correlation of mechanical stimulation in CFs phenotypic remodeling, which could be used to develop new preventive or therapeutic strategies for cardiac fibrosis.
Keywords: cardiac fibrosis, organ-on-a-chip, mechanical stimulation, hydrogel, transforming growth factor-β
1. Introduction
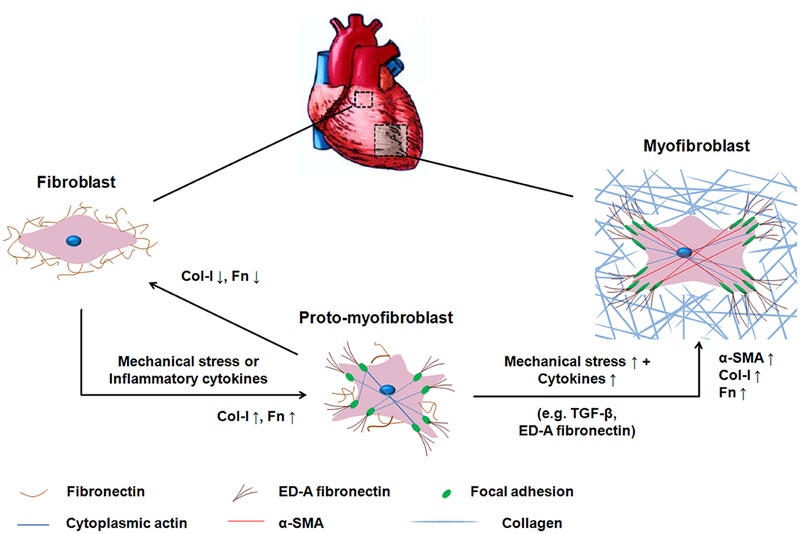
Cardiac fibroblasts (CFs), one of the major cell types in the heart, act a critical role in maintaining the function of a healthy heart.[1] Upon heart injury, e.g., myocardial infarction, due to the generation of pro-fibrotic and inflammatory cytokines, as well as the increased mechanical load,[2] the quiescent CFs are activated into proto-myofibroblasts.[3, 4] Through the formation of cytoplasmic actin-containing stress fibers in proto-myofibroblasts, tension is generated.[3] In a normal healing process, this activation of myofibroblasts is transient and reversible as ECM tension is restored after injury and takes over the mechanical load (Figure 1).[5, 6] However, in conditions of elevated mechanical load or chronic injury, persistent myofibroblast activation could lead to excessive ECM deposition, stiffening and contraction of cardiac tissue, resulting in impairment and dysfunction of cardiomyocytes and cardiac fibrosis. This is characterized by expressing alpha-smooth muscle actin (α-SMA).[4, 5, 7] The alteration of biochemical and biomechanical microenvironments in cardiac tissue accordingly determines the fate of CFs fibrotic remodeling. Since the mechanism by which fibrosis occurs is unclear, clarifying the underlying crosstalk between biochemical and biomechanical cues is paramount for the development of new therapeutic strategies for cardiac fibrosis.
Figure 1.
Activation of CFs towards cardiac myofibroblast phenotypes. Interstitial fibroblasts are characterized by the production of fibronectin and the absence of filamentous-actin, α-SMA, and ED-A fibronectin. Under mechanical stress or inflammatory cytokines, fibroblasts are activated into proto-myofibroblasts. The proto-myofibroblast produces ED-A fibronectin, contains stress fibers and focal adhesions, but does not yet contain the contractile α-SMA thus representing an immature myofibroblast. The increasing mechanical stress and TGF-β promote the modulation of proto-myofibroblasts into mature myofibroblasts. Mature myofibroblasts show abundant production of ED-A fibronectin and F-actin and are characterized by the presence of α-SMA. The transition from fibroblasts to proto-myofibroblasts is reversible.
In terms of biochemical cues, many soluble cytokines (e.g., transforming growth factor-β (TGF-β), angiotensin II, interleukin-6, endothelin-1, basic fibroblast growth factor) have been implicated in myofibroblast activation.[8] Among which, TGF-β acts a major role in the activation of myofibroblasts by promoting expression of α-SMA and synthesis of extra domain-A (ED-A) fibronectin.[9] Otherwise, the highly dynamic and contractile traits of cardiac tissue also possess very important biomechanical factors (e.g., ECM stiffness, mechanical strain) in the regulation of heart fibrosis and pathological remodeling.[10] Similarly, the fibroblasts/myofibroblasts are regulated by biomechanical conditions when cultured in vitro. For example, due to the resistance, it provides against cell traction force, stiff culture substrate induces myofibroblast activation.[6] Once the substrate stiffness is lowered to a level matching in vivo tissue, myofibroblast activation can be suppressed or reversed.[11] In the human heart, a mechanical strain is considered to be cyclic at roughly 1 Hz, ranging from 2% (loss of contractility)to 10% (physiological) to 15% (pathological) or even 20% (hyper-pathological) depending on pathological states.[12, 13] It has been established that the altered mechanical properties of the myocardium are associated with regulation of the phenotypic remodeling of CFs.[5] Various responses of myofibroblast activation to cyclic strains have been reported. In a combined mechanical strain and hypoxia in vitro model, compared with 2 % stretching strain (1 Hz), 8% stretching strain attenuated both CFs remodeling and pro-fibrotic responses taking place in hypoxia.[12] Cyclic stretch (4% and 8%, 1 Hz) of fibroblasts on silicone membranes was shown to increase the expression and production of collagen and fibronectin.[14] Cyclic biaxial stretch (10%, 1 Hz) enhanced the expression of α-SMA and collagen I (Col-I) in rat CFs.[15] A higher magnitude of 15% cyclic strain (1 Hz) was reported to either trigger cardiac fibrotic remodeling, or induce a fibroblastic phenotype over the myofibroblast phenotype.[16] However, different responses of myofibroblast activation with combined effects of TGF-β and mechanical strain were also observed. Cyclic stretch (10%, 1 Hz) attenuated the promoting effect of TGF-β1 in cardiac myofibroblast differentiation,[17] while cyclic strain (15%, 1 Hz) and TGF-β1 synergistically induced myofibroblastic phenotypes in aortic valve interstitial cells.[18] The conflicting findings shown may be attributed to different cell culture systems, mechanical strains, biological properties of substrate/matrix, or state of myofibroblast maturity, making data comparisons between studies difficult. Additionally, in order to clarify the impact of biomechanical cues on myofibroblast activation within different physiological and pathological states, the strain-response correlation between magnitudes of strain and modulation of phenotypes warrants further investigation.
The comprehensive correlations and spatiotemporal alterations of biochemical and biomechanical cues highly demand the development of in vitro cell culture systems which closely resemble the original cardiac physiological and biochemical microenvironment.[19, 20] A perspective in vitro bioreactor should involve, but is not limited to, the following characterizations: three-dimensional (3D) cell culture, resemblance to the biological properties of native cardiac ECM, substrate stiffness similar to cardiac ECM matrix, and cyclic strains.[6, 19] In our previous study, 3D cardiac fibrotic tissue was developed by stimulating a contractile 3D cardiac tissue with TGF-β1. The cardiac tissue was fabricated by using a mechanically tuned, ECM based hydrogel laden with cardiac cells to mimic the mechanical stiffness of native heart tissues.[21] This mechanically tuned hydrogel has mainly maintained the quiescent state of CFs compared with 2D stiff culture substrate that induced myofibroblast activation. After the TGF-β1 treatment, CFs have been attributed with the elevated deposition of ECM components that were presented to recapitulate a fibrogenic microenvironment. Therefore, biomimicked 3D matrixes are required to define cardiac remodeling.
A commercialized cyclic stretching device, Flexcell, is able to provide well-characterized strain profiles (sinusoidal wave, etc.) and tunable magnitudes of strain (from 0% to 30%), but is limited by only one magnitude of stretch which can be attained at any given time.[22] Similar limitations also occurred on other self-designed bioreactors.[23] A most recently developed bioreactor was able to achieve two magnitudes of stretching strains (2% and 8%) which were exerted on a 2-dimensional (2D) stretchable polydimethylsiloxane (PDMS) membrane.[12] It is still a challenge to integrate a wide range of mechanical strains due to the limited stretching strain of PDMS membrane, covering physiological and pathological levels (5–20%), into one bioreactor that could be used to investigate the relationship between strain and myofibroblast activation. In addition, most stretchable substrates with high stiffness fail to mimic the mechanical properties of native cardiac ECM, which might induce myofibroblast activation without any biochemical activation. Furthermore, most of the stretchable devices have difficulty holding and stretching cell-laden 3D matrices due to their weak mechanical properties, which are required to maintain proper cellular behaviors within the 3D matrix.
In this study, a bioreactor with the ability to accurately mimic the biomechanical properties of cardiac physiological and pathological microenvironments was developed based on our previous study,[24] which was modified by integrating inter-connectable dynamic compression bioreactors into one bioreactor. With the goal of recreating a more realistic in vivo cardiac environment, microfabrication was employed to enable incorporation of biochemical, biomaterial and biomechanical factors in one microdevice. PDMS-based microfluidic bioreactors or chips are good candidates to develop a bioreactor integrated with a mechanical stimulation component due to being easy and flexible to manipulate by lithography to create a range of compression strains. By integrating 3D culture and microfabrication, biochemical and biomechanical cues regarding fibrotic progress could be combined in this microdevice to study the mechanical regulation of myofibroblast activation and fibrotic remodeling, especially to uncover the strain-response relationship of mechanical cues and CFs phenotypic remodeling. So, this microdevice can apply cyclic compressions with gradient magnitudes onto 3D hydrogels laden with CFs. Although compression stimulation is usually applied for bone and cartilage tissue engineering,[24, 25] it has also been confirmed to favor formation of cardiac muscle in vitro.[26] Additionally, the hydrogel used for 3D cell culture can be deemed as incompressible. Therefore, vertical compression must be accompanied by horizontal expansion, placing the surrounding gel matrix in tension (Figure S1).[27] Chemically modified gelatin, which is desaturated collagen as an abundant ECM component of the heart, was used to resemble biological and mechanical properties (cellular behavior) of native heart ECM. Furthermore, TGF-β1 was applied to define the cardiac remodeling under various compression strains.
2. Results and Discussions
2.1. Microdevice characterization
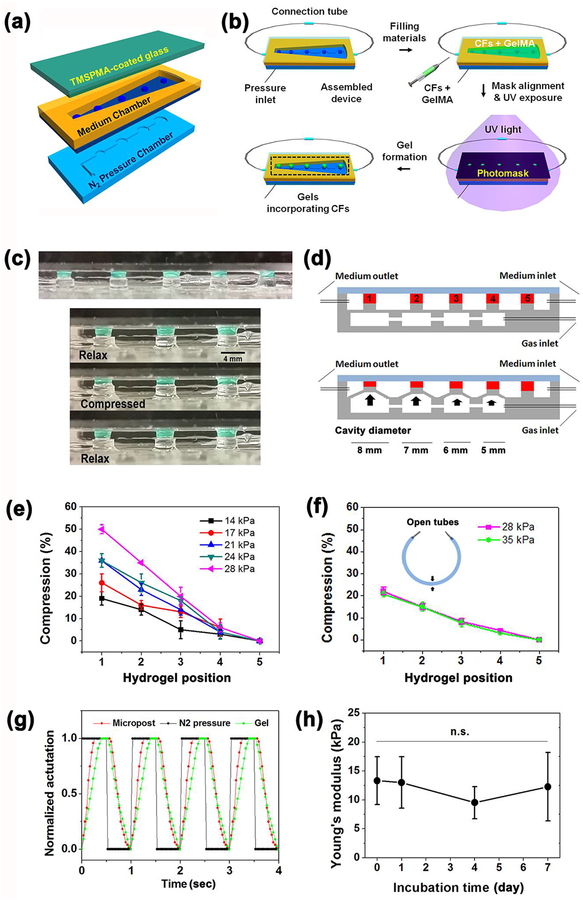
A microdevice was composed of an N2 pressure chamber on the bottom, a medium chamber in the middle and a cover glass on the top (Figure 2a). Similar to previous bioreactor designs,[24, 28] by varying the diameter of actuation cavities under each post (from post 1 to 4, the diameter was 8–5 mm), one single N2 injection was able to create a series of vertical displacements and form gradient magnitudes of compression (Figure 2c, d). Larger cavity diameters led to larger deformation of hydrogels. Compression strain of the hydrogel was defined as the percent of deformation and calculated by the following equation:
Figure 2.
(a) Schematic illustration of bioreactor fabrication consisting of N2 pressure chamber, PDMS membrane with pillars, medium chamber and TMSPMA coated glass from the bottom up. (b) Flowchart of bioreactor assembly, loading CFs with GelMA pre-polymer solution, UV exposure to cure cell-laden gels along PDMS pillars. (c) Side view of five hydrogels (Diameter: 3 mm) patterned on top of pillars in the bioreactor and sequential photographs showing the compression and relaxation of a patterned hydrogel under cyclic compression. Scale bar = 4 mm. Green dye was incorporated into the hydrogel to aid in visualization. (d) Schematic illustration of bioreactors in a static state (top) and actuated state (bottom) by injection of gas to generate a different strain of compression upon hydrogels, the hydrogels were normalized with numbers 1–5 indicating cavity diameter 8–5 mm of gas chambers. (e-f) Experimental data of compression strainas a function of hydrogel position and applied N2 pressures at 1 Hz in (e) air and (f) culture medium that filled inside the bioreactor leaving the medium tubes open (n=5). (g) Characterization of the dynamic compression response of a patterned hydrogel upon actuation of a post by applied pressure. (h) Young’s modulus of hydrogels after different incubation days of mechanical compression under 1 Hz actuation. (n=5)
| (1) |
The compression strain could also be modulated by the applied N2 pressure. As shown in Figure 2e, compression strain positively correlated with the cavity diameter of the gas chamber under each pillar and the applied gas pressure at 1 Hz. As previously reported, the compression strain applied to the hydrogels was not perfectly homogenous, which was highest near the bottom of the hydrogels but lowest near the top.[24] Relatively equal distribution of strain could be found in intermediate regions rather than the bottom or top surfaces of the hydrogels. The highest compression strain achieved was 50% under a pressure of 28 kPa with air filling in the medium chamber. As the medium chamber filled with cell culture medium, the presence of cell culture medium produced resistances that counteracted compressions to a certain extent. Resultantly, it could only generate as high as 20% strain even with a pressure of 35 kPa in a closed microdevice (FigureS2b). Notably, when the medium tubes were left open, the cell culture medium filled microdevice generated 20% strain and a similar slope with a lower pressure of 28 kPa (Figure 2f). The opening of the medium chamber provided a pathway to release the inner-pressure by expelling minor amounts of the medium into tubes in a compressed state, while the pressure would be gone in a relaxed state. Considering that the inner-pressure would accumulate inside the medium chamber in the closed case, opening the chamber without any connecting tubes was applied for following cyclic compression stimulations. Consequently, gradient compression strains of 0, 5, 10, 15 and 20% were obtained across the pillars, covering the physiological and pathological mechanical strain range of real cardiac tissue.[12, 13] The dynamic compression stimulation achieved a sinusoidal cyclic loading strain profile with a gradual increase in strain, which would be more favorable to the mechanical modulation of CFs (Figure 2g). A higher compression frequency of 2 Hz also worked on this microdevice but gave rise to strains less than 1 Hz (Figure S2a). The results certified that the microdevice was qualified to provide tunable cyclic mechanical stimulation with gradient strains to fulfill the dynamic mechanical demands of the in vitro 3D engineered cardiac tissue.
Besides the cyclic mechanical strains, the stiffness of the matrix is another key factor in creating a compliant microenvironment for the CFs to reside in the hydrogel. In our previous study, 7% GelMA hydrogel has been shown to have the best spreading and myofiber networking for the cardiac cells along with strong beating behavior.[21] Upon exposure to varying durations of UV crosslink, Young’s moduli of the 7% GelMA hydrogel after a 40 s UV exposure was 9.8±0.6 kPa, which closely resembled the stiffness of a healthy neonatal rat heart ECM ranging from 4 to 11 kPa (Figure S3a).[29] The encapsulation of CFs induced the stiffness to increase slightly to 13.3 ± 4.1 kPa. However, there were no significant differences compared to the pristine GelMA hydrogel. In addition, we found that cell-laden GelMA hydrogels showed a similar Young’s modulus during7 days culture in the microdevices along with cyclic compression under 37 °C (Figure 2h). Nonsignificant variation was also found in terms of thickness and diameter of the cell-laden hydrogels (Figure S3b, c), demonstrating the CF-laden GelMA hydrogels were mechanically and morphologically stable for 7 days of mechanical stimulation. Consequently, we observed negligible degradation of the cell-laden GelMA hydrogel. The results guarantee that the mechanical properties of the hydrogels would induce almost no impact on the following phenotypic remodeling of CFs during cyclic mechanical stimulation for up to 7 days.
2.2. Cell proliferation and spreading behaviors
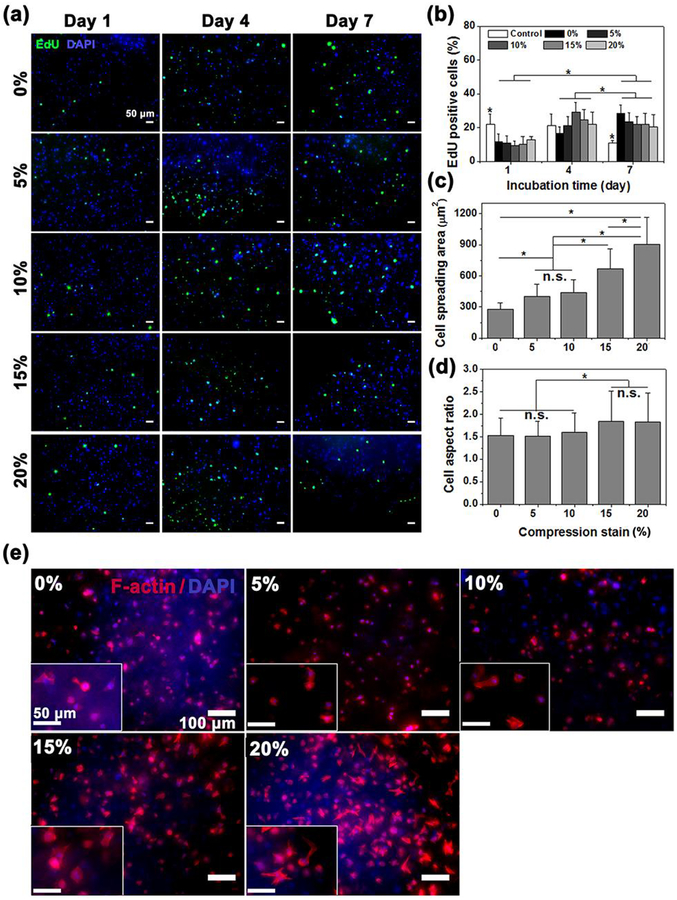
To assess the effect of cyclic compression with gradient magnitudes (0–20%) on cell proliferation, CF-laden 7% GelMA hydrogel was fabricated and incubated for 7 days. Fluorescent images of EdU labeled CFs in the GelMA hydrogel were analyzed on days 1, 4 and 7 of culture (Figure 3a). The quantification of the EdU positive cells demonstrated that the proliferation rate of CFs in the 7% GelMA hydrogel increased over7 days of culture (Figure 3b). Whereas varying compression strains had no significant impact on the proliferation of CFs, suggesting that the proliferation of CFs was independent of mechanical stimulation. For culture dimensions, 2D control exhibited a significantly higher growth rate than 3D cells on day 1, but the superiority disappeared on day 4 and was even reversed on day 7. Similar to the previous report, the proliferation of 2D cells showed higher rates initially but stopped once the cells reached confluence.[30] This is because the structure and biological properties of the scaffold are the major factors contributing to cell proliferation and attachment.[31] Since the optimized 3D hydrogels could provide larger space for cell attachment, migration and spread than a 2D culture plate, the cells in the 3D hydrogels were able to proliferate longer periods than that of the 2D culture.[32] Furthermore, dynamic disturbance of the medium occurred in the culture chamber due to cyclic compression, which was favorable for the diffusion of nutrients and oxygen through 3D scaffold for the proliferation of cells for extended culture periods.
Figure 3.
(a) Representative confocal images of EdU Click-iT labeling (green) of CF-laden 3D GelMA hydrogels with different compression strains at days 1, 4, and 7. (b) Representative quantification of proliferating cells inside CF-laden 3D GelMA hydrogels as determined by the percentage of EdU positive cells at days 1, 4, and 7 of culture (n=6). CFs cultured on 2D well plates were taken as control (n=6). (c and d) CFsspreading area and aspect ratio after 7 days of mechanical stimulation(n=8). (e) Representative confocal images of Actin/DAPI results under different compression strain from 0 to 20%. Data depict Mean ± Standard deviation. *p<0.05.
Cell spreading in response to cyclic compression was evaluated by F-actin staining of the CFs (Figure 3e). Markedly increased amounts of spreading were found with rising compression strain, and growing numbers of CFs varied from round shape to stellar morphology. Quantitative determination of area and aspect ratio covered by F-actin fibers confirmed a significant increase in cell spreading in the presence of compression (Figure 3c and d). Physiological strains of 10% and 5%created a similar spreading area, but a non-significant difference in aspect ratio when compared to 0%. However, the spreading area tripled under a physiological strain of 20%, compared to a strain of 0%. The higher strain also had a significantly varied cellular aspect ratio (Figure 3d). As compression was applied to the hydrogels, the cells were both vertically compressed and horizontally stretched, leading to a larger aspect ratio due to the increase in displacement along the long axis.[33] The results demonstrated that cyclic compression significantly enabled CFs to spread and vary their morphology in the hydrogels, both of which were shown to be dependent on compression strain. These cell behaviors were very similar to our previous study that showed human mesenchymal stem cells (hMSCs) under cyclic compression.[24]
2.3. Effect of myofibroblast maturity on the phenotypic remodeling of CFs upon dynamic mechanical strains
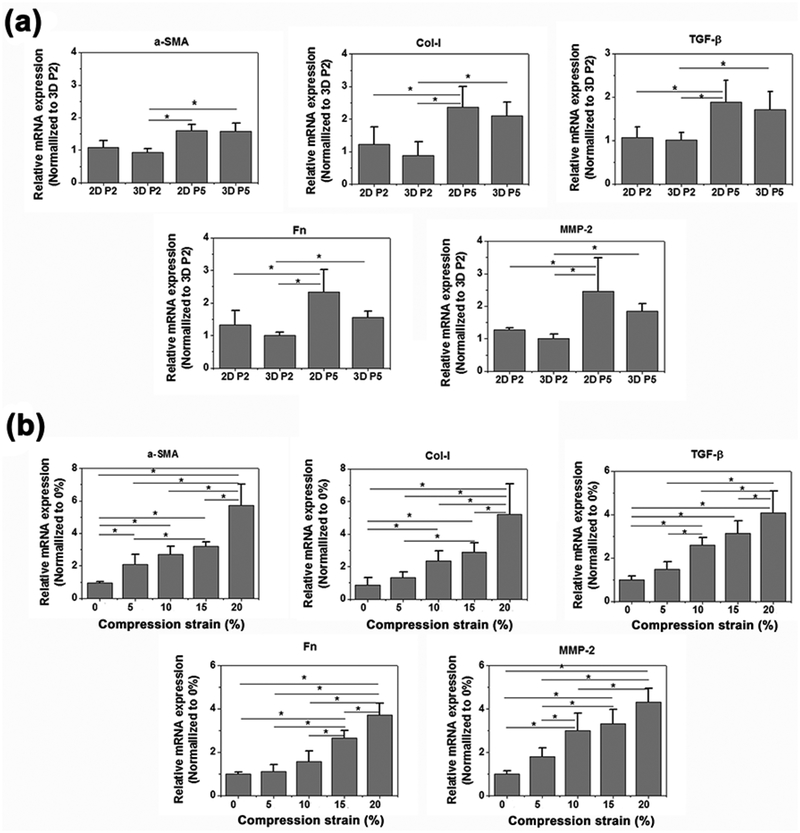
Based on previous studies implying that aging of the heart induces a phenotypic transition from CFs to myofibroblastic cells,[34] we questioned whether the passage number of CFs would influence the degree of fibrosis in CFs. In order to answer this question, CFs of passage 2 (P2) and P5 were cultured for 7 days on 2D well plates and in 3D hydrogel microdevices to compare their phenotype shifting. This was determined by using mRNA expression against widely used fibrosis biomarkers, α-SMA, Col-I, TGF-β, fibronectin (Fn) and MMP-2 (Figure 4a). Cells in the high passage (P5) displayed a significantly higher expression of the fibrotic markers than cells in the low passage (P2) regardless of culture form, which corresponded to previous studies demonstrating that quiescent CFs spontaneously differentiated into activated myofibroblasts with higher passage numbers.[34] In addition, the spatial dimension of cell culture also acts an important role in the phenotype shifting of CFs towards myofibroblastic phenotypes. Cells cultured in 3D hydrogels showed lower expression levels of the fibrotic markers when compared to those cultured on the 2D stiff substrate, which indicated that the mimicking of matrix elasticity in native cardiac tissues might help to maintain their CF phenotype. Together, these results demonstrated that CFs in the high passage might undergo a phenotypic transition to myofibroblastic cells showing higher myofibroblast maturity, and higher levels of the phenotypic transition may be promoted when the cells are cultured in 2D plastic substrates.
Figure 4.
RT-PCR of relative mRNA expression of α-SMA, Col-I, TGF-β, Fn and MMP-2 of (a) CFs of P2 or P5 which were cultured on 2D TCPS cell plate or in 3D hydrogels in microdevices; (b) CFs of P2 cultured in 3D GelMA hydrogels after 7 days cyclic compression stimulation of different strain from 0 to 20% (n=4). Data depicts fold-change ± standard deviation. *p<0.05.
To investigate the effect of myofibroblast maturity on the fibrotic remodeling of CFs under stimulation of dynamic compressions, P2 CFs in the GelMA hydrogel were cultured for 7 days under dynamic compressions. CFs displayed high mRNA expression of biomarkers towards the myofibroblast direction under higher mechanical strains (Figure 4b). Although there was no significant difference between strains of 5% and 10% for α-SMA, Col-I, and Fn, except for MMP-2, the maximal expression level was obtained under a strain of 20%, which was 4–6 times higher than that of 0%. TGF-β displayed a similar trend with the biomarkers, notably, with significant differences found even between strains of 5% and 10%.
It has been acknowledged that elevated mechanical stress is essential to the activation of fibroblastic cells towards the myofibroblastic phenotype.[3] In addition, TGF-β is a central mediator in the modulation of fibroblast phenotype and gene expression.[35] The crosstalk between mechanical tension and TGF-β thereby acts as a key role in fibrotic phenotype transition. In this experiment, the only difference among treatments was the level of mechanical stimulation. Intriguingly, the ECM of CFs provides a repository for various growth factors including TGF-β in a latent state, which can be exploited to enhance cell signaling by extracellular activation from its protective pro-peptide. Once CFs are traced upon exogenous mechanical tension, the latent TGF-β could be activated through traction-mediated activation. The amount of active TGF-β released depends on the mechanical strain applied to the ECM.[36] This theory confirms the finding that an increase in the expression of fibrotic markers would also increase the levels of TGF-β expression under higher mechanical compressions.
To gain insight into how phenotypic shifting mediated by myofibroblast maturity may exert an influence on the fibrotic remodeling of CFs upon dynamic mechanical strains, P5 CFs were also cultured for 7 days under stimulation of dynamic compression. Unlike the trend from P2 cells showing increased expression levels of fibrotic markers with increasing mechanical compressions, fibrotic makers showed magnitude dependent expression for P5 CFs after 7 days of treatment with dynamic compression (Figure S4). Cells under static 3D culture (0%) expressed higher levels of fibrotic markers compared to those under 5 and 10% mechanical compression (physiological strain), while CFs were activated again towards the myofibroblast direction under harsh mechanical tension (pathological strains, 15 and 20%) (Figure S4b). This may be attributed to the facts shown in the passage-number dependent levels of fibrotic marker expression. We showed that a high passage number of CFs (P5) did own high myofibroblast maturity and exhibited more obvious proto-myofibroblastic features (Figure S4a), which may lead to variable effects of cyclic strain on myofibroblast activation. In addition, a previous study showed that designing biomaterials which resemble native matrix elasticity would play an important role in the suppression or reversion of myofibroblast activation,[11] which corresponded to the findings that decreased degree of fibrosis was shown when proto-myofibroblastic phenotypes were cultured in a microenvironment mimicking the natural stiffness in vivo in combination with physiologically relevant strain (5 and 10%).
In the process of myofibroblast differentiation, three phenotypes are involved, resident fibroblasts, conversion to proto-myofibroblasts, and finally myofibroblasts. Proto-myofibroblasts serve as an intermediate in this process and are characterized by elevated levels of fibronectin expression, more specifically elevated levels of an alternatively spliced ED-A isoform, in response to mechanical tension.[37] The actin cytoskeleton of a proto-myofibroblast incorporates stress fibers generated by cytoplasmic β and γ actin microfilaments, which align parallel to the principal strain in a GelMA lattice (Figure 3e). As levels of mechanical strain increase, focal adhesions which appear at the ends of the stress fibers transform into larger and more mature adhesions, capable of detecting mechanical tension in the ECM and transmitting it to the cells as a contractile force. The further rise in mechanical tension, together with the activated TGF-β, induces differentiation into the myofibroblast phenotype, whose stress fibers are formed by α-SMA (Figure 1).[4, 38] The increased expression of α-SMA verified the activation of myofibroblasts, but it may not guarantee that the proto-myofibroblasts would be completely activated. This meant CFs might be plastic when cultured in a more realistic 3D microenvironment with the combination of hydrogels encapsulating cells and physiological mechanical compressions, both of which mimicked mechanical microenvironments in vivo. This also corresponded to the results from (1) P5 CFs displaying decreased expression levels of fibrotic markers under the physiological condition (5% and 10%) and (2) P2 CFs showing that there was no significant difference between strains of 5% and 10% for α-SMA, Col-I, and Fn. Future work exploring phenotypic remodeling with CFs in different passages, or a disease model that controls the level of proto-myofibroblasts may prove useful in discerning how physiologically relevant mechanical strains may guide reprogramming of proto-myofibroblasts into the fibroblast phenotype.
2.4. Mechanical and biochemical impacts on CFs phenotypic remodeling
As found above, mechanical compression enabled CFs with different myofibroblast maturity or phenotypes develop different proto-myofibroblastic features in response to gradient magnitudes. The latent TGF-β in CFs was activated and released by traction-mediated activation, which would trigger CFs phenotypic remodeling but was quite dependent upon exogenous mechanical stimulations. Methods to distinguish the impacts of biochemical and mechanical cues and how much they respectively contribute to CFs phenotypic remodeling still require additional research. Furthermore, what effects mechanical cues would play on an activated myofibroblast are still unknown. Answers to such questions would not only provide insight into the underlying mechanism of CFs phenotypic remodeling but would also be able to provide new preventive or therapeutic strategies for cardiac fibrosis.
Upon these considerations, a fibrosis model was established by treating CFs (P5) assembled in microdevices with 10 ng/mL TGF-β1 as model cytokine for 24 hours before mechanical stimulation, the dose of which was sufficient to trigger fully activated myofibroblasts according to a previous study.[39] Fibrotic markers displayed magnitude dependent expression for P5 CFs, rather than an increasing trend with elevating mechanical compressions, demonstrating that P5 CFs may be plastic within fibrosis and deserve to be clarified. In order to distinguish the roles of TGF-β and mechanical compression, 4 groups were set up: the first group receiving only mechanical stimulation, labeled M; the second group continuously treated with TGF-β inhibiting drug, Tranilast, and mechanical stimulation, labeled DM; the third group continuously treated with TGF-β1 and mechanical stimulation for 7 days, labeled TM (Figure S5a); and the last group without any mechanical compression or TGF-β1 treatment, labeled C. The expression of fibrosis markers α-SMA and Col-I were examined and the mRNA expression of Fn and MMP-2 were also assessed. Mechanical compression induced intermediate upregulation of α-SMA and Col-I expression, Tranilast and mechanical compression (DM) induced the lowest expression, while TM induced the highest level of expression (Figure 5a).
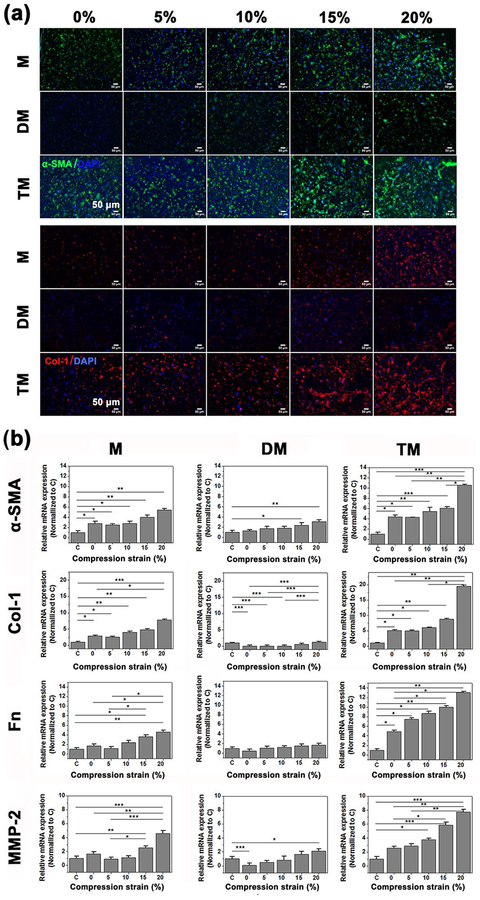
Figure 5.
A cardiac fibrosis model was established by treating CFs (P5) in the microdevices with 10 ng/mL TGF-β1 for 24 hours prior to compression stimulation, the first group receiving only mechanical stimulation, labeled M; the second group continuously treated with TGF-β inhibiting drug, Tranilast, and mechanical stimulation, labeled DM; the third group continuously treated with TGF-β1 and mechanical stimulation for 7 days, labeled TM; and the last group without any mechanical compression or TGF-β1 treatment, labeled C. (a) Representative confocal images of immunofluorescence stained α-SMA (green) and collagen-I (Col I, red) after 7 days cyclic compression stimulation with different strains from 0 to 20% (n=4). (b) RT-PCR of relative mRNA expression of α-SMA, Col-I, Fnand MMP-2 ofCFsafter 7 days cyclic compression stimulation of different strain from 0 to 20% (n=4). Data depicts fold-change ± standard deviation. *p<0.05,** p<0.01, *** p<0.001.
For M group, strain 0% was 2.8, 2.9, 1.7 and 1.6 times higher than the control for expressions of α-SMA, Col-I, Fn and MMP-2, respectively (Figure 5b). Strain 20% retained the highest level that was similar to the above results. It was noteworthy that a decrease was found in α-SMA and Col-I mRNA expression, although not significant, under strain 5%. The results were similar to the findings ahead.
Regarding DM, the overall mRNA expression of biomarkers was dramatically reduced. All experiments with strain 0% showed no difference with the control, implying that the myofibroblast phenotype in static hydrogels lost their fibrotic features and returned to the quiescent fibroblastic phenotype. Only strains of 15% and 20% displayed significantly higher expression of α-SMA than the control, which was 3.1 and 2.4, respectively. No statistical difference has been found between strain 20% and the control for Col-I and Fn. Especially, mRNA expression of Col-I was significantly downregulated under strains of 5% and 10%. The results were attributed to the treatment of TGF-β inhibitor, Tranilast, which was an anti-allergic drug and reported to have efficacy in attenuating TGF-β production and secretion,[40] suppressing TGF-β signaling pathway,[41] inhibiting TGF-β induced ECM production and collagen synthesis.[42] Although it was also reported to inhibit fibroblast proliferation, the 50 μM Tranilast applied in our study was nontoxic for cells according to a previous study.[43] The DM group thereby represented a bare effect of mechanical stimulation on CFs activation without the confounding influence of TGF-β.
For TM group, continuous treatment of TGF-β1 maintained the myofibroblastic phenotype. The quantitative mRNA expression of fibrogenic biomarkers displayed a significantly high level, with strain 0% being 4.3, 5.1, 4.9 and 2.5 times higher than the control group for expressions of α-SMA, Col-I, Fn and MMP-2, respectively. The overall mRNA expression levels were dramatically increased compared with M. Apparently, mechanical compression played a synergetic role to evolve fibrotic progress in a strain-dependent manner. As confirmed by RT-PCR data, the mRNA expression increased with compression magnitude and peaked under strain 20%. However, physiological strain levels, namely 5 and 10%, triggered no significant increase compared with 0% for α-SMA and Col-I, suggesting CFs could maintain their fibrotic status. The extent of α-SMA formation and collagen accumulation in Figure 5a also verified the results. In contrast, larger amounts of inter-connective stress fibers and collagen accumulation were observed under strains of 15 and 20%.
The above results suggested that we were able to distinguish the effects of mechanical cues, representative of compression, and biochemical cues, representative of TGF-β, and highlighted the strain-response correlation between dynamic mechanical compressions and myofibroblast activation. In a fibrosis model, mechanical compression exerted strain-dependent regulation of myofibroblast activation, which could maintain or attenuate the myofibroblastic phenotype under physiological strains (5% and 10%), and promote myofibroblast transition under pathological strains (15% and 20%). If the synthesis or action of TGF-β was inhibited, such as in group DM, the tendency of strain-mediated fibrotic response was unchanged, and mechanical compression alone could still activate the myofibroblastic phenotype under pathological strains (15% and 20%). When TGF-β1 was continuously supplied, such as in group TM, the highest fibrotic level was obtained and mechanical compression would synergistically promote the CFs phenotypic remodeling. Limited expression of TGF-β mRNA in the presence of exogenous TGF-β1 supplementation regardless of M, DM or TM also verified that the endogenous synthesis of TGF-β was inhibited (Figure S5b). At the same time, the activity of TGF-β was almost completely inhibited by Tranilast, as evidenced by the decrease of Col-I and Fn message expression to the control level even under strain 20%. On this point, it was suggested that promoting the phenotypic transition from fibroblast to myofibroblast may be a complicated process between the effects of mechanical tension and TGF-β. However, it seemed obvious that mechanical compression played a crucial role in promoting the phenotypic transition from fibroblast to myofibroblast with strain-dependent changes.
There is increasing evidence that TGF-β and mechanical stimulation are key regulators of the fibroblast transition towards myofibroblasts in the myocardium. Here, we indicated that CFs distinguished between strain magnitude dependence-induced and TGF-β induced phenotypic remodeling, though crosstalk occurs between the actions. The differential actions were determined by different myofibroblast maturity, and plasticity of CFs seeded in hydrogels. Without the exogenous addition of TGF-β, P2 CFs (P2) were activated towards the myofibroblast direction upon increased cyclic mechanical compression through traction-mediated activation of the latent TGF-β. While P5 CFs developed into proto-myofibroblasts, physiological strain (5% and 10%) could restore the ECM tension and reverse the proto-myofibroblastic phenotype into fibroblasts. Higher mechanical strains (15% and 20%) would again promote CFs activation towards the proto-myofibroblast direction. Although the mechanism of how TGF-β activates the expression of α-SMA is still not clearly understood, TGF-β bioactivity was confirmed by enhanced phosphorylation of Smad2, a downstream effector of the TGF-β signaling pathway.[44] In the case of mature myofibroblasts activated by exogenous TGF-β supplementation, mechanical compression would maintain or promote active myofibroblastic tendency rather than negate the effect of TGF-β. However, if the action of TGF-β was inhibited, CFs may be more plastic so that cells cultured under physiological strains (5–10%) would show similar or lower levels of fibrotic markers, while those cultured under larger mechanical load (15–20%) would prefer to express higher levels of those markers compared to the controls. The mechanism of such mechano-regulation is yet unknown. The results implied that mechanical compression and TGF-β played synergetic roles in inducing myofibroblast transition. Both of which could promote the transition, but the mechanical compression of physiological strains could maintain or attenuate the strain-mediated promotion, demonstrating that mechanical compression functioned as a means of protection in CFs phenotypic remodeling. However, the effect of mechanical compression was unable to dominate that of TGF-β.
Since cardiac tissue is highly dynamic and contractile, when a mild mechanical load or inflammation is applied, the activation of myofibroblasts could be attenuated via self-healing capacity from the cardiac physiological rhythm. In the case of pathological mechanical load elevation and chronic injury, drug intervention is necessary to reduce cytokine level and consequently modulate the mechanical stress in the diseased region. Any other strategies that could attenuate local mechanical tension could also be considered to alleviate the symptoms.
However, we recognized the limitations of this cardiac fibrotic remolding model, as it lacks the involvement of cardiomyocytes and their interplay with fibroblasts to recapitulate the native micro-environment in cardiac fibrosis remodeling. Moreover, characterization of fibrosis hallmarks and functional studies of the fibrotic tissues needed to be evaluated. Further study required to verify the effect of mechanical stimulations on fibrotic remodeling using a more physiologically biomimetic model in the future.
3. Conclusion
In this study, we developed a microdevice capable of accurately and simultaneously mimicking the biomechanical properties of the cardiac physiological and pathological microenvironment. The microdevice could apply 1 Hz cyclic compressions with gradient strain, ranging from 5% to 20%, onto 3D cultured CFs. CFs could proliferate within the cyclic mechanical compression applied on GelMA and displayed strain-mediated spreading behavior. Strain-dependent cardiac myofibroblastic activations were found after 7 days of mechanical compression, which also strongly relied on the myofibroblast maturity of the CFs seeded. Physiological strains (5–10%) could maintain or attenuate the myofibroblast activation, while pathological strains (15–20%) would significantly promote the phenotype transition towards myofibroblasts, demonstrating the crucial role which mechanical cue played in CFs phenotypic remodeling. Furthermore, mechanical compression and TGF-β played synergetic roles in CFs phenotypic remodeling, demonstrating close crosstalk lying between them. The strain-response of mechanical stimulation in cardiac myofibroblast activation may not only be favorable in the understanding of the mechanism of CFs phenotypic remodeling but would also provide new preventive or therapeutic strategies for cardiac fibrosis.
4. Experimental Section
4.1. Preparation of GelMA
GelMA was prepared following the method described previously.[45] Porcine skin gelatin (Type A, Sigma-Aldrich) was added in DPBS (Sigma-Aldrich) at a concentration of 10% (w/v) at 50°C. Then, methacrylic anhydride (Sigma-Aldrich) was added by drop while being stirred at 50°C with a concentration of 5% (v/v). After a 2hour period of reaction, the solution was dialyzed to remove the unreacted reagents using dialysis membrane tubes (molecular weight cut off:12–14 kDa) against deionized water for 7 days at 40°C. The obtained solution was freeze-dried for 5 days to obtain medium degree methacrylate (MM) GelMA.
4.2. Mechanical Properties of GelMA Hydrogels
7% MM GelMA and 0.25% (w/v) photoinitiator (Igracure 2595, Sigma-Aldrich) were dissolved in DPBS. In order to figure out the optimal mechanical stiffness, different UV exposure times were applied to photo-crosslink GelMA and subjected to Young’s modulus measurements. Disc-shaped hydrogels were fabricated by sandwiching the pre-polymer solution (200 μL) between 1 mm thick spacers. This pre-polymer solution was exposed to UV light (50 mW/cm2) for 30, 40, 50 or 60s. The crosslinked hydrogels were punched into small discs with diameters of 6 mm and detached from the glass slide. After removing excess liquid, the gels were compressed uniaxially on Instron (5542, USA) at 1mm/min. The Young’s modulus was then obtained by calculating the slope from 0% to 10% strain (n=5).
4.3. Microdevice Fabrication
A microdevice is composed of an N2 pressure chamber on the bottom, a medium chamber in the middle and a cover glass on the top (Figure 2a). The N2 pressure chamber consisted of aligned cylindrical cavities with varying diameters that were connected by channels (1.5 mm in width) for applying pressure from one gas inlet. The medium chamber was patterned with a lower part of 5 aligned posts (Diameter: 3.5 mm) and an upper part of sidewalls. The cavity was designed with one narrow end and another wide end to save the amount of hydrogel and cells afterward. The height of the posts (1.5 mm) was lower than that of the sidewalls (3 mm) to allow the formation of 3D hydrogels between the posts and the cover glass. The thickness of the medium chamber bottom membrane was 300 μm. The medium and N2 pressure chambers were prepared by pouring a mixture of polydimethylsiloxane (PDMS) base and curing agent into their individual poly(methyl methacrylate) (PMMA) molds. After a 1hourcuring period at 80 °C, they were carefully peeled off from the molds and adhered together using O2 plasma with the posts aligned to the gas chambers. Especially noted, the narrow end of the medium chamber had to be oriented towards the gas port with the fifth post located on the gas channel rather than the cavity (Figure 2d). The plasma bonded PDMS constructs were then incubated at 80 °C for 30 min. Fluidic and gas ports were first pierced by a needle and inserted with a stainless connector (1 mm in diameter). The PDMS construct was then combined with a 3-(trimethoxysilyl) propyl methacrylate (TMSPMA) coated glass through plasma treatment. The inlet and outlet ports were connected using tubes.
4.4. Cardiac Fibroblast Isolation and Culture in Microdevices
To obtain primary cardiac fibroblasts (CFs), we isolated hearts from neonatal Sprague Dawley rats (two-days old) following a protocol approved by the Institution’s Committee on Animal Care.[46] The extracted hearts were cut into small pieces, incubated in trypsin solution at 4 °C overnight and were subjected to collagenase digestions. After separating the cardiomyocytes, the attached CFs were cultured in Dulbecco’s Modified Eagle Medium (DMEM, Gibco, USA) along with 10% fetal bovine serum (Gibco, USA) and 1% penicillin/streptomycin (Gibco, USA) in an incubator at 37° C and 5% CO2. CFs were used in no more than 5 passages. CFs were trypsinized and resuspended in GelMA pre-polymer solutions at a ratio of 6 × 106 cells/mL. The GelMA pre-polymer solutions with cells were injected through the tube into the medium chamber of the microdevice. A photomask was then covered on the top of the microdevice and then UV light (50mW/cm2) was applied for 40 seconds to crosslink the cell-laden GelMA polymer solution above the posts. The uncrosslinked GelMA polymer solution was then withdrawn from the medium chamber and refilled with cell culture medium (Figure 2b). The microdevices with cell-laden GelMA hydrogels were cultured in an incubator at 37° C and 5% CO2. Using a WAGO controller and a customized MATLAB program to control solenoid valves, N2 gas was propelled into the pressure chamber via the gas inlet.[47] Cyclic compressions of 1 Hz were applied to the crosslinked hydrogel within the microdevices for 7 days of culture.
4.5. MicrodeviceCharacterization
The microdevice was subjected to varying dynamic compressive strains actuated by cyclic gas pressure. In order to quantify the compression strain of hydrogels in response to the cyclic loading pressure, time–sequential images were taken from a side view of hydrogel patterned microdevices. For each post, 10 cycles of compression were taken. ImageJ software (NIH) was employed to measure displacements (minimum3 measures per cycle and 5 cycles per post) and calculate the average strain. Parameters including actuation pressure (14–35 kPa), cyclic frequency (1–2 Hz), the fullness of the medium chamber (air or DMEM) and openness of connection tubes was considered for their impacts on the compression strains.
4.6. Cell Laden Hydrogels Degradation Assay
During the process of cell culture in the presence of cyclic compressions for7 days, GelMA hydrogels underwent degradation. In order to evaluate how much the degradation would affect the mechanical properties or the morphology of the hydrogels, hydrogels were detached from the cover glass at days 1, 4 and 7 and photos were taken for analysis using ImageJ. Dimensions (thickness and diameter) of hydrogels from each post were measured and then the Young’s modulus was determined on Instron (minimum 3 measures per post of 5 different devices).
4.7. Cell Proliferation Assay
Cell proliferation within 3D hydrogels (n=4) at days 1, 4 and 7were assessed qualitatively and quantitatively using Click-iT Plus EdU Alexa Fluor® 488 Imaging Kit (Thermo Fisher Scientific). Cell-laden hydrogels were incubated with 10 μM EdU at 37°C for 24 hours. The samples were then fixed with 4% paraformaldehyde (PFA) for 30 min and permeabilized with 0.5% Triton X-100 (Sigma-Aldrich) for 40 min. Click-iT Plus reaction cocktail was added into the samples and incubated for 40 min in the dark. After another rinse with 3% bovine serum albumin (BSA), the samples were stained using Hoechst 33342 (1:2000) at room temperature. CFs cultured on 2D well plates (5000 cells/well, n=4) were taken as control. A confocal microscope (Leica SP5 X MP, Germany) was used to take fluorescence images of the samples. Acquisition parameters were not changed throughout the imaging of each experiment. The amount of EdU positive cells and Hoechst positive cells were analyzed using the ImageJ program, respectively. The value obtained from the EdU positive cells divided by the total number of Hoechst positive cells was determined to be the percentage of cell proliferation.
4.8. Cell Spreading Analysis
Cell spreading of CFs in hydrogels in response to varying compression strains was assessed through visualization of the stained F-actin fibers inside cells. After 7 days of mechanical stimulation, the cell-laden hydrogels (n=4) were incubated at a dilution of 1:100 in 1% BSA under shaking conditions with Alexa Fluor 488 Phalloidin (Invitrogen) for 2 hr. To stain cell nuclei, 4’,6-diamidino-2-phenylindole dihydrochloride (DAPI; Vector Laboratories) at 1:100 dilution in 1% DPBS was used for 2 hours. Fluorescence images were taken by a confocal microscope and two random regions were chosen for quantitative evaluation of cell spreading (n=8). In total, no less than 100 cells in each hydrogel per condition were manually selected for cell area and respect ratio measurement using ImageJ software.
4.9. TGF-β Activation and Inhibition Assay
After the fabrication of microdevices patterned with cell-laden hydrogels, the CFs were treated with TGF-β1 (10 ng/mL) as model cytokine supplemented DMEM for 24 hr. Subsequently, the microdevices were divided into 3 groups (n=4) and subjected to cyclic compression with a frequency of 1 Hz for 7 days: (1) TGF-β1 activation + mechanical stimulation group (TM), CFs were treated with TGF-β1 supplemented culture medium; (2) TGF-β1 inhibiting drug + mechanical stimulation group (DM), CFs were treated with 50 μMTranilast (Sigma), an inhibitor of TGF-β1, supplemented culture medium; (3) mechanical stimulation group (M), CFs were treated with DMEM culture medium. CFs cultured in DMEM without any mechanical stimulation, or TGF-β1 treatment were taken as a control group (C). All the mediums for culture were changed every 48 hr. The samples were collected after 7 days and total RNA was extracted for RT-PCR analysis. Meanwhile, the fibrosis markers were immunostained to visualize their expression under different TGF-βlevels and varying cyclic compressions strains.
4.10. Immunofluorescence Staining
CFs in hydrogels, activated in response to varying compression strains, were immunostained for cardiac fibrotic biomarkers. The cell-laden hydrogels were fixed in 4% PFA for 30 min, permeabilized with 0.1% Triton X-100, and blocked with 1% BSA. The samples were incubated with 200 times diluted primary antibodies in 3% goat serum, monoclonal rabbit anti-α-SMA (Abcam, ab5694), monoclonal mouse anti-collagen I (Thermo Fisher, 1–26771), polyclonal rabbit anti-fibronectin (Abcam, 23751), and polyclonal rabbit anti-MMP-2 (Abcam, 37150) overnight at 4 °C while under shaking. The 200 times diluted secondary antibodies (goat anti-mouse AlexaFluor 594 (Invitrogen, 1107474), goat anti-rabbit Alexa Fluor 488(Life Technology, 1515529) or goat anti-rabbitCy5 (Abcam)) in 3% goat serum were applied to the samples for2 h at room temperature while under shaking. DAPI (1:100) was used to stain the nuclei for 2 h. Fluorescence images were obtained from the middle of each hydrogel by a confocal microscope to eliminate edge effects. Two randomly-selected regions in each hydrogel per condition and four samples were imaged (n=8). The images were processed with ImageJ software. For a better comparison of different groups, the intensities of detection channels (photomultipliers or PMTs, Leica) were optimized and kept unchanged throughout the imaging. CFs cultured on 2D well plates (5000 cells/well, n=4) were taken as control.
4.11. Real-Time Polymerase Chain Reaction
RNA was isolated from mechanically disrupted cell-laden hydrogels (n=4) using TRIzol reagent (Life Technologies). NanoDrop (Thermo Scientific) was used to measure the amount of RNA. Reverse transcription of RNA was carried out using the QuantiTect ® Reverse Transcription kit (Qiagen). RT-PCR was conducted using iTaq™ Universal SYBR® Green Supermix (Bio-Rad, USA) and predesigned KiCqStart™ SYBR® Green primers (Sigma) targeting the genes of fibrosis markers: TGF-β (catalog #KSPQ12012G), Collagen1A1 (catalog #KSPQ12012G), Fibronectin (catalog #KSPQ12012G),α-smooth muscle actin (catalog #KSPQ12012G), and Matrix metalloproteinase-2 (catalog #KSPQ12012G). Relative expressions were calculated by the ΔΔCt method and then normalized by glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene expression.
4.12. Statistical Analysis
We used a mean ± standard deviation for all quantitative data. One-way analysis of variance (ANOVA) was used for statistical analysis along with a Turkey significant difference post hoc test using GraphPad Prism 5.02(*p < 0.05, **p < 0.01, and ***p < 0.001).
Supplementary Material
Acknowledgments
The authors gratefully acknowledge Praveen Bandarufor his generous help in the design of microdevice, Hong Chen and Aiyun Wen for their generous support regarding laser scanning confocal microscopic observation, and Shreya Mehrotra for her work in RT-PCR conduction and analysis. The authors gratefully acknowledge funding from the National Institutes of Health (NIH) (AR066193 and CA214411). S.R.S. would like to recognize and thank Brigham and Women’s Hospital President Betsy Nabel, MD, and the Reny family, for the Stepping Strong Innovator Award through their generous funding.
References
- [1].Porter KE, Turner NA, Pharmacology & therapeutics 2009, 123, 255; C. A. Souders, S. L. Bowers, T. A. Baudino, Circ Res 2009, 105, 1164. [DOI] [PubMed] [Google Scholar]
- [2].Yokoyama T, Sekiguchi K, Tanaka T, Tomaru K, Arai M, Suzuki T, Nagai R, Journal of Cardiac Failure 1999, 5, 1968; A. Leask, Cardiovascular research 2007, 74, 207; R. Mazhari, J. H. Omens, J. W. Covell, A. D. McCulloch, Cardiovascular research 2000, 47, 284. [Google Scholar]
- [3].Tomasek JJ, Gabbiani G, Hinz B, Chaponnier C, Brown RA, Nature reviews. Molecular cell biology 2002, 3, 349. [DOI] [PubMed] [Google Scholar]
- [4].Hinz B, The Journal of investigative dermatology 2007, 127, 526. [DOI] [PubMed] [Google Scholar]
- [5].Herum K, Choppe J, Kumar A, Engler A, McCulloch A, Mol. Biol. Cell 2017, 28, 1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].van Putten S, Shafieyan Y, Hinz B, Journal of molecular and cellular cardiology 2016, 93, 133. [DOI] [PubMed] [Google Scholar]
- [7].Davis J, Molkentin JD, Journal of molecular and cellular cardiology 2014, 70, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Biernacka A, Dobaczewski M, Frangogiannis NG, Growth factors 2011, 29, 196; T. M. Hale, Journal of molecular and cellular cardiology 2016, 93, 125; G. P. Rossi, M. Cavallin, G. G. Nussdorfer, A. C. Pessina, Journal of cardiovascular pharmacology 2001, 38 Suppl 2, S49; L. Ronnov-Jessen, O. W. Petersen, Laboratory investigation; a journal of technical methods and pathology 1993, 68, 696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Ma Y, de Castro Bras LE, Toba H, Iyer RP, Hall ME, Winniford MD, Lange RA, Tyagi SC, Lindsey ML, Pflugers Archiv : European journal of physiology 2014, 466, 1113; G. Serini, M. L. Bochaton-Piallat, P. Ropraz, A. Geinoz, L. Borsi, L. Zardi, G. Gabbiani, The Journal of cell biology 1998, 142, 873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Hinz B, Journal of biomechanics 2010, 43, 146. [DOI] [PubMed] [Google Scholar]
- [11].Yeung T, Georges PC, Flanagan LA, Marg B, Ortiz M, Funaki M, Zahir N, Ming W, Weaver V, Janmey PA, Cell motility and the cytoskeleton 2005, 60, 24; A. M. Kloxin, J. A. Benton, K. S. Anseth, Biomaterials 2010, 31, 1. [DOI] [PubMed] [Google Scholar]
- [12].Ugolini GS, Pavesi A, 2017, 6.
- [13].Balachandran K, Sucosky P, Jo H, Yoganathan AP, American journal of physiology. Heart and circulatory physiology 2009, 296, H756. [DOI] [PubMed] [Google Scholar]
- [14].Guo Y, Zeng QC, Zhang CQ, Zhang XZ, Li RX, Wu JM, Guan J, Liu L, Zhang XC, Li JY, Wan ZM, International journal of medical sciences 2013, 10, 1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Dalla Costa AP , Clemente CF, Carvalho HF, Carvalheira JB, Nadruz W Jr., Franchini KG, Cardiovascular research 2010, 86, 421. [DOI] [PubMed] [Google Scholar]
- [16].Song J, Zhu Y, Li J, Liu J, Gao Y, Ha T, Que L, Liu L, Zhu G, Chen Q, Xu Y, Li C, Li Y, Journal of molecular and cellular cardiology 2015, 79, 145; A. S. Waxman, B. G. Kornreich, R. A. Gould, N. S. Moise, J. T. Butcher, Journal of veterinary cardiology : the official journal of the European Society of Veterinary Cardiology 2012, 14, 211. [DOI] [PubMed] [Google Scholar]
- [17].Watson CJ, Phelan D, Xu M, Collier P, Neary R, Smolenski A, Ledwidge M, McDonald K, Baugh J, Fibrogenesis & tissue repair 2012, 5, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Merryman WD, Lukoff HD, Long RA, Engelmayr GC Jr., Hopkins RA, Sacks MS, Cardiovascular pathology : the official journal of the Society for Cardiovascular Pathology 2007, 16, 268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Yong KW, Li Y, Huang G, Lu TJ, Safwani WK, Pingguan-Murphy B, Xu F, American journal of physiology. Heart and circulatory physiology 2015, 309, H532. [DOI] [PubMed] [Google Scholar]
- [20].Spinale FG, Frangogiannis NG, Hinz B, Holmes JW, Kassiri Z, Lindsey ML, Circ Res 2016, 119, 1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Sadeghi AH, Shin SR, Deddens JC, Fratta G, Mandla S, Yazdi IK, Prakash G, Antona S, Demarchi D, Buijsrogge MP, Sluijter JPG, Hjortnaes J, Khademhosseini A, Advanced healthcare materials 2017, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Balestrini JL, Billiar KL, Journal of biomechanical engineering 2009, 131, 051005. [DOI] [PubMed] [Google Scholar]
- [23].Gould RA, Chin K, Santisakultarm TP, Dropkin A, Richards JM, Schaffer CB, Butcher JT, Acta biomaterialia 2012, 8, 1710; P. A. Galie, J. P. Stegemann, Tissue engineering. Part C, Methods 2011, 17, 527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Seo J, Shin JY, Leijten J, Jeon O, Bal Ozturk A, Rouwkema J, Li Y, Shin SR, Hajiali H, Alsberg E, Khademhosseini A, ACS applied materials & interfaces 2018, 10, 13293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Orr DE, Burg KJ, Annals of biomedical engineering 2008, 36, 1228. [DOI] [PubMed] [Google Scholar]
- [26].Shachar M, Benishti N, Cohen S, Biotechnology progress 2012, 28, 1551. [DOI] [PubMed] [Google Scholar]
- [27].Visser J, Melchels FP, Jeon JE, van Bussel EM, Kimpton LS, Byrne HM, Dhert WJ, Dalton PD, Hutmacher DW, Malda J, Nature communications 2015, 6, 6933. [DOI] [PubMed] [Google Scholar]
- [28].Moraes C, Wang G, Sun Y, Simmons CA, Biomaterials 2010, 31, 577. [DOI] [PubMed] [Google Scholar]
- [29].Bhana B, Iyer RK, Chen WL, Zhao R, Sider KL, Likhitpanichkul M, Simmons CA, Radisic M, Biotechnol Bioeng 2010, 105, 1148. [DOI] [PubMed] [Google Scholar]
- [30].Xie Y, Yang ST, Kniss DA, Tissue engineering 2001, 7, 585. [DOI] [PubMed] [Google Scholar]
- [31].Ingber DE, Prusty D, Sun Z, Betensky H, Wang N, Journal of biomechanics 1995, 28, 1471. [DOI] [PubMed] [Google Scholar]
- [32].Ou KL, Hosseinkhani H, International journal of molecular sciences 2014, 15, 17938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Cui Y, Hameed FM, Yang B, Lee K, Pan CQ, Park S, Sheetz M, Nature communications 2015, 6, 6333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Biernacka A, Frangogiannis NG, Aging and disease 2011, 2, 158; J. J. Santiago, A. L. Dangerfield, S. G. Rattan, K. L. Bathe, R. H. Cunnington, J. E. Raizman, K. M. Bedosky, D. H. Freed, E. Kardami, I. M. Dixon, Developmental dynamics : an official publication of the American Association of Anatomists 2010, 239, 1573. [PMC free article] [PubMed] [Google Scholar]
- [35].Bujak M, Frangogiannis NG, Cardiovascular research 2007, 74, 184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Hinz B, Matrix biology : journal of the International Society for Matrix Biology 2015, 47, 54. [DOI] [PubMed] [Google Scholar]
- [37].Hinz B, Gabbiani G, Thrombosis and haemostasis 2003, 90, 993; B. Hinz, G. Gabbiani, Current opinion in biotechnology 2003, 14, 538. [DOI] [PubMed] [Google Scholar]
- [38].Meran S, Steadman R, International journal of experimental pathology 2011, 92, 158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Cucoranu I, Clempus R, Dikalova A, Phelan PJ, Ariyan S, Dikalov S, Sorescu D, Circ Res 2005, 97, 900. [DOI] [PubMed] [Google Scholar]
- [40].Martin J, Kelly DJ, Mifsud SA, Zhang Y, Cox AJ, See F, Krum H, Wilkinson-Berka J, Gilbert RE, Cardiovascular research 2005, 65, 694. [DOI] [PubMed] [Google Scholar]
- [41].Charng MJ, Wu CH, British journal of pharmacology 2006, 147, 117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Yamada H, Tajima S, Nishikawa T, Murad S, Pinnell SR, Journal of biochemistry 1994, 116, 892. [DOI] [PubMed] [Google Scholar]
- [43].Kubo M, Zhao Y, Moriguchi T, Archives of dermatological research 2012, 304, 745. [DOI] [PubMed] [Google Scholar]
- [44].Galie PA, Russell MW, Westfall MV, Stegemann JP, Experimental cell research 2012, 318, 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Nichol JW, Koshy ST, Bae H, Hwang CM, Yamanlar S, Khademhosseini A, Biomaterials 2010, 31, 5536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Khademhosseini A, Eng G, Yeh J, Kucharczyk PA, Langer R, Vunjak-Novakovic G, Radisic M, Biomedical microdevices 2007, 9, 149. [DOI] [PubMed] [Google Scholar]
- [47].Shin SR, Kilic T, Zhang YS, Avci H, Hu N, Kim D, Branco C, Aleman J, Massa S, Silvestri A, Kang J, Desalvo A, Hussaini MA, Chae S-K, Polini A, Bhise N, Hussain MA, Lee H, Dokmeci MR, Khademhosseini A, Advanced Science 2017. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.