ABSTRACT
Neisseria meningitidis, a commensal β-proteobacterium of the human nasopharynx, constitutes a worldwide leading cause of sepsis and epidemic meningitis. A recent genome-wide association study suggested an association of its type II-C CRISPR/Cas system with carriage and thus less invasive lineages. Here, we show that knock-out strains lacking the Cas9 protein are impaired in the adhesion to human nasopharyngeal cells which constitutes a central step in the pathogenesis of invasive meningococcal disease. Transcriptome sequencing data further suggest that meningococcal Cas9 does not affect the expression of surface adhesins but rather exerts its effect on cell adhesion in an indirect manner. Consequently, we speculate that the meningococcal CRISPR/Cas system exerts novel functions beyond its established role in defence against foreign DNA.
KEYWORDS: Neisseria meningitidis, CRISPR/Cas, Cas9, virulence, nasopharynx, RNA-seq, RIP-seq
1. Introduction
The discovery of clustered, regularly interspaced, short palindromic repeat (CRISPR)/CRISPR-associated protein (Cas)-based interference that confers adaptive, sequence-based immunity against viruses has been one of the most exciting advances in microbiology in the past decade [1]. While the role of CRISPR/Cas systems in host protection against mobile genetic elements is now well established, the understanding of their potential contribution to gene regulation and virulence is still in its infancy [2,3]. Of the six current main types of CRSIPR/Cas systems [4], Type II CRSIPR/Cas systems are associated with pathogenic bacteria including, amongst others, Campylobacter jejuni, Francisella novicida and Neisseria meningitidis [5]. These systems require a trans-activating crRNA (tracrRNA) and an endogenous RNase (RNase III) for maturation of the crRNAs encoded in the CRISPR array, as well as the endonuclease activity of the Cas9 protein.
The β-proteobacterium N. meningitidis is a genetically highly diverse, commensal pathogen that normally resides in the human nasopharynx but which also constitutes a worldwide leading cause of sepsis and epidemic meningitis [6]. The mechanisms that allow bacteria belonging to so-called hyperinvasive lineages to switch from the commensal to the pathogenic state and to cause invasive disease remain enigmatic yet [7]. Recent work by Sampson and co-workers [8] has suggested a potential contribution of the CRISPR-Cas system to meningococcal virulence. Specifically, they showed that deletion of the cas9 gene reduced the adhesion to, invasion of and replication in human adenocarcinoma alveolar basal epithelial cells. However, as meningococci only very rarely cause pneumonia in humans [9], the relevance of this finding for the pathogenesis of invasive meningococcal disease (IMD) remains unclear. Of more immediate relevance would be factors that alter the interaction of meningococci with human nasopharyngeal cells since such interactions are likely affect its propensity to cause IMD [10,11].
We have recently characterized the CRISPR/Cas system of N. meningitidis serogroup C strain 8013 [12,13] and showed that meningococci possess the most streamlined CRISPR/Cas system characterized to date [4]. In the meningococcal type II-C system crRNAs are transcribed from promoters that are embedded within each repeat and the degradation of foreign DNA is independent of RNase III [13]. Similar to findings in the unrelated Francisella genus [14], we further observed a significant association of the CRISPR/Cas locus with meningococcal carriage and therefore less pathogenic lineages but not with the hyperinvasive ones [15]. This suggests that the CRISPR/Cas systems might constitute an ‘anti-virulence’ factor in meningococci. In addition, there was a highly significant inverse association between the presence of a CRISPR/Cas locus and the meningococcal diseases associated prophage MDAΦ [15]. Bille and co-workers recently showed that this prophage increased host cell colonization and suggested that the increased invasiveness of strains lysogenized with this prophage might be a consequence of a high bacterial load at the site-of-entry. They further suggested that this in turn increases the chance of translocation of the bacteria in the bloodstream and/or the dissemination of the bacteria in a population and the number of carriers [10]. Therefore, one mechanism whereby the CRISPR/Cas system could affect meningococcal virulence may be by restricting lysogenization with the virulence-conferring MDAΦ. However, some isolates such as the serogroup C model strain 8013 belong to hyperinvasive lineages yet contain a CRISPR/Cas system but no MDAΦ [16]. This raises the question whether there is also a direct impact of the CRISPR/Cas system on the interaction of meningococci with the human host. Here, by using strain 8013 which also lacks any MDAΦ-specific crRNAs, we address the potentially direct impact of CRISPR/Cas on adhesion to human nasopharyngeal cells and thus on meningococcal virulence.
2. Results
2.1. The CRISPR/Cas system impacts meningococcal adhesion to human nasopharyngeal cells
To assess whether CRISPR/Cas genes impacted on adhesion to human nasopharyngeal cells, we generated deletion mutants and corresponding complemented strains for cas9, rnc (encoding RNase III), tracrRNA and the CRISPR array using standard cloning approaches [13]. We first assessed the growth of these strains in EMM+++ medium which is used in the cell adhesion assay, to exclude potential effects on bacterial fitness. As depicted in Figure 1, growth of the Δcas9 (panel A), Δrnc (panel B), ΔtracrRNA (panel C) and ΔCRISPR (panel D) mutant and corresponding complemented strains showed only slight impairment as compared to the wild-type strain. This showed that CRISPR/Cas system and RNase III are not essential for growth in a rich medium such as EMM+++.
Figure 1.
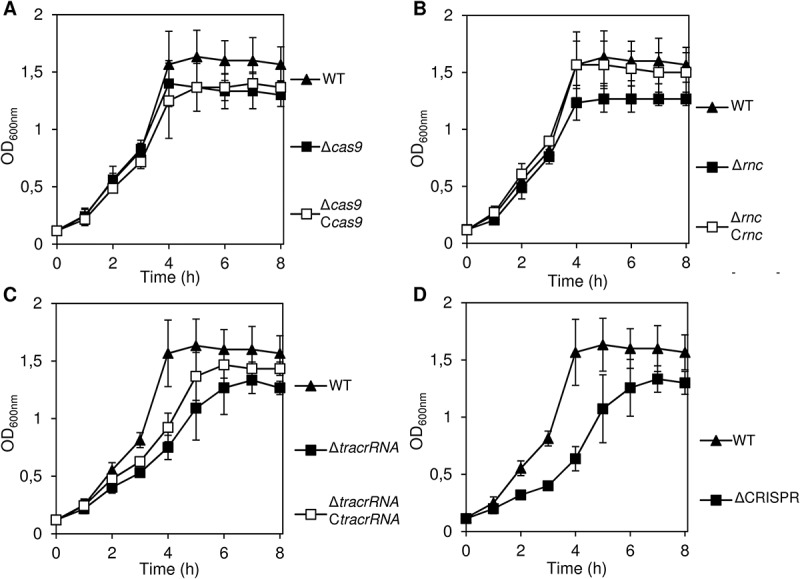
Growth curves of CRISPR/Cas wild-type and mutant strains in rich medium.
For each strain and time point, the mean OD600nm values are given from three individual measurements in cell culture medium EMEM+++ over a time course of 8 h at 37°C. For each strain, the strain designation is given in parentheses.
A. cas9 deletion (AH12) and complemented strain (AH61).
B. rnc deletion (B49) and complemented strain (AH15).
C. tracrRNA deletion (B45) and complemented strain (AH18).
D. CRISPR deletion strain (AH5).
Next, we tested the adhesion of these strains to human nasopharyngeal cells (Figure 2). We observed a significant reduction of the adhesion rates with the strains that carried individual deletions of cas9, tracrRNA or rnc. For two of these mutants – Δcas9 and Δrnc – wild-type adhesion rates could be restored by complementing the respective genes in trans via integration of rnc into the porA and of cas9 into the lctP and aspC locus, respectively (see Supplementary Materials and Methods for details). Notably, this confirms the previous conclusion by Sampson and co-workers [8] that Cas9 is required for meningococcal adhesion to human adenocarcinoma alveolar basal epithelial cells. Regarding our failure to complement the ΔtracrRNA mutant, we reason that deletion of tracrRNA is likely to exert a polar effect on cas9 expression due to the proximity of the tracrRNA and the cas9 promoter regions (Supplementary Figure 1).
Figure 2.
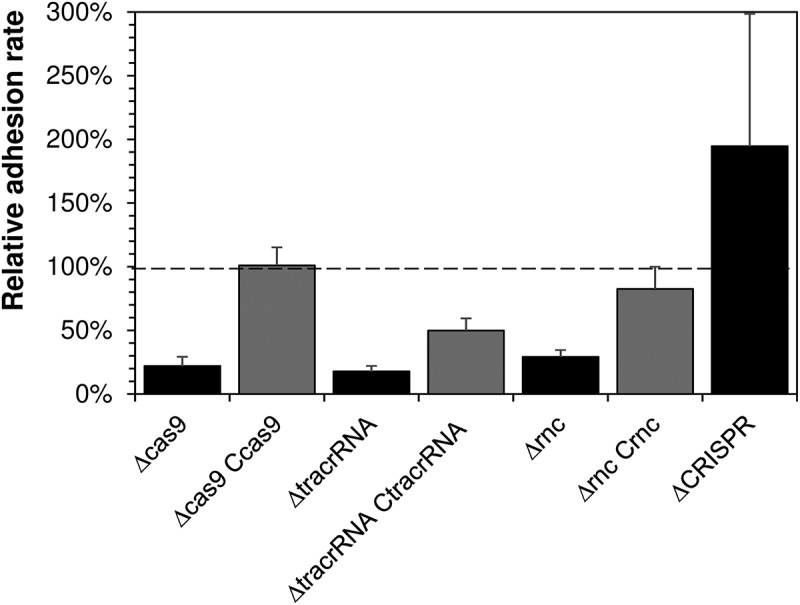
Adhesion of CRISPR/Cas wild-type and mutant strains to the human nasopharyngeal epithelial cell line Detroit562.
Depicted is the average relative adhesion rate of each mutant strain given on the abscissa to the wild-type strain (set to 100%) from three independent experiments each carried out in triplicate. The error bars give the standard error of the mean. For each strain, the adhesion rate was calculated as the number of colony forming units (CFU) recovered after 4 h of infection divided by the seeded CFU determined in parallel. The relative adhesion rate is the adhesion rate of the mutant divided by the adhesion rate of the wild-type strain. All adhesion experiments were performed with the same strains depicted in Figure 1.
Deletion of the CRISPR repeat array had no statistically significant effect on bacterial adhesion rates (Figure 2); if at all, it would promote adhesion, perhaps due to a hyperactivity of Cas9 in the absence of crRNAs. Together, these data provide experimental evidence that the cas9 and rnc genes (and possibly also that of tracrRNA) are required for adhesion to human epithelial cells.
2.2. Transcriptome-wide analysis of Cas9 derived mutants
To further analyse whether the CRISPR/Cas system affects the expression of genes involved in host-cell interaction, we compared the transcriptomes of the wild-type strain and the isogenic Δcas9 mutant in mid-log (OD600nm = 0.5) and early-stationary (OD600nm = 1.5) growth phase in rich medium (Figure 3) by transcriptome sequencing (RNA-seq). Altogether, 10 loci showed a greater than 2-fold expression difference between both strains with Benjamini-Hochberg corrected P-value of less than 0.1 (Table 1).
Figure 3.
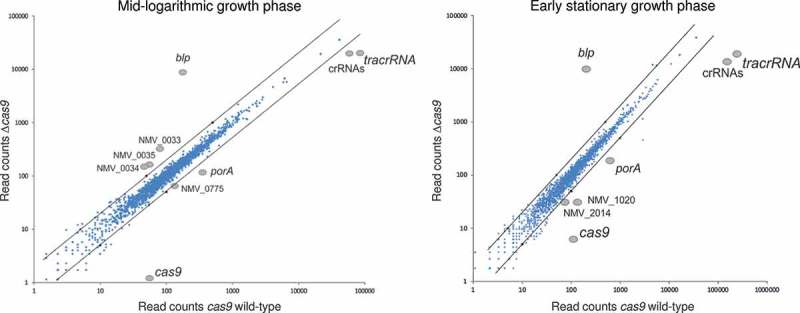
Scatter-plots comparing transcript abundances as determined by RNA-seq analyses in a Cas9 wild type and mutant strain in rich medium. Axes represent the number of cDNA read counts (abundance) in a Δcas9 mutant (y-axis) and the isogenic wild-type (x-axis) strain in mid logarithmic (OD600nm = 0.5) (left panel) and early stationary (OD600nm = 1.5) (right panel) growth phase in GC broth. Dots above the upper (below the lower) black line correspond to transcripts with an expression level difference greater than 2 (smaller than ½) in the Δcas9 mutant compared to the wild-type strain. Only transcripts with log2-fold expression level differences greater than 1 or smaller than −1 were considered to be specific and are highlighted in both figure panels.
Table 1.
Differentially expressed genes in a Δcas9 mutant compared to the wild-type.
| log2-fold expression difference |
|||
|---|---|---|---|
| Locus tag | Gene product* | mid logarithmic phase | early stationary phase |
| NMV_0031 | Hypothetical lipoprotein | 5.5 | 5.6 |
| NMV_0033 | Ferrous iron permease EfeU | 1.9 | - |
| NMV_0034 | Ferrous iron uptake system component EfeO | 1.5 | - |
| NMV_0035 | Deferrochelatase/peroxidase EfeB | 1.5 | - |
| NMV_0775 | Topoisomerase IV subunit A | −1.0 | - |
| NMV_0958 | Major outer membrane protein PorA | −1.5 | −1.5 |
| NMV_1020 | L-lactate dehydrogenase | - | −1.2 |
| NMNmisc_RNA1916856R | trans-activating crRNA (tracrRNA) | −2.0 | −3.6 |
| NMV_1993 | CRISPR/Cas system-associated endonuclease Cas9 | −5.6 | −4.1 |
| CRISPR RNA | CRISPR/Cas system repeat array | −1.2 | −3.4 |
*Gene product annotations were taken from the NeMeSys database[16]
The three protein coding genes NMV_0331, porA (NMV_0958) and cas9 (NMV_1993) as well as the genes for the tracrRNA and the crRNAs all showed differential expression independent of the growth phase. In addition to cas9, also tracrRNA and the crRNAs were constitutively less expressed in the Δcas9 mutant since these RNAs are less stable in the absence of the Cas9 protein. Furthermore, the mRNA of major outer membrane protein PorA showed constitutively reduced expression in the Δcas9 mutant. This porin protein is one of the most abundant outer membrane proteins in meningococci and an important protein antigen, although its exact role in the interaction between meningococci and host cells remains to be fully understood [17,18].
The three genes NMV_0033, NMV_0034 and NMV_0035 forming an operon that codes for an iron-uptake system as well as the topoisomerase IV subunit A gene were in turn all higher expressed in the Δcas9 mutant but only during the mid-log growth-phase while the gene NMV_1020 coding for a L-lactate dehydrogenase was less expressed in the mutant strain in the early stationary growth phase. So far, there are no experimental data that would support a role of one of these proteins in the adhesion to human epithelial cells [19–21].
While there surprisingly were no other ‘classical’ adhesins among the differentially expressed genes, we observed a strong upregulation of a hypothetical lipoprotein encoded by NMV_0031 in the Δcas9 mutant. Based on its nucleotide sequence the gene encodes a basic (pI 8.33), putative lipoprotein of 172 amino acids length with a calculated molecular mass 19.3 kDa; for convenience, we will refer to it as Blp (basic lipoprotein). BLASTP searches against the NCBI non-redundant protein database showed that Blp is highly conserved in meningococci. Of note, the promoter region of the blp gene contains a polyadenine stretch between its −10 and −35 box, indicating that its expression might also be phase-variable. Finally, an operon encoding an iron uptake system was higher expressed in the Δcas9 mutant in the mid logarithmic but not in the early stationary growth phase.
To further test whether blp is differentially expressed in cas9 wild-type, Δcas9 mutant and Δcas9 Ccas9 complemented strains, we performed Northern blot analyses of RNA samples from the same growth phases used for RNA-seq combined with SDS-PAGE analyses of total protein extracts (Figure 4). The Δcas9 mutant showed higher blp expression compared to the wild-type and the complemented strain that has the same polyadenine tract length in its blp promoter as the Δcas9 mutant.
Figure 4.
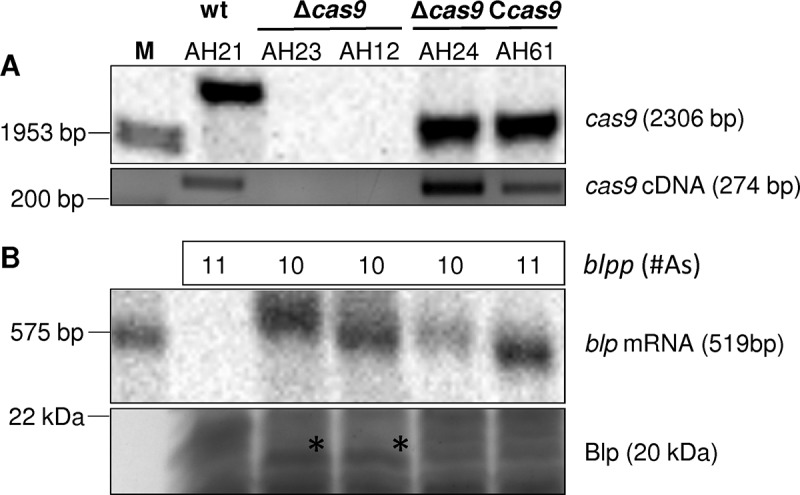
Analysis of Cas9 and Blp expression in wild-type, mutant and complemented strains.
A. Southern blot (upper panel) and RT-PCR (lower panel) for the detection of cas9 in wild-type, mutant as well as complemented strains. respectively, to confirm the cas9 genotype of the tested strains. The designations of different clones are given at the top of the upper panel.
B. Northern blot (upper panel) and SDS-PAGE (lower panel) assessing blp transcription and translation in the Cas9 wild-type, mutant and complemented strains. At the top of panel B the numbers give the length of the poly-A tract in the blp promoter region as determined by direct Sanger sequencing of specific PCR products. The asterisk in the lower panel depicts the Blp band as confirmed by mass spectrometry.
Importantly, however, targeted sequencing of the blp promoter region in different clones of the same Δcas9 mutant revealed variation in the length of the polyadenine tract (10 vs. 11 adenines). Northern blot analyses further showed that blp expression was somewhat higher in the cas9 complemented strain with a polyadenine tract length of 11 residues compared to one with 10 residues (Figure 4(B)). In contrast, there was no difference in blp expression between these two clones at the protein level as revealed by SDS-PAGE followed by mass spectrometry. Likewise, also the adhesion rates of both clones were almost identical (151 ± 49% for AH24 vs. 101 ± 14% for AH61) and thus similar to the wild-type. Together, these data suggest that, on a transcriptome-wide level, Cas9 has only a limited effect on gene expression. They further indicate that blp mRNA might be a potential target for post-transcriptional regulation by Cas9 superimposed on stochastic variation of blp expression due to phase variation at the transcriptional level. The complex interplay between both modes of regulation and its impact on host adhesion is subject to current experimental investigation.
2.3. Identification of alternative Cas9 RNA targets
In order to comprehensively identify target RNAs for binding by Cas9, co-immunoprecipitation (coIP) of Cas9 along with bound RNAs was performed followed by deep-sequencing of co-purified RNA from a strain expressing a chromosomally encoded Cas9-3xFLAG protein (RIP-Seq) (Figure 5). Total RNA was prepared from bacteria grown to either mid-logarithmic or early-stationary growth phase, and RNA-seq libraries of Cas9 coIP and control coIP data were compared with each other in terms of enrichment and abundance. Only cDNA reads enriched for more than two fold as indicated by the dashed line in Figure 5 and with more than 10 reads and a resulting Benjamini–Hochberg corrected P-value < 0.1 were considered significantly enriched. As shown in Figure 5 and Table 2, sequences recovered with Cas9 in both growth phases were predominantly derived from crRNAs and tracrRNAs. In the mid-logarithmic growth phase, there was also a slight enrichment of blp mRNA and of the recently described non-coding (nc)RNA NMnc0040 which was found to be associated with the CRISPR/Cas locus in N. meningitidis. Together, these data indicate that Cas9 binds only to a very limited number of non-canonical target-RNAs in meningococci, and there was in particular no enrichment of porA mRNA or of any ‘classical’ adhesin-encoding mRNA in the FLAG-tagged Cas9 RNA samples.
Figure 5.
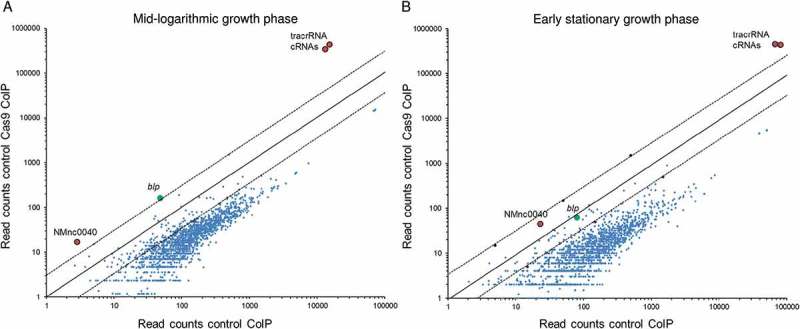
Scatter-plots comparing transcript abundances as determined by RNA-seq analysis after Cas9 co-immunoprecipitation (coIP) of total RNA in a wild type and in a mutant strain expressing 3xFLAG tagged Cas9 in rich medium. Axes represent the number of cDNA read counts (abundance) in a cas9::3xFLAG mutant (y-axis) and the wild-type (x-axis) strain in mid logarithmic (OD600nm = 0.5) (left panel) and early stationary (OD600nm = 1.5) (right panel) growth phase in GC broth. Dots above the upper (below the lower) dashed line correspond to transcripts that are enriched (depleted) greater than 2 (smaller than ½) in the cas9::3xFLAG mutant compared to the non FLAG-tagged wild-type strain. Only transcripts with log2-fold enrichment (depletion) greater than 1 or smaller than −1 were considered to be specific and are highlighted in both figure panels. This revealed crRNA and tracrRNAs to be associated with Cas9.
Table 2.
Genes enriched in a FLAG-tagged Cas9 mutant strain compared to the wild-type.
| OD600nm = 0.5 |
OD600nm = 1.5 |
||||
|---|---|---|---|---|---|
| Locus taga | Gene productb | Reads | LFE | Reads | LFE |
| NMV_0031 | Hypothetical lipoprotein | 47 | 1.8 | 80 | - |
| NMV_0503 | Glutamate racemase | 90 | 1.1 | 122 | - |
| NMNmisc_RNA1916856R | trans-activating crRNA (tracrRNA) | 12,930 | 4.8 | 79,699 | 2.5 |
| CRISPR RNA | CRISPR/Cas system repeat array | 14,925 | 4.9 | 66,472 | 2.8 |
aOnly Cas9-binding candidates with cDNA read-counts ≥ 10 and a log2-fold enrichment (LFE) ≥ 1 in the FLAG-tagged Cas9 mutant strain compared to the wild-type are listed.
bGene product annotations were taken from the NeMeSys database[16].
3. Discussion
In meningococci adhesion to human cells is primarily affected by the type IV pilus (tfp), the so called major adhesins Opa and Opc, a number of minor adhesins and the polysaccharide capsule [19,22,23]. Neisserial tfp are homopolymeric filaments composed of the major subunit pilin (PilE) with another 23 genes required for tfp biogenesis and/or function [16] and are of central importance for almost all aspects of meningococcal colonization and infection [24]. Additionally, the Opa and Opc outer membrane proteins constitute another class of major adhesins. However, Opc is missing in strain 8013, and the polymeric tracts in all four genes for different Opa alleles are in the OFF-state of phase-variable expression. Beyond these major adhesion, a number of so called minor adhesins have been identified including the autotransporters NhhA and App, respectively, and the Oca-type adhesin NadA [19] which are present in strain 8013. While the capsule sterically inhibits the interaction of most adhesin molecules with receptors on the host cell surface [22] it does not strongly inhibit tfp-mediated adhesion, and the tfp appears to be an indispensable colonization factor in the presence of a capsule [22]. Additionally but to a somewhat lesser extent, the presence of a capsule does also not interfere with the adherence mediated by NhhA, App, or NadA [19]. Since the expression of PilE and the serogroup C capsule was not different among the CRISPR/Cas mutants and the wild-type respectively complemented strains (data not shown), and because there is no experimental evidence that Cas9 acts itself as an adhesin, we hypothesize that the CRISPR/Cas system and in particular Cas9 interacts indirectly either with (i) the function and/or biogenesis of the meningococcal tfp, (ii) one or more of the minor adhesins App, NadA and/or Nhh, or (iii) directly affects composition of the meningococcal cell envelope thereby affecting adhesion to human airway epithelia. For example, in the ϵ-proteobacterium C. jejuni Cas9 was shown to somehow control the topology and/or composition of the bacterial cell envelope thereby affecting virulence [25]. In the γ-proteobacterium F. novicida a complex comprising Cas9, tracrRNA and another short non-coding CRSIPR-associated RNA termed scaRNA was shown to alter the stability of a mRNA encoding a small lipoprotein which in turn helps the bacterium to escape immune recognition by the host [8]. Furthermore, the Cas9-dependent CRISPR-Cas system was shown to be involved in enhancing envelope integrity in F. novicida through the regulation of this lipoprotein affecting resistance against numerous antibiotics [26].
Since we could not identify any adhesin-encoding mRNA in our Cas9-RIP-Seq data and given the small size of Blp the mechanism by which Blp affects adhesion is indirect and likely involves the tfp and/or other outer membrane components as explained above. Although the meningococcal Blp is about 100 AAs shorter than the small lipoprotein (FTN_1102) described in F. novicida [25] (172 vs. 282 amino acids) with only a low sequence identity of 27% it is yet tempting to speculate that, based on our data, Cas9 might affect the expression of Blp at the post-transcriptional level also in meningococci. In addition, the CRISPR/Cas locus harbors the small non-coding RNA NMnc0040 [12] that was found to be associated with Cas9 in the RIP-Seq-data. This situation therefore resembles the situation in F. novicida and suggests that both components might be involved in the regulation of meningococcal adherence to human cells by Cas9.
In conclusion, our data provide experimental evidence that the cas9 and rnc genes (and possibly also that of tracrRNA) are required for adhesion to human nasopharyngeal cells. Furthermore, our data on blp and the ncRNA NMnc0040 resemble to some extent the situation in F. novicida, but the exact molecular mechanism mediating the effect of Cas9 on host cell adhesion awaits further investigation.
4. Materials and methods
4.1. Bacterial strains, plasmids and oligonucleotides
A complete list of bacterial strains is provided in Supplementary Table S1. A complete list of all plasmids, as well as information on their construction, is given in Supplementary Table S2 and Supplementary Materials and Methods. A complete list of DNA oligonucleotides is given in Supplementary Table S3.
4.2. Bacterial growth and construction of mutant strains
Details about bacterial cultivation and the generation of Neisseria mutant strains are given in the Supplementary Materials and Methods.
4.3. Cell culture and cell adhesion experiments
The human nasopharyngeal epithelial cell line Detroit562 (ATCC® number CCL-138™) was cultivated in Eagle’s minimum essential medium (EMEM) (Lonza, Basel, Switzerland) with 10% feta calf serum (Thermo Fisher, Frankfurt/Main, Germany), 1% nonessential amino acids (Lonza) and 1% sodium pyruvate (Lonza) (EMM+++) at 37°C and 5% CO2. Cell adhesion assays are described in detail in the Supplementary Materials and Methods [27]. For each strain, the adhesion rate was calculated as the number of colony forming units (CFU) recovered after 4 h of infection divided by the seeded CFU determined in parallel. The relative adhesion rate is the adhesion rate of the mutant divided by the adhesion rate of the wild-type strain. All infection experiments were carried out in triplicate at a multiplicity of infection (MOI) of 20 in EMM+++ at 37°C and 5% CO2.
4.4. Sample collection for RNA-Seq analysis and RNA extraction
Samples for RNA-Seq were collected from bacterial cultures grown to optical densities (OD)600nm of 0.5 or 1.5, corresponding to the mid logarithmic and late logarithmic/early stationary growth phase, respectively. Samples were fixated by the addition of STOP Mix [95% (vol/vol) EtOH and 5% (vol/vol) phenol], frozen in liquid nitrogen and subsequently stored at −80°C until RNA preparation. Total RNA was extracted from bacterial lysates using the hot phenol method as described in ref.12.
4.5. Northern blot analysis
For Northern blot analysis, 5 μg of DNase I-treated total RNA was isolated from N. meningitidis cells grown to OD600nm = 0.5 as described above and separated on an 8% polyacrylamide gel containing 8.3 M urea. RNA was transferred onto Hybond-XL membranes, hybridized overnight at 42°C with γ32P-ATP end-labeled oligodeoxyribonucleotides (Supplementary Table S3) and visualized on a Phosphorimager (Typhoon FLA 7000, GE Healthcare).
4.6. Protein extraction and SDS-PAGE
For protein analysis, samples from N. meningitidis cells grown to OD600nm = 0.5 were collected by centrifugation at 16,100 × g at 4°C for 2 min and dissolved in 50 μl Laemmli loading buffer. After incubation for 5 min at 95°C, 0.1 OD600nm equivalents of samples were separated by 12% (vol/vol) SDS-polyacrylamide gel electrophoresis (PAGE) and stained with Coomassie 2250 (Serva, Heidelberg).
4.7. RNA co-immunoprecipitation (coIP) with Cas9 (RIP-seq)
N. meningitidis expressing either 3× FLAG-tagged- or wild-type (WT) Cas9 protein were grown in the presence of kanamycin until an OD600 of 0.5 and 1.5, respectively. For each strain cells equivalent to an OD600 of 50 were collected and subjected to Cas9 coIP and control coIP as described in detail in ref.12 and briefly in the Supplementary Materials and Methods.
4.8. Construction and sequencing of cDNA libraries for RNA-seq and Cas9 RIP-seq
cDNA libraries of RNA-seq samples were constructed by Vertis Biotechnology AG, Munich, Germany, exactly as described in ref.12.
cDNA libraries of Cas9 RIP-seq samples were constructed according to the instructions of the NEBnext Multiplex sRNA library Prep Set for Illumina according to the manufacter’s instruction. cDNA libraries were pooled and sequenced using either an Illumina MiSeq machine in the single-read mode. The raw de-multiplexed reads as well as the normalized coverage files of all cDNA libraries have been deposited in NCBI’s Gene Expression Omnibus [28] and are accessible through GEO Series accession number GSE110891 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE110891). A detailed description of the RNA-seq and RIP-seq read mappings and their normalization is given in refs [12,29]. and in the Supplementary Materials and Methods.
4.9. Analysis of Cas9 RIP-seq data
Read mapping and enrichment analyses were performed according to ref. 12 as described in the Supplementary Materials and Methods.
Funding Statement
This work was supported by the Deutsche Forschungsgemeinschaft (DFG) - Project Number 192503913 under Grant Vo875/7-2.
Acknowledgments
We would like to thank Barbara Conrad for expert technical assistance in constructing the mutants and conducting the cell culture experiments. We thank Andreas Schlosser for the mass spectrometric analysis of Blp and Lei Li und Thorsten Bischler for their help with the RNA-seq and RIP-seq data analysis.
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplementary material
Supplemental data for this article can be accessed here.
References
- 1.Marraffini LA. CRISPR-Cas immunity in prokaryotes. Nature. 2015;526:55–61. [DOI] [PubMed] [Google Scholar]
- 2.Sampson TR, Weiss DS. Alternative roles for CRISPR/Cas systems in bacterial pathogenesis. PLoS Pathog. 2013;9:e1003621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Westra ER, Buckling A, Fineran PC. CRISPR-Cas systems: beyond adaptive immunity. Nat Rev Microbiol. 2014;12:317–326. [DOI] [PubMed] [Google Scholar]
- 4.Koonin EV, Makarova KS, Zhang F. Diversity, classification and evolution of CRISPR-Cas systems. Curr Opin Microbiol. 2017;37:67–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Makarova KS, Haft DH, Barrangou R, et al. Evolution and classification of the CRISPR-Cas systems. Nat Rev Microbiol. 2011;9:467–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stephens DS. Conquering the meningococcus. FEMS Microbiol Rev. 2007;31:3–14. [DOI] [PubMed] [Google Scholar]
- 7.Harrison OB, Schoen C, Retchless AC, et al. Neisseria genomics: current status and future perspectives. Pathog Dis. 2017;75:ftx060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sampson TR, Saroj SD, Llewellyn AC, et al. A CRISPR/Cas system mediates bacterial innate immune evasion and virulence. Nature. 2013;497:254–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Winstead JM, McKinsey DS, Tasker S, et al. Meningococcal pneumonia: characterization and review of cases seen over the past 25 years. Clin Infect Dis. 2000;30:87–94. [DOI] [PubMed] [Google Scholar]
- 10.Bille E, Meyer J, Jamet A, et al. A virulence-associated filamentous bacteriophage of Neisseria meningitidis increases host-cell colonisation. PLoS Pathog. 2017;13:e1006495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Klughammer J, Dittrich M, Blom J, et al. Comparative genome sequencing reveals within-host genetic changes in Neisseria meningitidis during invasive disease. PLoS One. 2017;12:e0169892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heidrich N, Bauriedl S, Barquist L, et al. The primary transcriptome of Neisseria meningitidis and its interaction with the RNA chaperone Hfq. Nucleic Acids Res. 2017;45:6147–6167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang Y, Heidrich N, Ampattu BJ, et al. Processing-independent CRISPR RNAs limit natural transformation in Neisseria meningitidis. Mol Cell. 2013;50:488–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sampson TR, Weiss DS. Degeneration of a CRISPR/Cas system and its regulatory target during the evolution of a pathogen. RNA Biol. 2013;10:1618–1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Joseph B, Schwarz RF, Linke B, et al. Virulence evolution of the human pathogen Neisseria meningitidis by recombination in the core and accessory genome. PLoS One. 2011;6:e18441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rusniok C, Vallenet D, Floquet S, et al. NeMeSys: a biological resource for narrowing the gap between sequence and function in the human pathogen Neisseria meningitidis. Genome Biol. 2009;10:R110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Orihuela CJ, Mahdavi J, Thornton J, et al. Laminin receptor initiates bacterial contact with the blood brain barrier in experimental meningitis models. J Clin Invest. 2009;119:1638–1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Massari P, Ram S, Macleod H, et al. The role of porins in neisserial pathogenesis and immunity. Trends Microbiol. 2003;11:87–93. [DOI] [PubMed] [Google Scholar]
- 19.Carbonnelle E, Hill DJ, Morand P, et al. Meningococcal interactions with the host. Vaccine. 2009;27(Suppl 2):B78–89. [DOI] [PubMed] [Google Scholar]
- 20.Coureuil M, Join-Lambert O, Lecuyer H, et al. Pathogenesis of meningococcemia. Cold Spring Harb Perspect Med. 2013;3:a012393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Virji M. Pathogenic neisseriae: surface modulation, pathogenesis and infection control. Nat Rev Microbiol. 2009;7:274–286. [DOI] [PubMed] [Google Scholar]
- 22.Merz AJ, So M. Interactions of pathogenic neisseriae with epithelial cell membranes. Annu Rev Cell Dev Biol. 2000;16:423–457. [DOI] [PubMed] [Google Scholar]
- 23.Joseph B, Schneiker-Bekel S, Schramm-Gluck A, et al. Comparative genome biology of a serogroup B carriage and disease strain supports a polygenic nature of meningococcal virulence. J Bacteriol. 2010;192:5363–5377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Imhaus AF, Dumenil G. The number of Neisseria meningitidis type IV pili determines host cell interaction. EMBO J. 2014;33:1767–1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Louwen R, Horst-Kreft D, de Boer AG, et al. A novel link between Campylobacter jejuni bacteriophage defence, virulence and Guillain-Barre syndrome. Eur Jl Clin Microbiol Infect Dis. 2013;32:207–226. [DOI] [PubMed] [Google Scholar]
- 26.Sampson TR, Napier BA, Schroeder MR, et al. A CRISPR-Cas system enhances envelope integrity mediating antibiotic resistance and inflammasome evasion. Proc Natl Acad Sci U S A. 2014;111:11163–11168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Slanina H, Hebling S, Hauck CR, et al. Cell invasion by Neisseria meningitidis requires a functional interplay between the focal adhesion kinase, Src and cortactin. PLoS One. 2012;7:e39613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Edgar R, Domrachev M, Lash AE. Gene expression omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chao Y, Papenfort K, Reinhardt R, et al. An atlas of Hfq-bound transcripts reveals 3ʹ UTRs as a genomic reservoir of regulatory small RNAs. EMBO J. 2012;31:4005–4019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


