Abstract
The gut microbiota is crucial for our health, and well-balanced interactions between the host’s immune system and the microbiota are essential to prevent chronic intestinal inflammation, as observed in inflammatory bowel diseases (IBD). A variant in protein tyrosine phosphatase non-receptor type 22 (PTPN22) is associated with reduced risk of developing IBD, but promotes the onset of autoimmune disorders. While the role of PTPN22 in modulating molecular pathways involved in IBD pathogenesis is well studied, its impact on shaping the intestinal microbiota has not been addressed in depth. Here, we demonstrate that mice carrying the PTPN22 variant (619W mice) were protected from acute dextran sulfate sodium (DSS) colitis, but suffered from pronounced inflammation upon chronic DSS treatment. The basal microbiota composition was distinct between genotypes, and DSS-induced dysbiosis was milder in 619W mice than in WT littermates. Transfer of microbiota from 619W mice after the first DSS cycle into treatment-naive 619W mice promoted colitis, indicating that changes in microbial composition enhanced chronic colitis in those animals. This indicates that presence of the PTPN22 variant affects intestinal inflammation by modulating the host’s response to the intestinal microbiota.
Keywords: Gastroenterology
Keywords: Inflammatory bowel disease
Introduction
Our intestines are populated by trillions of bacteria, many of which are harmless commensals or even beneficial to our health. However, some species are potentially harmful and can drive pathogenesis under certain conditions, e.g., in immune-deficient patients or upon epithelial barrier defects (1, 2). Although the intestinal immune system is adapted to cope with the high bacterial load in the intestinal lumen, dysfunction of immunoregulatory mechanisms can result in an inadequate response to invading pathogens, or render the body hyperreactive toward harmless commensals and food antigens. Both events cause an increased susceptibility for (chronic) intestinal inflammation, as observed in inflammatory bowel diseases (IBD) (1, 3).
Apart from changes in intestinal microbiota composition, genetic and environmental factors contribute to the risk of developing IBD (4, 5). Among the over 240 genetic risk loci that affect the susceptibility to develop IBD, a variant in the gene encoding protein tyrosine phosphatase non-receptor type 22 (PTPN22) (6) is of particular interest. This variant has been associated with an increased risk of developing a broad number of autoimmune disorders, including rheumatoid arthritis, systemic lupus erythematodes, and type 1 diabetes (7–11), but protects from the onset of Crohn’s disease (CD) (6), one of the main subtypes of IBD. In contrast to other inflammatory disorders, which occur in organs that are typically sterile, IBD occurs in an organ with an exceptionally high bacterial load. This suggests that the protective effect of the PTPN22 variant observed in CD might result from an influence of bacteria, or bacteria-induced signaling cascades.
PTPN22 is exclusively expressed in immune cells (12) and acts as an important negative regulator of T and B cell activation (13), regulates type 1 as well as type 2 interferon signaling (14–16), and is involved in inflammasome activation (17). Further, we have previously shown that PTPN22 affects signaling pathways induced by the bacterial cell wall product muramyl-dipeptide (MDP), and loss of PTPN22 promotes autophagy, a cellular pathway not only involved in removing damaged proteins and organelles from the cytosol, but also essential for the clearance of invading bacteria (18). Defects in autophagy result in impaired goblet cell function (19, 20), and variants in genes involved in autophagy are associated with an enhanced risk of developing IBD (4, 21), underlining the role of autophagy in intestinal health. Despite its documented role in regulating pathways involved in bacterial handling, it is not known whether normal PTPN22 function is involved in shaping the composition of the intestinal microbiota. Since the microbiota is importantly involved in IBD pathogenesis, we investigated here how presence of the disease-associated variant in PTPN22 affects colitis development and consequently microbial composition in the intestine during health and disease.
Results
Presence of the CD-associated variant in PTPN22 results in attenuated colitis upon acute DSS treatment, but results in enhanced chronic DSS-induced colitis.
In order to understand how the autoimmunity-associated variant in PTPN22 affects the extent of intestinal inflammation, we subjected mice carrying either the WT or the disease-associated PTPN22 619W variant (the mouse ortholog of the human R620W variant, referred to as 619W mice) to acute dextran sulfate sodium–induced (DSS-induced) colitis (an overview of the timeline of the experiments and procedures can be found in Supplemental Methods; supplemental material available online with this article; https://doi.org/10.1172/JCI123263DS1). Since the functional consequence of the PTPN22 variant is still a matter of debate (22), we also included PTPN22-deficient (PTPN22–/–) mice in our studies. Consistent with previous reports (14, 17), PTPN22–/– mice suffered from enhanced colitis in response to acute DSS treatment, while 619W mice were protected from disease onset (Supplemental Figure 1).
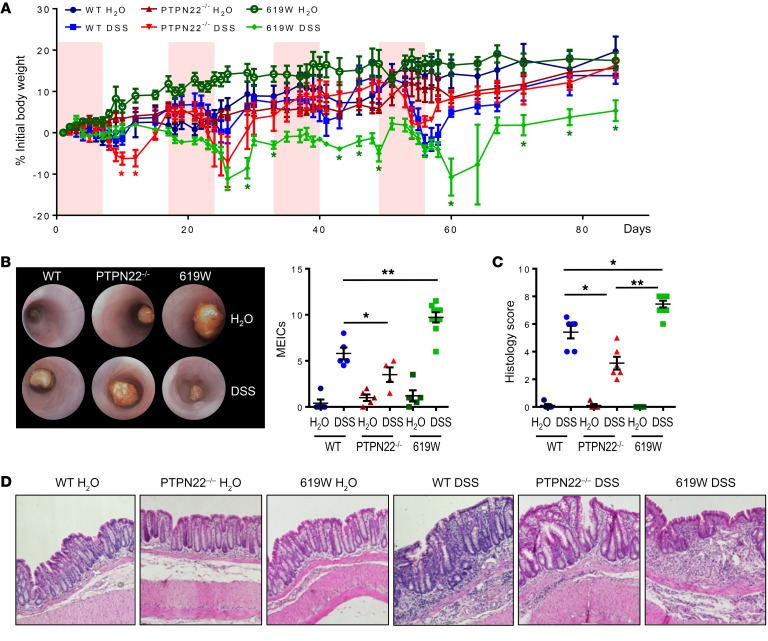
When PTPN22–/– and 619W mice were subjected to repeated DSS treatment, 619W mice were no longer protected from colitis induction. Although 619W mice lost significantly less weight during/after the first DSS cycle, they showed weight development similar to that of their WT littermates in the second cycle, and after the third and fourth cycle they even revealed a delayed recovery when compared with WT and PTPN22–/– mice (Figure 1A). Pronounced colitis in 619W mice upon 4 DSS cycles was also obvious in colonoscopy, and associated with pronounced colon shortening and increased histology scores at the end of the experiment (Figure 1, B–D).
Figure 1. Presence of the 619W variant promotes chronic colitis.
Chronic colitis was induced in PTPN22–/– and 619W mice as well as in their respective WT littermates via administration of 4 cycles of DSS (1.5% DSS in the drinking water for 7 days, followed by 10 days recovery, each cycle). (A–D) Weight development (A), representative pictures from colonoscopy (original magnification, ×10) and MEICS (B), quantification of colitis severity (C), and representative pictures from H&E-stained colon sections (original magnification, ×20) at day 85 (D) reveal that 619W mice show enhanced colitis from the second DSS cycle onward. Depicted are mean values and SEM (A–C). Data are representative of 1 of 2 independent experiments with n = 3–6 per experimental group, each experiment. (B–D) Each dot represents 1 mouse. Graph in A shows weight development from the same individuals as in B–D. *P < 0.05, **P < 0.01, Kruskal-Wallis test.
Presence of the 619W variant in PTPN22 results in decreased levels of Th1 cells in the inflamed intestine, while promoting Tregs.
PTPN22 negatively regulates molecules in the T cell receptor signaling pathway (13), and it has been demonstrated that it influences the development of Th cell subsets (23). Consistent with previous reports (23, 24), we found enhanced levels of regulatory T cells (Tregs) in PTPN22–/– mice undergoing acute and chronic DSS colitis (Supplemental Figure 2, A and B). Notably, a similar but less pronounced phenotype was observed in 619W animals (Supplemental Figure 2, A and B). This is in line with reports published by Dai et al. and Brownlie et al. (22, 23), and indicates that presence of the 619W variant has effects similar, with respect to T cell biology, to those of a deletion of Ptpn22. Further, we found reduced levels of Th1 cells in PTPN22–/– mice in both colitis models, while Th17 cells were not affected consistently (Supplemental Figure 2, C and D, and data not shown). Induction of effector Th cells was overall low in the DSS models, arguing against a prominent role of Th cells in DSS-mediated colitis (Supplemental Figure 2 and data not shown).
Inflammasome activation is enhanced in the inflamed intestine of 619W mice.
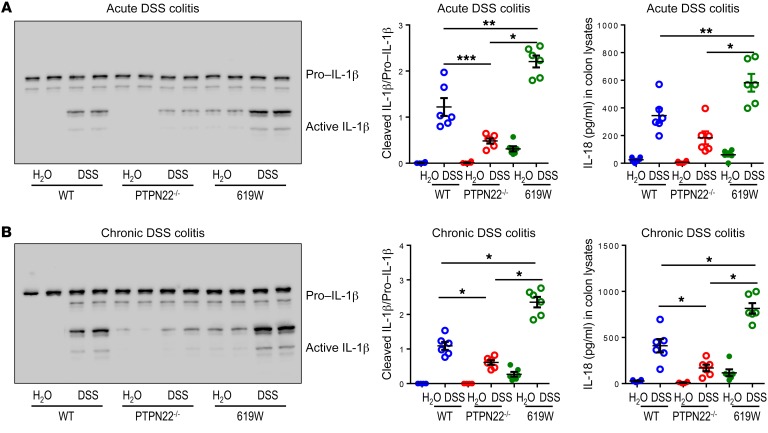
We have previously shown that loss of PTPN22 results in decreased NLRP3 inflammasome activation, while presence of the 619W variant promotes NLRP3-mediated IL-1β secretion (17, 25). Therefore, we next addressed whether inflammasome activation is altered within acute and chronic DSS colitis. Consistent with our previous findings, PTPN22–/– mice showed decreased inflammasome activity (as observed by reduced IL-1β cleavage and reduced levels of IL-18 in the colon) in acute and chronic DSS colitis, while it was enhanced in 619W mice (Figure 2, A and B).
Figure 2. Presence of the 619W variant in PTPN22 affects inflammasome activation and IL-18 secretion.
Colitis was induced in WT, 619W, and PTPN22–/– mice by (A) administration of DSS for 7 days; and (B) administration of 4 DSS cycles. Colon pieces were analyzed by Western blot and ELISA for IL-1β and IL-18, respectively. Each lane in the Western blot represents 1 individual mouse, and each dot in the graph represents 1 individual. Data are representative of 1 of 2 independent experiments with n = 3–6 per experimental group. *P < 0.05, **P < 0.01, ***P < 0.001, Kruskal-Wallis test. See complete unedited blots in the supplemental material.
Cohousing transfers the colitis phenotype from PTPN22–/– and 619W mice to WT mice.
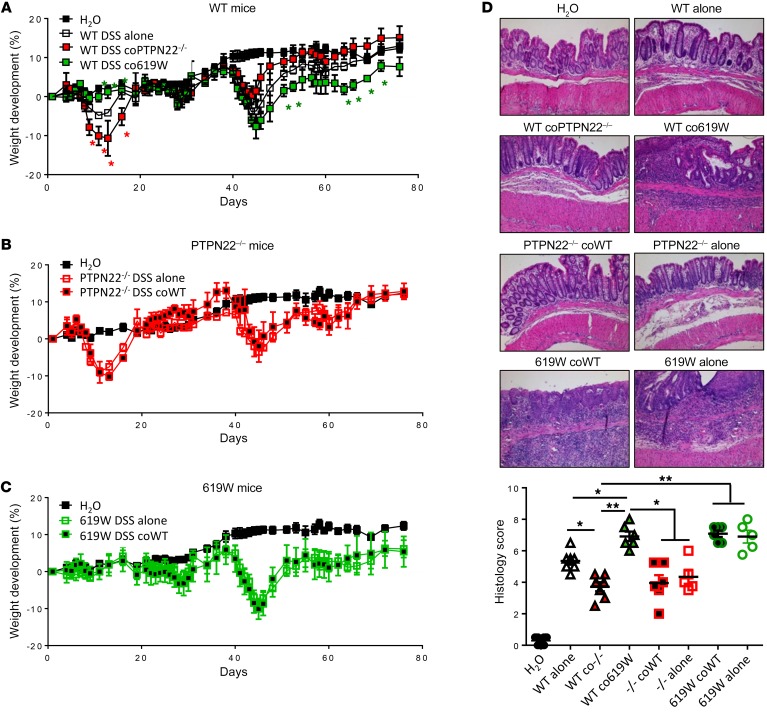
Though the observed changes in inflammasome activation and alterations in T cell subsets during colitis induction can partially explain the observed differences in colitis severity, they cannot explain why presence of the 619W variant in PTPN22 is protective during acute colitis, while promoting disease severity in chronic DSS colitis. Since aberrant T cell and inflammasome activity have both been linked to alterations in the intestinal microbiota, we hypothesized that the intestinal microbiota might be involved in the colitis phenotype in our mice. To address this, we first performed cohousing studies. When WT mice were cohoused with PTPN22–/– littermates after weaning and throughout the experiment, they showed a disease phenotype similar to that of PTPN22–/– counterparts, i.e., increased disease severity in acute colitis, but milder disease upon exposure to several DSS cycles (Figure 3A). Likewise, cohousing of WT mice with 619W mice throughout the experiment resulted in alleviated acute, but increased chronic, colitis (Figure 3, A and D), indicating that the microbiota at least partially contributes to this phenotype.
Figure 3. Cohousing transfers the phenotype from 619W and PTPN22–/– mice to WT littermates.
(A–C) Chronic colitis was induced in WT mice that were cohoused since birth with 619W or PTPN22–/– mice as indicated (A); PTPN22–/– mice that were cohoused since birth and throughout the experiment with WT mice, or mice that were housed only with PTPN22–/– mice after weaning and throughout the experiment (B); or 619W mice that were cohoused since birth and throughout the experiment with WT mice, or mice that were housed only with 619W mice after weaning and throughout the experiment (C). (D) Representative pictures (original magnification, ×10) and scoring of histologic colitis severity in H&E-stained sections of the terminal colon from mice in A–C. Data are representative of 1 of 2 independent experiments. Weight curves show mean ± SEM for each group; n = 3 for H2O group and n = 5 for all other groups. The other graphs show values and SEM, and each dot represents 1 individual mouse. –/–, PTPN22–/–. *P < 0.05, **P < 0.01, Kruskal-Wallis test.
Interestingly, cohousing of PTPN22–/– or 619W mice with WT mice had no effects on colitis severity (Figure 3, B–D), indicating that the phenotype is transferred from PTPN22–/– and 619W mice to their WT counterparts but not vice versa. A further indication that alterations in microbial composition promote enhanced colitis in 619W mice upon chronic DSS exposure is the fact that antibiotic treatment annulated this effect (Supplemental Figure 3), although the initial, more severe disease was still present in PTPN22–/– mice.
Altered microbiota in 619W and PTPN22–/– mice.
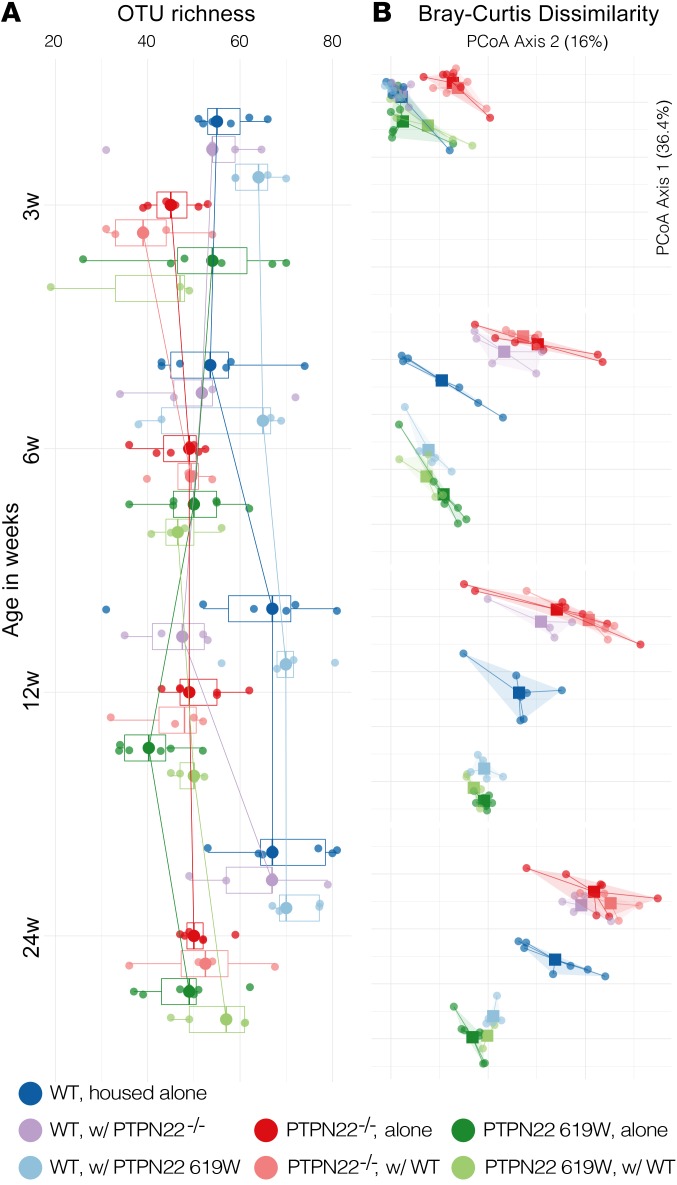
Having observed drastic effects of cohousing and microbiota depletion, we next analyzed the composition of the intestinal microbiota in the different mouse lines. In a first step we analyzed the microbiota in the 3 genotypes at the time of weaning (until that time, the PTPN22–/– and 619W mice were housed together with their respective WT littermates) and how it develops until week 24 (the same age as mice after the chronic DSS experiments). Starting at an age of 12 weeks, both PTPN22–/– and 619W mice harbored lower microbial richness than WT mice, though effects were overall minor (Figure 4A and Supplemental Table 1).
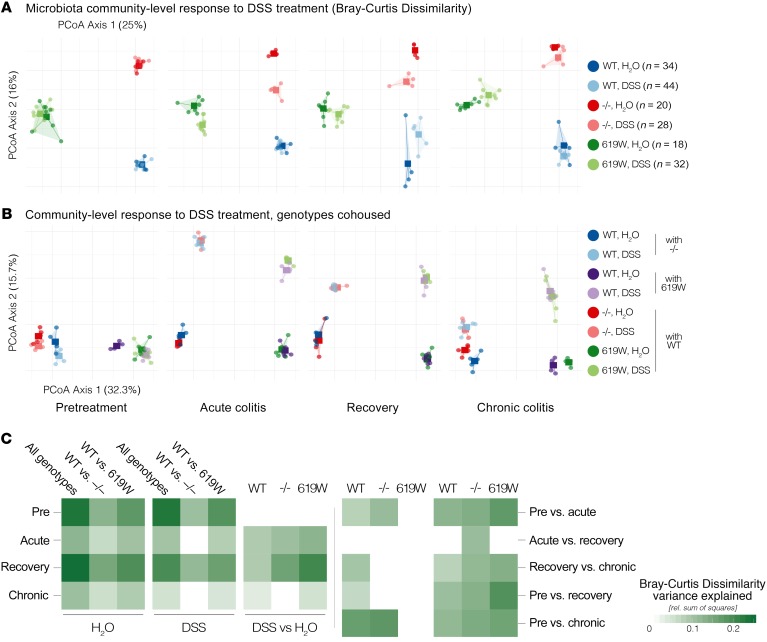
Figure 4. 619W and PTPN22–/– genotypes are associated with a distinct gut microbiota.
Relative to matched WT controls, both 619W and PTPN22–/– mice showed baseline differences in microbiota taxa richness (A) and overall community composition (B; principal coordinates analysis [PCoA] of Bray-Curtis dissimilarities) that were conferred to some extent to cohoused WT littermates, but not vice versa. See Supplemental Table 1 for full data and individual statistical test results.
Microbial community composition, in contrast, was clearly distinct between the 3 genotypes, as indicated by trends in Bray-Curtis dissimilarity (permutational multivariate ANOVA [PERMANOVA], P ≤ 10–4 in 9999 permutations; Figure 4B). These differences increased with age: while at the time of weaning, genotypes were already compositionally distinct (R2 = 0.284, P ≤ 10–4), microbiome maturation was characterized by more pronounced and genotype-specific compositional shifts (R2 > 0.6 at later time points). WT mice that were cohoused with 619W mice or PTPN22–/– mice after weaning developed a microbiota similar to that of their 619W or PTPN22–/– littermates. In contrast, PTPN22–/– and 619W mice cohoused with WT remained within genotype. This indicates that presence of the variant or deletion of PTPN22 clearly and characteristically influences microbial composition.
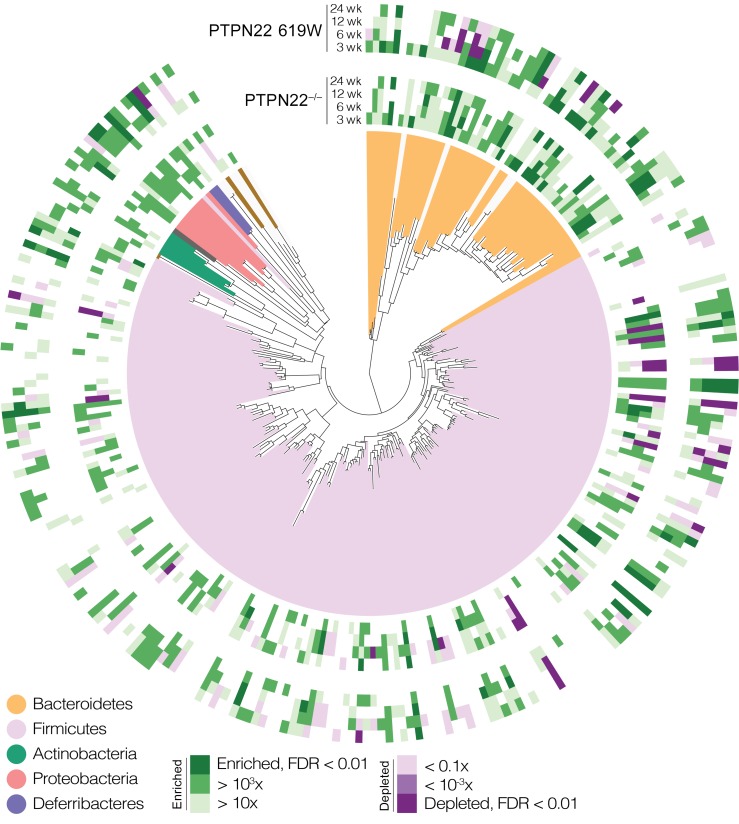
A total of 83 operational taxonomic units (OTUs) were differentially abundant in PTPN22–/– or 619W mice relative to WT baselines at any of the tested time points, representing the core murine gut bacterial phyla Bacteroidetes, Firmicutes, Proteobacteria, and Deferribacteres (Figure 5 and Supplemental Table 1 for a full list). While some univariate associations were shared between genotypes (e.g., Bacteroides OTU S8307 was enriched in both, whereas Lachnospiraceae OTU S56104 was depleted), others were genotype-specific; for example, 619W mice were characteristically enriched for OTUs classified as Lactobacillus, Helicobacter, and Sutterellaceae. Further, WT mice that were not separated from their PTPN22–/– or 619W littermates after weaning closely resembled their 619W or PTPN22–/– counterparts. This might reflect that the microbiota is transferred between individuals in the same cage (26).
Figure 5. Associations of individual taxa with 619W and PTPN22–/– genotypes.
Operational taxonomic units (OTUs) from various taxonomic groups showed distinct univariate trends of enrichment or depletion relative to WT controls, concordant or discordant between PTPN22–/– and 619W genotypes. See Supplemental Table 1 for full data and individual statistical test results.
Changes in microbiota during colitis induction.
Given these baseline differences, we next assessed the impact of DSS treatment on the microbiota in the different genotypes, testing at baseline, after 1 DSS cycle (acute colitis), after a recovery phase of 10 days, and after 4 repeated DSS cycles (chronic colitis). Microbiota community composition was most strongly associated with time point (Figure 6; PERMANOVA on Bray-Curtis dissimilarities across all time points, genotypes, and treatments; R2 = 0.08) and genotype (R2 = 0.063), with both factors dominating DSS treatment (R2 = 0.022) and interaction terms (all terms at P ≤ 10–4 in a global PERMANOVA test).
Figure 6. Genotype-specific effects of DSS treatment on the gut microbiota.
(A) Distinct compositional shifts in each genotype relative to H2O-treated controls were observed upon 1 cycle of DSS treatment (acute colitis), following a 10-day recovery phase, and upon 4 DSS cycles (chronic colitis). (B) These shifts were conferred to WT mice cohoused with PTPN22–/– or 619W mice, but not vice versa. Note that mice in A were not cohoused, whereas in B, WT mice were cohoused with PTPN22–/– or 619W, and vice versa. (C) Compositional shifts could be differentially attributed to the factors genotype, treatment, and time point, as indicated by R2 values in PERMANOVA tests on data subsets. Note that for illustration purposes, only 1 of several replicate experiments is shown in A; the total number of mice per group used in statistical tests is reported on the right and in Supplemental Table 2.
Acute DSS-induced colitis triggered a significant shift in community composition in all 3 genotypes (Figure 6, A–C; P ≤ 10–4), most pronounced in 619W mice (R2 = 0.127) and mildest in WT (R2 = 0.089), while characteristically directed for each genotype. These shifts were exacerbated during the recovery phase for 619W (DSS-treated vs. untreated mice, R2 = 0.205) and PTPN22–/– (R2 = 0.159) but not in WT mice, with genotypes becoming more compositionally dissimilar in treated (R2 = 0.248) and untreated (R2 = 0.197) mice. This indicates that while WT mice at least partially recovered from acute DSS colitis, treatment-induced compositional shifts increased in PTPN22–/– and 619W mice.
After 3 additional DSS cycles (inducing a chronic colitis), composition between treatment groups within each genotype remained significantly different, but shifts were moderate (P < 0.05; R2 ≤ 0.05). Interestingly, genotype-associated compositional variation was larger in the untreated group (R2 = 0.108) than in the DSS group (R2 = 0.512) after 4 DSS cycles, indicating that at least some aspects of the compositional response to chronic colitis were shared among genotypes.
These trends were surprisingly robust to effects of cohousing during the experiment (Figure 6B). As observed in untreated mice (Figure 4A), WT mice cohoused with PTPN22–/– or 619W mice after weaning were compositionally similar to their respective littermates, but not vice versa. This observation extended to DSS-treated mice: WT mice cohoused with PTPN22–/– or 619W mice followed the treatment-induced compositional shifts of their counterparts at all tested time points. This indicates that the genotype-specific compositional response to treatment was transferred to cohoused WT mice. As in non-cohoused mice, genotype effects dominated over treatment effects (R2 = 0.184 and 0.075, respectively; see also clustering along principal coordinates analysis axes 1 and 2 in Figure 6B, explaining 32.3% and 15.7% variation, respectively).
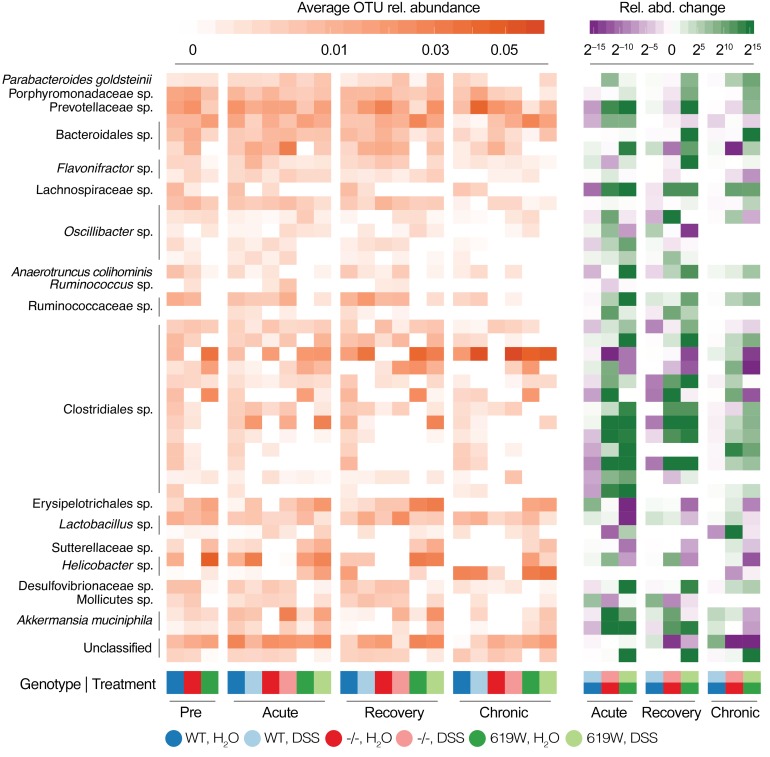
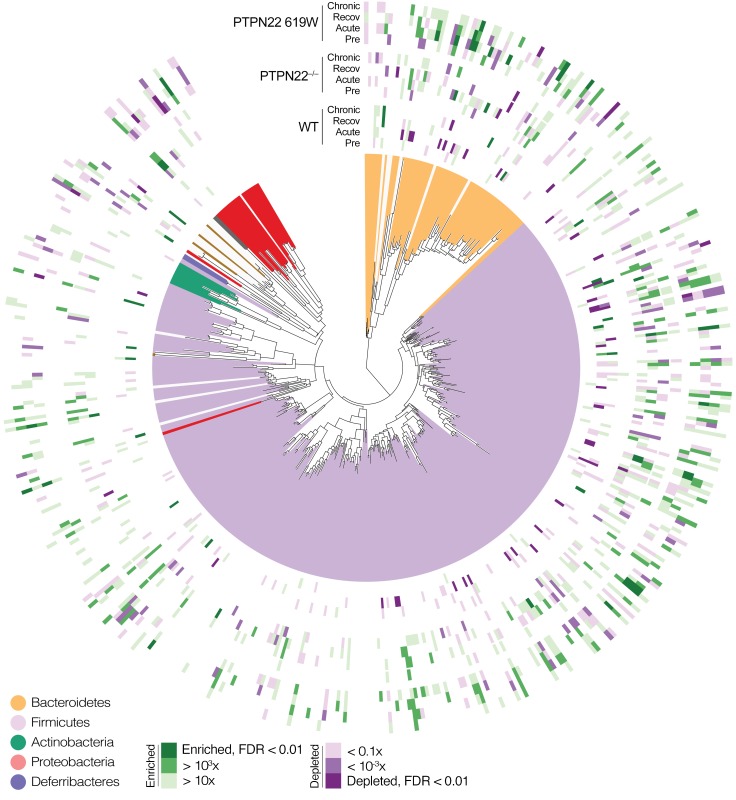
Genotype-specific trends in community-level variation were mirrored at the level of individual taxa, with a total of 164 OTUs across all major gut phyla differentially responding to DSS treatment among genotypes across tested time points (Figures 7 and 8 and Supplemental Table 2). In WT mice, acute colitis was associated with a depletion of several OTUs classified as Bacteroidales, Clostridiales, and Lactobacillus, accompanied by an enrichment of Erysipelotrichales (including Turicibacter sanguinis), Proteobacteria, and Mollicutes. Response to chronic colitis was more attenuated, with few significantly associated OTUs, in line with the more moderate trends in overall community composition.
Figure 7. Univariate associations of individual taxa with genotype and DSS treatment.
Average relative abundances (left) and relative fold changes (right) of selected taxa associated with genotype and DSS treatment response at various time points. See Supplemental Table 2 for full data on individual associations.
Figure 8. Phylogenetic representation of univariate genotype-specific taxa associations with DSS treatment.
Responders to DSS treatment (enrichment or depletion) across genotypes and time points were phylogenetically distributed, although some clades showed specific patterns consistent (or discordant) among genotypes (see main text and Figure 7). See Supplemental Table 2 for full data on individual associations.
In PTPN22–/– mice, Akkermansia muciniphila (among other OTUs, including Eubacterium and Lactobacillus spp.) were strongly enriched upon acute colitis, but depleted (relative to WT) in untreated mice. These associations generally did not persist at the recovery and chronic colitis stage, but were replaced by other (and fewer) univariate signatures, most prominently a strong enrichment of Flavonifractor spp.
Similarly, individual OTU signatures in 619W mice were specific to genotype and time point. Acute DSS colitis triggered a loss of Deferribacterales, Sutterellaceae, Helicobacter, and Turicibacter sanguinis, as well as an enrichment of several Bacteroidales and Clostridiales OTUs (including the butyrate producer Ruminococcaceae spp.). Most of these associations did not persist after recovery or during chronic colitis, which were characterized by few specific OTUs, and few consistently differential taxa. In contrast, untreated 619W were enriched for Akkermansia muciniphila, Sutterellaceae, and Helicobacter relative to WT controls at the recovery and chronic colitis time points, but depleted for several OTUs across phyla.
Thus, microbiota signatures in response to DSS treatment were highly genotype-specific, at the levels of both entire-community composition and the abundance of individual taxa, and each genotype showed characteristic changes at each tested experiment time point.
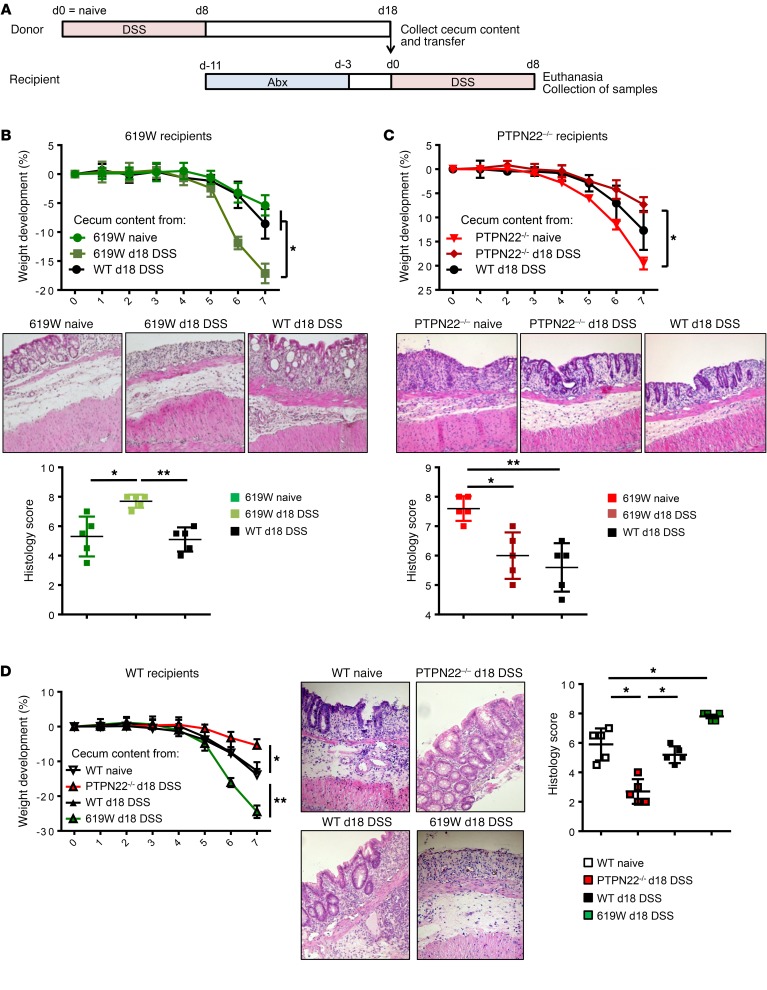
Transfer of microbiota from 619W mice after the first DSS cycle leads to enhanced colitis in treatment-naive 619W and WT mice.
To investigate whether the changes in microbial community after recovery from the first DSS cycle might be responsible for the altered disease outcome in the later disease course, we transferred microbiota from mice after recovery from the first cycle (day 18) into treatment-naive mice (Figure 9A). Notably, 619W mice that received cecum content from 619W mice collected at day 18 were no longer protected from acute DSS-induced colitis, but showed enhanced disease compared with mice that received cecum content from either WT mice at day 18 or treatment-naive 619W mice (Figure 9B). In PTPN22–/– mice, transfer of cecum content from PTPN22–/– mice obtained after recovery from the first DSS cycle (day 18) was sufficient to transmit the milder colitis that was observed in PTPN22–/– mice after several DSS cycles: PTPN22–/– mice that received stool collected from PTPN22–/– mice at day 18 showed decreased colitis severity upon DSS treatment compared with those that received cecum content from naive PTPN22–/– or WT mice at day 18 (Figure 9C). Likewise, transfer of cecum content from 619W mice at day 18 into WT mice promoted disease, while transfer of cecum content from WT mice did not affect disease severity and cecum content from PTPN22–/– mice resulted in milder disease (Figure 9D).
Figure 9. Transfer of stool after the first DSS cycle recapitulates the phenotype in chronic colitis.
Cecum content was collected from WT, PTPN22–/–, and 619W mice that were treated with DSS for 7 days and allowed to recover for 10 additional days (= day 18 DSS). Cecum content was then transferred into WT, PTPN22–/–, or 619W mice that had been treated with an antibiotic cocktail for 1 week to deplete the microbiota. (A) Schematic overview of the experimental setup. (B–D) Weight development, representative pictures (original magnification, ×10) from H&E-stained terminal colon sections, and scoring of histological damage of 619W mice that received cecum content of treatment-naive 619W mice, WT mice upon colitis induction, or 619W mice after colitis induction (B); PTPN22–/– mice that received cecum content of treatment-naive PTPN22–/– mice, WT mice upon colitis induction, or PTPN22–/– mice after colitis induction (C); and WT mice that received cecum content of treatment-naive WT mice or WT, 619W, or PTPN22–/– mice after colitis induction (D). Depicted are mean values and SEM; n = 5 for each experimental group. Data are representative of 1 of 2 independent experiments. Abx, antibiotic treatment. *P < 0.05, **P < 0.01, Kruskal-Wallis test.
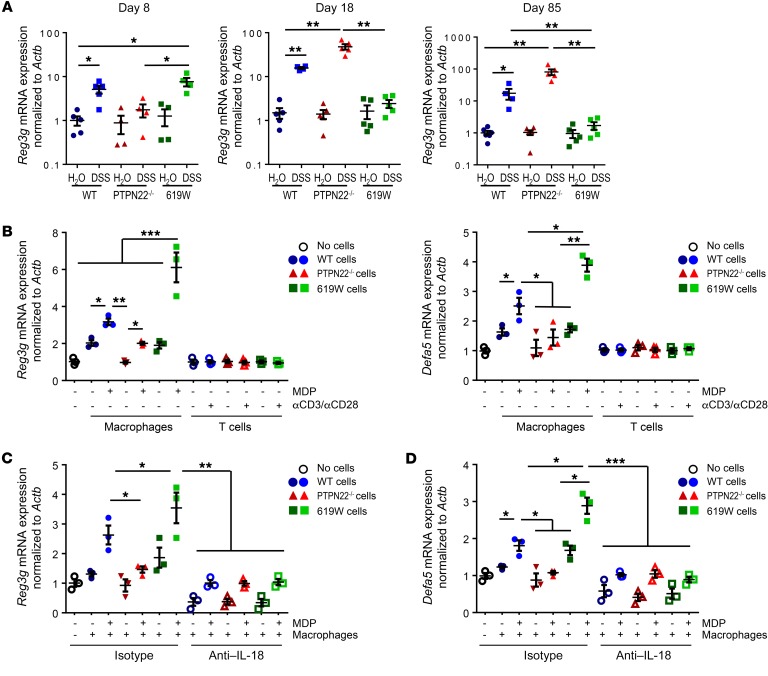
PTPN22 controls the expression of antimicrobial peptides via regulating IL-18 secretion from monocytes/macrophages.
PTPN22 is exclusively expressed in immune cells; hence we next addressed the mechanism of how loss or altered function of PTPN22 might affect the composition of the intestinal microbiota. Notably, mRNA expression of the antimicrobial proteins (AMPs) Reg3g and Defa5 was enhanced in intestinal epithelial cells from WT and 619W but not PTPN22–/– mice upon acute DSS treatment (Figure 10A and data not shown). After recovery from the first DSS cycle (day 18), however, expression of both of the AMPs decreased in 619W mice, but not in WT or PTPN22–/– mice, in which expression was further enhanced (Figure 10A and data not shown ). Similar results were obtained at the end of chronic DSS colitis induction (Figure 10A and data not shown).
Figure 10. PTPN22 affects expression of antimicrobial peptides in an IL-18–dependent manner.
(A) Reg3g mRNA expression in WT, PTPN22–/–, and 619W mice after the first DSS cycle (day 8), after recovery (day 18), or at the end of chronic colitis induction (day 85). (B) MC-38 cells were grown on inserts and cocultured with MDP-activated bone marrow–derived macrophages (BMDMs) or anti-CD3/CD28–activated T cells from WT, PTPN22–/–, or 619W mice, and mRNA levels of Reg3g were analyzed by quantitative PCR. (C and D) MC-38 cells were cocultured with MDP-activated BMDMs from WT, PTPN22–/–, or 619W mice in the presence of an isotype control or an anti–IL-18 antibody and analyzed for mRNA expression of Reg3g (C) and Defa5 (D). Data are representative of 1 of 2 independent experiments. Depicted are mean values and SEM, and each dot represents 1 individual mouse or experimental sample. *P < 0.05, **P < 0.01, ***P < 0.001, Kruskal-Wallis test.
Monocyte/macrophage-derived IL-18 induces the expression of antimicrobial peptides.
In order to determine how PTPN22 might affect the expression of AMPs, we next cocultured MC-38 intestinal epithelial cells (IECs) with bone marrow–derived macrophages (BMDMs) or T cells from WT, PTPN22–/–, or 619W mice. To induce production of factors that might influence the secretion of AMPs, we pulsed the BMDMs with MDP and stimulated T cells with anti-CD3/CD28 before coculture. Interestingly, we observed that coculture with BMDMs from PTPN22–/– mice resulted in moderate expression of Reg3g, while coculture with BMDMs from 619W mice promoted Reg3g expression (Figure 10B). On the other hand, coculture with T cells did not affect the expression of AMPs (Figure 10B).
It has been reported that IL-18 affects expression/secretion of AMPs in the intestine (27). In line with this observation, inhibition of IL-18 resulted in overall decreased induction of AMPs. Interestingly, the increase in AMP expression observed upon coculture with macrophages from 619W mice was no longer present when IL-18 was inhibited (Figure 10C). This strongly indicates that PTPN22 affects the expression of AMPs via modulating IL-18 secretion/signaling.
Discussion
Here we show that PTPN22 crucially affects colitis development, and that these effects are — at least in part — due to changes in the intestinal microbiota. In line with previous reports, we observe increased acute colitis in PTPN22–/– mice (14, 17), while mice carrying the autoimmunity-associated PTPN22 619W variant were protected from colitis upon acute DSS treatment. This protective effect of the variant is well in line with the notion that the variant protects from CD onset (6).
Notably, acute DSS colitis lacks a recovery phase. The natural disease course in IBD patients, however, is often characterized by phases of remission with few or no symptoms, intermitted by relapses with sometimes severe intestinal inflammation (3). Interestingly, when we subjected our mice to chronic DSS colitis — a model with several cycles of acute colitis followed by recovery, hence reflecting the varying disease course in IBD patients — mice carrying the PTPN22 variant were no longer protected. This is of great interest since it is similar to the human situation, in which the PTPN22 variant protects from CD onset, but in affected patients, presence of the variant does not correlate with a milder disease course or fewer complications (28).
When addressing T cell subsets or inflammasome activation, we observed phenotypes similar to those previously reported (17, 23), and moreover, these changes were consistent in acute and chronic colitis models, hence these changes alone cannot explain why the PTPN22 variant affects acute and chronic colitis differentially. Both inflammasome activation and altered T cell function have been reported to affect the intestinal microbiota, which might respond differentially in models with recovery.
Our results show that presence of the 619W variant in PTPN22 affects the composition of the intestinal microbiota per se, but even more importantly upon acute or chronic DSS treatment, which might crucially contribute to the observed alterations in colitis severity (protection in acute, but disease-promoting effects in chronic, DSS colitis with recovery phases). Cohousing of WT mice with 619W or PTPN22–/– mice showed that the phenotype of the PTPN22–/– and the 619W mice is transmissible, possibly mediated by the microbiota.
In line with this, we found profound differences within the intestinal microbiota, both at basal state and also after recovery from the first DSS cycle and after the induction of chronic DSS colitis: presence of the 619W variant resulted in increased levels of potentially beneficial bacteria upon acute DSS treatment, such as OTUs classified as Lactobacillus, and depletion of potential pathogens, such as Turicibacter sanguinis, which might explain the reduced disease severity at this stage. However, we also observed an increase of the potential pathogen Helicobacter at baseline and when mice were allowed to recover. However, the role of Helicobacter in IBD is controversial, and Helicobacter products might also exert antiinflammatory effects (29, 30). In contrast, PTPN22–/– mice showed a characteristic increase of Akkermansia and Ruminococcacea in acute colitis, which was not observed in 619W mice, but might account for the robust recovery and subsequent milder disease in later DSS cycles. Notably, Akkermansia also slightly increased in 619W mice upon acute DSS treatment, but was reduced upon recovery and in chronic colitis.
Akkermansia is a genus of Gram-negative mucus-degrading bacteria that has recently received increasing attention: its presence has been associated with better treatment response and recovery from colitis (31), and better treatment response in cancer patients treated with checkpoint inhibitors (32). The role of Akkermansia in intestinal health is controversial, and it is unclear whether its biological roles in the human and murine gut are equivalent: while some reports showed that Akkermansia might reduce inflammation (33), a recent study showed that presence of Akkermansia promotes colitis in IL-10–/– mice (34). Notably, this study also showed that defects in IL-18 production in IECs promote Akkermansia overgrowth, and supplementation of IL-18 was sufficient to reduce colitis in their model (34). This is of interest since we showed here that PTPN22 is involved in regulating IL-18 levels in the inflamed intestine. Hence, it might be that changes in IL-18 levels as observed in PTPN22–/– and 619W mice are responsible for the observed changes in Akkermansia abundance.
Another important group of bacteria involved in intestinal health are butyrate producers, which are drastically reduced in IBD patients (35, 36). Butyrate is an important energy source for IECs and promotes wound healing (37), while it dampens proliferation in colonic cancer cells (37). This is of interest since some butyrate producers, such as Lachnospiraceae and Ruminococcaceae, were increased in both PTPN22–/– and 619W mice during acute and chronic colitis, although disease severity was opposite. Nevertheless, some Clostridiales OTUs, which contain important butyrate producers, were clearly depleted specifically in 619W mice after chronic DSS colitis, possibly contributing to the enhanced colitis severity in chronic colitis observed in those mice.
Overall, while shifts in community composition and the enrichment or depletion of individual taxa merely correlated with genotype and differential treatment responses, the results of our transplantation and cohousing experiments provide orthogonal evidence suggesting a causal role of the microbiota in mediating genotype-specific effects.
Apart from changes in microbiota composition between the 3 genotypes, we found differences in the expression of AMPs. Notably, expression of Rag3g and Defa5 was initially increased in 619W mice, but significantly declined after the first recovery phase. Since AMPs are important factors to reduce the number of mucosa-adherent, potentially pathogenic bacteria (38), this might explain why 619W mice display increased disease in later DSS cycles.
AMPs were induced in intestinal epithelial cells upon coculture with macrophages that were pretreated with the bacterial cell wall product MDP, and this was dependent on IL-18 secretion. In line with our previous studies, inflammasome activation and IL-18 production are increased in the intestine of 619W mice. Further, it has been shown that IL-18 on one hand promotes AMP production (27), but on the other hand also promotes goblet cell loss during chronic DSS-induced colitis (39), resulting in decreased mucus and AMP production. Hence, enhanced IL-18 secretion in mice featuring the 619W variant in PTPN22 might promote antimicrobial peptide secretion in acute colitis, but in the chronic model might promote loss of goblet cells that in turn could drive the observed loss of an induction of AMPs and subsequently reduced levels of AMPs.
An additional mechanism of how PTPN22 might affect the secretion of AMPs is via its role in autophagy. We have shown that loss of PTPN22 results in enhanced autophagy (18), and recent reports demonstrated that in goblet cells, autophagy is crucial for secretion of lysozyme — especially in the presence of pathogenic bacteria (40). Therefore, it might be that PTPN22 affects AMP release via regulation of autophagy.
Taken together, our results demonstrate that PTPN22 is an important regulator of intestinal health via controlling the composition of the intestinal microbiota. Interestingly, induction of acute colitis changes the host response toward bacteria in 619W mice in a way that finally renders the 619W mice more susceptible to colitis induction. This is especially interesting, since it shows that the very same genetic variant can have both beneficial and disease-driving effects, depending on the timing of insults in the intestine. It might also explain why presence of the 619W variant protects from onset of CD but does not result in a milder disease course in affected patients (28).
Methods
Mice and colitis induction.
PTPN22-knockout (PTPN22–/–) mice in the C57BL/6 background (13) were initially obtained from A.C. Chan, Genentech, San Francisco, California, USA, and crossed with WT C57/B6 mice (The Jackson Laboratory) to obtain PTPN22+/– mice. A colony of PTPN22+/– mice was maintained in our vivarium and crossed with each other to obtain PTPN22–/– and WT littermates for experiments. Ten- to twelve-week-old female littermate mice were used for all studies. C57BL/6 mice carrying the 619W PTPN22 variant (619W mice) (22) were obtained from D. Rawlings, University of Washington School of Medicine, Seattle, Washington, USA, and crossed with WT mice to obtain mice heterozygous for the 619W allele. These mice were then crossed with each other to maintain a colony of mice heterozygous for the 619W allele and to obtain 619W and WT littermates for experiments. Unless otherwise stated, mice were separated according to genotype directly after weaning. For acute DSS colitis induction, 2.5% DSS was supplemented to the drinking water for 7 days. For induction of chronic DSS colitis, each mouse received 4 cycles of DSS treatment consisting of 7 days with 1.5% DSS in the drinking water followed by a 10-day recovery phase with normal drinking water. After the last DSS cycle, mice received normal drinking water for 4 weeks. For microbiota depletion using antibiotics, mice were treated with ampicillin (1 g/l; Sigma-Aldrich), neomycin sulfate (1 g/l; Sigma-Aldrich), metronidazole (1 g/l; Sigma-Aldrich), and vancomycin (500 mg/l; Sigma-Aldrich) in the drinking water starting 2 weeks before the first DSS cycle (chronic DSS colitis), or for 1 week followed by 3 days recovery before transfer of cecum content (transplantation studies).
Assessment of colitis severity and histological score.
Animals were anesthetized i.p. with 90–120 mg/kg body weight ketamine (Vétoquinol) and 8 mg/kg body weight xylazine (Bayer). Animals were examined as described previously (41). Recording was performed with the Karl Storz Tele Pack Pal 20043020 (Karl Storz Endoskope). Colonoscopy was scored using the murine endoscopic index of colitis severity (MEICS) scoring system as described previously (41) using the following 5 parameters: (a) transparency of the colon, (b) changes of the vascular pattern, (c) fibrin visible, (d) granularity of the mucosal surface, and (e) stool consistency. Histological scoring for inflammatory infiltration and epithelial cell damage was performed on H&E-stained sections of the most distal 1 cm of the mouse colon (41, 42).
Single-cell isolation and flow cytometry.
Single-cell suspensions were generated from spleen, mesenteric lymph nodes, and the colonic lamina propria as described previously (43). In brief, spleen and lymph nodes were smashed over a 70-μm cell strainer before red cell lysis using ACK buffer (1.5M NH4Cl; 100 mM KHC03, 10 mM EDTA). For isolation of lamina propria lymphocytes, intestinal tissue pieces were incubated for 30 minutes in HBSS supplemented with EDTA (2 nM) on a shaker (250 rpm) at 37°C, then vortexed vigorously, and intestinal epithelial cells were washed away with fresh HBSS. Tissue pieces were then incubated in digestion solution (RPMI containing 10% FCS, 2 mg/ml collagenase IV, 4 mg/ml dispase, 0.2 mg/ml DNase I) for 30 minutes. Cells were passed through a 70-μm strainer, washed with PBS, and used for FACS analysis. For intracellular cytokine staining, cells were treated with PMA (50 ng/ml; Sigma-Aldrich) and ionomycin (1 μg/ml; Sigma-Aldrich) in the presence of 10 μg/ml brefeldin A (Sigma-Aldrich) for 4 hours. Cells were harvested, washed in PBS, stained for surface molecules for 15 minutes, washed in FACS buffer (PBS, 2% FCS), and fixed in fixation/permeabilization buffer (BD) for 20 minutes. Cells were washed and resuspended in permeabilization buffer (BD) before addition of anti–IFN-γ and anti–IL-17 antibody for 30 minutes. Before analysis, cells were washed in permeabilization buffer and resuspended in FACS buffer. A FACS Canto II or an LSR Fortessa from BD was used for all analysis.
Western blotting.
Protein was extracted from colon specimens with M-PER lysis buffer (Thermo Fisher Scientific) using a GentleMACS (Miltenyi) tissue homogenizer. Cells (1 × 106) were lysed in 100 μl M-PER using a 26G needle. An aliquot of each lysate was mixed with loading buffer (NuPAGE 4× LDS Sample Buffer, Life Technologies) and 50 mM DTT and boiled for 5 minutes at 96°C. Proteins were separated by SDS-PAGE and transferred onto nitrocellulose membranes (GE Healthcare). Membranes were blocked with 1 % blocking solution, and primary antibody (anti–IL-1β: R&D Systems, ordering number AF-401-NA; anti–IL-18: Santa Cruz Biotechnology, ordering number sc-7954) was added in blocking buffer (3% BSA in washing buffer [Tris-buffered saline containing 1% Tween-20]). Membranes were washed for 30 minutes, HRP-labeled secondary anti-goat or anti-rabbit IgG antibody (Santa Cruz Biotechnology, ordering numbers sc-2020 and sc-2004, respectively) in blocking buffer was added for 30 minutes, and membranes were washed for 30 minutes with TBST. Immunoreactive proteins were detected using an ECL detection kit (Thermo Fisher Scientific). Densitometric analysis of Western blots was performed using NIH ImageJ software.
Real-time PCR.
Colon tissue was mechanically dissociated and lysed in RLT buffer (Thermo Fisher Scientific) using a GentleMACS tissue homogenizer (Miltenyi). Cells were disrupted in RLT buffer (Thermo Fisher Scientific) using a 26G needle. Total RNA was isolated using RNeasy Mini Kit (Qiagen) with a DNA removal step. RNA concentration was assessed by absorbance at 260 and 280 nm. cDNA synthesis was performed using a High-Capacity cDNA Reverse Transcription Kit (Thermo Fisher Scientific) following the manufacturer’s instructions. Commercially available TaqMan assays (Thermo Fisher Scientific) were used for real-time PCR on a Quant Studio 6 flex Thermocycler (Thermo Fisher Scientific).
16S sequencing.
Fecal samples were collected at different time points and DNA extracted using the PowerLyzer Soil kit (Qiagen, Thermo Fisher Scientific) according to the manufacturer’s instruction. Targeted amplification using the primer pair T515F × 806R described in the Earth Microbiome Project (44) and MiSeq sequencing of the 16S rRNA V4 region were performed by Microsynth. Raw sequence data are available from the European Nucleotide Archive, project accession PRJEB26758. Sample metadata and OTU count tables are available as Supplemental Tables 1 and 2. Relevant analysis code is available at https://git.embl.de/tschmidt/PTPN22_microbiota Demultiplexed paired-end reads were denoised, merged, and filtered for chimeric reads using DADA2 (45). Reads were then mapped to a preclustered reference database of full-length 16S rRNA sequences and taxonomically classified using MAPseq v1.0 (46). Reads that did not confidently map to 98% average linkage reference OTUs were aligned to a secondary-structure-aware bacterial 16S model using Infernal v1.1 (47) and hierarchically clustered into 98% average linkage OTUs using hpc-clust (48), as described previously (49). In short, we fitted negative binomial generalized linear models on dispersion-adjusted OTU count data, accounting for blocked designs involving repeated independent experiments, and conducted likelihood ratio tests on the resulting models, followed by test-wise FDR adjustment for multiple hypothesis testing with a significance level of FDR < 0.01 (see online analysis code for details).
Statistics.
Microbiome analyses were conducted using custom R scripts, relying on several external packages, including vegan, ggplot2, and edgeR. Hill diversities (50) were calculated on OTU counts rarefied to 10,000 reads. Community compositional distances were calculated as Bray-Curtis dissimilarity on nontransformed and sqrt-transformed counts, weighted and unweighted Jaccard distance, and weighted and unweighted TINA distances (51). Unless otherwise indicated, results are reported for Bray-Curtis dissimilarities on nontransformed counts, but findings were generally consistent between dissimilarity metrics. Differential abundance of individual OTUs was detected using edgeR with robust count dispersion estimation, as similarly reported previously (52).
For data on mouse and cell culture experiments, data were analyzed using GraphPad Prism software with the statistical test indicated in the respective figure legends. If not otherwise stated, data are presented as individual dots for each sample/mouse, a line for mean, and bars indicating SEM. Tests were always 2-sided where applicable, and a P value below 0.05 was considered significant.
Cell culture.
To generate BMDMs, bone marrow from WT, PTPN22–/–, or 619W mice was incubated for 7 days in DMEM supplemented with 10 IU M-CSF (Calbiochem), 10% FCS, and 1% penicillin/streptomycin solution (Thermo Fisher Scientific). MC-38 cells were obtained from ATCC and cultured in DMEM supplemented with 10% FCS. For coculture experiments, MC-38 cells (DSMZ) were seeded on inserts (Merck Millipore) and grown for 4 days. BMDMs were seeded in 6-well plates, pulsed for 8 hours with MDP (100 ng/ml; Invivogen), washed with PBS (Thermo Fisher Scientific), and incubated with fresh culture medium. Inserts with MC-38 cells were then transferred into the BMDC-containing wells. For coculture with T cells, 6-well plates were coated overnight with anti-CD3 (1 ng/ml; R&D Systems) and anti-CD28 (5 ng/ml; R&D Systems) antibody as described previously (43). CD3+ T cells were sorted from the spleen of WT, PTPN22–/–, and 619W mice as described previously (43), and 1.5 × 106 T cells were seeded per well. After 12 hours, inserts with MC-38 cells were transferred into the wells containing T cells. Cocultured MC-38 cells were harvested for mRNA isolation after 24 hours. mRNA isolation, cDNA synthesis, and quantitative PCR were performed as described previously (43). IL-18 was inhibited using 5 μg/ml rat anti–IL-18 IgG1 antibody (clone 93-10C; R&D Systems). Rat IgG1 isotype control antibody (clone MAB005; R&D Systems) was used as a control.
Study approval.
Animal experiments were performed according to Swiss animal welfare legislation and were approved by the local animal welfare commission (Veterinäramt Zürich; approval 255-2014 and 220-2016).
Author contributions
MRS, M Schwarzfischer, LH, KA, SL, and CG conducted mouse and cell culture experiments, and analyzed and interpreted data. AG, CL, TSBS, and CVM analyzed and interpreted microbiota data. XD, DJR, and ACC generated PTPN22–/– and 619W mice. GR, M Scharl, and TSBS critically revised the manuscript for intellectual content. MRS and M Scharl conceived, designed, and supervised the study.
Supplementary Material
Acknowledgments
This research has been supported by grants to M Scharl from the Stiftung Experimentelle Biomedizin, the Promedica Foundation, and the Swiss National Science Foundation (grant 314730-146204, grant 314730_166381/1, grant 314730_166381/2, grant 320030_184753/1, and grant CRSII3_154488/1).
Version 1. 05/20/2019
Electronic publication
Version 2. 06/03/2019
Print issue publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2019, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2019;129(6):2527–2541.https://doi.org/10.1172/JCI123263.
Contributor Information
Marlene Schwarzfischer, Email: Marlene.Schwarzfischer@usz.ch.
Larissa Hering, Email: Larissa.Hering@usz.ch.
Kirstin Atrott, Email: kirstin.atrott@usz.ch.
Silvia Lang, Email: silvia.lang@usz.ch.
Claudia Gottier, Email: claudia.gottier@usz.ch.
Annelies Geirnaert, Email: annelies.geirnaert@hest.ethz.ch.
Christophe Lacroix, Email: christophe.lacroix@hest.ethz.ch.
Xuezhi Dai, Email: tonyxuezhi.dai@seattlechildrens.org.
Christian von Mering, Email: mering@imls.uzh.ch.
Gerhard Rogler, Email: gerhard.rogler@usz.ch.
Michael Scharl, Email: michael.scharl@usz.ch.
References
- 1.Ni J, Wu GD, Albenberg L, Tomov VT. Gut microbiota and IBD: causation or correlation? Nat Rev Gastroenterol Hepatol. 2017;14(10):573–584. doi: 10.1038/nrgastro.2017.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Seo SU, et al. Distinct commensals induce interleukin-1β via NLRP3 inflammasome in inflammatory monocytes to promote intestinal inflammation in response to injury. Immunity. 2015;42(4):744–755. doi: 10.1016/j.immuni.2015.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cosnes J, Gower-Rousseau C, Seksik P, Cortot A. Epidemiology and natural history of inflammatory bowel diseases. Gastroenterology. 2011;140(6):1785–1794. doi: 10.1053/j.gastro.2011.01.055. [DOI] [PubMed] [Google Scholar]
- 4.Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447(7145):661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barrett JC, et al. Genome-wide association defines more than 30 distinct susceptibility loci for Crohn’s disease. Nat Genet. 2008;40(8):955–962. doi: 10.1038/ng.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Diaz-Gallo LM, et al. Differential association of two PTPN22 coding variants with Crohn’s disease and ulcerative colitis. Inflamm Bowel Dis. 2011;17(11):2287–2294. doi: 10.1002/ibd.21630. [DOI] [PubMed] [Google Scholar]
- 7.Dieudé P, Teixeira VH, Pierlot C, Cornélis F, Petit-Teixeira E. Testing for linkage and association with rheumatoid arthritis a ptpn22 promoter polymorphism reported to be associated and linked with type 1 diabetes in the Caucasian population. Ann Rheum Dis. 2008;67(6):900–901. doi: 10.1136/ard.2007.077180. [DOI] [PubMed] [Google Scholar]
- 8.Eliopoulos E, et al. Association of the PTPN22 R620W polymorphism with increased risk for SLE in the genetically homogeneous population of Crete. Lupus. 2011;20(5):501–506. doi: 10.1177/0961203310392423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kyogoku C, et al. Genetic association of the R620W polymorphism of protein tyrosine phosphatase PTPN22 with human SLE. Am J Hum Genet. 2004;75(3):504–507. doi: 10.1086/423790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Michou L, et al. Linkage proof for PTPN22, a rheumatoid arthritis susceptibility gene and a human autoimmunity gene. Proc Natl Acad Sci U S A. 2007;104(5):1649–1654. doi: 10.1073/pnas.0610250104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ramirez M, et al. The PTPN22 C1858T variant as a risk factor for rheumatoid arthritis and systemic lupus erythematosus but not for systemic sclerosis in the Colombian population. Clin Exp Rheumatol. 2012;30(4):520–524. [PubMed] [Google Scholar]
- 12.Arimura Y, Yagi J. Comprehensive expression profiles of genes for protein tyrosine phosphatases in immune cells. Sci Signal. 2010;3(137):rs1. doi: 10.1126/scisignal.2000966. [DOI] [PubMed] [Google Scholar]
- 13.Hasegawa K, Martin F, Huang G, Tumas D, Diehl L, Chan AC. PEST domain-enriched tyrosine phosphatase (PEP) regulation of effector/memory T cells. Science. 2004;303(5658):685–689. doi: 10.1126/science.1092138. [DOI] [PubMed] [Google Scholar]
- 14.Wang Y, et al. The autoimmunity-associated gene PTPN22 potentiates toll-like receptor-driven, type 1 interferon-dependent immunity. Immunity. 2013;39(1):111–122. doi: 10.1016/j.immuni.2013.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Holmes DA, et al. Autoimmunity-associated protein tyrosine phosphatase PEP negatively regulates IFN-α receptor signaling. J Exp Med. 2015;212(7):1081–1093. doi: 10.1084/jem.20142130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Spalinger MR, et al. Loss of protein tyrosine phosphatase nonreceptor type 22 regulates interferon-γ-induced signaling in human monocytes. Gastroenterology. 2013;144(5):978–988.e10. doi: 10.1053/j.gastro.2013.01.048. [DOI] [PubMed] [Google Scholar]
- 17.Spalinger MR, et al. NLRP3 tyrosine phosphorylation is controlled by protein tyrosine phosphatase PTPN22. J Clin Invest. 2016;126(5):1783–1800. doi: 10.1172/JCI83669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Spalinger MR, Lang S, Vavricka SR, Fried M, Rogler G, Scharl M. Protein tyrosine phosphatase non-receptor type 22 modulates NOD2-induced cytokine release and autophagy. PLoS One. 2013;8(8):e72384. doi: 10.1371/journal.pone.0072384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Patel KK, et al. Autophagy proteins control goblet cell function by potentiating reactive oxygen species production. EMBO J. 2013;32(24):3130–3144. doi: 10.1038/emboj.2013.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wlodarska M, et al. NLRP6 inflammasome orchestrates the colonic host-microbial interface by regulating goblet cell mucus secretion. Cell. 2014;156(5):1045–1059. doi: 10.1016/j.cell.2014.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Parkes M. Evidence from genetics for a role of autophagy and innate immunity in IBD pathogenesis. Dig Dis. 2012;30(4):330–333. doi: 10.1159/000338119. [DOI] [PubMed] [Google Scholar]
- 22.Dai X, et al. A disease-associated PTPN22 variant promotes systemic autoimmunity in murine models. J Clin Invest. 2013;123(5):2024–2036. doi: 10.1172/JCI66963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brownlie RJ, Miosge LA, Vassilakos D, Svensson LM, Cope A, Zamoyska R. Lack of the phosphatase PTPN22 increases adhesion of murine regulatory T cells to improve their immunosuppressive function. Sci Signal. 2012;5(252):ra87. doi: 10.1126/scisignal.2003365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Salmond RJ, Brownlie RJ, Morrison VL, Zamoyska R. The tyrosine phosphatase PTPN22 discriminates weak self peptides from strong agonist TCR signals. Nat Immunol. 2014;15(9):875–883. doi: 10.1038/ni.2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Spalinger MR, et al. PTPN22 regulates NLRP3-mediated IL1B secretion in an autophagy-dependent manner. Autophagy. 2017;13(9):1590–1601. doi: 10.1080/15548627.2017.1341453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Couturier-Maillard A, et al. NOD2-mediated dysbiosis predisposes mice to transmissible colitis and colorectal cancer. J Clin Invest. 2013;123(2):700–711. doi: 10.1172/JCI62236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ratsimandresy RA, Indramohan M, Dorfleutner A, Stehlik C. The AIM2 inflammasome is a central regulator of intestinal homeostasis through the IL-18/IL-22/STAT3 pathway. Cell Mol Immunol. 2017;14(1):127–142. doi: 10.1038/cmi.2016.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Spalinger MR, et al. Genotype-phenotype associations of the CD-associated single nucleotide polymorphism within the gene locus encoding protein tyrosine phosphatase non-receptor type 22 in patients of the Swiss IBD cohort. PLoS One. 2016;11(7):e0160215. doi: 10.1371/journal.pone.0160215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gravina AG, et al. Peptide Hp(2-20) accelerates healing of TNBS-induced colitis in the rat. United European Gastroenterol J. 2018;6(9):1428–1436. doi: 10.1177/2050640618793564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang H, et al. Helicobacter pylori colonization protects against chronic experimental colitis by regulating Th17/Treg balance. Inflamm Bowel Dis. 2018;24(7):1481–1492. doi: 10.1093/ibd/izy107. [DOI] [PubMed] [Google Scholar]
- 31.Shaw KA, et al. Dysbiosis, inflammation, and response to treatment: a longitudinal study of pediatric subjects with newly diagnosed inflammatory bowel disease. Genome Med. 2016;8(1):75. doi: 10.1186/s13073-016-0331-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Routy B, et al. Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science. 2018;359(6371):91–97. doi: 10.1126/science.aan3706. [DOI] [PubMed] [Google Scholar]
- 33.Kang CS, et al. Extracellular vesicles derived from gut microbiota, especially Akkermansia muciniphila, protect the progression of dextran sulfate sodium-induced colitis. PLoS One. 2013;8(10):e76520. doi: 10.1371/journal.pone.0076520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Seregin SS, et al. NLRP6 protects Il10–/– mice from colitis by limiting colonization of Akkermansia muciniphila. Cell Rep. 2017;19(10):2174. doi: 10.1016/j.celrep.2017.05.074. [DOI] [PubMed] [Google Scholar]
- 35.Weir TL, Manter DK, Sheflin AM, Barnett BA, Heuberger AL, Ryan EP. Stool microbiome and metabolome differences between colorectal cancer patients and healthy adults. PLoS One. 2013;8(8):e70803. doi: 10.1371/journal.pone.0070803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Baxter NT, Zackular JP, Chen GY, Schloss PD. Structure of the gut microbiome following colonization with human feces determines colonic tumor burden. Microbiome. 2014;2:20. doi: 10.1186/2049-2618-2-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hamer HM, Jonkers D, Venema K, Vanhoutvin S, Troost FJ, Brummer RJ. Review article: the role of butyrate on colonic function. Aliment Pharmacol Ther. 2008;27(2):104–119. doi: 10.1111/j.1365-2036.2007.03562.x. [DOI] [PubMed] [Google Scholar]
- 38.Mukherjee S, Hooper LV. Antimicrobial defense of the intestine. Immunity. 2015;42(1):28–39. doi: 10.1016/j.immuni.2014.12.028. [DOI] [PubMed] [Google Scholar]
- 39.Nowarski R, et al. Epithelial IL-18 equilibrium controls barrier function in colitis. Cell. 2015;163(6):1444–1456. doi: 10.1016/j.cell.2015.10.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bel S, et al. Paneth cells secrete lysozyme via secretory autophagy during bacterial infection of the intestine. Science. 2017;357(6355):1047–1052. doi: 10.1126/science.aal4677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Becker C, Fantini MC, Neurath MF. High resolution colonoscopy in live mice. Nat Protoc. 2006;1(6):2900–2904. doi: 10.1038/nprot.2006.446. [DOI] [PubMed] [Google Scholar]
- 42.Obermeier F, Kojouharoff G, Hans W, Schölmerich J, Gross V, Falk W. Interferon-gamma (IFN-γ)- and tumour necrosis factor (TNF)-induced nitric oxide as toxic effector molecule in chronic dextran sulphate sodium (DSS)-induced colitis in mice. Clin Exp Immunol. 1999;116(2):238–245. doi: 10.1046/j.1365-2249.1999.00878.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Spalinger MR, et al. PTPN2 controls differentiation of CD4+ T cells and limits intestinal inflammation and intestinal dysbiosis. Mucosal Immunol. 2015;8(4):918–929. doi: 10.1038/mi.2014.122. [DOI] [PubMed] [Google Scholar]
- 44.Thompson LR, et al. A communal catalogue reveals Earth’s multiscale microbial diversity. Nature. 2017;551(7681):457–463. doi: 10.1038/nature24621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJ, Holmes SP. DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods. 2016;13(7):581–583. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Matias Rodrigues JF, Schmidt TSB, Tackmann J, von Mering C. MAPseq: highly efficient k-mer search with confidence estimates, for rRNA sequence analysis. Bioinformatics. 2017;33(23):3808–3810. doi: 10.1093/bioinformatics/btx517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nawrocki EP, Eddy SR. Infernal 1.1: 100-fold faster RNA homology searches. Bioinformatics. 2013;29(22):2933–2935. doi: 10.1093/bioinformatics/btt509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Matias Rodrigues JF, von Mering C. HPC-CLUST: distributed hierarchical clustering for large sets of nucleotide sequences. Bioinformatics. 2014;30(2):287–288. doi: 10.1093/bioinformatics/btt657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schmidt TS, Matias Rodrigues JF, von Mering C. Limits to robustness and reproducibility in the demarcation of operational taxonomic units. Environ Microbiol. 2015;17(5):1689–1706. doi: 10.1111/1462-2920.12610. [DOI] [PubMed] [Google Scholar]
- 50.Hill MO. Diversity and evenness: a unifying notation and its consequences. Ecology. 1973;54(2):427–432. doi: 10.2307/1934352. [DOI] [Google Scholar]
- 51.Schmidt TS, Matias Rodrigues JF, von Mering C. A family of interaction-adjusted indices of community similarity. ISME J. 2017;11(3):791–807. doi: 10.1038/ismej.2016.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Becker E, et al. Effects of oral antibiotics and isotretinoin on the murine gut microbiota. Int J Antimicrob Agents. 2017;50(3):342–351. doi: 10.1016/j.ijantimicag.2017.03.017. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.