Abstract
Chronic glucocorticoid therapy has serious side effects, including diabetes and fatty liver. However, the molecular mechanisms responsible for steroid-induced diabetes remain largely enigmatic. Here, we show that hepatic Krüppel-like factor 9 (Klf9) gene expression is induced by dexamethasone and fasting. The overexpression of Klf9 in primary hepatocytes strongly stimulated Pgc1a gene expression through direct binding to its promoter, thereby activating the gluconeogenic program. However, Klf9 mutation abolished the stimulatory effect of dexamethasone on cellular glucose output. Adenovirus-mediated overexpression of KLF9 in the mouse liver markedly increased blood glucose levels and impaired glucose tolerance. Conversely, both global Klf9-mutant mice and liver-specific Klf9-deleted mice displayed fasting hypoglycemia. Moreover, the knockdown of Klf9 in the liver in diabetic mouse models, including ob/ob and db/db mice, markedly lowered fasting blood glucose levels. Notably, hepatic Klf9 deficiency in mice alleviated hyperglycemia induced by chronic dexamethasone treatment. These results suggest a critical role for KLF9 in the regulation of hepatic glucose metabolism and identify hepatic induction of KLF9 as a mechanism underlying glucocorticoid therapy–induced diabetes.
Keywords: Endocrinology, Metabolism
Keywords: Diabetes, Gluconeogenesis, Glucose metabolism
Introduction
In mammals, glucose homeostasis is maintained through the hormonal regulation of both hepatic glucose production (HGP) and peripheral glucose uptake. In the fasted state, the secretion of the pancreatic hormone glucagon and the adrenal hormone glucocorticoid (GC ) is induced to enhance HGP (gluconeogenesis) to maintain blood glucose levels within a narrow rang–e (1, 2). Glucagon triggers the gluconeogenic program in the liver through the cAMP/PKA/CREB/CRTC2 signaling pathway, where CREB indicates cAMP response element and CRTC2 indicates CREB-regulated transcriptional coactivator 2. The CREB/CRTC2 complex induces the expression of peroxisome proliferator-activated receptor γ coactivator 1 α (PGC1α), which in turn coactivates transcription factors, including hepatocyte nuclear factor 4 α (HNF4α), forkhead box O1 (FOXO1), and GC receptor (GR), to activate hepatic gluconeogenesis (3–6). However, the abnormal elevation of HGP contributes to fasting hyperglycemia in diabetes (7). In contrast, after a meal, circulating blood insulin levels increase, thereby inhibiting hepatic gluconeogenesis and stimulating glucose uptake and utilization by the peripheral tissues. Insulin inhibits hepatic gluconeogenesis by negatively regulating gluconeogenic transcriptional regulators, including FOXO1, PGC1α, and CRTC2 (4, 8–13).
Dexamethasone (Dex) and other GCs have been used clinically for decades and represent the most important and frequently used class of antiinflammatory drugs. Unfortunately, long-term GC therapy is associated with metabolic side effects, including hyperglycemia, hypertension, and hepatic steatosis (14, 15). Clinical studies show that GC is elevated in insulin-resistant and glucose-intolerant patients (16, 17). The endogenous overproduction of GCs characterized by Cushing’s syndrome is associated with glucose intolerance and the appearance of insulin resistance and altered lipid metabolism (18). In contrast, GC deficiency in humans is characterized by impaired gluconeogenesis, leading to hypoglycemia (19). Although it is well known that GCs induce the expression of hepatic gluconeogenic genes, leading to hyperglycemia, the underlying mechanism of the induction of hepatic gluconeogenesis by GCs remains largely unclear (15, 19).
Krüppel-like factor 9 (Klf9), also called basic transcription element–binding protein-1 (Bteb1), was originally isolated from a liver cDNA library (20). Current experimental evidence indicates that KLF9 plays a key role in development and is required for thyroid hormone–dependent actions during neurite extension and branching (21–23). Interestingly, a recent study suggests that KLF9 activates Pparg2 gene expression, thereby promoting preadipocyte differentiation (24). In addition to its expression in the brain, Klf9 is highly expressed in the liver (20); however, the physiological function of hepatic KLF9 remains unexplored.
In this study, we show that Dex and fasting induced the expression of hepatic Klf9, which in turn activated Pgc1a gene expression and the gluconeogenic program. Klf9 mutation in primary hepatocytes markedly compromised the Dex-mediated induction of the gluconeogenic program. Moreover, hepatic Klf9 deficiency alleviated Dex-induced hyperglycemia in mice.
Results
Hepatic Klf9 expression is controlled by Dex and nutritional status.
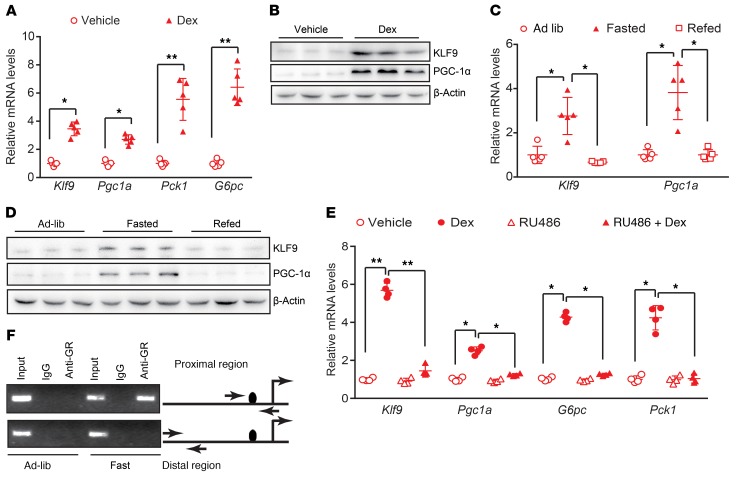
The molecular mechanism by which GCs activate hepatic gluconeogenesis remains largely unclear. To identify the mediator of the effects of GCs on hepatic glucose metabolism, we first performed RNA-Seq analysis of the livers of WT C57 BL/6J mice treated with Dex or saline (control) using Illumina/Solexa sequencing technology. Preliminary analysis of RNA-Seq data indicated that, as expected, Dex treatment induced the expression of gluconeogenic genes, including Pck1 and G6pc. Dex also altered the expression of genes involved in lipid metabolism and energy metabolism (Supplemental Figure 1, A–C; supplemental material available online with this article; https://doi.org/10.1172/JCI66062DS1). Notably, Dex treatment significantly induced the expression of Klf9, a transcription factor, and Pgc1a, a master regulator of hepatic gluconeogenesis (Supplemental Figure 1A). Quantitative PCR and Western blot analysis confirmed the Dex-mediated induction of the expression of Klf9 and gluconeogenic genes in intact liver (Figure 1, A and B, Supplemental Figure 1, D and E).
Figure 1. Hepatic Klf9 expression is induced by fasting and Dex.
(A) Quantitative PCR analysis of hepatic Klf9, Pgc1a, Pck1, and G6pc in C57BL/6J mice 12 hours after injection with saline or Dex (1 mg/kg) (n = 5/group). (B) Representative Western blot analysis of hepatic KLF9 and PGC1α in the mice described in A. (C) Quantitative PCR analysis of hepatic Klf9 and Pgc1a in mice under ad libitum–fed, 24 hour–fasted or 12 hour–refed conditions (n = 5/group). (D) Representative Western blot analysis of hepatic KLF9 and PGC1α in mice described in C. (E) Quantitative PCR analysis of Klf9, Pgc1a, Pck1, and G6pc in mouse primary hepatocytes treated with 100 nM Dex and/or 10 μM of the GR antagonist RU486 for 12 hours. (F) ChIP assays performed as described in Methods showing that fasting leads to the binding of endogenous GR proteins to GRE1/2 on the Klf9 promoter region, but not the distal region, which lacks GREs (negative control), as indicated. Data are shown as mean ± SEM. *P < 0.05; **P < 0.01, 2-tailed Student’s t test (A), 1-way ANOVA (C, E).
Under fasting conditions, glucagon and GCs are released into circulation to synergistically stimulate hepatic gluconeogenesis (15, 19). Thus, we further studied whether hepatic Klf9 expression could be regulated by nutritional status. First, preliminary analysis of microarray data indicated that fasting stimulated gluconeogenic genes, including Klf9 and Pgc1a (Supplemental Figure 1F). Moreover, real-time PCR and Western blotting analysis confirmed that fasting induced an increase in mRNA and protein levels of KLF9 and PGC1α in the liver and refeeding reversed this induction (Figure 1, C and D), which is a characteristic regulatory pattern for the genes involved in gluconeogenesis. Our results indicate a correlation between Klf9 expression and gluconeogenic potential in the liver.
GCs mediate their physiological effects through binding to GR, a member of the nuclear receptor superfamily of transcription factors. Indeed, treatment with the GR antagonist RU486 almost completely abolished the Dex-mediated induction of Klf9 and gluconeogenic genes in primary hepatocytes (Figure 1E). Although Dex treatment induced Klf9 gene expression, it did not significantly influence expression of Klf10 and Klf15, the 2 members of the Krüppel-like factor family that have been shown to regulate gluconeogenesis (25, 26) in hepatocytes (Supplemental Figure 1G). To further investigate the mechanism of the Dex-mediated activation of Klf9, the promoter region of Klf9 was cloned and fused to a luciferase reporter gene. The transfection of the GR expression plasmid into HepG2 cells caused marked activation of the Klf9 promoter-reporter gene. Our promoter deletion and mutation assays revealed that a potential GR response element half site (GRE1/2) in the Klf9 gene promoter mediated the stimulatory effect of Dex/GR (Supplemental Figure 1H). ChIP assays using liver extracts from ad libitum–fed and fasted C57BL/6J mice indicated that endogenous GR proteins in the fasted rather than the fed state were recruited to the proximal region, but not to the distal region, of the Klf9 promoter, suggesting that Klf9 is a direct target gene of GR in vivo (Figure 1F).
Klf9 activates the gluconeogenic program in primary hepatocytes.
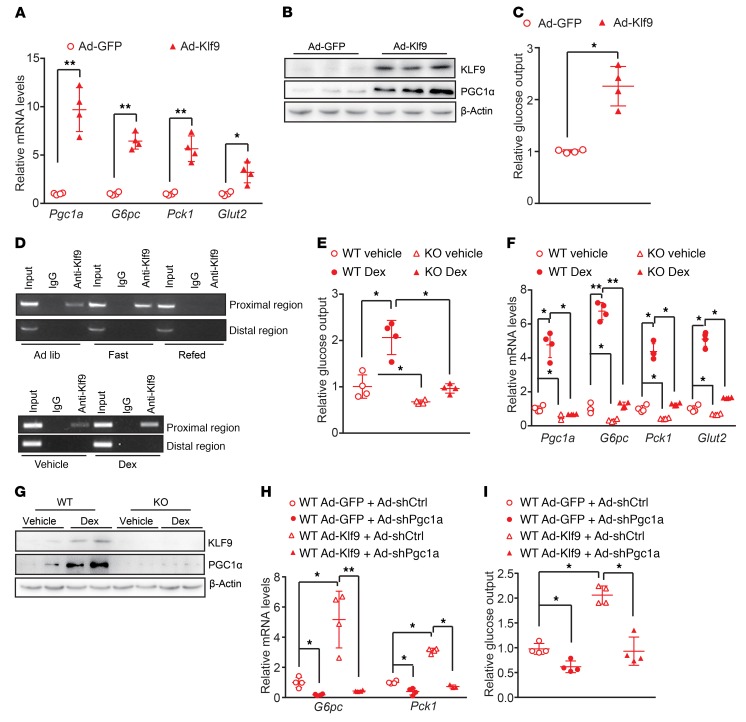
To explore the functional significance of the Dex-mediated induction of Klf9 expression, we first prepared an adenovirus expressing Klf9 (Ad-Klf9) and performed mRNA microarray analysis of primary hepatocytes infected with Ad-Klf9. The microarray data indicated that Klf9 overexpression stimulated the expression of Pgc1a and its downstream target genes involved in gluconeogenesis, fatty acid oxidation, and energy metabolism (Supplemental Figure 2, A–C). Furthermore, real-time PCR and Western blotting data confirmed that the induction of Klf9 expression by Ad-Klf9 in primary hepatocytes strongly activated the gluconeogenic program, subsequently promoting cellular glucose production (Figure 2, A–C). In contrast, Ad-Klf9 treatment of primary hepatocytes did not induce the expression of Pgc1b or other well-known factors involved in gluconeogenesis, including Foxo1, Hnf4a, Klf15, and Klf10 (Supplemental Figure 2D), indicating a specific stimulatory effect of KLF9 on PGC1α.
Figure 2. KLF9 activates the gluconeogenic program in primary hepatocytes through PGC1α.
(A) Quantitative PCR analysis showing mRNA levels of Pgc1a, G6pc, Pck1, and Glut2 in mouse primary hepatocytes infected with Ad-GFP or Ad-Klf9. Cells were harvested for further analysis 48 hours after infection. (B) Western blot analysis of KLF9 and PGC1α in primary hepatocytes treated as described in A. (C) Glucose output assay showing the effects of Klf9 overexpression on glucose production in primary hepatocytes as described in A. (D) ChIP assay performed as described in Methods, showing that both fasting (top panel) and Dex treatment (bottom panel) promote endogenous KLF9 binding to the proximal region of the Pgc1a gene promoter, but not the distal region (as a negative control). (E) Glucose output assay showing glucose production in primary hepatocytes isolated from global Klf9-mutant and WT C57BL/6 mice treated with Dex (100 nM) or saline for 12 hours. (F) Quantitative PCR analysis (left) of Pgc1a, G6pc, Pck1, and Glut2 in the primary hepatocytes described in E. (G) Representative Western blot analysis of hepatic KLF9 and PGC1α in the primary hepatocytes described in E. (H) Quantitative PCR analysis of G6pc and Pck1 in primary hepatocytes infected with the indicated adenoviruses. (I) Glucose output assay showing glucose production in the primary hepatocytes described in H. Data are represented as mean ± SEM. *P < 0.05; **P < 0.01, 2-tailed Student’s t test (A, C), 2-way ANOVA (E, F), or 1-way ANOVA (H, I).
Next, we determined whether KLF9 directly binds to and activates the Pgc1a gene promoter by examining the activity of the Pgc1a promoter. Mapping and mutation studies revealed that a potential KLF9-binding site in the Pgc1a promoter mediated the stimulatory effect of KLF9 on Pgc1a gene transcription (Supplemental Figure 2E). The occupancy of the KLF9 protein at this binding site on the Pgc1a promoter was confirmed using a ChIP assay (Figure 2D). Notably, fasting promoted the binding of endogenous KLF9 to the Pgc1a promoter in vivo, and refeeding reduced KLF9 occupancy on the Pgc1a promoter (Figure 2D). Moreover, Dex injection of mice enhanced hepatic KLF9 binding to the Pgc1a promoter (Figure 2D). These data suggest that KLF9 regulates PGC1α and the gluconeogenic program in a cell-autonomous manner.
To investigate the role of KLF9 in mediating Dex stimulation of glucose production in hepatocytes, primary hepatocytes were isolated from the livers of global Klf9-mutant mice and WT C57BL/6J mice, followed by treatment with Dex or saline (control). As a result, the treatment of WT primary hepatocytes with Dex resulted in a marked increase in cellular glucose output, whereas Klf9 mutation significantly attenuated the stimulatory effect of Dex (Figure 2E). Consistent with these effects, Klf9 mutation reduced the Dex-mediated induction of Pgc1a, Pck1, and G6pc expression (Figure 2, F and G). Of note, Klf9 mutation decreased expression of Pgc1a, Pck1, and G6pc, while the other factors involved in gluconeogenesis, including Foxo1, Hnf4a, and Klf15, were not altered in Klf9-mutant hepatocytes (Figure 2F and Supplemental Figure 2F). These data suggest an essential role of KLF9 in mediating the stimulatory effect of Dex on hepatocyte gluconeogenesis. To determine whether the stimulatory effects of KLF9 on PEPCK and G6Pase were PGC1α dependent, we generated an adenovirus encoding an shRNA specific to Pgc1a (Ad-shPGC1α). We found that the knockdown of Pgc1a significantly reduced the effects of KLF9 on the expression of G6pc and Pck1 and on cellular glucose production (Figure 2, H and I), confirming that PGC1α mediates the effects of KLF9 on cellular glucose production.
Overexpression of Klf9 in the liver stimulates gluconeogenesis in vivo.
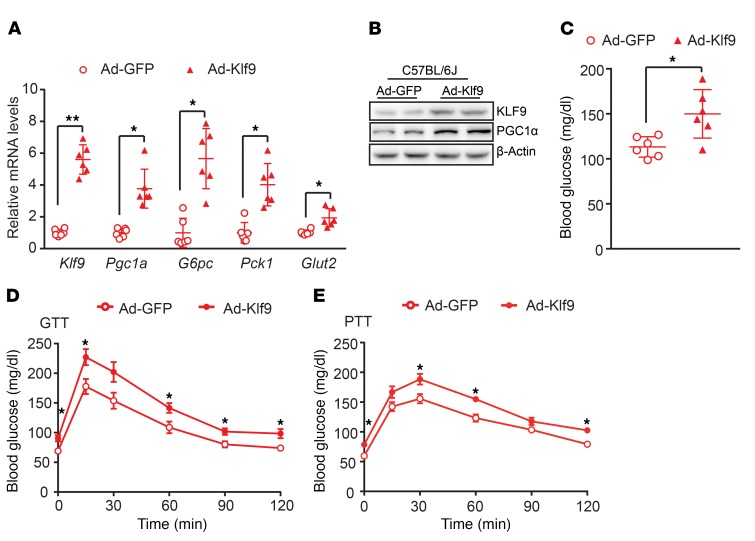
To explore the physiological function of KLF9 in the liver, we injected Ad-Klf9 into C57BL/6J mice via the tail vein. The induction of KLF9 in the liver using adenovirus markedly increased the expression of gluconeogenic genes, subsequently elevating blood glucose levels (Figure 3, A–C). Consistent with the previous study (4), endogenous PGC1α protein was recruited to the promoter regions of Pck1 and G6pc genes harboring a FOXO1-binding element. Of note, fasting enhanced PGC1α protein binding to the promoter of these genes (Supplemental Figure 3A). The glucose tolerance test (GTT) indicated that hepatic overexpression of Klf9 impaired glucose tolerance (Figure 3D). We also examined blood glucose levels in mice following i.p. injection of sodium pyruvate. The pyruvate tolerance test (PTT) indicated that hepatic gluconeogenesis was increased in Ad-Klf9–infected mice (Figure 3E). To further explore whether the effects of KLF9 on hepatic gluconeogenesis are PGC1α dependent in vivo, WT mice were injected with Ad-Klf9 in combination with Ad-shPGC1α or Ad-shCtrl (as a control). As a result, the knockdown of Pgc1a significantly reduced the stimulatory effects of KLF9 on hepatic gluconeogenic genes and blood glucose levels (Supplemental Figure 3, B and C), confirming that PGC1α is the major mediator of KLF9 action on hepatic gluconeogenesis.
Figure 3. Hepatic overexpression of Klf9 elevates blood glucose and impairs glucose tolerance in C57BL/6J mice.
(A) Quantitative PCR analysis of Klf9, Pgc1a, G6pc, Pck1, and Glut2 in the livers of C57BL/6J mice infected with Ad-GFP or Ad-Klf9 (n = 6/group). Seven days after infection, the 6 hour–fasted mice were sacrificed for further analysis. (B) Representative Western blot analysis of the hepatic KLF9 and PGC1α in mice treated as in A. (C) Blood glucose of 6 hour–fasted C57BL/6J mice treated as in A on day 7 after adenovirus injection. (D, E) Blood glucose during the GTT (D) and PTT (E) of C57BL/6J mice treated as in A (n = 6/group). Data are represented as mean ± SEM. *P < 0.05; **P < 0.01, 2-tailed Student’s t test (A, C-E).
Hepatic Klf9 deficiency in mice leads to fasting hypoglycemia.
Based on the above data, we hypothesized that endogenous KLF9 is required for maintaining blood glucose homeostasis. To test this hypothesis, we first employed global Klf9-mutant mice and WT littermates as controls (27). Klf9 mutation did not affect food intake or the weight or morphology of the liver under the feeding conditions used (data not shown). As expected, Western blot analysis failed to detect hepatic KLF9 protein in these mutant mice. The blood glucose levels in Klf9 mutant mice were comparable to those observed in the control mice on the normal chow diet (Supplemental Figure 4A). However, the hepatic mRNA and protein levels of gluconeogenic genes were decreased in Klf9 mutant mice compared with those in control mice in the fasted state (Supplemental Figure 4, B and C). Correspondingly, Klf9-mutant mice displayed lower fasting blood glucose levels (Supplemental Figure 4D). PTT experiments indicated that hepatic gluconeogenesis was decreased in Klf9-mutant mice compared with that in control mice (Supplemental Figure 4E).
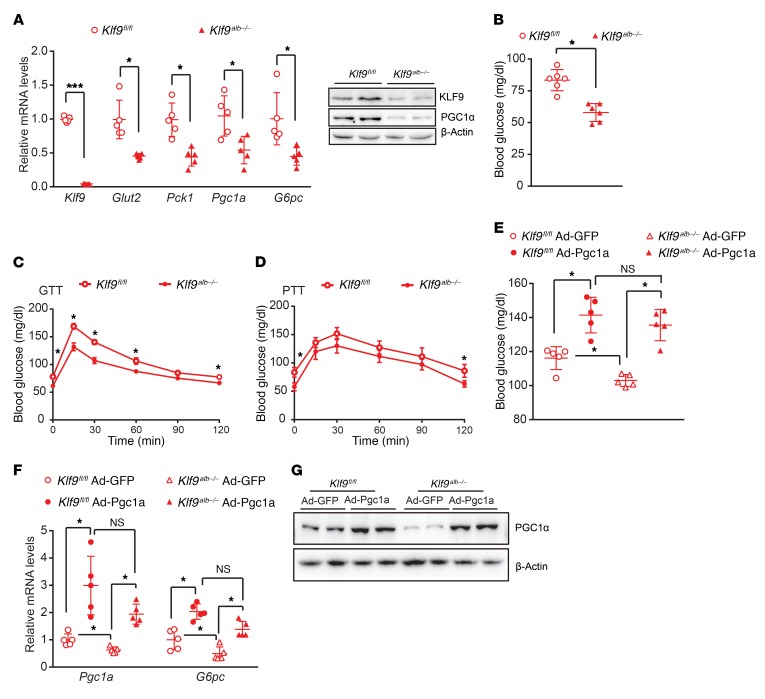
To further determine whether hepatic KLF9 deficiency contributes to fasting hypoglycemia, we first generated Klf9fl/fl mice with loxP sites flanking exon 1 of the Klf9 gene and obtained liver-specific Klf9-knockout mice by crossing Klf9fl/fl mice with Alb-Cre mice, which expressed the Cre-recombinase gene under the control of the albumin gene promoter. The resulting Alb-Cre; Klf9fl/fl mice are henceforth referred to as Klf9alb–/– mice (Supplemental Figure 5, A and B). Littermates lacking the Cre gene (homozygous Klf9fl/fl mice) were used as controls. Klf9 expression was specifically reduced in liver of Klf9alb–/– mice, while it remained unchanged in other tissues examined (Figure 4A and Supplemental Figure 5C). As a result, the Klf9alb–/– mice had decreased expression of gluconeogenic genes in the fasted state and displayed fasting hypoglycemia. GTT and PPT data indicated that the Klf9alb–/– mice had enhanced glucose tolerance and decreased hepatic gluconeogenesis (Figure 4, A–D). These data clearly suggest that hepatic KLF9 plays an essential role in regulating systemic glucose homeostasis. Furthermore, we also examined glucose metabolism of Klf9alb–/– mice fed a high-fat diet. Likewise, hepatic Klf9 deficiency also reduced expression of gluconeogenic genes in the fasted state, thereby decreasing fasting blood glucose levels, improving glucose tolerance, and lowering hepatic gluconeogenesis (Supplemental Figure 6).
Figure 4. Liver-specific Klf9-deficient mice display decreased fasting blood glucose and enhanced glucose tolerance.
(A) Quantitative PCR (left panel) and Western blot (right panel) of Klf9 and other gluconeogenic genes/proteins in the livers of 6 hour–fasted Klf9alb–/– mice and control littermates (Klf9fl/fl) at 8 to 9 weeks of age (n = 5/group). (B) Blood glucose in 6 hour–fasted Klf9fl/fl mice or Klf9alb–/– mice (n = 8/group). (C, D) Blood glucose during GTT (C) and PTT (D) of Klf9fl/fl mice or Klf9alb–/– mice (n = 6/group). (E) Blood glucose in 6 hour–fasted Klf9fl/fl mice and Klf9alb–/– mice injected with Ad-GFP or Ad-Pgc1a (n = 5/group). (F) Quantitative PCR analysis of hepatic Pgc1a and G6pc mRNA levels in mice treated as in E (n = 5/group). (G) Representative Western blot analysis of hepatic PGC1α protein levels in mice treated as in E. Data are represented as mean ± SEM. *P < 0.05; ***P < 0.001, 2-tailed Student’s t test (A-D) or 2-way ANOVA (E, F).
PGC1α lies downstream of KLF9, and hepatic Klf9 deficiency decreases Pgc1a expression. Thus, we also performed Pgc1a rescue experiments in Klf9alb–/– mice. As a result, adenovirus-mediated PGC1α overexpression increased blood glucose levels in both Klf9alb–/– and Klf9fl/fl mice, and the blood glucose levels in Ad-Pgc1a–infected Klf9alb–/– mice were comparable to those in Ad-Pgc1a–infected Klf9fl/fl mice (Figure 4E). Consistent with these effects, Pgc1a rescue reversed the decreased expression of G6pc genes in the livers of Klf9fl/fl mice (Figure 4F). These data further confirm that PGC1α mediates the stimulatory effects of KLF9 on hepatic glucose metabolism.
Mice with Klf9 deficiency in the liver display fasting-induced hepatic steatosis.
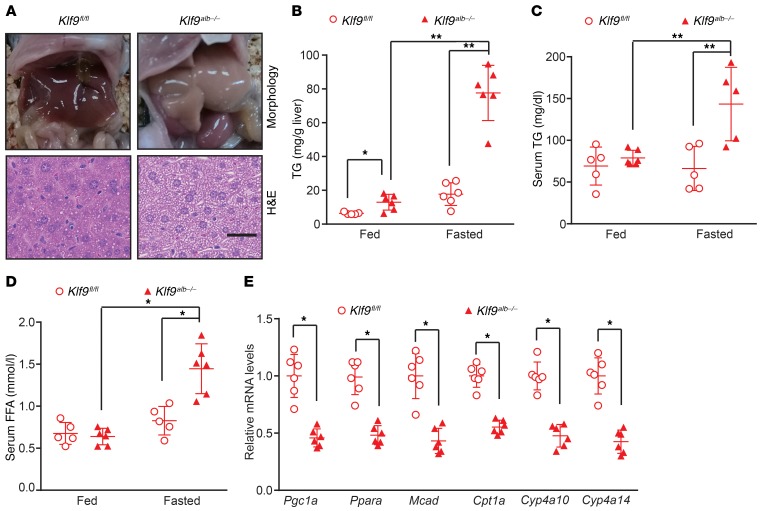
Given that PGC1α is a direct target gene of KLF9 and regulates hepatic fatty acid oxidation through coactivating PPARα in response to fasting (28–30), we next explored whether Klf9 deficiency affected hepatic lipid metabolism. Under feeding conditions, the livers of the Klf9alb–/– mice appeared grossly normal (Supplemental Figure 7A). However, after a prolonged fast (24 hours), the Klf9alb–/– mice displayed hepatic steatosis, as revealed by gross morphological changes and histological analysis (H&E staining) (Figure 5A). Biochemical analysis also revealed a significant increase in hepatic and serum triglycerides (TGs) (Figure 5, B and C). Additionally, serum free fatty acid (FFA) was increased in the Klf9alb–/– mice (Figure 5D). However, the hepatic and serum cholesterol remained unchanged (data not shown), indicating that KLF9 specifically regulated TG metabolism. We also studied the molecular mechanism responsible for the hepatic steatosis observed in the fasted Klf9alb–/– mice. We found that the mRNA levels of the target genes of Ppara, a master regulator of fatty acid oxidation, including Mcad, Cpt1a, Cyp4a10, and Cyp4a14, were decreased in the liver of the Klf9alb–/– mice (Figure 5E). Correspondingly, the ketone body levels in the serum of fasted Klf9alb–/– mice were reduced (Supplemental Figure 7B). Meanwhile, hepatic Klf9 deficiency decreased expression of lipogenic genes, including Srebp1c, Acc, and Scd1, in the fasted state (Supplemental Figure 7C). To further confirm that the decrease in Pgc1a contributes to hepatic steatosis observed in Klf9alb–/– mice, we performed Pgc1a rescue in livers of Klf9alb–/– mice. As expected, adenovirus-mediated Pgc1a overexpression reversed the decreased expression of genes involved in fatty acid oxidation, subsequently reducing hepatic TG levels in Klf9alb–/– mice (Supplemental Figure 7, D and E). We observed similar results in global Klf9-mutant mice (Supplemental Figure 8). These global Klf9-mutant mice also displayed fasting-induced hepatic steatosis and had increased TG levels in liver as well as increased serum TG and FFA levels (Supplemental Figure 8, A–D). Consistently, the expression of genes involved in fatty acid oxidation was also decreased in the Klf9-mutant mice (Supplemental Figure 8E). The changes in expression of these genes suggest that reduced fatty acid oxidation in hepatocytes may be a contributor to fasting-induced hepatic steatosis.
Figure 5. Liver-specific Klf9-deficient mice display fasting-induced hepatic steatosis.
(A) Representative gross morphology and H&E staining of livers from Klf9fl/fl mice and Klf9alb–/– mice fasted for 24 hours. (B–D) Biochemical analysis showing hepatic TG (B), serum TG (C), and FFA (D) content in ad libitum–fed or 24 hour–fasted Klf9fl/fl mice and Klf9alb–/– mice (n = 5/group). (E) Quantitative PCR analysis of Pgc1a, Ppara, Mcad, Cpt1a, Cyp4a10 and Cyp4a14 in the livers of 24 hour–fasted Klf9fl/fl mice and Klf9alb–/– mice (n = 5/group). Scale bars: 20 μm. Data are represented as mean ± SEM. *P < 0.05; **P < 0.01, 1-way ANOVA (B-D) or 2-tailed Student’s t test (E).
Hepatic Klf9 deficiency alleviates hyperglycemia induced by chronic Dex treatment.
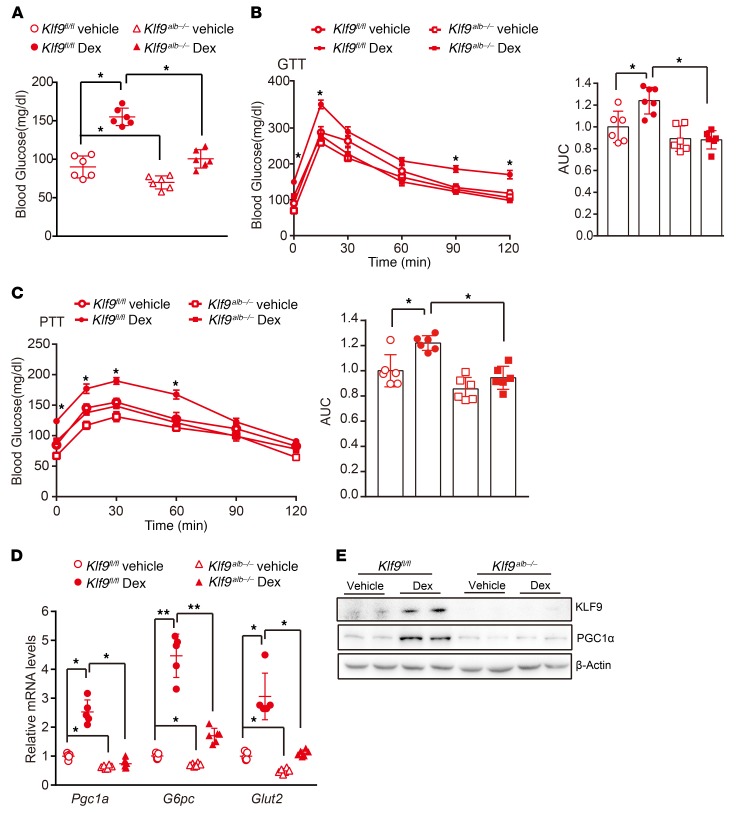
Long-term Dex therapy has serious side effects, including hyperglycemia and diabetes. To determine whether the induction of Klf9 by Dex/GR could account for this effect, Klf9alb–/– mice and Klf9fl/fl mice (control) were injected i.p. with Dex (1 mg/kg) or saline every other day for 2 months. Dex treatment did not significantly affect body weight gain or food intake (data not shown). However, long-term injection of Dex into control mice led to hyperglycemia, impaired glucose tolerance, and enhanced hepatic gluconeogenesis compared with saline treatment (Figure 6, A–C). Consistent with these findings, Dex treatment induced the expression of gluconeogenic genes in the livers of control mice (Figure 6, D and E). Strikingly, hepatic Klf9 deficiency significantly reduced the Dex-mediated induction of gluconeogenic gene expression and alleviated the glucose intolerance and hyperglycemia induced by Dex (Figure 6). These data clearly demonstrate that Klf9, at least in part, mediates the Dex/GR effects on hepatic glucose metabolism and hyperglycemia.
Figure 6. Hepatic Klf9 deficiency alleviates hyperglycemia induced by chronic Dex treatment.
(A)Blood glucose in the 16 hour–fasted Klf9fl/fl mice and Klf9alb–/– mice (n = 6/group) treated with saline or Dex (1 mg/kg) every other day for 2 months (n = 6/group). (B, C) Blood glucose during GTT (B) and PTT (C) of Klf9fl/fl mice and Klf9alb–/– mice treated as in A. (D) Quantitative PCR analysis of Pgc1a, G6pc, and Glut2 in livers of Klf9fl/fl mice and Klf9alb–/– mice treated as in A (n = 6/group). (E) Representative Western blot analysis of hepatic KLF9 and PGC1α in mice treated as in A. Data are represented as mean ± SEM. *P < 0.05; **P < 0.01, 2-way ANOVA (A-D).
Hepatic silencing of Klf9 alleviates hyperglycemia in other diabetic mice.
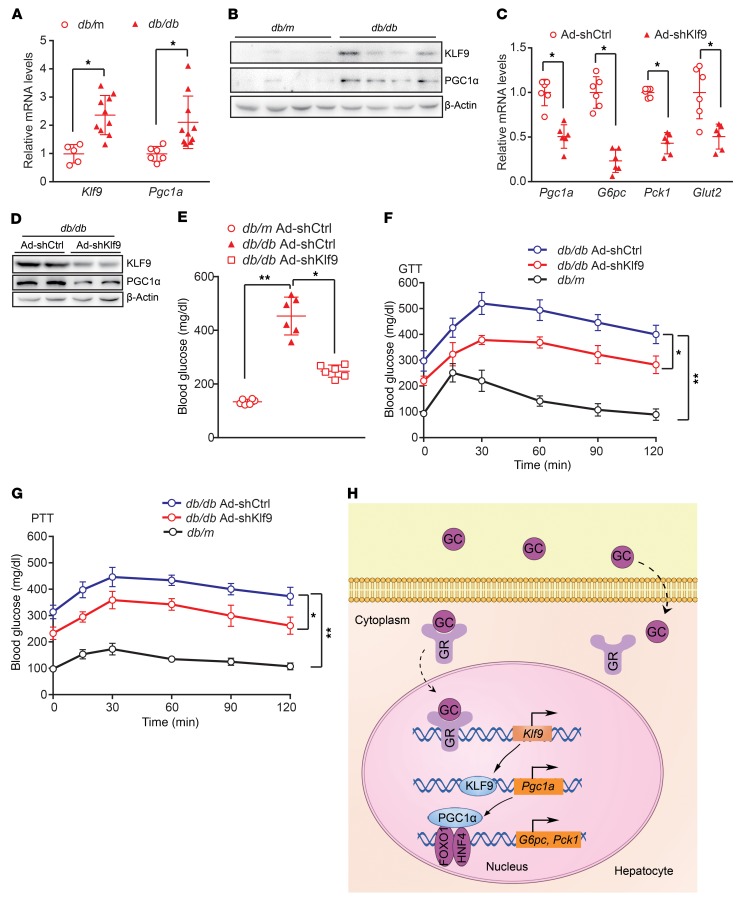
Increased hepatic gluconeogenesis contributes to hyperglycemia. Moreover, GCs are dramatically increased in various metabolic syndrome models, including db/db and ob/ob mice (31). Thus, we first examined Klf9 expression in the livers of db/db mice. Hepatic Klf9 expression was significantly increased in db/db mice compared with that in control mice (Figure 7, A and B). Next, we determined whether hepatic KLF9 also affected the diabetic phenotype in db/db mice. We generated an adenovirus encoding an shRNA specific to the Klf9 gene (Ad-shKlf9) and an adenovirus encoding an shRNA against luciferase as a control (Ad-shCtrl). Interestingly, the infusion of Ad-shKlf9 into db/db mice via the tail vein led to a decrease in the expression of Klf9 and its target genes in the liver (Figure 7, C and D), subsequently leading to a marked decrease in fasting blood glucose levels in obese db/db mice (Figure 7E). The GTT and PTT results showed that Klf9 knockdown led to an improvement in glucose intolerance and a decrease in hepatic gluconeogenesis in db/db mice (Figure 7, F and G).
Figure 7. Hepatic Klf9 knockdown decreases blood glucose in db/db mice.
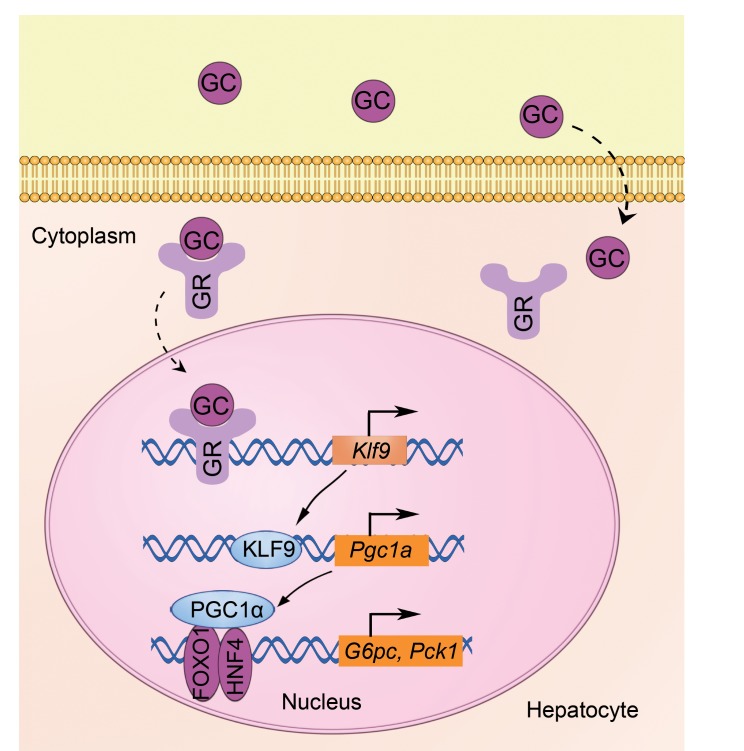
(A) Quantitative PCR analysis of hepatic Klf9 and Pgc1a in db/db or db/m mice (db/db, n = 10 mice; db/m, n = 6 mice). (B) Representative Western blot analysis of hepatic KLF9 and PGC1α in db/db or db/m mice. (C) Quantitative PCR analysis of hepatic Pgc1a, G6pc, Pck1, and Glut2 in db/db mice injected with Ad-shCtrl or Ad-shKlf9 (n = 6/group). Seven days after infection, 6 hour–fasted mice were sacrificed for further analysis. (D) Representative Western blot analysis of KLF9 and PGC1α in livers of db/db mice treated as in C. (E) Blood glucose of 6 hour–fasted db/db or db/m mice treated as in C (n = 6/group). (F–G) Blood glucose during GTT (F) and PTT (G) in db/db mice infected with Ad-shCtrl or Ad-shKlf9 or db/m mice (n = 6/group). (H) Proposed model of GC induction of hepatic gluconeogenesis and hyperglycemia. GCs are diffused into hepatocytes, where GCs bind to GRs in the cytosol. The GC/GR complex translocates into the nucleus to activate Klf9 gene transcription, which in turn promotes Pgc1a expression, thereby activating the hepatic gluconeogenic program and leading to hyperglycemia. Data are represented as mean ± SEM. *P < 0.05; **P < 0.01, 2-tailed Student’s t test (A, C) or 1-way ANOVA (E-G)
These experiments were broadened to ob/ob mice, and similar results were observed. The knockdown of Klf9 in the livers of ob/ob mice also significantly decreased the expression of Klf9 and its downstream genes, subsequently decreasing blood glucose levels (Supplemental Figure 9, A–C), improving glucose intolerance (GTT data), and decreasing hepatic gluconeogenesis (PTT data) (Supplemental Figure 9, D and E). These results suggest that hepatic Klf9 knockdown in diabetic mice reduces hepatic gluconeogenesis, eventually resulting in reduced blood glucose levels.
Based on previous reports and the results obtained in the present study, we propose a model for the regulation of hepatic gluconeogenesis by GCs. GCs promote hepatic gluconeogenesis through a GC/GR/KLF9/PGC1α/gluconeogenic gene signaling pathway (Figure 7H).
Discussion
GCs can induce hepatic gluconeogenesis, insulin resistance, and hyperglycemia (14, 32–34). However, the molecular mechanisms responsible for these effects of GCs are not well understood. Dex stimulates Pck1 gene expression (35–37). The GC response of the Pck1 gene is mediated through a complex GC response unit (GRU), which comprises a tandem array of 3 accessory factor elements (AF1–3; HNF4α binds to AF1 and AF3 element; FOXO1 binds to AF2) and 2 nonconsensus GR-binding elements (GR1–2) (3, 4, 31, 35). However, GR1 and GR2 have little intrinsic activity in the context of the Pck1 gene promoter or a heterologous promoter. Moreover, both of these nonconsensus elements bind to GR in vitro with low affinity (36–38). Notably, the mutation of any of the accessory factor elements results in at least a 50% reduction of GC-stimulated Pck1 gene transcription. Any combination of 2 mutations in AF elements nearly abolishes the response (36, 38, 39). These findings suggest that HNF4α and FOXO1 are required for the Dex-mediated induction of Pck1. Interestingly, all of these transcription factors, including FOXO1, HNF4α, and GR, can be coactivated by PGC1α (3, 4); thus, it is logical to speculate that PGC1α may mediate the effect of Dex on the induction of the Pck1 gene. However, no GRE is identified in the Pgc1a gene promoter (3, 40, 41), indicating that Dex induces PGC1α via an indirect mechanism. In this study, we show that Dex induces expression of the Klf9 gene, which in turn activates Pgc1a and gluconeogenic genes. Thus, our results clarify the molecular mechanism by which Dex induces the Pck1 gene and clearly explain why both FOXO1 and HNF4 are required for this process, although expression of FOXO1 and HNF4 are not altered by Klf9 overexpression or Dex treatment.
In this study, we also assessed the molecular mechanism of Dex-mediated activation of Klf9 expression and showed that Dex/GR activates Klf9 gene transcription through direct binding to the Klf9 gene promoter. Notably, we also observed that the mutation of the identified GRE1/2 in the Klf9 promoter did not completely abolish the stimulatory effects of Dex/GR on reporter activity, implying that more than one GRE exists in the Klf9 promoter or that other transcription factors mediate the effects of Dex/GR on Klf9 expression. We further confirmed that KLF9 directly bound to and activated the Pgc1a gene promoter. Based on previous reports and the data obtained in the present study, we propose that Dex activates gluconeogenic genes through the Dex/GR/KLF9/PGC1α signaling pathway.
In addition to KLF9, the other 2 members of the KLF family, KLF10 and KLF15, have been shown to regulate hepatic gluconeogenesis (25, 26). KLF10 was reported to repress gluconeogenesis (male global Klf10-KO mice displayed postprandial and fasting hyperglycemia [ref. 42], suggesting that KLF9 and KLF10 have the opposite role in the case of gluconeogenesis). Additionally, Gray et al. reported that KLF15 regulates gluconeogenesis (25). However, KLF15 regulates gluconeogenesis through modulating expression of genes encoding amino acid–degrading enzyme, such as alanine aminotransferase 1 (ALT1), proline dehydrogenase (ProDH), tryptophan 2,3-dioxygenase (TDO2), and others, to control gluconeogenic substrate availability. Remarkably, KLF15 does not regulate expression of gluconeogenic genes, including Pck1 and G6pc (25). Thus, although both KLF9 and KLF15 regulate hepatic gluconeogenesis, they exert their physiological function through completely different molecular mechanism. Asada et al. reported that Dex promotes adipogenic differentiation of mesenchymal stem cells and mouse embryonic fibroblasts via induction of Klf15 (42). However, in the present study, we did not observe that Dex treatment induces Klf15 significantly in primary hepatocytes, indicating that Dex induction of Klf15 is cell type specific. Notably, Dex still modestly induced expression of G6pc and Pck1 in Klf9-knockout hepatocytes, although the induction extent in Klf9-knockout hepatocytes is less than that in WT hepatocytes. These results suggest that KLF9 partially mediates Dex induction of these gluconeogenic genes and that other factors might also play a role in this physiological process. In addition to Dex/GR, Klf9, also known as Bteb1, might be regulated by other physiological processes, such as exercise (43). Likewise, Klf9 overexpression still modestly stimulated expression of these gluconeogenic genes and glucose production in the hepatocytes or liver with Pgc1a knockdown. These results indicate that factors other than PGC1α also mediate KLF9 effects on gluconeogenesis. The other possibility is that KLF9 might also directly activate transcription of G6pc and Pck1 genes. Further studies are required to clarify this question.
Morita et al. and Simmen et al. reported that Klf9 mutant mice exhibited deficits in development (27, 44). However, they did not examine the metabolic phenotype in these mutant mice. In the present study, we show that global Klf9-mutant mice display fasting hypoglycemia. Liver-specific Klf9-deleted mice also exhibit fasting hypoglycemia, and hepatic Klf9 deficiency alleviates Dex induction of hyperglycemia. These results clearly demonstrate that hepatic Klf9 deficiency leads to a defect in gluconeogenesis and contributes to fasting hypoglycemia, and hepatic KLF9 specifically mediates the stimulatory effects of Dex on gluconeogenesis. Interestingly, Opherk et al. reported that mice with hepatic deletion of the GR exhibited fasting hypoglycemia (32), a metabolic phenotype similar to that observed in our liver-specific Klf9-deleted mice, also suggesting that KLF9 acts as a mediator of GC/GR signaling. In this study, we show that Dex-induced KLF9 plays an important role in regulating hepatic gluconeogenesis. Of note, we studied glucose metabolism of mouse models used in the present study with traditional GTT and PTT experiments. Euglycemic-hyperinsulinemic clamp experiments are required to further confirm our results.
Although KLF9 inhibition would improve hyperglycemia in insulin-resistant and diabetic states, it might exacerbate fatty liver due to possibly reduced fatty acid oxidation and mitochondrial oxidative phosphorylation. Consistently, Satapati et al. also reported that inhibition of gluconeogenesis prevents fatty acid–induced oxidative metabolism and stress (45). Based on our study, we propose that GC stimulates Klf9 expression and hepatic gluconeogenesis, thereby leading to hyperglycemia. Theoretically, stimulation of Klf9 mediated by GC should alleviate fatty liver through promoting fatty acid oxidation. However, in reality, long-term treatment of mice with GC leads to fatty liver. One possible molecular mechanism is that GC induces hepatic steatosis via interference with Hes1 gene expression (33). Thus, GC induces hyperglycemia and fatty liver through distinct molecular mechanisms.
Additionally, our preliminary data (insulin tolerance test [ITT]) indicates that alteration of Klf9 expression in livers of WT mice or db/db mice led to a change in insulin sensitivity. Pgc1a, the direct target gene of KLF9, has been reported to remarkably promote insulin resistance in liver through the induction of TRB3 (28). Thus, we speculated that KLF9 might regulate insulin resistance through the KLF9/PGC1α/TRB-3 signaling pathway. More evidence is required to confirm this hypothesis. Notably, the adenovirus-mediated knockdown of hepatic Pgc1a in WT and db/db diabetic mice reduced fasting blood glucose and increased hepatic TGs (28). Consistent with these findings, Pgc1a-knockout mice displayed fasting hypoglycemia and hepatic steatosis (29, 46), which virtually phenocopied the Klf9-mutant mice, underscoring the role of PGC1α in mediating the effects of KLF9 on glucose and lipid metabolism. Although KLF9 activated PGC1α in the liver and both Klf9-deficient and Pgc1a-knockout mice displayed fasting hypoglycemia, the biological functions of KLF9 and PGC1α are not completely consistent. For example, Zucker et al. reported that KLF9 increases ROS levels in various types of cells and mouse tissues. Klf9-deficient mice display resistance to bleomycin-induced oxidative stress (47). In contrast, PGC1α protects neural cells from oxidative stress–mediated death. Pgc1a-null mice are much more sensitive to the neurodegenerative effects of oxidative stress (48). Moreover, despite having decreased mitochondrial function, Pgc1a-deficient mice are paradoxically lean, which is attributed to a profound hyperactivity of these mutant mice associated with lesions in the striatal region of the brain (46), although hyperactivity is not observed in the global Klf9-mutant mice. In sharp contrast, global Klf9-mutant mice become obese with age (our unpublished observations). The molecular mechanisms underlying these discrepancies warrant further investigation.
Aberrant hepatic gluconeogenesis contributes to insulin resistance and diabetes. Thus, the inhibition of gluconeogenesis is an attractive therapeutic strategy for the treatment of diabetes. Our data identify the hepatic activation of KLF9 as a mechanism underlying GC-induced hepatic gluconeogenesis and diabetes. KLF9 may serve as a target for the treatment of type 2 diabetes, especially GC therapy–driven diabetes.
Methods
Animal treatment.
Klf9fl/fl mice were generated by the CRISPR/Cas9 system at Biocytogen. Briefly, eight 5′ sgRNA and eight 3′ sgRNA were designed by the CRISPR design tool (http://crispr.mit.edu) to target a region upstream and downstream of exon 1, respectively, and then were screened for on-target activity using a Universal CRISPR Activity Assay (UCA) (Biocytogen). Based on an sgRNA activity assay, 2 sgRNAs (sgRNA3: 5′-TATGTACAATAGTGTATAGC-3′; sgRNA16: 5′-ACCACCAGAGCTAGTTGACC-3′) were selected. The T7 promoter sequence was added to the Cas9 or sgRNA template by PCR amplification in vitro. Circular targeting vector containing exon 1 flanked by 2 LoxP sites and 2 homologous arms was mixed with Cas9 mRNA and sgRNAs and then comicroinjected into the cytoplasm of 1-cell stage fertilized C57BL/6N eggs. The injected zygotes were transferred into oviducts of Kunming pesudopregnant females to generate F0 mice. F0 mice with the expected genotype confirmed by tail genomic DNA PCR and sequencing were mated with C57BL/6N mice to establish germline-transmitted F1 founders. F1 founders were genotyped by tail genomic PCR/DNA sequencing, and Southern blot examination was performed to further confirm correct genotype.
Albumin-Cre mice were provided by Hongbing Zhang (Institute of Basic Medical Sciences, Peking Union Medical College). To generate hepatic-specific Klf9-knockout mice, Klf9fl/fl mice were crossed with Albumin-Cre, Klf9fl/– mice to obtain Albumin-Cre, Klf9fl/fl mice (called Klf9alb–/–). The Klf9fl/fl littermates were used as controls. Global Klf9-mutant mice were obtained from the Jackson Laboratory (catalog 012909). Male 8- to 12-week-old C57BL/6J, ob/ob, and db/db mice were purchased from the Model Animal Research Center of Nanjing University (Nanjing, China) and were housed and maintained in 12-hour light/12-hour dark photoperiods. For the viral injections, 1 to 2 × 109 PFU per recombinant virus were administered by tail vein injection into the C57BL/6J, ob/ob, and db/db mice. For chronic Dex injections, 8-week-old Klf9fl/fl and Klf9alb–/– male mice received i.p. injections of Dex (1 mg/kg/every other day) or saline for 2 months.
Preparation of recombinant adenoviruses.
Adenoviruses expressing Klf9 and Pgc1a were prepared as previously described (49). Briefly, the full-length mouse Klf9 or Pgc1a gene was first cloned into the pAd-Track-CMV shuttle vector. The resultant plasmid was linearized by digesting with restriction endonuclease PmeI and subsequently transformed into competent cells, which were BJ5183 derivatives containing the adenoviral backbone plasmid pAdEasy-1. Recombinants were selected for kanamycin resistance, and recombination was confirmed by restriction endonuclease analyses. Overall, the confirmed recombinant adenovirus plasmids were digested with PacI to liberate both inverted terminal repeats and transfected into HEK-293A cells. Recombinant adenoviruses were typically generated within 14 to 20 days.
For RNA interference, oligonucleotides targeting mouse Pgc1a (5′-GGTGGATTGAAGTGGTGTAGA-3′) or luciferase (5′-CTTACGCTGAGTACTTCGA-3′) were annealed and cloned into the pAd-Track-U6 vector. Adenoviruses expressing shRNA against luciferase and Pgc1a were prepared as above. Adenoviruses were purified by the cesium and dialyzed against PBS buffer containing 10% glycerol.
Isolation and culture of mouse primary hepatocytes.
For hepatocyte preparation, mice were anesthetized with an i.p. injection of bromethol. Livers were subjected to 2-step collagenase perfusion through the portal vein, as previously described (50). First, 50 ml of Ca2+-free/Mg2+-free HBSS supplemented with 2 mM EGTA, 20 mM Hepes, and 10 mM NaHCO3, pH 7.4, was infused with a pump at a flow rate of 7 ml/min for 6 minutes. Then the livers were perfused with HBSS containing 0.05% (w/v) collagenase type II (MilliporeSigma) at the same flow rate for 5 minutes. The HBSS was preinfused with 95% O2 and 5% CO2 and kept in a 37°C water bath. After the 2-step perfusion, livers were excised and hepatocytes were flushed out and filtered through a 70 μm pore size mesh nylon filter. Cells were washed 3 times with ice-cold HBSS and centrifuged at 50 g for 2 minutes at 4°C. The cells were more than 80% viable, as evaluated by Trypan blue exclusion. Hepatocytes were seeded at 107 cells/10 cm2 on rat tail collagen-coated plates and cultured with RPMI 1640 medium (H10394; Invitrogen) containing 10% (v/v) FBS, 50 units/ml penicillin, and 50 μg/ml streptomycin. After 4 hours, unattached cells were washed away with PBS and fresh medium was added for an overnight incubation.
Adenoviral infections.
Six to twelve hours after attachment, primary hepatocytes were infected with indicated adenoviruses (1.0 × 107 infection particles per cell of a 6-well plate) for 48 hours in RPMI 1640 medium (containing 10%FBS, 50 units/ml penicillin, and 50 μg/ml streptomycin). Forty-eight hours after infection, cells were harvested for total RNA or protein extraction.
Tolerance tests.
For the GTT, the mice were fasted for 16 hours and injected i.p. with glucose (1 g/kg). For PTT, the mice were fasted for 16 hours and injected i.p. with 1.5 g/kg sodium pyruvate. The blood glucose concentrations were measured from the tail blood at the indicated times.
Metabolites.
Serum concentrations of TG, cholesterol, and FFAs were determined using an automated Monarch device (Peking Union Medical College Hospital). Blood glucose levels were monitored from tail vein blood using a glucose monitor (OneTouch; LifeScan Inc.). Hepatic concentrations of triacylglycerol and total cholesterol were measured using a colorimetric diagnostic kit (Applygen Technologies E1013). Serum ketone body concentrations were determined by EnzyChrom Ketone Body Assay Kit (BioAssay Systems, EKBD-100).
Quantitative PCR.
Total RNA from either the mouse livers or the primary hepatocytes was extracted using the TRIzol-based method (Invitrogen). cDNA was prepared using the Applied Biosystems High-Capacity cDNA Reverse-Transcription Kit. Quantitative PCR was performed using SYBR Green PCR Master <ix (Promega) on a Bio-Rad C1000 thermal cycler CFX96 Real-Time System. All PCR data were normalized to 36B4 expression in the corresponding samples. Sequences of primers are shown in Supplemental Table 1.
ChIP assay.
The mouse livers were homogenized and treated with 1% formaldehyde at room temperature for 10 minutes and lysed with ChIP cell lysis buffer (10 mM Tris-HCl, pH 8.0, 10 mM NaCl, 3 mM MgCl2, 0.5% NP-40, and protease inhibitor cocktail) and ChIP nuclear lysis buffer (50 mM Tris-HCl, pH 8.0, 5 mM EDTA, 1% SDS, and protease inhibitor cocktail). Cell lysates were sonicated to shear the chromatin and immunoprecipitated with antibodies specific for KLF9 (Abcam, catalog ab227920), GR (MilliporeSigma, catalog SAB4501309) or unspecific IgG (Santa Cruz Biotechnology Inc., catalog sc-2027). The immunoprecipitates were isolated using protein A/G–agarose beads (Invitrogen), and these were washed and eluted with1% SDS in 0.1 M NaHCO3. After reversing the crosslinks by incubation at 65°C overnight and using proteinase K digestion, the immunoprecipitated DNA fragments and input DNA were recovered by a PCR purification kit (QIAGEN). The purified DNA was used to amplify the GR and Klf9 regulatory element on the mouse Klf9 and Pgc1a promoters, respectively. ChIP assay was performed as described above. The ad libitum–fed or 24 hour–fasted liver lysates were immunoprecipitated with antibodies specific for PGC1α or unspecific IgG; subsequently, the purified DNA was used to amplify the mouse Pck1 and G6pc promoters, respectively. PCR products were visualized by electrophoresis on an agarose gel containing ethidium bromide under UV light. Primers directed at upstream or downstream of the binding site were used as negative controls. Specific primers used are shown in Supplemental Table 2.
Protein analysis.
Proteins were extracted from tissue samples or cultured hepatocytes in lysis buffer (20 mM Tris-Cl pH 7.5, 140 mM NaCl, 1 mM CaCl2 and MgCl2, 10 mM NaF, 1% NP-40, 10% glycerol, 2 mM Na-Vanadate, and 1 mM PMSF) supplemented with cOmplete Protease Inhibitor Cocktail (Roche). Protein extracts were subjected to 10% SDS-polyacrylamide gel and blotted on PVDF membranes. Immunoblotting was performed using the following primary antibodies: KLF9 (ABclonal, catalog A7196), PGC1α (MilliporeSigma, catalog AB3242), β-actin (ABclonal, catalog AC026).
Glucose output assay.
Primary hepatocytes were cultured in 6-well plates in RPMI 1640 media (Invitrogen) containing 10% FBS) After 6-12 hr of attachment, cells were treated with Dex (100 nM) or saline (control) for 12 hours, or infected with indicated Ad-Klf9 or Ad-GFP (control) for 48 hours. The medium was then replaced with 1.5 ml of glucose production buffer (GPB) consisting of glucose-free DMEM (pH 7.4) without phenol red, supplemented with 20 mM sodium lactate and 2 mM sodium pyruvate and incubated for 3 hours at 37°C. The GPB was collected and the glucose concentration was measured using a colorimetric glucose assay kit (MilliporeSigma). The total amount of the final glucose production was corrected for protein content.
Histology analysis.
For H&E staining, liver tissues were fixed in 10% neutral-buffered formalin, embedded in paraffin, and cut into 5 μm sections.
Preparation of the expression plasmids.
Full-length mouse Klf9 (GenBank NM_010638) was obtained by PCR amplification of the cDNA from mouse liver and cloned into pcDNA3.1+ with EcoRI/XhoI restriction sites. The full-length human GR (GenBank NM_000176) was cloned into pcDNA4-myc/His with BamHI/XhoI restriction sites. All constructs were confirmed using direct nucleotide sequencing, and the primers used for plasmid construction are shown in Supplemental Table 3.
Construction of the reporter gene plasmids.
The 5′ end of the mouse Klf9 gene extending from position −1771 bp (relative to the transcription start site) to +220 was cloned into the pGL3-Basic (Promega) luciferase reporter plasmid with the MluI/XhoI sites. The 5′ deletion and corresponding mutation constructs of Klf9 (−944Luc and mut) were prepared by PCR using −1771Luc as a template. The mouse Pgc1a gene promoter (−729 to +82 bp) was amplified using mouse genomic DNA and inserted into pGL3-Basic (−729Luc). A series of 5′ deletions and corresponding mutation constructs of Pgc1a(−464Luc, −174Luc, +68Luc, mut) were prepared by PCR using −729Luc as a template. The primers used for plasmid construction are shown in Supplemental Table 3.
Transient transfection and luciferase assays.
The HepG2 cells were grown in 24-well plates using DMEM (Gibco/BRL, Thermo Fisher Scientific, 37°C, 5% CO2) containing 10% FBS. The cells were cotransfected with each of the indicated luciferase reporter and expression plasmids using Lipofectamine TM 2000 (Invitrogen) according to the manufacturer’s recommendations. Cotranfections were performed with a constant amount of DNA by adding the empty pcDNA3.1. A Renilla luciferase expression vector, pCMV-RL-TK, was used as an internal control to adjust for the transfection efficiency. The luciferase activity was measured at 48 hours after transfection using the Dual Luciferase Reporter Assay System (Promega).
See complete unedited blots in the supplemental material.
Statistics.
The quantitative data are represented as the mean ± SEM of 3 independent experiments. In most of the cases for in vivo experiments in mice, n = 5 was the minimum amount used. Two-tailed, unpaired Student’s t test was used for pairwise comparison of genotypes or treatments. One-way ANOVA and 2-ANOVA were used when comparing 3 or more groups, as indicated in the figure legends and elsewhere. Analysis was performed using Microsoft Excel and/or GraphPad Prism. P < 0.05 was considered significant.
Study approval.
All animal experiments were conducted under protocols approved by the Institutional Animal Care, Use, and Welfare Committee, Institute of Basic Medical Sciences, Chinese Academy of Medical Sciences and Peking Union Medical College.
Author contributions
YC, AC, HF, QC, and JY designed research studies. AC, HF, Yinliang Zhang, and Yujie Zhang conducted experiments. AC, HF, Yinliang Zhang, Yujie Zhang, DN, SL, LS, YL, HZ, YX, and YC acquired data. AC, HF, Yinliang Zhang, Yujie Zhang, WM, JY, QC, YC, QL, QW, ZS, XX, and FF analyzed data. YC, ZS, and QW provided reagents. YC wrote the manuscript.
Supplementary Material
Acknowledgments
We thank Teresa C. Leone (Center for Cardiovascular Research, Washington University School of Medicine, St. Louis, Missouri, USA) for providing the Pgc1a-Myc plasmid. This work was supported by the National Natural Science Foundation of China (grants 81730024, 81825004, 81670749, 81570518, and 81471049) and the National Key Research and Development Program of China (2016YFC1304802).
Version 1. 04/29/2019
Electronic publication
Version 2. 06/03/2019
Print issue publication
Footnotes
AC’s present address is: Basic Medical College, Jining Medical University, Jining, China.
HF’s present address is: Institute of Human Stem Cells, General Hospital of Ningxia Medical University, Ningxia, China.
WM’s present address is: Central Laboratory, Navy General Hospital of PLA, Beijing, China.
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2019, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2019;129(6):2266–2278.https://doi.org/10.1172/JCI66062.
See the related Commentary at Taking KLF9 to “Cort” for crimes against metabolism.
Contributor Information
Anfang Cui, Email: caf911@126.com.
Heng Fan, Email: 80768383@qq.com.
Yinliang Zhang, Email: zhangyinliang8@foxmail.com.
Yujie Zhang, Email: 1269788276@qq.com.
Dong Niu, Email: pumcniudong@gmail.com.
Shuainan Liu, Email: liusn@imm.ac.cn.
Quan Liu, Email: popliu@imm.ac.cn.
Wei Ma, Email: langmawei@hsc.pku.edu.cn.
Zhufang Shen, Email: shenzhf@imm.ac.cn.
Lian Shen, Email: shenlian881@126.com.
Yanling Liu, Email: liuyanling938@126.com.
Huabing Zhang, Email: slzhang1977@163.com.
Yuan Xue, Email: xueyuan2010@gmail.com.
Ying Cui, Email: cuiying119@163.com.
Qinghua Wang, Email: wangq@smh.ca.
Xinhua Xiao, Email: Xiaoxh2014@vip.163.com.
Fude Fang, Email: fangfudepumc@126.com.
Jichun Yang, Email: yangj@bjmu.edu.cn.
Qinghua Cui, Email: cuiqinghua@bjmu.edu.cn.
Yongsheng Chang, Email: changy@ibms.pumc.edu.cn.
References
- 1.Saltiel AR, Kahn CR. Insulin signalling and the regulation of glucose and lipid metabolism. Nature. 2001;414(6865):799–806. doi: 10.1038/414799a. [DOI] [PubMed] [Google Scholar]
- 2.Flier JS. Obesity wars: molecular progress confronts an expanding epidemic. Cell. 2004;116(2):337–350. doi: 10.1016/S0092-8674(03)01081-X. [DOI] [PubMed] [Google Scholar]
- 3.Yoon JC, et al. Control of hepatic gluconeogenesis through the transcriptional coactivator PGC-1. Nature. 2001;413(6852):131–138. doi: 10.1038/35093050. [DOI] [PubMed] [Google Scholar]
- 4.Puigserver P, et al. Insulin-regulated hepatic gluconeogenesis through FOXO1-PGC-1alpha interaction. Nature. 2003;423(6939):550–555. doi: 10.1038/nature01667. [DOI] [PubMed] [Google Scholar]
- 5.Herzig S, et al. CREB regulates hepatic gluconeogenesis through the coactivator PGC-1. Nature. 2001;413(6852):179–183. doi: 10.1038/35093131. [DOI] [PubMed] [Google Scholar]
- 6.Koo SH, et al. The CREB coactivator TORC2 is a key regulator of fasting glucose metabolism. Nature. 2005;437(7062):1109–1111. doi: 10.1038/nature03967. [DOI] [PubMed] [Google Scholar]
- 7.Saltiel AR. New perspectives into the molecular pathogenesis and treatment of type 2 diabetes. Cell. 2001;104(4):517–529. doi: 10.1016/S0092-8674(01)00239-2. [DOI] [PubMed] [Google Scholar]
- 8.Li X, Monks B, Ge Q, Birnbaum MJ. Akt/PKB regulates hepatic metabolism by directly inhibiting PGC-1alpha transcription coactivator. Nature. 2007;447(7147):1012–1016. doi: 10.1038/nature05861. [DOI] [PubMed] [Google Scholar]
- 9.Brunet A, et al. Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell. 1999;96(6):857–868. doi: 10.1016/S0092-8674(00)80595-4. [DOI] [PubMed] [Google Scholar]
- 10.Nakae J, Park BC, Accili D. Insulin stimulates phosphorylation of the forkhead transcription factor FKHR on serine 253 through a Wortmannin-sensitive pathway. J Biol Chem. 1999;274(23):15982–15985. doi: 10.1074/jbc.274.23.15982. [DOI] [PubMed] [Google Scholar]
- 11.Dentin R, et al. Insulin modulates gluconeogenesis by inhibition of the coactivator TORC2. Nature. 2007;449(7160):366–369. doi: 10.1038/nature06128. [DOI] [PubMed] [Google Scholar]
- 12.Dong XC, et al. Inactivation of hepatic Foxo1 by insulin signaling is required for adaptive nutrient homeostasis and endocrine growth regulation. Cell Metab. 2008;8(1):65–76. doi: 10.1016/j.cmet.2008.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Matsumoto M, Pocai A, Rossetti L, Depinho RA, Accili D. Impaired regulation of hepatic glucose production in mice lacking the forkhead transcription factor Foxo1 in liver. Cell Metab. 2007;6(3):208–216. doi: 10.1016/j.cmet.2007.08.006. [DOI] [PubMed] [Google Scholar]
- 14.Bernal-Mizrachi C, et al. Dexamethasone induction of hypertension and diabetes is PPAR-alpha dependent in LDL receptor-null mice. Nat Med. 2003;9(8):1069–1075. doi: 10.1038/nm898. [DOI] [PubMed] [Google Scholar]
- 15.Vegiopoulos A, Herzig S. Glucocorticoids, metabolism and metabolic diseases. Mol Cell Endocrinol. 2007;275(1-2):43–61. doi: 10.1016/j.mce.2007.05.015. [DOI] [PubMed] [Google Scholar]
- 16.Phillips DI, et al. Elevated plasma cortisol concentrations: a link between low birth weight and the insulin resistance syndrome? J Clin Endocrinol Metab. 1998;83(3):757–760. doi: 10.1210/jcem.83.3.4634. [DOI] [PubMed] [Google Scholar]
- 17.Reynolds RM, Walker BR, Syddall HE, Whorwood CB, Wood PJ, Phillips DI. Elevated plasma cortisol in glucose-intolerant men: differences in responses to glucose and habituation to venepuncture. J Clin Endocrinol Metab. 2001;86(3):1149–1153. doi: 10.1210/jcem.86.3.7300. [DOI] [PubMed] [Google Scholar]
- 18.Rosmond R, Björntorp P. The hypothalamic-pituitary-adrenal axis activity as a predictor of cardiovascular disease, type 2 diabetes and stroke. J Intern Med. 2000;247(2):188–197. doi: 10.1046/j.1365-2796.2000.00603.x. [DOI] [PubMed] [Google Scholar]
- 19.Andrews RC, Walker BR. Glucocorticoids and insulin resistance: old hormones, new targets. Clin Sci. 1999;96(5):513–523. doi: 10.1042/cs0960513. [DOI] [PubMed] [Google Scholar]
- 20.Imataka H, et al. Two regulatory proteins that bind to the basic transcription element (BTE), a GC box sequence in the promoter region of the rat P-4501A1 gene. EMBO J. 1992;11(10):3663–3671. doi: 10.1002/j.1460-2075.1992.tb05451.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Denver RJ, Ouellet L, Furling D, Kobayashi A, Fujii-Kuriyama Y, Puymirat J. Basic transcription element-binding protein (BTEB) is a thyroid hormone-regulated gene in the developing central nervous system. Evidence for a role in neurite outgrowth. J Biol Chem. 1999;274(33):23128–23134. doi: 10.1074/jbc.274.33.23128. [DOI] [PubMed] [Google Scholar]
- 22.Cayrou C, Denver RJ, Puymirat J. Suppression of the basic transcription element-binding protein in brain neuronal cultures inhibits thyroid hormone-induced neurite branching. Endocrinology. 2002;143(6):2242–2249. doi: 10.1210/endo.143.6.8856. [DOI] [PubMed] [Google Scholar]
- 23.Bonett RM, Hu F, Bagamasbad P, Denver RJ. Stressor and glucocorticoid-dependent induction of the immediate early gene kruppel-like factor 9: implications for neural development and plasticity. Endocrinology. 2009;150(4):1757–1765. doi: 10.1210/en.2008-1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pei H, Yao Y, Yang Y, Liao K, Wu JR. Krüppel-like factor KLF9 regulates PPARγ transactivation at the middle stage of adipogenesis. Cell Death Differ. 2011;18(2):315–327. doi: 10.1038/cdd.2010.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gray S, et al. Regulation of gluconeogenesis by Krüppel-like factor 15. Cell Metab. 2007;5(4):305–312. doi: 10.1016/j.cmet.2007.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guillaumond F, et al. Kruppel-like factor KLF10 is a link between the circadian clock and metabolism in liver. Mol Cell Biol. 2010;30(12):3059–3070. doi: 10.1128/MCB.01141-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morita M, et al. Functional analysis of basic transcription element binding protein by gene targeting technology. Mol Cell Biol. 2003;23(7):2489–2500. doi: 10.1128/MCB.23.7.2489-2500.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koo SH, et al. PGC-1 promotes insulin resistance in liver through PPAR-alpha-dependent induction of TRB-3. Nat Med. 2004;10(5):530–534. doi: 10.1038/nm1044. [DOI] [PubMed] [Google Scholar]
- 29.Leone TC, et al. PGC-1alpha deficiency causes multi-system energy metabolic derangements: muscle dysfunction, abnormal weight control and hepatic steatosis. PLoS Biol. 2005;3(4):e101. doi: 10.1371/journal.pbio.0030101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vega RB, Huss JM, Kelly DP. The coactivator PGC-1 cooperates with peroxisome proliferator-activated receptor alpha in transcriptional control of nuclear genes encoding mitochondrial fatty acid oxidation enzymes. Mol Cell Biol. 2000;20(5):1868–1876. doi: 10.1128/MCB.20.5.1868-1876.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Anstee QM, Goldin RD. Mouse models in non-alcoholic fatty liver disease and steatohepatitis research. Int J Exp Pathol. 2006;87(1):1–16. doi: 10.1111/j.0959-9673.2006.00465.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Opherk C, et al. Inactivation of the glucocorticoid receptor in hepatocytes leads to fasting hypoglycemia and ameliorates hyperglycemia in streptozotocin-induced diabetes mellitus. Mol Endocrinol. 2004;18(6):1346–1353. doi: 10.1210/me.2003-0283. [DOI] [PubMed] [Google Scholar]
- 33.Lemke U, et al. The glucocorticoid receptor controls hepatic dyslipidemia through Hes1. Cell Metab. 2008;8(3):212–223. doi: 10.1016/j.cmet.2008.08.001. [DOI] [PubMed] [Google Scholar]
- 34.Bernal-Mizrachi C, et al. An afferent vagal nerve pathway links hepatic PPARalpha activation to glucocorticoid-induced insulin resistance and hypertension. Cell Metab. 2007;5(2):91–102. doi: 10.1016/j.cmet.2006.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stafford JM, Waltner-Law M, Granner DK. Role of accessory factors and steroid receptor coactivator 1 in the regulation of phosphoenolpyruvate carboxykinase gene transcription by glucocorticoids. J Biol Chem. 2001;276(6):3811–3819. doi: 10.1074/jbc.M009389200. [DOI] [PubMed] [Google Scholar]
- 36.Scott DK, Mitchell JA, Granner DK. The orphan receptor COUP-TF binds to a third glucocorticoid accessory factor element within the phosphoenolpyruvate carboxykinase gene promoter. J Biol Chem. 1996;271(50):31909–31914. doi: 10.1074/jbc.271.50.31909. [DOI] [PubMed] [Google Scholar]
- 37.Scott DK, Strömstedt PE, Wang JC, Granner DK. Further characterization of the glucocorticoid response unit in the phosphoenolpyruvate carboxykinase gene. The role of the glucocorticoid receptor-binding sites. Mol Endocrinol. 1998;12(4):482–491. doi: 10.1210/mend.12.4.0090. [DOI] [PubMed] [Google Scholar]
- 38.Imai E, Stromstedt PE, Quinn PG, Carlstedt-Duke J, Gustafsson JA, Granner DK. Characterization of a complex glucocorticoid response unit in the phosphoenolpyruvate carboxykinase gene. Mol Cell Biol. 1990;10(9):4712–4719. doi: 10.1128/MCB.10.9.4712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sugiyama T, Scott DK, Wang JC, Granner DK. Structural requirements of the glucocorticoid and retinoic acid response units in the phosphoenolpyruvate carboxykinase gene promoter. Mol Endocrinol. 1998;12(10):1487–1498. doi: 10.1210/mend.12.10.0187. [DOI] [PubMed] [Google Scholar]
- 40.Finck BN, Kelly DP. PGC-1 coactivators: inducible regulators of energy metabolism in health and disease. J Clin Invest. 2006;116(3):615–622. doi: 10.1172/JCI27794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Handschin C, Spiegelman BM. Peroxisome proliferator-activated receptor gamma coactivator 1 coactivators, energy homeostasis, and metabolism. Endocr Rev. 2006;27(7):728–735. doi: 10.1210/er.2006-0037. [DOI] [PubMed] [Google Scholar]
- 42.Asada M, et al. DNA binding-dependent glucocorticoid receptor activity promotes adipogenesis via Krüppel-like factor 15 gene expression. Lab Invest. 2011;91(2):203–215. doi: 10.1038/labinvest.2010.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zambon AC, et al. Time- and exercise-dependent gene regulation in human skeletal muscle. Genome Biol. 2003;4(10):R61. doi: 10.1186/gb-2003-4-10-r61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Simmen RC, et al. Subfertility, uterine hypoplasia, and partial progesterone resistance in mice lacking the Kruppel-like factor 9/basic transcription element-binding protein-1 (Bteb1) gene. J Biol Chem. 2004;279(28):29286–29294. doi: 10.1074/jbc.M403139200. [DOI] [PubMed] [Google Scholar]
- 45.Satapati S, et al. Mitochondrial metabolism mediates oxidative stress and inflammation in fatty liver. J Clin Invest. 2015;125(12):4447–4462. doi: 10.1172/JCI82204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lin J, et al. Defects in adaptive energy metabolism with CNS-linked hyperactivity in PGC-1alpha null mice. Cell. 2004;119(1):121–135. doi: 10.1016/j.cell.2004.09.013. [DOI] [PubMed] [Google Scholar]
- 47.Zucker SN, et al. Nrf2 amplifies oxidative stress via induction of Klf9. Mol Cell. 2014;53(6):916–928. doi: 10.1016/j.molcel.2014.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.St-Pierre J, et al. Suppression of reactive oxygen species and neurodegeneration by the PGC-1 transcriptional coactivators. Cell. 2006;127(2):397–408. doi: 10.1016/j.cell.2006.09.024. [DOI] [PubMed] [Google Scholar]
- 49.Luo J, et al. A protocol for rapid generation of recombinant adenoviruses using the AdEasy system. Nat Protoc. 2007;2(5):1236–1247. doi: 10.1038/nprot.2007.135. [DOI] [PubMed] [Google Scholar]
- 50.Wang R, et al. Sterol-regulatory-element-binding protein 1c mediates the effect of insulin on the expression of Cidea in mouse hepatocytes. Biochem J. 2010;430(2):245–254. doi: 10.1042/BJ20100701. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.