Abstract
Intrinsically disordered proteins (IDPs) are emerging as attractive drug targets by virtue of their prevalence in various diseases including cancer. Drug development targeting IDPs is challenging because IDPs have dynamic structure features and conventional drug design is not applicable. NUPR1 is an IDP that plays an important role in pancreatic cancer. We previously reported that trifluoperazine (TFP), an antipsychotic agent, was capable of binding to NUPR1 and inhibiting tumor growth. Unfortunately, TFP showed strong central nervous system side effects. In the present work, we undertook a multidisciplinary approach to optimize TFP based on the synergy of computer modeling, chemical synthesis, and a variety of biophysical, biochemical, and biological evaluations. A family of TFP-derived compounds was produced and the most active one, ZZW-115, showed a dose-dependent tumor regression with no neurological effects and an ability to induce cell death mainly by necroptosis. This study opens a new perspective for drug development against IDPs, demonstrating the possibility of successful ligand-based drug design for such challenging targets.
Keywords: Gastroenterology, Oncology
Keywords: Bioinformatics, Cell stress, Drug screens
Introduction
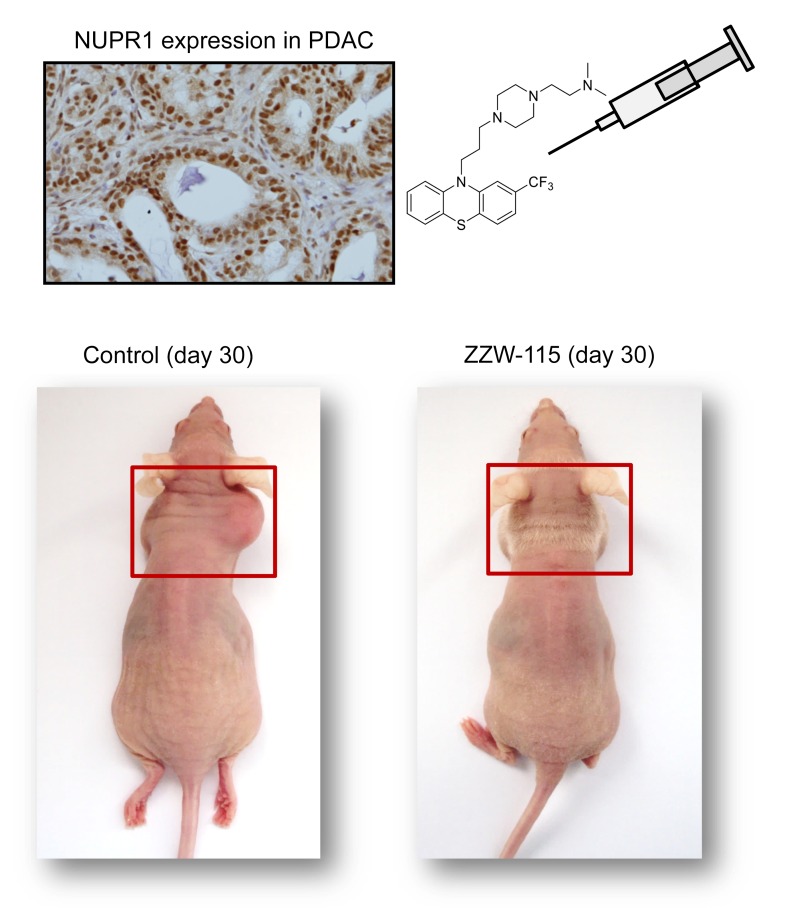
NUPR1 was first described as being activated in the exocrine pancreas in response to the cellular injury induced by pancreatitis (1). Subsequently, the inducible expression of Nupr1 was discovered to be a common response to many stresses (2, 3), including minimal ones (4), in almost all cells. Moreover, NUPR1 was found to be overexpressed in some, if not all, cancer tissues compared with healthy tissues, making NUPR1 an excellent target for cancer treatment. From a molecular point of view, NUPR1 binds to DNA in a manner similar to other chromatin proteins (5, 6) to control the expression of gene targets (7). At the cellular level, NUPR1 participates in many cancer-associated processes, including cell-cycle regulation, apoptosis (8, 9), senescence (6), cell migration and invasion (10), development of metastasis (11), and DNA repair responses (12). Indeed, NUPR1 has recently elicited significant attention for its role in promoting cancer development and progression in the pancreas (7, 13). Notably, NUPR1-dependent effects also mediate resistance to anticancer drugs (14–16). We previously showed that genetic inactivation of Nupr1 antagonizes the growth of pancreatic cancer (10, 17), and other laboratories have also shown that genetic inactivation of Nupr1 stops the growth of hepatocarcinoma (18), non–small cell lung cancer (19), cholangiocarcinoma (20), glioblastoma (21), multiple myeloma (22–23), and osteosarcoma (24), thereby supporting this protein’s role as a promising therapeutic target for developing new cancer therapies.
Structurally, NUPR1 is an intrinsically disordered protein (IDP) with an entirely disordered conformation (5, 25–28). Consequently, the target-based high throughput screening for drug selection toward this protein is highly challenging. In fact, drug-targeting IDPs is difficult due to their extremely dynamic nature, typically weak binding affinities with their natural partners, and the fact that many of them have several binding hotspots. Trying to use NUPR1 as a model IDP to be drug-targeted, we recently developed a combination of biophysical, biochemical, bioinformatic, and biological approaches for a molecular screening in vitro, in vivo, in silico, and in cellulo to select potential drug candidates against NUPR1. To this aim, we previously followed a bottom-up approach (29). We first characterized in vitro the interactions between NUPR1 and the potential ligands by using a collection of 1120 FDA-approved compounds. We employed a screening method based on fluorescence thermal denaturation (30), and identified the well-known antipsychotic agent trifluoperazine (TFP) and its structurally related fluphenazine hydrochloride as ligands inducing marked differences in the temperature denaturation profile for NUPR1. Phenotypic assays were carried out to assess the potential bioactivity of TFP, as selected from biophysical screenings. Cell viability assays in the presence of TFP have led to an IC50 of around 10 μM. Tests of TFP in vivo with human pancreatic cancer cell–derived xenografts implanted into immunocompromised mice have shown a tumor volume increase of only 50% compared with the control, whereas in mice treated with a higher dose of TFP the tumor growth was rapidly and almost completely stopped (29). Therefore, we previously successfully repurposed TFP as a possible cancer drug for treating pancreatic ductal adenocarcinoma (PDAC). Unfortunately, high doses of TFP also led to neurological effects on treated mice, such as strong lethargy and hunched posture. Although relatively efficient as an anticancer agent, the neurological effects observed in mice preclude the use of TFP to treat cancers in clinics. For this reason, in this work we developed a multidisciplinary approach to improve the compound by, on one hand, increasing its anticancer effect and, on the other hand, reducing its undesirable neurological side effects. In fact, a rational, in silico ligand design guided the organic synthesis of TFP-derived compounds, which showed a stronger affinity in vitro for NUPR1, as indicated by a combination of spectroscopic and biophysical studies. ZZW-115 showed evident antitumor activity through its interaction with NUPR1, therefore becoming a promising candidate for the treatment of PDAC and other cancers. We observed that this compound induced cell death by necroptotic and apoptotic mechanisms, with a concomitant mitochondrial metabolism failure that triggers lower production of ATP and overproduction of reactive oxygen species (ROS). The present work demonstrated how the repurposing of a drug can be used as a starting point to improve the design and efficiency of better drugs against cancer, even for challenging targets such as IDPs, and constitutes an innovative example of successful ligand-based (as opposed to structure-based) design of an inhibitor for an entirely unfolded protein.
Results
Ligand-based design and synthesis of TFP-derived compounds
Drug design via targeting NUPR1.
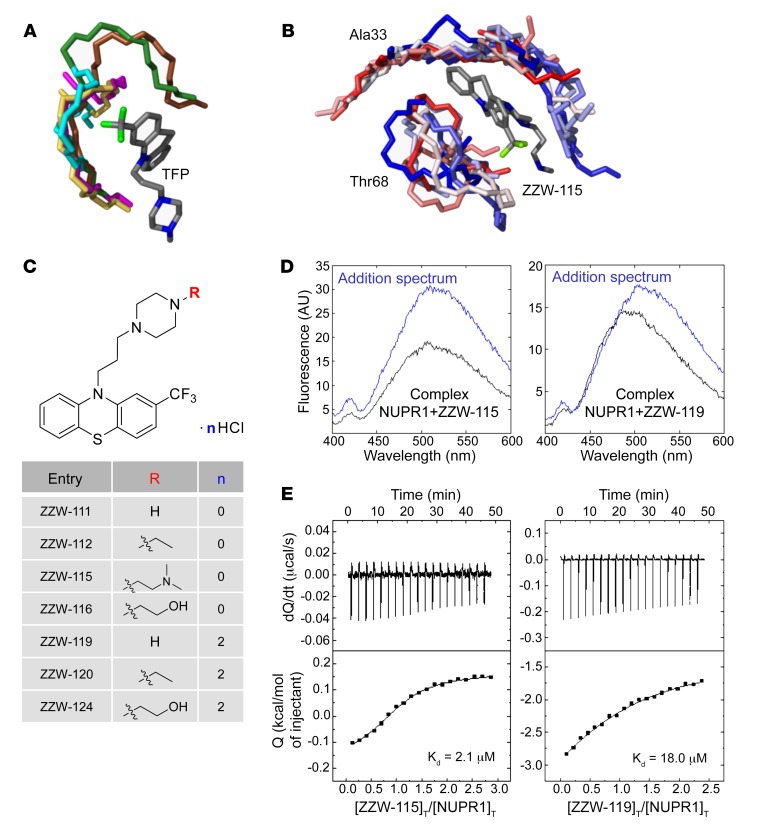
The first step in the development of TFP-derived compound was to carry out a ligand-based rational design in silico, also known as “indirect” drug design. In brief, starting from the structure of TFP, we defined models of the target regions of NUPR1 that are prone to coordinate this ligand, although in a context of high molecular flexibility. To this aim, molecular dynamic (MD) simulation of NUPR1 was performed, and short fragments of the protein (up to 7 amino acid residues) were used as receptors for molecular docking of TFP. These relatively small protein-ligand anchoring locations were completed with docking of additional fragments of NUPR1, and the resulting complexes were further refined by MD simulations. The complexes obtained (Figure 1A), which mimic highly flexible and possibly transient protein binding locations, were used as models to (a) identify the most important chemical moieties of TFP in the interaction with NUPR1 and (b) design modifications of the drug within this template ligand. The results showed that the phenothiazine and the attached trifluoromethyl group are important for TFP to maintain coordination with the protein, with average binding energy of –7.0 kcal/mol for the parent ligand. The rest of the ligand structure is progressively less critical in anchoring to NUPR1, and may provide entropic more than enthalpic contributions to the binding. On these bases, molecular modifications were explored in the propyl linker between the phenothiazine and piperazine ring, and in the methyl group attached to the latter (Figure 1B). Variations in the length of the linker were not significantly effective in improving the docking of TFP-derived compounds, due to a reduction in the enthalpic component of the binding. This left the methyl group as the most suitable region to modify for obtaining new, active analogs of TFP for inhibiting NUPR1, and we focused our synthetic effort on changing this methyl group. Improvements in the binding energies with the methyl group substitution were up to –0.5 kcal/mol, although the improvements depended on the specific modification and showed a typical uncertainty of the same order of the variation estimated.
Figure 1. Synthesis of the compounds, and docking and biophysical characterization of NUPR1 to the TFP-derived compounds.
(A) Docking of TFP to short fragments of NUPR1 sequence (in different colors), mimicking potential binding locations. The phenothiazine and trifluoromethyl groups of TFP are critical for anchoring, whereas the rest of the structure contributes less. (B) Example of simulation of ZZW-115 (a single representative structure is shown in gray) in complex with sequence segments of NUPR1 (ensembles are in gradients of color, from red to blue); the labels indicate the central protein residue for each of the two 7-residue fragments. (C) Structure of the synthesized TFP-derived compounds. (D) Fluorescence ANS spectra in the presence or absence of ZZW-115 (left) and ZZW-119 (right) for the complex between both molecules (black) and for that obtained from the addition of the spectra of both isolated molecules: NUPR1 and the TFP-derived compound (blue). Experiments were performed in 20 mM sodium phosphate pH 7.0 at 25°C. (E) Calorimetric titrations corresponding to the interaction of ZZW-115 (left) and ZZW-119 (right) with NUPR1. Thermograms (thermal power as a function of time) are shown on the top, and binding isotherms (ligand-normalized heat effects as a function of molar ratio) are shown on the bottom. Experiments were performed at 25°C in sodium phosphate 20 mM, pH 7, 2% DMSO, with 20 μM NUPR1 in the calorimetric cell and 200 μM compound in the titrating syringe, using an Auto-iTC200 instrument (MicroCal-Malvern Panalytical).
Compound synthesis.
The structures of the synthesized TFP-derived compounds ZZW-111, ZZW-112, ZZW-115, and ZZW-116 are presented in Figure 1C. All the target compounds can be prepared starting from 2-(trifluoromethyl)-10H-phenothiazine, which was coupled with 1,3-dibromepropane and further various piperazine analogs to afford the TFP analog bearing different functional groups in the piperazine moiety (Figure 1C). Moreover, the hydrochlorides of these compounds (ZZW-119, ZZW-120, and ZZW-124) were also synthesized to obtain the corresponding water-soluble candidates (Figure 1C).
Fluorescence, NMR, circular dichroism, and isothermal titration calorimetry studies of the candidate compounds
Fluorescence.
With the TFP-derived compounds predicted in silico in hand, we screened the selected compounds following a fluorescence-based strategy to identify the hits that could bind to NUPR1: (a) acquisition of the intrinsic fluorescence spectra of NUPR1 (which only contains 2 Tyr residues) either in the absence or presence of the TFP derivatives; and (b) acquisition of the fluorescence spectra of the probe ANS (8-anilinonaphtalene-1-sulfonic acid) when NUPR1 was either isolated in solution or in the presence of the compounds. In the 2 strategies we were trying to monitor whether there were changes in the environment around, respectively, the 2 Tyr residues in NUPR1, or around where ANS binds to NUPR1 (Figure 1D and Supplemental Figure 1A; supplemental material available online with this article; https://doi.org/10.1172/JCI127223DS1). By using both fluorescent approaches we were able to show that (a) the region around Tyr30 and Tyr36 of NUPR1 changed its conformation in the presence of any of the ZZW compounds (Supplemental Figure 1A); and (b) ANS fluorescence also varied in the presence of the TFP derivatives, indicating that ANS competes with the compounds for solvent-exposed hydrophobic patches of NUPR1. The above results revealed that NUPR1 bound to any of the compounds, and that binding occurred around the 30s region of the sequence and altered some solvent-exposed hydrophobic patches of NUPR1.
CD.
We also acquired far-UV circular dichroism (CD) experiments with the selected compounds ZZW-116 and ZZW-115. The presence of DMSO, used to prepare the stock solutions of the compounds, precluded the acquisition of many of the CD spectra (either that of the isolated compound or in the presence of NUPR1), as it absorbs strongly at short wavelengths. The aim of using CD as a spectroscopic probe was to determine whether NUPR1 modified its secondary structure in the presence of the compounds. The results with ZZW-115 and ZZW-116 (Supplemental Figure 1B) indicate that none of the compounds changed the secondary structure of the protein (as it is further confirmed by the NMR results).
NMR.
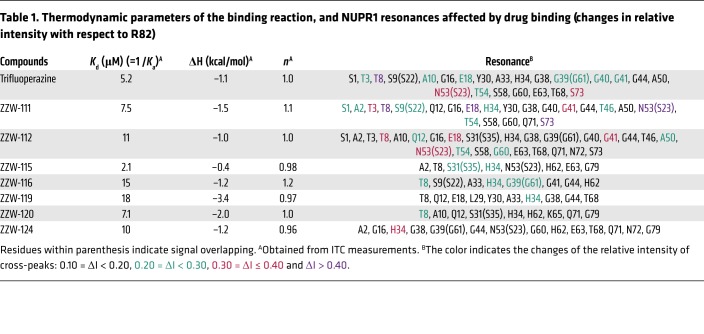
Once we had spectroscopically determined that there was binding between NUPR1 and the compounds, we aimed to verify whether the NUPR1 binding region was the same as that observed for TFP (29). Thus, 2D1H-15N-HSQC NMR experiments were carried out with 15N-labeled NUPR1. For all compounds, we found that there were no changes in the chemical shifts of the cross-peaks of NUPR1 (Supplemental Figure 1C), but rather variations in the intensities of the cross-peaks upon addition of any of the compounds (Supplemental Figure 1D). This indicated that NUPR1 remained disordered upon binding to the compounds and no new secondary or tertiary structures were acquired. The protein residues whose cross-peak intensities were affected upon addition of the ZZW compounds are indicated in Table 1. When compared with TFP, all compounds affected the intensities of the cross-peaks of a similar set of residues, namely those around the 20s region and those at the C terminus of the protein. Aside from TFP-derived compounds, ZZW-111 and ZZW-112 showed the largest number of protein residues of NUPR1 affected in the binding. Thus, we concluded that there was binding between the compounds and NUPR1, and that this binding was not unspecific, since only a subset of residues was affected.
Table 1. Thermodynamic parameters of the binding reaction, and NUPR1 resonances affected by drug binding (changes in relative intensity with respect to R82).
Isothermal titration calorimetry (ITC).
The interaction between TFP-derived compounds and NUPR1 was further studied by ITC, the gold-standard technique in binding-affinity determination. The affinity, enthalpy, and stoichiometry of binding were determined at 25°C. From the thermogram (thermal power as a function of time), the binding isotherm (ligand-normalized heats as a function of the molar ratio) was obtained through integration of the individual heat effect associated with injection of each ligand solution. We used a nonlinear least squares regression analysis employing a model that considered a single binding site in NUPR1 to estimate binding parameters such as dissociation constant, binding enthalpy, and stoichiometry. All compounds exhibited dissociation constants in the low micromolar range (Table 1). Among them, ZZW-115 was the strongest binder, with a dissociation constant close to 2 μM (Figure 1E).
Effect of TFP-derived compounds on pancreatic cancer cell growth
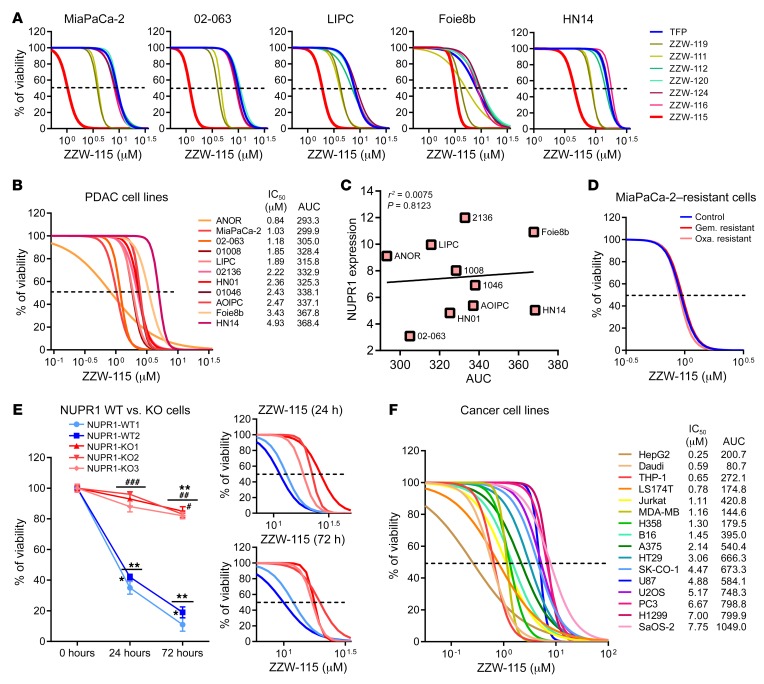
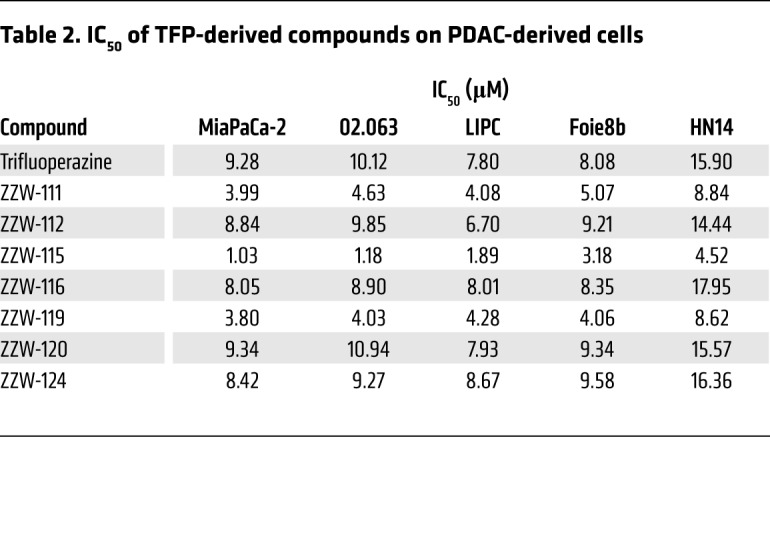
Treatment of MiaPaCa-2 (a traditional cell line), 02.063 and LIPC (basal subtype), Foie8b (derived from a liver metastasis), and HN14 (classical subtype) cells with compounds ZZW-112, ZZW-116, ZZW-120, and ZZW-124 showed an effect on cell growth close to that induced by TFP, whereas treatment with ZZW-111 and ZZW-119 was 2 to 3 times more efficient in decreasing cell growth. Remarkably, treatment with ZZW-115 was around 10 times more efficient than treatment with TFP (Figure 2A and Table 2). These data strongly suggested that (a) the effect of these TFP-derived compounds is independent of the subtype of pancreatic cancer cells; (b) ZZW-115 was the most efficient compound to kill the pancreatic cancer cells; (c) ZZW-111 and ZZW-119 had an intermediate activity compared with TFP; and (d) ZZW-112, ZZW-116, ZZW-120, and ZZW-124 had an antiproliferation activity similar to TFP. Moreover, ZZW-115 antitumor activity was tested on a panel of 11 primary PDAC-derived cells and it was found to be efficient in killing cancer cells, with an IC50 in the range of 0.84 μM (ANOR) to 4.93 μM (HN14), a very small therapeutic window (Figure 2B). These in vitro results are therefore in agreement with the ITC measurements, which showed that ZZW-115 induced the greatest affinity and resulted in the best anticancer activity. RNA-seq data showed that NUPR1 expression is systematically overexpressed in PDAC cells, but small differences among cell lines were founded. Finally, NUPR1 expression was plotted against the AUC after ZZW-115 treatment and no significant correlation was found (Figure 2C). In this regard, we cannot cluster cell lines in low/high expression or low/high sensibility.
Figure 2. TFP-derived compounds have antitumoral effect due to NUPR1 inhibition.
(A) Viability upon a 72-hour treatment with TFP-derived compounds of pancreatic cancer cells MiaPaCa-2, 02-063, LIPC, Foie8b, and HN14. (B) Viability of a panel of 11 additional pancreatic cancer cells upon a 72-hour treatment with ZZW-115. (C) NUPR1 expression values by RNA-seq are plotted against AUC. Data were statistically analyzed by simple linear regression and r2 and P values calculated. (D) Chemogram assays to test the sensitivity of the MiaPaCa-2–resistant cells to gemcitabine (Gem.) or oxaliplatin (Oxa.) after ZZW-115 treatment. (E) Viability of 2 NUPR1-WT clones and 3 NUPR1-KO clones PDAC cells upon a 24- or 72-hour treatment with 15 μM ZZW-115, or at increasing concentrations. (F) Viability of cell lines U87, A375, U2OS, SaOS-2, HT29, SK-CO-1, LS174T, H1299 and H358, HepG2, PC3, THP-1, Daudi, Jurkat and MDA-MB-231 at increasing concentrations of ZZW-115. AUC obtained from integration. Statistical significance: *P < 0.05 and **P < 0.01 compared with 0 hours of treatment; #P < 0.05, ##P < 0.01, and ###P < 0.001 compared with NUPR1-WT clones (2-way ANOVA, Bonferroni’s post hoc test). Data represent mean ± SEM, n = 3 (with technical triplicates).
Table 2. IC50 of TFP-derived compounds on PDAC-derived cells.
Since resistance to chemotherapy is a common issue that oncologists must face when treating patients with PDAC, we used the MiaPaCa-2 cell line, which has become resistant to the 2 chemotherapeutic agents oxaliplatin or gemcitabine, to assess whether resistance to them is also conferring resistance to ZZW-115. Remarkably, ZZW-115 treatment of resistant MiaPaCa-2 cells showed the same sensitivity as the parental cells, as demonstrated in Figure 2D. We therefore suggest that the antitumor effect of ZZW-115 is not influenced by resistance to other drugs and may affect some other intracellular pathways.
Finally, to verify that ZZW-115 displays its anticancer activity by targeting NUPR1, we incubated Panc1 cells with wild-type (NUPR1-WT) or inactivated NUPR1 (NUPR1-KO) in the presence of ZZW-115. As shown in Figure 2E, NUPR1-KO cells are significantly more resistant to ZZW-115 treatment at different time points after challenging them with increasing concentrations of the compound. For example, after 72 hours of treatment with 15 μM ZZW-115, only 15.01% ± 4.58% of NUPR1-WT cells survived compared with 84.60% ± 3.84% of NUPR1-KO cells. These results indicated that ZZW-115 was probably exerting its effect by binding to NUPR1. Although it is not unambiguously proved that NUPR1 is the only protein that ZZW-115 could target, our results showed that targeting NUPR1 may be the main mode of action of ZZW-115, and the ligand-protein binding contributed to its antitumor effect.
Altogether, the data showed that ZZW-115 was capable of efficiently killing the pancreatic tumor cell lines by targeting NUPR1, with no distinction regarding the PDAC subtype or even if the cells are resistant to the classical drugs currently used in clinical practice.
ZZW-115 inhibits the growth of other tumor-derived cells
Since NUPR1 is overexpressed in several (if not all) tumors, we evaluated the effect of the treatment in cellular lines derived from other different tumor at increasing concentrations of ZZW-115. Treatment of cells such as U87 (glioblastoma), A375 and B16 (melanoma), U2OS and SaOS-2 (osteosarcoma), HT29, SK-CO-1, and LS174T (colon cancer), H1299 and H358 (lung cancer), HepG2 (hepatocarcinoma), PC-3 (prostate cancer), THP-1 (acute monocytic leukemia), Daudi (lymphoma), Jurkat (acute T cell leukemia), and MDA-MB-231 (breast cancer) demonstrated that ZZW-115 was efficient to kill these tumor cells with an IC50 in the range of 0.42 μM (Hep2G cells) to 7.75 μM (SaOS-2 cells) (Figure 2F). These data showed that ZZW-115 could be potentially active for treating cancers from various tissues by targeting NUPR1.
ZZW-115 inhibits the growth of pancreatic xenografted tumors in vivo
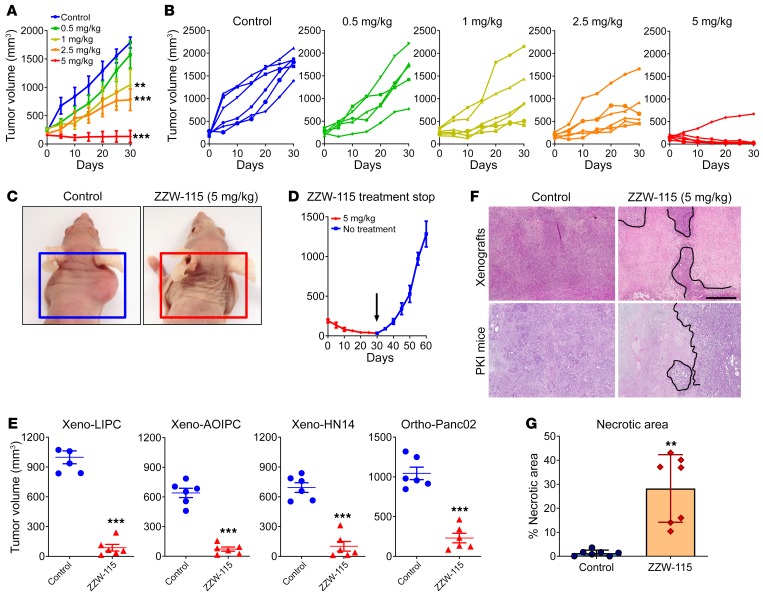
Because the compound ZZW-115 was the most efficient candidate for treating cancer cells in vitro, we selected it to treat MiaPaCa-2 cell–xenografted mice. We applied a protocol similar to that previously used for validating TFP in vivo (29). When tumors reached 200 mm3 we started a daily treatment for 30 days with 5, 2.5, 1.0, or 0.5 mg/kg ZZW-115.The control group received an equivalent volume of vehicle solution. As expected, tumor volumes increased in an exponential manner in control mice (from 217.7 ± 16.7 mm3 to 1790.7 ± 97.0 mm3 during the 30 days of observation). In contrast, when the mice were injected with 5 mg/kg ZZW-115, the tumors stopped growing a few days after treatment and their size decreased progressively, almost disappearing at the end of the treatment (from 205.9 ± 23.7 mm3 to 134.2 ± 106.8 mm3; 27.4 ± 4.6 mm3 when excluding the outlier mouse). In the outlier mouse the tumor showed a decrease in growth, but it did not disappear completely (Figure 3, A–C). Moreover, mice treated with 2.5 mg/kg showed a tumor growth from 191.9 ± 23.7 mm3 to 779.6 ± 189.8 mm3 at 30 days (603.9 ± 27.6 mm3 when excluding 1 outlier mouse), tumors from mice treated with 1.0 mg/kg grew from 269.5 ± 27.6 mm3 to 1045.4 ± 266.1 mm3, and those treated with 0.5 mg/kg grew from 238.6 ± 29.6 mm3 to 1574.6 ± 237.4 mm3. These data clearly show a dose-dependent effect with almost no significant outcome in animals treated with the lowest doses. Notably, contrary to what we observed for the TFP treatment (29), we did not find any appreciable neurological side effects, including in a 30-day treatment period using 10 mg/kg (data not shown). Moreover, in an independent group treated with 5 mg/kg, we stopped the treatment after 30 days and followed the tumor growth for an additional 30 days. The results presented in Figure 3D show that the growth arrest on the tumors was dependent on ZZW-115 administration, since the remaining tumor started growing again after suspension of the drug administration. Three additional primary PDAC-derived cells (LIPC, AOIPC, and HN14) were xenografted and treated for 30 days with 5 mg/kg ZZW-115 and we observed that tumor volume decreased (Figure 3E). Moreover, immunocompetent C57BL/6 mice were orthotopically implanted with Panc02 cells and treated with 5 mg/kg ZZW-115 for 30 days, and we observed that tumor size was almost unmeasurable in some cases (Figure 3C). Histological analysis of the tumors after 30 days of treatment with ZZW-115 showed obvious areas of necrosis, as shown in Figure 3F. Numerous apoptotic cells were also observed.
Figure 3. ZZW-115 has strong antitumoral effect in vivo.
NMRI-Foxn1nu/Foxn1nu mice (nude mice) xenografted with MiaPaCa-2 cells were separated into 5 groups of 6 mice and treated daily for 30 days with 0.5% DMSO in physiologic serum (control group) or 5, 2.5, 1.0, or 0.5 mg/kg ZZW-115 compound. Tumor volume was measured every 5 days. Mean of the volume of each treatment (A) and individual volume of each mouse (B) are shown. For each treatment, statistical significance is **P < 0.01 and ***P < 0.001 (1-way ANOVA, Tukey’s post hoc test). (C) Representative pictures of mice from control and 5 mg/kg ZZW-115 treatment groups. (D) Tumor volume of 5 mice treated with 5 mg/kg ZZW-115 measured after an additional 30 days without treatment; the arrow indicates the day the treatment was stopped. Student’s 2-tailed unpaired t test. (E) Tumor volume of nude mice xenografted with PDAC cells or C57BL/6 mice orthotopically implanted with Panc02 cells, and treated with 5 mg/kg ZZW-115 or vehicle. Tumor volume after 30 days of treatment (n = 6). (F) Representative pictures of histologic sections of the tumor xenografts or tumors from PDX1-Cre KrasG12D Ink4afl/fl, PKI mice (both from control or 5 mg/kg ZZW-115–treated mice). Scale bar: 500 μm. (G) Necrotic area in PKI mice, calculated for n = 9 mice. For each treatment, statistical significance is **P < 0.01 and ***P < 0.001 (Student’s 2-tailed unpaired t test).
We then evaluated the effect of ZZW-115 on the PDX1-Cre KrasG12D Ink4afl/fl mouse model. This animal model can spontaneously develop an aggressive PDAC that leads to death at the age of 9 to 10 weeks. We started the daily ZZW-115 treatment (5 mg/kg) at week 5 of life and then sacrificed the mice at the end of week 8. Importantly, the pronecrotic effect induced by ZZW-115 was still noticeable after 3 weeks of treatment in PDX1-Cre KrasG12D Ink4afl/fl mice (Figure 3F). The percentage of necrotic area in the tumors was calculated by histological analysis after H&E staining. We found a significant increase of necrosis in ZZW-115–treated mice, from 10% to 43% of the surface area in some tumors, whereas in the PDAC from the untreated mice these areas ranged from 0% to 3.5% (Figure 3G). We concluded that ZZW-115 could disrupt tumor growth (at low doses) and could reduce tumor size (at high doses) for the xenografted tumors, without obvious neurological side effects in the mice.
ZZW-115 induces pancreatic cell death by necrosis and apoptosis
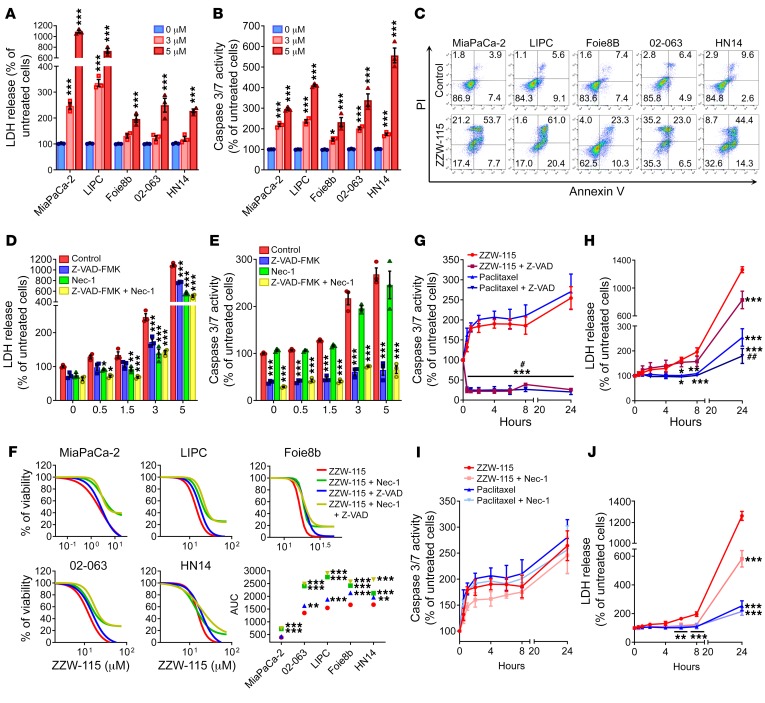
Although ZZW-115 treatment kills cancer cells and decreases the size of the xenografted pancreatic cancer tumors, details of its molecular mechanism are unknown. Prompted by our results described above, we measured necrotic and apoptotic effects throughout LDH release and caspase 3/7 activity, respectively, in several pancreatic primary cancer cells. Upon ZZW-115 treatment, we found that LDH release was significantly higher in ZZW-115–treated cells than in control cells in a concentration-dependent manner as presented in Figure 4A and Supplemental Figure 2A. Similarly, caspase 3/7 activity was also greater in ZZW-115–treated cells (Figure 4B and Supplemental Figure 2B). These experiments demonstrated that ZZW-115 exerted both pronecrotic and proapoptotic effects. We used flow cytometry after colabeling cells with annexin V and PI to measure apoptosis and necrosis, respectively, in ZZW-115–treated cells. Treatment with ZZW-115 increased necrotic and apoptotic events (Figure 4C and Supplemental Figure 3A), which agrees with the data concerning LDH release and caspase 3/7 activity, respectively. Moreover, analysis of the nuclear shape of PI-stained cells corroborates the combination of proapoptotic and pronecrotic effects of ZZW-115 treatment (Supplemental Figure 3B). Altogether, these findings indicate that ZZW-115 treatment induced cell death by both mechanisms: necrosis and apoptosis. Interestingly, pretreatment of MiaPaCa-2 cells with the pan-caspase inhibitor Z-VAD-FMK and/or the necrosis inhibitor Nec-1 prevented LDH release (Figure 4D). For example, in a 5-μM ZZW-115 treatment, the LDH was reduced to 70% with Z-VAD-FMK pretreatment, 52% with Nec-1 pretreatment, and 46% with the combination of both inhibitors. At the same concentration of ZZW-115, pretreatment with Z-VAD-FMK drastically inhibited the caspase 3/7 activity, whereas Nec-1 pretreatment did not affect it (Figure 4E). In line with these results, we performed rescue experiments on a panel of PDAC primary cells by incubating them with increasing concentrations of ZZW-115 in combination with either Z-VAD-FMK or Nec-1, or both. Our results show that both inhibitors can improve cell viability, the treatment with Nec-1 being more efficient, indicating that ZZW-115 induces necroptosis and apoptosis by independent mechanisms (Figure 4F).
Figure 4. ZZW-115 induces cell death by necrosis and apoptosis in vitro.
A panel of pancreatic cancer cell lines was incubated using 3 or 5 μM ZZW-115 for 24 hours and (A) LDH release and (B) caspase 3/7 activity were measured. Statistical significance is *P < 0.05 and ***P < 0.001 compared with untreated group (2-way ANOVA, Bonferroni’s post hoc test). (C) Flow cytometry analysis of annexin V/PI staining following 24 hours of treatment with 5 μM ZZW-115. A representative experiment of the dot plot profile of cells is shown (n = 3). MiaPaCa-2 cells were incubated from 0.5–5 μM ZZW-115 in the presence or absence of Z-VAD-FMK (20 μM) or/and Nec-1 (40 μM) for 24 hours, and (D) LDH release and (E) caspase 3/7 activity were measured. Statistical significance is *P < 0.05, **P < 0.01, and ***P < 0.001 compared with control group (2-way ANOVA, Bonferroni’s post hoc test). (F) Chemogram assays were done on pancreatic cancer cell lines with increasing concentrations of ZZW-115 in the presence or absence of Z-VAD-FMK (20 μM) or/and Nec-1 (40 μM) for 24 hours. AUC was calculated by integration. Statistical significance is **P < 0.01 and ***P < 0.001 compared with ZZW-115–treated cells (2-way ANOVA, Bonferroni’s post hoc test). MiaPaCa-2 cells were incubated in 5 μM ZZW-115 compound or 8 nM paclitaxel in the presence or absence of Z-VAD-FMK (20 μM), and (G) caspase 3/7 activity and (H) LDH release were measured. MiaPaCa-2 cells were incubated in 5 μM ZZW-115 or 8 nM paclitaxel in the presence or absence of Nec-1 (40 μM), and (I) caspase 3/7 activity and (J) LDH release were measured. Statistical significance is *P < 0.05, **P < 0.01, and ***P < 0.001 compared with ZZW-115–treated cells; #P < 0.05 and ##P < 0.01 compared with paclitaxel-treated cells (2-way ANOVA, Bonferroni’s post hoc test). Data represent mean ± SEM, n = 3.
We compared ZZW-115 to paclitaxel, a standard proapoptotic drug used in PDAC treatment, to elucidate whether their effects overlapped. To this end, we measured LDH release and caspase 3/7 activity at different times. Both drugs used alone induced similar caspase activity and, as expected, Z-VAD-FMK almost completely prevented caspase activity, showing that both compounds stimulated caspase-dependent apoptosis to a similar extent in PDAC cells (Figure 4G). Interestingly, LDH release was 5 times lower in paclitaxel-treated cells compared with ZZW-115–treated cells, and incubation with Z-VAD-FMK had a significant effect, reducing LDH release in both the paclitaxel and ZZW-115 treatments (Figure 4H). Remarkably, pretreatment with Nec-1 did not reduce the caspase activation in either the ZZW-115 or the paclitaxel treatment (Figure 4I). However, and most importantly, incubation with Nec-1 induced a significant rescue in ZZW-115–treated cells but not in paclitaxel-treated cells (Figure 4J). Our data support the idea that ZZW-115 treatment strongly induces programmed cell death by both necrosis and apoptosis with little, if any, interdependency between the 2 pathways.
ZZW-115 treatment induces metabolic failure
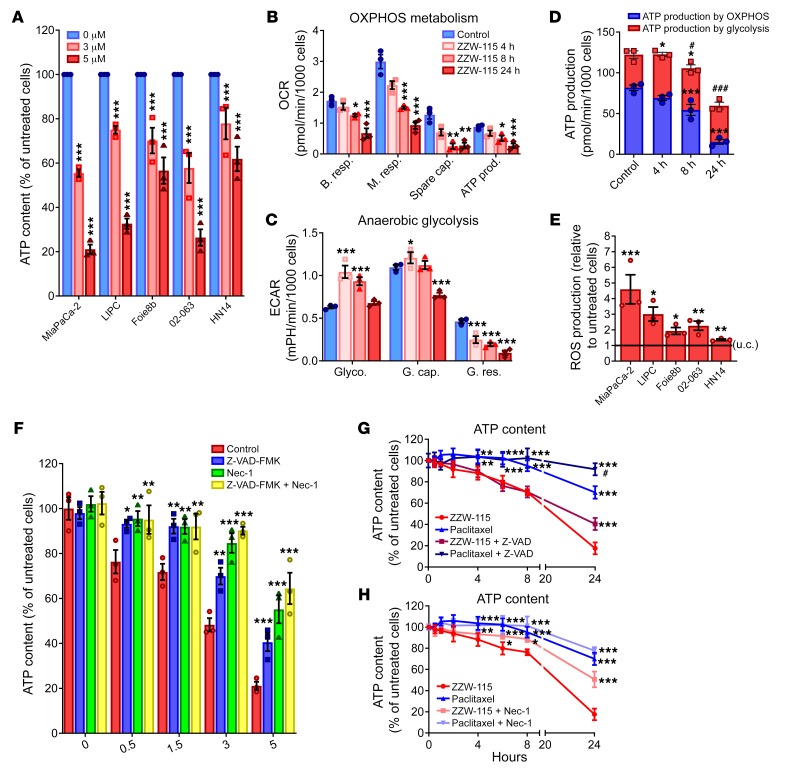
ATP production is essential for most cellular processes, and its monitoring is a good marker to understand the metabolic changes occurring during cell death. In this regard, we evaluated the ATP content in ZZW-115–treated cells. Remarkably, we found that the levels of ATP were significantly reduced in cells treated with ZZW-115 in a concentration-dependent manner on a panel of PDAC-derived primary cells (Figure 5A), as well as on other cancerous ones (Supplemental Figure 4A). Consequently, we investigated whether this decrease in the intracellular ATP content might be the result of reduced efficiency of the mitochondrial metabolism or anaerobic glycolysis. We evaluated different mitochondrial parameters such as basal respiration, maximal respiration, spare capacity, and ATP production by mitochondrial respiration, as reflected in the oxygen consumption rate (OCR). We measured the OCR in MiaPaCa-2 cells after 4, 8, and 24 hours in the presence of ZZW-115 (Figure 5B). When OCR in MiaPaCa-2 cells was compared with untreated cells, we observed a time-dependent variation in mitochondrial respiration, with a substantial decrease of all mitochondrial parameters after 8 hours of incubation with ZZW-115. Interestingly, the mitochondrial respiration failure induced by ZZW-115 treatment is not specific to the PDAC cells, as it was also found in other cancer cell lines (Supplemental Figure 4B). Since O2-dependent ATP production was also affected in ZZW-115–treated cells, we evaluated energy production by anaerobic glycolysis, measuring the extracellular acidification rate (ECAR) by the proton efflux into the media during lactate release. These experiments demonstrated that the cells shifted to anaerobic glycolytic metabolism between 4 and 8 hours after ZZW-115 treatment (Figure 5C), similar to the shift observed following a mitochondrial failure (31). This switch is transitory because cells rapidly metabolize their glycolytic reserve in a time-dependent manner, also losing their glycolytic capacity. Glycolytic capacity failure after ZZW-115 treatment was also confirmed in various pancreatic primary cancer cells, as well as in other cancer cell lines (Supplemental Figure 4C). By calculating the total ATP production by the 2 different pathways, we observed a significant time-dependent decrease of ATP production from mitochondrial metabolism, as well as a transient increase of glycolytic ATP production with a significant decrease of total ATP production after 8 hours of treatment (Figure 5D).
Figure 5. ZZW-115 treatment induces a decrease in ATP production and induces a ROS overproduction.
(A) ATP content measured in a panel of pancreatic cancer cell lines incubated at 3 or 5 μM ZZW-115 for 24 hours. (B) OXPHOS metabolism, reflected by oxygen consumption rate (OCR) levels for basal respiration (B. resp.), maximal respiration (M. resp.), spare capacity (Spare cap.), and ATP production (ATP prod.) and (C) anaerobic glycolytic metabolism reflected by extracellular acidification rate (ECAR) levels for glycolysis (Glyco.), glycolytic capacity (G. cap.), and glycolysis reserve (G. res.) were measured in MiaPaCa-2 cells treated with 3 μM ZZW-115 compound for 4, 8, or 24 hours. (D) ATP production by OXPHOS and anaerobic glycolysis were determined in MiaPaCa-2 cells treated with 3 μM ZZW-115 for 4, 8, or 24 hours. Statistical significance is *P < 0.05, **P < 0.01 and ***P < 0.001 for OXPHOS or glycolysis compared with control cells; #P < 0.05 and ###P < 0.001 for total ATP compared with control cells (2-way ANOVA, Bonferroni’s post hoc test). (E) ROS production was detected using MitoSOX Red by flow cytometry analysis on a panel of pancreatic cancer cell lines incubated with ZZW-115 at 3 μM. Statistical significance: *P < 0.05, **P < 0.01, and ***P < 0.001 compared with control cells (Student’s 2-tailed unpaired t test). (F) MiaPaCa-2 cells were incubated at a ZZW-115 concentration range of 0.5–5 μM in the presence or absence of Z-VAD-FMK (20 μM) or/and Nec-1 (40 μM) for 24 hours, and ATP content was measured. (G and H) MiaPaCa-2 cells were incubated with 5 μM ZZW-115 or 8 nM paclitaxel in the presence or absence of Z-VAD-FMK (20 μM) or Nec-1 (40 μM), and ATP content was measured. Statistical significance: *P < 0.05, **P < 0.01, and ***P < 0.001 compared with ZZW-115–treated cells; #P < 0.05 and ###P < 0.001 compared with paclitaxel-treated cells (2-way ANOVA, Bonferroni’s post hoc test). Data represent mean ± SEM, n = 3.
It is well-known that mitochondrial dynamics are related to the cell death pathways. Mitochondrial dysfunction, including decreased respiration and oxidative phosphorylation (OXPHOS), metabolic shift toward glycolysis, and increased mitochondrial ROS formation, leads to the activation of prosurvival or prodeath pathways, depending on the stimulus (32). We were interested in understanding whether mitochondrial ROS production was involved in cell death induced by ZZW-115. To answer this question we measured superoxide formation by using MitoSOX labeling and found a significant increase in the ROS formation (Figure 5E). Moreover, in the PDAC cell lines panel, ROS production is positively correlated with cell sensibility and inversely correlated with ATP production. To better understand the relation between cell death and metabolism failure, we performed experiments in ZZW-115–treated MiaPaCa-2 cells, in which we measured the ATP content after pretreatment with Z-VAD-FMK or/and Nec-1. Our results showed that the ATP content could be restored by inhibiting both necrosis and apoptosis (Figure 5F). Nec-1, which showed the most efficient rescue capacity after ZZW-115 treatment, also demonstrated a stronger ability to restore the ATP content. Moreover, Nec-1 pretreatment of ZZW-115–treated cells was able to increase anaerobic glycolysis, whereas no improvement was found in the mitochondrial metabolism (Supplemental Figure 5). Our results showed that Nec-1 is not able to rescue mitochondrial metabolic activity, meaning that the mitochondrial disruption takes place before the activation of the necroptosis. Consequently, RIP1 activation may not be upstream of mitochondrial dysfunction after ZZW-115 treatment. These data have been validated with siRNAs. We also compared paclitaxel treatment with ZZW-115 treatment to elucidate whether their effects on ATP content were similar. As expected, both isolated drugs induced a time-dependent decrease in the ATP content (Figure 5, G and H), the effect being greater with ZZW-115 and starting after 4 hours of treatment. More interestingly, both cell death inhibitors were capable of restoring the ATP content in ZZW-115–treated cells, whereas only the Z-VAD-FMK inhibitor was capable of increasing ATP content in paclitaxel-treated cells.
Taken together, these results strongly support a model in which, under ZZW-115 treatment, induced cell death is the consequence of a decrease of ATP production by the mitochondria accompanied by an increase of mitochondrial ROS production with a transient metabolic shift toward anaerobic glycolysis. In agreement with our previous data, inhibition of apoptosis, and mainly necrosis by Nec-1 incubation, can restore metabolic functions and ATP content (33).
Discussion
A TFP-derived compound decreases pancreatic tumor size by acting on NUPR1.
In this work we described the synthesis and characterization of a novel TFP-derived compound with a strong anticancer effect both in vitro and in vivo, due to its capability of inducing programmed necrosis and apoptosis. Based on a modeled structure of NUPR1 and of the selected fragments of its sequence involved in ligand binding, a collection of compounds was designed and synthesized with the aim of improving the affinity toward the protein target compared with the cognate drug. The biophysical and biochemical analyses of these compounds showed that ZZW-115 was bound to NUPR1 more favorably than to TFP (Table 1) and, in return, this compound exhibited a better anticancer effect in vitro on several primary PDAC cells, as well as on other cancer cell lines. Most importantly, ZZW-115 was a very efficient anticancer drug candidate in vivo on mice because, due mainly to necrosis, it was capable not only of stopping tumor growth but also of decreasing the tumor size until its disappearance.
TFP is an effective antipsychotic drug for people with schizophrenia (34), acting by blocking central dopamine receptors. In early attempts to repurpose the drug, TFP and some of its derived compounds were noted to have anticancer effects in some (35–38) but not all cancer models (39). However, TFP cannot be used in clinic for treating patients with cancer, due to the numerous undesirable side effects that occur at efficient anticancer doses. This is why it is important to use a rational approach to develop new TFP-derived compounds, with fewer side effects and higher anticancer activity. On the basis of our previous observation that TFP binds to NUPR1 and blocks its activity (29), leading to an anticancer effect in PDAC, we built models of the NUPR1-TFP complex, which could give us hints on how to improve the interaction with some novel derivatives. This constitutes an original ligand-based approach to the rational design of drugs against a totally unstructured protein, whereas previous pioneer attempts only included structure-based design against partly folded IDPs (40). The capacity of such new compounds to interact with NUPR1 was biophysically and biochemically validated using several spectroscopic and biophysical strategies. A stronger interaction with NUPR1 was found in particular for the compound ZZW-115 (Table 1), which resulted in improved anticancer activity both in vitro and in vivo. Noteworthy, and in contrast with TFP treatment, no apparent neurological effect was observed on mice, even at a dose of 10 mg/kg administrated daily for 30 days.
Inhibition of the NUPR1 activity as an anticancer strategy was evaluated and proven in pancreatic cancer (10, 17), but also in several other solid and liquid tumors such as hepatocarcinoma (18), non–small cell lung cancer (19), cholangiocarcinoma (20), glioblastoma (21), multiple myeloma (22, 23), and osteosarcoma (24). These findings indicate that ZZW-115 could be used for treating tumors from different origins with an IC50 range of 0.42–7.75 μM (Figure 2E). We believe that ZZW-115 is a promising agent for treating various cancers due to its effectiveness in targeting NUPR1, since the protein is commonly overexpressed in many cancerous tissues.
ZZW-115 acts at the cellular level by promoting programmed necrosis: insights into its cellular functions.
Histological study of PDAC mouse models, xenografts, and tumors from PDX1-Cre KrasG12D Ink4afl/fl mice demonstrate a strong pronecrotic effect of ZZW-115 treatment; this effect makes ZZW-115 an original drug candidate. In this regard, we noted that only a very small number of compounds rely on the capability of inducing necrosis as the main cause of their anticancer activity. For each novel candidate proposed to become an anticancer drug, such as ZZW-115, an essential prerequisite is to understand its mechanism of action. Moreover, understanding the cell death pathways elicited through a potential anticancer drug represents a milestone in preclinical studies. To clarify this aspect, we performed several functional studies and found that treatment with ZZW-115 of a whole panel of pancreatic cancer cells, as well as other cancer cells, induced in all cases a programmed cell death mainly by necrosis, but concomitantly also by apoptosis. In fact, cell rescue was possible using 2 distinct cell death inhibitors: Nec-1 for necroptosis and Z-VAD-FMK for apoptosis. Moreover, the combination of ZZW-115–induced necrosis and apoptosis offered an improvement in the treatment of cancers compared with exclusively proapoptotic drugs, because the former enhances their effect by providing 2 different mechanisms of cell death induction. Likewise, we have demonstrated that ZZW-115 can efficiently target cells that are already resistant to proapoptotic drugs commonly used in clinics. In this regard, a combination of the promotion of both apoptosis and necroptosis could improve treatment against cancer cells that very frequently develop drug resistance (41).
An important question that needs to be answered is why the cancer cells are sensitive to this strong pronecrotic compound, whereas normal cells, as observed in mice treated with high doses of up to 10 mg/kg/day for 30 days, do not show evidence of off-target effects. We suggest that this is due to the fact that cancer cells are more reliant than normal cells on NUPR1-dependent pathways. In fact, we hypothesize that since cancer cells are growing under hypoxia, with low contribution of nutrients, mechanic stress, and various other constrained conditions, these stress situations activate NUPR1 expression, and therefore NUPR1-dependent mechanisms help cells adapt to their environmental conditions. Cancer cells become highly dependent on the NUPR1 activity to live and grow under stress conditions. That is, cancer cells, but not healthy cells, are probably fully dependent on a NUPR1-adaptive mechanism.
It is also interesting to note that after ZZW-115 treatment we found a remarkable metabolic failure due to a mitochondrial metabolism rupture. This mitochondrial failure was accompanied by an increase of mitochondrial ROS production that raised cell instability and oxidative stress. Following mitochondrial respiration stress, cells shift toward anaerobic glycolytic metabolism to compensate for the lack of ATP (31). This anaerobic shift was observed between 4 and 8 hours after treatment with ZZW-115 (Figure 5C). Glycolysis is a less favorable ATP-producer pathway and cells rapidly consume glycolytic reserves, leading to ATP decrease and then to programmed cell death by both necroptosis and apoptosis. These results are in agreement with the fact that treating pancreatic cells with a siRNA against NUPR1 (33) or with TFP (our unpublished data) induces a similar metabolic disruption.
Methods
Computational modeling of a TFP-derived compound.
MD simulations of NUPR1 were carried out in the isobaric-isothermal ensemble using the GROMACS simulation package (42) in combination with the force fields AMBER ff99SB-ILDN (43). The protein was equilibrated in explicit TIP4P-D water (44) starting from an extended conformation, following a protocol previously described (26). Fragments of the simulated protein structure with a length of 5 or 7 amino acid residues were used as receptors for molecular docking of either TFP or TFP-derived compounds, by using the AutoDock Vina software (45). The choice of using short fragments of NUPR1 was motivated not only by the necessity of reducing its exceedingly large conformational space, but also by the fact that ligand binding to the protein is dictated by local hydrophobicity spanning up to 7 amino acid residues (27, 29). We previously proved that molecular docking is sufficiently accurate to reveal the binding locations of drugs to NUPR1 in a blind search (29). After docking of the ligand, the complex was considered as is or completed with addition of a second fragment of NUPR1, selected among those that had shown the most favorable affinity for the same ligand in the previous docking calculations. The resulting complexes were equilibrated by 1 ns of MD simulation performed using the GAFF force field (46) for the compounds. Simulation conditions including treatment of the electrostatics and van der Waals interactions, and reference values and coupling times for both the thermostat and barostat were as previously reported (26, 28). After the MD simulation runs, the binding affinity of the compounds was evaluated using the scoring function of AutoDock Vina (45), by performing a redocking of the ligand in the position occupied within the average structure of the simulated complex.
Synthesis of TFP-derived compounds.
The 2-(trifluoromethyl)-10H-phenothiazine (1204 mg), 1,3-dibromepropane (0.4 ml), and Cs2CO3 (150 mg) were dissolved in 1.6 ml dimethylformamide (DMF) and stirred at 65°C for 12 hours. After completion of the reaction, the solvent was removed. The residue was dissolved in ethyl acetate and washed with brine. The combined organic layer was dried over Na2SO4, filtered, and concentrated under reduced pressure to yield crude product which was further purified by column chromatography to generate the intermediate compound 2 (96 mg, 31.8%). Compound 2 and piperazine (or other piperazine analogs) were dissolved in DMF and stirred at 25°C overnight. After completion of the reaction, the solvent was removed and the residue was purified by column chromatography to generate the TFP-derived compounds. The yields of ZZW-111, ZZW-112, ZZW-115, and ZZW-116 were 68%, 77%, 64% and 76%, respectively. The TFP-derived compounds were completely dissolved in CH2Cl2 at 25°C. The anhydrous HCl was bubbled through the solution for 5 minutes. The solvent was removed and the residue was purified by column chromatography to generate the hydrochloride salt of the TFP-derived compound with a yield of 99%.
Protein expression and purification.
NUPR1 was produced and purified from transformed E. coli grown in lysogeny broth (LB) media as previously described (5). For the production of 15N-labeled samples, the cells were grown in M9 minimal media, with 1 g of 15NH4Cl per liter of media, supplemented with vitamins, and purified as the protein obtained from E. coli grown in LB media.
Fluorescence spectroscopy.
Fluorescence spectra were collected in a Cary Eclipse spectrofluorometer (Varian-Agilent Technologies) interfaced with a Peltier-thermostated multicell holder. The slit widths were 5 nm for both excitation and emission wavelengths. Excitation wavelength was 280 nm. Emission spectra from 300–400 nm were acquired at 25°C, in a 1-cm pathlength quartz cell (Hellma Analytics). The NUPR1 concentration was 8 μM in buffer in 20 mM sodium phosphate pH 7.0, and 50 μM of each TFP-derived compound were added in each experiment. For the experiments in the presence of ANS, the final concentration of the probe was 200 μM, with the same concentrations for the compound and NUPR1 as was used in the intrinsic fluorescence experiments. Excitation wavelength was 370 nm and emission spectra were collected from 400–600 nm. Slit widths were the same as before.
NMR spectroscopy.
The NMR data were acquired at 25°C, pH 4.5 (acetate buffer), on a Bruker Avance DRX-500 spectrometer equipped with a triple-resonance probe and z gradients. All spectra were referenced to external TSP (47). The 2D 1H-15N heteronuclear single-quantum coherence (HSQC) spectra were acquired either for isolated 15N-labeled NUPR1 (100 μM), or alternatively for 15N-labeled NUPR1 (100 μM) in the presence of the corresponding TFP-derived compound (400 μM). Frequency discrimination in the indirect dimensions was achieved by using the echo/antiecho-TPPI method (47). The spectra were acquired with 1024 complex points in the 1H dimension, 128 complex points in the 15N dimension, and 200 scans. The carrier of the 1H dimension was set at 8.00 ppm, and that of 15N was set at 120 ppm. The spectral widths used were 10 ppm and 35 ppm in the 1H and 15N dimensions, respectively. Water signal was suppressed with the WATERGATE sequence (48). Data were zero-filled to double the number of original points in both dimensions, apodized with shifted squared sine-bell functions in the 2 dimensions, and Fourier transformed with the program TopSpin 2.1. Assignments were taken from those previously reported for NUPR1 (25). The intensity of the signals from each row in the HSQC spectra was measured by using TopSpin 2.1 and taking into account, as an internal reference, the intensity of the last residue of NUPR1. Differences in intensity of a particular protein residue between spectra of the complex (NUPR1/compound) and that of isolated NUPR1 were considered significant only if greater than 10%.
Far-UV circular dichroism spectroscopy.
The far-UV CD experiments were acquired in a Jasco J815 spectropolarimeter with a thermostated cell holder, and interfaced with a Peltier unit at 25°C. The instrument was periodically calibrated with (+)-10-camphorsulphonic acid. A pathlength cell of 0.1 cm was used (Hellma). Spectra of the isolated molecules (NUPR1 or the corresponding compound) were acquired at a scan speed of 50 nm/min with a response time of 2 seconds and averaged over 6 scans with a bandwidth of 1 nm. Concentration was 20 μM of each molecule either in isolation or when forming the complex. The buffer used was phosphate buffer at pH 7.0 (50 mM).
ITC assays.
The binding of the TFP-derived ZZW compounds to NUPR1 was determined with a high sensitivity isothermal titration calorimeter Auto-iTC200 (MicroCal-Malvern Panalytical). Protein samples and solutions were properly degassed. Experiments were performed with freshly prepared protein solutions at 25°C. A solution of NUPR1 (20 μM, in sodium phosphate 20 mM, pH 7.0, 2% DMSO) in the calorimetric cell was titrated with a solution of each compound (200 μM, in sodium phosphate 20 mM pH 7.0, 2% DMSO). A standard protocol was employed: 19 titrant injections with 2 μl compound solution were programmed with a time spacing of 150 seconds, a stirring speed of 750 rpm, and a reference power of 10 μcal/s. The heat evolved after each ligand injection was obtained from the integral of the calorimetric signal. The heat due to the binding reaction was obtained as the difference between the reaction heat and the corresponding heat of injection, the latter estimated as a constant value throughout the experiment, and included as an adjustable parameter in the analysis. Control experiments (with ZZW compounds injected into buffer) were performed under the same experimental conditions. The association constant (Ka) and the enthalpy change (ΔH) of the binding reaction were obtained through nonlinear regression analysis of experimental data to a model assuming a single ligand binding site for the protein. Experiments were performed in replicate and data were analyzed using in-house–developed software implemented in Origin 7 (OriginLab).
Cell viability assays.
Cell viability after treatment was assessed in the following cancer cell lines: MiaPaCa-2 and Panc-1 (pancreatic cancer), U87 (glioblastoma), A375 and B16 (melanoma), U2OS and SaOS (osteosarcoma), HT29, SK-CO-1 and LS174T (colon cancer), H1299 and H358 (lung cancer), HepG2 (hepatocarcinoma), PC3 (prostate cancer), Jurkat (acute T cell leukemia), and MDA-MB-231 (breast cancer). Cells were obtained from ATCC and maintained in DMEM (Invitrogen) supplemented with 10% FBS at 37°C with 5% CO2. The primary pancreatic cancer–derived cells were cultured as previously described (49). Cells were plated in 96-well plates (5000 cells/well). Twenty-four hours later, the media were supplemented with 0–100 μM concentration of the compound to be tested, and were incubated for another additional 24 or 72 hours. Cell viability was estimated after addition of the CellTiter-Blue viability reagent (Promega) for 3 hours according to the protocol provided by the supplier. Cell viability was normalized with respect to untreated cell rates.
Animals.
Female NMRI-Foxn1nu/Foxn1nu mice (nude mice) were provided by Janvier Laboratories. Male C57BL/6 mice were obtained from Charles River Inc. Male Pdx1-Cre Ink4a/Arffl/fl LSL-KrasG12D mice were obtained by crossing the following strains: Pdx1-Cre Ink4a/Arffl/fl and LSL-KrasG12D. Mice were kept within the Experimental Animal House of the Centre de Cancérologie de Marseille, pôle Luminy (Centre de Recherche en Cancérologie de Marseille). Ten million MiaPaCa-2 cells were inoculated subcutaneously in nude mice (8 weeks old) and they were separated into 5 groups of 6 subjects each. Ten million LIPC, AOIPC, and HN14 cells were subcutaneously injected into nude mice. Mice were separated into 2 groups (treated and untreated). In C57BL/6 mice (9 weeks old), Panc02 cells (3 × 105) were injected with a 30-gauge needle in the tail of the pancreas, and the peritoneum and skin were closed with 5-0 prolene sutures. Mice were treated daily with 0.5% DMSO in physiologic serum (vehicle), and 5, 2.5, 1.0, or 0.5 mg/kg ZZW-115 when the tumor volume reached 200 mm3. Every 5 days, the mice were weighed and the tumor volumes were measured. Mice were sacrificed after 30 days of treatment. In an independent group of 6 mice, animals were xenografted and treated with 5 mg/kg ZZW-115 for 30 days as described above. Tumor growth was followed by an additional 30-day period without any additional treatment.
Necrosis evaluation.
To perform necrosis evaluation of the tumor, formalin-fixed samples were embedded in paraffin and 5-μm sections were stained with H&E. Percentage of necrotic area was calculated by the ImageJ program.
CRISPR-Cas9 clone development.
Cells were seeded and transfected with 1 μg NUPR1 double nickase plasmid or double control nickase plasmid (Santa Cruz Biotechnology) using Lipofectamine 3000 Transfection Reagent (Thermo Fisher Scientific) in each well, following the manufacturer’s protocol. Transfected cell selection was performed with puromycin, and after 72 hours single-cell colonies were isolated. Complete allelic knockouts were confirmed using Sanger sequencing.
siRNA transfection.
Cells were plated at 70% confluence and INTERFERin reagent (Polyplus-transfection) was used to perform siRNA transfections, according to the manufacturer’s protocol. Scrambled siRNA that targets no known gene sequence was used as a negative control. Assays were carried out 72 hours after transfection.
LDH assay, ATP production, and caspase 3/7 activity assay.
MiaPaCa-2 cells were seeded at a density of 10,000 cells/well in 96-well plates. Cells were allowed to attach overnight and treated the next day for 24 hours with different concentrations of ZZW-115. At the end of the experiment, LDH release, ATP production, and caspase 3/7 activity were monitored using CytoTox-ONE (Promega G7890), CellTiter-Glo (Promega G7571) and Caspase-Glo 3/7 (Promega G8091) assays, respectively. Data were normalized with respect to the cell number.
OXPHOS and glycolitic metabolism measurements.
Measurements were performed using a Seahorse Bioscience XF24 Extracellular Flux Analyzer. Cells were plated at 30,000 cells/well onto Seahorse 24-well plates 48 hours before the assay, and treated 24 hours later with ZZW-115. The OCR (pmol/min) was measured using the XF Cell Mito Stress Test Kit (Agilent) under basal conditions and in response to 1 μM oligomycin, 0.25 carbonylcyanide p-(trifluoro-methoxy)phenylhydrazone (FCCP), and rotenone and antimycin A (0.5 μM each) (MilliporeSigma). ECAR (mpH/min) was measured using the Seahorse XF Glycolysis Stress Test Kit (Agilent) at basal conditions and upon glucose (10 mM), oligomycin (1 μM), and 2-deoxyglucose (100 mM) addition (MilliporeSigma). The contribution of OXPHOS and glycolysis to ATP production was calculated using the OCR and proton production rate as previously described (50).
AnnexinV/PI staining.
Cells were collected after incubation for 24 hours of treatment with ZZW-115. Cells were washed and then detached with Accutase (Gibco, Life Technologies), and resuspended in annexin-binding buffer. Pacific-Blue annexin V (5 μl, BioLegend) was added to the cell suspension and incubated for 15 minutes. Before analysis by flow cytometry, propidium iodide (5 μl, Miltenyi Biotec) was added to the suspension. A MACSQuant-VYB (Miltenyi Biotec) was used to collect 10,000 events per sample. Data analysis was performed using FlowJo software.
Superoxide anion production by the mitochondria.
MitoSOX (Molecular Probes) was added to a final concentration of 5 μM for 10 minutes. After incubation, cells were washed with warm PBS, detached with Accutase, and resuspended in HBSS (Gibco, Life Technologies) for flow cytometry. 10,000 events per sample were collected in a MACSQuant-VYB, and data analysis was performed using FlowJo software.
Statistics.
Statistical analyses were performed by using the unpaired 2-tailed Student t test; either nonnormal distribution, 1-way ANOVA, or 2-way ANOVA with Bonferroni’s post hoc test was also used when appropriate. IC50 and AUC values were calculated by nonlinear regression curves with robust fit using GraphPad software. Values are expressed as mean ± SEM. Data are representative of at least 3 independent experiments with technical triplicates completed. A P value less than 0.05 was considered significant.
Study approval.
All experimental protocols were carried out in accordance with the nationally approved guidelines for the treatment of laboratory animals. All experimental procedures on animals were approved by the Comité d’éthique de Marseille numéro 14 (C2EA-14).
Author contributions
PSC, WL, ZZ, CH, PS, AVC, OA, BR and JLN conducted experiments. PSC, YX, LP, BR, JLN and JI contributed to study design and reviewed and revised the manuscript. PSC, JLN and JI analyzed and interpreted the data. PSC, YX, PS, BR, JLN and JI wrote the manuscript. JI designed and supervised the study.
Supplementary Material
Acknowledgments
This work was supported by La Ligue Contre le Cancer, INCa, Canceropole PACA and INSERM (to JI); Miguel Servet Program from Instituto de Salud Carlos III (CPII13/00017 to OA); Fondo de Investigaciones Sanitarias (PI15/00663 and PI18/00343 to OA); Spanish Ministry of Economy and Competitiveness (BFU2016-78232-P to AVC, CTQ2015-64445-R to JLN); Diputación General de Aragón (Protein Targets Group B89 to AVC, and Digestive Pathology Group B01 to OA); Generalitat Valenciana (Prometeo 018/2013 to JLN); Centro de Investigación Biomédica en Red en Enfermedades Hepáticas y Digestivas (CIBERehd); Fundación Alfonso Martín-Escudero and Fondation de France (to PSC); China Scholarship Council (to WL and CH); Programme XU GUANGQI (to YX and JI); National Natural Science Foundation of China (81502920) and the Fundamental Research Funds for the Central Universities (106112017CDJQJ468823) (both to YX). BR acknowledges hospitality from the European Magnetic Resonance Center (CERM), Sesto Fiorentino, Florence, Italy.
Version 1. 03/28/2019
In-Press Preview
Version 2. 05/13/2019
Electronic publication
Version 3. 06/03/2019
Print issue publication
Footnotes
Conflict of interest: PSC, YX, LP, AVC, OA, BR, JLN and JI have a pending patent entitled “NUPR1 Inhibition for Treating Cancer,” filed May 31, 2018 (European patent application no. EP18305672.0).
Copyright: © 2019, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2019;129(6):2500–2513.https://doi.org/10.1172/JCI127223.
Contributor Information
Patricia Santofimia-Castaño, Email: patricia.santofimia@inserm.fr.
Yi Xia, Email: yixia@cqu.edu.cn.
Wenjun Lan, Email: wenjun.lan@etu.univ-amu.fr.
Zhengwei Zhou, Email: zhouzhengwei610@126.com.
Can Huang, Email: can.huang@inserm.fr.
Ling Peng, Email: ling.peng@univ-amu.fr.
Philippe Soubeyran, Email: philippe.soubeyran@inserm.fr.
Bruno Rizzuti, Email: bruno.rizzuti@fis.unical.it.
Juan Iovanna, Email: juan.iovanna@inserm.fr.
References
- 1.Mallo GV, et al. Cloning and expression of the rat p8 cDNA, a new gene activated in pancreas during the acute phase of pancreatitis, pancreatic development, and regeneration, and which promotes cellular growth. J Biol Chem. 1997;272(51):32360–32369. doi: 10.1074/jbc.272.51.32360. [DOI] [PubMed] [Google Scholar]
- 2.Goruppi S, Iovanna JL. Stress-inducible protein p8 is involved in several physiological and pathological processes. J Biol Chem. 2010;285(3):1577–1581. doi: 10.1074/jbc.R109.080887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cano CE, Hamidi T, Sandi MJ, Iovanna JL. Nupr1: the Swiss-knife of cancer. J Cell Physiol. 2011;226(6):1439–1443. doi: 10.1002/jcp.22324. [DOI] [PubMed] [Google Scholar]
- 4.Garcia-Montero A, Vasseur S, Mallo GV, Soubeyran P, Dagorn JC, Iovanna JL. Expression of the stress-induced p8 mRNA is transiently activated after culture medium change. Eur J Cell Biol. 2001;80(11):720–725. doi: 10.1078/0171-9335-00209. [DOI] [PubMed] [Google Scholar]
- 5.Encinar JA, et al. Human p8 is a HMG-I/Y-like protein with DNA binding activity enhanced by phosphorylation. J Biol Chem. 2001;276(4):2742–2751. doi: 10.1074/jbc.M008594200. [DOI] [PubMed] [Google Scholar]
- 6.Grasso D, et al. Genetic inactivation of the pancreatitis-inducible gene Nupr1 impairs PanIN formation by modulating Kras(G12D)-induced senescence. Cell Death Differ. 2014;21(10):1633–1641. doi: 10.1038/cdd.2014.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hamidi T, et al. Nuclear protein 1 promotes pancreatic cancer development and protects cells from stress by inhibiting apoptosis. J Clin Invest. 2012;122(6):2092–2103. doi: 10.1172/JCI60144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Malicet C, Dagorn JC, Neira JL, Iovanna JL. p8 and prothymosin alpha: unity is strength. Cell Cycle. 2006;5(8):829–830. doi: 10.4161/cc.5.8.2686. [DOI] [PubMed] [Google Scholar]
- 9.Malicet C, Giroux V, Vasseur S, Dagorn JC, Neira JL, Iovanna JL. Regulation of apoptosis by the p8/prothymosin alpha complex. Proc Natl Acad Sci U S A. 2006;103(8):2671–2676. doi: 10.1073/pnas.0508955103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sandi MJ, et al. p8 expression controls pancreatic cancer cell migration, invasion, adhesion, and tumorigenesis. J Cell Physiol. 2011;226(12):3442–3451. doi: 10.1002/jcp.22702. [DOI] [PubMed] [Google Scholar]
- 11.Ree AH, Pacheco MM, Tvermyr M, Fodstad O, Brentani MM. Expression of a novel factor, com1, in early tumor progression of breast cancer. Clin Cancer Res. 2000;6(5):1778–1783. [PubMed] [Google Scholar]
- 12.Gironella M, et al. p8/nupr1 regulates DNA-repair activity after double-strand gamma irradiation-induced DNA damage. J Cell Physiol. 2009;221(3):594–602. doi: 10.1002/jcp.21889. [DOI] [PubMed] [Google Scholar]
- 13.Cano CE, et al. Genetic inactivation of Nupr1 acts as a dominant suppressor event in a two-hit model of pancreatic carcinogenesis. Gut. 2014;63(6):984–995. doi: 10.1136/gutjnl-2013-305221. [DOI] [PubMed] [Google Scholar]
- 14.Giroux V, et al. p8 is a new target of gemcitabine in pancreatic cancer cells. Clin Cancer Res. 2006;12(1):235–241. doi: 10.1158/1078-0432.CCR-05-1700. [DOI] [PubMed] [Google Scholar]
- 15.Tang K, Zhang Z, Bai Z, Ma X, Guo W, Wang Y. Enhancement of gemcitabine sensitivity in pancreatic cancer by co-regulation of dCK and p8 expression. Oncol Rep. 2011;25(4):963–970. doi: 10.3892/or.2011.1139. [DOI] [PubMed] [Google Scholar]
- 16.Palam LR, Gore J, Craven KE, Wilson JL, Korc M. Integrated stress response is critical for gemcitabine resistance in pancreatic ductal adenocarcinoma. Cell Death Dis. 2015;6:e1913. doi: 10.1038/cddis.2015.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vasseur S, Hoffmeister A, Garcia S, Bagnis C, Dagorn JC, Iovanna JL. p8 is critical for tumour development induced by rasV12 mutated protein and E1A oncogene. EMBO Rep. 2002;3(2):165–170. doi: 10.1093/embo-reports/kvf023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Emma MR, et al. NUPR1, a new target in liver cancer: implication in controlling cell growth, migration, invasion and sorafenib resistance. Cell Death Dis. 2016;7(6):e2269. doi: 10.1038/cddis.2016.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guo X, et al. Lentivirus-mediated RNAi knockdown of NUPR1 inhibits human nonsmall cell lung cancer growth in vitro and in vivo. Anat Rec (Hoboken) 2012;295(12):2114–2121. doi: 10.1002/ar.22571. [DOI] [PubMed] [Google Scholar]
- 20.Kim KS, Jin DI, Yoon S, Baek SY, Kim BS, Oh SO. Expression and roles of NUPR1 in cholangiocarcinoma cells. Anat Cell Biol. 2012;45(1):17–25. doi: 10.5115/acb.2012.45.1.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li J, et al. Knockdown of NUPR1 inhibits the proliferation of glioblastoma cells via ERK1/2, p38 MAPK and caspase-3. J Neurooncol. 2017;132(1):15–26. doi: 10.1007/s11060-016-2337-0. [DOI] [PubMed] [Google Scholar]
- 22.Zeng C, Yi B, Li X, Chen J. [Knockdown of nuclear protein 1 (NUPR1) gene inhibits proliferation and promotes apoptosis of human multiple myeloma U266 cells] Xi Bao Yu Fen Zi Mian Yi Xue Za Zhi. 2017;33(9):1240–1246. [PubMed] [Google Scholar]
- 23.Zeng C, et al. Knockdown of NUPR1 inhibits the growth of U266 and RPMI8226 multiple myeloma cell lines via activating PTEN and caspase activation‑dependent apoptosis. Oncol Rep. 2018;40(3):1487–1494. doi: 10.3892/or.2018.6544. [DOI] [PubMed] [Google Scholar]
- 24.Zhou C, et al. Long non-coding RNA FEZF1-AS1 promotes osteosarcoma progression by regulating miR-4443/NUPR1 axis. Oncol Res. 2018;26(9):1335–1343. doi: 10.3727/096504018X15188367859402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aguado-Llera D, et al. Deciphering the binding between Nupr1 and MSL1 and their DNA-repairing activity. PLoS One. 2013;8(10):e78101. doi: 10.1371/journal.pone.0078101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Neira JL, Rizzuti B, Iovanna JL. Determinants of the pKa values of ionizable residues in an intrinsically disordered protein. Arch Biochem Biophys. 2016;598:18–27. doi: 10.1016/j.abb.2016.03.034. [DOI] [PubMed] [Google Scholar]
- 27.Santofimia-Castaño P, et al. Intrinsically disordered chromatin protein NUPR1 binds to the C-terminal region of Polycomb RING1B. Proc Natl Acad Sci U S A. 2017;114(31):E6332–E6341. doi: 10.1073/pnas.1619932114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Santofimia-Castaño P, Rizzuti B, Abián O, Velázquez-Campoy A, Iovanna JL, Neira JL. Amphipathic helical peptides hamper protein-protein interactions of the intrinsically disordered chromatin nuclear protein 1 (NUPR1) Biochim Biophys Acta Gen Subj. 2018;1862(6):1283–1295. doi: 10.1016/j.bbagen.2018.03.009. [DOI] [PubMed] [Google Scholar]
- 29.Neira JL, et al. Identification of a drug targeting an intrinsically disordered protein involved in pancreatic adenocarcinoma. Sci Rep. 2017;7:39732. doi: 10.1038/srep39732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Velazquez-Campoy A, Sancho J, Abian O, Vega S. Biophysical screening for identifying pharmacological chaperones and inhibitors against conformational and infectious diseases. Curr Drug Targets. 2016;17(13):1492–1505. doi: 10.2174/1389450117666160201110449. [DOI] [PubMed] [Google Scholar]
- 31.Dickman KG, Mandel LJ. Differential effects of respiratory inhibitors on glycolysis in proximal tubules. Am J Physiol. 1990;258(6 Pt 2):F1608–F1615. doi: 10.1152/ajprenal.1990.258.6.F1608. [DOI] [PubMed] [Google Scholar]
- 32.Nagdas S, Kashatus DF. The interplay between oncogenic signaling networks and mitochondrial dynamics. Antioxidants (Basel) 2017;6(2):33. doi: 10.3390/antiox6020033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Santofimia-Castaño P, et al. Inactivation of NUPR1 promotes cell death by coupling ER-stress responses with necrosis. Sci Rep. 2018;8(1):16999. doi: 10.1038/s41598-018-35020-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Koch K, Mansi K, Haynes E, Adams CE, Sampson S, Furtado VA. Trifluoperazine versus placebo for schizophrenia. Cochrane Database Syst Rev. 2014;(1):CD010226. doi: 10.1002/14651858.CD010226.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pulkoski-Gross A, et al. Repurposing the antipsychotic trifluoperazine as an antimetastasis agent. Mol Pharmacol. 2015;87(3):501–512. doi: 10.1124/mol.114.096941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yeh CT, et al. Trifluoperazine, an antipsychotic agent, inhibits cancer stem cell growth and overcomes drug resistance of lung cancer. Am J Respir Crit Care Med. 2012;186(11):1180–1188. doi: 10.1164/rccm.201207-1180OC. [DOI] [PubMed] [Google Scholar]
- 37.Pinheiro T, et al. A chemical screen identifies trifluoperazine as an inhibitor of glioblastoma growth. Biochem Biophys Res Commun. 2017;494(3–4):477–483. doi: 10.1016/j.bbrc.2017.10.106. [DOI] [PubMed] [Google Scholar]
- 38.Zhelev Z, et al. Phenothiazines suppress proliferation and induce apoptosis in cultured leukemic cells without any influence on the viability of normal lymphocytes. Phenothiazines and leukemia. Cancer Chemother Pharmacol. 2004;53(3):267–275. doi: 10.1007/s00280-003-0738-1. [DOI] [PubMed] [Google Scholar]
- 39.Wen Y, Zhang Y, Li J, Luo F, Huang Z, Liu K. Low concentration trifluoperazine promotes proliferation and reduces calcium-dependent apoptosis in glioma cells. Sci Rep. 2018;8(1):1147. doi: 10.1038/s41598-018-19413-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ruan H, Sun Q, Zhang W, Liu Y, Lai L. Targeting intrinsically disordered proteins at the edge of chaos. Drug Discov Today. 2019;24(1):217–227. doi: 10.1016/j.drudis.2018.09.017. [DOI] [PubMed] [Google Scholar]
- 41.Trivedi R, Mishra DP. Trailing TRAIL resistance: novel targets for TRAIL sensitization in cancer cells. Front Oncol. 2015;5:69. doi: 10.3389/fonc.2015.00069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hess B, Kutzner C, van der Spoel D, Lindahl E. GROMACS 4: Algorithms for highly efficient, load-balanced, and scalable molecular simulation. J Chem Theory Comput. 2008;4(3):435–447. doi: 10.1021/ct700301q. [DOI] [PubMed] [Google Scholar]
- 43.Lindorff-Larsen K, et al. Improved side-chain torsion potentials for the Amber ff99SB protein force field. Proteins. 2010;78(8):1950–1958. doi: 10.1002/prot.22711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Piana S, Donchev AG, Robustelli P, Shaw DE. Water dispersion interactions strongly influence simulated structural properties of disordered protein states. J Phys Chem B. 2015;119(16):5113–5123. doi: 10.1021/jp508971m. [DOI] [PubMed] [Google Scholar]
- 45.Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31(2):455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang J, Wolf RM, Caldwell JW, Kollman PA, Case DA. Development and testing of a general amber force field. J Comput Chem. 2004;25(9):1157–1174. doi: 10.1002/jcc.20035. [DOI] [PubMed] [Google Scholar]
- 47. Cavanagh J, et al. Protein NMR spectroscopy: principles practice, 1st ed. San Diego, CA: Academic Press; 1996. [Google Scholar]
- 48.Piotto M, Saudek V, Sklenár V. Gradient-tailored excitation for single-quantum NMR spectroscopy of aqueous solutions. J Biomol NMR. 1992;2(6):661–665. doi: 10.1007/BF02192855. [DOI] [PubMed] [Google Scholar]
- 49.Lomberk G, et al. Distinct epigenetic landscapes underlie the pathobiology of pancreatic cancer subtypes. Nat Commun. 2018;9(1):1978. doi: 10.1038/s41467-018-04383-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wu H, Ying M, Hu X. Lactic acidosis switches cancer cells from aerobic glycolysis back to dominant oxidative phosphorylation. Oncotarget. 2016;7(26):40621–40629. doi: 10.18632/oncotarget.9746. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.