Abstract
Central activation of fibroblast growth factor (FGF) receptors regulates peripheral glucose homeostasis and reduces food intake in preclinical models of obesity and diabetes. The current work was undertaken to advance our understanding of the receptor expression, as sites of ligand action by FGF19, FGF21, and FGF1 in the mammalian brain remains unresolved. Recent advances in automated RNAscope in situ hybridization and droplet digital PCR (ddPCR) technology allowed us to interrogate central FGFR/beta klotho (Klb) system at the cellular level in the mouse, with relevant comparisons to non-human primate and human brain. FGFR1–3 gene expression was broadly distributed throughout the CNS in mus musculus, with FGFR1 exhibiting the greatest heterogeneity. FGFR4 expression localized only in the medial habenula and subcommissural organ of mice. Likewise, Klb mRNA was restricted to the suprachiasmatic nucleus (SCh) and select midbrain and hindbrain nuclei. ddPCR in the rodent hypothalamus confirmed that, although expression levels are indeed low for Klb, there is nonetheless a bonafide subpopulation of Klb+ cells in the hypothalamus. In NHP and human midbrain and hindbrain, Klb+ cells are quite rare, as is expression of FGFR4. Collectively, these data provide the most robust central map of the FGFR/Klb system to date, and highlight central regions that may be of critical importance to assess central ligand effects with pharmacological dosing, such as the putative interactions between the endocrine FGFs and FGFR1/Klb, or FGF19 with FGFR4.
Keywords: fibroblast growth factors, beta-klotho, in situ hybridization, type 2 diabetes, obesity, RRID: AB_2532109, RRID: AB_839504, RRID: AB_2109645
Graphical Abstract
We refined the central map of fibroblast growth factor receptor (FGFR) and beta klotho (Klb) expression using automated in situ hybridization in three mammalian species. FGFR4 and Klb expression were limited in the rodent CNS, and absent in primate and man, suggesting species-specific divergence in central FGF signaling.
1. Introduction
Rapidly rising global rates of obesity, diabetes, and metabolic disease highlight the need for new, more effective therapeutic interventions for these disorders. Three members of the fibroblast growth factor (FGF) family of ligands - FGF1, FGF19, and FGF21 - have emerged as promising therapeutic targets to reduce body weight, inhibit food intake, increase energy expenditure, and normalize circulating glucose and plasma lipid levels when administered at pharmacological doses. While endogenous FGF ligands play an important role in the development and function of a variety of tissues, mounting evidence indicates that the anti-obesity and anti-diabetic effects of these ligands can be driven by central action (Douris et al., 2015; Gasser, Moutos, Downes, & Evans, 2017; Marcelin et al., 2014; Morton et al., 2013; Rojas et al., 2015; Sarruf et al., 2010; Scarlett et al., 2016; Suh et al., 2014; Talukdar, Zhou, et al., 2016). Evidence in support of this assertion stems in part from the potent metabolic benefits induced by central administration of these ligands in preclinical animal models, but the specific FGF receptor (FGFR) systems involved in these central actions remains unclear. FGF19 and FGF21, behaving as endocrine FGFs, engage with the “III/c” isoforms of FGF receptors (FGFR) 1, 2, and 3, requiring both the FGFR and the plasma-membrane bound beta klotho (Klb) co-receptor (Agrawal et al., 2018; Itoh, Ohta, & Konishi, 2015; Kurosu et al., 2007). FGF19 is unique in that it can also bind to FGFR4 in both the presence and absence of Klb (Adams et al., 2012; X. Wu et al., 2010). In contrast, FGF1 is an autocrine FGF ligand that requires a heparan sulfate binding partner, but does not require Klb and can engage all known FGFR isoforms. A number of studies have sought to clarify the CNS expression patterns of the rodent FGFR-Klb system using radioisotopic in situ hybridization (ISH), PCR on tissue blocks and on microdissected brain tissue (Belluardo et al., 1997; Bookout et al., 2013; Fon Tacer et al., 2010; Itoh et al., 1994; Talukdar, Owen, et al., 2016; Yazaki et al., 1994). While these studies have provided an excellent overview of FGFR-Klb neuroanatomy at the gross level, their technical limitations preclude a clear understanding of the brain nucleus specificity, cellular resolution, and signal-to-noise in regions with sparse expression of FGFR and Klb. Therefore, we sought to map the central FGFR/Klb system using RNAscope ISH (Advanced Cell Diagnostics [ACD Bio], Newark, CA) to better resolve the central anatomy of this system at the cellular level, and extend these findings to higher-order mammalian species (cynomolgus monkey and human).
Fully automated RNAscope ISH (C. M. Anderson et al., 2016; Wang et al., 2012) provided cellular-level resolution of FGFR and Klb expression in three mammalian species. The methods we employed in this study allowed for rapid, robust, and replicable assessment of the central expression of the FGFR/Klb system. Traditional ISH methods are typically hampered by technical complexity, low resolution, high noise for rare transcripts, and in the case of radioisotopic ISH, extensive time constraints on the order of weeks between an ISH experiment and a result (Cassidy & Jones, 2014). Similar to the use of the RNAscope platform on a Leica Bond (C. M. Anderson et al., 2016), our automated ISH method allowed us to stain 300 slides in 10 batches on the Ventana Discovery ULTRA with minimal user input. Even the manual version of the RNAscope assay provides turnaround time to data in 1–2 work days for 20 slides (Heppner et al., 2017). Beyond the streamlined workflow, this platform allowed for cellular level resolution of ISH and immunohistochemical identification of different cell populations. The only corollary molecular technique that can provide this type of spatial resolution is single cell capture combined with qPCR, which is also considerably less-throughput than automated ISH.
In mice, we observed robust expression of FGFR1–3 throughout the entire brain, and FGFR1 in particular showed the most dynamic range across different brain regions. The relative abundance of FGFR2 was less than the observed ISH signal for FGFR3, and the morphology of the FGFR3 signal appears similar to that of non-neuronal cells. FGFR4 expression, on the other hand, was quite restricted in the rodent CNS: parallel to previous observations in rat (Itoh et al., 1994; Miyake & Itoh, 1996), FGFR4 ISH signal was only detected in the medial portion of the habenula and the subcommissural ependymal layer. Klb ISH signal was also highly restricted: we detected Klb in the suprachiasmatic nucleus (SCh), reticular thalamus, medial vestibular nucleus, medial trigeminal neurons (Me5), principal sensory nucleus of the trigeminal nerve (Pr5N), but not in the area postrema (AP) (Bookout et al., 2013). Dual ISH revealed that FGFR1 and Klb indeed co-localize within cells of the SCh, Me5 and Pr5N but not in the reticular thalamus or AP. We used PCR to validate our findings on hypothalamic Klb expression using droplet digital PCR (ddPCR). ddPCR measures single copies of RNA, allowing us to resolve the presence of rare RNA species that may be undetectable otherwise. Our ddPCR data validated our ISH findings in the rodent hypothalamus, indicating low copy numbers of Klb and robust expression of FGFR1. Taken together, these techniques highlight the power of high resolution in situ hybridization to interrogate rare RNA species and therefore cell phenotypes in the mouse brain.
In addition, we included non-human primate (NHP) and human tissue samples in our ISH assays to assess species differences specifically in midbrain/hindbrain FGFR/Klb distribution. We found ubiquitous expression of FGFR1–3 in midbrain and hindbrain regions, but only a handful of Klb+ neurons in the nucleus of the solitary tract (NTS) in human. Together this work provides the most comprehensive assessment of brain FGFR/Klb expression using RNAscope and ddPCR, and highlights sites of interest for ligand-receptor interaction including FGF19-FGFR4 in the habenula and subcommissural organ, and FGF21-FGFR1/Klb in the SCh, Me5, and Pr5N. Interestingly, Klb expression in these hindbrain regions in rodents were not observed in NHP and in only a few cells in human samples, suggesting that these ligands may drive divergent physiology in different mammalian species.
2. Materials & Methods
2.1. Animals.
All procedures were approved by the Institutional Animal Care and Use Committees and Institutional Review Board at Novo Nordisk Research Center Seattle, Novo Nordisk A/S, and the University of Washington, and followed the U.S. Public Health Service guidelines for the humane care and use of experimental animals. Adult male C57/Bl6 mice were used for all experiments. Central expression of FGF receptors by PCR and ddPCR and Klb was performed on brain blocks from wildtype animals. Transgenic animals expressing Fgfr1-eGFP (GP338Gsat/Mmucd; Mutant Mouse Regional Resource Center, University of California, Davis, CA) were used for all ISH experiments. Formalin-fixed, paraffin-embedded (FFPE) NHP samples from cynomolgus were procured from OHSU (Beaverton, OR), and FFPE human brain samples were procured from the Edinburgh Brain Bank (Edinburgh, Scotland). For each component of this study, 4–6 animals were used per experiment.
2.2. In situ hybridization.
Fgfr1-eGFP mice were anesthetized with ketamine and xylazine, and were perfused with PBS followed by 4% paraformaldehyde (PFA) in 0.1 M PBS. Brains were extracted, dissected into 3 mm-thick slabs in the coronal plane, and were paraffin embedded on an ASP300S tissue processor (Leica Biosystems, Buffalo Grove, IL) with the following sequence: Two 70% EtOH steps (1-hr), two 95% EtOH steps (1.5-hr), two 100% EtOH steps (1.5-hr), three xylene steps (1-hr), and three paraffin wax (Surgipath Paraplast, Leica) steps (1.5-hr each). Paraffin-infused tissues were embedded rostral side down in warm paraffin to form square blocks for sectioning. A series of 10 sections (5 μm/section) was collected every 200 μm along the rostro-caudal axis to sample the entire brain. Liver tissues were simultaneously prepared using the same paraffin-embedding technique and were sectioned at 5 μm. For cynomolgus and human samples, midbrain and hindbrain sections (4.5 μm thick) were used for all ISH experiments. All tissues were retrieved, hybridized and stained using a Ventana Discovery ULTRA or Discovery XT system (Ventana Medical Systems, Tucson, AZ) and the 2.5 VS RED reagent kit from Advanced Cell Diagnostics following the protocol outlined technical bulletin #322250-USM-ULT. We also performed dual ISH to co-localize Mm-Fgfr1c and Mm-Klb in mouse brains; these experiments used the same tissue preparation, but utilized RNAscope Universal reagents described in ACD Bio technical bulletin #323300-USM. Our assays followed the standard mRNA RED ULTRA protocol with software settings for each tissue type described in Table 1. Our total experiment time for uniplex or duplex ISH from start to finish is approximately 12–14 hours, 11–13 of which are fully automated. We list all steps and reagents for our assays in Table 1. A list of control and experimental probes used in this assay are outlined in Table 2. For every ISH run, the e. coli gene dapB was used as a negative control, and we used Mm-Polr2a, Mm-PPIB, and Hs-PECAM as housekeeping genes. All sections were counterstained with hematoxylin (Millipore-Sigma, St. Louis, MO) or QD DAPI (Ventana Medical Systems), dehydrated in xylenes (1 dip) and coverslipped using EcoMount (BioCare, Pacheco, CA).
Table 1. User input protocol parameters for Ventana Discovery ULTRA or Discovery XT for uniplex or duplex ISH.
This table outlines user-modifiable protocol steps for two different tissue types (liver, brain), as well as two ISH assay (uniplex, duplex). All other steps in these assays are hard-programmed by the Ventana Discovery software and are not modifiable by laboratory staff.
| ISH step | Uniplex ISH (liver) | Uniplex ISH (brain) | Duplex ISH (brain) |
|---|---|---|---|
| Baking | 32 min / 69°C | 32 min / 69°C | 32 min / 69°C |
| Deparaffinization | Selected | Selected | Selected |
| Cell Conditioning | 32 minutes / 97°C | 24 minutes / 97°C | 24 minutes / 97°C |
| Protease pretreatment | 24 minutes / 37°C | 12 minutes / 37°C | 12 minutes / 37°C |
| Probe application | 4 minutes / 37°C | 4 minutes / 37°C | 4 minutes / 37°C |
| Hybridization | 2 hours / 43°C | 2 hours / 43°C | 2 hours / 43°C |
| AMP 1 (duplex only) | - | - | 32 min / 39°C |
| AMP 2 (duplex only) | - | - | 32 min / 39°C |
| AMP 5 | 32 min / 39°C | 32 min / 39°C | 4 min / 39°C |
| AMP 8 (duplex only) | - | - | 4 min / 39°C |
Table 2. ISH probes for mouse, cynomolgus monkey (NHP), and human.
Control and experimental probes used for ISH experiments. All probes are annotated based on NCBI accession number and a defined target region. The “Z pairs” column denotes the size of the hybridization scaffold that builds on a given transcript for fast red staining. All FGFR probes are considered pan-isoform with the exception of Mm-FGFR1, which was specifically designed to identify Mm-FGFR1c and not Mm-FGFR1b.
| Probe | ACD Bio Cat. No. | Accession ID | Target Region | Z pairs | Use/Specificity |
|---|---|---|---|---|---|
| dapB | 312039 | EF191515 | 414–862 | 10 | Negative control (bacterial gene) |
| Mm-PPIB | 313919 | NM_011149.2 | 98–856 | 15 | Positive control (high copy number) |
| Mm-Polr2a | 312479 | NM_009089.2 | 2802–3678 | 20 | Positive control (low copy number) |
| Mm-NPY | 313329 | NM_023456.2 | 28–548 | 12 | Positive control probe (brain) |
| Mm-Pomc | 314089 | NM_008895.4 | 19–995 | 10 | Positive control probe (brain) |
| Mm-FGFR1-O1 | 454949 | NM_001079909.2 | 87–4624 | 20 | Designed for the 1c variant of FGFR1, and against the 1b variant #AF176552 |
| Mm-FGFR2 | 443509 | NM_010207.2 | 2–1677 | 18 | Pan-FGFR2 probe |
| Mm-FGFR3 | 440779 | NM_008010.5 | 511–1615 | 20 | Pan-FGFR3 probe |
| Mm-FGFR4 | 443519 | NM_008011.2 | 25–1953 | 17 | No human cross-reactivity |
| Mm-Klb-O1 | 481219 | NM_031180.2 | 2–1825 | 35 | Designed for maximum signal amplification |
| Hs-PECAM | 316729 | NM_001032378.1 | 915–1827 | 20 | Positive control (NHP and human) |
| Hs-FGFR1 | 310079 | NM_023110 | 811–1832 | 23 | Pan-FGFR1 for NHP and human |
| Hs-FGFR2 | 311179 | NM_000141.4 | 1743–3033 | 25 | Pan-FGFR2 for NHP and human |
| Hs-FGFR3 | 310799 | NM_000142.4 | 536–3245 | 28 | Pan-FGFR3 for NHP and human |
| Hs-FGFR4 | 506279 | NM_002011.4 | 151–1690 | 13 | No mouse cross-reactivity |
| Hs-Klb | 445159 | NM_175737.3 | 729–1680 | 20 | Klb for NHP and human |
2.3. ISH assay and probe validation.
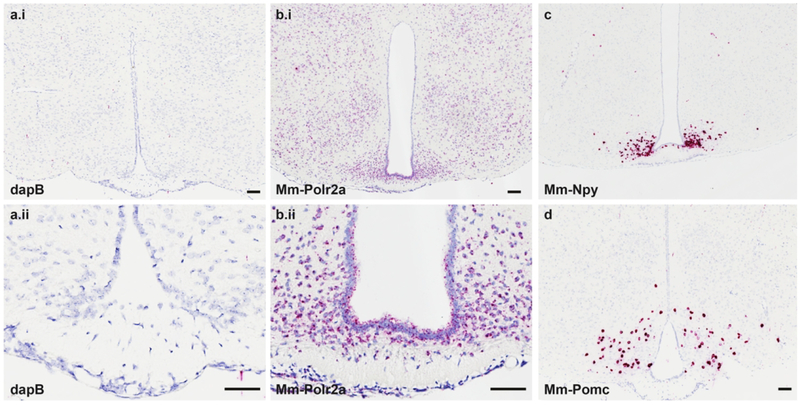
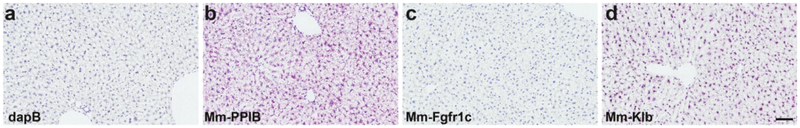
The central FGFR system has been classically described as containing FGFRs 1–3 across mammalian species, with a paucity or complete lack of central FGFR4 using radioisotopic ISH or laser capture/PCR techniques (Belluardo et al., 1997; Bookout et al., 2013; Fon Tacer et al., 2010; Itoh et al., 1994; Yazaki et al., 1994). We therefore aimed to refine this map using RNAscope ISH. To develop the RNAscope platform on our autostainer, we first sought to optimize retrieval conditions in FFPE mus musculus brain tissue. Optimal ISH staining retains crisp nuclear morphometry with well-defined borders, as well as ISH signal yielding ~2–10 red dots/cell cytoplasm using positive control probes. Brain tissues treated as described above and hybridized with the e. coli gene dapB (negative control gene) retained tissue morphology and was devoid of any red ISH signal (Fig. 1a), whereas the adjacent brain tissue hybridized with Mm-Polr2a (positive control gene) exhibited positive red signal in essentially every cell (Fig. 1b). To further test the specificity of this assay, we employed probes designed to identify Mm-NPY (Fig. 1c) and Mm-Pomc (Fig. 1d). In line with classic neuropeptide staining and transgenic reporter labeling of these cells (Broberger, Johansen, Johansson, Schalling, & Hokfelt, 1998; Lam et al., 2015; Luquet, Perez, Hnasko, & Palmiter, 2005; Mercer et al., 2014), the RNAscope ISH platform identified hypothalamic NPY+ cells that straddle the most mediobasal portion of the arcuate nucleus (ARC) along the third ventricle (3V), whereas POMC+ neurons are dispersed more laterally and uniformly throughout the ARC. These cells expressed such an abundance of NPY or POMC mRNA that we could not resolve single ISH dots, despite the fewer number of ZZ pairs that were complexed together to build the signal amplification tree (Table 1). We also validated the utility of our experimental probes by testing them in mouse liver samples (Fig. 2). Using the retrieval and protease preparation timing described in Methods, we assessed ISH expression of dapB (Fig. 2a), Mm-PPIB (Fig. 2b), Mm-Fgfr1 (Fig. 2c) and Mm-Klb (Fig. 2d) in mouse liver. By PCR methods, Mm-Fgfr1 is expressed at very low levels in the mouse liver, whereas Mm-Klb is expressed at relatively high levels in liver homogenates (Fisher et al., 2010). We paralleled these findings by RNAscope ISH, and observed only a low number of red dots in samples hybridized with the Mm-Fgfr1 probe, but near ubiquitous expression in liver sections hybridized with the Mm-Klb probe (Fig. 2d). Collectively, these validation studies showed the robust and high signal-to-noise capabilities of the RNAscope platform (dapB versus Mm-PPIB or Mm-Polr2a), neuroanatomical accuracy (Mm-Npy and Mm-Pomc), and probe set specificity (Mm-Fgfr1c and Mm-Klb in liver) to dissect the central FGFR/Klb system by ISH. Stable transfected Baf3 cells expressing the different receptors, i.e. human-Klb, human-FGFR1c, human-FGFR2, human-FGFR3, and human-FGFR4, were used as positive controls for the ISH analysis of NHP and human samples (data not shown).
Figure 1.
The negative control gene dapB (a1-a2) shows no RNAscope signal in the hypothalamus, whereas the positive control gene Mm-Polr2a (b1) exhibits near ubiquitous expression of 6–10 dots/cell in the mouse brain (b2). Arcuate (ARC) nucleus-specific gene expression of Mm-NPY (c) is clustered along the 3V in the most mediobasal portion of this nucleus, whereas Mm-Pomc (d) is expressed more laterally and dorsally in the ARC. Scale bars = 200 μm.
Figure 2.
dapB shows no expression in the liver using autostainer parameters outlined in Table 1 (a), whereas Mm-PPIB is expressed ubiquitously throughout this organ (b). Mm-Fgfr1c was only detected in a small number of cells (c), whereas Mm-Klb appears in most cells throughout the liver (d). Scale bars = 200 μm.
2.4. Antibody characterization.
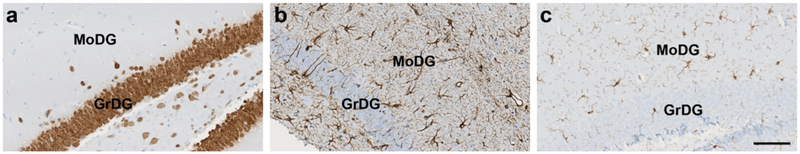
Rodent brains were phenotyped using antibodies directed against specific neuronal, microglia, and astrocyte markers. Neurons were identified using a rabbit monoclonal anti-NeuN antibody (#ab177847, RRID: AB_2532109, clone EPR17263; Abcam, Cambridge, UK) at a 1:500 dilution. The anti-NeuN antibody is raised against amino acids 1–100 of human NeuN, which exhibits 100% homology to mouse and rat NeuN. Microglia were labeled using a rabbit polyclonal anti-Iba1 antibody (#019–19741, RRID: AB_839504; FUJIFILM Wako Diagnostics, Mountain View, CA) at a 1:250 dilution. This antibody was raised against the c-terminus of synthetic Iba1 protein common to human, mouse, and rat Iba1. Astrocytes were identified with a polyclonal rabbit anti-GFAP antibody (#AB5804, RRID: AB_2109645; Sigma/EMD Millipore, St. Louis, MO) at a dilution of 1:1000. The anti-GFAP antibody was developed against purified bovine GFAP. Prior to our use in experiment 2.5, we validated the specificity of these antibodies using standard uniplex diaminobenzidine-based IHC methods (Fig. 3), as well as isotype antibody negative controls (data not shown), and obtained results similar to previously published data with regards to neuron (Saito et al., 2018), astrocyte (Talos, Fishman, et al., 2006; Talos, Follett, et al., 2006), and microglia (Homma, Li, Hayashi, Kawano, & Kawano, 2006; Yamada & Jinno, 2013) morphology.
Figure 3.
Uniplex IHC demonstrated the specificity of three antibodies to detect neurons, astrocytes, and glia in the hippocampus. The anti-NeuN antibody labeled neurons in the granular layer of the dentate gyrus (GrDG, a), whereas anti-GFAP and anti-Iba1 antibodies labeled astrocytes (b) and microglia (c), respectively, in the molecular layer of the dentate gyrus (MoDG). Scale bar = 100 μm.
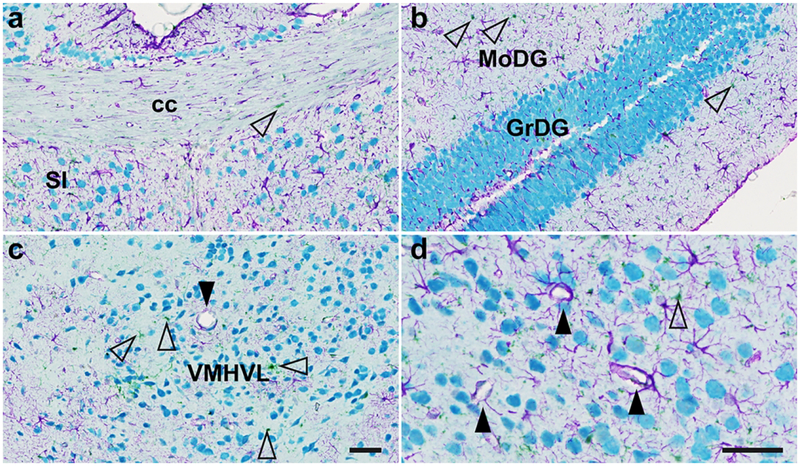
To further validate the specificity of these antibodies, we performed iterative chromogenic IHC with denaturing steps to multilabel mouse brains using all three antibodies (anti-GFAP, anti-Iba1, anti-NeuN). In this experiment, sections were deparaffinized, retrieved in sodium citrate (pH 6.0) for 16 min at 95C, and each antibody was incubated for 2 hr at RT. Primary antibody staining was followed by a 20 min incubation of OmniMap anti-rabbit HRP secondary antibody (#5269679001, Roche Diagnostics, Risch-Rotkreuz, Switzerland) and visualized with chromogenic substrates. We visualized the anti-NeuN antibody with teal-HRP substrate (#8254338001; Roche Diagnostics), the anti-GFAP antibody with purple HRP substrate (#7053983001; Roche Diagnostics), and the anti-Iba1 antibody with green HRP substrate (beta test product from Dr. A. Murillo, Ventana/Roche Diagnostics). Between antibody incubation and staining steps, a 16 min denaturing treatment using sodium citrate (pH 6.0) at 95C removed primary and secondary antibodies without perturbing chromogenic precipitates. This allowed us to triple label brains for astrocytes, glia, and neurons with negligible chromogen overlap between these distinct cell classes (Fig. 4). Any permutation of antibody/chromogen order revealed the same staining of astrocytes, glia, and neurons (data not shown), revealing a robust assay to multilabel brains using antibodies developed in the same species.
Figure 4.
Triple IHC for neurons (NeuN, teal), astrocytes (GFAP, purple), and glia (Iba1, green, open arrows) with three antibodies raised in rabbit. This experiment allowed us to demonstrate the distinct staining patterns for these cell classes simultaneously in the same sections. Representative images show lack of NeuN staining in classical white matter tracts that are nonetheless supported by glia and astrocytes (corpus callosum, cc, 3a), whereas the granular layer of the hippocampal dentate gyrus (GrDG) is packed with neural cell bodies (3b) interspersed with Iba1+ glia and GFAP+ astrocytes. In the ventromedial hypothalamus, ventrolateral portion (VMHVL), astrocytes supporting the blood-brain barrier around a capillary are shown in panel 3c (closed arrow). Panel 3d shows a higher magnification image of astrocyte-ensheathed blood vessels in the contralateral VMHVL (arrows). Scale bars = 25 μm.
2.5. Duplex fluorescent ISH (FISH) and immunofluorescence (IF) for cellular phenotyping.
To phenotype FGFR+ cells in the rodent brain, we combined HRP-based ISH (ACD Bio) to label FGFR mRNA with a rhodamine conjugate (#7259883001; Roche Diagnostics), followed by IF to identify common CNS cell populations using the antibody dilutions outlined in Methods 2.4. The ISH component of this assay followed the same pretreatment and cell conditioning parameters outlined in Table 1. IF was programmed sequentially after the ISH assay, and each primary antibody was incubated for 2 hr at RT on the Discovery ULTRA, followed by a 20 min application of OmniMap anti-rabbit HRP secondary antibody (#5269679001; Roche Diagnostics), and secondary antibodies were visualized using FITC-HRP substrate for 20 min (#7259212001, Roche Diagnostics). We did not perform multiplex IHC in combination with ISH, as the tissue denaturing steps (sodium citrate, pH 6.0 for 16 min at 95C) described in Methods 2.4 compromised antibody binding in our IF sequences if performed after ISH pretreatment/retrieval (data not shown).
2.5. Droplet digital PCR in mouse hypothalamus, cerebrum, and liver.
Five wildtype C57/Bl6 mice were euthanized with 2% CO2, and their brains and livers were rapidly extracted. The hypothalamus from each animal was sub sectioned and harvested as a ~17 mm3 block. Hypothalami were blocked coronally from the preoptic area (+0.26 mm from bregma) to the midbrain (−3.52 mm from bregma), sagitally from optic tract-to-optic tract (+/− 1.5 mm from midline), and horizontally through the ventral thalamus (1.5 mm from the ventral brain surface). RNA was extracted from tissues from 5 animals using an RNeasy mini kit (Qiagen, Hilden, Germany), and quality control for all samples was performed using an Agilent 2100 Bioanalyzer Instrument (Agilent, Santa Clara, CA) and RNA 6000 nano kit reagents (Agilent). All samples had an RNA integrity number exceeding 8.0 (on a 1–10 scale based on 28S and 18S ribosome electropherogram data) and were included for ddPCR assays. For these assays, 10–100 ng of cDNA was made using a high capacity cDNA reverse transcription kit (Applied Biosystems, Foster City, CA) or the iScript cDNA Synthesis Kit (Bio-Rad, Hercules, CA). All samples were assayed in triplicate. ddPCR was performed on a QX200 droplet digital PCR system (Bio-Rad), and data output for positive ddPCR read These assays utilized the same probe sets from Applied Biosystems outlined in Table 3.
Table 3. ddPCR probes.
All probes were purchased from Applied Biosystems
| Probe | Cat. No. | Accession ID | Exon Boundary | Assay Location | Amplicon Length |
|---|---|---|---|---|---|
| Mm-FGFR1 | Mm00438990-m1 | NM_001079909.2 | 9–10 | 1917 | 76 |
| Mm-Klb | Mm00473122-m1 | NM_031180.2 | 1–2 | 824 | 113 |
| Mm-NPY | Mm01410146-m1 | NM_023456.2 | 3–4 | 349 | 130 |
| Mm-POMC | Mm00435874-m1 | NM_008895.4 | 2–3 | 285 | 60 |
| Mm-HPRT | Mm03024075-m1 | NM_013556.2 | 2–3 | 276 | 131 |
2.6. Imaging.
Slides were imaged on an AxioScan.Z1 (Zeiss AG, Oberkochen, Germany) at 40x in brightfield mode or using Colibri 7 LED illumination (Zeiss) to visualize DAPI and rhodamine channels for post-hoc assessment. Rhodamine and DAPI were visualized with an exposure time of 200 ms and 50 ms, respectively. Stained slides with NHP and human brain tissue were scanned into digital images on an Olympus VS120 (Olympus, Hamburg, Germany). Qualitative assessment of FGFR or Klb ISH expression were based on hybridization signal strength from N = 5 animals. ++++: highest density (saturated ISH signal); +++: high density (10–12 dots/cell); ++: moderate density (6–10 dots/cell); +: low density (2–6 dots/cell); +/−: labeling inconsistently above background; −: no labeling. Duplex fluorescent FISH/IF images were captured on an upright DM500 microscope (Leica) at 40x equipped with a CTR 5500 epifluorescent light source (Leica) and a Photofluor LM-75 camera (89 North, Williston, VT). Fluorescent images were captured with exposure times of 50 ms, 500 ms, and 200 ms for DAPI, FITC, and rhodamine channels, respectively. All images were compiled post-hoc in Illustrator (Adobe Inc., San Jose, CA).
3. Results
3.1. Central mRNA expression of Mm-Fgfr1, Mm-Fgfr2, and Mm-Fgfr3 in mus musculus.
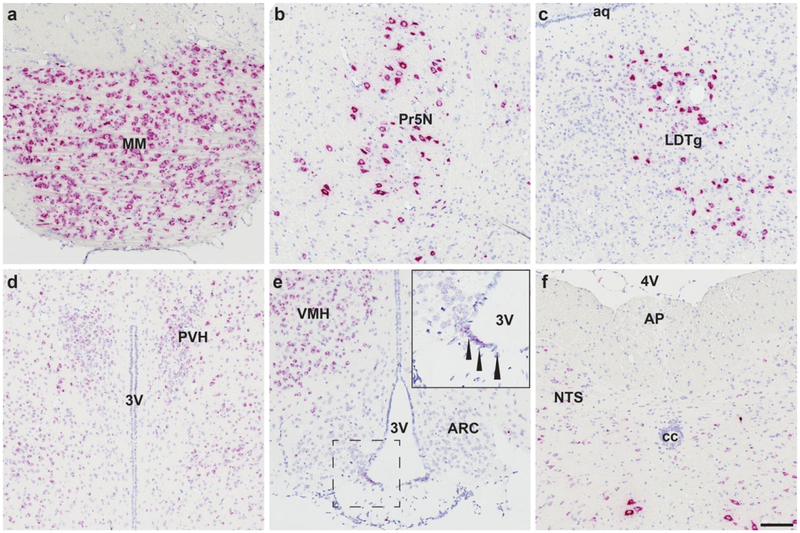
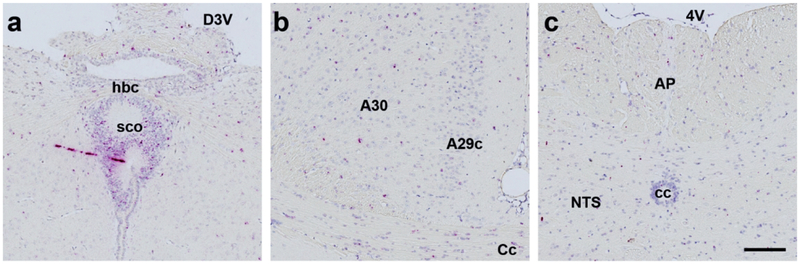
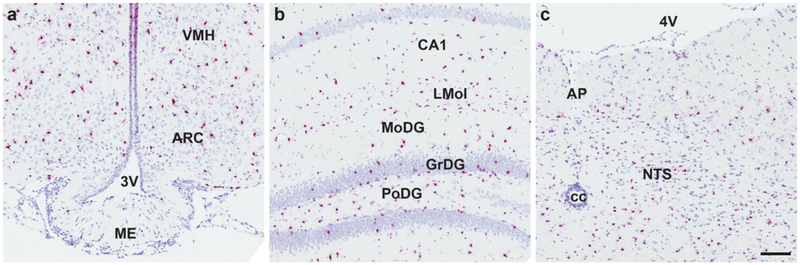
We found Mm-Fgfr1, Mm-Fgfr2, and Mm-Fgfr3 signal expressed throughout the entire mouse brain (representative low-power images are shown in Fig. 5). Fgfr1+ cells with the most robust expression were found in the MM (Fig. 5a), 5N (Fig. 5b), LDTg (Fig. 5c). Cells in these regions expressed enough RNAscope signal that, similar to our hypothalamic Mm-NPY and Mm-Pomc staining, we could not resolve individual red dots in the ISH assay, indicating that these cells strongly express Mm-Fgfr1. Nevertheless, there was differential ISH signal intensity that varied in a region-specific manner. In brain nuclei critical for the maintenance of energy homeostasis, we observed moderately strong staining in the PVH (Fig. 5d), and VMH (Fig. 5e), but fewer copies of Mm-Fgfr1c in the ARC (Fig. 5e), and hindbrain (AP/NTS, Fig. 5f). Mm-Fgfr2 and Mm-Fgfr3 both displayed much more consistent and ubiquitous CNS staining patterns: Signal for either ISH transcript was observed throughout the mouse brain, with highlights for Mm-Fgfr2 and Mm-Fgfr3 shown in Figs. 6 and 7, respectively. Interestingly, we noticed less regional variability with these two probes despite a similar probe design length (20 Z pairs, Table 2), suggesting that many cell populations throughout the CNS express similar amount of Fgfr2 and Fgfr3. Despite the relative dearth of Fgfr2 signal compared to Fgfr1 or Fgfr3, one notable region that showed higher ISH signal across all five mouse brains was the subcommissural organ (Fig. 6a).
Figure 5.
Mm-Fgfr1c is expressed throughout the mouse CNS, with the most qualitatively robust expression observed in the mammillary body (MM, a), principal sensory nucleus of the trigeminal nerve (5N, b), and laterodorsal tegmentum (LDTg, c). These cells express so much Fgfr1 that the ISH staining appears to fill cell bodies, giving them a neuronal-like appearance. In food intake/energy balance governing centers, we observed consistent medium-high expression in the paraventricular hypothalamus (PVN, d), ventromedial hypothalamus (VMH, e), β2-tanycytes (inset, e), and nucleus of the solitary tract (NTS, f), but lower expression in the arcuate nucleus (ARC, e) or area postrema (AP, f). Scale bar = 200 μm.
Figure 6.
While expressed in low levels in the entirety the mouse CNS (1–4 dots/cell), Mm-Fgfr2 appeared the most concentrated in the subcommissural organ (sco, a). Additional representative Mm-Fgfr2 ISH staining is shown in the medial cingulate cortex area 29 and area 30 (b) and hindbrain (c). Scale bar = 200 μm.
Figure 7.
Mm-Fgfr3 is robustly expressed in the mouse CNS, as highlighted in the hypothalamus (a), hippocampus (b), and hindbrain (c). Scale bar = 200 μm.
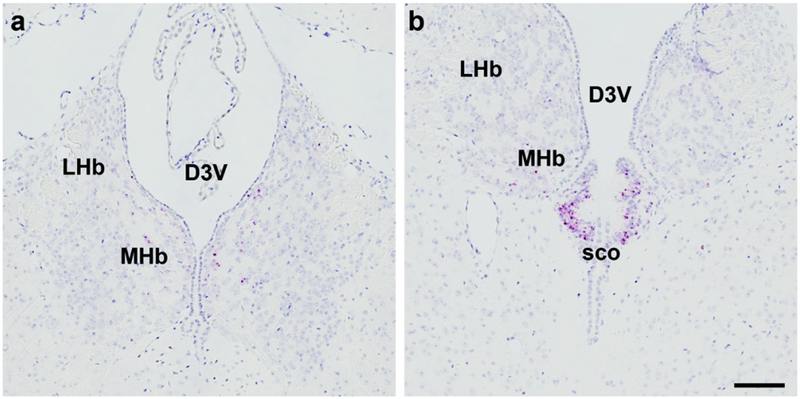
3.2. FGFR4 mRNA expression is restricted to the subcommissural organ (sco) and lateral habenula (LHb) of mus musculus.
Laser capture microdissection and PCR analysis has yielded little information on central expression of Fgfr4 (Fon Tacer et al., 2010). While radioisotopic ISH (Itoh et al., 1994) or digoxigenin ISH suggest small amounts of Fgfr4 in brain regions that lie lateroventral to the dorsal third ventricle, these techniques are hampered by high background relative to low-abundance mRNA targets (DIG) and less spatial resolution (radioisotopic). RNAscope allowed us to assess Mm-Fgfr4 expression in a much greater spatially sensitive manner: we detected Mm-Fgfr4 expression in the lateral habenula (LHb), medial habenula (MHb), and subcommissural organ (sco) (Fig. 8). It is notable that we could distinguish the sco, as this ependymal layer is quite small, extending ~125 μm lateral from midline, 250 μm in the rostrocaudal axis, with a thickness of ~100 μm (Paxinos, Franklin, & Franklin, 2001). Consistent with previous studies, we detected robust Mm-Fgfr4 mRNA in the liver using RNAscope (data not shown), suggesting that our probe is specific and sensitive, and that the rodent CNS only expresses Mm-Fgfr4 in the MHb, LHb, and sco. A complete qualitative comparison of central mouse Fgfr1–4 ISH expression is described in Table 4.
Figure 8.
The central site of Mm-Fgfr4 expression was found in the lateral and medial habenula (a) and more concentrated in the subcommissural organ (sco, b). Scale bar = 200 μm.
Table 4. Relative density of central FGFR1–4 and Klb expression in mus musculus brain.
Qualitative estimates of FGFR1–4 and Klb ISH expression were based on hybridization signal strength from N = 5 animals. ++++: highest density (saturated ISH signal); +++: high density (10–12 dots/cell); ++: moderate density (6–10 dots/cell); +: low density (2–6 dots/cell); +/−: labeling inconsistently above background; −: no labeling.
| Region | Nucleus | FGFR1 | FGFR2 | FGFR3 | FGFR4 | Klb |
|---|---|---|---|---|---|---|
| Cerebral Cortex | Claustrum | ++ | +/− | ++ | − | − |
| Dorsal endopiriform nucleus | ++ | +/− | +++ | − | − | |
| Insular, dysgranular (3, 4, and 6b) | ++ | +/− | ++ | − | − | |
| Insular, granular (3, 4, and 6b) | ++ | +/− | ++ | − | − | |
| Piriform | +++ | +/− | +++ | − | − | |
| Motor, primary (3, and 6b) | ++ | +/− | ++ | − | − | |
| Somatosensory, primary (3, 4, and 6b) | ++ | +/− | ++ | − | − | |
| Somatosensory, secondary (3, 4, and 6b) | ++ | +/− | ++ | − | − | |
| Retrosplenial, agranular (3, and 6b) | ++ | +/− | ++ | − | − | |
| Retrosplenial, granular (3, 4, and 6b) | ++ | +/− | ++ | − | − | |
| Perirhinal | + | +/− | ++ | − | − | |
| Ectorhinal | + | +/− | ++ | − | − | |
| Entorhinal | +++ | +/− | +++ | − | − | |
| Parasubiculum | ++ | +/− | +++ | − | − | |
| Lateral entorhinal | ++ | +/− | ++ | − | − | |
| Auditory, secondary (3, 4, and 6b) | ++ | +/− | ++ | − | − | |
| Temporal association (3, 4, and 6b) | ++ | +/− | ++ | − | − | |
| Forebrain | Nucleus accumbens core | ++ | +/− | + | − | − |
| Nucleus accumbens shell | +++ | +/− | ++ | − | − | |
| Caudate putamen | + | +/− | + | − | − | |
| Hippocampus and septum | Ammon’s horn CA1 | ++ | +/− | ++ | − | − |
| Ammon’s horn CA2 | ++ | +/− | ++ | − | − | |
| Ammon’s horn CA3 | ++ | +/− | ++ | − | − | |
| Dentate gyrus | +++ | + | + | − | − | |
| Lateral septal nucleus, intermediate | +++ | +/− | ++ | − | − | |
| Lateral septal nucleus, ventral | +++ | +/− | ++ | − | − | |
| Bed nucleus of the stria terminalis | ++ | − | +++ | − | − | |
| Thalamus | Paraventricular nucleus, anterior | ++ | +/− | +++ | − | − |
| Anteromedial nucleus | +/− | − | +++ | − | − | |
| Central medial nucleus | +/− | +/− | +++ | − | − | |
| Central lateral nucleus | +/− | + | +++ | − | − | |
| Interoanteromedial nucleus | +/− | + | +++ | − | − | |
| Paracentral nucleus | +/− | + | +++ | − | − | |
| Reticular thalamus | − | +/− | ++ | − | + | |
| Subparafasciucular nucleus | ++ | + | +++ | − | − | |
| Reuniens nucleus | +/− | +/− | +++ | − | − | |
| Submedial nucleus | +/− | +/− | +++ | − | − | |
| Ventral posterolateral nucleus | + | +/− | +++ | − | − | |
| Ventral posteromedial nucleus | +/− | +/− | +++ | − | − | |
| Lateral habenula | ++ | +/− | + | − | − | |
| Medial habenula | ++ | +/− | + | + | − | |
| Subcommissural organ | + | +++ | − | ++ | − | |
| Hypothalamus | Medial preoptic area | +++ | + | +++ | − | − |
| Median preoptic nucleus | ++ | + | +++ | − | − | |
| Lateral preoptic area | ++ | + | +++ | − | − | |
| Paraventricular nucleus | ++ | + | +++ | − | +/− | |
| Suprachiasmatic nucleus | ++ | + | +++ | − | + + + | |
| Supraoptic nucleus | ++ | + | +++ | − | − | |
| Anterior hypothalamic area, posterior | + | +/− | +++ | − | +/− | |
| Arcuate nucleus | + | +/− | +++ | − | − | |
| Median eminence | + | − | + | − | ||
| Ventromedial nucleus, central | ++ | +/− | +++ | − | +/− | |
| Ventromedial nucleus, dorsomedial | +++ | +/− | +++ | − | +/− | |
| Lateral hypothalamic area | ++ | + | +++ | − | − | |
| Dorsomedial nucleus, dorsal | +++ | +/− | +++ | − | +/− | |
| Dorsomedial nucleus, ventral | +++ | +/− | +++ | − | − | |
| Ventral premammillary nucleus | ++++ | + | +++ | − | − | |
| Posterior hypothalamic area | ++ | + | +++ | − | − | |
| Supramammillary nucleus | ++++ | + | ++++ | − | − | |
| β-tanycytes - rostral (−1.2 mm from bregma) | ++ | + | + | − | − | |
| β-tanycytes - mid (−1.6 mm from bregma) | ++++ | ++ | ++ | − | − | |
| β-tanycytes - caudal (−1.9 from bregma) | ++ | + | + | − | − | |
| α-tanycytes - rostral (−1.2 from bregma) | + | + | ++ | − | − | |
| α-tanycytes - mid (−1.6 from bregma) | + | + | ++ | − | − | |
| α-tanycytes - caudal (−1.9 from bregma) | + | ++ | ++++ | − | − | |
| Cerebellum | ++ | ++ | ++ | − | − | |
| Midbrain | Precommissural nucleus | ++ | + | +++ | − | − |
| Ventral tegmental area | ++ | + | ++++ | − | − | |
| Ventrolateral tegmental area | ++ | + | ++++ | − | − | |
| Substantia nigra, compact part | ++ | + | +++ | − | − | |
| Edinger Westphal nucleus | +++ | + | +++ | − | − | |
| Rostral linear nucleus | + | +/− | ++ | − | − | |
| Superior colliculus, deep gray | ++ | − | ++ | − | − | |
| Superior colliculus, superficial gray | ++ | − | ++ | − | − | |
| Deep mesencephalic nucleus | ++ | +/− | +++ | − | − | |
| Cuneiform nucleus | +++ | +/− | +++ | − | − | |
| Medial vestibular nucleus | + | + | +++ | − | + | |
| Lateral vestibular nucleus | − | + | +++ | − | − | |
| Lateral parabrachial nucleus | ++++ | + | +++ | − | − | |
| Medial parabrachial nucleus | ++ | + | +++ | − | − | |
| Medial trigeminal neurons | +++ | − | ++ | − | + | |
| Pontine reticular nucleus, caudal part | ++ | +/− | +++ | − | − | |
| Pontine reticular nucleus, oral part | ++ | +/− | +++ | − | − | |
| Supratrigeminal nucleus | +++ | + | ++ | − | − | |
| Principal sensory nucleus of the trigeminal nerve | ++++ | + | ++ | − | − | |
| Facial nucleus | +++ | − | +++ | − | − | |
| Hindbrain | Area postrema | +/− | +/− | + | − | +/− |
| Nucleus of the solitary tract | +++ | +/− | ++ | − | +/− | |
| Dorsal motor nucleus of the vagus | +++ | +/− | ++ | − | − | |
| Intercalate nucleus of the medulla | ++ | − | ++ | − | − | |
| Hypoglossal nucleus | ++++ | +/− | +++ | − | + | |
| Intermediate reticular nucleus | ++ | +/− | ++ | − | − | |
| Medullary reticular nucleus, ventral | +++ | +/− | ++ | − | − | |
| Medullary reticular nucleus, dorsal | ++++ | +/− | ++ | − | − |
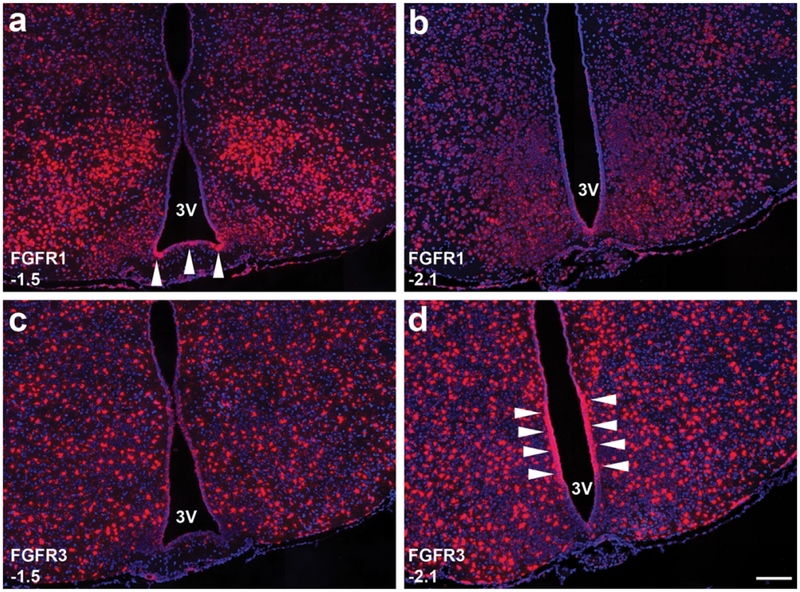
3.3. Heterogeneity of FGFR expression in mouse tanycytes.
Based on our sampling schematic (Methods) we were able to detect FGFR expression in tanycytes at 2–3 levels per animal (Fig. 9). At −1.5 mm from bregma (Fig. 9a), Mm-Fgfr1 appears much more abundantly expressed in the lateral β2-tanycytes, but appears much more homogenously expressed in the caudal ARC (−2.1 mm from bregma; Fig. 9b). Mm-Fgfr3, on the other hand, is homogenously expressed in the tanycyte layer at −1.5 mm from bregma (Fig 9c), but appears highly enriched in the β1-α2 tanycyte transition zone in the caudal ARC (Fig. 9d).
Figure 9.
Tanycytes lining the 3V within the hypothalamus exhibit heterogeneous expression of FGFR1 (a-b) and FGFR3 (c-d): At −1.5 mm from bregma, β2-tanycytes at the base of the 3V strongly express FGFR1 (arrows, a), whereas FGFR3 (C) is expressed much more homogenously in the entire tanycytic layer. In the caudal ARC (−2.1 from bregma), FGFR1 expression is much more uniform (b), whereas FGFR3 at this level is highly expressed in the β1-α2 tanycyte transition zone (arrows, d). Scale bar = 250 μm.
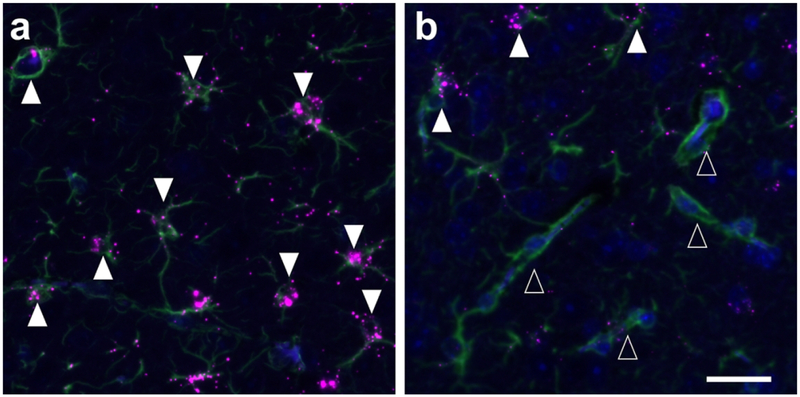
3.4. A phenotypic survey of FGFR+ cells in the mus musculus brain.
To assess the cellular identity of FGFR+ cells in the mouse brain, we performed duplex FISH/IF to visualize receptor mRNA in neurons, microglia, and astrocytes (Fig. 10). Hypothalamic (ARC, VMH, DMH) NeuN+ neurons showed occasional co-localization with FGFR1 ISH signal, whereas only a small subset of Iba1+ microglia co-expressed FGFR2 (data not shown). The consistent FGFR phenotype we observed in the rodent brain, encompassing both hypothalamic and non-hypothalamic sites, was that cells strongly expressing FGFR3 mRNA are GFAP+ astrocytes (Fig. 10a). However, a proportion of these GFAP+ astrocytes do not strongly express FGFR3+, and this subpopulation appears to be surrounding presumptive blood vessels (Fig. 10b).
Figure 10.
We observed occasional FGFR1+/NeuN+ neurons and FGFR2+/Iba1+ glia in the hypothalamus (ARC, VMH, DMH) and hippocampus, but these were a minority of observed total neuron or glial cell populations. GFAP+ astrocytes, on the other hand, consistently expressed high levels of FGFR3 (closed arrows, 8a), with the exception of astrocytes surrounding presumptive blood vessels (open arrows, 8b). Scale bar = 25 μm.
3.5. Klb ISH expression in mice is only found in a small subset of brain regions, and exhibits limited co-localization with Fgfr1c in the SCh, Me5 and Pr5N.
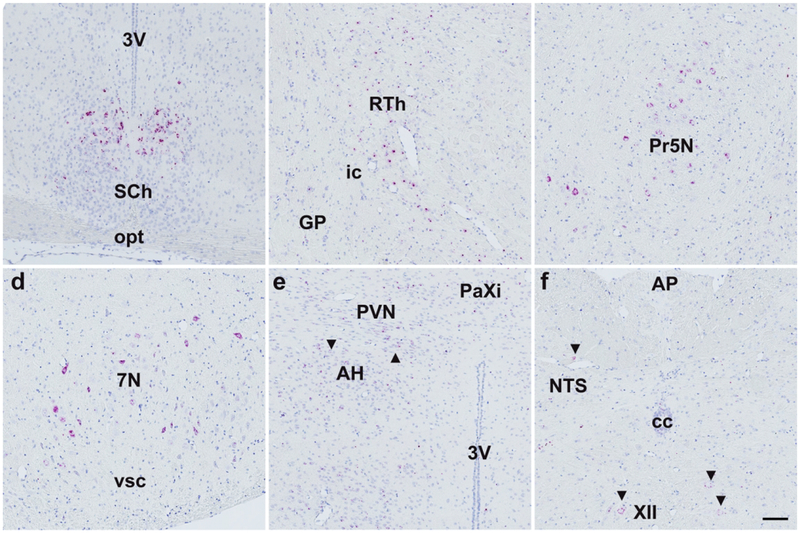
Similar to Fgfr4, central Mm-Klb expression was restricted to a limited number of brain regions: We detected the strongest Mm-Klb expression in the SCh (Fig. 11a), and Mm-Klb+ cells were observed in all animals in the reticular thalamus (RTh, Fig. 11b), principal sensory trigeminal nucleus (Pr5N, Fig. 11c), medial trigeminal neurons (Me5N, Fig. 11d), and hippocampal CA1-CA3 transition zone (data not shown). Klb expression was observed in a small number of cells in the hypothalamus (Fig. 11e) and hindbrain (Fig. 11f), with no apparent expression in the AP (Fig. 10f). We next assessed the degree to which Klb and Fgfr1 co-localize in Klb+ cells. Duplex ISH revealed limited co-expression between Fgfr1 (red, Fig. 12) and Klb (brown, Fig. 12) to cells in the SCh (Fig. 12a), Pr5N (Fig. 12b), Me5N (Fig. 12c), and hippocampal CA1-CA3 transition zone (data not shown). We detected no co-localization of Fgfr1 in Klb+ cells in the RTh, and contrary to previous results (Bookout et al., 2013); we observed sparse-to-no Klb in the AP (Fig 12d) or any other hindbrain regions.
Figure 11.
Klb ISH signal was found predominantly concentrated in the suprachiasmatic nucleus (SCh, a), reticular thalamus (RTh, b), principal sensory trigeminal nucleus (5N, c), and facial nerve nucleus (7N, d). Expression using this technique showed occasional Klb+ cells in the anterior hypothalamus (AH, e, black arrows), paraventricular hypothalamus (PVN, e, black arrows), nucleus of the solitary tract (NTS, f, black arrows), and motor nucleus (XII, f, black arrows), but negligible expression elsewhere in the hypothalamus or hindbrain. Scale bar = 200 μm.
Figure 12.
Mm-Fgfr1c (red) and Klb (brown) were found co-expressed in the suprachiasmatic nucleus (SCh, a), principal sensory nucleus of the trigeminal nerve (Pr5N, b), medial trigeminal neurons (Me5N, c), but not in the area postrema (AP, d). Pr5N and Me5N exhibited low expression of Klb (1–2 DAB+ dots/cell), but this expression pattern was consistent in all neurons assessed in these regions from N=3 animals. Scale bar = 200 μm.
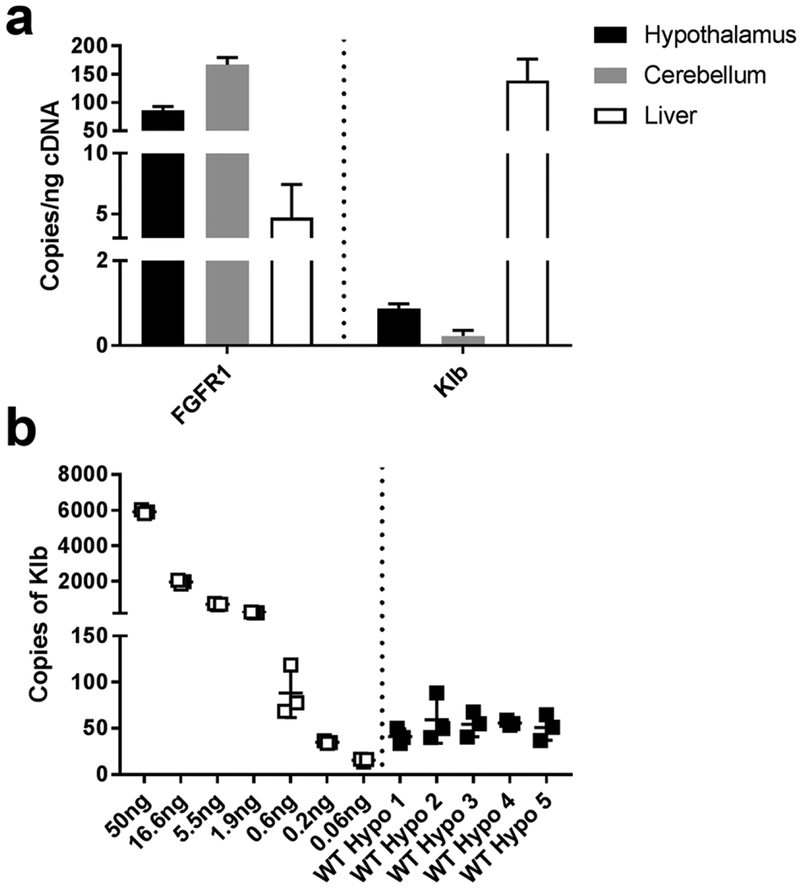
3.6. FGFR/Klb expression in mouse brain tissues by droplet digital PCR (ddPCR).
A disadvantage of qPCR is that low abundance mRNA transcripts (Mm-Klb), or mRNA transcripts with distinct regional expression patterns (Mm-Klb in the SCh, with little signal in the rest of the hypothalamus) are difficult to detect in a sizable piece of tissue. Additionally, qPCR results are relative to the expression of a housekeeping gene, which may vary region-to-region in the CNS. We therefore employed ddPCR (Bio-Rad) to probe fresh frozen tissues to validate our ISH data showing that the mouse hypothalamus does indeed contain Mm-Klb (Fig. 13). ddPCR uses technology that can make nanoliter-size lipid droplets for PCR reactions, allowing users to detect a single copy of RNA. Fgfr1 was robustly expressed in hypothalamus and cerebellum samples (86–166 copies/ng cDNA, Fig. 13a), and as predicted based on ISH data (Fig. 2), Fgfr1 levels in liver were on the order of 5±3 copies/ng cDNA. We measured low levels of Klb in the hypothalamus (1 copy/ng cDNA) and cerebellum (0.2 copies/ng cDNA), but high expression in the liver (139 copies/ng cDNA). To determine the dynamic range for Klb mRNA detection using the ddPCR system, we titrated cDNA from pooled liver mRNA libraries (starting at 50 ng cDNA/20 μl reaction). In this experiment, our lowest liver mRNA input (0.06 ng cDNA/20 μl reaction) yielded 0.837 Klb copies/ng mRNA (Fig. 13b). Comparatively, we measured ~1–2 copies of Klb mRNA per ng cDNA (Fig. 13b). Together, this indicates that our hypothalamic Klb measurements by ddPCR are above the detection limit of the system, and verify that, although few copies are observed in the hypothalamus, it nevertheless contains measureable Mm-Klb (1–2 copies per ng cDNA).
Figure 13.
a: Using ddPCR, we detected 86±6, 166±13, and 5±3 copies of FGFR1/ng cDNA in the hypothalamus (black), cerebellum (gray), and liver (black), respectively. From these same samples, we were able to measure 0.9±0.1, 0.2±0.1, and 139±38 copies of Klb/ng cDNA in the hypothalamus, cerebellum, and liver, respectively. b: To validate our findings that the hypothalamus does indeed contain low levels of Klb expression, we titrated 50 ng of cDNA from liver samples (white) and compared the Klb copy yield to triplicate 50 ng reactions from the hypothalamus (black). We detected a mean of 52.4 copies of Klb/50 ng cDNA reaction in the hypothalamus, which was 4-fold higher than the lower limit of detection in liver samples (15.59 copies of Klb/0.06 ng cDNA reaction).
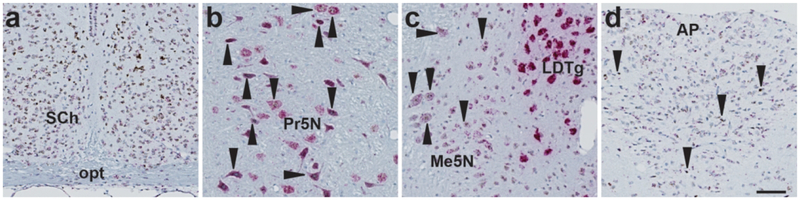
3.7. Human and NHP expression of Fgfr1c, Fgfr2, Fgfr3, Fgfr4, and Klb in the hindbrain and vestibular nuclei.
In order to evaluate the translatability of the mouse ISH data to higher species, automated ISH was performed on human and cynomolgous monkey brain tissue sections encompassing the AP, NTS, and the vestibular nuclei. Abundant Fgfr1c expression was observed in the cynomolgous monkey and human AP and NTS (Fig. 14). In contrast, Klb+ cells were observed in a small number of cells in the human NTS (Fig. 14). Scarce expression of Fgfr2 was detected in human and cynomolgous monkey AP, whereas Fgfr3 ISH signal was only observed in the NTS region of both species (data not shown). No expression of Fgfr4 was observed in any of the human or cynomolgous monkey hindbrain regions investigated (data not shown).
Figure 14.
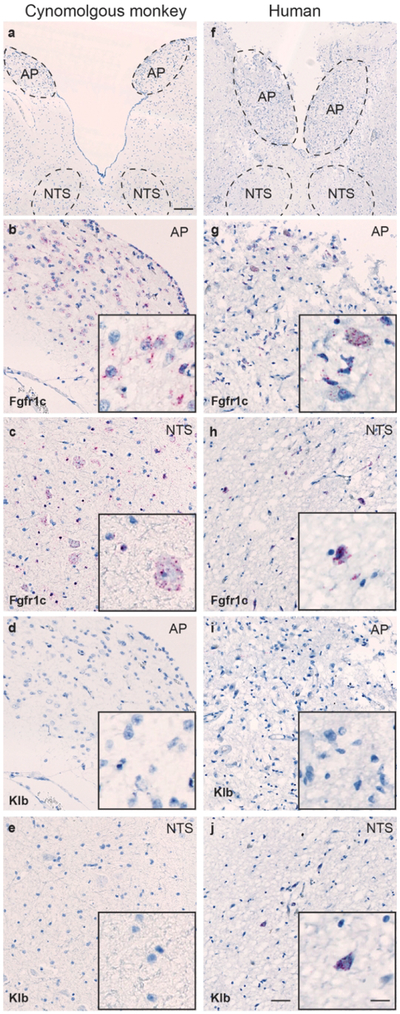
Overview of AP and NTS in human and cynomolgous monkey hindbrain (a and b, respectively. Scale = 250 μm). Evident Fgfr1c ISH signal was found in the AP area of the cynomolgous monkey (b) and human (g) hindbrain, and scant Fgfr1C expression was observed in the NTS of both species (c and h, respectively). Klb ISH signal was only observed in a few numbers of cells in the human NTS (j), whereas no signal for Klb was observed in the cynomoglous AP or NTS (scale = 100 μm). Inserts in figure b-e and g-j show higher magnification of ISH signal (scale = 20 μm).
Investigation of human and cynomolgous monkey tissue sections encompassing the vestibular nuclei revealed no ISH signal of Fgfr1c or Klb in this area (data not shown). Evident expression of both Fgfr2 and Fgfr3 was observed in the human and cynomolgous monkey vestibular nuclei area, whereas no ISH signal of Fgfr4+ cells were detected in this nucleus (data not shown). No ISH signal was detected after hybridization with the negative control probe dapB in any of the tissue sections investigated.
4. Discussion
4.1. Clarifying the central FGFR/Klb receptor system.
Using complementary neuroanatomical, molecular, and multi-species animal approaches, this work provides a comprehensive and systematic dissection of the central FGFR/Klb system. We were able to obtain cellular-level specificity using the RNAscope ISH platform. This methodology allowed us to validate previous works describing ubiquitous, widespread expression of FGFR1–3 (Belluardo et al., 1997; Fon Tacer et al., 2010; Itoh et al., 2015; Yazaki et al., 1994). We further defined these cell populations by dual FISH/IF labeling; our data suggest that cells containing high expression of FGFR3 mRNA are GFAP+ astrocytes. We noted subpopulations of neurons, glia, and astrocytes expressing FGFR1–2, but there was no consistent expression pattern observed in our initial phenotyping observations in the hypothalamus and hippocampus. These data are in line with published RNAseq databases in the rodent cerebral cortex (Zhang et al., 2014) and hypothalamus (Campbell et al., 2017), suggesting that further studies are needed to clarify the regional and phenotypic profiles of FGFR+ cells in the brain. Previous data suggest that FGFR4 is only expressed in the MHb of the rat (Itoh et al., 1994; Miyake & Itoh, 1996), and Klb expression in the mouse is predominantly restricted to the AP and SCh (Bookout et al., 2013; Fon Tacer et al., 2010; Talukdar, Owen, et al., 2016). Our findings further refine this map, indicating expression of FGFR4 in the MHb as well as the LHb and sco, and extend the Klb+ brain regions to include the SCh, Me5N, RTh, and 5N, as well as a few cells observed in the PVN and VMH. Unlike Bookout et al., we only identified a handful of low expressing Klb+ neurons in the NTS and AP despite employing a probe design for maximum signal amplification. With other RNAscope probe sets, we have observed non-specific signal in the choroid plexus using our brain retrieval conditions (K.H. & A.J.M., personal communication), and therefore the AP Klb+ signal observed in the choroid plexus of the fourth ventricle by Bookout et al. may be promiscuous ISH staining. A secondary explanation may be that central Klb expression, particularly in the AP, is regulated by the age, the circadian clock or other extrinsic factors, such as circulating signals or afferent input from visceral sensory fibers transmitted via the vagus nerve (Bookout et al., 2013; Diaz-Delfin et al., 2012; Jager et al., 2016; Kim et al., 2011). Our animals were all adults (>6 months of age) and were euthanized at mid-day, and it is conceivable that this may be a point at which hindbrain Klb expression is reduced. ddPCR experiments, on the other hand, substantiated the presence of Klb in the hypothalamus (Fig. 13). We postulate that the bulk of this signal comes from collecting the SCh as part of our larger hypothalamic blocks, and not from the few Klb+ cells we found in the PVH and VMH (Fig. 11e). Collectively, we were able to corroborate the previous data illustrating central expression of FGFR1–3 throughout the entirety of the rodent CNS, and pinpoint the rare populations of Klb+ and FGFR4+ cells in the mouse brain.
4.2. Species comparisons of FGFR and Klb expression.
Using three mammalian models, our findings indicate that FGFR1–3 are broadly expressed across the CNS of mice, and appear to have similar expression patterns in the hindbrain and midbrain in NHP and man. While FGFR4 and Klb were detected in discrete ependymal, hypothalamic, midbrain, and hindbrain CNS nuclei in mice, mRNA encoding these receptors was not detected in cynomolgus or human brain samples. The two most parsimonious explanations for these species differences are that 1) Klb+ and FGFR4+ cells are much rarer in the brain of higher-order mammals, or 2) mice have unique Klb and FGFR4 expression patterns that are different from primates. Ryan et al. report that FGFR4 is present in rat hypothalamus, providing further evidence of species-specific divergence in CNS FGFR4 and Klb expression (Ryan et al., 2013). We may also have more readily detected rare Klb transcripts in mice due to a more substantial ISH oligonucleotide probe (Table 2), but the total lack of Klb+ midbrain and hindbrain neurons in the Me5N and Pr5N across all primate samples indicates that these cells are quite rare. Interestingly, FGF21 analogs that require Klb for potency can nevertheless drive metabolically relevant decreases in food intake (Stanislaus et al., 2017; Talukdar, Zhou, et al., 2016) and/or body weight (Andersen et al., 2018) in both mice and NHPs. It remains to be determined, however, if these analogs drive metabolic changes through peripheral or central pathways, and notably if these effects are governed by cells and system we have identified that are Klb+.
4.3. Implications for central FGF ligand signaling and the control of peripheral energy homeostasis.
Our initial purpose in mapping the central FGFR/Klb receptor system in greater detail was to provide mechanistic insights into FGF19, FGF21, and FGF1 actions that ameliorate diabetes, obesity, and promote energy expenditure. With regards to endocrine FGFs (principally FGF19 and FGF21), physiological actions of these two ligands requires co-expression of Klb alongside FGFR1c or FGFR4 (Agrawal et al., 2018; Kurosu et al., 2007). By this logic, FGF19 and FGF21 signaling would be limited to sites of FGFR1c/Klb co-expression (Fig. 12). On the other hand, if FGF19 can stimulate FGFR4 in the absence of Klb (Adams et al., 2012; X. Wu et al., 2010), such an action would presumably be limited to the subcommissural organ and habenula (Fig 8). It has been reported that central actions alone of FGF19, FGF21, or an FGFR1/Klb activating antibody can drive improvements in body weight and metabolic state (Foltz et al., 2012; Kolumam et al., 2015; Lan et al., 2017; Morton et al., 2013; Sarruf et al., 2010; A. L. Wu et al., 2011), suggesting that cells in the SCh, Pr5N, and Me5N (Fig. 11–12) are critical for driving this physiological change. Administration of pharmacological doses of these compounds, however, may lessen the requirement of Klb for ligand-receptor-ERK signaling (Adams et al., 2012). In this scenario, ligands that drive responses via FGFR1 may have the capacity to act on many CNS sites, as has been reported for the effect of FGF19 to hyperpolarize hypothalamic AgRP neurons (Marcelin et al., 2014), more recently shown in electrophysiological preparations (A.F.B., personal communication). The possibility that such effects are mediated indirectly via projections from brain areas where both FGFR1 and Klb are expressed can also be considered. Nevertheless, our ISH data has highlighted key regions and cells that express FGFRs and Klb, which can be used to disentangle sites of action after pharmacologic treatment with endocrine FGFs. One limitation in our data set is that the ISH expression patterns identify cells likely to be responsive to FGF ligand stimulation, but do not identify potential non-somatic sites where FGF ligands are active in the brain. Techniques such as in situ ligand binding may reveal the pathways by which FGF19 and FGF21 elicit their putative central effects.
The central actions of FGF1, on the other hand, are complicated by its ligand-receptor biology: FGF1 is a pan-FGFR activator (Beenken & Mohammadi, 2009), and could therefore act on sites across the entire rostro-caudal axis of the CNS of all mammalian species that we have studied. Mechanistically, FGF1 appears to decrease food intake acutely and modulate peripheral blood glucose via hypothalamic astrocytes, tanycytes, and neurons (Ferguson & Johnson, 1991; Robins et al., 2013; Scarlett et al., 2016; Suzuki, Li, Akaike, & Imamura, 2001; Suzuki, Li, Ishisaki, et al., 2001). If FGF1 acts on FGFR3 in astrocytes, FGF1 may induce astrocyte activation, differentiation, and biosynthesis of apoE-HDL, with downstream consequences for local neural networks (G. Anderson & Maes, 2014; Ito et al., 2005; Kang, Lee, Han, Choi, & Song, 2014; Pringle et al., 2003). Within the tanycyte layer, we observed distinct and heterogeneous expression of FGFR1 and FGFR3 in mouse (Fig. 9). Tanycytes lining the third ventricle are regarded as central sensors of circulating metabolites and glucose (Elizondo-Vega et al., 2015; Lopez-Gambero, Martinez, Salazar, Cifuentes, & Nualart, 2018). Notably, the ventral β-tanycytes are implicated in this sensing process, and also appear to directly respond to FGF1 agonism, potentially contributing the mechanism by which central FGF1 application can induce diabetes remission (Scarlett et al., 2016). This divergent expression pattern raises a few interesting questions: Do the different FGFRs in tanycytes govern acute food intake reductions versus prolonged diabetes remission after FGF1 treatment? Does centrally-delivered FGF1 stimulate tanycytes equivalently, or does each tanycyte subclass have specific signaling cascades dependent on the density of FGFR expression? Additionally, rodent brains contain detectable levels of endogenous FGF1 (Chen et al., 2015; Lein et al., 2007; Miller, Ortega, Bashayan, Basch, & Basilico, 2000), but it is unclear if this depot plays a role in the control of energy and glucose homeostasis. Further studies on the acute versus chronic effects of FGF1 on the different astrocyte populations and tanycyte layers, as well as contributions from endogenous central FGF1, may reveal how these interactions contribute to type 2 diabetes remission.
4.4. Concluding remarks.
In summary, we have identified novel brain regions expressing FGFRs/Klb that are postulated to drive the anti-diabetic and anti-obesity actions of central FGF ligand action. While the mechanistic actions of central FGF ligand stimulation remains unresolved, our ISH mapping may provide more mechanistic insights into the receptor classes driving peripheral metabolic homeostasis. Further pharmacologic dissection of FGF ligand stimulation in the mammalian CNS alongside our FGFR/Klb map may shed additional light on this biological system and identify additional pharmacological targets for reducing the burden of obesity and type 2 diabetes.
Acknowledgements
The authors would like to acknowledge Pia Mortensen (Novo Nordisk A/S), Deanna Saxbe (Novo Nordisk Research Center Seattle, Inc.), and Jeanette Bannebjerg Johansen (Novo Nordisk A/S) for expert technical assistance. Dr. Jacob Hecksher-Sorenson (Gubra), Dr. Anna Secher (Novo Nordisk A/S), Dr. Birgitte Andersen (Novo Nordisk, A/S), Dr. Sebastian Parlee (Novo Nordisk Research Center Indianapolis, Inc.), and Dr. Alexei Kharitonenkov (Novo Nordisk Research Center Indianapolis, Inc.) provided many valuable discussions during the development and analysis of this project. We would also like to thank Dr. Paul Kievit (OHSU) for providing non-human primate samples, and the Edinburgh Brain and Tissue Bank for providing human brain tissue samples.
This research was funded by a collaborative partnership between Novo Nordisk (K.L.G, F.H.K.) and the University of Washington (M.W.S.), as well as NIH grants F32 DK104461 (J.M.S.), R01 DK083042 (M.W.S.), and R01 DK101997 (M.W.S.).
K.H., A.F.B., A.C., Y.Z., C.B.G.S., E.J.W., F.H.K., K.L.G., and A.J.M. are employees of Novo Nordisk, which develops, manufactures, and markets therapeutics for the treatment of diabetes and obesity. M.W.S. is a recipient of an early drug discovery grant from Novo Nordisk for the treatment of diabetes and obesity. J.M.S., J.B., and G.J.M. have no conflicts of interest.
Footnotes
Data Sharing
The authors are pleased to share any raw data generated from this study. Contact the senior author (A.J.M.; aomc@novonordisk.com) for inquiries into full slide scans or brain regions of interest. A.J.M. also takes responsibility for the integrity of the data and the accuracy of the data analysis.
References
- Adams AC, Coskun T, Rovira AR, Schneider MA, Raches DW, Micanovic R, … Kharitonenkov A (2012). Fundamentals of FGF19 & FGF21 action in vitro and in vivo. PLoS One, 7(5), e38438. doi: 10.1371/journal.pone.0038438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agrawal A, Parlee S, Perez-Tilve D, Li P, Pan J, Mroz PA, … DiMarchi RD (2018). Molecular elements in FGF19 and FGF21 defining KLB/FGFR activity and specificity. Mol Metab, 13, 45–55. doi: 10.1016/j.molmet.2018.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen B, Straarup EM, Heppner KM, Takahashi DL, Raffaele V, Dissen GA, … Kievit P (2018). FGF21 decreases body weight without reducing food intake or bone mineral density in high-fat fed obese rhesus macaque monkeys. Int J Obes (Lond). doi: 10.1038/s41366-018-0080-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson CM, Zhang B, Miller M, Butko E, Wu X, Laver T, … Ma XJ (2016). Fully Automated RNAscope In Situ Hybridization Assays for Formalin-Fixed Paraffin-Embedded Cells and Tissues. J Cell Biochem, 117(10), 2201–2208. doi: 10.1002/jcb.25606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson G, & Maes M (2014). Reconceptualizing adult neurogenesis: role for sphingosine-1-phosphate and fibroblast growth factor-1 in co-ordinating astrocyte-neuronal precursor interactions. CNS Neurol Disord Drug Targets, 13(1), 126–136. [DOI] [PubMed] [Google Scholar]
- Beenken A, & Mohammadi M (2009). The FGF family: biology, pathophysiology and therapy. Nat Rev Drug Discov, 8(3), 235–253. doi: 10.1038/nrd2792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belluardo N, Wu G, Mudo G, Hansson AC, Pettersson R, & Fuxe K (1997). Comparative localization of fibroblast growth factor receptor-1, −2, and −3 mRNAs in the rat brain: in situ hybridization analysis. J Comp Neurol, 379(2), 226–246. [PubMed] [Google Scholar]
- Bookout AL, de Groot MH, Owen BM, Lee S, Gautron L, Lawrence HL, … Kliewer SA (2013). FGF21 regulates metabolism and circadian behavior by acting on the nervous system. Nat Med, 19(9), 1147–1152. doi: 10.1038/nm.3249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broberger C, Johansen J, Johansson C, Schalling M, & Hokfelt T (1998). The neuropeptide Y/agouti gene-related protein (AGRP) brain circuitry in normal, anorectic, and monosodium glutamate-treated mice. Proc Natl Acad Sci U S A, 95(25), 15043–15048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell JN, Macosko EZ, Fenselau H, Pers TH, Lyubetskaya A, Tenen D, … Tsai LT (2017). A molecular census of arcuate hypothalamus and median eminence cell types. Nat Neurosci, 20(3), 484–496. doi: 10.1038/nn.4495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassidy A, & Jones J (2014). Developments in in situ hybridisation. Methods, 70(1), 39–45. doi: 10.1016/j.ymeth.2014.04.006 [DOI] [PubMed] [Google Scholar]
- Chen MS, Lin HK, Chiu H, Lee DC, Chung YF, & Chiu IM (2015). Human FGF1 promoter is active in ependymal cells and dopaminergic neurons in the brains of F1B-GFP transgenic mice. Dev Neurobiol, 75(3), 232–248. doi: 10.1002/dneu.22225 [DOI] [PubMed] [Google Scholar]
- Diaz-Delfin J, Hondares E, Iglesias R, Giralt M, Caelles C, & Villarroya F (2012). TNF-alpha represses beta-Klotho expression and impairs FGF21 action in adipose cells: involvement of JNK1 in the FGF21 pathway. Endocrinology, 153(9), 4238–4245. doi: 10.1210/en.2012-1193 [DOI] [PubMed] [Google Scholar]
- Douris N, Stevanovic DM, Fisher FM, Cisu TI, Chee MJ, Nguyen NL, … Maratos-Flier E (2015). Central Fibroblast Growth Factor 21 Browns White Fat via Sympathetic Action in Male Mice. Endocrinology, 156(7), 2470–2481. doi: 10.1210/en.2014-2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elizondo-Vega R, Cortes-Campos C, Barahona MJ, Oyarce KA, Carril CA, & Garcia-Robles MA (2015). The role of tanycytes in hypothalamic glucosensing. J Cell Mol Med, 19(7), 1471–1482. doi: 10.1111/jcmm.12590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson IA, & Johnson EM Jr. (1991). Fibroblast growth factor receptor-bearing neurons in the CNS: identification by receptor-mediated retrograde transport. J Comp Neurol, 313(4), 693–706. doi: 10.1002/cne.903130412 [DOI] [PubMed] [Google Scholar]
- Fisher FM, Chui PC, Antonellis PJ, Bina HA, Kharitonenkov A, Flier JS, & Maratos-Flier E (2010). Obesity is a fibroblast growth factor 21 (FGF21)-resistant state. Diabetes, 59(11), 2781–2789. doi: 10.2337/db10-0193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foltz IN, Hu S, King C, Wu X, Yang C, Wang W, … Li Y (2012). Treating diabetes and obesity with an FGF21-mimetic antibody activating the betaKlotho/FGFR1c receptor complex. Sci Transl Med, 4(162), 162ra153. doi: 10.1126/scitranslmed.3004690 [DOI] [PubMed] [Google Scholar]
- Fon Tacer K, Bookout AL, Ding X, Kurosu H, John GB, Wang L, … Kliewer SA (2010). Research resource: Comprehensive expression atlas of the fibroblast growth factor system in adult mouse. Mol Endocrinol, 24(10), 2050–2064. doi: 10.1210/me.2010-0142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasser E, Moutos CP, Downes M, & Evans RM (2017). FGF1 - a new weapon to control type 2 diabetes mellitus. Nat Rev Endocrinol. doi: 10.1038/nrendo.2017.78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heppner KM, Baquero AF, Bennett CM, Lindsley SR, Kirigiti MA, Bennett B, … Smith MS (2017). GLP-1R Signaling Directly Activates Arcuate Nucleus Kisspeptin Action in Brain Slices but Does not Rescue Luteinizing Hormone Inhibition in Ovariectomized Mice During Negative Energy Balance. eNeuro, 4(1). doi: 10.1523/ENEURO.0198-16.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homma A, Li HP, Hayashi K, Kawano Y, & Kawano H (2006). Differential response of arcuate proopiomelanocortin- and neuropeptide Y-containing neurons to the lesion produced by gold thioglucose administration. J Comp Neurol, 499(1), 120–131. doi: 10.1002/cne.21097 [DOI] [PubMed] [Google Scholar]
- Ito J, Nagayasu Y, Lu R, Kheirollah A, Hayashi M, & Yokoyama S (2005). Astrocytes produce and secrete FGF-1, which promotes the production of apoE-HDL in a manner of autocrine action. J Lipid Res, 46(4), 679–686. doi: 10.1194/jlr.M400313-JLR200 [DOI] [PubMed] [Google Scholar]
- Itoh N, Ohta H, & Konishi M (2015). Endocrine FGFs: Evolution, Physiology, Pathophysiology, and Pharmacotherapy. Front Endocrinol (Lausanne), 6, 154. doi: 10.3389/fendo.2015.00154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh N, Yazaki N, Tagashira S, Miyake A, Ozaki K, Minami M, … Kawasaki T (1994). Rat FGF receptor-4 mRNA in the brain is expressed preferentially in the medial habenular nucleus. Brain Res Mol Brain Res, 21(3–4), 344–348. [DOI] [PubMed] [Google Scholar]
- Jager J, Wang F, Fang B, Lim HW, Peed LC, Steger DJ, … Lazar MA (2016). The Nuclear Receptor Rev-erbalpha Regulates Adipose Tissue-specific FGF21 Signaling. J Biol Chem, 291(20), 10867–10875. doi: 10.1074/jbc.M116.719120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang K, Lee SW, Han JE, Choi JW, & Song MR (2014). The complex morphology of reactive astrocytes controlled by fibroblast growth factor signaling. Glia, 62(8), 1328–1344. doi: 10.1002/glia.22684 [DOI] [PubMed] [Google Scholar]
- Kim J, Eskiocak U, Stadler G, Lou Z, Kuro-o M, Shay JW, & Wright WE (2011). Short hairpin RNA screen indicates that Klotho beta/FGF19 protein overcomes stasis in human colonic epithelial cells. J Biol Chem, 286(50), 43294–43300. doi: 10.1074/jbc.M111.267641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolumam G, Chen MZ, Tong R, Zavala-Solorio J, Kates L, van Bruggen N, … Sonoda J (2015). Sustained Brown Fat Stimulation and Insulin Sensitization by a Humanized Bispecific Antibody Agonist for Fibroblast Growth Factor Receptor 1/betaKlotho Complex. EBioMedicine, 2(7), 730–743. doi: 10.1016/j.ebiom.2015.05.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurosu H, Choi M, Ogawa Y, Dickson AS, Goetz R, Eliseenkova AV, … Kuro-o M (2007). Tissue-specific expression of betaKlotho and fibroblast growth factor (FGF) receptor isoforms determines metabolic activity of FGF19 and FGF21. J Biol Chem, 282(37), 26687–26695. doi: 10.1074/jbc.M704165200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam DD, Attard CA, Mercer AJ, Myers MG Jr., Rubinstein M, & Low MJ (2015). Conditional expression of Pomc in the Lepr-positive subpopulation of POMC neurons is sufficient for normal energy homeostasis and metabolism. Endocrinology, 156(4), 1292–1302. doi: 10.1210/en.2014-1373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan T, Morgan DA, Rahmouni K, Sonoda J, Fu X, Burgess SC, … Mangelsdorf DJ (2017). FGF19, FGF21, and an FGFR1/beta-Klotho-Activating Antibody Act on the Nervous System to Regulate Body Weight and Glycemia. Cell Metab, 26(5), 709–718 e703. doi: 10.1016/j.cmet.2017.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lein ES, Hawrylycz MJ, Ao N, Ayres M, Bensinger A, Bernard A, … Jones AR (2007). Genome-wide atlas of gene expression in the adult mouse brain. Nature, 445(7124), 168–176. doi: 10.1038/nature05453 [DOI] [PubMed] [Google Scholar]
- Lopez-Gambero AJ, Martinez F, Salazar K, Cifuentes M, & Nualart F (2018). Brain Glucose-Sensing Mechanism and Energy Homeostasis. Mol Neurobiol. doi: 10.1007/s12035-018-1099-4 [DOI] [PubMed] [Google Scholar]
- Luquet S, Perez FA, Hnasko TS, & Palmiter RD (2005). NPY/AgRP neurons are essential for feeding in adult mice but can be ablated in neonates. Science, 310(5748), 683–685. doi: 10.1126/science.1115524 [DOI] [PubMed] [Google Scholar]
- Marcelin G, Jo YH, Li X, Schwartz GJ, Zhang Y, Dun NJ, … Chua S Jr. (2014). Central action of FGF19 reduces hypothalamic AGRP/NPY neuron activity and improves glucose metabolism. Mol Metab, 3(1), 19–28. doi: 10.1016/j.molmet.2013.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer AJ, Stuart RC, Attard CA, Otero-Corchon V, Nillni EA, & Low MJ (2014). Temporal changes in nutritional state affect hypothalamic POMC peptide levels independently of leptin in adult male mice. Am J Physiol Endocrinol Metab, 306(8), E904–915. doi: 10.1152/ajpendo.00540.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller DL, Ortega S, Bashayan O, Basch R, & Basilico C (2000). Compensation by fibroblast growth factor 1 (FGF1) does not account for the mild phenotypic defects observed in FGF2 null mice. Mol Cell Biol, 20(6), 2260–2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyake A, & Itoh N (1996). Rat fibroblast growth factor receptor-4 mRNA in the brain is preferentially expressed in cholinergic neurons in the medial habenular nucleus. Neurosci Lett, 203(2), 101–104. [DOI] [PubMed] [Google Scholar]
- Morton GJ, Matsen ME, Bracy DP, Meek TH, Nguyen HT, Stefanovski D, … Schwartz MW (2013). FGF19 action in the brain induces insulin-independent glucose lowering. J Clin Invest, 123(11), 4799–4808. doi: 10.1172/JCI70710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paxinos G, Franklin KBJ, & Franklin KBJ (2001). The mouse brain in stereotaxic coordinates (2nd ed.). San Diego: Academic Press. [Google Scholar]
- Pringle NP, Yu WP, Howell M, Colvin JS, Ornitz DM, & Richardson WD (2003). Fgfr3 expression by astrocytes and their precursors: evidence that astrocytes and oligodendrocytes originate in distinct neuroepithelial domains. Development, 130(1), 93–102. [DOI] [PubMed] [Google Scholar]
- Robins SC, Stewart I, McNay DE, Taylor V, Giachino C, Goetz M, … Placzek M (2013). alpha-Tanycytes of the adult hypothalamic third ventricle include distinct populations of FGF-responsive neural progenitors. Nat Commun, 4, 2049. doi: 10.1038/ncomms3049 [DOI] [PubMed] [Google Scholar]
- Rojas JM, Matsen ME, Mundinger TO, Morton GJ, Stefanovski D, Bergman RN, … Schwartz MW (2015). Glucose intolerance induced by blockade of central FGF receptors is linked to an acute stress response. Mol Metab, 4(8), 561–568. doi: 10.1016/j.molmet.2015.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan KK, Kohli R, Gutierrez-Aguilar R, Gaitonde SG, Woods SC, & Seeley RJ (2013). Fibroblast growth factor-19 action in the brain reduces food intake and body weight and improves glucose tolerance in male rats. Endocrinology, 154(1), 9–15. doi: 10.1210/en.2012-1891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K, Koike T, Kawashima F, Kurata H, Shibuya T, Satoh T, … Mori T (2018). Identification of NeuN immunopositive cells in the adult mouse subventricular zone. J Comp Neurol, 526(12), 1927–1942. doi: 10.1002/cne.24463 [DOI] [PubMed] [Google Scholar]
- Sarruf DA, Thaler JP, Morton GJ, German J, Fischer JD, Ogimoto K, & Schwartz MW (2010). Fibroblast growth factor 21 action in the brain increases energy expenditure and insulin sensitivity in obese rats. Diabetes, 59(7), 1817–1824. doi: 10.2337/db09-1878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarlett JM, Rojas JM, Matsen ME, Kaiyala KJ, Stefanovski D, Bergman RN, … Schwartz MW (2016). Central injection of fibroblast growth factor 1 induces sustained remission of diabetic hyperglycemia in rodents. Nat Med, 22(7), 800–806. doi: 10.1038/nm.4101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanislaus S, Hecht R, Yie J, Hager T, Hall M, Spahr C, … Xu J (2017). A Novel Fc-FGF21 With Improved Resistance to Proteolysis, Increased Affinity Toward beta-Klotho, and Enhanced Efficacy in Mice and Cynomolgus Monkeys. Endocrinology, 158(5), 1314–1327. doi: 10.1210/en.2016-1917 [DOI] [PubMed] [Google Scholar]
- Suh JM, Jonker JW, Ahmadian M, Goetz R, Lackey D, Osborn O, … Evans RM (2014). Endocrinization of FGF1 produces a neomorphic and potent insulin sensitizer. Nature, 513(7518), 436–439. doi: 10.1038/nature13540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki S, Li AJ, Akaike T, & Imamura T (2001). Intracerebroventricular infusion of fibroblast growth factor-1 increases Fos immunoreactivity in periventricular astrocytes in rat hypothalamus. Neurosci Lett, 300(1), 29–32. [DOI] [PubMed] [Google Scholar]
- Suzuki S, Li AJ, Ishisaki A, Hou X, Hasegawa M, Fukumura M, … Imamura T (2001). Feeding suppression by fibroblast growth factor-1 is accompanied by selective induction of heat shock protein 27 in hypothalamic astrocytes. Eur J Neurosci, 13(12), 2299–2308. [DOI] [PubMed] [Google Scholar]
- Talos DM, Fishman RE, Park H, Folkerth RD, Follett PL, Volpe JJ, & Jensen FE (2006). Developmental regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole-propionic acid receptor subunit expression in forebrain and relationship to regional susceptibility to hypoxic/ischemic injury. I. Rodent cerebral white matter and cortex. J Comp Neurol, 497(1), 42–60. doi: 10.1002/cne.20972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talos DM, Follett PL, Folkerth RD, Fishman RE, Trachtenberg FL, Volpe JJ, & Jensen FE (2006). Developmental regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole-propionic acid receptor subunit expression in forebrain and relationship to regional susceptibility to hypoxic/ischemic injury. II. Human cerebral white matter and cortex. J Comp Neurol, 497(1), 61–77. doi: 10.1002/cne.20978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talukdar S, Owen BM, Song P, Hernandez G, Zhang Y, Zhou Y, … Kliewer SA (2016). FGF21 Regulates Sweet and Alcohol Preference. Cell Metab, 23(2), 344–349. doi: 10.1016/j.cmet.2015.12.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talukdar S, Zhou Y, Li D, Rossulek M, Dong J, Somayaji V, … Calle RA (2016). A Long-Acting FGF21 Molecule, PF-05231023, Decreases Body Weight and Improves Lipid Profile in Non-human Primates and Type 2 Diabetic Subjects. Cell Metab, 23(3), 427–440. doi: 10.1016/j.cmet.2016.02.001 [DOI] [PubMed] [Google Scholar]
- Wang F, Flanagan J, Su N, Wang LC, Bui S, Nielson A, … Luo Y (2012). RNAscope: a novel in situ RNA analysis platform for formalin-fixed, paraffin-embedded tissues. J Mol Diagn, 14(1), 22–29. doi: 10.1016/j.jmoldx.2011.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu AL, Kolumam G, Stawicki S, Chen Y, Li J, Zavala-Solorio J, … Sonoda J (2011). Amelioration of type 2 diabetes by antibody-mediated activation of fibroblast growth factor receptor 1. Sci Transl Med, 3(113), 113ra126. doi: 10.1126/scitranslmed.3002669 [DOI] [PubMed] [Google Scholar]
- Wu X, Ge H, Lemon B, Vonderfecht S, Weiszmann J, Hecht R, … Li Y (2010). FGF19-induced hepatocyte proliferation is mediated through FGFR4 activation. J Biol Chem, 285(8), 5165–5170. doi: 10.1074/jbc.M109.068783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada J, & Jinno S (2013). Novel objective classification of reactive microglia following hypoglossal axotomy using hierarchical cluster analysis. J Comp Neurol, 521(5), 1184–1201. doi: 10.1002/cne.23228 [DOI] [PubMed] [Google Scholar]
- Yazaki N, Hosoi Y, Kawabata K, Miyake A, Minami M, Satoh M, … Itoh N (1994). Differential expression patterns of mRNAs for members of the fibroblast growth factor receptor family, FGFR-1-FGFR-4, in rat brain. J Neurosci Res, 37(4), 445–452. doi: 10.1002/jnr.490370403 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Chen K, Sloan SA, Bennett ML, Scholze AR, O’Keeffe S, … Wu JQ (2014). An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J Neurosci, 34(36), 11929–11947. doi: 10.1523/JNEUROSCI.1860-14.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]