Abstract
We report here the development of a suite of biocompatible SuFEx transformations from the SOF4-derived iminosulfur oxydifluoride-hub in aqueous buffer conditions. Thes biocompatible SuFEx reactions of iminosulfur oxydifluorides (R-N=SOF2) with primary amines give sulfamides (8 examples, up to 98%), while the reaction with secondary amines furnish sulfuramidimidoyl fluoride products (8 examples, up to 97%). Likewise, under mild buffered conditions, phenols react with the iminosulfur oxydifluorides (Ar-N=SOF2) to produce sulfurofluoridoimidates (13 examples, up to 99%), which can themselves be further modified by nucleophiles. These transformations open the potential for asymmetric and trisubstituted linkages projecting from the sulfur(VI) center, including versatile S-N and S-O connectivity (9 examples, up to 94%). Finally, the SuFEx bioconjugation of iminosulfur oxydifluorides to amine-tagged single-stranded DNA and to BSA protein demonstrate the potential of SOF4 derived SuFEx click chemistry in biological applications.
Keywords: SuFEx, Click Chemistry, Bioconjugation, Protein modification, DNA Encoded Libraries
Graphical Abstract
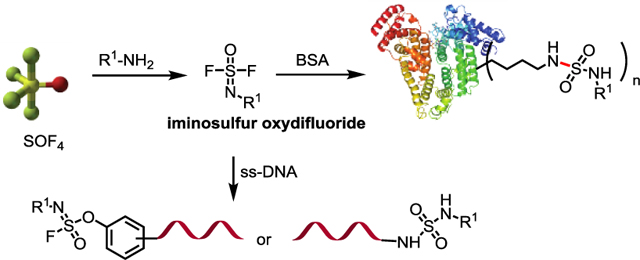
We describe the first high yielding and biocompatible SOF4 based SuFEx reaction methodology that operates at low concentration in aqueous solution under mild pH and temperature conditions. The biocompatible SuFEx reactions open the potential for asymmetric and trisubstituted products at the sulfur(VI) center with versatile S-N and S-O connectivity. The SuFEx bioconjugation of iminosulfur oxydifluorides to either BSA protein or to single-stranded DNA support the development of these useful reactions in bioconjugation applications.
Synthetic methods that allow the chemoselective derivatization of biomolecules, such as proteins, DNA, RNA, and carbohydrates etc., have enabled many important discovery-based investigations, and are essential tools for interrogating biomolecules in their native environment.[1]
The development of bio-orthogonal click chemistry over the past decade[2], and in particular CuAAC and the strain promoted copper-free Huisgen cycloaddition reactions have had enormous impact as tools for enabling chemical conjugations. Despite these major advances, there remains a growing demand for engineering fast, selective, and high yielding transformations that can be conducted in a complex biological milieu under physiological conditions. This is particularly pertinent in the burgeoning field of DNA-Encoded Chemical Libraries (DELs). Pioneered by Lerner and Brenner in 1992, DELs comprise individual organic molecules with distinctive DNA tags that serve as amplifiable identification bar codes to allow the construction and screening of combinatorial libraries of unprecedented size.[3]
To avoid damaging the DNA during the course of synthesizing the library, there are a number of requirements that a reaction must meet. For example: the chemistry must be compatible with aqueous media and proceed at moderate temperatures(< 90 °C) within pH 4–14; the reaction rate constant should be high enough to accommodate low concentrations of reactants (< 1 mM); and finally, the reactants should be inert to phosphates and nucleotides. Unsurprisingly, only a handful of reactions are available (Figure 1),[4] and there is an urgent need to expand the repertoire of DNA compatible transformations.
Figure 1.
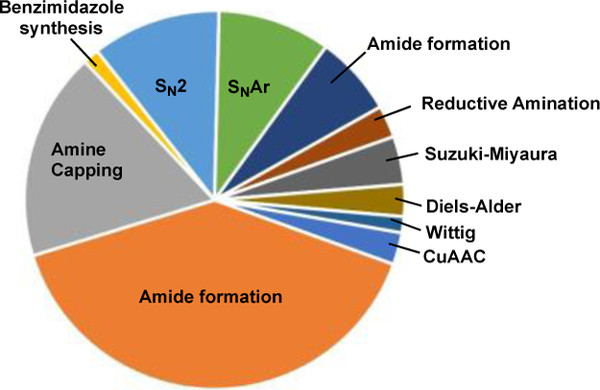
Reactions that are commonly used and compatible for DELs chemistry. [4f]
Sulfur-fluoride exchange (SuFEx) is a new generation of click chemistry that enables new connections to be formed through S-F exchange with a range of nucleophiles, to give stable S-N, S-O, and S-C covalent linkages. Much like nature’s own phosphate hubs, SuFEx connectors enable the modular construction of functional molecules through SuFExable hubs including: sulfuryl fluoride (SO2F2),[5] ethenesulfonyl fluoride (ESF),[6] and thionyl tetrafluoride (SOF4).[7]
A growing number of applications of SuFEx click chemistry have been reported since it was first introduced in 2014,[5] including new synthetic methodologies,[8] polymer preparation,[5,9] and bioconjugation.[10] However, it wasn’t until the evolution of Thionyl tetrafluoride (SOF4) as the first multidimensional SuFEx hub that the true power of polyvalent click chemistry was realized—opening the door to the 3-dimensional world and the domain of biological space.[7]
SOF4 is a reactive gas that under SuFEx catalysis undergoes S-F exchange with primary amines, including anilines and aliphatic primary amines, to afford tetrahedral iminosulfur oxydifluoride products I (Figure 2).[7a-b] The remaining two SuFExable S–F handles of I can themselves undergo sequential SuFEx reactions with N-, O-, and C- nucleophiles, to form a diverse array of sulfur fluoride compounds and downstream multi-substituted derivatives II–VII. Collectively, these transformations form the framework for creating SOF4 derived SuFEx platforms and libraries of extreme, hub-based special variety.[7]
Figure 2.
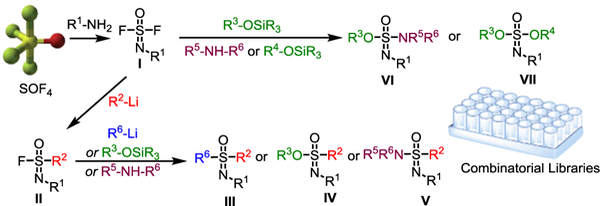
SuFEx connections from the polyvalent and multidimensional hub of SOF4.
SuFEx enabled “Schotten-Baumann-like”, linkage reactions with amines and phenols are nearly perfect at aqueous buffer interfaces, but better in that unwanted hydrolysis is even more suppressed. This interfacial enhanced mode of S-F reactivity is deemed perfect for bioconjugation endeavors where SuFEx activation can be achieved with nucleophilic catalysts (e.g. DBU (1,8-diazabicyclo[5.4.0]undec-7-ene)), and also by vigorous stirring with an immiscible buffered aqueous phase. The blending of water with miscible solvents such as THF or acetonitrile also aid the process, although we posit that most SuFEx reactions will benefit from two-phase conditions in which the aqueous phase contains bifluoride (KFHF). We anticipated that such conditions would be amenable for the development of biocompatible SuFEx chemistry, and report here the first example of SOF4-derived SuFEx reactions that proceed in dilute aqueous solutions under mild conditions of pH and temperature. We demonstrate the power of the new bioconjugation methodology by the joining of small molecules to amine-tagged single-stranded DNA and also to BSA-protein.[11]
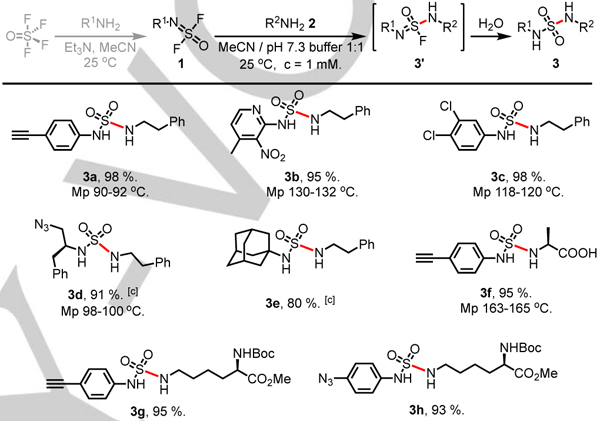
The p-ethynyl-aniline iminosulfur oxydifluoride 1a and 2-phenylethan-1-amine 2a were selected as model connective partners for the development of the SuFEx reactions in aqueous conditions (Table S1, see supporting information for condition optimization). Rigorous screening of conditions revealed the reaction of 2a and iminosulfur oxydifluoride 1a proceeded rapidly at low concentration (1 mM in pH 7.3 aq. PBS buffer/MeCN 1:1 mixture) at room temperature. Notably, the sulfuramidimidoyl fluoride intermediate 3’ was not detected in the reaction mixture—itself spontaneously hydrolyzing to furnish the sulfamide product 3 (Table 1).
Table 1.
 |
Reactions were performed with 1 (0.10 mmol), 2 (0.10 mmol), MeCN 50 mL, pH 7.3 aq. PBS buffer 50 mL, at 25 °C for 5 minutes unless otherwise stated.
Isolated yield.
For 3 d and 3 e, the reaction was carried out for 1 hour.
Under the optimized reaction conditions, the iminosulfur oxydifluorides 1 underwent the substitution-hydrolysis sequence with 2a to give the corresponding sulfamide products in excellent yield (3a-3h, 80–98%). The aliphatic amine iminosulfur oxydifluorides 2d and 2e were found to be slightly less reactive than the aniline counterparts, although the reactions were generally complete within 1 hour to giving products 3d (91%) and 3e (80%). It is noteworthy that the reaction of the amino acid L-alanine 2f and 1a gave product 3f in 95% yield, demonstrating that the SuFEx is compatible with free carboxylic acid functionality. Similarly, the long chain aliphatic primary amine N-Boc-lysine methyl ester 2g coupled successfully with both 1a and 1h to form the products 3g and 3h respectively.
Next, the substitution by secondary amines 4 under mild aqueous conditions were examined (Table 2). A slight increase of the reaction temperature (37 °C) and prolonged reaction time (12 h) were required to yield the corresponding sulfuramidimidoyl fluorides 5 in excellent yield (5a–h, 88–97%).
Table 2.
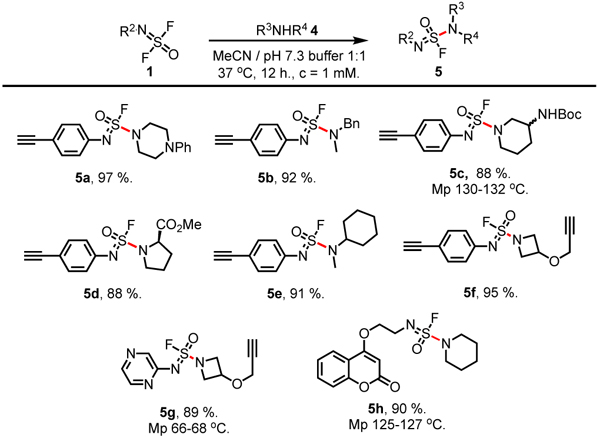 |
Reactions were performed with 1 (0.10 mmol), 4 (0.10 mmol), MeCN 50 mL, pH 7.3 aq. PBS buffer 50 mL, at 37 °C for 12 hours.
Isolated yield.
In the context of bioconjugation, the potential SuFEx chemistry of tyrosine was considered particularly important, since it is found abundantly in naturally occurring proteins. We then examined the reactivity of a number of phenols 6 with iminosulfur oxydifluorides 1 under buffered aqueous conditions (pH 8.0). As illustrated in table 3, the monosubstituted sulfurofluoridoimidates 7a–7l (95–99%) were readily obtained with stirring at 37 °C for 16 h in slightly basic pH 8.0 aq. PBS buffer/MeCN 1:1 mixtures. A wide selection of aromatic substituents on both the iminosulfur oxydifluorides 1 and the phenols 6 were well tolerated, although the reaction of the aliphatic iminosulfur oxydifluoride 1m and phenol produced 7m in only moderate yield (19%), even when a large excess of difluoride 1m (10 equiv.) was used. In this particular case, most of the starting material 1m was hydrolyzed to the (2-phenoxyethyl)sulfamic acid. Significantly, the products 7j-k, and 7l, derived from the reaction with N-Boc-tyrosine methyl ester and the N-Boc-tyrosine respectively, were isolated in excellent yield. The results support the notion that bioconjugation to protein tyrosine residues through the SOF4-derived iminosulfur oxydifluoride-hub is viable.
Table 3.
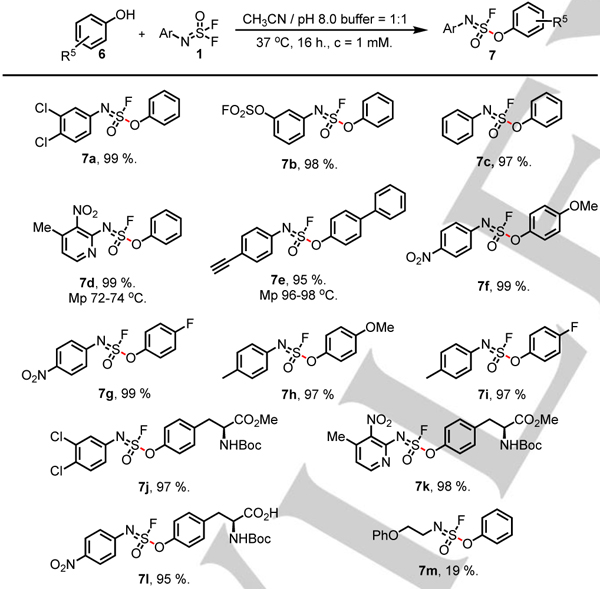 |
Reactions were performed with 1 (0.10 mmol), 6 (0.10 mmol), MeCN 50 mL, pH 8.0 aq. PBS buffer 50 mL, at 37 °C for 16 hours unless otherwise stated.
Isolated yield.
For 7 m, 1mmol (10 eq.) of 1m was used.
The stability of the monofluoride species 5 and 7 were next tested in 1mM aqueous buffered solutions as a function of pH (Figure S1 and S2). The compound 7a was found to be stable after agitation at 37 °C for 48 h from pH 6 to pH 8, but hydrolyzed to the sulfonate at more acidic or basic pH. On the other hand, compound 5a was more stable even after subjection to agitation at 37 °C for 96 h in pH 2 to pH 14 solutions. Based upon these results, we studied the reaction of monofluoride 7a and pyrrolidine 4j (Table S2), to implement a second SuFEx reaction.
Screening different reaction conditions revealed that a large excess of nucleophile (50 equiv.) under more extreme conditions (80 °C, pH 10) were required to achieve satisfactory conversion. The reaction with a range of secondary amines 4a-e gave the corresponding sulfonimidates 8a-e in good yields, whereas reaction with phenols 6a-d gave excellent conversion to the sulfurimidates 9a-d (86–94%) (Table 4).
Table 4.
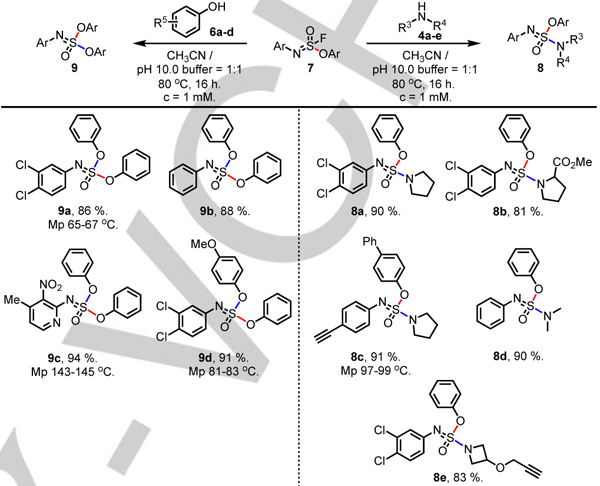 |
Reactions were performed with 7 (0.10 mmol), 4 or 6 (5 mmol, 50 eq), MeCN 50 mL, pH 10 aq. PB buffer 50 mL, at 80 °C for 16 hours.
Isolated yield.
In sharp contrast, efforts to substitute the S-F bond of the sulfuramidimidoyl fluoride 5 using ArOH or secondary amine as nucleophile were unsuccessful, even under forcing conditions (100 °C, pH 10, 48 h)—this is consistent with the increased stability imparted by the greater electron density on sulfur associated with two nitrogen-sulfur bonds.
With optimized conditions in hand, we next investigated the suitability of the new biocompatible SOF4 derived SuFEx conditions in the labelling of DNA, and hence as new technology DEL (Figure 3). The single-stranded DNA 11, comprising a long chain primary amine tail overhang and hairpin structure was selected as a suitable model substrate for conjugation reaction. Agitation of a reaction mixture comprising difluoride 1a and 11 at room temperature for 6 h resulted in successful conjugation and formation of the product 12 in 70% yield (determined by HPLC). The phenol-tailed single-stranded DNA 14 was then synthesized by DNA-amide condensation and desilylation with linker molecule 13. Coupling of 14 with the difluoride 1a gave the corresponding small molecule-DNA bioconjugate with the CuAAC-ready alkyne 15.[11g]
Figure 3.
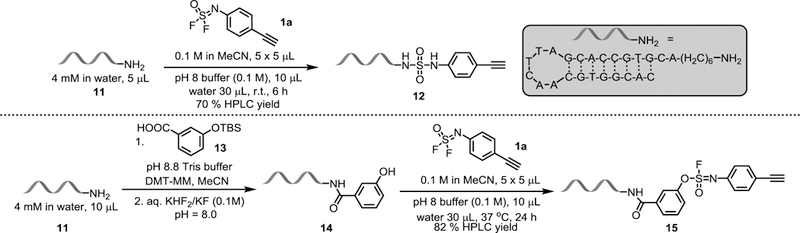
Bioconjugation of difluoride 1a to amine-tagged ss-DNA.
To explore the new SOF4-derived reagents for protein modification, bovine serum albumin (BSA) was chosen as a model protein to react with the alkyne-bearing difluoride 1a (1–100 equivalents). The resultant BSA-SuFEx bioconjugates were purified and analyzed using liquid chromatography-mass spectrometry (LC-MS) (Figure 4a). Using 1 molar equivalent of 1a to react with BSA (15 μM in pH 7.3 PBS) for 1 hour at 25 °C, a single modification of the target protein was observed. With 10 equivalents of 1a under the same condition, a triple modification was detected in the MS spectrum, and one modification site was confirmed as Lys548 by LC-MS/MS (Figure S11). When 100 equivalents of difluroride 1a were used, BSA was found to be modified by 8 molecules of 1a (Scheme S7–S9). Remarkably, in all cases, only lysine residues were found to be modified. The alkyne-modified BSA conjugates enabled further reaction with biotin-PEG3-azide via ligand-assisted CuAAC[12]. Likewise, the biotin-PEG3-amine difluoride 16 could be directly connected to BSA via an exposed amine such as a lysine residue (Figure 4b). However, due to lower reactivity of this aliphatic amine derived difluoride, only moderate SuFEx conversion of ~30% was observed (Scheme S12).
Figure 4.
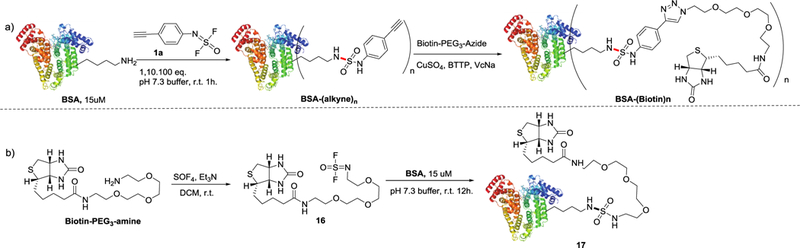
a) Sequential SuFEx bioconjugation and biotin CuAAC labeling of BSA protein.
b) Modification of BSA protein with biotin-PEG3-amine difluoride 16.
In conclusion, we have developed a series of high yielding and biocompatible SOF4 based SuFEx reactions that operate at low concentration, in aqueous solution under mild pH and temperature conditions. A diverse selection of chemical space was sampled through the polyvalent and multidimensional N- and O-nucleophilic substitutions from the SOF4 hub. The efficient labelling of BSA protein and amine-tagged single-stranded DNA was also accomplished, demonstrating the potential of the biocompatible SuFEx ligation sequence as a useful tool for bioconjugation applications. Collectively, the experimental results validate the suitability for aqueous phase combinatorial chemistry and we posit that SOF4 mediated SuFEx will prove to be a generally useful addition to the click chemistry toolbox with wide application in chemical biology, biophysics, bioengineering, and materials science.
Supplementary Material
Acknowledegements
The authors gratefully acknowledge financial support from the NIH (R01 GM117145 to K.B.S. R01GM093282 to P.W.), the ARC for supporting a Future Fellowship FT170100156 (J.E.M), and NSFC 21302126(F. L).
References
- [1].Sletten EM, Bertozzi CR, Angew. Chem. Int. Ed. 2009, 48, 6974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].a) Kolb HC, Finn MG, Sharpless KB, Angew. Chem. Int. Ed. 2001, 40, 2004. [DOI] [PubMed] [Google Scholar]; b) Baskin JM, Bertozzi CR, Qsar & Comb. Sci. 2007, 26, 1211. [Google Scholar]; c) Tornøe CW, Christensen C, Meldal M, J. Org. Chem. 2002, 67, 3057. [DOI] [PubMed] [Google Scholar]; d) Rostovtsev VV, Green LG, Fokin VV, Sharpless KB, Angew. Chem. Int. Ed. 2002, 41, 2596. [DOI] [PubMed] [Google Scholar]; e) Baskin JM, Prescher JA, Laughlin ST, Agard NJ, Chang PV, Miller IA, Lo A, Codelli JA, Bertozzi CR, Proc. Natl. Acad. Sci. 2007, 104, 16793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].a) Brenner S, Lerner RA, Proc. Natl. Acad. Sci. 1992, 89, 5381. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Brenner S, Lerner RA, Angew. Chem. Int. Ed. 2017, 56, 1164. [DOI] [PubMed] [Google Scholar]; c) Neri D, Lerner RA, Annu. Rev. Biochem. 2018, 87, 479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].a) Malone ML, Paegel BM, ACS Comb. Sci. 2016, 18, 182. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Nicholas N, Bassi G, Scheuermann J, Neri D, FEBS Lett. 2018, 592, 2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Dong J, Krasnova L, Finn MG, Sharpless KB, Angew. Chem. Int. Ed. 2014, 53, 9430. [DOI] [PubMed] [Google Scholar]
- [6].a) Krutak JJ, Burpitt RD, Moore WH, Hyatt JA, J. Org. Chem. 1979, 44, 3847; [Google Scholar]; b) Zheng Q, Dong J, Sharpless KB, J. Org. Chem. 2016, 81, 11360. [DOI] [PubMed] [Google Scholar]; c) Qin H, Zheng Q, Bare GAL, Wu P, Sharpless KB, Angew. Chem. Int. Ed. 2016, 55, 14155. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Chinthakindi PK, Govender KB, Kurnar AS, Kruger HG, Govender T, Naicker T, Arvidsson PI, Org. Lett. 2017, 19, 480. [DOI] [PubMed] [Google Scholar]; e) Zha G, Zheng Q, Leng J, Wu P, Qin H, Sharpless KB, Angew. Chem. Int. Ed. 2017, 56, 4849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].a) Li S, Wu P, Moses JE, Sharpless KB, Angew. Chem. Int. Ed. 2017, 56, 2903. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Gao B, Li S, Wu P, Moses JE, Sharpless KB, Angew. Chem. Int. Ed. 2018, 56, 2903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].a) Kende AS, Mendoza JS, J. Org. Chem. 1990, 55, 1125; [Google Scholar]; b) Chen W, Dong J, Li S, Liu Y, Wang Y, Yoon L, Wu P, Sharpless KB, Kelly JW, Angew. Chem. Int. Ed. 2016, 55, 1835. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Zhang E, Tang J, Li S, Wu P, Moses JE, Sharpless KB, Chem. Eur. J. 2016, 22, 5692. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Ren G, Zheng Q, Wang H, Org. Lett. 2017, 19, 1582. [DOI] [PMC free article] [PubMed] [Google Scholar]; e) Dondoni A, Marra A, Org. Biomol. Chem. 2017, 15, 1549. [DOI] [PubMed] [Google Scholar]
- [9].a) Dong J, Sharpless KB, Kwisnek L, Oakdale JS, Fokin VV, Angew. Chem. Int. Ed. 2014, 53, 9466. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Li S, Beringer LT, Chen S, Averick S, Polymer 2015, 78, 37. [Google Scholar]; c) Yatvin J, Brooks K, Locklin J, Chem. Eur. J. 2016, 22, 16348. [DOI] [PubMed] [Google Scholar]; d) Oakdale JS, Kwisnek L, Fokin VV, Macromolecules 2016, 49, 4473. [Google Scholar]; e) Wang H, Zhou F, Ren G, Zheng Q, Chen H, Gao B, Klivansky L, Liu Y, Wu B, Xu Q, Lu J, Sharpless KB, Wu P, Angew. Chem. Int. Ed. 2017, 56, 11203. [DOI] [PMC free article] [PubMed] [Google Scholar]; f) Brendel JC, Martin L, Zhang J, Perrier S, Polym. Chem. 2017, 8, 7475. [Google Scholar]; g) Wang H, Ren F, Zhou G, Zheng Q, Chen H, Gao B, Klivansky L, Liu Y, Wu B, Xu Q, Angew. Chem. Int. Ed. 2017, 56, 11203. [DOI] [PMC free article] [PubMed] [Google Scholar]; i) Zhu H, Chen D, Li N, Xu Q, Li H, He J, Wang H, Wu P, Lu J, Chemistry 2017, 23, 14712. [DOI] [PubMed] [Google Scholar]; j) Gao B, Zhang L, Zheng Q, Zhou F, Klivansky LM, Lu J, Liu Y, Dong J, Wu P, Sharpless KB, Nat. Chem. 2017, 9, 1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].a) Grimster NP, Connelly S, Baranczak A, Dong J, Krasnova LB, Sharpless KB, Powers ET, Wilson IA, Kelly JW, J. Am. Chem. Soc. 2013, 135, 5656. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Baranczak A, Liu Y, Connelly S, Du W, Greiner ER, Genereux JC, Wiseman RL, Eisele YS, Bradbury NC, Dong J, Noodleman L, Sharpless KB, Wilson IA, Encalada SE, Kelly JW, J. Am. Chem. Soc. 2015, 137, 7404. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Zelli R, Tommasone S, Dumy P, Marra A, Dondoni A, Eur. J. Org. Chem. 2016, 30, 5102. [Google Scholar]; d) Li S, Cohen-Karni D, Beringer LT, Wu C, Kallick E, Edington H, Passineau MJ, Averick S, Polymer 2016, 99, 7. [Google Scholar]; e) Hoppmann C, Wang L, Chem. Commun. 2016, 52, 5140. [DOI] [PubMed] [Google Scholar]; f) Chen W, Dong J, Plate L, Mortenson DE, Brighty GJ, Li S, Liu Y, Galmozzi A, Lee PS, Hulce JJ, Cravatt BF, Saez E, Powers ET, Wilson IA, Sharpless KB, Kelly JW, J. Am. Chem. Soc. 2016, 138, 7353. [DOI] [PMC free article] [PubMed] [Google Scholar]; g) Álvarez NH, van de Langemheen H, Brouwer AJ, Liskamp RM, Bioorganic & Medicinal Chemistry 2017, 25, 5055. [DOI] [PubMed] [Google Scholar]; i) Mortenson DE, Brighty GJ, Plate L, Bare G, Chen W, Li S, Wang H, Cravatt BF, Forli S, Powers ET, Sharpless KB, Wilson IA, Kelly JW, J. Am. Chem. Soc. 2018, 140, 200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].a) Hermanson GT, Bioconjugate Techniques (Academic Press, San Diego, CA, 1996). [Google Scholar]; b) Griffin BA, Adams SR, Tsien RY, Science, 1998, 281, 269. [DOI] [PubMed] [Google Scholar]; c) Wu P, Shui W, Carlson BL, Hu N, Rabuka D, Lee J, Bertozzi CR Proc. Natl. Acad. Sci. USA, 2009, 106, 3000. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Casi G, Huguenin-Dezot N, Zuberbuhler K, Scheuermann J, Neri D. J. Am. Chem. Soc. 2012, 134, 5887. [DOI] [PubMed] [Google Scholar]; e) MacDonald JI, Munch HK, Moore T, Francis MB. Nat. Chem. Biol. 2015, 11, 326. [DOI] [PubMed] [Google Scholar]; f) Zhang C, Welborn M, Zhu T, Yang NJ, Santos MS, Van Voorhis T, Pentelute BL, Nat. Chem. 2016, 8, 120. [DOI] [PMC free article] [PubMed] [Google Scholar]; g) Choi EJ, Jung D, Kim J, Lee Y, Kim BM, Chem. Eur. J. 2018, 24, 10948. [DOI] [PubMed] [Google Scholar]
- [12].Soriano del Amo D, Wang W, Jiang H, Besanceney C, Yan AC, Levy M, Liu Y, Marlow FL, Wu P, J. Am. Chem. Soc. 2010, 132, 16893. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


