Abstract
Lipid-conjugated small-interfering RNAs (siRNAs) exhibit accumulation and gene silencing in extrahepatic tissues, providing an opportunity to expand therapeutic siRNA utility beyond the liver. Chemically engineering lipids may further improve siRNA delivery and efficacy, but the relationship between lipid structure/configuration and siRNA pharmacodynamics is unclear. Here, we synthesized a panel of mono-, di-, and tri-meric fatty acid-conjugated siRNAs to systematically evaluate the impact of fatty acid structure and valency on siRNA clearance, distribution, and efficacy. Fatty acid valency significantly altered the physicochemical properties of conjugated siRNAs, including hydrophobicity and micelle formation, which affected distribution. Trivalent lipid-conjugated siRNAs were predominantly retained at the site of injection with minimal systemic exposure, whereas monovalent lipid-conjugated siRNAs were quickly released into the circulation and accumulated primarily in kidney. Divalent lipid-conjugated siRNAs showed intermediate behavior, and preferentially accumulated in liver with functional distribution to lung, heart, and fat. The chemical structure of the conjugate, rather than overall physicochemical properties (i.e. hydrophobicity), predicted the degree of extrahepatic tissue accumulation necessary for productive gene silencing. Our findings will inform chemical engineering strategies for enhancing the extrahepatic delivery of lipophilic siRNAs.
Keywords: Therapeutic oligonucleotides, RNA interference, Conjugated siRNAs, Branched lipid conjugate, Pharmacokinetics, siRNA delivery, Gene silencing in vivo
Introduction
Small interfering RNAs (siRNAs) are an emerging class of drugs [1] that target disease-causing messenger RNA (mRNA) for degradation in a sequence-specific manner [2]. Therapeutic siRNAs are potent, have a long duration of effect, and are able to target previously “undruggable” disease genes [3–5]. However, they exhibit poor in vivo stability and distribution. To overcome these limitations, siRNAs are first chemically modified, then conjugated with diverse chemical moieties [6, 7]. Fully chemically modified siRNAs conjugated to a trivalent N-acetylgalactosamine (GalNAc) show functional delivery to hepatocytes in humans [8–12], and represent a major breakthrough in the therapeutic oligonucleotide field.
Lipid conjugation has also emerged as a delivery platform for siRNAs [6]. Sterols, fatty acids, and vitamins (with or without a phosphocholine moiety) conjugated to siRNAs have been shown to impact both local and systemic siRNA distribution [7, 13–17]. Lipid-conjugated siRNAs primarily accumulate in clearance tissues (liver, kidney, and spleen). Less hydrophobic compounds preferentially bind to high density lipoprotein (HDL) in serum and accumulate in kidneys, whereas more hydrophobic siRNAs primarily bind low density lipoprotein (LDL) and distribute to the liver [17, 18]. Lipophilic siRNAs also distribute to extrahepatic and extrarenal tissues, which may be influenced by conjugate configuration [17, 19, 20].
Lipid engineering may be a useful strategy for further enhancing siRNA delivery to specific tissues. Indeed, lipid branching and multi-valency approaches are used in the development of lipid-based nanocarriers with advanced properties [21–25]. However, we currently have a limited understanding of how lipid chemical structure and configuration impact siRNA pharmacokinetic/pharmacodynamic behavior, tissue accumulation, and efficacy.
Here, we synthesized a panel of siRNAs conjugated to mono-, di- or tri-meric docosahexaenoic acid, eicosapentaenoic acid, or myristic acid to evaluate the impact of lipid structure and valency on siRNA clearance, distribution, and efficacy. Altering fatty acid valency significantly impacted siRNA hydrophobicity, resulting in different clearance profiles: (i) trivalent lipid-conjugated siRNAs were retained at the injection site with minimal systemic exposure; (ii) monovalent lipid-conjugated siRNAs were quickly released to the circulation and predominantly accumulated in kidney; and (iii) divalent lipid-conjugated siRNAs showed intermediate behavior, with preferential liver accumulation but wide distribution to other tissues (lung, heart, fat). In addition to hydrophobicity, conjugate structure also contributed to the degree of tissue accumulation required for productive silencing. Our study provides the first example of how rational lipid engineering may be used to fine-tune the properties of therapeutic siRNAs, and their extrahepatic delivery.
Materials and Methods
Oligonucleotide synthesis, deprotection, and purification
Oligonucleotides were synthesized following standard protocols on an Expedite ABI DNA/RNA synthesizer or a MerMade 12 (BioAutomation, Irving, TX, USA). Sense strands were synthesized at 1-μmole scales on synthesized fatty acid-functionalized CPG supports (Supplementary Schemes 1, 2 and 3). Antisense strands were synthesized at 10-μmole scales on CPG functionalized with Unylinker® (ChemGenes, Wilmington, MA). 2´-O-methyl phosphoramidites (ChemGenes, Wilmington, MA), 2´-fluoro phosphoramidites (BioAutomation, Irving, Texas), Cy3-labeled phosphoramidites (Gene Pharma, Shanghai, China), and custom synthesized (E)-vinylphosphonate phosphoramidites [26] were used for preparing oligonucleotides. Oligonucleotides were removed from CPG and deprotected using 40% aqueous methylamine, and purified by HPLC as described previously [15, 26]. Purified oligonucleotides were desalted by size-exclusion chromatography, and purity and identity were determined by Liquid Chromatography-Mass Spectrometry (LC-MS) on an Agilent 6530 accurate-mass Q-TOF LC/MS (Agilent technologies, Santa Clara, CA) (Supplementary Figure 1).
Physicochemical characterization of conjugated oligonucleotides
The relative hydrophobicities of conjugated siRNAs were determined by the retention time of each compound on an Agilent Prostar System equipped with a Water HxSil C18 column (75 × 4.6) in a gradient of 100% buffer A (0.1 M trimethylamine acetate in water) to 100% buffer B (0.1 M trimethylamine acetate in acetonitrile) at a flow rate of 1 ml/min for 16 min at 60°C.
Hydrodynamic diameters of siRNA were determined by dynamic light scattering (DLS) using a Zetasizer ZEN3600 (Malvern Instruments, UK). siRNAs solutions (10 nmol in 1 mL PBS) were analyzed at 25°C in triplicate. All the scattered photons were collected at a 173°- scattering angle. The scattering intensity data was processed using instrumental software to obtain the hydrodynamic diameter and the size distribution of each sample.
Injection of conjugated siRNAs into mice
Animal experiments were performed in accordance with animal care ethics approval and guidelines of University of Massachusetts Medical School Institutional Animal Care and Use Committee (IACUC, protocol number A-2411). Female FB/NJ mice (The Jackson Laboratory) 7- to 8-weeks old were injected subcutaneously with phosphate buffered saline (PBS controls) or with 20 mg/kg conjugated siRNA (or unconjugated control) suspended in PBS (160 μL). For pharmacokinetic studies, 6 mice per group were injected (n = 18). For distribution studies, 3 mice per conjugate were studied (n = 9 + 1 for PBS controls). For the efficacy studies, 6 mice per group were injected (n = 30, including non-targeting controls and PBS controls).
Pharmacokinetic studies
After injection, serial blood sampling was carried out using a previously described protocol [27]. Briefly, microsamples of blood were collected from the lateral saphenous vein at different time points. Sterile needles were used to puncture the vein, slight pressure was applied above the knee joint, and blood droplets were collected using Microvette CB300 K2R tubes (Sarstedt). Blood samples were stored at −80°C until analysis.
Peptide nucleic acid (PNA) hybridization assay
Blood and tissue concentrations of antisense strands were determined using a PNA hybridization assay [27, 28]. Blood samples (10 μL) were diluted in tissue lysis solution (MasterPure, EpiCentre) to a total of 200 μL containing 1 μL proteinase K (20 mg/mL) (Invitrogen). Tissues (15 mg) were placed in QIAGEN Collection Microtubes holding 3-mm tungsten beads and lysed in 300 μl tissue lysis solution containing 3 μl proteinase K using a QIAGEN TissueLyser II. For all samples, sodium dodecyl sulphate (SDS) was precipitated from lysates by adding 20–30 μL 3 M potassium chloride and centrifuging at 5000 × g for 15 minutes. Supernatants were then diluted in 150 μL of hybridization buffer (50 mM Tris 10% acetonitrile pH 8.8) containing 5 pmol of a Cy3-labeled PNA probe complementary to the antisense strand (PNABio, Thousand Oaks, CA, USA). Annealing was carried out by heating the samples at 90°C for 15 min and 50°C for 15 min. Samples were analyzed by HPLC (Agilent, Santa Clara, CA) over a DNAPac PA100 anion-exchange column (Thermo Fisher Scientific). Cy3 fluorescence was monitored and peaks integrated. Final concentrations were ascertained using calibration curves generated by spiking known quantities of conjugated siRNA into blood or tissue lysates from an untreated animal.
Fluorescence microscopy
At 48 hours post-injection, mice were euthanized and perfused with PBS. Tissues were collected and immersed in 10% formalin solution overnight at 4°C. Tissues were embedded in paraffin and sliced into 4-μm sections that were mounted on glass slides. Tissue sections on glass slides were deparaffinized by incubating twice in xylene for 8 min. Sections were rehydrated in an ethanol series from 100% to 95% to 80%, for 4 min each. Slides were then washed twice with PBS, 2 min each, incubated with DAPI (250 ng/mL, Molecular Probes) in PBS for 1 minute, and washed again in PBS for 2 minutes. Slides were mounted with PermaFluor mounting medium (Molecular Probes) coverslips, and dried overnight at 4°C. Sections were imaged at 5× and 40× using a Leica DM5500B microscope fitted with a DFC365 FX fluorescence camera.
mRNA silencing experiments
At 1-week post-injection, mice were euthanized. Tissues were collected and stored in RNAlater (Sigma) at 4°C overnight. mRNA was quantified using the QuantiGene 2.0 Assay (Affymetrix). Briefly, 1.5-mm punches (3 punches per tissue) were placed in QIAGEN Collection Microtubes holding 3-mm tungsten beads and lysed in 300 μl Homogenizing Buffer (Affymetrix) containing 0.2 mg/ml proteinase K (Invitrogen) using a QIAGEN TissueLyser II. Samples were then centrifuged at 1,000 × g for 10 min and incubated for 1 h at 55° to 60°C. Lysates and diluted probe sets (mouse Htt, or mouse Hprt) were added to the bDNA capture plate and signal was amplified and detected as described by Coles et al. [29]. Luminescence was detected on a Tecan M1000 (Tecan, Morrisville, NC, USA).
Statistical analysis
Data were analyzed using GraphPad Prism 7.01 software (GraphPad Software, Inc., San Diego, CA). For each independent efficacy experiment in mice, the level of silencing was normalized to the mean of the control (PBS) group. Data were analyzed using non-parametric one-way ANOVA with Dunnett’s test for multiple comparisons, with significance calculated relative to PBS controls.
Results
Synthesis of branched fatty acids conjugated to fully chemically stabilized siRNAs
To define the impact of fatty acid conjugate chemical structure and valency on siRNA pharmacokinetics, distribution, and efficacy in vivo, we synthesized a library of siRNAs conjugated to one, two, or three fatty acids with different carbon chain lengths and degrees of unsaturation: myristic acid (Myr, C14:0), docosahexaenoic acid (DHA, C22:6), and eicosapentaenoic acid (EPA, C20:5) (Figure 1).
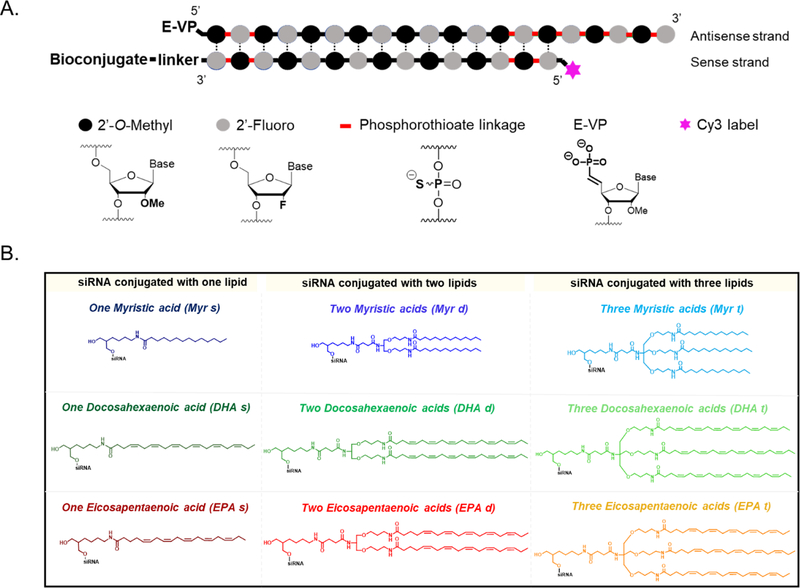
Figure 1.
Library of fatty acid-conjugated siRNAs (A.) Schematic of chemically modified siRNAs used in this study. The desired conjugate is attached at the 3’-end of the sense strand (B.) Structures of the conjugates attached to the siRNAs: monomeric (s); dimeric (d) and trimeric (t) fatty acids (Myristic acid, Myr; Docosahexaenoic acid, DHA; Eicosapentaenoic acid, EPA).
A fully chemically stabilized asymmetric siRNA was used as a scaffold (Figure 1A) [30]. Asymmetric siRNAs consist of a short duplex region (15 base-pairs) and a single-stranded fully phosphorothioate-modified tail that assists membrane association [31, 32]. All riboses are modified using an alternating 2´-O-methyl and 2´-fluoro modification pattern, which confers stability and minimizes innate immune activation [5, 33, 34]. The antisense strand is modified with a 5´-(E)-vinylphosphonate (E-VP) group that mimics the 5´-phosphate of the antisense strand to promote recognition by RISC (RNA-induced silencing complex) [35, 36] and provides stability against phosphatases and exonucleases [15, 37, 38]. The sense strand is labeled with Cy3 at the 5´-end, allowing for visualization of siRNA spatial distribution in tissues.
Fatty acids were covalently attached to the 3′ end of the siRNA sense strand (Figure 1B), which tolerates a range of covalent modifications [7, 39, 40]. All fatty acid conjugated siRNAs were synthesized using a functionalized solid support (Supplementary Schemes 1, 2 and 3) [16]. For the incorporation of either two or three fatty acids, synthetic dividers (Supplementary Schemes 2 and 3, compounds 9 and 16, respectively) were introduced on the solid support to minimize steric hindrance, followed by fatty acid conjugation (Supplementary Schemes 2 and 3, solid supports 11 and 18). Oligonucleotides were purified by High Performance Liquid Chromatography (HPLC) and characterized by mass spectrometry (Supplementary Figure 1).
The valency of fatty acid conjugate affects siRNA hydrophobicity and aggregation
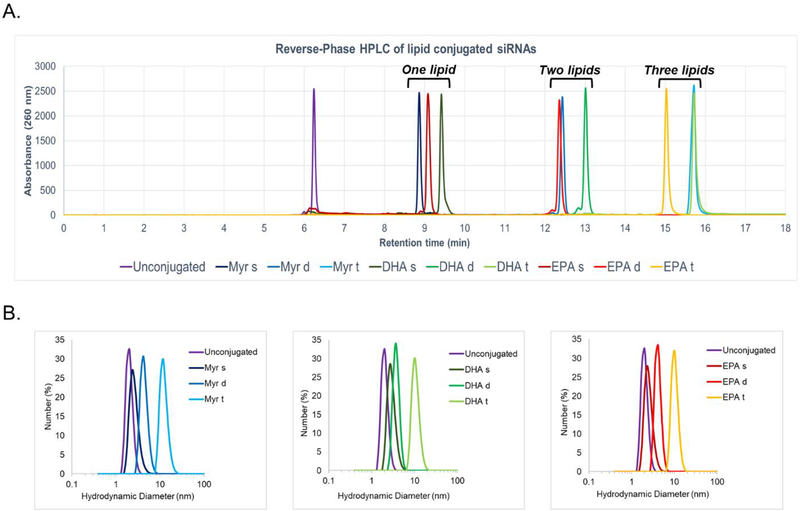
To evaluate the impact of conjugate chemical structure and valency on siRNA physicochemical properties, we measured the overall hydrophobicity and aggregation of each compound. The retention time in reversed-phase HPLC was used to determine overall hydrophobicity (increases with retention time) (Figure 2A) [41]. Monomeric fatty acid-conjugated siRNAs had the lowest retention time (8.8 to 9.5 min), followed by dimeric (12 to 13 min), and trimeric (15 to 16 min) fatty acids, respectively. The nature of the fatty acid carbon chain (Myr vs. DHA vs. EPA) had a relatively low impact on retention time. Our findings suggest that increasing the valency of the fatty acid conjugate increases overall compound hydrophobicity.
Figure 2.
The presence of fatty acids significantly impacts siRNA hydrophobicity and micelle formation (A.) High Performance Liquid Chromatography (HPLC) spectra of conjugated Cy3-siRNAHtt sense strands showing variation in retention time (hydrophobicity) due to the nature and number of fatty acids (B.) Hydrodynamic diameter of lipid-conjugated siRNAs determined by Dynamic Light Scattering. Mean diameter for: unconjugated = 2 nm; monomeric lipid = 2.3–2.7 nm; dimeric lipids = 3.6–4.2 nm; trimeric lipids = 10.1–11.7 nm.
Lipid-conjugated siRNAs can self-assemble in aqueous media [42]. To determine the impact of conjugate chemical structure and valency on aggregate size, hydrodynamic diameters of fatty acid-conjugated siRNAs were measured by Dynamic Light Scattering (DLS) (Figure 2B). The monomeric fatty acid-conjugated siRNAs did not aggregate, with a mean diameter of 2.5 nm, similar to unconjugated siRNA. By contrast dimeric and trimeric fatty acid-conjugated siRNAs did self-assemble into small aggregates and micelles. The average particle size was much larger for trimeric compared to dimeric compounds (11 versus 4 nm diameter, respectively). The nature of the fatty acid carbon chain (Myr vs DHA vs EPA) did not significantly affect the size of the aggregate formed. Our findings suggest that valency of the fatty acid conjugate impacts siRNA physicochemical properties.
Impact of fatty acid conjugate valency on siRNA tissue distribution profile.
Understanding clearance kinetics of engineered siRNAs is essential for further functional optimization [43]. Given that siRNA physicochemical properties were mainly affected by valency rather than the chemical nature of the conjugate carbon chain, we evaluated the impact of valency on clearance profile using one fatty acid—Myristic acid. The blood clearance profile for siRNAs conjugated with monomeric (Myr-s), dimeric (Myr-d), and trimeric (Myr-t) myristic acid were evaluated (Figure 3) by injecting siRNA into mice subcutaneously (n=6 per variant, 20 mg/kg dose), and collecting blood samples at different time points according to a previous method [27]. Pharmacokinetic profiles were determined by quantifying antisense strands in blood samples using a peptide nucleic acid (PNA) hybridization assay [28] (Figure 3A and 3B).
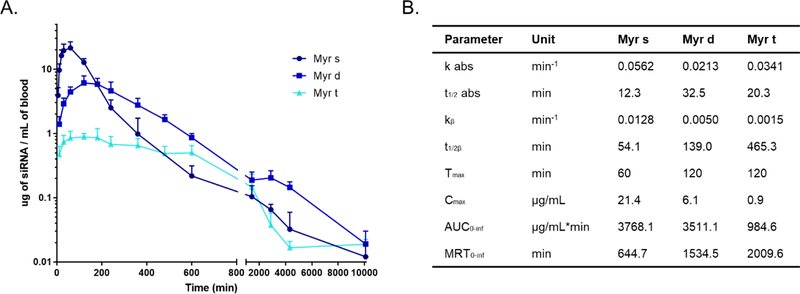
Figure 3.
Myr-s, Myr-d and Myr-t conjugated siRNAs exhibit distinct pharmacokinetic profiles. (A.) Mice were injected subcutaneously with Myr variant conjugated siRNAs (n = 6, 20 mg/kg). Serial blood samples were collected through the lateral saphenous vein at different time points after injection over a 7 day period. Antisense strands in blood samples were quantified using PNA hybridization assay (B.) Pharmacokinetic parameters for Myr variant conjugated siRNAs. AUC = area under the curve. MRT = mean residence time.
Myr-s, Myr-d, and Myr-t conjugated siRNAs showed distinct clearance profiles. Myr-s siRNAs were rapidly released into the blood from the site of injection (t1/2 abs = 12 min, Tmax = 60 min), whereas Myr-d exhibited slower release (t1/2 abs = 32 min, Tmax= 120 min). Although the areas under the curve (AUC) between Myr-s and Myr-d siRNAs were similar (3768 and 3511 μg/mL*min, respectively), the mean residence time (MRT) for Myr-d siRNAs was more than 2- fold higher than for Myr-s siRNAs (1543 and 644 min, respectively). These data suggest that Myr-d siRNAs stayed in the circulation longer than Myr-s siRNAs despite the two having comparable levels in the blood. By contrast, the majority of Myr-t siRNAs were not released from the site of injection (AUC of 984 μg/mL*min) even after one week. This is likely due to Myr-t siRNAs forming highly hydrophobic micelles (Figure 2A). Our results suggest that altering fatty acid conjugate valency generates siRNA compounds with substantially different pharmacokinetic profiles.
Fatty acid conjugate valency fundamentally defines siRNA tissue accumulation profiles
To evaluate the impact of fatty acid conjugate valency on siRNA tissue distribution, we injected mice subcutaneously with Myr-s, -d, or -t siRNA variants (20 mg/kg, n = 3 per variant). 48 hours post injection, we collected 15 tissues per mouse—liver, kidney, adrenal gland, lung, heart, thymus, spleen, pancreas, intestine, fallopian tube, bladder, fat (white fat from the belly), muscle, injection site, and skin—and evaluated spatial and quantitative siRNA tissue distribution in each sample. Spatial siRNA distribution was evaluated using fluorescence microscopy (Cy3 fluorophore attached to 5’ end of sense strand). We have previously shown that the presence of Cy3 has minimal impact on overall tissue distribution in the context of lipophilic siRNAs [17].
We quantified antisense strand accumulation using a PNA hybridization assay [28], which is not dependent on the presence of Cy3. Both methodologies generated overall consistent data.
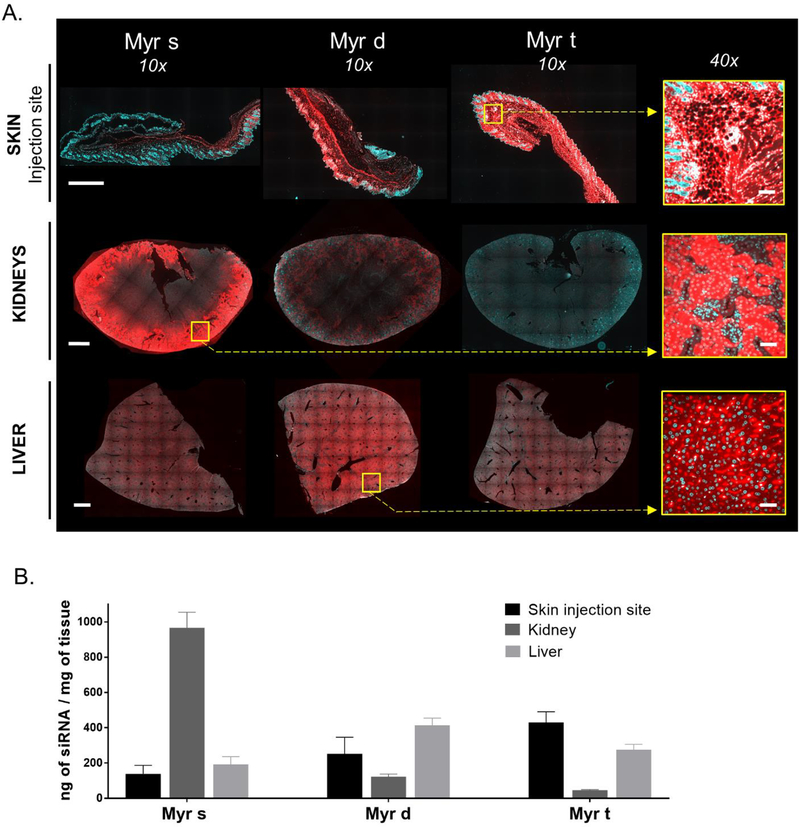
siRNA variants were mostly cleared from the blood (AUC0–48h > 85% of total AUC) at 48 hours post-injection, suggesting that tissue distribution profiles at this time point are representative of overall long-term delivery. Figure 4 shows siRNA distribution in primary sites—liver, kidney and site of infection, where compounds accumulate to the largest extent. In general, levels of siRNA accumulation in primary sites were at least 20-fold higher than in extra-hepatic tissues (Figure 4, Figure 5). Fatty acid conjugate valency had a profound impact on the liver-to-kidney-to skin (site of injection) distribution ratio (Figure 4). Representative fluorescent images (Figure 4A) and antisense strand quantification (Figure 4B) of liver, kidney, and skin (site of injection) show that Myr-s siRNAs preferentially accumulated in kidneys, Myr-d in liver, and Myr-t at the injection site. The difference in the liver-to-kidney distribution between Myr-s and Myr-d is consistent with our previous result that more hydrophobic compounds accumulate in liver and less hydrophobic compounds accumulate in kidney [17]. The preferential skin delivery and reduced liver accumulation of Myr-t is consistent with its limited release from the skin, and lower Cmax and AUC (Figure 3B).
Figure 4.
Lipid valency defines siRNA accumulation in skin (injection site), kidney, and liver. (A.) Representative fluorescence images of skin (injection site), kidney, and liver sections from mice injected subcutaneously (n=3, 20 mg/kg) with Myr variant Cy3-labeled siRNAs (red). Nuclei stained with DAPI (blue). Tissues were collected 48h after injection. Images taken at 10× and 40× magnifications and collected at the same laser intensity and acquisition time. Scale, 1 mm (10×) and 50 μm (40×) (B.) Bar graph showing siRNA quantification in skin (injection site), kidney (cortex), and liver measured by PNA hybridization assay (mean ± SD)
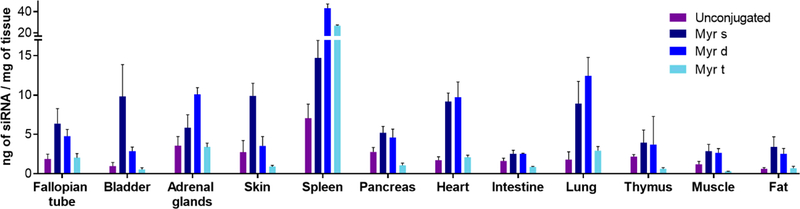
Figure 5.
siRNA accumulation is defined by the valency of lipids: monomeric and dimeric fatty acids enhance siRNA retention into extra-hepatic tissues. Bar graph (mean ± SD) showing the quantity of unconjugated, Myr-s, Myr-d, and Myr-t conjugated siRNAs in 12 tissues at 48h after a single subcutaneous injection (20 mg/kg) in mice (n=3). siRNA quantification was determined by PNA hybridization assay.
Measuring antisense strand accumulation in 15 tissues, which comprise most of the mouse body, allowed us to estimate the fraction of the injected siRNA dose that was retained after 48 hours (Supplementary Figure 2A, Supplementary Table 1). The mass of each tissue was experimentally defined, or approximated based on published mouse organ weights [44–46]. More than 85% of unconjugated siRNAs were cleared from the body, while the majority of the retained siRNA accumulated in kidneys (Supplementary Figure 2). The addition of fatty acid conjugates dramatically enhanced siRNA overall retention—approximately 100% of the injected dose for all three variants was accounted for. These findings indicate that fatty acid conjugates, regardless of valency status, reduce clearance into the urine compared to unconjugated siRNAs.
Figure 5 shows quantification of antisense strand accumulation in twelve tissues, including bladder, spleen, heart, lung, muscle, and fat. Myr-s and Myr-d siRNAs distributed to extra-hepatic tissues significantly more than unconjugated siRNAs and Myr-t. Overall tissue distribution profiles for Myr-s and Myr-d conjugated siRNAs were similar, with a few exceptions (Supplementary Table 1). Myr-s accumulated in bladder and skin (systemic, far from the site of injection) to a significantly higher degree than Myr-d. By contrast, Myr-d showed enhanced delivery to the lung. These findings indicate that modulating the valency of fatty acid conjugates can alter compound tissue distribution, and potentially enhance extra-hepatic delivery.
Fatty acid conjugated siRNAs enable functional gene silencing in several tissues
To evaluate the effect of conjugate valency on gene silencing, mice were injected subcutaneously with Myr-siRNA variants targeting Huntingtin (Htt) mRNA or controls (n = 6 per group, 20 mg/kg). We elected to target Htt mRNA because it is widely expressed in all tissues and has a validated siRNA sequence available [47]. Htt and Hprt (hypoxanthine-guanine phosphoribosyl transferase, a housekeeping gene) mRNA expression was measured one week post injection using the QuantiGene Assay. The efficacy of Htt targeting compounds with different Myr variants was evaluated in liver, kidneys, spleen, heart, lung, fat, bladder, and fallopian tube (Figure 6, Supplementary Figure 3). PBS, unconjugated siRNAs, and a non-targeting siRNA (Ntc, compound of identical chemical configuration but not targeting Htt mRNA) were used as controls.
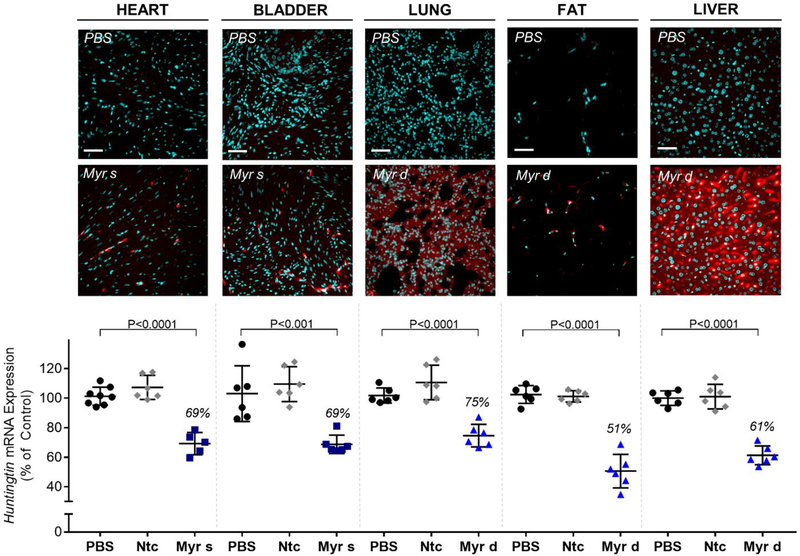
Figure 6.
The amounts of conjugated siRNA in several tissues are sufficient to induce mRNA silencing. Representative fluorescence images of heart, bladder, lung, fat, and liver sections from mice (n=3 per conjugate) injected subcutaneously with 20 mg/kg Cy3-labeled Myr variant conjugated siRNAs (red) or PBS. Nuclei stained with DAPI (blue). Tissues were collected 48h after injection. Images taken at 40× magnification and collected at the same laser intensity and acquisition time. Scale, 50 μm. For the measurement of mRNA, mice were injected subcutaneously with 20 mg/kg of conjugated siRNA (n = 6 per group, Ntc = non-targeting controls). The tissues were collected after 1 week, and Huntingtin (Htt) mRNA levels were measured using QuantiGene®, normalized to a housekeeping gene, Hprt (Hypoxanthine-guanine phosphoribosyl transferase), and presented as percent of PBS control (mean ± SD). Statistical analysis: One-way ANOVA with Bonferroni correction
Ntc was indistinguishable from PBS in all tissues tested, suggesting that any observed modulation of gene expression is target-specific and not related to overall oligonucleotide chemical configuration. Unconjugated compounds induced statistically significant silencing in kidneys (Supplementary Figure 3). This finding is consistent with previously reported data, and is likely due to the phosphorothioate modifications in our siRNA scaffold driving renal epithelia retention [31, 48].
Conjugation of fatty acids had a profound impact on siRNA efficacy, and statistically improved silencing in several tissues (compared to unconjugated, PBS and Ntc compounds) (Figure 6, Supplementary Figure 3). In general, there was a correlation between the degree of accumulation and the level of silencing within the same tissue. Myr-s conjugated siRNAs accumulated to a higher extent in bladder (~10 ng/mg, Figure 5), resulting in productive silencing (31% silencing, Figure 6), while Myr-d conjugated siRNA compounds show higher accumulation and more productive silencing in lung and spleen (8 ng/mg, 25% silencing; and 42 ng/mg, 29% silencing, respectively) (Figure 5, Figure 6, Supplementary Figure 3).
The levels of accumulation necessary for productive silencing differed significantly between tissues, likely due to differences in internalization pathways and intracellular trafficking. For example, Myr-d siRNAs accumulated to 3 ng/mg in fat (Figure 5), translating to ~50% silencing (P<0.0001) (Figure 6), whereas Myr-d kidney accumulation was >100 ng/mg (Figure 4B) and did not support functional silencing (Supplementary Figure 3). Figure 6 shows representative data for delivery and functional silencing in key tissues for the best-delivered siRNA per tissue—Myr-s siRNAs for heart, and bladder, and Myr-d siRNAs for lung, fat and liver. Myr-s conjugated siRNAs induced 31% silencing in both tissues, and Myr-d siRNA compounds induced 25%, 49% and 39% silencing in lung, fat, and liver respectively. All observed effects were statistically significant relative to PBS or Ntc (P<0.0001 or P<0.001, One-way ANOVA with Bonferroni correction). Our collective findings suggest that altering fatty acid conjugate valency affects tissue-specific siRNA accumulation for productive gene silencing.
siRNA hydrophobicity does not fully explain differences in siRNA distribution and efficacy
We and others have previously demonstrated that differences in conjugate-mediated siRNA tissue distribution are partially explained by changes in serum lipoprotein binding that are driven by hydrophobicity [14, 18, 19]. To evaluate how conjugate chemical structure impacts siRNA tissue distribution profiles independent of hydrophobicity, we compared tissue accumulation and efficacy of three conjugated siRNA: Myr-d, cholesterol (Chol), and α-tocopheryl succinate (TS) siRNAs.
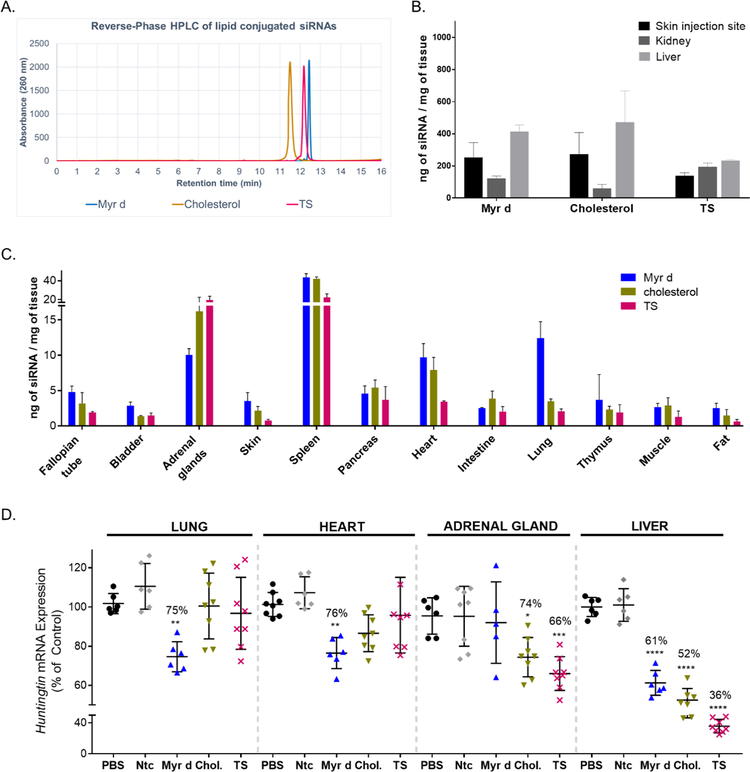
Figure 7A shows reverse-phase HPLC traces for the three conjugated siRNAs. All three compounds have similar retention time (within 11.5–12.5 minutes), indicating they have similar hydrophobicities. siRNA variants were then injected subcutaneously (20 mg/kg dose) into mice (n=3 per variant) and tissue accumulation was evaluated at 48 hours post injection (PNA hybridization assay). Figure 7B shows that accumulation in primary clearance tissues (liver/kidney/injection site) were similar between Myr-d and Chol but different for TS compounds. While Myr-d and Chol siRNAs mostly distributed to liver and site of injection, TS siRNAs accumulated equally between all three tissues. Next, we evaluated the impact of conjugate chemistry on extrahepatic/extrarenal delivery, and observed significant differences (Figure 7C). For example, Myr-d siRNAs accumulated to higher levels in lung (~13 ng/mg), and heart (~10 ng/mg), whereas TS siRNA clearly accumulated to a higher degree in adrenal gland (Figure 7C).
Figure 7.
Hydrophobicity does not fully explain differences in siRNA distribution and efficacy in vivo. (A.) High Performance Liquid Chromatography (HPLC) spectra of conjugated Cy3-siRNAHtt sense strands showing similar hydrophobicities between Myr-d, TS (tocopheryl succinate conjugate), and cholesterol-conjugated siRNA (B.) Bar graph showing siRNA quantification in skin (injection site), kidney (cortex), and liver for Myr-d, cholesterol and TS siRNAs, measured by PNA hybridization assay (3 mice per conjugate, mean ± SD) (C.) Bar graph showing the quantities of Myr d, cholesterol (chol.) and TS conjugated siRNAs present in 12 tissues at 48h after a single subcutaneous injection with 20 mg/kg (n=3 ± SD). siRNA quantification was measured by PNA hybridization assay. (D.) Measurement of Huntingtin mRNA levels in mice injected subcutaneously with 20 mg/kg of Myr d, cholesterol or TS conjugated siRNA (n = 6–8 per group, Ntc = non-targeting controls). The tissues were collected after 1 week and Huntingtin (Htt) mRNA levels were measured using QuantiGene®, normalized to a housekeeping gene, Hprt (Hypoxanthine-guanine phosphoribosyl transferase), and presented as percent of PBS control (mean ± SD). Statistical analysis: One-way ANOVA with Bonferroni correction, * = P<0.1, ** = P<0.01, *** = P<0.001.
Finally, we evaluated the impact of conjugate chemical structure on gene silencing in tissues where differences in accumulation were observed (n=6–8, compared to PBS and Ntc, 1-week post-injection, QuantiGene Assay). Myr-d siRNAs show statistically significant silencing in lung and heart (25% and 24% silencing respectively, P<0.01), whereas Chol and TS siRNAs did not. Similarly, TS and Chol siRNAs induced silencing in adrenal gland (34% and 26% silencing, P<0.001 and P<0.1 respectively), but Myr d siRNA compounds did not (Figure 7D). In liver, significant silencing is observed for all three compounds, which can be explained by the significant accumulation of all three siRNAs in this tissue (Figure 7B).
Our findings suggest that although the physicochemical properties (i.e. hydrophobicity) of a conjugate may affect siRNA clearance and distribution, the chemical nature/structure and self-association properties of conjugated-siRNA drive cellular internalization and functional silencing.
Discussion
The recent clinical success of GalNAc-conjugated siRNAs for liver-associated disorders demonstrates that the conjugation of chemically-stabilized siRNA is primordial for developing therapeutic oligonucleotides [8, 9, 11, 49]. Lipid conjugation of siRNAs supports broad tissue distribution beyond the liver [6, 17]. Advancements in lipid engineering may further refine siRNA delivery, but the relationship between conjugate structure/configuration and siRNA pharmacodynamic behavior must be more clearly defined. This paper is the first to report the effects of lipid conjugate valency on siRNA delivery. We demonstrate that modulating conjugate valency alters siRNA physicochemical properties, which directly affect clearance, distribution, and silencing. This study demonstrates that rational engineering of lipid conjugates may be used to enhance extrahepatic siRNA delivery and efficacy.
Unconjugated siRNAs are quickly cleared from the bloodstream [27, 50, 51] with less than ~15% body retention [17]. Primary retention is in the kidney epithelia due to the high phosphorothioate (PS) content of our siRNA scaffold (13 PS bonds within a 35-nucleotide scaffold). This retention mechanism is similar to that of short single-stranded PS-antisense oligonucleotides [31, 48, 52]. In general, lipid conjugates significantly improve siRNA overall body retention (almost 100%) and extra-hepatic/extra-renal tissue exposure. We and others have shown that the higher overall retention and tissue distribution of lipophilic siRNAs is primarily explained by changes in hydrophobicity that drive serum lipoprotein binding [14, 18, 19]. Less hydrophobic compounds preferentially bind HDL and accumulate in kidney; more lipophilic siRNAs preferentially bind LDL and accumulate in liver.
By manipulating lipid conjugate valency, we were able to modulate hydrophobicity such that siRNA pharmacokinetic behaviors and distribution to primary clearance tissues were altered. Trimeric fatty acid-conjugated siRNAs are highly hydrophobic, resulting in ineffective release from the injection site, low siRNA blood level and systemic exposure, and limited tissue accumulation. These poor pharmacokinetic properties may be due to micelle formation and non-productive entrapment in neighboring subcutaneous fat. The hydrophobicity of the trivalent conjugate appears to be well above the optimal range for siRNA distribution in vivo.
Monomeric fatty acid-conjugated siRNAs were rapidly released into the bloodstream and predominantly accumulated in kidney (Figure 3). Their overall behavior is similar to relatively less lipophilic conjugates like DHA, which preferentially bind HDL [18, 27]. Dimeric fatty acid-conjugated siRNAs are released from the injection site, but stayed in the blood longer than monomeric compounds, and accumulated predominantly in liver. Thus, divalent lipid-conjugated siRNAs act similar to LDL-binding compounds.
Although hydrophobicity primarily explains the effect of conjugate valency on the liver-to-kidney accumulation profile, the exact chemical nature/structure of the conjugate impacted the degree of siRNA accumulation in extrahepatic/extrarenal tissues. Myr-d siRNA showed lung accumulation levels ~3 times higher than other conjugated siRNAs of similar hydrophobicities, which resulted in productive silencing. The mechanism underlying enhanced lung delivery is unknown, but may be due to the impact of lipid branching on membrane fluidity, receptor interactions, and trafficking/endosomal escape.
Consistent with previous reports on siRNA accumulation/efficacy ratios for different tissues [17, 53], we found that the level of tissue accumulation sufficient to induce mRNA silencing is tissue-dependent. Liver and kidney require relatively high (>100 ng/mg, Figure 4B) siRNA accumulation, whereas lung, fat, and heart require relatively low siRNA accumulation (2–13 ng/mg, Figure 5) for productive silencing. This is likely related to tissue-specific internalization mechanisms. Liver and kidneys are responsible for eliminating metabolic bi-products from the bloodstream. Thus, oligonucleotides largely accumulate in those tissues as a byproduct of filtering and are trapped non-productively. By contrast, cellular internalization in non-filtering tissues results from active endocytosis.
This study reports that the valency of fatty acid conjugates is a strong determinant of the siRNA pharmacokinetic and distribution profiles in vivo. Thus, chemically engineering lipid-conjugated siRNA is a viable strategy for improving extrahepatic delivery and efficacy of therapeutic siRNAs.
Supplementary Material
Acknowledgements
We thank Khvorova lab members for insightful discussions and support, and Emily Mohn for helping with the manuscript writing and editing.
Funding
This work was supported by National Institutes of Health grants [RO1GM10880304, S10 OD020012].
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplementary data
Supplementary data are available online
Competing interest statement
A.K owns stock of RXi Pharmaceuticals and Advirna. Other authors declare no competing financial interest.
References
- 1.Zhou J; Shum K-T; Burnett JC; Rossi JJ Nanoparticle-based delivery of RNAi therapeutics: Progress and challenges. Pharmaceuticals 2013. 6, 85–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Scherman D; Rousseau A; Bigey P; Escriou V Genetic pharmacology: Progresses in si RNA delivery and therapeutic applications. Gene Ther. 2017. 24, 151–156. [DOI] [PubMed] [Google Scholar]
- 3.Nair JK; Willoughby JLS; Chan A; Charisse K; Alam MR; Wang Q; Hoekstra M; Kandasamy P; Kel’in AV; Milstein S; Taneja N; O’Shea J; Shaikh S; Zhang L; van der Sluis RJ; Jung ME; Akinc A; Hutabarat R; Kuchimanchi S; Fitzgerald K; Zimmermann T; van Berkel TJC; Maier MA; Rajeev KG; Manoharan M Multivalent N‑ acetylgalactosamine-conjugated siRNA localizes in hepatocytes and elicits robust RNAi-mediated gene silencing. JACS 2014. 136, 16958–16961. [DOI] [PubMed] [Google Scholar]
- 4.Zimmermann TS; Karsten V; Chan A; Chiesa J; Boyce M; Bettencourt BB; Hutabarat R; Nochur S; Vaishnaw A; Gollob J Clinical proof of concept for a novel hepatocyte-targeting galnac-siRNA conjugate. Mol. Ther 2017. 25, 71–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jackson AL; Burchard J; Leake D; Reynolds A; Schelter J; Guo J; Johnson JM; Lim L; Karpilow J; Nichols K; Marshall W; Khvorova A; Linsley PS Position-specific chemical modification of siRNAs reduces “off-target” transcript silencing. RNA 2006. 12, 1197–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Osborn MF; Khvorova A Improving siRNA delivery in vivo through lipid conjugation. Nucleic Acid Ther. 2018. 28, 128–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Soutschek J; Akinc A; Bramlage B; Charisse K; Constien R; Donoghue M; Elbashir S; Geick A; Hadwiger P; Harborth J; John M; Kesavan V; Lavine G; Pandey RK; Racie T; Rajeev KG; Rohl I; Toudjarska I; Wang G; Wuschko S; Bumcrot D; Koteliansky V; Limmer S; Manoharan M; Vornlocher H-P Therapeutic silencing of an endogenous gene by systemic administration of modified siRNAs. Nature 2004. 432, 173–178. [DOI] [PubMed] [Google Scholar]
- 8.Huang H Preclinical and clinical advances of GalNAc-decorated nucleic acid therapeutics. Mol. Ther. Nucleic Acids 2017. 6, 116–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rajeev KG; Nair JK; Jayaraman M; Charisse K; Taneja N; O’Shea J; Willoughby JLS; Yucius K; Nguyen T; Shulga-Morskaya s.; Milstein S; Liebow A; Querbes W; Borodovsky A; Fitzgerald K; Maier MA; Manoharan M Hepatocyte-specific delivery of siRNAs conjugated to novel non-nucleosidic trivalent N-acetylgalactosamine elicits robust gene silencing in vivo. ChemBioChem 2015. 16, 903–908. [DOI] [PubMed] [Google Scholar]
- 10.Matsuda S; Keiser K; Nair JK; Charisse K; Manoharan RM; Kretschmer P; Peng CG; Kel’in AV; Kandasamy P; Willoughby JLS; Liebow A; Querbes W; Yucius K; Nguyen T; Milstein S; Maier MA; Rajeev KG; Manoharan M siRNA conjugates carrying sequentially assembled trivalent N-acetylgalactosamine linked through nucleosides elicit robust gene silencing in vivo in hepatocytes. ACS Chem. Biol 2015. 10, 1181–1187. [DOI] [PubMed] [Google Scholar]
- 11.Prakash TP; Graham MJ; Yu J; Carty R; Low A; Chappell A; Schmidt K; Zhao C; Aghajan M; Murray HF; Riney S; Booten SL; Murray SF; Gaus H; Crosby J; Lima WF; Guo S; Monia BP; Swayze EE; Seth PP Targeted delivery of antisense oligonucleotides to hepatocytes using triantennary N-acetyl galactosamine improves potency 10-fold in mice. NAR 2014. 42, 8796–8807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tanowitz M; Hettrick L; Revenko A; Kinberger GA; Prakash TP; Seth PP Asialoglycoprotein receptor 1 mediates productive uptake of N-acetylgalactosamine-conjugated and unconjugated phosphorothioate antisense oligonucleotides into liver hepatocytes. Nucleic Acids Res. 2017. 45, 12388–12400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nishina K; Unno T; Uno Y; Kubodera T; Kanouchi T; Mizusawa H; Yokota T Efficient in vivo delivery of siRNA to the liver by conjugation of α-tocopherol. Mol. Ther 2008. 16, 734–740. [DOI] [PubMed] [Google Scholar]
- 14.Wolfrum C; Shi S; Jayaprakash KN; Jayaraman M; Wang G; Pandey RK; Rajeev KG; Nakayama T; Charrise K; Ndungo EM; Zimmermann T; Koteliansky V; Manoharan M; Stoffel M Mechanisms and optimization of in vivo delivery of lipophilic siRNAs. Nat. Biotechnol 2007. 25, 1149–1157. [DOI] [PubMed] [Google Scholar]
- 15.Nikan M; Osborn MF; Coles AH; Biscans A; Godinho BM; Haraszti RA; Sapp E; Echeverria D; DiFiglia M; Aronin N; Khvorova A Synthesis and evaluation of parenchymal retention and efficacy of a metabolically stable O-phosphocholine-N-docosahexaenoyl-L-serine siRNA conjugate in mouse brain. Bioconjugate Chem. 2017. 28, 758–1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nikan M; Osborn MF; Coles AH; Godinho BM; Hall LM; Haraszti RA; Hassler MR; Echeverria D; Aronin N; Khvorova A Docosahexaenoic acid conjugation enhances distribution and safety of siRNA upon local administration in mouse brain. Mol. Ther. Nucleic Acids 2016. 5, e344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Biscans A; Coles A; Haraszti R; Echeverria D; Hassler M; Osborn M; Khvorova A Diverse lipid conjugates for functional extra-hepatic siRNA delivery in vivo NAR 2018. 10.1093/nar/gky1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Osborn MF; Coles AH; Biscans A; Haraszti RA; Roux L; Davis S; Ly s.; Echeverria D; Hassler MR; Godinho BMDC; Nikan M; Khvorova A Hydrophobicity drives the systemic distribution of lipid-conjugated siRNAs via lipid transport pathways NAR 2018. 10.1093/nar/gky1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sarett SM; Werfel TA; Lee L; Jackson MA; Kilchrist KV; Brantley-Sieders D; Duvall CL Lipophilic siRNA targets albumin in situ and promotes bioavailability, tumor penetration, and carrier-free gene silencing. PNAS 2017. 114, E6490–E6497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Karaki S; Benizri S; Mejías R; Baylot V; Branger N; Nguyen T; Vialet B; Oumzil K; Barthélémy P; Rocchi P Lipid-oligonucleotide conjugates improve cellular uptake and efficiency of TCTP-antisense in castration-resistant prostate cancer. J. Control. Release 2017. 258, 1–9. [DOI] [PubMed] [Google Scholar]
- 21.Akinc A; Zumbuehl A; Goldberg M; Leshchiner ES; Busini V; Hossain N; Bacallado SA; N. ND; Fuller J; Alvarez R; Borodovsky A; Borland T; Constien R; de Fougerolles A; Dorkin JR; Jayaprakash KN; Jayaraman M; John M; Koteliansky V; Manoharan M; Nechev L; Racie T; Raitcheva D; Rajeev KG; Sah DWY; Soutschek J; Toudjarska I; Vornlocher H-P; Zimmermann TS; Langer R; Anderson DG A combinatorial library of lipid-like materials for delivery of RNAi therapeutics. Nat. Biotechnol 2008. 26, 561–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen S; Tam YYC; Lin PJC; Sung MMH; Tam YK; Cullis PR Influence of particle size on the in vivo potency of lipid nanoparticle formulations of siRNA. J. Control. Release 2016. 235, 236–244. [DOI] [PubMed] [Google Scholar]
- 23.Suhr OB; Coelho T; Buades Pouget, J.; Conceicao I; Berk J; Schmidt H; Waddington-Cruz; Campistol JM; Bettencourt BR; Vaishnaw A; Gollob J; Adams D Efficacy and safety of patisiran for familial amyloidotic polyneuropathy: A phase ii multi-dose study. Orphanet J. Rare Dis. 2015. 10, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dahlman JE; Barnes C; Khan O; Thiriot A; Jhunjunwala S; Shaw TE; Xing Y; Sager HB; Sahay G; Speciner L; Bader A; Bogorad RL; Yin H; Racie T; Dong Y; Jiang S; Seedorf D; Dave A; Sandu KS; Webber MJ; Novobrantseva T; Ruda VM; Lytton-Jean AKR; Levins CG; Kalish B; Mudge DK; Perez M; Abezgauz L; Dutta P; Smith L; Charisse K; Kieran MW; Fitzgerald K; Nahrendorf M; Danino D; Tuder RM; von Andrian UH; Akinc A; Schroeder A; Panigrahy D; Kotelianski V; Langer R; Anderson DG In vivo endothelial si RNA delivery using polymeric nanoparticles with low molecular weight. Nat. Nanotechnol 2014. 9, 648–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Whitehead KA; Dorkin JR; Vegas AJ; Chang PH; Veiseh O; Matthews J; Fenton OS; Zhang Y; Olejnik KT; Yesilyurt V; Chen D; Barros S; Klebanov B; Novobrantseva T; Langer R; Anderson DG Degradable lipid nanoparticles with predictable in vivo siRNA delivery activity. Nat. Commun 2014. 5, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haraszti RA; Roux L; Coles AH; Turanov AA; Alterman JF; Echeverria D; Godinho BM; Aronin N; Khvorova A 5΄-vinylphosphonate improves tissue accumulation and efficacy of conjugated siRNAs in vivo. Nucleic Acids Res. 2017. 45, 7581–7592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Godinho BMDC; Gilbert JW; Haraszti RA; Coles AH; Biscans A; Roux L; Nikan M; Echeverria D; Hassler M; Khvorova A Pharmacokinetic profiling of conjugated therapeutic oligonucleotides: A high-throughput method based upon serial blood microsampling coupled to peptide nucleic acid hybridization assay. Ncleic Acid Ther. 2017. 27, 323–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roehl I; Schuster M; Seiffert S Oligonucleotide detection method. US Patent US20110201006A1 2011, 1–9. [Google Scholar]
- 29.Coles AH; Osborn MF; Alterman JF; Turanov AA; Godinho BM; Kennington L; Chase K; Aronin N; Khvorova A A high-throughput method for direct detection of therapeutic oligonucleotide-induced gene silencing in vivo. Nucleic Acid Ther. 2016. 26, 86–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hassler MR; Turanov AA; Alterman JF; Haraszti RA; Coles AH; Osborn MF; Echeverria D; Nikan M; Salomon WE; Roux L; Godinho BMDC; Davis SM; Morrissey DV; Zamore PD; Karumanchi SA; Moore MJ; Aronin N; Khvorova A Comparison of partially and fully chemically-modified siRNA in conjugate-mediated delivery in vivo. Nucleic Acids Res. 2018. 46, 2185–2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Geary RS; Norris D; Yu R; Bennett CF Pharmacokinetics, biodistribution and cell uptake of antisense oligonucleotides. Adv. Drug. Deliv. Rev 2015. 87, 46–51. [DOI] [PubMed] [Google Scholar]
- 32.Ly S; Navaroli DM; Didiot MC; Cardia J; Pandarinathan L; Alterman JF; Fogarty K; Standley C; Lifshitz LM; Bellve KD; Prot M; Echeverria D; Corvera S; Khvorova A Visualization of self-delivering hydrophobically modified siRNA cellular internalization. Nucleic Acids Res. 2017. 45, 15–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Allerson CR; Sioufi N; Jarres R; Prakash TP; Naik N; Berdeja A; Wanders L; Griffey RH; Swayze EE; Bhat B Fully 2’-modified oligonucleotide duplexes with improved in vitro potency and stability compared to unmodified small interfering RNA. J. Med. Chem 2005. 48, 901–904. [DOI] [PubMed] [Google Scholar]
- 34.Nallagatla SR; Bevilacqua PC Nucleoside modifications modulate activation of the protein kinase pkr in an RNA structure-specific manner. RNA 2008. 14, 1201–1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ma JB; Yuan YR; Meister G; Pei Y; Tuschl T; Patel DJ Structural basis for 5’-end-specific recognition of guide RNA by the A. Fulgidus piwi protein. Nature 2005. 434, 666–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Frank F; Sonenberg N; Nagar B Structural basis for 5’-nucleotide base-specific recognition of guide RNA by human ago2. Nature 2010. 465, 818–822. [DOI] [PubMed] [Google Scholar]
- 37.Parmar R; Willoughby JLS; Liu J; Foster DJ; Brigham B; Theile CS; Charisse K; Akinc A; Guidry E; Pei Y; Strapps W; Cancilla M; Stanton MG; Rajeev KG; Sepp-Lorenzino L; Manoharan M; Meyers R; Maier MA; Jadhav V 5′-(e)-vinylphosphonate: A stable phosphate mimic can improve the RNAi activity of siRNA–GalNAc conjugates. ChemBioChem 2016. 17, 987–989. [DOI] [PubMed] [Google Scholar]
- 38.Lima WF; Prakash TP; Murray HM; Kinberger GA; Li W; Chappell AE; Li CS; Murray SF; Gaus H; Seth PP; Swayze EE; Crooke ST Single-stranded siRNAs activate RNAi in animals. Cell 2012. 150, 883–894. [DOI] [PubMed] [Google Scholar]
- 39.Morrissey DV; Blanchard K; Shaw L; Jensen K; Lockridge JA; Dickinson B; McSwiggen JA; Vargeese C; Bowman K; Shaffer CS; Polisky BA; Zinnen S Activity of stabilized short interfering RNA in a mouse model of hepatitis B virus replication. Hepatology 2005. 41, 1349–1356. [DOI] [PubMed] [Google Scholar]
- 40.Harbort J; Elbashir SM; Vandenburgh K; H. M; Scaringe SA; Weber K; Tuschl T Sequence, chemical, and structural variation of small interfering RNAs and short hairpin RNAs and the effect on mammalian gene silencing. Antisense Nucl. Acid Drug Dev. 2003. 13, 83–105. [DOI] [PubMed] [Google Scholar]
- 41.Smith M; Jungalwala FB Reversed-phase high performance iiquid chromatography of phosphatidylcholine: A simple method for determining relative hydrophobic interaction of various molecular species. J. Lipid Res. 1981. 22, 697–704. [PubMed] [Google Scholar]
- 42.Pokholenko O; Gissot A; Vialet B; Bathany K; Thiéry A; Barthélémy P Lipid oligonucleotide conjugates as responsive nanomaterials for drug delivery. J. Mater. Chem. B 2013. 1, 5329–5334. [DOI] [PubMed] [Google Scholar]
- 43.Liu B; Chang J; Gordon WP; Isbell J; Yingyao Z; Tuntland T Snapshot pk: A rapid rodent in vivo preclinical screening approach. Drug Discovery Today 2008. 13, 360–367. [DOI] [PubMed] [Google Scholar]
- 44.Reed DR; Bachmanov AA; Tordoff MG Forty mouse strain survey of body composition. Physiol. Behav 2007. 91, 593–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wanke R; Milz S; Rieger N; Ogiolda L; Renner-Müller I; Brem G; Hermanns W; Wolf E Overgrowth of skin in growth hormone transgenic mice depends on the presence of male gonads. J. Investig. Dermatol 1999. 113, 967–971. [DOI] [PubMed] [Google Scholar]
- 46.Taniguchi T; Miyauchi E; Nakamura S; Hirai M; Suzue K; Imai T; Nomura T; Handa T; Okada H; Shimokawa C; Onishi R; Olia A; Hirata J; Tomita H; Ohno H; Horii T; Hisaedaa H Plasmodium berghei anka causes intestinal malaria associated with dysbiosis. Sci. Rep 2015. 5, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Alterman JF; Hall LM; Coles AH; Hassler MR; Didiot M-C; Chase K; Abraham; Sottosanti E; Johnson E; Sapp E; Osborn MF; DiFiglia M; Aronin N; Khvorova A Hydrophobically modified siRNAs silence huntingtin mRNA in primary neurons and mouse brain. Mol. Ther. Nucleic Acids 2015. 4, e266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Oberbauer R; Schreiner GF; Meyer TW Renal uptake of an 18-mer phosphorothioate oligonucleotide. Kidney Int. 1995. 48, 1226–1232. [DOI] [PubMed] [Google Scholar]
- 49.Khvorova A Oligonucleotide therapeutics — a new class of cholesterol-lowering drugs. N. Engl. J. Med 2017. 376, 4–7. [DOI] [PubMed] [Google Scholar]
- 50.Solano EC; Kornbrust DJ; Beaudry A; Foy JW; Schneider DJ; Thompson JD Toxicological and pharmacokinetic properties of qpi-1007, a chemically modified synthetic siRNA targeting caspase 2 mRNA, following intravitreal injection. Nucleic Acid Ther. 2014. 24, 258–266. [DOI] [PubMed] [Google Scholar]
- 51.Thompson JD; Kornbrust DJ; Foy JW; Solano EC; Schneider DJ; Feinstein E; Molitoris BA; Erlich S Toxicological and pharmacokinetic properties of chemically modified siRNAs targeting p53 RNA following intravenous administration. Nucleic Acid Ther. 2012. 22, 255–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Geary RS Antisense oligonucleotide pharmacokinetics and metabolism. Expert Opin. Drug Metab. Toxicol 2009. 5, 381–391. [DOI] [PubMed] [Google Scholar]
- 53.Khan T; Weber H; DiMuzio J; Matter A; Dogdas B; Shah T; Thankappan A; Disa J; Jadhav V; Lubbers L; Sepp-Lorenzino L; Strapps WR; Tadin-Strapps M Silencing myostatin using cholesterol-conjugated siRNAs induces muscle growth. Mol. Ther. Nucleic Acids 2016. 5, e342. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.