Abstract
Brain arteriovenous malformation (bAVM) is an important risk factor for intracranial hemorrhage. The pathogenesis of bAVM has not been fully understood. Animal models are important tools for dissecting bAVM pathogenesis and testing new therapies. We have developed several mouse bAVM models using genetically modified mice. However, due to the body size, mouse bAVM models have some limitations. Recent studies identified somatic mutations in bAVM. To develop a feasible tool to create sporadic bAVM in rodent and animals larger than rodent, we tested using CRISPR/Cas9 technique to induce somatic gene mutations in mouse brain in situ. Two sequence-specific guide RNAs (sgRNAs) targeting mouse Alk1 exons 4 and 5 were cloned into pAd-Alk1e4sgRNA+e5sgRNA-Cas9 plasmid. These sgRNAs were capable to generate mutations in Alk1 gene in mouse cell-lines. After packaged into adenovirus, Ad-Alk1e4sgRNA+e5sgRNA-Cas9 was co-injected with an adeno-associated viral vector expressing vascular endothelial growth factor (AAV-VEGF) into the brains of wild-type C57BL/6J mice. Eight weeks after viral injection, bAVMs were detected in 10 of 12 mice. Compared to the control (Ad-GFP/AAV-VEGF-injected) brain, 13% of Alk1 alleles were mutated and Alk1 expression was reduced by 26% in the Ad-Alk1e4sgRNA+e5sgRNA-Cas9/AAV-VEGF-injected brains. Around Ad-Alk1e4sgRNA+e5sgRNA-Cas9/AAV-VEGF injected site, Alk1-null endothelial cells were detected. Our data demonstrated that CRISPR/Cas9 is a feasible tool for generating bAVM model in animals.
Keywords: Alk1, CRISPR/Cas9, brain arteriovenous malformation, somatic gene mutation
Introduction
Brain arteriovenous malformations (bAVMs) are tangles of abnormal vessels shunting blood directly from arteries into veins without passing the capillary bed [1]. Rupture of bAVM vessels causes life-threatening intracranial hemorrhage [1]. The pathogenesis of bAVM is currently unclear. An animal model is an important tool for dissecting bAVM pathogenesis and testing new therapies.
Brain AVM can be sporadic or inherited. Patients with hereditary hemorrhagic telangiectasia (HHT), an autosomal dominant disorder, are among those inherited cases. Majorities of HHT patients have gene mutations in endoglin (ENG) or activin receptor-like kinase 1 (ALK1 also known as ACVLR1). Using Alk1[2] or Eng [3] floxed mice, we have established several bAVM mouse models through conditional knockout of Eng or Alk1 in combination with focal angiogenic stimulation [4-6]. Using these models, we have identified several key factors involved in bAVM pathogenesis [7, 8] and tested new therapies [9-11].
Due to the body size, unlike human bAVMs [12], mouse bAVM vessels are small and have thin walls. Brain AVM models in animals larger than rodent are likely to have bAVM vessels that are closer to humans and could be used in studies that are difficult to perform on mice. However, current bAVM surrogate models of large animals do not have true AVM nidus in the brain [13-15]. Although large animal models with true AVM in the brain are needed, generating a genetically modified large animal model is challenging [16, 17].
The causal gene for sporadic bAVM is unknown. Recently, somatic mutations of genes in Ras-MAPK (mitogen-activated protein kinase) pathway, such as KRAS, MAP2K1 (dual specificity mitogen-activated protein kinase kinase 1) and BRAF, have been found in sporadic bAVMs and extra-neural AVMs [18-20]. The mutations resulted in gain of function of the affected genes. The causal effect of these mutations in bAVM formation needs yet to be validated. The traditional transgenic/knockout animals may not be suitable to create sporadic bAVM models. The ability to induce specific somatic mutations in situ in the brain would be an effective way.
The CRISPR/Cas9 system consists of Cas9 nuclease and a short gene-specific RNA sequence (single guide sequence [sgRNA]) [21]. The DNA target of Cas9 and sgRNA requires an immediate adjacent 5’-NGG sequence, also known as protospacer adjacent motif (PAM) [22]. When the sgRNA guides Cas9 to the targeted site, Cas9 generates a double-strand breakdown (DSB) in the target DNA locus. Damaged DNA is repaired by the host system through either the error-prone non-homologous end joining (NHEJ) or the high-fidelity homology-directed repair (HDR), which introduces non-specific insertion or deletion (indel) mutations or template-mediated specific mutation in the target locus. The Cas9 and sgRNA have been expressed by viral vectors to induce somatic gene mutation in normal mouse tissue, and to correct mutations in diseased tissues in situ [23-25].
In this study, we tested the feasibility of using CRISPR/Cas9 system to create bAVM models through induction of Alk1 somatic mutations in the mouse brain in situ. We showed that co-injection of an adenoviral vector expressing two sgRNAs that target Alk1 exons 4, 5 and Cas9 nuclease (Ad-Alk1e4sgRNA+e5sgRNA-Cas9) with an adeno-associated vector expressing vascular endothelial growth factor (AAV-VEGF) induced a bAVM phenotype in adult mice.
Method and Materials
Design mouse Alk1 sgRNA sequences and generation of Cas9sgRNA expressing vectors.
Using http://tools.genome-engineering.org, 2 sgRNAs that target mouse Alk1 exon 4 (e4, 5’-TCCCGCAGCCCCGATAGACC-3’) and exon 5 (e5, 5’- CTCTAGGCTTGTGGCGTGTC-3’) were chosen. The corresponding DNA sequences were synthesized (Elim Bio. Inc. Hayward, CA): e4-sense: 5'-ACCTCCCGCAGCCCCGATAGACC-3' and e4-antisense: 5'-AACGGTCTATCGGGGCTGCGGGA-3' (bold and underlined nucleotides are overhang sequence of SapI restrict enzyme site), e5-sense: 5'-ACCCTCTAGGCTTGTGGCGTGTC-3' and e5-antisense: 5'-AACGACACGCCACAAGCCTAGAG-3'. The sense and antisense sequences of each sgRNA were annealed and cloned into plasmid AAV-U6sgRNA-hSYN-GFP-KASH-bGH (PX552, #60958, Addgene, Cambridge, MA) at SapI site to generate AAV-U6-Alk1e4sgRNA and AAV-U6-Alk1e5sgRNA (Figure 1A)[22, 24]. Expression cassettes of U6-Alk1-e4sgRNA and U6-Alk1-e5sgRNA were PCR amplified using Phusion high fidelity DNA polymerase (Cat#: F530S, Fisher Scientific, Waltham, MA) using primers: 5’-AATTGGTACCGAGGGCCTATTTCCCATGATTC-3’ and 5’-TCGAGGTACCAAAAAAGCACCGACTCGGTGCCACTTTTTC-3’, and cloned into a pAd-shuttle-PKG-Cas9 vector (Plasmid #58253, Addgene, Cambridge, MA) [23] to generate pAd-Alk1sge4+sge5-Cas9 (Figure 2A). The sequences of U6-sgRNA expression cassettes were verified by sequencing. pAd-Alk1e4sgRNA+e5sgRNA-Cas9 was packaged into type 5 adenovirus by Vector Biolabs (Malvern, PA)
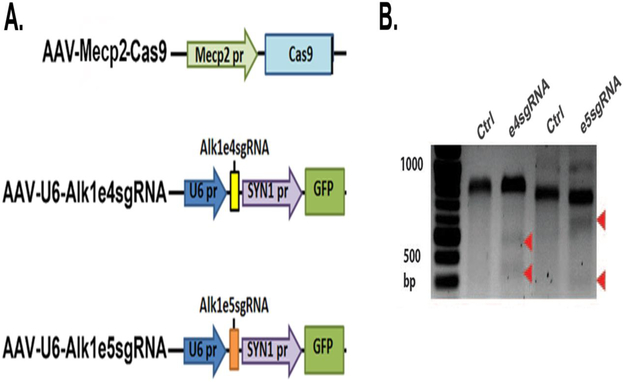
Figure 1. Alk1 e4sgRNA and e5sgRNA induced indel mutations in Alk1 gene in mouse cells.
A. Structure of vectors. Mecp2 pr: the promoter of a neural specific gene, Mecp2; SYN pr: the promoter of a neural specific gene, synapsin. B. Surveyor assay image. Indel mutations in Alk1 gene were detected in Neuro2A cells transfected with plasmids expressing Cas9 with e4sgRNA or e5sgRNA. For exon 4, wild-type band (Ctrl) is 761 base pairs (bp); mutant bands are 557 and 204 bps (arrows). For exon 5, wild-type band (Ctrl) is 830 bp; mutant bands are 480 and 350 bps (arrows).
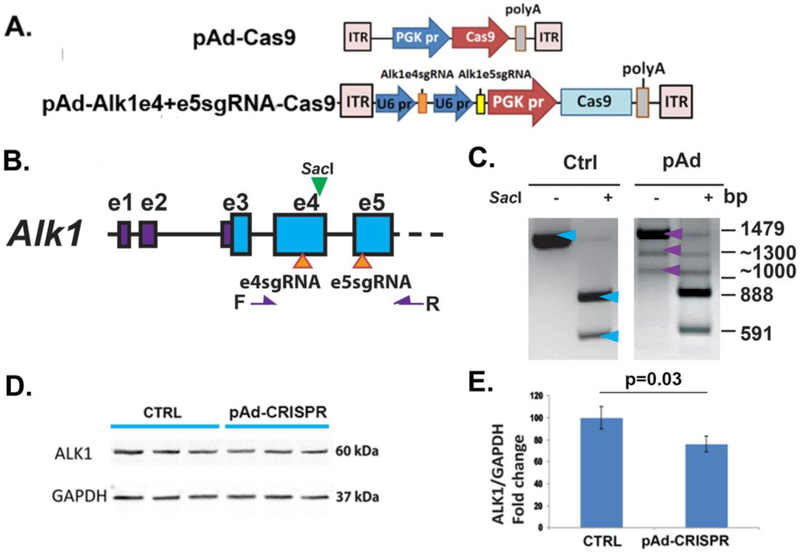
Figure 2. pAd-Alk1e4sgRNA+e5sgRNA-Cas9 induced deletion mutation in Alk1 gene and reduced Alk1 expression in mouse cells.
A. Structure of pAd-Cas9 and pAd-Alk1e4sgRNA+e5sgRNA-Cas9 vectors. pr: promoter. PGK: phosphoglycerate kinase. ITR: inverted terminal repeats. B. Location of e4sgRNA and e5sgRNA sites (arrows at the bottom) and primers F and R (purple arrows) for detection of deletions between e4sgRNA and e5sgRNA sites. SacI (green arrow at the top) is a restriction enzyme that cleaves wild-type PCR product. C. Deletion mutation in pAd-Alk1e4sgRNA+e5sgRNA-Cas9 transfected Neuro2A cells. A 1479 bp (top blue and purple arrows) fragment was amplified from pAd-Cas9 trensfected cells (Ctrl). SacI cut this fragment into 591 and 888 bp (bottom two blue two arrows). Two additional bands (1300 and 1000 bps, bottom two purple arrows) containing frameshift and deletion mutations were amplified from pAd-Alk1e4sgRNA+e5sgRNA-Cas9 transfected cells (pAd). D. A representative image of western blot using protein isolated from bEnd3 cells. CTRL: pAd-Cas9 transfect bEnd.3 cells; pAd-CRISPR: pAd-Alk1e4sgRNA+e5sgRNA-Cas9 transfected bENd.3 cells. E. Quantitative analysis of Alk1 expression in bEnd3 cells. N=3.
Transfection
Plasmid DNA was transfected in to Neuro2A mouse neuroblastoma cells (Sigma-Aldrich, St. Louis, MO) or bEnd.3 mouse brain endothelial cells using Lipofectamine LTX (Cat#15338100, Thermo Fisher Scientific, South San Francisco, CA) according to the manufacturer’s protocol. DNA, RNA or protein were extracted 48 h or 72 h after transfection for analyses.
Detection of Indel mutations
Site-specific mutagenesis of Alk1 e4 and e5 were tested in Neuro2A by co-transfection of AAV-U6-Alk1e4sgRNA or pAAV-U6-Alk1e5sgRNA plasmid with AAV-Mecp2-Cas9 plasmid, in which the expression of Cas9 nuclease is driven by a neuron specific promoter, Mecp2 promoter [24]. Since AAV-U6-Alk1e4sgRNA and pAAV-U6-Alk1e5sgRNA also express GFP driving by the promoter of synapsin 1 (SYN1, another neuron specific promoter) (Figure 1A), transfected cells were enriched by selecting GFP+ cells through cell sorting. Mutations in Alk1 exons 4 and 5 were detected through Surveyor assay according to the manufacturer’s instruction (Cat# 706020, Integrated DNA technologies, Redwood City, CA). Primers used for exon 4 Surveyor assay were F1: 5’-GCTGGAGAGGAACAGTAGTCC-3’ and R1: GGGCATGAGAAGAAAGCAGGG-3’. PCR amplification using F1 and R1 primers will yield a 761 bp fragment. If indel (insertion or deletion) mutation occurred, the 761 bp sequence will be cut into ~557 bp and ~204 bp fragments by Surveyor nuclease. Primers used for exon 5 Surveyor assay were: F2: GCAACCACAACGTGTCTCTG-3’ and R2: 5’-GTGTCTTACTCTGACCCAGGC-3’. PCR amplification using F2 and R2 primers will generate an 830 bp fragment. If indel mutation occurs, the 830 bp fragment will be cut into ~478 bp and ~352 bp fragments by Surveyor nuclease (Figure 1B).
Detection of Deletion Mutations
To analyze deletion mutations caused by co-expression of e4 and e5 sgRNAs, Ad-Alk1sge4+sge5-Cas9 that expresses e4 and e5 sgRNAs, and Cas9 nucleases was transfected into Neur2A cells. Genomic DNAs were isolated and were subjected to PCR amplification using F1 (F) and R2 (R) primers that were used for Surveyor assay (Figure 1B & 2B). Phusion high fidelity DNA polymerase (Cat#: F530S, Fisher Scientific, Waltham, MA) was used. A 1,479 bp PCR product was amplified from wild type DNA that has a SacI registration enzyme site located at 11 bp away from the 3’ end of e4 sgRNA (Figure 2B). SacI (Cat#: R0156S, NEB Biolabs, Ipswich, MA) cuts 1,479 bp PCR product into 888 bp and 591 bp fragments (Figure 2C). When deletion mutations occur between e4 and e5 targeted sites, smaller PCR fragments were amplified, which may not have SacI restriction enzyme site. All PCR products were cloned into a TA cloning vector, pCR2.1 (Cat # K202020, Thermo Fisher Scientific) and sequenced (GENEWIZ, South San Francisco).
RNA Extraction and Quantitative RT-PCR (qRT-PCR) Analyses
Total RNA was extracted from cells and tissues using RNAzol RT (Molecular Research Center, Cincinnati, OH) and reverse-transcribed into cDNA using Superscript III First-Strand Synthesis System (Invitrogen, Carlsbad, CA). Real-time PCR was performed using TaqMan Fast Advanced Master Mix (Applied Biosystems, Foster City, CA). Acvrl1 (Mm00437432_m1) and Gapdh (Mm99999915_g1) specific primers and probes were purchased from Applied Biosystems. All samples were run in triplicate. Relative gene expression was calculated using the comparative threshold cycle (CT) and normalized to Gapdh.
Animals
All experimental protocols and procedures of using laboratory animals were approved by the Institutional Animal Care and Use Committee (IACUC) at the University of California, San Francisco (UCSF). Animal husbandry was provided by the staff of the IACUC, under the guidance of supervisors who are certified Animal Technologists, and by the staff of the Animal Core Facility. Veterinary care was provided by IACUC faculty members and veterinary residents located on the San Francisco General Hospital campus. All mice were housed in a pathogen-free area in 421×316 cm2 cages with 12 hours light/dark cycle.
Equal number of eight weeks old male and female C57BL/6J mice were purchased from the Jackson Laboratory (Bar Harbor, ME) and were randomly assigned to different groups. A total of 85 mice were used.
Stereotactic injection of viral vectors into the basal ganglia
After being anesthetized through isoflurane (4%) inhalation, mice were placed in a stereotactic frame with a holder (David Kopf Instruments, Tujunga, CA). A burr hole, 1 mm lateral to the sagittal suture and 2 mm posterior to the coronal suture, was drilled in the pericranium. A 33-gauge needle attached to a Hamilton syringe was inserted 3 mm into the brain. A total of 2×109 genome copies (gcs) of AAV1-VEGF (AAV-VEGF vector packaged into AAV type 1 capsid) and 1×107 gcs of Ad-Alk1e4sgRNA+e5sgRNA-Cas9 or Ad-GFP (Cat#1060, Vector Biolabs, Malvern, PA) were stereotactically injected into the right basal ganglia at a rate of 0.2 μl per minute. The wound was closed with a suture after the injection.
Isolation of basal ganglia from injected brain and quantification of copies of Alk1 alleles with deletion mutation
Basal ganglia around the injected site was dissected from collected brain samples under a dissection scope. DNA was extracted using a PureLink Genomic DNA mini kit (K182001, Thermo Fisher Scientific) according to the manufacturer’s protocol. Real-time PCR was performed using power SYBR (Power SYBR Green Master Mix, Cat# 4367659, ThermoFisher) and primers flanking Alk1 e4 and e5 sgRNA targeted regions: e4F: 5’-GGTCTATCGGGGCTGCGGGAG-3’ and e5R: 5’-GACACGCCACAAGCCTAGAGC-3’. Primers flanking Alk1 exons 7 and 8 of were designed: e7F: 5’-GGTTCGTGGCATGGCGAGAG-3’ and e8R: 5’-GCAGAAAGTCATAGAGGGAGCCGTG-3’, to amplify a DNA fragment that has same size of the DNA fragment amplified by e4F and e5R, which is used as internal control (Figure 3B) for amplification efficacy.
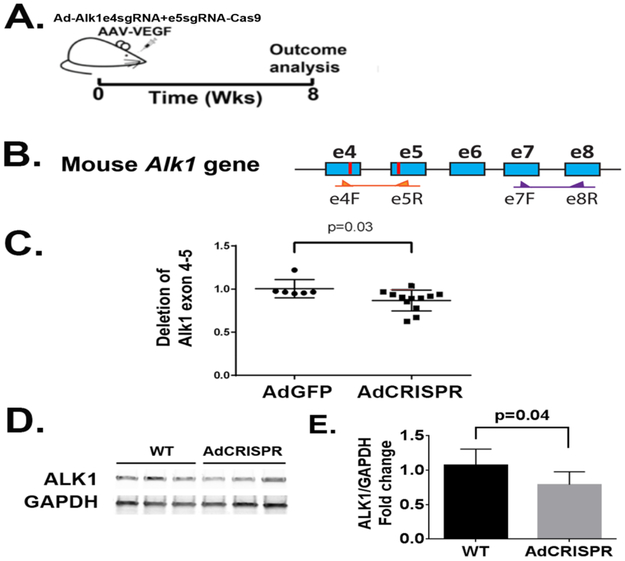
Figure 3. Ad-Alk1e4sgRNA+e5sgRNA-Cas9 induced deletion mutation in Alk1 gene and reduced Alk1 expression in the mouse brain.
A. Design for in vivo studies. Ad-Alk1e4sgRNA+e5sgRNA-Cas9 and AAV-VEGF were co-injected into basal ganglia of mouse brain. Eight weeks after virus injection, brain vessels were casted by latex to detect AVM phenotypes. Through intra-cardiac ventricle perfusion, latex dye enters vein only in the presence of arteriovenous shunt. Due to the particle size, latex cannot pass capillaries. Dysplasia vessels were quantified on immunostained sections. Alk1 gene mutation was analyzed by qPCR. Alk1 expression was analyzed by western blot. B. Schematic of mouse Alk1 exons 4 to 8 (e4-e8). Blue boxes are exons. The red lines are e4sgRNA and e5sgRNA target sites. The primers used for detecting deletion mutations between e4sgRNA and e5sgRNA sites and to amply sequence between exons 7 and 8 are indicated by arrows. C. Quantification of deletion mutations. N=6 for Ad-GFP group and N=12 for Ad-Alk1e4sgRNA+e5RNA-Cas9 (Ad-CRISPR) group. D. A western blot image. E. Quantification of Alk1 protein expression. WT: brain tissues of mice injected with Ad-GFP (N=7). Ad-CRISPR: brain tissues of mice injected with Ad-Alk1e4sgRNA+e5sgRNA-Cas9 (N=6).
Western blot
Basal ganglia around the injected site was dissected out as described above. Protein was extracted using a RIPA lysis and extraction buffer (Cat#89900, ThermoFisher) supplemented with Protease inhibitor cocktail (#8849, Sigma-Aldrich), 1mM NaF, 1mM sodium orthovandate and 1mM PMSF (#8553, Cell Signaling, Danvers, MA) and quantified by the Bradford method (Cat#: 500-0006, Bio-Rad, Hercules, CA) using a microplate reader (Emax, Molecular Devices, Sunnyvale, CA). Protein samples were loaded into 4–20% Tris-Glycine gels (Bio-Rad) and transferred onto nitrocellulose membranes (Bio-Rad). Immunoblotting were performed using primary antibodies specific to Alk1 (Abcam, ab108207) and Gapdh (Abcam, ab9485) and a goat anti-Rabbit secondary antibody (Cat#: IRDye800LT, Li-Cor, Lincoln, NE). Alk1 and Gapdh bands were detected by Li-Cor Quantitative western blot scanner and quantified using Li-Cor imaging software.
Latex perfusion
Latex dye was used to detected arteriovenous shunts (A-V shunts), a hallmark of AVM. Due to the particle size, latex dye enters veins after intra-left cardiac ventricle only when there is A-V shunt. Mice were anesthetized through isoflurane inhalation. Their abdominal and thoracic cavities were opened. After cutting off the left and right atria, latex dye (Blue latex, catalog BR80B; Connecticut Valley Biological Supply Co. Southampton, MA) was injected into the left ventricle using a 27-gauge needle attached to a 3 ml syringe. Brain samples were collected, washed briefly in PBS, fixed with 10 % formalin overnight, dehydrated using methanol series, and cleared with an organic solvent (benzyl alcohol/benzyl benzoate, 1:1; Sigma-Aldrich) [13].
Immunohistochemistry
After the mice were anesthetized with isoflurane inhalation, their brains were collected and frozen in dry ice. Brain samples were cut into 20-μm-thick coronal sections using a Leica RM2155 Microtome (Leica Microsystems, Wetzlar, Germany).
For quantification of vessel density, dysplastic vessels and vascular smooth muscle coverage, two sections per brain approximately 500 μm nostril or caudal to the injection site were co-stained with anti-CD31 (sc-18916, Santa Cruz Biotechnology, Santa Cruz, CA) antibody and anti-αSMA (α smooth muscle actin) antibody (1:1000, Cat #MA11547, Thermo Fisher Scientific). For analysis of Alk1 positive endothelial cells: two sections adjacent to the sections used for vessel quantification were co-stained with anti-CD31 antibody and anti-Alk1 antibody (1:50, AF770, R&D Systems, Minneapolis, MN). Sections were incubated with primary antibodies at 4°C overnight. Alexa Fluor 594-conjugated or Alexa Fluor 488-conjugated (1:250 or 1:500, Invitrogen, Carlsbad, CA) was used as secondary antibody to visualize positive stains. Vectashield antifade mounting medium containing 4′,6-diamidino-2-phenylindole (DAPI),Vector Laboratories, Cat. #: H-1200, Burlingame, CA) was used to stain the cell nuclei and mount the slide.
Brain sections were examined and imaged using a Keyence fluorescence microscopy under a 20X objective lens (Model BZ-9000, Keyence Corporation of America, Itasca, IL). A total of six images were taken and used for analysis for each animal brain, three from each brain section (to the right, to the left and below the injection site). Vascular density (number of vessels per mm2), dysplasia index (the number of vessels larger than 15 μm per 100 vessels), and vascular smooth muscle cell coverage (percentage of vessels with diameter larger than 15 μm completely covered or uncovered with α-SMA positive cells) [26] were quantified by at least two researchers blinded with the group assignment. The images were coded by a research who did not participate in the quantification.
Statistics
All data were analyzed using one-way ANOVA to compare the means of multiple groups followed by Turkey’s multiple comparisons or Student’s t test to compare the means of two groups using GraphPad Prism 6.0. Data are presented as mean ± standard deviation (SD). A p value < 0.05 was considered to be statistically significant. Sample sizes were N=6 unless otherwise stated.
Results
Alk1 e4sgRNA and e5sgRNA induced Alk1 gene mutation and reduced Alk1 expression in cell-lines
To test whether the sgRNA sequences we selected can induce site specific mutations in Alk1 gene, e4sgRNA or e5sgRNA was co-expressed with Cas9 in Neuro2A, a mouse neuroblastoma cell-line. The Neuro2A cell line was used because neuron specific promoters were used to drive the expression of the Cas9 and the GFP in sgRNA plasmids (Figure 1A). Transfected cells were enriched by selecting GFP+ cells through cell sorting. Indel mutations were detected at e4sgRNA and e5sgRNA targeted sites by Surveyor assay. Both sgRNAs have introduced indel mutations at their targeted sites. Fragments ~557 bp and ~204 bp were detected in pAAV-U6-Alk1e4sgRNA transfected cells, and fragments ~478 bp and ~352 bp were detected in pAAV-U6-Alk1e5sgRNA transfected cells (Figure 1B).
To test if co-expression of Alk1 e4sgRNA and e5sgRNA induces deletion mutations between their targeted sites, these sgRNAs and Cas9 were cloned into a plasmid to generate pAd-Alk1e4RNA+e5sgRNA-Cas9 (Figure 2A). In addition to the wild type band (1,474 bp), two additional bands (1,300 bp and 1,000 bp) were detected by PCR in the transfected Neuro2A cells (Figure 2B & C). Sanger sequencing showed that the 1,300 bp fragment contained frame shift mutations and the 1,000 bp band contained deletion mutations.
Compared to pAd-Cas9 transfected cells, Alk1 expression in pAd-Alk1e4sgRNA+e5sgRNA-Cas9 transfected bEnd.3 was reduced by 24% (Figure 2D & E).
Co-expression of Alk1 e4sgRNA and e5sgRNA with Cas9 and VEGF mutated the Alk1 gene, reduced Alk1 expression and induced AVM phenotypes in adult mouse brain
After packaging pAd-Alk1e4sgRNA+e5sgRNA-Cas9 into adenovirus, the vector was co-injected with AAV1-VEGF into mouse brain (Figure 3A). Compared to Ad-GFP injected brain, Ad-Alk1e4sgRNA+e5sgRNA-Cas9 injected brain showed 13% fewer of Alk1 wild-type alleles between e4sgRNA and e5sgRNA target sites (p=0.03, Figure 3B), and 26% less of Alk1 protein (p=0.037, Figure 3D & E).
Furthermore, by immunostaining, we detected Alk1 null endothelial cells in Ad-Alk1e4sgRNA+e5sgRNA-Cas9 injected brain (Figure 4A).
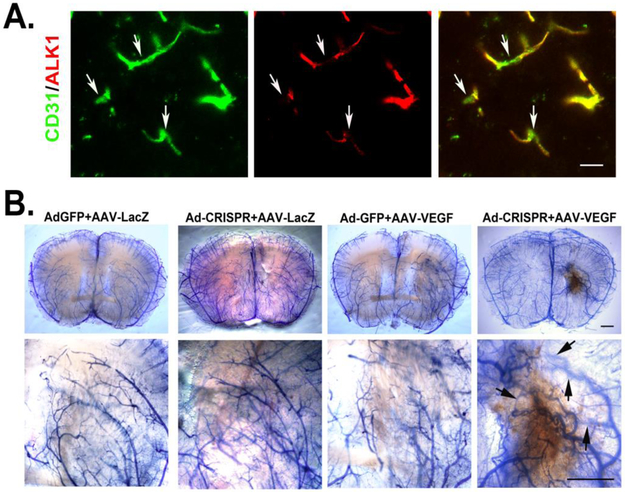
Figure 4. Ad-Alk1e4sgRNA+e5sgRNA-Cas9 silenced ALK1 expression in endothelial cells and induced bAVM in adult mouse.
A. Images show ALK1 null endothelial cells (arrows). Endothelial cells were stained by an anti-CD31 antibody (green). ALK1 expression was detected using an anti-ALK1 antibody (red). Scale bar: 20 μm. B. Representative images of latex perfused brains. Scale bar: 100 μm. The bottom pictures are zoom in images of the viral vector injected regions. Arrows indicate latex perfused veins.
AVMs were detected in 10 of 12 Ad-Alk1e4sgRNA+e5sgRNA-Cas9/AAV-VEGF injected mice through latex vascular casting 8 weeks after vector-injection. None AVM structure was detected in control mice injected with Ad-CMV-GFP/AAV-LacZ, Ad-Cas9-Alk1e4+e5sgRNA/AAV-LacZ, or Ad-CMV-GFP/AAV-VEGF (Figure 4B).
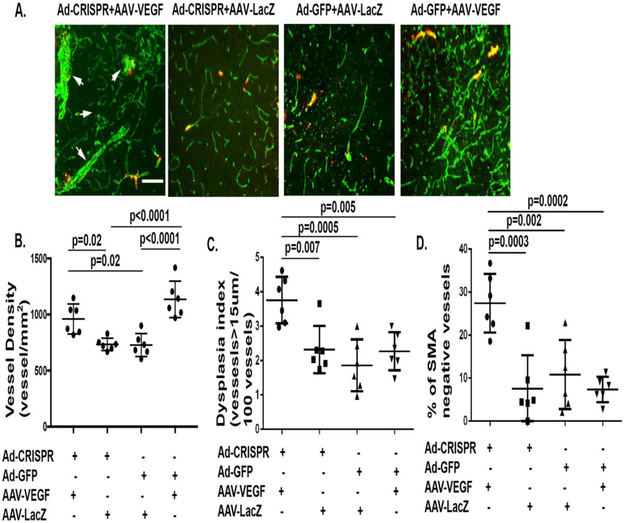
More vessels that are abnormal were found in the Ad-Alk1e4sgRNA+e5sgRNA-Cas9/AAV-VEGF injected brains compared to that of control groups (Figure 5). Vessels in the bAVM lesion of Ad-Alk1e4sgRNA+e5sgRNA-Cas9/AAV-VEGF injected mice have fewer smooth muscle coverage than controls (Figure 5).
Figure 5. Ad-Alk1e4sgRNA+e5sgRNA-Cas9 injected brain have more smooth muscle negative abnormal vessels.
A. Representative images of brain sections co-stained with antibodies specific to CD31 (green) and αSMA (red). Scale bar: 100 μm. B. Quantification of vessel densities (vessels/mm2). C. Quantification of dysplasia index (number of dysplasia vessels that are larger than 15 μm per 100 vessels). D. Quantification of αSMA negative vessels, Y axis shows % of vessels that were larger than 15 μm and αSMA negative. Ad-CRISPR: Ad-Alk1e4sgRNA+e5sgRNA-Cas9.
Discussion
In the present paper, we showed that CRISPR/Cas9 system is a feasible tool for inducing bAVM in animals. The major advantage of this system is that mutation of bAVM causal gene is induced somatically in the brain of adult animal in situ, which bypasses the challenges of germline modifications [27], such as, embryonic lethality, the time and cost that are needed for establishing, breeding and maintaining genetically modified animals.
We showed that CRISPR/Cas9 mediated Alk1 somatic mutation in wild-type mouse brain in situ induced AVM phenotype in the brain angiogenic region. Similar to the Alk1-deficient bAVM model induced through stereotactic injection of Ad-Cre and AAV-VEGF [4], bAVMs generated in this study were located at the vector injection site, where the Alk1 mutations occurred. The number of the abnormal vessels of this model and the Alk1-floxed mouse model are comparable [4]. By replacing the sgRNAs in Ad-Alk1e4sgRNA+e5sgRNA-Cas9, this technique is ready to be applied to induce bAVMs in animals that are larger than mouse, such as rabbit and pig, and to induce sporadic bAVM models.
. BAVM models of animals larger than mouse can be used to accomplish studies that are difficult to perform in mice. These studies include: (1) monitoring bAVM progression and regression in living animals, (2) using clinical criteria to evaluate the lesion severity [30, 31], (3) studying hemodynamic changes during lesion formation and progression, (4) analyzing flow induced changes in feeding arteries, vessels in the nidus and graining veins, and (5) testing the risk factors for AVM rupture. The bAVMs in large animal will also allow us to use microPET/CT scanning [32] to analyze cerebral metabolism during bAVM development and use MRI to detect acute and chronic hemorrhage in the bAVMs [33] without sacrificing the animals from time to time.
Somatic gene editing is also a suitable tool to induce sporadic bAVM animal models as recent studies indicated that somatic mutation instead of germline mutation could be the cause of sporadic bAVM as well as peripheral AVMs [18-20]. By replacing the sgRNAs in Ad-Alk1e4sgRNA+e5sgRNA-Cas9, this system can be used to mutate genes that have been identified in sporadic bAVMs.
In summary, we demonstrated the feasibility of using CRISPR/Cas9 system to induce bAVM in wild-type mouse. This technique can be easily adapted to create hereditary and sporadic bAVMs in other animals.
Acknowledgments
Funding Information
This study was supported by grants to Dr. Hua Su from NIH (R01 NS027713 and R01 HL122774), and the Michael Ryan Zodda Foundation.
Footnotes
Conflict of Interest
The authors declare that they have not conflict of interest.
Ethical Approval
All applicable international, national, and institutional guidelines for the care and use of animals were followed. The protocol and experimental procedures for using laboratory animals were approved by the Institutional Animal Care and Use Committee (IACUC) at the University of California, San Francisco. Animal husbandry and veterinary cares were provided by staffs in the IACUC and the Animal Core Facility.
Bibliography
- 1.Kim H, Su H, Weinsheimer S, Pawlikowska L, Young WL. Brain arteriovenous malformation pathogenesis: a response-to-injury paradigm. Acta Neurochir Suppl. 2011;111:83–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Park SO, Lee YJ, Seki T, Hong KH, Fliess N, Jiang Z, et al. ALK5- and TGFBR2-independent role of ALK1 in the pathogenesis of hereditary hemorrhagic telangiectasia type 2 (HHT2). Blood. 2008;111(2):633–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allinson KR, Carvalho RL, van den Brink S, Mummery CL, Arthur HM. Generation of a floxed allele of the mouse Endoglin gene. Genesis. 2007;45(6):391–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Walker EJ, Su H, Shen F, Choi EJ, Oh SP, Chen G, et al. Arteriovenous malformation in the adult mouse brain resembling the human disease. Ann Neurol. 2011;69(6):954–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Choi EJ, Chen W, Jun K, Arthur HM, Young WL, Su H. Novel brain arteriovenous malformation mouse models for type 1 hereditary hemorrhagic telangiectasia. PLoS One. 2014;9(2):e88511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen W, Sun Z, Han Z, Jun K, Camus M, Wankhede M, et al. De novo cerebrovascular malformation in the adult mouse after endothelial Alk1 deletion and angiogenic stimulation. Stroke. 2014;45(3):900–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang R, Han Z, Degos V, Shen F, Choi EJ, Sun Z, et al. Persistent infiltration and pro-inflammatory differentiation of monocytes cause unresolved inflammation in brain arteriovenous malformation. Angiogenesis. 2016;19(4):451–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ma L, Shen F, Jun K, Bao C, Kuo R, Young WL, et al. Integrin β8 deletion enhances vascular dysplasia and hemorrhage in the brain of adult Alk1 heterozygous mice. Transl Stroke Res. 2016;7(6):488–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Walker EJ, Su H, Shen F, Degos V, Amend G, Jun K, et al. Bevacizumab attenuates VEGF-induced angiogenesis and vascular malformations in the adult mouse brain. Stroke. 2012;43(7):1925–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhu W, Shen F, Mao L, Zhan L, Kang S, Sun Z, et al. Soluble FLT1 gene therapy alleviates brain arteriovenous malformation severity. Stroke. 2017;48(5):1420–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhu W, Chen W, Zou D, Wang L, Bao C, Zhan L, et al. Thalidomide Reduces Hemorrhage of Brain Arteriovenous Malformations in a Mouse Model. Stroke. 2018;49(5):1232–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pekmezci M, Nelson J, Su H, Hess C, Lawton MT, Sonmez M, et al. Morphometric characterization of brain arteriovenous malformations for clinical and radiological studies to identify silent intralesional microhemorrhages. Clin Neuropathol. 2016;35(3):114–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen W, Young WL, Su H. Induction of brain arteriovenous malformation in the adult mouse. Methods Mol Biol. 2014;1135:309–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Raj JA, Stoodley M. Experimental animal models of arteriovenous malformation: a review. Vet Sci. 2015;2(2):97–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Papagiannaki C, Clarencon F, Ponsonnard S, Couquet C, Maizeroi-Eugene F, Bresson D, et al. Development of an angiogenesis animal model featuring brain arteriovenous malformation histological characteristics. J Neurointerv Surg. 2016. [DOI] [PubMed] [Google Scholar]
- 16.Honda A, Hatori M, Hirose M, Honda C, Izu H, Inoue K, et al. Naive-like conversion overcomes the limited differentiation capacity of induced pluripotent stem cells. J Biol Chem. 2013;288(36):26157–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tachibana M, Sparman M, Ramsey C, Ma H, Lee HS, Penedo MC, et al. Generation of chimeric rhesus monkeys. Cell. 2012;148(1-2):285–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Couto JA, Huang AY, Konczyk DJ, Goss JA, Fishman SJ, Mulliken JB, et al. Somatic MAP2K1 mutations are associated with extracranial arteriovenous malformation. Am J Hum Genet. 2017;100(3):546–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nikolaev SI, Vetiska S, Bonilla X, Boudreau E, Jauhiainen S, Rezai Jahromi B, et al. Somatic Activating KRAS Mutations in Arteriovenous Malformations of the Brain. N Engl J Med. 2018;378(3):250–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Al-Olabi L, Polubothu S, Dowsett K, Andrews KA, Stadnik P, Joseph AP, et al. Mosaic RAS/MAPK variants cause sporadic vascular malformations which respond to targeted therapy. J Clin Invest. 2018;128(4):1496–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Platt RJ, Chen S, Zhou Y, Yim MJ, Swiech L, Kempton HR, et al. CRISPR-Cas9 knockin mice for genome editing and cancer modeling. Cell. 2014;159(2):440–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8(11):2281–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maggio I, Holkers M, Liu J, Janssen JM, Chen X, Goncalves MA. Adenoviral vector delivery of RNA-guided CRISPR/Cas9 nuclease complexes induces targeted mutagenesis in a diverse array of human cells. Sci Rep. 2014;4:5105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Swiech L, Heidenreich M, Banerjee A, Habib N, Li Y, Trombetta J, et al. In vivo interrogation of gene function in the mammalian brain using CRISPR-Cas9. Nat Biotechnol. 2015;33(1):102–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ruan GX, Barry E, Yu D, Lukason M, Cheng SH, Scaria A. CRISPR/Cas9-mediated genome editing as a therapeutic approach for Leber Congenital Amaurosis 10. Mol Ther. 2017;25(2):331–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen W, Guo Y, Walker EJ, Shen F, Jun K, Oh SP, et al. Reduced mural cell coverage and impaired vessel integrity after angiogenic stimulation in the Alk1-deficient brain. Arterioscler Thromb Vasc Biol. 2013;33(2):305–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maddalo D, Manchado E, Concepcion CP, Bonetti C, Vidigal JA, Han YC, et al. In vivo engineering of oncogenic chromosomal rearrangements with the CRISPR/Cas9 system. Nature. 2014;516(7531):423–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Park SO, Wankhede M, Lee YJ, Choi EJ, Fliess N, Choe SW, et al. Real-time imaging of de novo arteriovenous malformation in a mouse model of hereditary hemorrhagic telangiectasia. J Clin Invest. 2009;119(11):3487–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen W, Choi EJ, McDougall CM, Su H. Brain arteriovenous malformation modeling, pathogenesis and novel therapeutic targets. Transl Stroke Res. 2014;5(3):316–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Spetzler RF, Martin NA. A proposed grading system for arteriovenous malformations. J Neurosurg. 1986;65(4):476–83. [DOI] [PubMed] [Google Scholar]
- 31.Lawton MT, Kim H, McCulloch CE, Mikhak B, Young WL. A supplementary grading scale for selecting patients with brain arteriovenous malformations for surgery. Neurosurgery. 2010;66(4):702–13; discussion 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu NW, Ke CC, Zhao Y, Chen YA, Chan KC, Tan DT, et al. Evolutional Characterization of Photochemically Induced Stroke in Rats: a Multimodality Imaging and Molecular Biological Study. Transl Stroke Res. 2017;8(3):244–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guo D, Wilkinson DA, Thompson BG, Pandey AS, Keep RF, Xi G, et al. MRI Characterization in the Acute Phase of Experimental Subarachnoid Hemorrhage. Transl Stroke Res. 2017;8(3):234–43. [DOI] [PMC free article] [PubMed] [Google Scholar]