Abstract
The interaction between light and matter can be controlled efficiently by structuring materials at a length scale shorter than the wavelength of interest. With the goal to build optical devices that operate at the nanoscale, plasmonics has established itself as a discipline, where near-field effects of electromagnetic waves created in the vicinity of metallic surfaces can give rise to a variety of novel phenomena and fascinating applications. As research on plamonics has emerged from the optics and solid-state communities, most laboratories employ top-down lithography to implement their nanophotonic designs. In this review, we discuss the recent, successful efforts of employing self-assembled DNA nanostructures as scaffolds for creating advanced plasmonic architectures. DNA self-assembly exploits the base-pairing specificity of nucleic acid sequences and allows for the nanometer-precise organization of organic molecules but also for the arrangement of inorganic particles in space. Bottom-up self-assembly thus bypasses many of the limitations of conventional fabrication methods. As a consequence, powerful tools such as DNA origami have pushed the boundaries of nanophotonics and new ways of thinking about plasmonic designs are on the rise.
1. Introduction
The key component of plasmonics is metals. When light interacts with a metal nanoparticle, collective oscillations of conduction electrons known as particle plasmons are excited. As two metal nanoparticles are brought into proximity, their plasmon oscillations couple to each other, resulting in altered optical behavior that is highly distance dependent. 1–3 This arises from the fact that the plasmon resonance of a metal nanoparticle is influenced by other nanoparticles in its immediate environment. Such coupling effects have a profound impact on plasmonic assemblies for achieving novel phenomena and properties. 4–7
A prerequisite to build advanced plasmonic architectures, which possess well-defined configurations and tailored optical functionalities, is the ability to precisely control the arrangement of metal nanoparticles in space. To this end, DNA represents an ideal construction material owing to its unique sequence specificity.8 The almost infinite possibilities of designing individual sequences, which can be programmed to interact with each other, allow for an extremely rich versatility of DNA nanostructures.9,10 In general, DNA nanotechnology can be divided into two categories: structural and dynamic DNA nanotechnology. The former is often used to construct static two-dimensional (2D) and three-dimensional (3D) nano-objects of varying geometries and sizes,11 while the latter affords a remarkable capability to achieve dynamic nano-devices with desired reconfigurability12,13 and to perform computational tasks. 14–16
Over the last two decades, the utilization of DNA nanotechnology in plasmonics has led to unprecedented and unexpected novel research directions, which are mutually advantageous for both fields. When DNA nanotechnology meets plasmonics, it does not merely enable the realization of fascinating plasmonic architectures that were impossible to build before, 17,18 but also brings about tailored optical functionalities that are not possessed by DNA structures alone. 19,20 Thanks to the everlasting research efforts in this inter-disciplinary field, DNA nanotechnology-enabled plasmonics has significantly flourished and rendered possible a variety of interesting applications in biomolecular sensing, 21–26 surface-enhanced Raman and fluorescence spectroscopy, 27–41 diffraction-limited optics, 42, energy transfer 43,44 etc. In particular, the DNA route to build plasmonic architectures holds distinct advantages over top-down techniques due to its inherent molecular recognition and thus unprecedented nanoscale precision through well-controlled self-assembly processes. It also allows for large-scale and even bulk production of plasmonic nanostructures in a highly parallel manner. Importantly, it opens a new research pursuit towards reconfigurable and autonomous plasmonic nanosystems, which are beyond the state of the art of top-down nanofabrication techniques.
In this review, we aim to provide an overview of the recent progress in DNA-assembled advanced plasmonic architectures. It is noteworthy that we will focus on discrete, non-periodic plasmonic nanostructures as extensive reviews are available on periodic nanostructures such as plasmonic chains, arrays, and lattices assembled by DNA. 17,45 We will first describe the general experimental strategies that have been established for fabricating DNA-assembled plasmonic nanostructures. Then, we will introduce a diverse set of examples according to their characteristic optical properties and functionalities. Subsequently, we will discuss the inevitable evolution from static to dynamic plasmonic systems along with the fast development of this interdisciplinary field. Finally, possible future directions and perspectives on the remaining challenges and open opportunities will be elucidated.
2. Fabrication of DNA-Assembled Plasmonic Nanostructures
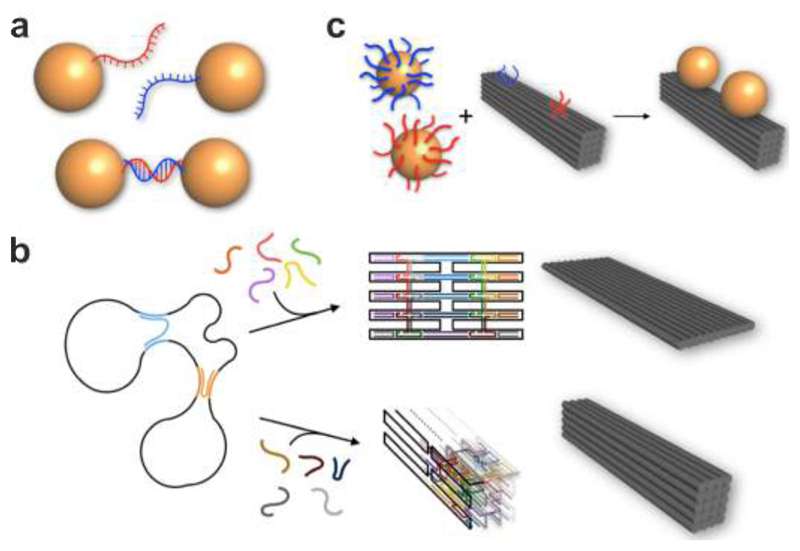
A general strategy to fabricate DNA-assembled plasmonic nanostructures relies on functionalization of metal nanoparticles, for example functionalization of gold nanoparticles (AuNPs) with thiol-modified single-stranded DNA (ssDNA) as programmable linkers (see Fig. 1a). 46–51 Unbound DNA can be removed by centrifugation and resuspension. For the formation of dimers, AuNPs modified with complementary ssDNA are added together for incubation. Through Watson-Crick base-pairing, AuNP dimers can be formed and subsequently purified from monomers as well as higher-order aggregates by gel electrophoresis. 47 Other than dimers, a variety of structures including homo/hetero dimers and trimers, 52–54 27,55 tetramers, 56 57 multimers, 58,59, chains, 60–62 lattices 63 64 65 etc. can also be achieved. Nevertheless, due to lack of structural rigidity, positioning of particles within individual assemblies cannot be accurately controlled. Also, due to lack of spatially directed organization, particle aggregates rather than discrete structures are often formed and still present after gel electrophoresis. To avoid such problems, a number of innovative protocols have been carried out to anisotropically functionalize AuNPs with DNA, e.g. through the use of Janus particles66 or geometric restrictions imposed by a solid substrate or other particles. 67,68 Noteworthy, Chad Mirkin, Oleg Gang and others have developed a wide variety of excellent crystalline assemblies using DNA-functionalized nanoparticles. 45,69–72 As in most of these examples the plasmonic properties are not examined, we recommend the reader to refer to the existing literature, while we limit our coverage to plasmonic studies.
Figure 1.
Fabrication of DNA-assembled plasmonic nanostructures. a. AuNPs with thiol-modified ssDNA can form a plasmonic dimer through Watson-Crick base-pairing. b. In DNA origami a long scaffold ssDNA strand, typically several thousand nucleotides long, is folded into arbitrary 2D and 3D shapes by hundreds of short staple strands. AuNPs functionalized with single or multiple DNA linkers can be assembled at designated binding sites through hybridization with their complementary DNA strands extending from the DNA origami template to form a wide variety of plasmonic architectures.
An alternative strategy to realize DNA-assembled plasmonic nanostructures is based on the DNA origami technique (see Fig. 1b). The concept of DNA origami was introduced by Rothemund in 2006 73 and relies on the folding of a long scaffold ssDNA strand – usually derived from the M13 phage genome – by hundreds of short staple strands into arbitrary 2D and 3D shapes. 74–78 Generally, nucleic acid nanostructures can also be assembled from oligonucleotides only, for instance, from RNA strands or from any combination of these and other building blocks. 10,79,80 DNA structures usually comprise bundles of interconnected double helices and have dimensions of a few up to several hundred nanometers. The bending rigidity of DNA bundles increases proportional to their area moment of inertia. Six-helix bundles, for example, have been reported to exhibit persistence lengths between 2 µm81,82 and 3 µm 83, which is ~ 50 times larger than that of an individual double strand (40 nm – 50 nm)84,85, and ten-helix bundles already have a persistence length of 17 µm. 83 Owing to high rigidities, high assembly yields and the inherent sequence-defined addressability86 of DNA origami, this approach is ideally suited to precisely organize metal nanoparticles in space for constructing plasmonic architectures with well-defined configurations. Metal nanoparticles functionalized with single or multiple DNA linkers can be assembled at designated binding sites through hybridization with their complementary DNA strands extended from DNA origami. 49,50,87 In particular, DNA origami offers positioning accuracy of a few nanometers, giving rise to superior molecular templates with nanoscale addressability. Pivotal for the emerging success of DNA origami and other DNA assembly methods is the development of open-access and easy-to-use computational tools for both designing and analyzing DNA structures. 88–90In parallel, growing interest of theoreticians in DNA assembly processes leads to important insights into the molecular and dynamic aspects. 91,92 After ten years of research efforts, a multitude of plasmonic architectures with different structural complexities and tailored optical functionalities have been accomplished using the DNA origami technique.
3. DNA-Assembled Static Plasmonic Nanostructures
DNA-assembled static plasmonic nanostructures refer to plasmonic DNA structures, which can yield well-defined optical properties, but do not exhibit dynamic behavior nor possess structural reconfigurability. Through specific spatial arrangements, such plasmonic nanostructures can exhibit interesting optical phenomena including plasmon hybridization, 53,93–96 Fano resonances, 97–99 magnetic resonances, 100 chiral response, 19,20,101–103 104 105, photonic crystal modes 106 etc. They may also serve as powerful plasmonic platforms for surface-enhanced Raman 30 31 32 33 107 34 35 and fluorescence spectroscopy 37 36 38,42 to understand fundamental light-matter interaction on the nanoscale.
3.1. Plasmon Hybridization
Plasmon hybridization is a theory developed by Nordlander to interpret coupling effects in complex plasmonic nanostructures. 93,94 It states that plasmons in assemblies of metal nanoparticles bear a direct resemblance to electrons in quantum molecular orbitals. Plasmons excited in adjacent metal nanoparticles interact, mix, and hybridize just like the electronic wave functions of atomic and molecular orbitals. The simplest case is plasmon hybridization in a dimer of AuNPs as shown in Fig. 2a. The near-field coupling between the two AuNPs in close proximity can result in the formation of bonding and antibonding modes, respectively. The antibonding mode is located at a higher frequency arising from its larger net dipole moment, when compared to the bonding mode.
Figure 2.
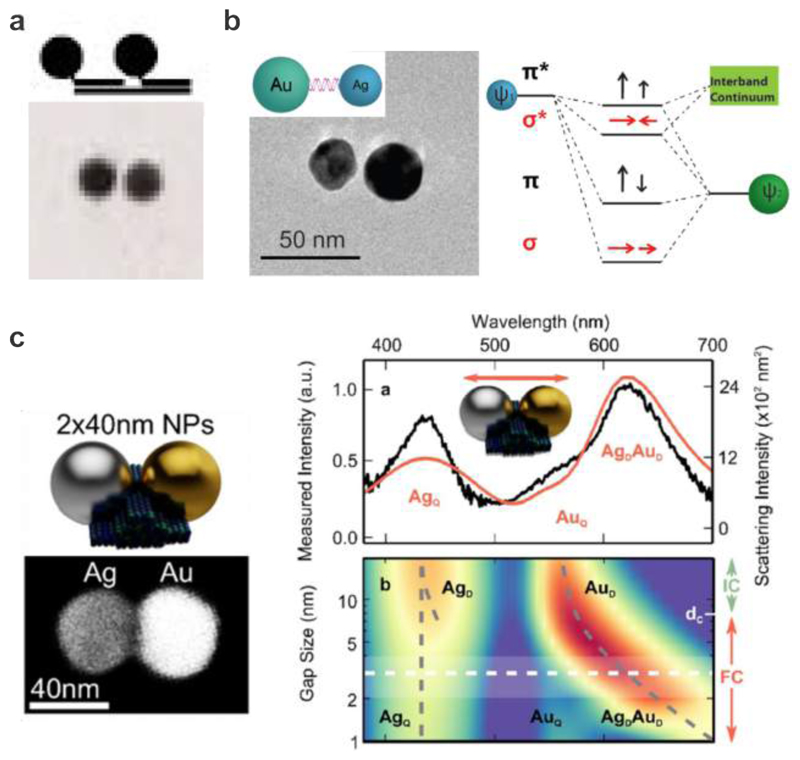
Plasmon hybridization in DNA-assembled plasmonic nanostructures. a. AuNP homodimer formation using DNA-monofunctionalized AuNPs. 52 Images reproduced with permission from ref 52. Copyright 1999 Wiley. 11b. Formation of a DNA-assembled Au-Ag heterodimer and the corresponding energy level diagram illustrating the plasmon hybridization. 53 Images reproduced from ref 53. Copyright 2010 American Chemical Society. c. Left: Au-Ag heterodimer hosted on a DNA origami template and TEM image of a representative structure. Right: Scattering response of the heterodimers under longitudinal polarization and color map for the scattering response as a function of the gap size. 95 Images reproduced from ref 95. Copyright 2016 American Chemical Society.
Alivisatos et al. first demonstrated the realization of discrete AuNP assemblies through DNA-monofunctionalization (see Fig. 2a). 47 AuNPs modified with ssDNA in a 1:1 ratio were achieved by taking advantage of the restricted surface-areas of the 1.4 nm particles. After monofunctionalization, the AuNPs were assembled with secondary short scaffolding strands into homodimers and homotrimers through Watson-Crick base-pairing followed by a gel electrophoresis purification step. Later, the same group reported the assembly of heterodimers and heterotrimers with a similar strategy using 5 nm and 10 nm AuNPs. 52 The optical characterization of the 10 nm homodimers was carried out using UV/Vis spectroscopy. A decrease in absorbance, spectral broadening, and a spectral shift to longer wavelengths were observed for the homodimers with respect to the monomers. These results exemplified the first experimental proof of plasmon hybridization in DNA-assembled plasmonic nanostructures.
The critical parameters that determine the optical properties of a complex plasmonic nanostructure include the sizes, shapes, material compositions of the individual nanoparticles, the interparticle spacing, as well as the overall configuration in space. Due to technical constraints, AuNPs utilized in the early work on DNA-assembled plasmonic nanostructures were rather small (≤ 10 nm) and the interparticle distances were often as large as or even larger than their actual sizes. The resulting small dipole moments and weak coupling strengths thus led to indiscernible spectral changes, when compared to the spectra of the individual AuNPs. This obstacle was overcome only later, thanks to successive methodological improvements in this field. One of the remarkable advancements was achieved by Sönnichsen et al., who demonstrated the first plasmon rulers composed of a 40 nm AuNP dimer or a silver (Ag) nanoparticle dimer, which exhibited distinct plasmon hybridization effect. 22 This landmark work will be discussed later in the section of DNA-assembled dynamic plasmonic nanostructures as it represents one of the very first dynamic plasmonic systems enabled by DNA nanotechnology.
Other than homodimers, heterodimers are also of great interest due to their complex optical response, offering rich information and profound insights into plasmonic coupling mechanisms. Asymmetries are often introduced by applying two particles of different sizes or shapes. As a matter of fact, asymmetry can also be created using different material compositions. As shown in Fig. 2b, Au-Ag heterodimers were assembled using DNA by Sheikholeslami et al. 53 Thiolated ssDNA molecules were conjugated on the Au (40 nm) and Ag (30 nm) particles, followed by passivation using thiolated polyethylene glycol to provide enhanced stability. The Au and Ag particles were then mixed in a 1:1 ratio and allowed to hybridize overnight. Optical measurements with polarized scattering spectroscopy revealed the excitation of both the bonding and antibonding plasmonic modes. Interestingly, the antibonding modes were red shifted relative to the resonance of the Ag particle. This anomalous shift was due to the coupling between the Ag particle resonance and the quasi-continuum of the interband transitions in Au, which did not occur in the cases of single-composition dimers presented in the same paper.
The fabrication of Au-Ag heterodimers could also be realized by utilization of DNA origami as shown in Fig. 2c. 95 The rigidity of DNA origami allowed for less variability in the interparticle spacing than the aforementioned cases. 40 nm Ag and 40 nm Au particles were assembled into a heterodimer on a DNA origami platform with a sub-5 nm gap. The different material compositions were verified using scanning electron microscopy and energy-dispersive X-ray characterization. Importantly, the authors also developed a new interparticle-spacing-dependent coupling model for heterodimers, very useful for controlling symmetry breaking in collective plasmon modes. Their study confirmed that direct plasmonic coupling in the Au-Ag dimer could be observed in both experiment and theory only for small gap sizes, as it required the Ag dipolar mode frequency to drop below the frequency of the Au interband transitions.
By placing a Ag NP between two remote AuNPs, Roller et al. introduced a concept for ultrafast and low-dissipative transfer energy. 44 With its excitation energy level being too high to be occupied, the Ag particle here served as a virtual transmitting state during the passage. As a result, no heat was generated in the Ag particle, while the two Au particles were still perfectly coupled. This behavior could be described by classical simulations and with a quantum mechanical model, both reproducing the experimental results. The remarkably short plasmon transfer times of the non-dissipative plasmon passage were on the femtosecond scale, opening the door towards a new class of plasmonic wave-guides.
3.2. Plasmonic Fano Resonances
Fano interference was originally studied in atomic and quantum mechanical systems. 97,99,108–110 In fact, it is also ubiquitous in classical systems, generally when energy transfer from an initial state to a final state takes place via two pathways that destructively interfere. Plasmonic systems can also support Fano interference, which often occurs in assemblies composed of closely-packed metal nanoparticles. Such Fano phenomena promise many useful applications in optical sensing, switching, nonlinear, and slow-light devices.
Plasmonic Fano interference in DNA-assembled particle clusters was first demonstrated by Fan et al. (see Fig. 3a). 111 Au nanospheres (74 nm) and slightly larger Au nanoshells (62.5 and 92.5 nm for the inner and outer shells, respectively) were functionalized separately with DNA strands that were complementary. They were mixed and incubated together to first form loosely bound structures. The clusters were then compressed into closely-packed pentamers through a drying process on a hydrophilic substrate. Scattered light from a single pentamer was collected using dark-field spectroscopy. The Fano resonance was identified as a narrow and asymmetric dip imposed on a broad spectral profile as shown in Fig. 3b. This effect resulted from the destructive interference between a superradiant bright mode, in which the dipolar plasmons of all the AuNPs oscillated in phase, and a subradiant dark mode, in which the small nanosphere capacitively coupled with two of the nanoshells (see Fig. 3b). In the quasistatic, nonretarded limit, the dark mode should possess nearly no net dipole moment and should be not easily excited by light. In the retarded limit, the bright and dark modes became supperadiant and subradiant, respectively. In other words, the coupling mediated by the plasmonic near-fields led to the interaction between the sub- and superradiant modes, introducing a Fano resonance in the superradiant continuum at the frequency of the subradiant mode. The Fano resonance depth is strongly dependent on the constituent particle sizes, shapes, interparticle spacing, the number of particles, as well as the cluster symmetry as demonstrated by a vast of follow-up experiments, mainly carried out using top-down nanotechniques. 112,113 114 115
Figure 3.
DNA-mediated assembly of plasmonic heteropentamers to generate Fano effects. a. Au nanospheres and nanoshells are functionalized with thiolated DNA and incubated together. After drying on a hydrophilic substrate, close-packed pentamers can be formed. A representative TEM image of the structure. b. Extinction spectrum and surface charge plots of the heteropentamer for the dark and bright modes. 111 Images reproduced from ref 111. Copyright 2011 American Chemical Society.
3.3. Plasmonic Magnetic Resonances
Plasmonic magnetic resonances have attracted a lot of attention due to the enthusiastic pursuits of artificial negative refractive index materials, which hold the promise for many fascinating phenomena including invisibility cloaking, super lensing, and negative refraction. 116,117 118 119 In general, simultaneous negative permittivity and negative permeability are required to occur in the same frequency region to effectively achieve a negative refractive index. While metals naturally possess negative permittivity at visible frequencies, negative permeability has to be created through an artificial magnetic response. Nevertheless, structural designs such as split-ring resonators, which are used to achieve negative permeability at lower frequencies, 120,121 cannot be easily transferred due to the so-called saturation effect of the magnetic response at visible frequencies. 122 To overcome this problem, Engheta et al. came up with an elegant design, in which metal nanoparticles are brought together in a ring geometry. 123 124 Such a structure can support a circulating displacement current due to plasmon excitations, which can lead to the occurrence of magnetic resonances at high frequencies. Indeed, several groups recorded magnetic resonances arising from planar ring arrangements 98,125 and from “raspberry”-shaped colloidal assemblies synthesized in solution.126,127
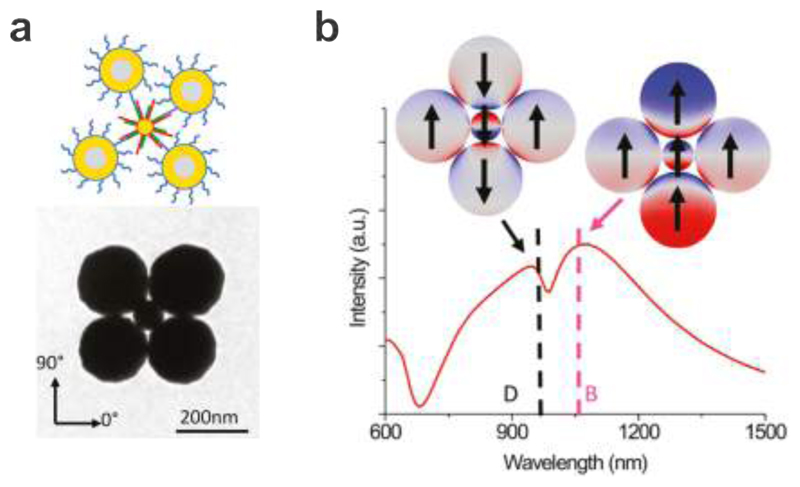
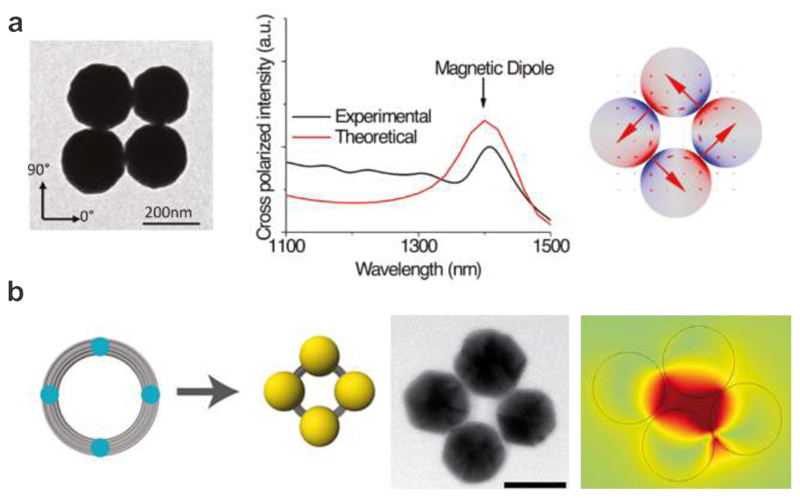
Fig. 4a shows the formation of the magnetic resonances in quadrumers demonstrated by Fan et al. 111 who assembled four Au nanoshell particles in a ring geometry using DNA as a linker molecule. The cross-polarized scattering spectrum of the quadrumer exhibited a clear and narrow peak near 1400 nm. The simulated mode plot at this resonance revealed a circulating current around the ring of the nanoshells, which identified the excitation of the magnetic resonance. Later, Roller et al. utilized DNA origami to assemble small nanoparticle rings, which exhibited magnetic resonances at visible frequencies as shown in Fig. 4b. 100 A DNA origami structure composed of 14 parallel arranged DNA helices of 200 nm length was designed to curve into a ring by insertion and deletion of bases at selected sites.78 The ring had a diameter of 62 nm and a cross-section of ~10 nm. Four AuNPs (40 nm) covered with ssDNA were then assembled on one origami ring through DNA hybridization. The individual plasmonic nanostructures immobilized on a glass substrate were characterized by dark-field spectroscopy. It was demonstrated that the magnetic resonance appeared only in four-particle rings with broken symmetry, which enabled a coupling channel between the magnetic mode and the far-field photons propagation perpendicular to the substrate. The simulated charge and magnetic field distributions confirmed the formation of circulating currents and a magnetic dipole in the particle ring structure, respectively, indicating the excitation of the magnetic resonance as shown in Fig. 4b.
Figure 4.
Magnetic resonances in DNA-assembled particle clusters. a. Left: TEM image of a DNA-assembled quadrumer. Experimental and theoretical spectra of the DNA-assembled quadrumer, which reveal narrow magnetic dipole peaks near 1400 nm. 111 Images reproduced from ref 111. Copyright 2011 American Chemical Society. b. Left: schematic illustration of the DNA origami template for assembly of four AuNPs to form a plasmonic quadrumer and a representative TEM image. Right: simulation of the surface charge distribution for the magnetic resonance. 100 Images reproduced from ref 100. Copyright 2015 American Chemical Society.
3.4. Plasmonic Chirality
A geometrical object is chiral, if its mirror image is non-superimposable with itself. Just like our two hands, which are non-superimposable to each other, many molecules can exist in left-handed (LH) and right-handed (RH) forms, the so-called enantiomers. Chirality is crucially important in pharmaceutics, as the biological activities of enantiomer molecules can be distinctly opposite. One form may be helpful, whereas the other form may be inactive or even toxic.
The commonly used spectroscopic technique to study chiral molecules is circular dichroism (CD), which is defined as the difference in extinction of LH and RH circularly polarized light. Natural chiral molecules such as proteins and DNA exhibit weak CD mostly in the UV spectral range. Govorov et al. theoretically proposed that it is possible to build plasmonic analogs of chiral molecules, which show much stronger CD than their molecular counterparts and possess large spectral tunability. 128
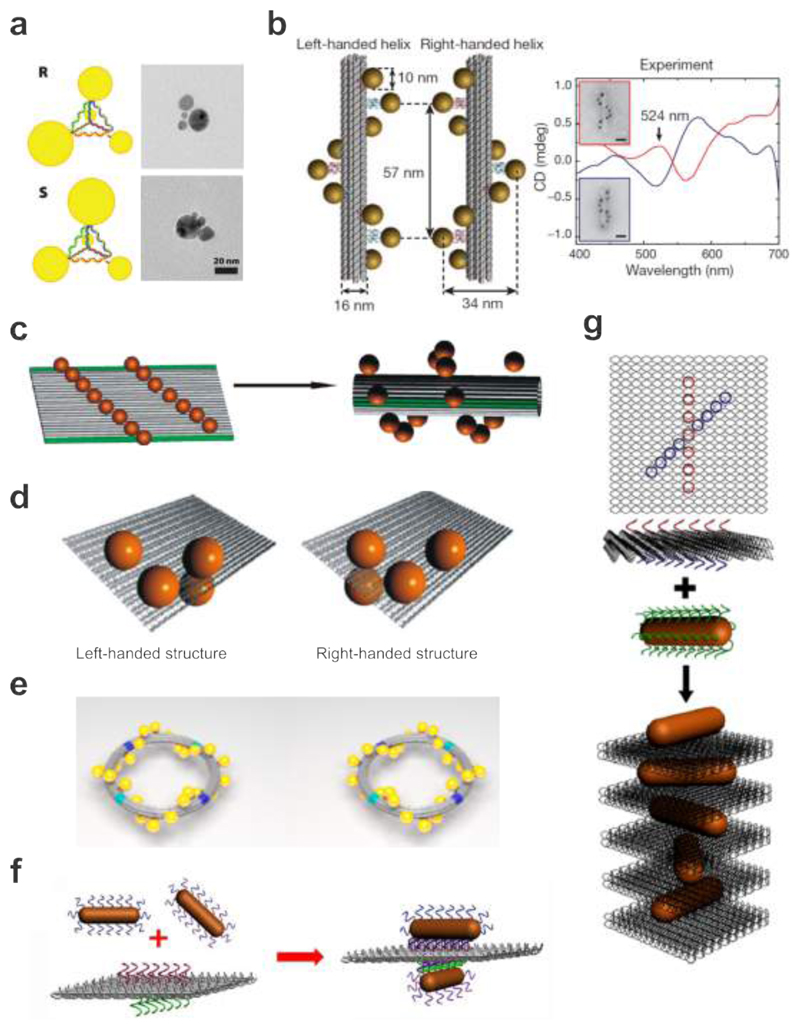
One of the first attempts toward DNA-assembled plasmonic chiral nanostructures was accomplished by Mastroianni et al. AuNP pyramids linked by DNA were carefully constructed as shown in Fig. 5a. 57 In an individual structure, each DNA strand was designed with a unique sequence, which allowed for positioning of a single AuNP at one of the pyramidal tips. Four AuNPs of different diameters (5, 10, 15, 20 nm) were arranged in one structure. The enantiomeric version was obtained by switching the positions of two of the AuNPs through conjugation with the opposite strands. Unfortunately, CD was not observed from these plasmonic assemblies mainly due to negligible coupling between the AuNPs of substantially different sizes and comparatively large inter-particle distances. In addition, the non-rigidity of the assemblies led to varying configurations over different nanostructures and therefore vanishing CD.
Figure 5.
DNA-assembled plasmonic chiral nanostructures. a. Schematic and TEM images of the DNA-AuNP pyramids in different handedness. 57 Images reproduced from ref 57. Copyright 2009 American Chemical Society. b. LH and RH nanohelices are formed by nine AuNPs that are attached to the surface of DNA origami bundles. AuNPs carry multiple thiol-modified DNA strands, which are complementary to the staple extensions on origami. Experimental CD spectra of the LH and RH helices of nine AuNPs, showing characteristic bisignate signatures in the visible. 20 Images reproduced with permission from ref 20. Copyright 2012 Macmillan Publishing Ltd. c. Fifteen AuNPs are assembled on a rectangular origami sheet. Addition of the folding strands leads to rolling and subsequent stapling of the 2D sheet into a hollow tube. As a consequence the AuNPs are arranged into a 3D helix. 19 Images reproduced from ref 19. Copyright 2011 American Chemical Society. d. Chiral plasmonic tetramers assembled on DNA origami sheets in different handedness. 104 Images reproduced from ref 104. Copyright 2013 American Chemical Society. e. Twenty four AuNPs are assembled in a helical fashion along an origami ring to form a LH or RH plasmonic toroidal structure. 103 Images reproduced from ref 103. Copyright 2016 American Chemical Society. f. Schematic of the bifacial DNA origami-directed assembly of 3D AuNR dimers. 105 Images reproduced from ref 105. Copyright 2013 American Chemical Society. g. Schematic of the self-assembly of RH-AuNR helices. This design enables one-pot assembly of AuNR helical superstructures. 131 Images reproduced from ref 131. Copyright 2014 American Chemical Society.
The power of structural DNA nanotechnology for constructing rigid plasmonic architectures was first demonstrated by Kuzyk et al. 20 The authors designed two 100 nm long DNA origami bundles of 24 DNA helices as templates to organize 10 nm AuNPs in LH and RH helical forms, respectively (see Fig. 5b). Capture strands were extended from nine binding sites on each of the two templates. Through DNA hybridization, AuNPs functionalized with complementary DNA were assembled around the DNA origami bundles in a staircase fashion, forming LH and RH AuNP helices, respectively. Due to the near-field interactions between the AuNPs in a 3D chiral order, these plasmonic assemblies gave rise to a bisignate CD response, which resembled that of chiral molecules, however, was orders of magnitude stronger and became apparent at visible frequencies. As predicted by theory, the LH and RH AuNP helices exhibited mirrored CD spectra. The authors also demonstrated the tunability of the CD response by Ag enhancement of the AuNP helices. In an approach by Shen et al., a rectangular DNA origami sheet dressed with thirteen AuNPs forming two parallel lines was rolled up into a tubular shape as shown in Fig. 5c. 19 As the repulsion between the particles favored the closure of the sheet toward one direction, AuNP helices of defined handedness were formed and shown to exhibit a measurable CD response in the visible.
As of today, a great variety of DNA origami-templated chiral plasmonic architectures have been realized. For example, Shen et al. also demonstrated DNA-assembled plasmonic tetramers (see Fig. 5d). 104 Each tetramer contained four identical 20 nm AuNPs assembled on a rectangular DNA origami template. In contrast to the aforementioned work of Mastroianni et al., the four AuNPs in the plasmonic tetramer could strongly couple due to their identical size. Also, the rigidity of the origami template and the utilization of multiple capture strands per AuNP contributed to the homogeneity of the structural configurations, allowing for experimental observation of distinct CD from the LH and RH plasmonic tetramers.
DNA origami is ideally suited to construct chiral systems with increasing complexities, for example, a plasmonic toroidal structure (see Fig. 5e), 103 which is extremely challenging to achieve using top-down nanotechniques. Four curved origami bundles were linked in a ring geometry. Twenty four AuNPs (13 nm) functionalized with complementary DNA strands compared to the capture strands extended from the origami ring were assembled in a helical fashion to form a LH or RH plasmonic toroidal structure. The assembled plasmonic toroids with designated handedness exhibited pronounced optical activity in the visible spectral range. In addition, the authors demonstrated that given the unique circular symmetry, distinct chiroptical response along the axial orientation of the toroids could be revealed by simple spin-coating of the structures on substrates. Such orientation self-alignment neither required surface functionalization nor introduced birefringence effects, which often arise from symmetry breaking.
The dominating role of Au nanospheres as key player for building DNA-assembled plasmonic nanostructures was terminated after the successful demonstration of well-controlled gold nanorod (AuNR) assemblies on origami by Pal et al. 129 Due to their strong optical response and anisotropic nature, AuNRs are excellent candidates for the realization of advanced plasmonic architectures with distinct and tailored optical functionalities. Soon after the work of Pal et al., Lan et al. realized crossed AuNR (11 nm × 37 nm) dimers templated by bifacial DNA origami as shown in Fig. 3f. 105,130 Importantly, the 3D spatial configuration of the dimer could be precisely tuned by rationally shifting the AuNR positions on origami. The two crossed AuNRs constituted a 3D plasmonic chiral object, which generated a theme of handedness when interacting with LH and RH circularly polarized light, giving rise to strong CD. As a matter of fact, the CD response from such AuNR dimers can be well described by the Born-Kuhn model, which was already used to interpret the phenomenon of optical activity in natural chiral molecules. A coupled AuNR dimer mimics the geometry of a molecule containing two chromophores, a situation commonly described as exciton-coupling in organic chemistry. Taking a step further, Lan et al. also created AuNR helices, which contained twisted AuNRs directed by DNA origami as shown in Fig. 5g 131. Capture strands in an ‘X’ pattern were arranged on the two sides of a 2D origami template. AuNRs functionalized with complementary DNA sequences were positioned on the origami and subsequently led to AuNR helices with the origami intercalated between neighboring AuNRs. LH and RH AuNR helices were conveniently accomplished by tuning the mirrored-symmetric ‘X’ patterns of the capture strands on the origami. These DNA-assembled plasmonic chiral nanostructures shed light on many interesting applications including tunable chiral fluids, enantiomer sensing, chiral signal amplification, and fluorescence-combined chiral spectroscopy.
3.5. Surface-Enhanced Raman Spectroscopy
Surface-enhanced Raman spectroscopy (SERS) is a surface sensitive technique that utilizes metal surfaces or nanostructures to enhance Raman scattering of adsorbed molecules. 132,133The ability to analyze specific chemical fingerprints of minute amounts of molecules on the nanoscale makes SERS extremely useful for applications in environmental analysis, pharmaceuticals, material sciences, drug and explosive detection, as well as food quality analysis. 134,135 However, SERS substrates often encounter poor reproducibility and performance in that the Raman scattering cross-sections of the probed molecules can be orders of magnitude smaller than fluorescence cross-sections. 136,137 Fortunately, SERS signals can be greatly enhanced by placing Raman-active molecules in plasmonic hot spots, where intense electromagnetic fields are highly localized.138,139 136,140 Such SERS intensity is proportional to the fourth power of the localized electric field with a possible enhancement factor beyond 1010.141,142 In general, the electromagnetic fields in plasmonic hot spots are enhanced as the nanogap size decreases, especially down to the 1 nm region. 29,36,107 Therefore, over the last decades continuous efforts have been exerted to synthetize various plasmonic nanostructures possessing narrow gaps for achieving strong and reliable SERS signals. 27 143
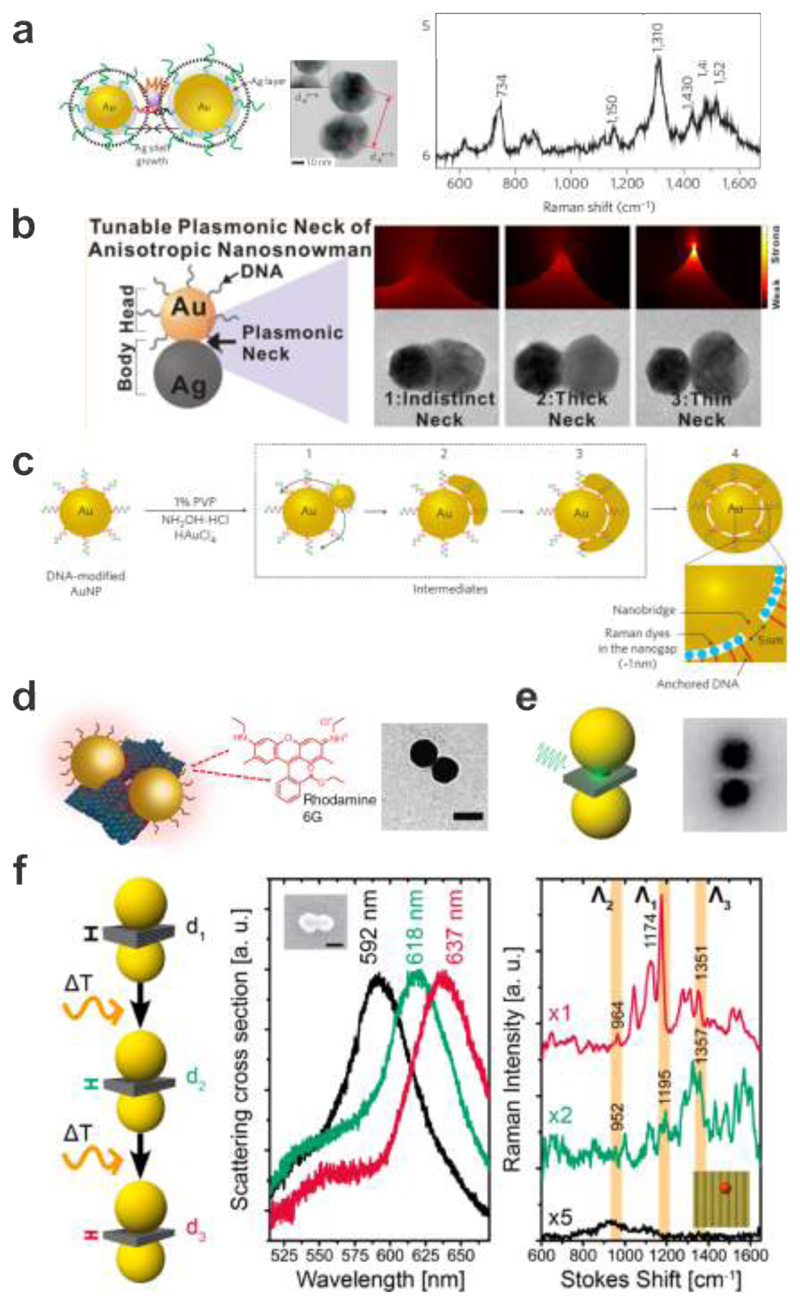
The successful synthesis of DNA-assembled Au core/Ag shell heterodimers was reported by Lim et al. 28 These particles could be used to detect Raman signals from single dye molecules placed in the dimer gaps as shown in Fig. 6a. AuNP (20 and 30 nm) heterodimers were first synthesized by means of DNA hybridization. A single Cy3 dye molecule was then conjugated between the two DNA-tethered AuNPs. Ag shell growth on the AuNPs was carefully controlled to generate gap-engineerable core-shell dimers. Such DNA-assembled heterodimers manifested remarkable single-molecule sensitivity with high structural reproducibility. When modified by other biomolecules such as proteins, these heterodimers could further serve as both in vitro and in vivo bio-labelling probes with ultrahigh sensitivity, quantification potentials, and multiplexing capability. Furthermore, another synthetic strategy has been developed by the same group for preparing Au-Ag nanosnowman structures with crevice nanogaps as shown in Fig. 6b. 40 nm AuNPs as seeds were modified with thiolated-DNA. 144 Ag precursors and other reagents were then added for the asymmetric growth of AgNPs on the DNA-AuNP surfaces. Such nanosnowman particles could generate strong SERS signals in that the narrow conductive junctions allowed for plasmonically enhanced electromagnetic fields and Raman-active molecules could be locally positioned by DNA in the hotspots. Notably, the synthetic yield of the nanosnowman particles was as high as 95%, greatly suitable for SERS-based chemical and biological sensing.
Figure 6.
Surface-enhanced Raman scattering from DNA-assembled plasmonic nanostructures. a. Ag-shell growth-based AuNP heterodimers using DNA modification. A single Raman-active Cy3 dye molecule is located between DNA-tethered AuNPs. TEM image of a representative structure and SERS spectrum taken from the sample showing characteristic Raman peaks for the Cy3 dye. 28 Images reproduced with permission from ref 28. Copyright 2009 Macmillan Publishing Ltd. b. Schematic, calculated electromagnetic field distributions, and TEM images of the Au-Ag nanosnowman structures with various neck junctions. 144 Images reproduced from ref 144. Copyright 2014 American Chemical Society. c. Synthetic scheme for the Au nanobridged particles using DNA-modified AuNPs as templates. Raman dyes are enclosed in the nanogaps. 29 Images reproduced with permission from ref 29. Copyright 2011 Macmillan Publishing Ltd. d. Schematic of the AuNP dimer assembled on a DNA origami platform with a thin layer of Rhodamine 6G adsorbed onto the dimer structure. TEM image of a representative structure. 31 Images reproduced with permission from ref 31. Copyright 2014 Macmillan Publishing Ltd. e. Schematic illustration and TEM image of a DNA origami assembled AuNP hybrid structure used for SERS measurements. A plasmonic hot spot is formed in the gap between the AuNPs for enhancing Raman signals. 33 Images reproduced from ref 33. Copyright 2014 American Chemical Society. f. Rayleigh and Raman scattering spectra of an individual AuNP dimer modified with a single Cy3.5 molecule placed at the hot spot before and after a first and a second round of laser excitation. 107 Images reproduced from ref 107. Copyright 2016 American Chemical Society.
It is worth mentioning that Lim et al. also synthesized a new class of plasmonic nanostructures termed Au nanobridged nanogap particles (Au-NNPs) as shown in Fig. 6c. 29 The Au-NNP had a hollow, 1 nm interior gap between a Au core and a Au shell. The process involved modification of AuNPs with DNA possessing Raman dyes and thiols. The thickness of a single strand of DNA is ~ 1nm, matching the interior gap size. The Au nanobridges that connected the core and the shell were mainly governed by the sequence of the thiolated DNA bases and the DNA grafting density.145 The precise and quantitative positioning of Raman dyes inside the narrow gap of the Au-NNP led to strong, highly reproducible SERS signals. Later, Kang et al. successfully applied such Au-NNPs, responding to near-infrared excitation (785 nm), in combination with high-speed confocal Raman microscopy for live cell Raman imaging. 146 These particles allowed for monitoring spatial particle distributions within their targeted sites such as cytoplasm, mitochondria, or nucleus without inducing significant damages on cells as well as for detecting rapidly changing cell morphologies induced by addition of highly toxic potassium cyanide to cells.
As DNA origami represents a reliable fabrication platform to create probes of strongly coupled plasmonic nanostructures with well-defined geometries, several research groups have already implemented DNA origami-templated AuNP assemblies for SERS. 30 32 As an extra benefit, the rigidity of the origami surface prevents samples from degradation and ensures stability during SERS measurements. The first Raman signals originating from a DNA origami platform were measured and reported by Prinz et al.30 In this work, TAMRA-modified DNA strands have been placed between 5 nm gold nanoparticles that were overgrown with additional Au by electroless deposition. Also, Pilo-Pais et al. used Au overgrowth on particles assembled in a tetramer on a DNA origami sheet.32 Subsequently, the structures were incubated with the Raman dye (4-aminobenzenethiol), which covalently attached to the Au surfaces and gave rise to clear signals. In particular, Thacker et al. demonstrated the assembly of 40 nm AuNP dimers with sub-5 nm gaps on a DNA origami platform as shown in Fig. 6d. 31 This design allowed for a strong coupling between the two AuNPs. Individual dimers were immobilized on a Au-coated silicon wafer and then briefly incubated in a Rhodamine 6G solution to form a monolayer on the dimer structures. The observed enhancement was highly sensitive to the polarization of the laser with respect to the dimer axis. Five molecules were estimated to contribute to the observed SERS signal. The calculated surface enhancement factors were between five and seven orders of magnitude. Almost at the same time, Kühler et al. also demonstrated AuNP dimers assembled by DNA origami for SERS applications (see Fig. 6e). 33 Their AuNP (40 nm) dimers were linked together by a three-layered DNA origami block at a separation distance of 6 nm to achieve plasmonic coupling and the formation of a plasmonic hot spot. Different from the work of Thacker et al., the authors located Raman-active molecules of interest by selectively incorporating SYBR Gold only in the DNA structure and thus in the hot spot region between the AuNPs. SYBR Gold is a minor groove-binding fluorescent nucleic acid stain that has a high affinity to double-stranded DNA (dsDNA). The measured SERS signal was estimated to originate from ~ 25 dye molecules localized in the hot spot region.
In order to push the sensing limit towards single molecule resolution, further reduction of the interparticle distance between two AuNPs to achieve higher field enhancements was carried out by Simoncelli et al. as shown in Fig. 6f.107 The authors used optothermal-induced shrinking of a DNA origami template to control the gap sizes between two 40 nm AuNPs in a range from 1 to 2 nm. Before laser irradiation, the Raman spectrum of a single Cy3.5 molecule placed in the center of the hotspot did not show any visible peaks. Upon irradiation with a continuous-wave 612 nm laser (60 kW/cm2) for 10 s, a red shift of the scattering peak was observed corresponding to reduction of the interparticle gap. Signature peaks for the single Cy3.5 dye were observable in the Raman spectrum. The gap sizes were inferred using Mie theory calculations according to the measured scattering spectra before and after laser irradiation. The numerical simulations showed that the gap size was reduced from ˜ 3.3 to 1.9 nm. The intensities of the Raman peaks were more pronounced after repeating the heating step for a second time. The scattering peak of the Au dimer was further red-shifted by about 30 nm. The gap was estimated to be further reduced from ~1.9 to 1.3 nm. This work, together with the work by Prinz et al., who used Ag overgrowth on DNA origami-assembled AuNPs to obtain SERS signals from single molecules, 34 outlines the path towards achieving reliable plasmonic platforms for single-molecule SERS detection.
3.6. Surface-Enhanced Fluorescence Spectroscopy
Quantifying the interplay between single emitters and plasmonic nanostructures provides important insight into the underlying physics of light-matter interaction on the nanoscale. Among different interaction mechanisms, surface-enhanced fluorescence holds tremendous potential for detection of single molecules, biosensing applications, and nanoscale light control. Plasmonic nanostructures, which on the one hand may efficiently alter the local electric fields in hot spots, can on the other hand directly affect the excitation rate of a fluorophore as well as influence its emission properties such as quantum yield and angular emission pattern. 37,147–150 All these enhancement effects are highly dependent on the plasmonic nanostructure’s size, geometry, material, and relative position to the fluorphore. In the past, significant efforts have been made to place single molecules into hot spots of plasmonic nanostructures. Unfortunately, most of the experimental schemes failed to provide nanoscale positioning accuracy. In contrast, DNA assembly intrinsically offers nanoscale docking sites that are fully addressable, opening a unique pathway to study light-matter interaction with unprecedented control.
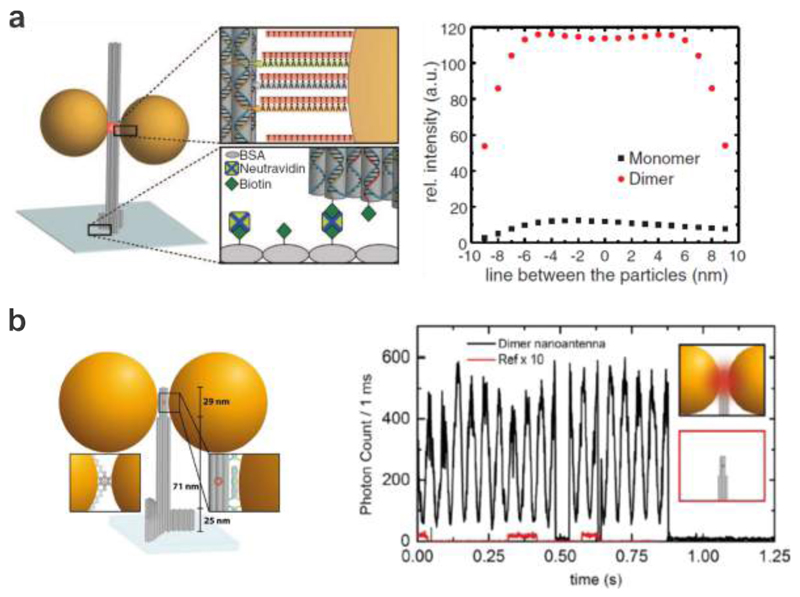
Pioneering work toward this direction was carried out by Acuna et al., who used a tower-shaped DNA origami structure as shown in Fig. 7a. 37 This origami platform allowed for precise assembly of a AuNP (100 nm) dimer with a defined interparticle distance and simultaneously provided a docking site for locating fluorescent dyes in the plasmonic hot spot. The 12-helix bundle tower had a height of 220 nm and a diameter of 15 nm. Three additional 6-helix bundles widened the base of the origami tower to ensure its stable immobilization on a cover slip through biotin-streptavidin conjugation. 82 Two DNA-functionalized AuNPs were hybridized to three capture strands each extending from the sides of the origami tower at pre-designed sites. A dye-labeled strand (ATTO647N) was incorporated in the origami at the center of the plasmonic hot spot during assembly such that the dye was aligned with respect to the electric field polarization. The authors observed a maximum of 117-fold fluorescence enhancement for the dye molecule positioned in the 23 nm gap between two 100 nm AuNPs. Later, the same group also reported optimized origami platforms (see Fig. 7b) by improving the structural robustness, reducing the interparticle distance, etc. Fluorescence enhancement of more than 5000-fold was achieved. 40 Evidently, such DNA origami platforms outperformed top-down lithographic ones. Also, the authors avoided the problem of bleaching the dye molecules by using short, dye-modified oligonucleotides that only transiently bound to an anchor sequence protruding from the position of interest on the origami. 151
Figure 7.
Surface-enhanced fluorescence from DNA-assembled plasmonic nanostructures. a. Left: schematic of a DNA origami pillar with two AuNPs forming a dimer. The dye is located between the NPs within the central bundle of the pillar. The DNA origami pillar is bound via biotins to a neutravidin-functionalized cover slip. Right: Numerical simulations of the fluorescence enhancement along the gap for a dye oriented in the radial direction. 36 Images reproduced with permission from ref 36. Copyright 2012 Association for the Advancement of Science (AAAS). b. Single-molecule fluorescence transients for a AuNP dimer (black line) and for a DNA origami structure without NPs (red line). 40 Images reproduced from ref 40. Copyright 2015 American Chemical Society.
In most of the aforementioned applications, the high-level control over the particle spacing and the resulting gap sizes is the essential feature of DNA assembly. Nevertheless, it is important to point out that heating in plasmonic hot spots, evaporation of solvents after the adsorption of the plasmonic devices on surfaces and other surface effects can lead to the deformation and even disassembly of the double helices that originally hold the particles in place. 107,152 On one hand, such effects are not only successfully applied for light-induced heating for controlled DNA release 153–155 but also for reversible DNA binding and unbinding as it is used in the polymerase chain reaction.156,157 On the other hand, these effects can lead to uncertainties of the particle positions and thus potentially influence the designed plasmonic effects and thus require careful considerations and further research.
4. DNA-Assembled Dynamic Plasmonic Nanostructures
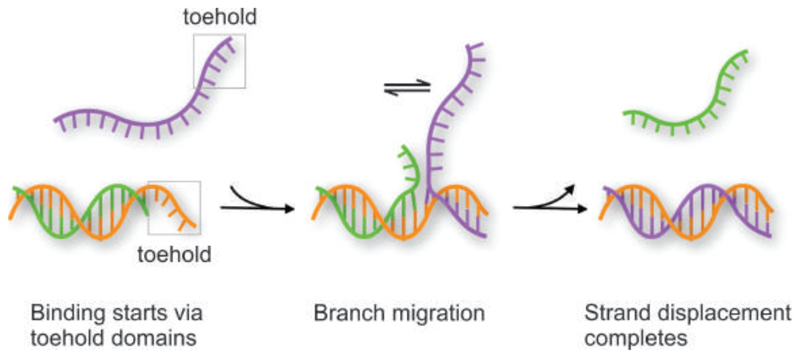
DNA-assembled dynamic plasmonic nanostructures refer to DNA-assembled plasmonic nanostructures, which not only yield well-defined optical properties but also possess desired structural reconfigurability. As a result of this reconfigurability and the chemical addressability of DNA, the optical properties of such plasmonic systems can be actively altered. Dynamic plasmonic devices hold great promise for applications in adaptable nanophotonic circuitry, artificial nanomachinery, as well as optical sensing of molecular binding and interaction activities. DNA structures generally allow for various ways to control their dynamic behavior. Basic schemes for structural reconfigurations of DNA structures can utilize DNA hybridization/dehybridization as well as pH and ion concentration stimuli. Probably, the most versatile and thus widespread approach is the so-called “toehold-mediated strand displacement” scheme as shown in Figure 8. In this process, a freshly added DNA sequence binds to a short extension, i.e., the toehold, on the DNA structure and replaces another sequence through branch migration. 12 More intriguing schemes include enzymatic cutting of pivotal connector strands or structural adaptions of aptamers to the presence of target molecules. 158 159 Also, photoresponsive molecules can be employed through incorporation with DNA to alter the strand’s hybridization kinetics in response to light stimuli. 160 161
Figure 8.
Scheme of the toehold-mediated strand displacement process. Note that after binding of the purple strand to the toehold region, the branching point can migrate randomly in both directions. This migration starts all over again when only the toehold region is bound but terminates once the branch has reached the left side and the purple strand has fully replaced the green strand.
4.1. Plasmon Rulers
Plasmon rulers are used to monitor distance changes with nanometer precision. The underlying mechanism can be explained by the plasmon hybridization theory: when two metal nanoparticles are placed into proximity, their plasmons couple to each other. 93 94 Depending on the interparticle separation, the resonance wavelength of the coupled system shifts sensitively. Importantly, plasmon rulers represent significant advantages over molecular rulers based on Förster Resonance Energy Transfer (FRET). 162 163 164 FRET often suffers from low and fluctuating signal intensities as well as photobleaching. In contrast, plasmon rulers neither blink nor bleach, offering unlimited observation time. In addition, FRET can only sense distance changes within 10 nm and the signal almost drops in a step-wise fashion, which often hampers quantitative interpretations. The range of accessible distances for plasmon rulers on the other hand can be up to several tens of nanometers, while the shifts of the plasmon resonances can occur more or less continuously. 3
The first plasmon rulers were created by Sönnichsen et al. using DNA-assembled 40 nm AuNP or AgNP dimers as shown in Fig. 9a. 22 The authors utilized surface-immobilized particles as anchors for ssDNA-functionalized particles. The ssDNA molecules were bound via thiol groups to the free AuNPs and had biotin at their other ends, allowing them to bind to the streptavidin-coated anchor particles. Upon binding, the coupled scattering centers suddenly changed color under dark-field illumination. The authors further demonstrated that DNA-linked AuNP dimers could be used to report interparticle distance changes in real time. They detected the hybridization of a complementary DNA oligonucleotide to the flexible ssDNA linker within a AuNP dimer through a significant blue-shift of the resonance position, which resulted from the stiff dsDNA pushing apart the AuNPs. As a result, the dynamics of DNA hybridization could be recorded in situ by monitoring the spectrum of a single AuNP dimer continuously.
Figure 9.
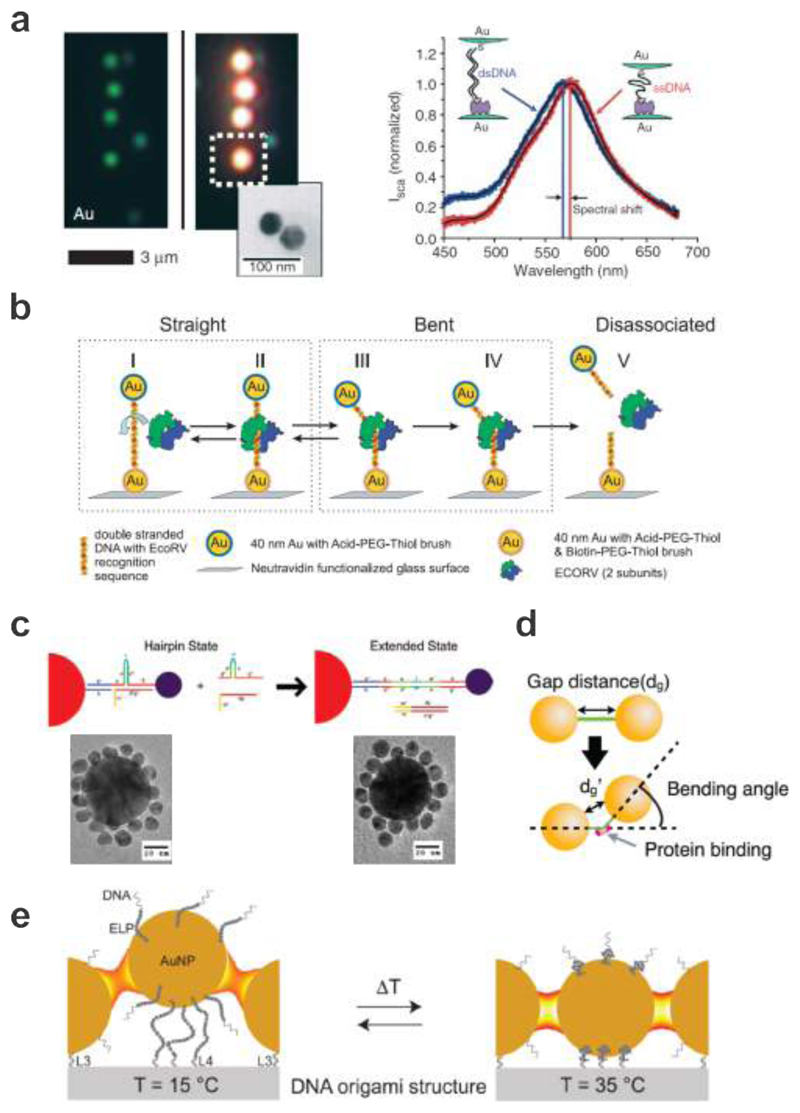
Plasmon rulers. a. Left: single AuNPs and DNA-assembled AuNP dimers appear blue and blue-green under dark field microscopy, respectively. Right: spectral shift of a AuNP dimer upon DNA hybridization. 22 Images reproduced with permission from ref 22. Copyright 2005 Macmillan Publishing Ltd. b. A plasmon ruler is immobilized with one particle to a glass surface through biotin-Neutravidin chemistry. The EcoRV enzyme binds to the DNA strand between the two AuNPs. It bends the DNA strand at the target site and then cuts it creating a blunt ended by phosphoryl transfer. 165 Images reproduced with permission from ref 165. Copyright 2007 National Academy of Sciences. c. Satellite particles are initially linked to a Au core particle with DNA strands incorporated with hairpin bridging strands. Reconfiguration occurs upon addition of strands complementary to the hairpin strands. Corresponding TEM images are shown for the two configurations. 168 Images reproduced from ref 168. Copyright 2008 American Chemical Society. d. AuNP dimer before and after DNA bending induced by SOX2. 169 Images reproduced from ref 169. Copyright 2013 American Chemical Society. e. Thermoresponsive switching of a plasmonic waveguide. With a temperature increase above the transition temperature, a thermo-sensitive polymer (ELP) can be transformed into the collapsed state leading to shortening of the particle spacing. 170 Images reproduced from ref 170. Copyright 2016 American Chemical Society.
After this pioneering work, a variety of plasmon rulers designed for different applications have been created. For example, Reinhard et al. applied a plasmon ruler to measure the dynamical biophysical processes during the cleavage of DNA by the restriction enzyme EcoRV on the single molecule level (see Fig. 9b).165 A plasmon ruler composed of two linked 40 nm AuNPs was immobilized on the surface of a glass flow chamber via a biotin-Neutravidin bond between the biotin-functionalized particle and surface. The color and intensity of the plasmon ruler were monitored in real time using dark field microscopy. Upon addition of the enzyme, some plasmon rulers exhibited sudden intensity drops due to enzymatic DNA cleavage. Intriguingly, the authors could also demonstrate that the observed intensity increase before the cut was due to DNA bending by the EcoRV restriction enzyme. In contrast to irreversible cleaving events, Lee et al. monitored the reversible binding and unbinding of single molecules to a DNA aptamer sequence – i.e. a sequence evolved to bind to a specific target molecule – bridging the gap between two AuNPs.166 These and many more remarkable results were only possible to obtain, given the unlimited lifetime, high temporal resolution, and high signal-to-noise ratio of the plasmon rulers, which substantiate a unique tool for studying conformation changes of molecular events and subsequent dynamic activities at the single molecule level and in complex environments.167
Plasmon rulers with more complex geometries have also been attempted. For example, Sebba et al. demonstrated reconfigurable core-satellite assemblies as molecularly-driven plasmonic switches (see Fig. 9c). 168 A 50 nm Au core particle and 13 nm Au satellite particles were linked together using DNA, which contained a hairpin-shaped secondary structure for modulating the duplex length. Reconfiguration of the core-satellite assemblies from the hairpin state to the extended state through strand displacement reactions led to a significant decrease of the scattered intensity and a restoration of the plasmon resonance position to that of isolated core particles. Another interesting work was demonstrated by Morimura et al., who utilized AuNP dimers to report DNA conformation changes induced by transcription factor binding (see Fig. 9d). 169 The AuNP dimers bound by SOX2 shifted the plasmon resonance position to a longer wavelength, indicating bending of the DNA duplex induced by SOX2 binding. When the SOX2 formed a ternary complex with PAX6 on DC5, a further resonance red-shift was observed, implying additional bending in the DC5 sequence.
Other than utilizing resonance positions to correlate nanoscale distance changes, AuNPs assembled with DNA in a chain geometry can particularly work as an energy transfer platform for guiding electromagnetic energy below the optical diffraction limit.4 Recently, Vogele et al. have developed a switchable plasmonic waveguide composed of AuNPs assembled on a DNA origami structure that allowed for simple spectroscopic excitation and readout (see Fig. 9e). 170 Multiple AuNPs were positioned along a line on a DNA origami structure. The central AuNP was functionalized with thermoresponsive elastin-like peptides (ELPs). ELPs are synthetic peptides that exhibit a fully reversible hydrophobic collapse upon a certain transition temperature. One outer AuNP was functionalized with fluorescein (FAM) and it worked as the excitation port. Near-field coupling among the adjacent AuNPs enabled the electromagnetic energy transfer along the particle chain. Evaluation of the waveguide performance was achieved by detecting the emission of an acceptor dye (Atto 532), which was located on the surface of the other outer AuNP. Below the transition temperature, the coupling between the AuNPs was weak as the highly solvated and thus swollen ELPs pushed the central AuNP out of line. With a temperature increase above the transition temperature, the ELPs were transformed into the collapsed state leading to shortening of the particle spacing. The resulting stronger plasmonic coupling among the AuNPs facilitated the energy transfer, reflected by a slight increase of the fluorescence signal. Owing to the reversibility of the ELP swelling and collapsing, the fluorescence signal of the waveguide could be repeatedly switched between two states by temperature cycling.
4.2. Dynamic Manipulation of Plasmonic Chirality
Controlling molecular chirality is of great importance in stereochemistry. Chirality of natural molecules can be manipulated by reconfiguring molecular structures through light, electric field, and thermal stimuli. However, such chirality regulation lacks efficiency as the CD response of natural molecules is very weak. In contrast, plasmonic chiral nanostructures assembled by DNA allow for dynamic manipulation of chirality and reversible switching of strong CD responses.
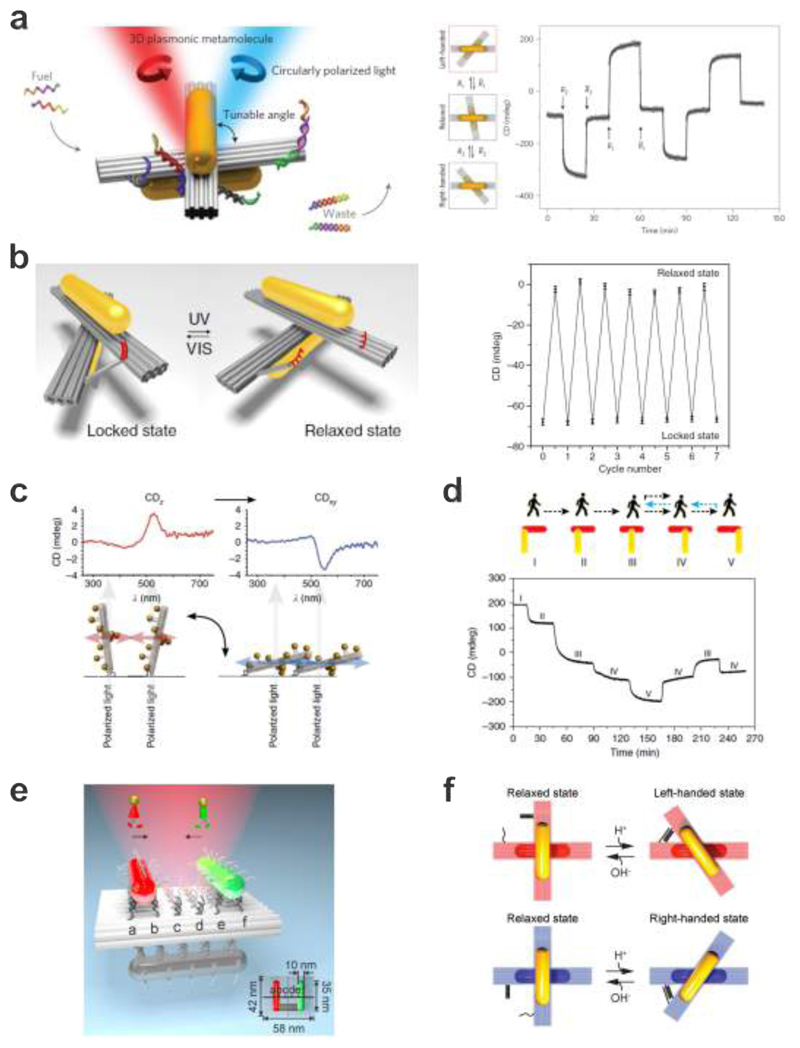
Kuzyk et al. reported the first reconfigurable 3D plasmonic nanostructures, which executed DNA-regulated conformational changes on the nanoscale (see Fig. 10a). 102 Two AuNRs were hosted on a reconfigurable DNA origami template, which consisted of two linked 14-helix bundles (80 nm × 16 nm × 8 nm). Twelve anchor strands were extended from each origami bundle for robust positioning of two AuNRs (40 nm × 10 nm, one on each bundle) functionalized with complementary DNA. Two DNA locks that were utilized to manipulate the configuration of the plasmonic nanostructure were extended from the sides of the DNA origami bundles. Each lock contained two DNA arms, which could be opened or closed through toehold-mediated strand displacement reactions upon addition of corresponding fuel strands. As a result, the configuration of the plasmonic nanostructure could be switched between LH, RH, and relaxed states. Importantly, the 3D plasmonic nanostructures worked as optical reporters, which transduced their configuration changes in situ into CD changes at visible frequencies.
Figure 10.
DNA-assembled plasmonic nanostructures for dynamic manipulation of strong chirality. a. Reconfigurable 3D plasmonic nanostructures consist of AuNRs hosted on switchable DNA origami templates. The relative angle between the AuNRs within the structure can be controlled using DNA locks, therefore giving rise to dynamic plasmonic chiral responses. 102 Images reproduced with permission from ref 102. Copyright 2014 Macmillan Publishing Ltd. b. Light-driven 3D plasmonic nanosystem reversibly regulated by UV and visible light for switching between a right-handed and a relaxed state. The dynamic function of the origami structure is enabled by introducing the azobenzene-modified DNA segment on the origami template. 172 Images reproduced with permission from ref 172. Copyright 2016 Macmillan Publishing Ltd. c. Chiral response switched by changing the helix orientation with respect to the light beam. 101 Images reproduced with permission from ref 101. Copyright 2013 Macmillan Publishing Ltd. d. A plasmonic walker that can perform stepwise walking on origami. The walking of the AuNR is enabled through toehold-mediated strand displacement. 173 Images reproduced with permission from ref 173. Copyright 2015 Macmillan Publishing Ltd. e. A plasmonic walker couple system, in which the two AuNR walkers can independently or simultaneously perform stepwise walking along the same DNA origami track. 174 Images reproduced from ref 174. Copyright 2015 American Chemical Society. f. pH regulation of DNA origami-based plasmonic chiral nanostructures. The plasmonic system can be switched between the relaxed and LH/RH state by opening/closing the pH-triggered DNA locks. 175 Images reproduced from ref 175. Copyright 2017 AAAS.
Introducing photon-switchable azobenzene-modified DNA segments into plasmonic particle systems allows for the reversible association and disassociation of DNA strands and consequently of nanoparticles.171 Kuzyk et al. also demonstrated a light-driven, reconfigurable 3D plasmonic DNA origami nanostructure that could translate molecular motion of azobenzene into reversible chiroptical function (see Fig. 10b). 172 The photoresponsive segment comprised two DNA branches, which were extended from the two origami bundles, respectively. One branch possessed a 20-base-pair-long dsDNA segment that was linked by a disulfide bond to a segment comprising azobenzene-modified nucleotides (Azo-ODN 1). The other branch contained the pseudocomplementary strand Azo-ODN 2. Upon UV light illumination, the azobenzene molecules in both Azo-ODNs were converted to the cis-form, resulting in dehybridization of the Azo-ODN duplex and opening of the origami template. In contrast, upon illumination with visible light, the azobenzene molecules were converted to the trans-form and the Azo-ODNs could hybridize again thus closing the origami template. The different conformation states could cyclically be induced and be read-out by probing light in real time. This system thus amplified the sub-nanometer conformation changes of azobenzene through large-scale changes of the host nanostructure and consequently translated the light-triggered molecular motion of azobenzene into strong and reversible plasmonic CD responses.
Dynamic CD switching has also been achieved by manipulating the orientations of plasmonic chiral nanostructures relative to the incident light direction. As shown in Fig. 10c, Schreiber et al. mounted DNA origami-assembled LH AuNP helices on a glass substrate through biotin-neutravidin binding. 101 CD measurements were then performed by placing the substrate perpendicular to the light beam inside a cuvette that was initially filled with buffer. The AuNP helices stood upright on the substrate and hence were aligned parallel to the probing light beam. Instead of a peak-dip CD of the LH helices isotropically dispersed in solution, an inverted dip-peak spectrum with a dominant peak was observed, which corresponded to the axial component of the averaged CD signal (CDz). In the next step, the sample was dried with nitrogen and placed again into the optical path. This resulted in the alignment of the AuNP helices perpendicular to the light beam and the recorded CD spectrum showed a peak-dip signal with a dominant dip, i.e., CDxy. Reversible switching between two alignment states and consequently switching between the signals of CDxy and CDz could be achieved by repeated flushing and drying processes without changing the orientation of the substrate.
The high sensitivity of CD on conformation changes also enables optically tracking of successive movements of plasmonic nanoparticles well below the diffraction limit. Zhou et al. demonstrated a dynamic plasmonic system, in which one AuNR could perform stepwise walking directionally and progressively on DNA origami (see Fig. 10d).173 A walker AuNR and a stator AuNR (an immobilized AuNR) were placed on two opposite surfaces of a rectangular DNA origami platform, forming a chiral geometry. While the stator was immobilized on one surface, the walker rod on the other surface could execute stepwise, directed movements. Along the track that was laid out on the origami surface, six parallel rows of footholds were utilized to establish five walking stations, evenly separated by 7 nm. The stepwise walking was powered by DNA hybridization and triggered by the addition of the respective blocking and removal strands. The dynamic walking process could be monitored in situ using a time-scan function of the CD spectrometer at a fixed wavelength. In the experiments, the plasmonic walker not only functioned as a walking element to carry out mechanical motion but also as an optical reporter, which could deliver its own translocation information through optical spectroscopy in real time. Later on, the same group demonstrated the implementation of two plasmonic walkers that could walk both individually and collectively on the same origami track (see Fig. 10e). 174 A sensitive plasmonic coupling scheme was introduced for in situ optically monitoring the dynamic walking of the two walkers with steps well below the diffraction limit. The two walkers and a stator were assembled on the two opposite surfaces of an origami template. The two walkers optically interact with the stator simultaneously. In particular, each walker and the stator constituted a chiral geometry. The chiroptical response of the entire system was jointly determined by the positions of both walkers relative to the stator. This rendered optical discrimination of the walking directions and the individual steps of the two walkers possible. In addition, the walker number and the optical response of the system could be correlated.
Very recently, selective control of different plasmonic nanostructure species coexisting within one ensemble has been demonstrated by Kuzyk et al. as shown in Fig. 10f. 175 Reconfigurable chiral plasmonic nanostructures were assembled using DNA origami in LH and RH states, respectively. In these structures, pH-sensitive DNA locks worked as active sites to trigger structural regulation over a wide pH range. Such locks exploited triplex DNA secondary structures that display pH-dependent behavior due to the presence of specific protonation sites. The selective reconfiguration was enabled by modulating the relative contents of TAT/CGC triplets in the DNA locks. To demonstrate the unprecedented enantioselectivity, the authors mixed LH and RH structures in equimolar amounts to form a quasi-racemic solution. Such plasmonic quasi-enantiomers, that is, enantiomers as plasmonic objects, but functionalized with DNA locks containing different TAT contents could be selectively activated upon pH tuning, elucidating an innovative approach to arbitrarily modulate chiroptical response at wish.
4.3. Dynamic Plasmonic Nanostructures for Sensing Applications
Due to the high sensitivity of plasmonic hot spots, the engineerable dynamic properties, and the unique biological specificity, DNA-assembled dynamic plasmonic architectures open the route for a new generation of optical sensors, which are vital for fundamental cellular studies and clinical diagnostics. Due to their ease of synthesis and sensitive read-out mechanisms, DNA-assembled plasmonic architectures are particularly attractive for analyte binding and molecular sensing.
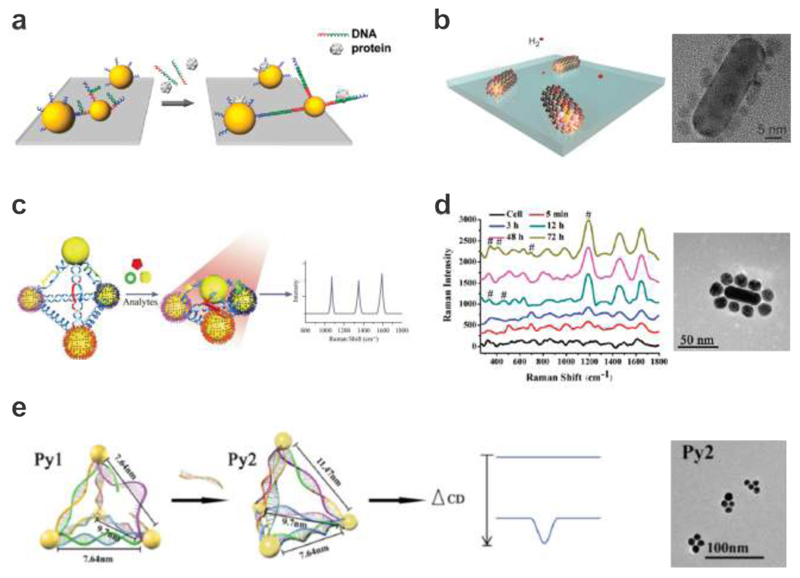
Chen et al. demonstrated that target-induced actuation of DNA-assembled AuNP dimers could be an effective mechanism for DNA molecule sensing based on the plasmon-hybridization scheme (see Fig. 11a).167 DNA-assembled plasmonic structures have also been used to sense gas molecules such as hydrogen. Li et al. reported DNA-assembled bimetallic plasmonic nanostructures composed of AuNRs and palladium nanoparticles (see Fig. 11b). 176 The DNA strands served both as linkers and seeding sites for the growth of the palladium nanoparticles and facilitated reliable positioning of palladium satellites around a AuNR with an ultrashort spacing of several nm. Upon hydrogen absorption, palladium underwent a phase transition to form palladium hydride, which then altered the neighboring environment of the centered AuNR. Using dark-field spectroscopy, these plasmonic superstructures were able to efficiently detect dynamic absorption and desorption of hydrogen on the single structure level.
Figure 11.
Dynamic plasmonic nanostructures for sensing applications. a. DNA-assembled AuNP dimer structures for identifying targets from nonspecific binding and for detecting targets in complex media. 167 Images reproduced from ref 167. Copyright 2010 American Chemical Society. b. DNA-assembled bimetallic hydrogen sensors, which consist of palladium nanoparticles linked to AuNRs using DNA strands. TEM image of a representative structure. 176 Images reproduced with permission from ref 176. Copyright 2014 Macmillan Publishing Ltd. c. SERS-encoded Ag-pyramids for detection of multiple biomarkers (PSA. thrombin, and mucin-1). 177 Images reproduced with permission from ref 177. Copyright 2015 Wiley. d. DNA-assembled plasmonic superstructures composed of AuNRs and spherical AuNPs for SERS-based in situ monitoring of intracellular metabolism. TEM image of a representative structure. 178 Images reproduced from ref 178. Copyright 2011 American Chemical Society. e. Chiral plasmonic sensors for DNA detection through CD response changes. TEM image of the plasmonic structures. 179 Images reproduced with permission from ref 179. Copyright 2014 Wiley.
More intriguingly, Xu et al. reported DNA-assembled Ag pyramids for simultaneous and ultrasensitive detection of multiple disease biomarkers such as prostate specific antigen, thrombin, and mucin-1 as shown in Fig. 11c. 177 Here a DNA frame holding aptamers specific to disease biomarkers was used to drive the self-assembly of AgNP pyramids dressed with Raman-active molecules. Upon binding of the designated disease markers to the aptamers, the 3D configuration of the pyramids could be altered. This process resulted in a shorter interparticle separation, which in turn caused Raman signal enhancement. Similarly, Kotov et al. produced DNA-assembled plasmonic superstructures composed of AuNRs and spherical AuNPs, which contained a varying number of plasmonic gaps (see Fig. 11d). 178 The authors applied these superstructures in live cells for SERS-based in situ monitoring of intracellular metabolism. Incubation of these superstructures with Hela cells indicated sufficient field enhancement to detect structural lipids of mitochondria and potentially small metabolites.
Chiral plasmonic nanostructures are a newly developed member in the family of optical sensors. One of the unique characters afforded by chiral plasmonic nanostructures is their high sensitivity on minute configuration changes. Also, their 3D nature renders possible sensing of analyte binding and other biochemical-related activities with extra degrees of freedom. For example, Yan et al. demonstrated a pyramidal sensor platform with reversible chiroptical signals for DNA detection (see Fig. 11e). 179 In the presence of target DNA, two types of nanoparticle pyramids underwent dynamic reconfiguration or a dissociation process, which functioned as an off-on or on-off switch towards CD signal changes. Target DNA could be detected at the attomolar level. Later, the same group also reported the realization of Au-upconversion nanoparticle pyramids to detect intracellular microRNA in real time. 180
5. Conclusion, Outlook, and Perspective
The benefits of self-assembled plasmonic systems are manifold and evident: literally nanometer-precise positioning of optical elements is paired with massive parallel assembly, which allows for the production of large quantities of custom-made meta-molecules. This enables plasmonic materials to be structured in all three dimensions on the nanoscale and they can thus perform in the visible frequency domain in the realm of possibility. Components of different types, e.g. AuNPs, AgNPs, quantum dots, organic dye molecules, analytes, etc., are readily combinable and can be assembled with high yields. Especially, considering the nanoscale positioning accuracy of DNA assembly, once short separations between the assembled entities are reached, classical electromagnetic interactions will fail and interesting quantum phenomena as well as nonlocal effects start to take place. This aspect certainly deserves more theoretical and experimental efforts for better understanding of the profound underlying physics. Furthermore, the crystalline quality of colloidal particles usually surpasses that of top-down deposited materials, which results in superior plasmonic performance of the devices. There is also a great potential for further exploring the material aspect beyond Au, Ag, etc.. Plasmonic nanoparticles suffer from absorption losses of metals at optical frequencies. A new promising route to overcome this hurdle has emerged from the discovery of electric and magnetic Mie-type resonances in high-permittivity all-dielectric nanoparticles made of Si, SiC, TiO2, and similar compounds 181 182. Therefore, research efforts on developing feasible and reliable functionalization protocols for assembly of such dielectric particles on DNA origami will be extremely valuable, as this would lead to novel low-loss nanophotonic devices at optical frequencies.
Finally, the development of dynamic plasmonic systems is just beginning and here DNA-based assembly opens ample opportunities for switchable systems that operate autonomously and react to chemical or optical cues. One of the futuristic directions using dynamic plasmonic systems could be sensing of molecular interactions 183 184 on the single molecule or single structure level and thereby readily convert the dynamic interaction behavior into optical signal changes in real time. Importantly, plasmonic signals do not suffer from blinking or bleaching, two effects that pose a severe hurdles to long-term single-molecule fluorescence experiments. A plasmonic approach is particularly beneficial for systems, in which large molecules or binding activities from multiple molecular entities are involved, as plasmonic interactions offer a much larger sensing distance range than FRET.
So far, DNA-assembled plasmonic devices have been either operating as individuals in solution or randomly distributed when attached to a surface. Outstanding exceptions are the works of Wallraff, Cha, Gopinath, Rothemund and others, who combined the benefits of DNA self-assembly with top-down methodologies by lithographically rendering surfaces into DNA binding patches surrounded by DNA repelling areas. 185 186 187 With this approach Gopinath et al. placed and oriented DNA origami triangles in photonic crystal cavities and beautifully illustrated the achievable control of light-matter interactions by re-painting van Gogh’s “The Starry Night” on their nanoscale canvas. 188 In the near future, we expect to see more of such combinations of nanotechnologies but also the combination of hierarchical and large-scale DNA assemblies with plasmonic metamolecules, which could this way be ordered into 2D arrays. In particular, a rapidly rising direction in nanoplasmonics is metasurfaces, which consist of 2D plasmonic nanostructure arrays with subwavelength periodicity.189 Due to its negligible thickness compared to the operating wavelength, a metasurface can function as an interface of discontinuity with abrupt changes in both the amplitude and phase of the incident light, enabling a variety of ultrathin functional devices. One interesting yet challenging avenue is to position DNA-assembled dynamic plasmonic structures in a well-defined 2D array using top-down technique for creating dynamic metasurfaces working in solution. As a result, the amplitude and phase of light could be dynamically controlled through manipulating the orientations of individual plasmonic nanostructures. This would lead to novel optical elements, for example dynamic lenses, vortex beam generators, and holography devices.
In this and other contexts, it is an important objective in the field of DNA nanotechnology to increase the mechanical stability of DNA structures and to enlarge the available surface area and mass of origami so that an increasing number of entities or larger particles can be hosted on for achieving more complex or more efficient devices. For static assemblies that are adsorbed on solids surfaces and are subsequently dried, usually no further treatment to increase their stability is necessary, as the configurations of the DNA templates and their guest entities are maintained on the substrates.100,190 Dynamic DNA structures, in contrast, operate in solutions and are susceptible to disassembly at temperatures above ~ 45°C or in the absence of salts. There have been successful attempts to render DNA structures stable under challenging conditions, which here means low salt concentrations, presence of enzymes or elevated temperatures.191–193 However, methods for site-directed mineralization of DNA structures or other strategies to stably maintain 3D shapes in air or vacuum have yet to be developed. Scaling up DNA origami, on the other hand, has already been achieved by employing biotechnological methods for enzyme-based production of ssDNA scaffolds and staples 194–196 and by using dsDNA sources as scaffold strands 197 198 199 Another approach, where several regular-sized origami components are linked together using again a long scaffolding strand is called ‘superorigami’ and successfully employs different levels of folding hierarchies.200 These and other strategies, including fractal growth and the assembly of giga-dalton scale superstructures,201–203 are currently developed further with the goal to build ever-larger DNA templates.
A more futuristic direction is to answer whether it is possible to build photonic molecular circuits or even nanofactories using DNA nanotechnology, taking a direct inspiration from natural organisms including living cells and photosynthesis systems.204 In such artificial circuits or nanofactories, plasmonic nanostructures serving as efficient energy funnels or transfer channels would be assembled together with other functional entities, for instance, light-harvesting complexes, enzymes, proteins, quantum dots, carbon nanotubes, etc.. Exploiting the assembly power of DNA origami, these constituents could thus work in a coordinated and cooperative fashion. In this scenario, dynamic plasmonic nanostructures would be ideally suited to regulate the relative distances and orientations between the plasmonic entities and other functional groups, which is critically important for optimized energy channeling efficiencies, reaction kinetics and resulting productivities.
The original motivation of Nadrian Seeman to assemble molecules in 3D lattices 9 has driven the field of DNA self-assembly for decades and recent achievements include the formation of rationally designed DNA crystals, large-area lattices built from DNA origami or ssDNA tiles. 205 206 207 208 Linking plasmonic architectures to such DNA lattices will help to establish order in the inherently non-ordered world of self-assembly and open the doors to studies on dynamic metasurfaces and metamaterials operating at visible wavelengths.
Acknowledement
Na Liu acknowledges support from the Sofja Kovalevskaja grant from the Alexander von Humboldt-Foundation, the Marie Curie CIG grant, the Volkswagen grant, and the European Research Council (ERC Dynamic Nano) grant. Tim Liedl acknowledges support from the Volkswagen Foundation and the European Research Council through the ERC Starting Grant ORCA (GA N°:336440).
Biographies
Na Liu is Professor at the Kirchhoff Institute for Physics at the University of Heidelberg. She received her Ph. D in Physics at the University of Stuttgart in 2009, working on 3D complex plasmonics at optical frequencies. In 2010, she worked as postdoctoral fellow at the University of California, Berkeley. From 2011 until 2012, she has worked at Rice University as Texas Instruments visiting professor. At the end of 2012, she obtained a Sofja Kovalevskaja Award from the Alexander von Humboldt Foundation and became an independent group leader at the Max-Planck Institute for Intelligent Systems in Stuttgart. She joined the University of Heidelberg in 2015. The research of Na Liu is multi-disciplinary. She works at the interface between nanoplasmonics, biology, and chemistry. Her group focuses on developing sophisticated and smart plasmonic nanostructures for answering structural biology questions as well as catalytic chemistry questions in local environments.
Tim Liedl is Professor for experimental physics at the Ludwig-Maximilians Universität since 2009. During his undergraduate studies he worked on the development of hydrophilic coatings for fluorescent semiconductor nanoparticles. In 2007 he obtained his Ph.D. in physics at the Ludwig-Maximilians Universität studying DNA-based nanodevices and switches driven by chemical oscillations. From spring 2007 till summer 2009 he visited Dana-Farber Cancer Institute / Harvard Medical School where he used the DNA-origami method to construct self-assembling two- and three-dimensional structures. The research of Tim Liedl is multi-disciplinary and exploratory positioned at the interface between nanoscience, plasmonics, synthetic- and cell-biology. Its current focus lies on the application of DNA-based nanostructures in biology and on self-assembled plasmonic materials.
Footnotes
Notes
The authors declare no competing financial interest.
References
- (1).Rechberger W, Hohenau A, Leitner A, Krenn JR, Lamprecht B, Aussenegg FR. Optical Properties of Two Interacting Gold Nanoparticles. Opt Commun. 2003;220:137–141. [Google Scholar]
- (2).Oubre C, Nordlander P. Optical Properties of Metallodielectric Nanostructures Calculated Using the Finite Difference Time Domain Method. J Phys Chem B. 2004;108:17740–17747. [Google Scholar]
- (3).Jain PK, Huang WY, El-Sayed MA. On the Universal Scaling Behavior of the Distance Decay of Plasmon Coupling in Metal Nanoparticle Pairs: A Plasmon Ruler Equation. Nano Lett. 2007;7:2080–2088. [Google Scholar]
- (4).Maier SA, Atwater HA. Plasmonics: Localization and Guiding of Electromagnetic Energy in Metal/Dielectric Structures. J Appl Phys. 2005;98 [Google Scholar]
- (5).Anker JN, Hall WP, Lyandres O, Shah NC, Zhao J, Van Duyne RP. Biosensing with Plasmonic Nanosensors. Nat Mater. 2008;7:442–453. doi: 10.1038/nmat2162. [DOI] [PubMed] [Google Scholar]
- (6).Schuller JA, Barnard ES, Cai WS, Jun YC, White JS, Brongersma ML. Plasmonics for Extreme Light Concentration and Manipulation (Vol 9, Pg 193, 2010) Nat Mater. 2010;9 doi: 10.1038/nmat2630. [DOI] [PubMed] [Google Scholar]
- (7).Gramotnev DK, Bozhevolnyi SI. Plasmonics Beyond the Diffraction Limit. Nat Photonics. 2010;4:83–91. [Google Scholar]
- (8).Watson JD, Crick FHC. Molecular Structure of Nucleic Acids - a Structure for Deoxyribose Nucleic Acid. Nature. 1953;171:737–738. doi: 10.1038/171737a0. [DOI] [PubMed] [Google Scholar]
- (9).Seeman NC. Nucleic-Acid Junctions and Lattices. J Theor Biol. 1982;99:237–247. doi: 10.1016/0022-5193(82)90002-9. [DOI] [PubMed] [Google Scholar]
- (10).Seeman NC. Nanomaterials Based on DNA. Annu Rev Biochem. 2010;79:65–87. doi: 10.1146/annurev-biochem-060308-102244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Seeman NC. Structural DNA Nanotechnology: Growing Along with Nano Letters. Nano Lett. 2010;10:1971–1978. doi: 10.1021/nl101262u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Yurke B, Turberfield AJ, Mills AP, Simmel FC, Neumann JL. A DNA-Fuelled Molecular Machine Made of DNA. Nature. 2000;406:605–608. doi: 10.1038/35020524. [DOI] [PubMed] [Google Scholar]
- (13).Gu HZ, Chao J, Xiao SJ, Seeman NC. A Proximity-Based Programmable DNA Nanoscale Assembly Line. Nature. 2010;465:202–U286. doi: 10.1038/nature09026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Adleman LM. Molecular Computation of Solutions to Combinatorial Problems. Science. 1994;266:1021–1024. doi: 10.1126/science.7973651. [DOI] [PubMed] [Google Scholar]
- (15).Jonoska N, Seeman NC. Computing by Molecular Self-Assembly. Interface Focus. 2012;2:504–511. doi: 10.1098/rsfs.2011.0117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Zhang DY, Seelig G. Dynamic DNA Nanotechnology Using Strand-Displacement Reactions. Nat Chem. 2011;3:103–113. doi: 10.1038/nchem.957. [DOI] [PubMed] [Google Scholar]
- (17).Tan SJ, Campolongo MJ, Luo D, Cheng WL. Building Plasmonic Nanostructures with DNA. Nat Nanotechnol. 2011;6:268–276. doi: 10.1038/nnano.2011.49. [DOI] [PubMed] [Google Scholar]
- (18).Chao J, Lin YF, Liu HJ, Wang LH, Fan CH. DNA-Based Plasmonic Nanostructures. Mater Today. 2015;18:326–335. [Google Scholar]
- (19).Shen XB, Song C, Wang JY, Shi DW, Wang ZA, Liu N, Ding BQ. Rolling up Gold Nanoparticle-Dressed DNA Origami into Three-Dimensional Plasmonic Chiral Nanostructures. J Am Chem Soc. 2012;134:146–149. doi: 10.1021/ja209861x. [DOI] [PubMed] [Google Scholar]
- (20).Kuzyk A, Schreiber R, Fan ZY, Pardatscher G, Roller EM, Hogele A, Simmel FC, Govorov AO, Liedl T. DNA-Based Self-Assembly of Chiral Plasmonic Nanostructures with Tailored Optical Response. Nature. 2012;483:311–314. doi: 10.1038/nature10889. [DOI] [PubMed] [Google Scholar]
- (21).Storhoff JJ, Elghanian R, Mucic RC, Mirkin CA, Letsinger RL. One-Pot Colorimetric Differentiation of Polynucleotides with Single Base Imperfections Using Gold Nanoparticle Probes. J Am Chem Soc. 1998;120:1959–1964. [Google Scholar]
- (22).Sonnichsen C, Reinhard BM, Liphardt J, Alivisatos AP. A Molecular Ruler Based on Plasmon Coupling of Single Gold and Silver Nanoparticles. Nat Biotechnol. 2005;23:741–745. doi: 10.1038/nbt1100. [DOI] [PubMed] [Google Scholar]
- (23).Guo LH, Jackman JA, Yang HH, Chen P, Cho NJ, Kim DH. Strategies for Enhancing the Sensitivity of Plasmonic Nanosensors. Nano Today. 2015;10:213–239. [Google Scholar]
- (24).Xu W, Xie XJ, Li DW, Yang ZQ, Li TH, Liu XG. Ultrasensitive Colorimetric DNA Detection Using a Combination of Rolling Circle Amplification and Nicking Endonuclease-Assisted Nanoparticle Amplification (Neana) Small. 2012;8:1846–1850. doi: 10.1002/smll.201200263. [DOI] [PubMed] [Google Scholar]
- (25).Taton TA, Mirkin CA, Letsinger RL. Scanometric DNA Array Detection with Nanoparticle Probes. Science. 2000;289:1757–1760. doi: 10.1126/science.289.5485.1757. [DOI] [PubMed] [Google Scholar]
- (26).Storhoff JJ, Lucas AD, Garimella V, Bao YP, Muller UR. Homogeneous Detection of Unamplified Genomic DNA Sequences Based on Colorimetric Scatter of Gold Nanoparticle Probes. Nat Biotechnol. 2004;22:883–887. doi: 10.1038/nbt977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Wustholz KL, Henry AI, McMahon JM, Freeman RG, Valley N, Piotti ME, Natan MJ, Schatz GC, Van Duyne RP. Structure-Activity Relationships in Gold Nanoparticle Dimers and Trimers for Surface-Enhanced Raman Spectroscopy. J Am Chem Soc. 2010;132:10903–10910. doi: 10.1021/ja104174m. [DOI] [PubMed] [Google Scholar]
- (28).Lim DK, Jeon KS, Kim HM, Nam JM, Suh YD. Nanogap-Engineerable Raman-Active Nanodumbbells for Single-Molecule Detection. Nat Mater. 2010;9:60–67. doi: 10.1038/nmat2596. [DOI] [PubMed] [Google Scholar]
- (29).Lim DK, Jeon KS, Hwang JH, Kim H, Kwon S, Suh YD, Nam JM. Highly Uniform and Reproducible Surface-Enhanced Raman Scattering from DNA-Tailorable Nanoparticles with 1-Nm Interior Gap. Nat Nanotechnol. 2011;6:452–460. doi: 10.1038/nnano.2011.79. [DOI] [PubMed] [Google Scholar]
- (30).Prinz J, Schreiber B, Olejko L, Oertel J, Rackwitz J, Keller A, Bald I. DNA Origami Substrates for Highly Sensitive Surface-Enhanced Raman Scattering. J Phys Chem Lett. 2013;4:4140–4145. [Google Scholar]
- (31).Thacker VV, Herrmann LO, Sigle DO, Zhang T, Liedl T, Baumberg JJ, Keyser UF. DNA Origami Based Assembly of Gold Nanoparticle Dimers for Surface-Enhanced Raman Scattering. Nat Commun. 2014;5 doi: 10.1038/ncomms4448. [DOI] [PubMed] [Google Scholar]
- (32).Pilo-Pais M, Watson A, Demers S, LaBean TH, Finkelstein G. Surface-Enhanced Raman Scattering Plasmonic Enhancement Using DNA Origami-Based Complex Metallic Nanostructures. Nano Lett. 2014;14:2099–2104. doi: 10.1021/nl5003069. [DOI] [PubMed] [Google Scholar]
- (33).Kuhler P, Roller EM, Schreiber R, Liedl T, Lohmuller T, Feldmann J. Plasmonic DNA-Origami Nanoantennas for Surface-Enhanced Raman Spectroscopy. Nano Lett. 2014;14:2914–2919. doi: 10.1021/nl5009635. [DOI] [PubMed] [Google Scholar]
- (34).Prinz J, Heck C, Ellerik L, Merk V, Bald I. DNA Origami Based Au-Ag-Core-Shell Nanoparticle Dimers with Single-Molecule Sers Sensitivity. Nanoscale. 2016;8:5612–5620. doi: 10.1039/c5nr08674d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Prinz J, Matkovic A, Pesic J, Gajic R, Bald I. Hybrid Structures for Surface-Enhanced Raman Scattering: DNA Origami/Gold Nanoparticle Dimer/Graphene. Small. 2016;12:5458–5467. doi: 10.1002/smll.201601908. [DOI] [PubMed] [Google Scholar]
- (36).Acuna GP, Moller FM, Holzmeister P, Beater S, Lalkens B, Tinnefeld P. Fluorescence Enhancement at Docking Sites of DNA-Directed Self-Assembled Nanoantennas. Science. 2012;338:506–510. doi: 10.1126/science.1228638. [DOI] [PubMed] [Google Scholar]
- (37).Acuna GP, Bucher M, Stein IH, Steinhauer C, Kuzyk A, Holzmeister P, Schreiber R, Moroz A, Stefani FD, Liedl T, et al. Distance Dependence of Single-Fluorophore Quenching by Gold Nanoparticles Studied on DNA Origami. ACS Nano. 2012;6:3189–3195. doi: 10.1021/nn2050483. [DOI] [PubMed] [Google Scholar]
- (38).Holzmeister P, Pibiri E, Schmied JJ, Sen T, Acuna GP, Tinnefeld P. Quantum Yield and Excitation Rate of Single Molecules Close to Metallic Nanostructures. Nat Commun. 2014;5 doi: 10.1038/ncomms6356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Pellegrotti JV, Acuna GP, Puchkova A, Holzmeister P, Gietl A, Lalkens B, Stefani FD, Tinnefeld P. Controlled Reduction of Photobleaching in DNA Origami-Gold Nanoparticle Hybrids. Nano Lett. 2014;14:2831–2836. doi: 10.1021/nl500841n. [DOI] [PubMed] [Google Scholar]
- (40).Puchkova A, Vietz C, Pibiri E, Wunsch B, Paz MS, Acuna GP, Tinnefeld P. DNA Origami Nanoantennas with over 5000-Fold Fluorescence Enhancement and Single-Molecule Detection at 25 Mu M. Nano Lett. 2015;15:8354–8359. doi: 10.1021/acs.nanolett.5b04045. [DOI] [PubMed] [Google Scholar]
- (41).Raab M, Vietz C, Stefani FD, Acuna GP, Tinnefeld P. Shifting Molecular Localization by Plasmonic Coupling in a Single-Molecule Mirage. Nat Commun. 2017;8 doi: 10.1038/ncomms13966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Heucke SF, Baumann F, Acuna GP, Severin PMD, Stahl SW, Strackharn M, Stein IH, Altpeter P, Tinnefeld P, Gaub HE. Placing Individual Molecules in the Center of Nanoapertures. Nano Lett. 2014;14:391–395. doi: 10.1021/nl401517a. [DOI] [PubMed] [Google Scholar]
- (43).Toppari JJ, Wirth J, Garwe F, Stranik O, Csaki A, Bergmann J, Paa W, Fritzsche W. Plasmonic Coupling and Long-Range Transfer of an Excitation Along a DNA Nanowire. ACS Nano. 2013;7:1291–1298. doi: 10.1021/nn304789w. [DOI] [PubMed] [Google Scholar]
- (44).Roller EM, Besteiro LV, Pupp C, Khorashad LK, Govorov AO, Liedl T. Hot Spot-Mediated Non-Dissipative and Ultrafast Plasmon Passage. Nat Phys. 2017;13:761–765. doi: 10.1038/nphys4120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Jones MR, Seeman NC, Mirkin CA. Programmable Materials and the Nature of the DNA Bond. Science. 2015;347 doi: 10.1126/science.1260901. [DOI] [PubMed] [Google Scholar]
- (46).Mirkin CA, Letsinger RL, Mucic RC, Storhoff JJ. A DNA-Based Method for Rationally Assembling Nanoparticles into Macroscopic Materials. Nature. 1996;382:607–609. doi: 10.1038/382607a0. [DOI] [PubMed] [Google Scholar]
- (47).Alivisatos AP, Johnsson KP, Peng XG, Wilson TE, Loweth CJ, Bruchez MP, Schultz P. Organization of 'Nanocrystal Molecules' Using DNA. Nature. 1996;382:609–611. doi: 10.1038/382609a0. [DOI] [PubMed] [Google Scholar]
- (48).Storhofff JJ, Elghanian R, Mirkin CA, Letsinger RL. Sequence-Dependent Stability of DNA-Modified Gold Nanoparticles. Langmuir. 2002;18:6666–6670. [Google Scholar]
- (49).Sharma J, Chhabra R, Andersen CS, Gothelf KV, Yan H, Liu Y. Toward Reliable Gold Nanoparticle Patterning on Self-Assembled DNA Nanoscaffold. J Am Chem Soc. 2008;130:7820. doi: 10.1021/ja802853r. + [DOI] [PubMed] [Google Scholar]
- (50).Ding BQ, Deng ZT, Yan H, Cabrini S, Zuckermann RN, Bokor J. Gold Nanoparticle Self-Similar Chain Structure Organized by DNA Origami. J Am Chem Soc. 2010;132:3248. doi: 10.1021/ja9101198. + [DOI] [PubMed] [Google Scholar]
- (51).Jones MR, Osberg KD, Macfarlane RJ, Langille MR, Mirkin CA. Templated Techniques for the Synthesis and Assembly of Plasmonic Nanostructures. Chem Rev. 2011;111:3736–3827. doi: 10.1021/cr1004452. [DOI] [PubMed] [Google Scholar]
- (52).Loweth CJ, Caldwell WB, Peng XG, Alivisatos AP, Schultz PG. DNA-Based Assembly of Gold Nanocrystals. Angew Chem Int Edit. 1999;38:1808–1812. doi: 10.1002/(SICI)1521-3773(19990614)38:12<1808::AID-ANIE1808>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- (53).Sheikholeslami S, Jun YW, Jain PK, Alivisatos AP. Coupling of Optical Resonances in a Compositionally Asymmetric Plasmonic Nanoparticle Dimer. Nano Lett. 2010;10:2655–2660. doi: 10.1021/nl101380f. [DOI] [PubMed] [Google Scholar]
- (54).Maye MM, Gang O, Cotlet M. Photoluminescence Enhancement in Cdse/Zns-DNA Linked-Au Nanoparticle Heterodimers Probed by Single Molecule Spectroscopy. Chem Commun. 2010;46:6111–6113. doi: 10.1039/c0cc00660b. [DOI] [PubMed] [Google Scholar]
- (55).Claridge SA, Goh SL, Frechet JMJ, Williams SC, Micheel CM, Alivisatos AP. Directed Assembly of Discrete Gold Nanoparticle Groupings Using Branched DNA Scaffolds. Chem Mater. 2005;17:1628–1635. [Google Scholar]
- (56).Fu AH, Micheel CM, Cha J, Chang H, Yang H, Alivisatos AP. Discrete Nanostructures of Quantum Dots/Au with DNA. J Am Chem Soc. 2004;126:10832–10833. doi: 10.1021/ja046747x. [DOI] [PubMed] [Google Scholar]
- (57).Mastroianni AJ, Claridge SA, Alivisatos AP. Pyramidal and Chiral Groupings of Gold Nanocrystals Assembled Using DNA Scaffolds. J Am Chem Soc. 2009;131:8455–8459. doi: 10.1021/ja808570g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Aldaye FA, Sleiman HF. Dynamic DNA Templates for Discrete Gold Nanoparticle Assemblies: Control of Geometry, Modularity, Write/Wrase and Structural Switching. J Am Chem Soc. 2007;129:4130. doi: 10.1021/ja070017i. + [DOI] [PubMed] [Google Scholar]
- (59).Xu XY, Rosi NL, Wang YH, Huo FW, Mirkin CA. Asymmetric Functionalization of Gold Nanoparticles with Oligonucleotides. J Am Chem Soc. 2006;128:9286–9287. doi: 10.1021/ja061980b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (60).Li HY, Park SH, Reif JH, LaBean TH, Yan H. DNA-Templated Self-Assembly of Protein and Nanoparticle Linear Arrays. J Am Chem Soc. 2004;126:418–419. doi: 10.1021/ja0383367. [DOI] [PubMed] [Google Scholar]
- (61).Deng ZX, Tian Y, Lee SH, Ribbe AE, Mao CD. DNA-Encoded Self-Assembly of Gold Nanoparticles into One-Dimensional Arrays. Angew Chem Int Edit. 2005;44:3582–3585. doi: 10.1002/anie.200463096. [DOI] [PubMed] [Google Scholar]
- (62).Beyer S, Nickels P, Simmel FC. Periodic DNA Nanotemplates Synthesized by Rolling Circle Amplification. Nano Lett. 2005;5:719–722. doi: 10.1021/nl050155a. [DOI] [PubMed] [Google Scholar]
- (63).Le JD, Pinto Y, Seeman NC, Musier-Forsyth K, Taton TA, Kiehl RA. DNA-Templated Self-Assembly of Metallic Nanocomponent Arrays on a Surface. Nano Lett. 2004;4:2343–2347. doi: 10.1021/nl0515495. [DOI] [PubMed] [Google Scholar]
- (64).Zhang JP, Liu Y, Ke YG, Yan H. Periodic Square-Like Gold Nanoparticle Arrays Templated by Self-Assembled 2d DNA Nanogrids on a Surface. Nano Lett. 2006;6:248–251. doi: 10.1021/nl052210l. [DOI] [PubMed] [Google Scholar]
- (65).Zheng JW, Constantinou PE, Micheel C, Alivisatos AP, Kiehl RA, Seeman NC. Two-Dimensional Nanoparticle Arrays Show the Organizational Power of Robust DNA Motifs. Nano Lett. 2006;6:1502–1504. doi: 10.1021/nl060994c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (66).Xing H, Wang ZD, Xu ZD, Wong NY, Xiang Y, Liu GLG, Lu Y. DNA-Directed Assembly of Asymmetric Nanoclusters Using Janus Nanoparticles. ACS Nano. 2012;6:802–809. doi: 10.1021/nn2042797. [DOI] [PubMed] [Google Scholar]
- (67).Huo FW, Lytton-Jean AKR, Mirkin CA. Asymmetric Functionalization of Nanoparticles Based on Thermally Addressable DNA Interconnects. Adv Mater. 2006;18:2304–2306. [Google Scholar]
- (68).Feng L, Dreyfus R, Sha RJ, Seeman NC, Chaikin PM. DNA Patchy Particles. Adv Mater. 2013;25:2779–2783. doi: 10.1002/adma.201204864. [DOI] [PubMed] [Google Scholar]
- (69).Park SY, Lytton-Jean AKR, Lee B, Weigand S, Schatz GC, Mirkin CA. DNA-Programmable Nanoparticle Crystallization. Nature. 2008;451:553–556. doi: 10.1038/nature06508. [DOI] [PubMed] [Google Scholar]
- (70).Lu F, Yager KG, Zhang YG, Xin HL, Gang O. Superlattices Assembled through Shape-Induced Directional Binding. Nat Commun. 2015;6 doi: 10.1038/ncomms7912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (71).Zhang YG, Pal S, Srinivasan B, Vo T, Kumar S, Gang O. Selective Transformations between Nanoparticle Superlattices Via the Reprogramming of DNA-Mediated Interactions. Nat Mater. 2015;14:840. doi: 10.1038/nmat4296. + [DOI] [PubMed] [Google Scholar]
- (72).O'Brien MN, Jones MR, Lee B, Mirkin CA. Anisotropic Nanoparticle Complementarity in DNA-Mediated Co-Crystallization. Nat Mater. 2015;14:833. doi: 10.1038/nmat4293. + [DOI] [PubMed] [Google Scholar]
- (73).Rothemund PWK. Folding DNA to Create Nanoscale Shapes and Patterns. Nature. 2006;440:297–302. doi: 10.1038/nature04586. [DOI] [PubMed] [Google Scholar]
- (74).Ke YG, Sharma J, Liu MH, Jahn K, Liu Y, Yan H. Scaffolded DNA Origami of a DNA Tetrahedron Molecular Container. Nano Lett. 2009;9:2445–2447. doi: 10.1021/nl901165f. [DOI] [PubMed] [Google Scholar]
- (75).Andersen ES, Dong M, Nielsen MM, Jahn K, Subramani R, Mamdouh W, Golas MM, Sander B, Stark H, Oliveira CLP, et al. Self-Assembly of a Nanoscale DNA Box with a Controllable Lid. Nature. 2009;459:73–U75. doi: 10.1038/nature07971. [DOI] [PubMed] [Google Scholar]
- (76).Douglas SM, Dietz H, Liedl T, Hogberg B, Graf F, Shih WM. Self-Assembly of DNA into Nanoscale Three-Dimensional Shapes. Nature. 2009;459:414–418. doi: 10.1038/nature08016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (77).Ke YG, Douglas SM, Liu MH, Sharma J, Cheng AC, Leung A, Liu Y, Shih WM, Yan H. Multilayer DNA Origami Packed on a Square Lattice. J Am Chem Soc. 2009;131:15903–15908. doi: 10.1021/ja906381y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (78).Dietz H, Douglas SM, Shih WM. Folding DNA into Twisted and Curved Nanoscale Shapes. Science. 2009;325:725–730. doi: 10.1126/science.1174251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (79).Ke Y, Ong LL, Shih WM, Yin P. Three-Dimensional Structures Self-Assembled from DNA Bricks. Science. 2012;338:1177–1183. doi: 10.1126/science.1227268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (80).Geary C, Rothemund PW, Andersen ES. Rna Nanostructures. A Single-Stranded Architecture for Cotranscriptional Folding of Rna Nanostructures. Science. 2014;345:799–804. doi: 10.1126/science.1253920. [DOI] [PubMed] [Google Scholar]
- (81).Liedl T, Hogberg B, Tytell J, Ingber DE, Shih WM. Self-Assembly of Three-Dimensional Prestressed Tensegrity Structures from DNA. Nat Nanotechnol. 2010;5:520–524. doi: 10.1038/nnano.2010.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (82).Kauert DJ, Kurth T, Liedl T, Seidel R. Direct Mechanical Measurements Reveal the Material Properties of Three-Dimensional DNA Origami. Nano Lett. 2011;11:5558–5563. doi: 10.1021/nl203503s. [DOI] [PubMed] [Google Scholar]
- (83).Schiffels D, Liedl T, Fygenson DK. Nanoscale Structure and Microscale Stiffness of DNA Nanotubes. ACS Nano. 2013;7:6700–6710. doi: 10.1021/nn401362p. [DOI] [PubMed] [Google Scholar]
- (84).Hagerman PJ. Flexibility of DNA. Annu Rev Biophys Biophys Chem. 1988;17:265–286. doi: 10.1146/annurev.bb.17.060188.001405. [DOI] [PubMed] [Google Scholar]
- (85).Baumann CG, Smith SB, Bloomfield VA, Bustamante C. Ionic Effects on the Elasticity of Single DNA Molecules. Proc Natl Acad Sci USA. 1997;94:6185–6190. doi: 10.1073/pnas.94.12.6185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (86).Voigt NV, Torring T, Rotaru A, Jacobsen MF, Ravnsbaek JB, Subramani R, Mamdouh W, Kjems J, Mokhir A, Besenbacher F, et al. Single-Molecule Chemical Reactions on DNA Origami. Nat Nanotechnol. 2010;5:200–203. doi: 10.1038/nnano.2010.5. [DOI] [PubMed] [Google Scholar]
- (87).Pal S, Deng ZT, Ding BQ, Yan H, Liu Y. DNA-Origami-Directed Self-Assembly of Discrete Silver-Nanoparticle Architectures. Angew Chem Int Edit. 2010;49:2700–2704. doi: 10.1002/anie.201000330. [DOI] [PubMed] [Google Scholar]
- (88).Douglas SM, Marblestone AH, Teerapittayanon S, Vazquez A, Church GM, Shih WM. Rapid Prototyping of 3d DNA-Origami Shapes with Cadnano. Nucleic Acids Res. 2009;37:5001–5006. doi: 10.1093/nar/gkp436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (89).Kim DN, Kilchherr F, Dietz H, Bathe M. Quantitative Prediction of 3D Solution Shape and Flexibility of Nucleic Acid Nanostructures. Nucleic Acids Res. 2012;40:2862–2868. doi: 10.1093/nar/gkr1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (90).Benson E, Mohammed A, Gardell J, Masich S, Czeizler E, Orponen P, Hogberg B. DNA Rendering of Polyhedral Meshes at the Nanoscale. Nature. 2015;523:441–444. doi: 10.1038/nature14586. [DOI] [PubMed] [Google Scholar]
- (91).Maffeo C, Yoo J, Aksimentiev A. De Novo Reconstruction of DNA Origami Structures through Atomistic Molecular Dynamics Simulation. Nucleic Acids Res. 2016;44:3013–3019. doi: 10.1093/nar/gkw155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (92).Snodin BE, Romano F, Rovigatti L, Ouldridge TE, Louis AA, Doye JP. Direct Simulation of the Self-Assembly of a Small DNA Origami. ACS Nano. 2016;10:1724–1737. doi: 10.1021/acsnano.5b05865. [DOI] [PubMed] [Google Scholar]
- (93).Prodan E, Radloff C, Halas NJ, Nordlander P. A Hybridization Model for the Plasmon Response of Complex Nanostructures. Science. 2003;302:419–422. doi: 10.1126/science.1089171. [DOI] [PubMed] [Google Scholar]
- (94).Nordlander P, Oubre C, Prodan E, Li K, Stockman MI. Plasmon Hybridizaton in Nanoparticle Dimers. Nano Lett. 2004;4:899–903. [Google Scholar]
- (95).Weller L, Thacker VV, Herrmann LO, Hemmig EA, Lombardi A, Keyser UF, Baumberg JJ. Gap-Dependent Coupling of Ag-Au Nanoparticle Heterodimers Using DNA Origami-Based Self-Assembly. ACS Photonics. 2016;3:1589–1595. [Google Scholar]
- (96).Reinhard BM, Siu M, Agarwal H, Alivisatos AP, Liphardt J. Calibration of Dynamic Molecular Rule Based on Plasmon Coupling between Gold Nanoparticles. Nano Lett. 2005;5:2246–2252. doi: 10.1021/nl051592s. [DOI] [PubMed] [Google Scholar]
- (97).Miroshnichenko AE, Flach S, Kivshar YS. Fano Resonances in Nanoscale Structures. Rev Mod Phys. 2010;82:2257–2298. [Google Scholar]
- (98).Fan JA, Wu CH, Bao K, Bao JM, Bardhan R, Halas NJ, Manoharan VN, Nordlander P, Shvets G, Capasso F. Self-Assembled Plasmonic Nanoparticle Clusters. Science. 2010;328:1135–1138. doi: 10.1126/science.1187949. [DOI] [PubMed] [Google Scholar]
- (99).Luk'yanchuk B, Zheludev NI, Maier SA, Halas NJ, Nordlander P, Giessen H, Chong CT. The Fano Resonance in Plasmonic Nanostructures and Metamaterials. Nat Mater. 2010;9:707–715. doi: 10.1038/nmat2810. [DOI] [PubMed] [Google Scholar]
- (100).Roller EM, Khorashad LK, Fedoruk M, Schreiber R, Govorov AO, Liedl T. DNA-Assembled Nanoparticle Rings Exhibit Electric and Magnetic Resonances at Visible Frequencies. Nano Lett. 2015;15:1368–1373. doi: 10.1021/nl5046473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (101).Schreiber R, Luong N, Fan ZY, Kuzyk A, Nickels PC, Zhang T, Smith DM, Yurke B, Kuang W, Govorov AO, et al. Chiral Plasmonic DNA Nanostructures with Switchable Circular Dichroism. Nat Commun. 2013;4 doi: 10.1038/ncomms3948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (102).Kuzyk A, Schreiber R, Zhang H, Govorov AO, Liedl T, Liu N. Reconfigurable 3d Plasmonic Metamolecules. Nat Mater. 2014;13:862–866. doi: 10.1038/nmat4031. [DOI] [PubMed] [Google Scholar]
- (103).Urban MJ, Dutta PK, Wang PF, Duan XY, Shen XB, Ding BQ, Ke YG, Liu N. Plasmonic Toroidal Metamolecules Assembled by DNA Origami. J Am Chem Soc. 2016;138:5495–5498. doi: 10.1021/jacs.6b00958. [DOI] [PubMed] [Google Scholar]
- (104).Shen XB, Asenjo-Garcia A, Liu Q, Jiang Q, de Abajo FJG, Liu N, Ding BQ. Three-Dimensional Plasmonic Chiral Tetramers Assembled by DNA Origami. Nano Lett. 2013;13:2128–2133. doi: 10.1021/nl400538y. [DOI] [PubMed] [Google Scholar]
- (105).Lan X, Chen Z, Dai GL, Lu XX, Ni WH, Wang QB. Bifacial DNA Origami-Directed Discrete, Three-Dimensional, Anisotropic Plasmonic Nanoarchitectures with Tailored Optical Chirality. J Am Chem Soc. 2013;135:11441–11444. doi: 10.1021/ja404354c. [DOI] [PubMed] [Google Scholar]
- (106).Park DJ, Zhang C, Ku JC, Zhou Y, Schatz GC, Mirkin CA. Plasmonic Photonic Crystals Realized through DNA-Programmable Assembly. Proc Natl Acad Sci USA. 2015;112:977–981. doi: 10.1073/pnas.1422649112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (107).Simoncelli S, Roller EM, Urban P, Schreiber R, Turberfield AJ, Liedl T, Lohmuller T. Quantitative Single-Molecule Surface Enhanced Raman Scattering by Optothermal Tuning of DNA Origami-Assembled Plasmonic Nanoantennas. ACS Nano. 2016;10:9809–9815. doi: 10.1021/acsnano.6b05276. [DOI] [PubMed] [Google Scholar]
- (108).Kobayashi K, Aikawa H, Katsumoto S, Iye Y. Tuning of the Fano Effect through a Quantum Dot in an Aharonov-Bohm Interferometer. Phys Rev Lett. 2002;88 doi: 10.1103/PhysRevLett.88.256806. [DOI] [PubMed] [Google Scholar]
- (109).Johnson AC, Marcus CM, Hanson MP, Gossard AC. Coulomb-Modified Fano Resonance in a One-Lead Quantum Dot. Phys Rev Lett. 2004;93 doi: 10.1103/PhysRevLett.93.106803. [DOI] [PubMed] [Google Scholar]
- (110).Hao F, Sonnefraud Y, Van Dorpe P, Maier SA, Halas NJ, Nordlander P. Symmetry Breaking in Plasmonic Nanocavities: Subradiant Lspr Sensing and a Tunable Fano Resonance. Nano Lett. 2008;8:3983–3988. doi: 10.1021/nl802509r. [DOI] [PubMed] [Google Scholar]
- (111).Fan JA, He Y, Bao K, Wu CH, Bao JM, Schade NB, Manoharan VN, Shvets G, Nordlander P, Liu DR, et al. DNA-Enabled Self-Assembly of Plasmonic Nanoclusters. Nano Lett. 2011;11:4859–4864. doi: 10.1021/nl203194m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (112).Lassiter JB, Sobhani H, Fan JA, Kundu J, Capasso F, Nordlander P, Halas NJ. Fano Resonances in Plasmonic Nanoclusters: Geometrical and Chemical Tunability. Nano Lett. 2010;10:3184–3189. doi: 10.1021/nl102108u. [DOI] [PubMed] [Google Scholar]
- (113).Mukherjee S, Sobhani H, Lassiter JB, Bardhan R, Nordlander P, Halas NJ. Fanoshells: Nanoparticles with Built-in Fano Resonances. Nano Lett. 2010;10:2694–2701. doi: 10.1021/nl1016392. [DOI] [PubMed] [Google Scholar]
- (114).Zhang SP, Bao K, Halas NJ, Xu HX, Nordlander P. Substrate-Induced Fano Resonances of a Plasmonic: Nanocube: A Route to Increased-Sensitivity Localized Surface Plasmon Resonance Sensors Revealed. Nano Lett. 2011;11:1657–1663. doi: 10.1021/nl200135r. [DOI] [PubMed] [Google Scholar]
- (115).King NS, Liu LF, Yang X, Cerjan B, Everitt HO, Nordlander P, Halas NJ. Fano Resonant Aluminum Nanoclusters for Plasmonic Colorimetric Sensing. ACS Nano. 2015;9:10628–10636. doi: 10.1021/acsnano.5b04864. [DOI] [PubMed] [Google Scholar]
- (116).Veselago VG. Electrodynamics of Substances with Simultaneously Negative Values of Sigma and Mu. Sov Phys Uspekhi. 1968;10:509. [Google Scholar]
- (117).Alu A, Engheta N. Achieving Transparency with Plasmonic and Metamaterial Coatings. Phys Rev E. 2005;72 doi: 10.1103/PhysRevE.72.016623. [DOI] [PubMed] [Google Scholar]
- (118).Liu YM, Zhang X. Metamaterials: A New Frontier of Science and Technology. Chem Soc Rev. 2011;40:2494–2507. doi: 10.1039/c0cs00184h. [DOI] [PubMed] [Google Scholar]
- (119).Soukoulis CM, Wegener M. Past Achievements and Future Challenges in the Development of Three-Dimensional Photonic Metamaterials. Nat Photonics. 2011;5:523–530. [Google Scholar]
- (120).Smith DR, Padilla WJ, Vier DC, Nemat-Nasser SC, Schultz S. Composite Medium with Simultaneously Negative Permeability and Permittivity. Phys Rev Lett. 2000;84:4184–4187. doi: 10.1103/PhysRevLett.84.4184. [DOI] [PubMed] [Google Scholar]
- (121).Schurig D, Mock JJ, Justice BJ, Cummer SA, Pendry JB, Starr AF, Smith DR. Metamaterial Electromagnetic Cloak at Microwave Frequencies. Science. 2006;314:977–980. doi: 10.1126/science.1133628. [DOI] [PubMed] [Google Scholar]
- (122).Zhou J, Koschny T, Kafesaki M, Economou EN, Pendry JB, Soukoulis CM. Saturation of the Magnetic Response of Split-Ring Resonators at Optical Frequencies. Phys Rev Lett. 2005;95 doi: 10.1103/PhysRevLett.95.223902. [DOI] [PubMed] [Google Scholar]
- (123).Alu A, Salandrino A. Negative Effective Permeability and Left-Handed Materials at Optical Frequencies. Opt Express. 2006;14:1557–1567. doi: 10.1364/oe.14.001557. [DOI] [PubMed] [Google Scholar]
- (124).Alu A, Engheta N. Dynamical Theory of Artificial Optical Magnetism Produced by Rings of Plasmonic Nanoparticles. Phys Rev B. 2008;78 [Google Scholar]
- (125).Shafiei F, Monticone F, Le KQ, Liu X-X, Hartsfield T, Alu A, Li X. A Subwavelength Plasmonic Metamoleculeexhibiting Magnetic-Based Optical Fano Resonance. Nat Nanotechnol. 2013;8:95–99. doi: 10.1038/nnano.2012.249. 2013, 95–99. [DOI] [PubMed] [Google Scholar]
- (126).Sheikholeslami SN, Alaeian H, Koh AL, Dionne JA. A Metafluid Exhibiting Strong Optical Magnetism. Nano Lett. 2013;13:4137–4141. doi: 10.1021/nl401642z. [DOI] [PubMed] [Google Scholar]
- (127).Qian Z, Hastings SP, Li C, Edward B, McGinn CK, Engheta N, Fakhraai Z, Park SJ. Raspberry-Like Metamolecules Exhibiting Strong Magnetic Resonances. ACS Nano. 2015;9:1263–1270. doi: 10.1021/nn5050678. [DOI] [PubMed] [Google Scholar]
- (128).Fan ZY, Govorov AO. Plasmonic Circular Dichroism of Chiral Metal Nanoparticle Assemblies. Nano Lett. 2010;10:2580–2587. doi: 10.1021/nl101231b. [DOI] [PubMed] [Google Scholar]
- (129).Pal S, Deng ZT, Wang HN, Zou SL, Liu Y, Yan H. DNA Directed Self-Assembly of Anisotropic Plasmonic Nanostructures. J Am Chem Soc. 2011;133:17606–17609. doi: 10.1021/ja207898r. [DOI] [PubMed] [Google Scholar]
- (130).Chen Z, Lan X, Chiu YC, Lu XX, Ni WH, Gao HW, Wang QB. Strong Chiroptical Activities in Gold Nanorod Dimers Assembled Using DNA Origami Templates. ACS Photonics. 2015;2:392–397. [Google Scholar]
- (131).Lan X, Lu XX, Shen CQ, Ke YG, Ni WH, Wang QB. Au Nanorod Helical Superstructures with Designed Chirality. J Am Chem Soc. 2015;137:457–462. doi: 10.1021/ja511333q. [DOI] [PubMed] [Google Scholar]
- (132).Willets KA, Van Duyne RP. Localized Surface Plasmon Resonance Spectroscopy and Sensing. Annu Rev Phys Chem. 2007;58:267–297. doi: 10.1146/annurev.physchem.58.032806.104607. [DOI] [PubMed] [Google Scholar]
- (133).Moskovits M. Surface-Enhanced Raman Spectroscopy: A Brief Retrospective. J Raman Spectrosc. 2005;36:485–496. [Google Scholar]
- (134).Laing S, Jamieson LE, Faulds K, Graham D. Surface-Enhanced Raman Spectroscopy for in Vivo Biosensing. Nat Rev Chem. 2017;1:0060. [Google Scholar]
- (135).Kneipp K, Kneipp H, Itzkan I, Dasari RR, Feld MS. Ultrasensitive Chemical Analysis by Raman Spectroscopy. Chem Rev. 1999;99:2957. doi: 10.1021/cr980133r. + [DOI] [PubMed] [Google Scholar]
- (136).Nie SM, Emery SR. Probing Single Molecules and Single Nanoparticles by Surface-Enhanced Raman Scattering. Science. 1997;275:1102–1106. doi: 10.1126/science.275.5303.1102. [DOI] [PubMed] [Google Scholar]
- (137).Aslan K, Gryczynski I, Malicka J, Matveeva E, Lakowicz JR, Geddes CD. Metal-Enhanced Fluorescence: An Emerging Tool in Biotechnology. Curr Opin Biotech. 2005;16:55–62. doi: 10.1016/j.copbio.2005.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (138).Su KH, Wei QH, Zhang X, Mock JJ, Smith DR, Schultz S. Interparticle Coupling Effects on Plasmon Resonances of Nanogold Particles. Nano Lett. 2003;3:1087–1090. [Google Scholar]
- (139).Giannini V, Fernandez-Dominguez AI, Heck SC, Maier SA. Plasmonic Nanoantennas: Fundamentals and Their Use in Controlling the Radiative Properties of Nanoemitters. Chem Rev. 2011;111:3888–3912. doi: 10.1021/cr1002672. [DOI] [PubMed] [Google Scholar]
- (140).Ding SY, Yi J, Li JF, Ren B, Wu DY, Panneerselvam R, Tian Q. Nanostructure-Based Plasmon-Enhanced Raman Spectroscopy for Surface Analysis of Materials. Nat Rev Mat. 2016;1:16021. [Google Scholar]
- (141).Le Ru EC, Etchegoin PG. Single-Molecule Surface-Enhanced Raman Spectroscopy. Annu Rev Phys Chem. 2012;63:65–87. doi: 10.1146/annurev-physchem-032511-143757. [DOI] [PubMed] [Google Scholar]
- (142).Novotny L, Hecht B. Principles of Nano-Optics. 2nd ed. Cambridge University Press; Cambridge: 2012. [Google Scholar]
- (143).Jiang J, Bosnick K, Maillard M, Brus L. Single Molecule Raman Spectroscopy at the Junctions of Large Ag Nanocrystals. J Phys Chem B. 2003;107:9964–9972. [Google Scholar]
- (144).Lee JH, You MH, Kim GH, Nam JM. Plasmonic Nanosnowmen with a Conductive Junction as Highly Tunable Nanoantenna Structures and Sensitive, Quantitative and Multiplexable Surface-Enhanced Raman Scattering Probes. Nano Lett. 2014;14:6217–6225. doi: 10.1021/nl502541u. [DOI] [PubMed] [Google Scholar]
- (145).Oh JW, Lim DK, Kim GH, Suh YD, Nam JM. Thiolated DNA-Based Chemistry and Control in the Structure and Optical Properties of Plasmonic Nanoparticles with Ultrasmall Interior Nanogap. J Am Chem Soc. 2014;136:14052–14059. doi: 10.1021/ja504270d. [DOI] [PubMed] [Google Scholar]
- (146).Kang JW, So PTC, Dasari RR, Lim DK. High Resolution Live Cell Raman Imaging Using Subcellular Organelle-Targeting Sers-Sensitive Gold Nanoparticles with Highly Narrow Intra-Nanogap. Nano Lett. 2015;15:1766–1772. doi: 10.1021/nl504444w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (147).Ruppin R. Decay of an Excited Molecule near a Small Metal Sphere. J Chem Phys. 1982;76:1681–1684. [Google Scholar]
- (148).Dulkeith E, Morteani AC, Niedereichholz T, Klar TA, Feldmann J, Levi SA, van Veggel FCJM, Reinhoudt DN, Moller M, Gittins DI. Fluorescence Quenching of Dye Molecules near Gold Nanoparticles: Radiative and Nonradiative Effects. Phys Rev Lett. 2002;89 doi: 10.1103/PhysRevLett.89.203002. [DOI] [PubMed] [Google Scholar]
- (149).Kuhn S, Hakanson U, Rogobete L, Sandoghdar V. Enhancement of Single-Molecule Fluorescence Using a Gold Nanoparticle as an Optical Nanoantenna. Phys Rev Lett. 2006;97 doi: 10.1103/PhysRevLett.97.017402. [DOI] [PubMed] [Google Scholar]
- (150).Anger P, Bharadwaj P, Novotny L. Enhancement and Quenching of Single-Molecule Fluorescence. Phys Rev Lett. 2006;96 doi: 10.1103/PhysRevLett.96.113002. [DOI] [PubMed] [Google Scholar]
- (151).Jungmann R, Steinhauer C, Scheible M, Kuzyk A, Tinnefeld P, Simmel FC. Single-Molecule Kinetics and Super-Resolution Microscopy by Fluorescence Imaging of Transient Binding on DNA Origami. Nano Lett. 2010;10:4756–4761. doi: 10.1021/nl103427w. [DOI] [PubMed] [Google Scholar]
- (152).Lee OS, Prytkova TR, Schatz GC. Using DNA to Link Gold Nanoparticles, Polymers and Molecules: A Theoretical Perspective. J Phys Chem Lett. 2010;1:1781–1788. doi: 10.1021/jz100435a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (153).Lee SE, Liu GL, Kim F, Lee LP. Remote Optical Switch for Localized and Selective Control of Gene Interference. Nano Lett. 2009;9:562–570. doi: 10.1021/nl802689k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (154).Jones MR, Millstone JE, Giljohann DA, Seferos DS, Young KL, Mirkin CA. Plasmonically Controlled Nucleic Acid Dehybridization with Gold Nanoprisms. ChemPhysChem. 2009;10:1461–1465. doi: 10.1002/cphc.200900269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (155).Do J, Schreiber R, Lutich AA, Liedl T, Rodriguez-Fernandez J, Feldmann J. Design and Optical Trapping of a Biocompatible Propeller-Like Nanoscale Hybrid. Nano Lett. 2012;12:5008–5013. doi: 10.1021/nl302775e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (156).Stehr J, Hrelescu C, Sperling RA, Raschke G, Wunderlich M, Nichtl A, Heindl D, Kurzinger K, Parak WJ, Klar TA, et al. Gold Nanostoves for Microsecond DNA Melting Analysis. Nano Lett. 2008;8:619–623. doi: 10.1021/nl073028i. [DOI] [PubMed] [Google Scholar]
- (157).Lee JH, Cheglakov Z, Yi J, Cronin TM, Gibson KJ, Tian B, Weizmann Y. Plasmonic Photothermal Gold Bipyramid Nanoreactors for Ultrafast Real-Time Bioassays. J Am Chem Soc. 2017;139:8054–8057. doi: 10.1021/jacs.7b01779. [DOI] [PubMed] [Google Scholar]
- (158).Bath J, Turberfield AJ. DNA Nanomachines. Nat Nanotechnol. 2007;2:275–284. doi: 10.1038/nnano.2007.104. [DOI] [PubMed] [Google Scholar]
- (159).Liedl T, Sobey TL, Simmel FC. DNA-Based Nanodevices. Nano Today. 2007;2:36–41. [Google Scholar]
- (160).Asanuma H, Takarada T, Yoshida T, Tamaru D, Liang XG, Komiyama M. Enantioselective Incorporation of Azobenzenes into Oligodeoxyribonucleotide for Effective Photoregulation of Duplex Formation. Angew Chem Int Edit. 2001;40:2671–2673. doi: 10.1002/1521-3773(20010716)40:14<2671::AID-ANIE2671>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- (161).Asanuma H, Liang X, Nishioka H, Matsunaga D, Liu M, Komiyama M. Synthesis of Azobenzene-Tethered DNA for Reversible Photo-Regulation of DNA Functions: Hybridization and Transcription. Nat Protoc. 2007;2:203–212. doi: 10.1038/nprot.2006.465. [DOI] [PubMed] [Google Scholar]
- (162).Forster T. *Zwischenmolekulare Energiewanderung Und Fluoreszenz. Ann Phys-Berlin. 1948;2:55–75. [Google Scholar]
- (163).Clegg RM. Fluorescence Resonance Energy Transfer. Curr Opin Biotechnol. 1995;6:103–110. doi: 10.1016/0958-1669(95)80016-6. [DOI] [PubMed] [Google Scholar]
- (164).Ha T, Enderle T, Ogletree DF, Chemla DS, Selvin PR, Weiss S. Probing the Interaction between Two Single Molecules: Fluorescence Resonance Energy Transfer between a Single Donor and a Single Acceptor. P Natl Acad Sci USA. 1996;93:6264–6268. doi: 10.1073/pnas.93.13.6264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (165).Reinhard BM, Sheikholeslami S, Mastroianni A, Alivisatos AP, Liphardt J. Use of Plasmon Coupling to Reveal the Dynamics of DNA Bending and Cleavage by Single Ecorv Restriction Enzymes. P Natl Acad Sci USA. 2007;104:2667–2672. doi: 10.1073/pnas.0607826104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (166).Lee SE, Chen Q, Bhat R, Petkiewicz S, Smith JM, Ferry VE, Correia AL, Alivisatos AP, Bissell MJ. Reversible Aptamer-Au Plasmon Rulers for Secreted Single Molecules. Nano Lett. 2015;15:4564–4570. doi: 10.1021/acs.nanolett.5b01161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (167).Chen JIL, Chen Y, Ginger DS. Plasmonic Nanoparticle Dimers for Optical Sensing of DNA in Complex Media. J Am Chem Soc. 2010;132:9600–9601. doi: 10.1021/ja103240g. [DOI] [PubMed] [Google Scholar]
- (168).Sebba DS, Mock JJ, Smith DR, LaBean TH, Lazarides AA. Reconfigurable Core-Satellite Nanoassemblies as Molecularly-Driven Plasmonic Switches. Nano Lett. 2008;8:1803–1808. doi: 10.1021/nl080029h. [DOI] [PubMed] [Google Scholar]
- (169).Morimura H, Tanaka SI, Ishitobi H, Mikami T, Kamachi Y, Kondoh H, Inouye Y. Nano-Analysis of DNA Conformation Changes Induced by Transcription Factor Complex Binding Using Plasmonic Nanodimers. ACS Nano. 2013;7:10733–10740. doi: 10.1021/nn403625s. [DOI] [PubMed] [Google Scholar]
- (170).Vogele K, List J, Pardatscher G, Holland NB, Simmel FC, Pirzer T. Self-Assembled Active Plasmonic Waveguide with a Peptide-Based Thermomechanical Switch. ACS Nano. 2016;10:11377–11384. doi: 10.1021/acsnano.6b06635. [DOI] [PubMed] [Google Scholar]
- (171).Yan Y, Chen JI, Ginger DS. Photoswitchable Oligonucleotide-Modified Gold Nanoparticles: Controlling Hybridization Stringency with Photon Dose. Nano Lett. 2012;12:2530–2536. doi: 10.1021/nl300739n. [DOI] [PubMed] [Google Scholar]
- (172).Kuzyk A, Yang YY, Duan X, Stoll S, Govorov AO, Sugiyama H, Endo M, Liu NA. Light-Driven Three-Dimensional Plasmonic Nanosystem That Translates Molecular Motion into Reversible Chiroptical Function. Nat Commun. 2016;7 doi: 10.1038/ncomms10591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (173).Zhou C, Duan XY, Liu N. A Plasmonic Nanorod That Walks on DNA Origami. Nat Commun. 2015;6 doi: 10.1038/ncomms9102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (174).Urban MJ, Zhou C, Duan XY, Liu N. Optically Resolving the Dynamic Walking of a Plasmonic Walker Couple. Nano Lett. 2015;15:8392–8396. doi: 10.1021/acs.nanolett.5b04270. [DOI] [PubMed] [Google Scholar]
- (175).Kuzyk A, Urban MJ, Idili A, Ricci F, Liu N. Selective Control of Reconfigurable Chiral Plasmonic Metamolecules. Science Advances. 2017;3:e1602803. doi: 10.1126/sciadv.1602803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (176).Li N, Tittl A, Yue S, Giessen H, Song C, Ding BQ, Liu N. DNA-Assembled Bimetallic Plasmonic Nanosensors. Light-Sci Appl. 2014;3 [Google Scholar]
- (177).Xu LG, Yan WJ, Ma W, Kuang H, Wu XL, Liu LQ, Zhao Y, Wang LB, Xu CL. Sers Encoded Silver Pyramids for Attomolar Detection of Multiplexed Disease Biomarkers. Adv Mater. 2015;27:1706–1711. doi: 10.1002/adma.201402244. [DOI] [PubMed] [Google Scholar]
- (178).Xu LG, Kuang H, Xu CL, Ma W, Wang LB, Kotov NA. Regiospecific Plasmonic Assemblies for in Situ Raman Spectroscopy in Live Cells. J Am Chem Soc. 2012;134:1699–1709. doi: 10.1021/ja2088713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (179).Yan WJ, Xu LG, Ma W, Liu LQ, Wang LB, Kuang H, Xu CL. Pyramidal Sensor Platform with Reversible Chiroptical Signals for DNA Detection. Small. 2014;10:4293–4297. doi: 10.1002/smll.201401641. [DOI] [PubMed] [Google Scholar]
- (180).Li S, Xu L, Ma W, Wu X, Sun M, Kuang H, Wang L, Kotov NA, Xu C. Dual-Mode Ultrasensitive Quantification of Microrna in Living Cells by Chiroplasmonic Nanopyramids Self-Assembled from Gold and Upconversion Nanoparticles. J Am Chem Soc. 2016;138:306–312. doi: 10.1021/jacs.5b10309. [DOI] [PubMed] [Google Scholar]
- (181).Fu YH, Kuznetsov AI, Miroshnichenko AE, Yu YF, Luk'yanchuk B. Directional Visible Light Scattering by Silicon Nanoparticles. Nat Commun. 2013;4 doi: 10.1038/ncomms2538. [DOI] [PubMed] [Google Scholar]
- (182).Kuznetsov AI, Miroshnichenko AE, Brongersma ML, Kivshar YS, Luk'yanchuk B. Optically Resonant Dielectric Nanostructures. Science. 2016;354 doi: 10.1126/science.aag2472. [DOI] [PubMed] [Google Scholar]
- (183).Funke JJ, Ketterer P, Lieleg C, Korber P, Dietz H. Exploring Nucleosome Unwrapping Using DNA Origami. Nano Lett. 2016;16:7891–7898. doi: 10.1021/acs.nanolett.6b04169. [DOI] [PubMed] [Google Scholar]
- (184).Funke JJ, Ketterer P, Lieleg C, Schunter S, Korber P, Dietz H. Uncovering the Forces between Nucleosomes Using DNA Origami. Science Advances. 2016;2:e1600974. doi: 10.1126/sciadv.1600974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (185).Kershner RJ, Bozano LD, Micheel CM, Hung AM, Fornof AR, Cha JN, Rettner CT, Bersani M, Frommer J, Rothemund PWK, et al. Placement and Orientation of Individual DNA Shapes on Lithographically Patterned Surfaces. Nat Nanotechnol. 2009;4:557–561. doi: 10.1038/nnano.2009.220. [DOI] [PubMed] [Google Scholar]
- (186).Hung AM, Micheel CM, Bozano LD, Osterbur LW, Wallraff GM, Cha JN. Large-Area Spatially Ordered Arrays of Gold Nanoparticles Directed by Lithographically Confined DNA Origami. Nat Nanotechnol. 2010;5:121–126. doi: 10.1038/nnano.2009.450. [DOI] [PubMed] [Google Scholar]
- (187).Gopinath A, Rothemund PWK. Optimized Assembly and Covalent Coupling of Single-Molecule DNA Origami Nano Arrays. ACS Nano. 2014;8:12030–12040. doi: 10.1021/nn506014s. [DOI] [PubMed] [Google Scholar]
- (188).Gopinath A, Miyazono E, Faraon A, Rothemund PWK. Engineering and Mapping Nanocavity Emission Via Precision Placement of DNA Origami. Nature. 2016;535:401–405. doi: 10.1038/nature18287. [DOI] [PubMed] [Google Scholar]
- (189).Yu N, Capasso F. Flat Optics with Designer Metasurfaces. Nat Mater. 2014;13:139–150. doi: 10.1038/nmat3839. [DOI] [PubMed] [Google Scholar]
- (190).Kim H, Surwade SP, Powell A, O'Donnel C, Liu H. Stability of DNA Origami Nanostructure under Diverse Chemical Environments. Chem Mater. 2014;26:5265–5273. [Google Scholar]
- (191).Mikkila J, Eskelinen AP, Niemela EH, Linko V, Frilander MJ, Torma P, Kostiainen MA. Virus-Encapsulated DNA Origami Nanostructures for Cellular Delivery. Nano Lett. 2014;14:2196–2200. doi: 10.1021/nl500677j. [DOI] [PubMed] [Google Scholar]
- (192).Cassinelli V, Oberleitner B, Sobotta J, Nickels P, Grossi G, Kempter S, Frischmuth T, Liedl T, Manetto A. One-Step Formation of “Chain-Armor”-Stabilized DNA Nanostructures. Angew Chem Int Edit. 2015;54:7795–7798. doi: 10.1002/anie.201500561. [DOI] [PubMed] [Google Scholar]
- (193).Ponnuswamy N, Bastings MMC, Nathwani B, Ryu JH, Chou LYT, Vinther M, Li WA, Anastassacos FM, Mooney DJ, Shih WM. Oligolysine-Based Coating Protects DNA Nanostructures from Low-Salt Denaturation and Nuclease Degradation. Nat Commun. 2017;8 doi: 10.1038/ncomms15654. 15654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (194).Pound E, Ashton JR, Becerril HA, Woolley AT. Polymerase Chain Reaction Based Scaffold Preparation for the Production of Thin, Branched DNA Origami Nanostructures of Arbitrary Sizes. Nano Lett. 2009;9:4302–4305. doi: 10.1021/nl902535q. [DOI] [PubMed] [Google Scholar]
- (195).Zhang HL, Chao J, Pan D, Liu HJ, Huang Q, Fan CH. Folding Super-Sized DNA Origami with Scaffold Strands from Long-Range Pcr. Chem Commun. 2012;48:6405–6407. doi: 10.1039/c2cc32204h. [DOI] [PubMed] [Google Scholar]
- (196).Praetorius F, Kick B, Behler KL, Honemann MN, Weuster-Botz D, Dietz H. Biotechnological Mass Production of DNA Origami. Nature. 2017;552:84–87. doi: 10.1038/nature24650. [DOI] [PubMed] [Google Scholar]
- (197).Hogberg B, Liedl T, Shih WM. Folding DNA Origami from a Double-Stranded Source of Scaffold. J Am Chem Soc. 2009;131:9154–9155. doi: 10.1021/ja902569x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (198).Marchi AN, Saaem I, Tian JD, LaBean TH. One-Pot Assembly of a Hetero-Dimeric DNA Origami from Chip-Derived Staples and Double-Stranded Scaffold. ACS Nano. 2013;7:903–910. doi: 10.1021/nn302322j. [DOI] [PubMed] [Google Scholar]
- (199).Nickels PC, Ke YG, Jungmann R, Smith DM, Leichsenring M, Shih WM, Liedl T, Hogberg B. DNA Origami Structures Directly Assembled from Intact Bacteriophages. Small. 2014;10:1765–1769. doi: 10.1002/smll.201303442. [DOI] [PubMed] [Google Scholar]
- (200).Zhao Z, Liu Y, Yan H. Organizing DNA Origami Tiles into Larger Structures Using Preformed Scaffold Frames. Nano Lett. 2011;11:2997–3002. doi: 10.1021/nl201603a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (201).Tikhomirov G, Petersen P, Qian L. Fractal Assembly of Micrometre-Scale DNA Origami Arrays with Arbitrary Patterns. Nature. 2017;552:67–71. doi: 10.1038/nature24655. [DOI] [PubMed] [Google Scholar]
- (202).Ong LL, Hanikel N, Yaghi OK, Grun C, Strauss MT, Bron P, Lai-Kee-Him J, Schueder F, Wang B, Wang P, et al. Programmable Self-Assembly of Three-Dimensional Nanostructures from 10,000 Unique Components. Nature. 2017;552:72–77. doi: 10.1038/nature24648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (203).Wagenbauer KF, Sigl C, Dietz H. Gigadalton-Scale Shape-Programmable DNA Assemblies. Nature. 2017;552:78–83. doi: 10.1038/nature24651. [DOI] [PubMed] [Google Scholar]
- (204).Simmel FC. DNA-Based Assembly Lines and Nanofactories. Curr Opin Biotechnol. 2012;23:516–521. doi: 10.1016/j.copbio.2011.12.024. [DOI] [PubMed] [Google Scholar]
- (205).Zheng JP, Birktoft JJ, Chen Y, Wang T, Sha RJ, Constantinou PE, Ginell SL, Mao CD, Seeman NC. From Molecular to Macroscopic Via the Rational Design of a Self-Assembled 3d DNA Crystal. Nature. 2009;461:74–77. doi: 10.1038/nature08274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (206).Liu WY, Zhong H, Wang RS, Seeman NC. Crystalline Two-Dimensional DNA-Origami Arrays. Angew Chem Int Edit. 2011;50:264–267. doi: 10.1002/anie.201005911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (207).Ke YG, Ong LL, Sun W, Song J, Dong MD, Shih WM, Yin P. DNA Brick Crystals with Prescribed Depths. Nat Chem. 2014;6:994–1002. doi: 10.1038/nchem.2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (208).Zhang T, Hartl C, Fischer S, Frank K, Nickels P, Heuer-Jungemann A, Nickels P, Liedl T. 3d DNA Origami Crystals. arXiv: 1706.06965. 2017 doi: 10.1002/adma.201800273. [DOI] [PubMed] [Google Scholar]