Abstract
In mammals, the emergence of totipotency after fertilization involves extensive rearrangements of the spatial positioning of the genome1,2. However, the contribution of spatial genome organization to the regulation of developmental programs is unclear3. Here, we have generated high-resolution maps in mouse pre-implantation embryos of genomic interactions with the nuclear lamina, a filamentous meshwork that lines the inner-nuclear membrane. We find that nuclear organisation is not inherited from the maternal germline but is instead established de novo rapidly after fertilisation. The two parental genomes establish lamina-associated domains (LADs4) with different features that converge after the 8-cell stage. We find that the mechanism of LAD establishment is unrelated to DNA replication. Instead, we show that paternal LAD formation in zygotes is prevented by ectopic expression of Kdm5b, suggesting that LAD establishment may be dependent upon remodelling of H3K4 methylation. Together, our data suggest a step-wise assembly model whereby early LAD formation precedes consolidation of Topologically Associating Domains (TADs).
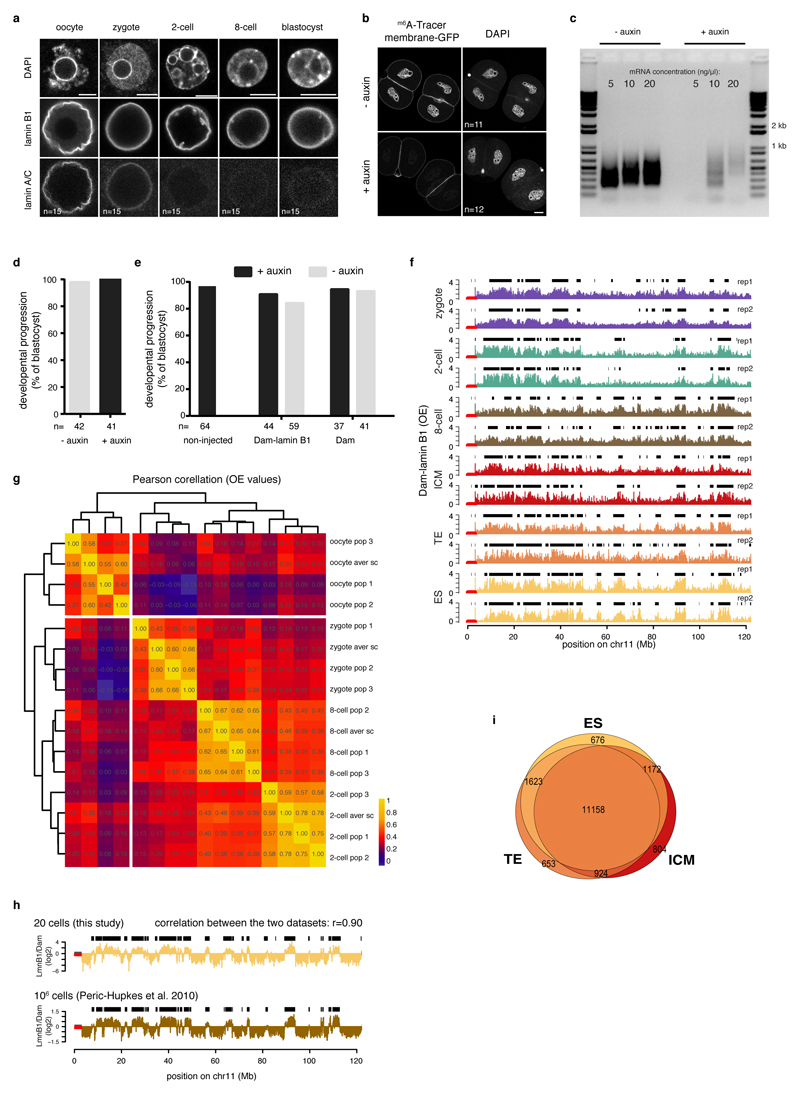
We established an experimental procedure to map LADs with DamID5 using lamin B1 and the untethered Dam enzyme as control. Lamin B1 is expressed throughout development and is therefore reliable to profile LADs (Extended Data Fig. 1a). Temporal control of Dam expression was ensured by the auxin inducible degron (AID) system6 (Extended Data Fig. 1b-c) and the m6ATracer7 enabled visualization of m6A-modified genome (Fig. 1a and Extended Data Fig. 1b). The experimental conditions did not interfere with embryonic development (Extended Data Fig. 1d-e).
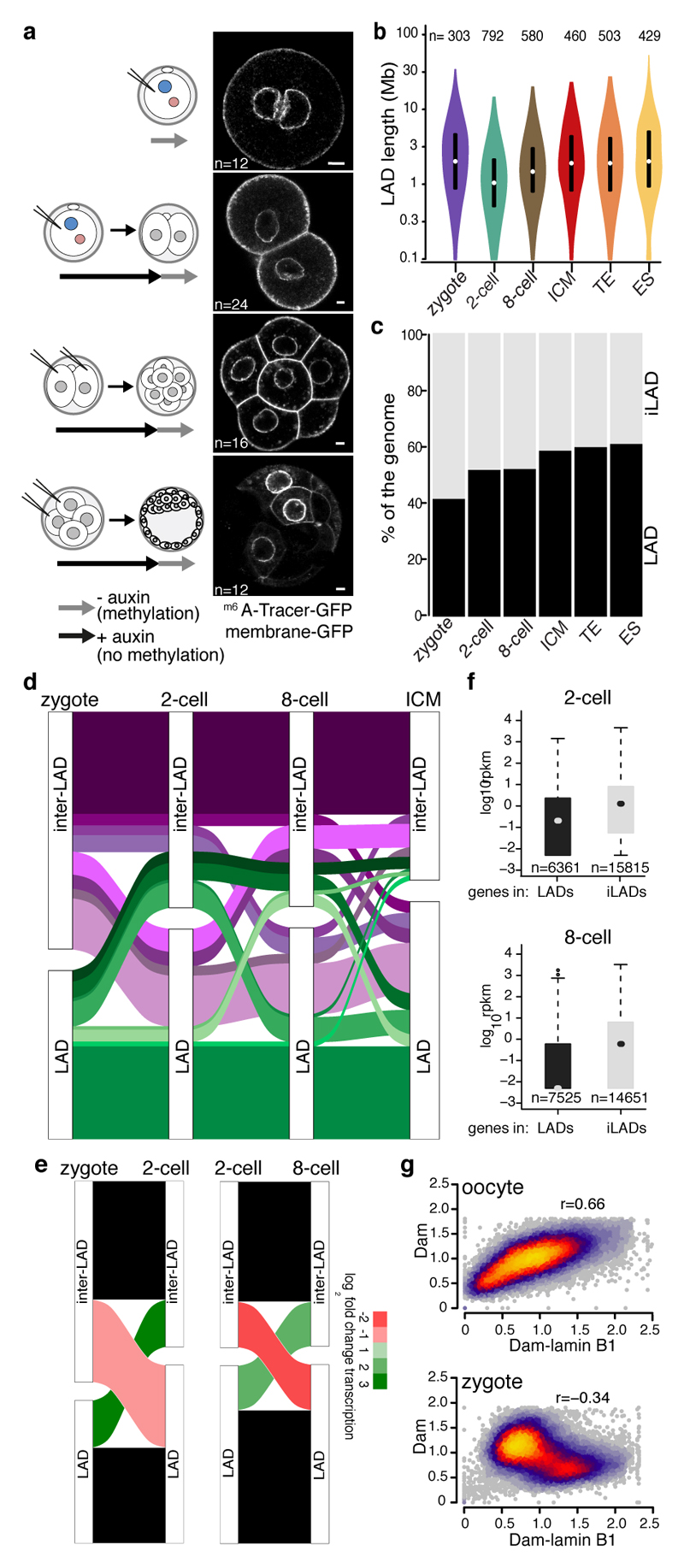
Figure 1. LADs establish de novo after fertilisation.
a, Experimental design. LAD methylation upon auxin removal, highlighted by GFP-m6ATracer. GAP43-EGFP expression marks cell membrane. Scale bar: 5 μm. Experiments were repeated at least five times. c, Distribution of LAD domain length. Violin plots show the 25th and 75th percentiles (black lines), median (circles) and the smallest/largest values at most 1.5 * IQR. n = number of LADs. d, Genomic LAD coverage. e, Alluvial plot showing LAD reorganisation during preimplantation development. f, Alluvial plot showing median log2 fold-change expression of genes20 for changing LADs between zygotes, 2-cell and 8-cell stages. g, RNAseq expression values20 of genes within LADs or iLADs. Box plots show the 25th and 75th percentiles (box), median (circles), the smallest/largest values at most 1.5 * IQR of the hinge (whiskers) and outliers (black circles). n = number of genes. h, Genome-wide scatter plots (100-kb bins) of Dam and Dam-lamin B1 scores in oocytes and zygotes. n = 3 biological independent samples.
We mapped LADs in fully-grown interphase oocytes (GV) arrested at the diplotene stage of prophase, zygotes, 2- and 8-cell embryos in populations and single-cell samples. The population replicates and single-cell average profiles displayed high concordance (Extended Data Fig. 1f-g). We also generated LAD profiles in trophectoderm (TE) and inner-cell-mass (ICM) cells, and in clonal mouse embryonic stem (ES) cells. LADs in ES cells correlate highly with previously published data (Extended Data Fig. 1g) and the similarity in LAD profiles between ICM and ES cell populations corresponds to the blastocyst origin of ES cells (Fig. 1b, Extended Data Fig. 1h). Genome-NL contacts on autosomes in zygotes, 2-cell, 8-cell and blastocysts stage embryos revealed broad continuous regions of m6A enrichment, characteristic of LADs in somatic cells (Extended Data Fig. 1f), which was vastly distinct from the Dam-injected embryos (Extended Data Fig. 2a). We conclude that the embryonic genome organises into LADs in zygotes.
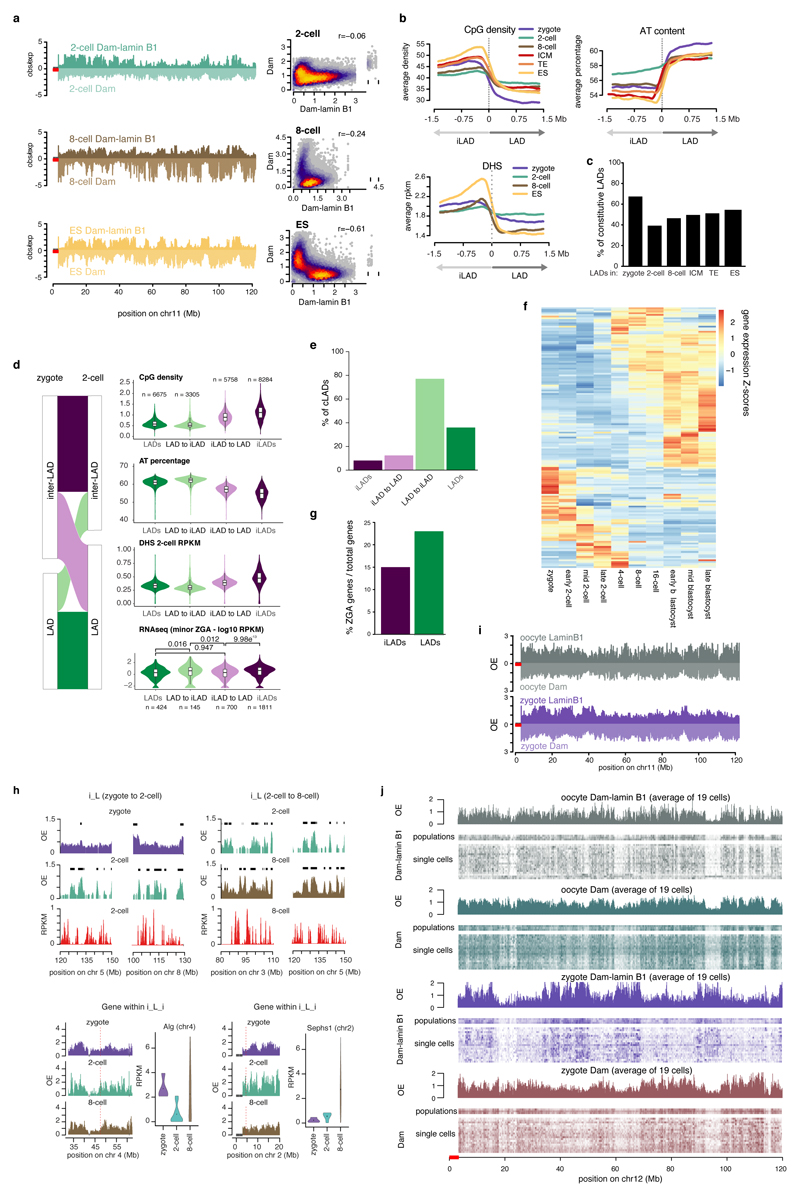
LADs in preimplantation development displayed broad domains with a median size between 1 Mb and 1.9 Mb and a genomic coverage between 42% and 61% (Fig. 1b and 1c). The 2- and 8-cell stages show more and smaller domains compared to the other stages (Fig. 1b and Extended Data Fig. 3). 42% of the zygotic LADs reposition to the nuclear interior at the 2- or 8-cell stage, but intriguingly 70% of these zygotic LADs, regain NL-association in blastocysts (Fig. 1d). Strikingly, LADs in zygotes overlap for 86% with the ICM and share a clear resemblance in associated genomic features (Extended Data Fig. 2b). Zygotic LADs are typified by high A/T content, low CpG density and a remarkable 67% overlap with previously identified cell-type invariable constitutive LADs (cLADs)8 (Extended Data Fig. 2c). The CpG density and A/T content is relatitvely low for LADs at the 2-cell stage. We postulate that this is the result of an exceptional reorganization of the genome at the 2-cell stage. Typical LADs in the zygote dislodge from the NL, while regions with intermediate LAD-features coincidently associate with the NL (Extended Data Fig. 2c). This reorganisation in 2-cell embryos involves large, typical LAD domains. Intriguingly, 77% of the dissociated LADs are cLADs, which further emphasizes the atypical nuclear positioning at the 2-cell stage (Extended Data Fig. 2e). Despite the unusual spatial rearrangements at the 2-cell stage, repositioning coincides with typical upregulation and downregulation of gene expression in de novo iLADs and LADs, respectively (Fig. 1e). 2-cell stage-specific LADs contain genes (n = 155) mainly expressed in the zygote and later stages of development, but are generally silent at the mid and late 2-cell stage (Extended Data Fig. 2f). The association between transcriptional changes and spatial repositioning at the 2-cell stage is further illustrated by the significantly stronger repression of minor zygotic genome activation (ZGA) genes in LADs (23 % minor ZGA gene-density), versus iLADs (15% minor ZGA gene-density) (Extended Data Fig. 2d and 2g). Between the 2- and 8-cell stage, differential gene expression also occurs in agreement with the spatial repositioning of genomic regions (Fig. 1e), and globally genes in LADs are lowly transcribed at the 2- and 8-cell stages compared to genes in iLADs (Fig. 1f). Examples of transitioning LADs at the 2-cell stage, and genes within them, are shown in Extended Data Fig. 2h. The less pronounced difference in gene activity between LADs and iLADs at the 2-cell compared to the 8-cell stage, may be attributed to a more open chromatin structure at the beginning of development9–11 (Extended Data Fig. 2b).
Remarkably, in fully-grown oocytes Dam-lamin B1 profiles appeared indistinguishable from the Dam controls (Fig. 1g and Extended Data Fig. 2i) and lack typical LADs for all oocyte samples including the single cells. In contrast, reproducible LAD-patterns are detected across all zygote samples (Extended Data Fig. 2j). These results suggest that fully-grown interphase oocytes are largely devoid of LADs. Taken together, our data uncover dynamic rearrangements of LADs in accordance with transcription changes, suggestive for a gene-regulatory role of LADs in the early embryo. Furthermore, because typical LADs are undetectable in oocytes we conclude that in the maternal pronucleus genome-lamina contacts are established de novo after fertilisation.
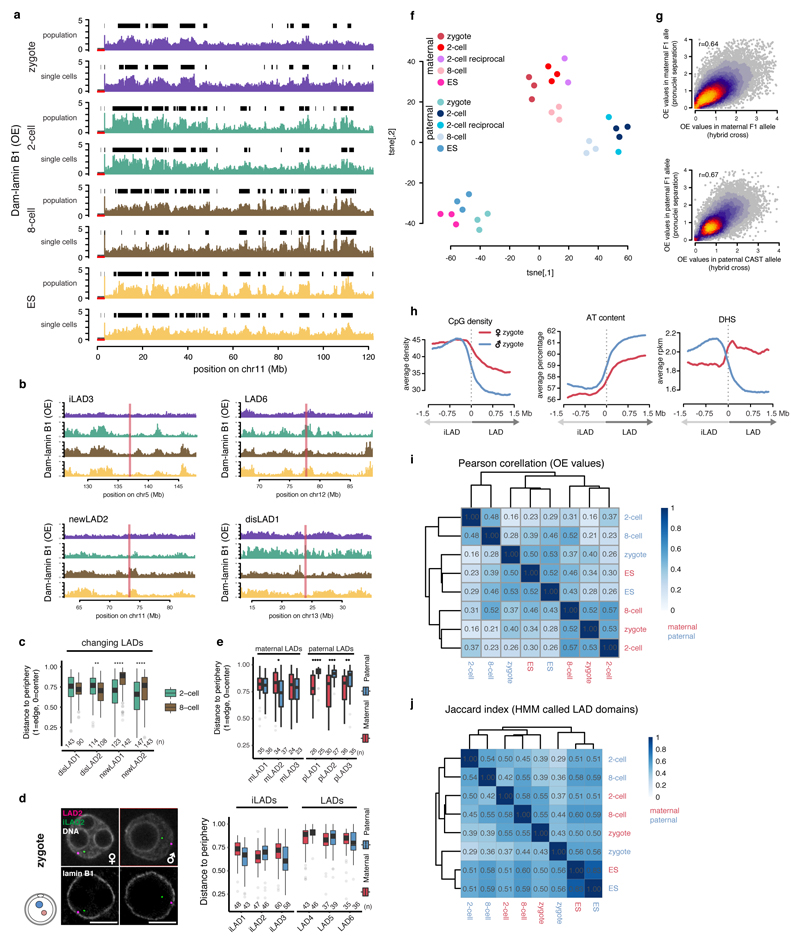
Intra-embryonic heterogeneity in mouse preimplantation embryos has been linked to cell fate decisions emerging in the blastocyst1. To address whether LADs display intra-embryonic heterogeneity, we converted single-cell DamID scores to binary contact frequency (CF) maps as previously described12. Single-cell and population average LAD profiles display high concordance (Extended Data Fig. 4a). We find that genome-NL contacts occur over a wide range of frequencies at all stages (Fig. 2a), yet between the individual cells of the three developmental stages, overall variability in CFs is comparable (Fig. 2b). Similarly, CF-distributions for zygotes, 2-cell and 8-cell embryos are consistent between variable and constant LADs (Fig. 2a). However, the spatial segmentation in ES cells appears more consistent as indicated by the lower coefficient of variation (CV) for LAD and iLADs (Fig. 2b). DNA-FISH coupled with 3D distance measurements on selected LADs and iLADs confirmed the DamID results (Fig. 2c and Extended Data Fig. 4b-c). Indeed, the mean distance values of DNA-FISH showed high positive correlation with the DamID CF scores (Fig. 2d). In addition, DNA-FISH confirmed the relocation of selected changing LADs between the 2- and 8-cell stage (Extended Data Fig. 4c). Thus, specific and robust NL-contacts form as early as the zygote and are largely maintained as development progresses.
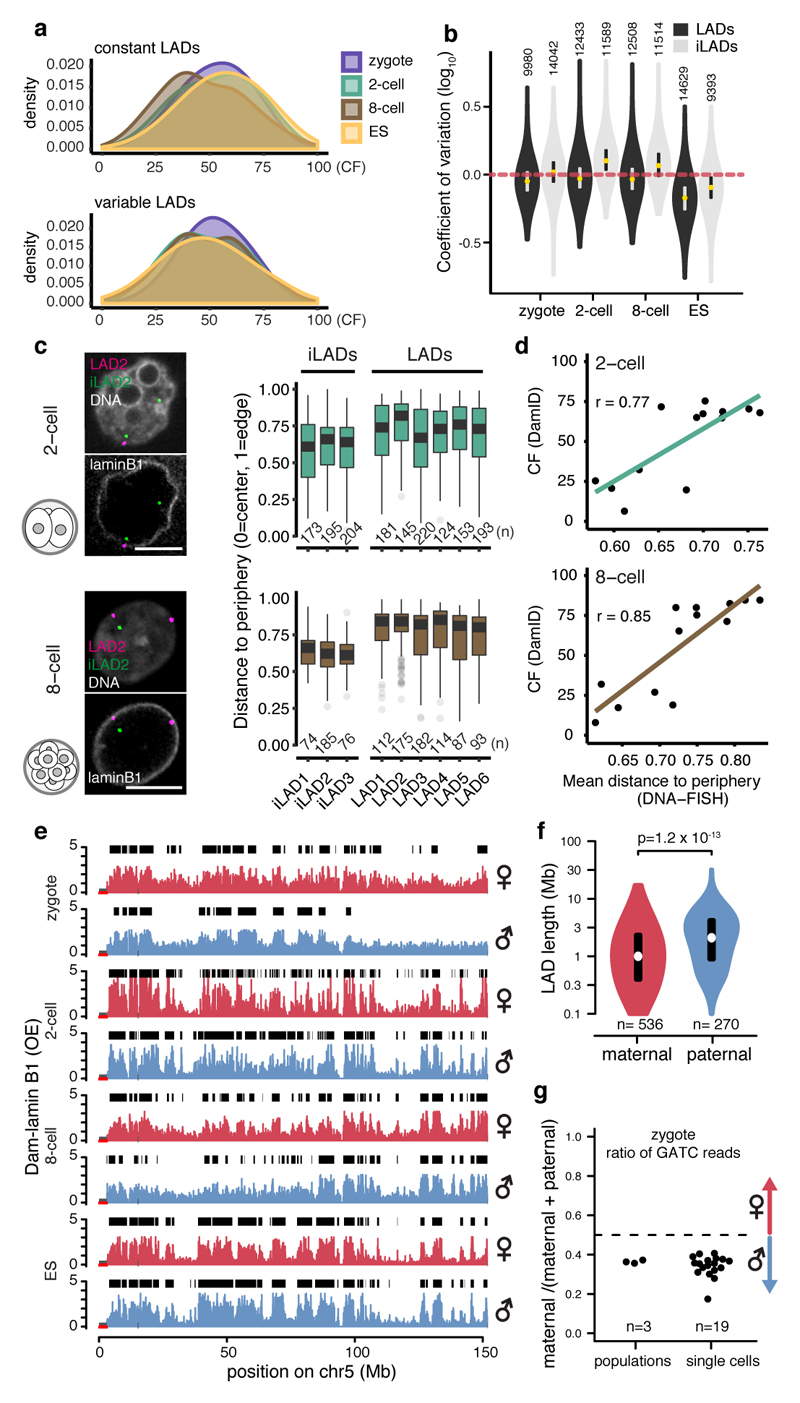
Figure 2. Single cells show consistent LAD patterning within the same developmental stages and LADs show parental differences.
a, Density distribution of contact frequencies (CF) across stages in constant (top) and variable (bottom) LADs. b, Coefficients of variation (CV) between LADs and iLADs. Violin plots show the 25th and 75th percentiles (black lines), median (circles) and the smallest/largest values at most 1.5 * IQR. n = number of 100-kb bins. c, 3D-DNA-FISH in 2-cell and 8-cell embryos, quantification shows position to the nuclear periphery. Scale bars: 5 μm. Box plots show the 25th and 75th percentiles (box), median (solid lines), the smallest/largest values at most 1.5 * IQR of the hinge (whiskers) and outliers (grey circles). n = number of DNA-FISH spots from at least 3 biological independent samples. d, Correlation between CF values from DamID and distance measurements from DNA-FISH. n = 13 DNA-FISH probes (black circles). r = Pearson’s r. e, Allelic Dam-lamin B1 profiles. Black boxes represent LAD domains. f, Distribution of allelic LAD domain length in zygotes. p = Wilcoxon rank-sum test p-value (two-sided). Violin plots show the 25th and 75th percentiles (black line), median (circles) and the smallest/largest values at most 1.5 * IQR. n = number of LADs. g, Allelic ratio of GATC counts in zygote Dam-lamin B1 samples.
Next, we used hybrid embryos to address whether parental differences exist in LADs. Notably, the paternal zygotic genome appears more defined, with broad domains as opposed to more fragmented patterns with fewer genome-NL contacts in the maternal pronucleus (Fig. 2e-g). LAD-regions determined by DamID are positioned with similar average distances to the nuclear periphery measured by DNA-FISH (Extended Data Fig. 4d). However, allele-specific DNA-FISH indicates that paternal-specific LADs associate with the NL more consistently than maternal-specific LADs (Extended Data Fig. 4e). Allelic differences persisted up until the 8-cell stage as revealed by t-SNE (Extended Data Fig. 4f), albeit much less pronounced than in the zygote. DamID on physically separated pronuclei from non-hybrid zygotes confirmed that our observations do not result from a genetic bias derived from different strains (Extended data Fig. 4f-g). Interestingly, unlike the paternal zygotic LADs, the maternal LADs are less enriched for typical LAD-features and even contain increased DNaseI hypersensitivity (Extended Data Fig. 4h). From the 2-cell stage onwards LADs showed reduced DNaseI hypersensitivity, which became more pronounced at the 8-cell stage, and in ES cells (Extended Data Fig. 2b). The Pearson correlation between the paternal zygote and 2-cell stage samples is 0.16; a marked contrast to the 0.53 for the maternal zygotic versus 2-cell stage (Extended Data Fig. 4i). Jaccard similarity coefficients confirmed these observations (Extended Data Fig. 4j). It therefore appears that the maternal genome-NL contacts initiate in the zygote and become reinforced at the 2-cell stage, while the paternal genome exhibits clearly defined LADs that are extensively rearranged during the zygote to 2-cell stage transition.
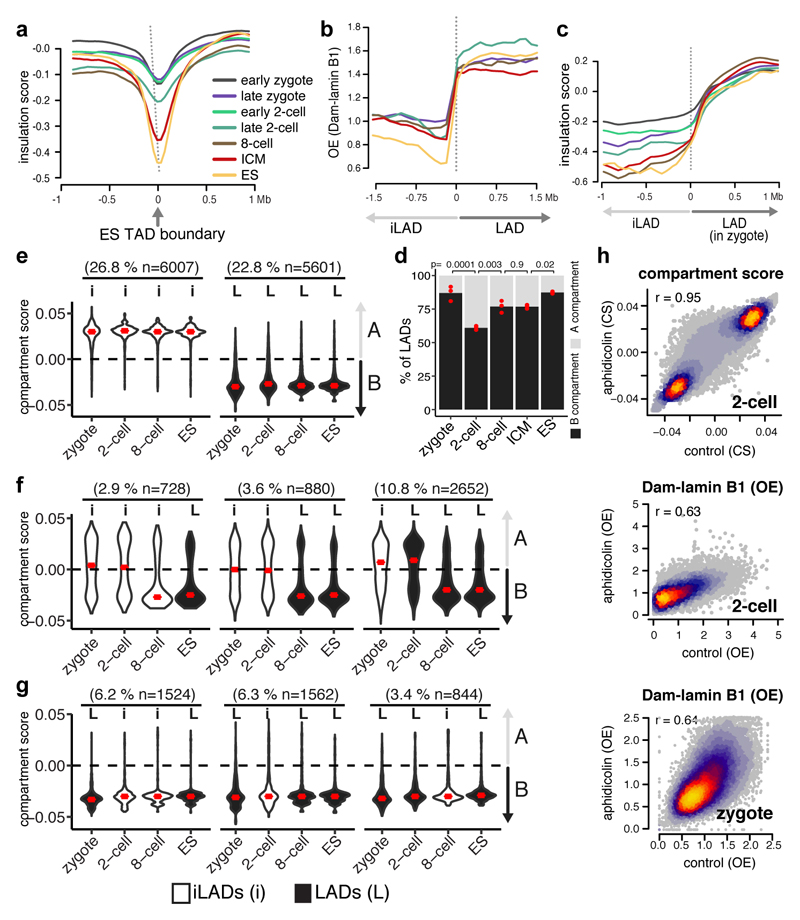
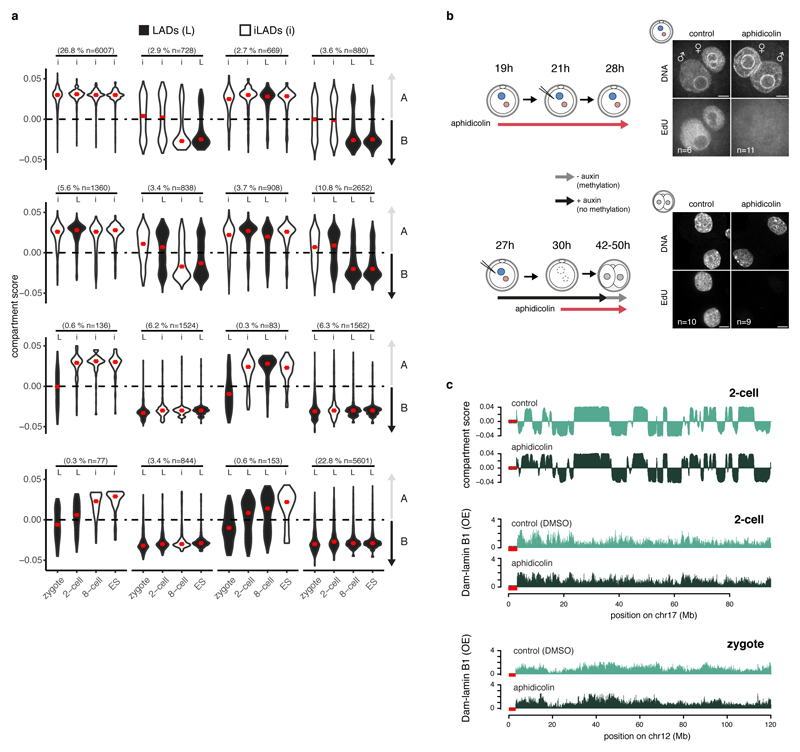
Whilst LADs are established immediately after fertilisation, TADs are largely absent in zygotes and gradually consolidate to form “mature” TADs at late cleavage stages13–14. To address the largely unexplored interdependency between spatial genome organisation and the establishment of chromatin topology, we analysed published Hi-C data13 and determined insulation scores in the zygote, 2-cell, 8-cell stage and ICM at TAD boundaries defined in ES cells. The insulation scores provide a measure of the overall level of contacts that occur over a given genomic region. Because only limited interactions occur between neighboring TADs, insulation scores are lowest at TAD boundaries15. As shown previously, TAD-boundaries become progressively insulated as development proceeds (Fig. 3a). In contrast, DamID-scores projected on per-stage LAD boundaries, reveals that LADs are already clearly defined in zygotes (Fig. 3b) and insulation scores progressively consolidate at the borders of zygotic LADs (Fig. 3c). These findings suggest that LADs may precede TAD establishment, and that chromatin scaffolding at the NL in the zygote may be directive for the formation of higher-order chromatin topology throughout early development. Unlike mature TADs, A and B compartments can be observed as early as the zygotic stage, albeit with higher compartment strength in the paternal genome13. The majority of LADs overlapped with B compartments in zygotes, 8-cell embryos, ICM and ES cells (Fig. 3d), in agreement with previous findings in ES cells16. However, unexpectedly, a significant portion of LADs in the 2-cell embryos occupies A compartments (39%). This prompted us to investigate whether LADs may help prime future B compartments. We determined compartments scores stratified for different patterns of LAD dynamics (Extended Data Fig. 5a). Constant iLADs and LADs respectively persist as A and B compartments throughout early development (Fig. 3e) which suggest that LAD and compartment formation occurs simultaneously for these regions (~50% genome coverage). In contrast, de novo 2-cell stage LADs that persist as LADs throughout early development (10.8% genome coverage), precede the establishment of B compartments (Fig. 3f). Thus, for certain genomic regions LADs may help prime the formation of B compartments, however, alternative scenarios exist (Fig. 3f). Notably, regions that constitute LADs in zygotes and dislodge from the NL at the 2- and/or 8-cell stage persist as B compartments throughout early development (Fig. 3g). This is intriguing, especially because NL-detachment of these regions during the zygote to 2-cell transition is associated with global transcriptional upregulation of these regions (Fig. 1e). Conversely, NL-associated regions at the 2-cell stage remain A-compartments, yet are associated with global gene-repression (Fig. 1e). These data suggest that LADs may be more directive in gene-regulation at this stage of development than the not yet fully consolidated TADs and compartments.
Figure 3. Replication unrelated formation of LADs is concordant with A/B compartments and precedes TAD structuring.
a, Insulation scores from Hi-C data plotted over ES TAD boundaries13. b, Dam-lamin B1 scores plotted over LAD boundaries. c, Insulation scores from Hi-C data plotted over zygotic LAD boundaries. d, Percentage of LADs overlapping A or B compartments. Bars represent the mean of three biological replicates indicated by red dots. p-values represent a post-hoc Tukey-test (two-sided). e, f, g, Violin plots of compartment scores for 100-kb genomic regions with different LAD dynamics. iLADs (‘I’) and LADs (‘L’) in the indicated stages and ES. Number in parentheses denote the percentage of the genome covered per category. Violin plots show the values at most 1.5 * IQR and median (red lines). Compartment scores are calculated based on 3 biological independent samples. n = number of bins. h, Genome-wide scatter plots of compartment scores (CS) and DamID scores in control and aphidicolin-treated zygotes and 2-cell embryos. n = 3 biological independent samples, r = Spearman’s rho.
Finally, we explored whether LADs, like TADs, are affected by blocking replication with aphidicolin13. Unlike TADs, zygotic and 2-cell stage LADs remained globally unaffected after aphidicolin treatment, although the patterns appeared less distinctive (Fig. 3h and Extended Data Fig. 5b-c). Similarly to LADs, analysis of published Hi-C data indicated that compartment formation is not affected by aphidicolin (Fig. 3h and Extended Data Fig. 5c). These results indicate that LADs and compartments are established independently of DNA replication. Collectively, our data support a model whereby scaffolding of the genome at the NL co-occurs with compartment formation but precedes TAD consolidation.
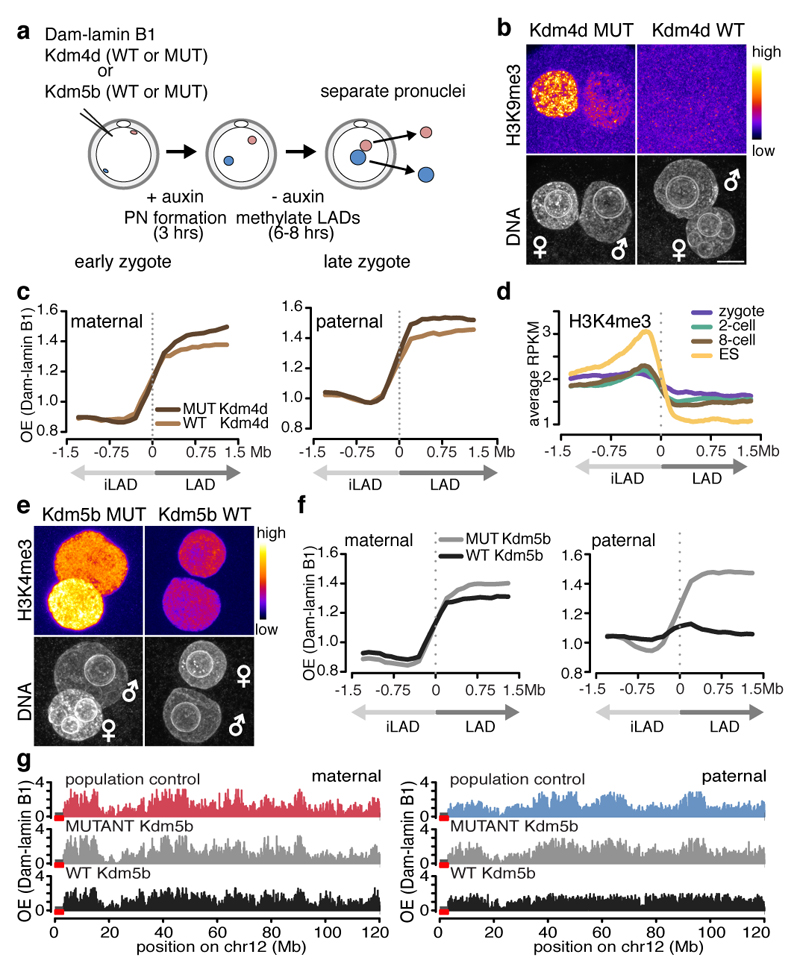
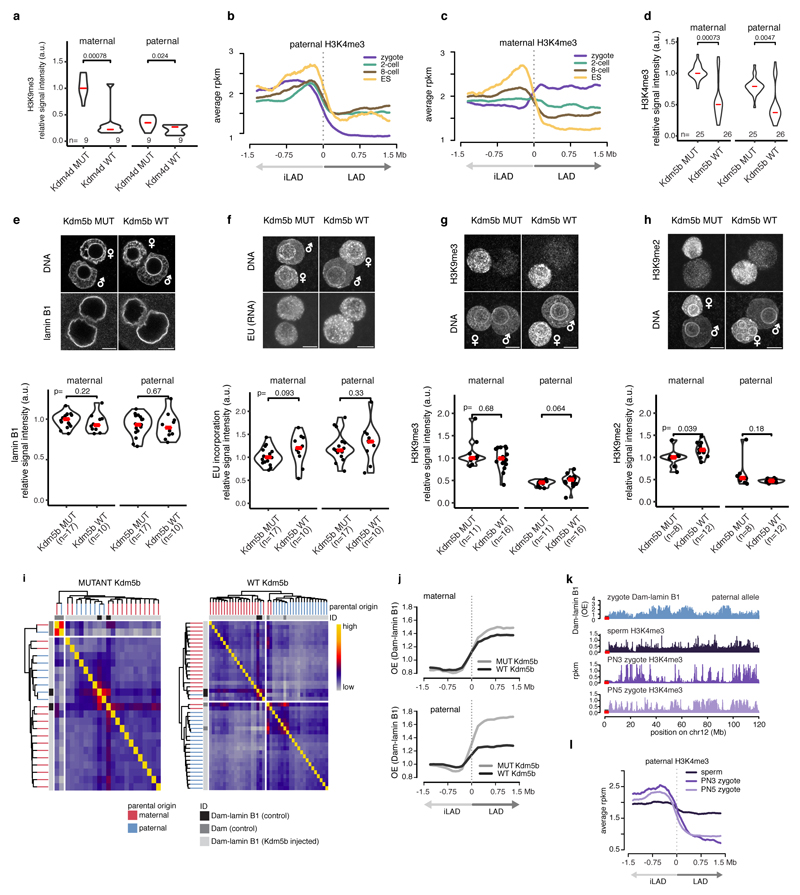
We next investigated the mechanism(s) underlying LAD formation in the zygote. Because LADs are enriched for H3K9me2/me3 in somatic cells7,17, we first asked if LAD establishment is dependent upon H3K9 methylation. For this, we expressed the H3K9 demethylase Kdm4d in zygotes and performed DamID (Fig. 4a). Remarkably, lowering global H3K9me2/3 levels via ectopic expression of Kdm4d (Fig. 4b and Extended Data Fig. 6a) had no gross effect on LAD structure (Fig. 4c).
Figure 4. Overexpression of Kdm5b histone demethylase abrogates paternal LAD establishment in the zygote.
a, Experimental design. b, H3K9me3 immunostaining in zygotes expressing wild-type and mutant KDM4D. c, Average Dam-lamin B1 signal at LAD boundaries in H3K9me3-manipulated embryos. Signal in maternal pronucei and paternal pronuclei. d, Average H3K4me3 signal at LAD boundaries. e, H3K4me3 Immunostaining of in zygotes expressing wild-type and mutant Kdm5b mRNA. Scale bars: 10 μm. f, Average Dam-lamin B1 signal at LAD boundaries. g, Dam-lamin B1 profiles on maternal and paternal alleles in non-manipulated controls or zygotes expressing KDM5BWT and KDM5B MUT.
The maternal chromatin harbours non-canonical ‘broad’ H3K4me3 domains, which are established during oocyte growth and persist until ZGA18–20. In contrast, paternal chromatin has significantly lower H3K4me3 levels at fertilisation21–22. LADs and iLADs are progressively demarcated by H3K4me3 levels as development proceeds (Fig. 4d), yet, only the paternal genome displays clear alternating patterns of H3K4me3 and LAD domains (Extended Data Fig. 6b-c). To address a potential role for H3K4me3 in LAD establishment, we expressed the H3K4me3 demethylase Kdm5b and performed DamID on physically isolated pronuclei (Fig. 4a). Kdm5b expression led to a pronounced reduction of H3K4me3 in both pronuclei compared to controls (Fig. 4e and Extended Data Fig. 6d). Importantly, lamin B1 localization, H3K9me2/me3, and global transcriptional activity were unaffected upon Kdm5b expression (Extended Data Fig. 6e-h). Remarkably, expression of the wild-type Kdm5b, but not the catalytically inactive mutant, resulted in a near complete erasure of LAD structure of the paternal genome, with little or no effect on maternal LADs (Fig. 4f-g). The erasure of LAD profiles was consistent across experiments and individual pronuclei analysed (Extended Data Fig. 6i). The same experiment performed in hybrid mice confirmed these findings (Extended Data Fig. 6j). We conclude that Kdm5b is critically involved in de novo establishment of LADs in the paternal pronucleus.
In summary, we show that LADs are established immediately after fertilisation without inheritance from the maternal germline. Paternal inheritance cannot be excluded, however, the de novo acquisition of H3K4me318,21 and the abrogation of LADs upon Kdm5b expression in the male pronucleus suggests LADs are formed de novo. The absence of H3K4me3 demarcation of LADs in sperms supports this (Extended Data Fig. 6k-l). De novo H3K4 methylation may support LAD formation or, an intact euchromatin compartment may be important for segregating heterochromatin to the NL. However, alternative mechanisms that are mediated by KDM5B demethylase activities cannot be formally excluded. Additional investigations are required, to further dissect the role of KDM5B and/or H3K4me3 in LAD organization. Our analyses indicate that LAD formation precedes TAD consolidation and may help prime B compartments for certain genomic regions. For the future, it will be important to further dissect the temporal and molecular interdependence of the different levels of nuclear organization. Overall, our work sheds light on the principles behind the establishment of nuclear organisation and higher-order chromatin structure during early mammalian development.
Methods
Oocyte and embryo collection and manipulation
Experiments with animals were carried out according to valid legislation in France and under the authorization of the Com'eth Institute of Genetics, Molecular and Cellular Biology ethical committee and in compliance with the local government (Government of Upper Bavaria). Preimplantation embryos were collected from 5-8 weeks old F1 (CBAxC57BL/6J) females mated with CAST/EiJ males for hybrid crosses and with F1 males for non-hybrid crosses. Ovulation was induced by injecting 10 IU PMSG (IDT Biologika Gmbh) and then hCG (MSD Animal Health) 46-48 hours later. GV oocytes were collected 44-48 hours after PMSG injection. Reciprocal crosses (CAST/EiJ females mated with F1 males) were performed without inducing ovulation. Oocytes were cultured in IBMX containing M16 while embryos were cultured in KSOM drops under paraffin oil (Sigma). The sample size was not chosen. In each experiment embryos from 4-8 female mice were pooled and randomly allocated to experimental groups. No blinding was done, since no manual assessment of images or experimets was performed. For DamID, an mRNA mixture containing 250 ng/μl TIR1, 50 ng/μl membrane-EGFP and embryonic stage dependent concentrations of AID-Dam-LaminB1 or AID-Dam were injected into the cytoplasm of oocytes and embryos. 50 ng/μl AID-Dam-lamin B1 mRNA injected from zygote to 2-cell stage – 5-fold higher mRNA concentration than otherwise used to map LADs at the 2-cell stage. To methylate LADs only at the stages of interest we washed the embryos into auxin-free media for 6-8 hours at the late-S, G2 phases of the cell cycle. Oocytes (48 hours post PMSG) and zygotes (21 hours post hCG) were isolated and injected with 5 ng/μl AID-Dam-LaminB1 or 20 ng/μl AID-Dam and kept in auxin free KSOM for 6-8 hours to methylate LADs or accessible regions, respectively. We performed additional DamID experiments in oocytes using 20 and 50 ng/μl of AID-Dam-LaminB1 mRNA, and obtained similar results to those obtained using only 5ng/μl (GSE112551). For DamID at 2-cell stage late-zygotes (27-28 hours post hCG) were isolated and injected with 10 ng/μl AID-Dam-LaminB1 or 40 ng/μl AID-Dam in auxin (500μM) containing media. Auxin was removed at 2-cell stage for 6-8 hours (from 42 to 48-50 hours post hCG). For DamID at 8-cell stage late-2-cell embryos (46-48 hours post hCG) were isolated and injected with 20 ng/μl AID-Dam-LaminB1 or 40 ng/μl AID-Dam in auxin containing media. Auxin was removed at 8-cell stage for 6-8 hours (from 66 to 72-74 hours post hCG). Afterwards, the zona pellucida was removed by treatment with 0.5% pronase in M2 at 37 °C and the polar bodies were mechanically separated from the embryos and discarded. Either pools of 20 blastomeres or single blastomeres were placed into 2 μl of DamID buffer (10mM TRIS acetate pH 7.5, 10mM magnesium acetate, 50mM potassium acetate) and stored at -80 °C until downstream processing. For the ICM/TE samples, 4-cell embryos were injected with 100ng/ul AID-Dam-lamin B1 or Dam mRNA at 60-62 hours post hCG injection. IAA was washed out at 90 hours post hCG when blastocysts started to cavitate. Embryos were treated with pronase at 96 hours and incubated in Fluorospheres (F8811 – Sigma) to mark TE cells for 2 minutes. Residual Fluorospheres were washed away and embryos were kept in Calcium-free M2 medium for 25 minutes followed by mechanical disaggregation. Fluorosphere positive TE cells were separated from negative ICM cells under a fluorescent microscope and placed into DamID buffer.
For DamID in embryos expressing Kdm5b or Kdm4d, early zygotes from non-hybrid crosses (18 hours post hCG) were injected with 2 μg/μl wild-type or mutant Kdm5b or 1.8 μg/μl wild-type or mutant Kdm4d, and 5 ng/μl AID-Dam-LaminB1, and 250 ng/μl TIR1 and 50 ng/μl membrane-EGFP coding mRNA. Zygotes were kept in auxin containing KSOM for 3 hours to prevent adenine methylation during pronucleus formation. At 21 hours post hCG auxin was removed for 6-8 hours to allow methylation. To separate pronuclei, the PN5 zygotes were transferred to M2 media containing 10 μg/ml cytochalasin B (Sigma-Aldrich). The zona pellucida was cut with a Piezo driven micromanipulator and one of the pronuclei was isolated into M2 drops. The pronuclei were distinguished based on their size and their relative position to the second polar body. The remaining embryos containing a single pronucleus were treated with pronase to remove the zona pellucida and the polar bodies were discarded. The karyoplasts and the single pronuclei containing embryos were frozen in DamID buffer as above. For DamID in replication inhibited 2-cell embryos, late zygotes (26-28 hours post hCG) from hybrid crosses (CBAxC57BL/6J females mated with CAST/EiJ males) were injected with 10 ng/μl AID-Dam-LaminB1, 250 ng/μl TIR1 and 50 ng/μl membrane-EGFP coding mRNA and kept in auxin containing media to prevent Dam activity in the zygote. Embryos were washed into aphidicoline (3 μg/ml) containing media when reaching the first metaphase. Auxin was removed from 42 to 48-50 hours post hCG to allow methylation of LADs in the late 2-cell stage.
Plasmid construction and mRNA production
The cDNA encoding wild-type Kdm5b was obtained from Addgene (86398)20. To generate a catalytically inactive version of Kdm5b the H499A mutation was introduced by site directed mutagenesis. The in vitro transcription plasmids containing the wild-type and mutant Kdm4d were obtained from Addgene (61553, 61554)25. mRNA was in vitro transcribed with T7 or T3 mMESSAGE mMACHINE kits (Ambion) and purified by LiCl precipitation. All plasmids generated in this study are available at Addgene under “Torres-Padilla lab plasmids”.
Immunofluorescence
Embryos were treated with 0.5% pronase in M2 to remove zona pellucida at 37 °C, washed in PBS and fixed in 4% PFA for 15 minutes at room temperature. After permeabilizing in 0.5% Triton-X 100 in PBS for 20 minutes, embryos were kept in blocking buffer (3% BSA in PBS) from one hour to overnight. Embryos were incubated overnight in primary antibody mixes (Supplementary Table 1) diluted in blocking buffer, washed three-times in PBS and stained with secondary antibodies conjugated with (Alexa 488, Alexa 568, Alexa 594 or Alexa 647) in blocking buffer for one hour. After washing three-times in PBS embryos were mounted in Vectashield containing DAPI. Lamin B1 is also known to be expressed in oocytes and blastocysts, as assessed by western blot23. For visualising global transcription, zygotes were pulsed with 50 μM EU for one hour (26-27 hours post hCG) and visualised with the Click-iT RNA Alexa Fluor 594 Imaging Kit according to the manufacturer’s instructions (Thermo Fischer). For visualising global DNA replication, zygotes or 2-cell embryos were pulsed with 10 μM EdU and visualised with the Click-iT EdU Alexa Fluor 594 Imaging Kit according to the manufacturer’s instructions (Thermo Fischer).
DNA-FISH
DNA-FISH was performed as described previously, using a protocol that preserves 3D information26. BACs were ordered from BACPAC or RIKEN DNABank (Supplementary Table 1) and purified with NucleoBond® Xtra Midi Plus kit (Macherey-Nagel). BACs were nick translated with 5-TAMRA, Atto594, Atto647N conjugated dUTPs according to the manufacturer’s instructions (Roche). To combine nuclear envelope staining with DNA-FISH, immunostaining was performed with mAb414 (1:1000) as described above, followed by postfixation in 2% PFA for 10 minutes. Next, embryos were washed in 0.5% Triton-X 100 for 10 minutes and treated with HCl solution (0.1N HCl, 0.5 Triton-X 100, and 1 mg/ml PVP in water) for 90 seconds, washed into prehybridization buffer (50 % formamide, 1 mg/ml PVP, 0.05% TritonX, 0.5 mg/ml BSA) and incubated in it at 37 °C for one hour. Embryos were transferred into drops of 0.2 µl hybridization buffer (prehybridization buffer containing 1 µg/µl mouse Cot-1 DNA) under mineral oil, denatured at 80 °C for 10 minutes and incubated at 37 °C for one hour. Embryos were transferred into drops of 0.2 µl hybridization buffer containing a mixture of three probes, each at 5 ng/μl which were previously denatured at 80 °C for 10 minutes under oil. After overnight hybridization at 37 °C, embryos were washed twice in 2x SSC, 0. Triton-X 100, 1 mg/ml PVP at RT followed by washing three times 10 minutes in in 2x SSC, 0. Triton-X 100, 1 mg/ml PVP at 55 °C and mounted in Vectashield containing DAPI on slides with spacers (Grace Bio-Labs SecureSeal) to preserve 3D structure.
Imaging and analysis
Microscopy images were acquired on a Leica SP8 confocal microscope equipped with a Plan Apochromat 63x/1.4 oil objective at 1.5 micron z steps for immunofluorescence and at 0.3 micron z steps for DNA FISH. H3K4me3, LaminB1 and EU intensity was quantified in the nuclei defined by creating masks on the DAPI staining using a custom made Icy protocol. DNA FISH spots were identified and their distance was measured relative to the DAPI mask periphery using another custom made Icy protocol. The centre of the DAPI mask was defined as 0 and the FISH spots location along the vector from the centre to the periphery defined as 1 was determined. Immunofluorescence signal intensities of all experimental groups were normalized to the median of the control group’s (KDM5B mutant) intensity separately for each biological replicate. The differences in signal intensity and FISH spot distance were subjected to Wilcoxon signed-rank test between groups or stages of development. For each stage between 71 to 220 FISH spots were analysed.
Hi-C data analysis
Hi-C data analysis. Data for untreated embryos were used from GSE82185, for the aphidicolin treated and their control 2-cell embryos from PRJCA000241. Raw files from all biological replicates were pooled and analyzed with HiC-Pro (version 2.10.0)27 as described in13 but aligning to the mm10 mouse reference genome. Compartments were called using the HiTC package27. ICE normalized 100-kb interaction matrices were binned with a bin size of 500-kb and a step size of 100-kb. Observed/expected matrices were used to generate correlation matrices and perform principal component analysis. A/B compartments were defined by the first principal component and gene density. TADs and insulation scores were calculated as described4.
Cell culture
F1 hybrid 129/Sv:Cast/Eij mouse embryonic stem cells27 were cultured at 37°C, 5% CO2 on primary mouse embryonic fibroblasts (MEFs), in Glasgows minimum essential medium (G-MEM; Gibco #21710025) supplemented with 10% fetal bovine serum (FBS; Sigma #F7524), 1% PEN/STREP (Gibco #15140122), 1% GlutaMAX (Gibco #35050038), 1% non-essential amino acids (Gibco #11140035), 1% sodium pyruvate (Gibco #11360039), 143 μM β-mercaptoethanol (Sigma #M6250) and 1:1000 human leukemia inhibitor factor (LIF; in-house production). F1 hybrid 129/Sv:Cast/Eij mouse embryonic stem cells were not authenticated; they were tested negative for mycoplasm
Generating cell lines
Stable clonal Dam and Dam-lamin B1 lines were created by transfection of EF1alpha-Tir1-neo with hPGK-AID-Dam-lamin B1 or hPGK-AID-Dam plasmids in a ratio of 1:5 plasmids with Effectene (Qiagen #301427). Clones were selected with 250 μg/ml G418 (Thermofisher #10131035) and selection of the clones was based on methylation levels as determined by DpnII-qPCR assays as previously described7. To reduce the background methylation levels in the presence of 1.0 mM indole-3-acetic acid (IAA; Sigma #I5148), we transduced the selected clones of both AID-Dam-lamin B1 and Dam with extra hPGK-Tir1-puro followed by selection with 0.8 μg/ml puromycin (Sigma #P8833-10mg). Positive clones were screened for IAA induction by DpnII-qPCR assays and DamID PCR products7.
DamID induction and harvesting
Expression of AID-Dam and AID-Dam-lamin B1 was suppressed by culturing the cells in the presence of 1.0 mM IAA for 48 hours. DamID was induced by IAA washout 12 hours prior to harvesting. 12 hours after IAA washout, cells were collected in G-MEM supplemented with 10% FBS and 1% PEN/STREP and stained with 10ug/ml Hoechst 34580 (Sigma #911004450) for 45 minutes at 37 °C. Single cell or 20-cell populations were sorted in 96-well plates at G2/M phase of the cell cycle based on the DNA content histogram.
Single cell DamID
We sequenced 3 independent population samples (of 15, 20 or 24 cells) for zygotes, 2- and 8-cell stage, respectively, and a total of 327 single cells for all stages. Single cells or populations of cells were manually sorted in 8-well PCR strips in 2 μl of DamID buffer (10mM TRIS acetate pH 7.5 (Sigma #T1258); 10mM magnesium acetate (Sigma #63052); 50mM potassium acetate (Sigma #95843); 2.01% Tween-20 (Sigma #P2287). 1 μl of lysis buffer with proteinase K (10mM TRIS acetate pH 7.5 (Sigma #T1258); 10mM magnesium acetate (Sigma #63052); 50mM potassium acetate (Sigma #95843); 2.01% Tween-20 (Sigma #P2287); 2.01% Igepal (Sigma #I8896) and 2.01mg/ml proteinase K (Roche #03115828001) was added to the samples, followed by proteinase K digestion at 42 °C for 12 hours in a thermoblock with heated lid. Proteinase K was inactivated by heating the samples for 20 minutes at 80 °C. In the following steps, reagents were added with an Eppendorf Multipipette Plus mounted with a 0.1 ml Combitip (Eppendorf #0030089405). The surface of the reaction volume was never touched by the pipette tip. Genomic DNA (gDNA) was digested for 8 hours by the addition of 7 μl of DpnI reaction mix (0.1 μl DpnI (10U/μl, New England Biolabs #R0176L); 0.7 μl 10x One-Phor-all-buffer plus (100mM TRIS acetate pH7.5; 100mM magnesium acetate; 500mM potassium acetate) and 6.2 μl nuclease free H2O) and incubation at 37 °C in a PCR machine, followed by heat inactivation at 80 °C for 20 minutes. Adaptor ligation was performed by the addition of 10 μl ligation mix (2 μl 2x T4 ligation buffer; 0.5 μl T4 ligase (5U/ul, Roche #10799009001); 0.05 μl 50μM double-stranded DamID adapter29 and 7.3 μl nuclease free H2O) and incubation in a PCR machine at 16 °C overnight. Heat inactivation at 65 °C for 10 minutes the next day was followed by PCR amplification by the addition of 30 μl PCR mix (10 μl 5x MyTaq Red reaction buffer (Bioline #25043), 1.25 μl PCR barcoded primer (50 µM) NNNNNNBARCODGTGGTCGCGGCCGAGGATC (Supplementary Table 2), 0.5 μl MyTaq DNA polymerase (Bioline #25043) and 18.25 μl nuclease free H2O). The PCR primer carries 6 random nucleotides at the 5' end to meet the Illumina software requirements of generating reads with diverse starting sequences and a 6 nucleotides sample barcode (Supplementary Table 2). The thermal cycling scheme is as follows:
| Step | Denature | Anneal | Extend | |
| 1 | 72 0C for 10 min | |||
| 2 | 94 0C for 1 min | 65 0C for 5 min | 72 0C for 15 min | |
| 3-6 | 94 0C for 1 min | 65 0C for 1 min | 72 0C for 10 min | |
| 7-35* | 94 0C for 1 min | 65 0C for 1 min | 72 0C for 2 min | |
33 for the population samples
Of the resulting PCR product 8 μl was used for standard 1% agarose gel electrophoresis for analytical purpose and estimation of DNA concentration. All samples were pooled and prepared for Illumina sequencing.
Single-cell DamID Illumina library preparation and sequencing
Of 300 ng purified PCR product the 3’ or 5’ overhanging ends were blunted in a 50 μl reaction following the manufacturer’s instructions (End-It DNA End-Repair Kit, Epicentre #ER81050). The blunted DNA samples were again purified using the PCR purification colomns of Qiagen and eluted with 26 μl nuclease free H2O. Next, a 3’ adenine was added by incubation for 30 minutes at 37 °C in a 50 μL reaction mix (1x New Engeland Biolabs restriction buffer 2, 200 μM dATP (Roche #11051440001) and 25 units of Klenow 3' → 5' exo– (New Engeland Biolabs #M0212M). After heat inactivation at 75 °C for 20 minutes, the DNA was purified with Agencourt AMPure XP beads (Beckman Coulter #A63881). A 1.8 x volume of beads over DNA sample was used, manufacturer’s instructions were followed and the DNA was eluted with 20 µl of nuclease free H2O. To the purified DNA the Illumina indexed Y-shaped adapters (TruSeq Nano DNA LT Library Prep Kit #FC-121-4402) were then ligated for two hours at room temperature in a 40 μl reaction mix (4μl 10x T4 ligation buffer, 0.5 μl T4 ligase (5U/ul) Roche #10799009001, 2.5 μl Illumina adapter, with nuclease-free H2O added to 40 μl final volume). Next, the T4 ligase reaction was heat inactivated at 65 °C for 10 minutes followed by 2 times DNA purification with 1.8 x volume followed by 1.2 x volume AMPure beads as described for the previous step. For the addition of the Illumina index primers a PCR reaction was performed with the DNA from the previous step in a 20 μl MyTaq red DNA polymerase PCR reaction mixture (10 μl 2x MyTaq reaction mixture (Bioline #BIO21110), 1 μl 2.5 μM Illumina oligo mix, nuclease-free H2O till a final volume of 20 μl). The DNA was amplified for 6-8 PCR amplification cycles (94 °C 1 minute; 94 °C 30 seconds, 58 °C 30 seconds and 72 °C for 30 seconds for 9 cycles and 72 °C for 2 minutes) after which 5 μl of each sample was analyzed by agarose gel electrophoresis. For Illumina multiplex sequencing typically 4 to 10 separate libraries of each 20-50 single cells were mixed in approximate equimolar ratios as judged from the agarose gel image. The pooled sample was subjected to a Qiagen PCR column purification and subsequent AMPure bead purification with 1.6 x volume of beads over DNA sample before it was used for sequencing.
Processing of single-cell DamID sequencing reads
Basecalling and filtering were performed using standard software of the Illumina HiSeq 2500. The total number of raw and final GATC reads are shown in Supplementary Table 3. Sequenced 151bp reads were parsed in order to obtain the gDNA for downstream analysis. When present, the first 6 random bases were discarded; subsequently the reads were demultiplexed and the 15 bp of adapter trimmed using custom scripts and cutadapt30. The pre-processed reads were then mapped to the mm10 (or alternative genome assemblies) using bwa (version bwa-0.7.12)31 aln with default parameters. Reads aligning to the genome with quality score below 25 were discarded. Please note that we applied a stringent threshold for the quality of the DamID datasets throughout all of the experiments, which we set at minimum 30.000 unique reads. Therefore, all the libraries that did not reach this QC were discarded from the analysis. The computation of Observed over Expected (OE) value per bin was carried out as described12. Briefly, reads that precisely flanked an annotated GATC site were associated with GATC-fragments and kept for downstream analysis. In order to compare population samples to single cell samples, multiple reads aligning to the same position were counted as one and subsequently aggregated in genomic segments of 100kb in order to determine the experimentally Observed value. This was then divided by the Expected value per 100kb bin. The expected value was generated by in-silico determining potential DamID-seq reads of the same length as the experimental data (~131 bp), aligning them to the mm10 genome assembly and selecting them based on the same filtering applied to the experimental data and aligned to GATC-fragments (see above). The final OE value per 100 kb bins was computed by dividing the ratio of the two counts (Observed, Expected) by the total number of observed reads per bin. LAD domain calling was performed on the average population replicates for each stage in (parental or non-allelic) OE values calculated using a two-state hidden Markov model (HMM)32, which allows the classification of each 100kb segment as LAD or iLADs.The computation of CF scores was carried out as described in12, by binarization of the OE values and subsequent summation of the CF score across the single cell samples per 100kb bin.
Identification of parental-specific reads
For hybrid samples C57BL/6 x CAST/EiJ or 129/Sv x CAST/EiJ, CAST/EiJ and 129/Sv genomes were de novo compiled by nucleotide substitution of strain specific SNPs using the SNPsplit_genome_preparation tool (version SNPsplit_v0.3.0/) (http://www.bioinformatics.babraham.ac.uk/projects/SNPsplit/) in the original mm10 genome assembly. The database of annotated SNPs between different mouse strains was obtained from ftp://ftp-mouse.sanger.ac.uk/current_snps/strain_specific_vcfs/. The reads were separately aligned to the parental (mm10 or hybrid) genomes using the above described parameters. The edit distance of the alignments of pre-filtered reads (quality score >= 25) was compared to the two genomes. The reads aligning with the lowest edit distance were assigned to the appropriate parental genome. The reads aligning with equal edit distance between the parental genomes were not assigned to the parental genomes but were kept for “non-allelic” profiles.
Comparative genomics to published datasets
Low-input ChIP data for H3K4me3 and DNA-hypersensitivity data were respectively obtained from Gene Expression Omnibus (GEO) accession numbers GSE71434 and GSE76642. Alignment was carried out as described for the DamID sequencing reads. Picard tools (version picard-tools-1.130) (http://picard.sourceforge.net) was used to remove PCR duplicates. Additionally, as the H3K4me3 arises from a mouse mixed genetic background C57BL/6N x PWK, the assignment of reads belonging to parental genomes was carried out as described for the DamID libraries. Normalisation in read per million (RPM) was then carried out in fixed genomic windows of 5kb or 100kb to allow direct comparison with DamID data. Gene expression data was obtained from GEO accession GSE71434. The samples were aligned to the mm10 genome assembly using hisat2 (version hisat2-2.0.3-beta33 with default parameters. Reads mapping with quality score lower than 200 were discarded. htseq-count (version 0.6.0)34 was then used to assign the mapped reads to a transcriptional model file (gencode.vM9.annotation.gtf) obtained from (https://www.gencodegenes.org). Only genes annotated in the refFlat (http://hgdownload.soe.ucsc.edu/goldenPath/mm10/database/) database were considered for downstream analysis. The reads per kilobase per million (RPKM) were calculated for each gene by normalising the total number of mapped reads per gene by the gene length in kb and sample size. A gene TSS was considered to be located within a LAD when the region surrounding its TSS (+/- 250 bp) was located within a LAD. edgeR35 was used to perform differential expression analysis between consecutive stages. A given gene was considered significantly changing if the associated FDR corrected p-values is lower or equal to 0.01. Maternally-specific genes as described by Park et al 201336 were depleted from this analysis. For the expression of minor zga genes, the datasets identified by Park 2013 was used (clusters 5,6,7,8). Minor zga genes undergo downregulation by the 4-cell stage. The minor zga gene dataset was ommitted for the differential expression analysis to compare the 2-cell stage and 8-cell stages in Figure 1g. For the heatmap in Extended Figure 2e, we defined the expression of the genes inside the LADs using the scRNA-Seq data sets of Deng, Q., et al 201437. The counts were quantified using the kallisto software (0.44.0)38. An RPKM normalization was applied to the raw counts using the R package edgeR, only those genes with counts >1 RPKM on average were considered as expressed. We calculated the mean expression of each gene per developmental stage and their z-scores were computed in order to visualize the relative gene expression between the stages, these values were plotted as a heatmap using the R package Pheatmap. For the comparison with previously obtained DamID data on LADs in ES cells24, the processed data was obtained from GEO accession GSM426758 in the form of log2 Dam-lamin B1 scores. In order to directly compare this dataset to the data generated in our study, average log2 Dam-lamin B1 scores were calculated for fixed genomic windows of 100-kb. HMM was then applied in order to call LADs in the different cell types obtained from GSM426758. 100-kb regions were finally defined as “cLADs” if they were classified as LADs in each of the individual cell types described in12. For Fig. 1g, genes were considered significantly changing if the associated FDR (false discovery rate) corrected p-values is lower or equal to 0.01. edgeR35 was used to perform differential expression analysis between consecutive stages. Only the log2 fold change of significantly differentially expressed genes between zygote and 2-cell, and 2-cell and 8-cell stages were included in the analysis. Fig. 1g displays the median log2 FC for changing LADs between zygote and 2-cell stage and 2-cell and 8-cell stage. Maternally-specific genes as defined by in reference 38, were removed from this analysis to prevent confounding effects due to mRNA degradation. For similar reasons, the minor ZGA gene-dataset based on reference 38 (Clusters 5, 6, 7 and 8) was omitted for the differential expression analysis between the 2-cell and 8-cell stages. This was decided because these mRNAs gradually decrease from the 2-cell to 8-cell transition but are still relatively highly abundant at the 2-cell stage compared to the 8-cell stage. Because of the unequal abundance of minor ZGA mRNA, gene-expression appears artificially inflated at the 2-cell stage. As a result, gene-expression at the 8-cell stage would appear overall downregulated compared to the 2-cell stage if not corrected for minor ZGA genes.
Statistical testing
Statistical tests were computed in order to test the correlation between datasets and/or the significance of specific features. The R programming language (versions R-3.1.2 and R-3.4.0)39 was widely used with this purpose. In general, before applying any test, the normality of the distributions was tested by the Anderson-Darling Normality test (R Package nortest). The computation of tests such as the Wilcoxon rank-sum test (two-sided, unless otherwise specified) or Pearson and Spearman correlation coefficients were carried out using the core statistics functions in R. Pair-wise Pearson correlation coefficients were calculated across conditions on the averaged, binned OE data and visualized as heatmaps with hierarchical clustering (Extended Data Fig. 4h).
Extended Data
Extended Data Figure 1. Establishment of DamID in mouse preimplantation embryos.
a, Immunostaining of lamin A/C and lamin B1 in oocytes, zygotes, 2-, 8-cell embryos and blastocysts. Scale bars represent 5 μm. Levels were quantified previously on Western-blot in23. Experiments were repeated at least 3 times. b, m6A-Tracer signals with/without auxin. Scale bars represent 20 μm. Experiments were repeated at least 3 times with similar results. c, PCR smears amplified from ten 2-cell embryos injected with varying amounts of Dam-lamin B1 mRNA and developed in the presence or absence of auxin. Experiments were repeated at least 3 times with similar results d, Development to the blastocyst in the absence/presence auxin. e, Development to the blastocyst of zygotes injected with Dam or Dam-lamin B1 mRNA in the absence or presence of auxin. f, Dam-lamin B1 profiles on chromosome 11. Black boxes represent LAD domains. g, Hierarchical clustering of Dam-lamin B1 population (n = 3) and average single cell profiles in oocyte (n = 56), zygote (n = 19), 2-cell (n = 47) and 8-cell stage (n = 42). n = number of biological independent samples. h, Comparison of genomic profiles obtained by DamID-seq (this study) to previous DamID on micro-arrays. Black boxes represent LAD domains called by Hidden-Markov modelling. r = Spearman’s rho. i, Venn diagram showing overlap between LADs in ES cells, ICM and TE.
Extended Data Figure 2. Features of embryonic LADs, analysis of gene expression and oocyte DamID.
a, Chromosome plots comparing Dam and Dam-lamin B1 DamID profiles in 2-, 8-cell embryos and ES cells. Genome-wide comparison scatter plots of Dam and Dam-lamin B1 scores per 100-kb bin in 2-, 8-cell embryos and ES cells (right panel). DamID scores were calculated based on n = 3 independent biological replicates. r = Spearman’s rho. b, Average CpG density, AT content and DNaseI hypersensitivity sites (DHS)10 over LAD boundaries. c, Percentage overlap of LADs in embryos with constitutive LADs (associating with the lamina in ES cells, astrocytes, NPCs and MEFs)8,24. d, CpG density, A/T content, DNase hypersensitivity (DHS) and minor ZGA gene expression at the 2-cell stage in genomic regions reorganising in respect to the nuclear periphery during the zygote to 2-cell stage transition. Violin plots show the 25th and 75th quantiles (white boxes), median (black horizontal line) and values at most 1.5 * IQR. For CpG density, A/T content and DHS n= number of bins; for minor ZGA expression n = number of minor ZGA genes. Wilcoxon rank-sum test p-values shown (two-sided). DHS data from ref.13, expression data from ref. 19. e, Percentage overlap of the de novo iLAD at the 2-cell stage with constitutive LADs (as defined for c). f, Heat map of gene expression Z-scores at different stages of early embryonic development, depicting only genes in LADs specific to the 2-cell stage. These genomic regions contain 957 genes of which the 155 expressed genes are presented in the heat map (see methods). g Percentage of minor ZGA genes of total number of genes in iLADs (purple) and LADs (green). h, Example regions of LAD dynamics between the zygote and 2-cell stage (left top) and the 2-cell and 8-cell stage (right top). For each region, the mRNA expression levels are depited in RPKM (red). The bottom panels display two examples of LAD dynamics between the zygote, 2-cell and 8-cell stages. The location of two genes in the 2-cell-specific LADs is indicated with a dotted line and the expression levels (RPKM) of the corresponding genes is displayed as violins plots. Violin plots show median (black points) and values at most 1.5 * IQR. n = biological independent samples, zygote (n = 4), 2-cell (n = 10) and 8-cell (n = 28). i, Chromosome plot comparing Dam and Dam-lamin B1 profiles in oocytes and zygotes. j, Chromosome plots comparing Dam and Dam-lamin B1 DamID population and single cell profiles in oocytes and zygotes.
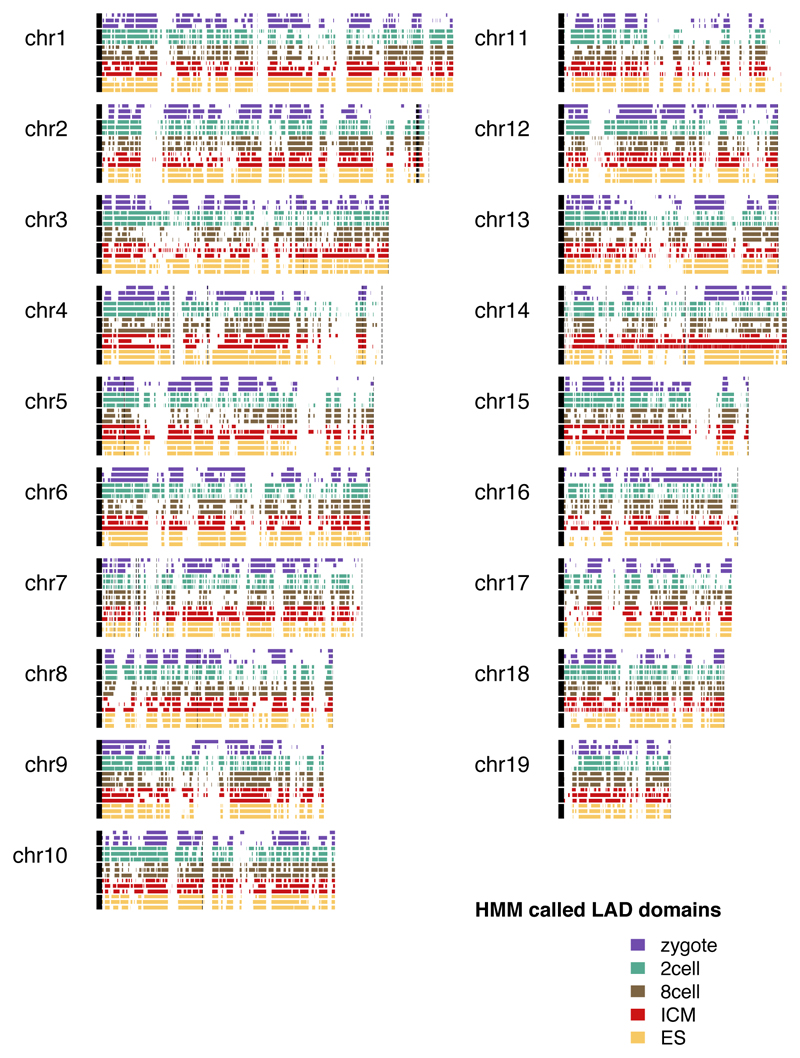
Extended Data Figure 3. Chromosome interaction profiles of Dam-lamin B1 triplicate samples.
Dam-lamin B1 chromosomal interaction maps from three biological replicate DamID population samples for each embryonic stage. Coloured blocks represent HMM-called LAD domains. Black bars on the left represent centromeres, red highlights indicate unmappable regions.
Extended Data Figure 4. Single cells show consistent LAD patterning within the same developmental stages but parental genomes display distinct features.
a, Chromosome profiles of population average (n = 3) and single-cell average Dam-lamin B1 signals. Black boxes represent LAD domains called by Hidden-Markov modelling. b, Genomic location of example DNA-FISH probes projected on Dam-lamin B1 chromosome profiles. c, Quantification of 3D preserved DNA-FISH spot distances to the nuclear periphery in 2-cell and 8-cell embryos of probes in regions that change LAD status from 2- to 8-cell stage according to DamID. Box plots show the 25th and 75th percentiles (box), median (solid lines), the smallest/largest values at most 1.5 * IQR of the hinge (whiskers) and outliers (grey circles). n = number of DNA-FISH spots from at least 3 biological independent samples. Wilcoxon rank-sum test p-values shown (two-sided). **: p <= 0.01, ****: p <= 0.0001. d, Images of 3D DNA-FISH in zygote pronuclei. Quantification shows distance to the nuclear periphery. Scale bars represent 10 μm. Box plots show the 25th and 75th percentiles (box), median (solid lines), the smallest/largest values at most 1.5 * IQR of the hinge (whiskers) and outliers (grey circles). n = number of DNA-FISH spots from at least 3 biological independent samples. e, Distance quantification of DNA-FISH probes in maternal and paternal specific LADs in zygotes. Box plots show the 25th and 75th percentiles (box), median (solid lines), the smallest/largest values at most 1.5 * IQR of the hinge (whiskers) and outliers (grey circles). n = number of DNA-FISH spots from at least 3 biological independent samples. Wilcoxon rank-sum test p-values shown (two-sided). *: p <= 0.05 **, : p <= 0.01, ***: p <= 0.001,****, : p <= 0.0001. f, t-SNE representation of the triplicate allelic Dam-lamin B1 population samples, including 2-cell embryos from reciprocal crosses. n = 3 biological independent samples. g, Scatter plot comparison between average Dam-lamin B1 signals obtained from pronucleus separated and hybrid zygote DamID. n = biological independent samples. Maternal pronucleus (n = 10), paternal pronucleus (n = 15) and hybrid zygote (n = 3). r = Spearman’s rho. h, Average CpG density, AT content and DHS sites over LAD boundaries ±1.5 Mb defined specifically for maternal and paternal alleles in zygotes. i, Allelic correlation matrix (Pearson) of OE scores form Dam-lamin B1 embryos. j, Jaccard indexes calculated over HMM called LAD domains.
Extended Data Figure 5. Compartment status of regions with different LAD dynamics during embryonic development.
a, Violin plots of compartment scores calculated for 100-kb genomic regions with different LAD dynamics. Number in parentheses represents the percentage of the genome covered by each of the 16 different categories of defined LAD reorganisations from zygote to ES. Violin plots show the values at most 1.5 * IQR and median (red lines). Compartment scores are calculated based on 3 biological independent samples. n = number of 100-kb bins b, Experimental schemes of DamID in aphidicolin treated zygotes and 2-cell embryos. Images show global DNA-replication measured by EdU incorporation in control and replication inhibited embryos. n = number of images of independent experiments. Scale bars represent 10 μm. c, Chromosome plots of compartment scores from Hi-C and DamID scores in control and aphidicoline treated zygotes and 2-cell embryos.
Extended Data Figure 6. Overexpression of Kdm5b histone demethylase abrogates paternal LAD establishment in the zygote.c.
a, Quantifications of immunostaining with H3K9me3 in zygotes injected with wild-type and mutant KDM4D. Violin plots show the values at most 1.5 * IQR and median (red lines). n = at least 3 biological independent samples. Wilcoxon rank-sum test p-values shown (two-sided). Scale bars represent 10 μm. b, Average paternal-specific H3K4me3 signal at paternal-specific LAD boundaries of each respective embryonic stage. c, Average maternal specific H3K4me3 signal at maternal specific LAD boundaries of each respective embryonic stage. d, Quantification as described for a but with H3K4me3. e, Immunostaining and quantification of lamin B1 localisation in the Kdm5b-expressing zygotes. Violin plots show the values at most 1.5 * IQR and median (red lines). n = at least 3 biological independent samples. Wilcoxon rank-sum test p-values shown (two-sided). Scale bars represent 10 μm. f, Global transcription detection (EU incorporation) and quantification in H3K4me3 in the Kdm5b-expressing zygotes. Violin plots show the values at most 1.5 * IQR and median (red lines). n = at least 3 biological independent samples. Wilcoxon rank-sum test p-values shown (two-sided). Scale bars represent 10 μm. g, Immunofluorescent staining of H3K9me3 in MUT and WT KDM5B-expressing embryos, violin plots show the quantified levels of H3K9me3. Violin plots show the values at most 1.5 * IQR and median (red lines). n = at least 3 biological independent samples. Wilcoxon rank-sum test p-values shown (two-sided). Scale bars represent 10 μm. h, Immunofluorescent staining of H3K9me2 in MUT and WT KDM5B injected embryos, violin plots show the quantified levels of H3K9me2. Violin plots show the values at most 1.5 * IQR and median (red lines). n = at least 3 biological independent samples. Wilcoxon rank-sum test p-values shown (two-sided). Scale bar represents 10 μm. i, Hierarchical clustering based on Pearson correlation of Dam-lamin B1 signal from single pronuclei of wild-type and mutant Kdm5b-injected zygotes (samples from 3 independent experiments). For comparison, population average Dam and Dam-lamin B1 signals are included as grey and black squares, respectively. j, Average Dam-lamin B1 signal at LAD boundaries in hybrid H3K4me3 manipulated embryos. Signal shown in maternal genome (top) and paternal genome (bottom). k, Chromosome profiles of H3K4me3 ChIP-seq20 signal from sperm, early-, and late-zygotes. l, Average H3K4me3 levels in sperm, early-and late-zygotes at paternal zygotic LAD borders.
Supplementary Material
Acknowledgements
We thank members of the JK and METP labs and T. Straub (Biomedical Center, Ludwig-Maximilians-University) for helpful discussions and comments on the manuscript, and I. Solovei, A. Scialdone and A. van Oudenaarden for critical reading of the manuscript and J.W. Jachowicz for embryo drawings. We thank R.Tepperino (Helmholtz Centre Munich) for support. We acknowledge funding by ERC-Stg EpiID (678423) to J.K. and EpiGeneSys NoE, ERC-Stg NuclearPotency (280840), the German Research Council (CRC 1064) and the Helmholtz Association to M.E.T.P. M.B. held a Boehringer Ingelheim Fonds Ph.D fellowship. The Oncode Institute is supported by KWF Dutch Cancer Society.
Footnotes
Data availability. The sequencing DamID data from this study are available from the Gene Expression Omnibus, accession number GSE112551 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE112551).
Code availability. Custom code generated to perform the analysis in this study are available upon request.
Contributions
M.B performed the embryo work, S.M.P managed the sequencing data and performed all bioinformatic analysis, except for the Hi-C data, which was analyzed by M.B. and T.S. and the expression analysis of Extended Data Fig. 1h, 2f and 2h by E.R.M. J.P. constructed the pipeline for 3D imaging analyses. K.L.d.L optimized the scDamID protocol. S.S.d.V. created and performed experiments with the mESC DamID lines. M.B. and S.M.P. contributed to experimental design and data interpretation. M.-E.T.-P. and J.K. conceived, designed and supervised the project.
Competing interest statement. The authors declare that no competing financial interest.
References
- 1.Burton A, Torres-Padilla ME. Chromatin dynamics in the regulation of cell fate allocation during early embryogenesis. Nat Rev Mol Cell Biol. 2014;15:723–734. doi: 10.1038/nrm3885. [DOI] [PubMed] [Google Scholar]
- 2.Xu Q, Xie W. Epigenome in Early Mammalian Development: Inheritance, Reprogramming and Establishment. Trends Cell Biol. 2018;28:237–253. doi: 10.1016/j.tcb.2017.10.008. [DOI] [PubMed] [Google Scholar]
- 3.Jachowicz J, et al. Heterochromatin establishment at pericentromeres depends on nuclear position. Genes Dev. 2013;27:2427–2432. doi: 10.1101/gad.224550.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van Steensel B, Belmont AS. Lamina-Associated Domains: Links with Chromosome Architecture, Heterochromatin, and Gene Repression. Cell. 2017;169:780–791. doi: 10.1016/j.cell.2017.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van Steensel B, et al. Chromatin profiling using targeted DNA adenine methyltransferase. Nat Genet. 2001;27:304–308. doi: 10.1038/85871. [DOI] [PubMed] [Google Scholar]
- 6.Nishimura K, et al. An auxin-based degron system for the rapid depletion of proteins in nonplant cells. Nat Methods. 2009;6:917–922. doi: 10.1038/nmeth.1401. [DOI] [PubMed] [Google Scholar]
- 7.Kind J, et al. Single-cell dynamics of genome-nuclear lamina interactions. Cell. 2013;153:178–192. doi: 10.1016/j.cell.2013.02.028. [DOI] [PubMed] [Google Scholar]
- 8.Meuleman W, et al. Constitutive nuclear lamina-genome interactions are highly conserved and associated with A/T-rich sequence. Genome Res. 2013;23:270–280. doi: 10.1101/gr.141028.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boskovic A, et al. Analysis of active chromatin modifications in early mammalian embryos reveals uncoupling of H2A.Z acetylation and H3K36 trimethylation from embryonic genome activation. Epigenetics. 2012;7:747–757. doi: 10.4161/epi.20584. [DOI] [PubMed] [Google Scholar]
- 10.Lu F, et al. Establishing Chromatin Regulatory Landscape during Mouse Preimplantation Development. Cell. 2016;165:1375–1388. doi: 10.1016/j.cell.2016.05.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu J, et al. The landscape of accessible chromatin in mammalian preimplantation embryos. Nature. 2016;534:652–657. doi: 10.1038/nature18606. [DOI] [PubMed] [Google Scholar]
- 12.Kind J, et al. Genome-wide maps of nuclear lamina interactions in single human cells. Cell. 2015;163:134–147. doi: 10.1016/j.cell.2015.08.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Du Z, et al. Allelic reprogramming of 3D chromatin architecture during early mammalian development. Nature. 2017;547:232–235. doi: 10.1038/nature23263. [DOI] [PubMed] [Google Scholar]
- 14.Ke Y, et al. 3D Chromatin Structures of Mature Gametes and Structural Reprogramming during Mammalian Embryogenesis. Cell. 2017;170:367–381 e320. doi: 10.1016/j.cell.2017.06.029. [DOI] [PubMed] [Google Scholar]
- 15.Crane E, et al. Condensin-Driven Remodeling of X-Chromosome Topology during Dosage Compensation. Nature. 2015;523:240–244. doi: 10.1038/nature14450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pope BD, et al. Topologically associating domains are stable units of replication-timing regulation. Nature. 2014;515:402–405. doi: 10.1038/nature13986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wen B, et al. Large histone H3 lysine 9 dimethylated chromatin blocks differentiated from embryonic stem cells. Nat Genet. 2009;41:246–250. doi: 10.1038/ng.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dahl JA, et al. Broad histone H3K4me3 domains in mouse oocytes modulate maternal-to-zygotic transition. Nature. 2016;537:548–552. doi: 10.1038/nature19360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu X, et al. Distinct features of H3K4me3 and H3K27me3 chromatin domains in pre-implantation embryos. Nature. 2016;537:558–562. doi: 10.1038/nature19362. [DOI] [PubMed] [Google Scholar]
- 20.Zhang B, et al. Allelic reprogramming of the histone modification H3K4me3 in early mammalian development. Nature. 2016;537:553–557. doi: 10.1038/nature19361. [DOI] [PubMed] [Google Scholar]
- 21.Lepikhov K, Walter J. Differential dynamics of histone H3 methylation at positions K4 and K9 in the mouse zygote. BMC Dev Biol. 2004;4:12. doi: 10.1186/1471-213X-4-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Torres-Padilla ME, et al. Dynamic distribution of the replacement histone variant H3.3 in the mouse oocyte and preimplantation embryos. Int J Dev Biol. 2006;50:455–461. doi: 10.1387/ijdb.052073mt. [DOI] [PubMed] [Google Scholar]
- 23.Houliston E, et al. Expression of nuclear lamins during mouse preimplantation development. Development. 1988;102:271–280. doi: 10.1242/dev.102.2.271. [DOI] [PubMed] [Google Scholar]
- 24.Peric-Hupkes D, et al. Molecular maps of the reorganization of genome-nuclear lamina interactions during differentiation. Mol Cell. 2010;38:603–613. doi: 10.1016/j.molcel.2010.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Matoba S, et al. Embryonic development following somatic cell nuclear transfer impeded by persisting histone methylation. Cell. 2014;159:884–895. doi: 10.1016/j.cell.2014.09.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miyanari Y, Torres-Padilla ME. Control of ground-state pluripotency by allelic regulation of Nanog. Nature. 2012;483:470–473. doi: 10.1038/nature10807. [DOI] [PubMed] [Google Scholar]
- 27.Servant N, et al. HiC-Pro: an optimized and flexible pipeline for Hi-C data processing. Genome Biol. 2015;16:259. doi: 10.1186/s13059-015-0831-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Monkhorst K, et al. X inactivation counting and choice is a stochastic process: evidence for involvement of an X-linked activator. Cell. 2008;132:410–421. doi: 10.1016/j.cell.2007.12.036. [DOI] [PubMed] [Google Scholar]
- 29.Vogel MJ, et al. Detection of in vivo protein-DNA interactions using DamID in mammalian cells. Nat Protoc. 2007;2:1467–1478. doi: 10.1038/nprot.2007.148. [DOI] [PubMed] [Google Scholar]
- 30.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnetjournal. 2011;17:10–12. [Google Scholar]
- 31.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler Transform. Bioinformatics. 2009;25:1754–60. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Filion GJ, et al. Systematic protein location mapping reveals five principal chromatin types in Drosophila cells. Cell. 2010;143:212–224. doi: 10.1016/j.cell.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim D, et al. HISAT: a fast spliced aligner with low memory requirements. Nature Methods. 2015;12:357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Anders S, et al. HTSeq — A Python framework to work with high-throughput sequencing data. bioRxiv. 2014 doi: 10.1101/002824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Robinson MD, et al. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Park SJ, et al. Inferring the choreography of parental genomes during fertilization from ultralarge-scale whole-transcriptome analysis. Genes Dev. 2013;27:2736–2748. doi: 10.1101/gad.227926.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Deng Q, et al. Single-cell RNA-seq reveals dynamic, random monoallelic gene expression in mammalian cells. Science. 2014;343:193–196. doi: 10.1126/science.1245316. [DOI] [PubMed] [Google Scholar]
- 38.Bray NL, et al. Near-optimal probabilistic RNA-seq quantification. Nat Biotechnology. 2016;34:525–527. doi: 10.1038/nbt.3519. [DOI] [PubMed] [Google Scholar]
- 39.R Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing; Vienna, Austria: 2017. URL https://www.R-project.org/ [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.