Abstract
CRISPR/Cas9-based therapies hold an important promise for the treatment of genetic diseases. Among these, Hutchinson-Gilford progeria syndrome (HGPS) – caused by a point mutation in the LMNA gene – stands out as a potential candidate. Here, we explore the efficacy of a CRISPR/Cas9-based approach that reverts several alterations in HGPS cells and mice by introducing frameshift mutations in the LMNA gene.
HGPS is a rare disease characterized by aging-like manifestations emerging in childhood1. Most cases (80-90%) result from a de novo point mutation in the LMNA gene - encoding the nuclear lamins A and C - which activates a cryptic splice site in exon 11 (c.1824C>T; p.Gly608Gly)2,3. This event leads to the expression of progerin, a truncated lamin A variant with an internal deletion of 50 amino acids, which remains farnesylated, inducing morphological and functional alterations of the nuclear envelope4. A mouse model - LmnaG609G/G609G- recapitulating the mutation and many of the clinical features of these children5, confirmed that HGPS is caused by progerin accumulation and not by the loss of normal lamin A5,6. Several approaches against this syndrome were tested in preclinical models7, including farnesyltransferase inhibitors, which provided clinical benefits to HGPS patients8,9.
CRISPR/Cas9 gene editing tools constitute promising alternatives for diseases such as Duchenne muscular dystrophy10, metabolopathies11 and deafness12. This system involves a Cas9 endonuclease directed by a single-guide RNA (sgRNA) that recognizes its target region plus a protospacer-adjacent motif (PAM). The nuclease generates double-strand breaks in the DNA, which are repaired mainly through non-homologous end-joining producing insertions and deletions (indels)13. The finding that mosaic mice with both normal and prelamin A-producing progeroid cells have a completely normal phenotype14 indicates that a partial reduction in the accumulation of farnesylated lamin A products could be sufficient for an important phenotype relief.
On this basis, we developed a CRISPR/Cas9-based strategy against HGPS aimed at blocking the accumulation of lamin A and progerin. The LMNA gene encodes lamin C (exons 1-10) and lamin A (exons 1-12) through alternative splicing and polyadenylation. Since lamin A appears to be dispensable5,6, our strategy would disrupt the last part of the LMNA gene, impeding lamin A/progerin production without affecting lamin C. We first designed an sgRNA (sgRNA-LCS1) with the 5’-NGG PAM sequence of Streptococcus pyogenes Cas9 to target LMNA exon 11 upstream of the HGPS mutation, in a region conserved across human and mouse (Fig. 1a).
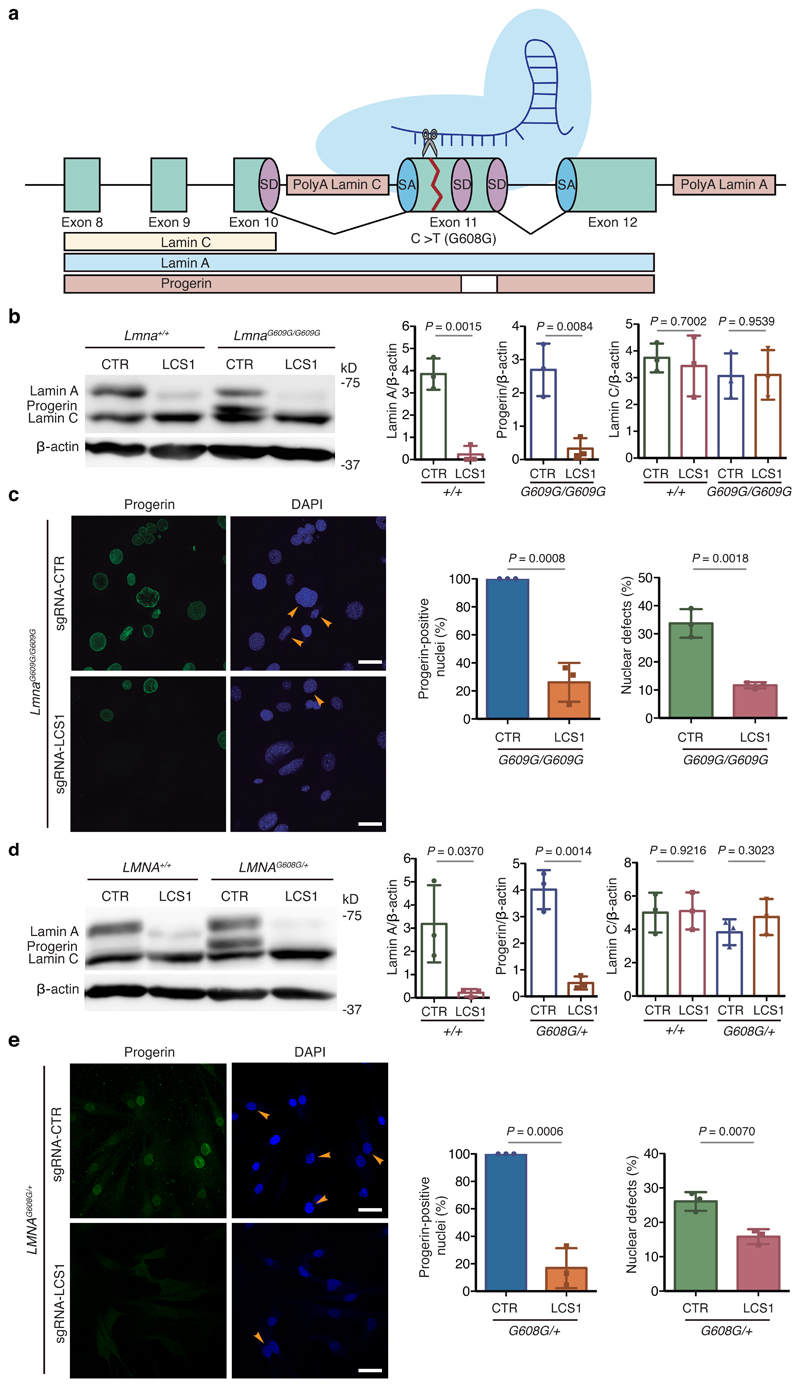
Figure 1. CRISPR/Cas9 testing in HGPS cellular models.
(a) sgRNA-LCS1 directs Cas9 nuclease against exon 11 of LMNA gene upstream of the HGPS mutation, disrupting lamin A and progerin without altering lamin C. (b) Cropped Western blot of lamin A, progerin and lamin C from wild-type and LmnaG609G/G609G mouse embryonic fibroblasts (MEFs) transduced with sgRNA-control or sgRNA-LCS1 (n=3 independent infections and MEF lines; two-tailed Student’s t-test). (c) Immunofluorescence analysis of progerin-positive nuclei and quantification of nuclear alterations by 4′,6-diamidino-2-phenylindole (DAPI) staining (n=3 independent infections and MEF lines; two-tailed Student’s t-test). Arrowheads indicate nuclear aberrations. (d) Cropped Western blot of lamin A, progerin and lamin C from wild-type and LMNAG608G/+ human fibroblasts transduced with sgRNA-control or sgRNA-LCS1 (n=3 independent infections; two-tailed Student’s t-test). (e) Progerin immunofluorescence and analysis of nuclear aberrations by DAPI staining (n=3 independent infections; two-tailed Student’s t-test). Arrowheads indicate blebbings and invaginations. Bar plots represent mean ± SD and individual values are overlaid. Scale bars, 40 μm. Uncropped blots are available as Source Data.
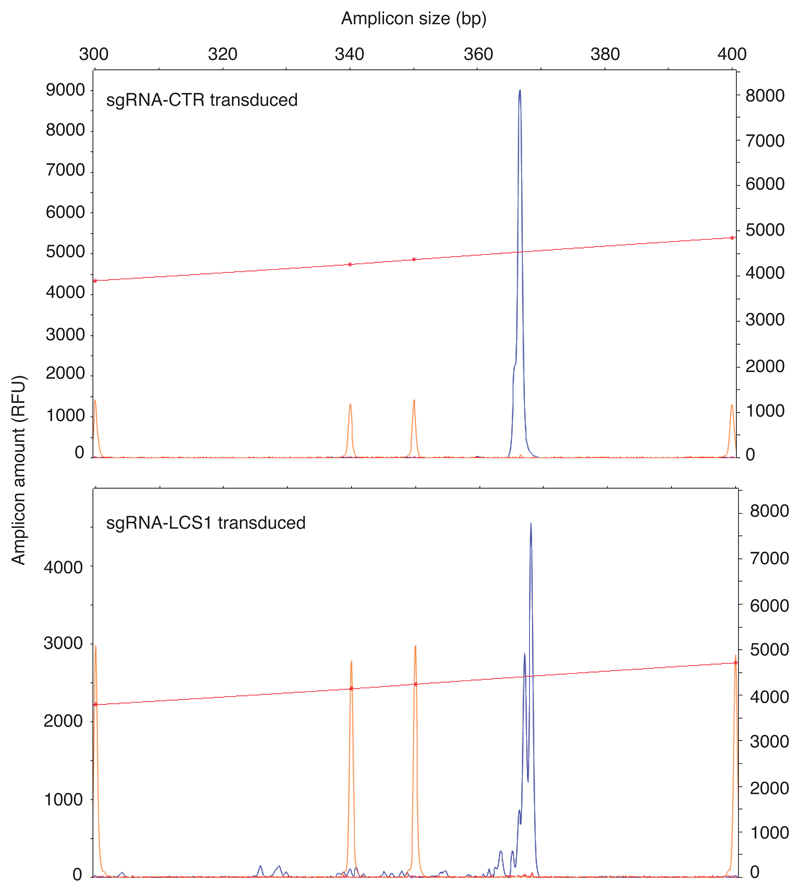
To test the efficacy of this approach, we cloned sgRNA-LCS1 or sgRNA-control in a lentiviral vector containing S. pyogenes Cas9 (lentiCRISPRv2) and used these to transduce Lmna+/+ and LmnaG609G/G609G murine fibroblasts. As a result, indels of variable lengths were produced in sgRNA-LCS1-transduced cells, as assessed by capillary electrophoresis-based fragment analysis (Extended Data 1). Western blot analysis showed a significant decrease in the accumulation of progerin and lamin A, while lamin C levels were not affected (Fig. 1b). Likewise, immunofluorescence analysis demonstrated that numbers of progerin-positive nuclei were reduced by 74% in sgRNA-LCS1-transduced cells compared to sgRNA-control-transduced cells (Fig. 1c). Accordingly, we found a 65% decrease in the number of nuclear alterations in LmnaG609G/G609G cells transduced with sgRNA-LCS1 compared to sgRNA-control-transduced cells (Fig. 1c).
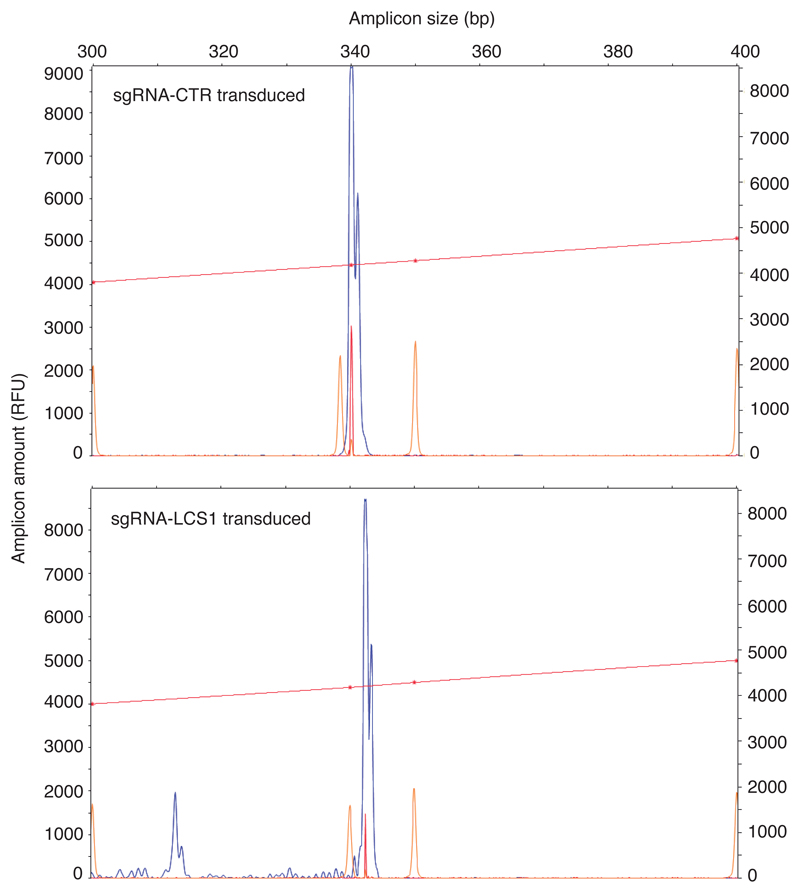
To test this system in human cells, we infected LMNAG608G/+ fibroblasts from HGPS patients and LMNA+/+ fibroblasts with these lentiviral vectors. Similar to mouse fibroblasts, we observed different indels in the DNA (Extended Data 2), a decrease in progerin and lamin A by Western blot (Fig. 1d), an 83% decrease in progerin-positive nuclei and a 39% reduction in the number of aberrant nuclei in sgRNA-LCS1- versus sgRNA-control-transduced HGPS cells (Fig. 1e).
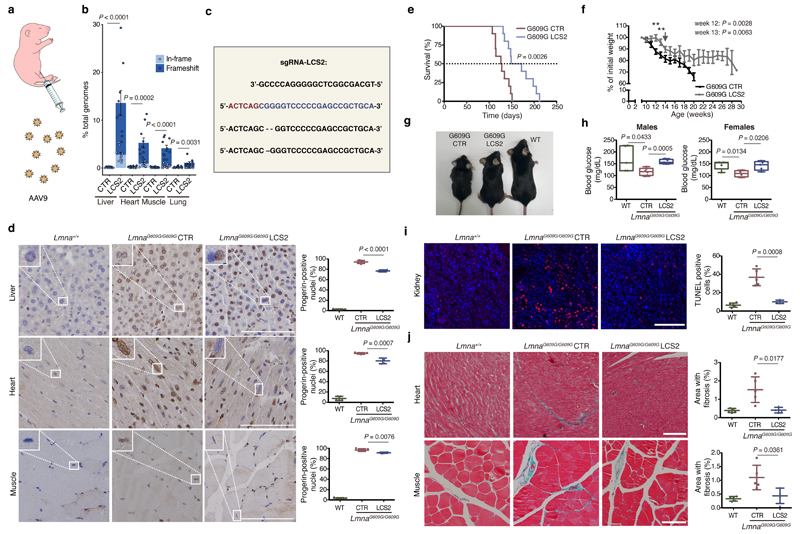
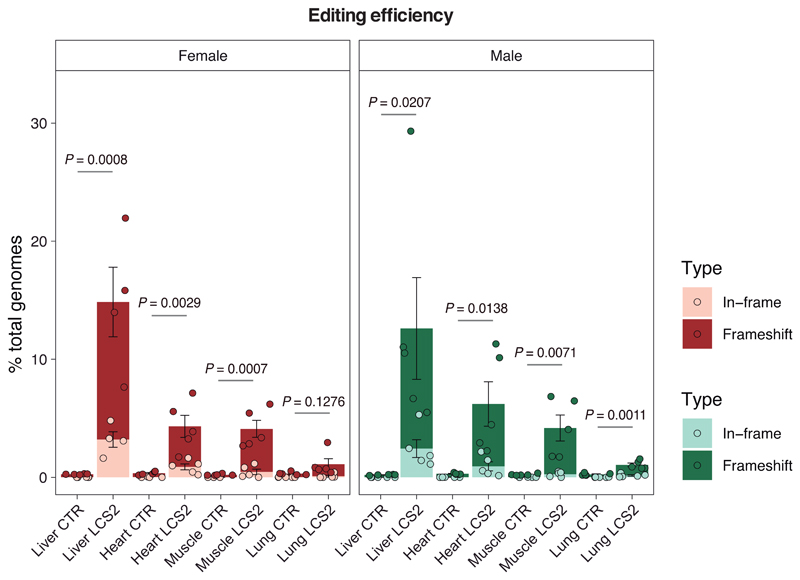
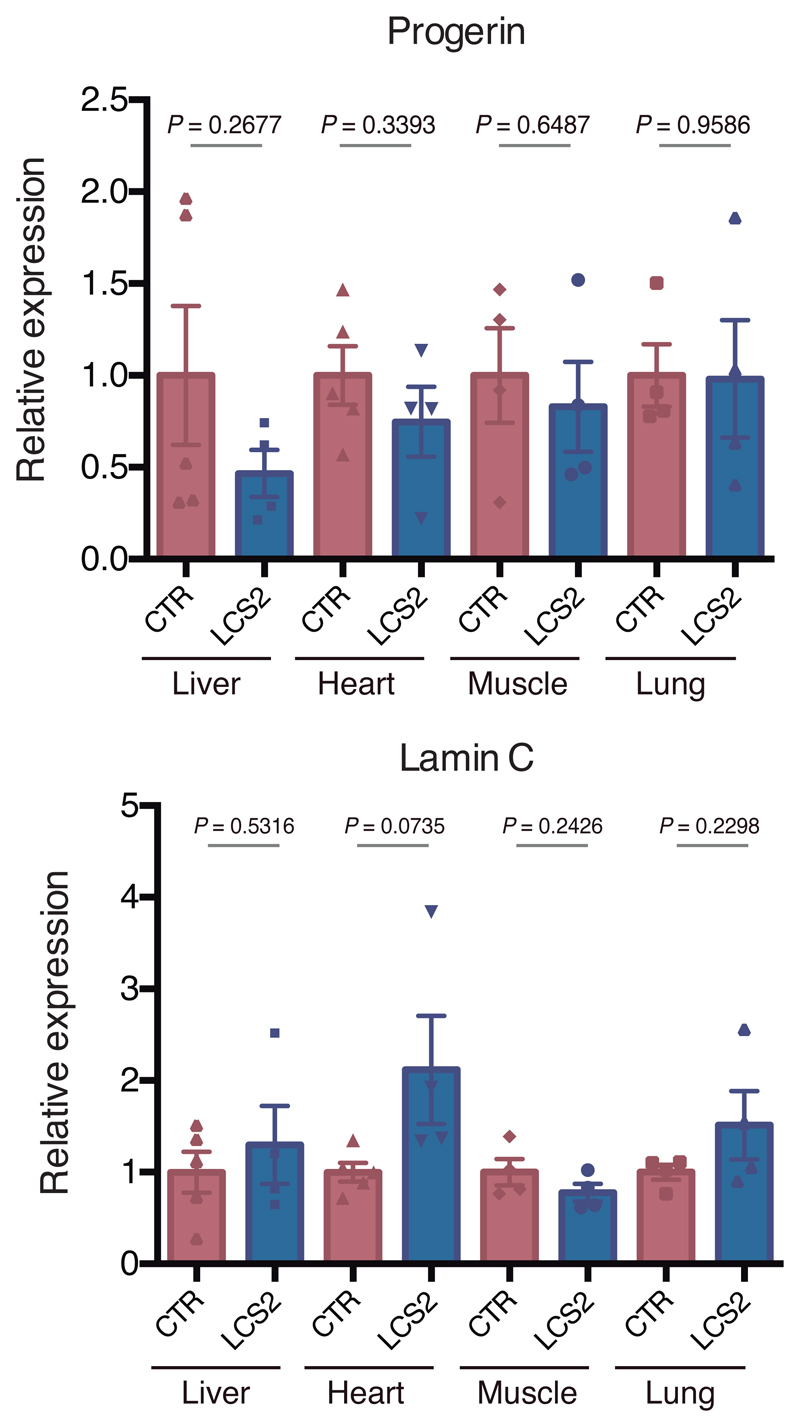
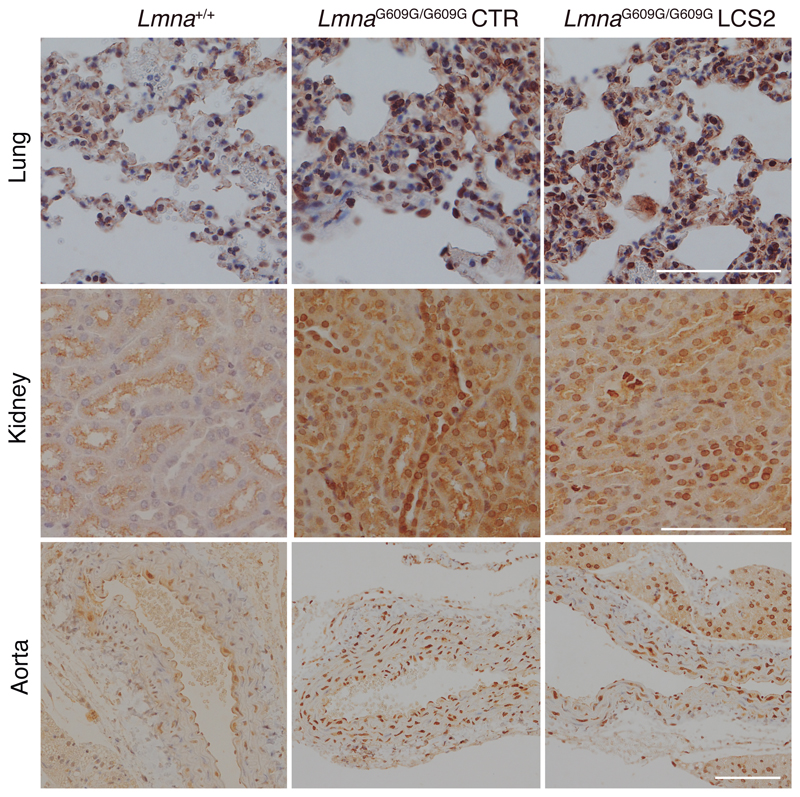
We next tested in vivo this editing approach using LmnaG609G/G609G mice as an HGPS animal model. We chose an adeno-associated virus 9 (AAV9) delivery vector due to its safety and broad tissue tropism. Given the packaging limit of these viruses (approximately 5 kb), we used Staphylococcus aureus Cas9 nuclease15 and designed a new sgRNA against the same region in exon 11 with the 5’-NNGRRT PAM sequence (sgRNA-LCS2). After packaging the vectors, with either sgRNA-LCS2 or the sgRNA-control, we injected intraperitoneally 2x1011 AAV9 genome copies in P3 LmnaG609G/G609G mice (Fig. 2a). To assess editing efficiency, we performed Illumina sequencing of the target region in DNA from AAV9 target organs – liver, heart, muscle and lung – of injected mice. Notably, Lmna contained indels in 13.6 ± 2.6% of the genome copies in liver, 5.3 ± 1.0% in heart, 4.1 ± 0.6% in muscle, and 1.1 ± 0.2% in lung (Fig. 2b,c; Extended Data 3; Supplementary Tables 1-4). Given the modest fraction of cells edited in vivo, the global decrease in progerin mRNA was too low to be reliably detected by RT-qPCR (Extended Data 4). However, immunohistochemical analysis revealed a significant reduction in progerin-positive nuclei in liver, heart and skeletal muscle from sgRNA-LCS2-transduced mice compared to sgRNA-control-transduced animals (Fig. 2d), which concurred with the DNA sequencing results. In lung, kidney and aorta, no reduction in the number of progerin-positive nuclei was observed, possibly due to the lower tropism of the AAV9 in these organs (Extended Data 5). Given the importance of vascular alterations in HGPS, the lack of noticeable direct effects on the aorta is a setback of the approach tested. Nevertheless, vascular pathologies characteristic of HGPS such as atherosclerosis are strongly influenced by systemic factors. Therefore, a reliable assessment of potential vascular benefits will require the use of susceptible mouse models carrying additional genetic alterations, such as Apoe or Ldlr inactivation.
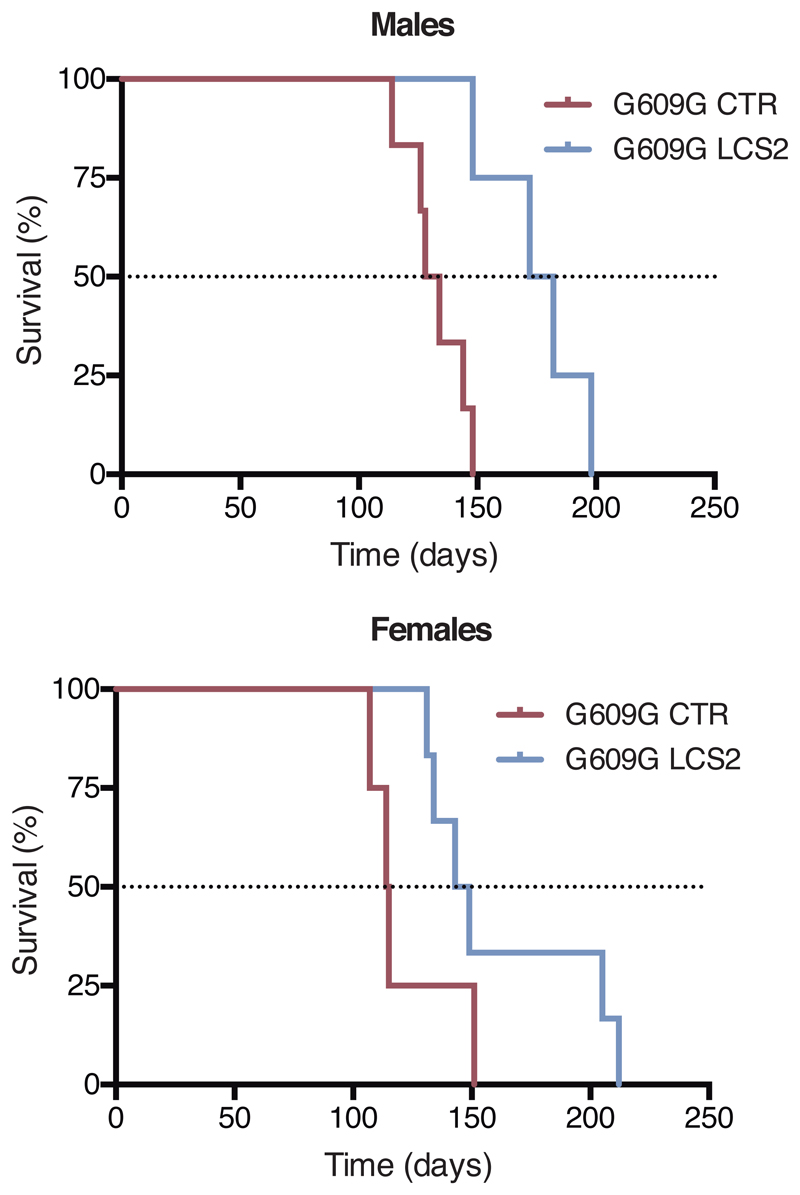
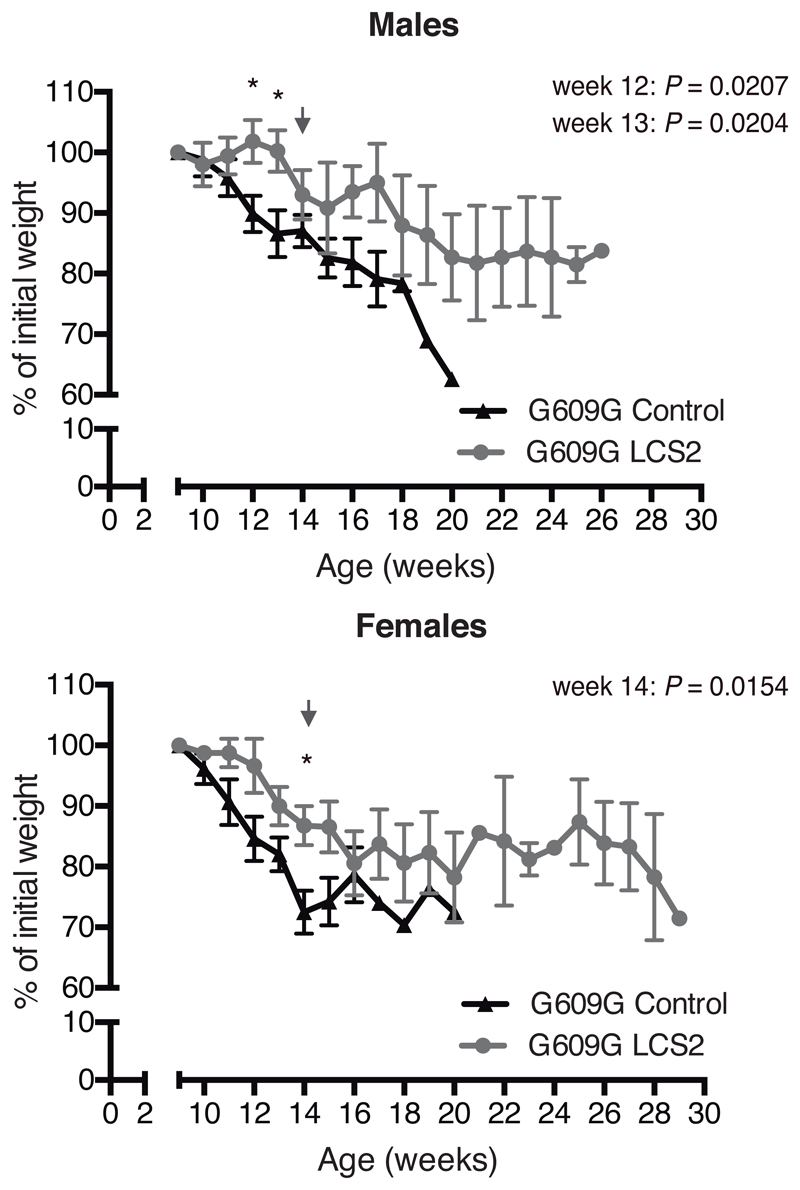
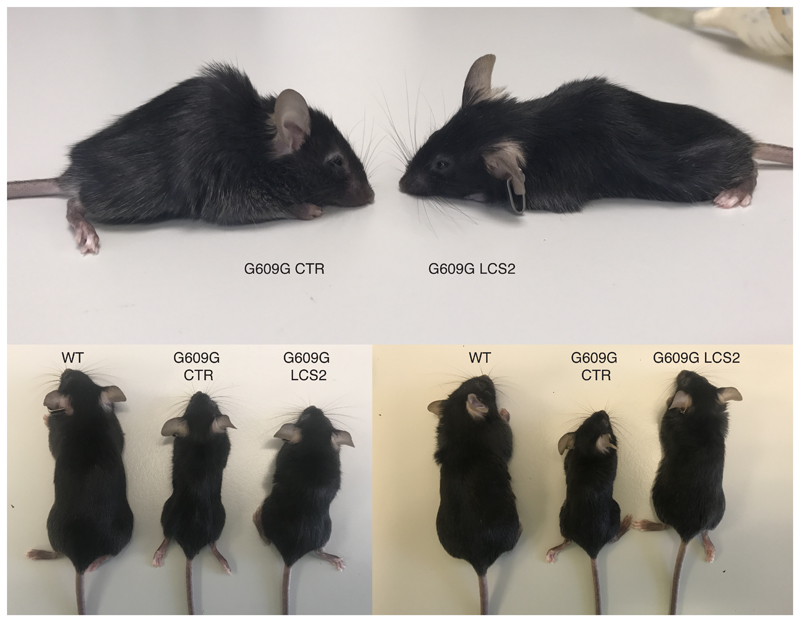
Figure 2. CRISPR/Cas9 delivery and phenotype amelioration in LmnaG609G/G609G mice.
(a) Intraperitoneal injection of AAV9 in P3 mice. (b) Percentage of in-frame and frameshift mutations at the Lmna target region in liver, heart, muscle and lung. Data are mean ± SEM (n=10 tissues per group, except n=9 sgRNA-LCS2-transduced liver; two-tailed Student’s t-test for total indels). (c) Alignment of the most common indels in sgRNA-LCS2-transduced mice. Blue, target sequence; red, PAM sequence. (d) Progerin immunohistochemistry of liver, heart and muscle from wild-type and LmnaG609G/G609G sgRNA-control-transduced or sgRNA-LCS2-transduced mice. Data are mean ± SD (n=5 wild-type and sgRNA-control-transduced mice; n=4 sgRNA-LCS2-transduced mice; two-tailed Student’s t-test). Insets, digital magnification of a selected area. (e) Kaplan-Meier survival plot of sgRNA-control- versus sgRNA-LCS2-transduced LmnaG609G/G609G mice (n=10 mice per group; two-sided Log-rank test). (f) Progression of body weight of mice transduced with sgRNA-control or sgRNA-LCS2, expressed as percentage of weight at 9 weeks. Vertical arrow, time point (3.5 months) at which the cohort destined for histological studies (4-5 mice per group) was sacrificed. Mean values ± SEM are represented (initial n=15 sgRNA-control-transduced mice; n=14 sgRNA-LCS2-transduced mice; two-tailed Student’s t-test). (g) Representative image of LmnaG609G/G609G sgRNA-control-transduced, sgRNA-LCS2-transduced and wild-type female mice at 3.5 months of age. (h) Glycemia in wild-type (males n=5; females n=5), sgRNA-control-transduced (males n=6; females n=4) and sgRNA-LCS2-transduced LmnaG609G/G609G mice (males, n=5; females, n=5). Data are represented by box plots and whiskers are minimum to maximum values (two-tailed Student’s t-test). (i) TUNEL assay in kidney of 3.5-month-old mice. Data are mean ± SD (n=5 wild-type and sgRNA-control-transduced mice; n=4 sgRNA-LCS2-transduced mice; two-tailed Student’s t-test). (j) Gomori staining in 3.5-month-old mouse tissues showing moderate perivascular and interstitial fibrosis in heart and quadriceps muscle of LmnaG609G/G609G mice (blue areas). Data are mean ± SD (n=5 wild-type and sgRNA-control-transduced mice; n=4 sgRNA-LCS2-transduced mice; two-tailed Student’s t-test). Scale bars, 100 μm (d, i, j).
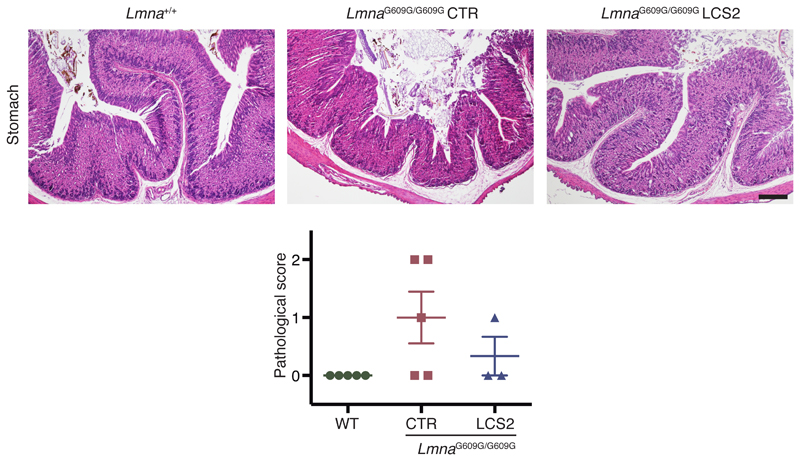
Importantly, progerin reduction in AAV9-sgRNA-LCS2-transduced mice was translated into an increase in their median survival of 33.5 days, from 127 to 160.5 days, compared to the sgRNA-control-transduced cohort, which represents a 26.4% lifespan increase (Fig. 2e; Extended Data 6). Mean survival was extended from 128.1 days (SD 15.73; 95% CI 116.8-139.4) to 167.4 days (SD 30.41; 95% CI 145.6-189.2). Likewise, maximum survival was extended from 151 to 212 days (P =0.0163; one-tailed Fisher exact test) (Fig. 2e). Phenotypically, sgRNA-LCS2-transduced LmnaG609G/G609G mice presented a healthier appearance, with retarded loss of grooming, slightly improved body weight and increased blood glucose levels, partially rescuing the hypoglycemia characteristic of these mice (Fig. 2f-h; Extended Data 7 and 8). Likewise, TUNEL analysis revealed that this group presented significantly less apoptotic cells in the kidney (Fig. 2i). Since progerin reduction was not detected in this organ, this result suggests an effect dependent on systemic factors. We also observed a slight decrease in gastric mucosa atrophy (Extended Data 9) and reduced focal and perivascular fibrosis in heart and quadriceps muscle in sgRNA-LCS2-transduced- compared to sgRNA-control-transduced mice, in accordance with the higher mobility of the former group (Fig. 2j; Supplementary Video).
Antisense oligonucleotides blocking the aberrant splicing of LMNA transcripts have been proposed for the treatment of progeria5,16. However, a longer-term therapy would be desirable. Here, we present a CRISPR/Cas9-based permanent genome editing approach that targets LMNA exon 11, specifically interfering with lamin A/progerin expression. Because the Cas9 endonuclease is not directed specifically against the p.Gly608Gly point mutation, this strategy could also be applicable to laminopathies caused by other LMNA mutations17. However, although lamin A is dispensable in cells and mice5,6, the consequences of abrogating its expression in humans remain unexplored. Therefore, alternative CRISPR/Cas9-based systems, such as base editors18, also need to be tested. The coexistence of progeroid and normal cells at a ratio of approximately 50/50 in mosaic mice resulted in a completely normal phenotype and lifespan14. In the current work, although the editing efficiency is lower, extensive phenotype amelioration and lifespan extension were obtained. The extent to which the modest editing efficiency attained is due to low delivery efficacy could not be determined reliably, as the packaging limit of the vector precluded the inclusion of a suitable reporter. Interestingly, the concurrent study by Beyret and colleagues19 describes another successful CRISPR/Cas9-based treatment of the LmnaG609G/G609G mouse model of HGPS. Nevertheless, further research on putative off-target events and adverse effects of the Cas9 nuclease will be needed to ensure the safety of this intervention. Regardless of these limitations, these two studies show preclinical efficacy of genome editing in a mouse model of progeria and pave the way for using CRISPR/Cas9 in HGPS and other currently incurable systemic diseases.
Methods
Plasmids and sgRNA cloning
All sgRNA-LCS were designed to target exon 11 of the LMNA gene by using the Benchling CRISPR Design tool. For the infections involving human and mouse fibroblasts, we used the lentiviral vector lentiCRISPRv2 (Addgene) in which we cloned the sgRNA-control (5’-GGAGACGGGATACCGTCTCT-3’) or the sgRNA-LCS1 (5’-AGCGCAGGTTGTACTCAGCG-3’). For AAV injection, we cloned the sgRNA-control or the sgRNA-LCS2 (5’-GTGCAGCGGCTCGGGGGACCCCG-3’) in the pX601-AAV vector from Addgene, containing the Cas9 nuclease from S. aureus.
Virus production
pX601-sgRNA-control or pX601-sgRNA-LCS2 plasmids were packaged as AAV serotype 9 by The Viral Vector Production Unit (UPV) of the Universitat Autònoma de Barcelona (Barcelona, Spain) followed by polyethylene glycol precipitation and iodixanol gradient purification. Aliquots of 2x1011 genome copies in 60 μL of PBS-MK were prepared for the injections.
Animal experiments
All animal experiments were performed in accordance with institutional guidelines and approved by the Committee of Animal Experimentation of University of Oviedo (Oviedo, Spain). For AAV injection, P3 LmnaG609G/G609G mice were injected intraperitoneally with either sgRNA-control- or sgRNA-LCS2-containing vectors. All gene-editing and phenotypic analyses were performed at 3.5 months of age. To analyze blood glucose, animals were starved overnight and glucose levels were measured with an Accu-Check glucometer (Roche Diagnostics) using blood from the tail vein.
Histological analysis and TUNEL staining
Tissues were collected in 4% paraformaldehyde in PBS and embedded in paraffin. Hematoxylin and eosin (H&E) staining was performed on stomach tissue, and atrophy of the gastric mucosa was blindly evaluated by a pathologist on three different sections per mouse, establishing a pathological score (0, normal; 1, mild; 2, moderate; 3, severe athrophy). TUNEL staining in mouse kidneys was done according to the manufacturer’s instructions (In-Situ Cell Death Detection Kit, TMR red, Roche). To determine the number of TUNEL-positive nuclei, ten random fields per mouse were blindly analyzed using ImageJ. H&E with Gomori’s trichrome staining was performed in heart and quadriceps muscle and five random fields per tissue were quantified with a FIJI plugin provided by A. M. Nistal (Servicios Científico-Técnicos, Universidad de Oviedo).
Illumina sequencing and bioinformatic analysis
MiSeq DNA sequencing was performed by Macrogen, using the Illumina 300bpPE. To prepare the library, DNA was isolated from liver, heart, muscle and lung from mice transduced with sgRNA-control- or sgRNA-LCS2-encoding AAVs. Next, we amplified the target region of the Cas9 nuclease with the Pfu DNA polymerase (Promega) adding the Illumina adapters by two PCRs: NGS1_fwd: ACACTCTTTCCCTACACGACGCTCTTCCGATCTNNNNTGTGACACTGGAGGCAGAAG and NGS1_rev: GTGACTGGAGTTCAGACGTGTGCTCTTCCGATCTCAAGTCCCCATCACTTGGTT for the first PCR, and NGS2_fwd: AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCT and NGS2_rev: CAAGCAGAAGACGGCATACGAGATXXXXXXGTGACTGGAGTTC for the second PCR. The N represents random bases and the X the sequence used for the index. Genomic reads in FASTQ format were aligned to the GRCm38.p6 assembly of the mouse genome using BWA v. 0.7.5a-r40520. Then, reads spanning the genomic region putatively affected by the CRISPR/Cas9 action (chr3: 88482555-88482615) were extracted with Samtools v. 1.3.121 and analyzed using in-house Perl scripts. Briefly, these scripts isolate the part of each read spanning the chosen region, highlight small insertions/deletions and output a count of each regional sequence. Then, we analyzed the percentage of the sequences showing regional differences in sgRNA-control- and sgRNA-LCS2-transduced mouse samples.
Capillary electrophoresis-based fragment analysis
We performed PCR amplification of the target region with the forward oligonucleotide labelled with 6FAM fluorophore in the 5’ position, allowing the fragment analysis of the resulting products by capillary electrophoresis. For human cells, we used the oligonucleotides HsLMNA_Fwd: [6FAM] GCACAGAACCACACCTTCCT and HsLMNA_Rev: TGACCAGATTGTCCCCGAAG, while for mouse cells we used MmLmna_Fwd: [6FAM] GTCCCCATCACTTGGTTGTC and MmLmna_Rev: TGACTAGGTTGTCCCCGAAG.
Cell culture, transfection and viral transduction
We maintained HEK-293T cell cultures in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum (FBS), 1% penicillin-streptomycin-L-glutamine and 1% antibiotic-antimycotic (Gibco) at 37 ºC in 5% CO2. In the case of human and mouse fibroblasts, 1x non-essential amino acids, 10 mM HEPES buffer, 100 μM 2-mercaptoethanol and 1x sodium pyruvate (Gibco) were also added to the previous medium and 15% FBS was used. For lentiviral infection, HEK-293T cells were transfected with lentiCRISPRv2 vector together with second-generation packaging plasmids using Lipofectamine reagent (Life Technologies). Supernatants were filtered through 0.45 μm polyethersulfone filters to collect the viral particles and added at 1:3 dilution to previously seeded human and mouse fibroblasts, supplemented with 0.8 μg/ml of polybrene (Millipore). Selection with puromycin (2 μg/ml) was performed two days after infection and the editing efficiency and nuclear aberrations were quantified one week later.
RNA preparation and RT-qPCR
Collected cells or tissues were homogenized in TRIzol reagent (Life Technologies) and RNA was extracted with the RNeasy Mini kit following the manufacturer’s instructions (QIAGEN). cDNA was synthesized with the QuantiTect Reverse Transcription kit (QIAGEN) using 1 μg of total RNA, and then RT-qPCR analysis of mouse tissues was performed. For progerin analysis, TaqMan PCR Universal Mastermix (Applied Biosystems) and the following oligonucleotides and probe were used: MmProgerin_fwd (5’-TGAGTACAACCTGCGCTCAC-3’), MmProgerin_rev (5’-TGGCAGGTCCCAGATTACAT-3’) and MmProgerin_probe (5’-CGGGAGCCCAGAGCTCCCAGAA-3’); using a β-actin (Applied Biosystems) as endogenous control. For lamin C analysis, we used SYBR green PCR Universal Mastermix (Applied Biosystems) and the oligonucleotides Lmnc_fwd (5’-CGACGAGGATGGAGAAGAGC-3’) and Lmnc_rev (5’-AGACTTTGGCATGGAGGTGG-3’) for lamin C; or Actb_Fwd (5’-CTGAGGAGCACCCTGTGCT-3’) and Actb_Rev (5’-GTTGAAGGTCTCAAACATGATCTG-3’) for β-actin as endogenous control.
Protein isolation and Western blot analysis
Cells were washed with 1x PBS and homogenized in RIPA lysis buffer containing 100 mM Tris pH 7.4, 150 mM NaCl, 10 mM EDTA pH 8, 1% sodium deoxycholate, 1% Triton X-100 and 0.1% SDS, supplemented with protease inhibitor cocktail (cOmplete, EDTA-free, Roche), and phosphatase inhibitors (PhosSTOP, Roche). Protein concentration was determined with the Pierce BCA Protein Assay Kit and 30 µg per lane were loaded onto 8% SDS-polyacrylamide gels. Gels were then transferred to nitrocellulose membranes, blocked with 5% nonfat dry milk in TBS-T buffer (20 mM Tris pH 7.4, 150 mM NaCl and 0.05% Tween 20) and incubated overnight at 4 ºC with primary antibodies: 1:500 mouse monoclonal anti-lamin A/C (MANLAC1, provided by G. Morris) for experiments involving mouse cells, 1:1,000 rabbit polyclonal anti-lamin A/C (sc-20681, Santa Cruz Biotechnology) for experiments involving human cells or 1:10,000 anti-β-actin (AC-15, Sigma) as an endogenous control. Finally, blots were incubated with 1:10,000 goat anti-mouse (Jackson ImmunoResearch) or 1:3,000 goat anti-rabbit horseradish peroxidase (HRP) (Cell Signalling) in 1.5% nonfat dry milk in TBS-T and washed with TBS-T. Immunoreactive bands were developed with Immobilon Western chemiluminescent HRP substrate (Millipore) in a LAS-3000 Imaging System (Fujifilm). Bands were quantified using ImageJ.
Immunofluorescence, immunohistochemistry and nuclear morphology analysis
For immunofluorescence assays, cells were fixed in 4% paraformaldehyde solution, rinsed in PBS and permeabilized with 0.5% Triton X-100. Afterwards, they were blocked with 15% goat serum solution and incubated overnight at 4 ºC with a rabbit polyclonal anti-progerin antibody in PBS (1:200). Next, slides were washed with TBS-T and incubated with 1:500 anti-rabbit secondary antibody Alexa Fluor 488 (Life Technologies). Nuclei were stained with 4’,6-diamidino-2-phenylindole (DAPI, Invitrogen). In the case of immunohistochemical analysis, tissues were fixed in 4% paraformaldehyde solution and incubated with Target Retrieval Solution at 95 ºC for 20 min, Peroxidase Blocking Solution for 5 min and Protein Block Serum Free (all from Dako) for 20 min before the incubation with the anti-progerin primary antibody (1:300 dilution) for 1 h. An HRP-conjugated polyclonal anti-rabbit was applied for 30 min and then 3,3'-diaminobenzidine for 10 min. Tissues were counterstained with hematoxylin (Dako) and visualized by light microscopy. Rabbit anti-progerin polyclonal antibody was generated using peptide immunogens and standard immunization procedures (S. Nourshargh et al., manuscript in preparation). The specificity of the antibody was confirmed by nuclear staining of LmnaG609G/G609G mouse-derived fibroblasts, which was negative in the case of wild-type cells. To determine the percentage of progerin-positive cells and nuclei with aberrations, five random fields per culture or tissue sample were blindly analyzed. For tissue samples, in each field a pre-established grid was used and five random areas were quantified.
Statistical analysis
Animals of the same age were used for comparisons between mice groups and no statistical method was used to predict sample size. For statistical analysis of differences between mouse cohorts, normality was assessed using the Shapiro-Wilk test in those cases where n>10. In the remaining cases, normality was assumed based on previous data and we performed two-tailed Student’s t-test to study the statistical significance. For survival comparisons, we used the Log-rank test and differences in maximum lifespan were calculated using the one-tailed Fisher exact test comparing the number of live sgRNA-control- and sgRNA-LCS2-transduced mice at the age corresponding to the 80th percentile of lifespan in the joint survival distribution. We used Microsoft Excel or GraphPad Prism software for the analysis and significant differences were considered when *P < 0.05, **P < 0.01, ***P < 0.001.
Extended Data
Extended Data 1. Representative capillary electrophoresis-based fragment analysis of sgRNA-control- and sgRNA-LCS1-transduced LmnaG609G/G609G mouse embryonic fibroblasts (n = 3 independent infections and MEF lines).
Red line and orange peaks correspond to size standards.
Extended Data 2. Representative capillary electrophoresis-based fragment analysis of sgRNA-control- and sgRNA-LCS1-transduced LMNAG608G/+ human fibroblasts (n = 3 independent infections).
Red line and orange peaks correspond to size standards.
Extended Data 3. Percentage of indels in LmnaG609G/G609G sgRNA-LC2-transduced male and female mouse tissues.
Data are mean ± s.e.m. (n = 5 tissues per group, except in sgRNA-LCS2-transduced female liver where n = 4; two-tailed Student’s t-test).
Extended Data 4. RT–qPCR analysis of progerin and lamin C in tissues from LmnaG609G/G609G sgRNA-control-transduced and LmnaG609G/G609G sgRNA-LCS2-transduced mice.
Data are mean ± s.e.m. (n = 4 tissues per group, except sgRNA-control-transduced liver and heart where n = 5; two-tailed Student’s t-test).
Extended Data 5.
Progerin immunohistochemistry in lung, kidney and aorta from WT, LmnaG609G/G609G sgRNA-control-transduced and LmnaG609G/G609G sgRNA-LCS2-transduced mice (lung and kidney, n = 5 for WT and sgRNA-control-transduced mice and n = 4 for sgRNA-LCS2-transduced mice; aorta, n = 2 for WT and n = 3 for sgRNA-control- and sgRNA-LCS2-transduced LmnaG609G/G609G mice). Scale bar, 100 μm.
Extended Data 6.
Kaplan–Meier survival plot of LmnaG609G/G609G male and female mice transduced with sgRNA-control (n = 6 males; n = 4 females) or sgRNA-LCS2 (n = 4 males; n = 6 females).
Extended Data 7. Progression of body weight of male and female mice transduced with sgRNA-control or sgRNA-LCS2, expressed as percentage of weight at 9 weeks.
Mean values ± s.e.m. are shown (for males, initial n = 9 sgRNA-control-transduced mice and n = 8 sgRNA-LCS2-transduced mice; for females, initial n = 6 mice per group; two-tailed Student’s t-test). Vertical arrow indicates the time point (3.5 months) at which the cohort destined for histological studies was sacrificed.
Extended Data 8.
Images of three sex- and age-matched mice transduced with the sgRNA-LCS2 compared to sgRNA-control-transduced animals.
Extended Data 9. H&E staining of gastric mucosa from WT, LmnaG609G/G609G sgRNA-controltransduced and LmnaG609G/G609G sgRNA-LCS2-transduced mice.
The graph shows atrophy quantification according to a pathological score as described in Methods. Data are mean ± s.e.m. (n = 5 for WT and sgRNA-control-transduced mice; n = 3 for sgRNA-LCS2-transduced mice).
Supplementary Material
Acknowledgements
We thank G. Velasco, R. Villa-Bellosta, C. Bárcena, A.P. Ugalde and X.M. Caravia for helpful comments and advice, and R. Feijoo, A. Moyano, D.A. Puente and S.A. Miranda for excellent technical assistance. We also acknowledge the generous support by J.I. Cabrera and Associazione Italiana Progeria Sammy Basso and the contribution of Dr. Matthew Golding to the generation of the anti-murine progerin antibody. The Instituto Universitario de Oncología del Principado de Asturias is supported by Fundación Bancaria Caja de Ahorros de Asturias. JMPF is supported by Ministerio de Economía y Competitividad (MINECO/FEDER: SAF2015-64157-R) and Gobierno del Principado de Asturias. CL-O is supported by grants from European Research Council (ERC-2016-ADG, DeAge), Ministerio de Economía y Competitividad (MINECO/FEDER: SAF2014-52413-R and SAF2017-87655-R), Instituto de Salud Carlos III (RTICC) and Progeria Research Foundation (PRF2016-66). OS-F is recipient of an FPU fellowship. ARF is recipient of a Ramón y Cajal fellowship. The generation of the progerin antibody was funded by the Wellcome Trust (098291/Z/12/Z to SN).
Footnotes
Reporting Summary. Further information on experimental design is available in the Life Sciences Reporting Summary attached to this paper.
Code availability. The in-house Perl scripts used for gene edition analysis can be freely downloaded at https://github.com/vqf/genotypes.
Data availability. The MiSeq data were deposited in the NCBI SRA (PRJNA505974). Other data from this study are available from the corresponding authors. Any materials that can be shared will be released via a Material Transfer Agreement.
Author contributions. F.G.O., J.M.P.F., and C.L.-O. conceived and designed experiments. O.S.-F., F.G.O., V.Q., F.R., S.B., D.M. and A.R.F. performed experiments and analyzed data. L.R., A.B. and S.N. provided reagents. O.S.-F., F.G.O., A.R.F., J.M.P.F., and C.L.-O. wrote the manuscript. All authors revised the manuscript.
Competing financial interest. The authors declare no competing interests.
References
- 1.Hennekam RC. Am J Med Genet A. 2006;140:2603–2624. doi: 10.1002/ajmg.a.31346. [DOI] [PubMed] [Google Scholar]
- 2.De Sandre-Giovannoli A, et al. Science. 2003;300:2055. doi: 10.1126/science.1084125. [DOI] [PubMed] [Google Scholar]
- 3.Eriksson M, et al. Nature. 2003;423:293–298. doi: 10.1038/nature01629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goldman RD, et al. Proc Natl Acad Sci U S A. 2004;101:8963–8968. doi: 10.1073/pnas.0402943101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Osorio FG, et al. Sci Transl Med. 2011;3 doi: 10.1126/scitranslmed.3002847. 106ra107. [DOI] [PubMed] [Google Scholar]
- 6.Fong LG, et al. J Clin Invest. 2006;116:743–752. doi: 10.1172/JCI27125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gordon LB, Rothman FG, Lopez-Otin C, Misteli T. Cell. 2014;156:400–407. doi: 10.1016/j.cell.2013.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gordon LB, et al. Circulation. 2014;130:27–34. doi: 10.1161/CIRCULATIONAHA.113.008285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gordon LB, et al. JAMA. 2018;319:1687–1695. doi: 10.1001/jama.2018.3264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Amoasii L, et al. Sci Transl Med. 2017;9 doi: 10.1126/scitranslmed.aan8081. eaan8081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang Y, et al. Nat Biotechnol. 2016;34:334–338. doi: 10.1038/nbt.3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gao X, et al. Nature. 2018;553:217–221. doi: 10.1038/nature25164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Doudna JA, Charpentier E. Science. 2014;346 doi: 10.1126/science.1258096. 1258096. [DOI] [PubMed] [Google Scholar]
- 14.de la Rosa J, et al. Nat Commun. 2013;4:2268. doi: 10.1038/ncomms3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ran FA, et al. Nature. 2015;520:186–191. doi: 10.1038/nature14299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Scaffidi P, Misteli T. Nat Med. 2005;11:440–445. doi: 10.1038/nm1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bar DZ, et al. J Med Genet. 2017;54:212–216. doi: 10.1136/jmedgenet-2016-104295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gaudelli NM, et al. Nature. 2017;551:464–471. doi: 10.1038/nature24644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Beyret E, et al. Nat Med. 2019;25:419–422. doi: 10.1038/s41591-019-0343-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li H, Durbin R. Bioinformatics. 2010;26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li H, et al. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.