Abstract
Histone H3 K27M mutation is the defining molecular feature of the devastating pediatric brain tumor, diffuse intrinsic pontine glioma (DIPG). The prevalence of histone H3 K27M mutations indicates a critical role in DIPGs, but the contribution of the mutation to disease pathogenesis remains unclear. We show that knockdown of this mutation in DIPG xenografts restores K27M-dependent loss of H3K27me3 and delays tumor growth. Comparisons of matched DIPG xenografts with and without K27M knockdown allowed identification of mutation-specific effects on the transcriptome and epigenome. The resulting transcriptional changes recapitulate expression signatures from K27M primary DIPG tumors, and are strongly enriched for genes associated with nervous system development. Integrated analysis of ChIP-seq and expression data showed that genes upregulated by the mutation are overrepresented in apparently bivalent promoters. Many of these targets are associated with more immature differentiation states. Expression profiles indicate K27M knockdown decreases proliferation and increases differentiation within lineages represented in DIPG. These data suggest that K27M-mediated loss of H3K27me3 directly regulates a subset of genes by releasing poised promoters, and contributes to tumor phenotype and growth by limiting differentiation. The delayed tumor growth associated with knockdown of H3 K27M provides evidence that this highly recurrent mutation is a relevant therapeutic target.
Keywords: Histone H3K27M, DIPG, glioma, epigenetic, H3K27me3, bivalent, oncohistone, isogenic knockdown, differentiation, stemness
INTRODUCTION
Diffuse intrinsic pontine gliomas (DIPGs) comprise approximately half of childhood high-grade gliomas (HGGs). There is a dire need for new therapeutic approaches, as 90% of DIPG patients succumb to the disease within two years of diagnosis [48]. DIPG incidence peaks between 6–8 years of age, and these deadly tumors are rarely found outside of the pediatric population. Although the histopathological features of DIPG match those of HGGs arising in other brain regions and across different age groups, there are unique molecular features pointing to a distinct pathogenesis. Nearly 80% of DIPGs contain histone H3 K27M somatic mutations [50]. While this mutation represents a unifying feature of DIPGs, there are a diverse array of other mutations found at much lower frequency in DIPGs, contributing to significant intertumoral heterogeneity [6, 16, 22, 30, 45, 51]. Outside the brainstem, H3 K27M mutations are also found in pediatric HGGs arising in other midline structures, predominantly the thalamus, but also cerebellum and spinal cord [41, 50]. In contrast, approximately 15% of HGGs arising in the cerebral hemispheres have alternate histone H3 mutations, G34R or G34V, which are found predominantly in older adolescents through young adulthood [22, 43]. This exquisitely specific association between the particular histone H3 mutation and spatiotemporal features of tumor origin highlights a critical connection between epigenetic dysregulation and brain development in which these mutations must confer a context-specific selective advantage.
H3 K27M causes a dominant and dramatic reduction in H3K27me3, a posttranslational modification (PTM) associated with transcriptional repression [5, 8, 27, 46]. While H3 K27M mutations are primarily restricted to pediatric brain tumors, alternative mutational mechanisms to dysregulate H3K27me3 are found in diverse cancers. For example, loss of function mutations in components of polycomb repressive complex 2 (PRC2), the complex that methylates H3K27, are found in multiple tumor types including early T-cell precursor acute lymphoblastic leukemia, myeloid malignancies, and head and neck squamous carcinomas [12, 35, 36, 42, 53]. However, gain of function mutations causing enhanced activity of EZH2, the histone methyltransferase component of PRC2, are found in non-Hodgkin lymphomas as well as other solid tumors [13]. Thus, perturbations resulting in increased or decreased levels of H3K27me3 can contribute to cancer in different contexts.
Several inhibitors of epigenetic regulators have shown promise in inhibiting DIPG growth in preclinical studies. Paradoxically, inhibition of H3K27me3 demethylases to attempt to restore levels of H3K27me3, or inhibition of EZH2, which drives further loss of H3K27me3, both inhibited DIPG growth [19, 32, 40]. HDAC inhibition, or transcriptional disruption through inhibiting BET domain proteins or CDK7 also inhibited DIPG growth [18, 34, 40]. While these findings offer possible new approaches for DIPG, the oncogenic mechanism of action downstream of H3 K27M remains unclear.
Understanding how effects of H3 K27M on the epigenome impact the transcriptome and oncogenic signaling is limited by the paucity of DIPG tumors with wild type H3 as a relevant reference, and further confounded by the intertumor genetic heterogeneity of DIPGs. Comparisons of signatures from midline and DIPG tumors or cell lines, which have K27M mutations, with cortical HGGs or cell lines, where K27M mutations are very rarely found, have provided important insights into the epigenetic and transcriptional signatures of these groups, demonstrating clear differences in genome-wide DNA methylation patterns that can reliably classify pediatric brain tumors [5, 7]. However, it is not possible to extricate the consequences of the H3 K27M mutation from the developmental signature of the tumor origin using primary tumors from different locations.
In this study, we knocked down expression of H3.3 K27M in three independent DIPG xenograft models to determine the biological contribution of the mutation to tumor growth. This approach also allowed us to identify K27M-dependent epigenetic and transcriptional signatures in the relevant tumor context using paired isogenic comparisons of DIPGs with or without histone H3.3 mutations. By removing the heterogeneity contributed by variations in age, location of tumor origin, differentiation state of cell of origin, and presence of different mutations that could confound identification of K27M signatures when comparing independent primary human tumors, we identified connections between the K27M-dependent epigenetic landscape and selective gene expression.
Materials and Methods
EXPERIMENTAL MODEL AND SUBJECT DETAILS
DIPG xenografts carrying H3F3A K27M mutations: SJDIPGX7 (H3F3AWT/K27M) and SJDIPGX37 (H3F3AK27M with loss of the wild type allele) were established by direct implantation of tumor cells from autopsy samples into recipient CD1-nude mice, 6 weeks or older. SJDIPGX7 also contains TP53 C176Y, PIK3CA E542K, and BCOR frameshift mutations. For SJDIPGX37, loss of chromosome 1q in the primary tumor implanted to generate the xenograft resulted in a hemizygous H3F3A K27M allele. It also contains an indel in PIK3R1 N453, and a PPM1D L513 frameshift. SJHGGX2 was established as an in vitro tumorsphere line from a surgical sample of a pediatric cerebellar glioblastoma, and subsequently implanted into CD1 nude mouse brain to generate a xenograft line that is passaged by serial transplants. It contains a PDGFRA N659K, TP53 C135W, but is wild-type for histone H3. Xenografts and cell lines were established with informed consent and approval from the St Jude Institutional Review Board. SUDIPG-VI, a gift from Dr. Michelle Monje (Stanford University), contains H3F3A K27M and TP53 mutations and was derived from an autopsy sample that was established and maintained as an in vitro cell line [18], and implanted into mouse brain for the in vivo tumorigenesis studies here. Each xenograft line was transduced with a lentivirus to express luciferase and yellow fluorescent protein (CL20-luc2aYFP), and maintained by successive implants of tumor cells in the brain cortex of CD-1 nude immunocompromised mice. Mice were maintained in an accredited facility of the Association for Assessment of Laboratory Animal Care in accordance with the NIH guidelines. The Institutional Animal Care and Use Committee of SJCRH approved all procedures in this study.
METHODS DETAILS
Transduction, ex vivo selection and implantation of xenograft cells
Tumor cells from SJDIPGX7 and SJDIPGX37 xenografts were dissected from the brain, mechanically dissociated with gentle pipetting, and kept as tumorspheres in Ultra Low Attachment flasks (Corning) under neural stem cell conditions (Neurobasal Medium, B-27, N-2, GlutaMAX, and growth factors hEGF, hFGF-2, PDGF-AA, hPDGF-BB at 20ng/mL each). Dissociated tumor cells were transduced for 3h with lentivirus and incubated for 48h prior to puromycin selection for 12 days. Tumorspheres for the pooled selected cells for each lentiviral transduction were mechanically disrupted to single cells before resuspension in matrigel and reimplantation into brain. For each experiment, a pool of acutely isolated PDX tumor cells was divided and transduced with control or knock-down viruses, and the same number of transduced cells was implanted for all mice in the cohort.
Intracranial implantations were completed as described [11]. Live animal imaging of tumor luciferase activity was carried out as in [33]. Radiance (photons/sec/cm2/steridian) was determined within regions of interest covering the head of each mouse using Xenogen IVIS-200 (Caliper). Mice were monitored daily for neurological and health symptoms and euthanized at a humane endpoint.
Lentiviral shRNA
Initial attempts to knockdown K27M mutant H3F3A without also targeting the wild type allele were unsuccessful. Therefore, we designed shRNAs highly specific for either H3F3A or H3F3B, the latter serving as a control for the knockdown of wild type H3.3. Sequences for specific shRNAs were cloned into the pGIPZ vector, which utilizes the backbone of miR30 for improved knockdown efficiency. We also adapted the pGIPZ vector to replace the miR30 backbone a miR-E backbone [14], and exchanged turboGFP with turboRFP (Evrogen). Specific shRNA sequences were either custom designed or obtained from the pGIPZ library of constructs, and adapted for cloning into the vector using the shRNA Retriever online tool. shRNA sequences are provided in [Supp. Table 1 (Online Resource 1)]. H3F3A specific shRNAs are referred to as shK27M when used in K27M mutant xenografts and shH3F3A when used in the H3 wild type xenograft. The notation shK27M refers to both H3F3A hairpins shK27M#1 and shK27M#2.
Immunofluorescence and Immunohistochemistry
Formalin-fixed paraffin-embedded tissue was sectioned and deparaffinized, and antigen retrieval was performed by microwave oven heating in sodium citrate buffer (10 mM, pH 6.0) for 10min. Primary antibodies (1:1000 except where noted) were turboGFP (Evrogen); MAG (1:2000) and Ki67 (Abcam); H3K27me3 (Cell Signaling); and Vimentin (1:200, Dako). Secondary antibodies (1:1000) were anti-rabbit Alexa 555, anti-mouse Alexa 488 and anti-rat Alexa 647 (ThermoFisher). Slides were stained with DAPI and mounted in Prolong Diamond (both ThermoFisher).
Western blotting
Histones were acid extracted using Histone Purification Mini Kit (Active Motif) from fresh frozen tumor tissue. Total protein stained with REVERT (LI-COR). Primary antibodies (1:1000 except where noted) were H3K27me3, H3K4me3 and H3K36me3 (Cell Signaling); H3K27ac (Active Motif); H3K27M (1:500) and H3.3 (Millipore Sigma); and H3 (Abcam). Secondary antibodies (1:10,000) were IRDye 800CW anti-rabbit and 680LTy anti-mouse (LI-COR). See also STAR methods. Blots imaged with Odyssey Fc and Image Studio software (LI-COR).
RNA and quantitative reverse transcription PCR
RNA was extracted using Trizol (Invitrogen) following the manufacturer’s protocol except for adding two additional 70% EtOH washes after precipitation. RNA quantity and purity were assessed by Nanodrop (Thermo Fisher). cDNA was generated by reverse transcription using Superscript III (Invitrogen). Gene expression was evaluated on a CFX-96 (Bio Rad) using QuantiTect (Qiagen) or iTaq (Bio Rad) SYBR. Primer sequences were designed with PrimerBLAST (NCBI) and are provided in [Suppl. Table 2 (Online Resource 1)]. Values shown on plots are normalized by GAPDH.
Chromatin Immunoprecipitation (ChIP)
ChIP and library construction for SJDIPGX7 xenografts (antibodies: H3K27me3 (Millipore) and H3K4me3 (Active Motif)) was performed by Active Motif. All other ChIP was performed at St. Jude using the following method. Xenograft tumors were snap frozen and ground to a powder before fixation. Single cell suspensions were generated from cultured cells then fixed. For histone ChIP, samples were fixed for 5min with 1% paraformaldehyde (PFA) in PBS at room temperature (RT). Fixation was quenched with 250mM Glycine, samples were pelleted and washed 2x with PBS. Pellets were resuspended in lysis buffer (50mM HEPES pH7.9, 140mM NaCl, 1mM EDTA pH8.0, 10% Glycerol, 0.5% Nonidet P-40, 0.25% Triton X-100) for 10min on ice for cell suspensions or for 20min with rotation at 4°C for tumors. Pellets of bare nuclei were washed 2x in wash buffer (10mM Tris HCl pH 8.0, 200mM NaCl, 1mM EDTA pH8.0, 0.5mM EGTA pH8.0), 2x shearing buffer (10mM Tris HCl pH8.0, 1mM EDTA pH8.0, 0.1% SDS) then resuspended in shearing buffer. Chromatin was sheared on a Covaris M220 ultrasonicator, then centrifuged remove cellular debris. For histone mark ChIPs, the reads arising from mouse cells entrapped within the xenograft were used to normalize the post ChIP samples to the human/mouse read ratio obtained for the pre-ChIP material (input library). For some experiments, chromatin from Drosophila S2 cells was also spiked in to chromatin before dividing sample for input control and ChIP and used to validate the use of mouse reads for normalization [21]. ChIP reactions were performed using a modified Upstate Biotechnology protocol. Briefly, sheared chromatin was diluted 1:10 with dilution buffer (21mM Tris HCL pH8.0, 1mM EDTA pH8.0, 167mM NaCl, 1.1% Triton X-100, 0.1% SDS), precleared with Protein A sepharose beads (GE Healthcare) and bovine serum albumin (BSA) at 4°C for 1–2h. The precleared chromatin was then rotated overnight at 4°C with the antibody of interest (see below), Protein A sepharose beads and BSA. The bead-bound chromatin was washed once each with low salt buffer (20mM Tris HCl pH8.0, 2mM EDTA pH8.0, 150mM NaCl, 1% Triton X-100, 0.1% SDS), high salt buffer (20mM Tris HCl pH8.0, 2mM EDTA pH8.0, 500mM NaCl, 1% Triton X-100, 0.1% SDS), LiCl buffer (10mM Tris HCl pH8.0, 1mM EDTA pH8.0, 250mM LiCl, 1% Nonidet P-40, 1% Deoxycholate) and twice with TE (10mM Tris HCl pH8.0, 1mM EDTA pH8.0). The chromatin was eluted off the beads with 0.1M NaHCO3 in 1% SDS by rotating at RT for 30min, the supernatant was incubated with 200nM NaCl overnight at 65°C, then incubated 2h at 37°C with 10µg Proteinase K, then cleaned up using a QiaQuick PCR Purification Kit (Qiagen) and eluted in 50μl 10mM Tris HCl pH8.5. Antibodies used were: H3K27me3 (Cell Signaling 9733), and H3K4me3 (Cell Signaling 9751).
Library Preparation and Sequencing
For RNA-seq, RNA quality was checked by 2100 Bioanalyzer RNA 6000 Nano assay (Agilent) or LabChip RNA Pico Sensitivity assay (Perkin Elmer) before library generation. Libraries were prepared from total RNA with the TruSeq Stranded Total RNA Library Prep Kit (Illumina). ChIP-seq libraries were prepared from 2–10ng of DNA using the NEBNext ChIP-Seq Library Prep Reagent Set for Illumina with NEBNext Q5 Hot Start HiFi PCR Master Mix according to the manufacturer’s instructions (New England Biolabs) with the following modifications: a second 1:1 Ampure cleanup was added after adaptor ligation. The Ampure size selection step prior to PCR was eliminated. Completed ChIP-seq libraries were analyzed for insert size distribution on a 2100 BioAnalyzer High Sensitivity kit (Agilent) or Caliper LabChip GX DNA High Sensitivity Reagent Kit (Perkin Elmer). All libraries were quantified using the Quant-iT PicoGreen dsDNA assay (Life Technologies), Kapa Library Quantification kit (Kapa Biosystems) or low pass sequencing on a MiSeq Nano v2 run (Illumina). One hundred cycle paired end sequencing (RNA-seq) or fifty cycle single end sequencing (ChIP-seq) was performed on an Illumina HiSeq 2500 or HiSeq 4000.
QUANTIFICATION AND STATISTICAL ANALYSIS
RNA-seq mapping and analysis
Total stranded RNA sequencing data was generated and mapped against human genome assembly GRCh37 using the StrongArm pipeline described previously [49] We used the XenoCP method to remove mouse reads misaligned to the human genome using (in preparation; Michael C Rusch, Xiang Chen, Liang Ding, Hongjian Jin, Gang Wu, Lei Wei, Michael A. Dyer, and Jinghui Zhang. Mouse read cleansing in xenograft sequencing data improves the accuracy of mutation discovery and gene quantification). The gene level read count values were obtained with HT-seq using human GENCODE annotation Release 19. Low expression genes that did not meet the threshold CPM>1 in at least 3 samples were excluded, and expression values normalized using TMM method and default settings (edgeR package). Differential expression analysis was performed with Voom and Limma methods (Limma package). Significantly up-and down-regulated genes by K27M were defined by at least 1.5 fold change with FDR adjusted p-value <0.05. A permutation procedure was used to evaluate enrichment of promoter bivalency status among differentially expressed genes. The Kruskal-Wallis test statistic was used to characterize differences in the distribution of the genes empirical Bayes t-statistics (Limma R package) according to their promoter bivalency status. The statistical significance of the Kruskal-Wallis test statistic (p value) was determined by repeating the differential expression analysis described above with permuted assignments of the treatment labels to expression profiles. In each analysis, the smaller of all possible or 1000 randomly selected permuted assignments were evaluated to compute the p value. SJDIPGX7, SJDIPGX37 and SU-DIPGVI all have the H3F3A K27M mutation, but show tumor-dependent differences in gene expression signatures that can obscure the effects of H3K27M knockdown. To remove this noise and clarify the K27M-dependent changes in gene expression, in combined analyses to identify up-regulated or down-regulated genes, the knockdown effects were adjusted for the difference between xenograft models by treating xenograft model as a covariate.
Gene expression signature analyses
Gene Set Enrichment Analysis (GSEA 3.0) analysis [44] was performed with 10,000 geneset permutations and default settings. Genes were ranked using signal-to-noise ratio, as provided by GSEA.
Single Sample GSEA (ssGSEA) was performed as in [2] through Gene Pattern server. RNA-seq data from human DIPG tumors was from published data [51].
This dataset was refined to only contain: confirmed H3.3 K27M bearing DIPGs (K27M) or DIPGs that were confirmed to be wild type for all genes encoding H3 histones (H3 WT); we also excluded all tumors showing genomic or transcriptomic evidence of MYC amplification, as our xenograft models did not include MYC amplified tumors (both K27M and H3 WT). We also excluded tumors (both K27M and H3 WT) that classified as tumor types distinct from diffuse midline glioma H3 K27M mutant in the DKFZ MolecularPathology.org tool [7].
Genesets used for GSEA and ssGSEA were: PRC2 targets in ES cells [4], Mature Oligo [54], Fetal Astro [55], OPC, Neural Tube Midline, [23], Housekeeping [52], single cell DIPG signatures [15]. Unsupervised scaled Principle Component Analysis (PCA) was performed on TMM normalized log2 CPM counts for all genes with FactoMineR package [26]. Gene Ontology term enrichment was performed with Panther Classification System [31]. Only transcripts annotated with HGNC Gene Symbol ID were used in the analysis.
Colors represent the difference of ssGSEA scores of K27M vs. shK27M samples, where the mean for each K27M/shK27M pair was set to zero (white).
ChIP-Seq Analysis
Mapping reads and visualizing data
Human, mouse and drosophila genomes were used in our study and we removed cross aligned reads by a competitive mapping method. Mouse chromosomes were renamed as “m.chr”, Drosophila chromosomes were renamed as “d.chr”, human chromosomes were not renamed and then the three fasta files (human, mouse and drosophila) were concatenated as one reference genome. We used BWA (version 0.7.12; default parameter) to align the reads simultaneously to the human, mouse and Drosophila melanogaster hybrid reference genome (hg19+mm9+dm3). We only used reads uniquely mapped to human or mouse or Drosophila with MAPQ >=1 in our analysis. We marked duplicated reads with Picard (version 1.65), with only nonduplicated reads kept by samtools (version 0.1.18, parameter ‘‘-q 1 -F 1024’’). Mapped human reads were extracted for downstream analysis. To control the quality of the data and estimate the fragment size, the nonduplicated version of SPP (version 1.11) was used to draw cross-correlation and calculate relative strand correlation value with support of R (version 2.14). Upon manually inspecting the cross-correlation plot generated by SPP, the smallest fragment size estimated by SPP was used to extend each read and to generate bigwig file for visualization on integrated genome viewer (IGV) (version 2.3.82 ). We scaled coverage according to spike-in normalization factor [37] to generate bigwig track for each sample. To show average of several replicates as a single track in the browser, the bigwig files were merged to a single average bigwig file BedGraph file using UCSC tools bigWigtoBedGraph, bigWigMerge and bedGraphToBigWig. The effect of shRNA treatment on the proportion of ChIP-Seq reads mapped to specific types of genetic loci (genic, enhancer, repeats, others) was statistically evaluated by fitting a linear model with animal model identifier and treatment as predictors of the logit-transformed ChIP-seq read proportions. The model estimates were then back-transformed to the original proportional abundance scale.
Spike-in normalization, differential analysis and peak annotation
ChIP-seq raw read counts were reported for each region/each sample using bedtools 2.25.0. Some samples had Drosophila chromatin spike-in added to the chromatin before separating samples for input and ChIP-seq. In some samples this was not available, so we used the amount of mouse DNA from entrapped mouse cells in the xenograft tumor for normalization. We used the samples with Drosophila spike-in to show that normalization with contaminating mouse gave comparable results. Then, for consistency, we used the mouse reads for normalization of all samples [21]. The spike-in normalization was performed by counting contaminating mouse reads and human reads in each IP sample and corresponding chromatin Input sample and using those counts to generate a spike-in normalization factor for each sample, which was calculated as (IP_mouse.reads/IP_human.reads)/(INPUT_mouse.reads/INPUT_human.reads). Raw read counts were voom normalized and statistically contrasted using the pipeline limma in R (version 3.30.13). Normalization factor defined above was used to modify human library size in edgeR (version 3.16.5) for counts per million (CPM) calculation and differential analysis. An empirical Bayes fit was applied to contrast K27M samples to shK27M samples and to generate log2 fold changes, p-values and false discovery rates for each peak region. For signal visualizations of ChIP-seq peaks and promoters, CPM values were log2 transformed. Binned counts for heat maps and histograms showing average ChIP-seq intensity around TSS (±2000bp) or across gene bodies were generated using annotatePeaks.pl, a program from HOMER suite (version 4.8.3, http://homer.salk.edu/homer/), or ngsplot (version 2.61), which were modified to allow library sizes to be adjusted according to their respective spike-in normalization factors.
Peak calling, promoter and enhancer characterization
MACS2 (version 2.1.1 20160309) was used to call narrow peaks (H3K4me3) with option “nomodel” and “extsize” defined as fragment size estimated above, FDR corrected p-value cutoff 0.01. For broad peaks (H3K27me3), SICER (version 1.1, with parameters of redundancy threshold 1, window size 200bp, effective genome fraction 0.86, gap size 600bp, FDR 0.00001 with fragment size defined above) was used for domain calling. Enriched regions were identified by comparing the IP library file to input library file. Peak regions were defined as the union of peak intervals in both K27M and shK27M samples. Promoters were defined as human RefSeq TSS ± 1000bp regions. For bivalent promoter analysis, MACS2 calls were used for both H3K27me3 and H3K4me3.
Retained H3K27me3 peaks/promoters in K27M:
Using the ChIP signal in the union of SICER H3K27me3 peaks between K27M and shK27M samples, H3K27me3 peaks retained in K27M were defined as those with K27M/shK27M log2FC > 0 and K27M signal > 0. Promoters with retained H3K27me3 peaks in K27M were defined as those with K27M/shK27M logFC > −0.15 in the ChIP signal in the 2kb surrounding the TSS and a MACS called H3K27me3 peak overlapping the promoter region in any K27M sample.
Analysis of Apparent Bivalency
Individual shK27M tumors (n=4 for SJDIPGX37 and n=3 each for SJDIPGX7 and SUDIPG-VI) that had both H3K27me3 and H3K4me3 ChIP-seq data were scored for the presence of MACScalled peaks overlapping the 2kb surrounding gene TSS, and promoters were binned into four categories: H3K27me3-H3K4me3-, H3K27me3+ H3K4me3-, H3K27me3-H3K4me3+, and H3K27me3+ H3K4me3+ (apparent bivalency). To be binned as H3K27me3+ H3K4me3+, both H3K27me3 and H3K4me3 peaks at a promoter had to co-occur in a single tumor.
RESULTS
H3.3 K27M knockdown delays tumor growth of DIPG-derived xenografts
To assess the role of the K27M mutation in oncogenesis, we utilized a lentiviral shRNA knockdown approach in three independent DIPG-derived lines bearing a K27M mutation in H3F3A; SJDIPGX7 and SJDIPGX37, both established from autopsy samples by direct implantation into mouse brain and passaged by serial implantation, and SUDIPG-VI, an autopsy sample established as an in vitro cell line [18] that we used for intracranial implantation in this study. Luciferase was introduced into all three lines by ex vivo lentiviral transduction to facilitate in vivo imaging of tumor growth. To knockdown H3F3A, xenograft cells were transduced with lentiviruses carrying shRNA, as well as a fluorescence marker and a puromycin resistance gene [Suppl. Fig. 1a (Online Resource 2)]. We tested the effects of two different shRNAs directed against H3F3A (shK27M) compared to two kinds of control: a non-silencing shRNA with no target in the human genome (referred to hereafter as K27M), or a shRNA targeting H3F3B (shH3F3B), the homolog of H3F3A that encodes the identical histone H3.3, but does not contain the K27M mutation. After selection ex vivo with puromycin, the transduced cells were reimplanted into the brain of immunocompromised mice and monitored for tumor progression by in vivo imaging of luciferase activity [Fig. 1a, and Suppl. Fig. 1b (Online Resource 2)].
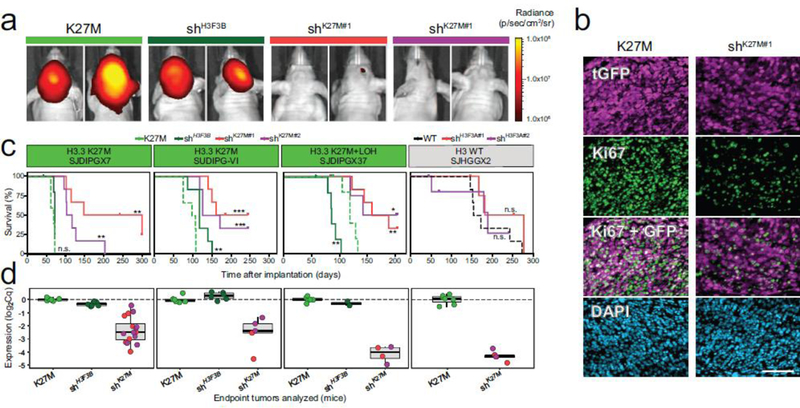
Figure 1. Knockdown of H3.3 K27M selectively delays tumor growth and extends survival of mice harboring H3.3 K27M mutant DIPG xenografts.
a. K27M mutant DIPG xenografts (SJDIPGX7, SUDIPG VI, and SJDIPGX37) expressing shRNAs targeting H3F3A (shK27M#1 and shK27M#2), control H3F3B (shH3F3B), or non-silencing shRNA (K27M) were implanted in brain of immunocompromised mice. Tumor growth was monitored by in vivo luciferase activity imaging. Equal numbers of cells for each construct were implanted on day 0, and representative images of SJDIPGX7 mice 42 days after implantation are shown. b. Tumors showed lower proliferation rate as measured by Ki67 staining upon H3F3A knockdown. Immunofluorescent detection of tGFP (magenta, marking shRNA transduced cells), Ki67 (green), and DAPI stain (blue) of nuclei is shown for sections of SJDIPGX7 xenografts. Scale bar is 100μm. c. Kaplan-Meier survival curves show significant delay in tumor growth upon knockdown with two H3F3A-specific shRNAs (shK27M#1, red and shK27M#2, purple) compared to non-silencing control (K27M, dotted light green line) in K27M mutant xenografts, SJDIPGX7, SUDIPG VI, and SJDIPGX37. H3F3B knockdown (shH3F3B, solid dark green line) did not consistently delay K27M mutant tumor growth. Knockdown of H3F3A (shH3F3A#1, red and shH3F3A#2, purple lines) did not significantly delay growth of an H3 wild type HGG xenograft (SJHGGX2) compared to non-silencing control (H3F3A WT, dotted black line). Non-silencing control shRNA is shown with a solid line on all plots. shH3F3A#1 and shK27M#1 are the same construct, as are shH3F3A#2 and shK27M#2. Mice were euthanized when showing clear neurological symptoms. Log-rank test p-values for comparisons to non-silencing controls are shown. n=6 for all groups except SJHGGX2 shH3F3A#1 where n=5. All shRNAs shown had a miR-E backbone. d. Potent H3F3A knockdown in endpoint tumors shown in panel C was measured by qRT-PCR. Expression of H3F3A relative to GAPDH is shown. * p<0.05, ** p<0.01, *** p<0.001, ns = not significant. See also [Suppl. Fig. 1 (Online Resource 2)].
We found that knockdown of H3.3 K27M with two different shRNAs, but not knockdown of H3F3B encoding wild type H3.3, resulted in slower tumor growth, as demonstrated by live tumor imaging (Fig. 1a). shK27M tumors also displayed a significantly lower proliferative index as measured by Ki67 expression compared to K27M tumors [p=0.003, Fig. 1b and Suppl. Figure 1c (Online Resource 2)]. Knockdown of H3F3B led to a slight extension of survival in SUDIPG-VI, but this was not observed in the other two PDXs, and was less significant than the change in survival induced by H3F3A shRNAs. H3F3B knockdown did not significantly change survival compared to the non-silencing control in SJDIPGX7. SJDIPGX37 contains an H3F3A K27M mutation, and loss of heterozygosity of chromosome 1q containing the second H3F3A allele. Strikingly, knockdown of H3F3B in this line, which would leave H3K27M as the predominantly expressed H3.3 in the cell, significantly accelerated tumor growth in SJDIPGX37. Ultimately, H3F3A knockdown using two different shRNAs significantly extended survival compared to the non-silencing control for all three DIPG xenograft models, while H3F3B knockdown did not consistently increase mouse survival (Fig. 1c and Suppl. Fig.1d (Online Resource 2)]. By contrast, in a cerebellar pHGG xenograft line with no histone H3 mutation (SJHGGX2; H3F3AWT/WT), efficient knockdown of H3F3A had no significant impact on tumor growth (Fig. 1c). Hematoxylin and eosin staining of K27M and shK27M xenograft tissue sections showed that both replicate the variable histopathologic features of primary DIPGs [Suppl. Fig. 1e–h (Online Resource 2)].
Delayed tumor growth showed that K27M depletion imparted a selective disadvantage; however, shK27M endpoint tumors showed robust knockdown as confirmed by quantitative real time reverse transcriptase polymerase chain reaction (qRT-PCR) (Fig. 1d). Thus, RNA interference generated K27M knockdown tumors for isogenic comparison with control tumors to identify the selective effects of K27M mutation. Together, these results indicate that the K27M mutation plays an ongoing role in the expansion of the tumor, rather than only setting a susceptible state for transforming effects of other mutations.
H3.3 K27M deprivation increases global H3K27me3 and reduces H3K27ac
The dominant negative effect of the K27M mutation on levels of H3K27me3 has been well documented and was also evident in our DIPG xenograft model system. The global reduction of H3K27me3 in the K27M tumors was alleviated in shK27M tumors as demonstrated by immunofluorescence (IF) (Fig. 2a).
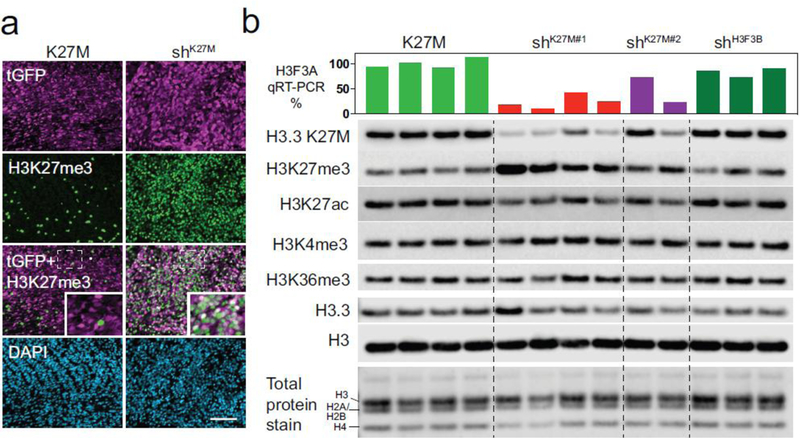
Figure 2. H3.3 K27M depletion restores global H3K27me3 and reduces H3K27ac.
a. Representative immunofluorescence image showing restoration of H3K27me3 in DIPG xenografts after knockdown of K27M. Non-silencing control (K27M) and H3F3A knockdown (shK27M) tumor cells stained for tGFP (magenta), H3K27me3 (green), and DAPI (blue). H3K27me3 positive, GFP negative cells in K27M tumors represent mouse cells entrapped within xenograft. Scale bar is 100μm. Area in dotted rectangle is enlarged in inset. b. Western blot of acid extracted histones from SJDIPGX7 xenografts with knockdown of H3F3A (shK27M#1 and shK27M#2) showing reduction of the H3.3 K27M protein detected with a mutantspecific antibody, (H3.3 K27M), and restoration of H3K27me3 compared to xenografts transduced with non-silencing control (K27M, leftmost four lanes) or shH3F3B control (rightmost 3 lanes). Level of H3.3 K27M protein loss and reciprocal H3K27me3 gain correlates with the decrease of H3F3A mRNA measured by qRT-PCR (shown as bar graph above western blot). H3K27ac is reduced in shK27M compared to K27M tumors, but H3K4me3 and H3K36me3 are not globally affected. Total H3 protein and total histone protein staining were used as loading controls.
The degree of H3K27me3 restoration correlated well with the extent of knockdown in tumors by western blot (Fig. 2b; bar graph above western indicates H3F3A RNA expression level for each tumor). At the global level, in addition to increased H3K27me3, shK27M tumors also showed reduced H3K27ac, while H3K4me3 and H3K36me3 levels were not impacted (Fig. 2b). Importantly, shH3F3B tumors did not show decrease in H3.3 K27M, or changes in the global levels of any histone H3 PTMs compared to K27M tumors (Fig. 2b), demonstrating that the alterations in H3K27 PTMs seen with H3F3A knockdown were a result of K27M reduction, not a change in the total amount of H3.3.
H3.3 K27M knockdown drives gene expression changes that recapitulate signatures found in primary DIPG tumors
One of the major problems in determining the effect of the K27M mutation on gene expression based on primary tumor data is the confounding impact of the extensive heterogeneity between tumors arising in different individuals. The isogenic comparison of K27M versus shK27M tumors for an individual xenograft model provides a means to address this issue. The extent of knockdown was somewhat variable between tumors. To maximize the clarity of the K27M-dependent signature, we only used tumors with 65%−94% knockdown of H3F3A for RNA-seq and ChIP-seq studies. Unsupervised clustering showed that the two different shRNAs for H3 K27M drove shared changes in gene expression in each of the three xenograft models [Suppl. Fig. 2a (Online Resource 2)]. An unsupervised Principal Component Analysis (PCA) of RNA-seq from tumors with or without K27M knockdown from all three DIPG xenograft models, shows that the majority of variation is represented by differences between the xenograft models (Fig. 3a, xenograft line identity dominates PC1 and PC2). The effect of K27M knockdown, while smaller than the variability between xenograft models is shown robustly in principal component 3 (Fig. 3a, PC3).
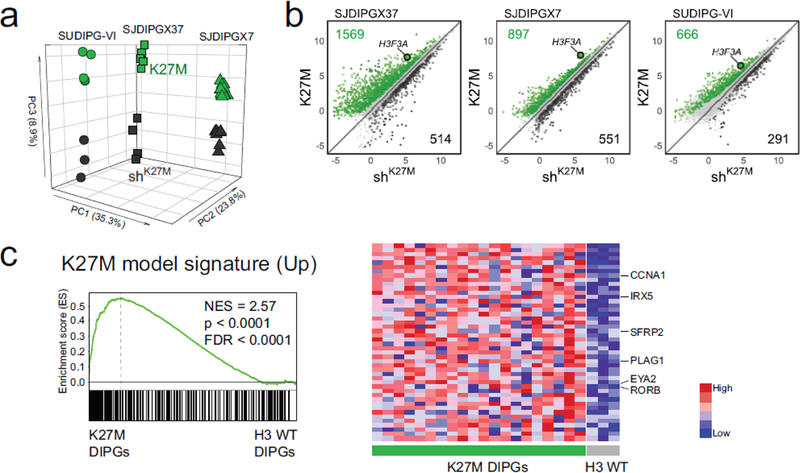
Figure 3. H3.3 K27M knockdown drives gene expression changes that recapitulate signatures found in primary DIPG tumors.
a. Unsupervised PCA for all three K27M xenograft models; SJDIPGX37 (squares), SJDIPGX7 (triangles) and SUDIPG VI (circles). PCA generated using all expressed genes from all samples. K27M (green) and shK27M (black). b. RNA-seq log2 CPM signal from K27M mutant xenografts transduced with non-silencing (K27M) or H3F3A shRNA (shK27M). Genes differentially regulated |FC|>1.5 and adjusted p-value <0.05 are marked; green are K27M-induced and black K27M-repressed. H3F3A is highlighted to show effect of knockdown. SJDIPGX7 K27M n=13, shK27M n=8; SJDIPGX37 K27M n=5, shK27M n=4; SUDIPG-VI K27M n=5, shK27M n=3. c. GSEA using the top 300 (ranked by limma p-value) K27M-induced genes derived from a combined analysis of K27M vs. shK27M for all three xenograft models applied to primary human DIPGs. K27M-induced genes (downregulated upon knockdown) are enriched in the primary K27M DIPG tumors in comparison to H3 WT DIPGs. Heat map shows the relative expression of the top 50 genes in the K27M (left side) and H3 WT (right side) DIPGs. See also Suppl. Fig. 2 (Online Resource 2) and Suppl. Tables 1 and 2 (Online Resource 1)].
While H3F3A knockdown produced global restoration of the repression-associated H3K27me3, the effect on gene expression for all three xenograft models was much more focused and resulted in differential expression of genes in both directions [(Fig. 3b and Suppl. Fig. 2b (Online Resource 2); at least 1.5 fold change (FC), adjusted p-value < 0.05]. As would be expected with a global decrease in the repression-associated PTM H3K27me3, there was a higher proportion of genes induced by K27M [Fig. 3b and Suppl. Fig. 2b (Online Resource 2), K27M-induced genes and their total number colored in green] compared to K27M-repressed genes (Fig. 3b and Suppl. Fig. 2b, colored in black (Online Resource 2)]. A comparison of the K27M downregulated gene lists between the three xenograft lines found 17 genes in common. This common downregulated gene list did not show significant enrichment for genes in Enrichr libraries: Epigenomics Roadmap HM ChIP-seq, ENCODE Histone Modifications 2015 and ENCODE TF ChIP-seq 2015. By contrast, the 78 genes in common between the three K27M upregulated gene lists resulted in 44 gene sets with significant adjusted p value in the same three Enrichr libraries, of which 40 were H3K27me3 or PRC2 component gene sets. This analysis is consistent with K27M upregulated genes being regulated directly by H3K27me3 loss, while K27M downregulated genes are indirectly regulated [Suppl Table 3 (Online Resource 3]. The ratio of K27M-induced to repressed genes varied by xenograft model but the ratio always favored K27M-induced genes. By comparison, while H3F3B knockdown produces a clear decrease in the H3F3B transcript, few other genes are differentially regulated [Suppl. Fig. 2c (Online Resource 2)].
The genes significantly upregulated by the K27M mutation differed considerably in between the three DIPG xenograft models [Suppl. Table 3 (Online Resource 3)]. Nonetheless, Gene Set Enrichment Analysis (GSEA) revealed that these three K27M-induced gene lists are cohesively and significantly enriched in the H3.3 K27M compared to H3 WT primary DIPGs [Suppl. Fig. 2d (Online Resource 2)]. We next performed a combined analysis of all three DIPG xenograft models to derive a K27M expression signature that was robust between the models [Suppl. Table 3, Combined (Online Resource3)]. Genes in the combined K27M-induced signature are collectively associated with the biological processes of neurogenesis (p=1.32E-20) and nervous system development (p=4.84E-23) (Gene Ontology (GO) [1]. Critically, GSEA showed that the top 300 genes from the combined K27M-induced signature were significantly enriched in the H3.3 K27M primary DIPGs [Fig. 3c and Suppl. Table 4 (Online Resource 4)]. As would be expected with loss of H3K27me3, a list of PRC2 target genes [4] was significantly enriched in K27M compared to shK27M xenografts by GSEA [Suppl. Fig. 2e (Online Resource 2)]. Thus, gene expression changes associated with H3F3A knockdown in xenografts recapitulate K27M signatures seen in primary patient DIPGs and these signatures show overlap with PRC2 targets.
H3.3 K27M knockdown produces global changes in the level of H3K27me3 without redistribution to different genomic regions
We next performed chromatin immunoprecipitation with high throughput sequencing (ChIP-seq) to investigate how the global effect of K27M on H3K27me3 impacted the epigenetic profile of these xenograft models in vivo. Normalized H3K27me3 ChIP-seq confirmed that knockdown of the K27M mutation reversed global loss of H3K27me3. Analysis of the union of H3K27me3 peaks called in either the K27M or shK27M tumors revealed that the global loss of H3K27me3 associated with the K27M mutation affects nearly every locus, with the exception of a very small fraction of locations maintaining similar levels or gaining H3K27me3 in the presence of K27M [(Fig. 4a and Suppl. Fig. 3a (Online Resource 2)]. For all three xenograft lines, more than 50% of H3K27me3 peaks that are retained in K27M (defined as having 90% or more of the signal in shK27M) fall into genic and active enhancer regions. None of the K27M retained H3K27me3 peak loci were in common between all three xenografts, but 29 peaks were in common between two xenografts, and these loci were enriched for transcription factors (GO DNA-binding transcription factor activity, RNA polymerase II-specific, p = 6.51E-10) [Suppl. Table 5 (Online Resource 5)].
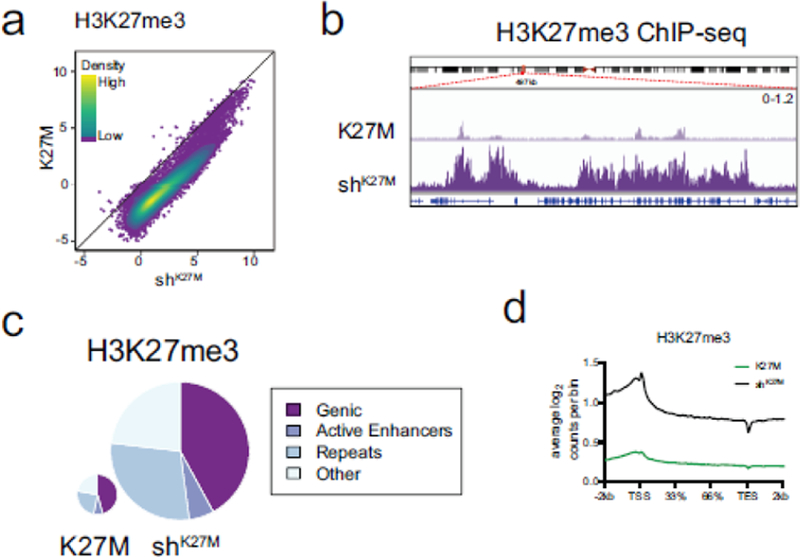
Figure 4. H3.3 K27M knockdown produces global changes in the level of H3K27me3 without redistribution to different genomic regions.
a. Spike-in normalized ChIP-seq peak calls for H3K27me3 (K27M n=4, shK27M n=4) display robust gain of H3K27me3 upon knockdown of K27M in SJDIPGX37. Colored overlay indicates density. b. Representative IGV tracks demonstrating restoration of H3K27me3 with knockdown of K27M. Region shown, chr3:57,097,000–57,600,000 was randomly selected. Note that H3K27me3 restoration occurs in both peak and “baseline” regions. Also, H3K27me3 peaks in K27M occur in regions that also have peaks in shK27M tumors, not novel loci. Scale of both tracks shown at right. c. H3K27me3 ChIP-seq read assignments to genomic categories in non-silencing (K27M) and H3F3A knockdown (shK27M) samples in SJDIPGX37. Pie charts are scaled by total read counts normalized to spike-in. Wilcoxson rank-sum test of scaled read counts between K27M and shK27M: H3K27me3 p=0.028. d. Distribution of H3K27me3 across the average of all gene bodies in K27M (green) and shK27M (black) in SJDIPGX37; 2kb upstream of the transcriptional start site (TSS) and downstream of the transcriptional end site (TES) is shown. Gene bodies are scaled by percentage to account for different lengths. See also Suppl. Fig. 3 (Online Resource 2).
A visual inspection of the genomic sequence tracks shows widespread depletion of H3K27me3 in K27M compared to shK27M tumors [(Fig. 4b and Suppl. Fig. 3b (Online Resource 2)]. Importantly, the H3K27me3 global landscape shows peaks that are present in K27M tumors are in the same loci in both conditions and do not reflect an altered pattern of deposition. The marked changes in the level of H3K27me3 associated with the K27M mutation are found across all types of genomic locations. Pie charts showing the amount (size of pie) and localization (wedges of pie) of H3K27me3 reads across the genome demonstrate that while the amount of this PTM changes between K27M and shK27M tumors, it remains similarly distributed across different types of genomic features including genes, enhancers, repeats and others [(Fig. 4c and Suppl. Fig. 3c; assessed across all three models, no feature showed a change of greater than 3%, although some changes were statistically significant (Online Resource 2)]. The similar distribution of H3K27me3 in K27M mutant cells was further supported by analysis of isogenic embryonic neural stem cells isolated from genetically engineered H3f3a K27M knock-in mice and wild-type controls [Suppl. Fig. 3d (Online Resource 2)] [24]. Further, although the level of H3K27me3 reads across gene bodies varies in K27M compared with shK27M tumors, the pattern of deposition across the gene body is the same [Fig. 4d and Suppl. Fig.3e (Online Resource 2)].
K27M-dependent global loss of H3K27me3 affects most promoters, but a subset is spared
The global increase of H3K27me3 seen in response to H3F3A knockdown is also reflected at promoters, but is not associated with a dramatic shift in distribution around the transcriptional start site (TSS) [Fig. 5a,b and Suppl. Fig. 4a,b (Online Resource 2)]. Interestingly, although most promoters have higher H3K27me3 signal in shK27M than K27M tumors, the difference in signal is not equal across the range of H3K27me3 levels [Fig. 5a,c and Suppl. Fig. 4a,c (Online Resource 2)]. A plot of the H3K27me3 signal in the 2kb surrounding the TSS [(Fig. 5d and Suppl. Fig. 4d, K27M in green, shK27M in black, same ranking as heat map (Online Resource 2)] demonstrates that the biggest difference in H3K27me3 signal between K27M and shK27M occurs at relatively high signal promoters in shK27M tumors. The distribution of K27M upregulated genes along the promoters ranked by H3K27me3 signal [Fig. 5e and Suppl. Fig. 4e, rug plots below dot plot (Online Resource 2)] shows that they are enriched at the promoters with relatively high signal in shK27M tumors and the biggest H3K27me3 differential between K27M and shK27M tumors. These promoters contrast with the majority of promoters where there is low signal in the shK27M tumors [(Fig. 5d and Suppl. Fig. 4d (Online Resource 2)]. Downregulated genes show some enrichment for promoters with low H3K27me3, although this is less consistent across xenograft lines [Fig. 5e and Suppl. Fig. 4e, rug plots below dot plot (Online Resource 2)].
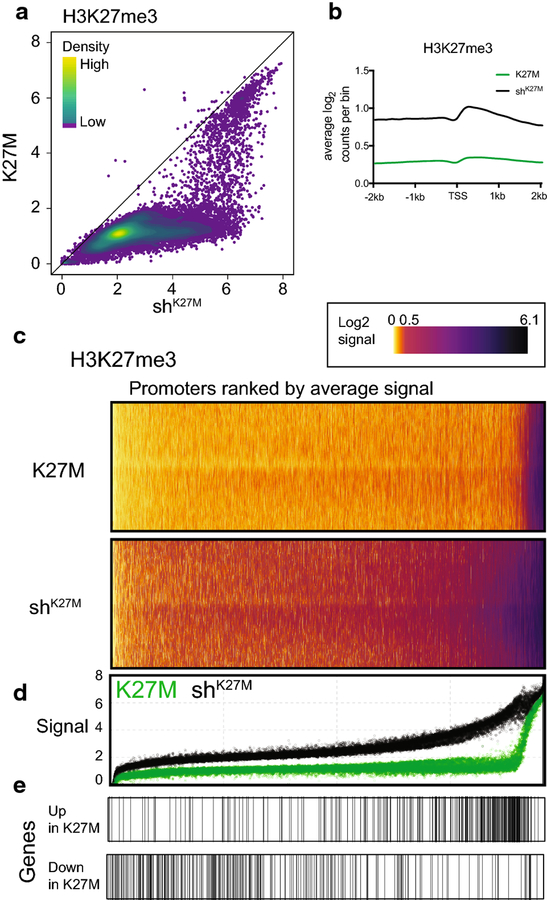
Figure 5. K27M-dependent global loss of H3K27me3 affects most promoters, but a subset is spared.
a. Scatter plot of H3K27me3 ChIP-seq signal at promoters (log2(CPM+1) in 2kb surrounding TSS) of K27M and shK27M SJDIPGX37 tumors. Colored overlay indicates density. K27M n=4, shK27M n=4. b. Distribution of H3K27me3 ChIP-seq log2 read counts across promoters (4kb centered on TSS) in K27M (green) and shK27M (black) xenografts. c. Heat maps showing distribution of H3K27me3 ChIP-seq log2 read counts binned across promoters (4kb centered on TSS) in K27M and shK27M tumors. Promoters ranked by average signal across all samples. d. Dot plot displaying H3K27me3 signal in 2kb promoter region with same ranking as heat maps for K27M (green) or shK27M (black) tumors. E. Distribution of the top 300 K27M up- and down- regulated gene promoters (ranked by limma p-value) along the H3K27me3 heat maps/dot plot (c,d). See also Suppl. Fig. 4 (Online Resource 2).
While most promoter regions have less H3K27me3 signal in K27M compared to shK27M tumors, there are a small collection of promoters that maintain similar levels of H3K27me3 in both conditions [Fig. 5a,c and Suppl. Fig. 4c; note band of very high signal promoters at right of the K27M H3K27me3 heat map (Online Resource 2)]. The CDKN2A locus, a well-known target of PRC1 recruitment and repression, encodes ARF and INK4A, both important tumor suppressors and regulators of cell cycle progression. The CDKN2A locus is one of the rare loci that retains H3K27me3 in K27M tumors, and at similar levels to that seen in shK27M tumors [Suppl. Fig. 4f (Online Resource 2)]. K27M-dependent repression of the INK4A transcript specifically has been reported to be a critical regulator of the oncogenic effect of the mutation (Cordero et al., 2017; Mohammad et al., 2017).
While INK4A expression was repressed by K27M in SJDIPGX7, the K27M effect was inconsistent between xenograft models [Suppl. Fig. 4g (Online Resource 2)]. Importantly, in both SJDIPGX7 and SJDIPGX37, the ARF transcript of CDKN2A was also significantly upregulated by K27M knockdown [Suppl. Fig. 4g (Online Resource 2)], indicating that K27M does not impact the INK4A transcript specifically in all K27M tumor models. In contrast, there was extremely low expression and no significant increase in expression of either CDKN2A transcript in response to K27M knockdown in SUDIPG-VI. This shows that there is a modeldependent component to the status of the CDKN2A locus as a K27M target, as previously suggested (Piunti et al., 2017). Only 6 of the promoter peaks with retained H3K27me3 signal were shared by two xenograft models none were shared by all three, and none of these are consistently associated with differential expression [Suppl. Table 5 (Online Resource 5)].
In contrast to H3K27me3, neither promoters with low nor high H3K4me3 signal show a consistent global shift with knockdown of H3F3A [Suppl. Fig. 4h (Online Resource 2)]. Together these results demonstrate that while promoters show the same effects on H3K27me3 that are seen globally, K27M up-regulated gene promoters are enriched for the most dramatic changes in H3K27me3, suggesting that the regulation of this subset of genes is directly correlated to H3K27me3 levels in a unique and meaningful way.
Combinatorial analysis of histone PTMs suggests that H3.3 K27M-induced genes are enriched for apparently bivalent promoters
Looking at the combination of H3K27me3 and H3K4me3, it becomes clear that the promoters of differentially regulated genes in both the K27M increased and repressed directions show epigenetic signatures consistent with their regulation. As expected from the global change in H3K27me3 the bulk of promoters show reduced H3K27me3 in the presence of the K27M mutation, regardless of whether their expression is increased, decreased, or unchanged [(Fig. 6a and Suppl. Fig. 5a (Online Resource 2)]. However, when the focus is narrowed to differentially expressed genes, promoters of K27M-induced genes show an overall wider range in the magnitude of H3K27me3 loss than K27M-repressed genes. For K27M upregulated genes, H3K4me3 changes follow the canonical model of increased H3K4me3 with increased gene expression [Fig. 6a and Suppl. Fig. 5a, red overlay (Online Resource 2)]. However, downregulated genes were not uniformly associated with loss of H3K4me3, but rather show little change in this PTM relative to promoters with no change in expression [Fig. 6a and Suppl. Fig. 5a -see histogram on right showing how downregulated (blue) genes are concentrated at same location as unchanged (gray) genes for H3K4me3 (Online Resource 2)]. In all, the epigenetic signature of these genes is consistent with their downregulation being an indirect effect of the K27M mutation.
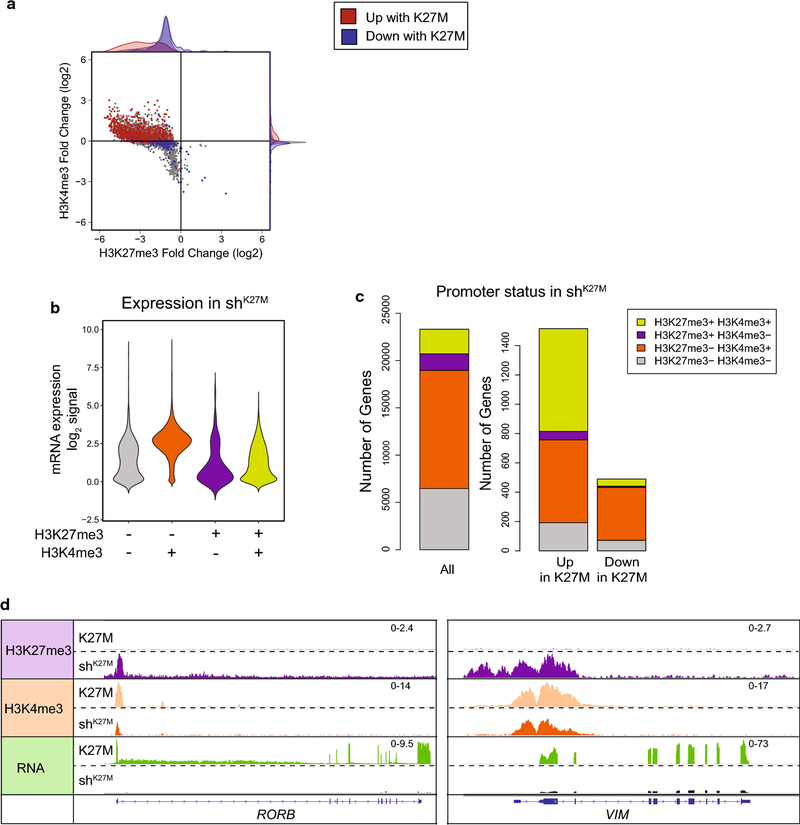
Figure 6. Combinatorial analysis of histone PTMs suggests that H3.3 K27M-induced genes are enriched for apparently bivalent promoters.
a. Fold change (FC) in ChIP-seq signal in SJDIPGX37 transduced with non-silencing (K27M) vs. H3F3A shRNA (shK27M) in 2kb surrounding TSS for H3K27me3 vs. H3K4me3. Colored dots represent promoters with differential gene expression FC>1.5 (red) or <−1.5 (blue) and an adjusted p-value<0.05. Marginal histograms display density, but are not quantitative between groups. b,c. Promoters binned by the presence or absence of MACS called H3K27me3 and H3K4me3 peaks inside the 2kb surrounding the TSS in shK27M SJDIPGX37 tumors. b. Violin plot showing log2 RPKM expression in the shK27M tumors for all genes binned by promoter type. Lime green represents promoters marked with H3K27me3 and H3K4me3 consistent with bivalency. c. Left bar graph shows H3K27me3 and H3K4me3 promoter status for all genes in shK27M tumors, while the bar graphs on the right show promoter status in shK27M tumors for genes that are differentially regulated (|FC|>1.5, adjusted p-value<0.05). d. IGV tracks showing representative loci scored as H3K27me3+H3K4me3+ (left tracks, RORB, right tracks, VIM) in shK27M tumors. Scale for pairs of tracks shown at right. See also Suppl. Fig. 5 (Online Resource 2)].
Using the presence or absence of a H3K27me3 and/or H3K4me3 peak in the 2 kb promoter region surrounding gene TSS in shK27M tumors, we binned promoters into four categories, including H3K27me3+ H3K4me3+ which could represent bivalency, a poised state where both PTMs co-occur [47]. Genes marked with both H3K27me3 and H3K4me3 in shK27M tumors (lime green) primarily had low to no expression as would be expected for a poised state [Fig. 6b and Suppl. Fig. 5b (Online Resource 2)]. The expression level and range was similar to genes marked with H3K27me3 only (purple) or with neither H3K4me3 nor H3K27me3 (gray) and differed from the H3K4me3 only marked genes (orange) that showed higher expression consistent with the activation-associated mark [Fig. 6b and Supp. Fig. 5b (Online Resource 2)]. This indicates that these apparently bivalent promoters likely represent a mixed population of poised and active promoters. When considering the promoters of all genes as a baseline in shK27M tumors where H3K27me3 is restored, 11% had both an H3K27me3 and an H3K4me3 peak in SJDIPGX37 shK27M tumors (7% for both SJDIPGX7 and SUDIPG-VI); however, for just K27M-induced gene promoters, the percentage significantly increased to 46% [48% for SJDIPGX7 and 32% for SUDIPG-VI; Fig. 6c and Suppl. Fig. 5c, p = 0.01 (Online Resource 2)]. By contrast, K27M-repressed genes showed no enrichment of H3K27me3+ H3K4me3+ (SJDIPGX37 10%, SJDIPGX7 11%, and SUDIPG-VI 8%). Retinoid-Related Orphan Receptor Beta (RORB) and Vimentin (VIM) are examples of K27M upregulated genes that have promoter PTMs consistent with a transition from an active state in K27M tumors to a bivalent state with K27M knockdown in shK27M tumors (Fig. 6d). The enrichment of promoters marked with both H3K27me3 and H3K4me3, consistent with a bivalent state among K27M-induced genes, represents a potential mechanism dictating which genes are targets of K27M upregulation.
Expression signatures driven by H3.3 K27M are primarily defined by NPC stemness
DIPG tumors show significant heterogeneity in their expression signatures [24, 38]. Filbin et al. recently used single cell RNA sequencing to identify the cellular hierarchy of DIPGs, which showed cells with cycling/stem-like, astrocyte-like, OPC-like and oligodendrocyte-like signatures [15]. Single sample GSEA (ssGSEA) reveals the enrichment of different expression signatures within bulk tumors, with primary human DIPGs and all three DIPG xenograft models showing strong enrichment with signatures from the DIPG cellular hierarchy identified by Filbin et al., and with signatures from developing, immature and differentiated normal brain cell types [23, 54, 55]. Within this framework of glioma expression signatures, the specific gene sets with greatest enrichment showed intertumoral variation among primary tumors and among the xenograft models [Suppl. Fig. 6a, b (Online Resource 2)]. ssGSEA further revealed effects of K27M on the differentiation hierarchy within the xenograft models. For all xenograft lines, gene sets representing different stages of neural progenitor cells, including human embryonic midline neural progenitors and astrocytic-lineage committed fetal cells were consistently more enriched in K27M compared with shK27M tumors [Fig. 7a, upper panel, Suppl. Fig. 7a (Online Resource 2)], indicating a decrease in normal stem/progenitor signatures with knockdown. Clear shifts in enrichment of signature gene sets from the DIPG cellular hierarchy were also apparent (Fig. 7a, middle panel). The astrocyte-like signature, which is most highly enriched in SJDIPGX37, showed the most dramatic loss in enrichment with K27M knockdown in this model. K27M knockdown decreased the enrichment of the cycling/stem-like and astrocyte-like DIPG signatures in SJDIPGX7 and SUDIPG-VI. As enrichment of those signatures decreased, in all three models, K27M knockdown induced increased enrichment of the oligodendrocyte-like gene signature found in DIPGs (Fig. 7a, middle panel). Increased oligodendrocyte differentiation in SJDIPGX7 was more pronounced, resulting in a clear increased enrichment in the expression signatures of normal mature oligodendrocytes [Fig. 7a, upper panel, Suppl. Fig. 7a (Online Resource 2)]. Importantly, neither a set of genes highly expressed throughout differentiation from progenitor to more differentiated midline neural cell types (“housekeeping”,[23]), nor a random collection of genes were substantially different between K27M and shK27M tumors (Fig. 7a, lower panel).
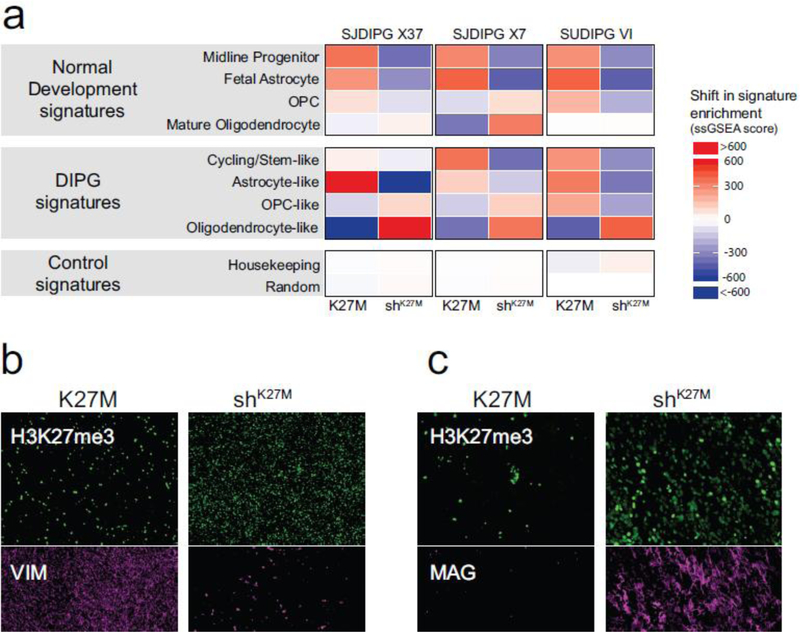
Figure 7. Expression signatures driven by H3.3 K27M are primarily defined by NPC stemness.
a. Heat map showing the shift in ssGSEA enrichment scores for xenograft models with and without knockdown of K27M. Colors represent the difference of ssGSEA scores of K27M vs. shK27M samples, where the mean for each K27M/shK27M pair was set to zero (white). Increased signature enrichment is colored in red, while loss of enrichment is colored in blue. b. IF showing decreased vimentin (VIM) expression (magenta) in shK27M SJDIPGX7 tumors. Scale bar is 200μm. c. IF showing increased MAG expression (magenta) in shK27M SJDIPGX7 tumors. Scale bar is 100μm. In b,c, H3K27me3 is shown in green and DAPI staining of nuclei is shown in blue. See also Suppl. Fig. 6 (Online Resource 2)].
Vimentin (VIM), which is normally expressed during embryonic development in mesenchymal cells, but was also associated with the astrocyte-like DIPG gene signature, was significantly decreased in shK27M compared to K27M tumors in SJDIPGX7 [Suppl. Fig. 7b (Online Resource 2)]. We confirmed by IF that vimentin protein was also lower in shK27M than K27M tumors [Fig. 7a, Suppl. Fig. 7c (Online Resource 2)]. Consistent with the increased oligodendrocyte signature in shK27M SJDIPGX7 tumors, we found that the expression of the oligodendrocyte marker, Myelin Associated Glycoprotein (MAG) was significantly upregulated in shK27M relative to K27M tumors [Suppl. Fig. 7d (Online Resource 2)] and MAG protein expression was also increased [Fig. 7c, Suppl. Fig. 7e (Online Resource 2)]. Interestingly, despite the increase in expression, the MAG promoter gains a small H3K27me3 peak at its promoter in response to K27M knockdown, suggesting it is regulated indirectly rather than by the change in epigenetic status [Suppl. Fig. 7f (Online Resource 2)]. Together, these data support a role for the K27M mutation in maintaining more immature and proliferative states in tumors and preventing differentiation.
DISCUSSION
The current study aimed to determine the biological, epigenetic and transcriptional contributions of H3.3 K27M mutation to DIPG pathogenesis by focusing on in vivo tumor growth of biologically relevant models. We used shRNA to knockdown H3F3A. An advantage of this approach is that we were able to use pooled populations and avoid artifacts of clonal heterogeneity that can complicate techniques such as selection of individual clonal lines targeted by CRISPR/CAS9. We used two different shRNAs to knockdown H3F3A and two different controls for specificity of the effects. However, shRNAs may introduce off-target effects which could complicate the analysis. Future studies to correct the H3 K27M mutation without altering expression of H3F3A would provide an important comparison. Knockdown of H3F3A, which encodes H3.3 K27M, significantly prolonged survival of mice with intracranial implantation of DIPG xenografts for three independent lines, showing that this mutation plays an important role in the ongoing growth of DIPG. Depleting the mutant protein reversed the dominant effects of H3.3 K27M mutation, restoring the balance of PTMs at K27. Integrated analysis of ChIP-seq and RNA-seq data also yielded insights into mechanisms connecting epigenetic disruption to the transcriptional consequences of K27M mutation.
Analysis of the three DIPG xenograft lines highlights the manner in which tumor heterogeneity can obscure K27M-dependent effects. Although the global change in H3K27me3 levels were consistent features across xenograft lines, K27M-driven effects on gene expression were far less pronounced than the expression signatures that distinguished the three xenograft lines from one another. This is likely a combined effect of differences in the precise developmental stage of the cell of origin, other somatic mutations, and individual genetic background. Paired isogenic comparisons using knockdown allowed identification of the K27M-dependent expression in each xenograft line, which was highly enriched in genes associated with neurogenesis and nervous system development.
DIPGs are found almost exclusively in children, indicating an intimate connection between development and disease etiology. Recent single cell RNA sequencing shows a cellular hierarchy recapitulating glial development in gliomas, with a rare population of stem/cycling cells, as well as populations with varying degrees of differentiation along the astrocyte and oligodendrocyte lineage in IDH mutant gliomas and IDH wild type adult glioblastomas. Single cell sequencing analysis of DIPGs revealed a less pronounced hierarchy, with cells resembling oligodendrocyte progenitor cells (OPCs) comprising the majority of the tumor, and cells with astrocyte- or oligodendrocyte-like signatures showing overall less differentiation, with some tumors showing little lineage differentiation [15]. Interestingly, proliferation of oligodendrocyte progenitors expressing Sox2 and Olig2 drive a massive early postnatal expansion of the developing pons, a developmental stage that is relevant to the pediatric incidence of DIPG [28]. Consistent with OPC-like cells as a major cellular constituent and potential cell of origin for DIPG, enhancer mapping in DIPG revealed active superenhancers for oligodendrocyte lineage genes [34]. Intriguingly, in our experiments, depletion of H3.3 K27M was associated with a reduction in enrichment of signatures of normal midline stem and progenitor cells and DIPG cycling/stem-like cells and an increased enrichment of the more differentiated oligodendrocyte-like signature. These results support the hypothesis that the H3 K27M mutation serves to maintain an epigenetic state of stem/progenitor cells. Loss of H3 K27M shifts the epigenetic state towards more differentiated cells. The cycling stem-like cells resemble OPCs, and may transition through the OPC-like state and progress to oligodendrocyte-like differentiation. The cellular hierarchy of DIPGs was shown by single cell sequencing to also include astrocyte-like cells in varying proportions, so there is intertumoral variation in plasticity of differentiation patterns within DIPGs, as illustrated by the strong astrocyte-like signature which is decreased as the oligodendrocyte-like signature increases in SJDIPGX37 [15]. The K27M-dependent extent of differentiation varied between xenograft lines, consistent with the variable proportion and extent of differentiated cells observed among primary DIPGs [15]. Based on the known cellular hierarchy in DIPG, each of these signatures would represent a different population of cells, which would explain why the expression changes observed upon K27M knockdown are relatively modest in magnitude.
The K27M-dependent genome-wide decrease in H3K27me3 may be expected to have similar consequences to loss of function in PRC2, which catalyzes this PTM and plays important roles in cell fate decisions and maintenance of cellular identity [9]. Deletion of PRC2 components Ezh2, Suz12, or Eed, impairs differentiation of embryonic stem cells and causes embryonic lethality in mice [25]. In developing brain, loss of PRC2 function disrupted normal transitions in cell fate and differentiation, with specific consequences varying between developmental stages. Ezh2 deletion in cortical progenitors at E9.5 reduced the neurogenic phase and induced a premature shift to gliogenesis [39]. However, Ezh2 deletion in neural progenitor cells at E13.5 disrupted a later cell fate transition, prolonging neurogenesis and delaying onset of gliogenesis [20]. Bivalent genes may show an enhanced susceptibility to deregulation in response to changes in H3K27me3. Deletion of Ezh2 or Eed in pancreatic β cells causes ectopic activation of bivalent genes, disrupting cell identity [29]. Conversely, gain of function mutations in EZH2, which increase levels of H3K27me3, drive the silencing of bivalent gene promoters, thus preventing differentiation in diffuse large B-cell lymphomas [3]. In this study, integrated analysis of ChIP-seq and RNA-seq from K27M and shK27M tumors showed that a substantial component of differential expression was attributable to the K27M-dependent reduction in H3K27me3 causing release of an apparent bivalent state at promoters and resulting in upregulated expression. Thus, a significant proportion of the K27M-dependent effects on transcription are attributable to a selective direct effect of the global H3K27me3 reduction. In a genetically engineered mouse model for spontaneous DIPG formation, expression of H3.3 K27M from the endogenous H3f3a promoter also induced upregulation of bivalent genes associated with neurodevelopment [24]. It is important to note that the high frequency of H3K27M mutations in DIPGs and other diffuse midline gliomas, and the absence of loss of function mutations in PRC2 [30] provides compelling evidence that different mutational mechanisms to reduce H3K27me3 are not functionally interchangeable in driving contextdependent tumorigenesis. Rather, it seems likely that the magnitude of change in H3K27me3 is critical. Further, H3K27M may have multiple additional functional consequences beyond its effects on H3K27me3 levels.
Identification of H3 K27M mutations in DIPG illuminated highly recurrent mutation of a novel oncogene in the context of extensive inter-and intra-tumoral heterogeneity, raising a new hope that there may a unifying therapeutic vulnerability for this incurable disease. H3.3 K27M alone enhances self-renewal of mouse neural stem cells [24], and when combined with mutated PDGFRα and loss of TP53, can also enhance self-renewal of human neural progenitor cells [17], suggesting that H3.3 K27M may contribute to DIPG formation by expanding the pool of cells susceptible to transformation. However, this could be a transient effect that no longer contributes to tumor growth after additional mutations are acquired. Importantly, our study shows that the effects of H3.3 K27M extend beyond a possible enhanced predisposition for transformation and contribute to in vivo tumor growth in the context of fully developed human DIPG tumors. Interfering RNA therapies have been developed for other neurological diseases such as spinal muscular atrophy [10], and RNAi-mediated depletion of H3 K27M could be considered as a potential therapeutic approach for DIPG. shK27M tumors grew with delayed kinetics and maintained knockdown of H3.3 K27M. This provided essential material for isogenic comparisons. The knockdown did not completely deplete the mutant message, and the pooled populations of knockdown cells had varying levels of knockdown efficiency. Therefore, it is possible that K27M mutation is required for DIPG growth or survival, and genetic deletion of this mutation could completely ablate tumorigenic activity. However, therapeutic intervention typically cannot achieve inhibition similar to complete deletion of a target. Thus, our results imply that effective therapy would require targeting the effects of H3.3 K27M, potentially through inhibition of self-renewal, in combination with approaches that target the other more canonical cancer mutations found in DIPG.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Michelle Monje for sharing SUDIPG-VI cells. Surgeries and preclinical imaging were performed by the Center for In Vivo Imaging and Therapeutics which is supported by SJCRH and NCI grants P30CA021765 and R50CA211481.
This work was supported by NIH grants CA096832 and CA188516 (SJB), R25CA23944 (RMN), the NCI Cancer Center Support Grant CA21765, the St Jude Children’s Research Hospital-Washington University Pediatric Cancer Genome Project, and ALSAC.
Footnotes
Publisher's Disclaimer: This Author Accepted Manuscript is a PDF file of an unedited peer-reviewed manuscript that has been accepted for publication but has not been copyedited or corrected. The official version of record that is published in the journal is kept up to date and so may therefore differ from this version.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- 1.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, and Sherlock G (2000) Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 25:25–29. 10.1038/75556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barbie DA, Tamayo P, Boehm JS, Kim SY, Moody SE, Dunn IF, Schinzel AC, Sandy P, Meylan E, Scholl C, Frohling S, Chan EM, Sos ML, Michel K, Mermel C, Silver SJ, Weir BA, Reiling JH, Sheng Q, Gupta PB, Wadlow RC, Le H, Hoersch S, Wittner BS, Ramaswamy S, Livingston DM, Sabatini DM, Meyerson M, Thomas RK, Lander ES, Mesirov JP, Root DE, Gilliland DG, Jacks T, and Hahn WC (2009) Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature 462:108–112. 10.1038/nature08460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beguelin W, Popovic R, Teater M, Jiang Y, Bunting KL, Rosen M, Shen H, Yang SN, Wang L, Ezponda T, Martinez-Garcia E, Zhang H, Zheng Y, Verma SK, McCabe MT, Ott HM, Van Aller GS, Kruger RG, Liu Y, McHugh CF, Scott DW, Chung YR, Kelleher N, Shaknovich R, Creasy CL, Gascoyne RD, Wong KK, Cerchietti L, Levine RL, Abdel-Wahab O, Licht JD, Elemento O, and Melnick AM (2013) EZH2 is required for germinal center formation and somatic EZH2 mutations promote lymphoid transformation. Cancer Cell 23:677–692. 10.1016/j.ccr.2013.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ben-Porath I, Thomson MW, Carey VJ, Ge R, Bell GW, Regev A, and Weinberg RA (2008) An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat Genet 40:499–507. 10.1038/ng.127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bender S, Tang Y, Lindroth AM, Hovestadt V, Jones DT, Kool M, Zapatka M, Northcott PA, Sturm D, Wang W, Radlwimmer B, Hojfeldt JW, Truffaux N, Castel D, Schubert S, Ryzhova M, Seker-Cin H, Gronych J, Johann PD, Stark S, Meyer J, Milde T, Schuhmann M, Ebinger M, Monoranu CM, Ponnuswami A, Chen S, Jones C, Witt O, Collins VP, von Deimling A, Jabado N, Puget S, Grill J, Helin K, Korshunov A, Lichter P, Monje M, Plass C, Cho YJ, and Pfister SM (2013) Reduced H3K27me3 and DNA hypomethylation are major drivers of gene expression in K27M mutant pediatric high-grade gliomas. Cancer Cell 24:660–672. 10.1016/j.ccr.2013.10.006 [DOI] [PubMed] [Google Scholar]
- 6.Buczkowicz P, Hoeman C, Rakopoulos P, Pajovic S, Letourneau L, Dzamba M, Morrison A, Lewis P, Bouffet E, Bartels U, Zuccaro J, Agnihotri S, Ryall S, Barszczyk M, Chornenkyy Y, Bourgey M, Bourque G, Montpetit A, Cordero F, Castelo-Branco P, Mangerel J, Tabori U, Ho KC, Huang A, Taylor KR, Mackay A, Bendel AE, Nazarian J, Fangusaro JR, Karajannis MA, Zagzag D, Foreman NK, Donson A, Hegert JV, Smith A, Chan J, Lafay-Cousin L, Dunn S, Hukin J, Dunham C, Scheinemann K, Michaud J, Zelcer S, Ramsay D, Cain J, Brennan C, Souweidane MM, Jones C, Allis CD, Brudno M, Becher O, and Hawkins C (2014) Genomic analysis of diffuse intrinsic pontine gliomas identifies three molecular subgroups and recurrent activating ACVR1 mutations. Nat Genet 46:451–456. 10.1038/ng.2936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Capper D, Jones DTW, Sill M, Hovestadt V, Schrimpf D, Sturm D, Koelsche C, Sahm F, Chavez L, Reuss DE, Kratz A, Wefers AK, Huang K, Pajtler KW, Schweizer L, Stichel D, Olar A, Engel NW, Lindenberg K, Harter PN, Braczynski AK, Plate KH, Dohmen H, Garvalov BK, Coras R, Holsken A, Hewer E, Bewerunge-Hudler M, Schick M, Fischer R, Beschorner R, Schittenhelm J, Staszewski O, Wani K, Varlet P, Pages M, Temming P, Lohmann D, Selt F, Witt H, Milde T, Witt O, Aronica E, Giangaspero F, Rushing E, Scheurlen W, Geisenberger C, Rodriguez FJ, Becker A, Preusser M, Haberler C, Bjerkvig R, Cryan J, Farrell M, Deckert M, Hench J, Frank S, Serrano J, Kannan K, Tsirigos A, Bruck W, Hofer S, Brehmer S, Seiz-Rosenhagen M, Hanggi D, Hans V, Rozsnoki S, Hansford JR, Kohlhof P, Kristensen BW, Lechner M, Lopes B, Mawrin C, Ketter R, Kulozik A, Khatib Z, Heppner F, Koch A, Jouvet A, Keohane C, Muhleisen H, Mueller W, Pohl U, Prinz M, Benner A, Zapatka M, Gottardo NG, Driever PH, Kramm CM, Muller HL, Rutkowski S, von Hoff K, Fruhwald MC, Gnekow A, Fleischhack G, Tippelt S, Calaminus G, Monoranu CM, Perry A, Jones C, Jacques TS, Radlwimmer B, Gessi M, Pietsch T, Schramm J, Schackert G, Westphal M, Reifenberger G, Wesseling P, Weller M, Collins VP, Blumcke I, Bendszus M, Debus J, Huang A, Jabado N, Northcott PA, Paulus W, Gajjar A, Robinson GW, Taylor MD, Jaunmuktane Z, Ryzhova M, Platten M, Unterberg A, Wick W, Karajannis MA, Mittelbronn M, Acker T, Hartmann C, Aldape K, Schuller U, Buslei R, Lichter P, Kool M, Herold-Mende C, Ellison DW, Hasselblatt M, Snuderl M, Brandner S, Korshunov A, von Deimling A and Pfister SM (2018) DNA methylation-based classification of central nervous system tumours. Nature 555:469–474. 10.1038/nature26000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chan KM, Fang D, Gan H, Hashizume R, Yu C, Schroeder M, Gupta N, Mueller S, James CD, Jenkins R, Sarkaria J, and Zhang Z (2013) The histone H3.3K27M mutation in pediatric glioma reprograms H3K27 methylation and gene expression. Genes Dev 27:985–990. 10.1101/gad.217778.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Conway E, Healy E, and Bracken AP (2015) PRC2 mediated H3K27 methylations in cellular identity and cancer. Curr Opin Cell Biol 37:42–48. 10.1016/j.ceb.2015.10.003 [DOI] [PubMed] [Google Scholar]
- 10.Crooke ST, Witztum JL, Bennett CF, and Baker BF (2018) RNA-Targeted Therapeutics. Cell Metab 27:714–739. 10.1016/j.cmet.2018.03.004 [DOI] [PubMed] [Google Scholar]
- 11.Endersby R, Zhu X, Hay N, Ellison DW, and Baker SJ (2011) Nonredundant functions for Akt isoforms in astrocyte growth and gliomagenesis in an orthotopic transplantation model. Cancer Res 71:4106–4116. 10.1158/0008-5472.CAN-10-3597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ernst T, Chase AJ, Score J, Hidalgo-Curtis CE, Bryant C, Jones AV, Waghorn K, Zoi K, Ross FM, Reiter A, Hochhaus A, Drexler HG, Duncombe A, Cervantes F, Oscier D, Boultwood J, Grand FH, and Cross NC (2010) Inactivating mutations of the histone methyltransferase gene EZH2 in myeloid disorders. Nat Genet 42:722–726. 10.1038/ng.621 [DOI] [PubMed] [Google Scholar]
- 13.Feinberg AP, Koldobskiy MA, and Gondor A (2016) Epigenetic modulators, modifiers and mediators in cancer aetiology and progression. Nat Rev Genet 17:284–299. 10.1038/nrg.2016.13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fellmann C, Hoffmann T, Sridhar V, Hopfgartner B, Muhar M, Roth M, Lai DY, Barbosa IA, Kwon JS, Guan Y, Sinha N, and Zuber J (2013) An optimized microRNA backbone for effective single-copy RNAi. Cell Rep 5:1704–1713. 10.1016/j.celrep.2013.11.020 [DOI] [PubMed] [Google Scholar]
- 15.Filbin MG, Tirosh I, Hovestadt V, Shaw ML, Escalante LE, Mathewson ND, Neftel C, Frank N, Pelton K, Hebert CM, Haberler C, Yizhak K, Gojo J, Egervari K, Mount C, van Galen P, Bonal DM, Nguyen QD, Beck A, Sinai C, Czech T, Dorfer C, Goumnerova L, Lavarino C, Carcaboso AM, Mora J, Mylvaganam R, Luo CC, Peyrl A, Popovic M, Azizi A, Batchelor TT, Frosch MP, Martinez-Lage M, Kieran MW, Bandopadhayay P, Beroukhim R, Fritsch G, Getz G, Rozenblatt-Rosen O, Wucherpfennig KW, Louis DN, Monje M, Slavc I, Ligon KL, Golub TR, Regev A, Bernstein BE, and Suva ML (2018) Developmental and oncogenic programs in H3K27M gliomas dissected by single-cell RNA-seq. Science 360:331–335. 10.1126/science.aao4750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fontebasso AM, Papillon-Cavanagh S, Schwartzentruber J, Nikbakht H, Gerges N, Fiset PO, Bechet D, Faury D, De Jay N, Ramkissoon LA, Corcoran A, Jones DT, Sturm D, Johann P, Tomita T, Goldman S, Nagib M, Bendel A, Goumnerova L, Bowers DC, Leonard JR, Rubin JB, Alden T, Browd S, Geyer JR, Leary S, Jallo G, Cohen K, Gupta N, Prados MD, Carret AS, Ellezam B, Crevier L, Klekner A, Bognar L, Hauser P, Garami M, Myseros J, Dong Z, Siegel PM, Malkin H, Ligon AH, Albrecht S, Pfister SM, Ligon KL, Majewski J, Jabado N, and Kieran MW (2014) Recurrent somatic mutations in ACVR1 in pediatric midline high-grade astrocytoma. Nat Genet 46:462–466. 10.1038/ng.2950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Funato K, Major T, Lewis PW, Allis CD, and Tabar V (2014) Use of human embryonic stem cells to model pediatric gliomas with H3.3K27M histone mutation. Science 346:1529–1533. 10.1126/science.1253799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grasso CS, Tang Y, Truffaux N, Berlow NE, Liu L, Debily MA, Quist MJ, Davis LE, Huang EC, Woo PJ, Ponnuswami A, Chen S, Johung TB, Sun W, Kogiso M, Du Y, Qi L, Huang Y, Hutt-Cabezas M, Warren KE, Le Dret L, Meltzer PS, Mao H, Quezado M, van Vuurden DG, Abraham J, Fouladi M, Svalina MN, Wang N, Hawkins C, Nazarian J, Alonso MM, Raabe EH, Hulleman E, Spellman PT, Li XN, Keller C, Pal R, Grill J, and Monje M (2015) Functionally defined therapeutic targets in diffuse intrinsic pontine glioma. Nat Med 21:827 10.1038/nm0715-827a [DOI] [PubMed] [Google Scholar]
- 19.Hashizume R, Andor N, Ihara Y, Lerner R, Gan H, Chen X, Fang D, Huang X, Tom MW, Ngo V, Solomon D, Mueller S, Paris PL, Zhang Z, Petritsch C, Gupta N, Waldman TA, and James CD (2014) Pharmacologic inhibition of histone demethylation as a therapy for pediatric brainstem glioma. Nat Med 20:1394–1396. 10.1038/nm.3716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hirabayashi Y, Suzki N, Tsuboi M, Endo TA, Toyoda T, Shinga J, Koseki H, Vidal M, and Gotoh Y (2009) Polycomb limits the neurogenic competence of neural precursor cells to promote astrogenic fate transition. Neuron 63:600–613. 10.1016/j.neuron.2009.08.021 [DOI] [PubMed] [Google Scholar]
- 21.Jin H, Silveira AB, Kasper LH, Wu G, Zhang J, Baker SJ, and Fan Y (In preparation) XenoScale – a Normalization Method for Quantifying Global Shift of Epigenetic Marks in Human Derived Xenograft Models Using Mouse Reads as Internal Controls
- 22.Jones C and Baker SJ (2014) Unique genetic and epigenetic mechanisms driving paediatric diffuse high-grade glioma. Nat Rev Cancer 14 10.1038/nrc3811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.La Manno G, Gyllborg D, Codeluppi S, Nishimura K, Salto C, Zeisel A, Borm LE, Stott SRW, Toledo EM, Villaescusa JC, Lonnerberg P, Ryge J, Barker RA, Arenas E, and Linnarsson S (2016) Molecular Diversity of Midbrain Development in Mouse, Human, and Stem Cells. Cell 167:566–580 e519. 10.1016/j.cell.2016.09.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Larson JD, Kasper LH, Paugh BS, Jin H, Wu G, Kwon CH, Fan Y, Shaw TI, Silveira AB, Qu C, Xu R, Zhu X, Zhang J, Russell HR, Peters JL, Finkelstein D, Xu B, Lin T, Tinkle CL, Patay Z, Onar-Thomas A, Pounds SB, McKinnon PJ, Ellison DW, Zhang J, and Baker SJ (2019) Histone H3.3 K27M Accelerates Spontaneous Brainstem Glioma and Drives Restricted Changes in Bivalent Gene Expression. Cancer Cell 35:1–16. 10.1016/j.ccell.2018.11.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Laugesen A and Helin K (2014) Chromatin repressive complexes in stem cells, development, and cancer. Cell Stem Cell 14:735–751. 10.1016/j.stem.2014.05.006 [DOI] [PubMed] [Google Scholar]
- 26.Le S, Josse J, and Husson F (2008) FactoMineR: An R package for multivariate analysis. J Stat Softw 25:1–18 [Google Scholar]
- 27.Lewis PW, Muller MM, Koletsky MS, Cordero F, Lin S, Banaszynski LA, Garcia BA, Muir TW, Becher OJ, and Allis CD (2013) Inhibition of PRC2 activity by a gain-of-function H3 mutation found in pediatric glioblastoma. Science 340:857–861. 10.1126/science.1232245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lindquist RA, Guinto CD, Rodas-Rodriguez JL, Fuentealba LC, Tate MC, Rowitch DH, and Alvarez-Buylla A (2016) Identification of proliferative progenitors associated with prominent postnatal growth of the pons. Nat Commun 7:11628 10.1038/ncomms11628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lu TT, Heyne S, Dror E, Casas E, Leonhardt L, Boenke T, Yang CH, Sagar, Arrigoni L, Dalgaard K, Teperino R, Enders L, Selvaraj M, Ruf M, Raja SJ, Xie H, Boenisch U, Orkin SH, Lynn FC, Hoffman BG, Grun D, Vavouri T, Lempradl AM, and Pospisilik JA (2018) The Polycomb-Dependent Epigenome Controls beta Cell Dysfunction, Dedifferentiation, and Diabetes. Cell Metab 27:1294–1308 e1297. 10.1016/j.cmet.2018.04.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mackay A, Burford A, Carvalho D, Izquierdo E, Fazal-Salom J, Taylor KR, Bjerke L, Clarke M, Vinci M, Nandhabalan M, Temelso S, Popov S, Molinari V, Raman P, Waanders AJ, Han HJ, Gupta S, Marshall L, Zacharoulis S, Vaidya S, Mandeville HC, Bridges LR, Martin AJ, Al-Sarraj S, Chandler C, Ng HK, Li X, Mu K, Trabelsi S, Brahim DH, Kisljakov AN, Konovalov DM, Moore AS, Carcaboso AM, Sunol M, de Torres C, Cruz O, Mora J, Shats LI, Stavale JN, Bidinotto LT, Reis RM, Entz-Werle N, Farrell M, Cryan J, Crimmins D, Caird J, Pears J, Monje M, Debily MA, Castel D, Grill J, Hawkins C, Nikbakht H, Jabado N, Baker SJ, Pfister SM, Jones DTW, Fouladi M, von Bueren AO, Baudis M, Resnick A, and Jones C (2017) Integrated Molecular Meta-Analysis of 1,000 Pediatric High-Grade and Diffuse Intrinsic Pontine Glioma. Cancer Cell 32:520–537 e525. 10.1016/j.ccell.2017.08.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mi H, Muruganujan A, Casagrande JT, and Thomas PD (2013) Large-scale gene function analysis with the PANTHER classification system. Nat Protoc 8:1551–1566. 10.1038/nprot.2013.092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mohammad F, Weissmann S, Leblanc B, Pandey DP, Hojfeldt JW, Comet I, Zheng C, Johansen JV, Rapin N, Porse BT, Tvardovskiy A, Jensen ON, Olaciregui NG, Lavarino C, Sunol M, de Torres C, Mora J, Carcaboso AM, and Helin K (2017) EZH2 is a potential therapeutic target for H3K27M-mutant pediatric gliomas. Nat Med 23:483–492. 10.1038/nm.4293 [DOI] [PubMed] [Google Scholar]
- 33.Murphy BL, Obad S, Bihannic L, Ayrault O, Zindy F, Kauppinen S, and Roussel MF (2013) Silencing of the miR-17~92 cluster family inhibits medulloblastoma progression. Cancer Res 73:7068–7078. 10.1158/0008-5472.CAN-13-0927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nagaraja S, Vitanza NA, Woo PJ, Taylor KR, Liu F, Zhang L, Li M, Meng W, Ponnuswami A, Sun W, Ma J, Hulleman E, Swigut T, Wysocka J, Tang Y, and Monje M (2017) Transcriptional Dependencies in Diffuse Intrinsic Pontine Glioma. Cancer Cell 31:635–652 e636. 10.1016/j.ccell.2017.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nikoloski G, Langemeijer SM, Kuiper RP, Knops R, Massop M, Tonnissen ER, van der Heijden A, Scheele TN, Vandenberghe P, de Witte T, van der Reijden BA, and Jansen JH (2010) Somatic mutations of the histone methyltransferase gene EZH2 in myelodysplastic syndromes. Nat Genet 42:665–667. 10.1038/ng.620 [DOI] [PubMed] [Google Scholar]
- 36.Ntziachristos P, Tsirigos A, Van Vlierberghe P, Nedjic J, Trimarchi T, Flaherty MS, Ferres-Marco D, da Ros V, Tang Z, Siegle J, Asp P, Hadler M, Rigo I, De Keersmaecker K, Patel J, Huynh T, Utro F, Poglio S, Samon JB, Paietta E, Racevskis J, Rowe JM, Rabadan R, Levine RL, Brown S, Pflumio F, Dominguez M, Ferrando A, and Aifantis I (2012) Genetic inactivation of the polycomb repressive complex 2 in T cell acute lymphoblastic leukemia. Nat Med 18:298–301. 10.1038/nm.2651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Orlando DA, Chen MW, Brown VE, Solanki S, Choi YJ, Olson ER, Fritz CC, Bradner JE, and Guenther MG (2014) Quantitative ChIP-Seq normalization reveals global modulation of the epigenome. Cell Rep 9:1163–1170. 10.1016/j.celrep.2014.10.018 [DOI] [PubMed] [Google Scholar]
- 38.Paugh BS, Broniscer A, Qu C, Miller CP, Zhang J, Tatevossian RG, Olson JM, Geyer JR, Chi SN, da Silva NS, Onar-Thomas A, Baker JN, Gajjar A, Ellison DW, and Baker SJ (2011) Genome-wide analyses identify recurrent amplifications of receptor tyrosine kinases and cell-cycle regulatory genes in diffuse intrinsic pontine glioma. J Clin Oncol 29:3999–4006. 10.1200/JCO.2011.35.5677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pereira JD, Sansom SN, Smith J, Dobenecker MW, Tarakhovsky A, and Livesey FJ (2010) Ezh2, the histone methyltransferase of PRC2, regulates the balance between self-renewal and differentiation in the cerebral cortex. Proc Natl Acad Sci U S A 107:15957–15962. 10.1073/pnas.1002530107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Piunti A, Hashizume R, Morgan MA, Bartom ET, Horbinski CM, Marshall SA, Rendleman EJ, Ma Q, Takahashi YH, Woodfin AR, Misharin AV, Abshiru NA, Lulla RR, Saratsis AM, Kelleher NL, James CD, and Shilatifard A (2017) Therapeutic targeting of polycomb and BET bromodomain proteins in diffuse intrinsic pontine gliomas. Nat Med 23:493–500. 10.1038/nm.4296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schwartzentruber J, Korshunov A, Liu XY, Jones DT, Pfaff E, Jacob K, Sturm D, Fontebasso AM, Quang DA, Tonjes M, Hovestadt V, Albrecht S, Kool M, Nantel A, Konermann C, Lindroth A, Jager N, Rausch T, Ryzhova M, Korbel JO, Hielscher T, Hauser P, Garami M, Klekner A, Bognar L, Ebinger M, Schuhmann MU, Scheurlen W, Pekrun A, Fruhwald MC, Roggendorf W, Kramm C, Durken M, Atkinson J, Lepage P, Montpetit A, Zakrzewska M, Zakrzewski K, Liberski PP, Dong Z, Siegel P, Kulozik AE, Zapatka M, Guha A, Malkin D, Felsberg J, Reifenberger G, von Deimling A, Ichimura K, Collins VP, Witt H, Milde T, Witt O, Zhang C, Castelo-Branco P, Lichter P, Faury D, Tabori U, Plass C, Majewski J, Pfister SM, and Jabado N (2012) Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature 482:226–231. 10.1038/nature10833 [DOI] [PubMed] [Google Scholar]
- 42.Stransky N, Egloff AM, Tward AD, Kostic AD, Cibulskis K, Sivachenko A, Kryukov GV, Lawrence MS, Sougnez C, McKenna A, Shefler E, Ramos AH, Stojanov P, Carter SL, Voet D, Cortes ML, Auclair D, Berger MF, Saksena G, Guiducci C, Onofrio RC, Parkin M, Romkes M, Weissfeld JL, Seethala RR, Wang L, Rangel-Escareno C, Fernandez-Lopez JC, Hidalgo-Miranda A, Melendez-Zajgla J, Winckler W, Ardlie K, Gabriel SB, Meyerson M, Lander ES, Getz G, Golub TR, Garraway LA, and Grandis JR (2011) The mutational landscape of head and neck squamous cell carcinoma. Science 333:1157–1160. 10.1126/science.1208130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sturm D, Bender S, Jones DT, Lichter P, Grill J, Becher O, Hawkins C, Majewski J, Jones C, Costello JF, Iavarone A, Aldape K, Brennan CW, Jabado N, and Pfister SM (2014) Paediatric and adult glioblastoma: multiform (epi)genomic culprits emerge. Nat Rev Cancer 14:92–107. 10.1038/nrc3655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, and Mesirov JP (2005) Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 102:15545–15550. 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Taylor KR, Mackay A, Truffaux N, Butterfield Y, Morozova O, Philippe C, Castel D, Grasso CS, Vinci M, Carvalho D, Carcaboso AM, de Torres C, Cruz O, Mora J, Entz-Werle N, Ingram WJ, Monje M, Hargrave D, Bullock AN, Puget S, Yip S, Jones C, and Grill J (2014) Recurrent activating ACVR1 mutations in diffuse intrinsic pontine glioma. Nat Genet 46:457–461. 10.1038/ng.2925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Venneti S, Garimella MT, Sullivan LM, Martinez D, Huse JT, Heguy A, Santi M, Thompson CB, and Judkins AR (2013) Evaluation of histone 3 lysine 27 trimethylation (H3K27me3) and enhancer of Zest 2 (EZH2) in pediatric glial and glioneuronal tumors shows decreased H3K27me3 in H3F3A K27M mutant glioblastomas. Brain Pathol 23:558–564. 10.1111/bpa.12042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Voigt P, Tee WW, and Reinberg D (2013) A double take on bivalent promoters. Genes Dev 27:1318–1338. 10.1101/gad.219626.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Warren KE (2012) Diffuse intrinsic pontine glioma: poised for progress. Front Oncol 2:205 10.3389/fonc.2012.00205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wu G, Barnhill RL, Lee S, Li Y, Shao Y, Easton J, Dalton J, Zhang J, Pappo A, and Bahrami A (2016) The landscape of fusion transcripts in spitzoid melanoma and biologically indeterminate spitzoid tumors by RNA sequencing. Mod Pathol 29:359–369. 10.1038/modpathol.2016.37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wu G, Broniscer A, McEachron TA, Lu C, Paugh BS, Becksfort J, Qu C, Ding L, Huether R, Parker M, Zhang J, Gajjar A, Dyer MA, Mullighan CG, Gilbertson RJ, Mardis ER, Wilson RK, Downing JR, Ellison DW, Zhang J, Baker SJ, and St. Jude Children’s Research Hospital-Washington University Pediatric Cancer Genome P (2012) Somatic histone H3 alterations in pediatric diffuse intrinsic pontine gliomas and non-brainstem glioblastomas. Nat Genet 44:251–253. 10.1038/ng.1102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wu G, Diaz AK, Paugh BS, Rankin SL, Ju B, Li Y, Zhu X, Qu C, Chen X, Zhang J, Easton J, Edmonson M, Ma X, Lu C, Nagahawatte P, Hedlund E, Rusch M, Pounds S, Lin T, Onar-Thomas A, Huether R, Kriwacki R, Parker M, Gupta P, Becksfort J, Wei L, Mulder HL, Boggs K, Vadodaria B, Yergeau D, Russell JC, Ochoa K, Fulton RS, Fulton LL, Jones C, Boop FA, Broniscer A, Wetmore C, Gajjar A, Ding L, Mardis ER, Wilson RK, Taylor MR, Downing JR, Ellison DW, Zhang J, and Baker SJ (2014) The genomic landscape of diffuse intrinsic pontine glioma and pediatric non-brainstem high-grade glioma. Nat Genet 46:444–450. 10.1038/ng.2938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zeisel A, Munoz-Manchado AB, Codeluppi S, Lonnerberg P, La Manno G, Jureus A, Marques S, Munguba H, He L, Betsholtz C, Rolny C, Castelo-Branco G, Hjerling-Leffler J, and Linnarsson S (2015) Brain structure. Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-seq. Science 347:1138–1142. 10.1126/science.aaa1934 [DOI] [PubMed] [Google Scholar]
- 53.Zhang J, Ding L, Holmfeldt L, Wu G, Heatley SL, Payne-Turner D, Easton J, Chen X, Wang J, Rusch M, Lu C, Chen SC, Wei L, Collins-Underwood JR, Ma J, Roberts KG, Pounds SB, Ulyanov A, Becksfort J, Gupta P, Huether R, Kriwacki RW, Parker M, McGoldrick DJ, Zhao D, Alford D, Espy S, Bobba KC, Song G, Pei D, Cheng C, Roberts S, Barbato MI, Campana D, Coustan-Smith E, Shurtleff SA, Raimondi SC, Kleppe M, Cools J, Shimano KA, Hermiston ML, Doulatov S, Eppert K, Laurenti E, Notta F, Dick JE, Basso G, Hunger SP, Loh ML, Devidas M, Wood B, Winter S, Dunsmore KP, Fulton RS, Fulton LL, Hong X, Harris CC, Dooling DJ, Ochoa K, Johnson KJ, Obenauer JC, Evans WE, Pui CH, Naeve CW, Ley TJ, Mardis ER, Wilson RK, Downing JR, and Mullighan CG (2012) The genetic basis of early T-cell precursor acute lymphoblastic leukaemia. Nature 481:157–163. 10.1038/nature10725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang Y, Chen K, Sloan SA, Bennett ML, Scholze AR, O’Keeffe S, Phatnani HP, Guarnieri P, Caneda C, Ruderisch N, Deng S, Liddelow SA, Zhang C, Daneman R, Maniatis T, Barres BA, and Wu JQ (2014) An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J Neurosci 34:11929–11947. 10.1523/JNEUROSCI.1860-14.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang Y, Sloan SA, Clarke LE, Caneda C, Plaza CA, Blumenthal PD, Vogel H, Steinberg GK, Edwards MS, Li G, Duncan JA 3rd, Cheshier SH, Shuer LM, Chang EF, Grant GA, Gephart MG, and Barres BA (2016) Purification and Characterization of Progenitor and Mature Human Astrocytes Reveals Transcriptional and Functional Differences with Mouse. Neuron 89:37–53. 10.1016/j.neuron.2015.11.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.