Abstract
Fusarium circinatum is a harmful pathogenic fungus mostly attacking Pinus species and also Pseudotsuga menziesii, causing cankers in trees of all ages, damping-off in seedlings, and mortality in cuttings and mother plants for clonal production. This fungus is listed as a quarantine pest in several parts of the world and the trade of potentially contaminated pine material such as cuttings, seedlings or seeds is restricted in order to prevent its spread to disease-free areas. Inspection of plant material often relies on DNA testing and several conventional or real-time PCR based tests targeting F. circinatum are available in the literature. In this work, an international collaborative study joined 23 partners to assess the transferability and the performance of nine molecular protocols, using a wide panel of DNA from 71 representative strains of F. circinatum and related Fusarium species. Diagnostic sensitivity, specificity and accuracy of the nine protocols all reached values >80%, and the diagnostic specificity was the only parameter differing significantly between protocols. The rates of false positives and of false negatives were computed and only the false positive rates differed significantly, ranging from 3.0% to 17.3%. The difference between protocols for some of the performance values were mainly due to cross-reactions with DNA from non-target species, which were either not tested or documented in the original articles. Considering that participating laboratories were free to use their own reagents and equipment, this study demonstrated that the diagnostic protocols for F. circinatum were not easily transferable to end-users. More generally, our results suggest that the use of protocols using conventional or real-time PCR outside their initial development and validation conditions should require careful characterization of the performance data prior to use under modified conditions (i.e. reagents and equipment). Suggestions to improve the transfer are proposed.
Subject terms: PCR-based techniques, Invasive species
Introduction
Fusarium circinatum Nelson Nirenberg & O’Donnell, formerly also known as Gibberella circinata Nirenberg & O’Donnell, is a harmful fungus causing pitch canker, a serious disease on pine trees. This pathogenic ascomycete attacks all Pinus species and Pseudotsuga menziesii (Mirb.) Franco, with varying levels of virulence1–3. All stages of pine are susceptible to the pathogen: seedling blight and damping-off, dieback of branches and stems on young and mature trees, where the most conspicuous symptoms are cankers accompanied by sometimes copious resin exudate (≪pitch canker≫)4. Dieback symptoms are also commonly observed in the crown due to the obstruction of water flow caused by the cankers and saturation of xylem by the excess resin produced by the tree. The multiplication of severe cankers on young or mature trees may lead to tree death5.
As for most tree pathogens, no economically and environmentally viable treatment is available to control or eradicate the fungus on a large scale. Management strategies are therefore focused on preventing the introduction of the pathogen, early detection and eradication of outbreaks in previously disease-free areas. Fusarium circinatum has been reported in different parts of the world, where it causes severe losses to the pine production industry (USA, South Africa, Korea, Japan, Spain) as well as in nurseries (Mexico, Haiti, Chile, Uruguay, Brazil, Colombia, South Africa) (EPPO Global database, https://gd.eppo.int/). In Europe, F. circinatum is officially present in Spain and Portugal in pine forests6,7, while it is also occasionally found in pine nurseries. The pathogen has also been found in French nurseries8, and in a public garden in Italy9, but it is currently considered as officially eradicated in both countries (EPP0 Global database, https://gd.eppo.int/). Since 2007, F. circinatum has been listed as a quarantine fungus for the EU, in order to prevent new introductions of infected material and further spread of the disease10. As a consequence of its quarantine status, a zero-tolerance policy is in force.
Management efforts should therefore focus on early detection of the pathogen in the different pathways of movement and introduction. Based on the pest risk assessment issued by the European Food and Environment Safety Agency (EFSA), the main pathways for the potential introduction of this fungus to disease-free areas are through contaminated pine seeds and seedlings8. To minimize the probability of an introduction and to reduce the cost associated with the eradication and control of this invasive pathogen, efficient management measures are needed11. For these reasons, the development of reliable diagnostic protocols is fundamental for the early and accurate detection of F. circinatum in pine-related commodities which can harbor and consequently spread the pathogen (i.e. substrates) such as seeds, seedlings, plants, young and mature trees. The diagnostic protocols should therefore be as specific and sensitive as possible10,12. False-negative detection results may lead to introduction of the pathogen in disease-free areas, while false-positive results may be responsible for unfair and inappropriate destruction of plant material, or a ban on trade with severe economic consequences.
Numerous diagnostic protocols targeting F. circinatum are currently available in the scientific literature and their use is suggested in international standard protocols by international bodies such as EPPO, ISTA or FAO-IPPC13–15. Despite most of them providing validation data supporting their accuracy, performance values are inconsistently reported or assessed in the original articles describing their development. Validation actions should be carried out to provide objective evidence that the test is suitable for the circumstances of use and can be considered for screening purposes16.
For instance, identification of F. circinatum may be achieved by isolation and morphological characterization of the pathogen, a common technique used in mycology. Fusarium circinatum displays several typical features, such as the presence of mono- and polyphialides as well as coiled sterile hyphae that aid diagnosis17,18. However, a recent study based on the phenotypical characteristics of isolates from a wide geographical range (Europe, America, Africa, and Asia) found that coiled sterile hyphae may not be a reliable morphological trait of F. circinatum, as previously reported19. Recent description of new Fusarium species in Colombia, which are also pine pathogens and can produce similar coiled hyphae in culture, further challenges the specificity of this morphology-based technique20. Additionally, morphological characterization is lengthy and requires considerable mycological expertise, whilst also not being efficient for the detection of quiescent forms of the pathogen that can be encountered in seedlings21 or in seeds.
A number of conventional and real-time polymerase chain reaction (PCR) assays targeting F. circinatum have been developed13,22–28, and another one is presented in the Supplementary Information section (Supplementary Information 1, hereafter referred to as Baskarathevan et al., unpublished). In the original articles, variable levels of validation are presented. For instance, the assessment of specificity and inclusivity used a more or less exhaustive range of Fusarium strains, depending on the availability of testing material, which included numbers of F. circinatum strains from different continents and newly described species, genetically related to F. circinatum. Considering the paramount importance of the reliability of a test when dealing with a quarantine pathogen, efforts should be focused on the continuous verification of the specificity of the protocols. This is particularly relevant when a Fusarium species is targeted, since this genus includes a steadily increasing number of newly described and cryptic species20,29,30. In addition, there is typically no data available to support the reliability of a given protocol when carried out in different laboratories, with different equipment and reagents than those described in the original papers. For instance, one may imagine that the specificity and the sensitivity of a test using conventional and real-time PCR may be altered by changing the brand of DNA polymerase enzyme, commercial ready-to-use master mix, or thermal-cycler/software. This in turn may affect the reliability of a diagnostic protocol targeting economically important pathogens such as F. circinatum31,32. Validation of diagnostic protocols is therefore a key element in establishing reference methods and to assess a laboratory’s competence and ability to produce reliable analytical data12.
The currently ongoing European COST action FP1406 “Pine Pitch Canker Strategies for Management of Gibberella circinata in Green Houses and Forests” (PINESTRENGTH) brings together 34 countries to establish a European-focused network dedicated to the Pine Pitch Canker pathogen. The main objectives are to increase knowledge on the biology, ecology and spread pathways of F. circinatum, to evaluate the potential for the development of effective and environmentally friendly prevention and mitigation strategies and to deliver these outcomes to stakeholders and policy makers. Early and accurate detection of F. circinatum is essential to achieve these goals. In this study, we compared for the first time performance of existing molecular tools targeting F. circinatum, with a wide range of DNA from target and non-target Fusarium species, in a large panel of 23 laboratories from European countries, South Africa and Chile. This collaborative study enabled us to provide useful data for the transferability of the different molecular-based diagnostic protocols. We propose recommendations for the preparation and use of future standard diagnostic protocols using conventional or real-time PCR assays.
Results
Indeterminate results
All 23 participants carried out the tests as requested. However, a few samples could not be tested by some of the participants due to loss or shortage of DNA template after multiple PCR run failures and were thus not considered in the analyses.
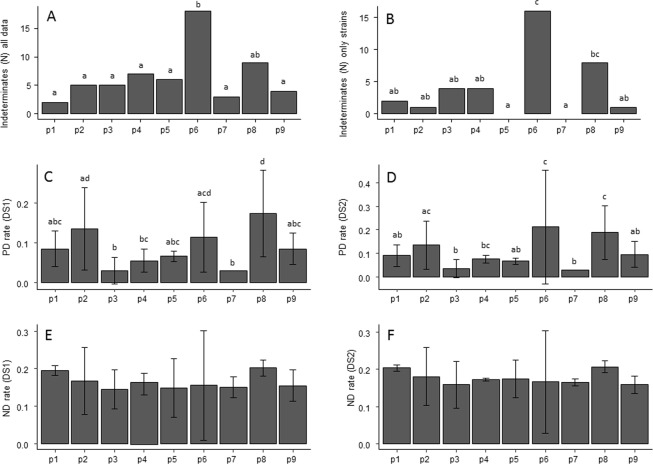
All nine protocols generated indeterminate results (Table 1, Fig. 1A). The total number of indeterminate results was 58 (1.5% of the total analysis) and ranged from 0.42% for protocol p1 to 4.58% for protocol p6 (Table 1). The participants reported several reasons for rating results as indeterminate. These included late mean cycle threshold (Ct) values or inconsistent Ct values from the replicates for hydrolysis probe-based protocols, melting temperatures (Tm) and melting peaks slightly different from the positive control, or late mean Ct values for SYBR Green based protocols. Generally, laboratories performing conventional PCR protocols did not provide an explanation for indeterminate results. However, one participant sequenced each amplicon and rated the results as indeterminate when the sequence was not readable. Comparison of indeterminate results by Fisher’s exact tests revealed significant differences (P < 0.001) between protocols (Table 1, Fig. 1A). In particular, protocol p6 based on SYBR Green real-time PCR, yielded significantly more indeterminate results than most of the other protocols. In this case, the melting peak analysis required by protocol p6 often revealed the presence of melting peaks slightly different from the positive control, the presence of double melting peaks, or only one out of two replicates were positive for some of the DNA extracts.
Table 1.
Performance values obtained during the collaborative study.
| Protocol | End point PCR | SYBR Green real-time PCR | Hydrolysis probe real-time PCR | Significance of the difference between protocols | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| p1 | p9 | p4 | p5 | p6 | p2 | p3 | p7 | p8 | ||
| Number of laboratories involved | 6 | 6 | 6 | 5 | 4 | 6 | 6 | 4 | 6 | |
| Number of samples analyzed and retained | 474 | 472 | 473 | 393 | 393 | 474 | 474 | 316 | 473 | |
| Number of indeterminate resultsa,b | 2 A (2 AB) | 4 A (2 AB) | 7 A (4 AB) | 6 A (0 A) | 18 B (16 C) | 5 A (1 AB) | 5 A (4 AB) | 3 A (0 AB) | 9 AB (8 BC) | P < 0.001 (P < 0.001) |
| Negative Accord (NA)b,c | 181 (180) | 183 (179) | 187 (183) | 154 (154) | 146 (130) | 171 (171) | 192 (191) | 128 (128) | 163 (161) | — |
| Positive Accord (PA)b,c | 223 (221) | 229 (231) | 230 (227) | 194 (189) | 192 (191) | 230 (226) | 236 (232) | 156 (153) | 221 (220) | — |
| Negative Deviation (ND)b,c | 53 (55) | 43 (44) | 45 (48) | 34 (40) | 35 (37) | 46 (50) | 40 (44) | 28 (31) | 55 (56) | — |
| Positive Deviation (PD)b,c | 17 (18) | 17 (19) | 11 (15) | 11 (11) | 19 (35) | 27 (27) | 6 (7) | 4 (4) | 34 (37) | — |
| Diagnostic Sensitivity % (SE)b,c | 80.8 (80.1) | 84.3 (83.6) | 83.6 (82.5) | 85.0 (82.8) | 84.0 (83.6) | 83.3 (81.9) | 85.5 (84.1) | 84.8 (83.2) | 80.1 (79.7) | P = 0.71 (P = 0.88) |
| Diagnostic Specificity % (SP)b,c | 91.4 ABCD (90.9 AB) | 92.4 ABCD (90.4 AB) | 94.4 AC (92.4 AB) | 93.3 ABC (93.3 AB) | 88.5 ABD (78.8 C) | 86.4 BD (86.4 AC) | 97.0 C (96.6 B) | 97.0 C (97.0 B) | 82.6 D (81.6 C) | P < 0.001 (P < 0.001) |
| Diagnostic accuracy % (AC)b,c | 85.2 AB (84.6 ABC) | 87.7 AB (86.4 AB) | 88.2 A (86.7 AB) | 88.5 A (87.3 ACD) | 85.9 AB (81.6 ABC) | 84.6 AB (83.8 ABC) | 90.3 A (89.2 B) | 89.9 A (88.9 D) | 81.2 B (80.5 CD) | P = 0.02 (P < 0.001) |
| Concordance level (%)b | 96.8 A | 93 C | 96.8 A | 93.7 C | 74.6 D | 88.5 B | 93.2 C | 97.7 A | 93.5 C | P < 0.001 |
| Positive deviation (PD)rate (%)b,c | 8.6 ABC (9.1 AB) | 8.5 ABC (9.6 AB) | 5.6 BC (7.6 AB) | 6.7 ABC (6.7 AB) | 11.5 ACD (21.2 C) | 13.6 AD (13.6 AC) | 3.0 B (3.5 B) | 3.0 B (3.0 B) | 17.3 D (18.7 C) | P < 0.001 (P < 0.001) |
| Negative deviation (ND) rate %b,c | 19.2 (19.9) | 15.7 (16.1) | 16.4 (17.5) | 14.9 (17.5) | 15.4 (16.2) | 16.7 (18.1) | 14.5 (15.9) | 15.2 (16.8) | 19.9 (20.3) | P = 0.716 (P = 0.89) |
aNumbers outside brackets are the total number of indeterminate results, while those between brackets indicate the number of indeterminate results considering only pure Fusarium strain DNA (i.e. no DNA extracts from inoculated seeds).
bValues followed by the same letter in a line are not significantly different (P > 0.05).
cNumbers outside brackets are estimations for the DS1 dataset and those between brackets are estimations for the DS2 dataset.
Figure 1.
(A) Total number of indeterminate results by protocol considering all data (strains and inoculated seeds); (B) Total number of indeterminate results by protocol considering only strains (i.e. no inoculated seed data); (C) PD rate mean values and standard deviation by protocol for DS1 dataset; (D) PD rate mean values and standard deviation by protocol for DS2 dataset. (E) ND rate mean values and standard deviation by protocol for DS1 dataset; (F) ND rate mean values and standard deviation by protocol for DS2 dataset. The x – axis in all graphs represents the nine protocols tested in this study, from p1 to p9. Please refer to Table 4 for details of each protocol. Different letters indicate values are significantly different, according to Fisher’s Exact Test, for count data with simulated P-values based on 1e + 05 replicates.
When indeterminate results were analyzed with only pure Fusarium strain DNA (no DNA extracts from inoculated seeds), 37 indeterminate results were reported (63.8% of the total number of indeterminate results). Fisher’s exact tests revealed significant differences between protocols for these indeterminate results (P < 0.001, Table 1, Fig. 1B), and the rate of indeterminate ranged from 0.0% (protocol p5 and protocol p7) to 4.6% (protocol p6). In general, these indeterminate results mostly concerned non-target species (88.9% of the Fusarium strain DNA material indeterminate results).
Comparisons of indeterminate results between laboratories by protocol revealed significant differences for p2 (indeterminate data by laboratory ranging from 0% to 5.1%, P = 0.02), p5 (indeterminate data by laboratory ranging from 0% to 7.6%, P = 0.002), p6 (indeterminate data by laboratory ranging from 0% to 16.5%, P < 0.001), p8 (indeterminate data by laboratory ranging from 0% to 7.6%, P < 0.001) and p9 (indeterminate data by laboratory ranging from 0% to 5.1%, P < 0.001).
Positive and negative deviation rates
All nine protocols exhibited positive deviations (or false positives) and negative deviations (or false negatives) regardless of the data set used (Table 1, Fig. 1C–F). Analysis of DS1 (the dataset for which an indeterminate result is ultimately determined to be the expected result, see methods section) revealed positive deviation rates (PD) ranging from 3.0% (protocol p7) to 17.3% (protocol p8) (Table 1, Fig. 1C). Comparison of PD by Fisher’s exact tests revealed significant differences (P < 0.001) between protocols (Table 1, Fig. 1C). Negative deviation rates (ND) in DS1 ranged from 14.5% (protocol p3) to 19.9% (protocol p8), but according to Fisher’s exact tests, the ND rates between protocols were not different (P = 0.71) (Table 1, Fig. 1E). Concerning DS2 (the dataset for which an indeterminate result is ultimately determined to be the contrary of the expected result, see methods section), PD ranged from 3.0% (protocol p7) to 21.2% (protocol p6) (Table 1, Fig. 1D). Significant differences between protocols for PD rates were observed according to Fisher’s exact tests (P < 0.001). ND in DS2 ranged from 15.9% (protocol p3) to 20.3% (protocol p8) (Table 1, Fig. 1F). ND comparisons between protocols using Fisher’s exact tests did not reveal any significant differences (P = 0.88).
Comparisons between laboratories using DS1 revealed significant PD rate differences only for p2 (PD rate by laboratory ranging from 3.0% to 27.3%, P = 0.007) and p8 (PD rate by laboratory ranging from 9.1% to 37.5%, P = 0.04). Concerning the ND rate in DS1, significant differences were observed in p2 (ND rate by laboratory ranging from 6.5% to 32.6%, P = 0.031) and p6 (ND rate by laboratory ranging from 4.3% to 40.9%, P < 0.001). When comparisons were performed using DS2, significant differences between laboratories were revealed for p2 (PD rate by laboratory ranging from 3.0% to 27.3%, P = 0.007) and p6 (PD rate by laboratory ranging from 6.1% to 63.6%, P < 0.001), for the PD rate, and for p6 (ND rate by laboratory ranging from 6.5% to 40.9%, P < 0.001) for the ND rate.
Pattern of cross-reactions with non-target species
As shown previously, all protocols exhibited different levels of positive deviations (Table 1, Fig. 1C,D). Cross-reactions with DNA from strains of non-target Fusarium species were encountered for all nine protocols, but inconsistently between participating labs (Table 2). For many strains, a cros-s reaction with the DNA extract was reported for only a single participant among the four, five, or six laboratories involved, which corresponds to a unique reagent/equipment/operator combination.
Table 2.
Detailed list of the false positive and false negative results obtained with the panel DNA from target and non-target species in this study.
| Protocol number | Cross-reactions* | Indeterminate or false negative result |
|---|---|---|
| (F. circinatum strain, originating country)** | ||
| p1 | F. marasasianum (5/6)a | NRRL26431, Japan (5/6) |
| F. pinninemorale (5/6)a | 310/061, Spain (1/6) | |
| F. sororula (5/6)a | ||
| F. temperatum (2/6)a | ||
| p2 | F. begoniae (6/6)b | LSVM216, France (1/6) |
| F. concentricum (1/6)b | LSVM1221, Spain (1/6) | |
| F. culmorum (1/6)b | FcCa01, Spain (1/6) | |
| F. fracticaudum (3/6)a | FcCa05, Spain (1/6) | |
| F. parvisorum (3/6)a | FcCa06, Spain (1/6) | |
| F. pininemorale (2/6)a | CSF-13, Spain (1/6) | |
| F. sororula (2/6)a | 2306 BASA, Chile (1/6) | |
| F. subglutinans (4/6)b | ||
| p3 | F. culmorum (1/6)b | NRRL25708, USA (1/6) |
| F. subglutinans (3/6)b | NRRL25333, S. Africa (1/6) | |
| FcCa06, Spain (1/6) | ||
| CSF-18, Spain (1/6) | ||
| p4 | F. marasasianum (1/6)a | |
| F. proliferatum (1/6)a | ||
| F. subglutinans (6/6)b | ||
| p5 | F. proliferatum (1/5)b | CSF8, Spain (1/5) |
| F. subglutinans (6/5)b | CSF10, Spain (1/5) | |
| CSF11, Spain (1/5) | ||
| p6 | F. acuminatum (1/5)a | NRRL25708, USA (1/5) |
| F. fracticaudum (1/5)a | NRRL25331, USA (1/5) | |
| F. graminearum (1/5)b | NRRL25333, S. Africa (1/5) | |
| F. incarnatum (1/5)b | FcCa02, Spain (1/5) | |
| F. mangiferae (1/5)b | FcCa05, Spain (1/5) | |
| F. marasasianum (3/5)a | FC042V, Spain (1/5) | |
| F. parviporum (1/5)a | FC035, Spain (1/5) | |
| F. pininemorale (1/5)a | CSF1, Spain (1/5) | |
| F. reticulatum (1/5)a | CSF2, Spain (1/5) | |
| F. sacchari (1/5)b | CSF3, Spain (1/5) | |
| F. sororula (1/5)a | CSF4, Spain (1/5) | |
| F. sporotrichioides (1/5)b | CSF7, Spain (1/5) | |
| F. subglutinans (4/5)b | CSF8, Spain (1/5) | |
| F. torulosum (1/5)a | CSF11, Spain (1/5) | |
| F. thapsinum (1/5)b | CSF12, Spain (1/5) | |
| F. tricinctum (2/5)b | CMW1219, S. Africa (1/5) | |
| F. verticillioides (1/5)b | ||
| p7 | F. temperatum (4/4)c | |
| p8 | F. fracticaudum (4/6)a | NRRL26431, Japan (6/6) |
| F. incarnatum (1/6)a | LSVM211, France (1/6) | |
| F. mangiferae (1/6)a | LSVM216, France (1/6) | |
| F. parvisorum (4/6)a | ||
| F. proliferatum (1/6)a | ||
| F. sacchari (1/6)a | ||
| F. sororula (4/6)a | ||
| F. subglutinans (6/6)a | ||
| F. temperatum (5/6)a | ||
| F. verticillioides (1/6)a | ||
| p9 | F. begoniae (1/6)b | |
| F. concentricum (2/6)b | ||
| F. culmorum (1/6)b | ||
| F. fracticaudum (5/6)a | ||
| F. fractiflexum (1/6)b | ||
| F. marasasianum (1/6)a | ||
| F. proliferatum (1/6)b | ||
| F. sororula (1/6)a | ||
| F. subglutinans (5/6)b | ||
| F. torulosum (1/6)a |
*Number of participants for which a cross reaction was observed/number of participants involved. Species names in bold indicate a frequent cross-reaction was observed for the protocol with the 0.5 ng µL−1 DNA extract used.
**Number of participants for which the 0.5 ng µL−1 DNA extract of the F. circinatum strain was not picked up by the protocol or yielded an indeterminate result/number of participants involved.
aNo reference sequence is available for this marker on GenBank for comparison with amplicon sequence, at the time of this study.
bComparison of the amplicon sequence with IGS F. circinatum reference sequences available on GenBank shows several polymorphisms in the region between primers.
cComparison of the amplicon sequence with TEF 1alpha F. circinatum reference sequences available on GenBank shows no polymorphism in the region between primers.
Fusarium subglutinans was the species that accounted for the most frequent cross-reactions, and its DNA yielded false-positive results for seven out of the nine protocols. Depending on the protocol, from three, up to six out of six laboratories observed cross-reaction with DNA of this species. Fusarium temperatum was the only, albeit consistent, cross-reaction observed with protocol p7. DNA from the newly-described species on pine from Colombia, i.e. F. marasasianum, F. pinninemorale, F. sororula, F. fracticaudum, and F. parvisorum also yielded frequent cross-reactions with four protocols (p1, p2, p8 and p9), and less frequently with protocol p6.
Reference sequences of target genes from species whose DNA cross-reacted with some of the PCR or real-time PCR tests were retrieved from GenBank and aligned with orthologous reference sequences of F. circinatum. The regions upstream and downstream of the forward and reverse PCR primers were removed. The alignments were manually scrutinized to check for the presence of interspecific polymorphism between the regions targeted by the primers. For some of the PCR or real-time PCR tests, it was shown that the presence of polymorphisms after sequencing the amplicon would be helpful to confirm the occurrence of cross-reactions, and possible false-positive results (Table 2).
Inclusivity of the different protocols
In this work, inclusivity is defined as the ability of each protocol to detect DNA of the target species, regardless of the host plant, mating type, geographical origin and year of collection. Different patterns of inclusivity were observed between protocols. DNA from some of the F. circinatum strains yielded inconsistent negative results. Protocols p4, p7 and p9 successfully picked up all the 38 F. circinatum strains of both mating types included in the panel, regardless of the equipment, reagents and operator, thus supporting their excellent level of inclusivity. By contrast, protocols p1 and p8 almost systematically failed to yield positive results with DNA from the Japanese strain of F. circinatum NRRL2643. However, the rest of the false negative results were not reproducible between laboratories, and were only reported for one participant out of the four, five, or six involved, meaning that they were only observed for some operator/reagent/equipment combinations. Except for the Japanese strain of F. circinatum NRRL2643, these false negative results were observed with DNA from different F. circinatum strains originating from Spain, France, Chile, USA, and South Africa without any obvious pattern.
Performance criteria and reproducibility
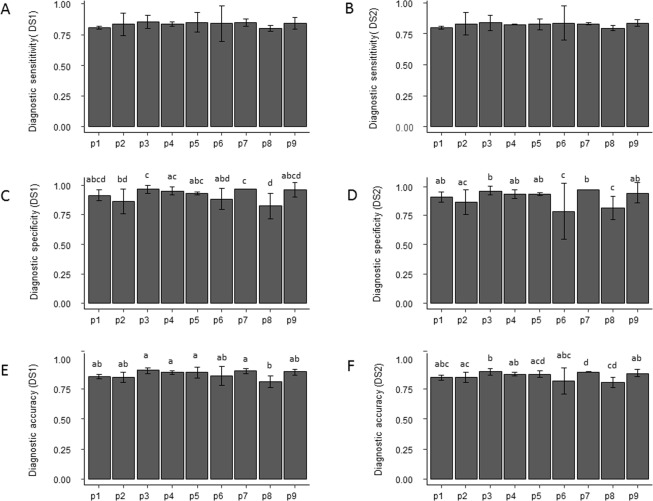
Diagnostic sensitivity (SE) ranged from 80.1% (protocol p8) to 85.5% (protocol p3) and from 79.7% (protocol p8) to 84.1% (protocol p3) using the DS1 and DS2 datasets, respectively. Fisher’s exact tests did not reveal significant differences for SE between the nine protocols either for DS1 (P = 0.72) or DS2 (P = 0.88) (Table 1, Fig. 2A,B). By contrast, diagnostic specificity (SP) differed significantly, using both datasets (both P-values < 0.001). SP ranged from 82.6% (protocol p8) to 97% (for both protocols p3 and p7) in DS1 (Table 1, Fig. 2C). When SP was assessed for DS2, it ranged from 78.8% (protocol p6) to 97% (protocol p7) (Table 1, Fig. 2D). Significant differences in Diagnostic accuracy (AC) were observed between protocols for both datasets (P = 0.002 and P < 0.001 for DS1 and DS2, respectively). AC ranged from 81.2% (protocol p8) to 90.3% (protocol p3) in DS1 and from 80.5% (protocol p8) to 89.2% (protocol p3) in DS2 (Table 1, Fig. 2E,F). Concordance ranged from 74.6% to 97.7% for p6 and p7, respectively. Fisher’s exact tests revealed significant differences between methods (P < 0.001, Table 1).
Figure 2.
(A) Diagnostic sensitivity and standard deviation by protocol for the DS1 dataset. (B) Diagnostic sensitivity and standard deviation by protocol for the DS2 dataset. (C) Diagnostic specificity and standard deviation by protocol for the DS1 dataset. (D) Diagnostic specificity and standard deviation by protocol for the DS2 dataset. (E) Diagnostic accuracy and standard deviation by protocol for the DS1 dataset. (F) Diagnostic accuracy and standard deviation by protocol for the DS2 dataset. The x – axis in all graphs represents the 9 protocols tested in this study, from p1 to p9. Please refer to Table 4 for details of each protocol. Different letters indicate values are significantly different, according to Fisher’s Exact Test, for count data with simulated P-values based on 1e + 05 replicates.
Analytical sensitivity
The analytical sensitivity was assessed for each protocol using serial dilutions of DNA from P. pinaster seeds spiked with F. circinatum conidia. However, inconsistent results were obtained with the serial dilutions. For example, when analyses were performed with the DS1 scenario, seven out of the nine evaluated protocols were able to detect at least one of the samples containing F. circinatum DNA in seeds at the highest concentration, i.e. 2 105 conidia/mL of ground seed homogenate: p2, p3, p4, p5, p6, p7 and p9. Only p6 was able to give a positive result for all samples of this concentration. Similarly, these inconsistent results were also observed in data from DS2, and only p2, p3, p5 and p6 were able to detect at least one of the samples of the highest concentration of F. circinatum in seeds. This inconsistent behavior of inoculated material was observed for all of the serial dilution samples, in both datasets (Supplementary Dataset 1).
Discussion
To establish an international surveillance network for the detection of outbreaks of pine pitch canker across Europe and other disease-free areas, harmonization of protocols for F. circinatum diagnosis is needed. To our knowledge, two international diagnostic protocols targeting this pathogen already exist. Nevertheless, one of them is a list of protocols that the international community has agreed are satisfactory, with minimal validation data available13,15 and the other relies solely on techniques such as mycological plating, that were selected because of their low cost and ease of implementation14. Most of the protocols targeting F. circinatum described in the literature lack a comprehensive evaluation of some basic performance criteria, such as specificity, inclusivity and sensitivity. Additionally, in most cases, specificity has not been re-evaluated in light of newly emerged or described Fusarium species occurring on Pinus spp20. In this study, we selected nine different detection protocols based on PCR and its variants. Their performance was assessed using a panel of 71 Fusarium strains and eight pine seed samples spiked with F. circinatum conidia. In order to achieve the best representation of F. circinatum strains (i.e. inclusivity), the panel included strains originating from six countries across four continents, and both mating types. Also included were recently-described Fusarium species isolated from pines, genetically related to F. circinatum, with overlapping morphological features, and some of them being pathogenic to Pinus20. This large panel of strains was used to assess the performance and transferability of the different protocols through an international collaboration involving a broad consortium of 23 partners. Each protocol was evaluated by a minimum of four, and up to six laboratories therefore ensuring a robust dataset. Practical and technical constraints led to an unbalanced number of laboratories involved per protocol, which means that some of the results should be read with care. In particular, although significant differences were observed for some criteria between participant laboratories and some of the protocols, caution is required before generalizing the results regarding non-significance of some of the statistical tests. Indeed, non-significant differences between protocols means non-identification of a difference rather than the absence of a difference altogether.
Our results showed that all protocols presented acceptable performance values in both datasets (>75%) for diagnostic accuracy, specificity and sensitivity, with some laboratories obtaining individual values close to 100% (Supplementary Dataset 1). Yet diagnostic specificity and accuracy differed considerably between protocols, irrespective of the technology involved (i.e. end-point PCR or real-time PCR using SYBR Green dye or specific hydrolysis probes). These differences were principally linked to cross-reactions with non-target species (positive deviations), and less commonly to consistent or erratic negative deviations with particular strains of F. circinatum.
The cross-reactions with DNA from non-target species observed in our study have not been reported in the original articles describing the protocols. We included a broader and more comprehensive panel of strains, revealing more information about the level of specificity of these protocols. However, our panel is not exhaustive and of course does not cover the entire biological range of genetically related Fusarium strains. Other unexpected cross-reactions may therefore occur, particularly with DNA from as yet undescribed Fusarium taxa. Cross-reactions were observed in all protocols, although at different levels, depending on the laboratory involved. Some of the erratic cross-reactions and false negative results may have occurred because of issues such as pipetting errors, DNA shearing, among others. However, some of the cross-reactions were more frequent and were due to lack of specificity/sensitivity of the molecular markers toward strains that had not been assessed during the original validation step of the original protocol by the authors, such as for genetically related F. temperatum and F. subglutinans. From a practical point of view, the presence of some of the Fusarium species whose DNA yielded false positive results are very unlikely on pine tissue, but the recent finding of Herron et al.20 showed that previously unknown species may be found on pine. In our experiment, a common DNA extraction procedure was followed for all the fungal strains, which sometimes differed from the original article describing each of the nine protocols. It cannot be ruled out that the DNA extraction procedure used in our study had an effect of PCR or real-time PCR specificity or inclusivity. Another aspect to consider is that for the sake of harmonization, a standard concentration of Fusarium DNA was used throughout the study (0.5 ng/µL). This may not always reflect the actual concentration that may occur when testing real pine samples contaminated with these Fusarium species, and the likelihood of cross-reactions with non-target DNA probably increases with higher concentrations. At the same time, certain strains of F. circinatum were ‘missed’ by some of the protocols (especially the F. circinatum strain from Japan), with false-negative results that were not reported in the original articles, except by Ramsfield et al. (2008) regarding protocol p1. This suggests that some of the F. circinatum strains travelling with plant material such as seeds might not be detected when using some of the protocols assessed here. Our data provide a first evaluation of the inclusivity of nine protocols, which can be useful for laboratories in charge of official analyses, by elucidating the level of uncertainty associated with some of the protocols used throughout the world. Although diagnostic specificity across all protocols was rather high (>75%), no protocol was 100% specific with the present panel of F. circinatum strains. These false-positives may not be acceptable when dealing with a pathogen subjected to strict phytosanitary regulations.
Concordance varied between protocols and ranged from 74.6 to 97.7%. Analysis of the differences between laboratories that tested the same protocol also showed that indeterminate results and negative and positive deviations differed significantly. These results suggest that molecular detection methods may not always be easily transferable. Basically, they clearly illustrate that deviations from the “original recipe”, i.e. the use of different equipment, consumables, but also operators, might compromise the stringency of the reactions, and therefore the specificity of the results. This is particularly important when dealing with a quarantine pathogen, with a zero-tolerance policy. Positive deviations may lead to the inappropriate destruction of goods, whereas negative deviations could fail to prevent introduction of the pathogen into disease-free areas.
We also showed that all the protocols exhibited some problems in result interpretation (indeterminate results), independently of the PCR technology used. However, end-point PCR generally yielded fewer indeterminate results, across all partners, probably linked to the simplicity of result interpretation, based on the observation of a band on an electrophoresis gel, with little room for doubt. Concerning the other PCR techniques, it can be suggested that interpretation of the melting curves was not straightforward when using the SYBR Green real-time PCR, and setting the fluorescence threshold for the calculation of the cycle threshold value when using a hydrolysis probe was sometimes done inconsistently between partners. In addition to this problem of result interpretation, some of the protocols required particular settings that may not have worked well under different conditions. For example, protocol p224 requires an unusually high hybridization temperature of 70 °C to ensure specificity, which seemed to cause a sensitivity problem when used in certain laboratories or with different reagents/equipment than the ones originally described. In this work, the statistical analysis of indeterminate results was enabled by processing data under two scenarios (DS1 and DS2). In all cases results from both datasets were consistent, leading consequently to the same conclusions. This is an important point because it means that differences in performance criteria between protocols were not influenced by indeterminate results, which represented less than 2% of the total results.
We did not evaluate and compare the sensitivity of a protocol based on mean Ct values, but rather on its ability to yield positive results with lower target concentrations. This approach enabled comparison of conventional PCR, for which no quantitative results are generated with real-time PCR protocols. Additionally, we chose not to provide cutoff values because the sensitivity of a test should not be dependent on Ct values, but rather on its ability to reliably amplify and detect a low concentration of target DNA33. Late Ct values may still be valid and confidently used if the test specificity has been correctly designed and evaluated34. Despite protocols p2, p3, p5 and p6 consistently yielding positive results for the highest conidia spiking quantity, the data of our study showed that positive results for lower concentrations were rarely obtained. This made it very difficult to compare the protocols to each other regarding analytical sensitivity with seeds. However, using protocols p2 and p6, successful detection of F. circinatum in naturally infected pine seeds has been reported by Ioos et al.24 and Dreaden et al.25, and protocol p2 has been used for years in ANSES for the interception of naturally infected imported seed lots (Guinet, C., ANSES, pers. Comm.). This suggests that the modified method used in our study for the preparation of artificially infected seed DNA was not able to provide samples with a sufficient level of contamination, probably inferior to what is expected with real-world samples. In this respect, a preliminary biologic enrichment of the seed in a broth of culture medium seems a very efficient method to improve detection of F. circinatum in seeds at low levels13.
One of the main recommendations resulting from the present study is that the transferability of a PCR or real-time PCR protocol should be thoroughly and continuously assessed before becoming a standard. In other words, the ability to yield accurate results when used under slightly different conditions should be checked. Indeed, it is very unlikely that all the specific brands of reagents and equipment described in the original scientific papers are available to end-users. In this study, partner laboratories were free to use their own real-time equipment and brand of reagents, such as DNA polymerases, PCR or real-time PCR master mixes. Discrepancies of results between laboratories regarding false positive and false negative rates, as well as the different analytical sensitivities confirm that changing the brand and type of DNA polymerase35 and equipment31,36 may affect the reliability of the results. Changing the DNA polymerase may also generate the amplification of non-specific amplicons, especially when working with symptomatic pine DNA extracts (Piškur, B., pers. comm.). This observation is in line with Bustin & Huggett33 who showed that the performance of a real-time PCR assay varied with different master mixes, probably due to differences in Mg2+ concentrations and the addition of undisclosed stabilizers to the buffer affecting primer and probe annealing.
Another parameter to be considered is that interpretation of the fluorescence levels yielded in the real-time PCR reactions requires the enforcement of decision rules, either by the operator, or the analysis software. In turn, the decision to rate a Ct value as a positive or indeterminate result may be influenced by internal rules, which are not the same between laboratories. In addition, slight variations induced by the operator or the equipment, such as pipetting errors, temperature drift or thermic heterogeneity of the thermal cycler block may have an effect on the stringency of the PCR reaction and thus, in turn may affect the analytical specificity and sensitivity33,37–39.
In line with other guidelines proposed for testing genetically modified organisms40, we therefore recommend that a preliminary assessment of the robustness and transferability of a new protocol should be carried out to provide an indication of its performance under different conditions than the ones used during its development. This assessment should be carried out in addition to the classical performance criteria assessed during the initial validation process. This may be achieved by, for instance, the organization of a collaborative study, using a large and representative panel of target and non-target taxa, and involving as many different reagent brands and thermal cycler types as possible. Therefore, the end users should bear in mind that the performance data of a conventional or real-time PCR protocol described in the original articles are intimately linked to the reagents, equipment and decision rules used. It is strongly suggested that individual laboratories should carry out their own characterization if these parameters are modified, and even if no parameter is modified at all. To this aim, a series of “reference samples” should be maintained and provided by a “reference laboratory” to any laboratory intending to establish and maintain an accurate diagnostic test41. The organization of training sessions by these reference laboratories would also help to share experience and knowledge about the use of a given protocol, and would harmonize the practices and decision rules. This is of paramount importance when targeting a quarantine pathogen, for which very strict regulations are enforced.
We also advocate the continuous verification of the specificity of published protocols, in order to consider new taxa that are continuously described in the literature. This can be achieved by in silico evaluation, by blasting the primer and probe sequences on international DNA databases such as GenBank on a regular basis, and by wet lab testing of newly described strains. Another suggestion to ensure the accuracy of the positive results is to analyze the amplicon sequence and/or to use additional tests targeting other loci in the genome of the target organism. Currently, the two international protocols for the diagnosis of F. circinatum13,15 recommend sequencing of the amplicon after a positive result via conventional or SYBR Green real-time PCR using the CIRC1A-CIRC4A primers22. However, our study suggests that a similar procedure should also apply for the other available protocols targeting F. circinatum, even for those using hydrolysis probe-based methods. It is advisable that such a complementary approach should be followed to verify results of particular importance such as first reports in disease-free areas. For some of the protocols, analysis of the amplicon sequence, trimmed from the primers’ sequences may help confirm the accuracy of the result. However, sequencing will not always be sufficient. Firstly, this is dependent on reference sequences being available in databases. Secondly, this approach will not work if undescribed species share a 100% match to F. circinatum (see Table 2 footnotes). Lastly, confirmation by sequencing is not always possible when the conventional or real-time PCR test targets a region of unknown function such as a Sequence Characterized Amplified Region (SCAR). Hence, no orthologous sequences for other Fusarium species are available for the SCAR targeted by protocol p1 and p8.
In addition, positive samples could be further processed in order to isolate the pathogen in pure culture, allowing the identification of the pathogen by both morphological and molecular features42. Combining molecular and morphological data would of course secure identification of the pathogen, particularly important for first reports, and will help to increase knowledge of the morphologic and genetic diversity of the pathogen. In this respect, it is necessary to establish protocols providing a representative sampling strategy, starting from plant tissue. With the exception of seeds, for which a strategy has been proposed43, there is to our knowledge no standard for sampling plants or adult trees, tackling for instance the minimum number of samples that should be taken for assuring the absence of the pathogen, irrespective of the analysis technique chosen (molecular or isolation).
Methods
Participants and selection of protocols
An official call for participation was issued in 2016 in the framework of the COST action FP1406 PINESTRENGTH. In all, 23 laboratories representing 18 countries participated in the study (Table 3). Only laboratories with sufficient experience in molecular biology-based detection techniques and appropriate equipment were involved. Samples were sent to each participant on June 7th, 2017. The analyses were to be completed and results returned to the organizer by the end of August 2017.
Table 3.
Partners involved in the collaborative project.
| Partner Label | Institute/laboratory | Description | Country, city |
|---|---|---|---|
| 1 | Sustainable Forest Management Institute | University of Valladolid | Spain, Palencia |
| 2 | Institute of Forestry and Rural Engineering | Estonian University of Life Sciences | Estonia, Tartu |
| 3 | Institute for National and International Plant Health | Julius Kühn-Institute | Germany, Braunschweig |
| 4 | Vokė Branch, Lab of Biotechnology | Lithuanian Research Centre for Agriculture and Forestry | Lithuania, Vilnius |
| 5 | Department for Innovation in Biological, Agro-food and Forest Systems (DIBAF), | University of Tuscia | Italy, Viterbo |
| 6 | Instituto Agroforestal Mediterráneo | Universitat Politècnica de València | Spain, Valencia |
| 7 | Laboratoire de la santé des végétaux | French Agency for food, environmental and occupational health safety (ANSES) | France, Malzéville |
| 8 | Faculty of Biology and Environmental Sciences | Cardinal Stefan Wyszynski University in Warsaw | Poland, Warsaw |
| 9 | Forest Health and Biotic Interactions/Phytopathology | Swiss Federal Institute for Forest, Snow and Landscape Research WSL | Switzerland, Birmensdorf |
| 10 | Instituto Nacional de Investigação Agrária e Veterinária I.P., | State Laboratory of the Ministry of Agriculture, Forests and Rural Development | Portugal, Oeiras |
| 11 | Institute for Sustainable Plant Protection | National Research Council | Italy, Florence |
| 12 | Forest Research | Forestry Commission | United Kingdom, Farnham |
| 13 | Centro de Biotecnología | Universidad de Concepción | Chile, Concepción |
| 14 | Department of Food, Environmental and Nutritional Sciences | University of Milan | Italy, Milan |
| 15 | Forestry and Agricultural Biotechnology Institute | University of Pretoria | South Africa, Pretoria |
| 16 | Department of Agricultural Sciences, Biotechnology and Food Science | Cyprus University of Technology | Cyprus, Limassol |
| 17 | Department of Biology | University of Aveiro | Portugal, Aveiro |
| 18 | Dipartimento di Scienze delle Produzioni Agroalimentari e dell’Ambiente (DISPAA) | University of Florence | Italy, Florence. |
| 19 | Phytophthora Research Centre | Mendel University in Brno | Czech Republic, Brno |
| 20 | Laboratory of Forest Protection | Slovenian Forestry Institute | Slovenia, Ljubljana |
| 21 | Forest Health Center of Calabazanos | Regional Government of Castilla y León (JCyL) | Spain, Palencia |
| 22 | Faculty of Forestry, Forest Pathology Laboratories | Applied Sciences University of Isparta | Turkey, Isparta |
| 23 | Department of Agriculture, Food and Environment | University of Catania | Italy, Catania |
At the time the project was started, a total of nine conventional or real-time PCR protocols targeting F. circinatum (p1 to p9) were available in the literature or were brought to our knowledge (Table 4). Protocols included several formats of PCR amplification and labeling. Protocols p1 and p9 use conventional or end-point PCR13,23, p4, p5, and p6 use SYBR Green-based real-time PCR22,25,26, and p2, p3, p7, and p8 use real-time PCR hydrolysis probe-based tests24,27,28. In order to balance the comparison among the protocols and to provide a sufficient amount of data to compute the performance criteria, each protocol was assessed by at least four different partners.
Table 4.
List of F. circinatum diagnostic protocols assessed during the collaborative study.
| Protocol number | Reference | Target in the F. circinatum genome | Type of assay |
|---|---|---|---|
| p1 | Ramsfield et al.23 | Two independent sequence characterized amplified regions (SCAR) | End-point PCR |
| p2 | Ioos et al.24 | rDNA Intergenic spacer (IGS) | Real-time PCR with hydrolysis probe |
| p3 | Lamarche et al.27 | rDNA Intergenic spacer (IGS) | Real-time PCR with hydrolysis probe |
| p4 | Schweigkofler et al.22 | rDNA Intergenic spacer (IGS) | Real-time PCR with SYBR Green staining |
| p5 | Fourie et al.26 | rDNA Intergenic spacer (IGS) | Real-time PCR with SYBR Green staining |
| p6 | Dreaden et al.25 | rDNA Intergenic spacer (IGS) | Real-time PCR with SYBR Green staining |
| p7 | Luchi et al.28 | Translation elongation factor 1-alpha gene (TEF) | Real-time PCR with hydrolysis probe |
| p8 | Baskarathevan et al. (Supplementary Information 1) | Sequence characterized amplified region (SCAR) | Real-time PCR with hydrolysis probe |
| p9 | EPPO13, appendix 4 | rDNA Intergenic spacer (IGS) | End-point PCR |
Protocols were conducted following the description in the original article, observing the amplification parameters (cycling conditions, temperatures settings) and reaction mixtures (primers and probe concentrations, reaction and DNA template volumes) indicated by the authors and summarized in a reference document that was sent to each participant along with the DNA samples (Supplementary Information 2). However, if not available in the participant laboratory, the DNA polymerase or commercial real-time PCR master mix described in the original articles were replaced by the reagents typically used by the participant laboratory (for further information refer to Supplementary Information 3). Each participating laboratory was free to use its own PCR equipment.
F. circinatum-specific primers and probes
Each partner provided the primers for the protocols using end-point PCR and SYBR Green real-time PCR, as described in the original articles. To cut down costs, the primers/probe combinations required for the different hydrolysis probe real-time PCR protocols were only purchased once by one of the partners and distributed to all the participants as ready-to-use aliquots of 30 µM (primers) or 10 µM (probe) solutions, in 1.5 mL amber microtubes. Primers/probe combinations for p2 and p7, p3, and p8 were custom made by Eurogentec (Seraing, Belgium), Integrated DNA technology (Skokie, Illinois), and Biosearch Technologies (Petaluma, California), respectively. Primers and probes were shipped at room temperature by a fast delivery service and kept in a freezer until used for testing.
Fungal strains and preparation of panels of DNA samples
A panel of 71 monosporic Fusarium spp. strains representing 29 distinct species was used (Table 5). Species identity was confirmed by EF1 alpha gene sequencing44, if the strain was not obtained from an international fungal collection. It included 38 F. circinatum strains from different geographical origins, mating types, host tree species and environments, thus covering as much of the genetic diversity of the pathogen as possible. Thirty-three other Fusarium strains were also included. They represented species that are either genetically close to F. circinatum, or inhabit the same ecological niche, i.e. pine woody tissue, pine seeds, pine roots, or soil. Also included were recently described species of Fusarium associated with pine cankers in Colombia, i.e. F. parvisorum, F. sororula, F. marasasianum, F. pininemorale, and F. fracticaudum20. The strains were sent from different providers, and were kept on agar slants at 5 °C before handling. As F. circinatum is considered a quarantine organism for the European Union (EU), all strains from this species were maintained and manipulated in level 3 biohazard containment facilities in ANSES Plant Health Laboratory (here named as ANSES), in Malzéville, France, in compliance with EU Directive 2008/61/EC. Taking into consideration that the Fusarium strains from Colombia are recently described species20 not found in the EU, it was decided to manipulate them under the same conditions as F. circinatum.
Table 5.
List of Fusarium spp. strains used in the collaborative study.
| Code | Species | Strain | Mating type | Host | Origin | Environment | Provider |
|---|---|---|---|---|---|---|---|
| F1 | F. circinatum | LSVM211 | MAT-1 | P. menziesii | France (Perpignan) | Private garden | R. Ioos |
| F2 | F. circinatum | LSVM216 | MAT-2 | P. radiata | France (Vendée) | Nursery | R. Ioos |
| F3 | F. circinatum | LSVM217 | MAT-2 | P. radiata | France (Côtes d’Armor) | Nursery | R. Ioos |
| F4 | F. circinatum | LSVM1221 | MAT-2 | P. radiata | Espagne (Basque country) | Forest | J. Aguayo |
| F5 | F. circinatum | NRRL26431 | MAT-1 | unkn. | Japan | unkn. | K. O’Donnell |
| F6 | F. circinatum | NRRL25708 | MAT-1 | unkn. | USA | unkn. | K. O’Donnell |
| F7 | F. circinatum | NRRL25331 | MAT-1 | unkn. | USA | unkn. | K. O’Donnell |
| F8 | F. circinatum | NRRL25333 | MAT-2 | unkn. | S-Africa | unkn. | K. O’Donnell |
| F11 | F. circinatum | FcCa01 | MAT-2 | P. radiata | Spain (Cantabria, Rionansa) | Forest | J. Diez |
| F12 | F. circinatum | FcCa02 | MAT-2 | P. radiata | Spain (Cantabria, Castrourdiales) | Forest | J. Diez |
| F13 | F. circinatum | FcCa05 | MAT-2 | P. radiata | Spain (Cantabria, Mazcuerras) | Forest | J. Diez |
| F14 | F. circinatum | FcCa06 | MAT-2 | P. radiata | Spain (Cantabria, Comillas) | Forest | J. Diez |
| F15 | F. circinatum | FC042v | MAT-2 | P. radiata | Spain (Cantabria, Cabezón de la Sal) | Forest | J. Diez |
| F16 | F. circinatum | FC035 | MAT-2 | P. radiata | Spain (Cantabria, Cabezón de la Sal) | Forest | J. Diez |
| F17 | F. circinatum | CSF-1 | MAT-1 | P. pinea | Spain (Burgos) | Reforestation seedling | A. Sanz-Ros |
| F18 | F. circinatum | CSF-2 | MAT-1 | P. radiata | Spain (León) | Insect (Brachyderes sp.) | A. Sanz-Ros |
| F19 | F. circinatum | CSF-3 | MAT-1 | P. radiata | Spain (León) | Seed (cones) | A. Sanz-Ros |
| F20 | F. circinatum | CSF-4 | MAT-1 | P. radiata | Spain (León) | Forest (twig) | A. Sanz-Ros |
| F22 | F. circinatum | CSF-6 | MAT-1 | P. radiata | Spain (León) | Forest (stem) | A. Sanz-Ros |
| F23 | F. circinatum | CSF-7 | MAT-2 | P. radiata | Spain (León) | Forest (stem) | A. Sanz-Ros |
| F24 | F. circinatum | CSF-8 | MAT-2 | P. nigra | Spain (Palencia) | Reforestation seedling | A. Sanz-Ros |
| F26 | F. circinatum | CSF-10 | MAT-1 | P. nigra | Spain (León) | Reforestation seedling | A. Sanz-Ros |
| F27 | F. circinatum | CSF-11 | MAT-1 | P. nigra | Spain (Valladolid) | Nursery | A. Sanz-Ros |
| F28 | F. circinatum | CSF-12 | MAT-1 | P. sylvestris | Spain (Valladolid) | Nursery | A. Sanz-Ros |
| F29 | F. circinatum | CSF-13 | MAT-2 | P. pinaster | Spain (Valladolid) | Seeds | A. Sanz-Ros |
| F30 | F. circinatum | 116 | MAT-2 | P. nigra | Spain (Galicia) | Nursery | M. Berbegal |
| F31 | F. circinatum | 164 | MAT-1 | P. sylvestris | Spain (Asturias) | Nursery | M. Berbegal |
| F32 | F. circinatum | 221 | MAT-2 | P. radiata | Spain (Cantabria) | Nursery | M. Berbegal |
| F33 | F. circinatum | 253 | MAT-1 | P. nigra | Spain (Galicia) | Nursery | M. Berbegal |
| F34 | F. circinatum | 822 | MAT-1 | P. pinaster | Spain (Galicia) | Seeds | M. Berbegal |
| F35 | F. circinatum | 07/0649 1b | MAT-1 | P. pinaster | Spain (Asturias) | Nursery | M. Berbegal |
| F36 | F. circinatum | 310/061 | MAT-1 | P. palustris | Spain (Asturias) | Nursery | M. Berbegal |
| F37 | F. circinatum | 2028 | MAT-2 | P. radiata | Chile | Nursery | R. Ahumada |
| F38 | F. circinatum | 2738 | MAT-2 | P. radiata | Chile | Nursery | R. Ahumada |
| F39 | F. circinatum | INV19 | MAT-2 | P. radiata | Chile | Nursery | R. Ahumada |
| F40 | F. circinatum | 2306 BASA | MAT-2 | P. radiata | Chile | Nursery | R. Ahumada |
| F41 | F. circinatum | CMW 1219 | MAT-2 | Pinus sp. | South Africa | unkn. | MJ. Wingfield (FABI) |
| F42 | F. circinatum | CMW 350 | MAT-1 | Pinus sp. | USA (California) | unkn. | MJ. Wingfield (FABI) |
| F51 | F. begoniae | LSVM293 | MAT-1 | Begonia elatior | France | R. Ioos | |
| F52 | F. concentricum | NRRL 25181 | unkn. | France | K. O’Donnell | ||
| F53 | F. fujikuroi | LSV667 | MAT-2 | Zea mays | France | R. Ioos | |
| F54 | F. mangiferae | NRRL25226 | MAT-2 | unkn. | unkn. | K. O’Donnell | |
| F55 | F. nygamai | NRRL13448 | unkn. | unkn. | K. O’Donnell | ||
| F56 | F. proliferatum | LSVM673 | MAT-2 | Populus sp. | France | R. Ioos | |
| F57 | F. sacchari | NRRL13999 | unkn. | unkn. | K. O’Donnell | ||
| F58 | F. subglutinans | LSVM869 | MAT-1 | Zea mays | France | R. Ioos | |
| F59 | F. subglutinans | LSVM704 | MAT-1 | Zea mays | France | R. Ioos | |
| F60 | F. temperatum | LSVM870 | MAT-2 | Zea mays | France | R. Ioos | |
| F61 | F. thapsinum | NRRL22045 | unkn. | unkn. | K. O’Donnell | ||
| F62 | F. verticillioides | LSVM873 | Zea mays | France | R. Ioos | ||
| F63 | F. verticillioides | 437-6 | Glycine max | Italy | M Pasquali | ||
| F64 | F. fractiflexum | NRRL28852 | MAT-2 | unkn. | unkn. | K. O’Donnell | |
| F65 | F. proliferatum | FGSC 7421 | MAT-2 | unkn. | unkn. | J.F. Leslie | |
| F66 | F. parvisorum | CMW 25267 | MAT-2 | Pinus patula | Columbia | Commerial nursery | MJ. Wingfield (FABI) |
| F67 | F. sororula | CMW 25254 | MAT-2 | Pinus spp. | Columbia | Commerial nursery | MJ. Wingfield (FABI) |
| F68 | F. marasasianum | CMW 25261 | MAT-2 | Pinus patula | Columbia | Commerial nursery | MJ. Wingfield (FABI) |
| F69 | F. pininemorale | CMW 25243 | MAT-1 | Pinus tecunumanii | Columbia | Plantation | MJ. Wingfield (FABI) |
| F70 | F. fracticaudum | CMW 25245 | MAT-2 | Pinus maximinoi | Columbia | Plantation | MJ. Wingfield (FABI) |
| F72 | F. avenaceum | Do_US_Nat_2_1 | seed of Douglasia sp. | USA | WSL - Phytopathology | ||
| F73 | F. incarnatum-equiseti species complex | Do_US_Nat_3_1 | seed of Douglasia sp. | USA | WSL - Phytopathology | ||
| F74 | F. sporotrichioides | Do_US_Nat_32_1 | seed of Douglasia sp. | USA | WSL - Phytopathology | ||
| F75 | F. tricinctum species complex | Do_US_Sno_49_1 | seed of Douglasia sp. | USA | WSL - Phytopathology | ||
| F76 | F. acuminatum | Do_US_VC_49_1 | seed of Douglasia sp. | USA | WSL - Phytopathology | ||
| F77 | F. torulosum | Do_US_VC_5_1 | seed of Douglasia sp. | USA | WSL - Phytopathology | ||
| F78 | F. graminearum | Do-Mur/17-1 | seed of D. menziesii | USA | WSL - Phytopathology | ||
| F79 | F. proliferatum | FI-BOS/13-1 | seed of Picea sp. | Switzerland | WSL - Phytopathology | ||
| F80 | F. reticulatum negundis | FI-BOS/14-1 | seed of Picea sp. | Switzerland | WSL - Phytopathology | ||
| F81 | F. redolens | Do-D/11-1 | seed of Douglasia sp. | Switzerland | WSL - Phytopathology | ||
| F82 | F. culmorum | CSF-14 | Pinus pinea | Spain (Palencia) | reafforestation seedling | A. Sanz-Ros | |
| F83 | F. torulosum | CSF-15 | Pinus nigra | Spain (León) | reafforestation seedling | A. Sanz-Ros | |
| F84 | F. oxysporum | CSF-16 | MAT-2 | Pinus pinea | Spain (Palencia) | reafforestation seedling | A. Sanz-Ros |
| F85 | P. pinaster seed spiked with 105 conidia of strain F7 | — | — | ||||
| F86 | P. pinaster seed spiked with 104 conidia of strain F7 | — | — | ||||
| F87 | P. pinaster seed spiked with 103 conidia of strain F7 | — | — | ||||
| F88 | P. pinaster seed spiked with 102 conidia of strain F7 | — | — | ||||
| F89 | P. pinaster seed spiked with 105 conidia of strain F11 | — | — | ||||
| F90 | P. pinaster seed spiked with 104 conidia of strain F11 | — | — | ||||
| F91 | P. pinaster seed spiked with 103 conidia of strain F11 | — | — | ||||
| F92 | P. pinaster seed spiked with 102 conidia of strain F11 | — | — | ||||
To avoid biases generated by the involvement of different operators and laboratories and to minimize the risk of moving around living F. circinatum strains, all participants worked with DNA extracts rather than with living cultures. All strains were first gathered in ANSES and kept on site. For DNA extraction, the strains were first cultured on potato dextrose liquid media (PD Broth, DIFCO™), for approximately 5 days, after which 100 to 200 mg of fresh mycelium was harvested. Genomic DNA (gDNA) was extracted using the DNeasy plant mini kit™ (Qiagen, Courtaboeuf, France) following the manufacturer’s guidelines, after grinding mycelium with a Lysis matrix A tube containing one 6-mm ceramic sphere and garnet matrix (MP Biomedicals, Santa Ana, CA, USA) and homogenized for 20 s at 6.5 U (m/sec) using a FASTprep 24 device (MP Biomedicals). DNA concentration was estimated using the Nanodrop 2000 Spectrophotometer™. For each strain, genomic DNA was produced from biological replicates and mixed/homogenized in order to obtain enough DNA to be tested in all of the different protocols and to supply to all of the partners. For each strain, the quality of the DNA extract was assessed by successful PCR amplification of the Internal Transcribed Spacer rDNA using the ITS1/ITS4 primer pair45, and the DNA concentration normalized to 0.5 ng µL−1 and distributed as 50-µL aliquots in individual 2-mL microtubes (F1 to F84, Table 5). In total, each laboratory received 38 F. circinatum DNA samples (target DNA) and 33 non-target DNA samples. All samples were anonymously labeled, shipped at room temperature by fast delivery service, and kept in a freezer until analysis.
A set of DNA from F. circinatum-artificially infected pine seeds was also prepared. Two strains of F. circinatum (F7 and F11) were cultured for 6 days at 22 °C under cool-white fluorescent lights with a 12-h light period on Spezieller Nährstoffarmer Agar (SNA) medium to allow macro- and microconidia production46. Microconidia were harvested by washing the surface of cultures with 10 mL of deionized sterile water with 0.01% Tween 20. The resulting suspension was diluted with sterile water to obtain a final concentration of 2.52 × 103 and 1.90 × 103 conidia µL−1 for F7 and F11, respectively, based on counts made using a hemocytometer. Healthy Pinus pinaster seeds were first incubated in liquid PD Broth media as described by Ioos et al.24, in order to simulate biological enrichment of natural samples, as is performed in routine detection analysis. Four replicates of one thousand healthy seeds each were incubated for 72 h at 22 °C in a sterile Easy flat flask containing 50 mL of PD Broth. After incubation, the contents of the Easy flask (healthy seeds + liquid medium) were aseptically transferred into a sterilized grinding bowl, and ground for 1 min using a Microtron MB 550 mixermill (Kinematica, Lucerne, Switzerland). Seventy subsamples of 500 µL of homogenate were collected using a micropipette and transferred into individual sterile 2-mL microtubes. For each of the two F. circinatum strains, 4 sets of 14 homogenized healthy seed subsamples were spiked with 1 × 105, 1 × 104, 1 × 103, and 1 × 102 conidia, respectively. One set was spiked with sterile water to be used as negative control for the seeds. Total DNA was extracted from each spiked homogenate as described by Ioos et al.24 using the Nucleospin Plant II miniprep (Macherey-Nagel) DNA extraction kit. For each level of contamination, all the DNA extracts were pooled, homogenized, and then distributed as 50 µL F. circinatum contaminated seed DNA to be used as template for PCR testing (F85 to F92, Table 5). In total, each laboratory received eight pine seed DNA extracts anonymously labeled, which were transported using a fast delivery service and kept in a freezer until analysis.
In total, each partner received an identical panel of 79 DNA extracts, to be tested in duplicate analysis for each protocol that was assessed.
Data generation and analysis
Indeterminate results
Data were processed anonymously, and no communication was allowed about the trials between partners before the end of the collaborative study. For each protocol, each participant tested all 79 DNA extracts in duplicate. For each sample, results of the tests were reported as either “positive” or “negative”, based on the duplicate analyses. With the exception of protocol p8 (where a Ct < 36 should be considered as a positive result), none of the five published real-time PCR protocols recommended a decision cut-off value. Therefore, the decision to rate a DNA sample as “positive” or “negative” was up to each participant, following the decision rules in force in the laboratory. However, in case of doubt or difficulty in interpretation of the results, “indeterminate result” could be reported. The participants were nevertheless encouraged to submit a brief description of the problem encountered. Indeterminate results between protocols, as reported by the participating laboratories, were compared by Fisher’s exact tests for count data.
Indeterminate results were also compared between laboratories by protocol using Fisher’s exact tests. Ideally, these comparisons would have been performed comparing all indeterminate results generated by laboratories, across all tests. However, it must be noted that not all participants implemented all nine protocols, and missing data for some partners exist in the datasets (for example laboratories that only participated in one test would not be included in the statistical test). Therefore, the decision was made to compare laboratories by protocol, in order to have an idea of potential differences that can exist for example between equipment, location or staff in charge of the tests. In both cases, Fisher’s exact tests were performed with simulated P-values based on 1 × 105 replicates.
Indeterminate results were then transformed following two scenarios as suggested by Chabirand et al.47 and Loreti, et al.48. The first scenario considered that an indeterminate result would be further assessed by the laboratory, and would always be rated “as expected” (i.e. a sample containing F. circinatum DNA would be rated as positive, and a sample not containing F. circinatum DNA would be rated as negative), so that the participant would always make the right decision, eventually. This dataset is here referred to as Dataset 1 (DS1). In the second scenario, an indeterminate result would always be rated “not as expected” (i.e. a sample containing F. circinatum DNA would be rated as negative, and a sample not containing F. circinatum DNA would be rated as positive), so that the participant would always make the wrong decision. This dataset is here referred to as Dataset 2 (DS2).
As the objective was to show the potential biases in the application of a protocol that may arise through differences between equipment, location or staff in charge of the analysis, identity of laboratories is not revealed and only the range of indeterminate results is shown.
Rates of false positive and false negative results
Results of the protocols were assessed by computing a number of parameters using both datasets: (i) PA, the number of positive accords or true positives, defined as the number of DNA samples from F. circinatum strains (or DNA from seed samples contaminated with F. circinatum) yielding positive results with the protocol; (ii) NA, the number of negative accords or true negatives, here defined as the number of DNA samples from other Fusarium species yielding negative results with the protocol; (iii) PD, the number of positive deviations or false positives, which takes into account the number of DNA samples from other Fusarium species (or DNA from seed samples not contaminated with F. circinatum) yielding positive results with the protocol; and iv) ND, the number of negative deviations or false negatives, which corresponds to the number of DNA samples from F. circinatum strains (or DNA from seed samples contaminated with F. circinatum) yielding negative results with the protocol.
Similarly, for both datasets, the performance of the protocols regarding specificity was assessed using the PD rate, computed as PD rate = 100 × (number of misclassified known positive samples/total number of known negative samples), and the ND rate, computed as, ND rate = 100 × (number of misclassified known negative samples/total number of known positive samples). As for indeterminate results, the PD and ND rates were compared between laboratories by protocol (in total nine comparisons) for both datasets.
All comparisons were performed using Fisher’s exact tests for count data with simulated P-values based on 1 × 105 replicates.
Other performance criteria
For each protocol and for each participant the results obtained for the blind samples were processed according to EN ISO 16140 standard49 and the PM7/98 (2) EPPO standard16. Three performance criteria were assessed: relative accuracy (AC), diagnostic specificity (SP) and diagnostic sensitivity (SE). AC represents the agreement between the expected results and the results obtained using the protocol. SE provides an estimation of the ability of the procedure to detect the target when it is present (presence of F. circinatum DNA). SP provides an estimation of the ability of the procedure not to detect the target when it is not present (no F. circinatum DNA present in the sample). AC, SP and SE were estimated using PA, NA, PD and ND, described in the previous sections, as follows:
Tests on the equality of SE, SP and AC between methods were performed using Fisher’s exact test.
Qualitative reproducibility or concordance (CO) was also estimated for each protocol. Concordance is the probability that two identical test materials sent to different laboratories will both provide the same results (i.e. both found positive or both found negative)50. Concordance for qualitative analyses is similar to reproducibility for quantitative analyses, and this performance criterion is a means to assess the ability of a protocol to provide consistent results with identical samples that are tested under different conditions: operator, equipment, master mix or DNA polymerase brand, location, time49. In order to have a reliable estimation of CO, it was calculated for each protocol using the original data reported by the participating laboratories. This means that positive, negative and indeterminate results were included. CO between protocols was compared for both datasets using Fisher’s exact tests for count data.
Statistical software for data analysis
All statistical tests were performed using the R statistical software version 3.4.051. Statistical tests were considered as significant for estimated P-values with a confidence of less than 5%. All figures were produced using the R package “ggplot2”52.
Supplementary information
Acknowledgements
This work was supported by COST action FP1406 “Pinestrength”. The authors want to thank Laura Hernández Escribano (Instituto Nacional de Investigación y Tecnología Agraria y Alimentaria, Centro de Investigación Forestal INIA-CIFOR, Ctra. La Coruña, Km.7.5, 28040, Madrid, Spain), Špela Jagodic (Slovenian Forestry Institute), Dr. S. Markovskaja and J. Švediene (Lithuanian Nature Research Centre), Victoria Rodríguez (Centro de Biotecnología, Universidad de Concepción), Dr. Eugénia Andrade INIAV I.P., Portugal, Prof. Tadeusz Malewski (Museum and Institute of Zoology, Warsaw, Poland), Tuğba Doğmuş Lehtijarvi (Applied Sciences University of Isparta, Faculty of Forestry, Isparta, Turkey), Victoria Avgitidou (University of Milan, Maria Evoli (Department of Agriculture, Food and Environment, University of Catania, Italy), Gema Pérez, Paula Zamora, Ana Martín, Juan Carlos Domínguez, Miriam Dueñas, Africa Miravalles, Jorge Miranda, Eva Mayor, Alejandro González and Beltrán Álvarez (Calabazanos Forest Health Centre, JCyL), Tobias Wille (JKI, Institute for National and International Plant Health)) and Michael Melek (Mendel university in Brno) for their technical contribution to this work. The work of the Estonian team was supported by the Estonian Science Foundation grants PSG136 and IUT21-04. The work of Portuguese team from INIAV was financed by INIAV I.P. Institute. The work at U. Aveiro (Portugal) was financed by European Funds through COMPETE and National Funds through the Portuguese Foundation for Science and Technology (FCT) to CESAM (UID/AMB/50017/2013 – POCI-01- 0145-FEDER-007638). The work of Slovenian team was financed through Slovenian Research Agency (P4-0107) and by the Slovenian Ministry of Agriculture, Forestry and Food (Public Forestry Service). The British work was financially supported by the Forestry Commission, UK. The French work was financially supported by the French Agency for Food, environmental and occupational health safety (ANSES). The work in New Zealand was funded by Operational Research Programmes, Ministry for Primary Industries, New Zealand.
Author Contributions
R.I. conceived the project, and designed the study. B.P., C.G. and F.A. prepared and standardized the samples for comparison. All authors performed the experiments. R.I., J.A. and M.M. wrote the main manuscript text with inputs from coauthors. J.A. carried out the statistical analyses and prepared figures. All authors reviewed the manuscript.
Data Availability
All data generated or analyzed during this study are included in this published article (and its Supplementary Information and Dataset Files).
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-44672-8.
References
- 1.Schmale DG, III, Gordon TR. Variation in susceptibility to pitch canker disease, caused by Fusarium circinatum, in native stands of Pinus muricata. Plant Pathol. 2003;52:720–725. doi: 10.1111/j.1365-3059.2003.00925.x. [DOI] [Google Scholar]
- 2.Gordon TR, Kirkpatrick SC, Aegerter BJ, Wood DL, Storer AJ. Susceptibility of Douglas fir (Pseudotsuga menziesii) to pitch canker, caused by Gibberella circinata (anamorph = Fusarium circinatum) Plant Pathol. 2006;55:231–237. doi: 10.1111/j.1365-3059.2006.01351.x. [DOI] [Google Scholar]
- 3.Martínez‐Álvarez P, Pando V, Diez JJ. Alternative species to replace Monterey pine plantations affected by pitch canker caused by Fusarium circinatum in northern Spain. Plant Pathol. 2014;63:1086–1094. doi: 10.1111/ppa.12187. [DOI] [Google Scholar]
- 4.Wingfield MJ, et al. Pitch canker caused by Fusarium circinatum - a growing threat to pine plantations and forests worldwide. Australas. Plant Path. 2008;37:319–334. doi: 10.1071/AP08036. [DOI] [Google Scholar]
- 5.Bezos D, Martinez-Alvarez P, Fernandez M, Diez JJ. Epidemiology and management of pine pitch canker disease in Europe - a review. Balt. For. 2017;23:279–293. [Google Scholar]
- 6.Landeras E, et al. Outbreak of pitch canker caused by Fusarium circinatum on Pinus spp. in Northern Spain. Plant Dis. 2005;89:1015. doi: 10.1094/PD-89-1015A. [DOI] [PubMed] [Google Scholar]
- 7.Bragança H, Diogo E, Moniz F, Amaro P. First report of pitch canker on pines caused by Fusarium circinatum in Portugal. Plant Dis. 2009;93:1079–1079. doi: 10.1094/PDIS-93-10-1079A. [DOI] [PubMed] [Google Scholar]
- 8.EFSA Risk assessment of Gibberella circinata for the EU territory and identification and evaluation of risk management options. EFSA Journal. 2010;8:1620. doi: 10.2903/j.efsa.2010.1620. [DOI] [Google Scholar]
- 9.Carlucci A, Colatruglio L, Frisullo S. First report of pitch canker caused by Fusarium circinatum on Pinus halepensis and P. pinea in Apulia (Southern Italy) Plant Dis. 2007;91:1683. doi: 10.1094/PDIS-91-12-1683C. [DOI] [PubMed] [Google Scholar]
- 10.Vettraino A, Potting R, Raposo R. EU legislation on forest plant health: an overview with a focus on Fusarium circinatum. Forests. 2018;9:568. doi: 10.3390/f9090568. [DOI] [Google Scholar]
- 11.Möykkynen T, Capretti P, Pukkala T. Modelling the potential spread of Fusarium circinatum, the causal agent of pitch canker in Europe. Annals of Forest Sciences. 2015;72:169–181. doi: 10.1007/s13595-014-0412-2. [DOI] [Google Scholar]
- 12.Bustin S. A., Benes V., Garson J. A., Hellemans J., Huggett J., Kubista M., Mueller R., Nolan T., Pfaffl M. W., Shipley G. L., Vandesompele J., Wittwer C. T. The MIQE Guidelines: Minimum Information for Publication of Quantitative Real-Time PCR Experiments. Clinical Chemistry. 2009;55(4):611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- 13.EPPO PM 7/91(1): Gibberella circinata. EPPO Bull. 2009;39:298–309. doi: 10.1111/j.1365-2338.2009.02317.x. [DOI] [Google Scholar]
- 14.ISTA. 7-009: Detection of Gibberella circinata on Pinus spp. (pine) and Pseudotsuga menziesii (Douglas-fir) seed. Validated Seed Health TestingMethods (2015).
- 15.IPPC. ISPM 27, Diagnostic protocols for regulated pests, DP 22: Fusarium circinatum (2017).
- 16.EPPO PM 7/98 (2) Specific requirements for laboratories preparing accreditation for a plant pest diagnostic activity. EPPO Bull. 2014;44:117–147. doi: 10.1111/epp.12118. [DOI] [Google Scholar]
- 17.Nirenberg HI, O’Donnell K. New Fusarium species and combinations within the Gibberella fujikuroi species complex. Mycologia. 1998;90:434–458. doi: 10.1080/00275514.1998.12026929. [DOI] [Google Scholar]
- 18.Britz H, Coutinho TA, Wingfield MJ, Marasas WFO. Validation of the description of Gibberella circinata and morphological differentiation of the anamorph Fusarium circinatum. Sydowia. 2002;54:9–22. [Google Scholar]
- 19.Mullett M, Pérez-Sierra A, Armengol J, Berbegal M. Phenotypical and molecular characterisation of Fusarium circinatum: correlation with virulence and fungicide sensitivity. Forests. 2017;8:458. doi: 10.3390/f8110458. [DOI] [Google Scholar]
- 20.Herron DA, et al. Novel taxa in the Fusarium fujikuroi species complex from Pinus spp. Stud. Mycol. 2015;80:131–150. doi: 10.1016/j.simyco.2014.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Storer G, Clark SL. Association of the pitch canker fungus, Fusarium subglutinans f.sp. pini, with Monterey pine seeds and seedlings in California. Plant Pathol. 1998;47:649–656. doi: 10.1046/j.1365-3059.1998.00288.x. [DOI] [Google Scholar]
- 22.Schweigkofler W, O’Donnell K, Garbelotto M. Detection and quantification of airborne conidia of Fusarium circinatum, the causal agent of pine pitch canker, from two California sites by using a real-time PCR approach combined with a simple spore trapping method. Appl. Environ. Microbiol. 2004;70:3512–3520. doi: 10.1128/AEM.70.6.3512-3520.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ramsfield TD, Dobbie K, Dick MA, Ball RD. Polymerase chain reaction-based detection of Fusarium circinatum, the causal agent of pitch canker disease. Molecular Ecology Resources. 2008;8:1270–1273. doi: 10.1111/j.1755-0998.2008.02188.x. [DOI] [PubMed] [Google Scholar]
- 24.Ioos R, Fourrier C, Iancu G, Gordon TR. Sensitive Detection of Fusarium circinatum in Pine Seed by Combining an Enrichment Procedure with a Real-Time Polymerase Chain Reaction Using Dual-Labeled Probe Chemistry. Phytopathology. 2009;99:582–590. doi: 10.1094/PHYTO-99-5-0582. [DOI] [PubMed] [Google Scholar]
- 25.Dreaden TJ, Smith JA, Barnard EL, Blakeslee G. Development and evaluation of a real-time PCR seed lot screening method for Fusarium circinatum, causal agent of pitch canker disease. For. Path. 2012;42:405–411. doi: 10.1111/j.1439-0329.2012.00774.x. [DOI] [Google Scholar]
- 26.Fourie G, et al. Culture-independent detection and quantification of Fusarium circinatum in a pine-producing seedling nursery. Southern Forests: a Journal of Forest Science. 2014;76:137–143. doi: 10.2989/20702620.2014.899058. [DOI] [Google Scholar]
- 27.Lamarche J, et al. Molecular detection of 10 of the most unwanted alien forest pathogens in Canada using Real-Time PCR. PLoS ONE. 2015;10:e0134265. doi: 10.1371/journal.pone.0134265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Luchi N, Pepori AL, Bartolini P, Ioos R, Santini A. Duplex real-time PCR assay for the simultaneous detection of Caliciopsis pinea and Fusarium circinatum in pine samples. Applied Microbiology and Biotechnology. 2018;102:7135–7146. doi: 10.1007/s00253-018-9184-1. [DOI] [PubMed] [Google Scholar]
- 29.Sandoval-Denis Marcelo, Swart Wijnand J., Crous Pedro W. New Fusarium species from the Kruger National Park, South Africa. MycoKeys. 2018;34:63–92. doi: 10.3897/mycokeys.34.25974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Steenkamp ET, Wingfield BD, Desjardins AE, Marasas WF, Wingfield MJ. Cryptic speciation in Fusarium subglutinans. Mycologia. 2002;94:1032–1043. doi: 10.1080/15572536.2003.11833158. [DOI] [PubMed] [Google Scholar]
- 31.Garcia-Benitez C, et al. Proficiency of real-time PCR detection of latent Monilinia spp. infection in nectarine flowers and fruit. Phytopathologia Mediterranea. 2017;56:242–250. [Google Scholar]
- 32.Ebentier DL, et al. Evaluation of the repeatability and reproducibility of a suite of qPCR-based microbial source tracking methods. Water Research. 2013;47:6839–6848. doi: 10.1016/j.watres.2013.01.060. [DOI] [PubMed] [Google Scholar]
- 33.Bustin S, Huggett J. qPCR primer design revisited. Biomolecular Detection and Quantification. 2017;14:19–28. doi: 10.1016/j.bdq.2017.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Grosdidier M, Aguayo J, Marçais B, Ioos R. Detection of plant pathogens using real-time PCR: how reliable are late Ct values? Plant Pathol. 2017;66:359–367. doi: 10.1111/ppa.12591. [DOI] [Google Scholar]
- 35.Al-Soud WA, Rådström P. Capacity of nine thermostable DNA polymerases to mediate DNA amplification in the presence of PCR-inhibiting samples. Applied and environmental microbiology. 1998;64:3748–3753. doi: 10.1128/aem.64.10.3748-3753.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Saunders GC, Dukes J, Parkes HC, Cornett JH. Interlaboratory study on thermal cycler performance in controlled PCR and random amplified polymorphic DNA analyses. Clinical chemistry. 2001;47:47–55. [PubMed] [Google Scholar]
- 37.Boutigny A-L, et al. Optimization of a real-time PCR assay for the detection of the quarantine pathogen Melampsora medusae f. sp. deltoidae. Fungal Biology. 2013;117:389–398. doi: 10.1016/j.funbio.2013.04.001. [DOI] [PubMed] [Google Scholar]
- 38.Guinet C, Fourrier-Jeandel C, Cerf-Wendling I, Ioos R. One-step detection of Monilinia fructicola, M. fructigena, and M. laxa on Prunus and Malus by a multiplex real-time PCR assay. Plant Dis. 2016;100:2465–2474. doi: 10.1094/PDIS-05-16-0655-RE. [DOI] [PubMed] [Google Scholar]
- 39.Aguayo J, et al. Development of a hydrolysis probe-based real-time assay for the detection of tropical strains of Fusarium oxysporum f. sp. cubense race 4. PLoS ONE. 2017;12:e0171767. doi: 10.1371/journal.pone.0171767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Broeders S, et al. Guidelines for validation of qualitative real-time PCR methods. Trends in Food Science & Technology. 2014;37:115–126. doi: 10.1016/j.tifs.2014.03.008. [DOI] [Google Scholar]
- 41.Pelloux H, et al. A second European collaborative study on polymerase chain reaction for Toxoplasma gondii, involving 15 teams. FEMS Microbiology Letters. 1998;165:231–237. doi: 10.1111/j.1574-6968.1998.tb13151.x. [DOI] [PubMed] [Google Scholar]
- 42.Leslie, J. F. & Summerell, B. A. The Fusarium laboratory manual. (Blackwell Publishing, 2006).
- 43.Ioos R, et al. Test performance study of diagnostic procedures for identification and detection of Gibberella circinata in pine seeds in the framework of a EUPHRESCO project. EPPO Bull. 2013;43:267–275. doi: 10.1111/epp.12037. [DOI] [Google Scholar]
- 44.Geiser DM. FUSARIUM-ID v. 1.0: a DNA sequence database for identifying Fusarium. Eur. J. Plant Pathol. 2004;110:473–479. doi: 10.1023/B:EJPP.0000032386.75915.a0. [DOI] [Google Scholar]
- 45.White, T. J., Bruns, T., Lee, S. & Taylor, J. In PCR protocols: a guide to method and applications (eds Gelfand, D. H., Innis M. A., Sninsky, J. J. and White, T. J.) 315–322 (Academic Press, 1990).
- 46.Nirenberg HI. A simplified method for identifying Fusarium spp. occurring on wheat. Canadian Journal of Botany. 1981;59:1599–1609. doi: 10.1139/b81-217. [DOI] [Google Scholar]
- 47.Chabirand Aude, Loiseau Marianne, Renaudin Isabelle, Poliakoff Françoise. Data processing of qualitative results from an interlaboratory comparison for the detection of “Flavescence dorée” phytoplasma: How the use of statistics can improve the reliability of the method validation process in plant pathology. PLOS ONE. 2017;12(4):e0175247. doi: 10.1371/journal.pone.0175247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Loreti Stefania, Cunty Amandine, Pucci Nicoletta, Chabirand Aude, Stefani Emilio, Abelleira Adela, Balestra Giorgio M., Cornish Deirdre A., Gaffuri Francesca, Giovanardi Davide, Gottsberger Richard A., Holeva Maria, Karahan Aynur, Karafla Charikleia D., Mazzaglia Angelo, Taylor Robert, Cruz Leonor, Lopez Maria M., Vanneste Joel L., Poliakoff Françoise. Performance of diagnostic tests for the detection and identification of Pseudomonas syringae pv. actinidiae (Psa) from woody samples. European Journal of Plant Pathology. 2018;152(3):657–676. doi: 10.1007/s10658-018-1509-5. [DOI] [Google Scholar]
- 49.International Standardization Organization. ISO 16140:2003 Microbiology of food and animal feeding stuffs - Protocol for the validation of alternative methods (2003).
- 50.Langton S, Chevennement R, Nagelkerke N, Lombard B. Analysing collaborative trials for qualitative microbiological methods: accordance and concordance. International Journal of Food Microbiology. 2002;79:175–181. doi: 10.1016/S0168-1605(02)00107-1. [DOI] [PubMed] [Google Scholar]
- 51.R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna (2014). R Foundation for Statistical Computing (2017).
- 52.Wickham, H. ggplot2:elegant graphics for data analysis. (Springer, 2016).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article (and its Supplementary Information and Dataset Files).