Abstract
PD-L1 is a 40 kDa trans-membrane protein of B7 family and an important T cell regulator. Binding of PD-L1 and PD-1 inhibits proliferation and activation of T cell results cell exhaustion. This phenomenon can be reversed by blocking PD-L1/PD-1 interactions with single chain variables fragment (scFv) fusion proteins and by direct inhibition of tumor cells with drug conjugates. The human phage-displayed scFv library was utilized to generate scFv against the PD-L1 antigen by affinity bio-panning. The positive clones were selected by continuous transfection of bacterial cells and sequence analysis. The binding affinity and specificity of the scFv and antibody fragments were determined by using surface plasmon resonance biosensor, western blot analysis, and immunofluorescence assay. After three rounds of panning selection, about 30% of clones have a binding affinity with targeted PD-L1 antigen. Eight positive clones with accurate sequences were isolated and analyzed for binding affinity with PD-L1 antigen. Three of those with accurate sequences and binding affinity were selected for the recombinant formation and soluble expression by Escherichia coli host machinery. The highly positive recombinant clones with the exact orientation of FR and CDR domains were developed and can be used as a drug carrier tools in ADC formation or direct inhibition of immune checkpoint in cancer immunotherapy. The conjugate achieved its initial potency and need efficient improvement to enhance direct tumor suppression and bio-therapeutics strategies enrichment.
Electronic supplementary material
The online version of this article (10.1007/s10616-019-00316-3) contains supplementary material, which is available to authorized users.
Keywords: Expression and purification, Immunotherapy, Bio-panning, PD-L1, scFv, Phage display technology
Background
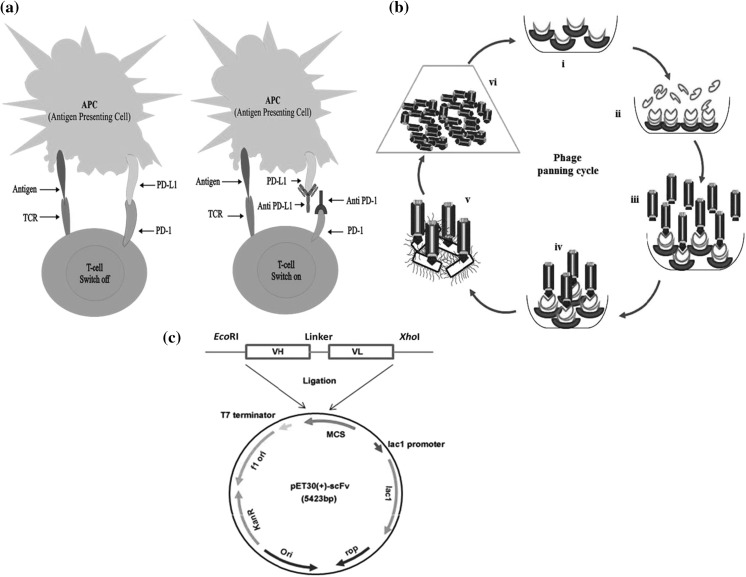
Programmed cell death ligand-1 (PD-L1) is a 40 kDa trans-membrane protein of B7 family that shares 40% homology with B7-DC/PD-L2 recorded more homologous to one another in this group (Dong et al. 1999; Latchman et al. 2001). The interactions of PD-L1 members result in down-regulation of T cell activation by decreasing interleukin-10, IL-4, IL-2 secretion, and interferon γ production via PD-1 receptors association (Carreno and Collins 2002; Freeman et al. 2000). Activated T cell and B cell result PD-1 expression while PD-L1 can be induced on macrophages and dendritic immune cells also by inflammatory cytokines. The T cell stress and down-regulation can be released by inhibition of PD-L1/PD-1 immune checkpoints via antibody therapies as shown in Fig. 1a. It was reported that immune checkpoint inhibition is most effective in pre-existing PD-L1 suppressed immunity patients and can be revived by antibody therapy (Herbst et al. 2014; Taube et al. 2014; Tumeh et al. 2014). Combination therapy of dabrafenib and trametinib with MEDI4736 antibody was also reported challenging anti-tumor activity by treating with dabrafenib and trametinib (Schreuer et al. 2017).
Fig. 1.
a Inhibition of PD-L1/PD-1 immune checkpoints with antibodies. Binding of PD-L1 and PD-1 inhibits proliferation and activation of T cell result cell exhaustion. This phenomenon can be reversed by blocking PD-L1/PD-1 binding with monoclonal antibodies. b Phage display bio-panning cycle. Phage bio-panning cycles: (i) immobilization of Ni–NTA sefinose beads on the solid surface. (ii) Binding of PD-L1 antigen with beads. (iii) Incubation of phage library. (iv) Removal of unbound phages via washing and selective phages are eluted out. (v). Infection of E. coli by rescued positive phages. (vi) Positive phage enrichment. c Genetic map of recombinant vector pET30 (+) scFv construction. The vector contains a Lac operon promoter region, gene encoding kanamycin resistance gene, the origin of replication fused with desired anti-PD-L1 scFv sequences including VH, VL and linker sequences loaded with EcoRI and XhoI restriction sites
The monoclonal antibodies are generally used for seizure antibody for their specificity and affinity but predictably time to consume and complex to achieve the desired monoclonal antibodies. Phage display technology makes it possible to generate single chain antibodies (scFv) with elevated specificity and high binding affinities (Li and Caberoy 2010; Pande et al. 2010). It was considered potent biological machinery for the screening of targeted ligands that bind to tumor surfaces, initially introduced in 1985 by Smith (1985). The simplicity of scFv can eradicate hurdles associated with whole antibodies, with a demonstration of superior penetration ability in tissues (Beckman et al. 2007; Chames et al. 2009; Yokota et al. 1992). The reduced complexity of scFv also allowed a desirable production in prokaryotic cells as compared to eukaryotic host machinery due to its lower maintenance and rapid growth (Makrides 1996). An scFv is actually a fusion protein of variable regions (VH and VL) connected by a short linker of 10–25 amino acids. These proteins possess the ability of both antigen binding capability and marker activity from hybridoma technology. The capabilities can be applied in the biological application as therapeutic gene delivery, biosensor and cancer treatment (Crivianu-Gaita and Thompson 2016). Additionally, for therapeutic purposes, the murine antibody can be humanized rapidly at the scFv production stage (Khantasup et al. 2015). Phage display technology is the most convenient method in terms of antibody production that designs a huge repertoire of peptides and proteins libraries (Clackson et al. 1991; McCafferty et al. 1990; Smith 1985). These libraries comprised hundreds of millions of molecules which display scFv fragments. Single chain variable fragments are made of variable regions of both heavy and light chains. These variable regions are joined by a flexible linker (Wen et al. 2010). Phage display technology shares tremendous advantage in antibody productions that it can be produced in a large amount and retains specific binding affinity to antigen deprived of immunization procedures (Dai et al. 2003; Weisser and Hall 2009). This system makes it possible to generate recombinant antibodies of elevated binding specificities and affinity with targeted antigens (Garet et al. 2010; Pande et al. 2010).
Current studies represent the novel approach of scFv production against the PD-L1 surface antigen from the recombinant human phage-displayed scFv library. The library was incubated initially with target proteins and unbound phages were excessively washed out. The high-affinity phages were eluted out and amplified by using host bacterial cells. These were further enriched in succeeding cycles. After panning rounds, clones were characterized by sequence analysis and ELISA to acquire the desired structural and functional information. In this study, we developed high-affinity phage environment and construct prokaryotic recombinant vector system to express the variable regions, a comprehensive foundation for further research in antibodies production and a carrier tool for loaded drug and chemicals.
Materials and methods
Cell lines and reagents
Tumor cell lines of lung cancer, A549, were obtained from ATCC and were maintained in RPMI-1640 supplemented with 10% fetal bovine serum in a humidified incubator containing 5% CO2 at 37 °C. M13KO7 helper phage (1.47 × 1012 pfu/ml, Lot: IG0286: Amersham Bioscience, USA). The human phage-displayed scFv library was previously established in our laboratory (Chinese Patent CN201210484448.6, 2015-02-25) and was used with 1.22 × 1012 concentrations. Escherichia coli strains were provided by Professor Jinbiao Zhan and were maintained under strict sterile conditions. Libraries were of high potency clones containing inserts that displayed as single chain fragments on pIII phage filaments. The scFv fragments were engineered in phagemid vector that comprised ampicillin resistant gene and single polypeptide chain with the variable region of heavy and light chains attached by Gly–Ser flexible linker. The PD-L1 extracellular domain was previously developed by our research group in gene and antibody engineering lab. Anti-PD-L1 IgG antibody and anti-6xHis Tag rabbit antibody (Cat No AB 10002) were from Life Science Production and Services, China. Rabbit anti-human IgG (H + L)-HRP (Cat No 6140-05; Lot No D2311-ZD51E) were from Southern Biotech USA) and goat anti-rabbit IgG-HRP (Cat No HA1001; Lot No G161011) were provided by Hangzhou HunAn Biotech Comp. China. All reagents, solutions, and buffers were maintained under high-grade purity and strict sterile condition.
Helper phage enrichment and library amplification
Escherichia coli TG1 was regenerated and incubated overnight at 37 °C into 5 ml 2 × YT tubes. Achieving logarithmic phase, M13KO7 helper phages (1.47 × 1012 pfu/ml) were added and incubated for 30 min at 37 °C followed by overnight incubation at 37 °C on 2 × YT culture plates supplemented with 50 µg/ml kanamycin. A single colony was transferred to E. coli TG1 at the logarithmic stage and incubated for 2 h at 37 °C. Bacterial culture was transferred to 200 ml 2 × YT in a conical flask and incubated for 1 h at 37 °C, 220 rpm followed by addition of 50 µg/ml kanamycin and incubated for 16 h at 30 °C and 220 rpm. Bacterial cells were pelleted out, the supernatant was collected at 7000 rpm for 20 min and phages were concentrated out with 20% PEG/2.5 M NaCl solution on ice for 4 h. The harvested pellets were dissolved in PBS and centrifuged at 12,000 rpm for 10 min to eliminate cell derbies. The supernatant was passed via a 0.22 µm syringe filter. The library was amplified with the same procedure by addition of enriched phages (1010) and concentrated with 20% polyethylene glycol (PEG8000) and 2.5 M NaCl solution and stored at − 80 °C with 15% glycerol.
Bio-panning screening and expression of positive scFv-PDL1 phages
Recombinant PD-L1-ECD was incubated overnight with Ni-sefinose beads at 4 °C. The mixture was vigorously washed with PBS and blocked with 5% BSA at 37 °C for 1 h. Amplified phages (100 µl) were added to tubes in blocking buffer at 37 °C for 2 h, followed by 10 min standing incubation. The liquid was discarded and washed five times with TBST and two times with dH2O to remove the unbound phages. The bound phage particles were eluted with elution buffer (200 mM imidazole, 0.5 M NaCl, 0.1 M PBS) followed by 0.2 M glycine–HCl (pH 1.7) at room temperature for 10 min and neutralized with 1 M Tris–HCl (pH 9.0). The eluted phages were subsequently used to infect E. coli TG1 at logarithmic stage (OD600nm = 0.4) for 30 min at 37 °C. Titers were also made and plated on 2 × YT plates supplemented with 100 µg/ml ampicillin and 1% glucose to calculate the phage infection. The infected E. coli TG1 was poured into 20 ml 2 × YT and incubated at 37 °C for 2–4 h. Helper phages (1010) were added and allowed to stand for 30 min at 37 °C. Cells were harvested at 2200xg for 15 min and suspended to new 200 ml 2 × YT medium supplemented with 100 μg/ml ampicillin and 50 μg/ml kanamycin and incubated for 16 h at 30 °C with regular shaking. The culture was harvested at 7000 rpm for 15 min and phages from the supernatant were concentrated by using PEG/NaCl solution and were dissolved in 2 ml PBS, centrifuged and filtered. These phages were further used for second and third rounds of panning. The schematic diagram of the bio-panning cycle is shown in Fig. 1b. The cycles of bio-panning were repeated twice to amplify and select the highly purified enrich phages. The positive colonies were selected from culture plates and 300 colonies were settled out for sequence analysis with Sangon Biotech Shanghai, China. Eight stronger clones were selected and were analyzed for binding affinity with PD-L1 by phage ELISA. Four samples out of eight were recorded of exact homology so were named as C36. The positive phages after ELISA and sequence determination were enriched further as described above. Positive samples were processed for soluble antibody production by using recombinant 6xHis-Tag pET 30(+) vector development using E. coli BL21 (DE3) host cell machinery. NCBI-BLAST tools and Clustal W programs were used to determine multiple sequence alignment and homologous sequence analysis with related peptides.
Phage ELISA
96 wells plates were coated with 100 µl per well PD-L1 antigen in PBS with 100 µg/ml concentration and incubated overnight at 4 °C. The liquid was discarded and wells were washed three times with PBS then blocked with 3% BSA (Bovine serum albumin) in PBS for 2 h at 37 °C. 10 µl positive phages in blocking buffer were added to each well and incubated for 1 h at 37 °C. Plates were washed five times with PBST (0.05% Tween 20 in PBS). The bound phages were detected by using 100 µl horseradish peroxidase-conjugated anti-M13 antibodies in each well with 1:5000 dilutions in blocking buffer and incubated for 1 h at 37 °C. 100 µl substrate TMB was added to develop color up to 30 min at 37 °C in dark and reaction was stopped by adding 2 M H2SO4 for 10 min at 37 °C. The binding affinity of phages and PD-L1 antigen were measured by using ELISA reader at 450 nm absorbance. All analyses were performed in triplicate.
Affinity determination of scFv by surface plasmon resonance (SPR)
The binding activity of anti-PD-L1 scFv to PD-L1 was assessed with Octet K2 bio-Layer Interferometry, BLI (Shuangtian Shengwu, China). The fully integrated SPR sensor used is highly stable and static. The emitted light by LED reaches polarizer and reflects by sensing the surface of gold. The detector measures the light and calculates the angle. Repeating the cycles at various concentrations, the association and dissociation constant can be a measure of the bio-molecular measure. Any change in molecule numbers of biosensor tips causes interference pattern shift that can be calculated in real time as shown in Table S1.
Positive phage enrichment
The selected positive phage clones, after ELISA and sequence analysis, were inoculated to 5 ml LB tubes and incubated overnight at 37 °C and 250 rpm. Next morning, 10 µl cultures were inoculated to 3 ml tubes and were allowed to grow until OD600 become 0.4. Then were poured into fresh 200 ml 2 × YT medium and 200 µl helper phages (1010) were added, followed by incubation for 30 min at 37 °C on standing position. The culture was allowed to regular shaking at 220 rpm for the next 30 min. Cells were pelleted out at 2200 rpm centrifugation for 15 min and transferred to a new 2 × YTAK medium (supplemented with 100 µg/ml ampicillin, 50 µg/ml kanamycin). After incubation at 30 °C and 250 rpm for 16 h, supernatants were collected at 7000 rpm centrifugation for 15 min and selected positive phages were precipitated out with 20% PEG/2.5 M NaCl on ice for 4 h. Phages were collected by centrifugation at 12,000 rpm and were dissolved in PBS. Cells debris were removed with 12,000 rpm centrifugation for 10 min and the supernatant was passed via 0.22 µm syringe filter. These phages were used for single-chain antibody expression.
PCR amplification of positive clones and construction of a recombinant vector
The DNA sequence of full-length VH and VL regions including linker copy of selective positive phages were amplified by using specific primers in a thermal cycler (Px2 Thermal Cycler, Thermo Hybaid Elect. Cooperation, USA). Three different sets of primers were design for the correct orientation of cloning and transformation of selected phages. The primers were also confirmed by NCBI database using Primer-BLAST tools. (http://www.ncbi.nlm.nih.gov/tools/primer-blast/) Two different sets of restriction enzymes sites EcoR1 (15U/µl, Lot K2703AA, TaKaRa) and XhoI (10 U/µl, Lot K2704AA, TaKaRa) was also added to facilitate the restriction digestion. PCR amplification was performed in 50 µl master mix containing 1 µl of phages, 0.5 pmol of each primer (TaKaRa), 2.5 mM concentration of each dNTPs Mg+ (Lot BJ1901A, TaKaRa), 5 U/µl of DNA polymerase (Lot KA1202DA, TaKaRa), 5 µl of 10X prime buffer and sterile ultrapure water. The thermal cycler condition was settled for one cycle denaturation for 5 min at 94 °C followed by 35 cycles of each denaturation for 45 s at 94 °C, annealing for 30 s at 58 °C, an extension for 45 s at 72 °C and final extension for 10 min at 72 °C. The final products were subjected to 0.8% agarose gel and visualized by addition of ethidium bromide and UV illuminator, and were recovered from the gel by using gel purification kit (TaKaRa).
The recovered PCR products were ligated to the pMD18-T cloning vector to generate the recombinant cloning vector and transferred to competent E. coli DH5α bacterial cell. The bacterial cells were plated on LB agar plates supplemented with 100 µg/ml ampicillin at final. The integrity of transformed plasmid was confirmed by PCR, restriction digestion and sequencing. The digested products were ligated into already digested vector pET 30(+) with EcoR1 and XhoI and transferred to competent E. coli BL21(DE3) as shown in Fig. 1c. Transformed cells were plated on LB agar plates supplemented with 50 µg/ml kanamycin, to ensure the exact transfer. The empty plasmid was also used as a control. Single colonies were selected and incubated overnight in LB tubes supplemented with 50 µg/ml kanamycin at 37 °C. Centrifuge tubes were made from the overnight culture and saved at − 80 °C in 15% glycerol also one tube was sent out for sequence analysis to determine the exact transfer of targeted sequences.
Expression and purification of recombinant scFv antibodies
The transformed bacterial strain was regenerated and transferred to 5 ml LB tubes supplemented with 50 µg/ml kanamycin and incubated overnight at 37 °C with 220 rpm shaking. The culture was poured to fresh 200 ml LB conical flask and allowed to grow to mid-logarithmic phase approximately for 3 h at 37 °C and 220 rpm. IPTG in a final concentration of 1 mM was added when OD600 reached 0.4–0.6 and incubated further at 30 °C for 5 h before being harvested. Aliquots were collected before and after addition of IPTG for SDS-PAGE analysis using standard protocols. After centrifugation at 4000 rpm for 10 min, pellets were dissolved in lysis buffer (20 mM Tris-Cl, 0.5 mM NaCl, 0.1% Triton-x100, 1 mM PMSF) and sonicated followed by centrifugation at 12,000 rpm for 15 min. Pellets were washed three times with washing buffer (50 mM TrisCl, 0.1 M NaCl, 1 mM EDTA, 0.1% T-x100) and one time with distilled water finally dissolved in 8 M urea buffer solution (8 M urea, 20 mM TrisCl, 0.5 mM NaCl). Both supernatant and pellets were saved and aliquots were collected to determine the exact band size position of expressed antibodies on SDS-PAGE.
Purification and refolding of recombinant scFv antibodies
SDS-PAGE detection indicated that maximum concentrations of expressed antibodies were found in the inclusive body. These were purified by affinity column chromatography on Ni–NTA gel matrix. The column of Ni–NTA nitrocellulose resin (BBI Life Science) was washed and equilibrated with washing urea buffer then cleared cell lysate (containing recombinant protein) were loaded onto the column. The recombinant antibodies were eluted out by elution buffer (8 M urea, 20 mM TrisCl, 0.5 mM NaCl, 20–150 mM imidazole) at different imidazole concentration as 20 mM, 50 mM, 100 mM, and 150 mM consecutively. The fractions were collected and examined by using 12% SDS-PAGE. Western blot analyses were also conducted to determine the purity of expressed antibodies. Purified protein were dialyzed against decreasing concentration of urea buffer (8 M, 4 M, 2 M) followed by buffer A (20 mM TrisCl, 0.5 mM NaCl, 5% glycerol, 5% sucrose, 1% arginine), B (20 mM TrisCl, 0.5 mM NaCl), and C (PBS 1X, 5% glycerol, 5% sucrose, 1% arginine) for 2 h at 4 °C using Economical Biotech Dialysis Membrane (BBI Life Science). Final dialysis run was carried out against PBS and products were examined by 12% SDS-PAGE and western blot analysis.
Western blot analysis
The final purified recombinant scFv products were separated by SDS-PAGE and electrophoretically transferred to polyvinylidene difluoride membrane (Bio-Rad Lab, Hercus, CA, USA) at 120 V for 90 min in a transfer buffer. Membranes were blocked by using 5% skim milk and then incubated with anti-6xHis Tag rabbit antibody (Cat No AB 10002, Life Science Production and Services China) at 1:500 dilutions for 2 h at room temperature. The membranes were washed with TBST, followed by goat anti-rabbit IgG-HRP (Cat No HA1001; Lot No G161011, Hangzhou HunAn Biotech Comp. China) at a dilution of 1:1000 for 1 h at room temperature. After washing with TBST bands were visualized using ECL detection reagents (Thermo Fisher Scientific).
Binding interaction of expressed recombinant antibodies with PD-L1 antigen
To investigate the binding affinity of expressed recombinant antibodies, the PD-L1 antigen with different concentrations (0.5, 0.1, 0.01, 0.001 and 0.0001 µg/ml) was employed to 96 wells ELISA plate and incubated overnight at 4 °C. After washing three times with PBST, plate wells were blocked in 3% BSA in PBS for 2 h at 37 °C. Wells were emptied and washed 5 times with PBST. Purified soluble antibodies were added to wells with same different concentrations and incubated for 1 h at 37 °C. Conjugated scFv antibodies were labeled with anti-His-HRP antibody and further incubated for 1 h at 37 °C. The remaining methodology was followed as described above. The binding affinities were measured by ELISA reader at 450 nm absorbance.
Immunofluorescence assay for phage and scFv antibodies localization
Cells were seeded onto glass coverslips at 2 × 104/ml in regular serum containing RPMI 1640 medium and incubated at 37 °C until 80% confluence. Cells were washed gently and incubated overnight at 4 °C with 1010 positive phages and 1 µg/ml expressed scFvs in separate experiments. Cells were washed three times (5 min for each wash) with ice-cold PBS and fixed with 4% paraformaldehyde. After washing three times, anti-M13 mouse monoclonal antibody (dilution 1:500) and anti-His rabbit antibody (dilution 1:1000) were added and incubated for 1 h at 37 °C. Coverslips were further washed 3 times with ice-cold PBS and FITC labeled goat anti-mouse antibody (dilution 1:500) and mouse anti-rabbit antibody (dilution 1:1000) were added for 1 h at 37 °C. DAPI (dilution 1:1000) were added after three times washing (5 min in each wash) for 10 min and coverslips were mounted in an inverted position on the slide and were visualized under 63 × oil immersion objective using Zeiss fluorescence microscope (Zeiss, Germany).
MTT analysis
Cell viability analysis was calculated by using MTT assay. The PD-L1 positive cell A549 and negative cell MDA453 were cultured in DMEM media supplemented with 10% FBS in 96 wells plate at a ratio of 1 × 104. These cells were exposed to scFv soluble proteins at different concentrations ranges from 1 to 0.0001 µg/ml. The cells were incubated for 72 h followed by addition of 50 µl MTT reagent. The plates were gently shaken and incubated up to 4 h. The supernatant was slowly removed and propanol was added to solubilize the formazan crystals. The absorbance was measured and growth inhibition was calculated using the formula:
whereas Ta was tested absorbance, Tb was blank absorbance and Tc was control absorbance.
Statistical analysis
One way analysis of variance by using GraphPad Prism 5 (GraphPad Software; Inc: La Jolla, CA) software was used to evaluate binding affinities of both phages and expressed scFvs. The significance level was set at p ≤ 0.05.
Results
Phage display bio-panning
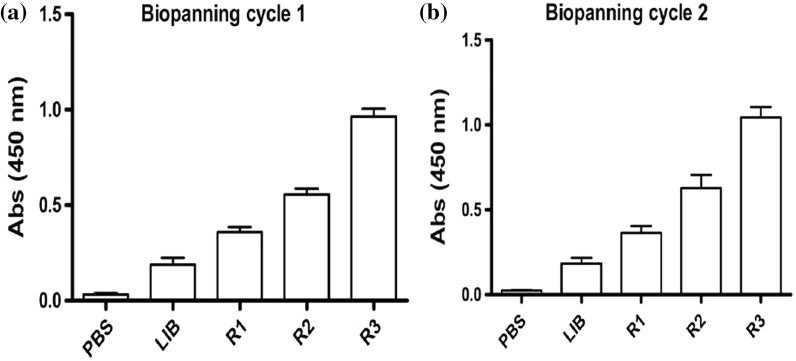
Panning is the in vitro binding activity of antibody fragments with targeted antigen bound to the solid support. Before panning, recombinant phages were separated from the soluble mixture in solution by using 20% PEG/2.5 M NaCl. The precipitated phages were then panned against the PD-L1 antigen. Equal or more than 1.22 × 1011 pfu of collected phages were used for each round of panning selection. The bio-panning cycles were repeated twice to select the high positive phages. The bio-panning intensity was elevated by increasing washing cycles with TBST for 10 and 15 times in each second and third round and successfully enrichment of phages were obtained as shown in Table 1. The unbound phages were eliminated during subsequent washing steps in each round. The rescued phages were allowed to infect the E. coli TG1 at log phase and plated onto 2 × YT plates supplemented with 100 µg/ml ampicillin. The single plaque was collected and inoculated to 2 × YT medium to the enriched production of targeted phages that were further precipitated out via PEG/NaCl solution. The output and input titers from each round of panning were calculated and phage enrichment intensity was accomplished in two different trial cycles as shown in Fig. 2. Results showed elevated output rate of 4.0 × 103 and 3.76 × 104 in both bio-panning cycles and the enrichment ratios were 22.56 and 1225.27, respectively. The second repeated cycle showed elevated enrichment of positive phages.
Table 1.
Phage enrichment rounds in the first cycle (a) and second cycle (b) of selection from a phage library
| Input (pfu) | Output (pfu) | Rate | Enrichment |
|---|---|---|---|
| (a) First cycle enrichment | |||
| 1.22 × 1012 | 4.0 × 103 | 3.27 × 10−9 | |
| 6.55 × 1012 | 3.827 × 105 | 5.8 × 10−8 | 17.7 |
| 4.56 × 1012 | 6.04 × 107 | 1.32 × 10−5 | 227.56 |
| Total yield | 4.0 × 103 | ||
| (b) Second cycle enrichment | |||
| 1.22 × 1012 | 8.0 × 103 | 5.92 × 10−9 | |
| 1.4 × 1012 | 2.56 × 105 | 1.82 × 10−7 | 30.74 |
| 2.7 × 1011 | 6.04 × 107 | 2.23 × 10−4 | 1225.27 |
| Total yield | 3.76 × 104 | ||
Fig. 2.
Selection of anti-PD-L1 scFv by phage panning technology. a First cycle of bio-panning, b the second cycle of bio-panning. Elevated enrichments were counted in the third round of selection in both repeated cycles
Phage specificity against the PD-L1 antigen by ELISA and sequence analysis
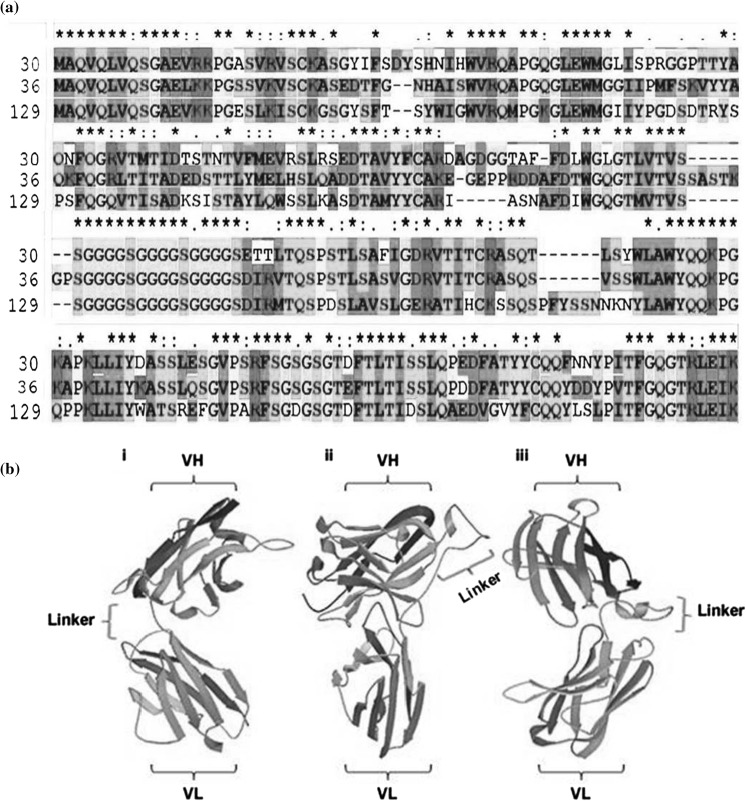
Hybridoma, spleen cells and B lymphocytes can provide mRNA construction sources by reversed transcription for the synthesis of scFv. The diverse and huge quantity of VH and VL gene can be created by using these methods (Marks et al. 1991). The phage display technique among affinity selection procedures was found the best to display scFv fragment on phage coat. The order of domains in scFv production can be VH-L-VL or VL-L-VH (Ahmad et al. 2012). The productions of the selected antibody from a specific hybridoma are essentially the same. After successful cloning and sequence analysis, the scFv fragments could be directly expressed in the mammalian or bacterial system. We purified positive phages from the human-phage recombinant library which was previously engineered in our lab and was evaluated by using phage ELISA. After the fourth round of bio-panning in both cycles, 300 clones were picked randomly and screened for sequence analysis to isolate the accurate functional framework of the heavy and light chain. Total of eight clones showed the exact orientation of targeted heavy and light chains FR and CDR regions without any shifted sequences due to point mutation. NCBI-BLAST tools and Clustal 2.1 programs were used to determine multiple sequence alignment and homologous sequence analysis with related peptides. Alignments results indicate highly conserved sequences which are shown in colors and star labeled as shown in Fig. 3a. Nucleotide sequence analyses of both heavy and light variable regions were determined. Coding sequences of positive samples B10, B30, B31, B36, B38, B46, B84, and B129 indicated high accuracy and binding affinity to the targeted PD-L1 antigen. The genetic makeups of these clones are shown in Table 2 that indicates the total calculated sequence analysis and also showed alignment results with human IgG. The Clustal 2.1 analysis tool of targeted positive sequences showed the alignment results and indicated that four samples were of 100% similarities as shown in Table 3, comprised of 363 bp VH gene and 321 bp of VL gene (Figures S1 and S2). These four similar samples were uniformly designated as C36 for further analysis. The high degree of mutation was calculated in VLCDR of the light chain and also point mutation were counted that directed to the generation of stop codon TGA in the functional framework of both heavy and light chain in remaining clones. These errors included clones were excluded out after sequence analysis. The five selected positive phages were chosen for single chain expression. Among these only three clones, B30, C36, and B129 could only express in E. coli BL21(DH3) system to produce highly soluble recombinant scFv proteins.
Fig. 3.
a Amino acid sequence alignment of positive phages by Clustal Omega. Different colors represent the physiochemical properties of amino acids. Red colors present small and hydrophobic, Blue presents Acidic, Magenta indicates Basic, Green shows Hydroxyl, sulfhydryl and amine residues. Grey colors indicate unusual amino acids. An asterisk (*) presents conserved residues in scFv. b Structural configuration of selected positive scFvs by protein modeling software Phyre2. Models show linkers connection with VH and VL regions of (i) B30, (ii) C36 and (iii) B129 scFv sequences
Table 2.
Total sequence analysis of positive phages
| Sample ID | Sequences (bp) | HC region | LC region |
|---|---|---|---|
| 38, 46, 36, 10 | 756 | humIGHV034 | humIGKV087 |
| 129 | 741 | humIGHV217 | humIGKV083 |
| 30 | 741 | humIGHV039 | humIGKV026 |
| 31 | 756 | humIGHV034 | humIGKV051 |
| 84 | 732 | humIGHV062 | humIGKV098 |
Table 3.
Alignment score of positive phages by Clustal 2.1 analysis
| Alignment score | Alignment score | Alignment score | Alignment score |
|---|---|---|---|
| Sequences (10:30): 65.83 | Sequences (30:31): 100 | Sequences (31:38): 100 | Sequences (36:129): 71.31 |
| Sequences (10:31): 65.83 | Sequences (30:36): 100 | Sequences (31:46): 92.46 | Sequences (38:46): 92.46 |
| Sequences (10:36): 65.83 | Sequences (30:38): 100 | Sequences (31:84): 73.06 | Sequences (38:84): 73.06 |
| Sequences (10:38): 65.83 | Sequences (30:46): 92.46 | Sequences (31:129): 71.31 | Sequences (38:129): 71.31 |
| Sequences (10:46): 65.42 | Sequences (30:84): 73.06 | Sequences (36:38): 100 | Sequences (46:84): 70.61 |
| Sequences (10:84): 63.49 | Sequences (30:129): 71.31 | Sequences (36:46): 92.46 | Sequences (46:129): 70.49 |
| Sequences (10:129): 63.18 | Sequences (31:36): 100 | Sequences (36:84): 73.06 | Sequences (84:129): 72.54 |
Structural prediction of scFv fragments
The phyre2 program was used to predict the function, mutation and structural analysis of recombinant antibodies (Kelley et al. 2015). These web portal analyses calculate ligands binding sites and analyze amino acids variants effects. All results showed 100.0% confidence by the highest scoring template. The tool SuSPect in the Phyre2 analysis has been used to calculate the possible mutation sites in query proteins. SuSPect results indicated a balanced accuracy of > 80% on the large benchmark test. Results indicated that 228 residues (90% of B30, C36) and 233 residues (90% of B129) have been modeled with 100.0% confidence by the single highest scoring template with balanced accuracy > 80% as shown in Fig. 3b, and indicated the exact position of the linker between VH and VL variable regions. Some researchers reported that scFv fragments possess low affinity as compared to parental whole antibody and form complexes of dimers, trimmers or aggregates (Borrebaeck et al. 1992). The linkers usually have 10–25 residues of amino acids but with 10–15 residues of amino acids showed no aggregates formation. If the linker length is further reduced one or two glycine residue, the formation of dimers will also be prevented and may show high potency (Atwell et al. 1999).
Selection of positive clone enrichment and affinity analysis
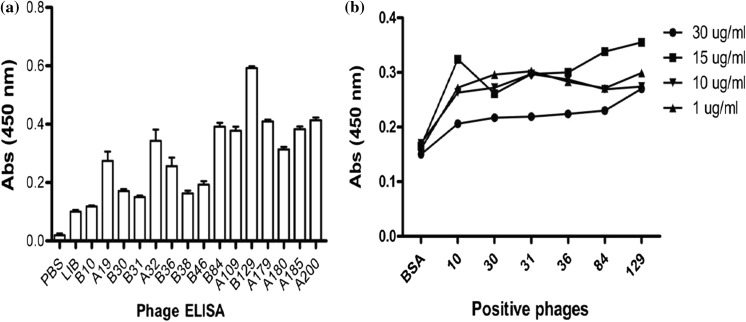
After three rounds of both bio-panning cycles and sequence analysis, selected clones were incubated in 200 ml of 2 × YT medium supplemented with 100 µg/ml ampicillin and phages were amplified as described in methodology. The enriched phages were tested by ELISA using anti-M13 antibody. ELISA results showed that rescued phages were of high affinity with PD-L1 protein as shown in Fig. 4a. The affinity constant was determined by using surface plasmon resonance as shown in Table S1. The recombinant phage dose was observed in the coated channel with the association (Ka), dissociation rate (Kd) and affinity constant (KD). The results indicated that phage scFv exhibited strong binding affinity and stability against the PD-L1 antigen. The phages were further processed for antibody production. At a different concentration of targeted antigen, the binding affinity was calculated for tested phages as shown in Fig. 4b. The maximum binding affinity was found at a concentration of 15 µg/ml of antigen through ELISA determination.
Fig. 4.
a ELISA results showed the binding specificity of phages and PD-L1 antigen. All tested samples have binding specificity but only eight clones (B10, B30, B31, B36, B38, B46, B84, and B129) were chosen after ELISA and sequence analysis. Three samples (B30, C36, and B129) were further processed for scFv expression after the successful construction of a recombinant vector. OD: optical density. b The binding affinity of eight positive phages with different concentration of PD-L1 by ELISA. PD-L1 protein was coated at different concentration in 96 wells plate and phages (1010) were added as the primary antibody. BSA was used as a control for non-specific binding
Recombinant formation and expression of scFv antibodies
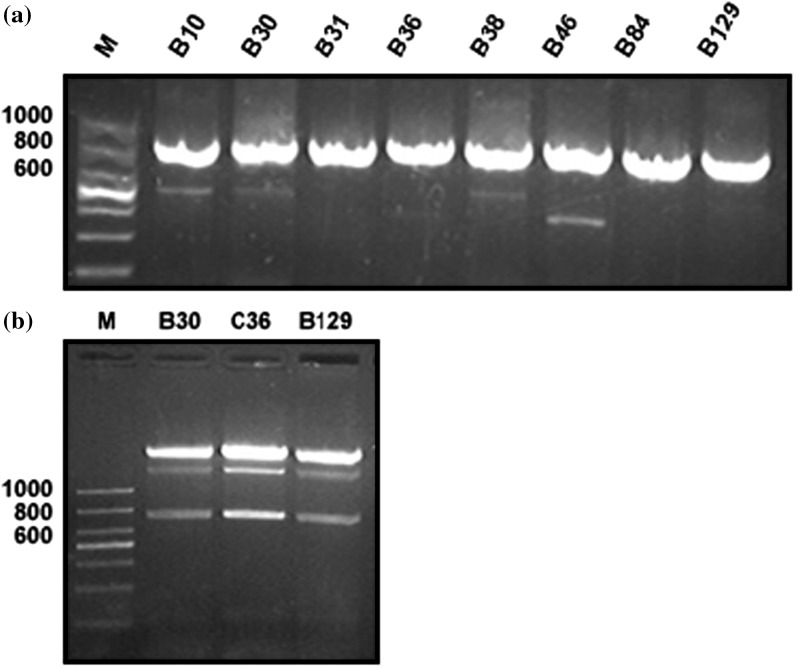
Primers were designed to maximize the cloning and recombinant formation of targeted sequences and evaluate their expression. The predicted band size of selected phages after amplification and recombinant formation was calculated and separated by using 1% agarose gel as shown in Fig. 5a, b. Single bacteria clone was picked and sequenced by Sangon Biotech. The recombinant expression vector was developed by insertion of targeted sequences into pET 30(+) vector and was confirmed by restriction enzyme digestion and sequence analysis. The sequences were further analyzed by using NCBI-BLAST tools. pET30 (+) vector comprised of 6x-His DNA marker that facilitates purification via Ni–NTA affinity chromatography (Hochuli et al. 1987, 1988). The recombinant expression vector pET30 (+) was transferred to E. coli BL21 (DE3) for expression. E. coli BL21 accomplished targeted protein expression by using T7 promoter and T7 RNA polymerases under control of lacUV5 promoter region. These are induced by using IPTG (Rosenberg et al. 1987; Studier and Moffatt 1986). After sonication and centrifugation, the products were loaded on polyacrylamide SDS-PAGE to determine the estimated molecular weight of about 33 kDa as shown in Fig. 6. Maximum expressions were recorded in inclusion bodies and were further purified by excessive washing with a urea solution to facilitate the removal of contamination. The soluble inclusion bodies were loaded to Ni–NTA column to elute the final pure products. It was found that maximum protein was eluted with 50 mM and 100 mM imidazole concentration. The eluted proteins were followed by refolding.
Fig. 5.
Gel electrophoresis of positive selective phages. a PCR polymerization results by primers using positive selective phages of interest. b Enzyme digestion products by using EcoR1 and Xho1 to produce cohesive ends. M. Molecular marker
Fig. 6.
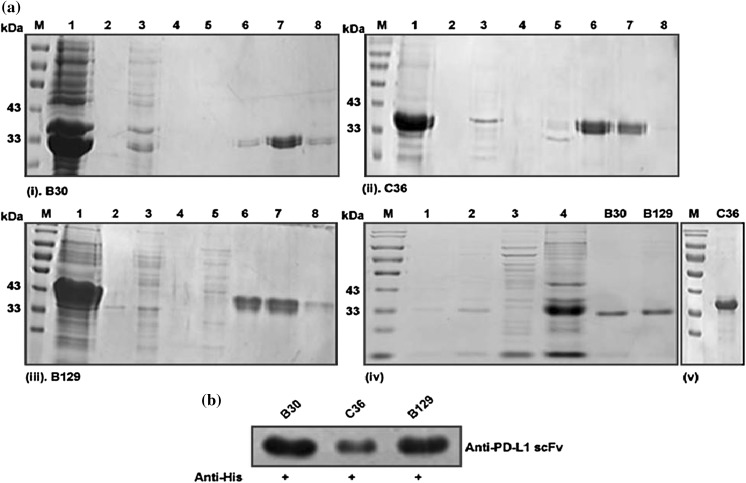
SDS-PAGE and western blot analysis of selected positive anti-PD-L1 scFvs expressed in E. coli BL21 (DE3). a (i) represents expression and purification of scFv-B30, (ii) shows the expression and purification of scFv-C36 and (iii) represents expression and purification of scFv-B129. M represents molecular marker, 1 indicates the expressed proteins after sonication and lyses, 2–4 show washing of column via urea solution, 5–8 represents eluted products at different concentrations of imidazole (20 mM, 50 mM, 100 mM, and 150 mM). a (iv) Represents the expression model of scFv. 1 represents expression without IPTG, 2 represents with IPTG, 3 shows expression in the supernatant, 4 represents expression in inclusive bodies. B30, B129, and C36 represent the final purified products after column purification and dialysis. b Represents bioactive confirmation of expressed purified scFvs with anti- 6xHis Tag Rabbit antibody
SDS-PAGE and western blot analysis
The anti-PD-L1 scFv purified above were identified by SDS-PAGE and western blot analysis. The gels were stained with Coomassie brilliant blue after gel electrophoresis and were photographed through a gel doc system. Purification analysis of positive scFv antibodies is indicated in Fig. 6a. The migration results of a single protein band on SDS-PAGE analysis predicted that purified products of the high purity of single chain recombinant antibodies with an exact molecular mass of about 30–34 kDa. Western blot analysis also showed the active bioactivity and verification of expressed proteins as shown in Fig. 6b. An apparent band signal was detected at appropriate molecular weight for expressed proteins. The yield was calculated by using a standard BCA kit and was estimated 1 mg/ml by absorbance of 450 nm.
However, it was also reported that a high level of protein expression in E. coli often obtained in inclusion bodies including insoluble aggregates (Moore et al. 1993). We further endure the purification, washing and refolding procedure to isolate targeted pure protein. We performed additional steps of washing the inclusion bodies with urea solution to eliminate the impurities. The soluble proteins after denaturation were applied to Ni–NTA column for additional purification. After column purification, the purified proteins were processed to refolding with continuous washing steps which are the most prevalent one (Amons and Schrier 1981; Wingfield et al. 2014; Winkler and Blaber 1986). Finally, the refolded anti-PD-L1 scFv was concentrated out by using 20000 PEG by yielding 2 mg/ml products and were filtered.
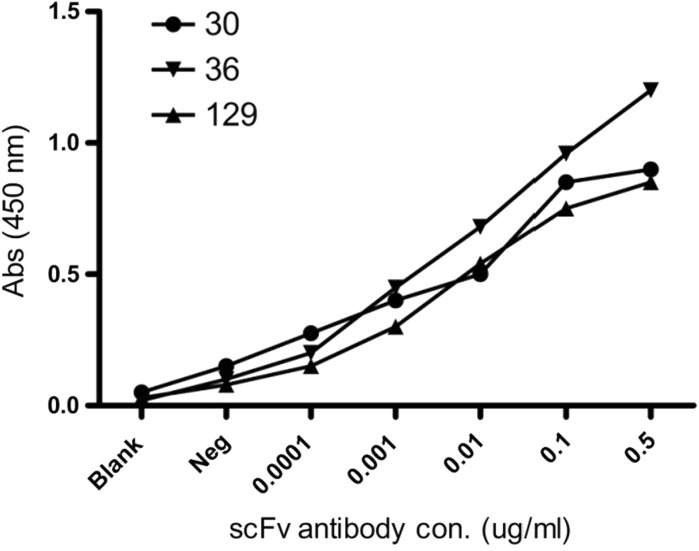
Binding affinity determination of expressed antibodies
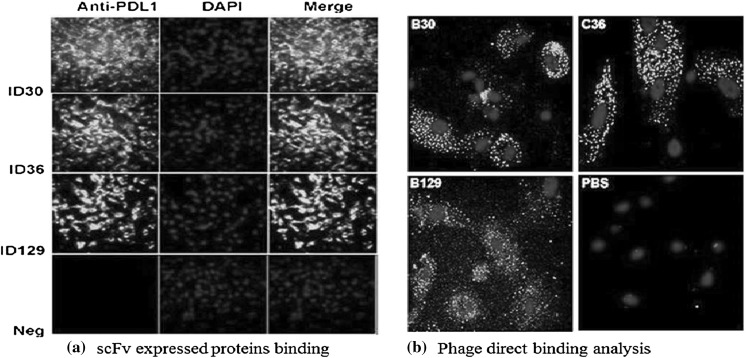
In order to examine the binding affinity and selectivity of expressed scFv for PD-L1 protein, we performed indirect competitive ELISA, western blotting, and immunofluorescence analysis to test the activity of scFv recombinant antibody with PD-L1. Results showed that scFv A30, C36, and A129 have high affinity with targeted antigen suggesting that these antibodies have high activity against PD-L1 positive tumor cells as reported in western blot analysis (Fig. 6b) and icELISA (Fig. 7). These results revealed the exact orientation of recombinant protein formation by using the pET system. According to the selection procedure, the most prominent positive clones were selected to perform the immunofluorescence assay against tumor cells to additionally confirm the binding specificity of expressed scFvs antibodies as shown in Fig. 8a. The fluorescent signal was detected on the surface of PD-L1 positive cells and no fluorescent was observed on surfaces of negative cell lines. The direct binding affinity of targeted phages was also calculated by confocal microscopy as shown in Fig. 8b.
Fig. 7.
Binding affinity of expressed proteins with PD-L1 antigen. ELISA results showed interaction between expressed scFvs with PD-L1 at different concentrations of both PD-L1 antigen and scFv expressed proteins (0.0001, 0.001, 0.01, 0.1, 0.5 µg/ml)
Fig. 8.
Immunofluorescence analysis of anti-PD-L1 scFv expressed proteins and phage particles. The figure shows confocal microscopy results of a newly generated anti-PDL1-ECD antibody that ratifies the bioactive conformation and specificity of recombinant anti-PD-L1scFv expressed proteins (a) and direct binding of positive phages (b)
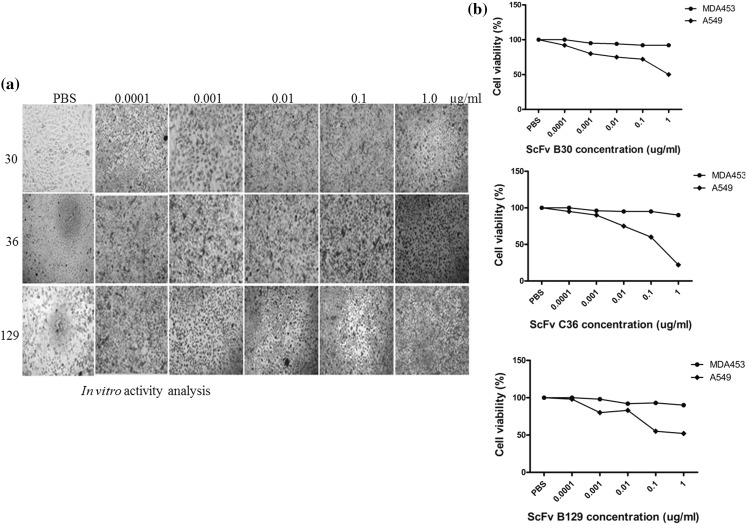
In vitro activity analysis
Cytotoxicity studies revealed that anti-PD-L1 scFvs have high toxicity against PD-L1 positive cancer cell lines A549 as compared to negative cells, MDA453. The expressed scFvs-PD-L1 was tested against different cancer cells and activities were checked by MTT analysis. The significant killing was recorded against A549 cells and less activity was found against MDA453 cells as shown in Fig. 9. The cytotoxicity was dose-dependent, and the percentage of growth inhibition was increased with the concentration of tested scFvs expressed proteins. The viability of A549 cancer cells was decreased after 72 h of incubation with anti-scFv (C36) as compared to two others scFvs with an increase of concentration up to 1 µg/ml. The toxicity effects against MDA453 were almost negligible and cells were allowed to grow freely in the presence of scFv. In this experiment, we found that blockage of PD-L1 by scFv proteins alone can cause the cell-growth arrest and cell death in PD-L1 positive A549 cells. This may be due to the intrinsic PD-L1 signals which exert oncogenic effects in cancer development. These antibody fragments may provide promises to enrich the efficacy of therapeutic PD-1/PD-L1 checkpoint inhibition alone or in combination with other immunotherapeutic approaches.
Fig. 9.
Cell cytotoxicity analysis of expressed scFv-PD-L1. a shows cell activity against A549 cells after 72 h of induction in 96 wells plate and b shows graphical analysis of cell viability by using three expressed scFv proteins (B30, C36, and B12) at different concentrations against A549 cells
Discussion
Immunotherapy has emerged a promising approach to detect and fight efficiently with cancers for several decades. These therapies include inflammatory agent injections, stem cell transplantation, vaccination, cytokines administration, immune checkpoint inhibitions, vaccinations, and others. However, until now efficacy of immunotherapy become restricted due to limited efficacy and elevated toxicities. Efforts are carried out to investigate agents that limit toxicity and selectively activate immune components that fight cancers. The monoclonal antibody has emerged as a promising approach for treating immune checkpoint inhibition in numerous malignancies (Muller et al. 2017; Robert et al. 2015; Vozy and Zalcman 2015). Monoclonal antibody production is more costly and acquired genetic maintenance of unstable hybridoma cell cultures. Additionally, these antibodies can lead to phagocytosis or fixation with the interaction of Fc domains by immune responses which create obstacles in investigational results and interfering with therapeutic effects (Chames et al. 2009; Hansel et al. 2010; Kalim et al. 2017b). It was examined to developed smaller antibody fragments to overcome related full-length antibodies problems (Holliger and Hudson 2005).
Recently, cancer targeted therapy has become one of the prominent hotlines in tumor eradication therapy. Highly specific carrier molecules are in practice to specifically deliver anti-tumor loaded drugs to target sites (Velasco-Velazquez et al. 2014). Small peptide molecules may act a most vital source of the carrier due to high specificity, structure simplicity, easy manipulation and solid penetration ability to tumor cells (Bastien et al. 2015). The most advantageous factor of phage display technology is that it recognizes the genotype and phenotypes binding and no need for its structural evidence in advance (Kaplan and Gershoni 2012; Silacci et al. 2005). So, it is conceivable to acquire the amino acid sequence indirectly by the recombinant formation and sequence analysis of positive phages clones followed by screening procedure. In the current study, we developed high phages enrichment environment and construct prokaryotic recombinant vector system to express the variable regions, a comprehensive foundation for further research in antibodies production and a carrier tool for loaded drug and chemicals. The expressed antibodies showed active binding affinity and also effective inhibition properties against PD-L1 positive cancer cell lines. The positively expressed scFv obtained presented the highly promising application in recognition of PD-L1 antigens in cancer therapy.
The binding affinity and assembly of anti-PD-L1 scFv by using human synthetic phage library were determined. The high-affinity titer was eluted by bio-panning and successively enriched by counter infection of E. coli TG1. From third rounds of both cycles, 300 clones were selected for sequence analysis. The precise and accurate genetic framework of selective positive clones were chosen and analyzed further for binding affinity with PD-L1 antigen. Eight clones (B10, B30, B31, B36, B38, B46, B84 and B129) showed the best activity. Four (B10, B36, B38, and B46) of these were of exact sequence similarities so named C36. Three samples B30, C36 and B129 were chosen for soluble expression. All these were successfully cloned by using recombinant technology techniques to develop recombinant vector and were transferred to E. coli BL21 (DE3) system. The maximum expression was recorded in inclusion bodies same as reported by others studies (Choi et al. 2004; Kalim et al. 2017a; Lombardi et al. 2005; Wang et al. 2010). Further washing steps successively decrease the impurities and purified by affinity column chromatography using Ni–NTA column. Single isolated bands by using SDS-PAGE predicted our results purity.
The functionality of recombinant scFv samples was evaluated by western blot analysis, direct ELISA and immunofluorescence detection to determine the prominent binding specificity and bioactivity. The generated scFv products were able to identify the receptor sites present positive results. The direct inhibition approach of scFv was also determined to evaluate the functional in vitro activities. In our experiment, we found that blockage of PD-L1 by scFv proteins alone can inhibit cell growth and cause cell death in A549 cells. So far, most studies have focused on PD-L1 and PD-1 interactions over T cell regulation but few have paid attention to intrinsic PD-L1 signals. It was suggested that intrinsic PD-L1 might exert an initial oncogenic effect in cancer development and plays an important regulatory role in cancer cell growth, proliferation, apoptosis and metastasis (Escors et al. 2018). Recently, Chen’s group demonstrated that overexpression of intrinsic PD-L1 promoted GBM cell proliferation via Ras/Erk/EMT mechanisms and the knockdown of PD-L1 abolished GBM development and invasion in vitro and in vivo (Qiu et al. 2018). It is reported that the intrinsic PD-L1 may also play important oncogenic roles in other types of cancer such as melanoma, gastric cancer, pancreatic cancer and lung adenocarcinoma (Clark et al. 2016).
Taken together, the growing evidence supports that PD-L1 itself can enhance tumor-cell proliferation besides it binds to PD-1 and results in T cell inactivation. The blockade of PD-L1 by scFv proteins can inhibit cell growth and eventually induce cell death. Various studies reported more complex methods to obtain scFv products and often functionally not sufficient (Visintin et al. 1999). Thus overall results of the expression, purification and refolding and binding strategies confirm the constancy of our expressed scFv. The utilization of expressed antibody by using phage display technology provides a basis for direct inhibition of tumor cells in cancer therapy.
Conclusions
It is still a bottleneck problem to prepare high-quality antibodies by immunoassay methods. In present studies with PD-L1 as a target antigen, scFv antibodies were successfully engineered and efficiently expressed in E. coli system. The inclusion bodies of anti-PD-L1 scFv were purified by using Ni–NTA affinity column chromatography in an effective manner. The synthetic scFv antibodies possessed appropriate antigen recognition sites for its bioactivity and sensitivity. The work suggested that these recombinant single chain antibodies can be utilized as a potent carrier to transport anti-tumor drugs and chemicals to cancer cells. These fragments can be utilized as a carrier source in designing of ADC to eradicate tumor or can be directly used to block the pathways in an immunoassay. The positively expressed scFv obtained presented the highly promising application in recognition of PD-L1 antigens in cancer therapy and can eradicate hurdles associated with other polyclonal antibodies of penetration in tissues and cells. The kinetic affinity of expressed scFv showed that these particles possess stable binding with the PD-L1 antigen. Therefore the scFv can be used as an alternative for monoclonal antibody and will serve as an effective carrier tool for various immunotherapy studies.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (Grant Nos. 81872784 and 81430081).
Abbreviations
- ADC
Antibody-drug conjugate
- ATCC
American Type Culture Collection
- CDR
Complementarity determining regions
- DAPI
4′,6-Diamidino-2-phenylindole
- ECD
Extracellular domain
- ELISA
Enzyme-linked immune-sorbent assay
- FITC
Fluorescein isothiocyanate
- FR
Framework region
- IL
Interleukin
- IPTG
Isopropyl β-d-1-thiogalactopyranoside
- Ni–NTA
Nickel–nitrilotriacetic acid
- PCR
Polymerase chain reaction
- PD-1
Programmed cell death protein-1
- PD-L1
Programmed cell death ligand-1
- PEG
Polyethylene glycol
- scFv
Single chain variable fragment
- SDS-PAGE
Sodium dodecyl sulfate–polyacrylamide gel electrophoresis
- TMB
3,3′,5,5′-Tetramethylbenzidine
- VH
Heavy variable region of Ig
- VL
Light variable region of Ig
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Ahmad ZA, Yeap SK, Ali AM, Ho WY, Alitheen NB, Hamid M. scFv antibody: principles and clinical application. Clin Dev Immunol. 2012 doi: 10.1155/2012/980250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amons R, Schrier PI. Removal of sodium dodecyl-sulfate from proteins and peptides by gel-filtration. Anal Biochem. 1981;116:439–443. doi: 10.1016/0003-2697(81)90385-7. [DOI] [PubMed] [Google Scholar]
- Atwell JL, Breheney KA, Lawrence LJ, McCoy AJ, Kortt AA, Hudson PJ. scFv multimers of the anti-neuraminidase antibody NC10: length of the linker between VH and VL domains dictates precisely the transition between diabodies and triabodies. Protein Eng. 1999;12:597–604. doi: 10.1093/protein/12.7.597. [DOI] [PubMed] [Google Scholar]
- Bastien JIL, McNeill KA, Fine HA. Molecular characterizations of glioblastoma, targeted therapy, and clinical results to date. Cancer. 2015;121:502–516. doi: 10.1002/cncr.28968. [DOI] [PubMed] [Google Scholar]
- Beckman RA, Weiner LM, Davis HM. Antibody constructs in cancer therapy—protein engineering strategies to improve exposure in solid tumors. Cancer. 2007;109:170–179. doi: 10.1002/cncr.22402. [DOI] [PubMed] [Google Scholar]
- Borrebaeck CA, Malmborg AC, Furebring C, Michaelsson A, Ward S, Danielsson L, Ohlin M. Kinetic analysis of recombinant antibody-antigen interactions: relation between structural domains and antigen binding. Bio/Technology. 1992;10:697–698. doi: 10.1038/nbt0692-697. [DOI] [PubMed] [Google Scholar]
- Carreno BM, Collins M. The B7 family of ligands and its receptors: New pathways for costimulation and inhibition of immune responses. In: Paul WE, Fathman CG, Glimcher LH, editors. Annual review of immunology. Cambridge: Genetics Institute/Wyeth Research; 2002. pp. 29–53. [DOI] [PubMed] [Google Scholar]
- Chames P, Van Regenmortel M, Weiss E, Baty D. Therapeutic antibodies: successes, limitations and hopes for the future British. J Pharmacol. 2009;157:220–233. doi: 10.1111/j.1476-5381.2009.00190.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi GH, et al. Cloning, expression, and characterization of single-chain variable fragment antibody against mycotoxin deoxynivalenol in recombinant Escherichia coli. Protein Expr Purif. 2004;35:84–92. doi: 10.1016/j.pep.2003.12.008. [DOI] [PubMed] [Google Scholar]
- Clackson T, Hoogenboom HR, Griffiths AD, Winter G. Making antibody fragments using phage display libraries. Nature. 1991;352:624–628. doi: 10.1038/352624a0. [DOI] [PubMed] [Google Scholar]
- Clark CA, et al. Tumor-intrinsic PD-L1 signals regulate cell growth, pathogenesis, and autophagy in ovarian cancer and melanoma. Cancer Res. 2016;76:6964–6974. doi: 10.1158/0008-5472.CAN-16-0258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crivianu-Gaita V, Thompson M. Aptamers, antibody scFv, and antibody Fab’ fragments: an overview and comparison of three of the most versatile biosensor biorecognition elements. Biosens Bioelectron. 2016;85:32–45. doi: 10.1016/j.bios.2016.04.091. [DOI] [PubMed] [Google Scholar]
- Dai HP, et al. Construction and characterization of a novel recombinant single-chain variable fragment antibody against white spot syndrome virus from shrimp. J Immunol Methods. 2003;279:267–275. doi: 10.1016/S0022-1759(03)00182-0. [DOI] [PubMed] [Google Scholar]
- Dong H, Zhu G, Tamada K, Chen L. B7-H1, a third member of the B7 family, co-stimulates T-cell proliferation and interleukin-10 secretion. Nat Med. 1999;5:1365–1369. doi: 10.1038/70932. [DOI] [PubMed] [Google Scholar]
- Escors D, Gato-Canas M, Zuazo M, Arasanz H, Garcia-Granda MJ, Vera R, Kochan G. The intracellular signalosome of PD-L1 in cancer cells. Signal Transduct Targeted Ther. 2018;3:26. doi: 10.1038/s41392-018-0022-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman GJ, et al. Engagement of the PD-1 immunoinhibitory receptor by a novel B7-family member leads to negative regulation of lymphocyte activation. Blood. 2000;96:810A–811A. doi: 10.1084/jem.192.7.1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garet E, Cabado AG, Vieites JM, Gonzalez-Fernandez A. Rapid isolation of single-chain antibodies by phage display technology directed against one of the most potent marine toxins: Palytoxin. Toxicon. 2010;55:1519–1526. doi: 10.1016/j.toxicon.2010.03.005. [DOI] [PubMed] [Google Scholar]
- Hansel TT, Kropshofer H, Singer T, Mitchell JA, George AJT. The safety and side effects of monoclonal antibodies. Nat Rev Drug Discov. 2010;9:325–338. doi: 10.1038/nrd3003. [DOI] [PubMed] [Google Scholar]
- Herbst RS, et al. Predictive correlates of response to the anti-PD-L1 antibody MPDL3280A in cancer patients. Nature. 2014;515:563–567. doi: 10.1038/nature14011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochuli E, Dobeli H, Schacher A. New metal chelate adsorbent selective for proteins and peptides containing neighboring histidine-residues. J Chromatogr. 1987;411:177–184. doi: 10.1016/S0021-9673(00)93969-4. [DOI] [PubMed] [Google Scholar]
- Hochuli E, Bannwarth W, Dobeli H, Gentz R, Stuber D. Genetic approach to facilitate purification of recombinant proteins with a novel metal chelate adsorbent. Bio-Technology. 1988;6:1321–1325. [Google Scholar]
- Holliger P, Hudson PJ. Engineered antibody fragments and the rise of single domains. Nat Biotechnol. 2005;23:1126–1136. doi: 10.1038/nbt1142. [DOI] [PubMed] [Google Scholar]
- Kalim M, et al. Construction of high level prokaryotic expression and purification system of PD-L1 extracellular domain by using Escherichia coli host cell machinery. Immunol Lett. 2017;190:34–41. doi: 10.1016/j.imlet.2017.06.004. [DOI] [PubMed] [Google Scholar]
- Kalim M, et al. Intracellular trafficking of new anticancer therapeutics: antibody-drug conjugates. Drug Design Dev Ther. 2017;11:2265–2276. doi: 10.2147/DDDT.S135571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan G, Gershoni JM. A general insert label for peptide display on chimeric filamentous bacteriophages. Anal Biochem. 2012;420:68–72. doi: 10.1016/j.ab.2011.08.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJE. The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protocols. 2015;10:845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khantasup K, Chantima W, Sangma C, Poomputsa K, Dharakul T. Design and generation of humanized single-chain Fv derived from mouse hybridoma for potential targeting application. Monoclon Antib Immunodiagn Immunother. 2015;34:404–417. doi: 10.1089/mab.2015.0036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latchman Y, et al. PD-L2 is a second ligand for PD-1 and inhibits T cell activation. Nat Immunol. 2001;2:261–268. doi: 10.1038/85330. [DOI] [PubMed] [Google Scholar]
- Li W, Caberoy NB. New perspective for phage display as an efficient and versatile technology of functional proteomics. Appl Microbiol Biotechnol. 2010;85:909–919. doi: 10.1007/s00253-009-2277-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombardi A, Sperandei M, Cantale C, Giacomini P, Galeffi P. Functional expression of a single-chain antibody specific for the HER2 human oncogene in a bacterial reducing environment. Protein Expr Purif. 2005;44:10–15. doi: 10.1016/j.pep.2005.05.013. [DOI] [PubMed] [Google Scholar]
- Makrides SC. Strategies for achieving high-level expression of genes in Escherichia coli. Microbiol Rev. 1996;60:512–538. doi: 10.1128/mr.60.3.512-538.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks JD, Hoogenboom HR, Bonnert TP, McCafferty J, Griffiths AD, Winter G. By-passing immunization. Human antibodies from V-gene libraries displayed on phage. J Mol Biol. 1991;222:581–597. doi: 10.1016/0022-2836(91)90498-U. [DOI] [PubMed] [Google Scholar]
- McCafferty J, Griffiths AD, Winter G, Chiswell DJ. Phage antibodies—filamentous phage displaying antibody variable domains. Nature. 1990;348:552–554. doi: 10.1038/348552a0. [DOI] [PubMed] [Google Scholar]
- Moore JT, Uppal A, Maley F, Maley GF. Overcoming inclusion body formation in a high-level expression system. Protein Expr Purif. 1993;4:160–163. doi: 10.1006/prep.1993.1022. [DOI] [PubMed] [Google Scholar]
- Muller M, Schouten RD, De Gooijer CJ, Baas P. Pembrolizumab for the treatment of non-small cell lung cancer. Expert Rev Anticancer Ther. 2017;17:399–409. doi: 10.1080/14737140.2017.1311791. [DOI] [PubMed] [Google Scholar]
- Pande J, Szewczyk MM, Grover AK. Phage display: concept, innovations, applications and future. Biotechnol Adv. 2010;28:849–858. doi: 10.1016/j.biotechadv.2010.07.004. [DOI] [PubMed] [Google Scholar]
- Qiu XY, et al. PD-L1 confers glioblastoma multiforme malignancy via Ras binding and Ras/Erk/EMT activation. Biochimica et Biophysica Acta (BBA) 2018;1864:1754–1769. doi: 10.1016/j.bbadis.2018.03.002. [DOI] [PubMed] [Google Scholar]
- Robert C, et al. Nivolumab in previously untreated melanoma without BRAF mutation. N Engl J Med. 2015;372:320–330. doi: 10.1056/NEJMoa1412082. [DOI] [PubMed] [Google Scholar]
- Rosenberg AH, Lade BN, Chui DS, Lin SW, Dunn JJ, Studier FW. Vectors for selective expression of cloned DNAs by T7 RNA-polymerase. Gene. 1987;56:125–135. doi: 10.1016/0378-1119(87)90165-X. [DOI] [PubMed] [Google Scholar]
- Schreuer M, Jansen Y, Planken S, Chevolet I, Seremet T, Kruse V, Neyns B. Combination of dabrafenib plus trametinib for BRAF and MEK inhibitor pretreated patients with advanced BRAF(V600)-mutant melanoma: an open-label, single arm, dual-centre, phase 2 clinical trial. Lancet Oncol. 2017;18:464–472. doi: 10.1016/S1470-2045(17)30171-7. [DOI] [PubMed] [Google Scholar]
- Silacci M, et al. Design, construction, and characterization of a large synthetic human antibody phage display library. Proteomics. 2005;5:2340–2350. doi: 10.1002/pmic.200401273. [DOI] [PubMed] [Google Scholar]
- Smith GP. Filamentous fusion phage: novel expression vectors that display cloned antigens on the virion surface. Science (New York, NY) 1985;228:1315–1317. doi: 10.1126/science.4001944. [DOI] [PubMed] [Google Scholar]
- Studier FW, Moffatt BA. Use of bacteriophage-T7 RNA-polymerase to direct selective high-level expression of cloned genes. J Mol Biol. 1986;189:113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- Taube JM, et al. Association of PD-1, PD-1 ligands, and other features of the tumor immune microenvironment with response to anti-PD-1 therapy. Clin Cancer Res. 2014;20:5064–5074. doi: 10.1158/1078-0432.CCR-13-3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tumeh PC, et al. PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature. 2014;515:568–571. doi: 10.1038/nature13954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velasco-Velazquez M, Xolalpa W, Pestell RG. The potential to target CCL5/CCR1 in breast cancer. Expert Opin Ther Targets. 2014;18:1265–1275. doi: 10.1517/14728222.2014.949238. [DOI] [PubMed] [Google Scholar]
- Visintin M, Tse E, Axelson H, Rabbitts TH, Cattaneo A. Selection of antibodies for intracellular function using a two-hybrid in vivo system. Proc Natl Acad Sci USA. 1999;96:11723–11728. doi: 10.1073/pnas.96.21.11723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vozy A, Zalcman G. Pembrolizumab for the treatment of non-small-cell lung cancer. Oncologie. 2015;17:407–408. [Google Scholar]
- Wang H, et al. Expression and purification of an anti-clenbuterol single chain Fv antibody in Escherichia coli. Protein Expr Purif. 2010;72:26–31. doi: 10.1016/j.pep.2010.03.001. [DOI] [PubMed] [Google Scholar]
- Weisser NE, Hall JC. Applications of single-chain variable fragment antibodies in therapeutics and diagnostics. Biotechnol Adv. 2009;27:502–520. doi: 10.1016/j.biotechadv.2009.04.004. [DOI] [PubMed] [Google Scholar]
- Wen S, Zhang X, Liu Y, Zhang Q, Liu X, Liang J. Selection of a single chain variable fragment antibody against Ivermectin from a phage displayed library. J Agric Food Chem. 2010;58:5387–5391. doi: 10.1021/jf904562x. [DOI] [PubMed] [Google Scholar]
- Wingfield PT, Palmer I, Liang S-M. Folding and purification of insoluble (inclusion body) proteins from Escherichia coli. Curr Protoc Protein Sci. 2014;78:6.5.1–6.5.30. doi: 10.1002/0471140864.ps0605s78. [DOI] [PubMed] [Google Scholar]
- Winkler ME, Blaber M. Purification and characterization of recombinant single-chain urokinase produced in Escherichia coli. Biochemistry. 1986;25:4041–4045. doi: 10.1021/bi00362a008. [DOI] [PubMed] [Google Scholar]
- Yokota T, Milenic DE, Whitlow M, Schlom J. Rapid tumor penetration of a single-chain FV and comparison with other immunoglobulin forms. Cancer Res. 1992;52:3402–3408. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.