Abstract
Antimicrobial peptides (AMPs) provide a promising strategy against infections involving multidrug-resistant pathogens. In previous studies, we designed a short 12 amino acid AMP DP7, using a machine-learning method based on an amino acid activity contribution matrix. DP7 shows broad-spectrum antimicrobial activities both in vitro and in vivo. Here, we aim to investigate the efficacy of DP7 against multidrug resistant Staphylococcus aureus (S. aureus) and reveal the potential mechanisms. First, by measuring the killing kinetics of DP7 against S. aureus and comparing these results with antibiotics with different antimicrobial mechanisms, we hypothesize that DP7, in addition to its known ability to induce cell wall cation damage, can also exert a full killing effect. With FITC-conjugated or biotin-labeled DP7, we tracked DP7’s attachment, membrane permeation and subsequent intracellular distribution in S. aureus. These results indicated that the possible targets of DP7 were within the bacterial cells. Transcriptome sequencing of S. aureus exposed to DP7 identified 333 differentially expressed genes (DEGs) influenced by DP7, involving nucleic acid metabolism, amino acid biosynthesis, cell wall destruction and pathogenesis, respectively, indicating the comprehensive killing efficacy of DP7. In addition, the genome sequencing results of the induced DP7 resistant strain S. aureus DP7-R revealed two-point mutations in the mprF and guaA gene. Moreover, in a murine model for MRSA blood stream infection, intravenously treating mice with DP7 showed a good protective effect on mice. In conclusion, DP7 is an effective bactericide for S. aureus, which deserves further study for clinical application and drug development.
Keywords: antimicrobial peptides DP7, MRSA, antimicrobial efficacy, blood stream infection, drug development
Introduction
The increasing frequency of multidrug-resistant bacterial infections is a rising threat to public health. Therefore, there is an urgent need to develop novel antimicrobial agents against infections induced by multidrug-resistant bacteria. Owing to the broad spectrum of antibacterial activity properties, multiple targets and hardly leading to induce bacterial resistance, AMPs are currently under evaluation as a potential defense against multidrug-resistant (MDR) pathogens (Fjell et al., 2012; Deslouches et al., 2013). AMPs are a diverse class of relatively short (12–100 amino acids) cationic peptides (with a net charge of +2 to +9) (Fjell et al., 2012). AMPs have been isolated from both single-cell and multi-cellular organisms; furthermore, it appears that AMPs are produced by all domains of life, acting as an ancient and non-specific innate immune system in defending the host against invading pathogenic organisms, including gram-negative and gram-positive bacteria, fungi, viruses and parasites (Brogden, 2005; Fjell et al., 2012). AMPs have been successfully deployed against bacterial infections in clinical and agricultural settings. Thus, AMPs could be effective antibacterial agents against MDR bacteria without inducing further spread of antimicrobial resistance, which is a common outcome of the use of traditional antibiotics.
Here, the antimicrobial peptide (AMP) DP7, a short 12 amino acid AMP with broad-spectrum antibacterial activities, was chosen to analyze its potential mechanisms for killing S. aureus. Compared with other AMPs against S. aureus ATCC 25923, such as LL-37 (37 amino acids, MIC = 14 mg/L), pexiganan (22 amino acids, MIC = 32 mg/L) and indolicidin (22 amino acids, MIC = 16 mg/L), DP7 (12 amino acids, MIC = 16 mg/L) was composed of less amino acids with similar or superior minimum inhibitory concentrations (MIC) (Table 1) (Falla et al., 1996; Subbalakshmi and Sitaram, 1998; Dorschner et al., 2001; Zhang et al., 2010; Shen et al., 2012). Compared to AMPs with similar amino acid composition, such as HH2 (12 amino acids, MIC = 64 mg/L), Bac2A (12 amino acids, MIC = 64 mg/L) and Hlf1-11 (11 amino acids, MIC = 64 mg/L), DP7 showed lower MIC values than S. aureus (Table 1) (Costa et al., 2014; Yao et al., 2016). Therefore, considering the synthesis cost and antimicrobial potency, DP7 was a potent drug against S. aureus in future clinical applications.
Table 1.
MIC of various AMPs against S. aureus.
| Peptide | Sequence | Number of amino acids | MIC for ATCC 25923 (mg/L) | MIC for ATCC 33591 (mg/L) | net + n at neutral pH | References |
|---|---|---|---|---|---|---|
| DP7 | VQWRIRVAVIRK | 12 | 16a | 16a | +4 | |
| LL-37 | LLGDFFRKSKEKIGKEFKRI VQRIKDFLRNLVPRTES | 37 | 14b | 28b | +6 | Shen et al., 2012 |
| Pexiganan | GIGKFLKKAKKFGKAFVKILKK | 22 | 32b | 16a | +9 | Zhang et al., 2010 |
| HH2 | VQLRIRVAVIRA | 12 | 64b | 64a | +3 | Wu et al., 2014 |
| Hlf1-11 | GRRRRSVQWCA | 11 | 64a | 64b | +3.95 | Costa et al., 2014 |
| Bac2A | RLARIVVIRVAR | 12 | 64b | 43.7a | +4 | Wu et al., 2014 |
| Indolicidin | ILPWKWPWWPWRR | 13 | 16b | 64a | +3 | Wu et al., 2014 |
aMICs measured in our experiments. bMICs obtained from the references.
In this study, we aimed to explore the potential mechanism of DP7 killing S. aureus. First, we compared the killing curves of DP7 with other antibiotics whose mechanisms are well-studied, confirming that DP7 presented killing effects. Next, we analyzed the effects of DP7 on nucleic acid metabolism, amino acid biosynthesis, cell wall and pathogenesis of S. aureus at the transcriptome level. Genome sequencing of the DP7-resistant S. aureus strain DP7-R showed two-point mutations in the mprF and guaA genes. Furthermore, DP7 showed a reduced inhibitory effect on S. aureus RN4220 (a natural sigB deletion strain) compared with S. aureus ATCC 25923. As we expected, DP7 showed favorable therapeutic efficacy in a lethal systemic MRSA infectious mouse model.
Materials and Methods
Bacterial Strains
Three S. aureus strains, the methicillin-sensitive strain ATCC 25923 (MSSA), the methicillin-resistant strain ATCC 33591 (MRSA), the sigB-defective strain RN4220 and Escherichia coli (ATCC 25922), as well as the Pseudomonas aeruginosa (PAO1) strain were all obtained from the American Type Culture Collection (ATCC, Rockville, MD, United States), and the clinical S. aureus isolates were obtained from Southwest Hospital of China (Sichuan, China). This experimental program was approved by the “West China Hospital Review Committee” and written informed consent was obtained from the patients, who volunteered.
Peptides and Reagents
DP7 (VQWRIRVAVIRK), DP8 (VQLRIRVCVIRK), DP10 (VQLRIRVCVIRK), HH2 (VQLRIRVCVIRK), pexiganan (GIGKFLKKAKKFGKAFVKILKK), FITC-labeled DP7 and biotin-labeled DP7 were synthesized by Shanghai Science Peptide Biological Technology (Shanghai, China) using fluorenylmethyloxycarbonyl chemistry. The synthesized peptides were purified by high performance liquid chromatography (HPLC) to 99% purity, and mass spectrometry was used to confirm their molecular weights. Vancomycin, gentamicin, noroxin, rifampicin, ampicillin and erythromycin were obtained from Dalian Medium Biological Technology Co. Ltd. (Dalian, China).
Susceptibility Testing and in vitro Time-Killing Assays
The susceptibility testing methods were performed as previously described (Wiegand et al., 2008). Briefly, the logarithmic growth-phase bacteria (ATCC 25923 and ATCC 33591) were treated with different drug concentrations at 37°C for 22 h, and the OD600 values were subsequently measured to determine the minimum inhibitory concentration (MIC). After MIC determination, the culture medium in all wells where no visible bacterial growth was observed was inoculated in a nutrient agar plate and cultured overnight at 37°C. The minimum bactericidal concentration (MBC) endpoint is the lowest concentration of antibacterial agent that kills > 99.9% of the initial bacterial population where no visible growth of bacteria was observed on nutrient agar plates. Peptides and antibiotics at concentrations corresponding to 1, 2, and 4× MIC (as determined above) were added to bacterial samples (106 CFU/mL) during early log phase to mid-log phase growth. Subsequently, the bacterial samples were counted at different time points (0, 1, 2, 4, 6, 8, 18, and 24 h).
Confocal Microscopy Assay
Exponential-phase bacteria (108 CFU/mL) were incubated with 1× MIC FITC-DP7 for 30 or 60 min at 37°C. Next, the unbound peptides were removed by extensive washing in PBS (three times) on a shaker. The imaging was carried out using a Zeiss LSM 800 confocal laser scanning microscope (CLSM, Carl Zeiss, Oberkochen, Germany) with a 63× oil objective. Bacteria treated with PBS was served as a control in this experiment.
Immunoelectron Microscopy
Mid-log phase bacterial cultures of S. aureus ATCC 25923 (1 × 108 CFU/mL) were treated with biotin-DP7 at 37°C in 10 mM sodium phosphate buffer (pH = 7.4, 5 mM of glucose, 150 mM of NaCl). After incubation for 1 h, the samples were washed three times with phosphate buffer, and then the S. aureus were fixed with 2.5% glutaraldehyde at 4°C. The S. aureus were then centrifuged and resuspended in water mixed with 2% agar. Small blocks of agar (approximately 2 Cubic millimeter) (Sigma-Aldrich) containing the samples were dehydrated by soaking in increasing concentrations of ethanol, infiltrated with LR White M acrylic resin (Sigma-Aldrich), and embedded in alginate capsules following procedures previously published (Podda et al., 2006). Ultra-thin sections (100 nm) were cut from the embedded samples, mounted on 300 mesh copper grids and incubated at room temperature for 1 h with drops of 0.1 M of Tris-buffered saline solution (TBS) with pH 7.4, containing 1% (w/v) BSA. After washing three times in PBS, the sections were incubated with a 1:50 dilution of gold-conjugated streptavidin (10 nm gold) (BoHua company, Shanghai, China) for 1 h, and washed three times with PBS to remove the free streptavidin. Non-DP7-treated cells acted as a control in this experiment. Finally, the samples were analyzed by transmission electron microscope (TEM) (Tecnai G2 F20 S-Twin; FEI).
RNA-seq Library Preparation and Analysis
The quality of the RNA was verified using a Bioanalyzer (Agilent Technologies, Santa Clara, CA, United States) and a NanoDrop spectrometer (Thermo, United States). The A260/280 absorbance ration was in the range of 2.0–2.2 and the A260/230 ratio was above 1.8. The RNA was sent to Shanghai Personal Biotechnology Co., Ltd. (Shanghai, China) for sequencing using an Illumina Nextseq500 system. We performed quality control analysis by FastQC1, and the fastx-toolkit was used to remove potential adapter sequences2. The RPKM (reads per kilobase per million reads) values were used to measure the expression level of each gene in the sample. Next, we used Rockhopper to compare the gene values and to analyze the differentially regulated genes. We screened the differentially expressed genes (DEGs) according to the differences in expression and the p-values. Finally, the gene functions in the eggNOG genome database (Evolutionary genealogy of genes: Non-supervised Orthologous Groups)3,4 were used to classify and analyze the gene functional distribution on the macroscopic level.
Gene Expression Pathway Analysis
Differentially expressed genes were identified by transcriptome sequencing, and genes with p < 0.05 were defined as displaying a significant expression difference. After enrichment analysis of the GO (Gene Ontology) and KEGG (Kyoto Encyclopedia of Genes and Genomes) pathways, the differential enrichment p-values were corrected by the Bonferroni method (Silva-Vargas et al., 2016). Corrected p-values less than 0.05 [-Ln (p-value) ≈3] indicated that a pathway was enriched in the DEGs.
Cell Treatment and RNA Extraction
Mid-log phage S. aureus ATCC 25923 (approximately OD600 = 0.4–0.6) cells were treated with 16 mg/L DP7 for 1 h and washed three times. The cells were then subjected to the saturated phenol-chloroform extraction method to obtain the total RNA. Untreated cells served as the control. Three independent repeats were performed for this experiment.
Quantitative Real-Time RT-PCR
According to a standard protocol, total RNA was reverse transcribed using 5 × All-In-One RT MasterMix (Abm, Nanjing, China). The experiment was performed using EvaGreen 2 × qPCR Master Mix (Takara, Dalian, China) and a Lightcycle 96 (Roche) with normal cycling parameters. The cycle threshold values were determined and the relative fold differences were calculated by the 2-ΔΔCT method, using gyrB as the reference gene. Three independent repeats were run in triplicate for every experiment.
Resistance Selection Experiment
According to a published protocol, we chose 4 replicate populations from S. aureus ATCC 25923, S. aureus ATCC 33591, and cultured them with supplementation with increasing concentrations of DP7 (starting with 1/8× MIC, 2 mg/L), and four control populations were propagated in the medium that lacked any DP7 (Habets and Brockhurst, 2012). For every 48 h, we transferred 2% of each culture to new flasks containing fresh medium with a twofold higher DP7 concentration. When growth was observed at the higher concentration, we used this culture for subsequent inoculation. Aliquots from each population were stored in 20% glycerol at -80°C. Every generation with a higher MIC was tested.
Whole Genome Sequencing
We extracted the genomic DNA of DP7-resistant bacteria and DP7-sensitive bacteria using the TIANamp Bacteria DNA Kit (QIAGEN) and conducted whole genome sequencing in Novogene (Beijing, China). The original image data files obtained from the high-throughput sequencing (Illumina HiSeq TM2000 and MiSeq sequencing platform) were transformed into the original sequencing sequences (Sequenced Reads) by CASAVA base-pair recognition (Base Calling) analysis. The sequencing data were filtered to remove the sequences containing adapter and low-quality data. The obtained clean data were used for subsequent analysis and the SNPs were detected using SAMTOOLS. Small insertions and deletions (InDels) with lengths less than 50 bp were analyzed. The SNPs/InDels variations in genomic functional regions were analyzed.
Experimental Murine Model of Blood Stream Infection
A well-characterized murine model of blood stream infection was employed to study the pharmacodynamics profiles of DP7 and compare them with those of vancomycin. Briefly, C57 female mice (body weight 17 g ± 1 g), purchased from Hua Fu Kang Biological Technology Co., Ltd (Beijing, China), were intravenously infected with 1 × 108 CFU/100 μL of MRSA strain ATCC 33591 in the logarithmic growth phase (recovery on defibrinated sheep blood agar, Oxoid, United Kingdom). One hour after infection, the mice were treated with various doses of DP7 (0.5, 1, and 2 mg/kg), while the mice in the control group were injected with the same volume of saline. Vancomycin at 10 mg/kg was used as the positive control. Survival and weight changes were observed every 24 h for 1 week. At 24 and 72 h, organs, including the heart, liver, spleen, lung, kidney, and blood were collected and the bacterial load was counted for each murine. At 72 h, the liver, spleen, lungs, and kidneys were fixed with 4% paraformaldehyde for 1 week, sectioned at 4 μm after processing for paraffin embedding, and then stained with hematoxylin and eosin (H&E). All animal procedures were approved and controlled by the Institutional Animal Care and Treatment Committee of Sichuan University and conducted according to the Animal Care and Use Guidelines of Sichuan University.
Statistical Analysis
All data comparisons were analyzed using Prism 6.0 with Student’s t-test. p-values ≤ 0.05 were considered statistically significant, and p-values ≤ 0.01 were considered markedly statistically significant.
Results
DP7 Showed an Efficient Bactericidal Activity
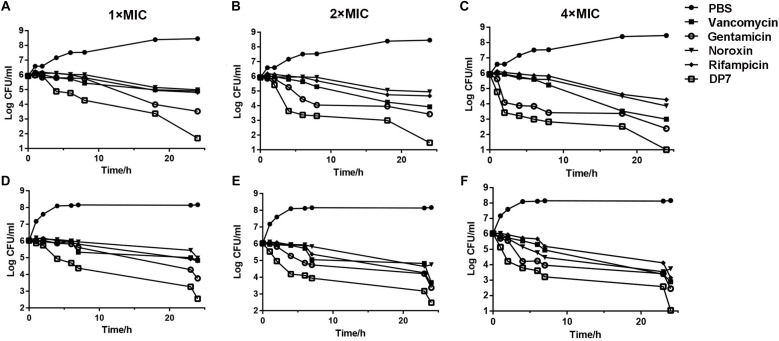
DP7 exhibited broad-spectrum antimicrobial properties (Table 2). We compared DP7’s killing kinetics with some clearly targeted antibiotics. Exposing bacteria to DP7 decreased the CFU/ml (DP7 treatment at 2× and 4× MIC), and the number of living cells decreased 3 logs within 24 h; furthermore, the number of bacteria did not increase over 2 h, indicating that DP7 was a bactericide. The trend in the DP7 killing curve was similar to that resulting from gentamicin exposure (Figure 1). Similarly, the killing curves resulting from exposure to 2× and 4× DP7 MIC showed significantly faster rates of bactericidal activity compared to the rates in bacteria treated with all of the other drugs at higher concentrations. At 4× MIC, DP7 was found to completely stop cell growth within 24 h. As a result, the features of DP7’s killing kinetics suggest a possible comprehensive bactericidal mechanism.
Table 2.
Antimicrobial assay of DP7.
| Strain | MIC (mg/L) | MBC (mg/L) |
|---|---|---|
| S. aureus (ATCC 25923) | 16 | 64 |
| S. aureus (ATCC 33591) | 16 | 64 |
| E. coli (ATCC 25922) | 16 | 64 |
| P. aeruginosa (PAO1) | 16 | 64 |
| S. aureus clinical isolates (MRSA) (SCQ1, SCQ3, S150320016, S3250016, S3396, S3750, S260029) S. aureus clinical isolates (MRSA) (S3768) | 16 32 | 128 256 |
FIGURE 1.
In vitro time-killing curves of DP7 compared with some other general antibiotics. Time-killing kinetics of DP7 (open squares) (MIC use here is 16 mg/L), vancomycin (squares) (MIC use here is 0.25 mg/L), gentamicin (open circles) (MIC use here is 0.125 mg/L), noroxin (inverted triangles) (MIC use here is 1 mg/L), and rifampicin (diamonds) (MIC use here is 0.125 mg/L) with untreated bacteria as a growth control (closed circles) at (A) 1× MIC, (B) 2× MIC, and (C) 4× MIC against S. aureus ATCC 25923 at 0, 1, 2, 4, 6, 8, 18, and 24 h. (n = 3); at (D) 1× MIC, (E) 2× MIC, and (F) 4× MIC against S. aureus ATCC 33591 at 0, 1, 2, 4, 6, 8, 18, and 24 h (n = 3).
Intracellular Localization of DP7 in S. aureus
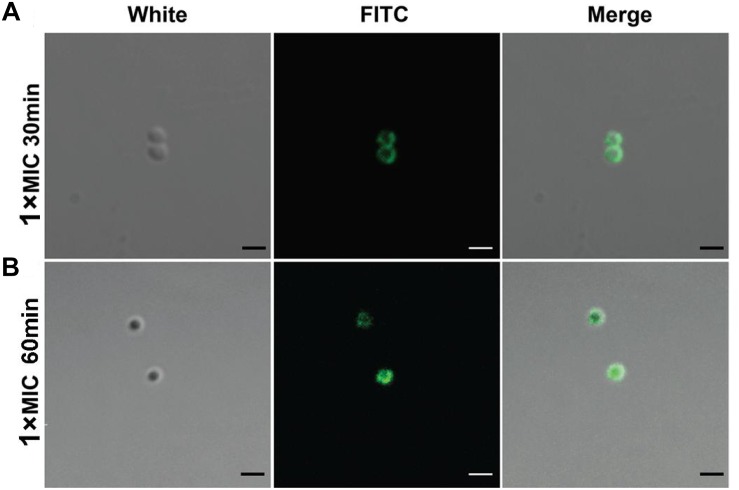
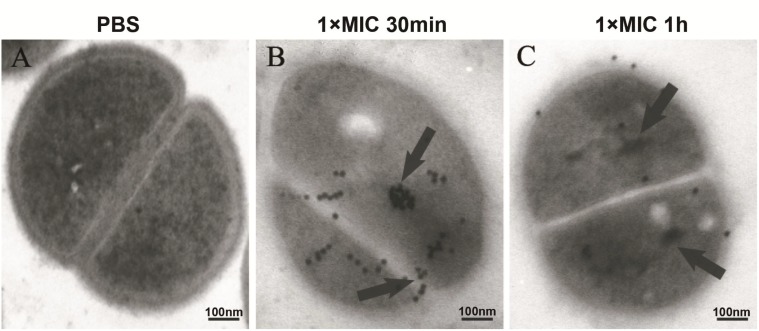
The intracellular localization of DP7 in S. aureus cells was examined with confocal laser scanning microscopy (CLSM) and FITC-labeled DP7 (MIC = 32 mg/L). S. aureus ATCC 25923 (1 × 107 CFU/mL) was incubated with 1× MIC of FITC-DP7 for 30 and 60 min. Any DP7 accumulation on or within the bacterial cells would result in a green fluorescent signal generation by the FITC that would be visible under the confocal microscope. As shown in Figure 2A, 30 min exposure to FITC-labeled DP7 resulted in an ambient green signal accumulation around the periphery of the cells, suggesting peripheral aggregation of DP7 on the surface of the bacteria (Figure 2A). When the bacteria were exposed to DP7 for 60 min (Figure 2B), the entire cell was encapsulated in green fluorescence, perhaps indicating that DP7 entered the cells (cells treated with PBS or FITC without conjugated DP7 were used as a negative control for this experiment). To further explore this possibility, transmission electron microscopy (TEM) was used. Based on the knowledge that gold-labeled streptavidin binds specifically to biotin, we assumed that the positions of the gold particles might indicate the location of the biotin-conjugated DP7 within the bacteria. Importantly, the control cells treated with PBS and incubated with streptavidin-gold particles showed no gold particles in the view (Figure 3A). However, after culturing bacteria with DP7 (1× MIC) for 30 min, the bacteria showed the accumulation of many bound gold particles (black dots) located in the bacteria, which suggested possible surface adhesion (Figure 3B). After treatment for 1 h, the original sharp black dots became fuzzy, suggesting DP7 penetrated into the cytoplasm. At this time, the cell wall was disrupted, and the inner membranes were obvious. After prolonged peptide incubation, the cell wall became obviously diffuse, thinner and the inner membrane became blurrier than that in the cells without peptide treatment. In the images of cells treated with DP7 for 1 h, there were black dots located in the membranes that looked as if they had “passed through” the cell wall and entered the cytoplasm, and this intracellular accumulation appeared to have a symmetrical distribution (Figure 3C).
FIGURE 2.
Confocal microscopic imaging shows DP7 aggregation on the cell surface and entry into cells. Confocal microscopic images of S. aureus ATCC 25923 after treatment with FITC-DP7 at 1× MIC for (A) 30 and (B) 60 min. The increasing fluorescence intensity from 30 to 60 min after treatment with the DP7 peptide may indicate progressive membrane damage and pore formation in S. aureus ATCC 25923, allowing increased uptake of the fluorescent probe. The scale bar in this figure is 1 μm.
FIGURE 3.
Localization of DP7 in S. aureus by transmission electron microscopy. S. aureus ATCC 25923 (approximately 1 × 108 CFU/mL) were incubated with PBS (A), or biotin-DP7 (1× MIC) for 30 min (B) or for 1 h (C). Fixed bacteria were then incubated with streptavidin (10 nm gold) for 1 h, and then observed under TEM. The length of the bars corresponds to 100 nm. (n ≥ 3).
Transcriptomic Analysis of DP7-Treated S. aureus ATCC 25923 and S. aureus ATCC 33591
In this study, S. aureus lab strains, methicillin-sensitive S. aureus (MSSA) ATCC 25923 and methicillin-resistant S. aureus (MRSA) ATCC 33591 were used as model species to determine the global gene expression change upon DP7 treatment. For S. aureus, there are 333 DEGs (250 upregulated genes) from a total of 2820 genes in DP7-treated S. aureus ATCC 25923 and 312 DEGs (147 upregulated genes) from a total of 2730 genes in DP7-treated S. aureus ATCC 33591, respectively (Supplementary Tables S1, S2). Gene annotations including statistical analysis of the enrichment pathways of S. aureus ATCC 25923 and S. aureus ATCC 33591 can be found in Supplementary Tables S3, S4.
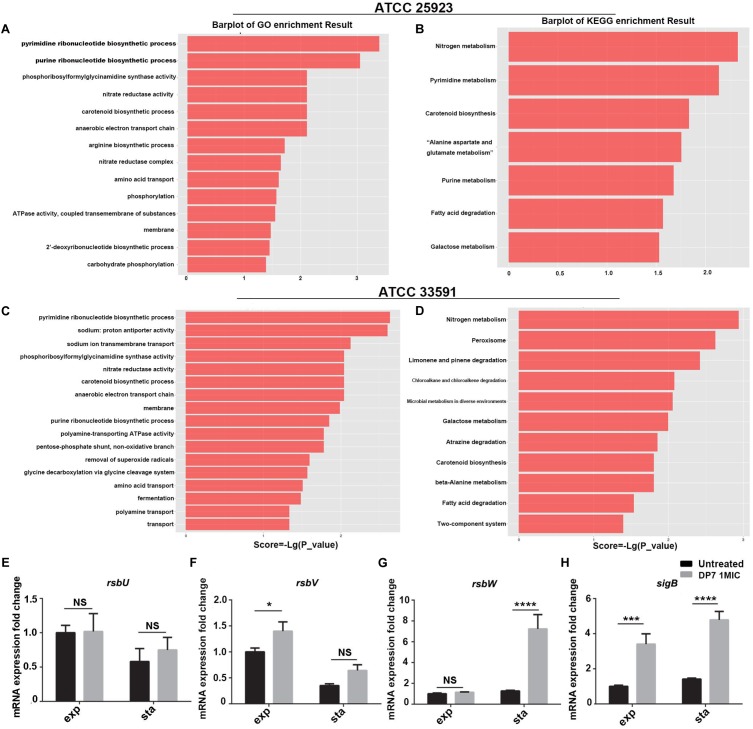
First, the impact of DP7 treatment on nucleic acid metabolism were analyzed, and the results showed that DNA and RNA biosynthetic process-associated genes exhibited significant differential expression after DP7 treatment on S. aureus, especially for pyrimidine and purine ribonucleotide biosynthetic process-related genes (Figure 4A–D). Notably, genes encoding DNA polymerase subunits, DNA gyrase subunits, DNA topoisomerase subunits and chromosome replication protein DnaD were differentially expressed. Some DNA-directed RNA polymerase submits were found to be differentially expressed after DP7 treatment, including alpha-subunit upregulation and beta, delta, omega-subunit downregulation in S. aureus ATCC 33591. This is accompanied by upregulation of RNA polymerase sigma factor sigB in both two S. aureus strains, and RNA polymerase sigma factor rpoD in S. aureus ATCC 33591. SigB was upregulated in DP7-treated cells in both S. aureus ATCC 25923 and S. aureus ATCC 33591, but there no differential expression was observed with DNA-binding RNA polymerase subunits, which indicated DP7 had no inhibitory activity on RNA polymerase. But RNA methyltransferase was influenced by DP7 in S. aureus ATCC 33591. Two of three translation-initiation factor-2 (IF-2) and IF-3 were differentially expressed after DP7 treatment. Downregulation of the A subunits in DNA topoisomerase IV was found in S. aureus ATCC 25923 and S. aureus ATCC 33591 after DP7-treatment. The expression of topoisomerase I and III were decreased in DP7-treated S. aureus ATCC 33591. In S. aureus ATCC 33591, DNA gyrase subunit A and IS1272 transposase were downregulated after being treated with DP7. In addition, carAB (Carbamoyl-phosphate synthase small chain) in the pyrimidine pathways was downregulated 17- and 10-fold lower than those of the control. Additionally, pyrC (Dihydroorotase) was upregulated 18.37-fold higher than that of normal control.
FIGURE 4.
Correlation pathway enrichment of differentially expressed genes after DP7 (1× MIC) treatment of S. aureus ATCC 25923 and ATCC 33591. (A) GO term enrichment results of DP7-treated ATCC 25923. (B) KEGG term enrichment results of DP7-treated ATCC 25923. (C) GO term enrichment results of DP7-treated ATCC 33591. (D) KEGG term enrichment results of DP7-treated ATCC 33591. (E–H) Expression of sigB-related genes in S. aureus ATCC 25923 (black bars), DP7 (16 mg/L)-treated S. aureus ATCC 25923 (gray bars) during exponential (exp) and stationary (sta) phases. Each single row in the heat map corresponds to a single gene. (n ≥ 3; ∗p < 0.05; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001).
The transcriptomic analysis of S. aureus ATCC 25923 and S. aureus ATCC 33591 showed that DP7 has effects on amino acid biosynthesis processes in S. aureus. From the gene enrichment analyses of DP7-treated S. aureus, the precursory pathways responsible for amino acid biosynthesis were noted. These included nitrogen, aromatic compound, amine acid, and carboxylic compound biosynthesis processes. The DEGs encoded for various amino acids were reported, including tryptophan, lysine, valine, leucine, isoleucine, arginine, proline, histidine, alanine, aspartate, glutamate, and tyrosine metabolism. The aromatic family amino acids tryptophan, the lysine in basic amino acids, and aliphatic family amino acids lysine and valine were affected significantly. The expression of DEGs resulted in new metabolisms of stress pathways being activated. Several genes were associated with arginine biosynthetic process pathways, nitrate reductase complex and amino acid transport, lyase, and chorismate’s metabolic process. Similar results were observed in DP7-treated S. aureus ATCC 33591. In addition, the generation of peptides/proteins on the downstream processes following influenced amino acids biosynthesis were affected by the treatments as well. Changes of RNA expressions were associated with tRNA ligase activity, 30S and 50S ribosomal proteins, and ribosomal small subunit assembly. The translation-initiation factors (IFs) IF2 and IF3 were downregulated after DP7 treatment in S. aureus ATCC 33591.
Next, the effects of DP7 treatment on the cell wall of S. aureus and pathogenesis were analyzed. Gene enrichment analyses highlighted that genes encoding for cell membrane and amino acid transport pathways were clearly impacted in DP7-treated S. aureus. More than 60 genes were differentially expressed in the two pathways and represented the largest gene sets as compared to other pathways. Moreover, DP7-treated S. aureus was reported with changes in the carotenoid biosynthetic process (GO:0016117), phosphoribosylformylglycinamidine syntheses activity (GO:0004642), nitrate reductase activity (GO:0008940), and anaerobic electron transport chain (GO:0019645). In DP7-treated S. aureus, two genes (KQ76_RS12545, KQ76_RS02320) associated with cell wall synthesis were downregulated in S. aureus ATCC 25923 and S. aureus ATCC 33591. In addition, DP7 treatment resulted in upregulation of the carotenoid biosynthetic pathway (GO:0016117)-related genes and lysostaplin resistance protein A (lyra, KQ76_RS13890) and toxin (KQ76_RS11290).
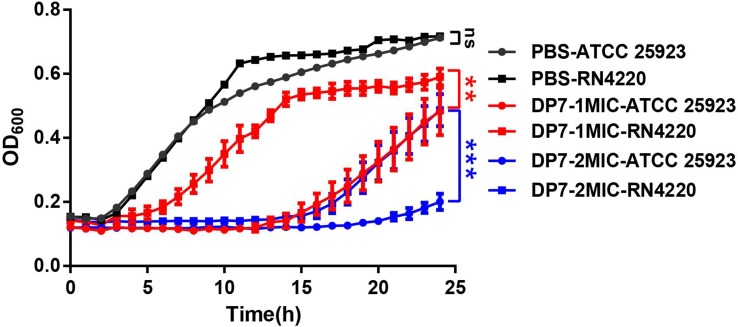
The Growth Inhibitory Rate of DP7 on S. aureus RN4220 Was Lower Than That in S. aureus ATCC 25923
In this section, we used RT-PCR to verify the consistency of sigB-related gene expression and the results of the transcriptome sequencing in DP7-treated S. aureus ATCC 25923 (Table 3) (Figure 4E–H). Primers were listed in Supplementary Table S5. A sigB-defective strain was used to further verify our results. S. aureus RN4220, with an 11-bp defect in rsbU, was described as a natural sigB-defective strain (Kullik et al., 1998; Rachid et al., 2000; Giachino et al., 2001). The growth curves of S. aureus ATCC 25923 and S. aureus RN4220 were tested in the presence or absence of DP7 to clarify whether DP7 killing S. aureus was related to the sigB synthesis regulon. Cells were grown in triplicate at 37°C with aeration using a Bioscreen C automated growth analysis system. In this experiment, the OD values were measured every 3 min, and only the hourly OD values were used for plotting. As seen in Figure 5, the growth inhibitory rate of DP7 on S. aureus RN4220 was lower than that in S. aureus ATCC 25923 at 1× MIC and 2× MIC, demonstrating that sigB plays an important role in the DP7 killing of S. aureus.
Table 3.
The fold changes determined by RNA-seq of some sigB-related genes.
| Gene | Description | Fold- |
|---|---|---|
| name | -change | |
| Rank upregulated | ||
| sigB | RNA polymerase sigma factor sigB | 5.37 |
| rsbU | Serine phosphatase | 1.20 |
| rsbV | Anti-sigma-B factor antagonist | 2.72 |
| rsbW | Serine-protein kinase rsbW | 3.70 |
| purR | LacI family transcriptional regulator | 1.53 |
| guaA | GNAT family acetyltransferase | 1.34 |
| graS | Sensor histidine kinase graS | 1.38 |
| graR | Response regulator protein graR | 1.66 |
| mprF | Phosphatidylglycerol lysyltransferase | 1.72 |
| agrA | Histidine kinase | 1.81 |
| agrB | Accessory gene regulator protein B | 1.21 |
| Rank downregulated | ||
| purF | Amidophosphoribosyltransferase | -3.83 |
| purN | Phosphoribosylglycinamide formyltransferase | -2.45 |
| purQ | Phosphoribosylformylglycinamidine synthase subunit purQ | -4.58 |
| purL | Phosphoribosylformylglycinamidine synthase subunit purl | -5.23 |
| purM | Phosphoribosylformylglycinamidine cyclo-ligase | -3.00 |
| purK | N5-carboxyaminoimidazole ribonucleotide synthase | -2.17 |
| purC | Phosphoribosylaminoimidazole-succinocarboxamide synthase | -2.72 |
| guaB | Inosine-5′-monophosphate dehydrogenase | -1.71 |
| guaC | GMP reductase | -2.24 |
| dnaN | DNA polymerase III subunit beta | -1.05 |
| rpoA | DNA-directed RNA polymerase subunit alpha | -1.36 |
FIGURE 5.
The growth inhibition rates of DP7 in S. aureus RN4220 were lower than in S. aureus ATCC 25923. Cells were cultured at 37°C with aeration using a Bioscreen C automated growth analysis system (n = 3). In this study, the OD values were recorded every 3 min, and the hourly OD values are plotted in this figure. ∗∗p < 0.01; ∗∗∗p < 0.001.
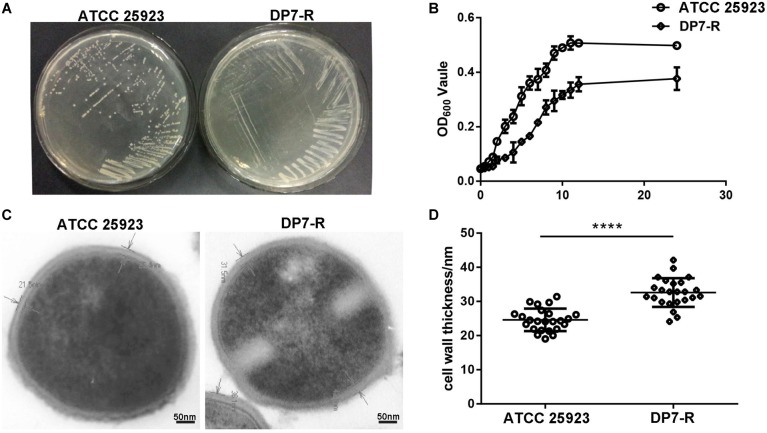
DP7-R (DP7-Resistant S. aureus Strain) Showed Point Mutations at mprF and guaA
After the 6 months of resistance selection experiments, the S. aureus strain DP7-R (MIC = 64 mg/L) (obtained from ATCC 25923 through DP7-induced drug resistance, while ATCC 33591 did not produce DP7-resistant strains) was selected from this experiment. After serial passage in normal medium, DP7-R was still resistant to DP7. S. aureus DP7-R carried two-point mutations in mprF (N353K and S567L) and one-point mutation in guaA (D450Y) in its genome sequence (Table 4). It was worth noting that S. aureus DP7-R had a slower growth rate compared with S. aureus ATCC 25923 (Figure 6A,B). Based on the upregulation of cell wall-related genes after DP7 treatment, we characterized the thickness of the cell wall in S. aureus ATCC 25923 and DP7-R. Consistent with the described physiology, the cell wall of S. aureus DP7-R was thicker than that of S. aureus ATCC 25923 (Figure 6C,D). We also tested the MICs of other AMPs against DP7-R, and the results revealed that DP7-R showed resistance to other peptides, such as HH2 (1× MIC to 4× MIC) (MIC = 64 mg/L), Pexiganan (1× MIC to 4× MIC) (MIC = 32 mg/L), DP8 (1× MIC to 4× MIC) (MIC = 16 mg/L), DP10 (1× MIC to 4× MIC) (MIC = 64 mg/L), and Hlf1-11 (1× MIC to 4× MIC) (MIC = 64 mg/L) (Table 5), while DP7-R was not resistant to some antibiotics, such as ofloxacin (1× MIC to 2× MIC) (MIC = 0.25 mg/L), moxifloxacin hydrochloride (1× MIC to 1× MIC) (MIC = 0.125 mg/L), norfloxacin (1× MIC to 2× MIC) (MIC = 1 mg/L), vancomycin (1× MIC to 1× MIC) (MIC = 0.25 mg/L), azithromycin (1× MIC to 1× MIC) (MIC = 0.125 mg/L), rifampin (1× MIC to 1× MIC) (MIC = 0.125 mg/L), but was resistant to gentamicin (1× MIC to 4× MIC) (MIC = 0.0625 mg/L).
Table 4.
Single nucleotide changes located in open reading frames.
| Strain and sitea | Base change | Amino acid change | Locus | MICb (mg/L) |
|---|---|---|---|---|
| DP7-R | ||||
| 1354274 | T→G | N353K | mprF | 64 |
| 1354918 | C→T | S567L | mprF | |
| 396702 | G→T | D450 Y | guaA |
aLocation of mutations in the S. aureus strain ATCC 25923 sequence. bThe MICs with the highest frequencies for each species. Values are averages from three independent experiments performed in triplicate.
FIGURE 6.
DP7-R showed a slower growth rate and thicker cell wall compared with wild-type DP7-sensitive strains. (A) Comparison of wild-type S. aureus ATCC 25923 and DP7-R. (B) DP7-R shows a slower growth rate than ATCC 25923. (C) The cell wall thicknesses of S. aureus ATCC 25923 and DP7-R. (D) Cell wall thickness statistics in different cells (approximately 20 cells) in several fields of view (n ≥ 3; ∗∗∗∗p < 0.0001).
Table 5.
Determination of MICs of a variety of antimicrobial peptides in ATCC 25923 and DP7-R.
| MIC (mg/L) |
||||||
|---|---|---|---|---|---|---|
| Strains | DP7 | HH2 | Pexiganan | DP8 | DP10 | Hlf1-11 |
| ATCC 25923 | 16 | 64 | 32 | 16 | 64 | 64 |
| DP7-R | 64 | 256 | 128 | 64 | 256 | 256 |
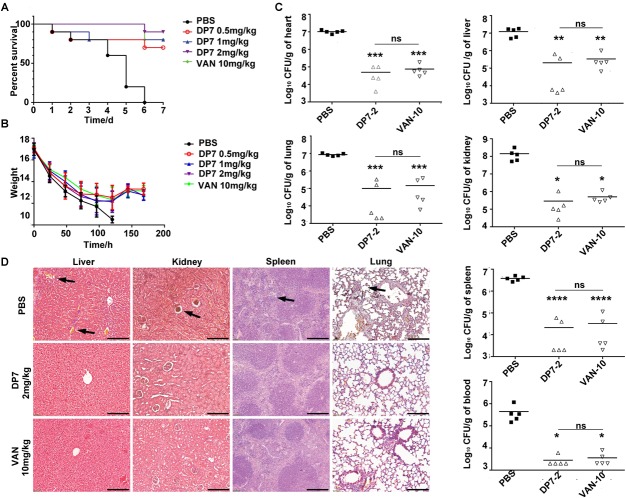
DP7 Showed Favorable Therapeutic Efficacy in a Mouse Model of MRSA Blood Stream Infection (BSI)
Blood stream infections (BSIs) have been an important cause of mortality in recent years, especially in S. aureus (20%) BSIs (Wisplinghoff et al., 2004). In this study, the therapeutic efficiency of DP7 was tested in a lethal model of MRSA BSI. The inoculums were optimized to produce lethal infection, with death occurring within 1-week post-infection. Typical physical and behavioral presentations associated with severe MRSA infection (such as decreased body weight, increased eye secretions, and decreased activity) were observed, and the infection was confirmed by testing MRSA in organ homogenates. The results showed that DP7 at doses of 0.5, 1, and 2 mg/kg led to 70, 80, and 90% reductions, respectively, in the lethality of the infection up to day 7 post-infection (Figure 7A). The protection level of DP7 at 1 mg/kg was equivalent to 10 mg/kg vancomycin in this infection model (Supplementary Figure S1). In addition, the survival of the systemically infected mice was significantly enhanced after DP7 treatment. Furthermore, DP7-treated infected mice showed slower weight loss than the control and showed recovery of weight 5 days post-infection, as occurs with vancomycin treatment (Figure 7B). The bacterial load in the heart, liver, spleen, lung, kidney, and blood was determined after 72 h of therapy. Compared with PBS-treated mice at the end of the therapy, 2 mg/kg DP7 and 10 mg/kg vancomycin led to statistically significant reductions in the bacterial load (Figure 7C). Organs from the infected control mice showed multiple lesions, damage to tissue structure, infiltration of erythrocytes and heavy alveolar congestion in the lungs (black arrow). This type of histological damage in DP7 (2 mg/kg)-treated mice was significantly minimal compared to the control mice (Figure 7D).
FIGURE 7.
Experimental murine model of blood stream infection. C57 female mice were intravenously injected with 1 × 108 CFU/100 μL ATCC 33591, and antimicrobial drugs were intravenously administered 1 h after injection. The treatments of DP7 doses were 0.5, 1, and 2 mg/kg. PBS and 10 mg/kg of vancomycin acted as negative and positive control, respectively. The bacterial loads at 72 h after infection were counted. Every symbol in Figure 1C represents the viable bacterial counts determined for each mouse, while the average values for each group is represented by the horizontal lines. (A) The survival rate and (B) weight changes after DP7 treatment (n = 10). (C) CFU counts/g of organs after treatment with 2 mg/kg DP7 or 10 mg/kg vancomycin at 24 h and 72 h (n = 5). (D) Histology of major organs of mice following lethal MRSA infection. The length of the bars corresponds to 100 μm.
Discussion
Due to serious drug resistance, conventional antibiotics are becoming increasingly ineffective. Thus, it is imperative to develop new antibacterial strategies. AMPs are used to treat multidrug-resistant bacteria because of their broad-spectrum antimicrobial activities, multi-targets and difficulty in inducing drug resistance. However, the synthetic cost of AMPs may limit their application. Therefore, it is still worth developing a more efficient and cost-effective broad-spectrum amplifier. Here, we are concerned with DP7—compared with other AMPs such as LL-37, pexiganan and indolicidin, DP7 has less amino acid composition and the MIC for S. aureus is not higher than these AMPs. Compared with Hlf1-11, Bac2A and HH2, DP7 has the same amino acid composition, and the MIC of DP7 to S. aureus is lower or equal to these AMPs. Therefore, considering antimicrobial activity and cost-effectiveness, DP7 deserves further study.
In the present study, we aimed to explore the DP7’s potential mechanisms for killing S. aureus. DP7 was designed based on the identified peptide HH2 and optimized with an amino acid-based prediction method by determining the contribution of each amino acid to the antimicrobial activity of the peptide (Wu et al., 2014). Our method, based on a novel peptide prediction method and determination of the quantitative structure-activity, was more accurate at determining the efficacy of the peptide but was not helpful in identifying potential targets (Ostberg and Kaznessis, 2005; Wu et al., 2014). Considering the amino sequence and lengths of DP7, the chances that it has a secondary structure is unlikely, and evidence of self-assembly is absent for DP7. Therefore, it is difficult to explain the uneven attachment and distribution of DP7 on the S. aureus cell surface. Furthermore, the similarities in the killing kinetics shared between DP7 and ribosome-targeted or nucleic synthesis inhibitory antibiotics (rather than cell wall-active antibiotics) prompted us to explore if there are additional DP7 targets (Schneider et al., 2010). Then we mapped the physiological changes after DP7 treatment via transcriptomic analysis. Through the whole genome sequencing analysis, we examined the changes in physiological functions in S. aureus before and after induced DP7 resistance. One of the goals of the present study was to outline the possible signaling pathways modulated by DP7 which could help to better understand its possible mode of action.
To analyze the global effects of DP7 in S. aureus, we performed comparative transcriptome analysis on MSSA strain S. aureus ATCC 25923 and the MRSA strain S. aureus ATCC 33591 with or without DP7 exposure. Interestingly, DP7 had drastic effects on S. aureus’s pyrimidine and purine biosynthesis metabolism. Specifically, carAB, a component of the pyrimidine pathway, was downregulated by 17- and 10-fold relative to the control. As a part of the arginine and pyrimidine pathways, the carAB gene encodes the enzyme carbamoylphosphate synthetase (CPS), which catalyzes the synthesis of carbamoylphosphate (Piette et al., 1984; Martinussen and Hammer, 1998; Nicoloff et al., 2001; Butcher et al., 2016). Carbamoylphosphate is a common intermediate in arginine and pyrimidine biosynthesis (Werner et al., 1985). Although the exact regulated mode of carAB in S. aureus is still unclear, similar studies performed on other gram-negative microbes suggest that arginine is likely involved in the regulation (Sekowska et al., 2001). Regulation of the carAB operon in P. aeruginosa is controlled by arginine and pyrimidines at the transcriptional level, possibly through an attenuation mechanism (Lu et al., 1997; Butcher et al., 2016). Further studies on E. coli identified an arginine box located upstream of the control region of carAB that served as the initiation site, but which was blocked upon arginine recognition (Piette et al., 1984; Crowell et al., 1987; Alwan et al., 2017). Due to the arginine-rich nature of DP7’s sequence, combined with the results of our earlier study proving that DP7 possesses no secondary structure and has a low chance of self-assembly, we speculated that the arginine box might be recognized by DP7 to inhibit carAB transcription. This possibility must be demonstrated through future experiments, as no evidence has suggested the existence of such arginine boxes in S. aureus or any other gram-positive species. Interestingly, while almost all the genes involved in the de novo synthesis of pyrimidines were downregulated, the expression of pyrC was upregulated by 18.37-fold compared to its level in the normal control. This result indicates that these pathways are tightly regulated and balanced in the cell. Further studies are warranted to investigate the mechanisms of nucleotide regulation in S. aureus, e.g., deciphering the role of end-product repression in these pathways.
During the induction of cell envelope stress, it is reasonable to assume that AMPs could bind to molecules located on the cell wall or membrane. Some of these molecules might act as global regulators that sense envelope stress and react as “effectors” after stimulation due to sustained exposure (Hu and Coates, 1999). The stress-activated sigma factor B (sigB) was reported to be involved in resistance to cell wall-active antibiotics such as beta-lactamase and vancomycin. There are two primary regulators of sigB, rsbV and rsbW. RsbW can bind to sigB protein, which renders sigB unavailable to RNA polymerase, while rsbV is the sigB release factor. Under stressful conditions, rsbW binds to non-phosphorylated rsbV and releases sigB (Voelker et al., 1996; Thompson et al., 2015). In this study, we observed that DP7 perturbed the rsbU-sigB transcription system, which could partly explain the strong effects of DP7 on gene expression, bacteria growth or infectivity. Related to the sigB regulon, DP7 increased the sigB transcriptional level by 5.37-fold in S. aureus ATCC 25923. The increased sigB positively regulates the purine operon repressor (purR), then inhibits the expression of the downstream pur genes, and downregulates the expression of the gua genes, thus affecting the synthesis of GMP. Inhibition of GMP synthesis inhibits the expression of DNA polymerase III subunit beta (dnaN) and DNA-directed RNA polymerase subunit alpha (rpoA), thereby inhibiting DNA and RNA synthesis (Table 3). Combined with the observation that DP7-R was found to develop a thicker cell wall upon ongoing DP7 exposure, our data were consistent with the fact that artificial overexpression of sigB in S. aureus resulted in a thickened cell wall (Mishra et al., 2009). Furthermore, other cell wall-targeted antibiotics have also been proven to alter S. aureus virulence by affecting sigB-regulated biofilm formation. Even though no specific DP7 target was identified in this study, the impaired killing effect of DP7 in the sigB-negative S. aureus strain RN4220 suggested that sigB might be one of the potential targets of the DP7.
DP7-R carried two-point mutations in mprF (N353K and S567L) and single point mutation in guaA (D450Y). The upregulation of mprF reduced the negative net charge of the cell membrane via the formation of lysyl-phosphatidylglycerol (Lys-PG), which limits the attraction to AMP (Ernst and Peschel, 2011; Rubio et al., 2012). The two point mutations in mprF have not been mentioned in previous studies, thus they may indicate a new cause of reduced membrane charge (Quinn et al., 2007). GuaA, known as an integrated hot spot for genetic islands, transposons and bacteriophages in bacterial species, has not been reported to be associated with peptide resistance (Mulhbacher et al., 2010; Wipf et al., 2015). We assumed that the point mutant in guaA may be related to a new mechanism of peptide resistance, which will be clarified in our future studies. Furthermore, DP7-R showed multi-peptide resistance, which may indicate that the activated sigB system may also enhance the tolerance of S. aureus to other peptides. In the case of MSI-78, which was previously studied by Habets and Brockhurst (2012), S. aureus could develop a stable 10- to 50-fold increase in the MIC. However, in our study, S. aureus showed, at most, an 8-fold increase in DP7 resistance, which may indicate that the development of DP7 resistance by bacteria may be difficult.
Proliferation of S. aureus in the blood represents one of the most dangerous stages of S. aureus infection. Considering the inhibitory effect of DP7, we examine the possible therapeutic efficacy of DP7 in S. aureus blood infection. We tested the potency of DP7 in a murine model of BSI by counting the survival of DP7-treated animals. Following intravenous administration, DP7 showed an effective protective effect on mice with blood infection. In summary, by conducting the murine model of BSI, we demonstrate that DP7 could be a promising strategy for fighting S. aureus infection in the future. These experiments demonstrated that DP7, a computer-designed AMP, may be effective in a more clinical scenario.
Ethics Statement
All animal procedures were approved and controlled by the Institutional Animal Care and Treatment Committee of Sichuan University and conducted according to the Animal Care and Use Guidelines of Sichuan University. In addition, we declare all animal experiments comply with the ARRIVE guidelines.
Author Contributions
RZ and ZW coordinated the whole project and wrote the article. YT and QY synthesized and purified some peptides. BZ, XC, and ML performed the bioinformatics analysis. XZ performed in vitro experiments for MIC. The manuscript was written with contributions from all the authors. All authors have given approval to the final version of the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding. This work was supported by the National Natural Science Foundation of China (31570927), The National Key Research and Development Program (No. 2016YFC1303502), and the 1.3.5 project for disciplines of excellence, West China Hospital, Sichuan University.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.01175/full#supplementary-material
References
- Alwan R., Bruel A. L., Da Silva A., Blanquet V., Bouhouche K. (2017). An siRNA-based screen in C2C12 myoblasts identifies novel genes involved in myogenic differentiation. Exp. Cell Res. 359 145–153. 10.1016/j.yexcr.2017.07.037 [DOI] [PubMed] [Google Scholar]
- Brogden K. A. (2005). Antimicrobial peptides: pore formers or metabolic inhibitors in bacteria? Nat. Rev. Microbiol. 3 238–250. 10.1038/nrmicro1098 [DOI] [PubMed] [Google Scholar]
- Butcher B. G., Chakravarthy S., D’Amico K., Stoos K. B., Filiatrault M. J. (2016). Disruption of the carA gene in Pseudomonas syringae results in reduced fitness and alters motility. BMC Microbiol. 16:194. 10.1186/S12866-016-0819-Z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa F., Maia S., Gomes J., Gomes P., Martins M. C. L. (2014). Characterization of hLF1-11 immobilization onto chitosan ultrathin films, and its effects on antimicrobial activity. Acta Biomater. 10 3513–3521. 10.1016/j.actbio.2014.02.028 [DOI] [PubMed] [Google Scholar]
- Crowell D. N., Reznikoff W. S., Raetz C. R. H. (1987). Nucleotide-sequence Of the Escherichia-Coli gene for lipid-a disaccharide synthase. J. Bacteriol. 169 5727–5734. 10.1128/jb.169.12.5727-5734.1987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deslouches B., Steckbeck J. D., Craigo J. K., Doi Y., Mietzner T. A., Montelaro R. C. (2013). Rational design of engineered cationic antimicrobial peptides consisting exclusively of arginine and tryptophan, and their activity against multidrug-resistant pathogens. Antimicrob. Agents Chemother. 57 2511–2521. 10.1128/Aac.02218-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorschner R. A., Pestonjamasp V. K., Tamakuwala S., Ohtake T., Rudisill J., Nizet V., et al. (2001). Cutaneous injury induces the release of cathelicidin anti-microbial peptides active against group A streptococcus. J. Investig. Dermatol. 117 91–97. 10.1046/j.1523-1747.2001.01340.x [DOI] [PubMed] [Google Scholar]
- Ernst C. M., Peschel A. (2011). Broad-spectrum antimicrobial peptide resistance by MprF-mediated aminoacylation and flipping of phospholipids. Mol. Microbiol. 80 290–299. 10.1111/j.1365-2958.2011.07576.x [DOI] [PubMed] [Google Scholar]
- Falla T. J., Karunaratne D. N., Hancock R. E. W. (1996). Mode of action of the antimicrobial peptide indolicidin. J. Biol. Chem. 271 19298–19303. 10.1074/jbc.271.32.19298 [DOI] [PubMed] [Google Scholar]
- Fjell C. D., Hiss J. A., Hancock R. E. W., Schneider G. (2012). Designing antimicrobial peptides: form follows function. Nat. Rev. Drug Discov. 11 124–124. [DOI] [PubMed] [Google Scholar]
- Giachino P., Engelmann S., Bischoff M. (2001). sigma(B) activity depends on RsbU in Staphylococcus aureus. J. Bacteriol. 183 1843–1852. 10.1128/Jb.183.6.1843-1852.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habets M. G., Brockhurst M. A. (2012). Therapeutic antimicrobial peptides may compromise natural immunity. Biol. Lett. 8 416–418. 10.1098/rsbl.2011.1203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J. M., Coates A. R. M. (1999). Transcription of two sigma 70 homologue genes, sigA and sigB, in stationary-phase Mycobacterium tuberculosis. J. Bacteriol. 181 469–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kullik I., Giachino P., Fuchs T. (1998). Deletion of the alternative sigma factor sigma(B) in Staphylococcus aureus reveals its function as a global regulator of virulence genes. J. Bacteriol. 180 4814–4820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C. D., Kwon D. H., Abdelal A. T. (1997). Identification of greA encoding a transcriptional elongation factor as a member of the carA-orf-carB-greA operon in Pseudomonas aeruginosa PAO1. J. Bacteriol. 179 3043–3046. 10.1128/jb.179.9.3043-3046.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinussen J., Hammer K. (1998). The carB gene encoding the large subunit of carbamoylphosphate synthetase from Lactococcus lactis is transcribed monocistronically. J. Bacteriol. 180 4380–4386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra N. N., Yang S. J., Sawa A., Rubio A., Nast C. C., Yeaman M. R., et al. (2009). Analysis of cell membrane characteristics of in vitro-selected daptomycin-resistant strains of methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 53 2312–2318. 10.1128/Aac.01682-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulhbacher J., Brouillette E., Allard M., Fortier L. C., Malouin F., Lafontaine D. A. (2010). Novel riboswitch ligand analogs as selective inhibitors of guanine-related metabolic pathways. PLoS Pathog. 6:e1000865. 10.1371/journal.ppat.1000865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicoloff H., Hubert J. C., Bringel F. (2001). Carbamoyl-phosphate synthetases (CPS) in lactic acid bacteria and other gram-positive bacteria. Lait 81 151–159. 10.1051/lait:2001119 [DOI] [Google Scholar]
- Ostberg N., Kaznessis Y. (2005). Protegrin structure-activity relationships: using homology models of synthetic sequences to determine structural characteristics important for activity. Peptides 26:197–206. 10.1016/j.peptides.2004.09.020 [DOI] [PubMed] [Google Scholar]
- Piette J., Nyunoya H., Lusty C. J., Cunin R., Weyens G., Crabeel M., et al. (1984). DNA-sequence of the cara gene and the control region of carab - tandem promoters, respectively controlled by arginine and the pyrimidines, regulate the synthesis of carbamoyl-phosphate synthetase in Escherichia-Coli K-12. Proc. Natl. Acad. Sci. U.S.A. 81 4134–4138. 10.1073/pnas.81.13.4134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podda E., Benincasa M., Pacor S., Micali F., Mattiuzzo M., Gennaro R., et al. (2006). Dual mode of action of Bac7, a proline-rich antibacterial peptide. Biochim. Biophys. Acta Gen. Subj. 1760 1732–1740. 10.1016/j.bbagen.2006.09.006 [DOI] [PubMed] [Google Scholar]
- Quinn B., Hussain S., Malik M., Drlica K., Zhao X. (2007). Daptomycin inoculum effects and mutant prevention concentration with Staphylococcus aureus. J. Antimicrob. Chemother. 60 1380–1383. 10.1093/jac/dkm375 [DOI] [PubMed] [Google Scholar]
- Rachid S., Ohlsen K., Wallner U., Hacker J., Hecker M., Ziebuhr W. (2000). Alternative transcription factor sigma(B) is involved in regulation of biofilm expression in a Staphylococcus aureus mucosal isolate. J. Bacteriol. 182 6824–6826. 10.1128/Jb.182.23.6824-6826.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio A., Moore J., Varoglu M., Conrad M., Chu M., Shaw W., et al. (2012). LC-MS/MS characterization of phospholipid content in daptomycin-susceptible and -resistant isolates of Staphylococcus aureus with mutations in mprF. Mol. Membr. Biol. 29 1–8. 10.3109/09687688.2011.640948 [DOI] [PubMed] [Google Scholar]
- Schneider T., Kruse T., Wimmer R., Wiedemann I., Sass V., Pag U., et al. (2010). Plectasin, a fungal defensin, targets the bacterial cell wall precursor lipid II. Science 328 1168–1172. 10.1126/science.1185723 [DOI] [PubMed] [Google Scholar]
- Sekowska A., Robin S., Daudin J. J., Henaut A., Danchin A. (2001). Extracting biological information from DNA arrays: an unexpected link between arginine and methionine metabolism in Bacillus subtilis. Genome Biol. 2 research0019.1–research0019.12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen J., Lu X. M., Jin X. B., Ding J., Li X. B., Mei H. F., et al. (2012). Expression of a novel dual-functional protein - The antimicrobial peptide LL-37 fused with human acidic fibroblast growth factor in Escherichia coli. Protein Expr. Purif. 81 119–125. 10.1016/j.pep.2011.09.007 [DOI] [PubMed] [Google Scholar]
- Silva-Vargas V., Maldonado-Soto A. R., Mizrak D., Codega P., Doetsch F. (2016). Age-dependent niche signals from the choroid plexus regulate adult neural stem cells. Cell Stem Cell 19 643–652. 10.1016/j.stem.2016.06.013 [DOI] [PubMed] [Google Scholar]
- Subbalakshmi C., Sitaram N. (1998). Mechanism of antimicrobial action of indolicidin. FEMS Microbiol. Lett. 160 91–96. 10.1016/S0378-1097(98)00008-1 [DOI] [PubMed] [Google Scholar]
- Thompson C. C., Griffiths C., Nicod S. S., Lowden N. M., Wigneshweraraj S., Fisher D. J., et al. (2015). The Rsb phosphoregulatory network controls availability of the primary sigma factor in chlamydia trachomatis and influences the kinetics of growth and development. PLoS Pathog. 11:e1005125. 10.1371/journal.ppat.1005125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voelker U., Voelker A., Haldenwang W. G. (1996). Reactivation of the bacillus subtilis anti-sigma(B) antagonist, RsbV, by stress- or starvation-induced phosphatase activities. J. Bacteriol. 178 5456–5463. 10.1128/jb.178.18.5456-5463.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner M., Feller A., Pierard A. (1985). Nucleotide-sequence of yeast gene cpa1 encoding the small subunit of arginine-pathway carboamoyl-phosphate synthetase - homology of the deduced amino-acid sequence to other glutamine amidotransferases. Eur. J. Biochem. 146 371–381. 10.1111/j.1432-1033.1985.tb08663.x [DOI] [PubMed] [Google Scholar]
- Wiegand I., Hilpert K., Hancock R. E. W. (2008). Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat. Protoc. 3 163–175. 10.1038/nprot.2007.521 [DOI] [PubMed] [Google Scholar]
- Wipf J. R., Schwendener S., Nielsen J. B., Westh H., Perreten V. (2015). The new macrolide-lincosamide-streptogramin B resistance gene erm(45) is located within a genomic island in Staphylococcus fleurettii. Antimicrob. Agents Chemother. 59 3578–3581. 10.1128/AAC.00369-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisplinghoff H., Bischoff T., Tallent M., Seifert H., Wenzel R. P., Edmond M. B. (2004). Nosocomial bloodstream infections in US hospitals: analysis of 24,179 cases from a prospective nationwide surveillance study. Clin. Infect. Dis. 39 1093–1093. [DOI] [PubMed] [Google Scholar]
- Wu X. Z., Wang Z. L., Li X. L., Fan Y. Z., He G., Wan Y., et al. (2014). In vitro and in vivo activities of antimicrobial peptides developed using an amino acid-based activity prediction method. Antimicrob. Agents Chemother. 58 5342–5349. 10.1128/Aac.02823-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Z. S., Han S. P., Jin R. Z. (2016). ROCK-I derived Bi-functional peptides with antibacterial infection activity. Int. J. Pept. Res. Ther. 22 407–412. 10.1007/s10989-016-9519-7 [DOI] [Google Scholar]
- Zhang Y., Zhao H., Yu G. Y., Liu X. D., Shen J. H., Lee W. H., et al. (2010). Structure-function relationship of king cobra cathelicidin. Peptides 31 1488–1493. 10.1016/j.peptides.2010.05.005 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.