Abstract
Epidermal growth factor receptor tyrosine kinase inhibitors (EGFR-TKIs) are a standard therapy for patients with non-small cell lung cancer (NSCLC) with sensitive mutations. However, acquired resistance emerges following a median of 6–12 months. Several studies demonstrated that EGFR-TKI-induced tumor microenvironment stresses and autophagy are important causes of resistance. The current review summarizes the molecular mechanisms involved in EGFR-mediated regulation of autophagy. The role of autophagy in EGFR-TKI treatment, which may serve a role in protection or cell death, was discussed. Furthermore, co-inhibiting EGFR and autophagy signaling as a rational therapeutic strategy in the treatment of patients with NSCLC was explored.
Keywords: epidermal growth factor receptor, autophagy, non-small-cell lung cancer, resistance
1. Introduction
Lung cancer is the most common cancer worldwide in 2014 (1). There are two broad types of lung cancer: Small cell lung cancer (SCLC) and non-small cell lung cancer (NSCLC). NSCLC accounts for 80% of lung cancers (2). NSCLC is further divided into three subtypes: Adenocarcinoma, squamous cell carcinoma and large cell carcinoma. For the majority of patients with advanced NSCLC, platinum-based chemotherapy was previously the standard first-line treatment (3).
Currently, epidermal growth factor receptor tyrosine kinase inhibitors (EGFR-TKIs) are efficacious in treating patients with NSCLC with sensitive EGFR mutations (4). These drugs have become the standard therapy for patients with advanced NSCLC with sensitive mutations (4). Compared with platinum-based chemotherapy, treatment with EGFR-TKI may improve the overall survival time of patients with NSCLC with EGFR exon 19 deletion and mutations in exon 21 (L858R), exon 18 (G719X) and exon 20 (S768I) (5,6). However, following 8–10 months of treatment, numerous patients who originally responded to EGFR-TKIs eventually develop drug resistance (7). Strategies designed to reverse primary or acquired resistance to EGFR-TKIs are required for the development of more effective treatments. Previous studies demonstrated that EGFR-TKI-induced tumor microenvironment stresses and autophagy are important causes of resistance (8,9).
The current review focused on the known molecular mechanisms of EGFR-mediated regulation of autophagy. The role of autophagy in EGFR-TKI treatment was also discussed. Furthermore, co-inhibiting EGFR and autophagy signaling as a novel strategy to improve the efficacy of EGFR-TKIs was explored.
2. EGFR and EGFR-TKI
The EGFR family consists of tyrosine transmembrane glycoproteins that are encoded by proto-oncogenes, including human epidermal growth factor receptor (HER)-1, HER-2, HER-3 and HR-4 (10). EGFR is mainly expressed in epithelial, mesenchymal and neuronal cells (10). Since its discovery in the 1980s, the EGFR signaling pathway has been implicated in organ development, including the mammary glands and epiphyseal cartilage (11,12). EGFR serves roles in tumor cell proliferation, differentiation, migration, adhesion, treatment resistance, survival and apoptosis (13–15).
At present, EGFR-targeted drugs consist of two types: Small molecule TKIs that inhibit the tyrosine kinase activity of the EGFR intracellular domain and artificially synthesized EGFR monoclonal antibodies that block the extracellular domain and inhibit the activation of the EGFR ligand binding domain (4,5). EGFR-TKI drugs have been used clinically for a relatively long time and are recommended by the National Comprehensive Cancer Network and European Society for Medical Oncology guidelines for the treatment of advanced NSCLC with sensitive mutations (16,17). EGFR targeting is an emerging strategy for treating other tumors, including anaplastic thyroid cancer and colorectal cancer (18,19).
The first-generation EGFR-TKIs, gefitinib, erlotinib and icotinib, are effective as first-line treatment for patients with advanced NSCLC harboring activating EGFR mutations (17,20). The second-generation EGFR-TKIs, afatinib and dacomitinib, irreversibly bind to the tyrosine kinase of EGFR and other ErbB-family members (4,21). Afatinib has been approved as a first-line treatment of patients with advanced NSCLC with sensitive mutations (21). Dacomitinib reduced lung cancer progression in patients with NSCLC exhibiting EGFR activating mutations compared with gefitinib in a phase III trial (22). Third-generation EGFR-TKIs, osimertinib (AZD9291), rociletinib (CO-1686), olmutinib (HM61713) and others (EGF816, ASP8273), suppress EGFR activating and resistance mutations (23,24). In clinical trials, the third-generation drugs demonstrated higher response rates among tumors with acquired EGFR T790M (25,26).
Although treatment efficacy has been reported in patients treated with EGFR-TKIs, drug resistance eventually develops following ~10 months (7). The mechanisms resulting in this resistance are various and not fully understood (27). The most common mechanism is the acquisition of secondary EGFR T790M mutations (28,29). Other mechanisms of resistance may include the following: Secondary mutations or amplification in EGFR, alternative pathway activation, histologic and phenotypic transformation, tumor growth factor β-dependent interleukin (IL)-6 secretion (30) or β2-adrenergic receptors (β2-ARs) activation (Fig. 1) (31). Previous studies demonstrated that EGFR-TKI-induced tumor microenvironment stresses and autophagy are important causes of resistance (8,9).
Figure 1.
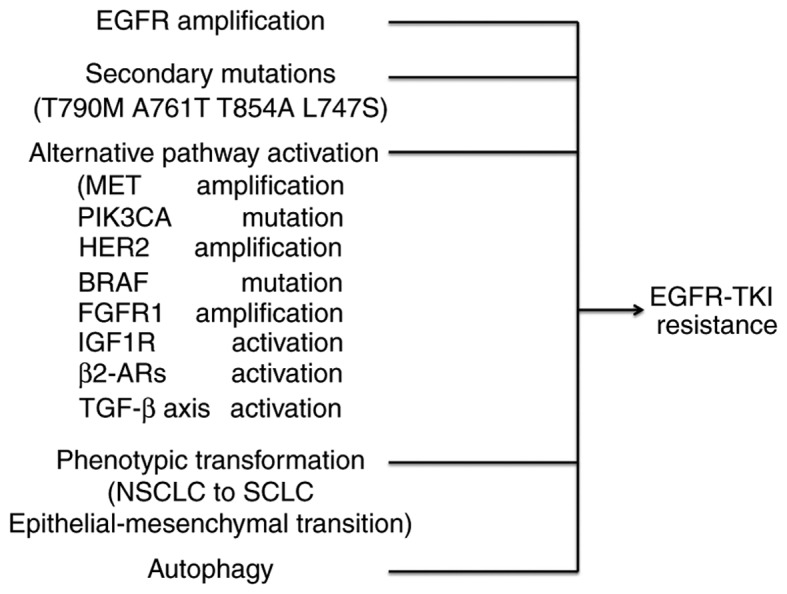
Main mechanisms involved in acquired resistance to EGFR TKIs. The main reported mechanisms of resistance to EGFR-TKIs involve EGFR gene amplification, new secondary mutations in EGFR, bypass or alternative pathway activation, histological and phenotypic transformation and autophagy. EGFR, epidermal growth factor receptor; TKIs, tyrosine kinase inhibitors; MET, MET proto-oncogene, receptor tyrosine kinase; PIK3CA, phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit α; HER2, human epidermal growth factor receptor 2; BRAF, B-Raf proto-oncogene, serine/threonine kinase; FGFR1, fibroblast growth factor receptor 1; IGF1R, insulin like growth factor 1 receptor; β2-ARs, β2-adrenergic receptors; TGF-β, transforming growth factor β, NSCLC, Non-small cell lung carcinoma; SCLC, small cell lung carcinoma.
3. Autophagy and its regulation
Autophagy is a conserved catabolic process (32). Cells digest and recycle their own cellular contents and dysfunctional organelles for cell survival (32,33). Autophagy can be divided into three types in eukaryotic cells: Macroautophagy, microautophagy and chaperone-mediated autophagy (CMA) (34). The term ‘autophagy’ usually indicates macroautophagy. In macroautophagy, the cytoplasm and organelles are engulfed into double-membrane bound structures, termed autophagosomes, and delivered to lysosomes (34). In microautophagy, cytosolic cargos are engulfed into lysosomes directly (32). In CMA, substrate proteins are delivered into the lysosome by molecular chaperones, including members of the heat shock protein 70 family (35).
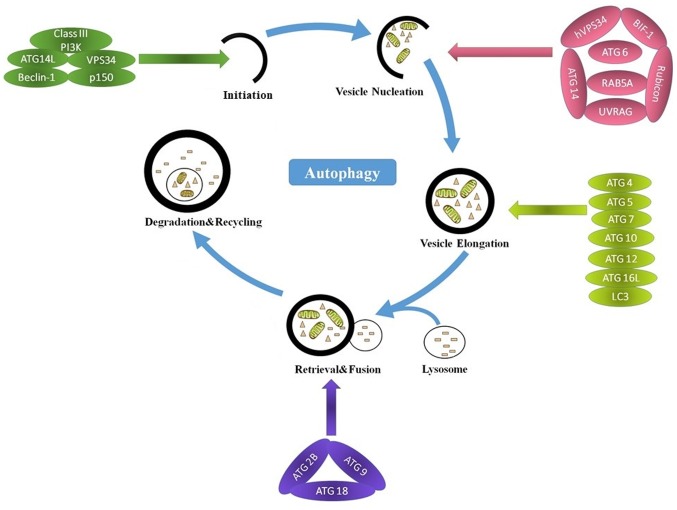
Autophagy can be divided into three main steps: Initiation, elongation and degradation (32). Autophagy is regulated by complex signaling pathways and autophagy-related genes (ATGs) (36,37). The main steps and molecular regulators of autophagy are presented in Fig. 2. Long non-coding RNA (lncRNA) has been implicated in the regulation of autophagy (38,39). microRNAs (miRNAs/miRs) have been reported to regulate certain ATGs at different stages of autophagy (40). Furthermore, the majority of lncRNAs serve a regulatory role by acting as sponges to sequester autophagy-associated miRNAs from their targets (41–43). Metastasis associated lung adenocarcinoma transcript 1 may upregulate the expression of the miR-101 targets stathmin 1, RAB5A, member RAS oncogene family and autophagy related 4D cysteine peptidase by acting as a sponge of miR-101, thus inducing autophagy (41). In anoxia/reoxygenation-induced autophagy of cardiomyocytes, the lncRNA autophagy promoting factor is upregulated to protect ATG7 from being degraded by miR-188-3p, and this promotes autophagic death of cardiomyocytes (42). The lncRNA HOXA transcript antisense RNA, myeloid-specific 1 regulates autophagy by sequestering miR-20a/106b and miR-125b and their targets unc-51 like autophagy activating kinase 1 (ULK1), E2F transcription factor 1 and DNA damage regulated autophagy modulator 2 (43).
Figure 2.
Main steps in autophagy and the molecular regulators involved. Autophagy occurs sequentially in five main steps: Initiation, vesicle nucleation, autophagosome elongation, retrieval and degradation. During the initiation, the formation of a Beclin 1, class III PI3K, VPS34, ATG14L and p150 complex contributes to the initiation and progression of autophagy. Vesicle nucleation is mediated by hVPS34, BIF-1, ATG6, ATG14, Rubicon, RAB5A and UVRAG. Autophagosome elongation is mediated by the ATG16L complex (ATG4, ATG5, ATG7, ATG10, ATG12 and ATG16L) and LC3. Retrieval is mediated by ATG2B, ATG9 and ATG18. In the last stage, engulfed contents are degraded following the fusion of the autophagosome with a lysosome, and the nutrients and energy are recycled. PI3K, phosphoinositide 3-kinase; VPS34, phosphatidylinositol 3-kinase catalytic subunit type 3; ATG, autophagy related; LC3, microtubule associated protein 1 light chain 3 α; RAB5A, RAB5A, member RAS oncogene family; BIF1, zinc finger and BTB domain containing 24; UVRAG, UV radiation resistance associated.
4. EGFR-mediated regulation of autophagy
The downstream pathways triggered by EGFR activation include the EGFR-Ras-rapidly accelerated fibrosarcoma (Raf)-c-Jun N-terminal (JNK), EGFR-PI3K/protein kinase B (AKT)/mechanistic target of rapamycin (mTOR), and EGFR-janus kinase (JAK)-signal transducer and activator of transcription 3 (STAT3) signaling pathways, all of which have regulatory effects on autophagy. The EGFR-Ras-Raf-JNK signaling pathway exerts a potent stimulatory effect on autophagy, while the EGFR-phosphoinositide 3-kinase (PI3K)/AKT/mTOR signaling pathway exerts a potent inhibitory effect on autophagy. The EGFR-JAK-STAT3 signaling pathway serves stimulatory and inhibitory roles in autophagy (44).
EGFR-Ras-Raf-JNK signaling pathway and autophagy
The RAS oncogene serves important roles in the regulation of cell survival and growth and is frequently activated in cancer. Following autophosphorylation, the adaptor protein growth factor receptor-bound protein 2 binds EGFR at the phosphorylated sites and activates son of sevenless (SOS), a guanosine triphosphate (GTP)-exchange factor for RAS. SOS then converts RAS-guanosine disphosphate into RAS-GTP (45–47). Previous studies demonstrated that autophagy is required for oncogenic Ras-induced malignant cell transformation (45). Increased autophagy in these K-ras mutation tumor cells is required for cell survival and transformation. Genetic inhibition of autophagy in RAS-transformed cells leads to decreased cell survival during starvation and abrogated tumorigenesis in mice (45). Alexandrova et al (46) revealed that oncogenic K-Ras expression upregulated autophagy through reactive oxygen species, p38, mitogen-activated protein kinase and JNK activation and subsequent upregulation of ATG5 and ATG7. Furthermore, JNK phosphorylates beclin-1 (BECN1) at three different tyrosine residues, T79, S60 and S87, as well as B-cell lymphoma 2 (Bcl-2), leading to the separation of BECN1 from the BECN1/Bcl-2 complex in response to starvation (47). The release of BECN1 results in autophagy activation (36).
EGFR-PI3K/AKT/mTOR signaling pathway and autophagy
PI3Ks are a family of lipid kinases, and class I are involved in tumorigenesis. Class I consists of a regulatory subunit p85 and a catalytic subunit p110. Class I kinases are often activated by growth factor stimulation through EGFR. The p85 regulatory subunit directly binds to phosphotyrosine residues on EGFR (48). This binding removes the intermolecular inhibition of the p110 catalytic subunit, allowing p110 to phosphorylate phosphatidylinositol-3,4 biphosphate into phosphatidylinositol-3,4,5 biphosphate (PIP3) (48). AKT is subsequently recruited to the plasma membrane by PIP3 and phosphorylated by pyruvate dehydrogenase kinase 1 at Thr308 and Ser473. AKT activates mTOR, relieving its negative effect on autophagy regulation (48). mTOR is a serine/threonine protein kinase that phosphorylates and inactivates unc-51-like autophagy activating kinase (ULK) 1/2 (49). The inhibited PI3K/AKT1 signaling upregulates the inhibitory activity of tuberous sclerosis complex 1/2 on Ras homolog, mammalian target of rapamycin complex-1 (mTORC1) binding, which is essential for mTOR activity in conditions of starvation or growth factor receptor inhibition. The decline in mTOR activity subsequently separates mTORC1 from the ULK1/2 complex [including ULK1/2, ATG13, ATG101, and RB1 inducible coiled-coil 1 (RB1CC1)] and thus activates ULK1/2. The activated ULK1/2 phosphorylates ATG13 and RB1CC1, two components of the ULK1/2 complex, which subsequently initiates the autophagy cascade (50).
EGFR-JAK-STAT3 signaling pathway and autophagy
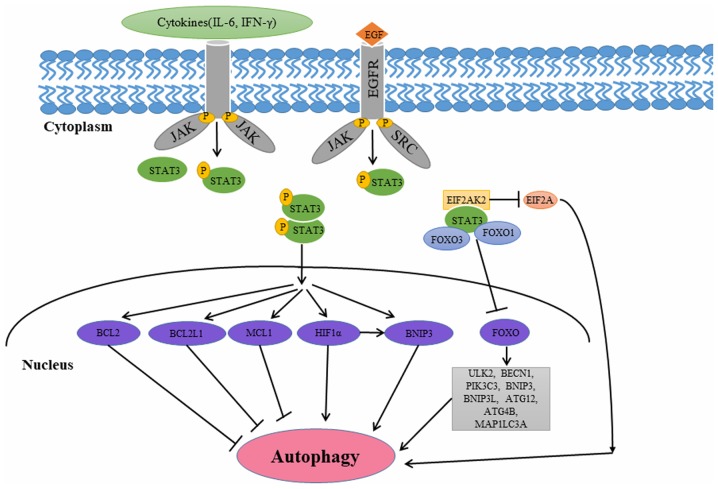
The JAK/STAT signaling pathway is a major pathway which is activated by EGFR family members (44). The STAT3 gene located on chromosome 17q21 encodes an 89 kDa protein. STAT3 belongs to a family of transcription factors that mainly exist in the cytoplasm (51). Growth-factor receptor tyrosine kinases, cytokine-receptor-associated kinases and nonreceptor tyrosine kinases phosphorylate conserved tyrosine residue 705 on STAT3, resulting in its activation and translocation from the cytoplasm to the nucleus (51). Unphosphorylated STAT3 can form dimers and translocate into the nucleus; however, tyrosine phosphorylation enhances STAT3 dimerization and translocation into the nucleus (52). Once in the nucleus, STAT3 regulates genes involved in cell proliferation, differentiation, survival and angiogenesis (52). STAT3 is involved in multiple aspects of autophagy (53–58). The different subcellular localization patterns of STAT3 affect autophagy in a transcriptional or nontranscriptional manner (Fig. 3).
Figure 3.
STAT3 in the regulation of autophagy. STAT3 has been implicated in multiple aspects of the autophagic process. The different subcellular localization patterns of STAT3 affect autophagy via transcriptional or nontranscriptional mechanisms. Cytoplasmic STAT3 inhibits autophagy by interacting with EIF2AK2, FOXO1 and FOXO3. Nuclear STAT3 inhibits autophagy by increasing the expression of several negative regulators of autophagy, including BCL2, BCL2L1 and MCL1. Nuclear STAT3 executes its pro-autophagic function by modulating HIF-1α and BNIP3. STAT3, signal transducer and activator of transcription 3; FOX, forkhead boxes; BCL2, BCL2 apoptosis regulator; JAK, janus kinase; IL-6, interleukin 6; SRC, SRC proto-oncogene, non-receptor tyrosine kinase; EIF2A, eukaryotic translation initiation factor 2A; ATF4, activating transcription factor 4; COX-2, cyclooxygenase 2; BNIP3, BCL2 interacting protein 3; EIF2AK2, eukaryotic translation initiation factor 2α kinase 2; HIF-1α, hypoxia-inducible factor 1α; MCL1, MCL1 apoptosis regulator, BCL2 family member; BCL2L1, BCL2 like 1; EGF, epidermal growth factor; EGFR, epidermal growth factor receptor; BNIP3L, BCL2 interacting protein 3 like; PIK3C3, phosphatidylinositol 3-kinase catalytic subunit type 3; BECN1, beclin 1; ULK2, unc-51 like autophagy activating kinase 2; MAP1LC3A, microtubule associated protein 1 light chain 3 α.
Regulation of autophagy by cytoplasmic STAT3
Several studies have revealed that cytoplasmic STAT3 suppresses autophagy by interacting with eukaryotic translation initiation factor 2 a kinase 2 (EIF2AK2), forkhead box protein (FOX) O1, and FOXO3 (53–56). Niso-Santano et al (53) demonstrated that cytoplasmic STAT3 may bind to the EIF2AK2 catalytic domain competitively, inhibiting its function in the process. Thus, STAT3 may function as a competitive inhibitor of EIF2AK2, inhibiting EIF2AK2 enzymatic activity and phosphorylation of EIF2A, a known autophagy activator (53). Eukaryotic translation initiation factor 2A-activating transcription factor 4 signaling pathway-mediated cyclooxygenase 2 overexpression may contribute to cadmium-induced autophagy in kidney (54). Furthermore, cytoplasmic STAT3 may interact with the autophagy-associated proteins FOXO1 and FOXO3 (55). FOXO3 can upregulate multiple autophagy-associated genes including ULK2, BECN1, phosphatidylinositol 3-kinase catalytic subunit type 3, Bcl-2 interacting protein 3 (BNIP3), Bcl-2 interacting protein 3 like, ATG12, ATG4B and microtubule associated protein 1 light chain 3 a following dephosphorylation and translocation to the nucleus. The active FOXO1 and FOXO3a exist exclusively in the nucleus of naïve T cells (56). In the cytoplasm of activated T cells, the inactive pFOXO1 and pFOXO3a integrate with unphosphorylated STAT3 (56). FOXO1/FOXO3a rapidly relocalize to the nucleus following IL-6 or IL-10 mediated pSTAT3 activation. STAT3 inhibitors completely inhibit cytokine-induced translocation of FOXO1/FOXO3a into the nucleus (56).
Regulation of autophagy by nuclear STAT3
Nuclear STAT3 increases the expression of negative regulators of autophagy including Bcl-2, Bcl-2 like 1 and MCL1 apoptosis regulator, Bcl-2 family member (MCL1), thus inhibiting autophagy (57,58). Following its activation, nuclear STAT3 increases Bcl-2 expression, leading to autophagy inhibition (57). In pancreatic ductal adenocarcinoma cells, miR506 triggers autophagic flux by direct inhibition of the STAT3-Bcl-2-BECN1 axis (59). Tai et al (60) demonstrated that sorafenib activated autophagy in a dose- and time-dependent manner through downregulation of phospho-STAT3 and MCL1 in hepatocellular carcinoma cell lines. The ectopic expression of MCL1 reversed the effect of sorafenib on autophagy. Nuclear STAT3 may inhibit autophagy through the downregulation of phosphatidylinositol 3-kinase catalytic subunit type 3 (60). The reduction of Vps34 protein levels in fiber type-specific regulation of autophagy and skeletal muscle atrophy occurs in a STAT3-dependent manner, which decreases ps34/p150/BECN1/Atg14 complex 1 (61).
Nuclear STAT3 may promote autophagy by modulating the hypoxic expression of hypoxia-inducible factor 1α (HIF-1α) and BNIP3 (62–65). STAT3 transcriptionally upregulates HIF-1α and inhibits its ubiquitination, which is mediated by von Hippel-Lindau tumor suppressor (62). Previous studies indicated that autophagy is closely associated with hypoxia (63). HIF-1α serves an essential role in various cellular responses; in particular, it promotes cell proliferation and survival (64). Hypoxia-induced autophagy serves an important role in HIF-1α-dependent general mechanisms of cell survival (64). BNIP3 and BNIP3L are the downstream targets of HIF-1α-dependent autophagy (65). BNIP3 is a conserved member of the BH3-only subfamily of the pro-apoptotic Bcl-2 family, and its expression is associated with the initiation of autophagy in different cell models (66). Furthermore, BNIP3 may dissociate BECN1 from the Bcl-2-BECN1complex (67). In addition, STAT3 regulates BNIP3 expression (68). Concanavalin A (Con A) induced autophagy through a STAT3-macrophage migration inhibitory factor-BNIP3-dependent signaling pathway (68). Pretreatment with epigallocatechin-3-gallate (EGCG) abrogated the upregulation of JAK1, JAK2, p-STAT3 and BNIP3 in a dose-dependent manner (69). The results obtained from the aforementioned studies indicated that ECGG decreases ConA-induced autophagy by downregulating the IL-6/JAKs/STAT/BNIP3-mediated signaling pathway (69).
5. Role of autophagy in EGFR-TKI treatment
Numerous studies demonstrated that initiation of autophagy may increase tumor resistance to anticancer therapies in different types of cancer cells (70). In lung cancer cells, autophagy may be activated by EGFR-TKIs, and coinhibiting EGFR signaling and autophagy demonstrates promising results in vitro (71,72). The autophagy induced by EGFR-TKIs executed a cytoprotective function in lung cancer cells. Autophagy inhibition by chloroquine (CQ) and small interfering RNAs targeting ATG5 and ATG7 increased the cytotoxic effect of the EGFR-TKIs gefitinib and erlotinib in vitro (71). Li et al (72) revealed that erlotinib induced apoptosis and autophagy in NSCLC cell lines with activating EGFR mutations (exon 19 deletion). Inhibition of autophagy may increase the sensitivity of NSCLC cell lines to erlotinib, suggesting that autophagy functions as a protective mechanism. Furthermore, the resistance to erlotinib may be reversed through autophagy inhibition (72). Wang et al (73) revealed that erlotinib induced autophagy in TKI-sensitive and TKI-resistant lung cancer cells. Inhibition of autophagy significantly increased sensitivity to erlotinib in TKI-resistant cancer cells via regulation of endoplasmic reticulum stress-induced apoptosis (73). These results indicated that cotargeting autophagy and EGFR signaling may present novel clinical strategies in the treatment of NSCLC. The resistance to erlotinib in wild-type EGFR-expressing NSCLC cells may be overcome through autophagy inhibition (74). Furthermore, a phase I clinical trial investigated the efficacy, safety and pharmacokinetics of the combination of hydroxychloroquine (HCQ) with erlotinib in patients with advanced NSCLC (75). Although this study revealed no significant improvement in survival time, the safety of adding HCQ to erlotinib was established (75). The recommended dose of HCQ was 1,000 mg when given in combination with 150 mg of erlotinib in a phase 2 study (75). However, CQ in combination with chemotherapy and radiation therapy increased the median survival time of patients with glioblastoma multiforme compared with controls in a randomized, double-blind, placebo-controlled trial (76). There are currently several ongoing clinical trials involving CQ or HCQ, and preliminary antitumor activity has been demonstrated in pancreatic adenocarcinoma, melanoma and glioblastoma (77–79).
EGFR-TKI may induce autophagic cell death (80–82). In gefitinib-resistant NSCLC cell lines, the addition of the mTOR inhibitor everolimus (RAD001) increased autophagy and cell death induced by gefitinib, and the synergistic effect was enhanced in cell lines with a high proliferative index and a short doubling time (80). In EGFR-TKI-resistant lung cancer cells with the T790M mutation, the combination of a protein kinase casein kinase 2 inhibitor and EGFR-TKI induced a high level of autophagy and promoted apoptosis (81). BEZ235, a dual inhibitor of PI3K and mTOR, overcame the EGFR-TKI resistance induced by hepatocyte growth factor in an EGFR mutant lung cancer model (82).
These observations demonstrated that autophagy may have context-dependent and even opposing effects on the behavior of cancer cells. The efficacy of autophagy inhibition in different types of human cancer, including pancreatic adenocarcinoma, melanoma, and glioblastoma have also varied widely (77–79). Autophagy may be stimulated or inhibited during cancer treatment, depending on the type of cell, the stress signals, such as chemotherapy, radiotherapy or target therapy, and other circumstances, such as hypoxia or starvation. The identification of novel biomarkers for evaluating dynamic changes in autophagy and new methods to evaluate autophagy in clinical samples may improve patient outcomes.
6. Conclusions and perspectives
EGFR-TKIs are effective for the treatment of patients with NSCLC with EGFR-sensitive mutations, although acquired resistance inevitably emerges (7). EGFR and its downstream signaling pathways serve an essential role in autophagy regulation (44). Autophagy exhibits complex, context-dependent roles in cancer, and interventions to enhance or inhibit autophagy have been proposed as an addition to EGFR-TKI-based therapies (71,80). Co-targeting autophagy signaling may be a novel therapeutic strategy for cancer treatment. However, elucidation of the mechanisms involved in autophagy is required prior to the use of such strategies. Autophagy inhibition may reduce the antitumor immune response (83). Rao et al (83) revealed that autophagy deficiency favored oncogenesis via changes in the tumor microenvironment that involved the regulatory T-cell-mediated inhibition of anticancer immunosurveillance. Autophagy also serves important roles in the survival of dormant cancer cells (84). A recent study using a Drosophila melanogaster tumor model demonstrated that dormant tumors from autophagy-deficient animals reactivated tumor growth when transplanted into autophagy-proficient animals (84). This suggested that autonomous autophagy in the surrounding nontumor cells of the microenvironment may be involved in the regrowth of dormant tumors.
The combination of EGFR-TKIs and autophagy inhibitors or inducers is attracting increased attention in NSCLC therapy. Understanding the mechanisms underlying the context-dependent effects of autophagy on cancer may provide a basis for making rational decisions on strategies to manipulate autophagy during cancer therapy.
Acknowledgements
Not applicable.
Funding
The present review was supported by a grant from the General Research Program of Zhejiang Provincial Health Department of China (grant no. 2017KY127).
Availability of data and materials
Not applicable.
Authors' contributions
XW, WL, NZ and XZ were responsible for the literature search and manuscript preparation. ZJ was responsible for manuscript co-writing and correction. All authors revised the article and approved the final version for publication.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. CA Cancer J Clin. 2014;64:9–29. doi: 10.3322/caac.21208. [DOI] [PubMed] [Google Scholar]
- 2.Mao Y, Yang D, He J, Krasna MJ. Epidemiology of lung cancer. Surg Oncol Clin N Am. 2016;25:439–445. doi: 10.1016/j.soc.2016.02.001. [DOI] [PubMed] [Google Scholar]
- 3.Al-Farsi A, Ellis PM. Treatment paradigms for patients with metastatic non-small cell lung cancer, squamous lung cancer: First, second, and third-line. Front Oncol. 2014;4:157. doi: 10.3389/fonc.2014.00157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Inal C, Yilmaz E, Piperdi B, Perez-Soler R, Cheng H. Emerging treatment for advanced lung cancer with egfr mutation. Expert Opin Emerg Drugs. 2015;20:597–612. doi: 10.1517/14728214.2015.1058778. [DOI] [PubMed] [Google Scholar]
- 5.Sharma SV, Bell DW, Settleman J, Haber DA. Epidermal growth factor receptor mutations in lung cancer. Nat Rev Cancer. 2007;7:169–181. doi: 10.1038/nrc2088. [DOI] [PubMed] [Google Scholar]
- 6.Pao W, Chmielecki J. Rational, biologically based treatment of EGFR-mutant non-small-cell lung cancer. Nat Rev Cancer. 2010;10:760–774. doi: 10.1038/nrc2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carrera S, Buque A, Azkona E, Aresti U, Calvo B, Sancho A, Arruti M, Nuño M, Rubio I, de Lobera AR, et al. Epidermal growth factor receptor tyrosine-kinase inhibitor treatment resistance in non-small cell lung cancer: Biological basis and therapeutic strategies. Clin Transl Oncol. 2014;16:339–350. doi: 10.1007/s12094-013-1143-9. [DOI] [PubMed] [Google Scholar]
- 8.Liu D, Yang Y, Zhao S. Autophagy facilitates the EGFR-TKI acquired resistance of non-small-cell lung cancer cells. J Formos Med Assoc. 2014;113:141–142. doi: 10.1016/j.jfma.2012.10.017. [DOI] [PubMed] [Google Scholar]
- 9.Sakuma Y, Matsukuma S, Nakamura Y, Yoshihara M, Koizume S, Sekiguchi H, Saito H, Nakayama H, Kameda Y, Yokose T, et al. Enhanced autophagy is required for survival in EGFR-independent EGFR-mutant lung adenocarcinoma cells. Lab Invest. 2013;93:1137–1146. doi: 10.1038/labinvest.2013.102. [DOI] [PubMed] [Google Scholar]
- 10.Lemmon MA, Schlessinger J, Ferguson KM. The egfr family: Not so prototypical receptor tyrosine kinases. Cold Spring Harb Perspect Biol. 2014;6:a020768. doi: 10.1101/cshperspect.a020768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang X, Zhu J, Li Y, Lin T, Siclari VA, Chandra A, Candela EM, Koyama E, Enomoto-Iwamoto M, Qin L. Epidermal growth factor receptor (EGFR) signaling regulates epiphyseal cartilage development through β-catenin-dependent and -independent pathways. J Biol Chem. 2013;288:32229–32240. doi: 10.1074/jbc.M113.463554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brisken C, O'Malley B. Hormone action in the mammary gland. Cold Spring Harb Perspect Biol. 2010;2:a003178. doi: 10.1101/cshperspect.a003178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee HC, Su MY, Lo HC, Wu CC, Hu JR, Lo DM, Chao TY, Tsai HJ, Dai MS. Cancer metastasis and EGFR signaling is suppressed by amiodarone-induced versican V2. Oncotarget. 2015;6:42976–42987. doi: 10.18632/oncotarget.5621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clapéron A, Mergey M, Nguyen Ho-Bouldoires TH, Vignjevic D, Wendum D, Chrétien Y, Merabtene F, Frazao A, Paradis V, Housset C, et al. EGF/EGFR axis contributes to the progression of cholangiocarcinoma through the induction of an epithelial-mesenchymal transition. J Hepatol. 2014;61:325–332. doi: 10.1016/j.jhep.2014.03.033. [DOI] [PubMed] [Google Scholar]
- 15.Masuda H, Zhang D, Bartholomeusz C, Doihara H, Hortobagyi GN, Ueno NT. Role of epidermal growth factor receptor in breast cancer. Breast Cancer Res Treat. 2012;136:331–345. doi: 10.1007/s10549-012-2289-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shi YK, Wang L, Han BH, Li W, Yu P, Liu YP, Ding CM, Song X, Ma ZY, Ren XL, et al. First-line icotinib versus cisplatin/pemetrexed plus pemetrexed maintenance therapy for patients with advanced EGFR mutation-positive lung adenocarcinoma (CONVINCE): A phase 3, open-label, randomized study. Ann Oncol. 2017;28:2443–2450. doi: 10.1093/annonc/mdx359. [DOI] [PubMed] [Google Scholar]
- 17.Shi Y, Zhang L, Liu X, Zhou C, Zhang L, Zhang S, Wang D, Li Q, Qin S, Hu C, et al. Icotinib versus gefitinib in previously treated advanced non-small-cell lung cancer (ICOGEN): A randomised, double-blind phase 3 non-inferiority trial. Lancet Oncol. 2013;14:953–961. doi: 10.1016/S1470-2045(13)70355-3. [DOI] [PubMed] [Google Scholar]
- 18.Antonelli A, Fallahi P, Ulisse S, Ferrari SM, Minuto M, Saraceno G, Santini F, Mazzi V, D'Armiento M, Miccoli P. New targeted therapies for anaplastic thyroid cancer. Anticancer Agents Med Chem. 2012;12:87–93. doi: 10.2174/187152012798764732. [DOI] [PubMed] [Google Scholar]
- 19.Koustas E, Karamouzis MV, Mihailidou C, Schizas D, Papavassiliou AG. Co-targeting of EGFR and autophagy signaling is an emerging treatment strategy in metastatic colorectal cancer. Cancer Lett. 2017;396:94–102. doi: 10.1016/j.canlet.2017.03.023. [DOI] [PubMed] [Google Scholar]
- 20.Costa DB, Nguyen KS, Cho BC, Sequist LV, Jackman DM, Riely GJ, Yeap BY, Halmos B, Kim JH, Jänne PA, et al. Effects of erlotinib in EGFR mutated non-small cell lung cancers with resistance to gefitinib. Clin Cancer Res. 2008;14:7060–7067. doi: 10.1158/1078-0432.CCR-08-1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu HA, Pao W. Targeted therapies: Afatinib-new therapy option for EGFR-mutant lung cancer. Nat Rev Clin Oncol. 2013;10:551–552. doi: 10.1038/nrclinonc.2013.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu YL, Cheng Y, Zhou X, Lee KH, Nakagawa K, Niho S, Tsuji F, Linke R, Rosell R, Corral J, et al. Dacomitinib versus gefitinib as first-line treatment for patients with EGFR-mutation-positive non-small-cell lung cancer (ARCHER 1050): A randomised, open-label, phase 3 trial. Lancet Oncol. 2017;18:1454–1466. doi: 10.1016/S1470-2045(17)30608-3. [DOI] [PubMed] [Google Scholar]
- 23.Wang S, Cang S, Liu D. Third-generation inhibitors targeting EGFR T790M mutation in advanced non-small cell lung cancer. J Hematol Oncol. 2016;9:34. doi: 10.1186/s13045-016-0290-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Russo A, Franchina T, Ricciardi GRR, Smiroldo V, Picciotto M, Zanghi M, Rolfo C, Adamo V. Third generation EGFR TKIs in EGFR-mutated NSCLC: Where are we now and where are we going. Crit Rev Oncol Hematol. 2017;117:38–47. doi: 10.1016/j.critrevonc.2017.07.003. [DOI] [PubMed] [Google Scholar]
- 25.Soria JC, Ohe Y, Vansteenkiste J, Reungwetwattana T, Chewaskulyong B, Lee KH, Dechaphunkul A, Imamura F, Nogami N, Kurata T, et al. Osimertinib in untreated EGFR-mutated advanced non-small-cell lung cancer. N Engl J Med. 2018;378:113–125. doi: 10.1056/NEJMoa1713137. [DOI] [PubMed] [Google Scholar]
- 26.Santarpia M, Liguori A, Karachaliou N, Gonzalez-Cao M, Daffinà MG, D'Aveni A, Marabello G, Altavilla G, Rosell R. Osimertinib in the treatment of non-small-cell lung cancer: Design, development and place in therapy. Lung Cancer (Auckl) 2017;8:109–125. doi: 10.2147/LCTT.S119644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Piotrowska Z, Sequist LV. Epidermal growth factor receptor-mutant lung cancer: New drugs, new resistance mechanisms, and future treatment options. Cancer J. 2015;21:371–377. doi: 10.1097/PPO.0000000000000147. [DOI] [PubMed] [Google Scholar]
- 28.Yu HA, Riely GJ, Lovly CM. Therapeutic strategies utilized in the setting of acquired resistance to EGFR tyrosine kinase inhibitors. Clin Cancer Res. 2014;20:5898–5907. doi: 10.1158/1078-0432.CCR-13-2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rotow J, Bivona TG. Understanding and targeting resistance mechanisms in NSCLC. Nat Rev Cancer. 2017;17:637–658. doi: 10.1038/nrc.2017.84. [DOI] [PubMed] [Google Scholar]
- 30.Yao Z, Fenoglio S, Gao DC, Camiolo M, Stiles B, Lindsted T, Schlederer M, Johns C, Altorki N, Mittal V, et al. TGF-beta IL-6 axis mediates selective and adaptive mechanisms of resistance to molecular targeted therapy in lung cancer. Proc Natl Acad Sci USA. 2010;107:15535–15540. doi: 10.1073/pnas.1009472107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nilsson MB, Sun H, Diao L, Tong P, Liu D, Li L, Fan Y, Poteete A, Lim SO, Howells K, et al. Stress hormones promote EGFR inhibitor resistance in NSCLC: Implications for combinations with β-blockers. Sci Transl Med. 2017;9(pii):eaao4307. doi: 10.1126/scitranslmed.aao4307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Levine B, Kroemer G. Autophagy in aging, disease and death: The true identity of a cell death impostor. Cell Death Differ. 2009;16:1–2. doi: 10.1038/cdd.2008.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang Z, Klionsky DJ. Eaten alive: A history of macroautophagy. Nat Cell Biol. 2010;12:814–822. doi: 10.1038/ncb0910-814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cuervo AM, Wong E. Chaperone-mediated autophagy: Roles in disease and aging. Cell Res. 2014;24:92–104. doi: 10.1038/cr.2013.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.He C, Klionsky DJ. Regulation mechanisms and signaling pathways of autophagy. Annu Rev Genet. 2009;43:67–93. doi: 10.1146/annurev-genet-102808-114910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang Z, Guo M, Zhao S, Xu W, Shao J, Zhang F, Wu L, Lu Y, Zheng S. The update on transcriptional regulation of autophagy in normal and pathologic cells: A novel therapeutic target. Biomed Pharmacother. 2015;74:17–29. doi: 10.1016/j.biopha.2015.06.003. [DOI] [PubMed] [Google Scholar]
- 38.Jing Z, Han W, Sui X, Xie J, Pan H. Interaction of autophagy with microRNAs and their potential therapeutic implications in human cancers. Cancer Lett. 2015;356:332–338. doi: 10.1016/j.canlet.2014.09.039. [DOI] [PubMed] [Google Scholar]
- 39.Li L, Chen H, Gao Y, Wang YW, Zhang GQ, Pan SH, Ji L, Kong R, Wang G, Jia YH, et al. Long noncoding RNA MALAT1 promotes aggressive pancreatic cancer proliferation and metastasis via the stimulation of autophagy. Mol Cancer Ther. 2016;15:2232–2243. doi: 10.1158/1535-7163.MCT-16-0008. [DOI] [PubMed] [Google Scholar]
- 40.Su Z, Yang Z, Xu Y, Chen Y, Yu Q. MicroRNAs in apoptosis, autophagy and necroptosis. Oncotarget. 2015;6:8474–8490. doi: 10.18632/oncotarget.3523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fu Z, Luo W, Wang J, Peng T, Sun G, Shi J, Li Z, Zhang B. Malat1 activates autophagy and promotes cell proliferation by sponging miR-101 and upregulating STMN1, RAB5A and ATG4D expression in glioma. Biochem Biophys Res Commun. 2017;492:480–486. doi: 10.1016/j.bbrc.2017.08.070. [DOI] [PubMed] [Google Scholar]
- 42.Wang K, Liu CY, Zhou LY, Wang JX, Wang M, Zhao B, Zhao WK, Xu SJ, Fan LH, Zhang XJ, et al. APF lncRNA regulates autophagy and myocardial infarction by targeting miR-188-3p. Nat Commun. 2015;6:6779. doi: 10.1038/ncomms7779. [DOI] [PubMed] [Google Scholar]
- 43.Chen ZH, Wang WT, Huang W, Fang K, Sun YM, Liu SR, Luo XQ, Chen YQ. The lncRNA HOTAIRM1 regulates the degradation of PML-RARA oncoprotein and myeloid cell differentiation by enhancing the autophagy pathway. Cell Death Differ. 2017;24:212–224. doi: 10.1038/cdd.2016.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Henson E, Chen Y, Gibson S. EGFR family members' regulation of autophagy is at a crossroads of cell survival and death in cancer. Cancers (Basel) 2017;9(pii):E27. doi: 10.3390/cancers9040027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nyfeler B, Eng CH. Revisiting autophagy addiction of tumor cells. Autophagy. 2016;12:1206–1207. doi: 10.1080/15548627.2016.1170265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Alexandrova AY, Kopnin PB, Vasiliev JM, Kopnin BP. ROS up-regulation mediates Ras-induced changes of cell morphology and motility. Exp Cell Res. 2006;312:2066–2073. doi: 10.1016/j.yexcr.2006.03.004. [DOI] [PubMed] [Google Scholar]
- 47.Zhou YY, Li Y, Jiang WQ, Zhou LF. MAPK/JNK signalling: A potential autophagy regulation pathway. Biosci Rep. 2015;35(pii):e00199. doi: 10.1042/BSR20140141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yip PY. Phosphatidylinositol 3-kinase-AKT-mammalian target of rapamycin (PI3K-Akt-mTOR) signaling pathway in non-small cell lung cancer. Transl Lung Cancer Res. 2015;4:165–176. doi: 10.3978/j.issn.2218-6751.2015.01.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jung CH, Seo M, Otto NM, Kim DH. ULK1 inhibits the kinase activity of mTORC1 and cell proliferation. Autophagy. 2011;7:1212–1221. doi: 10.4161/auto.7.10.16660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kim J, Kundu M, Viollet B, Guan KL. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol. 2011;13:132–141. doi: 10.1038/ncb2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ernst M, Putoczki TL. Stat3: Linking inflammation to (gastrointestinal) tumourigenesis. Clin Exp Pharmacol Physiol. 2012;39:711–718. doi: 10.1111/j.1440-1681.2011.05659.x. [DOI] [PubMed] [Google Scholar]
- 52.Demaria M, Camporeale A, Poli V. Stat3 and metabolism: How many ways to use a single molecule? Int J Cancer. 2014;135:1997–2003. doi: 10.1002/ijc.28767. [DOI] [PubMed] [Google Scholar]
- 53.Niso-Santano M, Shen S, Adjemian S, Malik SA, Mariño G, Lachkar S, Senovilla L, Kepp O, Galluzzi L, Maiuri MC, Kroemer G. Direct interaction between STAT3 and EIF2AK2 controls fatty acid-induced autophagy. Autophagy. 2013;9:415–417. doi: 10.4161/auto.22910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Luo B, Lin Y, Jiang S, Huang L, Yao H, Zhuang Q, Zhao R, Liu H, He C, Lin Z. Endoplasmic reticulum stress eIF2α-ATF4 pathway-mediated cyclooxygenase-2 induction regulates cadmium-induced autophagy in kidney. Cell Death Dis. 2016;7:e2251. doi: 10.1038/cddis.2016.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Oh HM, Yu CR, Golestaneh N, Amadi-Obi A, Lee YS, Eseonu A, Mahdi RM, Egwuagu CE. STAT3 protein promotes T-cell survival and inhibits interleukin-2 production through up-regulation of Class O Forkhead transcription factors. J Biol Chem. 2011;286:30888–30897. doi: 10.1074/jbc.M111.253500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Oh HM, Yu CR, Dambuza I, Marrero B, Egwuagu CE. STAT3 protein interacts with Class O Forkhead transcription factors in the cytoplasm and regulates nuclear/cytoplasmic localization of FoxO1 and FoxO3a proteins in CD4(+) T cells. J Biol Chem. 2012;287:30436–30443. doi: 10.1074/jbc.M112.359661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ray S, Zhao Y, Jamaluddin M, Edeh CB, Lee C, Brasier AR. Inducible STAT3 NH2 terminal mono-ubiquitination promotes BRD4 complex formation to regulate apoptosis. Cell Signal. 2014;26:1445–1455. doi: 10.1016/j.cellsig.2014.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kiprianova I, Remy J, Milosch N, Mohrenz IV, Seifert V, Aigner A, Kögel D. Sorafenib sensitizes glioma cells to the BH3 mimetic ABT-737 by targeting MCL1 in a STAT3-dependent manner. Neoplasia. 2015;17:564–573. doi: 10.1016/j.neo.2015.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sun L, Hu L, Cogdell D, Lu L, Gao C, Tian W, Zhang Z, Kang Y, Fleming JB, Zhang W. MIR506 induces autophagy-related cell death in pancreatic cancer cells by targeting the STAT3 pathway. Autophagy. 2017;13:703–714. doi: 10.1080/15548627.2017.1280217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tai WT, Shiau CW, Chen HL, Liu CY, Lin CS, Cheng AL, Chen PJ, Chen KF. Mcl-1-dependent activation of Beclin 1 mediates autophagic cell death induced by sorafenib and SC-59 in hepatocellular carcinoma cells. Cell Death Dis. 2013;4:e485. doi: 10.1038/cddis.2013.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yamada E, Bastie CC, Koga H, Wang Y, Cuervo AM, Pessin JE. Mouse skeletal muscle fiber-type-specific macroautophagy and muscle wasting are regulated by a Fyn/STAT3/Vps34 signaling pathway. Cell Rep. 2012;1:557–569. doi: 10.1016/j.celrep.2012.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nechemia-Arbely Y, Khamaisi M, Rosenberger C, Koesters R, Shina A, Geva C, Shriki A, Klaus S, Rosen S, Rose-John S, et al. In vivo evidence suggesting reciprocal renal hypoxia-inducible factor-1 upregulation and signal transducer and activator of transcription 3 activation in response to hypoxic and non-hypoxic stimuli. Clin Exp Pharmacol Physiol. 2013;40:262–272. doi: 10.1111/1440-1681.12064. [DOI] [PubMed] [Google Scholar]
- 63.Li M, Tan J, Miao Y, Lei P, Zhang Q. The dual role of autophagy under hypoxia-involvement of interaction between autophagy and apoptosis. Apoptosis. 2015;20:769–777. doi: 10.1007/s10495-015-1110-8. [DOI] [PubMed] [Google Scholar]
- 64.Abdul Rahim SA, Dirkse A, Oudin A, Schuster A, Bohler J, Barthelemy V, Muller A, Vallar L, Janji B, Golebiewska A, Niclou SP. Regulation of hypoxia-induced autophagy in glioblastoma involves ATG9A. Br J Cancer. 2017;117:813–825. doi: 10.1038/bjc.2017.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hsieh DJ, Kuo WW, Lai YP, Shibu MA, Shen CY, Pai P, Yeh YL, Lin JY, Viswanadha VP, Huang CY. 17β-estradiol and/or estrogen receptor β attenuate the autophagic and apoptotic effects induced by prolonged hypoxia through HIF-1α-mediated BNIP3 and IGFBP-3 signaling blockage. Cell Physiol Biochem. 2015;36:274–284. doi: 10.1159/000374070. [DOI] [PubMed] [Google Scholar]
- 66.Wilkinson S, Ryan KM. Growth factor signaling permits hypoxia-induced autophagy by a HIF1alpha-dependent, BNIP3/3L-independent transcriptional program in human cancer cells. Autophagy. 2009;5:1068–1069. doi: 10.4161/auto.5.7.9821. [DOI] [PubMed] [Google Scholar]
- 67.Karpathiou G, Sivridis E, Koukourakis M, Mikroulis D, Bouros D, Froudarakis M, Bougioukas G, Maltezos E, Giatromanolaki A. Autophagy and Bcl-2/BNIP3 death regulatory pathway in non-small cell lung carcinomas. APMIS. 2013;121:592–604. doi: 10.1111/apm.12026. [DOI] [PubMed] [Google Scholar]
- 68.Lai YC, Chuang YC, Chang CP, Yeh TM. Macrophage migration inhibitory factor has a permissive role in concanavalin A-induced cell death of human hepatoma cells through autophagy. Cell Death Dis. 2015;6:e2008. doi: 10.1038/cddis.2015.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li S, Xia Y, Chen K, Li J, Liu T, Wang F, Lu J, Zhou Y, Guo C. Epigallocatechin-3-gallate attenuates apoptosis and autophagy in concanavalin A-induced hepatitis by inhibiting BNIP3. Drug Des Devel Ther. 2016;10:631–647. doi: 10.2147/DDDT.S99420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Huang Z, Zhou L, Chen Z, Nice EC, Huang C. Stress management by autophagy: Implications for chemoresistance. Int J Cancer. 2016;139:23–32. doi: 10.1002/ijc.29990. [DOI] [PubMed] [Google Scholar]
- 71.Han W, Pan H, Chen Y, Sun J, Wang Y, Li J, Ge W, Feng L, Lin X, Wang X, et al. EGFR tyrosine kinase inhibitors activate autophagy as a cytoprotective response in human lung cancer cells. PLoS One. 2011;6:e18691. doi: 10.1371/journal.pone.0018691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Li YY, Lam SK, Mak JC, Zheng CY, Ho JC. Erlotinib-induced autophagy in epidermal growth factor receptor mutated non-small cell lung cancer. Lung Cancer. 2013;81:354–361. doi: 10.1016/j.lungcan.2013.05.012. [DOI] [PubMed] [Google Scholar]
- 73.Wang Z, Du T, Dong X, Li Z, Wu G, Zhang R. Autophagy inhibition facilitates erlotinib cytotoxicity in lung cancer cells through modulation of endoplasmic reticulum stress. Int J Oncol. 2016;48:2558–2566. doi: 10.3892/ijo.2016.3468. [DOI] [PubMed] [Google Scholar]
- 74.Zou Y, Ling YH, Sironi J, Schwartz EL, Perez-Soler R, Piperdi B. The autophagy inhibitor chloroquine overcomes the innate resistance of wild-type EGFR non-small-cell lung cancer cells to erlotinib. J Thorac Oncol. 2013;8:693–702. doi: 10.1097/JTO.0b013e31828c7210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Goldberg SB, Supko JG, Neal JW, Muzikansky A, Digumarthy S, Fidias P, Temel JS, Heist RS, Shaw AT, McCarthy PO, et al. A phase I study of erlotinib and hydroxychloroquine in advanced non-small-cell lung cancer. J Thorac Oncol. 2012;7:1602–1608. doi: 10.1097/JTO.0b013e318262de4a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sotelo J, Briceño E, López-González MA. Adding chloroquine to conventional treatment for glioblastoma multiforme: A randomized, double-blind, placebo-controlled trial. Ann Intern Med. 2006;144:337–343. doi: 10.7326/0003-4819-144-5-200603070-00008. [DOI] [PubMed] [Google Scholar]
- 77.Boone BA, Bahary N, Zureikat AH, Moser AJ, Normolle DP, Wu WC, Singhi AD, Bao P, Bartlett DL, Liotta LA, et al. Safety and biologic response of pre-operative autophagy inhibition in combination with gemcitabine in patients with pancreatic adenocarcinoma. Ann Surg Oncol. 2015;22:4402–4410. doi: 10.1245/s10434-015-4566-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rangwala R, Leone R, Chang YC, Fecher LA, Schuchter LM, Kramer A, Tan KS, Heitjan DF, Rodgers G, Gallagher M, et al. Phase I trial of hydroxychloroquine with dose-intense temozolomide in patients with advanced solid tumors and melanoma. Autophagy. 2014;10:1369–1379. doi: 10.4161/auto.29118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Rosenfeld MR, Ye X, Supko JG, Desideri S, Grossman SA, Brem S, Mikkelson T, Wang D, Chang YC, Hu J, et al. A phase I/II trial of hydroxychloroquine in conjunction with radiation therapy and concurrent and adjuvant temozolomide in patients with newly diagnosed glioblastoma multiforme. Autophagy. 2014;10:1359–1368. doi: 10.4161/auto.28984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.La Monica S, Galetti M, Alfieri RR, Cavazzoni A, Ardizzoni A, Tiseo M, Capelletti M, Goldoni M, Tagliaferri S, Mutti A, et al. Everolimus restores gefitinib sensitivity in resistant non-small cell lung cancer cell lines. Biochem Pharmacol. 2009;78:460–468. doi: 10.1016/j.bcp.2009.04.033. [DOI] [PubMed] [Google Scholar]
- 81.So KS, Kim CH, Rho JK, Kim SY, Choi YJ, Song JS, Kim WS, Choi CM, Chun YJ, Lee JC. Autophagosome-mediated EGFR down-regulation induced by the CK2 inhibitor enhances the efficacy of EGFR-TKI on EGFR-mutant lung cancer cells with resistance by T790M. PLoS One. 2014;9:e114000. doi: 10.1371/journal.pone.0114000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sano T, Takeuchi S, Nakagawa T, Ishikawa D, Nanjo S, Yamada T, Nakamura T, Matsumoto K, Yano S. The novel phosphoinositide 3-kinase-mammalian target of rapamycin inhibitor, BEZ235, circumvents erlotinib resistance of epidermal growth factor receptor mutant lung cancer cells triggered by hepatocyte growth factor. Int J Cancer. 2013;133:505–513. doi: 10.1002/ijc.28034. [DOI] [PubMed] [Google Scholar]
- 83.Rao S, Yang H, Penninger JM, Kroemer G. Autophagy in non-small cell lung carcinogenesis: A positive regulator of antitumor immunosurveillance. Autophagy. 2014;10:529–531. doi: 10.4161/auto.27643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Katheder NS, Khezri R, O'Farrell F, Schultz SW, Jain A, Rahman MM, Schink KO, Theodossiou TA, Johansen T, Juhász G, et al. Microenvironmental autophagy promotes tumour growth. Nature. 2017;541:417–420. doi: 10.1038/nature20815. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.