Short abstract
Advances in high-throughput sequencing have ushered in a new era of research into the gut microbiome and its role in human health and disease. However, due to the unique characteristics of microbiome survey data, their use for the detection of ecological interaction networks remains a considerable challenge, and a field of active methodological development. In this review, we discuss the landscape of existing statistical and experimental methods for detecting and characterizing microbial interactions, as well as the role that host and environmental metabolic signals play in mediating the behavior of these networks. Numerous statistical tools for microbiome network inference have been developed. Yet due to tool-specific biases, the networks identified by these methods are often discordant, motivating a need for the development of more general tools, the use of ensemble approaches, and the incorporation of prior knowledge into prediction. By elucidating the complex dynamics of the microbial interactome, we will enhance our understanding of the microbiome’s role in disease, more precisely predict the microbiome’s response to perturbation, and inform the development of future therapeutic strategies for microbiome-related disease.
Impact statement
This review provides a comprehensive description of experimental and statistical tools used for network analyses of the human gut microbiome. Understanding the system dynamics of microbial interactions may lead to the improvement of therapeutic approaches for managing microbiome-associated diseases. Microbiome network inference tools have been developed and applied to both cross-sectional and longitudinal experimental designs, as well as to multi-omic datasets, with the goal of untangling the complex web of microbe-host, microbe-environmental, and metabolism-mediated microbial interactions. The characterization of these interaction networks may lead to a better understanding of the systems dynamics of the human gut microbiome, augmenting our knowledge of the microbiome’s role in human health, and guiding the optimization of effective, precise, and rational therapeutic strategies for managing microbiome-associated disease.
Keywords: Microbiota, systems, statistics, experimental, models, gut
Introduction
By the time the initial phase of the Human Microbiome Project (HMP) drew to a close in 2014, it had become widely accepted that the human gut microbiome plays a dramatically underappreciated role in human health and disease. Similar international projects by the Beijing Genomics Institute, the American gut project, and the EU-funded MetaHIT have also punctuated growing worldwide interest in the human microbiome. The insights gained from these projects, along with advances in immunology, high-throughput metagenomic sequencing, and the development of statistical and computational tools for processing these data, have made large-scale analysis of microbial communities possible in a way they have never been before, spawning considerable excitement in the microbiome among scientists and nonscientists alike. Sometimes referred to as the “last organ”1 or the “forgotten organ,”2 the human microbiome could be considered one of the last active frontiers of human physiology.3
Recent work has drawn fascinating connections between changes in human microflora and a breadth of human diseases and conditions. Microbiota have been shown to play an important role in gastrointestinal and related diseases such as obesity,4,5 diarrhea,6 diabetes,7 irritable bowel syndrome (IBS),8 inflammatory bowel disease (IBD),9,10 and colorectal cancer.11 Even more surprising, researchers are beginning to identify unexpected associations between the gut microbiome and neurological disorders such as autism,12,13 schizophrenia,14 Parkinson’s disease,15 as well as depression.16 While in many cases, the mechanisms of such associations remain murky, there are indications that therapeutic interventions such as fecal transplants and probiotics may be effective in reducing the symptoms of many of these disorders.17–19 Yet in clinical trials, many probiotics have failed to produce positive results, for conditions including eczema,20 diarrhea,21 and gastroenteritis.22,23 Critically absent from the design of these clinical trials is an adequate understanding of how probiotic therapies affect the microbiome on a systems-level, which would ostensibly guide species selection, dosing regimens, and even the engineering of healthier gut microbiomes.
Put another way, our current knowledge of both commensal and pathogenic microbes remains primarily restricted to pairwise interactions. While the behaviors and mechanisms of specific organisms have become well documented, a thorough characterization of the multispecies interaction network and its dynamics remains elusive. In addition to providing valuable insight into the biological function and significance of specific species, a more comprehensive and quantitative map of microbiome interactions will lead to a more detailed and systemic understanding of the ways that shifts in the composition of the microbiome can shape human health. Such knowledge will facilitate the identification of novel therapeutic interventions and inform the rational design of treatment regimens. Ultimately, a complete and quantitative understanding of the gut microbiome’s interaction dynamics will allow more precise manipulations, with the ultimate goal of engineering healthcare solutions to microbiome-associated diseases. It is therefore important to define the behavior and function of the human microbiome using a systems-biology approach, by refocusing experimental and analytical strategies on multivariate interactions between species.
The mapping of many canonical human gene pathways was established through careful experimentation over several decades, and these have been validated through computational modeling, network inference, and other tools from systems biology. Such networks have been constructed effectively using a variety of established statistical approaches, such as Bayesian networks, neural networks, and graphical Gaussian models.24,25 Yet due to inherent differences in the way that microbial survey data are collected and reported, many of these strategies have proven inadequate or inapplicable in the context of microbial network inference. This is due in part to the fact that the number of reads identified by 16S or shotgun metagenomic sequencing varies independently of overall microbial abundance. As a result, microbiome data for a particular sample are typically presented as relative abundances, or “compositions” which sum to one.26,27 Additionally, because microbial content varies between samples and the abundance of some microorganisms is often below the limit of detection, microbiome sample data contain a large portion of zeros, and are therefore highly sparse. These characteristics of microbiome survey data mean that standard methods of analyzing multivariate data are likely to be ineffectual and statistically untenable.28–30
Although the microbiome field has seen experimental methods, computational tools, and available data proliferate enormously over the last decade, statistical and experimental methods for microbial network inference remain under active development. As these networks are developed, a more comprehensive understanding of the gut microbial ecosystems will emerge, providing new opportunities for precisely and predictably altering the human microbiome. In this mini-review, we will summarize various statistical and experimental approaches to mapping and analyzing microbial interaction networks. In doing so, we will discuss some of the prominent challenges and directions for improvement that must be considered as the field of systems microbiology develops.
Ecology meets network theory
Ecological relationships in the microbial interactome can be generalized using network theory, a set of mathematical concepts describing relationships between discrete entities. A network essentially consists of a set of “nodes,” which are interconnected by “edges.” Applied to microbial ecology, the nodes of a microbial interaction network represent species or operational taxonomic units (OTUs), while the edges denote functional interactions between them. Although the mechanisms of these microbial interactions can be extraordinarily complex, they can still be characterized using familiar ecological terminology.
The nature of an ecological relationship between microbes is typified by the harmful or beneficial growth-rate effect that each microbe has on its interaction partner. Microbes can have a net negative or positive impact on one another by producing or consuming resources, but also by manipulating their environment, such as through modulations in pH.31 Microbes competing for metabolites and macromolecules have a mutually negative effect on one another (competition), while interaction partners producing mutually beneficial metabolites or environmental conditions both benefit (mutualism). They can also exhibit opposite effects on one another, such as in predator–prey relationships, in which one interactor benefits, while the other suffers (parasitism). Lastly, interactions can occur in which one microbe is unaffected, while the other is exclusively helped (commensalism) or harmed (amensalism). Networks are useful ways to model these forms of ecological relationships between microorganisms.
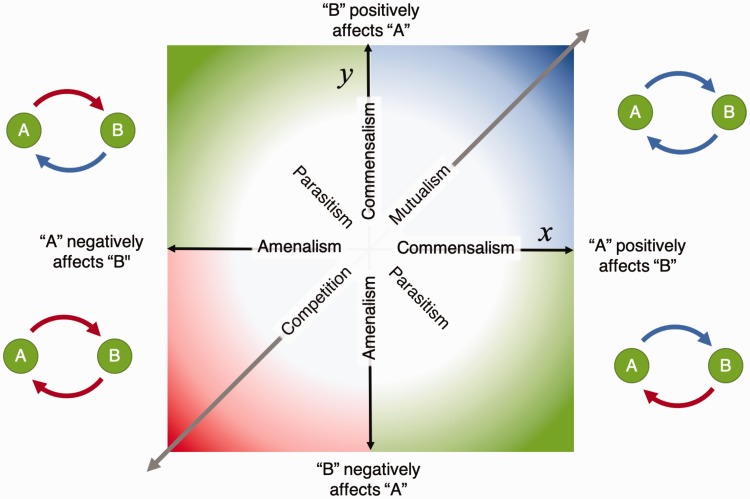
Typically, the ecological effect of one microorganism on another can be described by the sign of the interaction (e.g. positive, negative, or neutral) and the magnitude of the interaction (e.g. strong, weak). The bidirectional ecological relationship between microbes can thus be described using a coordinate pair (x, y) on a Cartesian grid (Figure 1), where x represents the net effect of microorganism A on microorganism B, and y represents the net effect of microorganism A on microorganism B. As reviewed by Faust et al.,32 this mathematical framework thus analogizes the five familiar ecological interaction mechanisms, wherein microbes exert mutual effects on one another: competition (–,–), mutualism (+,+), parasitism (+,–), commensalism (+,0), and amensalism (0,–). Each of these network formalisms have interpretable graphical representations, which are shown in Figure 1.
Figure 1.
The Cartesian plane of pairwise ecological interactions. Ecological interactions between pairs of microorganisms can be characterized by an (x, y) coordinate pair on a Cartesian plane, where x denotes the effect of microbe A on microbe B, and y characterize the effect of microbe B on microbe A. The sign of x and y denotes whether the effect of the interaction is positive or negative, while the magnitude denotes the strength of the effect. Classically defined ecological relationships of mutualism (+,+), competition (–,–), and parasitism (+,–) fall into the four quadrants, while amensalism (–,0) and commensalism (+,–), in which one organism is unaffected, lie along the axes. Weighted, undirected ecological networks only capture mutually positive or negative relationships such as competition and mutualism, and are therefore constrained along the diagonal, such that x = y.
These bidirectional ecological interactions fit nicely into mathematical framework of networks, allowing further characterization of network models of ecological interactions. In graph and network theory, networks are typified by the edges they contain. A network is called “weighted,” if we can quantify the strength or magnitude of a given interaction (Figure 2(a)), and “signed” if the weights can take on both positive and negative values (Figure 2(b)). A weighted, signed network is classified as “directed” if the relationships can be described in terms of source and target (or cause and effect) using the aforementioned coordinate pair (x, y). Directed edges are typically represented by arrows that designate the source and target of an interaction (Figure 2(c)). Undirected networks, however, merely describe mutually positive or negative ecological relationships such as mutualism or competition, but do not delineate the direction of causality for either interactor, rendering commensalism indistinguishable from mutualism, amensalism indistinguishable from competition, and the presence of parasitism largely ambiguous. Only directed networks that are weighted and signed are capable of describing all five forms of ecological interactions mentioned above.
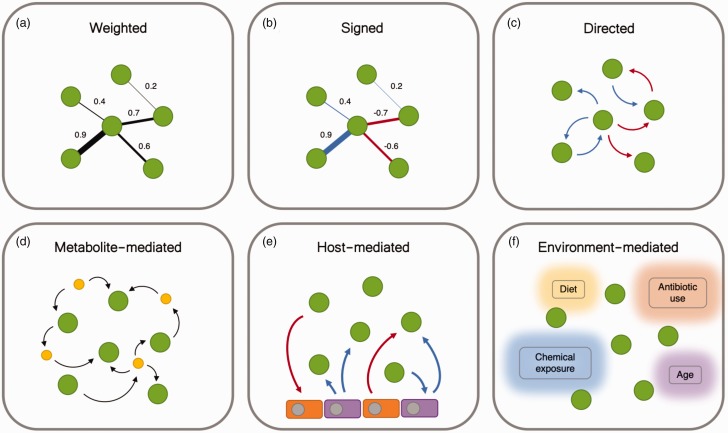
Figure 2.
Network abstractions of the microbial interactome. (a) Weighted networks characterize the strength of an interaction, but do not indicate whether the interaction is mutually positive or negative. Interactions characterized by non-linear relationships take this form. (b) Signed microbial interaction networks denote both the strength and direction of correlations between microbes, but do not indicate a causal relationship. Such networks are typically produced from cross-sectional data. (c) Directed microbial networks characterize source and target of an interaction, indicating a causal relationship. Such networks can be described using the ecological terms in Figure 1, and are typically produced from longitudinal (time-series) data. (d) Interactions between microorganisms are largely mediated by metabolites and macromolecules, which may be consumed or produced as a food source or waste. (e) Host cells play an important role in the microbial ecosystem. Host cells may affect the growth of microbes by secreting metabolites or antibiotics. Microbes break down and produce metabolites and macromolecules like short-chain fatty acids, which act as an energy source, and promote the differentiation of host cells in turn. (f) Environment-mediated microbial interaction networks contain context-dependent edges. These variables may conditionally alter the topology and dynamics of the microbial interaction networks.
While the concepts of ecological interactions are straightforward in principle, their precise detection of ecological interactions in experimental data remains a significant challenge in the field of microbial network inference.32 When network inference is the goal of microbial data acquisition, the experimental design depends largely on whether one’s objective is to construct a directed or undirected network. Approaches for identifying interactions in gut microbial ecosystems can thus be broadly classified by their underlying experimental design. Cross-sectional microbiome data, which consists of static snapshots of multiple individuals, can be used to detect or predict interactions, while longitudinal data, which involves repeated time-series measurements of one or more individuals, can be used to clarify the ecological mechanisms of such interactions. Broadly speaking, undirected, signed, and weighted microbial interaction networks can be inferred from cross-sectional sampling of metagenomic data, while directed network inference requires the collection of time-series, or longitudinal data.33,34
Cross-sectional methods for detecting microbial interactions
Undirected, weighted interaction networks, which may indicate positive or negative associations but not causal relationships, can be constructed using a variety of methods. Broadly speaking, these statistical methods are classified as parametric if they assume adherence to a particular statistical model, or non-parametric if they do not. The simplest and most familiar way to quantify the strength of interactions is using their correlation, and in most data analysis pipelines, the standard parametric statistic for calculating correlation is covariance. While the computation of covariance itself is straightforward under normal conditions, doing so for microbiome data remains a substantial challenge. Being compositional, data describing the proportions of species in microbial surveys are normalized such that the total abundance of a sample sums to a constant value. The result of this normalization is that an increase in the proportion of Firmicutes, all else held equal, is inherently coupled with an apparent decrease in the proportion of all other phyla, resulting in spurious negative correlations by many common statistical methods.35 Biases stemming from compositional effects and sparsity plague standard covariance metrics in these cases, motivating the development of alternative statistical methods for estimating covariance in compositional microbiome data.28,29 Since the overall microbial load of these populations cannot be measured directly, special statistical methods for estimating the covariance matrix must be used instead of calculating it directly.
A handful of parametric statistical methods for inferring the true covariance matrix in compositional microbiome data have been implemented as software programs, many of which are summarized in Table 1. One of the earliest such methods, SparCC,30 estimates the covariance using an iterative bootstrap selection procedure. Although designed to assume high data sparsity, it underperforms when sample diversity is high, and is prone to false negatives when the true number of true microbial interactions is large. Another approach, SPIEC-EASI,36 uses Aitchison’s centered log-ratio (CLR), a common compositional data transform, and performs well on datasets with high diversity. SPIEC-EASI has been expanded to allow for cross-domain associations, such as between bacteria and fungi.37 Other parametric methods use a regression method known as LASSO, a statistical technique that in this context, penalizes excessively complex microbial interaction networks. In particular, CCLasso38 and REBACCA39 show improved covariance estimation performance using this technique. Another LASSO-based method, BAnOCC,40 uses a Bayesian approach to estimate covariance, and thus has the benefit of providing uncertainty quantification for network predictions. Finally, MPLasso41 attempts to incorporate biological prior knowledge into its LASSO approach, by performing automated text-mining of PubMed abstracts to improve performance. Approaches that leverage the existing literature of microbe-microbe interactions are likely to be decisive in the construction and validation of microbial interaction networks.
Table 1.
Summary of statistical tools for inference of weighted, undirected microbial interaction networks from cross-sectional metagenomic data.
| SparCC | CCLasso | BAnOCC | MPLasso | SPIEC-EASI | CoNet | MIC | MENA | |
|---|---|---|---|---|---|---|---|---|
| Full name | Sparse correlations for compositional data | Correlation inference for compositional data through LASSO | Bayesian analysis of compositional covariance | Microbial prior LASSO | Sparse inverse covariance estimation for ecological association inference | Co-occurrence network analysis / renormalization and bootstrap | Mutual information coefficient | Molecular ecological network analysis / Random matrix theory |
| Implementation | Python | R | R | R | R | Cytoscape plugin | Java | Webapp |
| Description | Performs epirical covariance estimation using a bootstrapping procedure | Infers the correlation network for latent variables of compositional data using least squares L1 penalty | Estimates a sparse precision matrix through a LASSO prior and generates a posterior distribution by MCMC sampling | Integrates graphical LASSO of microbial co-occurrences with associations obtained from automated text mining of scientific literature | Infers underlying graphical model from conditional independence, using sparse neighborhood and inverse covariance selection | Uses an ensemble method based on multiple similarity measures in combination with generalized boosted linear models | A non-parametric method that detects various noisy distributions by data partitioning | Constructs ecological association networks through random matrix theory |
| Pros |
|
|
|
|
|
|
|
|
| Cons |
|
|
|
|
|
|
|
|
| References | Friedman and Alm30 | Fang and Deng,38 H. | Schwager and Huttenhower40 | Lo and Marculescu41 | Kurtz and Bonneau36 | Faust and Huttenhower47 | Reshef and Sabeti42 | Deng and Zhou46 |
Note: The table contains a brief description of the tool, the statistical methods underlying it, and some of the strengths and weaknesses of each approach.
Although these parametric methods for estimating covariance benefit from interpretability and utility for downstream data analysis, they, like direct covariance calculation methods, are only reliable for detecting linear dependencies between microbes.38 Other non-parametric strategies of identifying non-linear interactions between microbes have been proposed. For example, mutual information measures such as MIC42 rely on a measure of association borrowed from information theory to predict functional relationships between variables. A major advantage of MIC has over parametric methods is its ability to capture a broad range of non-linear microbial relationships. It does so by measuring the degree of noise present in potential interactions, rather than the shape of the interaction function itself. This approach revealed that co-exclusionary relationships represent a highly common association type between microbiota, as well as that many of the strongest non-linear relationships were dependent on external factors such as diet, sex, and collection method. Another non-parametric approach, LSA,43 was developed for identifying interactions among marine bacterioplankton, and is also capable of detecting non-linear dependencies between microbes. Although LSA was designed with a focus on time-series data, this method can generate undirected networks from static data if the time-delay parameter is set to zero. An expansion of this method, eLSA,44 was developed for time-series with replicates, as well as for approximating the statistical significance of its inferred relationships.45 Another non-parametric tool, MENA,46 was developed for characterizing microbial interactions in soil, and is highly robust to noise.34 MENA is based on methods from random matrix theory, a set of statistical tools borrowed from physics. Although non-parametric methods are capable of capturing noisy and highly non-linear microbial interactions, they do not necessarily indicate whether microorganisms are positively or negatively associated, and therefore may only capture the magnitude of an interaction. This means that while non-parametric methods may predict a broader range of microbial interactions, the nature of these interactions may be ambiguous or difficult to model.
While both parametric and non-parametric methods are capable of capturing broad ranges of ecological relationships, it is unlikely that any one method is general enough to detect them all, or even detect them with similar efficiency.42 Researchers must decide on which tool is right for their application, or otherwise rely on a combination of methods. A comprehensive meta-analysis of microbiome correlation metrics by Weiss et al.34 showed that precision of network inference could be dramatically improved by the use of ensembles. Because different statistical tools make different mistakes, relying on consensus across tools may be a powerful way to improve the accuracy of microbiome interaction networks. For example, CoNet47 combines multiple parametric and non-parametric similarity measures with generalized boosted linear models to predict microbial network interactions. As a result, it has a significantly lower false positive rate than other cross-sectional methods.34 CoNet has been used to identify interactions between species in the skin microbiome,48 as well as among pathobionts associated with cancer cachexia.49 Since it is integrated with the network visualization software Cytoscape, users quickly and easily construct and analyze microbial interaction networks
Despite the number of correlation methods available for inferring microbial interaction networks, there remains considerable room for improvement. The meta-analysis of eight such methods by Weiss et al.34 revealed an astonishing degree of variation in the sensitivity and precision of these tools. On average, methods shared less than a third of predicted interactions. Furthermore, they showed that precision depended greatly on the sequencing technology used, and that normalization choices have a significant impact on edge detection. Next, Weiss simulated several linear ecological relationships to compare tool detection performance. While nearly all methods surveyed were able to detect mutualism and commensalism, amensalism and parasitism were nearly undetectable by the majority of tools. Most concerning, however, was that precision was near zero for datasets with more than 50% sparsity. This suggests that abundance filtering is an important first step for detecting correlations between microbes, and that network inference for low-abundance OTUs remains an important area for improvement. Overall, they found that LSA, MIC, and SparCC were the most robust to distribution shape. SparCC performed best in cases where compositionality was high, while LSA did well at capturing both linear and non-linear ecological relationships, even under sparse conditions. LASSO-based methods were not tested, indicating that further work must be done to understand how the LASSO technique performs relative to previously described methods. However, the analysis did demonstrate that ensemble methods show promise in improving precision, particularly for highly sparse datasets. Until sufficiently general methods can be developed, combining methods with complementary strengths appears to be the best way to improve edge detection for microbial network inference.
Longitudinal methods for detecting microbial interactions
Fundamentally, all inference procedures for directed networks are concerned with determining causal relationships between discrete entities. Directed networks typically require longitudinal measurements to infer the source and target of a pairwise microbial relationship. Such methods are expected to be highly important for advancing dynamic models of microbial interactions,50 and may lead to highly precise manipulations of microbial ecosystems. Generally speaking, longitudinal data provide significantly more information on the dynamics of microbial interaction networks than cross-sectional methods. This is because even if the same number of replicates for a cross-sectional study is taken as timepoints for a longitudinal one, the ability to chronologically arrange discrete snapshots of microbial data allows the relational ordering of ecological events. Temporal tracking of blooms and busts in microbial populations thus facilitates the inference of directed microbial interaction networks, as time-delays can be used to indicate causal relationships. A summary of statistical tools for longitudinal network inference can be found in Table 2.
Table 2.
Summary of statistical tools for inference of directed microbial interaction networks from longitudinal metagenomic data.
| LSA | LIMITS | MetaMIS | MC-TIMME | MDSINE | TIME | |
|---|---|---|---|---|---|---|
| Full name | Local similarity analysis | Learning Interactions from microbial time series | Metagenomic microbial interaction simulator | Microbial counts and trajectories in infinite mixture model engine | Microbial dynamical systems inference engine | Temporal insights into microbial ecology |
| Implementation | R | Mathematica | Desktop app | Matlab | Matlab | Web app |
| Description | Detects complex, non-linear dependence associations between species and environmental factors without data reduction | Combines sparse linear regression with bootstrapping aggregation to infer a discrete-time Lotka–Volterra model | Uses an abundance-ranking strategy paired with partial least square regression to infer a discrete-time Lotka–Volterra model | Models time-varying counts of microbial taxa using an exponential relation process, coupled with adaptive Bayesian techniques | Provides a comprehensive toolbox for dynamical systems analysis of microbiota time-series to fit generalized Lotka–Volterra differential equations | Provides a workflows for time-series metagenomic data, including network inference using Granger LASSO causality |
| Pros |
|
|
|
|
|
|
| Cons |
|
|
|
|
|
|
| References | Ruan and Sun43 | Fisher and Mehta51 | Shaw and Wang52 | Gerber and Bry53 | Bucci and Gerber54 | Baksi and Mande55 |
Note: The table contains a brief description of the tool, how the statistical methods underlying it, and some of the strengths and weaknesses of each approach.
While true causality can only be determined using controlled experimentation,56 mathematical definitions of causality have been applied for predicting causal relationships from time-series data by comparing the histories of related entities.57,58 One statistical tool for causal inference is Granger causality,59 which was originally developed for economics but now been used extensively in neuroscience.56 For a pair of time series, X and Y, we say that X “Granger causes” Y if the histories X and Y together predict the current value of Y better than the history of Y alone. Popular in part due to its computational simplicity,60 this definition of causality has also proven helpful for inference of causal relationships in microbiome studies. TIME55 is a toolkit that provides a suite of analysis and visualization tools for microbial ecology analysis, and relies on a Granger-LASSO model to identify causal relationships.
Another way to construct directed microbial interaction networks from longitudinal data is to use goodness of fit to a defined model as evidence of causality. Perhaps the most straightforward model for time-dependent ecological modeling of microbiomes relies on generalized Lotka–Volterra (gLV) equations, which are commonly used to describe predator–prey interactions in ecology. This simple mathematical system was first developed by Lotka61 to describe autocatalytic chemical reactions, but was also derived independently by Volterra62 in early mathematical biology. Fundamentally, gLV defines the growth rate of a given organism as function of the abundances of all other organisms in a given ecosystem, producing a set of ordinary differential equations. Although these equations are best known for modeling macro-ecological systems, evidence suggests this framework may be applicable to microbiology as well. Here the network inference procedure involves deterministically estimating the interaction terms that determine the dynamics of pairwise ecological relationships, if any. One of the earliest gLV approaches to network inference of microbial time-series data came from a group using multilinear regression to identify interactions in cheese-making microbial communities.63 This inspired similar work to predict the gLV interaction terms of Clostridium difficile infection using regularized regression.64 Since then, a variety of gLV-based software tools have been developed for general applications to microbial network inference. For example, LIMITS51 uses sparse linear regression to determine the interaction coefficients for a gLV model of microbial interaction dynamics. To overcome the compositional effects associated with relative abundances, LIMITS uses a stepwise approach, iteratively adding edges that produce the lowest error. Another software platform, MetaMIS,52 relies on the use of a partial least square regression to identify the interaction terms, and is implemented as a graphical user interface with tools for network visualization. To maximize the identification of conserved interaction networks, MetaMIS uses an abundance-ranking strategy that prioritizes the identification of interactions between highly abundant microbes, although this strategy may overlook some novel interactions as a result. While gLV methods represent a powerful and ecologically relevant model, there are drawbacks to their use in microbiome network inference. The microbiome is subject to immigration of new species, spatial variability, and heterogeneity, characteristics that are not necessarily well modeled by gLV dynamics.65 Additionally, microbiota are understood to interact through a set of complex mechanisms, such as metabolic exchange, that may not be well modeled under this paradigm.66 And given that gLV models are occasionally unable to detect even pairwise interactions,67 the use of this framework may be questionable. Therefore, while the gLV equations are among the most well-characterized mathematical frameworks for modeling ecological interactions, network inference techniques based on this framework may not be able to fully capture the intricacies of gut microbiome dynamics. Indeed, there is a demonstrable need for network inference techniques that are able to reliably capture ecological relationships with more complex interaction mechanisms.
One promising area of development for network inference on time-series microbiome data involves the use of probabilistic time-series models. Like gLV, these tools are aimed at generating forecasting future behavior using a defined model but are typically better able to handle uncertainty. Such methods have been used extensively in genomics modeling,68–70 and have recently been expanded and adapted for the purpose of network inference on microbiome data. One such method is MC-TIMME,53 which uses a continuous-time dynamical model and a non-parametric Bayesian technique to identify interaction network. This tool performs OTU-level binning based on similarities in their temporal profiles, which allows improved estimations of the parameters regulating the dynamics of microbial interaction networks. Another probabilistic method, MDSINE,54 constitutes a comprehensive toolkit for dynamical systems inference, as multiple options for inference techniques provided. However, since MDSINE was designed for concentrations rather than relative abundances, it may give erroneous results in situations where overall microbial biomass does not remain constant, or cannot be otherwise measured. Many other approaches to probabilistic time-series modeling of microbial interactions use Dirichlet multinomial mixtures,71,72 which is a popular distribution for microbiome statistical analysis because it assumes that variables sum to certain value, and is therefore a natural way to model the compositional, relative abundances of microbes. Dynamic linear models, commonly used in commercial forecasting and control engineering,73 may also be useful for modeling microbiome dynamics. While both probabilistic time-series models and dynamic linear models are widely used techniques in other fields, they have only recently been applied to models of microbial dynamics. Hence, little information exists about the relative strengths of these approaches in the context of microbial modeling, demonstrating a need for analyses benchmarking the comparative performance of these tools against both synthetic and validated microbiome survey data. Additionally, the benefits of ensembles of time-series inference techniques should be evaluated, in light of the improvement Weiss et al.34 achieved by combining correlation measures with complementary strengths. As there is significant variation in the statistical approaches used to model longitudinal data, it is likely that ensemble approaches will similarly boost performance in the context of longitudinal models as well.
Indeed, dynamical systems and probabilistic models provide a powerful framework for inference of directed microbial interaction networks from longitudinal data.74–76 When applied correctly, such methods have significant advantages over cross-sectional or correlation-based approaches.30 By precisely defining the rules and parameters governing microbial interactions, inferred dynamical models are able to characterize the stability of an ecosystem in response to perturbation, and predict future behavior by simulating hypothetical ecosystems.58,77,78 Yet design of longitudinal microbiome studies is not without pitfalls: researchers must strike balance between the cost of data collection and the duration and frequency of sampling. This requires insight into the time scales in which microbial ecosystems fluctuate.78 Relative to macro-ecological systems, the time scale of microbial interactions is expected to be small,79 making acquisition of usable time-series data challenging or uninformative.80 Typically, longitudinal ex vivo studies sample participants on the order of days, while both in vivo and in vitro studies demonstrate that microbial dynamics likely operate on the scale of hours.81–83 Furthermore, given that metagenomic measurements are discrete and likely to capture steady-state behavior only,84 it may not be possible to capture microbiomes during state-transition, complicating network inference procedures on longitudinal data. While more development of statistical methods for time-series microbiome data is needed, optimization of the frequency and duration of sampling for longitudinal microbiome studies may be equally important.
The expanded universe of microbial interactions
While the inference of microbe-microbe interaction networks is a crucial step towards understanding the dynamics of the human microbiome, it somewhat abstracts the true mechanisms by which these interactions occur. Like macro-ecological systems, relationships between microbial species are largely dictated by their food source. While some bacteria, such as ciliates, consume other bacteria, most microbes consume metabolic byproducts excreted by their neighbors, giving rise to some of the ecological dynamics described in Figure 1. Mutualistic relationships such cross-feeding, or competition for metabolic resources are largely driven by the import and export of these compounds.85 Microbial interactions are mediated by metabolites and macromolecules that are broadly derived from three sources: (1) other endogenous microbiota, (2) endogenous host cells, and (3) exogenous environmental exposures, including dietary intake86,87 and chemical exposure.88 Graphical network representations of metabolite-mediated, host-mediated, and environment-mediated interactions are illustrated in Figure 2(d) to (f)).
The best approach to simultaneous measurement of the gut microbiome and the gut metabolic state is not obvious. The metabolic activity of microbiota can be assessed indirectly using shotgun metagenomic sequencing, by identifying marker genes associated with metabolic functions.89 A variety of software tools have been developed towards this end. For example, MicrobiomeAnalyst90 allows metabolic network visualization from metagenomic sequencing. Another network-based tool, PMRT,91 was developed to predict community metabolic functions from metagenomic data, and HUMAnN92 can also be used to infer the functional and metabolic potential of microbial metagenomes. If only 16S data are available, tools such as PICRUST,93 Tax4Fun,94 and PiPhillin95 can be used in combination with KEGG metabolic gene annotations to infer the metabolic state of a community. These approaches are limited, however, as meta-omics analyses have shown that marker genes for metabolism identified in metagenomic data may not be expressed,96,97 and genes inferred from 16S sequencing may not be present at all. Of course, the metabolic state of the gut can be measured directly, using methods such as nuclear magnetic resonance (NMR) spectroscopy and mass spectrometry (MS), which can then be integrated with microbiome survey data. Such multi-omics approaches, such as the one employed by Perez-Cobas et al.98 have potential to dramatically improve our understanding of metabolite-mediated microbial interactions. Methods for integrating such datasets, however, remain in their infancy. The aforementioned PRMT was extended to include integrated analysis of metagenomic and metabolomic data, using a tool called MIMOSA.99 Otherwise, multivariate correlation methods such as two-way orthogonal partial least squares (O2-PLS),100 canonical correlation analysis (CCA),101 and co-inertia analysis (CIA),102 may be applied to identify interactions between and within omics datasets.102,103 Identification of microbiome-metabolite interactions may also be guided by the literature. For example, NJS16,104 perhaps one of the most comprehensive maps of microbial-metabolite-host interaction networks, was constructed using an exhaustive review of experimental data and existing biological knowledge from the literature. As meta-omics experimental approaches improve, statistical tools for inferring microbiome-metabolite interaction networks must also be developed, integrated with prior knowledge, and tailored to the unique statistical properties of microbiome data.
Incorporation of host-microbiome dynamics into microbial interaction networks represents an even greater challenge. However, this will be an important step for the development of a comprehensive model of gut ecosystem dynamics. Microbes ferment short chain fatty acids, such as butyrate, which play important roles in host cell function, both as an energy source, and by regulating host gene expression and inflammatory response.105,106 Gut microbiota may also produce toxic metabolic byproducts, such as reactive oxygen species, that impair host cell function and promote disease.107 Conversely, host cells affect the metabolism of resident microorganisms. For example, goblet cells secrete mucin, while hepatocytes mediate glycine- and taurine-conjugated bile acid export.104 Microbe-interacting host cells are therefore influential components of the gut ecosystem, and models of microbial interaction networks will become more comprehensive with their inclusion. Although the metabolic dynamics of host cells has been reconstructed extensively108,109 in a number of model organisms,110 simultaneous measurement of microbial composition and host gene expression or host metabolism in humans is a challenge to perform non-invasively. One potential approach employed by Knight et al.111 involved extraction of RNA from infant stool samples containing both microbial populations and exfoliated epithelial cells. An in vitro approach using “artificial gut” microfluidics-based human-microbial co-culture systems such as HuMiX112 may also be a promising avenue for exploring host-microbiome interactions.113 Alternatively, model organisms may be used. In a review of this topic, Kostic et al.113 suggest the use of bobtail squid, Drosophila, zebrafish, and mice as alternatives to human models for the interrogation of host-microbiome dynamics. However, the clinical relevance of host-microbiome interaction networks developed using non-human experimental models remains circumspect. Even as technology for simultaneous measurement of human and microbial cellular populations improves, statistical approaches must continue to be developed for integrating omics datasets.
Lastly, it is well known than environmental factors shape microbial communities. Microbiome composition varies significantly according to age,114 geography,114,115 ethnicity,115 diet,116 social networks,117 and chemical exposure.88 Consequently, inferred microbial interaction networks are likely be significantly influenced by their environmental contexts. Diet in particular is an important environmental variable, as it strongly influences the metabolic environment of the gut microbiome.118 Identifying interactions between environmental characteristics and microbiome composition can be done using aforementioned multivariate approaches like CCA, O2-PLS, and CIA.119 Understanding how and when environmental factors will conditionally influence the topology of microbial interaction networks will be necessary not only to control for these factors, but also to understand the degree to which microbiome dynamics are context-dependent.
Discussion
Significant challenges remain in efforts to catalogue microbial interaction networks. The gut microbiome is composed of up to a thousand unique species, among which there is significant variation in metabolite consumption and production, in growth rate and conditions, and in their effects on and in response to host. The human microbiome forms a dense web of complex interactions, many of which are likely to be context-dependent. While a number of tools exist for inferring both undirected and directed interaction networks from cross-sectional and longitudinal studies, the predictions they generate are concerningly inconsistent. More work must be done to understand how and why these inconsistencies occur, as well as the design principles that drive them. There remains demand for sufficiently general tools that are designed with the unique statistical characteristics of microbiome data in mind. Furthermore, existing tools must be made sufficiently accessible to the research community, particularly those without a substantial background in systems biology or statistics.
Despite the apparent inconsistencies between networks drawn by the available inference tools, ensemble approaches show encouraging results for increasing the accuracy of predicted microbial interactions. Because different methods for inferring microbial interaction networks make different mistakes, precision may be improved greatly by the combined use of these tools. Additionally, it is likely that inference of the microbial interactome will be substantially improved by integrating existing statistical techniques with existing biological knowledge from the literature. As the field of microbiome research progresses, the manual curation of interaction networks, similar to KEGG for gene networks, will be highly beneficial. Finally, as we approach a more detailed understanding of microbial interaction dynamics, including the ways in which host-related, environmental, and metabolic factors influence these dynamics, it will become possible to make confident predictions about the effects of compositional changes to the microbial ecosystem, paving the way for novel and highly precise interventions for engineering healthier microbiomes.
ACKNOWLEDGEMENTS
The authors would like to thank Dr. Albert Siryaporn and Amanda Everett for their valuable comments and discussion during the preparation of this manuscript.
Authors’ contributions
AD researched and wrote the manuscript. XS contributed to the writing and advised on structure and content.
DECLARATION OF CONFLICTING INTERESTS
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
FUNDING
This work was supported by National Institutes of Health [R35GM122465]; and the National Science Foundation [DGE 1545220].
References
- 1.Baquero F, Nombela C. The microbiome as a human organ. Clin Microbiol Infect 2012; 18:2–4 [DOI] [PubMed] [Google Scholar]
- 2.O'Hara AM, Shanahan F. The gut flora as a forgotten organ. EMBO Rep 2006; 7:688–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hattori M, Taylor TD. The human intestinal microbiome: a new frontier of human biology. DNA Res 2009; 16:1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Perry RJ, Peng L, Barry NA, Cline GW, Zhang DY, Cardone RL, Petersen KF, Kibbey RG, Goodman AL, Shulman GI. Acetate mediates a microbiome-brain-beta-cell axis to promote metabolic syndrome. Nature 2016; 534:213–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, Sogin ML, Jones WJ, Roe BA, Affourtit JP, Egholm M, Henrissat B, Heath AC, Knight R, Gordon JI. A core gut microbiome in obese and lean twins. Nature 2009; 457:480–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pop M, Walker AW, Paulson J, Lindsay B, Antonio M, Hossain MA, Oundo J, Tamboura B, Mai V, Astrovskaya I, Bravo HC, Rance R, Stares M, Levine MM, Panchalingam S, Kotloff K, Ikumapayi UN, Ebruke C, Adeyemi M, Ahmed D, Ahmed F, Alam MT, Amin R, Siddiqui S, Ochieng JB, Ouma E, Juma J, Mailu E, Omore R, Morris JG, Breiman RF, Saha D, Parkhill J, Nataro JP, Stine OC. Diarrhea in young children from low-income countries leads to large-scale alterations in intestinal microbiota composition. Genome Biol 2014; 15:R76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gulden E, Wong FS, Wen L. The gut microbiota and type 1 diabetes. Clin Immunol 2015; 159:143–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jalanka-Tuovinen J, Salojarvi J, Salonen A, Immonen O, Garsed K, Kelly FM, Zaitoun A, Palva A, Spiller RC, de Vos WM. Faecal microbiota composition and host-microbe cross-talk following gastroenteritis and in postinfectious irritable bowel syndrome. Gut 2014; 63:1737–45 [DOI] [PubMed] [Google Scholar]
- 9.Morgan XC, Tickle TL, Sokol H, Gevers D, Devaney KL, Ward DV, Reyes JA, Shah SA, LeLeiko N, Snapper SB, Bousvaros A, Korzenik J, Sands BE, Xavier RJ, Huttenhower C. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol 2012; 13:R79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pascal V, Pozuelo M, Borruel N, Casellas F, Campos D, Santiago A, Martinez X, Varela E, Sarrabayrouse G, Machiels K, Vermeire S, Sokol H, Guarner F, Manichanh C. A microbial signature for Crohn's disease. Gut 2017; 66:813–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yu YN, Fang JY. Gut microbiota and colorectal cancer. Gastrointest Tumors 2015; 2:26–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mulle JG, Sharp WG, Cubells JF. The gut microbiome: a new frontier in autism research. Curr Psychiat Rep 2013; 15:337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li QR, Han Y, Dy ABC, Hagerman RJ. The Gut Microbiota and Autism Spectrum Disorders. Front Cell Neurosci 2017; 11:120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dickerson F, Severance E, Yolken R. The microbiome, immunity, and schizophrenia and bipolar disorder. Brain Behav Immun 2017; 62:46–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sampson TR, Debelius JW, Thron T, Janssen S, Shastri GG, Ilhan ZE, Challis C, Schretter CE, Rocha S, Gradinaru V, Chesselet MF, Keshavarzian A, Shannon KM, Krajmalnik-Brown R, Wittung-Stafshede P, Knight R, Mazmanian SK. Gut microbiota regulate motor deficits and neuroinflammation in a model of Parkinson's disease. Cell. 2016; 167:1469–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Winter G, Hart RA, Charlesworth RPG, Sharpley CF. Gut microbiome and depression: what we know and what we need to know. Rev Neurosci 2018; 29:629–43 [DOI] [PubMed] [Google Scholar]
- 17.Xu MQ, Cao HL, Wang WQ, Wang S, Cao XC, Yan F, Wang BM. Fecal microbiota transplantation broadening its application beyond intestinal disorders. World J Gastroenterol 2015; 21:102–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kang DW, Adams JB, Gregory AC, Borody T, Chittick L, Fasano A, Khoruts A, Geis E, Maldonado J, McDonough-Means S, Pollard EL, Roux S, Sadowsky MJ, Lipson KS, Sullivan MB, Caporaso JG, Krajmalnik-Brown R. Microbiota transfer therapy alters gut ecosystem and improves gastrointestinal and autism symptoms: an open-label study. Microbiome 2017; 5:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Evrensel A, Ceylan ME. Fecal microbiota transplantation and its usage in neuropsychiatric disorders. Clin Psychopharmacol Neurosci 2016; 14:231–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Allen SJ, Jordan S, Storey M, Thornton CA, Gravenor MB, Garaiova I, Plummer SF, Wang DL, Morgan G. Probiotics in the prevention of eczema: a randomised controlled trial. Arch Dis Child 2014; 99:1014–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Allen SJ, Wareham K, Wang DL, Bradley C, Hutchings H, Harris W, Dhar A, Brown H, Foden A, Gravenor MB, Mack D. Lactobacilli and bifidobacteria in the prevention of antibiotic-associated diarrhoea and Clostridium difficile diarrhoea in older inpatients (PLACIDE): a randomised, double-blind, placebo-controlled, multicentre trial. Lancet 2013; 382:1249–57 [DOI] [PubMed] [Google Scholar]
- 22.Schnadower D, Tarr PI, Charles CT, Gorelick MH, Dean MJ, O'Connell KJ, Mahajan P, Chun TH, Bhatt SR, Roskind CG, Powell EC, Rogers AJ, Vance C, Sapien RE, Gao F, Freedman SB. Randomised controlled trial of Lactobacillus rhamnosus (LGG) versus placebo in children presenting to the emergency department with acute gastroenteritis: the PECARN probiotic study protocol. BMJ Open 2017; 7:e018115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Freedman SB, Williamson-Urquhart S, Schuh S, Sherman PM, Farion KJ, Gouin S, Willan AR, Goeree R, Johnson DW, Black K, Schnadower D, Gorelick MH. Pediatric Emergency Research Canada Gastroenteritis Study G. Impact of emergency department probiotic treatment of pediatric gastroenteritis: study protocol for the PROGUT (probiotic regimen for outpatient gastroenteritis utility of treatment) randomized controlled trial. Trials 2014; 15:170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.De Smet R, Marchal K. Advantages and limitations of current network inference methods. Nat Rev Microbiol 2010; 8:717–29 [DOI] [PubMed] [Google Scholar]
- 25.Veiga DFT, Dutta B, Balazsi G. Network inference and network response identification: moving genome-scale data to the next level of biological discovery. Mol Biosyst 2010; 6:469–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lovell D, Muller W, Taylor J, Zwart A, Helliwell C. Proportions, percentages, ppm: do the molecular biosciences treat compositional data right? Composit Data Anal 2011;1:193–207 [Google Scholar]
- 27.Tsilimigras MCB, Fodor AA. Compositional data analysis of the microbiome: fundamentals, tools, and challenges. Ann Epidemiol 2016; 26:330–5 [DOI] [PubMed] [Google Scholar]
- 28.Aitchison J. The statistical-analysis of compositional data. J R Stat Soc B Met 1982; 44:139–77 [Google Scholar]
- 29.Li HZ. Microbiome, metagenomics, and high-dimensional compositional data analysis. Annu Rev Stat Appl 2015; 2:73–94 [Google Scholar]
- 30.Friedman J, Alm EJ. Inferring correlation networks from genomic survey data. Plos Comput Biol 2012; 8:e1002687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ilhan ZE, Marcus AK, Kang DW, Rittmann BE, Krajmalnik-Brown R. pH-mediated microbial and metabolic interactions in fecal enrichment cultures. mSphere. 2017; 2:e00047-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Faust K, Raes J. Microbial interactions: from networks to models. Nat Rev Microbiol 2012; 10:538–50 [DOI] [PubMed] [Google Scholar]
- 33.Xiao YD, Angulo MT, Friedman J, Waldor MK, Weiss ST, Liu YY. Mapping the ecological networks of microbial communities. Nat Commun 2017; 8:2042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weiss S, Van Treuren W, Lozupone C, Faust K, Friedman J, Deng Y, Xia LC, Xu ZZ, Ursell L, Alm EJ, Birmingham A, Cram JA, Fuhrman JA, Raes J, Sun F, Zhou J, Knight R. Correlation detection strategies in microbial data sets vary widely in sensitivity and precision. ISME J 2016; 10:1669–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pawlowsky-Glahn V, Egozcue JJ, Tolosana-Delgado R. Modeling and analysis of compositional data introduction. Stat Pract 2015;1:1–7 [Google Scholar]
- 36.Kurtz ZD, Muller CL, Miraldi ER, Littman DR, Blaser MJ, Bonneau RA. Sparse and compositionally robust inference of microbial ecological networks. PLoS Comput Biol 2015; 11:e1004226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tipton L, Muller CL, Kurtz ZD, Huang L, Kleerup E, Morris A, Bonneau R, Ghedin E. Fungi stabilize connectivity in the lung and skin microbial ecosystems. Microbiome 2018; 6:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fang H, Huang C, Zhao H, Deng M. CCLasso: correlation inference for compositional data through Lasso. Bioinformatics 2015; 31:3172–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ban Y, An L, Jiang H. Investigating microbial co-occurrence patterns based on metagenomic compositional data. Bioinformatics 2015; 31:3322–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schwager E, Mallick H, Ventz S, Huttenhower C. A Bayesian method for detecting pairwise associations in compositional data. PLoS Comput Biol 2017; 13:e1005852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lo C, Marculescu R. MPLasso: inferring microbial association networks using prior microbial knowledge. Plos Comput Biol. 2017; 13:e1005915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reshef DN, Reshef YA, Finucane HK, Grossman SR, McVean G, Turnbaugh PJ, Lander ES, Mitzenmacher M, Sabeti PC. Detecting novel associations in large data sets. Science 2011; 334:1518–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ruan Q, Dutta D, Schwalbach MS, Steele JA, Fuhrman JA, Sun F. Local similarity analysis reveals unique associations among marine bacterioplankton species and environmental factors. Bioinformatics 2006; 22:2532–8 [DOI] [PubMed] [Google Scholar]
- 44.Ki BM, Ryu HW, Cho KS. Extended local similarity analysis (eLSA) reveals unique associations between bacterial community structure and odor emission during pig carcasses decomposition. J Environ Sci Health A Tox Hazard Subst Environ Eng 2018; 53:718–27 [DOI] [PubMed] [Google Scholar]
- 45.Xia LC, Ai D, Cram J, Fuhrman JA, Sun F. Efficient statistical significance approximation for local similarity analysis of high-throughput time series data. Bioinformatics 2013; 29:230–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Deng Y, Jiang YH, Yang Y, He Z, Luo F, Zhou J. Molecular ecological network analyses. BMC Bioinform 2012; 13:113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Faust K, Sathirapongsasuti JF, Izard J, Segata N, Gevers D, Raes J, Huttenhower C. Microbial co-occurrence relationships in the human microbiome. PLoS Comput Biol 2012; 8:e1002606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cusco A, Belanger JM, Gershony L, Islas-Trejo A, Levy K, Medrano JF, Sanchez A, Oberbauer AM, Francino O. Individual signatures and environmental factors shape skin microbiota in healthy dogs. Microbiome 2017; 5:139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Potgens SA, Brossel H, Sboarina M, Catry E, Cani PD, Neyrinck AM, Delzenne NM, Bindels LB. Klebsiella oxytoca expands in cancer cachexia and acts as a gut pathobiont contributing to intestinal dysfunction. Sci Rep 2018; 8:12321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Caporaso JG, Lauber CL, Costello EK, Berg-Lyons D, Gonzalez A, Stombaugh J, Knights D, Gajer P, Ravel J, Fierer N, Gordon JI, Knight R. Moving pictures of the human microbiome. Genome Biol 2011; 12:R50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fisher CK, Mehta P. Identifying keystone species in the human gut microbiome from metagenomic timeseries using sparse linear regression. PLoS One 2014; 9:e102451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shaw GT, Pao YY, Wang D. MetaMIS: a metagenomic microbial interaction simulator based on microbial community profiles. BMC Bioinform 2016; 17:488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gerber GK, Onderdonk AB, Bry L. Inferring dynamic signatures of microbes in complex host ecosystems. PLoS Comput Biol 2012; 8:e1002624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bucci V, Tzen B, Li N, Simmons M, Tanoue T, Bogart E, Deng L, Yeliseyev V, Delaney ML, Liu Q, Olle B, Stein RR, Honda K, Bry L, Gerber GK. MDSINE: microbial dynamical systems inference engine for microbiome time-series analyses. Genome Biol 2016; 17:121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Baksi KD, Kuntal BK, Mande SS. ‘ TIME': a web application for obtaining insights into microbial ecology using longitudinal microbiome data. Front Microbiol 2018; 9:36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gourevitch B, Bouquin-Jeannes RL, Faucon G. Linear and nonlinear causality between signals: methods, examples and neurophysiological applications. Biol Cybern 2006; 95:349–69 [DOI] [PubMed] [Google Scholar]
- 57.Gibbons SM, Kearney SM, Smillie CS, Alm EJ. Two dynamic regimes in the human gut microbiome. PLoS Comput Biol 2017; 13:e1005364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sugihara G, May R, Ye H, Hsieh CH, Deyle E, Fogarty M, Munch S. Detecting causality in complex ecosystems. Science 2012; 338:496–500 [DOI] [PubMed] [Google Scholar]
- 59.Granger CWJ. Investigating causal relations by econometric models and cross-spectral methods. Econometrica 1969; 37:424–38 [Google Scholar]
- 60.Eichler M. Causal inference with multiple time series: principles and problems. Philos Trans R Math Phys Eng Sci 2013; 371:20110613. [DOI] [PubMed] [Google Scholar]
- 61.Lotka AJ. Contribution to the theory of periodic reactions. J Phys Chem 1910; 14:271–4 [Google Scholar]
- 62.Volterra V. Fluctuations in the abundance of a species considered mathematically. Nature 1926; 118:558–60 [Google Scholar]
- 63.Goerges S, Mounier J, Rea MC, Gelsomino R, Heise V, Beduhn R, Cogan TM, Vancanneyt M, Scherer S. Commercial ripening starter microorganisms inoculated into cheese milk do not successfully establish themselves in the resident microbial ripening consortia of a South German red smear cheese. Appl Environ Microbiol 2008; 74:2210–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Stein RR, Bucci V, Toussaint NC, Buffie CG, Ratsch G, Pamer EG, Sander C, Xavier JB. Ecological modeling from time-series inference: insight into dynamics and stability of intestinal microbiota. Plos Comput Biol 2013; 9:e1003388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gonze D, Coyte KZ, Lahti L, Faust K. Microbial communities as dynamical systems. Curr Opin Microbiol 2018; 44:41–9 [DOI] [PubMed] [Google Scholar]
- 66.Moree WJ, Phelan VV, Wu CH, Bandeira N, Cornett DS, Duggan BM, Dorrestein PC. Interkingdom metabolic transformations captured by microbial imaging mass spectrometry. Proc Natl Acad Sci U S A 2012; 109:13811–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Momeni B, Xie L, Shou WY. Lotka-Volterra pairwise modeling fails to capture diverse pairwise microbial interactions. Elife 2017; 6:e25051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bar-Joseph Z, Gitter A, Simon I. Studying and modelling dynamic biological processes using time-series gene expression data. Nat Rev Genet 2012; 13:552–64 [DOI] [PubMed] [Google Scholar]
- 69.Bonneau R, Reiss DJ, Shannon P, Facciotti M, Hood L, Baliga NS, Thorsson V. The Inferelator: an algorithm for learning parsimonious regulatory networks from systems-biology data sets de novo. Genome Biol 2006; 7:R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Aijo T, Butty V, Chen Z, Salo V, Tripathi S, Burge CB, Lahesmaa R, Lahdesmaki H. Methods for time series analysis of RNA-seq data with application to human Th17 cell differentiation. Bioinformatics 2014; 30:i113–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Aijo T, Muller CL, Bonneau R. Temporal probabilistic modeling of bacterial compositions derived from 16S rRNA sequencing. Bioinformatics 2018; 34:372–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chen J, Li H. Variable selection for sparse dirichlet-multinomial regression with an application to microbiome data analysis. Ann Appl Stat 2013; 7:418-442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.West M, Harrison J. Bayesian forecasting and dynamic models. 2nd ed New York: Springer, 1997, pp.xiv–680 [Google Scholar]
- 74.Stein RR, Bucci V, Toussaint NC, Buffie CG, Ratsch G, Pamer EG, Sander C, Xavier JB. Ecological modeling from time-series inference: insight into dynamics and stability of intestinal microbiota. Plos Comput Biol 2013; 9:e1003388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Marino S, Baxter NT, Huffnagle GB, Petrosino JF, Schloss PD. Mathematical modeling of primary succession of murine intestinal microbiota. Proc Natl Acad Sci U S A 2014; 111:439–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Buffie CG, Bucci V, Stein RR, McKenney PT, Ling L, Gobourne A, No D, Liu H, Kinnebrew M, Viale A, Littmann E, van den Brink MR, Jenq RR, Taur Y, Sander C, Cross JR, Toussaint NC, Xavier JB, Pamer EG. Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature 2015; 517:205–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bucci V, Xavier JB. Towards predictive models of the human gut microbiome. J Mol Biol 2014; 426:3907–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gerber GK. The dynamic microbiome. FEBS Lett 2014; 588:4131–9 [DOI] [PubMed] [Google Scholar]
- 79.Datta MS, Sliwerska E, Gore J, Polz MF, Cordero OX. Microbial interactions lead to rapid micro-scale successions on model marine particles. Nat Commun 2016; 7:11965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Angulo MT, Moreno JA, Lippner G, Barabasi AL, Liu YY. Fundamental limitations of network reconstruction from temporal data. J R Soc Interf 2017; 14:pii: 20160966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Liang X, Bushman FD, FitzGerald GA. Rhythmicity of the intestinal microbiota is regulated by gender and the host circadian clock. Proc Natl Acad Sci U S A 2015; 112:10479–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Trosvik P, Rudi K, Naes T, Kohler A, Chan KS, Jakobsen KS, Stenseth NC. Characterizing mixed microbial population dynamics using time-series analysis. ISME J 2008; 2:707–15 [DOI] [PubMed] [Google Scholar]
- 83.Trosvik P, Rudi K, Straetkvern KO, Jakobsen KS, Naes T, Stenseth NC. Web of ecological interactions in an experimental gut microbiota. Environ Microbiol 2010; 12:2677–87 [DOI] [PubMed] [Google Scholar]
- 84.Huttenhower C, Gevers D, Knight R, Abubucker S, Badger JH, Chinwalla AT, Creasy HH, Earl AM, FitzGerald MG, Fulton RS, Giglio MG, Hallsworth-Pepin K, Lobos EA, Madupu R, Magrini V, Martin JC, Mitreva M, Muzny DM, Sodergren EJ, Versalovic J, Wollam AM, Worley KC, Wortman JR, Young SK, Zeng QD, Aagaard KM, Abolude OO, Allen-Vercoe E, Alm EJ, Alvarado L, Andersen GL, Anderson S, Appelbaum E, Arachchi HM, Armitage G, Arze CA, Ayvaz T, Baker CC, Begg L, Belachew T, Bhonagiri V, Bihan M, Blaser MJ, Bloom T, Bonazzi V, Brooks JP, Buck GA, Buhay CJ, Busam DA, Campbell JL, Canon SR, Cantarel BL, Chain PSG, Chen IMA, Chen L, Chhibba S, Chu K, Ciulla DM, Clemente JC, Clifton SW, Conlan S, Crabtree J, Cutting MA, Davidovics NJ, Davis CC, DeSantis TZ, Deal C, Delehaunty KD, Dewhirst FE, Deych E, Ding Y, Dooling DJ, Dugan SP, Dunne WM, Durkin AS, Edgar RC, Erlich RL, Farmer CN, Farrell RM, Faust K, Feldgarden M, Felix VM, Fisher S, Fodor AA, Forney LJ, Foster L, Di Francesco V, Friedman J, Friedrich DC, Fronick CC, Fulton LL, Gao HY, Garcia N, Giannoukos G, Giblin C, Giovanni MY, Goldberg JM, Goll J, Gonzalez A, Griggs A, Gujja S, Haake SK, Haas BJ, Hamilton HA, Harris EL, Hepburn TA, Herter B, Hoffmann DE, Holder ME, Howarth C, Huang KH, Huse SM, Izard J, Jansson JK, Jiang HY, Jordan C, Joshi V, Katancik JA, Keitel WA, Kelley ST, Kells C, King NB, Knights D, Kong HDH, Koren O, Koren S, Kota KC, Kovar CL, Kyrpides NC, La Rosa PS, Lee SL, Lemon KP, Lennon N, Lewis CM, Lewis L, Ley RE, Li K, Liolios K, Liu B, Liu Y, Lo CC, Lozupone CA, Lunsford RD, Madden T, Mahurkar AA, Mannon PJ, Mardis ER, Markowitz VM, Mavromatis K, McCorrison JM, McDonald D, McEwen J, McGuire AL, McInnes P, Mehta T, Mihindukulasuriya KA, Miller JR, Minx PJ, Newsham I, Nusbaum C, O'Laughlin M, Orvis J, Pagani I, Palaniappan K, Patel SM, Pearson M, Peterson J, Podar M, Pohl C, Pollard KS, Pop M, Priest ME, Proctor LM, Qin X, Raes J, Ravel J, Reid JG, Rho M, Rhodes R, Riehle KP, Rivera MC, Rodriguez-Mueller B, Rogers YH, Ross MC, Russ C, Sanka RK, Sankar P, Sathirapongsasuti JF, Schloss JA, Schloss PD, Schmidt TM, Scholz M, Schriml L, Schubert AM, Segata N, Segre JA, Shannon WD, Sharp RR, Sharpton TJ, Shenoy N, Sheth NU, Simone GA, Singh I, Smillie CS, Sobel JD, Sommer DD, Spicer P, Sutton GG, Sykes SM, Tabbaa DG, Thiagarajan M, Tomlinson CM, Torralba M, Treangen TJ, Truty RM, Vishnivetskaya TA, Walker J, Wang L, Wang ZY, Ward DV, Warren W, Watson MA, Wellington C, Wetterstrand KA, White JR, Wilczek-Boney K, Wu YQ, Wylie KM, Wylie T, Yandava C, Ye L, Ye YZ, Yooseph S, Youmans BP, Zhang L, Zhou YJ, Zhu YM, Zoloth L, Zucker JD, Birren BW, Gibbs RA, Highlander SK, Methe BA, Nelson KE, Petrosino JF, Weinstock GM, Wilson RK, White O. Consortiu HMP. Structure, function and diversity of the healthy human microbiome. Nature 2012; 486:207–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Blaut M. Ecology and physiology of the intestinal tract. Curr Top Microbiol Immunol 2013; 358:247–72 [DOI] [PubMed] [Google Scholar]
- 86.Mackie RI, White BA, Isaacson RE. Gastrointestinal microbiology. New York: Chapman & Hall, 1997 [Google Scholar]
- 87.Phelan VV, Liu WT, Pogliano K, Dorrestein PC. Microbial metabolic exchange-the chemotype-to-phenotype link. Nat Chem Biol 2012; 8:26–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Rosenfeld CS. Gut dysbiosis in animals due to environmental chemical exposures. Front Cell Infect Microbiol 2017; 7:396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Chong J, Xia J. Computational approaches for integrative analysis of the metabolome and microbiome. Metabolites 2017; 7:pii: E62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Dhariwal A, Chong J, Habib S, King IL, Agellon LB, Xia JG. MicrobiomeAnalyst: a web-based tool for comprehensive statistical, visual and meta-analysis of microbiome data. Nucl Acids Res 2017; 45:W180–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Larsen PE, Collart F, Meyer F, Gilbert JA. Predicted relative metabolomic turnover predicting changes in the environmental metabolome from the metagenome. Bioinformatics 2011;1:337–45 [Google Scholar]
- 92.Abubucker S, Segata N, Goll J, Schubert AM, Izard J, Cantarel BL, Rodriguez-Mueller B, Zucker J, Thiagarajan M, Henrissat B, White O, Kelley ST, Methe B, Schloss PD, Gevers D, Mitreva M, Huttenhower C. Metabolic reconstruction for metagenomic data and its application to the human microbiome. Plos Comput Biol 2012; 8:e1002358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Langille MGI, Zaneveld J, Caporaso JG, McDonald D, Knights D, Reyes JA, Clemente JC, Burkepile DE, Thurber RLV, Knight R, Beiko RG, Huttenhower C. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol 2013; 31:814–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Asshauer KP, Wemheuer B, Daniel R, Meinicke P. Tax4Fun: predicting functional profiles from metagenomic 16S rRNA data. Bioinformatics 2015; 31:2882–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Iwai S, Weinmaier T, Schmidt BL, Albertson DG, Poloso NJ, Dabbagh K, DeSantis TZ. Piphillin: improved prediction of metagenomic content by direct inference from human microbiomes. PLoS One 2016; 11:e0166104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Franzosa EA, Morgan XC, Segata N, Waldron L, Reyes J, Earl AM, Giannoukos G, Boylan MR, Ciulla D, Gevers D, Izard J, Garrett WS, Chan AT, Huttenhower C. Relating the metatranscriptome and metagenome of the human gut. Proc Natl Acad Sci U S A 2014; 111:E2329–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Verberkmoes NC, Russell AL, Shah M, Godzik A, Rosenquist M, Halfvarson J, Lefsrud MG, Apajalahti J, Tysk C, Hettich RL, Jansson JK. Shotgun metaproteomics of the human distal gut microbiota. ISME J 2009; 3:179–89 [DOI] [PubMed] [Google Scholar]
- 98.Perez-Cobas AE, Gosalbes MJ, Friedrichs A, Knecht H, Artacho A, Eismann K, Otto W, Rojo D, Bargiela R, von Bergen M, Neulinger SC, Daumer C, Heinsen FA, Latorre A, Barbas C, Seifert J, dos Santos VM, Ott SJ, Ferrer M, Moya A. Gut microbiota disturbance during antibiotic therapy: a multi-omic approach. Gut 2013; 62:1591–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Noecker C, Eng A, Srinivasan S, Theriot CM, Young VB, Jansson JK, Fredricks DN, Borenstein E. Metabolic model-based integration of microbiome taxonomic and metabolomic profiles elucidates mechanistic links between ecological and metabolic variation. Msystems 2016; 1:e00013–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Trygg J, Wold S. O2-PLS, a two-block (X-Y) latent variable regression (LVR) method with an integral OSC filter. J Chemometrics 2003; 17:53–64 [Google Scholar]
- 101.Hotelling H. Relations between two sets of variates. Biometrika 1936; 28:321–77 [Google Scholar]
- 102.Doledec S, Chessel D. Co-inertia analysis – an alternative method for studying species environment relationships. Freshwater Biol 1994; 31:277–94 [Google Scholar]
- 103.Chong J, Xia JG. Computational approaches for integrative analysis of the metabolome and microbiome. Metabolites 2017; 7:pii:E62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Sung J, Kim S, Cabatbat JJT, Jang S, Jin YS, Jung GY, Chia N, Kim PJ. Global metabolic interaction network of the human gut microbiota for context-specific community-scale analysis. Nat Commun 2017; 8:15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Donohoe DR, Garge N, Zhang X, Sun W, O'Connell TM, Bunger MK, Bultman SJ. The microbiome and butyrate regulate energy metabolism and autophagy in the mammalian colon. Cell Metab 2011; 13:517–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Furusawa Y, Obata Y, Fukuda S, Endo TA, Nakato G, Takahashi D, Nakanishi Y, Uetake C, Kato K, Kato T, Takahashi M, Fukuda NN, Murakami S, Miyauchi E, Hino S, Atarashi K, Onawa S, Fujimura Y, Lockett T, Clarke JM, Topping DL, Tomita M, Hori S, Ohara O, Morita T, Koseki H, Kikuchi J, Honda K, Hase K, Ohno H. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature 2013; 504:446–50 [DOI] [PubMed] [Google Scholar]
- 107.Louis P, Hold GL, Flint HJ. The gut microbiota, bacterial metabolites and colorectal cancer. Nat Rev Microbiol 2014; 12:661–72 [DOI] [PubMed] [Google Scholar]
- 108.Thiele I, Swainston N, Fleming RM, Hoppe A, Sahoo S, Aurich MK, Haraldsdottir H, Mo ML, Rolfsson O, Stobbe MD, Thorleifsson SG, Agren R, Bolling C, Bordel S, Chavali AK, Dobson P, Dunn WB, Endler L, Hala D, Hucka M, Hull D, Jameson D, Jamshidi N, Jonsson JJ, Juty N, Keating S, Nookaew I, Le Novere N, Malys N, Mazein A, Papin JA, Price ND, Selkov E, Sr., Sigurdsson MI, Simeonidis E, Sonnenschein N, Smallbone K, Sorokin A, van Beek JH, Weichart D, Goryanin I, Nielsen J, Westerhoff HV, Kell DB, Mendes P, Palsson BO. A community-driven global reconstruction of human metabolism. Nat Biotechnol 2013; 31:419–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Thiele I, Palsson BO. A protocol for generating a high-quality genome-scale metabolic reconstruction. Nat Protoc 2010; 5:93–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Oberhardt MA, Palsson BO, Papin JA. Applications of genome-scale metabolic reconstructions. Mol Syst Biol 2009; 5:320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Knight JM, Davidson LA, Herman D, Martin CR, Goldsby JS, Ivanov IV. Donovan SM, Chapkin RS. Non-invasive analysis of intestinal development in preterm and term infants using RNA-Sequencing. Sci Rep 2014; 4:5453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Shah P, Fritz JV, Glaab E, Desai MS, Greenhalgh K, Frachet A, Niegowska M, Estes M, Jager C, Seguin-Devaux C, Zenhausern F, Wilmes P. A microfluidics-based in vitro model of the gastrointestinal human-microbe interface. Nat Commun 2016; 7: 11535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kostic AD, Howitt MR, Garrett WS. Exploring host-microbiota interactions in animal models and humans. Genes Dev 2013; 27:701–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, Magris M, Hidalgo G, Baldassano RN, Anokhin AP, Heath AC, Warner B, Reeder J, Kuczynski J, Caporaso JG, Lozupone CA, Lauber C, Clemente JC, Knights D, Knight R, Gordon JI. Human gut microbiome viewed across age and geography. Nature 2012; 486:222–2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Gaulke CA, Sharpton TJ. The influence of ethnicity and geography on human gut microbiome composition. Nat Med 2018; 24:1495–6 [DOI] [PubMed] [Google Scholar]
- 116.David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, Ling AV, Devlin AS, Varma Y, Fischbach MA, Biddinger SB, Dutton RJ, Turnbaugh PJ. Diet rapidly and reproducibly alters the human gut microbiome. Nature 2014; 505:559–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Tung J, Barreiro LB, Burns MB, Grenier JC, Lynch J, Grieneisen LE, Altmann J, Alberts SC, Blekhman R, Archie EA. Social networks predict gut microbiome composition in wild baboons. Elife 2015; 4:e05224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Zhang CH, Zhang MH, Wang SY, Han RJ, Cao YF, Hua WY, Mao YJ, Zhang XJ, Pang XY, Wei CC, Zhao GP, Chen Y, Zhao LP. Interactions between gut microbiota, host genetics and diet relevant to development of metabolic syndromes in mice. ISME J 2010; 4:312–3 [DOI] [PubMed] [Google Scholar]
- 119.Wang XH, Eijkemans MJC, Wallinga J, Biesbroek G, Trzcinski K, Sanders EAM, Bogaert D. Multivariate approach for studying interactions between environmental variables and microbial communities. PLoS One 2012; 7:e50267. [DOI] [PMC free article] [PubMed] [Google Scholar]