Short abstract
Gut bacterial microbiota is altered in patients with advanced renal disease and those on dialysis. However, it is not clear yet what bacterial composition changes are due to the renal insufficiency per se, and what are in result of the accompanying interventions and comorbid conditions. Most studies analyzed diabetic nephropathy, hypertensive nephropathy, and glomerulonephritis patients which might have directly influenced the microbiome regardless of alterations in renal function. We present in this report changes in gut bacterial microbiota in a highly selected group of patients with strict inclusion criteria to eliminate the effects of the confounding factors on the microbiome composition. We conducted multiple analysis approaches according to participants’ renal function to further understand microbiome alteration in different degrees of renal insufficiency. An interesting group of bacteria showed a step-wise change in relative abundance in response to the three groups’ analysis. These bacteria either decreased or increased from mild, moderate to severe renal insufficiency indicating strong and direct effects of the uremic milieu on its relative abundance. We also ran a sensitivity analysis that took into account an assembly of the significant taxa observed in an approach to investigate whether these taxa can fully explain the separation noted between the groups. We determined the projected metabolic pathways altered according to the gut microbiota composition changes. This report not only delineates with a higher certainty the effects of alteration in renal function on the microbiome, but also explores the possible role of dysbiosis on comorbid conditions through alterations in the projected metabolic pathways.
Impact statement
The heterogeneity of the renal disease, therapeutic interventions, and the original cause of the renal failure, all directly affect the microbiota. We delineate in this report the direct effect of decreased renal function on the bacterial composition following stringent criteria to eliminate the possibilities of other confounding factors and dissect the direct effects of the uremic milieu. We analyzed the microbiome following three different approaches to further evaluate the effects of mild, moderate and advanced renal insufficiency on the microbiome. We also present here a detailed functional analysis of the projected altered pathways secondary to changes in the microbiome composition.
Keywords: Polycystic kidney disease, gut bacterial microbiota, uremic milieu, PICRUSt analysis, renal insufficiency, hemodialysis
Introduction
The microbes residing within the human intestines have the ability to influence numerous aspects of human biology. Alterations in the function and composition of the gut microbial flora (microbiota) play a major role in the pathogenesis of diverse human illnesses such as chronic inflammation,1,2 diabetes mellitus,3–5 and cardiovascular diseases.6–8 Gut microbes provide protection against pathogenic organisms, contribute to energy metabolism, serve a clear role in the development and modulation of the human gut immune system, and participate in nitrogen and micronutrient homeostasis by synthesizing amino acids and various vitamins.9–14
We and other researchers have recently shown alterations in the bacterial gut microbiome in experimental animals models of chronic kidney disease (CKD)15,16 and patients with various degrees of renal insufficiency.17,18 It has been established that protein absorption in the small intestine is impaired in CKD.19 As a consequence, an increased amount of dietary protein becomes available to the colon. The reduction in the colonic carbohydrate-to-protein ratio may favor a shift from a saccharolytic to a proteolytic fermentation pattern, a shift accentuated by several other processes in CKD. Patients and animal models with advanced renal impairment will exhibit elevations in blood urea nitrogen (BUN) due to the decline in overall renal clearance of nitrogenous waste.20 Although the mechanisms are still unclear, increased BUN will result in increased intestinal urea content.15 This upsurge has been proposed to lead to high ammonia production from bacteria-mediated urea hydrolysis, which can raise luminal pH values and foster overgrowth of proteolytic species.16,21–23 A second CKD-associated process leading to dysbiosis may be prolonged colonic transit times.24,25 As a result, a larger part of the colon becomes carbohydrate-deprived, which will induce an expansion of proteolytic species.26,27 Earlier studies have confirmed the increased carbohydrate absorption and decreased carbohydrate fecal content in patients with increased colonic transit time.28–30
The challenge with identifying dysbiotic causality in CKD is the presence of many co-morbidities and therapeutic interventions. For example, diabetes and hypertension are the top two leading causes of CKD and end stage renal disease (ESRD) worldwide.31–36 Additionally, the vast majority of therapeutic strategies and dietary restrictions provided to patients with CKD can independently affect the composition of the gut microbiota.37–41 In an attempt to examine the independent impact of renal failure on the composition of gut microbiota, we studied patients with polycystic kidney disease (PKD). PKD is a hereditary disease involving a mutation in the gene encoding polycystin 1 and 2, which results in progressive cystic formation and structural damage to the kidneys. Compared to patients with renal failure due conditions such as diabetic nephropathy or glomerulonephritis, patients with PKD have less major co-morbid medical conditions or associated medical interventions (i.e. antimicrobial or anti-inflammatory therapies) that could potentially alter the gut microbiota. Therefore, we conducted a cross-sectional pilot study to evaluate colonic microbiome changes in PKD patients with varying stages of renal impairment.
Materials and methods
Study participants
This was an observational cross-sectional pilot study to evaluate the differences in gut bacterial microbiota composition in patients with PKD with varying degrees of renal insufficiency. The study was approved by the Icahn School of Medicine at Mount Sinai Institutional Review Board (IRB, GCO#13–1798). Patients with PKD who visited the Mount Sinai Medical Center between 2010 and 2015 were identified using the Icahn School of Medicine database registry. Full study protocol is available at the U.S. National Library of Medicine ClinicalTrials.gov (NCT02142101). Screening, IRB approval, and feasibility were first initiated on December 2013. Patient recruitment took place on May 2014 and was completed on January 2016.
Recruitment procedures, inclusion and exclusion criteria
This pilot observational, cross-sectional study was designed to evaluate changes in gut bacterial microbiota in patients with PKD and estimated glomerular filtration rate (eGFR) > 60 mL/min (group 1, CKD I–II), between 15–60 mL/min (group 2, CKD III–IV) and < 15 mL/min or on hemodialysis (HD) (group 3, CKD V-ESRD). GFR was estimated using the modification of diet in renal disease (MDRD) equation.42 Groups were enrolled to achieve a final participant ratio of 1:1:1. Patients who visited Mount Sinai Medical Center between 2010 and 2015 with a PKD diagnosis were contacted via phone and invited to participate in the study. A total of 1962 patients were identified. Due to the strict inclusion/exclusion criteria employed in this project, and to maintain the 1:1:1 ratio, we first identified and recruited PKD patients with CKD V-ESRD and then recruited the remaining two groups following the proposed ratio. Patients with advanced liver, cardiac, autoimmune disease or those with history of intra-abdominal surgery, small or large intestine resection, small bowel obstruction, colon cancer or gastrointestinal bleeding were all excluded. Patients who received oral multivitamins, vitamin D analogues, probiotics, prebiotics, antibiotics, immunosuppressive medications, steroids, or chemotherapy in the last three months prior to enrollment were also excluded. Patients receiving intravenous or oral iron supplementation, laxatives, or resins such as kayexalate in the last month before enrollment were excluded. Patients receiving phosphate binders were not excluded. Patients on peritoneal dialysis were not included in this study. Patient with mild polycystic liver changes with no liver dysfunction as measured by abnormal transaminase, elevated international normalized ratio (INR) were not excluded.
Stool sample collection and DNA extraction
Stool sample collections by the participants (and study personal) were conducted in the office. Participants were given a stool collection hat placed at the top of the toilet. Stool sample were collected using sterile tools, placed immediately in 50 mL conical tubes, and snap frozen in liquid nitrogen. Tubes were stored in −80°C until sample processing. DNA was extracted using PowerFecal® DNA isolation kits, according to the manufacturer protocol (MOBIO Laboratory Inc., QIAGEN Company, CA, USA). Fecal materials were obtained from three grossly firm stool areas absent of undigested food, mixed and homogenized on ice, then placed immediately in extraction tubes.
16S sequencing and annotation
16S sequencing and annotation was performed as described before.15,17 Briefly, metagenomic DNA was amplified using the 16S V3 (341F) forward and V4 (805R) reverse primer pairs with Illumina adapter overhang. Sequencing was performed using the MiSeq Reagent V3 Kit (Illumina) with 2 × 300 bp paired-end sequencing. Reads were joined using the default parameters in Paired-End reAd mergeR (PEAR v0.9.6),43 and filtered to retain sequences in which at least 90% of the bases have a quality score greater than Q30 using the FASTX Toolkit v0.0.13.44 Closed reference OTU picking was performed against the Green Genes v13.8 database.45
Laboratory measurements and evaluation of uremic symptoms
All measurements and clinical laboratory tests were obtained as standard of care during the clinic visits. Uremic symptoms were assessed by a modified Pittsburgh Symptom Score Index (PSSI) that included 10 physical symptoms checklist questionnaire (fatigue, trouble sleeping, difficulty concentrating, restless leg, changes in taste, loss of appetite, nausea or vomiting, pruritus, bone pain, muscle pain, or weakness).46–48 The PSSI gives a score from 0 (not at all) to 5 (very much).
Statistical analysis
To explore detailed changes in microbiota according to renal function, we performed multiple analyses using different eGFR cutoffs for groups’ allocations. We first performed all analyses on the previously mentioned three groups: eGFR > 60 mL/min, eGFR between 15–60 mL/min, and eGFR < 15 mL/min or on HD. Secondly, we divided the cohort into two groups based on the median eGFR and reran the analyses. Thirdly, we performed all analyses on HD patients in comparison to those not on HD. Operational taxonomic units (OTUs) tables were collapsed to the phyla, genera, and species levels and center log-ratio transformed for downstream analysis. OTU analysis was performed as described previously.15 In brief, ordination analyses and diversity measures were calculated using Python v3.5,49 and the SciKit-Bio library (Scikit-Bio (2015) Scikit-Bio. http://scikit-bio.org.). Data manipulation and statistical analysis was performed with Pandas v0.20.2,50 and numerical measures calculated using NumPy v1.12.1,51 and SciPy v0.19.0.52 Dimension reduction through principal component analysis (PCA) was performed using Scikit-Learn v0.18.1.53 To evaluate the significance and quantification of cluster separation in PCA plots, we used the Mahalanobis metric to quantify the distance between group centroids, and calculated a two-sample Hotelling's T2 statistic from the data. Then, an F-test is applied to determine the significance of the centroid separation.54 Clinical data are expressed as mean ± standard deviation (M ± SD). Microbiome data are presented as the mean of the center-log ratio (CLR) transformation. Two-sided unpaired t-tests were used to analyze data between two groups after determination of data distributions and variance. The ANOVA with Bonferroni correction was used when more than two groups were present. GraphPad Prism software was used for statistical analysis. P values less than 0.05 were considered to be statistically significant.
Functional profile of the microbial community
To evaluate the functional profile of the microbiome composition in PKD patients according to their renal function, the functional predictions were made against the Kyoto Encyclopedia of Genes and Genomes (KEGG).55,56 Orthology was assessed using Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (PICRUSt) version 1.1.13.57 Predictions were center-log ratio transformed to account for the compositional structure of the data, and significant orthologs were identified as those with a Student’s t-test P value < 0.05.58,59 Orthologs were subsequently amalgamated into functional pathways, which were similarly transformed and significance tested.
Results
Participants’ characteristics
We first contacted and recruited PKD patients with eGFR < 15 mL/min. Six patients fulfilled the inclusion criteria and agreed to participate in the study. We then contacted candidates in batches consisting of 50 PKD patients with eGFR between 15–60 mL/min, and 50 patients with eGFR >60 mL/min at any given time. We continued this process until we achieved the desired distribution of 1:1:1 between the groups (n = 6 each group). We recruited the first comers (fulfilling the inclusion/exclusion criteria) regardless of their age, gender and race. This resulted in a younger group 1 (eGFR > 15 mL/min) (Table 1). All group 3 participants (eGFR < 15 mL/min or on HD) were already on HD. The median eGFR in the entire cohort was 45 mL/min. Due to the small sample size, we were not able to detect/analyze the differences in PKD related polycystic liver disease events. However, no patient with abnormal liver function test as measured by abnormal transaminase, elevated international normalized ratio (INR) was recruited.
Table 1.
Participants characteristics according to their renal function.
| Group 1 | Group 2 | Group 3 | |
|---|---|---|---|
| Age (years) | 36.3 ± 8.9 | 56.5 ± 7.1** | 52.5 ± 5.4** |
| Height (cm) | 169 ± 6.7 | 167 ± 6.7 | 161 ± 3.4* |
| Weight (kg) | 72.7 ± 12.3 | 70.2 ± 15 | 70.6 ± 5.9 |
| BMI (kg/m2) | 25.1 ± 3.2 | 24.8 ± 4.3 | 27.2 ± 2.3 |
| Creatinine (mg/dL) | 1 ± 0.23 | 2.1 ± 0.85* | 7.7 ± 2.3*** |
| eGFR (mL/min) | 78.5 ± 16.4 | 32.8 ± 11.4*** | 6.8 ± 1.9*** |
| PSSI | 10.5 ± 9.5 | 6.8 ± 5.6 | 7.3 ± 4.3 |
| Male/Female (n) | 3/3 | 5/1 | 5/1 |
| Race (n) (AA/W/H) | 1/5/0 | 2/2/2 | 2/2/2 |
| HTN (yes/no) | 4/2 | 4/2 | 6/0 |
| DM (yes/no) | 0/6 | 0/6 | 0/6 |
| HD (yes/no) | 0/6 | 0/6 | 6/0 |
Group 1, with estimated glomerular filtration rate (eGFR) >60 mL/min are younger when compared to group 2 (eGFR 15–60 mL/min), or group 3 (eGFR < 15 mL/min or on dialysis). No differences noted in uremic symptoms between the groups using the Pittsburgh Symptoms Score Index (PSSI), or in body mass index (BMI) between the groups.
HTN: Hypertension, DM: Diabetes mellitus, HD: hemodialysis, AA: African American, W: While, H: Hispanic.
*P < 0.05, **P < 0.01, ***P < 0.001 in comparison to group 1.
Step-wise microbiome changes detected according to renal function
Patients on HD showed a non-statistically significant trend towards decreased α diversity when compared with either group (1 and 2) or to both combined (Figure 1(a) to 1(d)). For genera and species level comparisons between the three groups, we reported all OTUs with at least one significant difference between any of the three groups (Figure 2). No differences were observed on the phyla level. A group of 17 OTUs on the species level (Supplementary Table 1) and 11 OTUs on the genera level (Supplementary Table 2) were identified to have a significant difference between at least two of the groups. However, we focused on OTUs that followed a step-wise pattern across the groups (i.e. systematic increases or decreases across groups 1, 2 through 3). Table 2 summarizes OTUs on the genera and species levels that showed a step-wise change in expression according to renal function. We next performed a sensitivity analysis to explore whether more distinct bacterial composition is present after selecting the OTUs with the statistically significant changes. This was accomplished by utilizing heat-map, hierarchical clustering, and PCA first on selected OTUs, followed by the identical analyses on the remaining groups minus the selected OTUs (Supplementary Figures 1 and 2). Improved but yet poor separation and clustering was noted in the selected OTUs compared to the remaining.
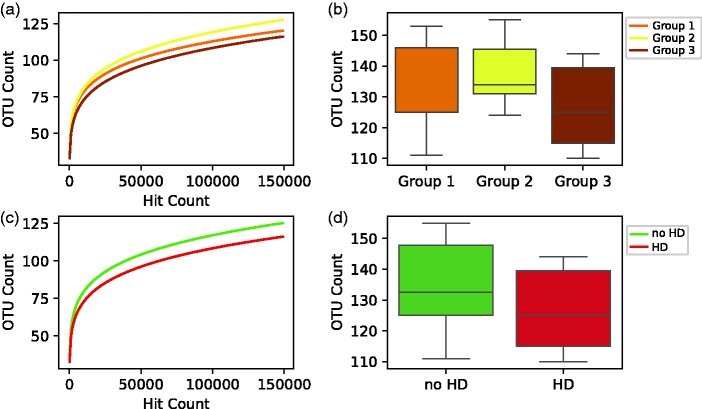
Figure 1.
Alpha diversity. (a, b) Patients with end stage renal disease on HD, (Group 3) showed a trend towards decreased α diversity when compared to Group 1 (patients with chronic kidney disease and eGFR > 60 mL/min) or Group 2 (patients with chronic kidney disease and eGFR between 15–60 mL/min). (c, d) In a separate analysis, patients on HD showed a decreased α diversity when compared to patients not on HD. HD: hemodialysis, OTUs: operational taxonomic units, eGFR: estimated glomerular filtration rate. (A color version of this figure is available in the online journal.)
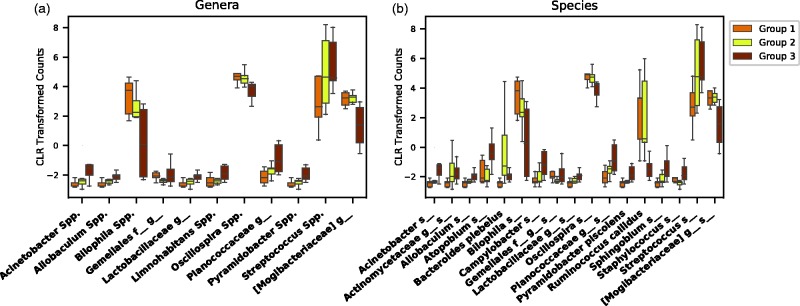
Figure 2.
Genera and species three groups’ analysis. Operational taxonomic units (OTUs) with at least one statistically significant comparison between two of the three groups are shown on genera (a) and species (b) levels. Significant differences were seen in 11 genera, and 17 species. Of note, many OTUs followed a stepwise increased/decreased relative abundance according to changes in renal function. CLR: center log-ratio. (A color version of this figure is available in the online journal.)
Table 2.
Operational taxonomic units (OTUs) with a step-wise change in relative abundance that follows changes in renal function.
| OTUs with increased relative abundance |
| Acinetobacter Spp.* |
| Allobacukum Spp.* |
| Campylobacter s_ |
| Lactobacillaceae g_ s_* |
| Plancoccaceae g_ s_* |
| Pyramidobacter Piscolens |
| Pyramidobacter Spp. |
| Sphingobium s_ |
| Streptococcus Spp.* |
| Limnohabitans Spp. |
| OTUs with decreased relative abundance |
| Bilophila Spp.* |
| Oscillospira Spp.* |
A significantly different changes in relative abundance was noted in multiple OTUs, mostly increased in relative abundance with worsening renal failure. Only Bilophila and Oscillospira genera showed a nominal decrease in relative abundance with worsening renal function.
*Significant change in a non-cultured species along with the changes noted on the genera level.
**Changes seen in a non-cultured species from the corresponding family.
Microbiome changes according to the median eGFR
To overcome the small sample size in the three groups’ analysis, we evaluated the microbial changes according to the cohort median eGFR (45 mL/min). On the phyla level, no major differences were noted when performing PCA. However, patients with eGFR < 45 mL/min showed a significantly lower relative abundance of the Bacteria Tenericutes phylum (Figure 3). Species and genera level analysis revealed a statistically significant relative abundance in 14 species (Supplementary Table 3) and 9 genera (Supplementary Table 4). Lactobacillus iners relative abundance was increased in patients with eGFR < 45 mL/min, while Prevotella stercorea, Ruminococcus callidus, Eubacterium biforme relative expression was decreased. PCA analysis resulted in a much better separation (Supplementary Figure 3(B) and 3(C)) than seen in the three groups’ analysis. Heat-map, hierarchical clustering (Supplementary Figure 4) on the species level showed a strong separation between the two groups when the 14 species were utilized, indicating a robust association between these OTUs and median eGFR.
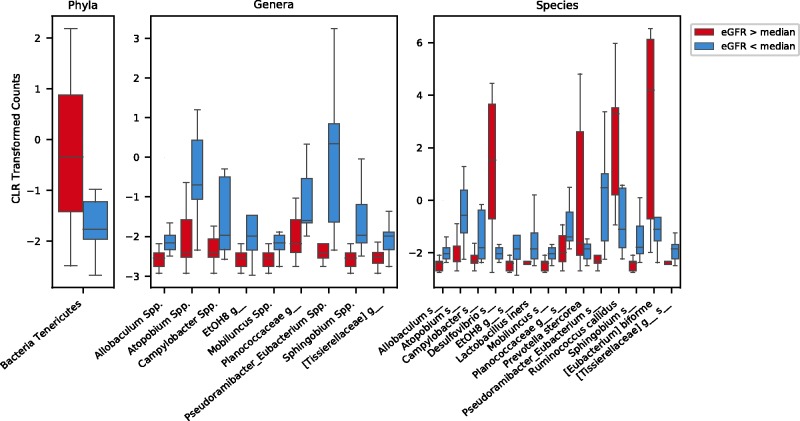
Figure 3.
Phyla, genera and species taxa analysis according to the median estimated glomerular filtration rate (eGFR). When OTUs were compared according to above or below the median eGFR, Bacteria Tenericutes phylum showed a decreased relative abundance in the lower eGFR group when compared to those above the median eGFR. All taxa with statistically significant differences on the genera level are increased in the lower eGFR group. At the species level, most of the significantly different bacteria were uncultivated (denoted by the s_) with the exception of Lactobacillus iners, Prevotella stercorea, Ruminococcus callidus, [Eubacterium] biforme. CLR: Center log-ratio. (A color version of this figure is available in the online journal.)
Microbiome changes in HD versus no HD groups
Metabolic changes in renal failure might be subtle in moderate renal insufficiency.60–67 Thus, we elected to study the HD group (n = 6) against the merged other two groups (groups 1 and 2, n = 12). This approach enabled us to identify OTUs with the greatest correlation to renal function as the previous two approaches might have diluted the effect size. This approach may also provide a better idea of the gut microbiota changes in dialysis patients.68–70 Patients on HD expressed lower α diversity when compared to the rest (Figure 1). The relative abundance of 21 species (Supplementary Table 5) and 18 genera (Supplementary Table 6) were differentially expressed between the two groups. Clostridium saccharogumia, Pyramidobacter piscolens, Streptococcus infantis, and Streptococcus lutecia relative abundances were increased in HD patients, while Ruminococcus callidus expression was decreased (Figure 4). PCA analysis and relative expression distribution between the HD and non-dialysis groups along with the sensitivity analysis after selecting the statistically significant OTUs on the genera and species levels are shown in Supplementary Figure 5. Heat-map and hierarchical clustering on the species level are shown in Supplementary Figure 6.
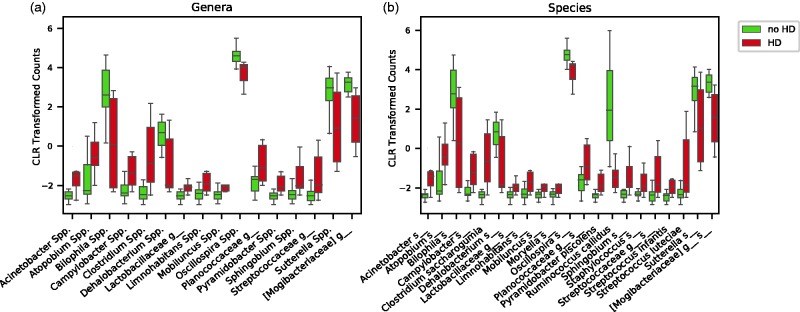
Figure 4.
Genera and species taxa analysis according to HD status. Patients were grouped based on whether or not they were undergoing hemodialysis. Statistical analysis was then performed, and statistically significant differences were observed in 16 genera (a), and 21 species (b). HD: hemodialysis, CLR: center log-ratio. (A color version of this figure is available in the online journal.)
Functional profiling of microbial communities prediction
To predict the metagenomic functional content using the bacterial 16S rRNA data, we used the Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (PICRUSt) software package. We first divided the two groups based on the median eGFR, then followed this with another comparison between those on HD and those who were not. The analysis based on median eGFR revealed 84 (Supplementary Table 7) pathways with statistically significant differences, while the analysis based on HD grouping revealed 153 (Supplementary Table 8) pathways with a statistically significant difference between the groups. It is of great interest to note that changes seen are mainly metabolic pathways. Alterations in taurine-pyruvate aminotransferase (K03851), tryptophan, glyoxylate and dicarboxylate, carbon, purine, and pyrimidine metabolism and biosynthesis of secondary metabolites (K03781, K01119, K00148), and biosynthesis of amino acids (K00619) pathways were noted. Changes seen in purine metabolism (K00230) are directly associated with the bacterial capacity to metabolize urea through possessing urease subunit gamma/beta (K00230-K14048, urea amidohydrolase EC 3.5.1.5).
Discussion
To our knowledge, this is the first report evaluating changes of the bacterial gut microbiome in patients with PKD and varying degrees of renal insufficiency. We excluded patients with other co-morbid conditions or therapeutic interventions that could have potentially influenced the composition of the gut microbiota. This was not a simple task when it comes to ESRD patients on HD, which was reflected in the small sample size. We present a group of OTUs that showed a stepwise change in relative abundance according to the eGFR values. This worked to strengthen the findings and helped confirm the strong effects of renal function alterations on the relative abundance of these taxa.
There are several lines of evidence to suggest that the gut microbiome is likely altered in patients with CKD. A distinct gut microbiome with decreases in both Lactobacillaceae and Prevotellaceae families has been associated with CKD.16 In contrast to these reports, we have previously shown an increase in the relative abundance of Lactobacillus Spp. in experimental CKD.15 In this report we present Lactobacillaceae family, uncultured genus and species from this family along with Lactobacillus iners are all increased in patients with decreased eGFR. This discrepancy may be attributed to the confounding effects of the comorbid conditions associated with CKD or the original disease, such as diabetes. The design of the current study was established to identify the sole impact of uremia on the gut bacterial microbiota by selecting PKD patients with virtually no other comorbid conditions or major therapeutic interventions. In comparison to our previous results in mice with experimental CKD, both PKD patients and CKD mice expressed decreased relative abundance of Oscillospira Spp.15
Byproducts of protein and amino acid catabolism are known to contribute to medical conditions. For example, gut microbe-mediated protein catabolism produces P-cresol from tyrosine and phenylalanine,71 and indole from tryptophan which later is converted by the liver to indoxyl sulfate.72 P-cresol sulfate and indoxyl sulfate are considered uremic toxins because they contribute to adverse cardiac outcomes, and serum concentrations of these compounds are elevated in patients with advanced kidney disease.73–75 Thus, we elected to conduct PICRUSt analysis to establish a dataset for future metabolic analyses. This will enable the scientific community to explore the potential pathways associated with microbiome alterations in CKD populations and discover new candidate metabolites for further studies.
Although the detailed evaluation of the PICRUSt analyzed pathways was not the focus of the current study, it is important to note that most of the highly significant pathways altered in advanced renal disease (HD and eGFR< median groups) were metabolic pathways. Of these pathways, interestingly we noted changes seen in purine metabolism (K00230) and pathways directly associated with the bacterial capacity to metabolize urea through possessing urease subunit gamma/beta (K00230-K14048, urea amidohydrolase EC 3.5.1.5). This, along with other previous reports, strengthens the role of intestinal urea content in shaping the gut bacterial microbiota in CKD.16,21,22,76,77
It is still unclear, however, if increased intestinal urea content is the major contributor to these changes or not. As we have recently shown an increased abundance of bacteria with the ability to metabolize urea in experimental CKD model.15 Changes in the bacterial composition after oral urea supplementation differed from those with the experimental model.15 This of course does not diminish the role of urea in CKD associated dysbiosis but indicates that it may take more than just a change in intestinal urea concentration to induce CKD-associated dysbiotic changes. CKD itself is accompanied with numerous biochemical and bio-physiological changes that most likely play a supporting dysbiotic role. Such changes may include increased colonic transit time, alterations in intestinal pH, and increased ammonia content.24,78–82
Here, we report changes in gut bacterial microbiota according to changes in renal function. We attempted to mitigate the effects on the gut microbiota by excluding patients exhibiting multiple confounding factors including CKD associated co-morbid conditions, therapeutic intervention, or originating disease that resulted in CKD development (e.g. diabetes). Our report also descriptively presented changes in functional pathways associated with the alteration in microbiota noted in this population. Another strength of the current work is the methodical approach in sample collection and downstream steps. Most previous analyses fell short in implementing the gold standard stool collection (fresh in office) and used multiple approaches including transporting samples on ice, room temperature, or utilizing the commercially available stool kits. This report, however, needs to be approached with the knowledge of multiple limitations. First and foremost, the small sample size. Following the strict inclusion/exclusion criteria, it was not feasible to achieve a larger sample size in the HD group. Secondly, dietary intake was not appropriately recorded by most of the participants which might have affected the results. Thirdly, the group with eGFR >60 mL/min was younger compared to other two groups. Despite these limitations, our analysis resulted in multiple findings that will further enable us and other researchers to better understand the CKD-associated dysbiosis through metagenomics PICRUSt analysis and detailed presentation of the OTUs associated with the observed step-wise changes.
Supplemental Material
Supplemental material, Supplemental Figure2 for Fecal microbiota analysis of polycystic kidney disease patients according to renal function: A pilot study by Rabi Yacoub, Girish N Nadkarni, Daniel I McSkimming, Lee D Chaves, Sham Abyad, Mark A Bryniarski, Amanda M Honan, Shruthi A Thomas, Madan Gowda, John C He and Jaime Uribarri in Experimental Biology and Medicine
Supplemental Material
Supplemental material, Supplemental Figure3 for Fecal microbiota analysis of polycystic kidney disease patients according to renal function: A pilot study by Rabi Yacoub, Girish N Nadkarni, Daniel I McSkimming, Lee D Chaves, Sham Abyad, Mark A Bryniarski, Amanda M Honan, Shruthi A Thomas, Madan Gowda, John C He and Jaime Uribarri in Experimental Biology and Medicine
Supplemental Material
Supplemental material, Supplemental Figure4 for Fecal microbiota analysis of polycystic kidney disease patients according to renal function: A pilot study by Rabi Yacoub, Girish N Nadkarni, Daniel I McSkimming, Lee D Chaves, Sham Abyad, Mark A Bryniarski, Amanda M Honan, Shruthi A Thomas, Madan Gowda, John C He and Jaime Uribarri in Experimental Biology and Medicine
Supplemental Material
Supplemental material, Supplemental Figure5 for Fecal microbiota analysis of polycystic kidney disease patients according to renal function: A pilot study by Rabi Yacoub, Girish N Nadkarni, Daniel I McSkimming, Lee D Chaves, Sham Abyad, Mark A Bryniarski, Amanda M Honan, Shruthi A Thomas, Madan Gowda, John C He and Jaime Uribarri in Experimental Biology and Medicine
Supplemental Material
Supplemental material, Supplemental Figure6 for Fecal microbiota analysis of polycystic kidney disease patients according to renal function: A pilot study by Rabi Yacoub, Girish N Nadkarni, Daniel I McSkimming, Lee D Chaves, Sham Abyad, Mark A Bryniarski, Amanda M Honan, Shruthi A Thomas, Madan Gowda, John C He and Jaime Uribarri in Experimental Biology and Medicine
Supplemental Material
Supplemental material, Supplemental Table1 for Fecal microbiota analysis of polycystic kidney disease patients according to renal function: A pilot study by Rabi Yacoub, Girish N Nadkarni, Daniel I McSkimming, Lee D Chaves, Sham Abyad, Mark A Bryniarski, Amanda M Honan, Shruthi A Thomas, Madan Gowda, John C He and Jaime Uribarri in Experimental Biology and Medicine
Supplemental Material
Supplemental material, Supplemental Table2 for Fecal microbiota analysis of polycystic kidney disease patients according to renal function: A pilot study by Rabi Yacoub, Girish N Nadkarni, Daniel I McSkimming, Lee D Chaves, Sham Abyad, Mark A Bryniarski, Amanda M Honan, Shruthi A Thomas, Madan Gowda, John C He and Jaime Uribarri in Experimental Biology and Medicine
Supplemental Material
Supplemental material, Supplemental Table3 for Fecal microbiota analysis of polycystic kidney disease patients according to renal function: A pilot study by Rabi Yacoub, Girish N Nadkarni, Daniel I McSkimming, Lee D Chaves, Sham Abyad, Mark A Bryniarski, Amanda M Honan, Shruthi A Thomas, Madan Gowda, John C He and Jaime Uribarri in Experimental Biology and Medicine
Supplemental Material
Supplemental material, Supplemental Table4 for Fecal microbiota analysis of polycystic kidney disease patients according to renal function: A pilot study by Rabi Yacoub, Girish N Nadkarni, Daniel I McSkimming, Lee D Chaves, Sham Abyad, Mark A Bryniarski, Amanda M Honan, Shruthi A Thomas, Madan Gowda, John C He and Jaime Uribarri in Experimental Biology and Medicine
Supplemental Material
Supplemental material, Supplemental Table5 for Fecal microbiota analysis of polycystic kidney disease patients according to renal function: A pilot study by Rabi Yacoub, Girish N Nadkarni, Daniel I McSkimming, Lee D Chaves, Sham Abyad, Mark A Bryniarski, Amanda M Honan, Shruthi A Thomas, Madan Gowda, John C He and Jaime Uribarri in Experimental Biology and Medicine
Supplemental Material
Supplemental material, Supplemental Table6 for Fecal microbiota analysis of polycystic kidney disease patients according to renal function: A pilot study by Rabi Yacoub, Girish N Nadkarni, Daniel I McSkimming, Lee D Chaves, Sham Abyad, Mark A Bryniarski, Amanda M Honan, Shruthi A Thomas, Madan Gowda, John C He and Jaime Uribarri in Experimental Biology and Medicine
Supplemental Material
Supplemental material, Supplemental Table7 for Fecal microbiota analysis of polycystic kidney disease patients according to renal function: A pilot study by Rabi Yacoub, Girish N Nadkarni, Daniel I McSkimming, Lee D Chaves, Sham Abyad, Mark A Bryniarski, Amanda M Honan, Shruthi A Thomas, Madan Gowda, John C He and Jaime Uribarri in Experimental Biology and Medicine
Supplemental Material
Supplemental material, Supplemental Table8 for Fecal microbiota analysis of polycystic kidney disease patients according to renal function: A pilot study by Rabi Yacoub, Girish N Nadkarni, Daniel I McSkimming, Lee D Chaves, Sham Abyad, Mark A Bryniarski, Amanda M Honan, Shruthi A Thomas, Madan Gowda, John C He and Jaime Uribarri in Experimental Biology and Medicine
Supplemental Material
Supplemental material, Supplemental Figure1 for Fecal microbiota analysis of polycystic kidney disease patients according to renal function: A pilot study by Rabi Yacoub, Girish N Nadkarni, Daniel I McSkimming, Lee D Chaves, Sham Abyad, Mark A Bryniarski, Amanda M Honan, Shruthi A Thomas, Madan Gowda, John C He and Jaime Uribarri in Experimental Biology and Medicine
Authors’ contributions
All authors contributed substantially to the manuscript preparation and editing, read and approved the final version before submission. JCH, JU and RY conceived the study and were responsible of the study design. RY and GNN recruited the participants. LDC, SA, AMH, SAT, MAB, and MG performed experiments and contributed to data analysis and presentation. DIM performed and advised on 16S sequencing, data annotation and analysis.
Authors’ note
Data will be made available upon request.
DECLARATION OF CONFLICTING INTERESTS
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article
FUNDING
The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: Research reported in this publication was supported by the National Center for Advancing Translational Sciences of the National Institutes of Health under award number UL1TR001412 to the University at Buffalo and by the NIH. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. This was also supported by University at Buffalo Genome, Environment and Microbiome (GEM), and department fund for RY and LDC. Innovative Micro-Programs Accelerating Collaboration in Themes (IMPACT) for RY.
References
- 1. Candela M, Turroni S, Biagi E, Carbonero F, Rampelli S, Fiorentini C, Brigidi P. Inflammation and colorectal cancer, when microbiota-host mutualism breaks. World J Gastroenterol 2014; 20:908–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Belkaid Y, Hand TW. Role of the microbiota in immunity and inflammation. Cell 2014; 157:121–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Sanz Y, Moya PA. Microbiota, inflammation and obesity. Adv Exp Med Biol 2014; 817:291–317 [DOI] [PubMed] [Google Scholar]
- 4.Mao K, Baptista AP, Tamoutounour S, Zhuang L, Bouladoux N, Martins AJ, Huang Y, Gerner MY, Belkaid Y, Germain RN. Innate and adaptive lymphocytes sequentially shape the gut microbiota and lipid metabolism. Nature 2018; 554:255–9 [DOI] [PubMed] [Google Scholar]
- 5.Forslund K, Hildebrand F, Nielsen T, Falony G, Le Chatelier E, Sunagawa S, Prifti E, Vieira-Silva S, Gudmundsdottir V, Pedersen HK, Arumugam M, Kristiansen K, Voigt AY, Vestergaard H, Hercog R, Costea PI, Kultima JR, Li J, Jorgensen T, Levenez F, Dore J, Meta HITc Nielsen HB, Brunak S, Raes J, Hansen T, Wang J, Ehrlich SD, Bork P, Pedersen O. Disentangling type 2 diabetes and metformin treatment signatures in the human gut microbiota. Nature 2015; 528:262–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Koeth RA, Wang Z, Levison BS, Buffa JA, Org E, Sheehy BT, Britt EB, Fu X, Wu Y, Li L, Smith JD, DiDonato JA, Chen J, Li H, Wu GD, Lewis JD, Warrier M, Brown JM, Krauss RM, Tang WH, Bushman FD, Lusis AJ, Hazen SL. Intestinal microbiota metabolism of L-carnitine, a nutrient in red meat, promotes atherosclerosis. Nat Med 2013; 19:576–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tang WH, Wang Z, Levison BS, Koeth RA, Britt EB, Fu X, Wu Y, Hazen SL. Intestinal microbial metabolism of phosphatidylcholine and cardiovascular risk. N Engl J Med 2013; 368:1575–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tang WH, Hazen SL. The contributory role of gut microbiota in cardiovascular disease. J Clin Invest 2014; 124:4204–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peterson CT, Sharma V, Elmen L, Peterson SN. Immune homeostasis, dysbiosis and therapeutic modulation of the gut microbiota. Clin Exp Immunol 2015; 179:363–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kabat AM, Srinivasan N, Maloy KJ. Modulation of immune development and function by intestinal microbiota. Trends Immunol 2014; 35:507–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yacoub R, Jacob A, Wlaschin J, McGregor M, Quigg RJ, Alexander JJ. Lupus: the microbiome angle. Immunobiology 2018; 223:460–5 [DOI] [PubMed] [Google Scholar]
- 12.Fellows R, Denizot J, Stellato C, Cuomo A, Jain P, Stoyanova E, Balazsi S, Hajnady Z, Liebert A, Kazakevych J, Blackburn H, Correa RO, Fachi JL, Sato FT, Ribeiro WR, Ferreira CM, Peree H, Spagnuolo M, Mattiuz R, Matolcsi C, Guedes J, Clark J, Veldhoen M, Bonaldi T, Vinolo MAR, Varga-Weisz P. Microbiota derived short chain fatty acids promote histone crotonylation in the colon through histone deacetylases. Nat Commun 2018; 9:105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ohira H, Tsutsui W, Fujioka Y. Are short chain fatty acids in gut microbiota defensive players for inflammation and atherosclerosis? J Atheroscler Thromb 2017; 24:660–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morrison DJ, Preston T. Formation of short chain fatty acids by the gut microbiota and their impact on human metabolism. Gut Microbes 2016; 7:189–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chaves LD, McSkimming DI, Bryniarski MA, Honan AM, Abyad S, Thomas SA, Wells S, Buck MJ, Sun Y, Genco RJ, Quigg RJ, Yacoub R. Chronic kidney disease, uremic milieu, and its effects on gut bacterial microbiota dysbiosis. Am J Physiol Renal Physiol 2018; 315:F487–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vaziri ND, Wong J, Pahl M, Piceno YM, Yuan J, DeSantis TZ, Ni Z, Nguyen TH, Andersen GL. Chronic kidney disease alters intestinal microbial flora. Kidney Int 2013; 83:308–15 [DOI] [PubMed] [Google Scholar]
- 17.Yacoub R, Nugent M, Cai W, Nadkarni GN, Chaves LD, Abyad S, Honan AM, Thomas SA, Zheng W, Valiyaparambil SA, Bryniarski MA, Sun Y, Buck M, Genco RJ, Quigg RJ, He JC, Uribarri J. Advanced glycation end products dietary restriction effects on bacterial gut microbiota in peritoneal dialysis patients: a randomized open label controlled trial. PLoS One 2017; 12:e0184789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu KY, Xia GH, Lu JQ, Chen MX, Zhen X, Wang S, You C, Nie J, Zhou HW, Yin J. Impaired renal function and dysbiosis of gut microbiota contribute to increased trimethylamine-N-oxide in chronic kidney disease patients. Sci Rep 2017; 7:1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bammens B, Verbeke K, Vanrenterghem Y, Evenepoel P. Evidence for impaired assimilation of protein in chronic renal failure. Kidney Int 2003; 64:2196–203 [DOI] [PubMed] [Google Scholar]
- 20.Isakova T, Nickolas TL, Denburg M, Yarlagadda S, Weiner DE, Gutierrez OM, Bansal V, Rosas SE, Nigwekar S, Yee J, Kramer H. KDOQI US Commentary on the 2017 KDIGO Clinical Practice Guideline Update for the diagnosis, evaluation, prevention, and treatment of chronic kidney disease-mineral and bone disorder (CKD-MBD). Am J Kidney Dis 2017; 70:737–51 [DOI] [PubMed] [Google Scholar]
- 21.Kang JY. The gastrointestinal tract in uremia. Dig Dis Sci 1993; 38:257–68 [DOI] [PubMed] [Google Scholar]
- 22.Macfarlane GT, Macfarlane S. Fermentation in the human large intestine: its physiologic consequences and the potential contribution of prebiotics. J Clin Gastroenterol 2011;45 Suppl:S120–7 [DOI] [PubMed] [Google Scholar]
- 23.Felizardo RJ, Castoldi A, Andrade-Oliveira V, Camara NO. The microbiota and chronic kidney diseases: a double-edged sword. Clin Trans Immunol 2016; 5:e86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu MJ, Chang CS, Cheng CH, Chen CH, Lee WC, Hsu YH, Shu KH, Tang MJ. Colonic transit time in long-term dialysis patients. Am J Kidney Dis 2004; 44:322–7 [DOI] [PubMed] [Google Scholar]
- 25.Lefebvre HP, Ferre JP, Watson AD, Brown CA, Serthelon JP, Laroute V, Concordet D, Toutain PL. Small bowel motility and colonic transit are altered in dogs with moderate renal failure. Am J Physiol Regul Integr Comp Physiol 2001; 281:R230–8 [DOI] [PubMed] [Google Scholar]
- 26.Cummings JH, Hill MJ, Bone ES, Branch WJ, Jenkins DJ. The effect of meat protein and dietary fiber on colonic function and metabolism. II. Bacterial metabolites in feces and urine. Am J Clin Nutr 1979; 32:2094–101 [DOI] [PubMed] [Google Scholar]
- 27.Cummings JH, Hill MJ, Jivraj T, Houston H, Branch WJ, Jenkins DJ. The effect of meat protein and dietary fiber on colonic function and metabolism. I. Changes in bowel habit, bile acid excretion, and calcium absorption. Am J Clin Nutr 1979; 32:2086–93 [DOI] [PubMed] [Google Scholar]
- 28.Stephen AM, Wiggins HS, Cummings JH. Effect of changing transit time on colonic microbial metabolism in man. Gut 1987; 28:601–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tottey W, Feria-Gervasio D, Gaci N, Laillet B, Pujos E, Martin JF, Sebedio JL, Sion B, Jarrige JF, Alric M, Brugere JF. Colonic transit time is a driven force of the gut microbiota composition and metabolism: in vitro evidence. J Neurogastroenterol Motil 2017; 23:124–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roager HM, Hansen LB, Bahl MI, Frandsen HL, Carvalho V, Gobel RJ, Dalgaard MD, Plichta DR, Sparholt MH, Vestergaard H, Hansen T, Sicheritz-Ponten T, Nielsen HB, Pedersen O, Lauritzen L, Kristensen M, Gupta R, Licht TR. Colonic transit time is related to bacterial metabolism and mucosal turnover in the gut. Nat Microbiol 2016; 1:16093. [DOI] [PubMed] [Google Scholar]
- 31.Tilg H, Moschen AR. Microbiota and diabetes: an evolving relationship. Gut 2014; 63:1513–21 [DOI] [PubMed] [Google Scholar]
- 32.Lv Y, Zhao X, Guo W, Gao Y, Yang S, Li Z, Wang G. The relationship between frequently used glucose-lowering agents and gut microbiota in type 2 diabetes mellitus. J Diabetes Res 2018; 2018:1890978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Burcelin R, Serino M, Chabo C, Blasco-Baque V, Amar J. Gut microbiota and diabetes: from pathogenesis to therapeutic perspective. Acta Diabetol 2011; 48:257–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jose PA, Raj D. Gut microbiota in hypertension. Curr Opin Nephrol Hypertens 2015; 24:403–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang T, Santisteban MM, Rodriguez V, Li E, Ahmari N, Carvajal JM, Zadeh M, Gong M, Qi Y, Zubcevic J, Sahay B, Pepine CJ, Raizada MK, Mohamadzadeh M. Gut dysbiosis is linked to hypertension. Hypertension 2015; 65:1331–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mell B, Jala VR, Mathew AV, Byun J, Waghulde H, Zhang Y, Haribabu B, Vijay-Kumar M, Pennathur S, Joe B. Evidence for a link between gut microbiota and hypertension in the Dahl rat. Physiol Genomics 2015; 47:187–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dostal A, Lacroix C, Bircher L, Pham VT, Follador R, Zimmermann MB, Chassard C. Iron modulates butyrate production by a child gut microbiota in vitro. MBio 2015; 6:e01453–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shanmugam NK, Trebicka E, Fu LL, Shi HN, Cherayil BJ. Intestinal inflammation modulates expression of the iron-regulating hormone hepcidin depending on erythropoietic activity and the commensal microbiota. J Immunol 2014; 193:1398–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vieira SM, Pagovich OE, Kriegel MA. Diet, microbiota and autoimmune diseases. Lupus 2014; 23:518–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moraes C, Fouque D, Amaral AC, Mafra D. Trimethylamine N-oxide from gut microbiota in chronic kidney disease patients: focus on diet. J Ren Nutr 2015; 25:459–65 [DOI] [PubMed] [Google Scholar]
- 41.Montemurno E, Cosola C, Dalfino G, Daidone G, De Angelis M, Gobbetti M, Gesualdo L. What would you like to eat, Mr CKD Microbiota? A Mediterranean diet, please! Kidney Blood Press Res 2014; 39:114–23 [DOI] [PubMed] [Google Scholar]
- 42.Levey AS, Coresh J, Greene T, Stevens LA, Zhang YL, Hendriksen S, Kusek JW, Van Lente F, Chronic Kidney D, Epidemiology C. Using standardized serum creatinine values in the modification of diet in renal disease study equation for estimating glomerular filtration rate. Ann Intern Med 2006; 145:247–54 [DOI] [PubMed] [Google Scholar]
- 43.Zhang J, Kobert K, Flouri T, Stamatakis A. PEAR: a fast and accurate Illumina Paired-End reAd mergeR. Bioinformatics 2014; 30:614–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gordon A, Hannon G. Fastx-toolkit. FASTQ/A short-reads preprocessing tools (unpublished), http://hannonlab.cshl.edu/fastx_toolkit (2010, accessed 25 November 2018).
- 45.DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol 2006; 72:5069–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Weisbord SD, Fried LF, Arnold RM, Rotondi AJ, Fine MJ, Levenson DJ, Switzer GE. Development of a symptom assessment instrument for chronic hemodialysis patients: the Dialysis Symptom Index. J Pain Symptom Manage 2004; 27:226–40 [DOI] [PubMed] [Google Scholar]
- 47.Novak MJ, Sheth H, Bender FH, Fried L, Piraino B. Improvement in Pittsburgh Symptom Score index after initiation of peritoneal dialysis. Adv Perit Dial 2008; 24:46–50 [PubMed] [Google Scholar]
- 48.Hong E, Bernardini J, Fried L, Samsonov M, Pirain B. The relationship between symptoms, depression, and quality of life in peritoneal dialysis patients. Adv Perit Dial 2006; 22:83–7 [PubMed] [Google Scholar]
- 49.Van Rossum G, Drake FL. Python language reference manual. Release 2.3. Network Theory Limited, 2003. [Google Scholar]
- 50.McKinney W. (ed). Data structures for statistical computing in python. In: Proceedings of the 9th python in science conference, 2010, SciPy Austin, TX, USA.
- 51.Walt Svd Colbert SC, Varoquaux G. The NumPy array: a structure for efficient numerical computation. Comput Sci Eng 2011; 13:22–30 [Google Scholar]
- 52.Jones E, Oliphant T, Peterson P. {SciPy}: open source scientific tools for {Python}, http://www.scipy.org. 2014.
- 53.Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, Blondel M, Prettenhofer P, Weiss R, Dubourg V. Scikit-learn: machine learning in python. J Mach Learn Res 2011; 12:2825–30 [Google Scholar]
- 54.Goodpaster AM, Kennedy MA. Quantification and statistical significance analysis of group separation in NMR-based metabonomics studies. Chemometr Intell Lab Syst 2011; 109:162–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kanehisa M, Sato Y, Morishima K. BlastKOALA and GhostKOALA: KEGG tools for functional characterization of genome and metagenome sequences. J Mol Biol 2016; 428:726–31 [DOI] [PubMed] [Google Scholar]
- 56.Kanehisa M, Sato Y, Kawashima M, Furumichi M, Tanabe M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res 2016; 44:D457–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Langille MG, Zaneveld J, Caporaso JG, McDonald D, Knights D, Reyes JA, Clemente JC, Burkepile DE, Vega Thurber RL, Knight R, Beiko RG, Huttenhower C. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol 2013; 31:814–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Aitchison J. The statistical analysis of compositional data. J R Stat Soc Series B Stat Methodol 1982;139–77 [Google Scholar]
- 59.Pincus R, Aitchison, J. The statistical analysis of compositional data. Chapman and Hall, London‐New York 1986, XII, 416 pp., £25, 00. Biom J 1988; 30:794 [Google Scholar]
- 60.Ketteler M, Block GA, Evenepoel P, Fukagawa M, Herzog CA, McCann L, Moe SM, Shroff R, Tonelli MA, Toussaint ND, Vervloet MG, Leonard MB. Executive summary of the 2017 KDIGO chronic kidney disease-mineral and bone disorder (CKD-MBD) Guideline Update: what's changed and why it matters. Kidney Int 2017; 92:26–36 [DOI] [PubMed] [Google Scholar]
- 61.Warnock DG. Uremic acidosis. Kidney Int 1988; 34:278–87 [DOI] [PubMed] [Google Scholar]
- 62.Bailey JL. Metabolic acidosis: an unrecognized cause of morbidity in the patient with chronic kidney disease. Kidney Int Suppl 2005; 68:S15–23 [DOI] [PubMed] [Google Scholar]
- 63.Widmer B, Gerhardt RE, Harrington JT, Cohen JJ. Serum electrolyte and acid base composition. The influence of graded degrees of chronic renal failure. Arch Intern Med 1979; 139:1099–102 [PubMed] [Google Scholar]
- 64.Franch HA, Raissi S, Wang X, Zheng B, Bailey JL, Price SR. Acidosis impairs insulin receptor substrate-1-associated phosphoinositide 3-kinase signaling in muscle cells: consequences on proteolysis. Am J Physiol Renal Physiol 2004; 287:F700–6 [DOI] [PubMed] [Google Scholar]
- 65.Ordonez FA, Santos F, Martinez V, Garcia E, Fernandez P, Rodriguez J, Fernandez M, Alvarez J, Ferrando S. Resistance to growth hormone and insulin-like growth factor-I in acidotic rats. Pediatr Nephrol 2000; 14:720–5 [DOI] [PubMed] [Google Scholar]
- 66.Kraut JA, Kurtz I. Metabolic acidosis of CKD: diagnosis, clinical characteristics, and treatment. Am J Kidney Dis 2005; 45:978–93 [DOI] [PubMed] [Google Scholar]
- 67.Kopple JD, Kalantar-Zadeh K, Mehrotra R. Risks of chronic metabolic acidosis in patients with chronic kidney disease. Kidney Int 2005; 67:S21–7 [DOI] [PubMed] [Google Scholar]
- 68.Zoccali C, Vanholder R, Massy ZA, Ortiz A, Sarafidis P, Dekker FW, Fliser D, Fouque D, Heine GH, Jager KJ, Kanbay M, Mallamaci F, Parati G, Rossignol P, Wiecek A, London G, European Renal and Cardiovascular Medicine Working Group of the European Renal Association – European Dialysis Transplantation Association. The systemic nature of CKD. Nat Rev Nephrol 2017; 13:344–58 [DOI] [PubMed] [Google Scholar]
- 69.Hocher B, Adamski J. Metabolomics for clinical use and research in chronic kidney disease. Nat Rev Nephrol 2017; 13:269–84 [DOI] [PubMed] [Google Scholar]
- 70.Aron-Wisnewsky J, Clement K. The gut microbiome, diet, and links to cardiometabolic and chronic disorders. Nat Rev Nephrol 2016; 12:169–81 [DOI] [PubMed] [Google Scholar]
- 71.De Smet R, Glorieux G, Hsu C, Vanholder R. p-cresol and uric acid: two old uremic toxins revisited. Kidney Int Suppl 1997; 62:S8–11 [PubMed] [Google Scholar]
- 72.Deguchi T, Ohtsuki S, Otagiri M, Takanaga H, Asaba H, Mori S, Terasaki T. Major role of organic anion transporter 3 in the transport of indoxyl sulfate in the kidney. Kidney Int 2002; 61:1760–8 [DOI] [PubMed] [Google Scholar]
- 73.Bammens B, Evenepoel P, Keuleers H, Verbeke K, Vanrenterghem Y. Free serum concentrations of the protein-bound retention solute p-cresol predict mortality in hemodialysis patients. Kidney Int 2006; 69:1081–7 [DOI] [PubMed] [Google Scholar]
- 74.Meijers BK, Claes K, Bammens B, de Loor H, Viaene L, Verbeke K, Kuypers D, Vanrenterghem Y, Evenepoel P. p-Cresol and cardiovascular risk in mild-to-moderate kidney disease. Clin J Am Soc Nephrol 2010; 5:1182–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Vanholder R, De Smet R, Hsu C, Vogeleere P, Ringoir S. Uremic toxicity: the middle molecule hypothesis revisited. Semin Nephrol 1994; 14:205–18 [PubMed] [Google Scholar]
- 76.Vaziri ND, Dure-Smith B, Miller R, Mirahmadi MK. Pathology of gastrointestinal tract in chronic hemodialysis patients: an autopsy study of 78 cases. Am J Gastroenterol 1985; 80:608–11 [PubMed] [Google Scholar]
- 77.Hatch M, Vaziri ND. Enhanced enteric excretion of urate in rats with chronic renal failure. Clin Sci (Sci) 1994; 86:511–6 [DOI] [PubMed] [Google Scholar]
- 78.Kes P. Serum gastrin concentration in chronic renal failure. Acta Med Croatica 1992; 46:47–58 [PubMed] [Google Scholar]
- 79.Quintero E, Ohning GV, Del Rivero M, Wong HC, Walsh JH, Guth PH. Gastrin mediates the increase in gastric cell growth in uremic rats. Am J Physiol 1995; 268:G586–91 [DOI] [PubMed] [Google Scholar]
- 80.Quintero E, Kaunitz J, Nishizaki Y, De Giorgio R, Sternini C, Guth PH. Uremia increases gastric mucosal permeability and acid back-diffusion injury in the rat. Gastroenterology 1992; 103:1762–8 [DOI] [PubMed] [Google Scholar]
- 81.Lee YT. Urea concentration in intestinal fluids in normal and uremic dogs. J Surg Oncol 1971; 3:163–8 [DOI] [PubMed] [Google Scholar]
- 82.Vaziri ND, Yuan J, Norris K. Role of urea in intestinal barrier dysfunction and disruption of epithelial tight junction in chronic kidney disease. Am J Nephrol 2013; 37:1–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, Supplemental Figure2 for Fecal microbiota analysis of polycystic kidney disease patients according to renal function: A pilot study by Rabi Yacoub, Girish N Nadkarni, Daniel I McSkimming, Lee D Chaves, Sham Abyad, Mark A Bryniarski, Amanda M Honan, Shruthi A Thomas, Madan Gowda, John C He and Jaime Uribarri in Experimental Biology and Medicine
Supplemental material, Supplemental Figure3 for Fecal microbiota analysis of polycystic kidney disease patients according to renal function: A pilot study by Rabi Yacoub, Girish N Nadkarni, Daniel I McSkimming, Lee D Chaves, Sham Abyad, Mark A Bryniarski, Amanda M Honan, Shruthi A Thomas, Madan Gowda, John C He and Jaime Uribarri in Experimental Biology and Medicine
Supplemental material, Supplemental Figure4 for Fecal microbiota analysis of polycystic kidney disease patients according to renal function: A pilot study by Rabi Yacoub, Girish N Nadkarni, Daniel I McSkimming, Lee D Chaves, Sham Abyad, Mark A Bryniarski, Amanda M Honan, Shruthi A Thomas, Madan Gowda, John C He and Jaime Uribarri in Experimental Biology and Medicine
Supplemental material, Supplemental Figure5 for Fecal microbiota analysis of polycystic kidney disease patients according to renal function: A pilot study by Rabi Yacoub, Girish N Nadkarni, Daniel I McSkimming, Lee D Chaves, Sham Abyad, Mark A Bryniarski, Amanda M Honan, Shruthi A Thomas, Madan Gowda, John C He and Jaime Uribarri in Experimental Biology and Medicine
Supplemental material, Supplemental Figure6 for Fecal microbiota analysis of polycystic kidney disease patients according to renal function: A pilot study by Rabi Yacoub, Girish N Nadkarni, Daniel I McSkimming, Lee D Chaves, Sham Abyad, Mark A Bryniarski, Amanda M Honan, Shruthi A Thomas, Madan Gowda, John C He and Jaime Uribarri in Experimental Biology and Medicine
Supplemental material, Supplemental Table1 for Fecal microbiota analysis of polycystic kidney disease patients according to renal function: A pilot study by Rabi Yacoub, Girish N Nadkarni, Daniel I McSkimming, Lee D Chaves, Sham Abyad, Mark A Bryniarski, Amanda M Honan, Shruthi A Thomas, Madan Gowda, John C He and Jaime Uribarri in Experimental Biology and Medicine
Supplemental material, Supplemental Table2 for Fecal microbiota analysis of polycystic kidney disease patients according to renal function: A pilot study by Rabi Yacoub, Girish N Nadkarni, Daniel I McSkimming, Lee D Chaves, Sham Abyad, Mark A Bryniarski, Amanda M Honan, Shruthi A Thomas, Madan Gowda, John C He and Jaime Uribarri in Experimental Biology and Medicine
Supplemental material, Supplemental Table3 for Fecal microbiota analysis of polycystic kidney disease patients according to renal function: A pilot study by Rabi Yacoub, Girish N Nadkarni, Daniel I McSkimming, Lee D Chaves, Sham Abyad, Mark A Bryniarski, Amanda M Honan, Shruthi A Thomas, Madan Gowda, John C He and Jaime Uribarri in Experimental Biology and Medicine
Supplemental material, Supplemental Table4 for Fecal microbiota analysis of polycystic kidney disease patients according to renal function: A pilot study by Rabi Yacoub, Girish N Nadkarni, Daniel I McSkimming, Lee D Chaves, Sham Abyad, Mark A Bryniarski, Amanda M Honan, Shruthi A Thomas, Madan Gowda, John C He and Jaime Uribarri in Experimental Biology and Medicine
Supplemental material, Supplemental Table5 for Fecal microbiota analysis of polycystic kidney disease patients according to renal function: A pilot study by Rabi Yacoub, Girish N Nadkarni, Daniel I McSkimming, Lee D Chaves, Sham Abyad, Mark A Bryniarski, Amanda M Honan, Shruthi A Thomas, Madan Gowda, John C He and Jaime Uribarri in Experimental Biology and Medicine
Supplemental material, Supplemental Table6 for Fecal microbiota analysis of polycystic kidney disease patients according to renal function: A pilot study by Rabi Yacoub, Girish N Nadkarni, Daniel I McSkimming, Lee D Chaves, Sham Abyad, Mark A Bryniarski, Amanda M Honan, Shruthi A Thomas, Madan Gowda, John C He and Jaime Uribarri in Experimental Biology and Medicine
Supplemental material, Supplemental Table7 for Fecal microbiota analysis of polycystic kidney disease patients according to renal function: A pilot study by Rabi Yacoub, Girish N Nadkarni, Daniel I McSkimming, Lee D Chaves, Sham Abyad, Mark A Bryniarski, Amanda M Honan, Shruthi A Thomas, Madan Gowda, John C He and Jaime Uribarri in Experimental Biology and Medicine
Supplemental material, Supplemental Table8 for Fecal microbiota analysis of polycystic kidney disease patients according to renal function: A pilot study by Rabi Yacoub, Girish N Nadkarni, Daniel I McSkimming, Lee D Chaves, Sham Abyad, Mark A Bryniarski, Amanda M Honan, Shruthi A Thomas, Madan Gowda, John C He and Jaime Uribarri in Experimental Biology and Medicine
Supplemental material, Supplemental Figure1 for Fecal microbiota analysis of polycystic kidney disease patients according to renal function: A pilot study by Rabi Yacoub, Girish N Nadkarni, Daniel I McSkimming, Lee D Chaves, Sham Abyad, Mark A Bryniarski, Amanda M Honan, Shruthi A Thomas, Madan Gowda, John C He and Jaime Uribarri in Experimental Biology and Medicine