Abstract
Survival rates decrease significantly when localized prostate cancer (CaP) becomes metastatic, emphasizing the need for improved targeted therapies. DDX3, an RNA helicase, has widespread functions in RNA regulation, in both the nucleus and cytoplasm. Although DDX3 has been implicated as a prognostic marker for many cancers, including primary CaP, its expression, localization, and function in metastatic CaP have not been investigated. Analysis of metadata and cell line models found increased DDX3 expression in metastatic versus primary CaP and benign prostate. Quantification of DDX3 expression in 320 human prostate samples, representing different stages of CaP progression, revealed an increase in epithelial whole cell, cytoplasmic, and nuclear DDX3 in primary CaP compared with benign prostate. In metastatic tissues, cytoplasmic DDX3 remained highly expressed, whereas nuclear DDX3 significantly decreased compared with primary CaP, suggesting a potential role for cytoplasmic DDX3 in metastatic CaP. Genetic and pharmacologic loss of function for DDX3 in metastatic CaP produced a significant decrease in cell viability, proliferation, and motility but did not affect apoptosis. The data suggest that cytoplasmic DDX3 is highly expressed in metastatic CaP and that inhibition of DDX3 affects metastatic growth by decreasing proliferation and motility. These findings introduce a novel role for cytoplasmic DDX3 in CaP progression and provide a foundation for clinically targeting DDX3 in metastatic CaP.
Prostate cancer (CaP) is the second most prevalent cancer diagnosed among men in the United States, with 164,690 new cases expected in 2018, corresponding to 9.5% of all new cancer cases.1 Of these diagnoses, 29,430 men are expected to die of CaP, equating to 4.8% of all cancer-related deaths.1 Importantly, although the 5-year survival rate for localized and regional CaP is >99%, patients with distant metastases have only a 30% 5-year survival rate.1 Treatment for metastatic CaP includes endocrine-based therapies, which despite initial success inevitably result in advanced disease progression.2, 3 After endocrine therapy failure, taxane-based chemotherapy is prescribed to nonspecifically target dividing cells, which improves median survival for metastatic CaP by 2 to 3 months.3, 4, 5 Although advances in immunotherapies have improved survival times to approximately 4.5 months,6, 7 identifying features of aggressive (metastatic) CaP is imperative to ascertain improved targeted therapies and patient survival.
DDX3 (DEAD-box helicase 3 X-linked) is an ATP-dependent RNA helicase that is known to play a role in nearly all stages of RNA processing in both the nucleus and cytoplasm. In the nucleus, DDX3 has roles in transcription, splicing, and nuclear export. As a transcriptional regulator, DDX3 i) interacts with SP1 transcription factor to up-regulate p21waf1/cip1 and KRAS, ii) directly binds the interferon β promoter to induce activation, and iii) regulates transcription of E-cadherin as both a repressor and an activator.8, 9, 10, 11, 12, 13 DDX3 has also been identified as a component of the spliceosome14, 15 and interacts with the nuclear export proteins chromosomal maintenance 1 and transporter associated with antigen processing to assist in mRNA nuclear export.16, 17 These functions of nuclear DDX3 suggest it may be involved in context-dependent growth induction or suppression, immune response, motility, and/or invasion.
In the cytoplasm, DDX3 acts as a regulator of translation. Under normal conditions, DDX3 binds mRNA as part of the translation initiation complex with other components, including the poly(A)-binding protein and eukaryotic initiation factors 2a, 3, and 4E/F.16, 18, 19, 20 Here, DDX3 hydrolyzes ATP to resolve the secondary structure of target mRNA, allowing translation initiation.16, 20, 21 Alternatively, under stress conditions, such as heat shock, oxidative stress, ischemia, or viral infection, DDX3 localizes to ribonucleoprotein granules known as stress granules to transiently prevent translation of target mRNA.19, 22, 23 Identified targets of DDX3-mediated translational regulation include Ras family small GTPase 1 and cyclin-E1, suggesting that cytoplasmic DDX3 as a translational regulator may influence cell division and/or cell growth.24, 25
DDX3 has recently been implicated as a prognostic marker in several cancer types.26 Nuclear DDX3 was identified as a negative prognostic factor in breast cancer and glioblastoma, whereas increased cytoplasmic DDX3 is considered a negative prognostic factor in breast, lung, gallbladder, head and neck, and skin cancers.26 Recently, increased DDX3 protein expression has been associated with triple negative breast cancer and advanced Gleason scores in primary CaP.11, 27, 28 RK33, a small molecule inhibitor specific to DDX3, has positive combinatorial therapeutic effects with radiotherapy in lung and primary CaP, where co-treatment decreased proliferation and increased apoptosis.27, 29 Given the correlation between DDX3 localization and primary CaP with high Gleason scores, it is expected that DDX3 localization and expression are also increased in CaP metastases, which has yet to be evaluated.
A better understanding of DDX3 in metastatic CaP could elucidate mechanisms of disease progression and provide a foundation for development of improved therapeutics. The various roles of DDX3, both nuclear and cytoplasmic, have been explored in a multitude of cancers; however, the role of DDX3 in metastatic CaP remains unstudied. Therefore, identifying the expression and localization of DDX3 in this context could provide insight into its function in advanced disease. Here, patient tissues and cell line models for CaP progression, as well as publicly available gene expression data, were used to investigate the expression, localization, and function of DDX3 in CaP metastases.
Materials and Methods
Meta-Analysis
Oncomine data were accessed online at https://www.oncomine.org (registration required).30 Data sets were analyzed for CaP using the cancer versus normal analysis for gene DDX3X.30 The reporter accession numbers for each data set are as follows: 212514_x_at,31 IMAGE: 745188,32 A_24_P126060,33 39744_at,34 and 826_at.35 Gene Expression Omnibus (GEO, https://www.ncbi.nlm.nih.gov/geo) data set GDS1439 was analyzed with the GEO2R analysis tool version 2013 for gene DDX3X (1558120_at).36
Patient Samples and Immunohistochemistry
The CaP progression tissue microarray was created by a board-certified pathologist (W.H.) and contains 336 cores of 0.6 mm from 168 patients (two cores per patient) cut into in 5-μm sections. This array includes 96 cores (48 patients) of benign prostatic tissue (BPT), 50 cores (25 patients) of high-grade prostatic intraepithelial neoplasia (HGPIN), 146 cores (73 patients) of CaP, and 44 cores (22 patients) of metastatic tissue (METS).37 Diagnosis of cores was determined by histologic findings that represented most of the core (<5% intermixed gland diagnoses) and confirmed by the board-certified pathologist (W.H.) at 10-section intervals. Cores that contained >5% intermixed glands, that contained <100 total cells, or were significantly damaged during processing were excluded from the analysis, resulting in total sample sizes of: 46 patients with BPT, 24 patients with HGPIN, 71 patients with CaP, and 19 patients with METS. Immunohistochemical analysis using a commercially available antibody against DDX3 (1:250 dilution, catalog number A300-474A; Bethyl Laboratories Inc, Montgomery, TX) was detected using diaminobenzidine chromogen (catalog number 8059; Cell Signaling Technology, Danvers, MA). Mayer's hematoxylin was used as the nuclear counterstain.
Automated Image Acquisition and Analysis
IHC quantification was performed as previously described.37, 38 Briefly, the progression tissue microarray slide was scanned and imaged at ×20 magnification using a Vectra2 quantitative pathology imaging system. inForm Cell Analysis software version 2.1 (PerkinElmer, Waltham, MA) was used to create spectral libraries of diaminobenzidine and hematoxylin staining, which allows for separation of the two stains. With the use of the Tissue Finder function of inForm software version 2.1 (PerkinElmer), tissue types (stroma, epithelia) and cell compartments (nuclear, cytoplasmic) were segmented and quantified.37 The mean optical density (OD) was calculated for each core, and the two cores from the same patient were averaged to create a single mean OD value for each patient. For each core, epithelial mean OD was normalized to stromal mean OD to control for background staining. Statistical change was analyzed using a one-way analysis of variance with Tukey's multicomparison test.
Cell Culture
RWPE-1, PC3, and DU145 cells were obtained from ATCC (Manassas, VA). BPH1 and BHPrE cells were generously provided by Dr. Simon Hayward at Northshore University Health Systems. LNCaP, C42, and C42B cells were obtained from collaborators at University of Wisconsin, Madison. Benign CaP (BCaP) cell lines were created in the Ricke laboratory as described previously.39, 40, 41 All cell lines were maintained in RPMI 1640 medium and 2.05 mmol/L l-glutamine media (Hyclone, Logan, UT) supplemented with 5% fetal bovine serum (Hyclone), 2.5% HEPES (Hyclone), 1% penicillin/streptomycin (Hyclone), and 0.2% normacin (Invivogen, San Diego, CA) and routinely passaged.
Protein Expression
Cells were seeded in tissue culture grade petri dishes and grown to confluence. Western blot protocol was performed as previously described,42 using BioRad precast gels and nitrocellulose membrane transfer packages (BioRad, Hercules, CA). Antibodies specific to DDX3 (catalog number A300-474A, Bethyl Laboratories Inc) and α-tubulin (α-tub) (catalog number 2125; Cell Signaling Technology) were diluted in 5% bovine serum albumin/Tris-buffered saline/Tween-20 (DDX3 1:2000; α-tub 1:1000) and incubated overnight at 4°C. Species-specific horseradish peroxidase–conjugated secondary antibodies (Bethyl Laboratories Inc) were incubated at room temperature for 1 hour and visualized using chemiluminescent substrate DURA (Thermo Fisher Scientific, Waltham, MA). Band intensity was quantified with ImageJ version 1.49v43 (NIH, Bethesda, MD; http://imagej.nih.gov/ij).and normalized to total protein, which was quantified using stain-free imaging as previously described.43 α-Tub was used as a visual loading control.
Cell Viability Assay
BCaPM1 or C42 cells were seeded at a density of 30,000 cells per well. After adherence, DDX3 inhibition was achieved by transfection with 25 nmol/L siRNA targeting DDX3 or treatment with 2 μM RK33. SMARTpool ON-TARGETplus Human DDX3X siRNA [siRNA targeting DDX3 (siDDX3)] was acquired from Dharmacon (catalog number L-006874-02-0005), with the associated siRNA nontargeting scramble control (siSCBL) (catalog number D-001810-10-05). Cells were transiently transfected with 25 nmol/L siDDX3 or siSCBL control using the Mirus Bio TransIT X2 Dynamic Delivery System (catalog number MIR 6005), according to Mirus' protocol. After 12 hours, transfection reagent was removed, and cells continued to grow in RPMI 1640 medium for 0, 1, 2, or 4 days after transfection. Alternatively, DDX3 was inhibited pharmacologically by treatment with 2 μM RK33 (catalog number 1070773-09-9; SelleckChem, Houston, TX), diluted in dimethyl sulfoxide (DMSO). RK33, or DMSO control, and added to media on day 0, and cells were counted on days 0, 2, 4, and 6 after treatment. Cell viability was assessed at each time point by staining with trypan blue and counted using a TC-20 automated cell counter (BioRad, Hercules, CA).
Immunofluorescence
Immunofluorescence was performed according to Abcam's protocol (Abcam, Cambridge, UK). Briefly, cells were fixed, permeabilized, and then incubated with primary antibodies DDX3 (diluted 1:250; catalog number A300-474A; Bethyl Laboratories Inc), Ki-67 (diluted 1:200; catalog number ab15580; Abcam), or cleaved caspase (cCASP)-3 (diluted 1:250; catalog number 9661; Cell Signaling Technology), followed by anti-rabbit conjugated to AlexaFluor 488 (catalog number A-21206; Thermo Fisher Scientific). For the gain-of-function experiments, BCaPM1 or C42 cells were grown on coverslips and treated as described above with either siRNA for 12 hours or RK33 for 48 hours before fixation. All cells were counterstained with DAPI nuclear stain. Images were taken at ×20 magnification. Fluorescence intensity (measured in arbitrary units) and percent positivity were calculated using ImageJ version 1.49v.44
Motility Assay
BCaPM1 or C42 cells were plated at a seeding density of 50,000 cells per well. After adherence, cells were treated with a DDX3 inhibitor or control. DDX3 was inhibited as described above by transfection with 25 nmol/L siRNA targeting DDX3 or treatment with 2 μM RK33. A total of 48 hours after treatment, a 20-μL pipette tip was used to create a scratch through the cell monolayer in the center of each well, as previously described.40 Images were taken at 0, 6, 12, and 24 hours, and the area of the scratch was quantified with ImageJ version 1.49v. Percentage of scratch closure was calculated by dividing the area of the scratch at each time point by the area of the scratch at 0 hour for each condition and subtracting the resulting percentage from 100. Statistics reflect 3 biological replicates for each treatment.
Statistical Analysis
Data reflect the mean of samples within each category or treatment, and error bars reported indicate SEM. GraphPad/Prism software version 7.05 (GraphPad Software, Inc., La Jolla, CA) was used for all statistical analysis, with a Welch t-test for 3 replicates or a one-way analysis of variance with Tukey's test for multicomparison statistics. P ≤ 0.05 was considered to be statistically significant.
Results
DDX3 Expression Increases in Meta-Analysis of CaP Progression
The expression of DDX3 in advanced/metastatic CaP has not been investigated. To assess DDX3 expression in CaP progression, data-mining platform Oncomine and other publicly available data sets were used. Oncomine data revealed a significant increase in DDX3 expression in primary CaP versus healthy samples in the Wallace et al31 and Tomlins et al33 data sets (P = 0.0002 and P = 0.046, respectively)30 (Table 1). However, other data sets found a trend toward significance (P = 0.054 in the study by Grasso et al32 and P = 0.072 in the study by Singh et al34) or no significant change (P = 0.164 in the study by LaTulippe et al35) (Table 1).30 To assess the expression of DDX3 (associated gene noted DDX3X) in primary versus metastatic CaP, the publicly available data set GDS1439 was used.36 Here, DDX3 was significantly increased in metastatic CaP samples (n = 6) versus primary CaP (n = 7) (P = 0.023) (Table 1). These data suggest DDX3 gene expression can be high in metastatic CaP.
Table 1.
DDX3 Expression Increases in Prostate Cancer (CaP) Progression
| Data set | P value | Fold change | Comparison | n |
|---|---|---|---|---|
| Wallace et al31 | 0.000184 | 2.154 | Primary CaP versus healthy | 89 |
| Tomlins et al33 | 0.046 | 1.302 | Primary CaP versus healthy | 101 |
| Grasso et al32 | 0.054 | 1.106 | Primary CaP versus healthy | 122 |
| Singh et al34 | 0.072 | 1.565 | Primary CaP versus healthy | 102 |
| LaTulippe et al35 | 0.164 | 1.309 | Primary CaP versus healthy | 35 |
| Varambally et al36 | 0.023 | 2.216 | Metastatic versus primary CaP | 13 |
Cytoplasmic DDX3 Is Highly Expressed in Metastatic CaP Tissue
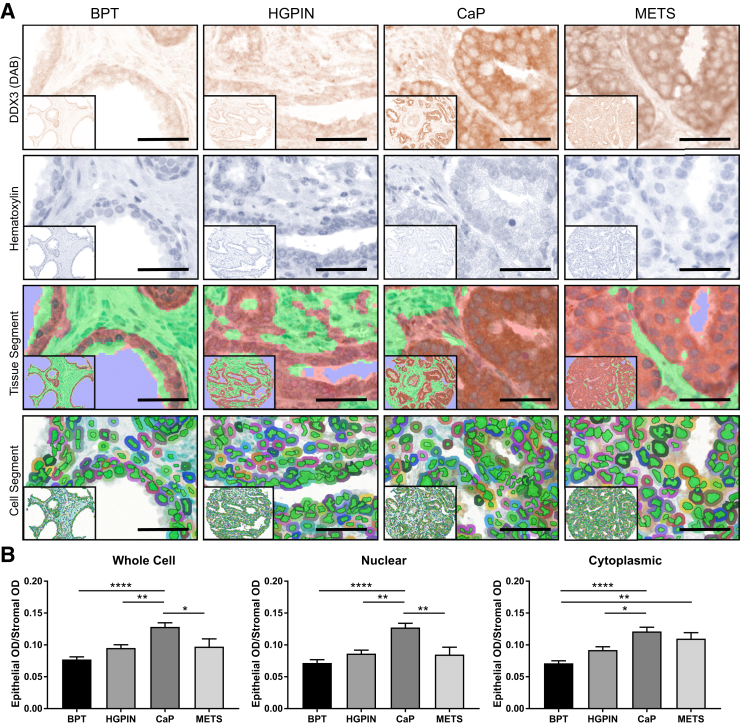
DDX3 is a multifunctional protein that plays a role in nuclear and cytoplasmic RNA processing. Therefore, analyzing the relative expression and localization of DDX3 in CaP progression may provide insight into its functional significance. DDX3 staining was detected in a human prostate tissue microarray that contained cores histologically categorized as BPT (n = 46), HGPIN (n = 24), CaP (n = 71), or METS (n = 19) (Figure 1A).37 inForm software version 2.1 was trained to segment prostate epithelia versus stroma (Figure 1A).45, 46 The mean OD of the epithelial staining was normalized to background by dividing it by the mean OD of the stroma for each core. The staining was then quantified by cell compartment (nuclear, cytoplasmic, whole cell) and compared between disease stages (Figure 1A). Both whole cell and nuclear DDX3 expression was significantly increased in CaP compared with BPT (P < 0.0001) and HGPIN (P = 0.021 for whole cell and P = 0.001 for nuclear) but significantly decreased in METS compared with CaP (P = 0.035 for whole cell and P = 0.002 for nuclear) (Figure 1B). There was no significant change in whole cell or nuclear DDX3 expression between BPT and METS (whole cell P = 0.143 and nuclear P = 0.713). Interestingly, cytoplasmic DDX3 was significantly increased in CaP versus BPT (P < 0.0001) and CaP versus HGPIN (P = 0.021) and remained significantly increased in METS versus BPT (P = 0.005) (Figure 1B). No significant difference was found in cytoplasmic DDX3 in CaP versus METS (P = 0.728) (Figure 1B). Taken together, these data indicate that cytoplasmic, not nuclear, DDX3 is highly expressed in CaP metastases, and investigation of the role of DDX3 in the cytoplasm of these cells may be valuable.
Figure 1.
Cytoplasmic DDX3 is highly expressed in metastatic prostate cancer (CaP) tissue. A: DDX3 expression (brown) was analyzed in a CaP progression tissue microarray (pTMA) that contained cores from patients with benign prostate tissue (BPT; n = 46), high-grade prostatic intraepithelial neoplasia (HGPIN; n = 24), CaP (n = 71), and metastatic tissue (METS; n = 19) (first row). Hematoxylin (blue) was used to counterstain nuclei (second row). Machine learning software (inForm Cell Analysis software version 2.1; PerkinElmer, Waltham, MA) was trained to segment prostate epithelia (red) versus stroma (green) (third row). Cell segmentation analysis distinguished nuclear (green) and cytoplasmic (multicolored) staining (fourth row). B: Quantification of DDX3 expression in the pTMA shows significantly increased mean optical density (OD) in all cell compartments (whole cell, nuclear, cytoplasmic) for CaP versus BPT and CaP versus HGPIN. DDX3 expression was increased in METS versus BPT in the cytoplasm only. DDX3 expression was decreased in METS versus CaP in whole cell and nuclear but not significantly different in the cytoplasm. Mean OD in the epithelia was normalized to stromal mean OD within each core to control for background staining. ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗∗P < 0.0001 (one-way analysis of variance with Tukey multicomparison test). Scale bars = 25 μm. Original magnification, ×20 (insets).
Cytoplasmic DDX3 Expression Is Highly Expressed in Cell Line Models of CaP Metastasis
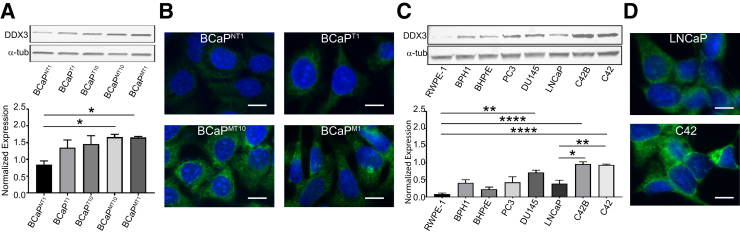
To address the role of DDX3 in the cytoplasm of metastatic CaP cells, both novel and established cell line models of CaP progression were used. First, DDX3 expression and localization was assessed in the BCaP model of CaP progression.40 The BCaP model was derived from the BPH1 cell line, which was immortalized from a nontumorigenic human prostate epithelial cell.41 When BPH1 tissue recombinants that contained rodent urogenital mesenchyme were grown in male athymic mice under steroid hormone treatment, epithelial cells in these grafts underwent malignant transformation as previously described.39, 40, 47 Cells isolated from tissue recombinants at different time points represent distinct stages of CaP progression: nontumorigenic (BCaPNT1), small indolent tumors (BCaPT1), large aggressive tumors (BCaPT10), and lymph node metastases (BCaPMT10 and BCaPM1).39, 40 In this model, DDX3 expression was significantly increased in both metastatic cell lines (BCaPMT10, BCaPM1) compared with nontumorigenic line BCaPNT1 (P = 0.038 and P = 0.041, respectively) (Figure 2A). Localization of DDX3 in BCaP cell lines found a DDX3 expression primarily in the cytoplasm (Figure 2B). In addition, these data confirmed the expression data from Figure 2A because DDX3 was more highly expressed in BCaPMT10 and BCaPM1 cell lines compared with cell lines representing earlier stages of carcinogenesis BCaPNT1 and BCaPT1.
Figure 2.
Cytoplasmic DDX3 expression is highly expressed in cell line models of prostate cancer (CaP) metastasis. A: Quantification of DDX3 expression by Western blot shows a significant increase in the metastatic lines BCaPMT10 and BCaPM1 compared with nontumorigenic line BCaPNT1. Bands were quantified by densitometry in ImageJ version 1.49v.and normalized to total protein. α-Tubulin (α-tub) was used as a visual loading control. B: Localization of DDX3 in BCaP cell lines shows high cytoplasmic DDX3 expression (green) in metastatic cell lines BCaPMT10 and BCaPM1 compared with nontumorigenic cell line BCaPNT1 and early primary tumor cell line BCaPT1. There is minimal nuclear staining. DAPI (blue) was used as the nuclear counterstain. C: Quantification of DDX3 expression in a panel of benign (RWPE-1, BPH1, BHPrE) and CaP (PC3, DU145, LNCaP, C42B, C42) cell lines shows increased DDX3 in DU145, C42B, and C42 compared with nontumorigenic prostate RWPE-1 cell lines. DDX3 expression is significantly increased in C42B and C42 compared with LNCaP. Bands were quantified by densitometry in ImageJ version 1.49v and normalized to total protein. α-Tub was used as a visual loading control. D: Localization of DDX3 in LNCaP parental and derivative (C42) cell lines shows that DDX3 (green) primarily localizes to the cytoplasm in both models, with higher expression in C42 (bottom panel) compared with LNCaP (top panel). DAPI (blue) was used as the nuclear counterstain. n = 3 Western blots (A and C). ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗∗P < 0.0001 (Welch t-test). Scale bars = 5 μm. Original magnification, ×20.
DDX3 expression was analyzed in a panel of established CaP cell lines, including LNCaP and the advanced LNCaP derivatives C42B and C42.48, 49 Although LNCaP is lowly metastatic in vivo, its derivative, C42, is a model for metastatic CaP because it readily metastasizes in vivo.50 A significant increase was seen in DDX3 expression in the CaP cell lines DU145, C42B, and C42 compared with nontumorigenic cell lines RWPE-1 (P = 0.0027, P < 0.0001, and P = 0.0001, respectively) (Figure 2C). Importantly, expression of DDX3 was significantly increased in advanced LNCaP derivative cell lines C42B and C42 compared with parental LNCaP (P = 0.007 and P = 0.0103, respectively) (Figure 2C). Localization of DDX3 in LNCaP and C42 revealed DDX3 primarily in the cytoplasm (Figure 2D). Moreover, DDX3 was more robustly expressed in the advanced LNCaP-derivative C42 versus parental LNCaP (Figure 2C and D). These data provide evidence that CaP cell line models represent similar trends in DDX3 expression because patient data increased expression in advanced/metastatic CaP and localization to the cytoplasm. Therefore, these models are appropriate to investigate the role of DDX3 in the cytoplasm of metastatic CaP.
Genetic Knockdown of DDX3 Decreases Proliferation in CaP Metastasis Models
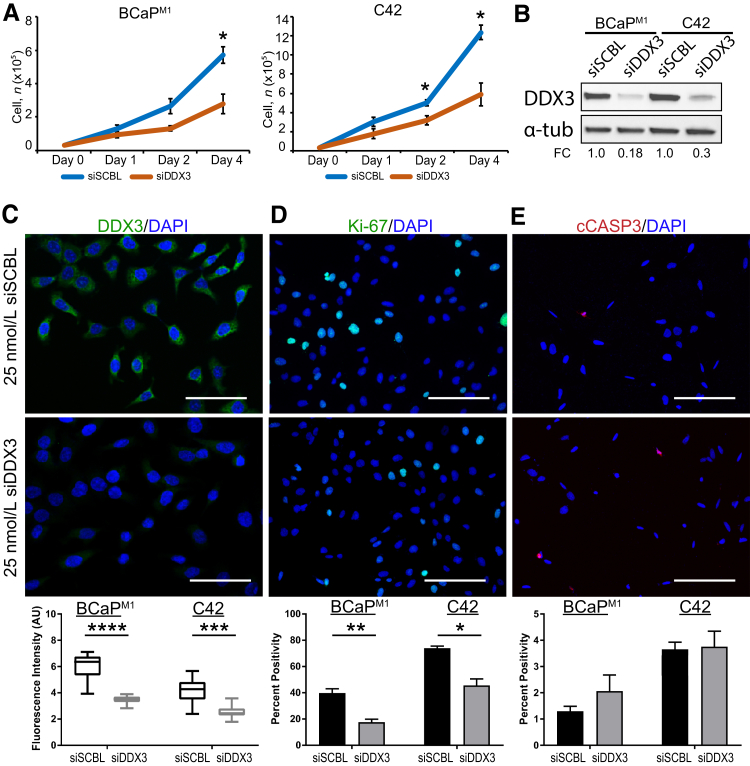
To determine functional implications of DDX3 in the cytoplasm of CaP metastases, DDX3 was genetically knocked down in BCaPM1 and C42 cells. To ascertain the effect of DDX3 knockdown on general cell viability, cells were transiently transfected at day 0 with 25 nmol/L of either siDDX3 or siSCBL and counted using trypan blue at days 1, 2, and 4 after transfection. In BCaPM1, at days 1 and 2, there was no significant change in cell number between siSCBL and siDDX3 transfected cells (P = 0.223 and P = 0.085, respectively). At day 4, a statistically significant decrease of cell number in siDDX3-transfected cells was found compared with control (P = 0.021) (Figure 3A). In C42, no significant difference in cell number at day 1 was found (P = 0.081), but a significant decrease in cell number at days 2 and 4 in the DDX3 knockdown cells was found compared with control (P = 0.012 and P = 0.035, respectively) (Figure 3A). A significant reduction of DDX3 was found in siDDX3-transfected cells compared with control by immunoblot [BCaPM1 fold change (FC) = 0.18, C42 FC = 0.3] and immunofluorescence (BCaPM1 P < 0.0001, C42 P = 0.0003), suggesting the siRNA was targeted and efficient (Figure 3, B–C). Similar to human CaP metastases, Figure 3C confirms DDX3 primarily in the cytoplasm of BCaPM1 cells, with similar results seen in C42 cells (images not shown). Two possible mechanisms through which DDX3 could decrease cell viability are slowing proliferation or inducing apoptosis. To differentiate between these two modalities, the expression of proliferation marker Ki-67 and apoptosis marker cCASP3 was quantified. A statistically significant reduction was seen in Ki-67 positivity in siDDX3-transfected cells compared with siSCBL [BCaPM1 P = 0.007, C42 (images not shown), P = 0.021] (Figure 3D). A change in cCASP3 staining between siSCBL and siDDX3 cells was not seen [BCaPM1 P = 0.341, C42 (images not shown), P = 0.890] (Figure 3E). Taken together, these data suggest that DDX3 plays a role in cell proliferation but not apoptosis in metastatic CaP.
Figure 3.
Genetic knockdown (KD) of DDX3 decreases proliferation in prostate cancer (CaP) metastasis models. A: Cell viability is significantly decreased with siRNA DDX3 (siDDX3) KD compared with control siRNA nontargeting scramble control (siSCBL) for the metastatic cell lines BCaPM1 and C42. Cells were transiently transfected at day 0 with 25 nmol/L siDDX3 or siSCBL and counted using trypan blue at days 1, 2, and 4 after transfection. In BCaPM1, there is no significant change in cell number between DDX3 KD and control at days 1 or 2 but a significant decrease with DDX3 KD at day 4. In C42, there is no significant difference in cell number at day 1 but a significant decrease with DDX3 KD at days 2 and 4. B: Western blot analysis shows a decrease of DDX3 protein with DDX3 KD by siRNA in both BCaPM1 and C42 cells. Expression of DDX3 was normalized to total protein, and α-tubulin (α-tub) was used as a visual loading control. Densitometry was quantified in ImageJ version 1.49v and represented as fold change (FC) over control. In BCaPM1 KD cells, DDX3 expression decreases to 0.18-fold compared with scramble control. In C42 KD cells, DDX3 expression decreases to 0.3-fold compared with scramble control. C: DDX3 protein expression (green) is significantly decreased in siDDX3-transfected cells compared with siSCBL (representative images shown for BCaPM1; images for C42 not shown). DAPI nuclear stain is shown in blue. Fluorescence intensity was quantified in arbitrary units (AU) using ImageJ . D: Staining with the proliferation marker Ki-67 (green) shows a statistically significant reduction in Ki-67 positivity in siDDX3-transfected cells compared with siSCBL (representative images shown for BCaPM1 ;images for C42 not shown). DAPI nuclear stain is shown in blue. Percent positivity was calculated in ImageJ version 1.49v. E: Staining with apoptosis marker cleaved caspase (cCASP)-3 (red) does not show a statistically significant difference in positivity between siDDX3 and siSCBL-transfected cells (representative images shown for BCaPM1; images not shown for C42). DAPI nuclear stain is shown in blue. Percent positivity was calculated in ImageJ. n = 3. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001, and ∗∗∗∗P < 0.0001 versus siDDX3 treatment (Welch t-test). Scale bars: 25 μm (C); 50 μm (D and E).
Pharmacologic Inhibition of DDX3 Decreases Proliferation in CaP Metastasis Models
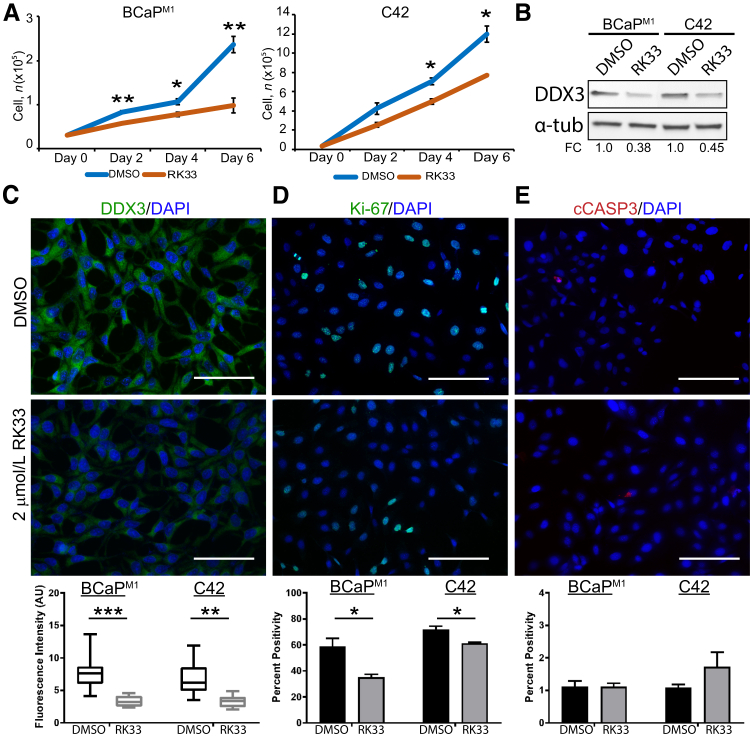
Given that DDX3 genetic knockdown is sufficient to decrease proliferation in metastatic CaP, it is possible that pharmacologic inhibition of DDX3 could produce similar results that are clinically relevant. RK33 is a small molecule inhibitor that specifically binds the ATP-binding domain of DDX3 and prevents its helicase activity.29 To determine the effect of DDX3 pharmacologic inhibition on cell viability, BCaPM1 and C42 cells were treated with 2 μM RK33 or DMSO control at day 0. Cells were collected and counted using trypan blue at days 2, 4, and 6 afer treatment. At all time points assessed, there was a statistically significant decrease of cell number in RK33-treated BCaPM1 cells compared with DMSO control (day 2 P = 0.004, day 4 P = 0.031, and day 6 P = 0.005) (Figure 4A). In C42, cell number was significantly decreased with RK33 treatment at days 4 (P = 0.0119) and 6 (P = 0.0355) (Figure 4A). Similar to genetic knockdown experiments, RK33-treated cells have lower expression of DDX3 protein by immunoblot (BCaPM1 FC = 0.38, C42 FC = 0.45) and immunofluorescence [BCaPM1 P = 0.001, C42 (images not shown), P = 0.003) (Figure 4, B and C). To address the antiproliferative effect of DDX3 inhibition, Ki-67 expression was assessed and a significant reduction was seen in Ki-67 positivity in RK33-treated cells compared with control [BCaPM1 P = 0.039, C42 (images not shown), P = 0.044] (Figure 4D). There was no significant change in cCASP3 staining between RK33 and DMSO [BCaPM1 P = 0.720, C42 (images not shown), P = 0.291] (Figure 4E). Taken together, these data indicate that pharmacologic inhibition of DDX3 in metastatic CaP effectively decreases proliferation but does not induce apoptosis.
Figure 4.
Pharmacologic inhibition of DDX3 decreases proliferation in prostate cancer (CaP) metastasis models. A: Cell viability is significantly decreased with 2 μM RK33 treatment compared with dimethyl sulfoxide (DMSO) control. BCaPM1 and C42 cells were treated at day 0 with 2 μM RK33 or DMSO control and counted using trypan blue at days 2, 4, and 6 after treatment. In BCaPM1, there is a significant decrease in cell viability of RK33-treated cells compared with DMSO control at days 2, 4, and 6. In C42, cell number was significantly decreased with RK33-treatment at days 4 and 6 but not day 2. B: Western blot analysis shows a decrease of DDX3 protein with DDX3 inhibited by RK33 in both BCaPM1 and C42 cells. Expression of DDX3 was normalized to total protein, and α-tubulin (α-tub) was used as a visual loading control. Densitometry was quantified in ImageJ version 1.49v and represented as fold change (FC) over control. In BCaPM1 cells treated with RK33, DDX3 expression decreased to 0.38-fold compared with DMSO control. In C42 cells treated with RK33, DDX3 expression decreased to 0.45-fold compared with DMSO control. C: DDX3 protein expression (green) is significantly decreased in RK33-treated cells compared with DMSO control (representative images are shown for BCaPM1;images for C42 are not shown). DAPI nuclear stain is shown in blue. Fluorescence intensity was quantified in arbitrary units (AU) using ImageJ. D: Staining with proliferation marker Ki-67 (green) shows a statistically significant reduction in Ki-67 positivity in RK33-treated cells compared with DMSO control (representative images for BCaPM1 are shown; images for C42 are not shown). DAPI nuclear stain is shown in blue. Percent positivity was calculated in ImageJ. E: Staining with apoptosis marker cleaved caspase (cCASP)-3 (red) does not show a statistically significant difference in positivity between RK33 and DMSO-treated cells (representative images for BCaPM1are shown; images for C42 are not shown). DAPI nuclear stain is shown in blue. Percent positivity was calculated in ImageJ. n = 3. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001, and ∗∗∗∗P < 0.0001 versus RK33 treatment (Welch t-test). No significance reflects P > 0.05. Scale bars = 50 μm (C–E).
Inhibition of DDX3 Decreases Cell Motility in Metastasis Models
In addition to proliferation and apoptosis, the effect of genetic and pharmacologic DDX3 inhibition was assessed on cell motility (Table 2). A significant decrease was seen in the percentage of scratch closure with siDDX3 at 12 and 24 hours compared with scramble control for both BCaPM1 (12 hours P = 0.013; 24 hours P = 0.009) and C42 (12 hours P = 0.042; 24 hours P = 0.016) (Table 2). No significant change in motility occurred between siDDX3 and siSCBL at 6 hours for either cell line (BCaPM1 P = 0.357; C42 P = 0.445) (Table 2). Similarly, there was a significant decrease in percentage of scratch closure with RK33 treatment at 12 and 24 hours compared with DMSO control for BCaPM1 (12 hours P = 0.025; 24 hours P = 0.003) and C42 (12 hours P = 0.02; 24 hours P = 0.024) (Table 2). There was no significant change in motility between RK33 and DMSO at 6 hours for either cell line (BCaPM1 P = 0.986; C42 P = 0.912) (Table 2). Representative images for the motility assays are shown in Supplemental Figure S1. Taken together, these data suggest that genetic and pharmacologic inhibition of DDX3 is sufficient to decrease cell motility. These findings provide a foundation for potential clinical implications of targeting DDX3 in metastatic CaP.
Table 2.
Cell Motility Decreases with DDX3 Inhibition
| Time, hours | BCaPM1 |
C42 |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| DMSO | RK33 | P value | siSCBL | siDDX3 | P value | siSCBL | siDDX3 | P value | DMSO | RK33 | P value | |
| 6 | 14.01 ± 2.15 | 13.93 ± 3.25 | 0.986 | 17.04 ± 4.58 | 10.94 ± 3.61 | 0.357 | 14.43 ± 5.63 | 7.56 ± 5.86 | 0.445 | 14.72 ± 2.27 | 13.82 ± 4.35 | 0.912 |
| 12 | 60.38 ± 5.25 | 35.79 ± 4.6 | 0.025 | 46.07 ± 2.26 | 31.04 ± 0.89 | 0.013 | 36.9 ± 4.32 | 19.01 ± 4.27 | 0.042 | 50.45 ± 4.25 | 29.53 ± 2.99 | 0.02 |
| 24 | 88.13 ± 5.31 | 43.18 ± 3.71 | 0.003 | 79.24 ± 3.44 | 56.29 ± 3.33 | 0.009 | 58.04 ± 5.44 | 29.4 ± 3.89 | 0.016 | 65.46 ± 5.59 | 36.62 ± 2.07 | 0.024 |
Data are expressed as percentage of scratch closure ± SEM. Bold values signify statistical significance between control and DDX3 inhibition at each time point.
DMSO, dimethyl sulfoxide; siDDX3, siRNA targeting DDX3; siSCBL, siRNA nontargeting scramble control.
Discussion
Metastatic CaP remains a prevalent pathologic condition with low survival rates in part because of a dearth of knowledge about molecular changes associated with advanced disease.1 Thus, identifying and developing more effective targeted therapies for metastatic CaP are necessary to improve patient survival. DDX3 has been implicated in many cancer types as both a positive and negative prognostic marker.26 In primary CaP, increased DDX3 is associated with CaP with a high Gleason score.27 Importantly, preclinical studies of RK33, a small molecule inhibitor specific to DDX3, have found a positive therapeutic value of targeting DDX3 in combination with radiotherapy in both lung and primary CaP27, 29; however, DDX3 expression in CaP metastases had not been assessed before the current study. Here, although DDX3 expression was increased in some meta-analyses of CaP versus healthy samples, it was not significantly changed in others (Table 1). Importantly, there was a significant increase of DDX3 expression in a meta-analysis of metastatic versus primary CaP (Table 1), suggesting that increased DDX3 expression is associated with metastatic CaP and warranting further investigation of the function of DDX3 in this context.
As a helicase, DDX3 plays roles in both nuclear and cytoplasmic RNA processing. DDX3 acts as a translational regulator in the cytoplasm, whereas it contributes to mRNA transcription, splicing, and export in the nucleus; these established roles for DDX3 suggest that its localization provides insight into its function. To quantify DDX3 expression and localization in prostate tissue, an objective machine-learning approach with inForm software version 2.1 was used (Figure 1A). By assessing subcellular localization of DDX3 in progression, an increase of whole cell, nuclear, and cytoplasmic epithelial DDX3 expression was observed in primary CaP compared with BPT (Figure 1B), supporting previously published data.27 In metastatic CaP, these data indicate that nuclear DDX3 was significantly decreased compared with primary CaP and similar to levels expressed in BPT. Interestingly, cytoplasmic DDX3 remained significantly increased in metastatic CaP compared with BPT (Figure 1B). These data suggest that cytoplasmic, not nuclear, DDX3 in the epithelial compartment is a characteristic of metastatic CaP and may have a functional significance in advanced disease. Alternatively, it is possible that the decrease in nuclear DDX3 is contributing to disease progression because loss of DDX3 is a negative prognostic marker in other cancer types, including lung, colon, and head and neck cancers.26 This finding may reveal an undescribed role for DDX3 in CaP progression, and future studies evaluating nuclear and cytoplasmic gain or loss of function may be informative.
To assess the loss of function of cytoplasmic DDX3 in metastatic CaP, DDX3 was inhibited in cell line models for metastatic CaP in two different ways: genetic knockdown with siRNA and pharmacologic inhibition with small molecule inhibitor RK33. These cell line models of CaP progression had similar trends for cytoplasmic DDX3 expression as metadata and clinical data, precipitating their use as models for assessing potential functional implications (Figure 2). With both genetic and pharmacologic loss of function, DDX3 expression was significantly decreased in metastatic CaP cells (Figures 3, B and C, and 4, B and C). Another study found a similar reduction of DDX3 protein after RK33 treatment,29 and Bol et al26 suggested that the mechanism for this decrease in DDX3 protein occurs through DDX3 degradation. Significantly, genetic and pharmacologic inhibition was sufficient to decrease cell viability compared with control in two different models (Figures 3A and 4A). By staining with markers for proliferation or apoptosis, it was concluded that DDX3 may contribute to metastatic growth through proproliferative, not antiapoptotic, effects, because DDX3 loss of function resulted in decreased proliferation but no change in apoptosis (Figures 3, D and E, and 4, D and E). In addition, DDX3 inhibition decreased cell motility in both models (Table 2). Extrapolating from these key findings, we believe it is possible that targeting DDX3 therapeutically may slow metastasis or metastatic cell proliferation; because RK33 is well tolerated in preclinical models in vivo, this may be a useful strategy to prevent or treat metastatic growth. Moreover, inhibiting cytoplasmic DDX3 may affect the profile of translated mRNA because of its role as a translational regulator, and characterization of this metastatic translation profile may reveal additional targetable agents for metastatic CaP.
The implications for targeting DDX3 in CaP have only been assessed once previously and serve as the basis of our current understanding; nuclear DDX3 was highly expressed in primary CaP versus benign tissue, and combination treatment with RK33 and radiotherapy decreased proliferation and increased apoptosis.27 Here, we addressed DDX3 expression and localization in metastatic CaP, differing from previous publication because of our focus on advanced stages of disease.27 This context is significant because of the low survival rates for men with metastatic CaP; RK33 is a nontoxic drug in vivo and if effective against metastatic CaP could be transitioned into the clinic quickly. DDX3 expression and localization in patient tissues were objectively quantified using a machine learning–based approach, implicating cytoplasmic, not nuclear, DDX3 in metastatic CaP. This method is unique because it does not rely on traditional and subjective scoring methods, requiring less time for pathologists and decreasing interobserver variability to foster National Institutes of Health–mandated rigor and reproducibility.37 Finally, RK33 treatment alone was sufficient to decrease proliferation and motility but not induce apoptosis. In previous studies, RK33 treatment alone was not sufficient to affect proliferation or apoptosis in vitro or in vivo, and motility was not assessed.27, 29 This discrepancy could be attributable to the difference in CaP models used in studies or the difference of DDX3 localization in models. To confirm results from previous publication,27 DU145 and PC3 cells were treated with RK33 alone, and a significant change in cell viability, proliferation, or apoptosis was not observed in vitro (data not shown). Although this study did not address combination treatments with RK33 (eg, radiotherapy), perhaps this therapeutic scheme would have even more significant effects than RK33 alone in metastatic CaP and should be explored in future studies.
In summary, cytoplasmic DDX3 is highly expressed in metastatic CaP, and inhibiting DDX3 in this context genetically and pharmacologically was sufficient to attenuate proliferation and motility. These analyses i) provide insight into expression and localization changes related to metastasis, ii) implicate cytoplasmic DDX3 as a feature of metastatic CaP, iii) introduce a previously undescribed role of cytoplasmic DDX3 in CaP progression, and iv) provide a foundation for potential clinical implications of targeting DDX3 in metastatic CaP. Consequently, further investigation into the role of cytoplasmic DDX3 in metastatic CaP may elucidate mechanisms underlying progression to advanced disease that result in reduced patient survival.
Acknowledgments
We thank the University of Wisconsin Translational Research Initiatives in Pathology Laboratory for use of its facilities and services; Dr. Glen Leverson for assistance with statistical analysis; and Drs. Teresa Liu, Samuel Thomas, Dalton McLean, Kristen Uchtmann, and Christian Ortiz-Hernandez for manuscript editing.
Footnotes
Supported by NIH grants DK104310 (W.A.R.), CA009135 (J.E.V.), and CA014520 (University of Wisconsin Carbone Cancer Center, H.B.). J.E.V. is a trainee in the Cancer Biology Graduate Program at the University of Wisconsin-Madison.
Disclosures: None declared.
Supplemental material for this article can be found at https://doi.org/10.1016/j.ajpath.2019.02.011.
Supplemental Data
Inhibition of DDX3 decreases motility in prostate cancer metastasis models. A: Representative images show scratch closure with siRNA DDX3 (siDDX3) knockdown compared with control siRNA nontargeting scramble control (siSCBL) for the metastatic cell lines BCaPM1 and C42. Cells were transiently transfected with 25 nM siDDX3 or siSCBL, and a scratch was made 48 hours after transfection. B: Representative images show cell motility is significantly decreased with RK33 treatment compared with dimethyl sulfoxide (DMSO) control for metastatic cell lines BCaPM1 and C42. Cells were treated with 2 μM RK33, then scratched 48 hours later. Images were taken at 0, 6, and 12 hours after scratching, and scratch boundaries were pseudo-colored in red. Original magnification, ×10 (A and B).
References
- 1.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67:7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 2.Kirby M., Hirst C., Crawford E. Characterising the castration-resistant prostate cancer population: a systematic review. Int Journal Clin Pract. 2011;65:1180–1192. doi: 10.1111/j.1742-1241.2011.02799.x. [DOI] [PubMed] [Google Scholar]
- 3.Sumanasuriya S., De Bono J. Treatment of advanced prostate cancer—A review of current therapies and future promise. Cold Spring Harb Perspect Med. 2018;8:a030635. doi: 10.1101/cshperspect.a030635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Petrylak D.P., Tangen C.M., Hussain M.H., Lara P.N., Jr., Jones J.A., Taplin M.E., Burch P.A., Berry D., Moinpour C., Kohli M. Docetaxel and estramustine compared with mitoxantrone and prednisone for advanced refractory prostate cancer. N Engl J Med. 2004;351:1513–1520. doi: 10.1056/NEJMoa041318. [DOI] [PubMed] [Google Scholar]
- 5.Berthold D.R., Pond G.R., Soban F., de Wit R., Eisenberger M., Tannock I.F. Docetaxel plus prednisone or mitoxantrone plus prednisone for advanced prostate cancer: updated survival in the TAX 327 study. J Clin Oncol. 2008;26:242–245. doi: 10.1200/JCO.2007.12.4008. [DOI] [PubMed] [Google Scholar]
- 6.Small E.J., Schellhammer P.F., Higano C.S., Redfern C.H., Nemunaitis J.J., Valone F.H., Verjee S.S., Jones L.A., Hershberg R.M. Placebo-controlled phase III trial of immunologic therapy with sipuleucel-T (APC8015) in patients with metastatic, asymptomatic hormone refractory prostate cancer. J Clin Oncol. 2006;24:3089–3094. doi: 10.1200/JCO.2005.04.5252. [DOI] [PubMed] [Google Scholar]
- 7.Higano C.S., Schellhammer P.F., Small E.J., Burch P.A., Nemunaitis J., Yuh L., Provost N., Frohlich M.W. Integrated data from 2 randomized, double-blind, placebo-controlled, phase 3 trials of active cellular immunotherapy with sipuleucel-T in advanced prostate cancer. Cancer. 2009;115:3670–3679. doi: 10.1002/cncr.24429. [DOI] [PubMed] [Google Scholar]
- 8.Botlagunta M., Vesuna F., Mironchik Y., Raman A., Lisok A., Winnard P., Jr., Mukadam S., Van Diest P., Chen J., Farabaugh P. Oncogenic role of DDX3 in breast cancer biogenesis. Oncogene. 2008;27:3912. doi: 10.1038/onc.2008.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chao C.-H., Chen C.-M., Cheng P.-L., Shih J.-W., Tsou A.-P., Lee Y.-H.W. DDX3, a DEAD box RNA helicase with tumor growth–suppressive property and transcriptional regulation activity of the p21waf1/cip1 promoter, is a candidate tumor suppressor. Cancer Res. 2006;66:6579–6588. doi: 10.1158/0008-5472.CAN-05-2415. [DOI] [PubMed] [Google Scholar]
- 10.Soulat D., Bürckstümmer T., Westermayer S., Goncalves A., Bauch A., Stefanovic A., Hantschel O., Bennett K.L., Decker T., Superti-Furga G. The DEAD-box helicase DDX3X is a critical component of the TANK-binding kinase 1-dependent innate immune response. EMBO J. 2008;27:2135–2146. doi: 10.1038/emboj.2008.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van Voss M.R.H., Schrijver W.A., Ter Hoeve N.D., Hoefnagel L.D., Manson Q.F., van der Wall E., Raman V., van Diest P.J., Dutch Distant Breast Cancer Metastases Consortium The prognostic effect of DDX3 upregulation in distant breast cancer metastases. Clin Exp Metastasis. 2017;34:85–92. doi: 10.1007/s10585-016-9832-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu D.-W., Lin P.-L., Cheng Y.-W., Huang C.-C., Wang L., Lee H. DDX3 enhances oncogenic KRAS-induced tumor invasion in colorectal cancer via the β-catenin/ZEB1 axis. Oncotarget. 2016;7:22687. doi: 10.18632/oncotarget.8143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu D., Lee M., Wang J., Chen C., Cheng Y., Lee H. DDX3 loss by p53 inactivation promotes tumor malignancy via the MDM2/Slug/E-cadherin pathway and poor patient outcome in non-small-cell lung cancer. Oncogene. 2014;33:1515. doi: 10.1038/onc.2013.107. [DOI] [PubMed] [Google Scholar]
- 14.Merz C., Urlaub H., Will C.L., Lührmann R. Protein composition of human mRNPs spliced in vitro and differential requirements for mRNP protein recruitment. Rna. 2007;13:116–128. doi: 10.1261/rna.336807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou Z., Licklider L.J., Gygi S.P., Reed R. Comprehensive proteomic analysis of the human spliceosome. Nature. 2002;419:182. doi: 10.1038/nature01031. [DOI] [PubMed] [Google Scholar]
- 16.Lai M.-C., Lee Y.-H.W., Tarn W.-Y. The DEAD-box RNA helicase DDX3 associates with export messenger ribonucleoproteins as well astip-associated protein and participates in translational control. Mol Biol Cell. 2008;19:3847–3858. doi: 10.1091/mbc.E07-12-1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yedavalli V.S., Neuveut C., Chi Y.-H., Kleiman L., Jeang K.-T. Requirement of DDX3 DEAD box RNA helicase for HIV-1 Rev-RRE export function. Cell. 2004;119:381–392. doi: 10.1016/j.cell.2004.09.029. [DOI] [PubMed] [Google Scholar]
- 18.Lee C.-S., Dias A.P., Jedrychowski M., Patel A.H., Hsu J.L., Reed R. Human DDX3 functions in translation and interacts with the translation initiation factor eIF3. Nucleic Acids Res. 2008;36:4708–4718. doi: 10.1093/nar/gkn454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shih J.-W., Wang W.-T., Tsai T.-Y., Kuo C.-Y., Li H.-K., Lee Y.-H.W. Critical roles of RNA helicase DDX3 and its interactions with eIF4E/PABP1 in stress granule assembly and stress response. Biochem J. 2012;441:119–129. doi: 10.1042/BJ20110739. [DOI] [PubMed] [Google Scholar]
- 20.Soto-Rifo R., Rubilar P.S., Limousin T., De Breyne S., Decimo D., Ohlmann T. DEAD-box protein DDX3 associates with eIF4F to promote translation of selected mRNAs. EMBO J. 2012;31:3745–3756. doi: 10.1038/emboj.2012.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oh S., Flynn R.A., Floor S.N., Purzner J., Martin L., Do B.T., Schubert S., Vaka D., Morrissy S., Li Y. Medulloblastoma-associated DDX3 variant selectively alters the translational response to stress. Oncotarget. 2016;7:28169. doi: 10.18632/oncotarget.8612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Anderson P., Kedersha N. Stressful initiations. J Cell Sci. 2002;115:3227–3234. doi: 10.1242/jcs.115.16.3227. [DOI] [PubMed] [Google Scholar]
- 23.Buchan J.R., Parker R. Eukaryotic stress granules: the ins and outs of translation. Mol Cell. 2009;36:932–941. doi: 10.1016/j.molcel.2009.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen H., Yu H., Cho W., Tarn W. DDX3 modulates cell adhesion and motility and cancer cell metastasis via Rac1-mediated signaling pathway. Oncogene. 2015;34:2790. doi: 10.1038/onc.2014.190. [DOI] [PubMed] [Google Scholar]
- 25.Lai M.-C., Chang W.-C., Shieh S.-Y., Tarn W.-Y. DDX3 regulates cell growth through translational control of cyclin E1. Mol Cell Biol. 2010;30:5444–5453. doi: 10.1128/MCB.00560-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bol G.M., Xie M., Raman V. DDX3, a potential target for cancer treatment. Mol Cancer. 2015;14:188. doi: 10.1186/s12943-015-0461-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xie M., Vesuna F., Tantravedi S., Bol G.M., Van Voss M.R.H., Nugent K., Malek R., Gabrielson K.L., Van Diest P.J., Tran P.T. RK-33 radiosensitizes prostate cancer cells by blocking the RNA helicase DDX3. Cancer Res. 2016;76:6340–6350. doi: 10.1158/0008-5472.CAN-16-0440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Epstein J.I., Allsbrook W.C., Jr., Amin M.B., Egevad L.L., Committee I.G. The 2005 International Society of Urological Pathology (ISUP) consensus conference on Gleason grading of prostatic carcinoma. Am J Surg Pathol. 2005;29:1228–1242. doi: 10.1097/01.pas.0000173646.99337.b1. [DOI] [PubMed] [Google Scholar]
- 29.Bol G.M., Vesuna F., Xie M., Zeng J., Aziz K., Gandhi N., Levine A., Irving A., Korz D., Tantravedi S. Targeting DDX3 with a small molecule inhibitor for lung cancer therapy. EMBO Mol Med. 2015;7:648–669. doi: 10.15252/emmm.201404368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rhodes D.R., Yu J., Shanker K., Deshpande N., Varambally R., Ghosh D., Barrette T., Pander A., Chinnaiyan A.M. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6:1–6. doi: 10.1016/s1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wallace T.A., Prueitt R.L., Yi M., Howe T.M., Gillespie J.W., Yfantis H.G., Stephens R.M., Caporaso N.E., Loffredo C.A., Ambs S. Tumor immunobiological differences in prostate cancer between African-American and European-American men. Cancer Res. 2008;68:927–936. doi: 10.1158/0008-5472.CAN-07-2608. [DOI] [PubMed] [Google Scholar]
- 32.Grasso C.S., Wu Y.-M., Robinson D.R., Cao X., Dhanasekaran S.M., Khan A.P., Quist M.J., Jing X., Lonigro R.J., Brenner J.C. The mutational landscape of lethal castration-resistant prostate cancer. Nature. 2012;487:239. doi: 10.1038/nature11125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tomlins S.A., Mehra R., Rhodes D.R., Cao X., Wang L., Dhanasekaran S.M., Kalyana-Sundaram S., Wei J.T., Rubin M.A., Pienta K.J. Integrative molecular concept modeling of prostate cancer progression. Nat Genet. 2007;39:41. doi: 10.1038/ng1935. [DOI] [PubMed] [Google Scholar]
- 34.Singh D., Febbo P.G., Ross K., Jackson D.G., Manola J., Ladd C., Tamayo P., Renshaw A.A., D'Amico A.V., Richie J.P. Gene expression correlates of clinical prostate cancer behavior. Cancer Cell. 2002;1:203–209. doi: 10.1016/s1535-6108(02)00030-2. [DOI] [PubMed] [Google Scholar]
- 35.LaTulippe E., Satagopan J., Smith A., Scher H., Scardino P., Reuter V., Gerald W.L. Comprehensive gene expression analysis of prostate cancer reveals distinct transcriptional programs associated with metastatic disease. Cancer Res. 2002;62:4499–4506. [PubMed] [Google Scholar]
- 36.Varambally S., Yu J., Laxman B., Rhodes D.R., Mehra R., Tomlins S.A., Shah R.B., Chandran U., Monzon F.A., Becich M.J. Integrative genomic and proteomic analysis of prostate cancer reveals signatures of metastatic progression. Cancer Cell. 2005;8:393–406. doi: 10.1016/j.ccr.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 37.Huang W., Hennrick K., Drew S. A colorful future of quantitative pathology: validation of Vectra technology using chromogenic multiplexed immunohistochemistry and prostate tissue microarrays. Hum Pathol. 2013;44:29–38. doi: 10.1016/j.humpath.2012.05.009. [DOI] [PubMed] [Google Scholar]
- 38.Bauman T.M., Vezina C.M., Ricke E.A., Halberg R.B., Huang W., Peterson R.E., Ricke W.A. Expression and colocalization of β-catenin and lymphoid enhancing factor-1 in prostate cancer progression. Hum Pathol. 2016;51:124–133. doi: 10.1016/j.humpath.2015.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hayward S.W., Wang Y., Cao M., Hom Y.K., Zhang B., Grossfeld G.D., Sudilovsky D., Cunha G.R. Malignant transformation in a nontumorigenic human prostatic epithelial cell line. Cancer Res. 2001;61:8135–8142. [PubMed] [Google Scholar]
- 40.Liu T.T., Ewald J.A., Ricke E.A., Bell R., Collins C., Ricke W.A. Modeling human prostate cancer progression in vitro. Carcinogenesis. 2018:bgy185. doi: 10.1093/carcin/bgy185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hayward S., Dahiya R., Cunha G., Bartek J., Deshpande N., Narayan P. Establishment and characterization of an immortalized but non-transformed human prostate epithelial cell line: BPH-1. In Vitro Cell Dev Biol Anim. 1995;31:14–24. doi: 10.1007/BF02631333. [DOI] [PubMed] [Google Scholar]
- 42.Towbin H., Staehelin T., Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci U S A. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Colella A.D., Chegenii N., Tea M.N., Gibbins I.L., Williams K.A., Chataway T.K. Comparison of Stain-Free gels with traditional immunoblot loading control methodology. Anal Biochem. 2012;430:108–110. doi: 10.1016/j.ab.2012.08.015. [DOI] [PubMed] [Google Scholar]
- 44.Schneider C.A., Rasband W.S., Eliceiri K.W. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Levenson R. Putting the “more” back in morphology: spectral imaging and image analysis in the service of pathology. Arch Pathol Lab Med. 2008;132:748–757. doi: 10.5858/2008-132-748-PTMBIM. [DOI] [PubMed] [Google Scholar]
- 46.Mansfield J.R. Cellular context in epigenetics: quantitative multicolor imaging and automated per-cell analysis of miRNAs and their putative targets. Methods. 2010;52:271–280. doi: 10.1016/j.ymeth.2010.10.001. [DOI] [PubMed] [Google Scholar]
- 47.Ricke W.A., Ishii K., Ricke E.A., Simko J., Wang Y., Hayward S.W., Cunha G.R. Steroid hormones stimulate human prostate cancer progression and metastasis. Int J Cancer. 2006;118:2123–2131. doi: 10.1002/ijc.21614. [DOI] [PubMed] [Google Scholar]
- 48.Thalmann G.N., Anezinis P.E., Chang S.-M., Zhau H.E., Kim E.E., Hopwood V.L., Pathak S., von Eschenbach A.C., Chung L.W. Androgen-independent cancer progression and bone metastasis in the LNCaP model of human prostate cancer. Cancer Res. 1994;54:2577–2581. [PubMed] [Google Scholar]
- 49.McLean D.T., Strand D.W., Ricke W.A. Prostate cancer xenografts and hormone induced prostate carcinogenesis. Differentiation. 2017;97:23–32. doi: 10.1016/j.diff.2017.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sato N., Gleave M.E., Bruchovsky N., Rennie P.S., Beraldi E., Sullivan L.D. A metastatic and androgen-sensitive human prostate cancer model using intraprostatic inoculation of LNCaP cells in SCID mice. Cancer Res. 1997;57:1584–1589. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Inhibition of DDX3 decreases motility in prostate cancer metastasis models. A: Representative images show scratch closure with siRNA DDX3 (siDDX3) knockdown compared with control siRNA nontargeting scramble control (siSCBL) for the metastatic cell lines BCaPM1 and C42. Cells were transiently transfected with 25 nM siDDX3 or siSCBL, and a scratch was made 48 hours after transfection. B: Representative images show cell motility is significantly decreased with RK33 treatment compared with dimethyl sulfoxide (DMSO) control for metastatic cell lines BCaPM1 and C42. Cells were treated with 2 μM RK33, then scratched 48 hours later. Images were taken at 0, 6, and 12 hours after scratching, and scratch boundaries were pseudo-colored in red. Original magnification, ×10 (A and B).