Abstract
The liver contains diploid and polyploid hepatocytes (tetraploid, octaploid, etc.), with polyploids comprising ≥90% of the hepatocyte population in adult mice. Polyploid hepatocytes form multipolar spindles in mitosis, which lead to chromosome gains/losses and random aneuploidy. The effect of aneuploidy on liver function is unclear, and the degree of liver aneuploidy is debated, with reports showing aneuploidy affects 5% to 60% of hepatocytes. To study relationships among liver polyploidy, aneuploidy, and adaptation, mice lacking E2f7 and E2f8 in the liver (LKO), which have a polyploidization defect, were used. Polyploids were reduced fourfold in LKO livers, and LKO hepatocytes remained predominantly diploid after extensive proliferation. Moreover, nearly all LKO hepatocytes were euploid compared with control hepatocytes, suggesting polyploid hepatocytes are required for production of aneuploid progeny. To determine whether reduced polyploidy impairs adaptation, LKO mice were bred onto a tyrosinemia background, a disease model whereby the liver can develop disease-resistant, regenerative nodules. Although tyrosinemic LKO mice were more susceptible to morbidities and death associated with tyrosinemia-induced liver failure, they developed regenerating nodules similar to control mice. Analyses revealed that nodules in the tyrosinemic livers were generated by aneuploidy and inactivating mutations. In summary, we identified new roles for polyploid hepatocytes and demonstrated that they are required for the formation of aneuploid progeny and can facilitate adaptation to chronic liver disease.
Hepatocytes are characterized by variations in polyploidy and aneuploidy. Polyploidy refers to an increase in the number of chromosome sets and, in hepatocytes, is regulated by the number of nuclei per cell (typically one or two) and the DNA content of each nucleus (diploid, tetraploid, octaploid, etc.).1, 2 At birth, hepatocytes are exclusively diploid, containing two copies of each chromosome. During postnatal development in rodents, a subset of proliferating diploid hepatocytes divides by acytokinetic mitosis to generate binucleate tetraploid daughters that contain two diploid nuclei.3 These daughter cells can then produce a pair of mononucleate tetraploid daughter cells with a round of cell cycling that includes mitosis, followed by successful cytokinesis. The cycle of acytokinetic mitosis, followed by traditional mitosis, continues to generate binucleate and mononucleate octaploids as well as hepatocytes with even higher ploidy.4, 5 More than 50% of adult human hepatocytes are polyploid, and, in adult mice ≥90% are polyploid.6, 7 The polyploid state is also reversible. Proliferating polyploid hepatocytes form multipolar mitotic spindles, and, in some cases, cell divisions produce three or more daughter cells with reduced ploidy states. Chromosome segregation errors are common during multipolar cell division and lead to random and unbiased whole chromosome gains and/or losses (aneuploidy).6, 7, 8 The degree of aneuploidy has been reported to be as low as 5% or as high as 60% in the liver.6, 7, 9, 10 Although aneuploidy is strongly correlated with cancer,11, 12 spontaneous liver cancer is extremely rare,13 suggesting that hepatocyte aneuploidy is a normal part of liver homeostasis. Taken together, the adult liver is comprised of a heterogeneous mixture of diploid and polyploid hepatocytes that are either euploid or aneuploid. Despite our understanding of the mechanisms that influence ploidy and aneuploidy in the liver, we are only just beginning to appreciate how these chromosomal alterations affect function and disease.
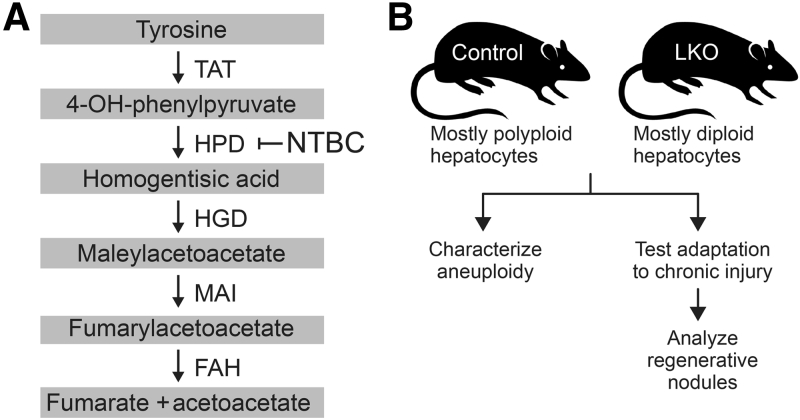
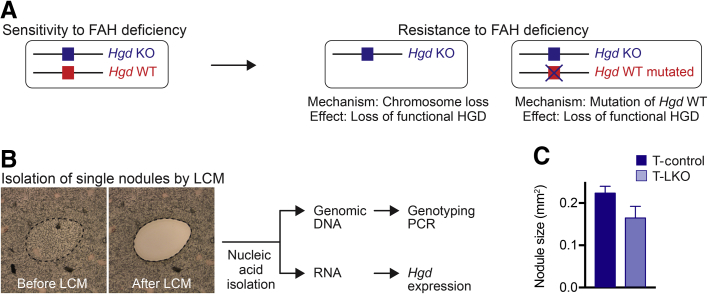
We previously investigated the role of hepatic aneuploidy in chronic liver injury with the use of Fah–/– mice, a model of hereditary tyrosinemia type 1.14, 15 The Fah–/– mouse model has been used in a variety of contexts to study liver function and disease.16, 17 Fumarylacetoacetate hydrolase (FAH) is expressed in the kidney and liver and catalyzes a distal step in the tyrosine catabolic pathway (Figure 1A). Mice lacking FAH accumulate genotoxic intermediates maleylacetoacetate and fumarylacetoacetate in hepatocytes. Cell cycle arrest/death is induced in the hepatocytes, which leads to liver failure and ultimately death of the animal. Blocking the tyrosine pathway upstream of maleylacetoacetate production prevents accumulation of both maleylacetoacetate and fumarylacetoacetate and ameliorates tyrosinemia.18, 19 Treatment with the drug 2-(2-nitro-4-trifluoro-methylbenzoyl)-1,3-cyclo-hexanedione (NTBC), which inhibits the upstream enzyme 4-hydroxyphenylpyruvate dioxygenase (HPD), prevents liver injury and allows Fah–/– mice to be bred and maintained in a healthy state.14 Genetic deletion and inactivation of homogentisate 1,2-dioxygenase (HGD) in the liver has a similar effect.15, 19 Earlier, chronic tyrosinemia was induced in Hgd+/– Fah–/– mice by removal of NTBC. Over 3 months, livers were spontaneously repopulated by subsets of healthy endogenous hepatocytes. These hepatic subsets were resistant to injury by HGD loss of function, which provided them with a competitive advantage, compared with injured hepatocytes, and permitted clonal expansion. It was estimated that half of the regenerating nodules were derived from aneuploid hepatocytes with whole or partial loss of chromosome 16 (Chr 16), containing the Hgd wild-type (WT) locus.15 We, therefore, concluded that aneuploid hepatocytes have the potential to play a major role in liver regeneration and adaptation to long-term injury.
Figure 1.
Diagram of the tyrosine catabolic pathway and experimental design. A: Loss of fumarylacetoacetate hydrolase (FAH) led to accumulation of fumarylacetoacetate and toxic metabolites. Fah deficiency was ameliorated by 2-(2-nitro-4-trifluoro-methylbenzoyl)-1,3-cyclo-hexanedione (NTBC) treatment or loss of homogentisate 1,2-dioxygenase (HGD). B: Schematic of experiments presented herein. HPD, 4-hydroxyphenylpyruvate dioxygenase; LKO, liver-specific E2f7/E2f8 double knockout; MAI, maleylacetoacetate isomerase; TAT, tyrosine aminotransferase.
Transcription factors E2F7 and E2F8, atypical members of the E2F family,20 are highly expressed between weeks 1 and 7 in the developing liver when hepatocyte polyploidization is maximal.21 Recent studies showed liver-specific E2f7/E2f8 double knockout (LKO) mice have a significant reduction of polyploid hepatocytes.21, 22, 23 E2F7 and E2F8 negatively regulate cytokinesis genes, and loss of E2F7 and E2F8 promotes successful cytokinesis of proliferating mononucleate diploid hepatocytes.21 LKO livers have normal development, zonation, and function and exhibit normal regeneration and recovery of mass in response to two-thirds partial hepatectomy.21, 22 Moreover, LKO livers respond to 3,5-diethoxycarbonyl-1,4-dihydrocollidine diet21 and high-fat diet22 similar to control mice. It has also been shown that livers deficient in E2f7 and/or E2f8 have increased susceptibility to spontaneous tumorigenesis and diethylnitrosamine-induced liver cancer.24, 25 Strikingly, hepatocytes from LKO mice are more proliferative in culture and during liver repopulation and partial hepatectomy–induced regeneration. The increased proliferation of LKO hepatocytes is predominately due to the enriched diploid phenotype, as corresponding studies with WT mice demonstrated that diploids are more proliferative than polyploid hepatocytes.22 Because LKO livers are functionally normal and exhibit increased proliferation consistent with diploid enrichment, LKO mice are a robust polyploidy knockout model that can be used to study how loss of polyploidy affects liver aneuploidy and adaptation.
It has been challenging to determine whether diploid and polyploid hepatocytes have similar roles in liver function and homeostasis because hepatocytes readily increase or decrease their ploidy. Here, LKO mice were used to investigate the role of diploids and polyploids in chronic liver injury (Figure 1B). In agreement with previous studies, LKO livers have a significant reduction in polyploid hepatocytes21, 22, 23 and remain predominantly diploid after extensive liver repopulation.22 In addition, cytogenetic studies revealed that LKO hepatocytes are mostly euploid, unlike control hepatocytes that are highly aneuploid with chromosome gains/losses occurring in a widespread and unbiased fashion. To test the idea that reduced polyploidy and aneuploidy in LKO mice would impair their ability to adapt to environmental stresses, LKO mice were bred onto the Hgd+/– Fah–/– background, and NTBC was withdrawn to induce tyrosinemia.15, 19 Survival was significantly reduced in tyrosinemic LKO mice, compared with control mice, but they maintained the ability to form regenerative nodules. Overall, we identify new roles for polyploid hepatocytes—they are required for the formation of aneuploid progeny and are essential mediators that promote adaptation to chronic liver disease.
Materials and Methods
Mouse Strains
The Institutional Care and Use Committee of the University of Pittsburgh approved all of the experiments that used mice. Optimice cages (AnimalCare Systems, Centennial, CO) with Sani-Chip Coarse bedding (P.J. Murphy, Montville, NJ) were used for housing the mice, which were provided ad libitum access to water and standard mouse chow (Purina ISO Pro Rodent 3000; LabDiet, St. Louis, MO) in a standard 12-hour light and 12-hour dark cycle. Huts and running wheels were provided for enrichment. All animals were sacrificed between 9 AM and noon daily. LKO mice were a kind gift from Alain deBruin (Utrecht University, Utrecht, the Netherlands) and Gustavo Leone (The Ohio State University, Columbus, OH).21, 23 LKO mice, which were a mixed background but predominantly FVB, contained floxed E2f7 and E2f8 alleles, a Rosa26-lacZ reporter (R26R-lacZ) and Cre recombinase driven by the albumin promoter (Alb-Cre): E2f7loxP/loxP E2f8 loxP/loxP R26RlacZ/lacZ Alb-CreTg/0. Control mice were negative for Alb-Cre: E2f7loxP/loxP E2f8 loxP/loxP R26RlacZ/lacZ. Fah–/– Rag2–/– IL-2 common γ chain–/– Nod background (FRGN) mice were used for liver repopulation flow cytometry and cytogenetic studies (Yecuris, Inc., Tualatin, OR).26, 27 Hgd+/– Fah–/– mice were acquired from Markus Grompe (Oregon Health and Science University, Portland, OR)15 and bred with the E2f7/E2f8 liver-specific control and knockout mice to generate Tyrosinemia-control (T-control) and Tyrosinemia-LKO (T-LKO) mice, respectively. T-control, T-LKO, and FRGN mice were maintained on 8 mg/L NTBC in their drinking water (Ark Pharm, Libertyville, IL). Male mice were used for experiments unless otherwise stated.
Hepatocyte Isolation
Primary hepatocytes were isolated with the use of a two-step collagenase perfusion.22, 28 Briefly, after general anesthesia induction, a catheter was inserted into the portal vein or inferior vena cava, and 0.3 mg/mL collagenase II (Worthington, Lakewood, NJ) was circulated through the liver. Collagenase-digested livers were placed in Dulbecco's modified Eagle's medium (DMEM)-F12 with 15 mmol/L HEPES (Corning, Corning, NY) + 5% fetal bovine serum (FBS; Sigma-Aldrich, St. Louis, MO), filtered with a 70-μm cell strainer and washed twice using low-speed centrifugation (55 × g for 2 minutes) to remove nonparenchymal cells. Hepatocyte viability, determined by trypan blue staining, was typically >80%. Hepatocytes from FRGN transplant recipients were isolated as described above in this section, except the collagenase II concentration was increased to 1 mg/mL.
Ploidy Analysis
To identify hepatocyte ploidy populations, primary hepatocytes were isolated by a two-step collagenase perfusion, washed with phosphate buffered saline (PBS), adjusted to a density of 1 to 2 million cells/mL and incubated on ice with 2 μL/mL Fixable Viability Dye (FVD) eFluor 780 (65-0865; eBioscience, San Diego, CA). After two PBS washes, hepatocytes were fixed with 2% paraformaldehyde, washed an additional two times with PBS, and incubated with permeabilization buffer (0.1% saponin and 0.5% bovine serum albumin in PBS) + 15 μg/mL Hoechst 33342 (H3570; Life Technologies, Carlsbad, CA). Cells were then washed twice and stored in PBS until flow cytometric analysis. Ploidy populations identified by flow cytometry are indicated with chromatid number c (2c, 4c, 8c). A mixture of quiescent and proliferating cells comprises the gated cells, even though >99% of adult hepatocytes in mice are quiescent (eg, a 4c cell is either a diploid in G2/M phase or a tetraploid in G0/G1 phase).
Liver Repopulation Using LKO or Control Hepatocytes
Primary hepatocytes were isolated from 4-month–old male control and LKO mice with the use of a two-step collagenase perfusion. Control or LKO donor (FAH+) hepatocytes (n = 300,000) were injected into the spleens of 2-month–old female FRGN (FAH–) recipients. The FRGN mice that received a transplant were maintained on 8 mg/L NTBC for 4 days after transplantation and then were cycled on/off NTBC for several cycles to stimulate donor cell (FAH+) expansion. Hepatocytes were isolated from repopulated mice, those whose body weight had stabilized to approximately 100% of the initial weight, using a two-step collagenase perfusion as previously described.22 To identify donor cells, the isolated hepatocytes were washed with PBS, adjusted to a density of 1 to 2 million cells/mL, and incubated on ice with 2 μL/mL FVD eFluor 780 (eBioscience). After two PBS washes, cells were incubated with rabbit anti-FAH primary antibody (a gift from Markus Grompe; Oregon Health and Science University), washed twice with permeabilization buffer (0.5% Saponin in PBS), and incubated with donkey anti-rabbit phosphatidylethanolamine secondary antibody (12-4739-81; eBioscience, San Diego, CA). After two washes in permeabilization buffer, cells were incubated with permeabilization buffer + 15 μg/mL Hoechst 33342, washed again with permeabilization buffer, and stored in 0.5% bovine serum albumin in PBS until flow cytometric analysis.
Cytogenetics
LKO or control hepatocytes from adult mice or fully repopulated FRGN recipients were isolated with the use of a two-step collagenase perfusion. Because >99% of hepatocytes in adult livers are quiescent, the freshly isolated cells were not actively proliferating. To induce proliferation 300,000 viable cells were seeded in 10-cm Primaria Cell Culture dishes (Corning) in seeding media: DMEM-F12 with 15 mmol/L HEPES (Corning), 5% FBS (Atlanta Biologicals, Atlanta, GA), and Antibiotic-Antimycotic Solution (Corning). After 4 hours, seeding medium was replaced with growth medium: DMEM-F12 with 15 mmol/L HEPES (Corning), 0.5% FBS (Atlanta Biologicals), Antibiotic-Antimycotic Solution (Corning), and ITS Supplement (containing 1 μg/mL insulin, 0.55 μg/mL transferrin, and 0.67 ng/mL sodium selenite; Gibco by Life Technologies, Carlsbad, CA). Primary mouse hepatocytes are highly proliferative in vitro under these and similar conditions as demonstrated through live cell imaging, 5′-bromo-2′-deoxyuridine incorporation, and cyclin gene expression.7, 22, 29, 30 After 44 hours in culture, KaryoMAX Colcemid solution (30 ng/mL) was added to the media for 3 hours to arrest dividing cells in metaphase. Proliferating cells must be arrested in metaphase to perform G-banding chromosome staining. After metaphase arrest, the cells were trypsinized, washed in PBS, incubated in prewarmed hypotonic swelling solution (56 mmol/L KCl in 5% FBS) for 10 minutes, and fixed in methanol/acetic acid (3:1 ratio). For each mouse, chromosome G-band staining and karyotype preparation were performed on 14 to 20 metaphase arrested hepatocytes, using a standard trypsin/Wright's staining protocol by the Oregon Health and Science University. For each cell karyotyped, the overall aneuploidy status was determined and all chromosome gains, losses, and other abnormalities (deletions, inversions, etc.) were noted.
NTBC Cycling
T-control and T-LKO mice were maintained on 8 mg/L NTBC until ages 2 to 3 months, at which time it was withdrawn. To induce partial liver repopulation, mice were cycled off NTBC for 1 to 2 weeks and placed on NTBC for 5 days for approximately 2 to 3 periods. Livers were harvested from T-control and T-LKO mice when the mice dropped <80% body weight and expressed high morbidity that could not be improved with placement back on NTBC.
General Processing of Paraffin-Embedded Tissue
Livers from mice were isolated, fixed in 10% neutral buffered formalin, embedded in paraffin, and sectioned at a thickness of 4 μm. Sections were deparaffinized with xylene and graded ethanol (100% to 95%) washes and incubated in citric-acid–based antigen retrieval (Vector, Burlingame, CA).
Immunofluorescence Staining
After paraffin embedding, sectioning, and antigen retrieval, liver sections were stained with Ki-67 and β-catenin to identify regenerating nodules and to quantify the mononuclear and binucleate hepatocytes within the nodules and surrounding tissue. Liver sections were blocked with 2% bovine serum albumin and stained with rabbit anti–Ki-67 primary antibody (ab15580; Abcam Cambridge, MA) and mouse anti–β-catenin primary antibody (sc-7963; Santa Cruz Biotechnology, Dallas, TX), followed by species-specific secondary antibodies: goat anti-rabbit Alexa Fluor 594 and goat anti-mouse Alexa Fluor 488 secondary antibody (A11012 and A11029, respectively; both from Life Technologies). The sections were counterstained with Hoechst 33342 to visualize nuclei and mounted with Fluoromount-G (SouthernBiotech, Birmingham, AL). A minimum of 100 hepatocytes/sample was counted to determine the percentage of mononucleate and binucleate hepatocytes in the nodules and surrounding tissue.
Immunohistochemistry
To identify regenerating nodules, tissue sections were first stained for Ki-67 (G1/S/G2/M phases) because regenerating nodules contained proliferating hepatocytes. After antigen retrieval, deparaffinized tissue sections were treated with 3% hydrogen peroxide to quench endogenous peroxidase and blocked with Avidin/Biotin blocking solution (SP-2001; Vector). Sections were stained with rabbit anti–Ki-67 primary antibody (ab15580; Abcam), followed by biotinylated goat anti-rabbit secondary antibody (BA-1000; Vector) and Avidin-conjugated peroxidase (PK-6100; Vector ABC kit); staining was visualized with ImmPACT DAB Peroxidase Substrate (SK-4105; Vector). Sections were counterstained with hematoxylin, dehydrated with ethanol and xylene washes, and mounted with Permount (Thermo Fisher Scientific, Pittsburgh, PA). Regenerating nodules were also identified with hematoxylin and eosin staining, which was performed by the Development Laboratory in the University of Pittsburgh Department of Pathology.
Laser Capture Microdissection
For laser capture microdissection (LCM), livers were harvested, washed in PBS, and frozen. Tissues were cut at a thickness of 8 μm, and individual nodules were isolated directly from unstained tissue sections with the use of an LMD 6500 laser microdissection system (Leica Microsystems, Buffalo Grove, IL).31
DNA and RNA Isolation and PCR
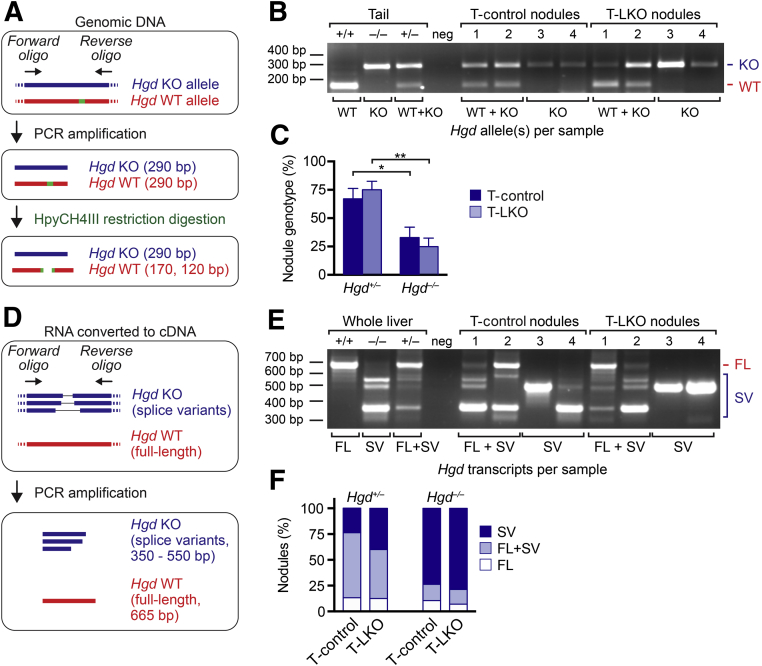
Total DNA and RNA were extracted from LCM-isolated nodules with the use of an AllPrep DNA/RNA Micro Kit (Qiagen, Germantown, MD). The Hgd genotype of the nodules was determined by performing an Hgd genotyping PCR reaction (94°C × 3 minutes; 45 cycles of 94°C × 30 seconds, 56°C × 30 seconds, 72°C × 45 seconds; final extension 72°C × 7 minutes) on the total DNA with the use of the forward primer 5′-TTTAGTCGCTGCATCACC-3′ and reverse primer 5′-CATTTTCACCGTGCTGAC-3′, incubating the PCR product with the restriction enzyme HpyCH4III (Thermo Fisher Scientific) for 4 hours at 37°C and then running the digested products on a 4% agarose gel. The restriction enzyme HpyCH4III cuts the 290-bp PCR product from the WT allele to generate fragments that are 170 bp and 120 bp long, but the KO allele has a one base mismatch at the HpyCH4III recognition site that prevents cleavage. Therefore, nodules that produce three bands after enzyme digestion (290 bp, 170 bp, and 120 bp) were considered Hgd+/–, and nodules that had lost the Hgd WT allele produced a single 290-bp band and were considered Hgd null (Hgd–/–).15, 19
For Hgd transcript analysis, total RNA was reverse-transcribed into cDNA, using Iscript Reverse Transcription Supermix (Bio-Rad, Hercules, CA), and a nested PCR reaction was performed to determine the presence or absence of full-length and splice variant Hgd transcripts in individual nodules. Briefly, the cDNA was run in the first PCR reaction (94°C × 5 minutes; 20 cycles of 90°C × 30 seconds, 62°C × 30 seconds, 72°C × 1 minute; final extension 72°C × 10 minutes), using the forward primer 5′-TTTGAGGAGACCAGGGGCTA-3′ and reverse primer 5′-TCAATTACAGTAGAGGGCTCCAGTC-3′, and after a 50-fold dilution, subjected to a second PCR reaction (94°C × 5 minutes; 40 cycles of 90°C × 30 seconds, 62°C × 30 seconds, 72°C × 1 minute; final extension 72°C × 10 minutes), using the forward primer 5′-GCCTGGTATGAAGATCGTCGAG-3′ and reverse primer 5′-TCAATTACAGTAGAGGGCTCCAGTC-3′. The final products of the second PCR were analyzed on a 2% agarose gel. The Hgd WT allele produces a full-length (FL) transcript corresponding to a 665-bp PCR product. The point mutation within the Hgd KO allele produces shorter splice variants (SVs) approximately 350 bp and 500 to 550 bp in length that form truncated and defective proteins.19, 32 The nodules were classified according to their Hgd genotype (Hgd+/– or Hgd–/–) in combination with their Hgd transcript profile (FL only, FL + SV, or SV only).
Flow Cytometry
All fluorescence-activated cell sorting (FACS) analyses were performed with a FACS Aria II-SORP (BD Biosciences, San Jose, CA) equipped with a 130-μm nozzle and running DiVa software version 8.0.2 (BD Biosciences). FACS plots were generated with FlowJo software version 9.9.6 (TreeStar, Ashland, OR).
Microscopy
Fluorescent images were acquired with a Nikon (Melville, NY) TiU fluorescent microscope equipped with a monochrome camera. Nonfluorescent images were acquired with a Nikon TS100 microscope equipped with a color camera. Gross morphologic images were taken with a Nikon D3100 DSLR Digital Camera with 18-55 mm f/3.5-5.6 Auto Focus-S Nikkor Zoom Lens (Nikon). Images were processed with Nikon NIS Elements Basic Research software version 4.13.00, Nikon NIS Elements Freeware software version 4.00.00, ImageJ software version 1.49 (NIH, Bethesda, MD; https://imagej.nih.gov/ij), and Photoshop CC software version 20.0.2 (Adobe Systems, Mountain View, CA).
Statistical Analysis
Statistical significance was calculated by Prism 7.0.a (GraphPad Inc., San Diego, CA) or Excel 16.16.6 (Microsoft, Redman, WA), using two-tailed t-test except for the Kaplan-Meier curve in which a log-rank (Mantel-Cox) test was performed. P values < 0.05 were considered significant.
Results
Livers Deficient for E2f7 and E2f8 Are Depleted of Polyploid Hepatocytes
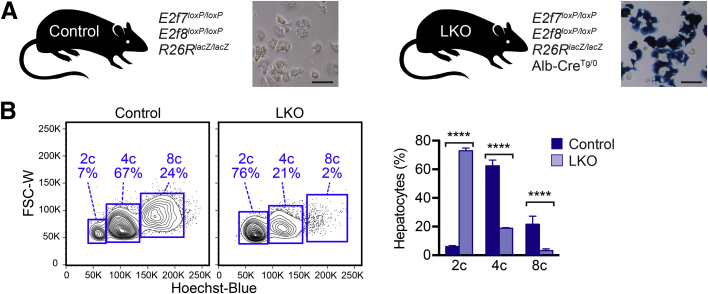
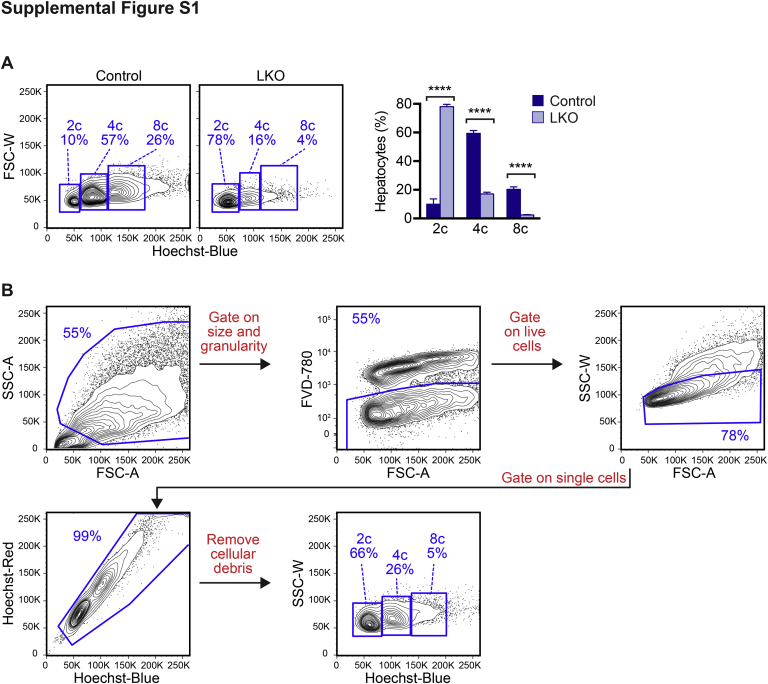
To confirm the reduced polyploidy phenotype in mice that lack E2F7 and E2F8, LKO mice that contained floxed E2f7, floxed E2f8, and a Rosa26-lacZ reporter (R26R-lacZ) were obtained.21, 22, 23 Genes were specifically deleted in LKO livers by Cre recombinase driven by the liver-specific Albumin promoter (Alb-Cre).33 Cre activity was highly efficient as indicated by expression of the β-galactosidase reporter in LKO hepatocytes and its absence in control cells (Figure 2A). Hepatocytes were isolated by two-step collagenase perfusion, stained with the viability marker FVD-780, and loaded with the DNA dye Hoechst 33342. Because >99% of adult hepatocytes were quiescent, cell cycle analysis by flow cytometry provided a precise readout of cellular ploidy. Consistent with previous observations, livers from LKO mice at 3 to 4 weeks and at 4 months were enriched with diploid hepatocytes and depleted of polyploids (Figure 2B, Supplemental Figure S1A).21, 22, 23 For example, 4-month–old LKO livers contained 73.0% ± 1.8% diploids, 18.8% ± 0.4% tetraploids, and 3.2% ± 1.3% octaploids, whereas control livers contained 6.1% ± 0.6% diploids, 62.4% ± 4.0% tetraploids, and 21.7% ± 5.6% octaploids (Figure 2B).
Figure 2.
Diploid hepatocytes were increased and polyploid hepatocytes were depleted in E2f7/E2f8-deficient livers. A: Control mice contained floxed alleles of E2f7/E2f8 and a Rosa26-lacZ reporter (R26RlacZ/lacZ) but did not contain Cre recombinase. E2f7/E2f8 liver-specific knockout mice (LKO) also contained Cre recombinase driven by the albumin promoter (Alb-Cre), a liver-specific driver of Cre, which deleted E2f7/E2f8 and activated the reporter in the liver. Hepatocytes isolated from 2.5-month–old control and LKO livers were stained with X-gal, a substrate for β-galactosidase (blue). The presence of blue hepatocytes confirmed Cre expression in LKO livers. B: Hepatocytes isolated from control and LKO male mice were stained with the viability dye Fixable Viability Dye (FVD)–780 and the nuclear dye Hoechst. The distribution of live hepatic ploidy populations is shown for 4-month–old adult mice. The full gating strategy is shown in Supplemental Figure S1B. Diploid (2c), tetraploid (4c), and octaploid (8c) hepatocytes are indicated on representative flow cytometric plots (left panels), and the percentage of each population is summarized (right panel). Data are expressed as means ± SEM (B). n = 3 livers per genotype (A); n = 4 mice per genotype (B); ∗∗∗∗P < 0.0001. Scale bar = 100 μm. FSC-W, forward scatter width.
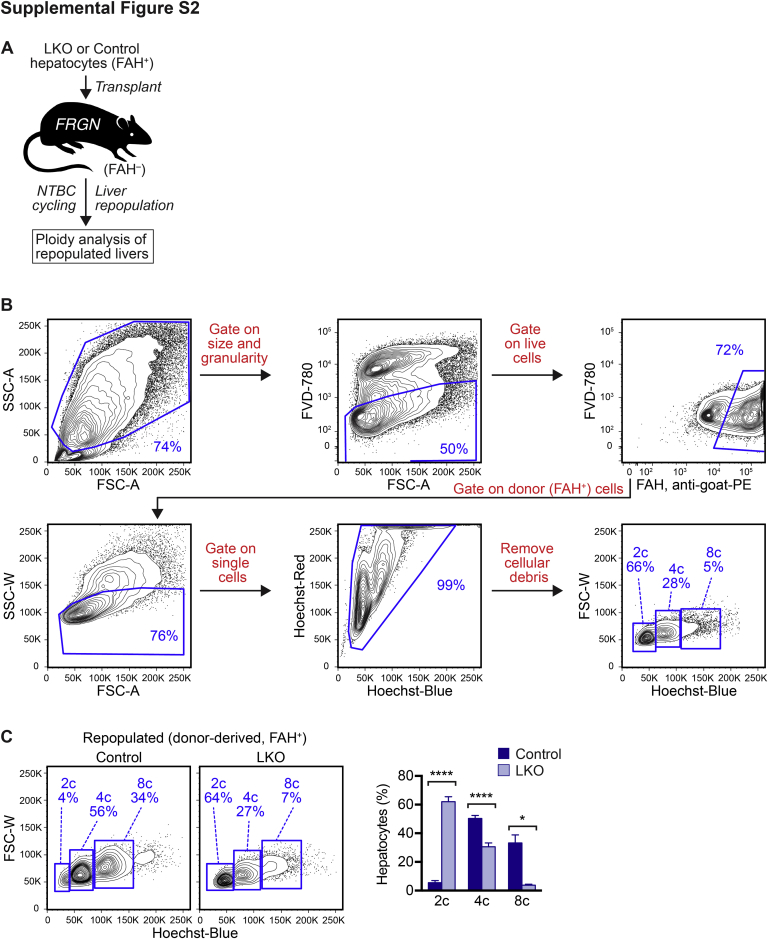
To determine whether the diploid-enriched phenotype in LKO mice remained stable during liver repopulation, the FAH repopulation model in which transplanted donor cells could expand 500- to 1000-fold was used.22, 28 Hepatocytes were isolated from control or LKO mice that were FAH+ and transplanted into Fah–/– recipients (Supplemental Figure S2A). Because control and LKO mice were a mixed background, immune-deficient recipients (FRGN) were used to prevent graft rejection.26, 27 Four days after transplantation, NTBC was withdrawn to promote proliferation of donor hepatocytes. When the body weight dropped to 80%, the mice were provided NTBC for 5 days and NTBC was subsequently removed again. After several rounds of NTBC cycling (approximately 3 months), livers maintained 100% of body weight in the absence of NTBC, an indicator of complete liver repopulation. FAH+ control and LKO donor hepatocytes had a survival advantage over the FAH– FRGN host hepatocytes and were able to proliferate and repopulate host livers over the course of NTBC withdrawal. After liver repopulation, hepatocytes were isolated from the repopulated livers and analyzed for ploidy. To determine the ploidy distribution, donor-derived hepatocytes were isolated from repopulated livers and stained for FAH (expressed only by donor cells), FVD-780, and Hoechst. The ploidy of FAH+ donor-derived hepatocytes was specifically analyzed. Repopulated livers contained approximately 75% FAH+ cells, indicating high-level repopulation (Supplemental Figure S2B). Similar to the ploidy distribution seen in adults, LKO hepatocytes were predominantly diploid and depleted of polyploid hepatocytes (Supplemental Figure S2C). Thus, the diploid-enriched phenotype in LKO mice was stable during normal aging and liver repopulation.
E2f7/E2f8-Deficient Livers Have Reduced Aneuploidy and Are Enriched with Euploid Hepatocytes
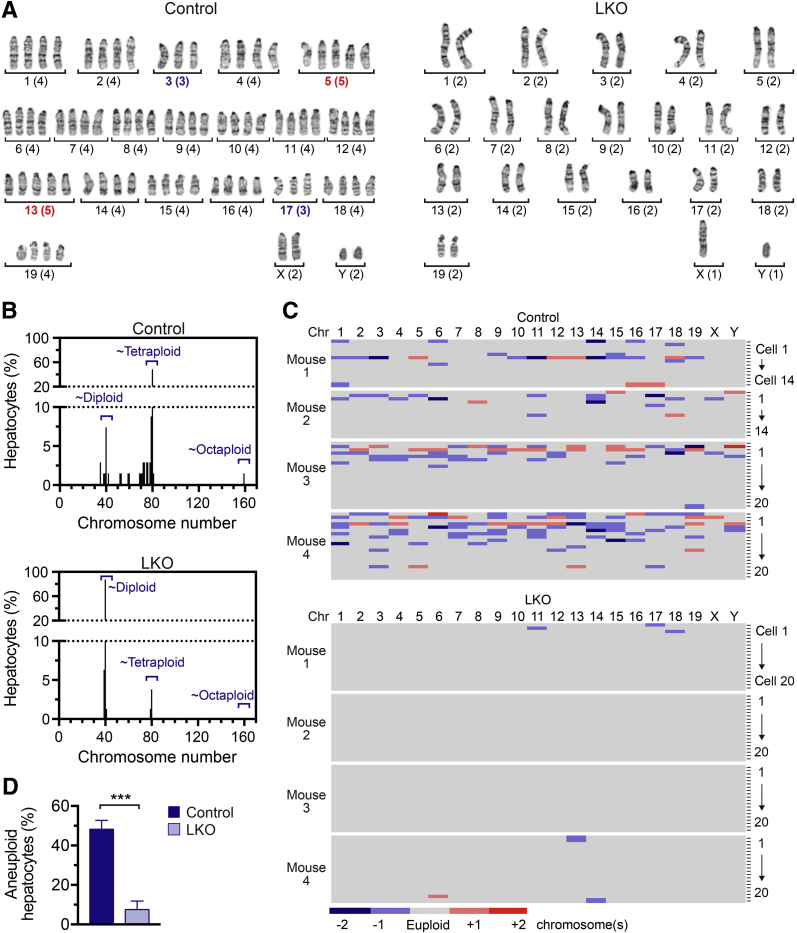
Proliferating polyploid hepatocytes undergo chromosome segregation errors to generate aneuploid daughter cells that have gained/lost ≥1 chromosomes.6, 7 It was rationalized that LKO hepatocytes, which are depleted of polyploids, would also have fewer aneuploid cells. To test this idea, hepatocytes were isolated from 4-month–old control and LKO mice, and individual hepatocytes were karyotyped with the use of standard metaphase cytogenetic analysis. Briefly, freshly isolated hepatocytes were cultured for 44 hours and treated with colcemid to arrest proliferating cells in metaphase. Subsequently, metaphase spreads were prepared and chromosomes were visualized by G-banding. Consistent with ploidy analysis by flow cytometry, most control hepatocytes were tetraploid or near-tetraploid with approximately 80 chromosomes, whereas diploid or near-diploid hepatocytes with approximately 40 chromosomes were uncommon (Figure 3, A and B). Aneuploid hepatocytes were identified by the presence or absence of chromosomes, leading to chromosome numbers close to, but not exactly, 40, 80, or 160 (Figure 3B). Aneuploid hepatocytes with a balanced number of gains and losses were also detected. For example, Figure 3A shows a karyogram for a tetraploid control hepatocyte that contained 80 chromosomes with four homologs of most autosomes; however, this cell contained 3 homologs of chromosomes 3 and 17 (indicating chromosome losses) and 5 homologs of chromosomes 5 and 13 (indicating chromosome gains). As a whole, chromosome losses outnumbered chromosome gains, and all chromosomes were affected, which was consistent with the random aneuploidy previously observed in WT hepatocytes (Figure 3C and Supplemental Table S1).7 In contrast to control cells, most hepatocytes from LKO mice were diploid and contained exactly 40 chromosomes, and aneuploid karyotypes were rarely observed (Figure 3, A–C, Supplemental Table S2). Taken together, 51.8% ± 4.5% of control hepatocytes were aneuploid compared with 7.5% ± 4.3% of LKO hepatocytes (Figure 3D).
Figure 3.
Hepatocytes from E2f7/E2f8-deficient livers were predominantly euploid. Hepatocytes isolated from control or liver-specific E2f7/E2f8 double knockout (LKO) mice (male) were cultured briefly to stimulate cell division, proliferating hepatocytes were arrested in metaphase, and chromosomes from individual cells were identified by G-banding. A: Representative karyograms are shown for control and LKO hepatocytes. Chromosome name (ie, 1 to 19, X, and Y) and number (listed in parentheses) are indicated. Chromosome losses are marked in blue, and chromosome gains are shown in red. The example control hepatocyte was tetraploid and aneuploid with 80 chromosomes (four copies of most autosomes and two copies of each sex chromosome) but lost one copy of chromosomes 3 and 17 and gained one copy of chromosomes 5 and 13. The example LKO hepatocyte was diploid and euploid (two copies of all autosomes and one copy of each sex chromosome). B: Chromosome number is shown for control and LKO hepatocytes. Skewed chromosome counts (eg, hypodiploid, hypotetraploid, and hypertetraploid) were frequent in control but not LKO hepatocytes. C: Chromosome (Chr) gains and losses were summarized relative to the euploid number of homologous chromosomes. Every row marks an individual cell, and each column corresponds to a different chromosome. Detailed karyotypes are provided in Supplemental Tables S1 and S2. D: The percentage of aneuploid hepatocytes is indicated. Data are expressed as means ± SEM (D). n = 4 mice per genotype (B–D); n = 14 to 20 cells per mouse (B–D); n = 68 control hepatocytes (B–D); n = 80 LKO hepatocytes (B–D). ∗∗∗P < 0.001.
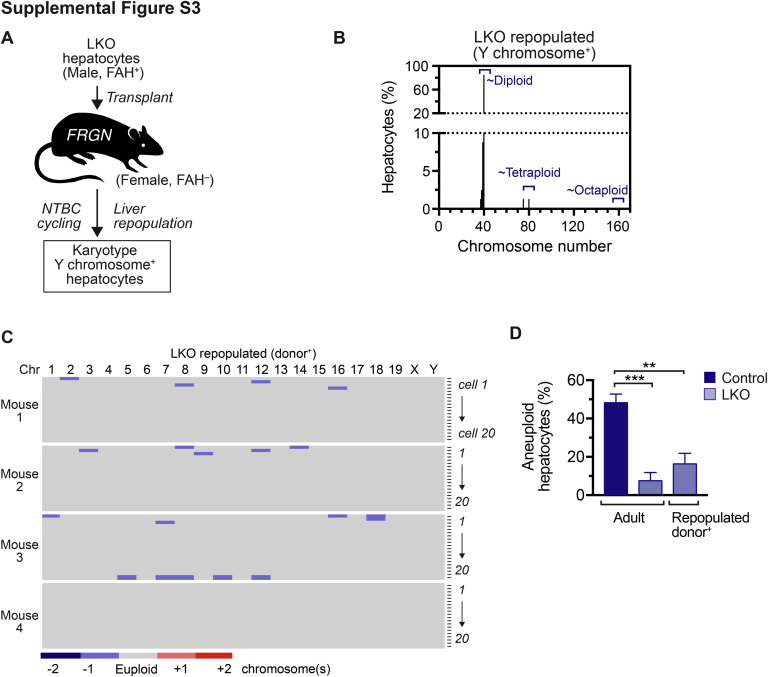
To determine whether LKO hepatocytes remained diploid and euploid after extensive proliferation, the FAH repopulation model was used again (Supplemental Figure S3A). LKO hepatocytes from 4-month–old male mice were transplanted into female FRGN recipients. Mice were cycled off and on NTBC, and, on complete liver repopulation, hepatocytes were isolated for cytogenetic analysis. The FAH+ LKO donor hepatocytes had a survival advantage over the FAH– FRGN host hepatocytes and were able to proliferate and repopulate the FRGN host livers over the course of NTBC withdrawal. After liver repopulation, hepatocytes were isolated and prepared for metaphase cytogenetic analysis. To measure aneuploidy among donor LKO hepatocytes in the repopulated livers, metaphase spreads that were positive for the Y chromosome, a marker for donor-derived cells, were studied. Most donor-derived hepatocytes contained 40 chromosomes and were euploid (Supplemental Figure S3, B and C, and Supplemental Table S3), although there was a trend toward increased aneuploidy in LKO hepatocytes after repopulation (16.3% ± 5.5%) compared with those harvested directly from 4-month–old mice (7.5% ± 4.3%) (Supplemental Figure S3D). Taken together, the data showed that E2f7/E2f8 deficiency markedly altered the chromosomal composition of the liver; LKO livers were highly enriched with diploid and euploid hepatocytes, whereas control livers were predominantly polyploid and aneuploid.
E2f7/E2f8-Deficient Mice Are Highly Sensitive to Chronic Tyrosinemia-Induced Liver Injury
Chronic tyrosinemia liver injury in Hgd+/– Fah–/– mice resulted in spontaneous liver repopulation by subsets of hepatocytes.15 In this model, loss of functional HGD blocked the tyrosine catabolic pathway upstream of maleylacetoacetate and provided resistance to injury (Figure 1A). It was estimated that half of the tyrosinemia-resistant clones acquired inactivating mutations in the Hgd WT allele, and the other regenerating nodules originated from aneuploid hepatocytes that lacked the Hgd WT locus through whole or partial loss of Chr 16.15, 19 The data strongly suggested that preexisting chromosomal variations were required for adaptation to chronic liver injury with the use of an aneuploidy mechanism. E2f7/E2f8-deficient mice, with markedly reduced polyploidy and aneuploidy, provided a unique opportunity to test this idea. It was, therefore, hypothesized that LKO mice with predominantly diploid and euploid livers would be incapable of generating injury-resistant aneuploid clones in the tyrosinemia model of chronic injury, resulting in defective liver adaptation and regeneration as well as reduced survival.
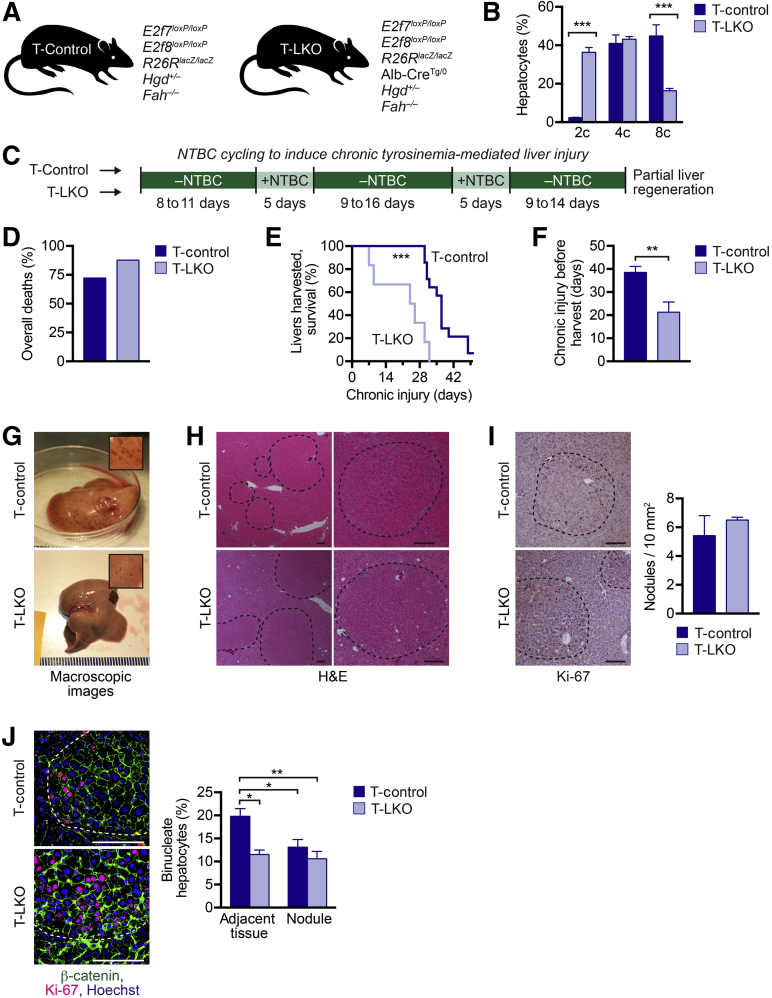
Hgd+/– Fah–/– tyrosinemia mice were bred with control and LKO models to generate Tyrosinemia-control (T-control) and Tyrosinemia-LKO (T-LKO) mice, respectively (Figure 4A). As expected, livers from T-LKO mice contained 16-fold more diploids and 1.5-fold fewer polyploid hepatocytes than T-control mice (Figure 4B). Surprisingly, mice bred to the Hgd+/– Fah–/– tyrosinemia background contained more polyploid hepatocytes than nontyrosinemic mice. For example, <25% of adult LKO hepatocytes were polyploid, whereas nearly 60% of T-LKO hepatocytes were polyploid (Figures 2B and 4B).
Figure 4.
Liver regeneration by control and liver-specific E2f7/E2f8 double knockout (LKO) livers during tyrosinemia-induced chronic injury. A: Control and LKO mice were mated with Hgd+/–Fah–/– mice to generate tyrosinemia models: Tyrosinemia-control (T-control; E2f7loxP/loxPE2f8loxP/loxPR26RlacZ/lacZHgd+/–Fah–/–) and Tyrosinemia-LKO (T-LKO; E2f7loxP/loxPE2f8loxP/loxPR26RlacZ/lacZAlb-CreTg/0Hgd+/–Fah–/–). B: Hepatocytes were isolated from T-control and T-LKO male mice and stained with the viability dye Fixable Viability Dye (FVD)–780 and the nuclear dye Hoechst. The distribution of live hepatic ploidy populations is shown for 4-month–old T-control and T-LKO mice. C: At 2.5 months of age, mice were cycled off/on 2-(2-nitro-4-trifluoro-methylbenzoyl)-1,3-cyclo-hexanedione (NTBC) three times, as indicated, to induce chronic injury and to promote liver regeneration by proliferation of disease-resistant hepatic clones. D: Overall survival during injury for T-control and T-LKO mice. E and F: Kaplan-Meier curve for harvested mice (E) and duration of injury before harvest (F) for T-control and T-LKO mice. G: Tyrosinemia-injured livers contained red macroscopic regenerating nodules. Representative images for T-control and T-LKO livers harvested after 37 and 42 days off NTBC, respectively. Insets depict magnified liver images. H and I: Microscopic nodules are visible by hematoxylin and eosin (H&E) staining (H; low magnification, left column; high magnification, right column) and presence of proliferating Ki-67+ hepatocytes (I, dark brown); the number of nodules is summarized. J: Mononucleate and binucleate hepatocytes were detected. β-catenin (green) marked cell membranes, Hoechst (blue) marked nuclei, and Ki-67 (red) marked proliferating hepatocytes within nodules. The percentage of binucleate cells within nodules and in the tissue adjacent to the nodules is indicated. Dashed lines in H–J indicate nodule boundaries. Data are expressed as means ± SEM (B, F, I, and J) or means only (D). n = 3 to 4 mice per genotype (B); n = 59 T-control mice (D); n = 47 T-LKO mice (D); n = 14 T-control mice (E and F); n = 6 T-LKO mice (E and F); n = 3 to 4 mice per genotype (I). ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.001. Scale bars: 100 μm (H–J). 2c, diploid hepatocytes; 4c, tetraploid hepatocytes; 8c, octaploid hepatocytes.
T-control and T-LKO mice were subjected to NTBC withdrawal for up to 7 weeks to induce tyrosinemia and chronic liver injury (Figure 4C). Experimental mice were monitored throughout the injury period and supplemented with NTBC after 8 to 11 days. Mice were harvested when body weight dropped 20% below the starting weight or exhibited severe morbidity that could not be alleviated with NTBC. The percentage of T-LKO mice that died before harvest (87%) was higher than T-control mice (approximately 72%) (Figure 4D). Moreover, among the mice that were harvested (Figure 4E), T-LKO livers were isolated much earlier (21.3 ± 4.4 days) than livers from T-control mice (38.5 ± 2.6 days) (Figure 4F). Livers harvested from T-LKO and T-control mice were pale, an indicator of liver damage. Most livers were decorated with red macroscopic regenerating nodules, but nodule size was independent of the duration of chronic injury (Figure 4G and Supplemental Figure S4). At the microscopic level, regenerating nodules were apparent in hematoxylin and eosin stains by the appearance of large healthy-looking cells with consistent eosin staining and absence of inflammatory cells (Figure 4H). The healthy and proliferative nature of the nodules was confirmed by the presence of Ki-67+ hepatocytes (Figure 4I). This was expected because hepatocytes within nodules were resistant to tyrosinemia-induced injury and were able to proliferate, whereas hepatocytes in the adjacent tissue were susceptible to tyrosinemia-induced injury and remained quiescent. Nodules were heterogeneous in size and equivalent in number in T-control and T-LKO livers. It has been demonstrated that binucleate hepatocytes could proliferate and that proliferating binucleates generated mononucleate daughters.7, 34 In agreement with these studies, the regenerating nodules in both T-control and T-LKO livers contained fewer binucleate hepatocytes than adjacent tissue, indicating the nodules were predominantly comprised of proliferating mononucleate cells (Figure 4J). Together, the data demonstrated that, although both T-control and T-LKO livers were able to develop healthy regenerating nodules in response to NTBC withdrawal, T-LKO mice were more susceptible to morbidity and death associated with chronic tyrosinemia-induced liver injury.
Aneuploidy and Inactivating Mutations Drive Formation of Regenerating Nodules during Chronic Tyrosinemia-Induced Injury
T-control and T-LKO mice developed regenerating nodules in response to NTBC withdrawal, and these nodules were likely generated by either aneuploidy or gene mutation15 (Figure 5A). In terms of aneuploidy, the nodules could arise from preexisting aneuploid hepatocytes that lost the chromosome containing the Hgd WT allele. Hgd is located on Chr 16 at position qB3, and it was previously shown that loss of the Hgd WT allele occurred by whole or partial loss of Chr 16.15 Because Hgd+/– mice contained only one Hgd WT allele per diploid genome, cells that have lost this allele lacked HGD activity and were, therefore, resistant to tyrosinemia-induced injury (Figure 1A). During chronic injury, these cells have a regenerative advantage and proliferate to restore liver function. The nodules could also be generated by genetic mutation.19 Here, chronic tyrosinemia caused by Fah deficiency led to accumulation of toxic metabolites that could create inactivating mutations in the Hgd WT allele (Figure 1A).19 Once again, these cells became resistant to injury and proliferated to restore liver function. We hypothesized that regenerating nodules from T-control livers were derived from a combination of aneuploid hepatocytes that lost the Hgd WT allele and hepatocytes with a mutated WT allele. In contrast, because LKO hepatocytes were euploid, it was hypothesized that regenerating nodules from T-LKO livers were derived exclusively from hepatocytes with the mutated WT allele.
Figure 5.
Approach to identify the origin of proliferating nodules. A:Hgd+/–Fah–/– tyrosinemic mice are sensitive to 2-(2-nitro-4-trifluoro-methylbenzoyl)-1,3-cyclo-hexanedione (NTBC) withdrawal. Hepatocytes that lose the Hgd wild-type (WT) allele become resistant to tyrosinemia and can proliferate during tyrosinemia-induced injury. Loss of the Hgd WT allele occurred by either loss of chromosome (Chr) 16, which contained the Hgd locus, or by inactivating mutations in the Hgd WT allele. B: Individual regenerating nodules were isolated by laser capture microdissection (LCM), and genomic DNA and RNA were prepared from each nodule for downstream analysis. Dashed lines indicate a representative nodule. C: The size of nodules in harvested Tyrosinemia-control (T-control) and Tyrosinemia–liver-specific E2f7/E2f8 double knockout (T-LKO) livers subjected to LCM was equivalent. n = 4 mice per genotype; mixed sex (C). FAH, fumarylacetoacetate hydrolase; HGD, homogentisate 1,2-dioxygenase; KO, knockout.
To determine the mechanism of nodule formation, individual nodules were isolated by LCM from T-control and T-LKO livers (Figure 5B). Unstained tissues were used because discrete nodules were easily identified within liver sections. Because nodule size and quantity are independent of the duration of injury (Supplemental Figure S4), livers with equivalent sized regenerative nodules were studied from both groups (Figure 5C). Although not statistically significant, there was a trend toward smaller nodules in T-LKO livers, compared with T-control livers, and this was attributed to the smaller size of LKO hepatocytes.21, 22, 23 LCM-isolated nodules were directly placed into lysis buffer, and DNA and RNA were isolated simultaneously to permit analysis of the Hgd locus and transcript. First, the DNA was analyzed to determine the Hgd genotype of the nodules (Figure 6A). The Hgd locus was PCR-amplified, and the resulting 290-bp product was digested with the restriction enzyme HpyCH4III. This enzyme cut the 290-bp product for the WT allele to generate fragments that were 170 bp and 120 bp long. The KO allele has a one base mismatch at the HpyCH4III recognition site that prevented cleavage of the 290-bp product. Hence, nodules that produced three bands after enzyme digestion (290 bp, 170 bp, and 120 bp) were considered Hgd+/–, and nodules that have lost the Hgd WT allele produce a single 290-bp band and were considered Hgd null (Hgd–/–).15, 19 Analysis of nodules from T-control livers revealed that 67.1% ± 9.2% of nodules were Hgd+/–, whereas 32.9% ± 9.2% were Hgd–/– (Figure 6, B and C, and Supplemental Table S4). Surprisingly, a similar distribution was found in nodules from T-LKO livers, and 75.1% ± 15.1% were Hgd+/– and 24.9% ± 15.1% were Hgd–/–.
Figure 6.
The Hgd wild-type (WT) allele was lost by similar mechanisms in equal frequencies by regenerating nodules from Tyrosinemia-control (T-control) and Tyrosinemia–liver-specific E2f7/E2f8 double knockout (T-LKO) livers. A–C: Genomic DNA analysis for each nodule. Strategy for detecting Hgd WT and knockout (KO) alleles in genomic DNA by PCR. Genomic DNA was amplified by PCR and was digested with restriction enzyme HpyCH4III (A). Tail tissue DNA isolated from Hgd+/+, Hgd–/–, and Hgd+/– mice illustrate the WT (170 bp) and KO (290 bp) bands. The 120-bp WT band is not shown. Neg is a negative control that did not contain template DNA. Representative nodules from T-control and T-LKO livers depict WT + KO alleles (nodules 1 and 2) and KO alleles (nodules 3 and 4; B). Nearly 25% of regenerating nodules from T-control and T-LKO livers lost the Hgd WT allele and are effectively Hgd–/– (C). D–F: Transcriptomic analysis of Hgd transcripts in each nodule. Strategy for detecting full-length (FL) transcript from the Hgd WT allele or splice variants (SVs) from the Hgd KO allele (D). Reverse transcription-PCR with the use of RNA from whole livers isolated from Hgd+/+, Hgd–/–, and Hgd+/– mice illustrate FL and SV banding patterns. Neg is a negative control that did not contain template cDNA. Representative nodules from T-control and T-LKO livers depict FL + SV Hgd transcripts (nodules 1 and 2) and SV Hgd transcripts (nodules 3 and 4; E). The percentage of Hgd transcripts is indicated for nodules genotyped as Hgd+/– and Hgd–/–. Hgd transcripts were expressed similarly in nodules from T-control and T-LKO mice, and, consistent with loss of the WT Hgd allele, up to 80% of Hgd–/– nodules contained Hgd SVs only (F). Data are expressed as means ± SEM (C) or means only (F). n = 57 nodules from 4 T-control mice; n = 54 nodules from 4 T-LKO mice. ∗P < 0.05, ∗∗P < 0.01.
To validate the genotyping approach, nodules were interrogated for Hgd mRNA expression. Nodules that lost the Hgd WT locus were expected to have also lost the FL Hgd transcript. mRNA was converted to cDNA and Hgd was PCR-amplified with primers directed to the 3′ end of the gene (Figure 6D). The Hgd WT allele produced a FL transcript that corresponded to a 665-bp PCR product. The point mutation within the Hgd KO allele produced shorter SVs approximately 350 bp and 500 to 550 bp in length that formed truncated and defective proteins.19, 32 Most regenerating nodules from T-LKO (50%) and T-control (65%) livers genotyped as Hgd+/– contained the FL transcript with or without SV transcripts (Figure 6, E and F, and Supplemental Table S4). Manning et al19 previously demonstrated that FL transcripts within these nodules acquired gene inactivating alterations (eg, deletion of 1 to 31 nucleotides, insertion of up to 27 nucleotides, missense mutations), which resulted in HGD loss of function. Most regenerating nodules genotyped as Hgd–/– contained SV only (T-control, 73%; T-LKO, 78%), which was consistent with complete loss of the Hgd locus (Figure 6, E and F, and Supplemental Table S4). Thus, as expected, the data suggested that regenerating nodules from T-controls originated from either aneuploid hepatocytes that lost the Hgd WT allele or by hepatocytes that acquired inactivating mutations within the WT gene. Although T-LKO nodule formation was expected to be driven exclusively by Hgd mutation, the data showed that T-LKO nodules were remarkably similar to T-control nodules because they originated by both the aneuploidy and gene mutation mechanisms.
Discussion
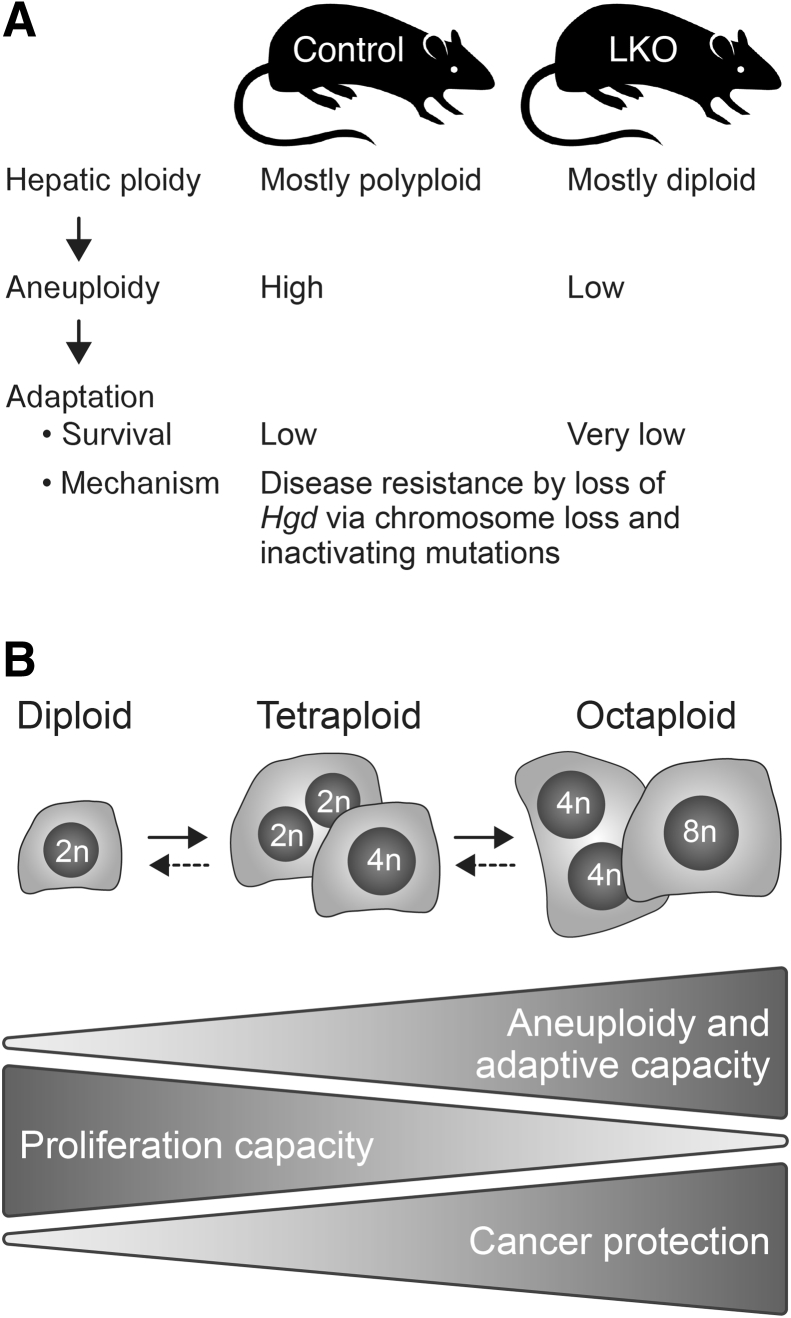
The ploidy conveyor model integrates hepatic polyploidization, ploidy reversal, and aneuploidy in the liver, generating widespread genetic heterogeneity and contributing to liver adaptation in response to stress and injury. Here, our goal was to determine whether loss of liver polyploidy leads to reduced aneuploidy among hepatocytes and impairs the liver's ability to adapt to chronic injury. LKO mice whose livers exhibit reduced hepatic polyploidy were used.21, 22, 23 In agreement with other studies, hepatocyte polyploidization was disrupted in LKO mice because their livers had a fourfold reduction of polyploid hepatocytes and a 10-fold enrichment of diploid hepatocytes. Moreover, the polyploidization defect was stable because LKO hepatocytes remained predominantly diploid after extensive proliferation during liver repopulation. Mice deficient in miR-122, another mouse model with a polyploidization defect, had reduced liver polyploidy and few aneuploid hepatocytes.29 To determine whether reduced liver polyploidy caused by E2f7/E2f8 deletion led to decreased hepatic aneuploidy, hepatocytes were karyotyped from LKO and control adult mice. Cytogenetic analysis revealed that almost all hepatocytes from LKO mice were euploid, whereas nearly 50% of control hepatocytes were aneuploid. LKO hepatocytes remained mostly euploid even after extensive proliferation during liver repopulation. The low-degree aneuploidy of LKO hepatocytes was within the range of background noise attributed to chromosome loss that occurs during slide preparation, and for the Oregon Health and Science University Cytogenetics Research Service Laboratory, this affects <15% of cells. Overall, a concomitant reduction of polyploidy and aneuploidy was observed in mice deficient in miR-122 and LKO mice, polyploidy knockout models, suggesting that polyploid hepatocytes are required for the generation of aneuploid daughter cells (Figure 7A).
Figure 7.
Impact of hepatic polyploidy on liver function. A: Liver-specific E2f7/E2f8 double knockout (LKO) mice with reduced polyploidy were mostly euploid and highly sensitive to tyrosinemia-induced chronic liver injury. In contrast, control hepatocytes were predominantly polyploid, enriched with aneuploid karyotypes, and were less sensitive to injury. B: Model summarizing unique capabilities of diploid and polyploid hepatocytes, incorporating current and recently published findings. Solid arrows indicate polyploidization. Dashed arrows indicate ploidy reversal. Polyploid hepatocytes are protected from tumorigenesis associated with tumor suppressor loss25 and, as demonstrated here, are essential for adaptation to chronic liver injury. In contrast, proliferation capacity was highest among diploid hepatocytes.22 2n, 2 chromosome sets; 4n, 4 chromosome sets; 8n, 8 chromosome sets.
Aneuploidy is common among hepatocytes in WT mice, in which 50% to 60% of adult mouse hepatocytes are aneuploid, and in humans at various ages.6, 7, 15 In 2014, Knouse et al10 subjected hepatic nuclei from one mouse and two human patients to single-cell DNA sequencing and observed aneuploidy among approximately 5% of the sequenced hepatocytes. A follow-up study in 2018 observed low levels of aneuploidy in hepatocytes with normal tissue organization and cell polarity, but increased chromosome segregation errors in hepatocytes dividing in vitro.9 The researchers hypothesized that disrupted tissue architecture increased the chance for merotelic microtubule-kinetochore errors to proceed unchecked in dividing polyploid hepatocytes, ultimately leading to aneuploid daughter cells.9 The idea that normal tissue architecture ensures chromosome segregation fidelity in dividing WT hepatocytes could explain why the metaphase cytogenetic analyses, where hepatocytes are cultured briefly in vitro, reveal a higher percentage of aneuploid hepatocytes. Thus, although the actual frequency of aneuploidy could range from 5% to 60%,7, 9, 10 even the most conservative aneuploidy estimate of 5% indicates that millions of hepatocytes in mice and humans contain random chromosome gains and/or losses, thus highlighting the necessity to understand the role of aneuploidy in liver disease pathology and treatment.
Previously, Manning and colleagues19 induced chronic tyrosinemia injury in Hgd+/– Fah–/– mice, a model of tyrosinemia, and found livers were spontaneously repopulated by disease-resistant hepatic nodules through complete loss of functional HGD, an enzyme located upstream of FAH in the tyrosine catabolic pathway (Figure 1A). Nearly one-half of the nodules were derived from hepatocytes that contained loss-of-function mutations in the Hgd WT allele, with the others originating from aneuploid hepatocytes with whole or partial loss of Chr 16 containing the Hgd WT locus.15 Here, we determined whether polyploidy facilitates adaptation to chronic injury by breeding LKO mice (reduced polyploidy and reduced aneuploidy) onto a tyrosinemia background (Hgd+/– Fah–/–), rationalizing that T-LKO mice would be more sensitive to tyrosinemia-induced injury because of a decreased ability to adapt by the aneuploidy mechanism. Indeed, T-LKO mice were more susceptible to morbidities and death associated with tyrosinemia, and it was necessary to harvest T-LKO livers earlier than T-control livers for analysis. Surprisingly, however, livers from both T-control and T-LKO mice contained healthy regenerating nodules that were comparable in size and frequency on harvest. The percentage of regenerating nodules from T-LKO livers that lost the Hgd WT locus by aneuploidy was equivalent to nodules from T-control livers.
One-half as many T-LKO mice survived tyrosinemia-induced liver injury compared with T-control mice, indicating T-LKO mice were much more susceptible to death and morbidities associated with tyrosinemia-induced liver failure (Figure 4D). It was hypothesized that T-LKO mice were more susceptible because their livers had reduced hepatic polyploidy and, as a consequence, decreased ability to generate aneuploid hepatocytes that could resist the disease. However, other mechanisms may have contributed to their increased susceptibility. First, gene expression differences associated with loss of E2f7 and E2f8 and between diploid and polyploid hepatocytes could have affected their survival. Several studies have found that E2f7 and E2f8 deficiency does not disrupt liver architecture or alter gene expression in mice between weeks 3 and 16.21, 22, 25 Moreover, Lu et al35 found gene expression was similar between diploid and polyploid hepatocytes, suggesting ploidy populations are equivalent at the RNA level. A second possibility is functional differences between diploid and polyploid hepatocytes. Kreutz et al36 found diploids had higher substrate-induced metabolism, measured through esterase activity, and a greater affinity for insulin, indicating that underappreciated functional differences could specifically affect diploid or polyploid hepatocytes during long-term injury. Thus, the increased susceptibility to tyrosinemia-induced liver injury by T-LKO mice may be attributed to multiple mechanisms, including decreased polyploidy and aneuploidy.
Our finding that a similar percentage of regenerating nodules in T-control and T-LKO livers originated by the aneuploidy mechanism was unexpected but could potentially be explained by a combination of factors. First, when LKO mice were bred onto the tyrosinemia background, LKO livers became more polyploid. Specifically, approximately 40% of the hepatocytes in T-LKO mice were diploid and the remaining approximately 60% were polyploid (Figure 4B), compared with approximately 75% being diploid and approximately 25% being polyploid in LKO mice (Figure 2B). The change in ploidy distribution is likely because of strain differences between the LKO and tyrosinemia models. Consistent with this, control mice (containing E2f7 and E2f8) bred onto the tyrosinemia background also exhibited an increased percentage of polyploid hepatocytes. Note that the ploidy differences are unrelated to hepatic injury because T-LKO and T-control mice were maintained on NTBC continuously (ie, no tyrosinemia) before ploidy analysis. T-LKO hepatocytes were not karyotyped because Fah-deficient cells die quickly in vitro and proliferate poorly, making it difficult to obtain metaphases for G-banding. Although the possibility that T-LKO hepatocytes have unique aneuploid karyotypes that make them more susceptible to tyrosinemia-induced morbidities and death cannot be excluded, the aneuploidy data in Figure 3 suggest they are highly euploid and exhibit little to no chromosome gains/losses. Second, only a small number of T-LKO mice were able to be harvested and analyzed, thus potentially biasing the selection for T-LKO livers. We speculate that analysis of only the surviving T-LKO mice inadvertently selected for a unique subset that, randomly, adapted to injury through a combination of aneuploidy and gene inactivation mechanisms. We predict that analysis of T-LKO mice that died and were not able to be harvested would have revealed a failure to generate regenerative nodules by the aneuploidy mechanism that entailed loss of the Hgd WT allele by Chr 16 aneuploidy.
Recent studies have shed new light on the role of hepatic ploidy in liver function and disease. First, Gentric and colleagues30 examined hepatic ploidy in fatty liver disease. With the use of flow analysis and tissue microscopy, they showed that mouse models of fatty liver disease exhibited increased levels of polyploid hepatocytes and a significant increase in mononucleate cells with high nuclear content. Moreover, patients diagnosed with nonalcoholic fatty liver disease who had undergone liver resections for hepatocellular carcinoma displayed similar phenotypes.30 Second, Zhang et al25 showed that polyploid hepatocytes protect against liver cancer. In a series of elegant experiments (supported by other studies22, 24), polyploid hepatocytes were found to buffer against tumor suppressor loss. Inactivation of one tumor suppressor copy in a diploid cell leads to loss of heterozygosity and increased potential for transformation. However, in polyploid hepatocytes, the additional chromosome sets effectively provide backup tumor suppressor copies, which reduce the likelihood of transformation. Third, compared with polyploids, diploid hepatocytes have a proliferative advantage.22 In response to proliferative stimuli, they had faster cell cycle entry and progression than polyploids, suggesting that diploid hepatocytes are immediate drivers of liver regeneration and tumor growth. Finally, multiple studies in patients and animal models have shown that hepatocellular carcinomas are enriched with diploid hepatocytes.37, 38, 39 Thus, taken together, the polyploid state could provide protection from tumorigenesis by providing extra copies of tumor suppressor genes and by restricting hepatocyte proliferation. The present work identifies another specialized role for hepatocytes based on ploidy. Polyploid hepatocytes perform an essential role in adaptation to tyrosinemia-induced chronic liver injury, with one adaptive mechanism involving production of aneuploid progeny. Thus, together with other studies,3, 34, 40 hepatocyte capabilities are directly influenced by chromosomal content (diploid versus polyploid; aneuploid versus euploid), which can dramatically alter cell fate and liver function (Figure 7B).
Future studies should focus on elucidating the conditions that drive polyploids to generate aneuploid hepatocytes in vivo and how this aneuploidy affects liver function and adaptation in homeostasis and disease. For example, transient cell polarity defects are common during liver regeneration induced by partial hepatectomy,41, 42 posing potential aneuploidy-related issues for liver resections and living donor liver transplants. In a cell therapy context, donor hepatocyte isolation involves complete disruption of tissue architecture and cell polarity, which must be reestablished after transplantation in recipients. Although the kinetics of normal architecture and polarity establishment by donor hepatocytes in a new microenvironment are unknown, it was previously demonstrated that donor hepatocyte aneuploidy increased up to twofold in liver repopulation after transplantation.7 In addition, hepatitis c infection has been shown to cause the formation of macroscopic nodules in cirrhotic livers. Of interest, 25% to 50% of these nodules have been shown to be monoclonal,43, 44, 45 which suggests they were derived from single cells with survival advantages because of unique aneuploid karyotypes. Researchers are only beginning to understand the relationship between polyploidy and aneuploidy in the liver; therefore, future studies are warranted to determine how hepatic polyploidy and aneuploidy can affect liver function and adaptation to injury and stress.
Acknowledgments
We thank Gustavo Leone (The Ohio State University, Columbus, OH) and Alain deBruin (Utrecht University, Utrecht, the Netherlands) for sharing the liver-specific E2f7/E2f8 knockout mice; Markus Grompe (Oregon Health and Science University, Portland, OR) for the Hgd+/– Fah–/– mice; Lynda Guzik (McGowan Institute Flow Cytometry Core at University of Pittsburgh, Pittsburgh, PA) for flow cytometry assistance; and Nichole Owen, Muhsen Al-Dhalimy, and Susan Olson (Cytogenetics Research Service Laboratory at Oregon Health and Science University, Portland, OR) for mouse karyotypes.
A.W.D. is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Footnotes
Supported by the NIH grant R01 DK103645 and the Commonwealth of Pennsylvania (A.W.D.).
Disclosures: None declared.
Supplemental material for this article can be found at http://doi.org/10.1016/j.ajpath.2019.02.008.
Supplemental Data
Supplemental Figure S1.
Diploid hepatocytes were increased and polyploid hepatocytes were depleted in E2f7/E2f8-deficient livers. A: Single cell suspensions of liver-specific E2f7/E2f8 double knockout (LKO) and control hepatocytes were stained with Fixable Viability Dye (FVD)–780 (a viability dye) + Hoechst dye and analyzed by flow cytometry. The distribution of live hepatic ploidy populations is shown for 3- to 4-week–old mice. Diploid (2c), tetraploid (4c), and octaploid (8c) hepatocytes are indicated on representative flow cytometry plots (left panels), and the percentage of each population is summarized (right panel). B: The fluorescence-activated cell sorting (FACS) gating strategy for identification of hepatic ploidy populations is shown. Cells were gated on the basis of size/granularity and viability. Subsequently, single cells were selected and cellular debris removed. Hepatic ploidy populations were determined by separating the cells based on incorporation of Hoechst, using the Hoechst-blue channel. Ploidy populations are marked with chromatid number c because they contain a mixture of cycling and quiescent cells (although >99% are quiescent). Representative FACS plots are shown for a 4-month–old LKO mouse. Data are expressed as means ± SEM (A). n = 4 to 7 mice per genotype (A). ∗∗∗∗P < 0.0001. FSC-A, forward scatter area; FSC-W, forward scatter width; SSC-A, side scatter area; SSC-W, side scatter width.
Supplemental Figure S2.
Hepatocytes from E2f7/E2f8-deficient livers remained predominantly diploid during extensive in vivo proliferation. A: Approximately 300,000 hepatocytes from control and liver-specific E2f7/E2f8 double knockout (LKO) mice (males, 4 months old) were transplanted into FRGN (Fah–/–Rag2–/– IL-2 common γ chain–/– Nod background) recipient mice (females, 2 months old) and subjected to 2-(2-nitro-4-trifluoro-methylbenzoyl)-1,3-cyclo-hexanedione (NTBC) cycling to promote liver repopulation by transplanted donor cells. After completed repopulation, livers that contained a mixture of fumarylacetoacetate hydrolase positive (FAH+) donor hepatocytes and FAH– host hepatocytes were isolated and stained with Fixable Viability Dye (FVD)–780, Hoechst, and FAH antibody (donor marker). B: Cells were first gated on the basis of size/granularity and viability. Single cells were then selected and cellular debris was removed. Donor-derived cells (FAH+) were selected and hepatic ploidy populations was determined. Representative fluorescence-activated cell sorting (FACS) plots are shown for an FRGN mouse repopulated by LKO hepatocytes. C: The distribution of hepatic ploidy populations of live FAH+ donor-derived hepatocytes from mice repopulated with control or LKO hepatocytes are shown in representative flow cytometric plots and summarized in the graph. Data are expressed as means ± SEM (C). n = 4 mice repopulated with control hepatocytes (C); n = 4 mice repopulated with LKO hepatocytes (C). ∗P < 0.05, ∗∗∗∗P < 0.0001. 2c, diploid hepatocytes; 4c, tetraploid hepatocytes; 8c, octaploid hepatocytes; FSC-A, forward scatter area; FSC-W, forward scatter width; SSC-A, side scatter area; SSC-W, side scatter width.
Supplemental Figure S3.
Liver-specific E2f7/E2f8 double knockout (LKO) hepatocytes remained predominantly euploid after repopulation of FRGN (Fah–/–Rag2–/– IL-2 common γ chain–/– Nod background) livers. A: Approximately 300,000 hepatocytes from LKO mice (males, 4 months old) were transplanted into FRGN recipient mice (females, 2 months old) and subjected to 2-(2-nitro-4-trifluoro-methylbenzoyl)-1,3-cyclo-hexanedione (NTBC) cycling to promote liver repopulation by transplanted donor cells. After completed repopulation, livers that contained a mixture of fumarylacetoacetate hydrolase positive (FAH+) donor hepatocytes and FAH– host hepatocytes were isolated. Hepatocytes were cultured briefly to stimulate cell division, proliferating hepatocytes were arrested in metaphase, and chromosomes from individual cells were identified by G-banding. Karyotypes were determined for donor-derived, male LKO hepatocytes that were Y chromosome+. B: Chromosome number is shown for LKO-derived hepatocytes. Skewed chromosome counts (eg, hypodiploid, hypotetraploid) were infrequent. C: Chromosome (Chr) gains and losses were summarized relative to the euploid number of homologous chromosomes. Every row marks an individual cell, and each column corresponds to a different chromosome. Detailed karyotypes are provided in Supplemental Table S3. D: The percentage of aneuploid LKO hepatocytes was compared with the degree of aneuploidy in freshly isolated 4-month–old adult control and LKO hepatocytes, which were originally reported in Figure 3D. Data are expressed as means ± SEM (D). n = 4 mice; 20 cells karyotyped per mouse (A); n = 80 LKO-derived hepatocytes (B). ∗∗P < 0.01, ∗∗∗P < 0.001.
Supplemental Figure S4.
The size of macroscopic regenerating nodules in tyrosinemia-injured livers was independent of the length of injury. Tyrosinemia-control (T-control) and Tyrosinemia–liver-specific E2f7/E2f8 double knockout (T-LKO) livers were cycled off/on 2-(2-nitro-4-trifluoro-methylbenzoyl)-1,3-cyclo-hexanedione (NTBC) as described in Figure 4C. Most livers contained macroscopic regenerating nodules, but nodule size and number varied. Representative images depict liver heterogeneity from T-control and T-LKO livers that were harvested. Livers are arranged from least (left) to most (right) nodules, and the duration of tyrosinemia-induced liver injury (caused by NTBC withdrawal) is indicated beneath each image.
References
- 1.Gentric G., Desdouets C. Polyploidization in liver tissue. Am J Pathol. 2014;184:322–331. doi: 10.1016/j.ajpath.2013.06.035. [DOI] [PubMed] [Google Scholar]
- 2.Pandit S.K., Westendorp B., de Bruin A. Physiological significance of polyploidization in mammalian cells. Trends Cell Biol. 2013;23:556–566. doi: 10.1016/j.tcb.2013.06.002. [DOI] [PubMed] [Google Scholar]
- 3.Celton-Morizur S., Merlen G., Couton D., Margall-Ducos G., Desdouets C. The insulin/Akt pathway controls a specific cell division program that leads to generation of binucleated tetraploid liver cells in rodents. J Clin Invest. 2009;119:1880–1887. doi: 10.1172/JCI38677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Duncan A.W. Aneuploidy, polyploidy and ploidy reversal in the liver. Semin Cell Dev Biol. 2013;24:347–356. doi: 10.1016/j.semcdb.2013.01.003. [DOI] [PubMed] [Google Scholar]
- 5.Margall-Ducos G., Celton-Morizur S., Couton D., Bregerie O., Desdouets C. Liver tetraploidization is controlled by a new process of incomplete cytokinesis. J Cell Sci. 2007;120:3633–3639. doi: 10.1242/jcs.016907. [DOI] [PubMed] [Google Scholar]
- 6.Duncan A.W., Hanlon Newell A.E., Smith L., Wilson E.M., Olson S.B., Thayer M.J., Strom S.C., Grompe M. Frequent aneuploidy among normal human hepatocytes. Gastroenterology. 2012;142:25–28. doi: 10.1053/j.gastro.2011.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Duncan A.W., Taylor M.H., Hickey R.D., Hanlon Newell A.E., Lenzi M.L., Olson S.B., Finegold M.J., Grompe M. The ploidy conveyor of mature hepatocytes as a source of genetic variation. Nature. 2010;467:707–710. doi: 10.1038/nature09414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Duncan A.W., Hickey R.D., Paulk N.K., Culberson A.J., Olson S.B., Finegold M.J., Grompe M. Ploidy reductions in murine fusion-derived hepatocytes. PLoS Genet. 2009;5:e1000385. doi: 10.1371/journal.pgen.1000385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Knouse K.A., Lopez K.E., Bachofner M., Amon A. Chromosome segregation fidelity in epithelia requires tissue architecture. Cell. 2018;175:200–211.e13. doi: 10.1016/j.cell.2018.07.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Knouse K.A., Wu J., Whittaker C.A., Amon A. Single cell sequencing reveals low levels of aneuploidy across mammalian tissues. Proc Natl Acad Sci U S A. 2014;111:13409–13414. doi: 10.1073/pnas.1415287111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ganem N.J., Storchova Z., Pellman D. Tetraploidy, aneuploidy and cancer. Curr Opin Genet Dev. 2007;17:157–162. doi: 10.1016/j.gde.2007.02.011. [DOI] [PubMed] [Google Scholar]
- 12.Thompson S.L., Bakhoum S.F., Compton D.A. Mechanisms of chromosomal instability. Curr Biol. 2010;20:R285–R295. doi: 10.1016/j.cub.2010.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leenders M.W., Nijkamp M.W., Borel Rinkes I.H. Mouse models in liver cancer research: a review of current literature. World J Gastroenterol. 2008;14:6915–6923. doi: 10.3748/wjg.14.6915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grompe M., Lindstedt S., al-Dhalimy M., Kennaway N.G., Papaconstantinou J., Torres-Ramos C.A., Ou C.N., Finegold M. Pharmacological correction of neonatal lethal hepatic dysfunction in a murine model of hereditary tyrosinaemia type I. Nat Genet. 1995;10:453–460. doi: 10.1038/ng0895-453. [DOI] [PubMed] [Google Scholar]
- 15.Duncan A.W., Hanlon Newell A.E., Bi W., Finegold M.J., Olson S.B., Beaudet A.L., Grompe M. Aneuploidy as a mechanism for stress-induced liver adaptation. J Clin Invest. 2012;122:3307–3315. doi: 10.1172/JCI64026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grompe M. Fah knockout animals as models for therapeutic liver repopulation. Adv Exp Med Biol. 2017;959:215–230. doi: 10.1007/978-3-319-55780-9_20. [DOI] [PubMed] [Google Scholar]
- 17.Nakamura K., Tanaka Y., Mitsubuchi H., Endo F. Animal models of tyrosinemia. J Nutr. 2007;137:1556S–1560S. doi: 10.1093/jn/137.6.1556S. discussion 1573S–1575S. [DOI] [PubMed] [Google Scholar]
- 18.Endo F., Kubo S., Awata H., Kiwaki K., Katoh H., Kanegae Y., Saito I., Miyazaki J., Yamamoto T., Jakobs C., Hattori S., Matsuda I. Complete rescue of lethal albino c14CoS mice by null mutation of 4-hydroxyphenylpyruvate dioxygenase and induction of apoptosis of hepatocytes in these mice by in vivo retrieval of the tyrosine catabolic pathway. J Biol Chem. 1997;272:24426–24432. doi: 10.1074/jbc.272.39.24426. [DOI] [PubMed] [Google Scholar]
- 19.Manning K., Al-Dhalimy M., Finegold M., Grompe M. In vivo suppressor mutations correct a murine model of hereditary tyrosinemia type I. Proc Natl Acad Sci U S A. 1999;96:11928–11933. doi: 10.1073/pnas.96.21.11928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moon N.S., Dyson N. E2F7 and E2F8 keep the E2F family in balance. Dev Cell. 2008;14:1–3. doi: 10.1016/j.devcel.2007.12.017. [DOI] [PubMed] [Google Scholar]
- 21.Pandit S.K., Westendorp B., Nantasanti S., van Liere E., Tooten P.C., Cornelissen P.W., Toussaint M.J., Lamers W.H., de Bruin A. E2F8 is essential for polyploidization in mammalian cells. Nat Cell Biol. 2012;14:1181–1191. doi: 10.1038/ncb2585. [DOI] [PubMed] [Google Scholar]
- 22.Wilkinson P.D., Delgado E.R., Alencastro F., Leek M.P., Roy N., Weirich M.P., Stahl E.C., Otero P.A., Chen M.I., Brown W.K., Duncan A.W. The polyploid state restricts hepatocyte proliferation and liver regeneration in mice. Hepatology. 2019;69:1242–1258. doi: 10.1002/hep.30286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen H.Z., Ouseph M.M., Li J., Pecot T., Chokshi V., Kent L., Bae S., Byrne M., Duran C., Comstock G., Trikha P., Mair M., Senapati S., Martin C.K., Gandhi S., Wilson N., Liu B., Huang Y.W., Thompson J.C., Raman S., Singh S., Leone M., Machiraju R., Huang K., Mo X., Fernandez S., Kalaszczynska I., Wolgemuth D.J., Sicinski P., Huang T., Jin V., Leone G. Canonical and atypical E2Fs regulate the mammalian endocycle. Nat Cell Biol. 2012;14:1192–1202. doi: 10.1038/ncb2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kent L.N., Rakijas J.B., Pandit S.K., Westendorp B., Chen H.Z., Huntington J.T., Tang X., Bae S., Srivastava A., Senapati S., Koivisto C., Martin C.K., Cuitino M.C., Perez M., Clouse J.M., Chokshi V., Shinde N., Kladney R., Sun D., Perez-Castro A., Matondo R.B., Nantasanti S., Mokry M., Huang K., Machiraju R., Fernandez S., Rosol T.J., Coppola V., Pohar K.S., Pipas J.M., Schmidt C.R., de Bruin A., Leone G. E2f8 mediates tumor suppression in postnatal liver development. J Clin Invest. 2016;126:2955–2969. doi: 10.1172/JCI85506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang S., Zhou K., Luo X., Li L., Tu H.C., Sehgal A., Nguyen L.H., Zhang Y., Gopal P., Tarlow B.D., Siegwart D.J., Zhu H. The polyploid state plays a tumor-suppressive role in the liver. Dev Cell. 2018;44:447–459.e5. doi: 10.1016/j.devcel.2018.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wilson E.M., Bial J., Tarlow B., Bial G., Jensen B., Greiner D.L., Brehm M.A., Grompe M. Extensive double humanization of both liver and hematopoiesis in FRGN mice. Stem Cell Res. 2014;13:404–412. doi: 10.1016/j.scr.2014.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Azuma H., Paulk N., Ranade A., Dorrell C., Al-Dhalimy M., Ellis E., Strom S., Kay M.A., Finegold M., Grompe M. Robust expansion of human hepatocytes in Fah-/-/Rag2-/-/Il2rg-/- mice. Nat Biotechnol. 2007;25:903–910. doi: 10.1038/nbt1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Overturf K., Al-Dhalimy M., Tanguay R., Brantly M., Ou C.N., Finegold M., Grompe M. Hepatocytes corrected by gene therapy are selected in vivo in a murine model of hereditary tyrosinaemia type I. Nat Genet. 1996;12:266–273. doi: 10.1038/ng0396-266. [DOI] [PubMed] [Google Scholar]
- 29.Hsu S.H., Delgado E.R., Otero P.A., Teng K.Y., Kutay H., Meehan K.M., Moroney J.B., Monga J.K., Hand N.J., Friedman J.R., Ghoshal K., Duncan A.W. MicroRNA-122 regulates polyploidization in the murine liver. Hepatology. 2016;64:599–615. doi: 10.1002/hep.28573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gentric G., Maillet V., Paradis V., Couton D., L'Hermitte A., Panasyuk G., Fromenty B., Celton-Morizur S., Desdouets C. Oxidative stress promotes pathologic polyploidization in nonalcoholic fatty liver disease. J Clin Invest. 2015;125:981–992. doi: 10.1172/JCI73957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yovchev M.I., Locker J., Oertel M. Biliary fibrosis drives liver repopulation and phenotype transition of transplanted hepatocytes. J Hepatol. 2016;64:1348–1357. doi: 10.1016/j.jhep.2016.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Manning K., Fernandez-Canon J.M., Montagutelli X., Grompe M. Identification of the mutation in the alkaptonuria mouse model. Mutations in brief no. 216. Hum Mutat. 1999;13:171. doi: 10.1002/(SICI)1098-1004(1999)13:2<171::AID-HUMU15>3.0.CO;2-W. Online. [DOI] [PubMed] [Google Scholar]
- 33.Postic C., Magnuson M.A. DNA excision in liver by an albumin-Cre transgene occurs progressively with age. Genesis. 2000;26:149–150. doi: 10.1002/(sici)1526-968x(200002)26:2<149::aid-gene16>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 34.Miyaoka Y., Ebato K., Kato H., Arakawa S., Shimizu S., Miyajima A. Hypertrophy and unconventional cell division of hepatocytes underlie liver regeneration. Curr Biol. 2012;22:1166–1175. doi: 10.1016/j.cub.2012.05.016. [DOI] [PubMed] [Google Scholar]
- 35.Lu P., Prost S., Caldwell H., Tugwood J.D., Betton G.R., Harrison D.J. Microarray analysis of gene expression of mouse hepatocytes of different ploidy. Mamm Genome. 2007;18:617–626. doi: 10.1007/s00335-007-9048-y. [DOI] [PubMed] [Google Scholar]
- 36.Kreutz C., MacNelly S., Follo M., Waldin A., Binninger-Lacour P., Timmer J., Bartolome-Rodriguez M.M. Hepatocyte ploidy is a diversity factor for liver homeostasis. Front Physiol. 2017;8:862. doi: 10.3389/fphys.2017.00862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Saeter G., Schwarze P.E., Nesland J.M., Juul N., Pettersen E.O., Seglen P.O. The polyploidizing growth pattern of normal rat liver is replaced by divisional, diploid growth in hepatocellular nodules and carcinomas. Carcinogenesis. 1988;9:939–945. doi: 10.1093/carcin/9.6.939. [DOI] [PubMed] [Google Scholar]
- 38.Schwarze P.E., Saeter G., Armstrong D., Cameron R.G., Laconi E., Sarma D.S., Preat V., Seglen P.O. Diploid growth pattern of hepatocellular tumours induced by various carcinogenic treatments. Carcinogenesis. 1991;12:325–327. doi: 10.1093/carcin/12.2.325. [DOI] [PubMed] [Google Scholar]
- 39.Nagasue N., Kohno H., Chang Y.C., Yamanoi A., Kimoto T., Takemoto Y., Nakamura T. DNA ploidy pattern in synchronous and metachronous hepatocellular carcinomas. J Hepatol. 1992;16:208–214. doi: 10.1016/s0168-8278(05)80117-5. [DOI] [PubMed] [Google Scholar]
- 40.Sigal S.H., Rajvanshi P., Gorla G.R., Sokhi R.P., Saxena R., Gebhard D.R., Jr., Reid L.M., Gupta S. Partial hepatectomy-induced polyploidy attenuates hepatocyte replication and activates cell aging events. Am J Physiol. 1999;276:G1260–G1272. doi: 10.1152/ajpgi.1999.276.5.G1260. [DOI] [PubMed] [Google Scholar]
- 41.Odin P., Obrink B. Dynamic expression of the cell adhesion molecule cell-CAM 105 in fetal and regenerating rat liver. Exp Cell Res. 1986;164:103–114. doi: 10.1016/0014-4827(86)90458-1. [DOI] [PubMed] [Google Scholar]
- 42.Odin P., Obrink B. The cell-surface expression of the cell adhesion molecule cellCAM 105 in rat fetal tissues and regenerating liver. Exp Cell Res. 1988;179:89–103. doi: 10.1016/0014-4827(88)90351-5. [DOI] [PubMed] [Google Scholar]
- 43.Ochiai T., Urata Y., Yamano T., Yamagishi H., Ashihara T. Clonal expansion in evolution of chronic hepatitis to hepatocellular carcinoma as seen at an X-chromosome locus. Hepatology. 2000;31:615–621. doi: 10.1002/hep.510310311. [DOI] [PubMed] [Google Scholar]
- 44.Paradis V., Dargere D., Bonvoust F., Rubbia-Brandt L., Ba N., Bioulac-Sage P., Bedossa P. Clonal analysis of micronodules in virus C-induced liver cirrhosis using laser capture microdissection (LCM) and HUMARA assay. Lab Invest. 2000;80:1553–1559. doi: 10.1038/labinvest.3780165. [DOI] [PubMed] [Google Scholar]
- 45.Paradis V., Laurendeau I., Vidaud M., Bedossa P. Clonal analysis of macronodules in cirrhosis. Hepatology. 1998;28:953–958. doi: 10.1002/hep.510280409. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.