Key Points
Question
Are there common genetic variants contributing to progression of geographic atrophy secondary to age-related macular degeneration?
Findings
This pooled analysis combines data from 4 independent studies assessing progression of geographic atrophy secondary to age-related macular degeneration. The correlation between square root–transformed geographic atrophy growth rate and more than 7 million genetic variants was estimated, and 2 gene loci with genome-wide significance were identified as geographic atrophy progression–associated candidates: protein arginine methyltransferase 6 (chromosome 1) and lanosterol synthase (chromosome 21).
Meaning
Gene prioritization within 2 identified loci as geographic atrophy progression–associated candidates, providing a basis for future functional studies exploring mechanisms of atrophic lesion growth.
This pooled analysis of data from 4 cohort studies assesses the contribution of common genetic variants to geographic atrophy lesion growth secondary to age-related macular degeneration in older adults.
Abstract
Importance
Age-related macular degeneration (AMD) is a common threat to vision loss in individuals older than 50 years. While neovascular complications in AMD are treatable, there is currently no therapy for geographic atrophy secondary to AMD. Geographic atrophy lesion progression over time shows considerable interindividual variability, but little is known about prognostic factors.
Objective
To elucidate the contribution of common genetic variants to geographic atrophy lesion growth.
Design, Setting, and Participants
This pooled analysis combined 4 independent studies: the Fundus Autofluorescence Imaging in Age-Related Macular Degeneration (FAM) study, the Directional Spread in Geographic Atrophy (DSGA) study, the Age-Related Eye Disease Study (AREDS), and the Geographic Atrophy Treatment Evaluation (GATE) study. Each provided data for geographic atrophy lesion growth in specific designs. Patients with geographic atrophy secondary to AMD were recruited to these studies. Genotypes were retrieved through the database of Genotypes and Phenotypes (for AREDS) or generated at the Cologne Center for Genomics (for FAM, DSGA, and GATE).
Main Outcomes
The correlation between square root–transformed geographic atrophy growth rate and 7 596 219 genetic variants passing quality control was estimated using linear regression. The calculations were adjusted for known factors influencing geographic atrophy growth, such as the presence of bilateral geographic atrophy as well as the number of lesion spots and follow-up times.
Main Outcomes and Measures
Slopes per allele, 95% CIs, and P values of genetic variants correlated with geographic atrophy lesion growth.
Results
A total of 935 patients (mean [SD] age, 74.7 [7.8] years; 547 female participants [59.0%]) were included. Two gene loci with conservative genome-wide significance were identified. Each minor allele of the genome-wide associated variants increased the geographic atrophy growth rate by a mean of about 15% or 0.05 mm per year. Gene prioritization within each locus suggests the protein arginine methyltransferase 6 gene (PRMT6; chromosome 1; slope, 0.046 [95% CI, 0.026-0.066]; P = 4.09 × 10−8) and the lanosterol synthase gene (LSS; chromosome 21; slope, 0.105 [95% CI, 0.068-0.143]; P = 4.07 × 10−7) as the most likely progression-associated genes.
Conclusions and Relevance
These data provide further insight into the genetic architecture of geographic atrophy lesion growth. Geographic atrophy is a clinical outcome with a high medical need for effective therapy. The genes PRMT6 and LSS are promising candidates for future studies aimed at understanding functional aspects of geographic atrophy progression and also for designing novel and targeted treatment options.
Introduction
Age-related macular degeneration (AMD) is a degenerative disease of the central retina and the most common cause of irreversible blindness in elderly individuals.1 Late-stage disease presents either as neovascular complications or as atrophic processes (geographic atrophy [GA]). While disease progression in neovascular AMD can be slowed by anti–vascular endothelial growth factor treatment, the development and progression of GA is currently untreatable.2,3,4,5
Both growth rate and appearance of GA lesions are highly variable among individual patients, although the 2 eyes of an individual generally reveal a high degree of concordance.6 The mean increase in GA lesions is between 1.3 mm2 and 2.6 mm2 per year, with several independent features contributing to progression variability. Specifically, the presence of GA in both eyes or neovascular AMD in the fellow eye; the location, appearance, and pattern of GA lesions; and an increased number of lesion spots significantly increase GA lesion growth.6
In recent years, major advances have been made to delineate the genetic landscape of AMD risk.7,8,9,10 Little is known, however, about genetic factors that influence disease progression once late stage features have become manifest.6,11 Here, we aimed to identify novel genetic variants conferring risk for GA progression by extending previous studies to a genome-wide undirected search in a well-powered genome-wide association study.
Methods
Ethics Statement
The study followed the tenets of the Declaration of Helsinki and was approved by the local ethics review boards at the University of Bonn, Bonn, Germany, and the US National Institutes of Health for all studies, as well as central and local country-specific institutional review boards for the Geographic Atrophy Treatment Evaluation (GATE) study. Informed written consent was obtained from each patient after explanation of the nature and implications of the study.
Study Characteristics
Inclusion and exclusion criteria were reported previously11 for the Fundus Autofluorescence Imaging in Age-Related Macular Degeneration (FAM),12 Directional Spread in Geographic Atrophy (DSGA),13 Age-Related Eye Disease Study (AREDS),14 and GATE3 studies. Briefly, FAM and DSGA are both longitudinal natural history studies of AMD, with DSGA representing an extension of FAM at the Department of Ophthalmology of the University of Bonn. The FAM study was a multicenter observational study with patient recruitment at multiple sites in Germany. The AREDS was a randomized, masked, multicenter clinical trial designed to evaluate the efficacy of dietary supplementation on preventing AMD and cataract. Finally, GATE was a controlled, masked, randomized, multicenter, phase 3 clinical trial designed to determine the efficacy and safety of a medication called AL-8309B. The treatment was shown to have no effect on GA lesion enlargement, rendering GATE a natural history study of GA progression.3
Classification of Geographic Atrophy
The classification of GA was previously described for FAM, DSGA,15 and AREDS.16 The GATE trial largely followed the definitions of the FAM study.3
GA Area Measurements and Calculation of GA Progression Rate
In FAM, DSGA, and GATE, fundus autofluorescence images were analyzed by RegionFinder Software (Heidelberg Engineering GmbH) at the GRADE Reading Center in Bonn, Germany.3,17 For AREDS, we directly measured the area of GA from color fundus photographs16 retrieved through the database of Genotypes and Phenotypes (accession number: phs000001.v3.p1).18 In brief, a trained ophthalmologist (C.B.) and medical student (S.H.) used the paintbrush tool implemented in ImageJ version 1.49v (National Institutes of Health and the Laboratory for Optical and Computational Instrumentation)19 to trace the extent of GA lesions. The pixel area (in pixels squared) was converted to millimeters squared using a conversion factor of 0.004506 millimeters per pixel, estimated from the mean vertical diameter of the optic nerve (426.1 pixels).20
The individual GA area measurement was square root transformed,6,21 which meant it was essentially representing the equivalent of the diameter or the circumference of the atrophic area. The progression rate for each eye was determined in millimeters per year by calculating the absolute square progression between the most distant evaluation points in time. The resulting linear progression rate per eye6,22,23 was summarized for each individual by computing the mean of the growth rates for the 2 eyes (if available). The GA lesion growth was inverse normal transformed24 and used as the main phenotype in the association analysis.
Clinical and Demographic Variables
Sex, age at first examination (in years), baseline square root–transformed lesion size in 1 eye or the mean of both eyes at first examination (in millimeters) as well as the follow-up time (in years) of each patient were recorded. We included the number of GA lesions, determined as a mean over both eyes for each participant to account for unifocal and multifocal GA lesions and the presence of GA in both eyes.
Genotyping and Imputation
The AREDS genotypes were retrieved through the database of Genotypes and Phenotypes (accession number: phs000001.v3.p1). Genome-wide genotyping in FAM, DSGA, and GATE was performed at the Cologne Center for Genomics in Cologne, Germany. Briefly, genomic DNA was genotyped on the Precision Medicine Research Array (ThermoFisher Scientific). The genotypes were called with the Axiom Analysis Suite version 3.0.1.4 (ThermoFisher Scientific) and the Best Practice Workflow (implemented in the Axiom Analysis Suite). Typed variants were removed if the call rate was less than 98% or the allele frequency was deviated significantly from Hardy-Weinberg equilibrium (P < 1.00 × 10−6).
The nonimputed genotypes of individual studies were phased with ShapeIt2 version 2.r904 (University of Oxford).25 Untyped genotypes were imputed with IMPUTE2 version 2.3.2 (University of Oxford)26 using the 1000 Genomes Release 1 version 3 reference database,27 transformed into the variant call format 4.2 by applying the QCTool28 version 2 and merged into a single file with BCFTools29 version 1.4.1-10-gc5be287 (Samtools). A total of 835 530 genotyped and 7 596 219 imputed variants with an imputation quality greater than 0.3 were used in a genome-wide association study (GWAS). The first 2 principal components of genetic variation were calculated from the imputed genotypes with R package SNPRelate (Bioconductor). Genetically related individuals as well as non-European individuals were excluded from further analysis, as described previously.8
Statistical Analyses and Visualization
Linear regression models were fit as implemented in R version 3.3.3 (R Foundation for Statistical Computing) and, unless noted otherwise, were univariate. A GWAS was performed with rvtest,30 conditioned on the first 2 principal components calculated from the imputed genotypes, the number of GA lesion spots, the presence of GA in the fellow eye, and the clinical follow-up time. The GWAS was calculated on the expected alternate allele counts of the imputed variants, and missing genotypes were imputed to the mean by rvtest. The quantile-quantile and Manhattan plots were generated with the qqman library in R, and the genomic inflation factor λ was calculated with the GenABEL library version 1.8-0,31 implemented in R. The boundaries of an associated genomic locus were defined by the genomic position of the most distant variants that were in moderate linkage disequilibrium (R>0.5, estimated in European participants from the 1000 Genomes Project with LDLink [National Cancer Institute]32) from the lead variant, including an additional 500-kbp buffer region. For each lead variant, we also investigated the association with AMD risk by using the summary statistics provided by the International AMD Genomics Consortium.8 The 95% and 99% credible set of associated variants were calculated, as described previously.33,34 Summary statistics of the study findings will be available in LD Hub (Broad Institute)35 and GWAS Central.
Estimating the Heritability of GA Progression
Heritability was estimated with the genomic restricted maximum likelihood estimation method implemented in genome-wide complex trait analysis36 with settings adjusted for the same covariates as described for the GWAS analysis. Furthermore, the heritability was estimated by linkage disequilibrium score regression35 from GWAS summary statistics.
Pathway Enrichment and Gene Prioritization
Pathway scoring algorithm (PASCAL)37 identified significantly enriched Reactome or Kyoto Encyclopedia of Genes and Genomes pathways based on gene scores computed from the association signals within genes while adjusting for linkage disequilibrium structure (estimated from the 1000 Genomes phase 3 haplotypes). The raw P values were adjusted per Benjamini and Hochberg,38 controlling the false discovery rate at less than 5%. In addition, we used g:Profiler (BioInformatics and Information Technology Research Group)39 with standard settings to assess the enrichment of Kyoto Encyclopedia of Genes and Genomes or Reactome pathways in candidate genes from genome-wide significant loci. To prioritize genes in associated loci, we applied a transcriptome-wide association study and identified significant correlations between expression quantitative trait loci in associated regions and the lead variant. We queried the International Mouse Phenotyping Consortium database to identify knockout mice of candidate genes underlying an abnormal retinal phenotype. Gene expression of candidate genes in retina and retinal pigment epithelium (RPE) or choroid was analyzed with Hierarchical Indexing for Spliced Alignment of Transcripts 2 (Johns Hopkins University)40 and ballgown implemented in R40 from the tuxedo suite. Publicly available gene expression data were retrieved from Gene Expression Omnibus41 (accession number: GSE94437).42 Genes were ranked by increasing the priority by 1 in cases in which one of the following features was present: nominally significant differential gene expression between locations in either the retina or the RPE or choroid, presence of at least 5 (of 43) significant transcriptome-wide association study signals, a nominally significant enrichment of associated variants estimated by PASCAL, or a mouse model with a retinal phenotype.
Results
Study characteristics are given in Table 1. A total of 935 samples were included; 547 of 935 (59.0%) were from female participants. Approximately 1 of every 4 participants (227 of 935 [25.6%]) had a single GA lesion. The mean (SD) number of lesions in the full sample was 2.7 (2.1) lesions per participant.
Table 1. Study Characteristics.
| Characteristic | Mean (SD) | |||
|---|---|---|---|---|
| AREDS | FAM/DSGA | GATE | Combined | |
| Samples, No. | 349 | 164 | 422 | 935 |
| Baseline square root geographic atrophy area, mm | 2.31 (1.42) | 2.14 (1.09) | 2.60 (0.84) | 2.41 (1.14) |
| Square root–transformed geographic atrophy growth rate, mm/y | 0.294 (0.278) | 0.298 (0.175) | 0.296 (0.16) | 0.295 (0.214) |
| Age at blood sampling, y | 70.0 (5.3) | 75.9 (7.1) | 78.1 (7.7) | 74.7 (7.8) |
| Female, No. (%) | 195 (55.9) | 106 (64.6) | 246 (58.3) | 547 (58.5) |
| Follow-up time within study, y | 5.6 (3.2) | 3.3 (2.4) | 1.9 (0.5) | 3.6 (2.8) |
| Geographic atrophy lesions, No. | 1.7 (1.1) | 2.7 (2.3) | 3.6 (2.3) | 2.7 (2.1) |
| Participants with a single geographic atrophy lesion, No. (%) | 151 (43.3) | 51 (31.1) | 35 (8.3) | 237 (25.4) |
| Age-related macular degeneration genetic risk scorea | 41.36 (5.19) | 41.76 (5.32) | 41.47 (5.03) | 41.48 (5.14) |
| Individuals with geographic atrophy in both eyes, No. (%) | 206 (59.0) | 95 (57.9) | 291 (69.0) | 592 (63.3) |
Abbreviations: AREDS, Age-Related Eye Disease Study; DSGA, Directional Spread in Geographic Atrophy; FAM, Fundus Autofluorescence Imaging in Age-Related Macular Degeneration; GATE, Geographic Atrophy Treatment Evaluation.
The effect size (log odds) weighted age-related macular degeneration genetic risk score was calculated from 37 condition-associated, well-imputed variants normalized by the mean log odds of all 37 variants.8
Influence of Covariates on GA Progression Rate
First, we evaluated the association of various covariates with GA progression. In agreement with previous reports,6 the presence of bilateral GA (slope, 0.233 [95% CI, 0.100-0.367]; P < .001) and the number of GA lesions (slope, 0.159 [95% CI, 0.128-0.191]; P = 2.0×10−23) were significantly correlated with an increased progression rate (Table 2). In addition, the review period was correlated with GA growth (slope, −0.045 [95% CI, −0.074 to −0.016] per year; P = .002), particularly in the AREDS cohort (slope, −0.035 [95% CI, −0.068 to −0.002] per year; P = .04) and the FAM and DSGA cohort (slope, −0.070 [95% CI, −0.133 to −0.006] per year; P = .03) and explained around 1% of the variation (Table 2), with a negative slope indicating that the growth rate slows down with longer periods of observation. A multivariable regression model including follow-up time, bilateral GA status, and number of GA lesions explained 9.7% of the trait variability. The first 2 principal components calculated from the imputed genotypes as well as age and the square root–transformed baseline GA size failed to reach significance.
Table 2. Univariate Correlation Between Clinical and Demographic Variables and Geographic Atrophy Growth Rates.
| AREDS | FAM/DSGA | GATE | Combined | |||||
|---|---|---|---|---|---|---|---|---|
| Slope (95% CI) | P Value | Slope (95% CI) | P Value | Slope (95% CI) | P Value | Slope (95% CI)a | P Value | |
| Baseline square root geographic atrophy area, per mm | −0.082 (−0.156 to −0.008) | .03 | 0.006 (−0.137 to 0.148) | .93 | 0.001 (−0.114 to 0.115) | .99 | −0.047 (−0.104 to 0.010) | .10 |
| Age, per y | −0.003 (−0.023 to 0.017) | .77 | 0.012 (−0.009 to 0.034) | .27 | 0.002 (−0.011 to 0.014) | .75 | −1.574 (−3.660 to 0.512) | .14 |
| Female | 0.117 (−0.095 to 0.328) | .28 | 0.062 (−0.266 to 0.389) | .71 | 0.081 (−0.115 to 0.276) | .42 | 0.091 (−0.04 to 0.223) | .19 |
| First principal componentb | −4.046 (−8.735 to 0.643) | .09 | −2.225 (−14.46 to 10.01) | .72 | −0.915 (−3.296 to 1.466) | .45 | 0.003 (−0.007 to 0.012) | .54 |
| Second principal componentb | 1.749 (−8.165 to 11.66) | .73 | 2.283 (−1.245 to 5.811) | .20 | −0.911 (−5.240 to 3.417) | .68 | 1.062 (−1.566 to 3.690) | .43 |
| Geographic atrophy in both eyes | 0.204 (−0.009 to 0.417) | .06 | 0.189 (−0.123 to 0.501) | .24 | 0.281 (0.074 to 0.487) | .008 | 0.233 (0.100 to 0.367) | .001 |
| Follow-up time within study period, per y | −0.035 (−0.068 to −0.002) | .04 | −0.070 (−0.133 to −0.006) | .03 | −0.157 (−0.356 to 0.043) | .12 | −0.045 (−0.074 to −0.016) | .002 |
| Geographic atrophy lesions, per lesion | 0.277 (0.192 to 0.363) | 6 × 10−10 | 0.161 (0.095 to 0.227) | 3 × 10−06 | 0.134 (0.095 to 0.173) | 6 × 10−11 | 0.159 (0.128 to 0.191) | 2 × 10−23 |
| Age-related macular degeneration genetic risk score,c per allele | 0.025 (0.006 to 0.044) | .02 | −0.015 (−0.043 to 0.013) | .21 | 0.016 (−0.003 to 0.034) | .09 | 0.014 (0.002 to 0.027) | .03 |
Abbreviations: AREDS, Age-Related Eye Disease Study; DSGA, Directional Spread in Geographic Atrophy; FAM, Fundus Autofluorescence Imaging in Age-Related Macular Degeneration; GATE, Geographic Atrophy Treatment Evaluation.
Adjusted for study.
Principal components calculated from the genotypes.
Effect size (log odds) weighted age-related macular degeneration genetic risk score calculated from 37 condition-associated, well-imputed variants, normalized by the mean log odds of all 37 variants.8
The AMD genetic risk score,10 computed from 37 AMD-associated risk variants8,43 was significantly correlated, although with a weak slope (slope, 0.014 [95% CI, 0.002-0.027]; P = .03). Furthermore, 2 of the 37 AMD risk variants (rs12357257 and rs183281136) were nominally significant for a correlation with GA lesion growth (eAppendix and eTable 1 in the Supplement), but none remained significant after adjustment for multiple testing.
GWAS Analysis
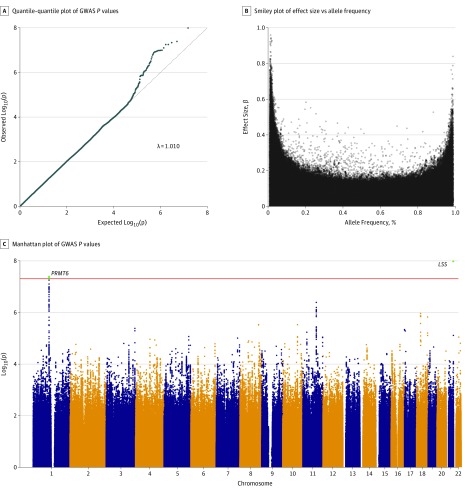
The P values from the GWAS revealed no substantial evidence for population stratification (λ, 1.010; Figure, A), while there was an excess of small P values exceeding expectation. Allele frequencies and effect sizes conform largely to the expected distribution (Figure, B). Overall, 2 genome-wide significant loci were identified, one on chromosome 1p13.3 (rs11184959 within the locus for protein arginine methyltransferase 6 [PRMT6]; slope, 0.046 [95% CI, 0.026-0.066]; P = 4.09 × 10−8) and the other on chromosome 21q22.3 (rs2839127 within the locus for lanosterol synthase [LSS]; slope, 0.059 [95% CI, 0.034-0.085]; P = 1.01 × 10−8; Table 3). In addition, a locus on chromosome 11 showed suggestive evidence for association (slope, 0.105 [95% CI, 0.068-0.143]; P = 4.07 × 10−7; Table 3). Additional adjustment for age at baseline, sex, and study did not change the observed association (data not shown). The 2 genome-wide significant loci at PRMT6 and LSS jointly explain 6.05% of the observed variability of GA lesion growth and are independently correlated with the trait without evidence for epistasis or interaction. Neither lead variant in PRMT6 or LSS was associated with AMD risk according to data provided by the International AMD Genomics Consortium. Each minor allele of the genome-wide associated variants increased the growth rate by a mean of about 0.05 mm per year. Thus, a person homozygous for a major allele would be expected to have a growth rate reduction of about 30% compared with a person homozygous for the minor allele.
Figure. Results of Genome-Wide Association Study (GWAS).
A, Quantile-quantile plot of GWAS P values; little evidence for population stratification was found (λ = 1.01). B, Smiley plot of effect size vs allele frequency. C, Manhattan plot of GWAS P values. Two regions with genome-wide significant variations are highlighted.
Table 3. Genome-Wide Significant and Suggestive Lead Variants Associated With Geographic Atrophy Lesion Growth.
| Varianta | Chr | Position (hg19) |
Reference Allele | Alternative Allele | Alternative Allele Frequency | Slope per Allele (95% CI)b | P Value | 95% of Credible Setc | 99% of Credible Setc | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AREDS | FAM/ DSGA |
GATE | All | Root Transformed, mm/y |
Inverse Normal Transformed |
||||||||
| rs11184959 | 1 | 107 213 976 | G | A | 0.375 | 0.340 | 0.338 | 0.352 | 0.046 (0.026- 0.066) |
0.246 (0.155- 0.338) |
4.09 × 10−8 | 24 | 37 |
| rs2839127 | 21 | 47 573 550 | G | A | 0.115 | 0.159 | 0.174 | 0.145 | 0.059 (0.034- 0.085) |
0.346 (0.221- 0.471) |
1.0109 × 10−8 | 1 | 1 |
| rs145146260d | 11 | 90 194 708 | CT | C | 0.079 | 0.046 | 0.096 | 0.070 | 0.105 (0.068- 0.143) |
0.441 (0.270- 0.612) |
4.0709 × 10−7 | 32 | 47 |
Abbreviations: AREDS, Age-Related Eye Disease Study; Chr, chromosome; DSGA, Directional Spread in Geographic Atrophy; FAM, Fundus Autofluorescence Imaging in Age-Related Macular Degeneration; GATE, Geographic Atrophy Treatment Evaluation.
Per dbSNP.
Adjusted for the number of geographic atrophy lesions, follow-up time, presence of bilateral geographic atrophy, and the first 2 principal components of ancestry.
Number of variants in the 95% or 99% credible set of association, containing the true variant with 95% or 99% probability, respectively.
Suggestive evidence for association (P < 1.00 × 10−6).
Pathway Enrichment Analysis
No significant pathway enrichment was observed for the 13 genes located within the 2 genome-wide significant loci (Table 4). In identifying significantly enriched pathways after adjustment for multiple testing, PASCAL software44 revealed the glycosphingolipid metabolism Reactome pathway to be statistically significant (Reactome accession number: R-HSA-1660662; false discovery rate <0.05). Kyoto Encyclopedia of Genes and Genomes pathways showed no statistically significant enrichment, although several pathways were nominally significant for an association with GA growth (eAppendix and eTable 2 in the Supplement).
Table 4. Gene Prioritization Within Genome-Wide Significant Loci.
| Gene | Score | Expression FPKM, Mean (SD) | TWASa | PASCAL P Value | Mouse Model, P Valueb | |||
|---|---|---|---|---|---|---|---|---|
| Retina, Macular | Retina, Peripheral | Retinal Pigment Epithelium or Choroid, Macular | Retinal Pigment Epithelium or Choroid, Peripheral | |||||
| rs11184959 on chromosome 1:106 777 024-107 851 213 | ||||||||
| PRMT6c | 2 | 3.57 (0.77) | 3.61 (0.96) | 1.41 (0.86)d | 2.39 (0.46)d | 0 | 0.599 | 5.37 × 10−5 |
| NTNG1 | 0 | 1.95 (0.79) | 1.66 (0.73) | 0.72 (0.24) | 0.97 (0.27) | 0 | 0.628 | NA |
| rs2839127 on chromosome 21:47 099 280-48 073 550 | ||||||||
| LSSd | 4 | 8.11 (2.44)e | 2.36 (1.02)e | 3.19 (0.88) | 3.51 (1.1) | 6f | 1.44E-03g | 8.13 × 10−6 |
| PCNTd | 2 | 2.43 (0.36)d | 1.59 (0.43)e | 1.84 (0.84) | 1.26 (0.19) | 1 | 0.010g | NA |
| SPATC1L | 2 | 1.23 (0.49) | 1.23 (0.34) | 2.24 (0.5) | 2.07 (0.6) | 30f | 2.98E-04g | NA |
| YBEY | 2 | 2.59 (1.12) | 2.13 (0.25) | 2.27 (0.61) | 2.18 (0.67) | 37f | 0.015g | NA |
| PCBP3c | 2 | 6.75 (1.17) | 5.92 (1.78) | 0.51 (0.26)d | 0.24 (0.13)c | 0 | 0.010g | NA |
| MCM3AP | 1 | 3.75 (0.51) | 3.11 (0.76) | 3.01 (0.56) | 2.81 (0.56) | 2 | 0.021g | NA |
| C21ORF58 | 1 | 0.49 (0.14) | 0.49 (0.15) | 0.14 (0.13) | 0.11 (0.08) | 1 | 0.010g | NA |
| FTCD | 1 | 0.04 (0.04) | 0.01 (0.02) | 0 (0.01) | 0 (0) | 0 | 2.93E-04g | NA |
| DIP2A | 1 | 1.8 (0.48) | 1.74 (0.53) | 0.67 (0.18) | 0.61 (0.11) | 6f | 0.059 | >.99 |
| PRMT2 | 1 | 4.92 (0.93) | 4.17 (0.39) | 4.39 (0.88) | 4.32 (1.14) | 5f | 0.197 | NA |
| COL6A1d | 1 | 4.62 (1.38)e | 2.76 (1.24)e | 24.76 (10.71) | 36.51 (14.39) | 0 | 0.701 | NA |
Abbreviations: C21ORF58, chromosome 21 open reading frame 58; COL6A1, collagen 3 type VI alpha 1 chain; DIP2A, disco interacting protein 2 homolog A; FPKM, fragments/reads per thousand base-pairs of transcript per million reads of the respective gene; FTCD, formimidoyltransferase 1 cyclodeaminase; LSS, lanosterol synthase; MCM3AP, minichromosome maintenance complex component 3 associated protein; NA, not applicable; NTNG1, netrin G1; PASCAL, Pathway scoring algorithm; PCBP3, poly(RC) binding protein 3; PCNT, pericentrin; PRMT2, protein arginine methyltransferase 2; PRMT6, protein arginine methyltransferase 6; SPATC1L, spermatogenesis and centriole associated 1–like; TWAS, transcriptome-wide association study; YBEY, YbeY metalloendoribonuclease.
Number of nominally significant TWAS results of 43 total possible results for gene over all tissues in Genotype-Tissue Expression project; number of tissues with significant local expression quantitative trait loci according to the expression quantitative trait loci.
Combined Retinal Morphology phenotype P value according to the International Mouse Phenotyping Consortium (http://www.mousephenotype.org/); NA indicates that no knockout mouse model is available or investigated for the respective gene.
Nominally significant differential expression between the central and peripheral retinal pigment epithelium or choroid.
Nominally significant differential expression between the central and peripheral retinal pigment epithelium.
Nominally significant differential expression between the central and peripheral retina.
More than 5 nominally significant TWAS results for gene over all tissues in Genotype-Tissue Expression project (https://www.gtexportal.org/home/).
Significant PASCAL gene score; statistically significant combined retinal morphology phenotype P value according to the International Mouse Phenotyping Consortium (http://www.mousephenotype.org/).
Gene Prioritization in Genome-Wide Significant Loci
None of the variants in linkage disequilibrium with the lead variants (R2> 0.5) or in the 95% or 99% credible set of associated variants were located within exons of known genes. To prioritize candidate genes, we used a transcriptome-wide association study, a method that correlates the slope and the respective effect allele of associated variants with gene expression from the Genotype-Tissue Expression project. Notably, the spermatogenesis and centriole associated 1–like (SPATC1L) and YbeY metalloendoribonuclease (YBEY) genes were significantly regulated by the lead variant rs2839127 in the LSS locus (Table 4), while expression of candidate genes in the PRMT6 locus showed no significant correlation with markers associated with GA lesion growth. In addition, several genes in the LSS locus, but not in the PRMT6 locus, revealed significant gene scores computed by PASCAL (Table 4).
Next, we queried the International Mouse Phenotyping Consortium database for knockout mice in genes located within the PRMT6 and LSS loci. Knockout mouse lines deficient for PRMT6 and LSS were reported to display an abnormal retinal phenotype (Table 4).
Finally, for each gene, we report the mean expression in macular (central) and peripheral retina as well as macular and peripheral RPE or choroid (Table 4). All genes except the formimidoyltransferase cyclodeaminase gene (FTCD) were expressed in either the RPE or choroid or the retina. We found that LSS, the pericentrin gene (PCNT), and the collagen type VI alpha 1 chain gene (COL6A1) were differentially regulated between peripheral and macular retina, while PRMT6 and poly(RC) binding protein 3 gene (PCBP3) expression was significantly different between the peripheral and central RPE or choroid. By ranking the genes in both chromosomal loci according to the number of significant features, the highest gene priority within the PRMT6 locus was PRMT6 and within the LSS locus was LSS.
Heritability Estimate of GA Progression
To assess the heritability of GA growth attributed to imputed variants, the genomic restricted maximum likelihood estimation method was used to estimate the heritability of GA growth at 28.0% (SE, 28.4%). We observed a high standard error, rendering the estimate to be the same as 0 (P = .14). Linkage-disequilibrium score regression was performed on the summary statistics of the GWAS results. This demonstrated that the sample size of the current study was too small to compute significant heritability estimates.
Discussion
This study presents a first GWAS for lesion growth in GA secondary to AMD and reports 2 genome-wide significant loci: the PRMT6 gene on chromosome 1p13.3 and the LSS gene on chromosome 21q22.3. The progression of GA lesions varies to a large extent between individuals but shows a high degree of similarity between twins and between both eyes of an individual.6 These findings suggest that genetic predisposition plays a role in this process. Furthermore, this GWAS revealed an excess of small P values greater than were expected by chance, indicating that genetic association signals are present. Consequently, this GWAS identified 2 genome-wide significant genetic variants, as well as additional suggestive variants implying that the heritability of GA progression is different from zero and quantifiable. Indeed, both genome-wide significant loci at PRMT6 and LSS jointly explain 6.05% of the observed variation in GA lesion progression. Together, genetic and clinical parameters explain 15.7% of disease variability underscoring the need to consider these findings in future clinical trials targeting GA growth rates. In addition, the findings may also point toward processes that are involved in other retinal degenerations accompanied by atrophic changes.
Within the PRMT6 and LSS loci, a total of 13 local genes could be implicated by the respective association signals. Since none of the lead variants nor any correlated variant (R2>0.5) nor any variant in the 95% or 99% credible set were located in the protein-coding region of the genes in question, we developed a prioritization scheme with a ranking of favored situations. First, genes are differentially expressed between the central and peripheral retina or the RPE or choroid45 (because such genes may be of interest as GA growth rates vary between the macula and the peripheral retina). Second, genes show a significant differential regulation in multiple tissues by the alleles of the lead variant46; since no expression quantitative trait loci data are available for the retinal tissue or the RPE or choroid, this approach is at present a best guess on the true underlying genomic regulation in the disease relevant tissues. Third, a significant enrichment of association signals are measured as a gene score in the gene body of candidate genes.47 Lastly, genes deleted in knockout mouse lines present with phenotypes relevant to the RPE or retinal tissues.48,49 We found that 1 gene at each locus (PRMT6 and LSS) indeed revealed a retinal phenotype in the respective homozygous knockout mice. While homozygous LSS deficiency has various abnormal changes in the retinal vasculature, the PRMT6 knockout mouse appears to be mainly affected by abnormal eye morphology.50 This finding is interesting because individuals with neovascular AMD in the fellow eye also present with higher GA enlargement rates.15,51 Taken together, we propose that both LSS and PRMT6 are likely functionally relevant genes for GA growth rates identified in this study.
The list of candidate genes within the PRMT6 and LSS loci showed no significant pathway enrichment. Nevertheless, the genome-wide analysis revealed a potential role of the glycosphingolipid metabolism in GA progression. However, neither LSS nor PRMT6 are part of this pathway, and thus, we advise caution in the interpretation of the pathway enrichment results without further experimental exploration and confirmation.
We found no evidence that genetic variants associated with complement genes or other AMD risk–associated pathways were significantly involved in GA lesion progression. This indicates different pathways in risk and progression of GA secondary to AMD. Although we and others have previously implicated AMD risk–associated variants in GA lesion progression (complement factor B,52 complement C3,11 and age-related maculopathy susceptibility 2,16 as summarized in Fleckenstein et al6), this study failed to confirm such connections with genome-wide statistical significance.
In this study, we have adjusted the GWAS by the number of GA lesions to identify genetic factors that only influence GA lesion growth but not new lesion initiation. In case the AMD risk variants are preferentially increasing the number of GA lesions and thus indirectly increasing the growth rate, we would not expect to find a significant association of these variants with GA growth rate after adjustment. Nevertheless, this study confirmed previous factors associated with GA growth, such as the number of lesions and the presence of bilateral geographic atrophy. These 2 factors accounted for a greater degree of the variability for GA growth than the 2 genetic loci that were identified in this study.
The strengths of the current study are several. They include its relatively large number of individuals with available genetic data, the concurrent and standardized genotype and phenotype processing, and the wide range of methods to assess lesion growth.
Limitations
This study is limited by the lack of a large replication cohort. Thus, it requires reliance on the conservative genome-wide significance threshold to account for potential false positive findings, which creates the potential of missing additional association signals.
Conclusions
In conclusion, the present study is a large genetic study on GA lesion progression, combining 4 studies with different inclusion and exclusion criteria and imaging platforms. The observed conservative genome-wide significant associations at the PRMT6 and LSS loci should be robust and reproducible. The data provide a basis for future studies investigating the role of the identified candidate genes and pathways on GA lesion growth.
eAppendix. Legends to eTables
eTable 1. Correlation of known AMD risk variants with GA lesion growth
eTable 2. Nominally significant pathways involved in GA lesion growth
References
- 1.Wong WL, Su X, Li X, et al. Global prevalence of age-related macular degeneration and disease burden projection for 2020 and 2040: a systematic review and meta-analysis. Lancet Glob Health. 2014;2(2):e106-e116. doi: 10.1016/S2214-109X(13)70145-1 [DOI] [PubMed] [Google Scholar]
- 2.Li J, Xu J, Chen Y, Zhang J, Cao Y, Lu P. Efficacy comparison of intravitreal anti-VEGF therapy for three subtypes of neovascular age-related macular degeneration: a systematic review and meta-analysis. J Ophthalmol. 2018;2018:1425707. doi: 10.1155/2018/1425707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jaffe GJ, Schmitz-Valckenberg S, Boyer D, et al. Randomized trial to evaluate tandospirone in geographic atrophy secondary to age-related macular degeneration: the GATE study. Am J Ophthalmol. 2015;160(6):1226-1234. doi: 10.1016/j.ajo.2015.08.024 [DOI] [PubMed] [Google Scholar]
- 4.Yaspan BL, Williams DF, Holz FG, et al. ; MAHALO Study Investigators . Targeting factor D of the alternative complement pathway reduces geographic atrophy progression secondary to age-related macular degeneration. Sci Transl Med. 2017;9(395):eaaf1443. doi: 10.1126/scitranslmed.aaf1443 [DOI] [PubMed] [Google Scholar]
- 5.Heier JS. Lampalizumab phase 3 trial for geographic atrophy secondary to AMD: the Spectri topline results. Paper presented at: American Academy of Ophthalmology Annual Meeting; November 11, 2017; New Orleans, LA. [Google Scholar]
- 6.Fleckenstein M, Mitchell P, Freund KB, et al. The progression of geographic atrophy secondary to age-related macular degeneration. Ophthalmology. 2018;125(3):369-390. doi: 10.1016/j.ophtha.2017.08.038 [DOI] [PubMed] [Google Scholar]
- 7.Grassmann F, Ach T, Brandl C, Heid IM, Weber BHF. What does genetics tell us about age-related macular degeneration? Annu Rev Vis Sci. 2015;1(1):73-96. doi: 10.1146/annurev-vision-082114-035609 [DOI] [PubMed] [Google Scholar]
- 8.Fritsche LG, Igl W, Bailey JNC, et al. A large genome-wide association study of age-related macular degeneration highlights contributions of rare and common variants. Nat Genet. 2016;48(2):134-143. doi: 10.1038/ng.3448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grassmann F, Fauser S, Weber BHF. The genetics of age-related macular degeneration (AMD)—novel targets for designing treatment options? Eur J Pharm Biopharm. 2015;95(Pt B):194-202. doi: 10.1016/j.ejpb.2015.04.039 [DOI] [PubMed] [Google Scholar]
- 10.Grassmann F, Kiel C, Zimmermann ME, et al. ; International AMD Genomics Consortium (IAMDGC) . Genetic pleiotropy between age-related macular degeneration and 16 complex diseases and traits. Genome Med. 2017;9(1):29. doi: 10.1186/s13073-017-0418-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grassmann F, Fleckenstein M, Chew EY, et al. Clinical and genetic factors associated with progression of geographic atrophy lesions in age-related macular degeneration. PLoS One. 2015;10(5):e0126636. doi: 10.1371/journal.pone.0126636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dreyhaupt J, Mansmann U, Pritsch M, Dolar-Szczasny J, Bindewald A, Holz FG. Modelling the natural history of geographic atrophy in patients with age-related macular degeneration. Ophthalmic Epidemiol. 2005;12(6):353-362. doi: 10.1080/09286580591005723 [DOI] [PubMed] [Google Scholar]
- 13.Lindner M, Bezatis A, Czauderna J, et al. Choroidal thickness in geographic atrophy secondary to age-related macular degeneration. Invest Ophthalmol Vis Sci. 2015;56(2):875-882. doi: 10.1167/iovs.14-14933 [DOI] [PubMed] [Google Scholar]
- 14.Age-Related Eye Disease Study Research Group . The Age-Related Eye Disease Study (AREDS): design implications, AREDS report No. 1. Control Clin Trials. 1999;20(6):573-600. doi: 10.1016/S0197-2456(99)00031-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fleckenstein M, Schmitz-Valckenberg S, Adrion C, et al. ; FAM Study Group . Progression of age-related geographic atrophy: role of the fellow eye. Invest Ophthalmol Vis Sci. 2011;52(9):6552-6557. doi: 10.1167/iovs.11-7298 [DOI] [PubMed] [Google Scholar]
- 16.Klein ML, Ferris FL III, Francis PJ, et al. Progression of geographic atrophy and genotype in age-related macular degeneration. Ophthalmology. 2010;117(8):1554-1559, 1559.e1. doi: 10.1016/j.ophtha.2009.12.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Scholl HPN, Fleckenstein M, Fritsche LG, et al. CFH, C3 and ARMS2 are significant risk loci for susceptibility but not for disease progression of geographic atrophy due to AMD. PLoS One. 2009;4(10):e7418. doi: 10.1371/journal.pone.0007418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tryka KA, Hao L, Sturcke A, et al. NCBI’s database of genotypes and phenotypes: dbGaP. Nucleic Acids Res. 2014;42(database issue):D975-D979. doi: 10.1093/nar/gkt1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schindelin J, Arganda-Carreras I, Frise E, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9(7):676-682. doi: 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tasman W, Jaeger EA. Anatomy of the visual sensory system. In: Duane’s Ophthalmology. New York, NY: Lippincott Williams and Wilkins; 2006:614-618. [Google Scholar]
- 21.Feuer WJ, Yehoshua Z, Gregori G, et al. Square root transformation of geographic atrophy area measurements to eliminate dependence of growth rates on baseline lesion measurements: a reanalysis of age-related eye disease study report No. 26. JAMA Ophthalmol. 2013;131(1):110-111. doi: 10.1001/jamaophthalmol.2013.572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lindblad AS, Lloyd PC, Clemons TE, et al. ; Age-Related Eye Disease Study Research Group . Change in area of geographic atrophy in the Age-Related Eye Disease Study: AREDS report number 26. Arch Ophthalmol. 2009;127(9):1168-1174. doi: 10.1001/archophthalmol.2009.198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thavikulwat AT, Jacobs-El N, Kim JS, et al. Evolution of geographic atrophy in participants treated with ranibizumab for neovascular age-related macular degeneration. Ophthalmol Retina. 2017;1(1):34-41. doi: 10.1016/j.oret.2016.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goh L, Yap VB. Effects of normalization on quantitative traits in association test. BMC Bioinformatics. 2009;10(1):415. doi: 10.1186/1471-2105-10-415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.O’Connell J, Gurdasani D, Delaneau O, et al. A general approach for haplotype phasing across the full spectrum of relatedness. PLoS Genet. 2014;10(4):e1004234. doi: 10.1371/journal.pgen.1004234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5(6):e1000529. doi: 10.1371/journal.pgen.1000529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Abecasis GR, Auton A, Brooks LD, et al. ; 1000 Genomes Project Consortium . An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491(7422):56-65. doi: 10.1038/nature11632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.QCtool v2. QCtool v2: a tool for quality control and analysis of gwas datasets. https://www.well.ox.ac.uk/~gav/qctool_v2/index.html. Published 2014. Accessed April 18, 2019.
- 29.BCFTools . Samtools/bcftools. https://github.com/samtools/bcftools. Published 2015. Accessed April 18, 2019.
- 30.Zhan X, Hu Y, Li B, Abecasis GR, Liu DJ. RVTESTS: an efficient and comprehensive tool for rare variant association analysis using sequence data. Bioinformatics. 2016;32(9):1423-1426. doi: 10.1093/bioinformatics/btw079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Aulchenko YS, Ripke S, Isaacs A, van Duijn CM. GenABEL: an R library for genome-wide association analysis. Bioinformatics. 2007;23(10):1294-1296. doi: 10.1093/bioinformatics/btm108 [DOI] [PubMed] [Google Scholar]
- 32.Machiela MJ, Chanock SJ. LDassoc: an online tool for interactively exploring genome-wide association study results and prioritizing variants for functional investigation. Bioinformatics. 2018;34(5):887-889. doi: 10.1093/bioinformatics/btx561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maller JB, McVean G, Byrnes J, et al. ; Wellcome Trust Case Control Consortium . Bayesian refinement of association signals for 14 loci in 3 common diseases. Nat Genet. 2012;44(12):1294-1301. doi: 10.1038/ng.2435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Grassmann F, Heid IM, Weber BHF; International AMD Genomics Consortium (IAMDGC) . Recombinant haplotypes narrow the ARMS2/HTRA1 association signal for age-related macular degeneration. Genetics. 2017;205(2):919-924. doi: 10.1534/genetics.116.195966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zheng J, Erzurumluoglu AM, Elsworth BL, et al. ; Early Genetics and Lifecourse Epidemiology (EAGLE) Eczema Consortium . LD Hub: a centralized database and web interface to perform LD score regression that maximizes the potential of summary level GWAS data for SNP heritability and genetic correlation analysis. Bioinformatics. 2017;33(2):272-279. doi: 10.1093/bioinformatics/btw613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet. 2011;88(1):76-82. doi: 10.1016/j.ajhg.2010.11.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lamparter D, Marbach D, Rueedi R, Kutalik Z, Bergmann S. Fast and rigorous computation of gene and pathway scores from SNP-based summary statistics. PLoS Comput Biol. 2016;12(1):e1004714. doi: 10.1371/journal.pcbi.1004714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yekutieli D, Benjamini Y. The control of the false discovery rate in multiple testing under dependency. Ann Stat. 2001;29(4):1165-1188. doi: 10.1214/aos/1013699998 [DOI] [Google Scholar]
- 39.Reimand J, Arak T, Adler P, et al. g:Profiler—a web server for functional interpretation of gene lists (2016 update). Nucleic Acids Res. 2016;44(W1):W83-9. doi: 10.1093/nar/gkw199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pertea M, Kim D, Pertea GM, Leek JT, Salzberg SL. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat Protoc. 2016;11(9):1650-1667. doi: 10.1038/nprot.2016.095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Barrett T, Wilhite SE, Ledoux P, et al. NCBI GEO: archive for functional genomics data sets–update. Nucleic Acids Res. 2013;41(database issue):D991-D995. doi: 10.1093/nar/gks1193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li M, Jia C, Kazmierkiewicz KL, et al. Comprehensive analysis of gene expression in human retina and supporting tissues. Hum Mol Genet. 2014;23(15):4001-4014. doi: 10.1093/hmg/ddu114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Grassmann F, Fritsche LG, Keilhauer CN, Heid IM, Weber BHF. Modelling the genetic risk in age-related macular degeneration. PLoS One. 2012;7(5):e37979. doi: 10.1371/journal.pone.0037979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fabregat A, Jupe S, Matthews L, et al. The reactome pathway knowledgebase. Nucleic Acids Res. 2018;46(D1):D649-D655. doi: 10.1093/nar/gkx1132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schmitz-Valckenberg S, Sahel J-A, Danis R, et al. Natural history of geographic atrophy progression secondary to age-related macular degeneration (Geographic Atrophy Progression Study). Ophthalmology. 2016;123(2):361-368. doi: 10.1016/j.ophtha.2015.09.036 [DOI] [PubMed] [Google Scholar]
- 46.Flutre T, Wen X, Pritchard J, Stephens M. A statistical framework for joint eQTL analysis in multiple tissues. PLoS Genet. 2013;9(5):e1003486. doi: 10.1371/journal.pgen.1003486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee S, Miropolsky L, Wu M. SKAT: SNP-set (sequence) kernel association test. https://cran.r-project.org/web/packages/SKAT/index.html. Published 2013. Accessed April 12, 2019.
- 48.Dickinson ME, Flenniken AM, Ji X, et al. ; International Mouse Phenotyping Consortium; Jackson Laboratory; Infrastructure Nationale PHENOMIN, Institut Clinique de la Souris (ICS); Charles River Laboratories; MRC Harwell; Toronto Centre for Phenogenomics; Wellcome Trust Sanger Institute; RIKEN BioResource Center . High-throughput discovery of novel developmental phenotypes. Nature. 2016;537(7621):508-514. doi: 10.1038/nature19356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kitsios GD, Tangri N, Castaldi PJ, Ioannidis JPA. Laboratory mouse models for the human genome-wide associations. PLoS One. 2010;5(11):e13782. doi: 10.1371/journal.pone.0013782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sunness JS, Margalit E, Srikumaran D, et al. The long-term natural history of geographic atrophy from age-related macular degeneration: enlargement of atrophy and implications for interventional clinical trials. Ophthalmology. 2007;114(2):271-277. doi: 10.1016/j.ophtha.2006.09.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Neault M, Mallette FA, Vogel G, Michaud-Levesque J, Richard S. Ablation of PRMT6 reveals a role as a negative transcriptional regulator of the p53 tumor suppressor. Nucleic Acids Res. 2012;40(19):9513-9521. doi: 10.1093/nar/gks764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Caire J, Recalde S, Velazquez-Villoria A, et al. ; Spanish Multicenter Group on AMD . Growth of geographic atrophy on fundus autofluorescence and polymorphisms of CFH, CFB, C3, FHR1-3, and ARMS2 in age-related macular degeneration. JAMA Ophthalmol. 2014;132(5):528-534. doi: 10.1001/jamaophthalmol.2013.8175 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
eAppendix. Legends to eTables
eTable 1. Correlation of known AMD risk variants with GA lesion growth
eTable 2. Nominally significant pathways involved in GA lesion growth