Abstract
Ribosome biogenesis (RiBi) is an extremely energy intensive process that is critical for gene expression. It is thus highly regulated, including through the tightly coordinated expression of over 200 RiBi genes by positive and negative transcriptional regulators. We investigated RiBi regulation as cells initiated meiosis in budding yeast and noted early transcriptional activation of RiBi genes, followed by their apparent translational repression 1 hour (h) after stimulation to enter meiosis. Surprisingly, in the representative genes examined, measured translational repression depended on their promoters rather than mRNA regions. Further investigation revealed that the signature of this regulation in our data depended on pre-treating cells with the translation inhibitor, cycloheximide (CHX). This treatment, at 1 h in meiosis, but not earlier, rapidly resulted in accumulation of RiBi mRNAs that were not translated. This effect was also seen in with CHX pre-treatment of cells grown in media lacking amino acids. For NSR1, this effect depended on the –150 to –101 region of the promoter, as well as the RiBi transcriptional repressors Dot6 and Tod6. Condition-specific RiBi mRNA accumulation was also seen with translation inhibitors that are dissimilar from CHX, suggesting that this phenomenon might represent a feedback response to global translation inhibition.
INTRODUCTION
The synthesis of ribosomes, the ribonucleoprotein machines that mediate translation of proteins from mRNA templates, is a tightly regulated process that has been well studied in the budding yeast Saccharomyces cerevisiae. Eighty ribosomal proteins (RPs) and four ribosomal RNAs make up the ribosome, and their processing and assembly requires an additional suite of more than 200 proteins that mediate ribosome biogenesis (RiBi; (1); reviewed in (2)). RiBi genes are tightly coordinated in their transcription, which typically involves at least one general factor (Abf1, Reb1, Rap1 and Tbf1) in concert with other positive transcriptional activators (reviewed in (3)), which are typically recruited to RiBi gene promoters by the nutrient-activated kinases Tor and PKA ((4–12); reviewed in (3,13)). Therefore, under conditions in which amino acids and glucose are plentiful, RiBi gene transcription is activated, which increases the synthesis and assembly of ribosomes. Negative transcriptional regulators—including Stb3, Dot6, and Tod6—also participate in regulation of the RiBi genes by binding their promoters and recruiting the histone deacetylase Rpd3L ((14–16), reviewed in (13)). Tor and/or PKA activity is known to relieve this repression via phosphorylation of these negative regulators, resulting in their removal from RiBi promoters and subsequent nuclear export (14,16). Robust expression of RiBi genes thus requires coordinated de-repression as well as active induction. Transcriptional repression of RiBi genes under non-inducing conditions appears to be important, as it has been shown that cells lacking Dot6/Tod6-based repression are unable to properly adapt to nutrient-limiting conditions (16). This is presumably due to leaky RiBi gene production that depletes cellular energy reserves required for cellular adaptation.
Much of what we currently know about RiBi regulation comes from studies of yeast in either rich laboratory growth conditions or under conditions of acute amino acid starvation. It is also known that RiBi and RP genes are modulated during other cellular conditions in yeast, including during the meiotic program, the conserved process by which gametes (spores in yeast) are created (17–19). RP and RiBi genes are known to be transcriptionally downregulated prior to meiotic entry relative to vegetative exponentially growing cells (18), and upregulated following the meiotic divisions (17). We previously found that translation levels and apparent ribosome number increase shortly after cells enter the meiotic program, and also that RPs are actively degraded following the meiotic divisions, concomitant with transcriptional upregulation of RP and RiBi genes (19). Given these interesting patterns of regulation and the central importance of the ribosome to gene expression, we investigated the regulation of RiBi genes in meiosis in greater depth. Here, we report our observation of apparent translational repression of RiBi genes ∼1 h after cells were stimulated to enter meiosis. We find that this apparent repression is limited to a narrow time window between 45 and 75 minutes (min) after transfer of cells into sporulation medium (SPO). Surprisingly, this is not a natural part of the meiotic program, but rather a response to the drug-based inhibition of translation that we employed to collect samples. We report that global translation inhibition by CHX resulted in transcriptional upregulation of RiBi genes that is partially dependent on Tod6 and Dot6. We find that a similar effect can be seen following CHX treatment during conditions of amino acid starvation, another context in which RiBi genes are normally actively repressed. We propose that this conditional transcriptional de-repression of RiBi genes may represent a feedback response to global translation inhibition, as it is seen in response to several distinct drug-based mechanisms of general translation inhibition.
MATERIALS AND METHODS
Yeast strains
All experiments were performed using S. cerevisiae strains of the SK1 background. Strains were fully prototrophic (LEU URA TRP LYS HIS). All strains are diploid (MATa/alpha) except for 4401, which is haploid (MATa). All the reporters are integrated at TRP1 locus with functional TRP1 gene as selection marker.
| Strain number | Genotype |
|---|---|
| 4401 | WT |
| 4484 | WT |
| 11324 | pNSR1-NSR1-3V5 |
| 11582 | pSAR1-NSR1-3V5 |
| 11584 | pSAR1-NSR1-3V5 (TSS2) |
| 11326 | pNIP7-NIP7-3V5 |
| 11586 | pSAR1-NIP7-3V5 |
| 13064 | tod6::NatMX tod6::NatMX |
| 13070 | dot6::NatMX dot6::NatMX |
| 13074 | tod6::NatMX tod6::NatMX dot6::NatMX dot6::NatMX |
| 14203 | stb3::NatMX stb3::NatMX |
| 15325 | pSAR1-SAR1-3V5 |
| 15328 | pNSR1(-100 to -51 swapped to pSAR1 sequence)-NSR1-3V5 |
| 15329 | pNSR-(-150 to -101 swapped to pSAR1 sequence)-NSR1-3V5 |
| 15330 | pNSR-(-200 to -151 swapped to pSAR1 sequence)-NSR1-3V5 |
| 15331 | pNSR-(-728 to -201 swapped to pSAR1 sequence)-NSR1-3V5 |
| 15332 | pNSR-(-50 to -1 swapped to pSAR1 sequence)-NSR1-3V5 |
| 16268 | pNSR1-SAR1-3V5 |
| 16272 | pNSR1(ΔRRPE-1 ΔRRPE-2 ΔPAC)-NSR1-3V5 |
Yeast growth conditions
Vegetative samples were collected after growth of 300 ml (sequencing and polysome fractionation experiments) or 15 ml (total RNA qPCR experiments) culture in YPD from OD600 0.1 to OD600 0.6 at 30°C with shaking. For meiotic samples, cells were grown in YEPD for 24 h, diluted to OD600 0.25 in buffered YTA, and grown for 16 h. Cells were washed in water, resuspended at OD600 1.9 in 300 ml (sequencing and polysome fractionation experiments) or 15 ml (total RNA qPCR experiments) sporulation media supplemented with 0.02% raffinose, and incubated at 30°C with shaking. For amino acid starvation experiments, cells were grown in YPD overnight, diluted to OD600 0.1 in fresh YPD, grown to OD600 0.6, resuspended in equal volume of SD media without amino acids, and incubated at 30°C with shaking.
Cell extract preparation
CHX (100 μg/ml final concentration) or equal volume of vehicle solvent (ethanol) was added to the cells 1.5 min prior to harvesting. Cells were collected by filtration and flash freezing in liquid nitrogen. Around 10% of the cells were stored in a separate tube for mRNA-seq. 3 ml frozen buffer (20 mM Tris pH 8, 140 mM KCl, 1.5 mM MgCl2, 100 μg/ml cycloheximide, 1% Trition X-100) were added to the cell pellets. Samples were lysed by Retsch mixermilling (3 × 3-minute rounds at 15 Hz). The powder was thawed at 30°C, spun at 4°C for 5 min at 3000 rcf. The supernatant was collected and spun at 20 000 rcf at 4°C for 10 min. The supernatant was aliquoted in 200 μl portions and flash frozen in liquid nitrogen. Identical extracts were used for ribosome profiling and polysome gradients.
Ribosome profiling and mRNA-seq data
Ribosome profiling and mRNA-seq data were analyzed from (17) and are deposited at NCBI GEO with accession number GSE34082. Cluster 3.0 and Treeview were used for cluster analyses and visualization (20,21). Mochiview was used for genome browsing visualization (22).
LTM profiling
Cells were harvested by filtration following treatment at a final concentration of 50 uM LTM in DMSO and 5 min incubation with shaking. Extract was prepared following mixermilling and 200 μl of cell extract were incubated with 15 U RNase I (Ambion) per A260 unit of extract for 1 h at room temperature. Samples were loaded onto 10–50% sucrose gradients and spun at 35 000 rpm for 3 h at 4°C in a SW41Ti rotor (Beckman). The gradients were then fractionated using a Gradient Station (BioComp) and the 80S/monosome peaks were collected. RNA was extracted with hot acid phenol, size selected from a polyacrylamide gel, dephosphorylated with PNK (NEB), polyA-tailed with Escherichia coli polyA polymerase (NEB), subjected to rRNA subtraction by antisense oligos, reverse transcribed with Superscript III (Thermo), circularized with Circ Ligase (Epicentre), and PCR amplified with Phusion polymerase (NEB). The Oligo oCJ200-oligodT was used for reverse transcription, and oNTI231 and aatgatacggcgaccaccgagatcggaagagcacacgtctgaactccagtcac-barcode-cgacaggttcagagttc index primers was used for PCR. The barcodes are six nucleotides in length. Sequencing was done using standard Illumina oligos. All samples were sequenced on an Illumina HiSeq 2500, 50SRR, with multiplexing, at the UC-Berkeley Vincent Coates QB3 Sequencing facility.
LTM sequencing data analysis
LTM ribosome profiling data were analyzed by Bowtie2-based alignment exactly as standard ribosome profiling data in (23–25). Mochiview was used for genome browsing visualization (22).
Polysome analysis
The sucrose gradients were performed exactly as in (32), except without RNAse I treatment. In short, 200 ul of extract was loaded on 10–50% sucrose gradients and samples were centrifuged in a Beckman XL-70 Ultracentrigue, using a Sw-Ti41 rotor for 3 h at 35 000 rpm at 4°C. Tube was loaded on a Bio-Comp Gradient Station and analyzed for absorbance at 260 nm. Eight fractions were collected per gradient and flash frozen in liquid nitrogen. 300, 500 and 700 ul of DEPC water was added to the fractions 6, 7 and 8, respectively, to bring their density below the density of phenol/chloroform mix prior to hot acid phenol extraction.
Cell harvesting for RT-qPCR
All YPD samples were harvested at OD600 0.6 in YEPD medium. All amino acid starvation samples were harvested 20 min after transferring from YEPD to SD medium without amino acids, except for the experiment showed in Figures 3E and Supplementary Figure S5C, in which the cells were harvested at the indicated timepoints.
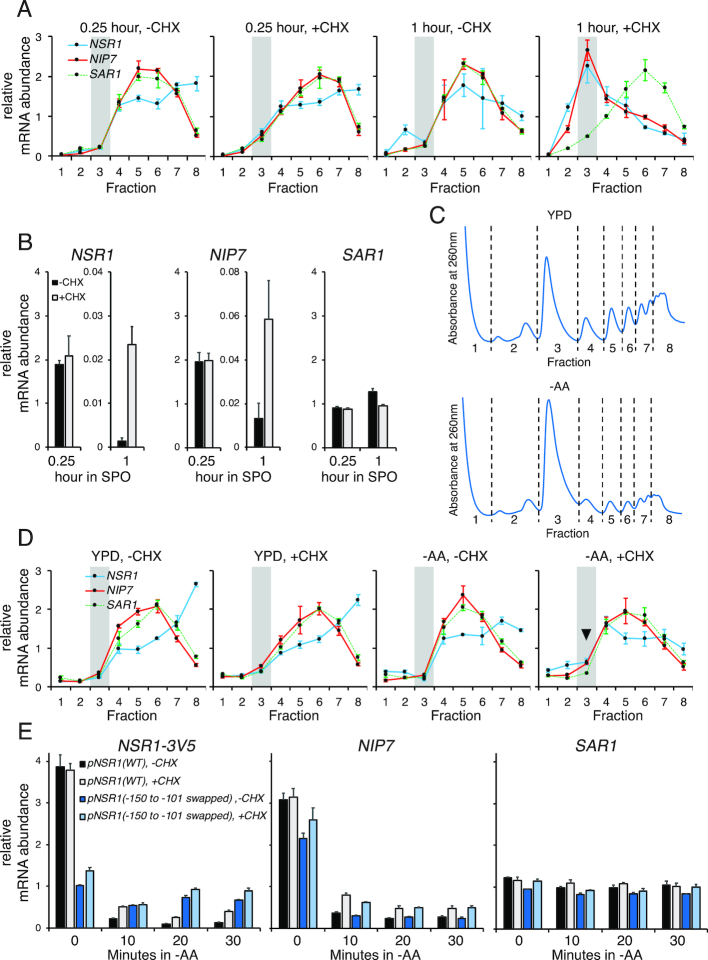
Figure 3.
CHX treatment causes accumulation of RiBi transcripts, explaining the apparent translational repression at 1 h in SPO. Throughout this figure, mRNA is quantified by RT-qPCR and error bars represent measurement variability as determined by three qPCR replicates. (A) Distributions of RiBi mRNAs (NSR1 and NIP7) and a non-RiBi mRNA (SAR1) in polysome gradients without or with CHX treatment at 0.25 h or 1 h in SPO. The highlighted region represents monosome/80S fraction. (B) mRNA levels of NSR1 (left), NIP7 (middle), and SAR1 (right) without or with 1.5-minute CHX treatment at 0.25 or 1 h in SPO. (C) Sucrose gradient absorbance profiles of exponentially growing cells (OD6000.6) in YPD (top) and after 20 min of amino acid starvation (bottom). (D) Distributions of RiBi mRNAs (NSR1 and NIP7) and a non-RiBi mRNA (SAR1) in polysome gradients without or with CHX treatment in rich medium (YPD) or at 20 min after amino acid starvation (–AA). The highlighted region represents monosome/80S fraction and arrowhead emphasizes the reproducible difference observed in –AA conditions. (E) mRNA levels of NSR1 (left), NIP7 (middle), and SAR1 (right) in response to CHX treatment following amino acid starvation. The first timepoint (0 min) was collected prior to the media switch from YPD. The CHX-dependent mRNA increase is greatly diminished in the NSR1 reporter with its –150 to –101 promoter region swapped to the corresponding SAR1 promoter sequence. Note that the increase in NSR1 mRNA seen with CHX in –AA conditions is statistically significant (Student's t-test P-value 0.0014 at 20 min) when comparing to the biological replicate data in Supplementary Figure S5C. This difference is no longer significant in the –150 to –101 promoter mutant.
1 ml of cells was mixed with drugs or solvent in a 2 ml screw-cap tube. The samples were kept shaking for 1.5 min at 30°C, spun at 20 000 rcf for 30 s, and flash frozen in liquid nitrogen.
The final drug concentrations were: CHX: 100 μg/ml; LTM: 10 μM; sordarin: 100 μg/ml; anisomycin: 20 μM. For controls, an equal volume of solvent was added to the cells (ethanol for CHX and sordarin, DMSO for LTM and anisomycin).
In the TOR inhibition experiment, rapamycin was added to the cells at 0.2 μg/ml final concentration, 10 min prior to CHX addition. An equal volume of the solvent, DMSO, was added to the control samples.
RT-qPCR
RNA was isolated from the gradient fraction samples or the cell pellets by hot acid phenol extraction, DNase-treated with Turbo DNase (Invitrogen), and purified with phenol extraction. The RNA samples were adjusted to similar concentration and 150–250 ng of RNA was used in reverse transcription with Superscript III (Thermo). Transcript levels were quantified on a StepOnePlus Real-Time PCR system using the SYBR green PCR mix (ThermoFisher), and normalized to the total RNA concentrations. CT values were first transformed to fold changes (according to the standard curve of the primer pair). Values were then normalized to total RNA amount in reverse transcription reaction of each fraction, and for gradient analyses, also normalized to the average of the eight fractions for each strain.
RT-qPCR analysis
For figures, to enable visualization of trends between transcripts of different expression levels, the values plotted are relative to the average values for that transcript in all fractions or conditions analyzed in the plot. Values are also relative to the total RNA in that fraction.
The qPCR primers are listed below:
NSR1-forward 5′ GTTCAATTCTCCAACATGGAGGACG 3′
NSR1-reverse 5′ CCATCGTTGTTTGGTCTTGGAGAAG 3′
NIP7-forward 5′ AGGCACATGTGGGTAAAATGTCTG 3′
NIP7-reverse 5′ ACCGAACCCTAATGGCACATC 3′
SAR1-forward 5′ TCAAGCTCGTCGTTTATGGAAGG 3′
SAR1-reverse 5′ TCTTTCAGGGTCAGCAGCATC 3′
REC8-forward 5′ TCTAACAGGTTCGAGCTTCATGGG 3′
REC8-reverse 5′ CATCAACGGGAATTTCATCCAGTGG 3′
CCW14-forward 5′ CCTCTACCAAGGCTTCTTCCAGT 3′
CCW14-reverse 5′ GTGGAAGAAGCCTTGCTAGAAGATG 3′
NSR1-3V5-forward 5′ CGCTTCTTTCGCTGGTTCAAAG 3′
NIP7-3V5-forward 5′ GTGAGTATTTGAGAGATGAAGACACCTTG 3′
SAR1-3V5-forward 5′ TAGAGGCGTTCCAATGGTTATCTC 3′
Common-3V5-reverse 5′ TGGTATTGGTTTTCCATCTAGTCCC 3′
5′ RACE
cDNA was synthesized from phenol extracted total RNA using GeneRacer kit with SuperScript III RT (Invitrogen), following the manufacture's protocol. cDNA was then amplified using Q5 Hot Start High-Fidelity DNA polymerase (NEB) with adapter primer in the GeneRacer kit and gene specific primers to generate 5′ end for individual genes. The 5′ ends were incubated with 1 U Taq polymerase (Invitrogen) to add 3′ A-overhangs, purified using agarose gel extraction, cloned into vector using TOPO TA Cloning Kit for Sequencing (Invitrogen), and transformed into E. coli. Around twenty clones per gene were sequenced. Only clones with intact adapter sequence were analyzed.
The gene specific primers are listed below
NSR1 5′ GATTCGGAGGAAGAGGAAGAGAC 3′
NIP7 5′ GCTTAGCCAATACTGTCAAAGAAGT 3′
SAR1 5′ TGACCACCCAAATCGAAAGTTGT 3′
RESULTS
RiBi mRNAs show signatures of translational repression shortly after stimulation of cells to enter meiosis
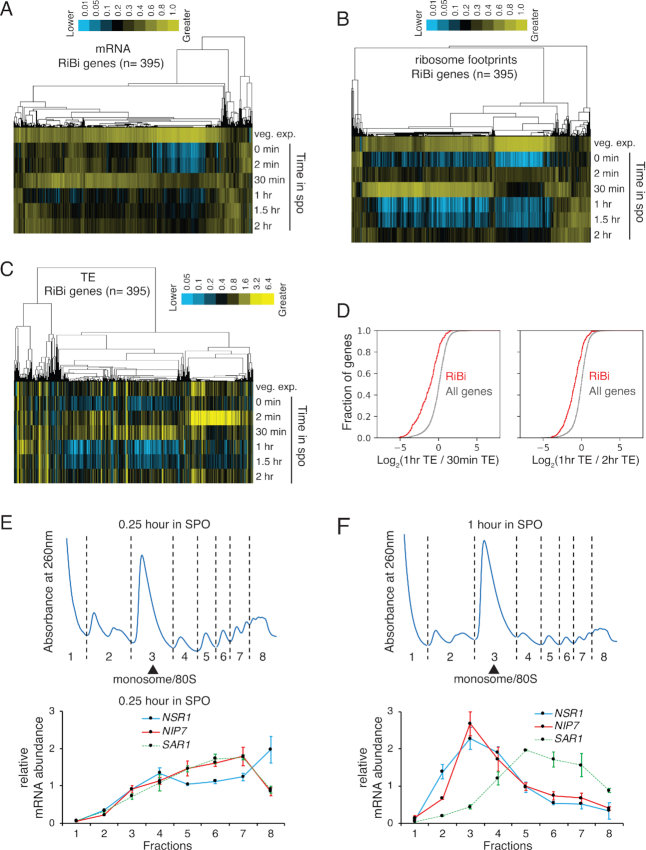
To begin investigating the regulation of RiBi genes in meiosis, we re-analyzed a previously generated dataset from our lab, which included mRNA abundance and translation measurements made by mRNA-seq and ribosome profiling, respectively, during a high-resolution timecourse of the budding yeast meiotic program (17). We found that, as expected, most annotated RiBi genes (325/395; ‘ribosome biogenesis’ at http://amigo.geneontology.org/amigo; Pol II-dependent transcripts only; includes RP genes with roles in RiBi) were coordinately regulated over all timepoints. mRNA levels of RiBi genes were very low prior to meiotic entry, but were rapidly increased within 30 min in SPO, followed by their rapid decline (Figure 1A). Our ability to observe this brief burst of transcription depended on the high-resolution of our timepoints relative to previous studies. We collected denser timepoints early in the meiotic program and found that RiBi transcripts peak at 15 min and decline by 45 min, timepoints that were not included in our original large-scale dataset (Supplementary Figure S1A). Translation levels, as assayed by ribosome footprint density over ORFs, showed a roughly similar trend as mRNA levels, which fit with our expectations (Figure 1B). Surprisingly, however, when we calculated the translation efficiency (TE, ribosome footprint abundance normalized to mRNA abundance) for RiBi mRNAs, we noted that there was apparent translational repression at ∼1 h after transfer of cells to SPO, shortly after transcript levels had plummeted (Figure 1C, D, Supplementary Figure S1A) and after expression of the early meiosis-specific gene, REC8, was detected, indicating commitment to the meiotic differentiation program (Supplementary Figure S1A). This translational repression was strong and consistent for RiBi genes (Figure 1D, Supplementary Figure S1B and C).
Figure 1.
RiBi genes show translational repression at 1 h in sporulation medium. (A–C) mRNA-seq, ribosome profiling, and TE values of RiBi genes from a high-resolution timecourse of the budding yeast meiotic program (17). Data are clustered by similar expression patterns of genes (columns) across timepoints in sporulation medium (rows). (D) Left: a cumulative histogram showing log2 values of the ratio between 1 h and 30 min TEs. The red line represents RiBi genes and the gray line represents all genes. A Kolmogerov–Smirnov (K–S) test was used to determine significance. P = 7.99E–54. Right: a cumulative histogram showing log2 values of the ratio between 1 and 2 h TEs. The red line represents RiBi genes and the gray line represents all genes. K–S test P = 1.75E–48. (E and F) Distributions of RiBi mRNA (NSR1 and NIP7) and non-RiBi mRNA (SAR1) in polysome gradients as determined by RT-qPCR. Error bars represent measurement variability as determined by three qPCR replicates. The sample shown in panel E was collected at 0.25 h in SPO, and the sample shown in panel F was collected at 1 h in SPO. Polysome gradients for these timepoints are shown above in each case.
To independently confirm the translational repression that we measured by global approaches, we analyzed the distribution of RiBi mRNAs in polysome gradients at 15 min and 1 h after transfer to SPO, timepoints prior to and during the period in which translational repression was measured, respectively (Figure 1C, E, F). Comparison of the mRNA distribution for representative RiBi genes NSR1 and NIP7 to non-RiBi gene SAR1 revealed that all were abundant in heavy polysome fractions at 15 min after transfer to SPO, suggesting high levels of translation, which was consistent with TE measurements from the global meiotic study (Figure 1C, E). Extract from cells 1 h after transfer to SPO showed a specific shift to the monosome/80S fraction for NSR1 and NIP7, but not SAR1, again consistent with our analysis of the global study (Figure 1C, F). To identify the regulatory regions of NSR1 and NIP7 responsible for this apparent translational repression, we mapped the 5′ ends of these mRNAs by 5′ RACE, along with that of SAR1 (Supplementary Figure S2A–C). Because disruption of NSR1, NIP7 or SAR1 would have likely resulted in secondary cellular consequences based on the critical cellular roles for these genes, we constructed strains carrying reporter constructs that did not alter the wild-type NSR1, NIP7 or SAR1 loci. To do this, we added a reporter construct, integrated at the TRP1 locus, which expressed a 3' 3V5-tagged version of each of these genes. We collected fractions from polysome gradients performed with cell extract of strains carrying these reporter constructs, and performed RT-qPCR against the 3V5 region in these fractions to distinguish our reporters from the endogenous genes. We found that the reporter constructs recapitulated the regulation seen at the endogenous locus for all three genes at 1 hr after transfer to SPO (Figure 2A–D, left panels).
Figure 2.
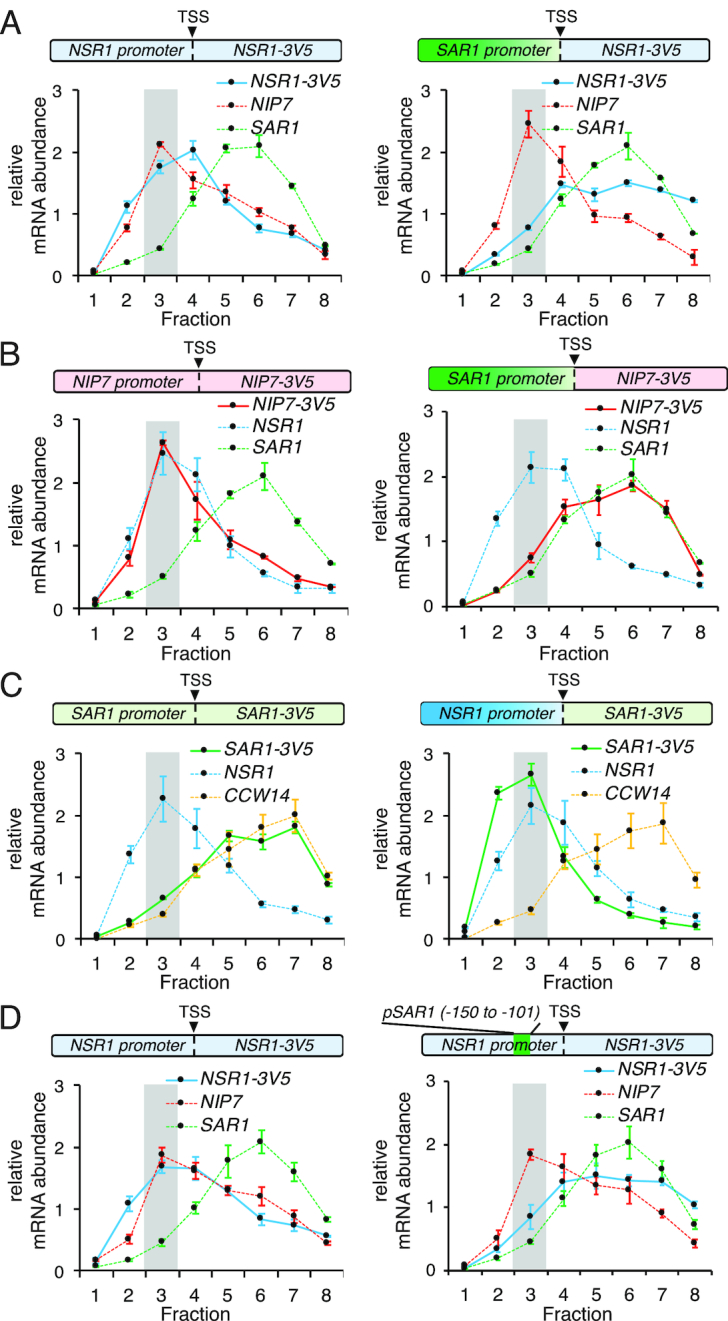
RiBi promoters are necessary and sufficient for their translational repression. (A–D) RT-qPCR was used to quantify mRNAs in polysome fractions at 1 h in SPO. The highlighted regions represent monosome/80S fractions. Error bars represent measurement variability as determined by three qPCR replicates. (A) Distribution of the mRNAs for the WT NSR1 reporter (left) and the NSR1 reporter under the SAR1 promoter (right) in polysome gradients at 1 h in SPO. (B) Distribution of the mRNAs for the WT NIP7 reporter (left) and the NIP7 reporter under the SAR1 promoter (right) in polysome gradients at 1 h in SPO. (C) Distribution of the mRNAs for the WT SAR1 reporter (left) and the SAR1 reporter under the NSR1 promoter (right) in polysome gradients at 1 h in SPO. (D) Distribution of the mRNAs for the WT NSR1 reporter (left) and the NSR1 reporter with its –150 to –101 promoter region swapped to the corresponding SAR1 promoter sequence (right) in polysome gradients at 1 h in SPO.
An interval of the NSR1 promoter is necessary and sufficient for its apparent translational repression
We next sought to identify the cis-regulatory regions responsible for this regulation. Interestingly, replacing the NSR1 promoter alone with that of SAR1 resulted in the retention of NSR1 –3V5 in heavy polysome fractions at 1 h in SPO (Figure 2A, Supplementary Figure S2D, S3A). A similar effect was seen when the SAR1 promoter was used to drive NIP7 with its 5′ UTR intact (Figure 2B). These results were a surprise because most translational regulation is thought to be mediated through untranslated regions (UTRs) of mRNAs and, although it has been reported that promoter regions can affect downstream processes including mRNA translatability and decay (26–29), this type of regulation had not been reported for RiBi genes, to our knowledge. We further explored this promoter-based RiBi regulation, finding that the NSR1 promoter was not only required for the shift of the reporter mRNA to the monosome fraction at 1 h in SPO, it was also sufficient for this shift, as a SAR1–3V5 reporter driven by the NSR1 promoter was seen in the monosome fraction at 1 h (Figure 2C). This was not the case for another non-RiBi mRNA, CCW14 at the same timepoint, suggesting that the effect seen was transcript-specific rather than a global perturbation of translation (Figure 2C). To identify the specific promoter region that was required for translational repression, we swapped out intervals of the NSR1 promoter with the corresponding region of the SAR1 promoter and found that one such interval replacement, for the region between -150 and -101 bp upstream of the NSR1 transcription start site (‘TSS’ in Supplementary Figure S2A), was necessary for the shift of the NSR1 reporter into the monosome/80S fraction at 1 h in SPO (Figure 2D, Supplementary Figure S3A and B). This region of the NSR1 promoter contains binding sites for the three known RiBi transcriptional repressors, Dot6, Tod6, and Stb3 (Supplementary Figure S3C; (30)). Deletion of any one of these proteins did not result in a loss of the observed translational repression at 1 h in SPO (Supplementary Figure S3D). Because Dot6 and Tod6 are paralogs and both bind the same DNA element (PAC motif; (30,31)), we attempted to assay a dot6Δtod6Δ double mutant, but found that these cells showed markedly reduced ability to undergo meiosis, obscuring our ability to observe normal RiBi regulation timing.
Cycloheximide treatment leads to context-dependent de-repression of RiBi mRNAs
The rapid decline in TE that we observed in meiotic cells was reminiscent of translational repression seen by the same ribosome profiling protocol applied to yeast cells that were transferred from rich media to media lacking amino acids (32). In addition, we became aware that a similar type of time-interval-specific apparent translational repression was detected by another group during amino acid starvation and metabolic cycling in yeast, and that it appeared to be dependent on the translation inhibitor, CHX, due to a resultant increase in RiBi mRNAs (personal communication, Dan Santos; reported in accompanying study (33)). CHX is a chemical that is frequently used to pre-treat cells prior to harvesting for ribosome profiling (32,34). In our study, CHX pre-treatment occurs for less than 2 min prior to harvesting of cells by filtration and flash freezing in liquid nitrogen. CHX pre-treatment has been reported to affect the distribution of ribosome footprints within mRNAs but has been reported to have a minimal effect on gene-by-gene quantification in most contexts examined (32,35–40). Because of this, and because harvesting with filtration without CHX pre-treatment results in loss of ribosomes from 5′ regions of ORFs (32), CHX is typically included in our protocols for harvesting cells for translation assays, including both ribosome profiling and polysome analyses. It seemed possible that this could explain the apparent translational repression that we observed.
Indeed, when we performed polysome analyses without CHX pre-treatment, we no longer observed a shift of NSR1 and NIP7 to the monosome/80S fraction at 1 h in SPO (Figure 3A). We assayed NSR1, NIP7, and SAR1 mRNA abundances by RT-qPCR at 15 min and 1 h in SPO with and without CHX pre-treatment and found that the high level of all of these mRNAs seen at 15 min in SPO was unaffected by CHX (Figure 3B, Supplementary Figure S4A). The total level of NSR1 and NIP7 mRNA was much lower at 1 h than at 15 min in SPO with or without CHX pre-treatment, but it was higher with CHX pre-treatment at 1 h in SPO than without CHX pre-treatment, a result that was not seen for SAR1 (Figure 3B, Supplementary Figure S4A). This change, while small in absolute terms, represented a 4- to 16-fold increase in NIP7 and NSR1, respectively (Figure 3B, Supplementary Figure S4A). This is approximately the degree of TE shift that we measured by ribosome profiling for these genes at 1 h in SPO (Figure 1C). Because TE is calculated by dividing the ribosome footprint RPKM (reads per kilobase per million mapped reads) by mRNA RPKM, and because CHX treatment inhibits new translation (42,43), we concluded that the apparent translational effect that we observed for RiBi genes at 1 h in SPO (Figure 1F) was an artifact of the time-window-specific mRNA induction of these genes in response to translation inhibition by CHX. In support of this model, we observed a build-up of ribosome footprints at start codons of RiBi genes specifically at ∼1 h in SPO in an experiment in which CHX-pretreatment was used (Supplementary Figure S4B and C). This type of build-up has been previously observed more globally in ribosome profiling data collected from cells under stress conditions (35) and has been thought to represent the fact that CHX inhibits post-initiation ribosomes (42). CHX treatment does not prevent new translation initiation, but does prevent elongation, which results in a build-up of ribosomes at start codons. This effect can be seen to some extent under all conditions, but is most extreme in cases in which many ribosomes are free to re-initiate, which appears to be the case following acute stress conditions that results in general inhibition of new translation.
Because the TE effect that we observed early in meiosis was similar to that observed previously in amino acid-starved cells (26), we next compared polysome fractions from vegetative cells grown in rich media (YPD) or shifted to media lacking amino acids (-AA), and observed a small but specific and reproducible CHX-dependent shift of NSR1 to the monosome fraction (Figure 3C, D and Supplementary Figure S5A). As expected, much lower levels of ribosomes in polysome relative to monosome fractions was observed in cells shifted to media lacking amino acids (Figure 3C). The CHX- dependent shift to the monosome fraction of NSR1 and NIP7, was modest compared to 1 h in SPO (Figure 3A, D and Supplementary Figure S5A). Consistently, a much more subtle CHX-dependent mRNA increase was seen following amino acid starvation than the one that we observed at 1 h in SPO (2.6-fold versus 17.3-fold for NSR1, 1.9-fold versus 4.8-fold for NIP7; Figure 3B, 3E, Supplementary Figures S4A, S5B-D). Despite the reduced degree of the effect, the specific mechanism of this mRNA induction appeared to be similar in meiosis and under conditions of amino acid limitation, as the CHX-dependent fold-increase in mRNA seen after transfer to media lacking amino acids was substantially reduced in the NSR1 reporter construct mutated for the –150 to –101 interval defined based on meiotic regulation (Figures 2D, 3E and Supplementary Figure S5B-D). The muted CHX-dependent fold increase in NSR1 mRNA was because removal of this NSR1 promoter region resulted in an increase in NSR1 mRNA whether or not CHX pre-treatment was used for cell harvesting (Figure 3E and Supplementary Figure S5B–D). This result suggested that the CHX-dependent mRNA increases that we observed could be a result of faulty RiBi repression at 1 h in SPO and under amino acid deplete conditions.
Loss of negative regulators of RiBi transcription, Dot6 and Tod6, ablates CHX-dependent RiBi de-repression
We previously noted that the –150 to –101 promoter region of NSR1 contained binding sites for repressors Stb3, Dot6, and Tod6 (Supplementary Figure S3C). The lack of binding of a repressor in this region was consistent with the CHX-independent de-repression that was seen in our reporter experiments (Figure 3B, E, Supplementary Figure S5B–D). While single deletions of STB3, DOT6 or TOD6 in meiosis did not ablate the effect (Supplementary Figure S3D), amino acid-depleted conditions did not rely on the type of sensitive time window that was seen during meiosis and allowed us to assay double-mutants without confounding effects on meiotic entry that previously stymied our meiotic dot6Δtod6Δ experiment. We found that cells deleted for both DOT6 and TOD6 no longer showed a CHX-dependent shift away from polysomes or an increase in either NSR1 or NIP7 mRNA in media lacking amino acids, suggesting that the CHX-dependent mRNA increase in RiBi genes is mediated through these two factors (Figure 4A, Supplementary Figure S6A–D). Akin to the reporter experiment in which the –150 to –101 promoter region was deleted (Figure 3E), the loss of CHX-dependent mRNA increase is associated with an elevated absolute level of RiBi mRNA relative to wild-type cells without CHX-treatment, and these levels do not increase more in cells harvested with CHX (Figure 4A, Supplementary Figure S6B–D). As in the meiotic experiments (Supplementary Figure S3D), mutation of STB3 had no effect on the CHX-dependent regulation (Figure 4A, Supplementary Figure S6B–D). Mutation of the consensus binding sites for STB3, DOT6 and TOD6, within the –150 to –101 interval, however, did not recapitulate this effect, suggesting that Dot6 and/or Tod6 may have additional cryptic binding sites in this region or they are recruited by another factor (Supplementary Figure S7A–D).
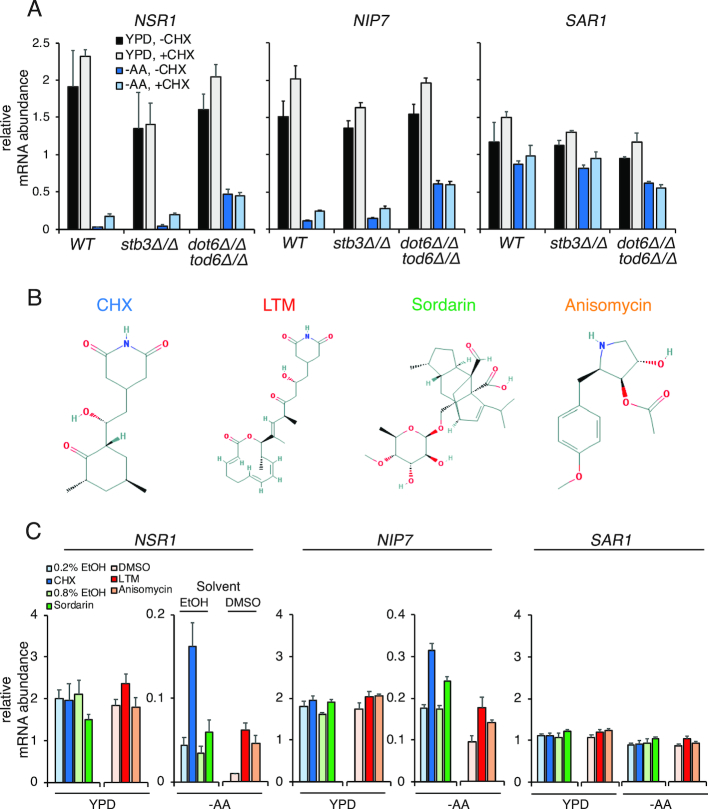
Figure 4.
The increase of RiBi transcript levels in response to CHX is mediated by Dot6/Tod6 and is caused by global translation inhibition. Throughout this figure, mRNA is quantified by RT-qPCR and error bars represent measurement variability as determined by three qPCR replicates. (A) mRNA levels of NSR1 (left), NIP7 (middle), and SAR1 (right) in response to CHX treatment. Deletion of DOT6 and TOD6 eliminates the CHX-induced accumulation of RiBi transcripts following amino acid starvation. Note that the increase in NSR1 in –AA conditions with CHX is statistically significant (Student's t-test P-value 0.0331) when comparing to the biological replicate data in Supplementary Figure S6C. This difference is no longer significant in cells lacking DOT6 and TOD6. (B) The structures of the translation inhibitors used in panel C are shown, from NCBI PubChem. (C) mRNA levels of NSR1 (left), NIP7 (middle), and SAR1 (right) in response to CHX (100 μg/ml), LTM (10 μM), anisomycin (20 μM), and sordarin (100 μg/ml). Stock solutions of CHX and sordarin were prepared in EtOH, and stock solutions of LTM and anisomycin were prepared in DMSO. Note that LTM and anisomycin treatment together with their vehicle control (DMSO) was done in a separate experiment from sordarin and CHX treatment and their vehicle controls (EtOH). Also note that sordarin-based inhibition of cell growth is less complete than other drugs (Supplementary Figure S9).
The known regulation of RiBi genes, which involves transcriptional activation and removal of transcriptional repression, along with the time-interval specificity of CHX-based mRNA induction during both meiotic and amino acid-starvation conditions and the low absolute levels of mRNAs for these genes seen during both of these time intervals (Figure 3B and E), suggested a possible mechanism for the effect that we observed. It seemed that the condition-specificity of this response might be based on whether RiBi transcriptional activation is occurring. If it is not, de-repression of transcription through Dot6 and Tod6 by a CHX-based mechanism could explain the modest absolute mRNA increases that resulted in large fold-change increases, and thus the measured TE decreases that we observed. We tested this hypothesis by treating cells in either rich media or under conditions of amino acid starvation with rapamycin, which inhibits activity of the Tor kinase (44), a major driver of RiBi transcriptional activation (6,45). This treatment resulted in lower overall mRNA levels for RiBi genes NSR1 and NIP7 in rich (YPD) media (Supplementary Figure S8A), as expected. This also resulted in a robust CHX-dependent increase in mRNA levels under these conditions, which was not observed without Tor inhibition (Supplementary Figure S8A and B). A similar effect was not seen for non-RiBi gene SAR1. In the case of amino-acid-starved cells, rapamycin treatment resulted in a more modest decrease in the levels of mRNA without CHX treatment, consistent with lower Tor activity in these conditions (46), but a CHX-dependent mRNA level increase was still evident (Supplementary Figure S8A and B). We note that the CHX-dependent mRNA increase remained in the presence of rapamycin in the prototrophic strain background used for all of our experiments, but that this was not seen in the auxotrophic strain background used for the accompanying study (33). The reason for this difference is unclear, but is consistent with the finding from Santos et al. that auxotrophies can modify the CHX-dependent effect on RiBi mRNAs (33).
RiBi mRNA de-repression is a response to global inhibition of translation
Our results to this point could be explained by two general models. By the first model, CHX results in de-repression of RiBi mRNA accumulation through Tod6 and Dot6 independent of its role in inhibiting translation. By the second model, it is inhibition of translation itself that causes RiBi mRNA accumulation. To distinguish between these possibilities, we treated cells grown in YPD and those starved for amino acids with three other drugs that rapidly inhibit translation: lactimidomycin (LTM), sordarin, and anisomycin ((42,47,48); (Figure 4B, Supplementary Figure S9A and B)). CHX inhibits ribosome translocation through E-site binding (49). LTM shows some structural similarity to CHX, but is larger, which causes its preferential binding to empty E-sites and thus preferential inhibition of the first ribosome translocation event on an mRNA (49). LTM is known to specifically inhibit translation initiation at 50 uM in mammalian cells (50), but we find that it inhibits both translation initiation and elongation at this concentration in yeast (Supplementary Figure S10A). Sordarin inhibits translation elongation by inhibiting elongation factor 2 activity (48). Anisomycin has been reported to partially inhibit translation elongation by binding at the peptidyl transfer center, but also results in a number of other cellular effects, including mitogen activated kinase pathway activation (47,51,52). Importantly, sordarin and anisomycin have structures that are dissimilar from CHX (Figure 4B), and thus would be likely to have different off-target cellular effects. All drugs, however, resulted in an increase on NSR1 mRNA abundance in conditions of amino acid starvation that were independent of their vehicle solvents (Figure 4C; Supplementary Figures S10B–D, S11A–D). Effects on NIP7 abundance were similar to NSR1 for CHX, LTM, and sordarin, but somewhat variable in the case of anisomycin for reasons that we do not yet understand. Nonetheless, these results suggested to us that condition-specific RiBi mRNA accumulation may represent a cellular response to inhibited translation rather than an artifact of treatment with CHX.
DISCUSSION
Ribosome biogenesis is a conserved, critical, and extremely energy intensive process that is thus highly regulated (53). Hundreds of RiBi genes are coordinately transcriptionally controlled and constitute targets of all three RNA polymerases (Pols I, II and III). Robust induction of protein-coding Pol II RiBi target genes requires both activation of a gas pedal (Tor-activated binding of activating transcription factors) and release of a brake (Tor- or PKA-based removal of repressive RiBi promoter-bound factors Dot6, and Tod6). Our study shows that inhibition of translation results in an increase in RiBi mRNA accumulation that is modest in absolute quantity but represents a high fold change early in meiosis. The de-repression of mRNA seen with CHX and other translation inhibitors could, in principle, be due to de-repression of transcription of RiBi genes or inhibition of degradation or RiBi mRNAs. We strongly favor the former model for two reasons. First, the CHX-based RiBi gene mRNA accumulation is dependent on Dot6 and Tod6, which are known to bind to RiBi promoters and repress their transcription. Second, this mRNA accumulation requires an interval in the promoter of NSR1 that is known to bind Tod6 and Dot6. The accompanying paper (33) reports that CHX-dependent RiBi mRNA accumulation is also associated with nuclear to cytoplasmic shuttling of Dot6, Tod6, and Stb3, which also supports a model of transcriptional de-repression.
We were unable to assay the effects of DOT6 and TOD6 loss on RiBi transcript abundance at 1 h in SPO because cells deleted for both of these genes could not efficiently complete meiosis (15% sporulation compared to 91% for WT cells). This defect suggests importance to RiBi transcriptional repression during meiosis, which is also consistent with the rapid and dramatic transcript regulation that naturally occurs as cells enter the meiotic program (Figure 1A, Supplementary Figure S1A). Prior to transfer of budding yeast cells to SPO, cellular ribosome and translation levels are extremely low because pre-sporulation medium lacks a non-fermentable carbon source and added amino acids, and meiotic entry requires low PKA activity (54). SPO is at least as nutrient-poor as pre-sporulation medium, yet within 30 min of transfer to SPO, ribosome and translation levels increase dramatically (17), which is consistent with the rapid transcript induction of RiBi genes that we observe by 15 min in SPO (Supplementary Figure S1A) and the fact that Tor activity is required for meiotic entry (54). Why is this transcriptional burst so brief? Although ribosome synthesis is likely required for cells to translate the proteins needed for meiotic progression, the low nutrient conditions that are also necessary for meiosis to occur in yeast may mean that cells must carefully balance their ribosome number to retain resources to make other proteins. Consistently, the wave of early genes that are transcribed in meiosis, including REC8, rise concomitant with a drop in RiBi transcript levels (Supplementary Figure S1A). A related explanation invokes splicing regulation. Most transcripts in budding yeast are not spliced, but the spliced set is highly enriched for RP genes and meiotic genes. Because RP transcripts and meiotic transcripts compete for splicesome components (55,56), very early transcriptional induction followed repression of RiBi/RP gene transcription may be required for meiotic yeast cells to properly process many of the transcripts that drive early meiotic events. By either model, rapid RiBi gene induction in conditions that favor sporulation may have evolved to enable the increase in protein synthesis capacity that is needed for meiotic commitment, and the low RiBi transcript levels seen soon after may enable expression of transcripts required for meiotic progression and, for the purposes of our study, represent a sensitized condition in which CHX-dependent de-repression is unmasked.
An important lesson of our study is that rapid secondary cellular responses can result from inhibition of core gene expression machinery. We were alerted to investigate the RiBi-specific CHX-dependent effect by authors of the accompanying study (33). We were surprised to observe the same effect our context, due to both on the speed of our cell harvesting—within 2 min of CHX treatment—and the specificity of the effect in time and scale. CHX-dependent mRNA increases were only seen for a subset of genes, strongly enriched for RiBi function, and only for a period of <1 h during the meiotic program, as discussed above. This effect is also observed following amino acid starvation of vegetatively growing cells. In both conditions, RiBi mRNA levels are low and this effect can be mimicked in cells grown in rich media in Tor-inhibited conditions, when RiBi mRNAs are also resultantly low. Interestingly, in meiosis, low RiBi mRNA levels do not appear sufficient to see this response. NSR1 mRNA levels, for example, are low during the meiotic divisions and the very low TE measurement that is the signature of this effect is not seen at this time, suggesting additional determinants that enable the effect to be observed. The rare conditions in which the CHX-dependent effect is seen and its isolation to specific gene subsets caused this artifact to appear to represent specific and temporally modulated translational control. This highlights a major challenge of measuring translation in vivo. Because translation is a dynamic process, ideal harvesting conditions should capture cells instantaneously without perturbation. Unfortunately, no such conditions have been reported, to our knowledge. Flash freezing cells in the absence of CHX is one excellent alternative to CHX pretreatment, but because this requires a human to transfer cells from a filter membrane to liquid nitrogen, ribosomes would be expected to move at least several codons along mRNAs during the process, which might also result in specific artifacts, and does result in general loss of ribosome density from the 5′ ends of ORFs. Confirmation of results from translation experiments using complementary harvesting conditions may be the best solution to avoid the type of prolonged data misinterpretation that was the basis for our study.
Does the unexpected artifact of our methodology offer insight into a real biological response? We argue that this is likely, based on the similarity of the response seen with treatment of cells by four different translation inhibitors. While these drugs all inhibit translation elongation, they do not share any particular structural feature and thus it does not seem likely that they would all share an off-target effect on RiBi mRNAs. Rather, these results are consistent with at least two biological models. First, it is possible that acute translation inhibition increases intracellular amino acid pools, which may mimic addition of nutrients and thus activate ribosome biogenesis through Tor activation. While this type of effect has been seen in mammalian cells treated with translation inhibitors (57) and consistent results exist in yeast (58), those experiments employed much longer inhibitor treatment times than ours (at least 30 min compared to less than 2 min), which may reveal downstream effects rather than the initial cellular response. Consistently, we did not find inhibition of Tor activity by rapamycin treatment to abolish CHX-dependent RiBi gene transcription in our prototrophic strain background. Alternatively, a feedback loop may exist in cells such that low or stalled translation leads to an increase in ribosome synthesis. Given the direct relationship between active ribosome number and cellular growth (53,59), it may not be surprising for such a failsafe to exist to enable synthesis of more ribosomes when a condition of reduced translation is encountered. It is possible, in fact, that one advantage of the evolution of dual RiBi gene regulation by gas pedals and brakes is that regulated brake release allows leaky ribosome production so that cells in poor nutrient conditions can maintain just enough translation to survive until conditions improve. It will be interesting to determine whether physiological conditions can be identified in which this type of regulation can be seen.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dan Santos and Jonathan Weissman for generous communication of results prior to publication. We thank Emily Powers and Tina Sing for helpful comments on this manuscript and all members of the Brar and Ünal lab for helpful project feedback.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health (NIH) [DP2-GM-119138]; investigator awards from the Alfred P. Sloan Foundation [FG-2016–6229] and Pew Charitable Trusts [00029624]; UC-Berkeley start-up funding, including from the Bowes Foundation (to G.A.B.). Funding for open access charge: NIH.
Conflict of interest statement. None declared.
REFERENCES
- 1. Wade C.H., Umbarger M.A., McAlear M.A.. The budding yeast rRNA and ribosome biosynthesis (RRB) regulon contains over 200 genes. Yeast Chichester Engl. 2006; 23:293–306. [DOI] [PubMed] [Google Scholar]
- 2. Klinge S., Woolford J.L.. Ribosome assembly coming into focus. Nat. Rev. Mol. Cell Biol. 2018; 20:116–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bosio M.C., Fermi B., Dieci G.. Transcriptional control of yeast ribosome biogenesis: a multifaceted role for general regulatory factors. Transcription. 2017; 8:254–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Berger A.B., Decourty L., Badis G., Nehrbass U., Jacquier A., Gadal O.. Hmo1 is required for TOR-dependent regulation of ribosomal protein gene transcription. Mol. Cell Biol. 2007; 27:8015–8026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hall D.B., Wade J.T., Struhl K.. An HMG protein, Hmo1, associates with promoters of many ribosomal protein genes and throughout the rRNA gene locus in Saccharomyces cerevisiae. Mol. Cell Biol. 2006; 26:3672–3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Jorgensen P., Rupes I., Sharom J.R., Schneper L., Broach J.R., Tyers M.. A dynamic transcriptional network communicates growth potential to ribosome synthesis and critical cell size. Genes Dev. 2004; 18:2491–2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Martin D.E., Soulard A., Hall M.N.. TOR regulates ribosomal protein gene expression via PKA and the Forkhead transcription factor FHL1. Cell. 2004; 119:969–979. [DOI] [PubMed] [Google Scholar]
- 8. Rudra D., Zhao Y., Warner J.R.. Central role of Ifh1p-Fhl1p interaction in the synthesis of yeast ribosomal proteins. EMBO J. 2005; 24:533–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Schawalder S.B., Kabani M., Howald I., Choudhury U., Werner M., Shore D.. Growth-regulated recruitment of the essential yeast ribosomal protein gene activator Ifh1. Nature. 2004; 432:1058–1061. [DOI] [PubMed] [Google Scholar]
- 10. Wade J.T., Hall D.B., Struhl K.. The transcription factor Ifh1 is a key regulator of yeast ribosomal protein genes. Nature. 2004; 432:1054–1058. [DOI] [PubMed] [Google Scholar]
- 11. Xiao L., Grove A.. Coordination of ribosomal protein and ribosomal RNA gene expression in response to TOR signaling. Curr. Genomics. 2009; 10:198–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhao Y., McIntosh K.B., Rudra D., Schawalder S., Shore D., Warner J.R.. Fine-structure analysis of ribosomal protein gene transcription. Mol. Cell Biol. 2006; 26:4853–4862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Broach J.R. Nutritional control of growth and development in yeast. Genetics. 2012; 192:73–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Huber A., French S.L., Tekotte H., Yerlikaya S., Stahl M., Perepelkina M.P., Tyers M., Rougemont J., Beyer A.L., Loewith R.. Sch9 regulates ribosome biogenesis via Stb3, Dot6 and Tod6 and the histone deacetylase complex RPD3L. EMBO J. 2011; 30:3052–3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Liko D., Slattery M.G., Heideman W.. Stb3 binds to ribosomal RNA processing element motifs that control transcriptional responses to growth in Saccharomyces cerevisiae. J. Biol. Chem. 2007; 282:26623–26628. [DOI] [PubMed] [Google Scholar]
- 16. Lippman S.I., Broach J.R.. Protein kinase A and TORC1 activate genes for ribosomal biogenesis by inactivating repressors encoded by Dot6 and its homolog Tod6. Proc. Natl. Acad. Sci. U.S.A. 2009; 106:19928–19933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Brar G.A., Yassour M., Friedman N., Regev A., Ingolia N.T., Weissman J.S.. High-resolution view of the yeast meiotic program revealed by ribosome profiling. Science. 2012; 335:552–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chu S., DeRisi J., Eisen M., Mulholland J., Botstein D., Brown P.O., Herskowitz I.. The transcriptional program of sporulation in budding yeast. Science. 1998; 282:699–705. [DOI] [PubMed] [Google Scholar]
- 19. Eisenberg A.R., Higdon A., Keskin A., Hodapp S., Jovanovic M., Brar G.A.. Precise Post-translational tuning occurs for most protein complex components during meiosis. Cell Rep. 2018; 25:3603–3617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. de Hoon M.J.L., Imoto S., Nolan J., Miyano S.. Open source clustering software. Bioinforma. Oxf. Engl. 2004; 20:1453–1454. [DOI] [PubMed] [Google Scholar]
- 21. Saldanha A.J. Java Treeview–extensible visualization of microarray data. Bioinforma. Oxf. Engl. 2004; 20:3246–3248. [DOI] [PubMed] [Google Scholar]
- 22. Homann O.R., Johnson A.D.. MochiView: versatile software for genome browsing and DNA motif analysis. BMC Biol. 2010; 8:49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Brar G.A., Yassour M., Friedman N., Regev A., Ingolia N.T., Weissman J.S.. High-resolution view of the yeast meiotic program revealed by ribosome profiling. Science. 2012; 335:552–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ingolia N.T., Brar G.A., Rouskin S., McGeachy A.M., Weissman J.S.. The ribosome profiling strategy for monitoring translation in vivo by deep sequencing of ribosome-protected mRNA fragments. Nat. Protoc. 2012; 7:1534–1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Langmead B., Salzberg S.L.. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012; 9:357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Trcek T., Larson D.R., Moldón A., Query C.C., Singer R.H.. Single-molecule mRNA decay measurements reveal promoter- regulated mRNA stability in yeast. Cell. 2011; 147:1484–1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bregman A., Avraham-Kelbert M., Barkai O., Duek L., Guterman A., Choder M.. Promoter elements regulate cytoplasmic mRNA decay. Cell. 2011; 147:1473–1483. [DOI] [PubMed] [Google Scholar]
- 28. Enssle J., Kugler W., Hentze M.W., Kulozik A.E.. Determination of mRNA fate by different RNA polymerase II promoters. Proc. Natl. Acad. Sci. U.S.A. 1993; 90:10091–10095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zid B.M., O’Shea E.K.. Promoter sequences direct cytoplasmic localization and translation of mRNAs during starvation in yeast. Nature. 2014; 514:117–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhu C., Byers K.J.R.P., McCord R.P., Shi Z., Berger M.F., Newburger D.E., Saulrieta K., Smith Z., Shah M.V., Radhakrishnan M. et al.. High-resolution DNA-binding specificity analysis of yeast transcription factors. Genome Res. 2009; 19:556–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Badis G., Chan E.T., van Bakel H., Pena-Castillo L., Tillo D., Tsui K., Carlson C.D., Gossett A.J., Hasinoff M.J., Warren C.L. et al.. A library of yeast transcription factor motifs reveals a widespread function for Rsc3 in targeting nucleosome exclusion at promoters. Mol. Cell. 2008; 32:878–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ingolia N.T., Ghaemmaghami S., Newman J.R.S., Weissman J.S.. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009; 324:218–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Santos D.A., Shi L., Tu B.P., Weissman J.S.. Cycloheximide can distort measurements of mRNA levels and translation efficiency. Nucleic Acids Res. 2019; doi:10.1093/nar/gkz205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ingolia N.T., Brar G.A., Rouskin S., McGeachy A.M., Weissman J.S.. The ribosome profiling strategy for monitoring translation in vivo by deep sequencing of ribosome-protected mRNA fragments. Nat. Protoc. 2012; 7:1534–1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gerashchenko M.V., Gladyshev V.N.. Translation inhibitors cause abnormalities in ribosome profiling experiments. Nucleic Acids Res. 2014; 42:e134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hussmann J.A., Patchett S., Johnson A., Sawyer S., Press W.H.. Understanding biases in ribosome profiling experiments reveals signatures of translation dynamics in yeast. PLoS Genet. 2015; 11:e1005732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lareau L.F., Hite D.H., Hogan G.J., Brown P.O.. Distinct stages of the translation elongation cycle revealed by sequencing ribosome-protected mRNA fragments. eLife. 2014; 3:e01257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Requião R.D., de Souza H.J.A., Rossetto S., Domitrovic T., Palhano F.L.. Increased ribosome density associated to positively charged residues is evident in ribosome profiling experiments performed in the absence of translation inhibitors. RNA Biol. 2016; 13:561–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Duncan C.D.S., Mata J.. Effects of cycloheximide on the interpretation of ribosome profiling experiments in Schizosaccharomyces pombe. Sci. Rep. 2017; 7:10331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Duncan C.D.S., Rodríguez-López M., Ruis P., Bähler J., Mata J.. General amino acid control in fission yeast is regulated by a nonconserved transcription factor, with functions analogous to Gcn4/Atf4. Proc. Natl. Acad. Sci. U.S.A. 2018; 115:E1829–E1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Schneider-Poetsch T., Ju J., Eyler D.E., Dang Y., Bhat S., Merrick W.C., Green R., Shen B., Liu J.O.. Inhibition of eukaryotic translation elongation by cycloheximide and lactimidomycin. Nat. Chem. Biol. 2010; 6:209–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Siegel M.R., Sisler H.D.. Inhibition of protein synthesis in vitro by cycloheximide. Nature. 1963; 200:675–676. [DOI] [PubMed] [Google Scholar]
- 44. Heitman J., Movva N.R., Hall M.N.. Targets for cell cycle arrest by the immunosuppressant rapamycin in yeast. Science. 1991; 253:905–909. [DOI] [PubMed] [Google Scholar]
- 45. Powers T., Walter P.. Regulation of ribosome biogenesis by the rapamycin-sensitive TOR-signaling pathway in Saccharomyces cerevisiae. Mol. Biol. Cell. 1999; 10:987–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Cardenas M.E., Cutler N.S., Lorenz M.C., Di Como C.J., Heitman J.. The TOR signaling cascade regulates gene expression in response to nutrients. Genes Dev. 1999; 13:3271–3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Grollman A.P. Inhibitors of protein biosynthesis. II. Mode of action of anisomycin. J. Biol. Chem. 1967; 242:3226–3233. [PubMed] [Google Scholar]
- 48. Justice M.C., Hsu M.J., Tse B., Ku T., Balkovec J., Schmatz D., Nielsen J.. Elongation factor 2 as a novel target for selective inhibition of fungal protein synthesis. J. Biol. Chem. 1998; 273:3148–3151. [DOI] [PubMed] [Google Scholar]
- 49. Garreau de Loubresse N., Prokhorova I., Holtkamp W., Rodnina M.V., Yusupova G., Yusupov M.. Structural basis for the inhibition of the eukaryotic ribosome. Nature. 2014; 513:517–522. [DOI] [PubMed] [Google Scholar]
- 50. Lee S., Liu B., Lee S., Huang S.-X., Shen B., Qian S.-B.. Global mapping of translation initiation sites in mammalian cells at single-nucleotide resolution. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:E2424–E2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Barros L.F., Young M., Saklatvala J., Baldwin S.A.. Evidence of two mechanisms for the activation of the glucose transporter GLUT1 by anisomycin: p38(MAP kinase) activation and protein synthesis inhibition in mammalian cells. J. Physiol. 1997; 504:517–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Mailliot J., Garreau de Loubresse N., Yusupova G., Meskauskas A., Dinman J.D., Yusupov M.. Crystal structures of the uL3 mutant Ribosome: Illustration of the importance of ribosomal proteins for translation efficiency. J. Mol. Biol. 2016; 428:2195–2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Warner J.R. The economics of ribosome biosynthesis in yeast. Trends Biochem. Sci. 1999; 24:437–440. [DOI] [PubMed] [Google Scholar]
- 54. Weidberg H., Moretto F., Spedale G., Amon A., van Werven F.J.. Nutrient control of yeast gametogenesis is mediated by TORC1, PKA and energy availability. PLoS Genet. 2016; 12:e1006075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Munding E.M., Shiue L., Katzman S., Donohue J.P., Ares M.. Competition between pre-mRNAs for the splicing machinery drives global regulation of splicing. Mol. Cell. 2013; 51:338–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Venkataramanan S., Douglass S., Galivanche A.R., Johnson T.L.. The chromatin remodeling complex Swi/Snf regulates splicing of meiotic transcripts in Saccharomyces cerevisiae. Nucleic Acids Res. 2017; 45:7708–7721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Beugnet A., Tee A.R., Taylor P.M., Proud C.G.. Regulation of targets of mTOR (mammalian target of rapamycin) signalling by intracellular amino acid availability. Biochem. J. 2003; 372:555–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Urban J., Soulard A., Huber A., Lippman S., Mukhopadhyay D., Deloche O., Wanke V., Anrather D., Ammerer G., Riezman H. et al.. Sch9 is a major target of TORC1 in Saccharomyces cerevisiae. Mol. Cell. 2007; 26:663–674. [DOI] [PubMed] [Google Scholar]
- 59. Marr A.G. Growth rate of Escherichia coli. Microbiol. Rev. 1991; 55:316–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.