Abstract
Eukaryotic translation initiation requires unwinding of secondary structures in the 5′-untranslated region of mRNA. The DEAD-box helicase eIF4A is thought to unwind structural elements in the 5′-UTR in conjunction with eIF4G and eIF4B. Both factors jointly stimulate eIF4A activities by modulation of eIF4A conformational cycling between open and closed states. Here we examine how RNA substrates modulate eIF4A activities. The RNAs fall into two classes: Short RNAs only partially stimulate the eIF4A ATPase activity, and closing is rate-limiting for the conformational cycle. By contrast, longer RNAs maximally stimulate ATP hydrolysis and promote closing of eIF4A. Strikingly, the rate constants of unwinding do not correlate with the length of a single-stranded region preceding a duplex, but reach a maximum for RNA with a single-stranded region of six nucleotides. We propose a model in which RNA substrates affect eIF4A activities by modulating the kinetic partitioning of eIF4A between futile, unproductive, and productive cycles.
INTRODUCTION
The initiation of translation of eukaryotic mRNAs is a tightly regulated process that requires the coordinated interplay of several translation initiation factors (eIFs). In yeast, the first step of the canonical, cap-dependent pathway of translation initiation is the recognition of the 5′-m7G cap of the mRNA by eIF4F, a heterotrimeric complex comprising the initiation factors eIF4A, eIF4G, and eIF4E (1,2). eIF4E binds to the 5′-cap (3,4), eIF4G is a scaffold protein that contacts both eIF4E and eIF4A (1,5) as well as RNA, and eIF4A is an RNA helicase of the DEAD-box family (1,6). The cap-bound eIF4F complex then recruits the 43S pre-initiation complex (PIC), formed by the 40S ribosomal subunit, the initiation factor eIF2·GTP, and the initiator tRNA Met-tRNAi, as well as eIF1, eIF1A, and eIF3. The 43S PIC scans the 5′-untranslated region (5′-UTR) of the mRNA toward the start codon (7). eIF4A has been implicated in the recruitment of mRNAs to the PIC (8). The helicase activity of eIF4A is thought to be required to disrupt secondary structures in the 5′-UTR and to displace bound proteins in this process (9,10). When the start codon is reached, binding of the 60S ribosomal subunit leads to formation of the elongation-competent 80S ribosome and the start of protein biosynthesis. In mammalians and plants, several alternative mechanisms of translation with different requirements for translation initation factors have been characterized (reviewed in (11)), including cap-dependent initiation on internal repeat expansions using non-AUG start codons (12), and cap-independent initiation mediated by internal ribosome entry sites (13) or cap-independent translation enhancers (14). The prevalence and role of such alternative initiation mechanisms in yeast is still unclear.
The DEAD-box helicase eIF4A consists of a canonical helicase core, formed by two RecA domains (15) (Figure 1A). Its RNA unwinding activity is coupled to a conformational cycle in which eIF4A alternates between an open conformation with a wide cleft between its two RecA domains (15–17), and a closed conformation in the presence of ATP and RNA, in which the two RecA domains interact with each other and with bound ATP and RNA (18,19). Formation of the closed state is linked to duplex destabilization (19,20). The translation initiation factors eIF4B and eIF4G (Figure 1A) jointly stimulate the weak intrinsic RNA-dependent ATPase and ATP-dependent RNA helicase activities of yeast and human eIF4A (21–24) through modulation of the eIF4A conformational cycle (19,25,26). In the presence of eIF4G, eIF4A alternates between a half-open conformation, stabilized by binding of eIF4G to both RecA domains of eIF4A (27), and the closed state (25). eIF4G accelerates opening and closing, but the effect on closing is larger (25), which leads to an increased population of the closed state in equilibrium (19). eIF4B binds to eIF4A through its 7-repeats domain (26). Binding of eIF4B to eIF4A further accelerates closing when eIF4G is present, and thus causes an additional shift of the conformational equilibrium of eIF4A toward the closed state (19,25).
Figure 1.
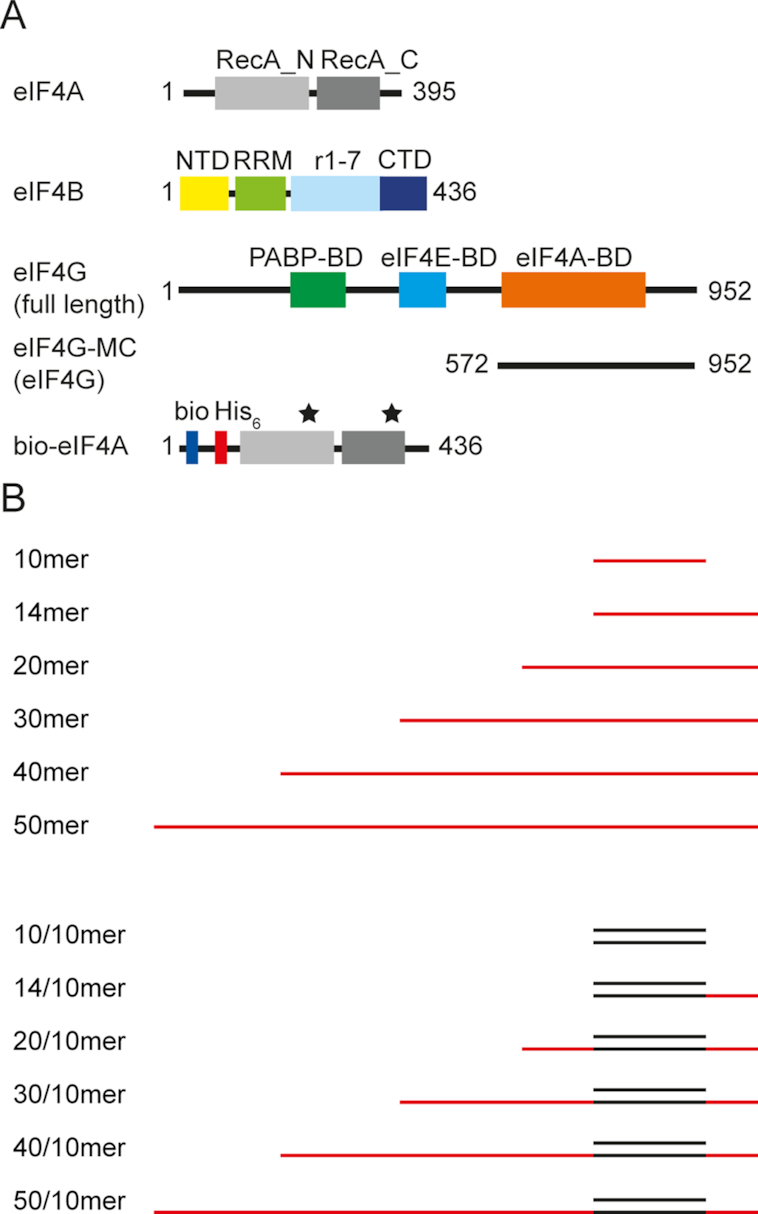
Constructs and RNA substrates used in this study. (A) Experiments were performed with full-length S. cerevisiae eIF4A and eIF4B, and a deletion variant of full-length eIF4G comprising the middle and C-terminal domains (eIF4G-MC, amino acids 572–952, referred to as eIF4G throughout). For smFRET experiments, the biotinylated eIF4A_Q186/G370C variant was used (18,19,25). bio: Biotin, His6: hexa-Histidine-tag. The black stars mark the positions of cysteines for fluorescent labeling. (B) Single-stranded RNA substrates of different lengths (10mer, 14mer, 20mer, 30mer, 40mer, 50mer) and double-stranded RNA substrates with a 10 bp duplex flanked by 5′-single-stranded regions of different lengths (10/10mer, 14/10mer, 20/10mer, 30/10mer, 40/10mer, 50/10mer). See Materials and Methods for sequences.
We have previously shown that the conformational dynamics of yeast eIF4A depends on the nature of the RNA substrate (25). poly-U RNA is frequently used as a model RNA to study the interaction of DEAD-box proteins with RNA. However, the rate constants for opening and closing of eIF4A differ substantially in the presence of poly-U RNA or a 50mer RNA of random sequence: While opening and closing occur with similar rate constants in the presence of poly-U RNA, kclose is larger than kopen with the 50mer RNA (25). The rate constants for eIF4A conformational changes are again different in the presence of RNA substrates containing a duplex that is unwound by eIF4A (25), and vary between two unwinding substrates which either contain a long single-stranded 5′-extension or lack single-stranded regions, respectively. In the presence of a 50/10mer, containing a 10mer duplex flanked by a 5′-single-stranded region, kclose is larger than kopen, whereas kclose is smaller than kopen in presence of a 32/9mer containing a 9 bp duplex flanked by a hairpin (25). We therefore set out to investigate the effect of the length of single-stranded RNAs and of 5′-single-stranded regions flanking a 10 bp duplex on eIF4A ATPase and helicase activities and on its conformational dynamics. We show that eIF4A undergoes about 3–4 conformational cycles per ATP hydrolyzed in the presence of single-stranded RNAs, independent of the RNA length. The behavior of eIF4A in the presence of a 10 bp-RNA duplex, flanked by 5′-single-stranded regions of different lengths, is dependent on the length of the 5′-extension. The length dependence of ATPase activity, RNA unwinding, and conformational cycling is different, however. eIF4A is predominantly in the open state when the 5′-single-stranded region is six nucleotides or shorter, and mostly in the closed state when it comprises 16 nucleotides or more. Similarly, the ATPase activity of eIF4A is low in the presence of RNAs with short single-stranded regions, and high in presence of RNAs with long single-stranded regions. In contrast, the rate constant of unwinding shows a maximum for an RNA with a 6-nucleotide single-stranded regions adjacent to the duplex, and unwinding is slower for RNAs with shorter or longer 5′-extensions. Overall, these results demonstrate that 5′-UTRs modulate eIF4A activity, and thus ribosome scanning and translation initiation, in an unprecedented and hitherto unappreciated fashion.
MATERIALS AND METHODS
Cloning, protein production and purification
Yeast eIF4A, N-terminally biotinylated eIF4A_Q186C/G370C (eIF4A), eIF4B and eIF4G comprising the middle domain and the C-terminal tail (eIF4G-MC, amino acids 572–952; Figure 1A) were produced recombinantly in Escherichia coli and purified to >95% purity according to Coomassie-stained SDS-PAGE as previously described (18,19,25).
RNA substrates
RNA oligonucleotides (Figure 1B) were purchased from Purimex (Grebenstein, Germany; PAGE-purified) and Axolabs (Kulmbach, Germany; HPLC-purified). Sequences were: 5′-ACCGUAAAGC-3′ (10mer), 5′-ACCGUAAAGC ACGC-3′ (14mer); 5′-ACUAGCACCGUAAAGCACGC-3′ (20mer), 5′-GGGGAGAAAA ACUAGCACCGUAAAGCACGC-3′ (30mer), 5′-GGGGAGAAAA ACAAAACAAA ACUAGCACCGUAAAGCACGC-3′ (40mer), 5′-GGGGAGAAAA ACAAAACAAA ACAAAACAAA ACUAGCACCGUAAAGCACGC-3′ (50mer). A second 10mer of the sequence 5′-GCUUUACGGU-3′ was annealed to the single-stranded RNAs to generate an unwinding substrate; the underlined part indicates the region of duplex formation.
Fluorescence anisotropy titrations
Dissociation constants (Kd) for protein/RNA-complexes were determined in fluorescence anisotropy titrations of 50 nM RNA containing a 10mer modified with 6-FAM (fluorescein) at the 5′-end with eIF4B, eIF4G, eIF4A/eIF4G or eIF4A/eIF4B/eIF4G in 30 mM HEPES/KOH, pH 7.4, 100 mM KOAc, 3 mM Mg(OAc)2, 2 mM DTT and 5 mM ATP (if present) at 25°C. In titrations in the presence of ATP, 23 μg/ml pyruvate kinase and 1 mM phosphoenolpyruvate were added to ensure ATP regeneration. Fluorescence was excited at 495 nM (2 nM bandwidth) and detected at 520 nM (5 nM bandwidth). Kd values were determined by describing the experimental data with a 1:1 binding model using non-linear regression, taking into account the change in quantum yield of the dye upon complex formation, as previously described (28). Measurements were performed at least in duplicate.
Steady-state ATPase activity
The steady-state ATPase activity of eIF4A in the absence and presence of eIF4G, eIF4B, and different RNAs was determined in a coupled enzymatic assay containing pyruvate kinase and lactate dehydrogenase that couples ATP hydrolysis to the oxidation of NADH (29). Concentrations were 1 μM eIF4A, 2 μM eIF4B, 2 μM eIF4G, 15 μM RNA, 2 mM ATP, 200 μM NADH, 13 μg/ml lactate dehydrogenase, 23 μg/ml pyruvate kinase, 1 mM phosphoenolpyruvate. Experiments were performed in quartz cuvettes (Hellma analytics, Germany) in an Ultrospec 2100 pro UV/VIS Spectrophotometer (Amersham Biosciences, Germany) as previously described (19). To determine kcat and Kapp,RNA, data were described with the Michaelis–Menten equation.
RNA unwinding
Duplex RNA (25 μM) for unwinding reactions was prepared by annealing a 10mer RNA, labeled with FAM at the 5′-end, with a two-fold excess of a complementary 10mer, or a 14mer, 20mer, 30mer, 40mer or 50mer RNA in 25 mM HEPES/KOH, pH 7.4. The mixture was heated to 96°C for 2 min, slowly cooled to room temperature and incubated on ice for 15 min. Experiments were performed in 30 mM HEPES/KOH, pH 7.4, 100 mM KOAc, 3 mM Mg(OAc)2, 2 mM DTT at 25°C with 500 nM duplex RNA, a 10-fold molar excess of unlabeled 10mer to ensure single turnover conditions, and 5 μM of eIF4A in the presence of 5 μM eIF4G and eIF4B. Unwinding reactions were initiated by the addition of 3 mM ATP, and stopped at different time points by adding an equal volume of a 2-fold stop solution (2% (v/v) SDS, 100 mM DTT, 10% glycerol, 0.1 mg/ml proteinase K). Substrate and products were separated by native gel electrophoresis. The fraction unwound at each time point was calculated from band intensities quantified by densitometry, and data were described by single-exponential functions to determine the rate constant of unwinding, kunwind.
Fluorescent labeling and single-molecule experiments
smFRET experiments were performed on a Picoquant Microtime 200 confocal microscope or an Olympus total internal reflection microscope as previously described (19,25,30). For confocal microscopy, biotinylated eIF4A_Q186C/G370C was labeled with Alexa488-maleimide (A488, donor) and Alexa546-maleimide (A546, acceptor); for TIRF experiments, biotinylated eIF4A_Q186C/G370C was labeled with Alexa555-maleimide (A555, donor) and Alexa647-maleimide (A647, acceptor), and immobilized on streptavidin-functionalized coverslips as described (19,25). All single-molecule measurements were performed in 50 mM Tris/HCl, pH 7.5, 80 mM KCl, 2.5 mM MgCl2, 1 mM DTT and 1% glycerol in the presence of 3 mM ATP and 15 μM RNA (if not stated otherwise) at 25°C. eIF4B and eIF4G concentrations were 10 μM. Measured donor and acceptor fluorescence intensities were corrected for donor cross-talk into the acceptor channel, acceptor cross-talk into the donor channel, different quantum yields and detection efficiencies of donor and acceptor fluorescence, and direct excitation of the acceptor as previously described (19,25,30), and FRET histograms were created using Origin. FRET time traces were idealized by hidden Markov modelling using vbFRET (31). Rate constants for opening and closing were extracted from cumulative dwell time histograms.
Electrophoretic mobility shift assays
Duplex RNA (10 μM) was prepared by annealing equimolar concentrations of 30mer or 20mer with 10mer RNA labeled with FAM at the 5′-end as described above. 10 μl samples containing 100 nM RNA, equimolar concentrations of the translation initiation factors (as indicated; 1% of eIF4G was labeled with Alexa647 to enable fluorescence detection), 4 units of RNase inhibitor (Roche), 5 mM ATP, 23 μg/ml pyruvate kinase and 1 mM phoshoenolpyruvate in 30 mM HEPES/KOH, pH 7.4, 100 mM KOAc, 3 mM Mg(OAc)2, 2 mM DTT were incubated for 10 min at 25 °C. After addition of 2 μl of 80% glycerol, the samples were applied to an 8% native polyacrylamide gel, and separated by gel electrophoresis for 3 h at 170 V. The fraction of RNA bound at each protein concentration was determined by densitometry, using fluorescein as a probe. eIF4G was detected using Alexa647 fluorescence as a probe.
Kinetic model for opening and closing in the presence of (short) double-stranded RNAs
For details on the kinetic models involving inter-conversion between the open state of eIF4A and one, two, or three closed states, and for the calculation of microscopic rate constants for individual steps in these models from the observed rate constants kopen, kclose,n, and the apparent equilibrium constant Kapp, see Supplementary Methods.
RESULTS
Effect of RNA length on the RNA-dependent ATPase activity of eIF4A
Poly-U RNA is often used as a model substrate for DEAD-box helicases. Poly-U RNA is a heterogeneous mixture of oligonucleotides with different lengths, ranging from 300 to 3000 nucleotides. To investigate the effect of the length of single-stranded RNA on the RNA-dependent ATPase activity of eIF4A, we therefore used a series of unstructured RNAs (22) with defined lengths of 10, 14, 20, 30, 40, and 50 nucleotides (Figure 1B). ATPase activities of eIF4A were determined under steady-state conditions in the absence of other translation initiation factors, in the presence of eIF4G-MC (comprising amino acids 572–952, Figure 1A, referred to as eIF4G), and in the presence of eIF4G and eIF4B (Figure 2A). The ATPase activity of eIF4A is reported relative to the activity in the presence of poly-U RNA, eIF4B, and eIF4G (measured as an internal control in each set of experiments and set to 100%). Turnover numbers in the presence of the 10mer, 14mer, 20mer, 30mer, 40mer, or 50mer single-stranded RNAs were increased approximately 8–10-fold compared to the low intrinsic ATPase activity of eIF4A in the absence of RNA. This stimulation is similar to the effect of poly-U RNA (6-fold). In the presence of eIF4G, the ATPase activity was increased approx. 2-fold compared to the intrinsic ATPase activity of eIF4A/G when the 10mer RNA was present. With the 14mer and 20mer RNAs, it increased further to 3- and 5-fold of the unstimulated ATPase activity, respectively. For the longer 30mer, 40mer, and 50mer RNAs, the stimulation saturated at 5–6-fold compared to the intrinsic hydrolysis rate, suggesting that a maximal turnover number was reached. The stimulation in the presence of the 50mer is similar to the stimulation by poly-U RNA (6.4-fold), supporting the notion that this value reflects a maximum. When eIF4B was also present (Figure 2A), the ATPase activity of eIF4A also increased linearly with the length of the RNA for the 10mer, 14mer, and 20mer, but now a further increase from the 20mer to the 30mer (7.8-fold to 9.8-fold) was observed. The stimulation remained at this level with the 40mer and 50mer RNAs. Again, the stimulation in the presence of the 50mer (9.6-fold) was similar to the value in the presence of poly-U RNA (9.2-fold), indicating that a maximum is reached. Thus, the presence of eIF4B shifts the turnover point where the maximum ATPase activity is reached from the 20mer to the 30mer.
Figure 2.
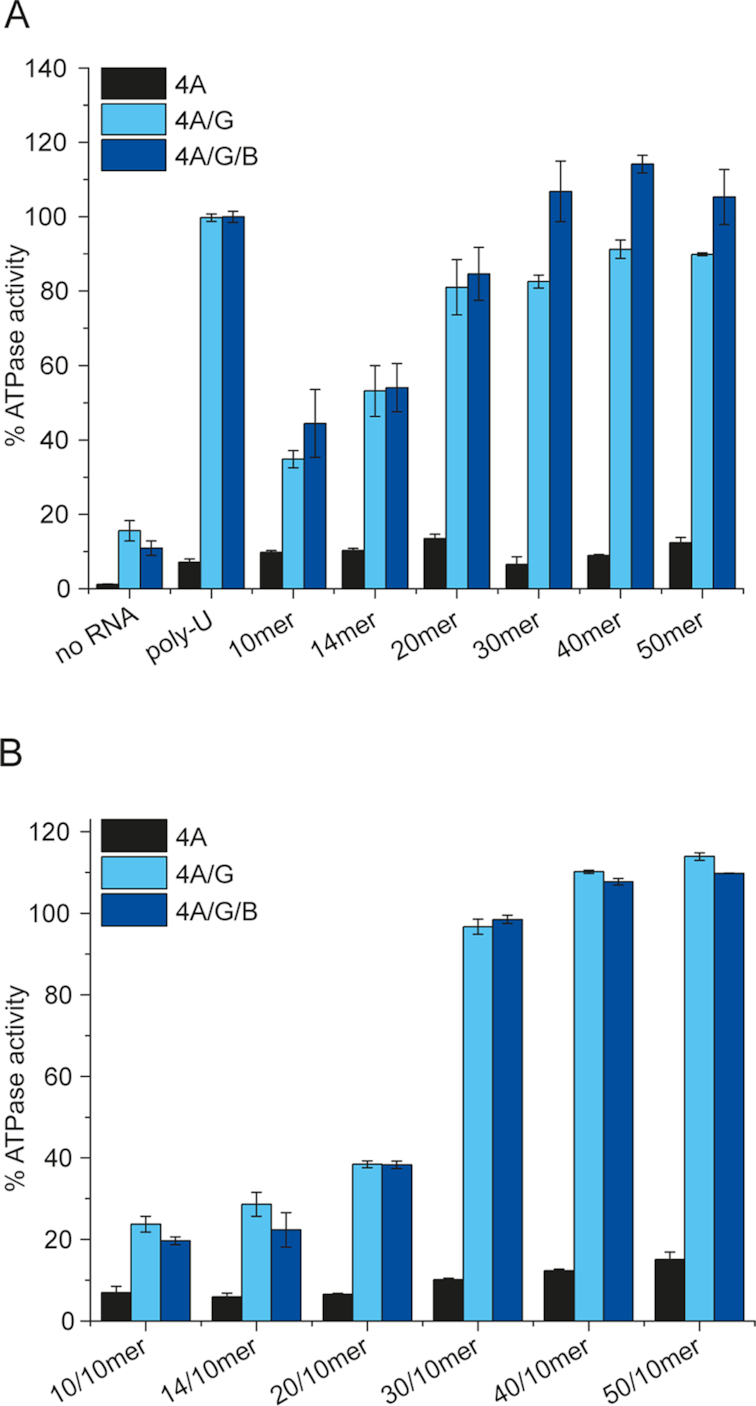
Dependence of the eIF4A steady-state ATPase activity on the length of the RNA substrate and of single-stranded 5′-tails. (A) Rate constant of ATP hydrolysis as a function of length of single-stranded RNA, relative to the ATP hydrolysis in the presence of poly-U RNA (set to 100%, typical values for the rate of ATP hydrolysis in the presence of poly-U RNA range between 70 and 100 × 10−3 s−1). Experiments were performed with 1 μM eIF4A, 2 μM eIF4B and eIF4G, 15 μM RNA (molecular concentration), 2 mM ATP in 30 mM HEPES/KOH, pH 7.4, 100 mM KOAc, 3 mM Mg(OAc)2, 2 mM DTT at 25°C. (B) Rate constant of ATP hydrolysis as a function of length of the 5′-single-stranded region flanking a 10 bp duplex. Black: eIF4A, light blue: eIF4A/eIF4G, dark blue: eIF4A, eIF4B, and eIF4G. Rate constants are given relative to the turnover number in the presence of poly-U RNA (see panel A), which was set to 100%. Experiments were performed at least twice; error bars reflect the standard error of the mean (n = 2) or the standard deviation (n > 2).
Effect of the length of 5′-single-stranded regions on the RNA-dependent ATPase activity of eIF4A
We next tested the effect of 5′-single-stranded regions of different lengths, flanking a 10 bp duplex, on the steady-state ATPase activity of eIF4A, using a 10/10mer, 14/10mer, 20/10mer, 30/10mer, 40/10mer, and 50/10mer RNA (Figure 2B). The 50/10mer was used previously to characterize eIF4A (19,22,25). All of these RNAs are unwound by eIF4A (see Figure 3). These RNAs stimulated the intrinsic ATP hydrolysis by eIF4A between 5- and 13-fold in the absence of other translation initiation factors, with a linear dependence on the length of the single-stranded region at the 5′-end. In the presence of eIF4G, the 10/10mer, 14/10mer and 20/10mer RNAs stimulated the ATPase activity 1.5- to 2.5-fold, and thus less than the corresponding single-stranded RNAs (10mer, 14mer, and 20mer). For the 30/10mer, 40/10mer, and 50/10mer RNAs, the stimulation increased further, to 6–7-fold. The most pronounced change occurred from the 20/10mer (6-nucleotide-overhang; 2.5-fold increase) to the 30/10mer (16-nucleotide-overhang; 6.2-fold increase). The turnover number did not increase much further when the single-stranded overhang was extended to 26 (40/10mer) or 36 nucleotides (50/10mer), and again leveled off at values similar to the turnover numbers in the presence of poly-U RNA (6.4-fold). Addition of eIF4B led to a slight overall increase in ATPase stimulation. Again, the largest change in stimulation was observed from the 20/10mer (3.5-fold) to the 30/10mer (9.0-fold).
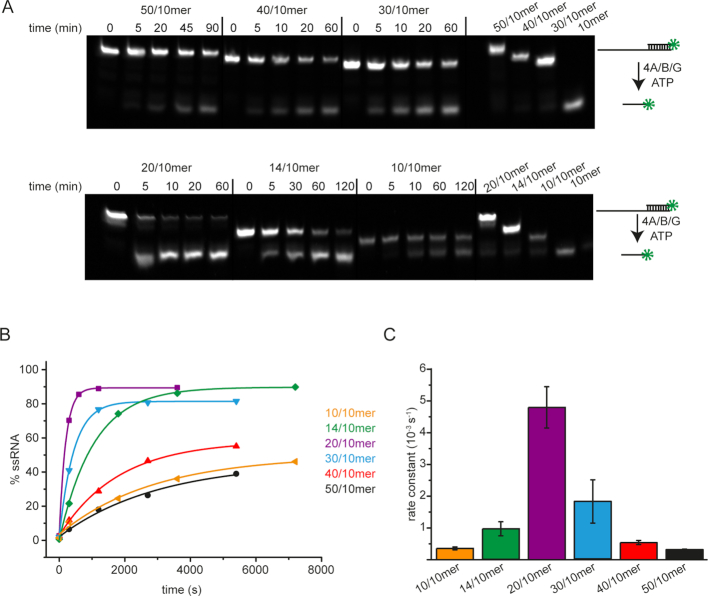
Figure 3.
Unwinding of the 10/10mer, 14/10mer, 20/10mer, 30/10mer, 40/10mer, and 50/10mer RNAs. (A) Unwinding of the 10/10mer, 14/10mer, 20/10mer, 30/10mer, 40/10mer, and 50/10mer RNAs (0.5 μM) by 5 μM eIF4A in the presence of 5 μM eIF4B and eIF4G in 30 mM HEPES/KOH, pH 7.4, 100 mM KOAc, 3 mM Mg(OAc)2, 2 mM DTT at 25°C. 5 μM of 10mer RNA was added as a trap to ensure single-turnover conditions. Reactions were started by addition of of 3 mM ATP, and stopped at indicated time points, followed by separation of substrate and product by native gel electrophoresis (see Methods). The 10/10mer, 14/10mer, 20/10mer, 30/10mer, 40/10mer, and 50/10mer RNAs in the absence of proteins were included as a control. The green star marks the position of the FAM label used for fluorescence detection of the RNA. (B) Time traces of unwinding, obtained by densitometric quantification of double-stranded substrate and single-stranded product. (C) Rate constants of RNA unwinding for the different RNAs. Rate constants were obtained by analyzing time traces (see B) with single-exponential functions. Error bars depict the standard deviation from at least three independent experiments.
Unwinding of RNA substrates with 5′-single-stranded regions of different lengths
The effect of the length of single-stranded regions on the eIF4A ATPase rate suggested that the 10/10mer 14/10mer, and 20/10mer might be unwound rather slowly, whereas the 30/10mer, 40/10mer, and 50/10mer RNAs that led to a large stimulation of the eIF4A ATPase activity might be unwound more rapidly. We followed unwinding of these RNAs in single-turnover experiments (Figure 3A). While the blunt-ended 10/10mer RNA was unwound only slowly (kunwind = 0.36 ± 0.05 × 10−3 s−1), the rate constant of unwinding increased for the 14/10mer (kunwind = 0.96 ± 0.22 × 10−3 s−1). This slight increase must be related to the presence of the four single-stranded nucleotides at the 3′-end of the duplex in the 14/10mer. Unwinding of the 20/10mer was 5-fold faster (kunwind = 4.80 ± 0.65 × 10−3 s−1). Instead of a further increase with increasing lengths of the 5′-single-stranded region for the 30/10-, 40/10-, and 50/10mer, however, we observed a decrease in the rate constant of unwinding (to 1.8 ± 0.68 × 10−3 s−1 for the 30/10mer). The 40/10mer and 50/10mer RNAs were again unwound slowly (kunwind = 0.54 ± 0.07 10−3 s−1 for the 40/10mer, and 0.32 ± 0.01 × 10−3 s−1 for the 50/10mer), with a very similar rate constant to the 10/10mer (Figure 3B, C). Any differences in rate constants for unwinding should be related to the 5′-single-stranded tail, as the duplex length, sequence, and stability are identical between these RNAs (ΔG = -67 kJ mol−1; ref. (32)). Thus, the 5′-single-stranded region of 6 nucleotides in the 20/10mer appears to be optimal for unwinding by eIF4A when eIF4B and eIF4G are present (Figure 3C).
Conformational dynamics of eIF4A in the presence of RNAs of different lengths
We previously analyzed the conformational dynamics of eIF4A in single-molecule FRET (smFRET) experiments and noticed that the rate constants for opening and closing are different in the presence of different RNAs (25). To test if the length of single-stranded RNA or of the 5′-single-stranded region flanking a 10 bp duplex had an effect on the conformational cycling of eIF4A, we immobilized donor/acceptor-labeled, N-terminally biotinylated eIF4A (carrying cysteines for fluorescent labeling at positions 186 and 370) on streptavidin-functionalized coverslips (25), and followed opening and closing of eIF4A as changes in FRET efficiency with time by total internal reflection fluorescence (TIRF) microscopy.
Effect of eIF4B and eIF4G
eIF4A rapidly inter-converted between a low- and a high-FRET state in the presence of eIF4B, eIF4G, ATP, and different single-stranded RNAs (Figure 4A), as observed previously (25). These states reflect the half-open and closed states, confirming that these are the relevant conformational states of eIF4A in presence of eIF4G and the different RNAs. We did not observe any excursions to the open state, which indicates that eIF4G remains bound to eIF4A on the timescale of these experiments, in agreement with the nanomolar affinity of eIF4A for eIF4G (18,27). The interaction between eIF4A and eIF4B is weaker, with a Kd on the order of 15 μM (in the presence of RNA and ADPNP; see (26)). We therefore suspected that eIF4B may not be saturating in these experiments, and tested the conformational dynamics of eIF4A in the presence of eIF4G and the 50mer RNA as a function of the concentration of eIF4B (0, 2.5, 5, 10, 30 μM; Supplementary Figure S1). The conformational equilibrium of eIF4A was shifted from the half-open state towards the closed state with increasing concentrations of eIF4B (Supplementary Figure S1A), and switching between these states became more frequent (Supplementary Figure S1B). Cumulative dwell time histograms (CDHs) gave a rate constant for opening of 0.31 s−1 that was independent of eIF4B (up to concentrations of 10 μM; Supplementary Figure S1C). The CDH in the presence of 30 μM eIF4B was double-exponential (kopen1 = 0.22 s−1, 38% amplitude, kopen2 = 0.79 s−1, 62% amplitude, Supplementary Figure S1D), which may be related to eIF4B oligomerization or aggregation. The CDHs for closing in the presence of different concentrations of eIF4B all exhibited two phases, and were best described by double-exponential functions (Supplementary Figure S1E). From a global analysis (excluding the data for 30 μM eIF4B), we obtained kclose1 = 0.17 s−1 and kclose2 = 0.69 s−1. The amplitude of the fast phase increased with increasing concentrations of eIF4B (0%, 45%, 90%, 96% at 0, 2.5, 5, and 10 μM eIF4B, respectively). Thus, we can conclude from these control experiments that opening of eIF4A is eIF4B-independent, but closing is eIF4B-dependent. The fast phase reflects closing of eIF4A in an eIF4A/B/G complex (Kclose = 1.9; rapid closing), the slow phase reflects closing of eIF4A that is part of an eIF4A/G complex (Kclose = 0.55; slow closing). At a concentration of eIF4B of 10 μM, the amplitude of the fast phase and thus the saturation of eIF4A with eIF4B was 96%. This value is higher than the saturation estimated from the Kd value determined in supernatant depletion assays in the presence of RNA and ADPNP (15 μM, (26), corresponding to a saturation of ∼40%), presumably because the interaction between eIF4A and eIF4B is favored in the presence of ATP. In agreement with this hypothesis, an apparent Kd of ∼2 ± 1.7 μM can be determined from the amplitudes of the fast phase of closing as a function of the concentration of eIF4B; a value of 2.9 ± 1.2 μM is obtained from the fraction closed in smFRET histograms; Supplementary Figure S1F). Overall, we can thus conclude that the concentration of eIF4B in single-molecule experiments is nearly saturating, but slower phases (of closing) with amplitudes <5% may occur due to the slower conformational cycling of a small population of eIF4/G complexes.
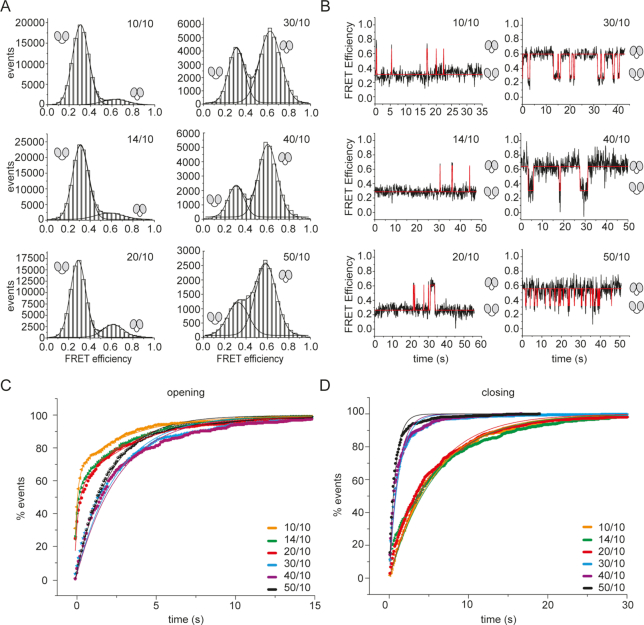
Figure 4.
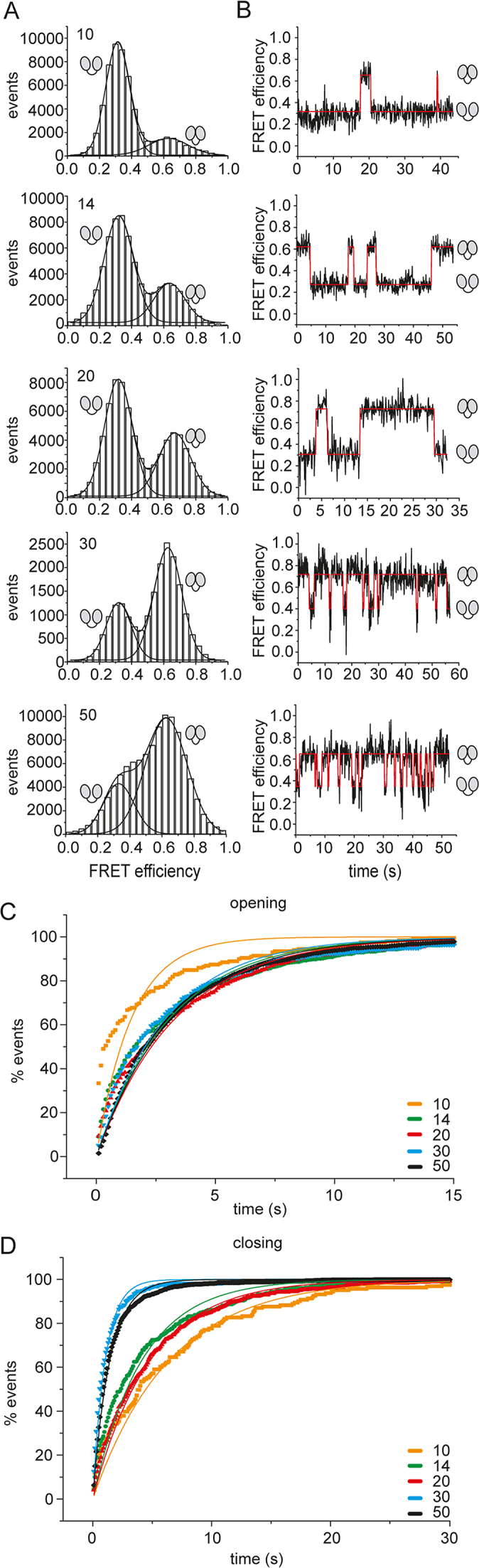
Effect of single-stranded RNAs of different lengths on the eIF4A conformational cycle. (A) FRET histograms for eIF4A in the presence of eIF4B, eIF4G, and the 10mer, 14mer, 20mer, 30mer, 40mer, or 50mer single-stranded RNAs. Experiments were performed with biotinylated, donor/acceptor-labeled, surface-immobilized eIF4A in the presence of 10 μM eIF4B and eIF4G, and 15 μM of the 10mer, 14mer, 20mer, 30mer, 40mer, or 50mer RNA, and 3 mM ATP in 50 mM Tris/HCl, pH 7.5, 80 mM KCl, 2.5 mM MgCl2, 1 mM DTT and 1% glycerol at 25°C. (B) Representative FRET time traces. (C) Normalized cumulative dwell time histograms for opening and single-exponential fits. See Supplementary Figure S2A for analyses with single-exponential functions and the respective residuals, Supplementary Figure S3A for analyses with double-exponential functions and residuals, and Supplementary Table S1 for corrected R2-values. (D) Normalized cumulative dwell time histograms for closing and single-exponential fits. See Supplementary Figure S2B for single-exponential fits and respective residuals, Supplementary Figure S3B for double-exponential fits and residuals, and Supplementary Table S1 for corrected R2-values. The cartoons indicate the low-FRET (half-)open conformation and the high-FRET closed state of eIF4A.
The effect of the length of single-stranded RNAs
FRET histograms for eIF4A in the presence of eIF4B, eIF4G, and the 10mer, 14mer, 20mer, 30mer, or 50mer single-stranded RNAs (Figure 4A, B) showed a shift in the conformational equilibrium of eIF4A from the half-open state towards the closed state with increasing length of the RNA. The RNAs fall into two classes: For the shorter (10mer, 14mer, and 20mer) RNAs, eIF4A is predominantly in the half-open state. In contrast, the closed state is predominantly populated in presence of the longer RNAs (30mer, 50mer). Rate constants for opening and closing were determined from the analysis of CDHs using single-exponential functions (Figure 4C, D, Supplementary Figure S2; Table 1). Double-exponential fits did not lead to a significant improvement of the quality of the fit (as judged by corrected R2 values and the systematic behavior of the residuals; Supplementary Figure S3A,B). An independent analysis of the single-molecule data for the 20mer with the program SMACKS (33) that uses a maximum-likelihood approach and ranks different models according to the Bayesian information criterion also favored a model with one open and one closed state (Supplementary Table S2), in agreement with the single-exponential behavior of opening and closing. The fact that the CDHs for closing were described satisfactorily by single-exponential functions confirms that eIF4B is saturating for eIF4A in these experiments. Rate constants of opening were similar, and varied between 0.31 s−1 and 0.39 s−1 in the presence of the 14mer, 20mer, 30mer, and 50mer RNAs; only in the presence of the 10mer a higher rate constant of kopen = 0.67 s−1 was determined, but in this case, the single-exponential function does not describe the data very well (Supplementary Figure S2). The rate constants for closing ranged between 0.16 and 0.26 s−1 in the presence of shorter RNAs (10mer, 14mer, 20mer), but jumped to 0.99 and 0.71 s−1 with the 30mer and 50mer RNAs. As a consequence, the rate constant for opening is higher than the rate constant for closing with the 10mer, 14mer, and 20mer RNAs, in agreement with the higher population of eIF4A in the open state with these RNAs (Table 1). With the 30mer and the 50mer, the rate constant of closing is higher than the rate constant for opening, in agreement with the higher population of eIF4A in the closed state with these RNAs as observed in FRET histograms.
Table 1.
Observed rate constants for opening (kopen or kopen,1, kopen,2) and closing (kclose) of eIF4A in the presence of different RNAs
| RNA | k open(,1) (s−1) | k open,2 (s−1) | k close (s−1) |
|---|---|---|---|
| 10mer | 0.67 | 0.16 | |
| 14mer | 0.32 | 0.24 | |
| 20mer | 0.30 | 0.21 | |
| 30mer | 0.34 | 0.95 | |
| 50mer | 0.31 | 0.69 | |
| 10/10mer | 0.30 (33%)a | 5.8 (67%)a | 0.17 |
| 14/10mer | 0.29 (43%)a | 6.1 (57%)a | 0.17 |
| 20/10mer | 0.26 (44%)a | 4.4 (56%)a | 0.19 |
| 30/10mer | 0.21 (48%)a | 0.86 (52%)a | 0.82 |
| 40/10mer | 0.19 (42%)a | 0.72 (58%)a | 0.91 |
| 50/10mer | 0.20 (18%)a | 0.59 (82%)a | 1.3 |
| 30/10mer | 0.39b | 0.82 | |
| 40/10mer | 0.37b | 0.91 | |
| 50/10mer | 0.47b | 1.3 |
aValues in brackets are the amplitudes of the slow and fast phases from double-exponential fits.
bRate constant for opening from single-exponential analysis of the CDHs in the presence of 30/10mer, 40/10mer and 50/10mer RNAs.
The switch of eIF4A from slow conformational dynamics and a higher fraction in the half-open state in the presence of short RNAs to faster conformational cycling and a larger fraction in the closed state in the presence of longer RNAs might be related to different interactions of eIF4A with eIF4B or could result from a direct effect of the length of these RNAs on eIF4A conformation and dynamics. We have shown previously that eIF4B does not bind to eIF4A in the presence of the 10mer RNA, but complex formation does occur in the presence of the 20mer RNA (26) (Supplementary Figure S4). Thus, if different binding of eIF4B were the reason for the change in conformational dynamics, a switch in eIF4A behavior would be expected from the 10mer (or 14mer) to the 20mer, rather than between the 20mer and 30mer RNAs. The observation of the switch from the 20mer to the 30mer indicates that the length of the RNA is responsible for the altered dynamics of eIF4A.
Conformational dynamics of eIF4A in presence of duplex RNA substrates with 5′-single-stranded regions of different lengths
We next analyzed the conformational dynamics of eIF4A in the presence of double-stranded RNAs with 5′-single-stranded overhangs of different lengths (Figure 5). FRET histograms (Figure 5A) showed that these RNAs also fall into two classes: With the 10/10mer, 14/10mer, and 20/10mer, the conformational equilibrium of eIF4A is on the side of the low-FRET half-open state, whereas the population of the closed state is dominant in presence of the 30/10mer, 40/10mer, and 50/10mer RNAs. In agreement with the conformational equilibrium, the rate constants kclose for closing, determined from the analysis of CDHs with single-exponential functions (Figure 5B, D, Supplementary Figure S5), are low for the 10/10mer, 10/14mer, and 20/10mer (0.17, 0.17, and 0.19 s−1), and higher for the 30/10mer, 40/10mer, and 50/10mer (0.82, 0.91, and 1.3 s−1, respectively; Figure 5B, D, Supplementary Figure S5). Inclusion of a second phase did not significantly improve the quality of the fits (Supplementary Figure S5). CDHs for opening in the presence of the 30/10mer, 40/10mer, and 50/10mer were also single-exponential, with rate constants kopen of 0.39, 0.37, and 0.47 s−1 (Figure 5C; see Supplementary Figure S6 for double-exponential fits), although the inclusion of a second phase improved the quality of the fit for the 30/10mer and 40/10mer (Supplementary Figure S6, Table 1). In contrast, CDHs for opening in the presence of the shorter RNAs showed significant deviations from single-exponential behavior (Supplementary Figure S5), but were well-described by double-exponential functions (Figure 5C, Supplementary Figure S6). From the slower phase, rate constants similar to the rate constants for opening in presence of the longer RNAs were obtained (0.30, 0.29, and 0.26 s−1, respectively; Table 1). The lifetimes τ of the faster phase (rate constants 5.8 s−1(τ = 170 ms), 6.1 s−1 (τ = 160 ms), and 4.4 s−1 (τ = 230 ms)) are close to the limit of the time-resolution (100 ms) in these experiments, and these rate constants therefore constitute lower limits. The occurrence of one observed rate constant for closing and two observed rate constants for opening is consistent with a model in which eIF4A inter-converts between a single open state and two alternative, non-inter-converting closed states. Analysis of the data with the program SMACKS (33) also favored a model with one open and two non-interconverting closed states in the presence of the 14/10mer, 20/10mer, and 30/10mer RNAs. In contrast, the data in the presence of the 50/10mer were more consistent with a model with one open and one closed state (Supplementary Table S2), in agreement with the single-exponential behavior of opening.
Figure 5.
Effect of 5′-single-stranded regions of different lengths on the eIF4A conformational cycle. (A) FRET histograms for eIF4A in the presence of 10 μM eIF4B and eIF4G and 15 μM of the 10/10mer, 14/10mer, 20/10mer, 30/10mer, 40/10mer, or 50/10mer RNA. Experiments were performed in 50 mM Tris/HCl, pH 7.5, 80 mM KCl, 2.5 mM MgCl2, 1 mM DTT, and 1% glycerol in the presence of 3 mM ATP at 25°C. (B) Representative FRET time traces. (C) Cumulative dwell time histograms for opening, double-exponential (10/10mer, 14/10mer, 20/10mer) and single-exponential fits (30/10mer, 40/10mer, 50/10mer). (D) Cumulative dwell time histograms for closing and single-exponential fits. See Supplementary Figure S4 for analyses with single-exponential functions and residuals obtained, Supplementary Figure S5 for analyses with double-exponential functions and residuals. The corrected R2-values are summarized in Supplementary Table S1.
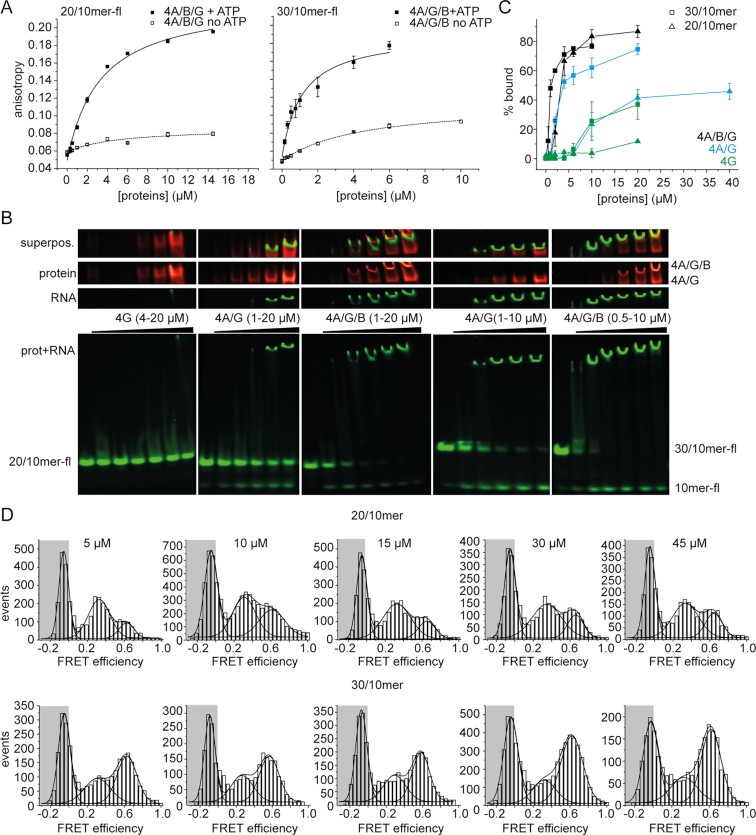
Interaction of 20/10mer and 30/10mer RNAs with eIF4A and its complexes with eIF4G and eIF4B
Single-molecule experiments with eIF4A in the presence of duplex RNAs with 5′-single-stranded extensions of different lengths clearly showed a switch in eIF4A conformational dynamics and the conformational equilibrium from the 20/10mer to the 30/10mer (6-nucleotide versus 16-nucleotide 5′-single-stranded region). The same threshold behavior was observed for the stimulation of ATP hydrolysis by these RNAs. To rule out that the pronounced change between the behavior of eIF4A in presence of the 20/10mer and 30/10mer is merely an effect of different affinities of eIF4A/B/G for these RNAs, we monitored binding of the 20/10mer and 30/10mer RNAs in fluorescence anisotropy titrations (Figure 6A). Titration of a fluorescein-labeled 20/10mer with eIF4A in presence of eIF4B, eIF4G, and ATP gave rise to an increase in anisotropy. From the binding isotherm, an apparent Kd value of 3.5 (±0.5) μM was determined. The same experiment with the fluorescein-labeled 30/10mer showed similar results, with an apparent Kd of 1.2 (±0.3) μM. In the absence of ATP, the observed increase in anisotropies was smaller for both RNAs, and the apparent Kd values were increased to 6.7 (±3.5) μM (20/10mer) and 4.0 (±0.6) μM (30/10mer; Figure 6A). eIF4A does not bind RNA in the absence of ATP, and these values should thus reflect the interaction of the respective RNAs with eIF4B and eIF4G as part of the eIF4A/B/G complex. eIF4B alone bound weakly to both RNAs (Kd = 9–10 μM; Supplementary Figure S7A). eIF4G alone also bound weakly to the 30/10mer, whereas no increase in anisotropy was detected when eIF4G was titrated to the 20/10mer (Supplementary Figure S7B). The eIF4A/G complex caused no increase in anisotropy when titrated to the 20/10mer (in the presence or absence of ATP), or to the 30/10mer (in the absence of ATP). In the presence of ATP, the apparent affinity of eIF4A/G for the 30/10mer was 1.6 (±0.5) μM. The final anisotropy reached in titrations with eIF4A/G was lower than the saturating value in the titration with eIF4A/B/G, which is consistent with eIF4A, eIF4B, and eIF4G jointly binding to the RNA.
Figure 6.
Interaction of eIF4A with 20/10mer and 30/10mer RNA. (A) Anisotropy titrations of fluorescein-labeled 20/10mer and 30/10mer RNAs with eIF4A/B/G in the absence (open squares) and presence of 5 mM ATP (filled squares). Titrations were performed in 30 mM HEPES/KOH, pH 7.4, 100 mM KOAc, 3 mM Mg(OAc)2, and 2 mM DTT at 25°C. Error bars depict the standard error of the mean from two independent experiments. (B) Electrophoretic mobility shift assay of 100 nM fluorescein-labeled 20/10- and 30/10mer with increasing concentrations of eIF4G, eIF4A/G and eIF4A/B/G in 30 mM HEPES/KOH, pH 7.4, 100 mM KOAc, 3 mM Mg(OAc)2, 2 mM DTT in the presence of 5 mM ATP and 0.4 U/μl of RNase inhibitor, incubated for 10 min at 25°C. To regenerate ATP, 23 μg/ml pyruvate kinase and 1 mM phoshoenolpyruvate were added. 1% of eIF4G is labeled with Alexa647 (red), the 10mer of the RNA substrate is labeled with fluorescein (fl; green). A representative gel from two independent EMSAs is shown. Note that these epxeriments were performed with an excess of translation initiation factors, such that under saturation all of the RNA is protein-bound, but only a small fraction of the translation factors is bound to RNA. See Supplementary Figure S8 for control experiments with labeled eIF4A and labeled eIF4B. (C) Quantification of RNA-bound complexes of eIF4G, eIF4A/G, and eIF4A/B/G from B. Error bars depict either the error of the mean from two independent experiments (20/10mer, eIF4G; 20/10mer, eIF4A, eIF4G, eIF4B) or the standard deviation from at least three independent experiments (all other experiments). (D) smFRET experiments in presence of different concentrations of 20/10mer and 30/10mer RNAs. 100 pM biotinylated eIF4A_Q186C/G370C, labeled with Alexa488-maleimide (A488, donor) and Alexa546-maleimide (A546, acceptor), in 50 mM Tris/HCl, pH 7.5, 80 mM KCl, 2.5 mM MgCl2, 1 mM DTT, and 1% glycerol in the presence of 3 mM ATP, 10 μM eIF4B and eIF4G, and 5, 10, 15, 30, and 45 μM RNA at 25°C.
As it is not possible to distinguish individual complexes in fluorescence equilibrium titrations, we also performed electrophoretic mobility shift assays with fluorescein-labeled 20/10mer and 30/10mer in the presence of eIF4A, eIF4B, Alexa647-labeled eIF4G, and ATP (Figure 6B, C). Thereby, we could distinguish between RNA bound to eIF4A/G complexes and eIF4A/B/G complexes through their different electrophoretic mobilities. The 20/10mer showed no shift in electrophoretic mobility in the presence of eIF4G alone in the concentration range tested (<20 μM), in agreement with the anisotropy measurements. In contrast, a substantial shift in electrophoretic mobility of the 20/10mer RNA was observed in the presence of eIF4A and eIF4G, indicating binding of this RNA to eIF4A/G. In the additional presence of eIF4B, the 20/10mer RNA showed a super-shift, consistent with binding to the eIF4A/B/G complex (and not to eIF4A/G) at higher protein concentrations. Both in presence and absence of eIF4B, the signals for RNA and eIF4G do not superimpose precisely (see also Supplementary Figure S8). This may be the consequence of the large excess of protein in these experiments, with only a small fraction of the proteins co-migrating with the RNA. Alternatively, it may point to the transient character of the protein/RNA complex. Possibly, ATP hydrolysis by eIF4A and formation of nucleotide-free eIF4A with low RNA affinity leads to dissociation of RNA during the experiment. The 30/10mer also bound to eIF4A/G, as evidenced by a large shift in electrophoretic mobility (Figure 6B, C). In the presence of eIF4B, the RNA shows a reduced electrophoretic mobility corresponding to the complex with eIF4A/G at lower protein concentrations. At higher protein concentrations, the shift is larger, pointing to preferential binding to eIF4A/B/G under these conditions. Control experiments with labeled eIF4A or labeled eIF4B are also consistent with the presence of all three factors in the RNA complex with the lowest electrophoretic mobility (Supplementary Figure S8). Altogether, the electrophoretic mobility shift assays thus indicate that both the 20/10mer and the 30/10mer are bound to the eIF4A/G/B complex at the concentrations of eIF4B and eIF4G used in single-molecule experiments (10 μM). However, eIF4A is present at much lower (picomolar) concentrations in these experiments. To unambiguously test whether eIF4A is bound to the 20/10mer and 30/10mer RNAs under these conditions, we therefore also repeated the smFRET experiments as a function of concentration of 20/10mer and 30/10mer RNAs (5, 10, 15, 30, and 45 μM; Figure 6D). The fraction of eIF4A in the closed conformation did not show a systematic increase with increasing RNA concentrations, and remained lower (∼30%) in presence of the 20/10mer RNA than in the presence of the 30/10mer (∼70%) at all RNA concentrations tested. Opening of eIF4A in the presence of 45 μM RNA was still biphasic (Supplementary Figure S7C), indicating that the appearance of a second phase for opening of eIF4A not caused by a lack of saturation with RNA at the lower concentration. Hence, the difference in eIF4A behavior in the presence of the 20/10mer and 30/10mer RNAs must be related to qualitatively different interactions with these RNAs, not by a lower saturation of eIF4A/B/G with the 20/10mer.
DISCUSSION
The length of single-stranded regions in mRNAs determines the conformational equilibrium and the velocity of conformational cycling of eIF4A
Here we have shown that the length of single-stranded RNA and the length of the 5′-single-stranded region preceding a 10 bp RNA duplex determine the conformational equilibrium and the velocity of conformational cycling of eIF4A. In the presence of RNAs with no or only short single-stranded regions (10mer, 14mer, 20mer; 10/10mer, 14/10mer, 20/10mer), opening of eIF4A is faster than closing, and the majority of eIF4A is in the open conformation. In the presence of RNAs with longer single-stranded regions (30mer, 50mer; 30/10mer, 40/10mer, 50/10mer), closing is faster than opening, and eIF4A is predominantly in the closed conformation. The switch in the rate-limiting step of the conformational cycle from closing (up to the 20mer or 20/10mer) to opening (from the 30mer or the 30/10mer on) leads to an overall faster conformational cycling of eIF4A in the presence of the longer RNAs and RNAs with longer single-stranded 5′-regions. This switch is not correlated with the dependence of the interaction between eIF4A and eIF4B on the length of the RNA. The switch in behavior is also not caused by different levels of saturation of eIF4A with the 20/10mer and the 30/10mer, as both RNAs are bound to the eIF4A/B/G complex under our experimental conditions. Hence, the observed threshold behavior is related to a different functional interaction of the eIF4A/B/G complex with shorter and longer RNAs.
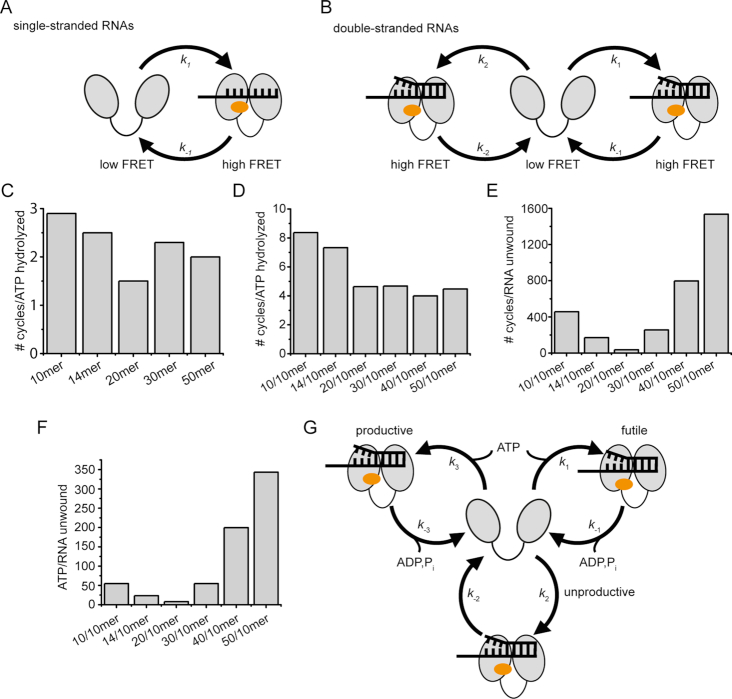
A kinetic model for eIF4A conformational cycling in the presence of single- and double-stranded RNAs
Closing and opening of eIF4A in the presence of all single-stranded RNAs show single-exponential behavior, in agreement with a simple one-step inter-conversion of open and closed states (Figure 7A). In the presence of RNAs with single-stranded extensions at the 5′-end of the duplex, closing is also single-exponential. Opening shows single-exponential behavior in the presence of double-stranded RNAs with long 5′-single stranded regions (30/10mer, 40/10mer, 50/10mer). With these RNAs, eIF4A inter-converts thus between a single open and closed state, similar to the behavior observed in the presence of single-stranded RNAs (Figure 7A). With RNAs that contain no or only a short 5′-single-stranded overhang (10/10mer, 14/10mer, 20/10mer), a second, rapid phase of opening is observed. The kinetic behavior observed is consistent with a model in which eIF4A can undergo two alternative conformational cycles, starting from a common open state and passing through two kinetically distinct closed states (Figure 7B). We have excluded a significant population of eIF4A/G complexes as the reason for the appearance of this second phase. A model where eIF4A undergoes three independent conformational cycles would lead to one kinetic phase for closing, and three kinetic phases for opening, which is not in agreement with the observations (see Supplementary Methods).
Figure 7.
Kinetic models for conformational cycling of eIF4A, and comparison of conformational dynamics, ATP hydrolysis, and RNA unwinding. (A) Kinetic model for eIF4A conformational changes in the presence of single-stranded RNAs and RNAs with long 5′-single-stranded regions. (B) Kinetic model for conformational cycling in the presence of RNAs with no or short 5′-single-stranded regions. (C) Coupling of conformational cycling to ATP hydrolysis in the presence of single-stranded RNAs. The number of conformational cycles per ATP hydrolyzed is independent of the length of the RNA. (D) Number of conformational cycles per ATP hydrolyzed in the presence of double-stranded RNAs. (E) Number of conformational cycles per RNA unwound. In the presence of the 20/10mer RNA, eIF4A undergoes the minimal number of approx. 37 conformational cycles per RNA unwound. (F) Coupling of ATP hydrolysis to RNA unwinding. In the presence of the 20/10mer RNA, eIF4A hydrolyzes the minimal number of 8 ATP per RNA unwound. (G) Model linking eIF4A conformational cycling with ATP hydrolysis and RNA unwinding. eIF4A can undergo futile cycles (ATP hydrolysis, but no RNA unwinding), unproductive cycles (no ATP hydrolysis, no RNA unwinding), and productive cycles (ATP-dependent RNA unwinding). In productive cycles the first strand of an RNA duplex dissociates from eIF4A closed state, prior to ATP hydrolysis. In futile cycles the duplex dissociates from eIF4A without unwinding, and ATP is then hydrolyzed. In unproductive cycles, eIF4A undergoes conformational changes without ATP hydrolysis and unwinding. The kinetic competition of the individual cycles determines the coupling of conformational cycling, ATP hydrolysis, and RNA unwinding. The cartoons reflect the conformation of eIF4A in the presence of the translation initiation factors eIF4B and eIF4G. Note that for simplicity the closed states in the different conformational cycles in panels B and G are depicted identically, but reflect different physical states with different functional properties.
For a model with two alternative conformational cycles, the observed rate constant for closing, kclose, determined from dwell times in FRET time traces (see Table 1), corresponds to the sum of the two microscopic rate constants for closing in the model, k1 and k2 (Figure 7B). The two observed rate constants for opening, kopen,1 and kopen,2 (see Table 1), are identical to the microscopic rate constants k-1 and k-2 (see Materials and Methods). The apparent equilibrium constant Kapp for closing determined from FRET histograms is the sum of K1 and K2 (see Supplementary Methods). From these considerations, the entire set of microscopic rate constants can be calculated (see Materials and Methods, Supplementary Methods; Table 2). While the two high-FRET closed states are similar in their global conformation, they must represent different functional states of eIF4A. Mechanistically, the two alternative conformational cycles could represent eIF4A interacting with either the single-stranded or the duplex part of the RNA substrate, or eIF4A undergoing conformational cycles that are or are not associated with ATP hydrolysis. The different kinetic models and their biological interpretation will be discussed in more detail below.
Table 2.
Microscopic rate constants for opening (k-1, k-2) and closing (k1, k2) of eIF4A in the presence of double-stranded RNAs, and individual equilibrium constants K1, and K2. See Figure 7B and Supplementary Methods for the definitions of rate constants
| RNA | k 1 (s−1) | k -1 (s−1) | K 1 | k 2 (s−1) | k -2 (s−1) | K 2 |
|---|---|---|---|---|---|---|
| 10/10mer | 0.024 | 0.30 | 0.18 | 0.15 | 5.8 | 0.025 |
| 14/10mer | 0.031 | 0.29 | 0.17 | 0.14 | 6.1 | 0.023 |
| 20/10mer | 0.049 | 0.26 | 0.24 | 0.14 | 4.4 | 0.032 |
| 30/10mer | 0.22 | 0.21 | 1.5 | 0.60 | 0.86 | 0.70 |
| 40/10mer | 0.39 | 0.19 | 1.9 | 0.52 | 0.72 | 0.73 |
| 50/10mer | 0.35 | 0.20 | 2.6 | 0.95 | 0.59 | 1.6 |
Coupling of ATPase and unwinding activities to eIF4A conformational dynamics
A comparison of conformational dynamics of eIF4A with rate constants for ATP hydrolysis and for RNA unwinding of duplex RNAs provides insight into coupling of ATP hydrolysis to RNA unwinding, and of ATP hydrolysis and RNA unwinding to conformational cycling of eIF4A (Figure 7C–F).
Coupling of ATP hydrolysis to conformational cycling in the presence of ssRNAs
The turnover number for ATP hydrolysis should be limited by the slowest step in the conformational cycle of eIF4A. In the presence of the 10mer, 14mer, and 20mer, closing of eIF4A is rate-limiting, with a rate constant of ∼0.20 s−1, whereas opening becomes rate-limiting in the presence of the 30mer and the 50mer (kopen ≈ 0.33 s−1). The overall conformational cycling of eIF4A is thus ∼1.7-fold faster in the presence of longer single-stranded RNAs. Rate constants for ATP hydrolysis increase with the length of the RNA present, and reach a maximum value of ∼0.1 s−1 for the 30mer, 40mer, and 50mer RNAs (see Figure 2). These findings are in reasonable agreement with previous studies on rabbit eIF4A that reported a minimal size of RNA of 12–18 nucleotides required for maximal ATPase stimulation (34). Conformational cycling is faster than ATP hydrolysis in the presence of all RNAs investigated: The number of conformational cycles of eIF4A per ATP hydrolyzed varies between 1.6 (in the presence of the 20mer RNA) and 2.9 (in the presence of the 10mer RNA), with an average value of 2.2 (±0.5) conformational cycles per ATP hydrolyzed. There is no evident correlation with the length of the RNA (Figure 7C).
Average coupling of ATP hydrolysis to conformational cycling in the presence of double-stranded RNAs
In the presence of double-stranded RNAs, rate constants for ATP hydrolysis increase with the length of the 5′-single-stranded overhang, and reach a maximum of ∼0.1 s−1 in the presence of the 30/10mer. Further increase in the length of the 5′-single-stranded extension does not lead to a significant increase in the turnover number. The presence of two alternative conformational cycles of eIF4A with different rate constants precludes a straightforward analysis of coupling in this case. However, the transit times for both conformational cycles can be determined by adding the inverse of the rate constants for opening and closing for each cycle. From the two transit times, the total number of conformational cycles per second can be calculated (see Supplementary Methods). Comparison of these values with the rate constants for ATP hydrolysis yields the average coupling over both cycles. In the presence of the 10/10mer and 14/10mer RNAs, this calculation gives a value of 7–8 conformational cycles per ATP hydrolyzed, whereas 4.0–4.6 conformational cycles are required per ATP hydrolysis event in the presence of the 20/10mer, 30/10mer, 40/10mer, and 50/10mer RNAs (Figure 7D). The average coupling for the longer RNAs is similar to the coupling observed for single-stranded RNAs, indicating that the activation of the ATPase activity may partly result from the interaction of eIF4A/B/G with the single-stranded part.
Altogether, the data show that conformational cycling of eIF4A is not rate-limiting for ATP hydrolysis. Conformational cycling that is faster than the chemical step of the reaction catalyzed is not unprecedented: a recent study correlated the conformational dynamics of adenylate kinase with the chemical step of phosphoryl transfer and found that opening and closing of the enzyme are two orders of magnitude faster than the reaction catalyzed (35).
Average coupling of conformational cycles and RNA unwinding
For mammalian eIF4A (in the presence of eIF4H), a previous smFRET study reported that conformational changes are rate-limiting and strictly coupled to unwinding of an RNA hairpin (16), which is clearly not the case for yeast eIF4A. In contrast to the saturation behavior of the ATPase activity with increasing length of the RNA, rate constants for RNA unwinding first increase with increasing length of the 5′-single stranded region, reach a maximum for the 20/10mer, and then decrease dramatically when the 5′-tail increases further in length (30/10mer, 40/10mer, 50/10mer). Consequently, the average number of conformational cycles per RNA unwound first decreases from the 10/10mer (∼460 cycles/RNA unwound) to the 14/10mer (170 cycles/RNA unwound), and reaches a minimum of approx. 37 conformational cycles per unwinding event for the 20/10mer (Figure 7E). For RNAs with longer 5′-single-stranded regions, the number of conformational cycles per RNA unwound increases again, to 260 (30/10mer), 800 (40/10mer), and 1500 (50/10mer; Figure 7E). It is important to note that the considerations on coupling are independent of whether opening in the presence of RNAs with longer single-stranded regions is described by a single- or double-exponential function. In contrast to ATP hydrolysis, RNA unwinding is thus less tightly coupled to conformational cycling of eIF4A in the presence of RNAs with longer 5′-single stranded regions. This may also reflect a predominant interaction of eIF4A/B/G with the single-stranded regions flanking the duplex in these RNAs, and the resulting competition of the single-stranded regions with duplex unwinding. In contrast, the lower coupling between conformational cycling and RNA unwinding with RNAs containing short single-stranded extensions may result from the enforced direct interaction with the duplex. Interaction of eIF4A with single-stranded regions is in agreement with the suggested role of eIF4A in binding of all mRNAs, not only those with secondary structure elements, and in recruitment of these mRNAs to the PIC (8,36).
Average coupling of ATP hydrolysis and RNA unwinding
Due to the different dependencies of rate constants for ATP hydrolysis and RNA unwinding on the length of the 5′-single-stranded region, coupling of ATP hydrolysis to RNA unwinding is maximal for the 20/10mer, with 0.12 unwinding events per ATP hydrolyzed (corresponding to approx. 8 ATP hydrolyzed per RNA unwound; Figure 7F). The 20/10mer offers 6 nucleotides for interaction with the eIF4A/B/G complex, which is less than the binding site of approx. 13 nucleotides estimated for rabbit eIF4A (37). Human eIF4A in the presence of eIF4B and eIF4G shows a strong increase in unwinding velocity for a 12 bp-duplex with a 20-nucleotide 5′-tail compared to a 10-nucleotide tail (24), which is in agreement with the size of the RNA protected by rabbit eIF4A. For yeast eIF4F, the strongest increase in unwinding rate was observed from an RNA with a 15-nucleotide 5′-single-stranded region to an RNA with a 30-nucleotide single-stranded region (38). The optimal length of the 5′-single-stranded tail exceeds that of the binding site of rabbit eIF4A, which may be caused by the presence of eIF4G and eIF4E and their binding to the RNA. It should be noted that the eIF4G-MC construct we used here contains only one of the three RNA binding domains of eIF4G (39). It can thus not be excluded that the unwinding optimum might shift to longer 5′-single-stranded regions in the presence of full-length eIF4G.
A certain length of the single-stranded region flanking an RNA duplex is necessary to provide a binding platform for the productive assembly of eIF4A, eIF4G, and eIF4B on the RNA, with eIF4B and eIF4G contacting the single-stranded region, and eIF4A positioned to unwind an adjacent duplex. For RNAs with longer single-stranded extensions, the entire eIF4A/B/G complex appears to preferentially interact with the single-stranded region, which competes with binding at the single-strand/double-strand junction and with productive duplex destabilization and unwinding by eIF4A. Rapid unwinding of secondary structures in the 5′-UTRs flanked by short single-stranded regions also makes sense in a physiological context: examination of yeast mRNAs (40), limited to the first 84 nucleotides (the average length of a 5′-UTR), revealed that single-stranded regions preceding double-stranded regions are very often short, with a length between 6 and 16 nucleotides in 69% of all mRNAs mapped. Thus, the 20/10mer with its 6-nucleotide single-stranded region is a suitable model system that mimics a typical structural feature of yeast 5′-UTRs.
Towards a quantitative kinetic model linking conformational cycling, ATP hydrolysis, and RNA unwinding by eIF4A
With double-stranded RNA, eIF4A can in principle undergo three different reactions, productive, unproductive, and futile cycles (Figure 7G). Productive cycles lead to ATP hydrolysis and RNA unwinding, unproductive cycles occur without ATP hydrolysis and RNA unwinding, and in futile cycles ATP is hydrolyzed without RNA unwinding. From these considerations, we would expect eIF4A to undergo three alternative conformational cycles in the presence of double-stranded RNAs (Figure 7G, Supplementary Figure S9).
Futile cycling has been observed previously for helicases of the superfamily 1 that couple ATP hydrolysis to translocation on single-stranded DNA, such as PcrA (41). However, it is unclear if futile and productive cycles are connected to differences in conformational dynamics of the enzyme. How can we reconcile the three different types of reaction catalyzed by eIF4A with the two conformational cycles observed in the presence of double-stranded RNAs? The appearance of two observed rate constants for opening suggests that two of the three reactions proceed through the same closed state. In this case, the observed rate constants for opening and closing in this cycle would be the sum of the two microscopic rate constants for opening and closing of these two reactions (k1+k3, k-1+k-3; see Supplementary Methods). There are two possible hypotheses: Either the two ATP-dependent processes, i.e. futile and productive cycles, pass through the same closed state (Supplementary Methods, Case 1), or the two reactions that occur without RNA unwinding, i.e. futile and unproductive cycles, pass though the same closed state (Supplementary Methods, Case 2). Upon closer inspection, the rate constants k1 and k-1 (see Table 2) in presence of the double-stranded RNAs are very similar (within 2-fold) to the rate of ATP hydrolysis observed, suggesting that this cycle might reflect the ATP-dependent processes. Consequently, k2 and k-2 would reflect unproductive cycles without ATP hydrolysis. From the three observed rate constants and the apparent equilibrium constant Kapp, k1+k3, k2, k-1+k-3, and k-2 can be calculated (see Supplementary Methods; Supplementary Table S3A). An alternative model in which one of the conformational cycles reflects futile and unproductive cycles passing through the same closed state, and the second conformational cycle reflects productive cycles, gives negative rate constants and can be discarded.
As only the sum of k1 and k3 or k-1 and k-3 can be determined from the data, we cannot a priori distinguish the fraction of eIF4A going through futile cycles, and the fraction undergoing productive cycles, which precludes a correlation of the conformational cycles with the unwinding behavior observed with different RNAs. Only if we assume that both processes that depend on ATP hydrolysis occur with the same rate constant (k1 = k3, k-1 = k-3), can we extract all six microscopic rate constants (Supplementary Methods; Supplementary Table S3). The net rate constant for productive and futile cycles does not show a maximum for the 20/10mer RNA (Supplementary Table S3B), although this RNA is unwound with the highest rate constant. However, it is conceivable that the contributions of k1 and k3 or k-1 and k-3 to the observed rate constants for opening and closing in futile and productive cycles are different for different RNAs, leading to the observed maximum of the unwinding rate constant, and optimal coupling of ATP hydrolysis and RNA unwinding with the 20/10mer RNA.
Kinetic partitioning fine-tunes eIF4A activity to preferentially unwind duplexes flanked by short single stranded regions that are prevalent in yeast 5′-UTRs
Our three-cycle model (Figure 7G) describes eIF4A activities as the result of a kinetic partitioning (42) between futile, unproductive, and productive cycles. The coupling of conformational cycling to ATP hydrolysis and RNA unwinding is determined by the kinetic balance of these cycles. The rate constant of unwinding is highest for RNA duplexes flanked by short single-stranded regions. In the presence of these RNAs, neither conformational cycling nor ATPase activity are fully stimulated, again pointing to the importance of balancing the rate constants for these processes rather than maximally accelerating all steps. Timing is key to achieve efficient ATP-driven duplex separation: In productive cycles, the first strand of the RNA duplex has to dissociate from the closed state of eIF4A before ATP is hydrolyzed, resulting in tight coupling of ATP hydrolysis to RNA unwinding. In futile cycles, ATP is hydrolyzed before the first strand of the duplex is released, and the intact duplex dissociates from eIF4A, leading to ATP hydrolysis without unwinding (uncoupled cycle). The fine-tuned balance of rate constants for all processes involved, modulated by other translation initiation factors and the 5′-UTR itself, results in efficient unwinding of duplex regions flanked by short single-stranded regions, which is an abundant structural feature in yeast 5′-UTRs, and thus tailors eIF4A to its physiological task.
Kinetic partitioning has been recognized decades ago as the underlying principle that ensures the fidelity in the biosynthesis of DNA and proteins (43). Although the functional implications for eIF4A still need to be unraveled, the kinetic partitioning model provides a valuable framework for future work aimed at understanding eIF4A activity and regulation in the context of translation initiation.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Andreas Heuer, University of Muenster, for discussions on HMM analysis and for simulating FRET time traces, Jochen Reinstein, Max-Planck Institute for Medical Research, for stimulating discussions, Sonja Schmid, Delft University of Technology, for advice on the SMACKS program, and Daniela Schlingmeier for technical assistance.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Deutsche Forschungsgemeinschaft [KL1153/7-1, 7-2]. The open access publication charge for this paper has been waived by Oxford University Press - NAR Editorial Board members are entitled to one free paper per year in recognition of their work on behalf of the journal.
Conflict of interest statement. None declared.
REFERENCES
- 1. Grifo J.A., Tahara S.M., Morgan M.A., Shatkin A.J., Merrick W.C.. New initiation factor activity required for globin mRNA translation. J. Biol. Chem. 1983; 258:5804–5810. [PubMed] [Google Scholar]
- 2. Neff C.L., Sachs A.B.. Eukaryotic translation initiation factors 4G and 4A from Saccharomyces cerevisiae interact physically and functionally. Mol. Cell Biol. 1999; 19:5557–5564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Sonenberg N., Morgan M.A., Merrick W.C., Shatkin A.J.. A polypeptide in eukaryotic initiation factors that crosslinks specifically to the 5′-terminal cap in mRNA. Proc. Natl. Acad. Sci. U.S.A. 1978; 75:4843–4847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Altmann M., Handschin C., Trachsel H.. mRNA cap-binding protein: cloning of the gene encoding protein synthesis initiation factor eIF-4E from Saccharomyces cerevisiae. Mol. Cell Biol. 1987; 7:998–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Goyer C., Altmann M., Lee H.S., Blanc A., Deshmukh M., Woolford J.L. Jr, Trachsel H., Sonenberg N.. TIF4631 and TIF4632: two yeast genes encoding the high-molecular-weight subunits of the cap-binding protein complex (eukaryotic initiation factor 4F) contain an RNA recognition motif-like sequence and carry out an essential function. Mol. Cell Biol. 1993; 13:4860–4874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Linder P., Slonimski P.P.. An essential yeast protein, encoded by duplicated genes TIF1 and TIF2 and homologous to the mammalian translation initiation factor eIF-4A, can suppress a mitochondrial missense mutation. Proc. Natl. Acad. Sci. U.S.A. 1989; 86:2286–2290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Pestova T.V., Kolupaeva V.G.. The roles of individual eukaryotic translation initiation factors in ribosomal scanning and initiation codon selection. Genes Dev. 2002; 16:2906–2922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yourik P., Aitken C.E., Zhou F., Gupta N., Hinnebusch A.G., Lorsch J.R.. Yeast eIF4A enhances recruitment of mRNAs regardless of their structural complexity. Elife. 2017; 6:e31476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Svitkin Y.V., Pause A., Haghighat A., Pyronnet S., Witherell G., Belsham G.J., Sonenberg N.. The requirement for eukaryotic initiation factor 4A (elF4A) in translation is in direct proportion to the degree of mRNA 5′ secondary structure. RNA. 2001; 7:382–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Jankowsky E., Gross C.H., Shuman S., Pyle A.M.. Active disruption of an RNA-protein interaction by a DExH/D RNA helicase. Science. 2001; 291:121–125. [DOI] [PubMed] [Google Scholar]
- 11. Jackson R.J., Hellen C.U., Pestova T.V.. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat. Rev. Mol. Cell Biol. 2010; 11:113–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Green K.M., Linsalata A.E., Todd P.K.. RAN translation-What makes it run. Brain Res. 2016; 1647:30–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hellen C.U., Sarnow P.. Internal ribosome entry sites in eukaryotic mRNA molecules. Genes Dev. 2001; 15:1593–1612. [DOI] [PubMed] [Google Scholar]
- 14. Shatsky I.N., Terenin I.M., Smirnova V.V., Andreev D.E.. Cap-Independent Translation: What's in a Name. Trends Biochem. Sci. 2018; 43:882–895. [DOI] [PubMed] [Google Scholar]
- 15. Caruthers J.M., Johnson E.R., McKay D.B.. Crystal structure of yeast initiation factor 4A, a DEAD-box RNA helicase. Proc. Natl. Acad. Sci. U.S.A. 2000; 97:13080–13085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sun Y., Atas E., Lindqvist L.M., Sonenberg N., Pelletier J., Meller A.. Single-Molecule kinetics of the eukaryotic initiation factor 4AI upon RNA unwinding. Structure. 2014; 22:941–948. [DOI] [PubMed] [Google Scholar]
- 17. Theissen B., Karow A.R., Kohler J., Gubaev A., Klostermeier D.. Cooperative binding of ATP and RNA induces a closed conformation in a DEAD box RNA helicase. Proc. Natl. Acad. Sci. U.S.A. 2008; 105:548–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hilbert M., Kebbel F., Gubaev A., Klostermeier D.. eIF4G stimulates the activity of the DEAD box protein eIF4A by a conformational guidance mechanism. Nucleic Acids Res. 2011; 39:2260–2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Andreou A.Z., Klostermeier D.. eIF4B and eIF4G jointly stimulate eIF4A ATPase and unwinding activities by modulation of the eIF4A conformational cycle. J. Mol. Biol. 2014; 426:51–61. [DOI] [PubMed] [Google Scholar]
- 20. Sengoku T., Nureki O., Nakamura A., Kobayashi S., Yokoyama S.. Structural basis for RNA unwinding by the DEAD-Box protein drosophila vasa. Cell. 2006; 125:287–300. [DOI] [PubMed] [Google Scholar]
- 21. Rogers G.W. Jr, Richter N.J., Lima W.F., Merrick W.C.. Modulation of the helicase activity of eIF4A by eIF4B, eIF4H, and eIF4F. J. Biol. Chem. 2001; 276:30914–30922. [DOI] [PubMed] [Google Scholar]
- 22. Rogers G.W. Jr, Richter N.J., Merrick W.C.. Biochemical and kinetic characterization of the RNA helicase activity of eukaryotic initiation factor 4A. J. Biol. Chem. 1999; 274:12236–12244. [DOI] [PubMed] [Google Scholar]
- 23. Nielsen K.H., Behrens M.A., He Y., Oliveira C.L., Sottrup Jensen L., Hoffmann S.V., Pedersen J.S., Andersen G.R.. Synergistic activation of eIF4A by eIF4B and eIF4G. Nucleic Acids Res. 2011; 15:67–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ozes A.R., Feoktistova K., Avanzino B.C., Fraser C.S.. Duplex unwinding and ATPase activities of the DEAD-Box helicase eIF4A are coupled by eIF4G and eIF4B. J. Mol. Biol. 2011; 412:674–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Harms U., Andreou A.Z., Gubaev A., Klostermeier D.. eIF4B, eIF4G and RNA regulate eIF4A activity in translation initiation by modulating the eIF4A conformational cycle. Nucleic Acids Res. 2014; 42:7911–7922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Andreou A.Z., Harms U., Klostermeier D.. eIF4B stimulates eIF4A ATPase and unwinding activities by direct interaction through its 7-repeats region. RNA Biol. 2016; 14:113–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Schutz P., Bumann M., Oberholzer A.E., Bieniossek C., Trachsel H., Altmann M., Baumann U.. Crystal structure of the yeast eIF4A-eIF4G complex: An RNA-helicase controlled by protein-protein interactions. Proc. Natl. Acad. Sci. U.S.A. 2008; 105:9564–9569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Samatanga B., Klostermeier D.. DEAD-box RNA helicase domains exhibit a continuum between complete functional independence and high thermodynamic coupling in nucleotide and RNA duplex recognition. Nucleic Acids Res. 2014; 42:10644–10654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Adam H. Methoden der enzymatischen Analyse. 1962; Weinheim: Bergmeyer, H.U.(Hrsg.), Verlag Chemie; 573–577. [Google Scholar]
- 30. Andreou A.Z., Klostermeier D.. Conformational changes of DEAD-Box helicases monitored by single molecule fluorescence resonance energy transfer. Methods Enzymol. 2012; 511:75–109. [DOI] [PubMed] [Google Scholar]
- 31. Bronson J.E., Fei J., Hofman J.M., Gonzalez R.L. Jr, Wiggins C.H.. Learning rates and states from biophysical time series: a Bayesian approach to model selection and single-molecule FRET data. Biophys. J. 2009; 97:3196–3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003; 31:3406–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Schmid S., Gotz M., Hugel T.. Single-molecule analysis beyond dwell times: demonstration and assessment in and out of equilibrium. Biophys. J. 2016; 111:1375–1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Abramson R.D., Dever T.E., Lawson T.G., Ray B.K., Thach R.E., Merrick W.C.. The ATP-dependent interaction of eukaryotic initiation factors with mRNA. J. Biol. Chem. 1987; 262:3826–3832. [PubMed] [Google Scholar]
- 35. Aviram H.Y., Pirchi M., Mazal H., Barak Y., Riven I., Haran G.. Direct observation of ultrafast large-scale dynamics of an enzyme under turnover conditions. Proc. Natl. Acad. Sci. U.S.A. 2018; 115:3243–3248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sokabe M., Fraser C.S.. A helicase-independent activity of eIF4A in promoting mRNA recruitment to the human ribosome. Proc. Natl. Acad. Sci. U.S.A. 2017; 114:6304–6309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Goss D.J., Woodley C.L., Wahba A.J.. A fluorescence study of the binding of eucaryotic initiation factors to messenger RNA and messenger RNA analogues. Biochemistry. 1987; 26:1551–1556. [DOI] [PubMed] [Google Scholar]
- 38. Rajagopal V., Park E.H., Hinnebusch A.G., Lorsch J.R.. Specific domains in yeast eIF4G strongly bias the RNA unwinding activity of the eIF4F complex towards duplexes with 5′-overhangs. J. Biol. Chem. 2012; 287:20301–20312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Berset C., Zurbriggen A., Djafarzadeh S., Altmann M., Trachsel H.. RNA-binding activity of translation initiation factor eIF4G1 from Saccharomyces cerevisiae. RNA. 2003; 9:871–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kertesz M., Wan Y., Mazor E., Rinn J.L., Nutter R.C., Chang H.Y., Segal E.. Genome-wide measurement of RNA secondary structure in yeast. Nature. 2010; 467:103–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Dillingham M.S., Wigley D.B., Webb M.R.. Demonstration of unidirectional single-stranded DNA translocation by PcrA helicase: measurement of step size and translocation speed. Biochemistry. 2000; 39:205–212. [DOI] [PubMed] [Google Scholar]
- 42. Cleland W.W. Partition analysis and the concept of net rate constants as tools in enzyme kinetics. Biochemistry. 1975; 14:3220–3224. [DOI] [PubMed] [Google Scholar]
- 43. Hopfield J.J. Kinetic proofreading: a new mechanism for reducing errors in biosynthetic processes requiring high specificity. Proc. Natl. Acad. Sci. U.S.A. 1974; 71:4135–4139. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.