Abstract
We report a novel system for efficient and specific targeted delivery of large nucleic acids to and into cells. Plasmid DNA and core histones were assembled to chromatin by salt gradient dialysis and subsequently connected to bispecific antibody derivatives (bsAbs) via a nucleic acid binding peptide bridge. The resulting reconstituted vehicles termed ‘plasmid-chromatin’ deliver packaged nucleic acids to and into cells expressing antigens that are recognized by the bsAb, enabling intracellular functionality without detectable cytotoxicity. High efficiency of intracellular nucleic acid delivery is revealed by intracellular expression of plasmid encoded genes in most (∼90%) target cells to which the vehicles were applied under normal growth/medium conditions in nanomolar concentrations. Specific targeting, uptake and transgene expression depends on antibody-mediated cell surface binding: plasmid chromatin of identical composition but with non-targeting bsAbs or without bsAbs is ineffective. Examples that demonstrate applicability, specificity and efficacy of antibody-targeted plasmid chromatin include reporter gene constructs as well as plasmids that enable CRISPR/Cas9 mediated genome editing of target cells.
INTRODUCTION
Addressing acquired or inherited diseases by providing gene products or by modifying the genetic setup of patients is the primary concept of gene therapy (1–4). In general, the manifold particular gene therapy concepts can be divided in ex vivo or in vivo approaches (5). During an ex vivo gene therapy cells of interest are isolated from the patient for subsequent treatment with the therapeutic gene followed by re-administration of the genetically modified cells (5–7). The in vivo approach on the contrary is based on direct local or systemic injection of a gene delivery system to treat the target cells or tissue (5,8). The common goal for both approaches is the efficient transfer of the genetic material over the cell membrane and finally into the nucleus (9,10). To mediate successful gene transfer, current clinical trials are dominated by two strategies, namely nucleic acid delivery by viral vectors or synthetic chemical systems (11,12). Viral gene delivery is highly efficient by nature but safety concerns due to random integration of the transgene into the host genome or potential immunogenicity issues limit their applicability (13–15). In addition a labour and cost intensive manufacturing comprising difficult to standardize processes are further issues for drug development (16–20). Synthetic chemical systems, most often composed of cationic lipids or polymers, are easier to manufacture and face minor concerns of biosafety/immunogenicity. Nevertheless, so far viral systems are favoured for the major fraction of current clinical trials, as non-viral systems are less efficient and their mode of action bear the risk for toxicity issues (21,22). Both systems, chemical as well as virus-derived entities, are also prone to unspecific uptake, i.e. deliver of nucleic acids to non-target cells. This can affect/decrease efficacy because uptake into non-target cells increases clearance, it may also elicit undesired effects in the non-target tissues (5,23,24). The significance of these issues is fortified by the fact that to date no systemic gene delivery approach succeeded phase III clinical trials to be approved for market access (25). All in all, this emphasizes the need for alternative systems for efficient and specific nucleic acid delivery to realize systemic gene therapy.
To develop a gene delivery system that is not aided by viral entities or synthetic transfection reagents, important characteristics of these systems have to be pointed out and taken into consideration. One common feature of most delivery systems is the protection of DNA to avoid degradation by nucleases (26). Furthermore, viral as well as synthetic nucleic acid delivery systems condense the large nucleic acid to reduce the exposed negative charge and size with the aim to form a compact particle for facilitated cellular uptake (27–31). Moreover, the DNA interaction is most often non-covalent to enable de-compaction and access of the transcription machinery inside the nucleus and to avoid chemical modification influencing gene expression (32–35). Finally, every system comprises a particular mechanism that enables DNA membrane translocation (36–38).
In principle, one inherent mechanism that meets the above mentioned criteria is the assembly of core histones on DNA. The assembly into chromatin is a highly conserved mechanism in eukaryotes to organize genomic DNA inside the nucleus by reducing its size and charge (39). Furthermore, previous studies demonstrated that all four core histone proteins contain protein transduction domains and compatibility of histones for gene delivery has been shown by several studies reviewed by Han et al. (40–44). However, the majority of histone based delivery systems comprise unspecific DNA complexation of core histones, single histone proteins or domains or peptides derived from them and most often combined with synthetic or viral entities (45–52). Wagstaff and co-workers demonstrated that plasmid DNA assembled into chromatin can be delivered into the nucleus, using modified histone H2B protein (53).
The objective of our work was to develop an efficient chromatin-based nucleic acid delivery system that does not contain any virus-derived components. In addition, the delivery system shall (in contrast to applying histones and/or chromatin for nonspecific DNA delivery) introduce nucleic acids only into desired target cells without addressing non-target cells. To achieve these objectives, we used purified histones for packaging DNA into plasmid chromatin (this avoids viral components). In contrast to approaches described above, however, these histones were deliberately kept as ‘wildtype proteins’, i.e. not mutated/modified and therefore exhibited a very low spontaneous delivery potential (53). We then analysed if we can convert such inactive plasmid chromatin to targeted plasmid chromatin with intracellular delivery functionality by adding antibody-derived cell surface targeting entities.
MATERIALS AND METHODS
In-vitro chromatin reconstitution of plasmid DNA
Calf thymus Histones for assembly were kindly provided by Prof. Dr Gernot Längst (University of Regensburg). A ∼4000 bp plasmid DNA encoding EGFP (pEGFP) was amplified and used for assembly of histones via salt gradient dialysis (54). To set up the assembly reaction we mixed DNA and histones in a 1:2 mass ratio in a reaction mix of 2 M NaCL 200 ng/ml BSA and 200 ng/ml BSA, 1 fold low salt buffer (10 mM Tris–HCl pH 7.6, 50 mM NaCl, 1 mM EDTA, 0.05% w/v Igepal CA-630), 2 M NaCl and histone octamer in a 1:2 DNA:histone weight ratio. The reaction mixture was transferred into 3.5 kDa MWCO mini dialysis devices (Thermo Fisher Scientific) equilibrated for 15 min in high salt buffer (10 mM Tris–HCl pH 7.6, 2 M NaCl, 1 mM EDTA, 0.05% w/v Igepal CA-630). Afterwards a 4 l beaker was prepared with 300 ml high salt buffer containing 1 mM beta-mercaptoethanol and a second beaker with 3 l 1-fold low salt buffer containing 1 mM beta-mercaptoethanol. A floater with the dialysis devices and a magnetic stir bar were added into the beaker with high salt buffer. The salt gradient dialysis was performed over night at 4°C. Therefore, the beaker was placed on a magnetic stirrer to allow slow mixing and a peristaltic pump was set to transfer the 3 l of low salt buffer into the beaker containing high salt buffer with a velocity of ∼300 ml/h. After buffer dilution, chromatin samples were purified and buffer was exchanged to PBS via size exclusion chromatography using Sephacryl S-1000 GE Superfine (Sigma Aldrich) matrix.
Antibody chromatin complex preparation
Hapten binding bispecific antibodies and TriFabs were generated and purified as previously described (55). Haptenylated CPXM2 peptide was synthesized by Biosynthan GmbH (Berlin). To prepare DNA binding antibody constructs, biotinylated peptide and biotin binding antibody was pre-incubated in PBS for 30 min in a ratio of two peptides per antibody for the bivalent biotin binding bsAb and one peptide per antibody for monovalent biotin binding TriFabs. Subsequently, constructs were added to chromatin and incubated for at least 30 min for antibody-peptide association at the DNA backbone.
Microscale thermophoresis
Microscale thermophoresis experiments, data processing and determination of KD values was performed by 2bind GmbH (Regensburg). Antibody and peptide were diluted in PBS and pre-incubated for 30 min at RT with a 1:1 or 1:2 molar ratio for TriFab: peptide or 2 + 2 bsAb:peptide, respectively. A serial dilution of the ligand was prepared in a way to match the final buffer conditions in the reaction mix (1× PBS, 0.05% Tween-20). 5 μl of each dilution step were mixed with 5 μl of fluorescent labelled plasmid chromatin. The final reaction mixture, which was filled in capillaries, contained a respective amount of ligand and constant 0.25 nM fluorescent molecule. The samples were analysed on a Monolith NT.115 Pico at 25°C, with 10% LED power and 60% Laser power. Fluorescence values were normalized and data were displayed according the analysed peptide concentration (56). KD values were determined, if normalized fluorescence values allowed a proper curve fit.
Analytic MNase digestion
For nuclease sensitivity assays, 2 μg of DNA assembled with chromatin was diluted in EX-80 buffer (10 mM Tris–HCl pH 7.6, 80 mM KCl, 10% v/v glycerol, 1.5 mM MgCl2, 1 mM DTT) and 1 μl BSA to a final volume of 50 μl. To stop the reaction, 1.5 ml tubes were prepared with 4 μl stop-buffer (100 mM EDTA, 4% w/v sodium dodecyl sulfate). The nuclease reaction was started by addition of 50 μl micrococcal nuclease (MNase) mix (6 mM CaCl2, 200 ng/μl BSA and 40 U MNase). After the indicated time-points, 30 μl of the reaction mix were transferred to the tubes containing stop-buffer. The DNA was de-proteinized by addition of 1 μl Proteinase K and incubation for 1 h at 50°C. The DNA was purified by ethanol precipitation and analysed by agarose gel-electrophoresis.
Flow cytometry
To generate fluorescent plasmid DNA and plasmid chromatin, Cy5 fluorescent dye was chemically conjugated to plasmid–DNA applying the Label IT® Nucleic Acid Labelling kit (Mirus) according to the manufacturer's specification. To generate fluorescent plasmid chromatin, assembly was performed with Cy5 labelled plasmid as described above. Cy3 labelling of antibody was performed via maleimide conjugation after partial antibody reduction with TCEP.
Previous to cell treatment, antibody chromatin complexes were formed as described above. 200,000 MCF7 cells per well were seeded in 96 well plates and treated with complexes with final concentration 1.6 μg/ml plasmid DNA bevor or after chromatin assembly, 50 nM antibody and 100 nM peptide for 1 h at 37°C. Single colour flow cytometry with unlabelled antibodies was performed with a FACScanto II (BD Biosciences). For dual colour flow cytometry, Cy3 labelled antibodies were used instead of unlabelled antibodies. Colour compensation was performed with single stained controls. Dual color flow cytometry was performed with an LSRFortessa (BD Biosciences).
Reporter gene expression and cytotoxicity assay
80 000 MCF7 cells per well were seeded in 12-well plates for reporter gene expression assays and 10 000 MCF7 cells per well were seeded in 96-well plates for LDH cytotoxicity assays. 24 h after seeding, cells were treated with complexes containing 8 μg/ml plasmid DNA bevor or after chromatin assembly, 250 nM antibody and 500 nM peptide prepared as described above or single components at the same concentration when indicated. The cells were exposed to complexes or single components for 48 h in the presence of serum. After 48 h gene expression or cytotoxicity was analysed. For gene expression analysis cells were washed, detached and the ratio of GFP positive cells was determined by flow cytometry with a FACScanto II (BD Biosciences). For cytotoxicity analysis, culture supernatant was removed and LDH activity was quantified with the Cytotoxicity Detection Kit (LDH) (Sigma-Aldrich) according to the manufacturer's protocol.
Confocal microscopy
For live cell imaging, MCF-7 cells (NCI) were cultured in phenolred-free RMPI medium supplemented with 10% fetal calf serum (FCS) and 100 U/ml penicillin and 100 μg/ml streptomycin. 20 000 cells/well were seeded into 8-well chamber slides (Lab-Tek™, Thermo Fisher Scientific, Braunschweig, Germany) and allowed to adhere overnight. Glass surfaces had been coated with 30 μg/ml fibronectin in PBS for 1 h at 37°C. Antibody plasmid DNA-Cy5 and Antibody-plasmid chromatin-Cy5 complexes were formed as described in example 7. Samples were added to MCF7 at a final concentration of 4 μg/ml plasmid DNA, 250 nM peptide and 125 nM antibody. 4 and 72 h after addition, internalization of antibody–chromatin complexes and GFP expression were followed by live cell fluorescence microscopy carried out on a Leica SP5 laser scanning confocal microscope using a 63×/1.2NA water immersion objective lens (Leica, Mannheim, Germany). Temperature, CO2 level and humidity were maintained at 37°C and 5% CO2 using a stage-top incubation chamber (Oko-touch, Okolab, Ottaviano, Italy). Sequential scans were performed using white light laser excitation at 488 nm, (561 nm) and 633 nm. Fluorescence emission was detected at 495–548 nm (GFP), 570–628 nm (Cy3) and 647–732 nm (Cy5) using HyD detectors. Images were processed with ImageJ (NIH, Bethesda, MD, USA). Immunocytochemistry was performed as previously described (57).
CRISPR/Cas9 targeting and knock-out quantification
It has previously been described that gene-editing mediated inactivation of DPH1, combined with assessment of cellular sensitivity towards Diphtheria Toxin (DT), can be used to quantify efficacy of gene editing (58). Inactivation of all cellular copies of DPH1 (as consequence of gene editing) in turn renders cells resistant to DT. This generates a very robust readout which can be quantified by counting DT-resistant colonies following gene editing. To prove targeting specificity and efficacy of the delivery system with plasmids encoding a therapeutically more relevant gene product, targeted delivery complexes were prepared as described above with CISPR/Cas9 ‘plasmid chromatin’ instead of pEGFP plasmid chromatin. Afterwards the complexes were added to MCF7 cells seeded in a 12-well plate (4000 cells/well 24 h before treatment) to a maximal final concentration of 8 μg/ml plasmid DNA assembled to Chromatin, 500 nM peptide and 250 nM antibody. After incubation of the complexes in normal serum containing cell culture medium for 72h, medium was removed and cells were exposed to the same medium containing DT at a final concentration of 4 nM. DT exposure was continued for 2 weeks with medium exchange every 3 to 4 days. After this period, cells were stained with methylene blue and efficiency of intracellular delivery and expression of the editing components was assessed by determination of DT-resistant colonies as previously described.
Statistics
Unpaired, two-tailed Student's t-tests were performed for single comparisons between two treatments. Multiple comparisons were statistically analyzed via one-way ANOVA, followed by Tukey's honestly different significance (HDS) post hoc tests. Significant differences were defined by P-values of < 0.05. The level of significance determined using Student's t-test or Tukey's HDS test is indicated in graphs by asterisks. One, two or three asterisks are defined by P < 0.05, P < 0.01 and P < 0.001, respectively.
RESULTS
In-vitro chromatin reconstitution of plasmid DNA by salt gradient dialysis
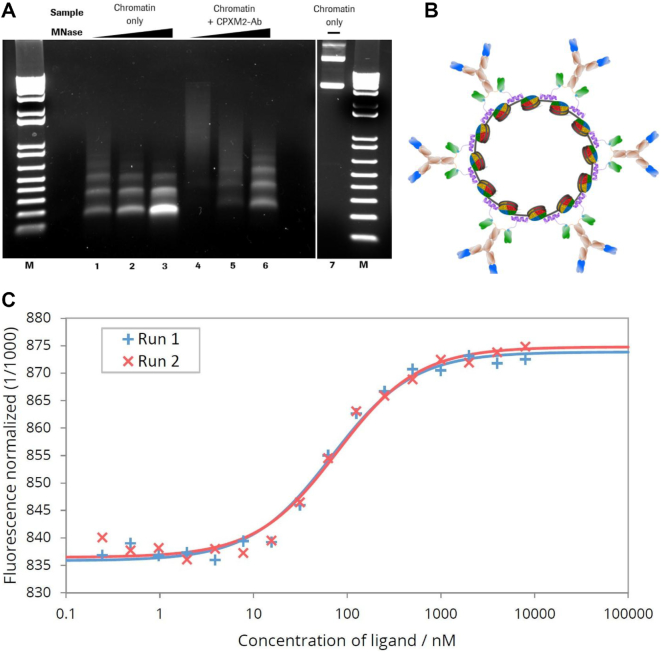
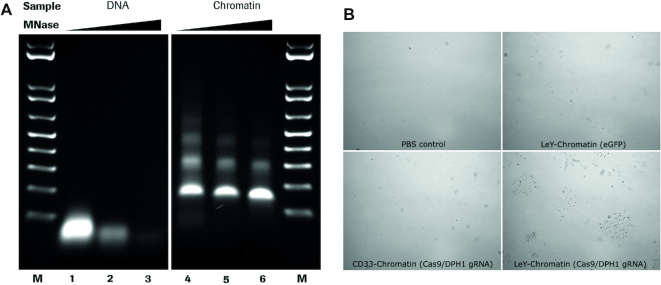
Chromatin can be efficiently reconstituted from DNA and histones by the salt gradient dialysis methods (54,59,60). Using supercoiled plasmid DNA and purified histone octamers, nucleosomes are formed that consist of the histone octamer and 147 bp of DNA wrapped ∼1.65 turns around the octamer (61). The salt gradient dialysis method gives rise to nucleosomal arrays on DNA that are separated by short DNA linkers with a size ∼15 bp. Fine titration of histone to DNA ratios results in plasmid chromatin fully covered by nucleosomes that are qualitatively evaluated by micrococcal nuclease (MNase) hydrolysis of DNA. The endonuclease MNase does preferentially hydrolyse DNA in the linker region between the nucleosomes, giving rise to an MNase ladder of DNA when partially hydrolyzing chromatin (62). We applied this method to generate plasmid chromatin with an eGFP expression plasmid. The quality of the reconstituted chromatin was determined by nuclease hydrolysis and subsequent agarose gel-electrophoresis (Figure 1A) (54). The partial DNA hydrolysis of assembled chromatin generates DNA fragments of multiples of 160 base pairs, suggesting that arrays of nucleosomes were formed on the plasmid DNA. Furthermore, the clear pattern of the nucleosomal ladder and the absence of DNA fragments shorter than 147 bp (sub-nucleosomal DNA), suggested the efficient reconstitution of the plasmid DNA into chromatin (Figure 1A).
Figure 1.
MNase digestion of antibody–chromatin complexes and antibody-complexation with plasmid-chromatin. (A) Agarose gel electrophoresis of chromatin without (lane 1–3) and in presence of biotin-CPXM2 ∼ anti biotin 2+2 bsAb constructs (lane 4–6) after partial DNA hydrolysis by MNase with increasing incubation time (20s, 80s and 270s); Chromatin without MNase treatment (lane 7) is shown as control. Mononucleosomal DNA bands (147bp) indicate complete digestion in contrast to higher molecular weight bands. In presence of biotin-CPXM2 ∼ anti biotin 2+2 bsAb constructs, chromatin is more nuclease resistant as the 147 bp DNA band only occurs at late time-points of Nuclease treatment in comparison to the chromatin only sample. (B) Scheme of antibody–chromatin complexes with plasmid DNA reconstituted into nucleosomes and associated antibody-peptide constructs. Variable regions against cell surface antigen (blue) faces outwards and anti biotin scFv (green) is bound at biotin-CPXM2 peptide (purple) associated at the DNA backbone. (C) MST runs for Chromatin + biotin-CPXM2 ∼ anti biotin 2+2 bsAb interaction. Ligand concentration refers to biotin–CPXM2 peptide (twice as much as the respective anti biotin 2+2 bsAb concentration). Exp 1 (blue) and Exp 2 (red) are independent experiments of the same construct with the respective curve fit for KD determination.
Antibody - chromatin complexes with improved nuclease resistance are formed via DNA binding peptide CPXM2
To capture plasmid DNA or plasmid chromatin via charge interaction with the negatively charged DNA backbone, we used a nucleic acid binding peptide (CPXM2 peptide) identified by Haas et al. and derived from human carboxypeptidase-like protein X2 (CPXM2 protein) (63). To enable binding of CPXM2 peptide to antibodies, we used a biotinylated version of CPXM2 peptide (biotin CPXM2 peptide) and biotin binding (anti biotin) bispecific antibodies (Figure 1B). Affinity of antibody-peptide constructs to chromatin was determined by microscale thermophoresis (MST). With this method affinity data were generated in solution without the need to capture antibody or peptide as this would affect affinity in this system due to avidity effects. To identify the most suitable antibody format, we compared monovalent biotin binding TriFabs with bivalent biotin binding bispecific antibodies (anti biotin 2+2 bsAb) towards affinity and potential aggregation due to crosslinking of the molecules (55,64). Affinity of biotin CPXM2 peptide ∼ anti biotin TriFab constructs to chromatin was in the three digits nanomolar range (300nM). The biotin CPXM2 peptide ∼ anti biotin 2+2 bsAb constructs demonstrated further stabilization (two-digit nM affinity) most likely due to avidity effects as two CPXM2 peptides can be bound by one antibody (Figure 1C). In addition, no aggregation was observed with the biotin CPXM2 peptide ∼ anti biotin 2+2 bsAb construct, indicating that no severe crosslinking occurs with this antibody format (Supplementary Figure S1). Specificity was proven by respective controls without peptide. The MST data set of the individual runs (Supplementary Figure S2) is summarized in Table 1. As the strongest interaction was observed when the peptide was coupled to anti biotin 2+2 bsAb, we used this antibody format for further studies. As the antibody peptide construct interacts with the negatively charged DNA backbone, we checked whether this interaction disrupts the nucleosomes, or alters nuclease resistance of plasmid chromatin after antibody-peptide assembly. After incubation of chromatin with antibody and peptide and subsequent nuclease digestion, the pattern of partially hydrolyzed DNA after 270s was similar to the pattern of nuclease treated chromatin alone after 20s (Figure 1A). This data clearly demonstrates that addition of the antibody-peptide reduces the nuclease accessibility, by probably binding to the accessible DNA linker, but the nucleosomal arrays remain intact.
Table 1.
Affinity between chromatin and antibody or antibody-peptide constructs; Interaction between chromatin and antibody or antibody-peptide constructs was determined by MST. Affinity value for bio-CPXM2 ∼ anti biotin 2+2 bsAb refers to biotin-CPXM2 peptide concentration (2-fold higher than antibody concentration as one antibody can bind two peptides); affinity values with respective SEM were determined by two independent measurements
| Construct | K D (nM) | SEM (nM) |
|---|---|---|
| Chromatin + biotin-CPXM2 ∼ anti biotin TriFab | 300.0 | 36.0 |
| Chromatin + biotin-CPXM2 ∼ anti biotin 2+2 bsAb | 73.1 | 3.4 |
| Chromatin + anti biotin TriFab only | no interaction | n.a. |
| Chromatin + anti biotin 2+2 bsAb only | no interaction | n.a. |
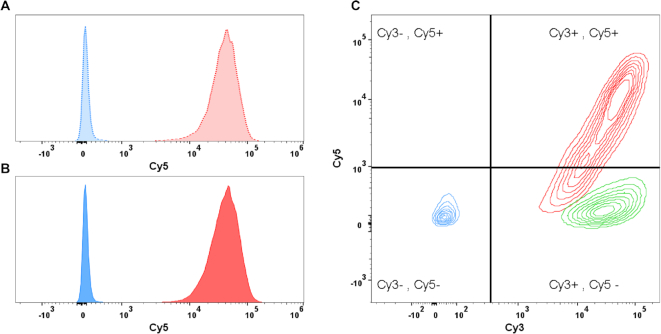
DNA as well as chromatin is specifically and efficiently delivered via CPXM2-antibody constructs
In addition to specific formation and prolonged nuclease resistance of the antibody–chromatin complex, we investigated DNA delivery to the cell surface via the associated antibodies. To determine delivery efficacy and specificity, anti-biotin 2+2 bsAbs with a second specificity against Lewis Y or CD33 were compared on MCF7 cells (LeY+++/CD33-). Furthermore, plasmid DNA labelled with Cy5 fluorophore was used to enable quantification of plasmid DNA on cells by flow cytometry 1h after cell treatment. To elaborate the influence of chromatin assembly on delivery specificity and efficacy, we applied the delivery system for plasmid DNA before and after chromatin assembly. Figure 2A shows Cy5 signal of MCF7 cells after treatment with plasmid DNA before chromatin assembly complexed with anti LeY (dotted red) and anti CD33 (dotted blue) antibody. A distinct fluorescence signal was detected after treatment with anti LeY–DNA–Cy5 complexes demonstrating that DNA delivery is highly efficient. In contrast, application of anti CD33–DNA–Cy5 complexes did not result in Cy5 positive cells (as MCF7 do not express CD33). This demonstrates that DNA delivery is mediated by the antibody and payload is delivered only to cells that express the cognate target antigen. After chromatin assembly, plasmid delivery efficacy and specificity was not affected as the same distinct fluorescence signal was observed after treatment with anti LeY-chromatin-Cy5 complexes (solid red) and no Cy5 signal was detected with anti CD33-chromatin-Cy5 complexes (solid blue) (Figure 2B). To confirm the presence of the antibody in our delivery system, we used anti CD33 and anti LeY antibodies labelled with Cy3 fluorophore together with Cy5 labelled chromatin. As displayed in figure 2C, MCF7 cells treated with anti CD33–Cy3–chromatin–Cy5 complexes did not show an elevated Cy5 as well as Cy3 signal (blue contours) demonstrating that neither antibody nor chromatin is present at the cell surface. In contrast, anti LeY–Cy3–chromatin–Cy5 treatment results in distinct fluorescence signals for Cy3 and Cy5 (red contours), proving antibody at the cell surface and confirming the successful delivery of chromatin (with somehow reduced efficiency compared to unlabelled antibody). Finally, we checked the second specificity of our targeting antibody against biotin. Therefore, we compared our targeted chromatin delivery system comprising biotinylated–CPXM2 peptide with a targeting system where the biotinylated peptide was exchanged against a peptide with the wrong hapten (digoxigenin instead of biotin). Figure 2 C highlights that both complexes (blue contours with biotin–CPXM2 peptide and green contours with digoxigenin-CPXM2 peptide) generate a distinct Cy3 fluorescent signal on MCF7 cells, whereas Cy5 signal was only detected after treatment with biotin–CPXM2 peptide comprising complexes. This clearly demonstrated that despite the cell surface specificity, also the second specificity against the hapten is necessary for chromatin and therefore plasmid DNA delivery without unspecific interaction between antibody and peptide/chromatin.
Figure 2.
Flow cytometric determination of delivery specificity; Binding and uptake of antibody-Cy3 and DNA-Cy5 (before and after chromatin assembly) was analysed by flow cytometry after incubation for 1 h. (A) Histogram of MCF7 cells after treatment with targeted (anti LeY; dotted red) and untargeted (anti CD33; dotted blue) DNA-Cy5 complexes. Cy5 signal was detected only after treatment with the targeted DNA-Cy5 construct. (B) Histogram of MCF7 cells after treatment with targeted (red) and untargeted (blue) chromatin-Cy5 complexes. Results are comparable to results after DNA–Cy5 delivery. (C) Contours plot of MCF7 Cy3 (x-axis) and Cy5 (y-axis) signals after treatment with various antibody chromatin complexes comprising Cy3 labelled antibody and Cy5 labelled DNA. Cells were treated with complexes comprising antibody without specificity against cell surface antigen but against CPXM2 peptide do neither show Cy3 nor Cy5 signals (blue). Cells treated with complexes comprising digoxigenin CPXM2 peptide (instead of biotin CPXM2 peptide) display Cy3 signal but no Cy5 signal, demonstrating that antibody but not chromatin is present at the cell surface (green). Cells treated with complexes comprising antibody with specificity against the cell surface and CPXM2 peptide display Cy3 signal and Cy5 signal, demonstrating that antibody as well as chromatin is present at the cell surface (red).
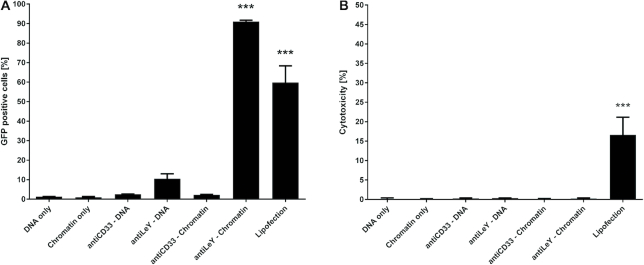
Targeted chromatin efficiently mediates transgene expression without cytotoxicity
After determination of delivery efficiency and specificity to the target cells, we addressed the nuclear delivery efficacy by quantifying GFP reporter gene expressing cells via flow cytometry. With this assay we can also directly depict the influence of chromatin assembly on intracellular plasmid DNA delivery as on cell DNA delivery is equally efficient with and without chromatin assembly. To address reporter gene-expression, we have treated MCF7 cells with different constructs for 48 h and subsequently identified GFP expressing cells via flow cytometry. The ratio of GFP positive cells was determined by comparison with respective vehicle or antibody only control. Incubation of MCF7 cells in presence of DNA or chromatin did not generate cells expressing detectable levels of GFP, indicating no unspecific nuclear uptake of plasmid DNA before and after chromatin assembly (Figure 3A). Moreover, association of antibody-peptide constructs did not generate GFP positive cells when the antibody does not bind the cell surface as shown for anti CD33–DNA as well as anti CD33–chromatin complexes (confirming the data of Figure 2 were no unspecific uptake of antibody-DNA and antibody–chromatin was detected). Targeting of plasmid DNA by associated antibody–peptide constructs generated single GFP positive cells (as observed under the microscope) but not to a significant extent despite efficient delivery to the cell surface as shown in Figure 2A. In contrast, antibody-peptide constructs targeting chromatin raised the ratio of GFP positive cells from single exceptions to the vast major population (>90% positive cells). Finally, Lipofection was used as a positive control, resulting in about 60% reporter gene expressing cells. Next, we addressed the cytotoxicity of the different treatments by LDH release relative to vehicle control and complete cell lysis. Lipofection mediated cytotoxicity to a certain extent (∼15% to lysis control), usual for most transfection reagents. None of the other treatments showed detectable cytotoxic effects (Figure 3B).
Figure 3.
Efficiency and cytotoxicity of gene delivery systems. (A) Delivery efficiency of EGFP expression plasmids was addressed by determination of GFP positive cells via flow cytometry 48 h after treatment with targeting complexes being present throughout that time. Significant numbers of GFP positive cells were achieved with Lipofection and anti LeY-chromatin complexes for MCF-7 cells. (B) Cytotoxicity was addressed by quantification of LDH release. Significant LDH release after 48 h was only observed with lipofection. Cells were exposed to respective treatment for the whole incubation period in normal (serum containing) cell culture medium. Mean values + SEM are shown (n = 3); P-values < 0.001 are indicated by three asterisks.
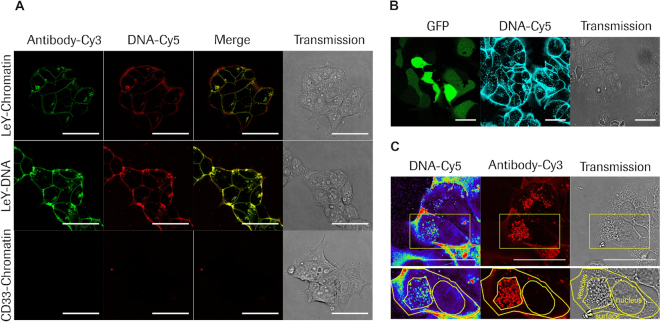
Chromatin is specifically delivered to target cells by bispecific antibodies followed by internalization into the vesicular system
As the impact of chromatin assembly on functional plasmid DNA delivery was surprisingly high, we addressed intracellular distribution of antibody and DNA after treatment with different complexes by confocal microscopy. Figure 4A highlights the distribution of antibody-Cy3 (green) and DNA-Cy5 (red) in living cells 4 h after treatment with targeted (LeY–) chromatin, targeted (LeY–) DNA or untargeted (CD33-) chromatin. Antibody as well as DNA was present at the cell surface as well as the vesicular system after targeting of chromatin (top row of images) as well as DNA (middle row of images). Overlay of both fluorescence signals indicates that most of antibody and DNA is co-localized and not separated. These data clearly demonstrate that targeted DNA gets delivered to the cell surface and internalized via the targeting antibody irrespective of assembled into chromatin or not. Specificity of the targeting system was confirmed by confocal microscopy of MCF7 cells after incubation with the untargeted chromatin complex, as neither antibody nor DNA was detected at the cell surface as well as inside vesicles (bottom row of images). Figure 4B shows that chromatin targeting does not only result in strong DNA accumulation at the cell surface and inside the vesicular system (cyan) but also in GFP expression (green). For imaging of GFP signal after 3days, unlabelled antibody was used as labelling reduced chromatin delivery efficacy. To image treated cells with higher resolution and in more detail, cells were fixed and the antibody was subsequently counterstained with anti-human IgG Cy3 antibody (Figure 4C). Confocal microscopy of cells that received targeted plasmid chromatin revealed strong above-background signals of the targeting antibodies on cell surfaces and vesicular compartments but not in nuclei. Cy5-labeled plasmid payload was found together with the antibody on cell surfaces and vesicular compartments but was also clearly detectable in nuclei. These observations are in line with previous findings that (i) noncovalent hapten-coupled payloads separate from targeting antibodies after internalization and become routed independently from the antibody (64,65) and (ii) that most antibodies bind to cells and internalize in an effective manner but by themselves have very low propensity to escape from vesicular compartments and enter the cytoplasm or nucleus (66).
Figure 4.
Confocal microscopy analysis of antibody mediated plasmid DNA or chromatin delivery and intracellular routing. (A) Live cell imaging of MCF7 cells 4h after treatment with targeted (LeY) chromatin (top row), targeted (LeY) DNA (middle row) and untargeted (CD33) chromatin (bottom row); Antibody-Cy3 signal is displayed in green and DNA-Cy5 level is displayed in red; Overlay of fluorescent images are shown in the ‘Merge’ column and the right column shows respective transmission images. (B) Live cell imaging of MCF7 cells 3 days after treatment with targeted (LeY) chromatin complexes comprising unlabelled antibody and Cy5 labelled DNA. Left panel shows GFP signal in green, middle panel DNA–Cy5 in cyan and right panel transmission, respectively. (C) Imaging of fixed MCF7 cells 3 days after treatment with targeted (LeY) chromatin. Left panel displays DNA Cy5 signal in pseudocolor, middle panel shows antibody signal generated by counterstaining with anti human IgG Cy3 antibody in red and the right panel represents the transmission image. Cell surface, vesicular compartments and nuclear envelope are marked by yellow contours. Scale bars: 50 μm.
Targeted chromatin delivery enables specific and efficient CRISPR/Cas9 mediated genome editing
Next, we addressed if chromatin delivery can be applied with plasmid DNA of larger size and with more complex function. Therefore we used a plasmid encoding a CRISPR/Cas9 knock-out system against Diphthamide synthesis gene 1 (DPH1) and performed the previously published Diphtheria toxin (DT) based assay for quantification of CRISPR/Cas9 mediated gene editing (58). This assay utilizes DT resistance mediated by homozygous DPH1 knock-out for identification of cell clones in which gene editing by Cas9 was successful. As a result, only homozygous DPH1 knock-out cells survive and display colony formation after 2 weeks of continuous DT selection. First of all, we transferred the chromatin assembly on the DPH1 gRNA Cas9 expression plasmid (Figure 5A). The delivery system was applied for of Cas9 DPH1 gRNA encoding ‘plasmid chromatin’ in the same manner as for GFP encoding ‘plasmid chromatin’. After treatment of MCF7 cells with targeted (LeY–) Cas9 DPH1 gRNA encoding ‘plasmid chromatin’, untargeted (CD33-) Cas9 DPH1 gRNA ‘plasmid chromatin’ and targeted (LeY–) GFP ‘plasmid chromatin’ and incubation for 3 days, cells were exposed to DT for two weeks. Finally, cells were fixed and colonies were counted under the microscope. Representative microscopic images are shown in Figure 5B and the ratio of colony number and number of initially seeded cells are summarized in Table 2 as percentage of DT resistant colonies and therefore percentage of clones with homozygous gene knock-out. Targeted delivery of Cas9 DPH1 gRNA chromatin results in almost 4% DT resistant clones whereas targeted delivery of GFP control chromatin does not result in any resistant colony, confirming that colony formation can only occur by expression of the CRISPR/Cas9 editing system. In line with the specificity of the chromatin targeting system as shown above for GFP, MCF7 treatment with untargeted (CD33–) Cas9 DPH1 gRNA chromatin does also not result in formation of DT resistant colonies. Compared to the absolute CRISPR/Cas9 knock-out frequencies and colony numbers from our previous experiments, the determined percentage of DT resistant clones would equal to >60% of Cas9 expressing cells (Table 2).
Figure 5.
Antibody-chromatin delivery with plasmid DNA encoding a CRISPR/Cas9 system. (A) Agarose gel electrophoresis of DPH1 gRNA/Cas9 plasmid DNA (lanes 1–3) and chromatin (lanes 4–6) after partial DNA hydrolysis by MNase with increasing incubation time (20s, 80s and 270s). (B) Representative microscopic images of DT resistant MCF7 cell clones after treatment with vehicle (PBS control), targeted control plasmid chromatin (LeY-Chromatin (eGFP)), non-targeted Cas9/DPH1 gRNA plasmid chromatin (CD33-Chromatin (Cas9/DPH1 gRNA)), targeted Cas9/DPH1 gRNA plasmid chromatin (LeY-Chromatin (Cas9/DPH1 gRNA)); DT resistant colonies were only observed after treatment with targeted DPH1 gRNA/Cas9 plasmid chromatin.
Table 2.
Transfection efficiencies with Cas9/DPH1 gRNA expression plasmids based on DPH1 editing efficiencies in MCF7 cells; Calculated cell transfection efficiencies (a) are based on determined Cas9/DPH1 gRNA mediated homozygous DPH1 knock-out (DPH1 k.o.) efficiencies (b) as previously published (58). DT-resistant DPH1 k.o. cells (c) are indicated as ratio between counted DT resistant colonies and initially seeded cells. Data of first row are derived from previous publication (58); data of second and third row are based on mean values of colony numbers obtained after respective treatments as described in B (n = 3)
| Treatment | a Transfection efficiency (% of seeded cells) | b DPH1 k.o. efficiency of Cas9/DPH1 gRNA (% transfected cells) | c DT-resisitant DPH1 k.o. cells (% of seeded cells) |
|---|---|---|---|
| Lipofection (Killian et al. SciRep 2017) | 40% measured | 6.3% calculated c/a | 2.5% measured |
| Targeted (LeY) Cas9/gDPH1 chromatin | 59% calculated c/b | 6.3% same as above | 3.7% measured |
| Non-targeted (CD33) Cas9/gDPH1 chromatin | 0% calculated c/b | 6.3% same as above | 0% measured |
DISCUSSION
The development of a targeted gene delivery system faces multiple challenges as it must overcome several hurdles and therefore needs well-balanced properties (67,68). For example, a DNA delivery system must have affinity to the target cells to efficiently mediate the uptake of DNA but in parallel must not interact with serum components or the cell membrane of other cells and tissues to minimize loss of DNA and avoid off-target effects along the delivery route (34,69,70). Furthermore, DNA has to be translocated over the membrane barrier to enter the cytosol and finally reach the nucleus to enable transgene expression (71,72). As a consequence, the required membrane interaction for DNA translocation has to be efficient but at the same time gentle enough to avoid cell cytotoxicity (70).
Our goal was to generate a highly flexible and modular gene delivery system to outline the influence of every component along the gene delivery route. We made use of the hapten-binding bsAb technology comprising antibody derivatives that are able to simultaneously bind cell surface antigens and to small molecule haptens like biotin or digoxigenin via antibody-antigen interactions57. This technology enables delivery of diverse hapten bearing molecules (payload) to the target cell surface and its broad applicability including nucleic acid delivery has already been demonstrated. The fact that hapten-binding bsAbs are available in different formats covering various sizes, geometries and stoichiometries (55,65,73,74) enhances their versatility as modules for targeted nucleic acid delivery. The non-covalent attachment of payload to hapten-binding bsAbs enables separation of payloads from targeting vehicles inside vesicular compartments. The latter is important for delivery of molecules with intracellular functionality such as nucleic acids. For those, non-covalent hapten coupling is advantageous compared to covalent conjugation strategies were payload release frequently needs to be optimized for example by introduction of cleavable linkers (64,65,75).
Functional plasmid DNA delivery can be observed in about 90% of treated cells without cytotoxicity therefore providing a mechanism for efficient but gentle DNA membrane translocation. Such high efficacies are comparable to viral gene delivery systems (24,76–80). However, the overall objective of our work was not only to achieve high efficacy, but also to combine that with targeting to specific cells. Attaching targeting entities to delivery vehicles to selectively address desired cell types is similar to that of next-generation viral or virus-like particle (VLP)- based delivery systems. Entities that confer targeting specificity can be added to VLPs by conjugating or fusing them to VLPs. Such specificity-enhancing entities that support enrichment on desired sites can be antibody-based or other protein domains or peptide derivatives (81–83). True specificity, however, requires not only addition of specific binding entities, but also reduction or elimination of non-targeted transfection activity. Targeted plasmid-chromatin described here fulfils targeting requirements and intracellular activity as well as reduction of nonspecific uptake without applying any virus-derived modules.
Hapten-binding bsAbs combined with the DNA binding CPXM2 peptide mediate efficient and specific delivery of plasmid chromatin to and into cells. Thus, while other delivery systems show target preference (84–87), this novel approach has the potential to reach very high specificity. Moreover, our data clearly show that the major component facilitating DNA membrane translocation is the organization of plasmid DNA into plasmid chromatin with naturally occurring histones, as we can deliver plasmid DNA with and without chromatin assembly to target cells with comparable efficiency and specificity but only plasmid chromatin mediates high ratios of transgene expressing cells. In contrast to previous observations, we could not observe that histone assembly affects DNA uptake by unspecific membrane binding and we could demonstrate that plasmid chromatin facilitates membrane translocation and nuclear DNA transport without further engineering of histone proteins (42,53). As we could not observe major differences in antibody mediated DNA or chromatin cell surface binding and internalization, DNA transduction at the cell surface as well as a specific vesicular escape mechanism cannot be excluded and might also not be the only mechanism behind the improved nuclear translocation in line with previous suggestions (41,42,53). Furthermore, DNA compaction and charge reduction may contribute to the facilitated DNA membrane translocation and also the transition from the cytoplasm into the nucleus might be altered by histones as previously suggested (88,53,89,90). Further studies like measuring the cytoplasmic to nuclear transition, as performed for oligonucleotides for example, are necessary to uncover the exact role of Histone assembly on plasmid DNA and this modular system can contribute to its further understanding (91).
Beside the fact of being non-toxic and highly specific, the developed gene delivery system exclusively consists of proteins and peptides of mammalian origin. Thus, the concerns about safety and the risk of immunogenicity are expected to be rather low. However, one concern of systemic application and exposure to the immune system is the chromatin itself. It was shown that plasmid DNA with bacterial DNA sequences might be recognized by immune cells via Toll like receptor interaction (89,92). But this hurdle might be solved via plasmid DNA engineering like the production of mini-circles or mini-vectors containing only a minor portion of bacterial sequences (93). In addition, extracellularly occurring histones are elevated in various autoimmune diseases but are also considered in potentially mediating inflammatory diseases (94). On the other hand extracellular chromatin release is an active mechanism of neutrophils to bind bacteria and therefore serve as a trap for gram positive as well as gram negative bacteria (95). Furthermore, in our system histones might not be completely exposed to the immune system as they are shielded by the wrapped DNA as well as the associated antibody to some extent. All in all, further studies have to be performed to investigate the potential risk of systemic chromatin delivery.
In conclusion, we have developed a novel system to deliver plasmid DNA with viral-like efficiency, high specificity and without cytotoxicity exclusively by mammalian entities. However, further studies are necessary for example to understand the exact mechanism of nuclear chromatin and in particular the translocation mechanism over the membrane barrier. Nevertheless, antibody mediated chromatin targeting is a novel approach for specific gene delivery with the potential of being a viable alternative to existing targeted gene delivery systems.
Supplementary Material
ACKNOWLEDGEMENTS
Tobias Killian is a member of the Regensburg International Graduate School of Life Sciences (RIGeL), support and valuable discussions with Prof. Dr Gernot Längst and with Prof. Dr Reinhard Sterner as supervisor are gratefully acknowledged.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Roche Postdoc Fund (RPF – Targeted Therapy) position (to A.B., in part). Funding for open access charge: Roche Pharma Research & Early Development, Large Molecule Research, Roche Innovation Center Munich, Penzberg, Germany [VAT de.143.840.627].
Conflict of interest statement. T.K., A.B., T.H., H.S., O.M. and U.B. are employed by Roche Pharma Research and Early Development. Roche is interested in targeted therapies.
REFERENCES
- 1. Mulligan R.C. The basic science of gene therapy. Science. 1993; 260:926–932. [DOI] [PubMed] [Google Scholar]
- 2. Maeder M.L., Gersbach C.A.. Genome-editing technologies for gene and cell therapy. Mol. Ther. 2016; 24:430–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kumar S.R., Markusic D.M., Biswas M., High K.A., Herzog R.W.. Clinical development of gene therapy: results and lessons from recent successes. Mol. Ther. Methods Clin. Dev. 2016; 3:16034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Dunbar C.E., High K.A., Joung J.K., Kohn D.B., Ozawa K., Sadelain M.. Gene therapy comes of age. Science. 2018; 359:eaan4672. [DOI] [PubMed] [Google Scholar]
- 5. Kaufmann K.B., Buning H., Galy A., Schambach A., Grez M.. Gene therapy on the move. EMBO Mol. Med. 2013; 5:1642–1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gowing G., Svendsen S., Svendsen C.N.. Ex vivo gene therapy for the treatment of neurological disorders. Prog. Brain Res. 2017; 230:99–132. [DOI] [PubMed] [Google Scholar]
- 7. Hernandez-Alcoceba R., Poutou J., Ballesteros-Briones M.C., Smerdou C.. Gene therapy approaches against cancer using in vivo and ex vivo gene transfer of interleukin-12. Immunotherapy. 2016; 8:179–198. [DOI] [PubMed] [Google Scholar]
- 8. Xue H.Y., Zhang X., Wang Y., Xiaojie L., Dai W.J., Xu Y.. In vivo gene therapy potentials of CRISPR-Cas9. Gene Ther. 2016; 23:557–559. [DOI] [PubMed] [Google Scholar]
- 9. Yao J., Fan Y., Li Y., Huang L.. Strategies on the nuclear-targeted delivery of genes. J. Drug Target. 2013; 21:926–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Johnson-Saliba M., Jans D.A.. Gene therapy: optimising DNA delivery to the nucleus. Curr. Drug Targets. 2001; 2:371–399. [DOI] [PubMed] [Google Scholar]
- 11. Nayerossadat N., Maedeh T., Ali P.A.. Viral and nonviral delivery systems for gene delivery. Adv. Biomed. Res. 2012; 1:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yin H., Kanasty R.L., Eltoukhy A.A., Vegas A.J., Dorkin J.R., Anderson D.G.. Non-viral vectors for gene-based therapy. Nat. Rev. Genet. 2014; 15:541–555. [DOI] [PubMed] [Google Scholar]
- 13. Kay M.A. State-of-the-art gene-based therapies: the road ahead. Nat. Rev. Genet. 2011; 12:316–328. [DOI] [PubMed] [Google Scholar]
- 14. Nayak S., Herzog R.W.. Progress and prospects: immune responses to viral vectors. Gene Ther. 2010; 17:295–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Thomas C.E., Ehrhardt A., Kay M.A.. Progress and problems with the use of viral vectors for gene therapy. Nat. Rev. Genet. 2003; 4:346–358. [DOI] [PubMed] [Google Scholar]
- 16. Li H., Malani N., Hamilton S.R., Schlachterman A., Bussadori G., Edmonson S.E., Shah R., Arruda V.R., Mingozzi F., Wright J.F. et al.. Assessing the potential for AAV vector genotoxicity in a murine model. Blood. 2011; 117:3311–3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wright J.F. Manufacturing and characterizing AAV-based vectors for use in clinical studies. Gene Ther. 2008; 15:840–848. [DOI] [PubMed] [Google Scholar]
- 18. Yang N. Nonviral gene delivery system. Int. J. Pharm. Investig. 2012; 2:97–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mingozzi F., High K.A.. Therapeutic in vivo gene transfer for genetic disease using AAV: progress and challenges. Nat. Rev. Genet. 2011; 12:341–355. [DOI] [PubMed] [Google Scholar]
- 20. Lock M., McGorray S., Auricchio A., Ayuso E., Beecham E.J., Blouin-Tavel V., Bosch F., Bose M., Byrne B.J., Caton T. et al.. Characterization of a recombinant adeno-associated virus type 2 reference standard material. Hum. Gene Ther. 2010; 21:1273–1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Boulaiz H., Marchal J.A., Prados J., Melguizo C., Aranega A.. Non-viral and viral vectors for gene therapy. Cell Mol. Biol. (Noisy-le-grand). 2005; 51:3–22. [PubMed] [Google Scholar]
- 22. Kizewski A., Ilies M.A.. Efficient and synergetic DNA delivery with pyridinium amphiphiles-gold nanoparticle composite systems having different packing parameters. Chem. Commun. (Camb.). 2016; 52:60–63. [DOI] [PubMed] [Google Scholar]
- 23. Waehler R., Russell S.J., Curiel D.T.. Engineering targeted viral vectors for gene therapy. Nat. Rev. Genet. 2007; 8:573–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Munch R.C., Janicki H., Volker I., Rasbach A., Hallek M., Buning H., Buchholz C.J.. Displaying high-affinity ligands on adeno-associated viral vectors enables tumor cell-specific and safe gene transfer. Mol. Ther. 2013; 21:109–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ginn S.L., Amaya A.K., Alexander I.E., Edelstein M., Abedi M.R.. Gene therapy clinical trials worldwide to 2017: an update. J. Gene Med. 2018; 20:e3015. [DOI] [PubMed] [Google Scholar]
- 26. Grigsby C.L., Leong K.W.. Balancing protection and release of DNA: tools to address a bottleneck of non-viral gene delivery. J. R. Soc. Interface. 2010; 7(Suppl. 1):S67–S82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mann A., Thakur G., Shukla V., Singh A.K., Khanduri R., Naik R., Jiang Y., Kalra N., Dwarakanath B.S., Langel U. et al.. Differences in DNA condensation and release by lysine and arginine homopeptides govern their DNA delivery efficiencies. Mol. Pharm. 2011; 8:1729–1741. [DOI] [PubMed] [Google Scholar]
- 28. Speir J.A., Johnson J.E.. Nucleic acid packaging in viruses. Curr. Opin. Struct. Biol. 2012; 22:65–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Huang L., Li S.. Liposomal gene delivery: a complex package. Nat. Biotechnol. 1997; 15:620–621. [DOI] [PubMed] [Google Scholar]
- 30. Morachis J.M., Mahmoud E.A., Sankaranarayanan J., Almutairi A.. Triggered rapid degradation of nanoparticles for gene delivery. J. Drug Deliv. 2012; 2012:291219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yang Y., Zhao H., Jia Y., Guo Q., Qu Y., Su J., Lu X., Zhao Y., Qian Z.. A novel gene delivery composite system based on biodegradable folate-poly (ester amine) polymer and thermosensitive hydrogel for sustained gene release. Sci. Rep. 2016; 6:21402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lee D., Lee Y.M., Kim J., Lee M.K., Kim W.J.. Enhanced tumor-targeted gene delivery by bioreducible polyethylenimine tethering EGFR divalent ligands. Biomater. Sci. 2015; 3:1096–1104. [DOI] [PubMed] [Google Scholar]
- 33. Mann K., Kullberg M.. Trastuzumab-targeted gene delivery to Her2-overexpressing breast cancer cells. Cancer Gene Ther. 2016; 23:221–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Moffatt S., Papasakelariou C., Wiehle S., Cristiano R.. Successful in vivo tumor targeting of prostate-specific membrane antigen with a highly efficient J591/PEI/DNA molecular conjugate. Gene Ther. 2006; 13:761–772. [DOI] [PubMed] [Google Scholar]
- 35. Ge Z., Chen Q., Osada K., Liu X., Tockary T.A., Uchida S., Dirisala A., Ishii T., Nomoto T., Toh K. et al.. Targeted gene delivery by polyplex micelles with crowded PEG palisade and cRGD moiety for systemic treatment of pancreatic tumors. Biomaterials. 2014; 35:3416–3426. [DOI] [PubMed] [Google Scholar]
- 36. Xiao P.J., Samulski R.J.. Cytoplasmic trafficking, endosomal escape, and perinuclear accumulation of adeno-associated virus type 2 particles are facilitated by microtubule network. J. Virol. 2012; 86:10462–10473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Xu Y., Szoka F.C. Jr. Mechanism of DNA release from cationic liposome/DNA complexes used in cell transfection. Biochemistry. 1996; 35:5616–5623. [DOI] [PubMed] [Google Scholar]
- 38. Liang W., Lam J.K.W.. Ceresa B. Molecular Regulation of Endocytosis. 2012; IntechOpen; 429–456. [Google Scholar]
- 39. Peterson C.L., Laniel M.A.. Histones and histone modifications. Curr. Biol. 2004; 14:R546–R551. [DOI] [PubMed] [Google Scholar]
- 40. Rosenbluh J., Hariton-Gazal E., Dagan A., Rottem S., Graessmann A., Loyter A.. Translocation of histone proteins across lipid bilayers and Mycoplasma membranes. J. Mol. Biol. 2005; 345:387–400. [DOI] [PubMed] [Google Scholar]
- 41. Rosenbluh J., Singh S.K., Gafni Y., Graessmann A., Loyter A.. Non-endocytic penetration of core histones into petunia protoplasts and cultured cells: a novel mechanism for the introduction of macromolecules into plant cells. Biochim. Biophys. Acta. 2004; 1664:230–240. [DOI] [PubMed] [Google Scholar]
- 42. Wagstaff K.M., Glover D.J., Tremethick D.J., Jans D.A.. Histone-mediated transduction as an efficient means for gene delivery. Mol. Ther. 2007; 15:721–731. [DOI] [PubMed] [Google Scholar]
- 43. Hariton-Gazal E., Rosenbluh J., Graessmann A., Gilon C., Loyter A.. Direct translocation of histone molecules across cell membranes. J. Cell Sci. 2003; 116:4577–4586. [DOI] [PubMed] [Google Scholar]
- 44. Han H., Yang J., Chen W., Li Q., Yang Y., Li Q.. A comprehensive review on histone-mediated transfection for gene therapy. Biotechnol. Adv. 2018; 37:132–144. [DOI] [PubMed] [Google Scholar]
- 45. Alipour M., Hosseinkhani S., Sheikhnejad R., Cheraghi R.. Nano-biomimetic carriers are implicated in mechanistic evaluation of intracellular gene delivery. Sci. Rep. 2017; 7:41507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Cheraghi R., Nazari M., Alipour M., Majidi A., Hosseinkhani S.. Development of a targeted anti-HER2 scFv chimeric peptide for gene delivery into HER2-Positive breast cancer cells. Int. J. Pharm. 2016; 515:632–643. [DOI] [PubMed] [Google Scholar]
- 47. Dai F.H., Chen Y., Ren C.C., Li J.J., Yao M., Han J.S., Gong Y., Yang S.L., Zhu J.D., Gu J.R.. Construction of an EGF receptor-mediated histone H1(0)-based gene delivery system. J. Cancer Res. Clin. Oncol. 2003; 129:456–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Puebla I., Esseghir S., Mortlock A., Brown A., Crisanti A., Low W.. A recombinant H1 histone-based system for efficient delivery of nucleic acids. J. Biotechnol. 2003; 105:215–226. [DOI] [PubMed] [Google Scholar]
- 49. Balicki D., Reisfeld R.A., Pertl U., Beutler E., Lode H.N.. Histone H2A-mediated transient cytokine gene delivery induces efficient antitumor responses in murine neuroblastoma. Proc. Natl. Acad. Sci. U.S.A. 2000; 97:11500–11504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Schneeweiss A., Buyens K., Giese M., Sanders N., Ulbert S.. Synergistic effects between natural histone mixtures and polyethylenimine in non-viral gene delivery in vitro. Int. J. Pharm. 2010; 400:86–95. [DOI] [PubMed] [Google Scholar]
- 51. Jung H.J., Hwang D.S., Wei Q.D., Cha H.J.. Carassius auratus-originated recombinant histone H1 C-terminal peptide as gene delivery material. Biotechnol. Prog. 2008; 24:17–22. [DOI] [PubMed] [Google Scholar]
- 52. Hatefi A., Karjoo Z., Nomani A.. Development of a recombinant multifunctional biomacromolecule for targeted gene transfer to prostate cancer cells. Biomacromolecules. 2017; 18:2799–2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wagstaff K.M., Fan J.Y., De Jesus M.A., Tremethick D.J., Jans D.A.. Efficient gene delivery using reconstituted chromatin enhanced for nuclear targeting. FASEB J. 2008; 22:2232–2242. [DOI] [PubMed] [Google Scholar]
- 54. Rhodes D., Laskey R.A.. Assembly of nucleosomes and chromatin in vitro. Methods Enzymol. 1989; 170:575–585. [DOI] [PubMed] [Google Scholar]
- 55. Mayer K., Baumann A.L., Grote M., Seeber S., Kettenberger H., Breuer S., Killian T., Schafer W., Brinkmann U.. TriFabs–Trivalent IgG-Shaped bispecific antibody derivatives: Design, generation, characterization and application for targeted payload delivery. Int. J. Mol. Sci. 2015; 16:27497–27507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Jerabek-Willemsen M., André T., Wanner R., Roth H.M., Duhr S., Baaske P., Breitsprecher D.. MicroScale Thermophoresis: Interaction analysis and beyond. J. Mol. Struct. 2014; 1077:101–113. [Google Scholar]
- 57. Metz S., Haas A.K., Daub K., Croasdale R., Stracke J., Lau W., Georges G., Josel H.P., Dziadek S., Hopfner K.P. et al.. Bispecific digoxigenin-binding antibodies for targeted payload delivery. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:8194–8199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Killian T., Dickopf S., Haas A.K., Kirstenpfad C., Mayer K., Brinkmann U.. Disruption of diphthamide synthesis genes and resulting toxin resistance as a robust technology for quantifying and optimizing CRISPR/Cas9-mediated gene editing. Sci. Rep. 2017; 7:15480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Dyer P.N., Edayathumangalam R.S., White C.L., Bao Y., Chakravarthy S., Muthurajan U.M., Luger K.. Reconstitution of nucleosome core particles from recombinant histones and DNA. Methods Enzymol. 2004; 375:23–44. [DOI] [PubMed] [Google Scholar]
- 60. Langst G., Bonte E.J., Corona D.F., Becker P.B.. Nucleosome movement by CHRAC and ISWI without disruption or trans-displacement of the histone octamer. Cell. 1999; 97:843–852. [DOI] [PubMed] [Google Scholar]
- 61. Patterton H.G., von Holt C.. Negative supercoiling and nucleosome cores. I. The effect of negative supercoiling on the efficiency of nucleosome core formation in vitro. J. Mol. Biol. 1993; 229:623–636. [DOI] [PubMed] [Google Scholar]
- 62. Langst G. Preparation of chromatin templates to study RNA polymerase I transcription In Vitro. Methods Mol. Biol. 2016; 1455:109–119. [DOI] [PubMed] [Google Scholar]
- 63. Haas A.K., Maisel D., Adelmann J., von Schwerin C., Kahnt I., Brinkmann U.. Human-protein-derived peptides for intracellular delivery of biomolecules. Biochem. J. 2012; 442:583–593. [DOI] [PubMed] [Google Scholar]
- 64. Schneider B., Grote M., John M., Haas A., Bramlage B., Ickenstein L.M., Jahn-Hofmann K., Bauss F., Cheng W., Croasdale R. et al.. Targeted siRNA delivery and mRNA knockdown mediated by bispecific Digoxigenin-binding antibodies. Mol. Ther. Nucleic Acids. 2012; 1:e46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Dengl S., Sustmann C., Brinkmann U.. Engineered hapten-binding antibody derivatives for modulation of pharmacokinetic properties of small molecules and targeted payload delivery. Immunol. Rev. 2016; 270:165–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Marschall A.L., Zhang C., Frenzel A., Schirrmann T., Hust M., Perez F., Dubel S.. Delivery of antibodies to the cytosol: debunking the myths. MAbs. 2014; 6:943–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Ibraheem D., Elaissari A., Fessi H.. Gene therapy and DNA delivery systems. Int. J. Pharm. 2014; 459:70–83. [DOI] [PubMed] [Google Scholar]
- 68. Mann A., Shukla V., Khanduri R., Dabral S., Singh H., Ganguli M.. Linear short histidine and cysteine modified arginine peptides constitute a potential class of DNA delivery agents. Mol. Pharm. 2014; 11:683–696. [DOI] [PubMed] [Google Scholar]
- 69. Schatzlein A.G. Targeting of Synthetic gene delivery systems. J. Biomed. Biotechnol. 2003; 2003:149–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Varkouhi A.K., Scholte M., Storm G., Haisma H.J.. Endosomal escape pathways for delivery of biologicals. J. Control Release. 2011; 151:220–228. [DOI] [PubMed] [Google Scholar]
- 71. Cervia L.D., Chang C.C., Wang L., Yuan F.. Distinct effects of endosomal escape and inhibition of endosomal trafficking on gene delivery via electrotransfection. PLoS One. 2017; 12:e0171699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Sanders N., Rudolph C., Braeckmans K., De Smedt S.C., Demeester J.. Extracellular barriers in respiratory gene therapy. Adv. Drug. Deliv. Rev. 2009; 61:115–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Dengl S., Hoffmann E., Grote M., Wagner C., Mundigl O., Georges G., Thorey I., Stubenrauch K.G., Bujotzek A., Josel H.P. et al.. Hapten-directed spontaneous disulfide shuffling: a universal technology for site-directed covalent coupling of payloads to antibodies. FASEB J. 2015; 29:1763–1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Kontermann R.E., Brinkmann U.. Bispecific antibodies. Drug Discov. Today. 2015; 20:838–847. [DOI] [PubMed] [Google Scholar]
- 75. McCombs J.R., Owen S.C.. Antibody drug conjugates: design and selection of linker, payload and conjugation chemistry. AAPS J. 2015; 17:339–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Veldwijk M.R., Fruehauf S., Schiedlmeier B., Kleinschmidt J.A., Zeller W.J.. Differential expression of a recombinant adeno-associated virus 2 vector in human CD34+ cells and breast cancer cells. Cancer Gene Ther. 2000; 7:597–604. [DOI] [PubMed] [Google Scholar]
- 77. Seth P., Brinkmann U., Schwartz G.N., Katayose D., Gress R., Pastan I., Cowan K.. Adenovirus-mediated gene transfer to human breast tumor cells: an approach for cancer gene therapy and bone marrow purging. Cancer Res. 1996; 56:1346–1351. [PubMed] [Google Scholar]
- 78. Parker L.P., Wolf J.K., Price J.E.. Adenoviral-mediated gene therapy with Ad5CMVp53 and Ad5CMVp21 in combination with standard therapies in human breast cancer cell lines. Ann. Clin. Lab. Sci. 2000; 30:395–405. [PubMed] [Google Scholar]
- 79. Krishnamachary B., Glunde K., Wildes F., Mori N., Takagi T., Raman V., Bhujwalla Z.M.. Noninvasive detection of lentiviral-mediated choline kinase targeting in a human breast cancer xenograft. Cancer Res. 2009; 69:3464–3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Lucas A., Kremer E.J., Hemmi S., Luis J., Vignon F., Lazennec G.. Comparative transductions of breast cancer cells by three DNA viruses. Biochem. Biophys. Res. Commun. 2003; 309:1011–1016. [DOI] [PubMed] [Google Scholar]
- 81. May T., Gleiter S., Lilie H.. Assessment of cell type specific gene transfer of polyomavirus like particles presenting a tumor specific antibody Fv fragment. J. Virol. Met. 2002; 105:147–157. [DOI] [PubMed] [Google Scholar]
- 82. Rhodes C.A., Pei D.. Bicyclic peptides as next-generation therapeutics. Chemistry. 2017; 23:12690–12703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Galaway F.A., Stockley P.G.. MS2 Viruslike particles: a robust, semisynthetic targeted drug delivery platform. Mol. Pharmaceutics. 2013; 10:59–68. [DOI] [PubMed] [Google Scholar]
- 84. Wolff J.A., Rozema D.B.. Breaking the bonds: non-viral vectors become chemically dynamic. Mol. Ther. 2008; 16:8–15. [DOI] [PubMed] [Google Scholar]
- 85. McCaskill J., Singhania R., Burgess M., Allavena R., Wu S., Blumenthal A., McMillan N.A.. Efficient biodistribution and gene silencing in the lung epithelium via intravenous liposomal delivery of siRNA. Mol. Ther. Nucleic Acids. 2013; 2:e96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Novo L., Mastrobattista E., van Nostrum C.F., Lammers T., Hennink W.E.. Decationized polyplexes for gene delivery. Expert Opin. Drug Deliv. 2015; 12:507–512. [DOI] [PubMed] [Google Scholar]
- 87. Mokhtarzadeh A., Parhiz H., Hashemi M., Ayatollahi S., Abnous K., Ramezani M.. Targeted gene delivery to MCF-7 cells using Peptide-Conjugated polyethylenimine. AAPS PharmSciTech. 2015; 16:1025–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Hariton-Gazal E., Rosenbluh J., Graessmann A., Gilon C., Loyter A.. Direct translocation of histone molecules across cell membranes. J. Cell Sci. 2003; 116:4577–4586. [DOI] [PubMed] [Google Scholar]
- 89. Demirhan I., Hasselmayer O., Chandra A., Ehemann M., Chandra P.. Histone-mediated transfer and expression of the HIV-1 tat gene in Jurkat cells. J. Hum. Virol. 1998; 1:430–440. [PubMed] [Google Scholar]
- 90. Balicki D., Putnam C.D., Scaria P.V., Beutler E.. Structure and function correlation in histone H2A peptide-mediated gene transfer. Proc. Natl. Acad. Sci. U.S.A. 2002; 99:7467–7471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Buntz A., Killian T., Schmid D., Seul H., Brinkmann U., Ravn J., Lindholm M., Knoetgen H., Haucke V., Mundigl O.. Quantitative fluorescence imaging determines the absolute number of locked nucleic acid oligonucleotides needed for suppression of target gene expression. Nucleic Acids Res. 2019; 47:953–969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Spies B., Hochrein H., Vabulas M., Huster K., Busch D.H., Schmitz F., Heit A., Wagner H.. Vaccination with plasmid DNA activates dendritic cells via Toll-like receptor 9 (TLR9) but functions in TLR9-deficient mice. J. Immunol. 2003; 171:5908–5912. [DOI] [PubMed] [Google Scholar]
- 93. Hardee C.L., Arevalo-Soliz L.M., Hornstein B.D., Zechiedrich L.. Advances in Non-Viral DNA vectors for gene therapy. Genes (Basel). 2017; 8:65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Chen R., Kang R., Fan X.G., Tang D.. Release and activity of histone in diseases. Cell Death Dis. 2014; 5:e1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Brinkmann V., Reichard U., Goosmann C., Fauler B., Uhlemann Y., Weiss D.S., Weinrauch Y., Zychlinsky A.. Neutrophil extracellular traps kill bacteria. Science. 2004; 303:1532–1535. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.