Abstract
Biomedical applications of nucleic acid aptamers are limited by their rapid degradation in biological fluids and generally demand tedious post-selection modifications that might compromise binding. One possible solution to warrant biostability is to directly evolve chemically modified aptamers from xenobiotic nucleic acids (XNAs). We have isolated fully modified 2′-O-methyl-ribose–1,5-anhydrohexitol nucleic acid (MeORNA–HNA) aptamers targeting the rat vascular endothelial growth factor 164 (rVEGF164). Three sequences have been identified that interact with the target protein with affinities in the low-nanomolar range and HNA modifications appeared to be mandatory for their tight binding. The evolution of these XNA aptamers was accomplished using an in vitro selection procedure starting from a fully sugar-modified library containing a 20mer 2′-OMe-ribonucleotide region followed by a 47mer HNA sequence. The high binding affinity and selectivity of the selected aptamers were confirmed by several methods including gel-shift, fluorescence polarisation, and enzyme-linked oligonucleotide assays. The isolated HNA ligands exhibited higher specificity to the rVEGF164 and human VEGF165 isoforms compared to rat VEGF120, while very low binding efficiencies were observed to streptavidin and thrombin. Furthermore, it was clearly demonstrated that the resulting aptamers possessed a superior stability to degradation in human serum and DNase I solutions.
INTRODUCTION
Aptamers are short nucleic acid molecules capable of binding with high affinity to a specific target. Over the last three decades, they have been proven as useful tools in different research fields, such as nanotechnology, medicinal chemistry, bioanalysis, and synthetic biology (1–3). Aptamer selection is performed using an in vitro screening method known as Systematic Evolution of Ligands by EXponential enrichment (SELEX) (1,2). The simplicity of this method has allowed for the generation of a myriad of RNA or DNA aptamers against different targets ranging from ions and small molecules to proteins and even whole cells (3).
However, despite the widespread production of aptamers, their effective use as drugs and biosensors still faces a major challenge with regard to their rapid degradation by cellular nucleases (4,5). The introduction of suitable chemical variations in the sugar-phosphate backbone of natural biopolymers has been suggested to prevent enzymatic hydrolysis. These include for instance various modifications at the 2′-OH position of RNA, which led to the anti-human VEGF165 aptamer Pegaptanib (or Macugen), whose backbone alternates 2′-F-pyrimidines and 2′-OMe-purine nucleotides added as in- and post-SELEX modifications, respectively (6,7). The first direct in vitro selection of fully 2′-OMe-modified aptamers binding to human neutrophil elastase has been reported by using engineered thermostable DNA polymerases for the synthesis and transcription of 2′-OMe-modified libraries (8).
One more recent approach that has been introduced for the selection of enzymatically stable aptamers starts from xenobiotic nucleic acid (XNA) libraries (9). Since XNAs significantly differ in their backbone structure from natural nucleic acids, they are inherently more resistant to cellular nucleases. The enzymatic production and enrichment of XNA libraries has become viable owing to the directed evolution of specialized XNA polymerases able to synthesize XNA sequences from a DNA template and then transcribe it back into DNA (10–12).
However, so far only a limited number of chemistries have been demonstrated to be compatible with the in vitro evolution of XNA ligands by the direct in-SELEX method. Specifically, a 2′-deoxy-2′-fluoroarabino nucleic acid (FANA)-based aptamer has been described with affinity for HIV-1 reverse transcriptase (13) along with threose nucleic acid (TNA) aptamers to human thrombin (14), HIV-1 reverse transcriptase (15) as well as the small-molecule ochratoxin A (16). Another highly promising example of XNA molecule compatible with in-SELEX was obtained by replacing the furanose ring of DNA with a six-membered carbohydrate sugar moiety. The resulting 1,5-anhydrohexitol (hexitol or HNA, Figure 1) nucleic acid can form stable duplexes with DNA and RNA, while possessing a remarkable nuclease (Turbo DNase I, 37°C, 2 h), acid (pH 1, 40°C, 3 h), and alkaline (pH 13.9, 65°C, 1 h) stability in vitro as well as low toxicity of the monomers in cell cultures (9,10,17–19). Two HNA aptamers against the HIV trans-activation response RNA (TAR) and hen egg lysozyme (HEL) have been previously selected using partial and entirely random HNA libraries with dissociation constants (KD) in the nanomolar range (10).
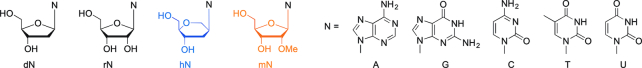
Figure 1.
Chemical structures of the natural deoxyribo (dN) and ribo (rN) nucleosides as well as modified nucleosides (hN – hexitol nucleosides, mN - 2′-OMe-ribonucleosides) investigated in this study.
In this study, we successfully accomplished the selection of three structurally unique 2′-O-methyl-ribose–1,5-anhydrohexitol nucleic acid (MeORNA–HNA) aptamers against the rat VEGF164 target protein. These ligands were demonstrated to bind to rVEGF164 with low-nanomolar affinity by virtue of the HNA/2′-OMe modifications. Herein, we describe in detail the successful SELEX process for the screening of high-affinity aptamers from a completely random sugar-modified MeORNA-HNA pool (HNA-SELEX), including their synthesis, specificity, binding, and affinity measurements by enzyme-linked oligonucleotide (ELONA), electrophoretic mobility shift (EMSA), and fluorescence polarisation (FP) assays. These HNA aptamers are the first examples of anti-VEGF aptamers with an alternative sugar unit in their backbone displaying a remarkable stability against DNase I and in human serum. Furthermore, one of the selected MeORNA–HNA aptamers, i.e. aptamer 2–21, exhibited moderate inhibitory activity as shown by its ability to diminish VEGF-induced tissue factor expression in human umbilical vein endothelial cells (HUVEC). The results of this study are expected to guide the future design and development of aptamer-based diagnostics, biosensors, and therapeutics using sugar modified XNAs as both in vitro and in vivo biomolecular tools.
MATERIALS AND METHODS
Materials
Deoxyribonucleoside triphosphates (dNTPs), Taq DNA polymerase, 10× ThermoPol buffer, MgSO4, Lambda exonuclease, 10× Lambda exonuclease reaction buffer, 10× DNase I reaction buffer and 6× purple gel loading dye were purchased from New England Biolabs. Turbo DNase I, magnetic beads (Dynabeads M-280 Streptavidin), SYBR Gold nucleic acid gel stain, CloneJet PCR cloning kit, Micro BCA protein assay kit, the biotinylation reagent EZ-Link Sulfo-NHS-LC-Biotin, Biotin Quantitation Kit (HABA, Pierce) as well as the chemiluminescent (SuperSignal West Pico or Femto) and calorimetric (1-Step Ultra TMB ELISA) HRP substrates were also purchased from ThermoFisher Scientific. The PCR purification kit was obtained from Macherey-Nagel (NucleoSpin Gel and PCR clean-up kit). Antibodies used in the immunodetection assays were a polyclonal rabbit anti-VEGFA antibody (Abcam), HRP-conjugated polyclonal anti-FITC antibody (ThermoFisher Scientific), HRP-conjugated streptavidin, and HRP-conjugated polyclonal anti-rabbit IgG (Jackson Immuno Research). All unmodified oligodeoxyribonucleotides and 2′-OMe-Pr2 were purchased from Integrated DNA Technologies (IDT, Leuven). Pol6G12_521L HNA polymerase and Pol521L_664K HNA reverse transcriptase expression vectors were kindly provided by Dr P. Holliger (MRC Laboratory of Molecular Biology, Cambridge, UK) and Dr V. Pinheiro (KU Leuven, Belgium) (10). The polymerases were expressed and stored in aliquots at −80°C. Human umbilical vein endothelial cells (HUVEC) were provided by the Laboratory of Virology and Chemotherapy (KU Leuven, Belgium). The rVEGF164 target was expressed in the lab of Prof. P. Carmeliet (VIB, KU Leuven, Belgium). Streptavidin and human α-thrombin were purchased from ThermoFisher Scientific. Human VEGF165 and rat VEGF120 were obtained from Bio-Connect Life Science. All other chemicals were obtained either from VWR or Sigma-Aldrich.
Methods
Hexitol nucleoside triphosphate (hNTP) synthesis as well as oligonucleotide and rVEGF164 target preparation are described in detail in the Supplementary Data section.
MeORNA–HNA library synthesis and preparation
The synthesis of MeORNA–HNA oligonucleotides was performed using a 67-nucleotide randomized DNA template (Lib25) and 2′-OMe-FAM-riboligonucleotide primer (2′-OMe-Pr2, Supplementary Table S1). Each reaction (100 μl) contained 1 μl (41.7 μM) of pol6G12_521L HNA polymerase, 3 μM of primer, 1 μM of DNA template, 125 μM of each hNTP, 4 mM of MgSO4 (final concentration), and 1× ThermoPol reaction buffer. Primer and template were melted at 95°C for 5 min, followed by annealing upon slow cooling to room temperature (RT) prior to addition of HNA polymerase, hNTPs, and MgSO4. The reaction mixtures were first incubated at 40°C for 10 min and then at 65°C for 1–24 h (9 h was optimal). Several reactions were performed to obtain a sufficient amount of material for the in vitro selection procedure. After synthesis, 2.5 μl (5 units) of Turbo DNase I and 11 μl of 10× DNase I reaction buffer were added to the reactions, which were incubated at 37°C for 2 h (0–12 h for optimization), followed by inactivation of Turbo DNase I at 80°C for 15 min. The reaction mixtures were subsequently purified using a NucleoSpin clean-up kit according to the manufacturer's recommendations and quantified by 15% PAGE with fluorescein-tagged primer standards (0.156–10 pmol of 2′-OMe-Pr2).
MeORNA–HNA aptamer synthesis and preparation
The synthesis of the selected MeORNA–HNA aptamers was performed as described above, with the exception that 1 μM of the complementary DNA templates (Supplementary Table S1 in the Supplementary Data) was used with either FAM-tagged 2′-OMe-Pr2 (ELONA1, FP and EMSA) or biotin-tagged Bio-2′-OMe-Pr2 (ELONA2) primers. The reaction mixtures were incubated first at 40°C for 10 min, then at 65°C for 3 h, and finally at 85°C for 2 min. The thermocycle was repeated three times. After hydrolysis by Turbo DNase I, the reaction mixtures were purified by 15% denaturing PAGE as described above in order to isolate the aptamer sequences.
Preparation of the selection and negative selection matrixes
For the selection matrix preparation, 100 μl (1 mg) of streptavidin-coated magnetic beads (MB) were resuspended and transferred to a new 1.5 ml Eppendorf tube, washed three times with 100 μl of 1× PBS, and incubated with 12 μl of bio-rVEGF164 for 1 h at RT under shaking at 1000 rpm (Supplementary Table S2). Subsequently, the beads were washed 4–5 times with 100 μl of PBS-T and blocked with 100 μl of PBS-T–5% BSA for 30 min at RT and 1000 rpm. Finally, the magnetic beads were washed three times with 100 μl PBS-T, and the resulting material was then used for the in vitro selection. For the negative selection matrix preparation, 100 μl of magnetic beads was blocked by 100 μl of PBS-T–5% BSA buffer and washed several times with 100 μl of PBS-T, as described above, without incubation with the rVEGF164 protein.
Selection of MeORNA–HNA aptamers
For anti-rVEGF164 aptamer selection, the MeORNA–HNA library was dissolved in the selection buffer (SB buffer, Supplementary Table S2) and renatured (95°C for 5 min followed by slow cooling to 8°C and equilibration to RT). For the first two rounds of selection 500 pmol of the MeORNA–HNA library in 100 μl SB was incubated with 100 μl (1 mg) of negative selection matrix (magnetic beads in SB without target) for 30 min at RT and 1100 rpm; bead-binding sequences were discarded, and the supernatant was used for the next selection step with ∼160 pmol of biotinylated rVEGF164 targets immobilized on magnetic beads (selection matrix). The MeORNA–HNA library was incubated with the selection matrix for 1 h at RT upon shaking at 1100 rpm, followed by two washing steps with 100 μl of WB and shaking for 5 min at RT and 1100 rpm after each washing step. The binding sequences were eluted with 100 μl of elution buffer (EB, Supplementary Table S2) upon incubation of the magnetic beads for 10 min at 90°C and shaking at 1100 rpm. The eluted aptamers were purified from the salt, target, and buffer components by using a NucleoSpin clean-up kit (with 400 μl NTC buffer and then two times elution from the column with 25 μl H2O). The samples were analysed after each step of each selection round by 15% denaturing PAGE using FAM-labelled 2′-OMe-Pr2 as standard. The gel was scanned using the Typhoon 9500 imaging system (Cy2-channel) and quantified by the ImageQuant TL v8.1 software (both from GE Healthcare Life Science). Selection stringency was increased by increasing the number, volume, and timing of the washing steps as well as the molar ratio of the MeORNA–HNA library to the target from ∼3:1 (1–2 rounds, 2 washing steps) to 6:1 (3–5 rounds, two washing steps) and 9:1 (6–7 rounds, three washing steps).
The eluted MeORNA–HNA sequences were lyophilized and reverse transcribed into DNA using 1 μM of Cy5-labeled DNA primer (Cy5-Pr1), 500 μM of dNTPs, approximately 10–100 nM of the selected MeORNA–HNA aptamers, and 0.5 μl of pol521L_664K reverse transcriptase (10) in 20 μl of 1 × ThermoPol buffer with 4 mM of MgSO4. Primer and MeORNA–HNA template were annealed before addition of the reverse transcriptase at 95°C for 5 min, followed by slow cooling to RT and the mixture was incubated at 65°C for 6–9 h (normally 9 h). Each 0.5–1 μl of reverse transcription products was PCR amplified in 20 μl of 1× ThermoPol buffer with 0.5 μM of primers (5′-phosphorylated reverse P-Pr2 primer and Pr1), 200 μM of dNTPs, and 25 U/mL of Taq DNA polymerase. The samples were first denatured at 95°C for 1 min, followed by 10 cycles of repetitive denaturation at 95°C for 30 s, annealing at 60°C for 30 s, extension at 68°C for 30 s, and final extension at 68°C for 5 min. The PCR reactions were repeated 1–2 times to reach 500–1000 pmol of dsDNA product (≤200 fmol of library per PCR tube). The final PCR product was collected and each 200 μl PCR was digested by 100 U/ml of lambda exonucleases (λexo) in 1× Lambda Exonuclease Reaction Buffer at 37°C for 1 h, followed by λexo inactivation at 80°C for 15 min. The reactions were ethanol precipitated, diluted with 100 μl H2O, and purified by using a NucleoSpin clean-up kit and 50 μl of water (2 × 25 μl). The samples were quantified, and the quality was checked by 15% denaturing PAGE. The resulting sequences were either used for further synthesis as described above followed by a selection round (up to seventh SELEX round) or used for cloning, sequencing, and analysis.
The selected oligonucleotides, recovered from the seventh round, were cloned into the pJET1.2 plasmid using the CloneJet PCR cloning kit according to the manufacturer's recommendations. The plasmid DNAs were isolated using the MiniPrep DNA isolation kit and sequenced. The aptamer sequences were analysed and grouped according to their conservative motifs.
Sequence analysis
The sequence analysis was performed using both the Jalview and EMBL-EBI software with either the Clustal W or Clustal Omega sequence alignment algorithm. The folding into secondary structures was generated using the online Mfold program and the default settings for RNA or DNA at 37°C. The appropriate DNA oligonucleotide templates for some recovered sequences were ordered and the synthesis of MeORNA–HNA material was performed as described above.
Magnetic beads-based binding test of the selected aptamers
Members of groups 1, 2 and 4–6 (Supplementary Table S3) were analysed after MeORNA–HNA synthesis and purification. Thus, 20 pmol of each renatured FAM-labelled individual sample in 10 μl of SB buffer was incubated with 10 μl of negative selection matrix for 30 min at RT and 1100 rpm. Then, the supernatant was incubated with 10 μl of the selection matrix (with 18 pmol target) for 30 min at RT and 1100 rpm. The resulting magnetic beads were washed two times with 10 μl of WB buffer for 5 min at RT and 1100 rpm; the samples were then eluted with 10 μl of EB buffer after 10 min incubation at 90°C and 1100 rpm. All samples were loaded on 15% PAGE using the FAM-labeled 2′-OMe-Pr2 standard (0.078–10 pmol). Gels were scanned as described above with the Cy2 channel. Aptamer binding was determined according to the relation [El/I0]aptamer/[El/I0]initial Lib25, where El and I0 are the amounts of eluted and initial material, respectively, measured for each aptamer and normalized to the values of the initial MeORNA–HNA library (Lib25). Next, the best 10 sequences were chosen for subsequent analysis by EMSA, FP, and ELONA assays.
Electrophoretic mobility shift assay (EMSA)
In this assay, 50 μl of the total reaction volume containing 25 nM of renatured FAM-labelled MeORNA–HNA aptamers or control solutions without or with 100 nM of rVEGF164 in SB was incubated for 2 h at RT upon shaking at 300 rpm. After incubation, 10 μl of 6× purple gel loading dye was added to the samples and 12 μl of the final solutions was run on a 6% native polyacrylamide gel with 0.5× TBE (running buffer) at 150 V and ∼18°C until the purple tracking dye reached  of the gel (∼ 3.5 h). The gel was scanned using the Typhoon 9500 imaging system (Cy2-channel) and quantified with the ImageQuant TL v8.1 software (both from GE Healthcare Life Science).
of the gel (∼ 3.5 h). The gel was scanned using the Typhoon 9500 imaging system (Cy2-channel) and quantified with the ImageQuant TL v8.1 software (both from GE Healthcare Life Science).
Fluorescence polarisation assay (FP)
In this assay, 20 nM of FAM-tagged aptamer and control solutions was first renatured in SB and then 25 μl was added to each well (black plates) containing either 25 μl of 800 nM of rVEGF164 in SB (400 nM final concentration in 50 μl total volume) or 25 μl of SB only (S0). The plate was sealed with an aluminium adhesive foil seal and the samples were incubated for 2 h at RT upon shaking at 300 rpm. After incubation, the plate was scanned on a ClarioStar microplate reader (BMG Labtech, Isogen Life Science) using the 482–16, LP504 and 530–40 filters. Focus and gain were adjusted before measurements (using a sample without target). During scanning, 100 flashes with 0.2 s setting time were applied to each well. Wells containing 50 μl of SB were used as background (BGR). The relative aptamer binding abilities were calculated as follows: Binding = [(F482-16 aptamer – F482-16 BGR)/(F530-40 aptamer – F530-40 BGR)] – [(F482-16 S0 – F482-16 BGR)/(F530-40 S0 – F530-40 BGR)] and normalized by the binding of the initial MeORNA–HNA Lib25.
Enzyme-linked oligonucleotide assay (ELONA)
To measure the specificity and binding affinity of the selected aptamers to proteins by ELONA, two different approaches were used (ELONA1 and 2, see Supplementary Table S4 for more details). In ELONA1, the target rVEGF164 and different competitors (thrombin, streptavidin, rVEGF120, and hVEGF165) were immobilized on microtiter plates via adsorption. The microplates covered by 50 μl (per well) of 0.5 or 1 μg/ml protein solutions in PBS were incubated overnight on ice upon gentle rotation, washed 4 times with 125 μl of WB, and blocked by 125 μl of BB for 2 h at RT upon gentle rotation. The wells were then washed 4 times with 125 μl of WB for 5 min, and 50 μl of the renatured FAM-aptamer or control solutions was added, followed by incubation for 1 h or overnight on ice upon gentle rotation. For specificity tests, 25 nM of aptamer/control samples was used. For KD measurements by ELONA, serial dilutions of 200 or 1000 nM aptamers/controls were employed. After incubation, the plates were washed five times with 125 μl of WB for 5 min at RT upon gentle rotation. The anti-FITC HRP-conjugated antibody was applied (1:3333 dilution in WB) for 2 h at RT upon rotation. The microplates were washed as described above, dried on paper, and 50 μl of 1-Step Ultra TMB ELISA was added to each well and incubated for 30 min at RT in the dark. The reactions were stopped by addition of 50 μl of 2 M H2SO4 and the protein-bound-aptamer complexes were quantified by measuring the absorbance at 450 and 570 nm (background) using ClarioStar (BMG Labtech, Isogen Life Science).
In ELONA2, pre-blocked NeutrAvidin coated (deglycosylated form of avidin) plates were used. The plates were pre-washed four times with 200 μl of WB for 5 min at RT upon gentle rotation. Biotinylated aptamers and controls were renatured, then 100 μl of 50 nM of each oligonucleotide sample was added to the wells. The plates were incubated overnight on ice upon gentle rotation and then washed four times as described above. Serial dilutions of 1000 nM rVEGF164 (100 μl) were added to the corresponding well, and the target solutions were incubated overnight at RT upon gentle rotation. After extensive washing, 100 μl of the rabbit anti-VEGFA antibody (1:500 dilution) together with the HRP-conjugated anti-rabbit-IgG antibody (1:2500) was added. Each antibody was incubated for 1 h upon rotation at RT and washed four times with WB for 5 min. The 1-Step Ultra TMB ELISA HRP substrate (100 μl) was used for detection as described above for ELONA1.
Stability of unmodified and modified aptamers in whole human serum
The 5′-FAM-MeORNA–HNA aptamers, their unmodified counterparts, as well as V7t1 and Macugen control aptamers (0.1 μM each) were incubated in 95% human serum at 37°C with a 100 μl total reaction volume. Aliquots (10 μl) of each reaction were removed at different time intervals (0, 1, 3, 6, 24, 30, 48 and 72 h or up to 7 days for Macugen and MeORNA-HNA 2–21), quenched by adding 20 μl of 2× gel loading dye, denatured at 95°C for 10 min, and kept at −20°C before analysis by denaturing 15% PAGE. Samples were visualized using the Typhoon 9500 imaging system (Cy2-channel).
Cellular inhibitory assay
The inhibitory effect of aptamers was evaluated by a VEGF-induced tissue factor (TF) expression assay as previously described (20,21). Specifically, HUVECs (100 000 cells/mL per well) were cultured in EBM-2 (Endothelial Cell Basal Medium, LONZA) in 24-well plates (Costar, Corning) at 37°C, under a 5% CO2 atmosphere. The cells were washed twice with OPTI-MEM (Gibco), and pre-incubated in OPTI-MEM (0.5 ml/well) for 2 h. The medium was then aspirated, and 0.5 ml of OPTI-MEM containing 0.3 nM hVEGF165 and constant 0.6 (or 1 nM) or a range of aptamer concentrations (0.1–100 nM) was added to the corresponding well. After 1 h incubation, the total RNA was isolated by using a RNeasy Mini Kit (QIAGEN). The obtained cDNAs were prepared by reverse transcription, using OligoT primer (Supplementary Table S1), M-MLV Reverse Transcriptase (Life Technology), and 100 ng of each mRNA sample. The reverse transcription products (1 μl) were used as templates for end-point PCR with Taq DNA polymerase and Real-Time PCR analysis with PowerUp SYBR Green Master Mix qPCR kit (Applied Biosystems) using 0.5 μM TF or actin primers specific for human TF or β-actin mRNAs, respectively (Supplementary Table S1). The expression of β-actin was used as reference, and the relative TF mRNA expression level was quantified using the 2−ΔΔCt comparative method. In each plate, the experiment was repeated two or four times under the same conditions, and the relative TF mRNA expression was calculated by comparison with that obtained in the presence of hVEGF165 and without any aptamer (100% expression). The mean values with standard deviations were taken as the final result.
RESULTS
MeORNA–HNA library synthesis
To define the functional potential of XNAs, we focused our interest on the in vitro evolution of sugar-modified hexitol nucleic acid (HNA, Figure 1) (22) due to its chemical and enzymatic stability along with the ability to form stable duplexes and quadruplexes (23,24).
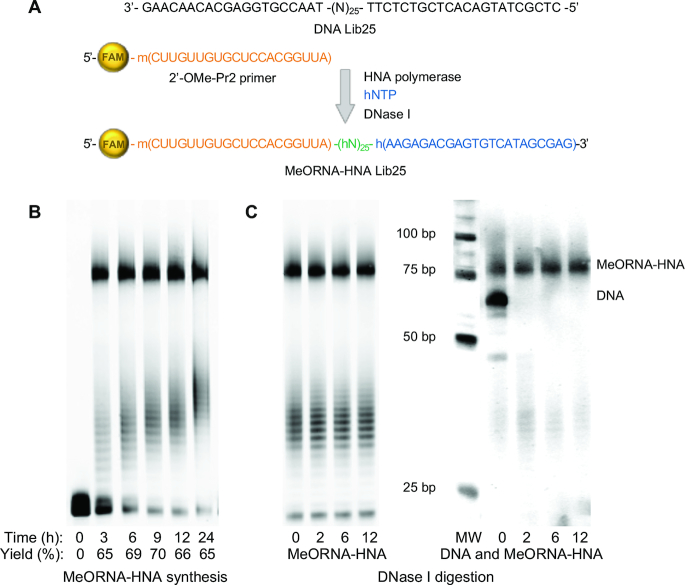
The MeORNA–HNA hybrid library used for this study was synthesized using primer extension reactions directed by a DNA template (Lib25) containing a random region made of 25 nucleotides (nt) annealed to a FAM-labeled 2′-OMe-RNA primer, in the presence of hexitol nucleoside triphosphates (hNTPs) containing the four natural nucleobases. A specialized variant of the TgoT DNA polymerase from Thermococcus gorgonarius, namely Pol6G12_521L HNA polymerase, was used as catalyst (10). This genetically engineered DNA-dependent HNA polymerase is able to generate full-length HNA products using DNA templates and either DNA or 2′-OMe-RNA primers. The resulting FAM-labelled MeORNA–HNA library consisted of 20 2′-OMe-ribonucleotides at the 5′ end, a central 25 mer random hexitol sequence, and 22 fixed hexitol nucleotides at the 3′ end (Figure 2A).
Figure 2.
MeORNA–HNA hybrid library synthesis. (A) A DNA oligonucleotide containing a 25 nt random region flanked by two fixed sequences at the 5′ (22 nt) and 3′ (20 nt) ends was used as template. The MeORNA–HNA hybrid library was produced using a 2′-OMe-Pr2 primer, hNTPs, and specialised DNA-depended HNA polymerase (TgoT Pol6G12_521L). The initial DNA template was digested by DNase I and the resulting ss MeORNA–HNA library was purified and subjected to the in vitro selection process. (B) Final MeORNA–HNA products after synthesis resolved in 15% PAGE. (C) Products after incubation with DNase I. After 15% PAGE scanned with Cy2 channel, only the FAM-labelled MeORNA–HNA product is visible (left); after staining with SYBR Gold, both the MeORNA–HNA hybrid and DNA are visible (right). MW stands for molecular weight marker.
The library synthesis conditions were optimized to gain the maximum yield of the MeORNA–HNA product, which is necessary to avoid the loss of enriched molecules throughout the in vitro selection (Figure 2B). The optimal reaction time and temperature were determined to be 9 h and 65°C, respectively, thus yielding 70% of the MeORNA–HNA product.
It has been reported that synthetic HNA molecules are completely resistant to degradation by DNA nucleases (17,18). Therefore, it was envisaged that the synthesized MeORNA–HNA hybrid library would not undergo detectable enzymatic degradation, allowing for an expedient and simple method to achieve the separation of the desired MeORNA–HNA product from the initial DNA template. Accordingly, it was found that upon treatment with highly processive Turbo DNase I, HNA remained stable even after 12 h incubation, while DNA was completely hydrolysed within 2 h (Figure 2C).
MeORNA–HNA aptamer selection procedure
For this study, we chose the rat vascular endothelial growth factor 164 (rVEGF164) as model target protein, since anti-rVEGF164 aptamers can be employed in well-established in vitro and in vivo assays. Furthermore, this protein has a high similarity with the therapeutically relevant human VEGF165, for which various DNA and RNA aptamers have been successfully selected (Supplementary Table S5, (6,7,25–27)). During the selection process, bound and unbound sequences were separated by exploiting the streptavidin-biotin interaction using streptavidin-coated magnetic beads (28,29). This method requires the biotinylation of the target protein, which was performed using a 3-molar excess of biotinylation reagent to rVEGF164. The success of the biotinylation reaction was confirmed by western blot and HABA assays (Supplementary Figure S1).
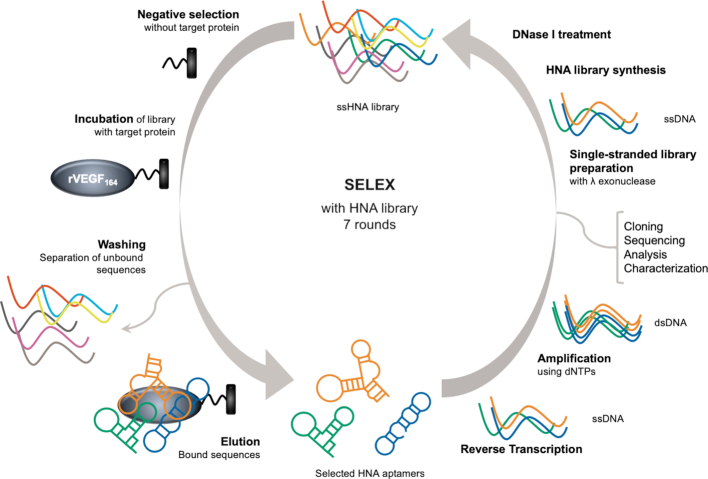
The previously prepared MeORNA–HNA library composed of approximately 1014 sequences was used as starting point for the selection process. In order to prevent unspecific interactions of the library with the streptavidin and magnetic beads support, we introduced a step of negative selection before each round of SELEX (Figure 3). During this step, the streptavidin-magnetic beads without immobilized protein were incubated with the MeORNA–HNA library, and the supernatant was then added to the selection matrix with the immobilized biotinylated rVEGF164. After extensive washing steps, the selected sequences were eluted from the magnetic beads. The amount of eluted MeORNA–HNA material was estimated using as standards the FAM-labelled 2′-OMe primer loaded in the gel together with the library after each step of the selection round (Supplementary Figure S2). The eluted sequences were further transcribed back into DNA by a specific HNA-dependent DNA polymerase (Pol521L_664K reverse transcriptase) (10). Transcripts were PCR amplified using natural DNA primers and deoxynucleoside triphosphates. The PCR was repeated 1–2 times to obtain a sufficient amount (0.5–1 μmol) of DNA library for the next selection round.
Figure 3.
Schematic representation of the in vitro selection of MeORNA–HNA aptamers. The initial library is incubated with streptavidin-coated magnetic beads first without and later with the immobilized biotinylated rVEGF164. Unbound sequences are removed by extensive washing, while the bound molecules are eluted, reverse transcribed, PCR amplified, and subjected to the next selection round.
For the preparation of the single-stranded DNA templates required for HNA synthesis, we performed the hydrolysis of the 5′-phosphorylated strand with Lambda exonuclease (λexo) (30,31). In order to avoid unspecific degradation of the non-phosphorylated strand, the optimal incubation time of the λexo enzyme with the library was determined (Supplementary Figure S3). Full digests were already obtained after 60 min in the presence of 0.2 U/μl of λexo with no unspecific hydrolysis of the non-phosphorylated single-stranded sequence occurring under these conditions. The resulting ss DNA sequences were used once again as templates in HNA synthesis reactions in the presence of the specific HNA polymerase and subjected to the next selection round (Figure 3).
Aptamers binding
After seven rounds of selection, the enriched MeORNA–HNA library was transcribed, amplified, and used for cloning and sequencing. After recovering 54 clones, the aptameric sequences were classified in 12 groups (including an unsorted and a G-rich group, Supplementary Table S3), according to the sequence analysis of the random regions and conserved motifs. Some sequences underwent several deletions or insertions in the random region during the selection process. It is likely that such errors are due to the use of genetically engineered HNA polymerase and reverse transcriptase with high error rates (10). The random regions of the oligonucleotides contained 17–27 nt instead of 25 nt, and only one sequence appeared more than once (clones 3–17 and 3–23).
Five aptamer groups showing sequence similarity (Group No. 1, 2, 4–6, for a total of 36 sequences) were chosen for the subsequent analysis of their target binding properties. For this purpose, the corresponding DNA templates of the MeORNA–HNA aptameric sequences were chemically synthesized and employed for aptamer synthesis using FAM-labeled 2′-OMe-Pr2.
To determine the affinity of the selected aptamers, we first used a magnetic bead binding assay. The selected MeORNA–HNA sequences were incubated with an equimolar ratio of immobilized rVEGF164, washed, and the amount of eluted sequences was then analysed. This procedure is similar to the in vitro selection from the random library described above and the same selection buffers were employed. The eluted amount of material was compared with that of the initial random pool and the sequences binding more efficiently (10 sequences, Table 1 (MB)) were further investigated.
Table 1.
Binding analysis of the selected MeORNA–HNA aptamers
| Bindingb (%) | |||||||
|---|---|---|---|---|---|---|---|
| Aptamer | Random region sequencea | Size of random | Group | MB | EMSA | FP | ELONAc |
| 1–18 | AAATGCGGGGGTGGCTAGTGTGTGTGC | 27 | 2 | 114 | 63 | 148 | 95 |
| 1–32 | TGATGAATGTGGGTGTGAATTATGGT | 26 | 2 | 121 | 39 | 156 | 63 |
| 2–8 | ATCCACACACAAACTAATCAGCATG | 25 | 6 | 114 | 197 | 140 | 133 |
| 2–15 | CACACGTGATACGGATATGGTTAG | 24 | 4 | 116 | 153 | 182 | 125 |
| 2–18 | GTAACCGACACGGATTGAGTTGATG | 25 | 4 | 115 | 167 | 79 | 61 |
| 2–21 | ACTAACGAGCGTACATGTGCATTG | 24 | 2 | 131 | 308 | 108 | 127 |
| 3–5 | ATATGCGTGTTTATGTGTAAGTGT | 24 | 5 | 105 | 77 | 134 | 70 |
| 3–7 | TGAAACGCAATCTAGAGAGAAT | 22 | 6 | 101 | 58 | 130 | 53 |
| 4–6 | TATTGCGGCATTACTGGTACCATG | 24 | 2 | 120 | 268 | 115 | 131 |
| 4–19 | ATGTATACGAGCCGCAATGACCT | 23 | 2 | 107 | 80 | 131 | 121 |
| Lib25 | NNNNNNNNNNNNNNNNNNNNNNNNN | 25 | - | 100 | 100 | 100 | 100 |
| V7t1d | TGTGGGGGTGGACGGGCCGGGTAGA | 25 | - | - | 50 | 81 | 192 |
aAll aptameric sequences can be found in Supplementary Table S3.
bMean values of the relative binding abilities of the selected aptamers compared to that of the initial MeORNA–HNA library (Lib25, taken as 100%). MB - magnetic bead binding assay; EMSA – electrophoretic mobility shift assay; FP – fluorescence polarization; ELONA – enzyme-linked oligonucleotide assay.
cELONA1 analysis was performed with rVEGF164 immobilized on a plate.
dAptamer to human VEGF165 from (32) with a KD of 1.4 nM as measured by SPR.
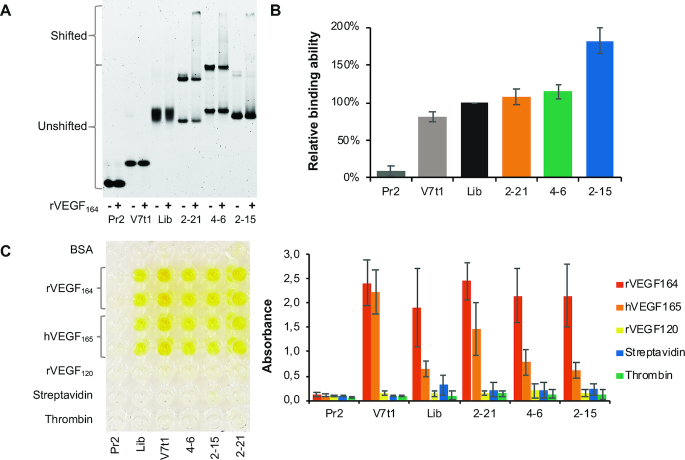
Specifically, we analysed the 10 best binding sequences by EMSA; this assay allowed to separate the free FAM-aptamers from the aptamer–rVEGF164 complexes by native PAGE (Supplementary Figure S4 and Table 1). The binding ability of these MeORNA–HNA aptamers was compared with those of the initial library, 2′-OMe-RNA primer (Pr2), and unmodified DNA aptamer V7t1 with a known nM-affinity to hVEGF165 (32). From Figure 4A, it is evident that MeORNA–HNA chimeras 2–21, 4–6 and 2–15 formed stable dimeric structures, as indicated by the presence of two distinguishable bands on the native gel and possessed a higher binding to rVEGF164 than the original library. Furthermore, the binding abilities of the 10 selected aptamers were confirmed by fluorescence polarisation studies (Figure 4B, Supplementary Figure S5, and Table 1). The FP of each fluorescently labelled aptamer was measured in the absence or presence of rVEGF164 and compared to the FP values of the library, Pr2 and V7t1. All aptamers except for 2–18 and V7t1 exhibited a higher binding affinity to the target protein than the initial library.
Figure 4.
Binding analysis of the selected MeORNA–HNA aptamers. (A) Electrophoretic gel mobility analysis (EMSA) of the FAM-labelled MeORNA–HNA aptamers (2–21, 4–6, and 2–15) and control sequences, i.e., 2′-OMe-Pr2, DNA V7t1, and MeORNA–HNA Lib25 at a 25 nM concentration after 2 h incubation without (−) or with (+) 100 nM rVEGF164 followed by separation by 6% native PAGE. The positions of the shifted and upshifted material are indicated. (B) Fluorescence polarisation assay (FP) with 10 nM FAM-labelled aptamers after 2 h incubation with 400 nM rVEGF164. The analysis was performed 6 times. (C) Determination of the specificity of anti-rVEGF164 aptamers using ELONA. An example of ELONA specificity experiment (left) and the binding abilities of the aptamers (right) are shown. The target rVEGF164 as well as the protein competitors (BSA, hVEGF165, rVEGF120, streptavidin and thrombin) at the same concentration of 0.5 μg/mL were incubated with the 25 nM FAM-labeled aptamers and control oligonucleotide sequences. The oligonucleotide-protein complexes were detected using the polyclonal anti-FAM HPR conjugated antibody with a calorimetric HPR substrate. The absorbance was measured at 450 nm. Each experiment was performed 3–6 times. See Supplementary Figures S4–S7 for further details.
Aptamers specificity
Next, the selected aptamers were also examined with regard to their selectivity by using the immunoassay ELONA. This analysis was performed with the non-biotinylated rVEGF164 target as well as alternative competitors (e.g. streptavidin, human α-thrombin, human VEGF165 and rat VEGF120) and a negative control (selection buffer with 0.1% of BSA). Figure 4C and Supplementary Figure S7 show that aptamers 2–21, 4–6 and 2–15 demonstrated a tighter binding to the target protein rVEGF164 than the initial MeORNA–HNA library. These aptamers were also found to bind to the human VEGF165, while they did not interact with rVEGF120 or unrelated proteins such as streptavidin, thrombin and BSA. The capacity of binding both rVEGF164 and hVEGF165 but not the rVEGF120 isoform of the VEGFA protein suggested that the selected aptamers primarily recognise the heparin-binding domain (HBD) of the protein since rVEGF120 lacks HBD.
To evaluate the importance of the HNA backbone on the binding of the selected aptamers, the fully natural DNA variants of MeORNA–HNA aptamers 2–21, 4–6 and 2–15 were chemically synthesized and subjected to the ELONA assay together with their MeORNA–HNA counterparts (Supplementary Figure S8). In all cases, the DNA aptamer variants exhibited a 4- to 6-fold lower binding activity to rVEGF164 than their parent MeORNA–HNA aptamers. These results confirm that the HNA modification significantly contributes to the tight binding of the resulting XNA aptamers to their biological target.
Aptamer affinity
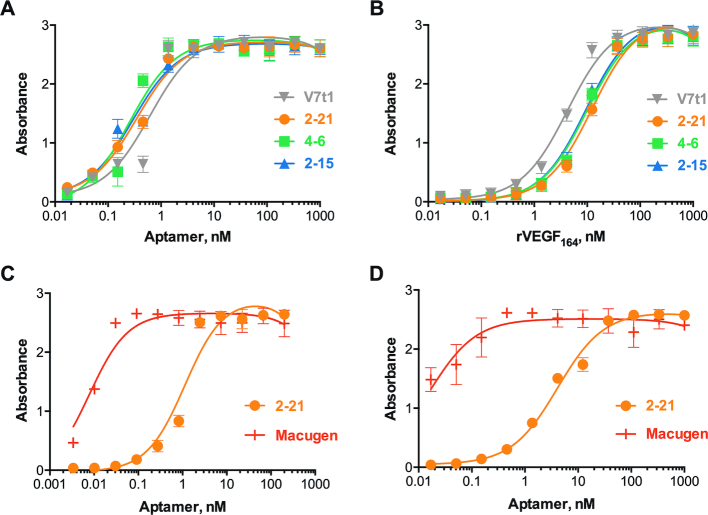
The three aptamers that demonstrated enhanced specificity and binding ability compared to the random pool, e.g., 2–21, 4–6 and 2–15, were further studied with regard to their affinity using different immunoassays. In particular, two ELONA-based assays were performed. One featured rVEGF164-coated plates with different concentrations of FAM-labeled aptamers (ELONA1), while the other made use of avidin-coated plates with immobilized biotinylated aptamers and different concentrations of the rVEGF164 target (ELONA2). The resulting binding affinities are presented in Figure 5A, B and Table 2. As reflected by these KD values, all three aptamers demonstrated affinities in the nanomolar range, i.e., 0.3–0.4 and 10–13 nM as measured by ELONA1 and ELONA2, respectively.
Figure 5.
Enzyme-linked oligonucleotide assays (ELONA) to determine the binding affinity of the selected aptamers. (A) ELONA1 assay was performed with rVEGF164 (1 μg/mL) coated plates and different concentrations of FAM-labeled aptamers. (B) ELONA2 assay was performed using avidin coated plates with immobilized biotinylated aptamers (50 nM) and different concentrations of rVEGF164 target. (C, D) Binding affinity comparison of MeORNA–HNA 2–21 and Macugen aptamers to either rVEGF164 (C) or hVEGF165 (D) at 1 μg/mL concentration. For more details see Supplementary Figures S9 and S11.
Table 2.
Affinity and activity of selected aptamers and controls
| K D measurements, nM | ||||
|---|---|---|---|---|
| ELONA1 | ELONA2 | |||
| Aptamer | rVEGF164 | hVEGF165 | rVEGF164 | IC50, nM |
| 2–15 | 0.29 ± 0.04 | 1.09 ± 0.09 | 9.6 ± 1.1 | n.d. |
| 2–21 | 0.35 ± 0.04 | 0.31 ± 0.03 | 12.6 ± 1.5 | 23.4 ± 7.8 |
| 4–6 | 0.25 ± 0.05 | 0.23 ± 0.03 | 10.0 ± 1.2 | n.d. |
| V7t1 | 0.62 ± 0.14 | 0.77 ± 0.17 | 4.3 ± 0.5 | n.d. |
| Macugena | 0.008 ± 0.01 | 0.016 ± 0.003 | n.d. | 15.7 ± 2.8 |
aAptamer to hVEGF165 from (6) with a KD = 50 pM.
We then explored the affinities of aptamers V7t1, 2–21, 4–6 and 2–15 to the human isoform of VEGF165 by ELONA1. The dissociation constant values of the selected aptamers to hVEGF165 resulted to be close to those measured against rVEGF164, and particularly ranging from 0.2 to 1.1 nM (Supplementary Figure S10). For comparison, we also tested the well-known 2′-OMe and 2′-F-substituted RNA aptamer Macugen, which has been shown to tightly bind hVEGF165 with a KD of 50 pM (6). The affinities of MeORNA–HNA 2–21 or Macugen to the rat and human VEGF targets were determined to be 1.1 and 4.1 nM or 8 and 16.4 pM, respectively (Figure 5C, D and Table 2).
Aptamers structure
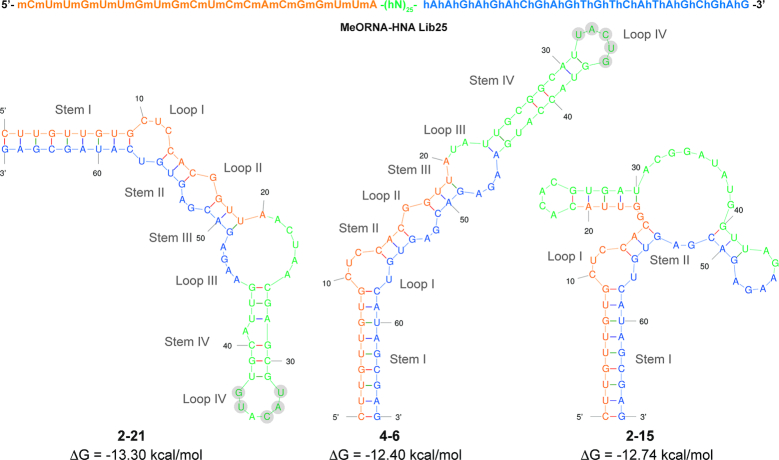
The potential for secondary structure formation of MeORNA–HNA sequences 2–21, 4–6 and 2–15 was assessed using the Mfold software according to pre-defined conditions (37°C). Since no software is currently available to predict the folding of HNA or any other XNA, we chose RNA as the closest folding form to that of the HNA cognate molecule. In fact, it was previously demonstrated that HNA sequences preferentially adopt RNA-like A-form helical conformations (23). However, it should be noted that the structures produced by Mfold may not be highly accurate and behave differently under our selection conditions. Figure 6 illustrates the predicted secondary structures of all four aptamers based on their lowest free energy (ΔG). As it can be seen, these aptamers displayed complex hairpin stem-loop secondary structures including 2–4 stem-loop regions. In particular, the structures of aptamers 2–21 and 4–6 shared analogous stem-loop patterns with a high similarity in loop IV. All possible structures for the selected aptamers obtained by using either the RNA or DNA folding form can be found in Supplementary Figures S12 and S13, respectively.
Figure 6.
Predicted secondary structures of the selected full-length MeORNA–HNA aptamers using the Mfold software. The RNA folding method was applied under standard conditions (37°C). The first 20 nt at the 5′-end correspond to the 2′-OMe-RNA primer sequence (orange) followed by a 24 nt random HNA region (green) and 22 nt 3′-HNA primer region (blue). Nucleotides in the random region (Loop IV) exhibiting high similarity among the different aptamers are highlighted in light grey circles. All possible structures are shown in Supplementary Figure S12 (RNA folding) and Supplementary Figure S13 (DNA folding).
Stability of MeORNA–HNA aptamers in human serum
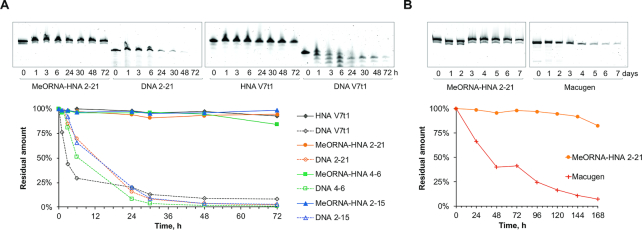
Subsequently, we tested the stability of the selected MeORNA–HNA aptamers and their corresponding DNA variants to nuclease degradation. As expected from the results obtained by previous DNase I enzymatic tests, the MeORNA–HNA aptamers did not show signs of nuclease digestion after 72 h of incubation at 37°C in 95% human serum (84–98% of the aptamers remained intact), while their DNA cognates as well as the V7t1 DNA aptamer were rapidly degraded within 24 h under the same conditions (Figure 7A). Furthermore, aptamer 2–21 was compared with Macugen by carrying out a stability assay in 95% whole human serum for up to 7 days at 37°C (Figure 7B). After seven days of incubation, 2′F/2′OMe-RNA Macugen was almost completely degraded (7% of full-length aptamer was detected), whereas aptamer MeORNA–HNA 2–21 was nearly intact (83%). It was reasoned that the partial hydrolysis (17%) of aptamer 2–21 might be due to the digestion of the FAM-2′-OMe-RNA region of the aptamer. These results are consistent with the known biological stability of HNA relative to DNA and RNA, which proves the significance of HNA biomolecules as convenient scaffolds for the development of biologically stable nucleic acid drugs (10,18).
Figure 7.
Nuclease resistance of aptamers in 95% human serum at 37°C. (A) Stability of MeORNA–HNA and DNA aptamer variants up to 72 h. (B) Comparison of the stability of MeORNA-HNA 2–21 and 2′F-/2′-OMe-RNA Macugen aptamers up to 7 days (168 h). Complete figures are shown in Supplementary Figures S14 and S15.
Inhibitory assay
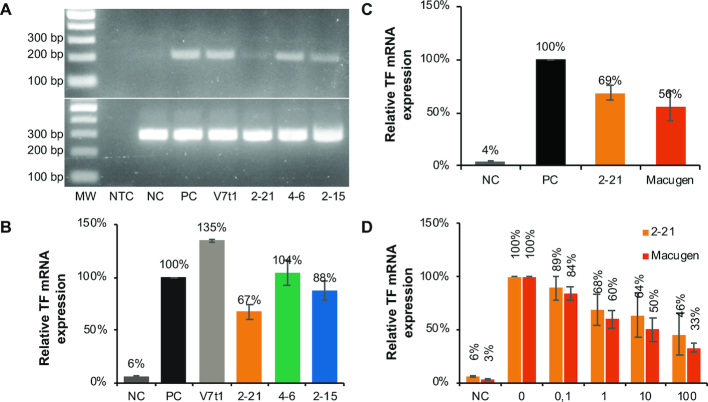
The inhibitory activity of MeORNA–HNA 2–21, 4–6 and 2–15 was evaluated in comparison with that of DNA V7t1 aptamer by a VEGF-induced tissue factor (TF) expression assay using HUVEC cells (20,21). It was found that MeORNA–HNA 2–21 and 2–15 hybrid sequences were able to reduce the mRNA expression level of TF by 33 ± 7% and 12 ± 9%, respectively, at a concentration of 0.6 nM (Figure 8A,,B). On the other hand, the MeORNA–HNA 4–6 and V7t1 aptamers slightly increased TF mRNA expression corresponding to 135 ± 1% and 104 ± 12%, respectively. We also determined the inhibitory effect of MeORNA–HNA 2–21 on TF mRNA expression in comparison with that of Macugen, which was found to be 44 ± 14% under identical conditions (Figure 8C).
Figure 8.
Inhibition of the interaction between hVEGF165 and its receptors by a range of aptamers. (A) PCR results after aptamer TF mRNA expression inhibition. The upper gel shows the detection of TF mRNA by PCR amplification, while the lower gel illustrates the β-actin mRNA expression (positive control). NTC stands for no template control, i.e., PCR without cDNA template. MW represents the 2-Log molecular weight ladder. (B) Relative TF mRNA expression levels in HUVEC treated with hVEGF165 in the presence or absence of each MeORNA–HNA aptamer and control V7t1 aptamer (0.6 nM). (C) Comparison of relative TF mRNA expression levels between MeORNA–HNA 2–21 and Macugen aptamers (1 nM). (D) Concentration-dependent inhibitory effect of different concentrations (0, 0.1, 1, 10 and 100 nM) of MeORNA-HNA 2–21 and Macugen aptamers added to HUVEC treated with hVEGF165. NC indicates the sample without hVEGF165 and aptamers, while PC is the sample with hVEGF165 (0.3 nM) but without aptamers.
Dose-response studies were further conducted using the same TF expression assay. Both MeORNA-HNA 2–21 and Macugen inhibited VEGF165-induced tissue factor mRNA expression in HUVECs in a concentration-dependent manner. Particularly, the concentration of each aptamer required to inhibit 50% of the hVEGF165 binding (IC50) was 23.4 ± 7.8 and 15.7 ± 2.8 nM for MeORNA-HNA 2–21 and Macugen, respectively (Figure 8D, Table 2).
DISCUSSION
Fully modified MeORNA–HNA hybrid aptamers against rVEGF164 have been evolved upon seven rounds of an in vitro selection procedure starting from an entirely modified library composed of two fixed 2′-OMe-RNA (20 nt) and HNA (22 nt) regions at the 5′- and 3′-end, respectively, together with a central random 25 mer HNA sequence. Their binding affinity to the target was measured by different methods, including homogenous (FP, EMSA) and heterogenous (ELONA) assays, with either the target or library immobilized on a support. Overall, these binding studies revealed that three aptamers among the selected functional sequences exhibited high affinity in the nanomolar range to the rVEGF164 molecule. Specifically, the KDs of the best aptamers fell in the 0.3–0.4 nM range, as determined by enzyme-linked oligonucleotide assay (ELONA1). The binding affinities of the selected XNA aptamers were compared to those of established unmodified DNA and 2′-substituted RNA aptamers, i.e., V7t1 and Macugen, respectively. The KD of V7t1 was comparable with those of the selected aptamers, while the KD of Macugen indicated a relatively higher binding affinity. On the other hand, the selected MeORNA–HNA chimeric aptamers showed a far superior biostability in human serum up to 7 days, whereas Macugen underwent complete degradation under the same experimental conditions. These findings suggest that the use of HNA aptamers might be particularly advantageous in direct assays and procedures involving biological fluids, cells, and serum, as well as for conducting in vivo experiments.
Notably, HNA modifications appear to be a prerequisite for high aptamer affinity. The target interactions and high-affinity binding was significantly decreased upon replacement of the unnatural MeORNA–HNA by a fully natural DNA sequence. The selected aptamers also demonstrated high specificity to the rat and human isoforms of VEGF, while no binding occurred to other targets such as rVEGF120, thrombin, streptavidin and BSA proteins. The biological effect of the selected aptamers on the target was further analysed in a cell culture assay by determining the variation of VEGF-stimulated TF mRNA expression in primary HUVECs. While aptamer MeORNA–HNA 2–21 reduced the VEGF activity by 33 ± 7%, the selected candidate MeORNA–HNA 4–6 led to an increased activity despite possessing closed structural elements and similar hVEGF165 binding activities. Strong binding aptamers may therefore modulate the activity of the target in both directions. Moreover, the results of this cell inhibitory assay suggested that under analogous conditions, MeORNA-HNA 2–21 inhibits TF mRNA expression in a dose-dependent manner and its inhibitory effect is comparable with that of Macugen (IC50 is 23.4 against 15.7 nM for Macugen).
In summary, with the present study we have established an efficient in vitro selection approach of HNA-based aptamers, which focuses on a direct selection from a fully modified library. Such HNA-SELEX led to the isolation of high-affinity aptamers characterized by low nanomolar KDs, high selectivity, and cellular activity. The resulting MeORNA–HNA sequences proved to be exceptionally nuclease-resistant (10,17,18). Strong sugar-modified binders with dissociation constants in the pM-nM range against alternative targets have been evolved from completely modified XNA libraries, which represent the HNA, TNA and FANA families of aptamers (10,13–16). Given the growing interest in XNA oligonucleotides as highly stable tools for diagnostics and therapy, the current study encourages to further investigate the evolution of XNA aptamers. The direct selection of HNA aptamers offers in fact a clear advantage over the post-SELEX modification of DNA and RNA aptamers.
The selected MeORNA–HNA hybrid aptamers represent the first example of XNA molecules targeting the therapeutically relevant VEGF protein, which is known for playing an important role in several pathological processes such as tumor growth, rheumatoid arthritis, and age-related macular degeneration (33,34). Several successful examples of anti-human VEGF165 aptamers have been generated in the past by other groups (Supplementary Table S5). However, all these aptamers are made either of 2′-substituted-RNA or DNA molecules, and no anti-VEGF aptamer comprising a sugar moiety other than ribose in the backbone structure has been reported so far. In view of the concurrent notable affinity and stability exhibited by the selected aptamers, HNA ligands could be finely tuned and ultimately developed into unique tools for use as therapeutics or biosensors. Thus, future work will focus on the optimisation of aptamer sequence and structure to create shorter and structurally more stable XNA aptamers for in vivo testing with improved affinity to VEGF. Furthermore, with a suitable HNA-SELEX protocol in hand (HNA library synthesis, reverse transcription, library optimization, detection, etc.), the selection of HNA-based aptamers will be extended to undruggable, toxic, and nonimmunogenic targets that are known to be inaccessible molecules for antibody generation.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Prof. Peter Carmeliet and Katleen Brepoels for providing the rVEGF164 target protein, Dr Philipp Holliger and Dr Vitor Pinheiro for providing the HNA polymerase and reverse transcriptase expression vectors. We would like to acknowledge the help of Erik Martens with the immunoassays. Authors are indebted to Prof. Jef Rozenski for mass spectrometry analysis, Dr Piotr Leonczak and Guy Schepers for the chemical synthesis of oligonucleotides and Chantal Biernaux for editorial help.
Notes
Present address: Lia Margamuljana, Agilent Technologies, Technologielaan 3, 3001 Heverlee, Belgium.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Fonds Wetenschappelijk Onderzoek (FWO, Flanders Research Foundation) [G.078014N to P.H., 12Q8619N to E.E.]; Research Fund KU Leuven [OT/14/128]; European Research Council under the European Union's Seventh Framework Program (FP7/2007–2013)/ERC [ERC-2012-ADG 20120216/320683]. Funding for open access charge: FWO [G.078014N and 12Q8619N].
Conflict of interest statement. None declared.
REFERENCES
- 1. Tuerk C., Gold L.. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990; 249:505–510. [DOI] [PubMed] [Google Scholar]
- 2. Ellington A.D., Szostak J.W.. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990; 346:818–822. [DOI] [PubMed] [Google Scholar]
- 3. Dunn M.R., Jimenez R.M., Chaput J.C.. Analysis of aptamer discovery and technology. Nat. Rev. Chem. 2017; 1:0076. [Google Scholar]
- 4. Taylor A.I., Arangundy-Franklin S., Holliger P.. Towards applications of synthetic genetic polymers in diagnosis and therapy. Curr. Opin. Chem. Biol. 2014; 22:79–84. [DOI] [PubMed] [Google Scholar]
- 5. Maier K.E., Levy M.. From selection hits to clinical leads: progress in aptamer discovery. Mol. Ther. Methods Clin. Dev. 2016; 5:16014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ruckman J., Green L.S., Beeson J., Waugh S., Gillette W.L., Henninger D.D., Claesson-Welsh L., Janjic N.. 2′-Fluoropyrimidine RNA-based aptamers to the 165-amino acid aorm of vascular endothelial growth factor (VEGF 165). J. Biol. Chem. 1998; 273:20556–20567. [DOI] [PubMed] [Google Scholar]
- 7. Ng E.W.M., Shima D.T., Calias P., Cunningham E.T., Guyer D.R., Adamis A.P.. Pegaptanib, a targeted anti-VEGF aptamer for ocular vascular disease. Nat. Rev. Drug Discov. 2006; 5:123–132. [DOI] [PubMed] [Google Scholar]
- 8. Liu Z., Chen T., Romesberg F.E.. Evolved polymerases facilitate selection of fully 2′-OMe-modified aptamers. Chem. Sci. 2017; 8:8179–8182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Herdewijn P., Marlière P.. Toward safe genetically modified organisms through the chemical diversification of nucleic acids. Chem. Biodivers. 2009; 6:791–808. [DOI] [PubMed] [Google Scholar]
- 10. Pinheiro V.B., Taylor A.I., Cozens C., Abramov M., Renders M., Zhang S., Chaput J.C., Wengel J., Peak-Chew S.-Y., McLaughlin S.H. et al.. Synthetic genetic polymers capable of heredity and evolution. Science. 2012; 336:341–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dunn M.R., Otto C., Fenton K.E., Chaput J.C.. Improving polymerase activity with unnatural substrates by sampling mutations in homologous protein architectures. ACS Chem. Biol. 2016; 11:1210–1219. [DOI] [PubMed] [Google Scholar]
- 12. Peng C.G., Damha M.J.. Polymerase-directed synthesis of 2′-deoxy-2′-fluoro-beta-D-arabinonucleic acids. J. Am. Chem. Soc. 2007; 129:5310–5311. [DOI] [PubMed] [Google Scholar]
- 13. Alves Ferreira-Bravo I., Cozens C., Holliger P., DeStefano J.J.. Selection of 2′-deoxy-2′-fluoroarabinonucleotide (FANA) aptamers that bind HIV-1 reverse transcriptase with picomolar affinity. Nucleic Acids Res. 2015; 43:9587–9599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yu H., Zhang S., Chaput J.C.. Darwinian evolution of an alternative genetic system provides support for TNA as an RNA progenitor. Nat. Chem. 2012; 4:183–187. [DOI] [PubMed] [Google Scholar]
- 15. Mei H., Liao J., Jimenez R.M., Wang Y., Bala S., McCloskey C., Switzer C., Chaput J.C.. Synthesis and evolution of a threose nucleic acid aptamer bearing 7-deaza-7-substituted guanosine residues. J. Am. Chem. Soc. 2018; 140:5706–5713. [DOI] [PubMed] [Google Scholar]
- 16. Rangel A.E., Chen Z., Ayele T.M., Heemstra J.M.. In vitro selection of an XNA aptamer capable of small-molecule recognition. Nucleic Acids Res. 2018; 46:8057–8068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Taylor A.I., Pinheiro V.B., Smola M.J., Morgunov A.S., Peak-Chew S., Cozens C., Weeks K.M., Herdewijn P., Holliger P.. Catalysts from synthetic genetic polymers. Nature. 2014; 518:427–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Taylor A.I., Beuron F., Peak-Chew S.-Y., Morris E.P., Herdewijn P., Holliger P.. Nanostructures from synthetic genetic polymers. ChemBioChem. 2016; 17:1107–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Verheggen I., Van Aerschot A., Toppet S., Snoeck R., Janssen G., Balzarini J., De Clercq E., Herdewijn P.. Synthesis and antiherpes virus activity of 1,5-anhydrohexitol nucleosides. J. Med. Chem. 1993; 36:2033–2040. [DOI] [PubMed] [Google Scholar]
- 20. Kimoto M., Nakamura M., Hirao I.. Post-ExSELEX stabilization of an unnatural-base DNA aptamer targeting VEGF165 toward pharmaceutical applications. Nucleic Acids Res. 2016; 44:7487–7494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lee J.-H., Canny M.D., De Erkenez A., Krilleke D., Ng Y.-S., Shima D.T., Pardi A., Jucker F.. A therapeutic aptamer inhibits angiogenesis by specifically targeting the heparin binding domain of VEGF165. Proc. Natl. Acad. Sci. U.S.A. 2005; 102:18902–18907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Vastmans K., Rozenski J., Van Aerschot A., Herdewijn P.. Recognition of HNA and 1,5-anhydrohexitol nucleotides by DNA metabolizing enzymes. Biochim. Biophys. Acta - Protein Struct. Mol. Enzymol. 2002; 1597:115–122. [DOI] [PubMed] [Google Scholar]
- 23. Herdewijn P. Nucleic acids with a six-membered ‘carbohydrate’ mimic in the backbone. Chem. Biodivers. 2010; 7:1–59. [DOI] [PubMed] [Google Scholar]
- 24. Zhou J., Abramov M., Liu F., Amrane S., Bourdoncle A., Herdewijn P., Mergny J.-L.. Effects of six-membered carbohydrate rings on structure, stability, and kinetics of G-quadruplexes. Chem. - A Eur. J. 2013; 19:14719–14725. [DOI] [PubMed] [Google Scholar]
- 25. Kimoto M., Yamashige R., Matsunaga K., Yokoyama S., Hirao I.. Generation of high-affinity DNA aptamers using an expanded genetic alphabet. Nat. Biotechnol. 2013; 31:453–457. [DOI] [PubMed] [Google Scholar]
- 26. Kanakaraj I., Chen W.-H., Poongavanam M., Dhamane S., Stagg L.J., Ladbury J.E., Kourentzi K., Strych U., Willson R.C.. Biophysical characterization of VEGF-aHt DNA aptamer interactions. Int. J. Biol. Macromol. 2013; 57:69–75. [DOI] [PubMed] [Google Scholar]
- 27. Potty A.S.R., Kourentzi K., Fang H., Jackson G.W., Zhang X., Legge G.B., Willson R.C.. Biophysical characterization of DNA aptamer interactions with vascular endothelial growth factor. Biopolymers. 2009; 91:145–156. [DOI] [PubMed] [Google Scholar]
- 28. Stoltenburg R., Reinemann C., Strehlitz B.. FluMag-SELEX as an advantageous method for DNA aptamer selection. Anal. Bioanal. Chem. 2005; 383:83–91. [DOI] [PubMed] [Google Scholar]
- 29. Mayer G. In vitro selection of ssDNA aptamers using biotinylated target proteins. Methods Mol. Biol. 2009; 535:19–32. [DOI] [PubMed] [Google Scholar]
- 30. Avci-Adali M., Paul A., Wilhelm N., Ziemer G., Wendel H.P.. Upgrading SELEX technology by using lambda exonuclease digestion for single-stranded DNA generation. Molecules. 2009; 15:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Marimuthu C., Tang T.-H., Tominaga J., Tan S.-C., Gopinath S.C.B.. Single-stranded DNA (ssDNA) production in DNA aptamer generation. Analyst. 2012; 137:1307–1315. [DOI] [PubMed] [Google Scholar]
- 32. Nonaka Y., Sode K., Ikebukuro K.. Screening and improvement of an Anti-VEGF DNA aptamer. Molecules. 2010; 15:215–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Carmeliet P. Angiogenesis in life, disease and medicine. Nature. 2005; 438:932–936. [DOI] [PubMed] [Google Scholar]
- 34. Jellinek D., Green L.S., Bell C., Janjic N.. Inhibition of receptor binding by high-affinity RNA ligands to vascular endothelial growth factor. Biochemistry. 1994; 33:10450–10456. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.