Abstract
Oxidation of the guanine (G) heterocycle to 8-oxo-7,8-dihydroguanine (OG) in mammalian gene promoters was demonstrated to induce transcription. Potential G-quadruplex forming sequences (PQSs) in promoters have a high density of G nucleotides rendering them highly susceptible to oxidation and possible gene activation. The VEGF PQS with OG or an abasic site were synthesized at key locations in the SV40 or HSV-TK model promoters to determine the location dependency in the gene expression profile in human cells. The PQS location with respect to the transcription start site (TSS) and strand of occupancy (coding versus non-coding strand) are key parameters that determine the magnitude and direction in which gene expression changes with the chemically modified VEGF PQS. The greatest impact observed for OG or F in the PQS context in these promoters was within ∼200 bp of the TSS. Established PQSs found to occur naturally in a similar location relative to the TSS for possible oxidation-induced gene activation include c-MYC, KRAS, c-KIT, HIF-1α, PDGF-A and hTERT. The studies provide experimental constraints that were used to probe bioinformatic data regarding PQSs in the human genome for those that have the possibility to be redox switches for gene regulation.
INTRODUCTION
Oxidative stress is characterized as a shift of the redox state toward oxidation that can result from increased production of reactive oxygen species (ROS) (1). The free radicals generated during oxidative stress are considered detrimental to biological processes by oxidizing biomolecules leading to mutations and inappropriate cellular activity (2–4); however, these findings represent only one piece of the overall picture regarding ROS. A growing body of evidence has found that ROS can function as signaling molecules and guide the cellular response to oxidative stress and inflammation (5–10). These free radicals may play an important signaling role for regulating cellular activity during oxidative or inflammatory stress to prevent long-term damage. This burgeoning field of study has many remaining questions for further inquiry.
One domain of this field has identified that ROS function to regulate cellular phenotype by invoking oxidative modification of promoter DNA sequences that lead to changes in mRNA levels (11–16). In an early study, Perillo et al. demonstrated that chromatin remodeling by LSD1 can activate transcription by delivering H2O2 to the promoter regions of estrogen-induced genes to effect oxidation of a guanine (G) heterocycle (14). Base excision repair (BER) of the oxidized G initiated activation of transcription. Their studies identified a coupling of oxidative modification of the genome yielding DNA repair-mediated induction of transcription.
Further developments in the field of promoter DNA oxidation and gene regulation have been made using the tools of biology. The Gillespie laboratory identified that hypoxia-induced oxidation of G within the G-rich control sequence of the VEGF promoter induces gene expression (15). Their proposed activation pathway also harnessed BER in the process. The Tell laboratory found that H2O2-mediated oxidation of the SIRT1 gene promoter led to gene activation and required the BER pathway (13). The proposed mechanism from Tell's laboratory identified a key role in the process for the BER endonuclease APE1 that serves a dual function as a DNA repair enzyme and as a trans-activating factor for transcription (13). Likewise, the Xodo laboratory documented that oxidation of a G nucleotide in the potential G-quadruplex forming sequence (PQS) in the KRAS gene promoter recruits a transcription factor, a protein factor, and DNA repair enzymes for gene induction (16). Lastly, Ba, Boldogh, and co-workers showed that oxidation of G in the TNF-α promoter near an NF-κB binding site recruits the DNA repair glycosylase OGG1 and transcription factors for gene activation during inflammatory stress (12,17). Collectively these studies indicate that oxidation of G in promoters of a number of genes can upregulate transcription, although the proposed mechanisms in each study differ. These observations stand in contrast to experiments that have found oxidatively modified G nucleotides in the coding region of a gene generally diminish gene expression (18,19).
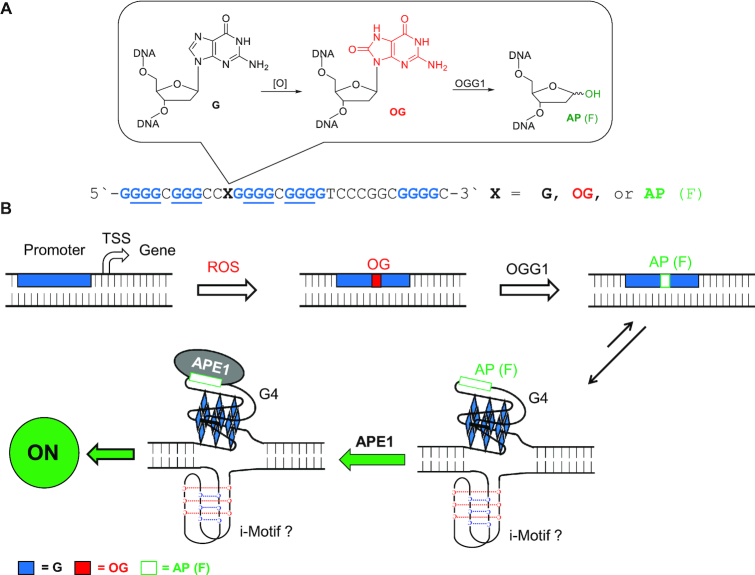
Our laboratory mapped a possible mechanism for gene activation as a result of G oxidation using chemical tools in a biological context. We demonstrated a possible mechanism by which the VEGF gene is activated by oxidation of the G-rich PQS in the promoter (11). We synthetically installed the G oxidation product 8-oxo-7,8-dihydroguanine (OG) at a hotspot for oxidation in the VEGF promoter PQS (Figure 1A) (20). Selection of OG was based on a collection of in vivo and in vitro studies demonstrating this modification is a major two-electron oxidation product of G under oxidative stress conditions inside cells (3,20–22). The OG-modified promoter was then placed in the context of the VEGF PQS replacing the TATA box of the simian vacuolating virus 40 (SV40) early promoter that drove a nearly 3-fold increase in expression of a luciferase gene upon transfection into mammalian cells (Figure 1B) (11). Our chemically defined approach allowed the claim that the oxidatively modified G heterocycle OG is a candidate modification to DNA that has regulatory potential (6,11). Additional experiments found activation was dependent on the BER pathway in which the glycosylase OGG1 functions as the initial reader protein to remove OG and yield an abasic site (AP). The AP results from OGG1 primarily functioning as a monofunctional glycosylase in vivo (18). The AP destabilizes the duplex state driving a shift in equilibrium favoring a G-quadruplex (G4) fold because the AP can reside in a loop region that does not impact the stability of the structure (11,20). Having the AP in a G4 loop stalls the endonuclease activity of APE1 processing its substrate (23) allowing the protein to function in its trans-acting factor capabilities for upregulation of transcription by likely interacting with the transcriptional activator proteins AP-1 and/or HIF-1α (15,24). Thus, APE1 is the second and pivotal reader protein in the regulatory process modulating gene expression. Support for G4 formation in the process was derived from studies with G4-negative sequences leading to a null result when located in the coding strand, and the roles for OGG1 and APE1 were established using either knockout cells or siRNA knockdown studies (11).
Figure 1.
(A) The VEGF promoter PQS and scheme of G oxidation to OG, as well as (B) the proposed APE1-dependent pathway for transcriptional modulation. The previous studies from our laboratory placed the G-rich sequence between –16 to –64 relative to the transcription start site of the SV40 promoter in the coding strand, and controls conducted with a G4-negative sequence support the non-canonical structure participates in gene modulation (11,25); however, the currently available data cannot speak to the role of the i-motif fold in the process.
Our proposed pathway addresses molecular details regarding how G oxidation to OG can activate transcription and provides an explanation for why APE1 stalls on the DNA substrate to function as a trans-acting regulator of transcription (6,11). Additional studies in our laboratory found a strand bias in gene modulation by OG in the PQS context when replacing the TATA box of the SV40 early promoter for luciferase expression. When the VEGF PQS is located between -16 to -64 relative to the TSS, placement in the coding strand (i.e., non-transcribed strand) leads to gene activation in an APE1-dependent process; in contrast, gene suppression occurs when placement of the same sequence is in the template strand in an APE1-independent process (Figure 1B) (25). Thus, OG in the context of a PQS can function as an on/off transcriptional switch when replacing the TATA box of the SV40 promoter sequence. The impact on gene expression for oxidative modification of G to OG was also demonstrated in the NTHL1, RAD17, and PCNA PQSs in the same plasmid system (11,26,27). In the complementary strand to the PQS is a potential i-motif forming sequence that could possibly fold and aid in the gene regulation process (28). At present, we know that DNA repair proteins are required to guide gene regulation that occurs on the G-rich strand because this is where the modified bases were synthesized (11); however, whether proteins interact with the i-motif fold in addition to the DNA repair proteins is not currently known, but future studies will aid in better understanding this step of the proposed activation process.
A number of laboratories have now documented that oxidative modification of a gene promoter results in modulation of gene expression (5–7,9,16). Although, the spatial constraints within a gene promoter by which OG in the PQS context can impact transcription are not known. How important is location? The previous luciferase experiments in our laboratory replaced the TATA box of the SV40 early promoter and enhancer driving Renilla luciferase (Rluc) expression with OG in the VEGF PQS context in the coding strand. In the present report, we moved the five-track VEGF PQS with and without OG or an abasic site analog (tetrahydrofuran, F; Figure 1A) to other critical control regions of the SV40 early promoter driving Rluc expression. These studies begin to shed light on the importance of PQS location relative to the transcription start site (TSS) to alter transcription when the G-rich sequence is oxidatively modified. At each location selected for study, the PQS was placed in both the coding or template strands in parallel experiments. The data collected from these studies found that the VEGF PQS in the wild-type and modified states uniquely modulate gene expression with dependency on the location in the promoter and the strand of occupancy (i.e., coding versus template). These findings would not have been predicted on the basis of our previous studies (11,25). Lastly, the experimental findings provide an initial window of sequence space to inspect literature data using bioinformatic tools for identification of genes that possibly can be turned on or off by oxidative modification of a promoter PQS, assuming genomic promoters function similar to the model promoters studied in the present work.
MATERIALS AND METHODS
Plasmid synthesis to install the VEGF PQS with and without modifications
The plasmids were constructed from the native psiCHECK2 plasmid (Promega) that contains coding sequences for the Renilla luciferase (Rluc) and firefly luciferase (luc) genes. The luc gene is regulated by the HSV-TK promoter that was generally not modified and harnessed as the internal standard to conduct a dual-glo luciferase assay (Promega). The Rluc gene was originally regulated by the SV40 early enhancer/promoter, which we modified by replacing key regulatory sequences of interest with the VEGF PQS flanked by Nt.BspQ1 nicking endonuclease recognition sequences following a PCR-based method previously described by our laboratory (11). Insertion of site-specific modifications into the plasmids was achieved following a literature protocol that we adapted (29), in which the native sequence was clipped out using the nicking endonuclease Nt.BspQ1 and replaced with a synthetic oligodeoxynucleotide containing the modification of interest. To confirm that the DNA modifications were introduced into the plasmid, a sequencing protocol for the modifications in DNA was applied (30). Complete details of the process can be found in the Supporting Information.
Cellular studies
The cells were purchased from ATCC. All cells were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 20 μg/mL gentamicin, 1× glutamax and 1× non-essential amino acids. The cells were grown at 37°C with 5% CO2 at ∼80% relative humidity and were split when they reached 70–80% confluence. The transfection experiments were conducted in white, 96-well plates by seeding 3 × 104 cells per well and then allowing them to grow for 24 h. After 24 h, the cells were transfected with 250–500 ng of plasmid using X-tremeGene HP DNA transfection agent (Roche) following the manufacturer's protocol in Opti-MEM media. Following 48 h incubation after transfection, a dual-glo luciferase (Promega) assay was conducted following the manufacturer's protocol. All transfection experiments were conducted at least four times.
The luminescence data obtained for luciferase expression were analyzed by converting the luminescence values measured into normalized relative response ratios (RRR), which is the luminescence of Rluc divided by the luminescence of luc (i.e. RRR = Rluc/luc). To obtain the normalized expression values reported, each RRR with the modified plasmid was divided by the RRR for the wild-type SV40-containing plasmid in that data set, for example, normalized expression = RRRX/RRRSV40. This data analysis approach allows control for the different expression levels of the two luciferase genes that occur in different cell lines. The error bars represent 95% confidence intervals obtained from the quadruplicate data sets collected. The actual values reported in the bar charts in the text are provided in Supplementary Table S1.
Bioinformatic analysis
The PQSs distributed throughout the human genome were previously identified by many laboratories using a PQS search algorithm (31–35). The data presented in the present work is from our previous analysis of the human genome (hg38) using the Quadparser algorithm (31). The RNA pol II ChiP-Seq data from U87 cells were taken from the ChIP-Atlas peak browser with a significance level of 200 (http://chip-atlas.org/). The PQSs and their distance from the TSS were calculated and plotted with an in-house Python script (https://github.com/dychangfeng/PQS_bacterial). Statistical overrepresentation tests of gene ontology terms were conducted with PANTHER (36).
RESULTS AND DISCUSSION
Experimental design for movement of the VEGF PQS within the context of the SV40 early enhancer/promoter
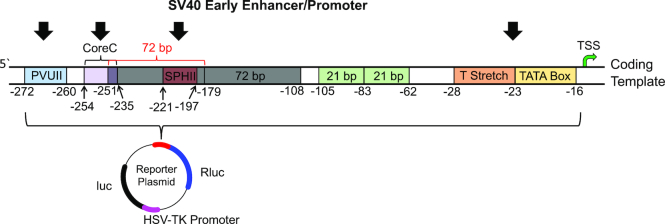
The SV40 early promoter and enhancer sequence is 347 base pairs (bp) long and is comprised of many established functional sequence elements (Figure 2 and Supplementary Figure S1) (37–43). All element positions are relative to the TSS. The TATA box is the region in which the RNA pol II preinitiation complex forms and is located between –16 and –23; additionally, this region has an upstream T stretch that extends to position -29 that aids in preinitiation of transcription. Next, there are two 21-bp boxes positioned between –62 to –83 and –84 to –105, and these two regions are essential for transcription to occur from the SV40 promoter in mammalian cell culture; thus, as discussed below, these regions were not altered in our studies. Each 21-bp box has two copies of the sequence 5′-CCGCCC-3′ on the coding strand that are recognition sites for the SP1 transcription factor; this is noteworthy because the VEGF PQS located in the promoter in mammalian cells has been established to be bound by the SP1 factor (44). Further upstream of the TSS are two 72-bp repeats located from positions –108 to –179 and –180 to –251. These two 72-bp repeats function independently. The upstream-most 72-bp repeat contains two unique enhancer elements, SPHII located from –197 to –221 and CoreC located from –235 to –251 that are not found in the downstream 72-bp repeat. The downstream-most 72-bp box was documented to be important for gene expression in mammalian cells, and therefore, this region was not modified in the studies described below. Next is the P-motif located from –253 to –259 that is another essential region for expression to occur in mammalian cell culture; thus, this region was not modified with the VEGF PQS. Lastly, the PVUII enhancer element is located in the promoter from positions –260 to –272. The regions outlined were found to be bound by specific protein factors that guide the magnitude and timing of gene expression from this promoter in mammalian cell culture (37–43).
Figure 2.
Diagrams of the SV40 promoter to illustrate the location of functional sequences and the plasmid containing the two luciferase genes studied. The black arrows depict sites where the VEGF PQS was studied in the promoter.
The goal of the present study was to selectively replace key SV40 promoter elements with the VEGF PQS in the wild-type and chemically modified states for regulation of Rluc expression. By making changes to the promoter and not the transcribed portion of the genes, the effects monitored result from changes in initiation of transcription, while all other processes in gene expression (i.e. mRNA processing and translation) were kept constant throughout the studies. This study allows the understanding of how the position of a PQS that can adopt a well-defined parallel-stranded G4 (45) in a promoter can impact gene expression. We measured the effect on gene expression in mammalian cells in which the wild-type VEGF PQS replaced the TATA box and T stretch, SPHII, CoreC or PVUII within regions of the SV40 promoter in the coding or template strands (Figures 2 and 3A). Following these initial control experiments, the oxidatively modified G nucleotide OG or an AP (i.e. F), that is the initial product of OG repair, was synthetically incorporated into one of the previously identified hotspots for oxidation in the VEGF sequence (18,20). The chemical modifications were studied at each location in which the PQS was placed in the SV40 promoter. The modulation of Rluc expression for each sequence trial was normalized against the luc expression levels derived from the gene located on the same reporter plasmid using a dual-glo luciferase assay to determine a relative response ratio (RRR = Rluc/luc). The luc expression level was harnessed as an internal standard that provides greater ability to control for experimental errors and biological variability. The assays were performed 48 h post transfection because our previous work on this system found this analysis time gave expression levels for the WT, OG- or F-modified VEGF PQS plasmids in U87 cells that could be maximally differentiated (25).
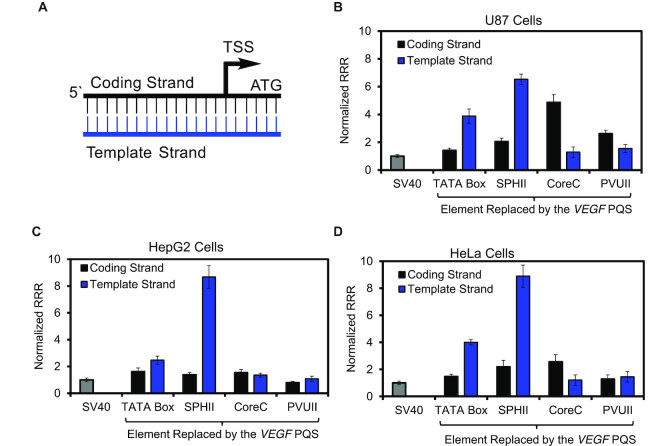
Figure 3.
Changes in Rluc expression observed as the VEGF PQS with no base modifications replaced key sequence elements in the SV40 promoter. (A) Promoter diagram to identify the color key for the plots. Relative response ratios (RRR = Rluc/luc) measured when the VEGF PQS replaced key elements of the SV40 promoter in plasmids that were transfected in (B) U87, (C) HepG2 or (D) HeLa cell lines. The values were normalized to the native SV40 promoter for comparison (normalized RRR = RRRexpt/RRRSV40). The luciferase expression levels were determined using a dual-glo luciferase assay conducted 48 h post transfection. The RRRs measured were normalized to the native psiCHECK2 plasmid RRR in the same cell line to account for cell-specific differences in expression.
VEGF PQS location dependency in the SV40 on Rluc expression
The modified plasmids with the TATA box and T stretch replaced with the VEGF PQS in either the coding or template strand were transfected in U87 (Figure 3B), HepG2 (Figure 3C), or HeLa (Figure 3D) cells. In all three cell lines, having the PQS in the template strand at this site resulted in greater expression than having the PQS in the coding strand. The increase in expression observed for a template strand versus coding strand VEGF PQS was 2.7-fold more in U87 cells, 1.5-fold more in HepG2 cells, and 2.7-fold more in HeLa cells (Figure 3B–D). When the TATA box that was the original site of RNA pol II binding for preinitiation of mRNA synthesis was replaced with the VEGF PQS, transcription was enhanced for both orientations of the G-rich sequence studied when compared to the native SV40 promoter in the cell lines studied.
Replacement of the enhancer elements in the SV40 promoter (SPHII, CoreC or PVUII) with the VEGF PQS on the coding or template strand resulted in strand orientation, positional, and cell line differences in expression between the studies (Figure 3B–D). Replacement of the SPHII element with the PQS, in general, provided an increase in expression relative to the native SV40 promoter in all three cell lines regardless of the strand the G-rich sequence resided. An interesting observation occurred when the VEGF PQS was located in the template strand at the SPHII site, wherein a dramatic increase in expression was observed relative to the native promoter (∼6-fold for U87, ∼8-fold for HepG2, and ∼9-fold for HeLa cells). When the CoreC element was replaced with the VEGF PQS in either strand, the expression levels measured were generally greater for the coding strand PQS than the template strand. For example, in U87 cells 3.7-fold more expression was observed for the coding strand VEGF PQS, 1.1-fold more in HepG2 cells, and 2.1-fold more in HeLa cells. The VEGF PQS at the CoreC position generally increased Rluc expression more than the native SV40 early promoter. Lastly, replacement of the enhancer element furthest from the TSS, PVUII, with the VEGF PQS in either strand provided similar expression levels between the strands; further, the expression levels measured were similar to the wild-type SV40 promoter in all three cell lines reported. Nearly identical findings for all the sequence permutations studied were found in a mouse embryonic fibroblast (MEF) cell line, supporting the conclusion that these observations are not unique to human cells (Supplementary Figure S2). These observations in their entirety suggest when a PQS is in an enhancer region of a promoter, the impact on expression is highly dependent on location, which includes both position relative to the TSS and location in the coding versus template strands.
The data collected from moving the VEGF PQS through the SV40 promoter on the coding or template strand while monitoring expression of the Rluc gene in the mammalian cells provide new insights regarding promoter PQSs. Previous studies to determine the impact of a promoter PQS on expression, as well as the strand bias, have produced mixed findings (34,46–56). These reports support promoter PQSs can either enhance or diminish expression depending on interactions with RNA pol II, transcription factors, or the folded G4 can function to maintain the open conformation of the DNA duplex for transcription bubbles to progress. Each report studied a different PQS or inspected these sequences on the genome-scale at different locations leading to mixed collective interpretations regarding how the G-rich sequences guide transcription. In the present studies, we systematically moved the same PQS to different locations on either the coding or template strand. This approach allowed us to rule out any PQS sequence effects, and by maintaining the same overall promoter (i.e. SV40), long-range promoter sequence effects were also controlled. We recognize these studies have taken the VEGF PQS outside of its native context and the SV40 promoter is of viral origin; however, moving the same PQS around a promoter region allows direct comparisons to be made.
The cell-line dependent expression levels observed are consistent with previous studies with the native SV40 promoter regulating a reporter gene (Figure 3) (57,58); thus, the differences observed were expected. Replacement of the TATA box with the VEGF PQS while still maintaining promoter activity was at first glance surprising. The TATA box is the sequence that RNA pol II loads for preinitiation of transcription (59); however, a TATA box is not essential for preinitiation of transcription that is exemplified by >75% of human genes possessing TATA-less promoters (60). Additionally, increasing the GC content of a promoter was previously demonstrated to enhance the activity of a promoter on the genome-wide scale (61), and the present findings are generally similar in that replacement of the TATA box (0% GC) with the VEGF PQS (97% GC) resulted in greater gene expression; however, the underlying sequence- and possible structural-context features leading to the observed changes are not known. Overall, the observation that Rluc expression was maintained when the TATA box was replaced with the VEGF PQS is not surprising.
The differences detected between the coding versus template strand PQS when replacing the TATA box were significant (1.1- to 3.7-fold; Figure 3B, C, and D). To determine whether the impact observed with the TATA box replacement was unique to the SV40 promoter, the experiments were repeated utilizing the same plasmid while replacing the TATA box of the HSV-TK promoter driving the luc gene with the VEGF PQS. These additional studies in U87 cells found a similar pattern in which the PQS in the template furnished greater expression than the one in the coding strand (Supplementary Figure S3). Next, placement of the VEGF PQS at enhancer element sites in the promoter identified a strand bias for the G-rich sequence with dependency on the location of the element replaced (Figure 3B–D). The observations made in these studies indicate that the impact of a PQS on gene expression within a promoter is location dependent with respect to both the distance from the TSS and its presence in the coding versus template strand; furthermore, there is not a clear trend for predicting the outcome from placement of the VEGF PQS, and likely any other PQS, on gene expression. A similar location dependency of a PQS on gene expression has been observed in prokaryotic cells (62). These findings identify location (i.e., distance from the TSS and coding versus template) strand are two important parameters that will impact how a PQS effects gene expression, beyond the sequence and G4 fold of a PQS.
A few final observations regarding the data were made. First, the VEGF PQS can replace any of the key enhancer elements of the SV40 promoter and gene expression was still retained (Figure 3). The expression observed was greatest for the VEGF PQS on the template strand at the location of the SPHII enhancer that suggests this location is critical for amplifying gene expression by a PQS. Interestingly, this element is of intermediate distance from the TSS (position -197) that suggests an optimal distance from the TSS provides the maximal increase in gene expression. A similar strand bias was generally observed for the VEGF PQS at the position of the further downstream CoreC element (position -235). As the PQS was moved further from the TSS, the changes observed relative to the wild-type SV40 promoter were found to diminish, as did the strand bias effect that was observed for the PQS at closer positions. In an additional study, the VEGF PQS was placed just upstream of the established SV40 early promoter (i.e., outside the promoter) to assay the impact on expression (Supplementary Figure S4). In this final study, we found no change on Rluc expression when the VEGF PQS was in either the coding or template strand in comparison to the native SV40 promoter. This control experiment confirms that when a PQS is located far enough from the TSS, it will not impact transcription.
In these studies, the VEGF PQS was always maintained in the same 5′ to 3′ orientation, and therefore, moving from the coding to the template strand reoriented the G-rich sequence relative to the TSS. A previous structural study found the inversion of G4 sequences exerts a substantial effect on the structure (63); therefore, to avoid this additional complication, we maintained the native VEGF PQS orientation in these studies. Moving the PQS from the coding to the template strand will reorient proteins that bind the sequence, and on the basis of these data, this change may best explain the impact on gene expressions observed when the same sequence was studied in the two strands at the same site.
Identification of the VEGF PQS functioning to enhance transcription from the coding or template strand is consistent with the findings from the Balasubramanian laboratory in which ∼10,000 G4s in the human genome on either strand can increase gene expression (51). Our results provide further confirmation of the previous studies and identify within a well-defined promoter such as SV40 that PQS strand orientation and distance from the TSS play complementary roles to establish the magnitude of gene expression. The present results do not address how the PQS interacts with other elements in the same promoter to set the basal expression level; nonetheless, the data establish the background levels of expression prior to analyzing the impact of oxidation of a G nucleotide on gene expression at each of the locations studied.
Oxidative modification of the PQS modulates transcription
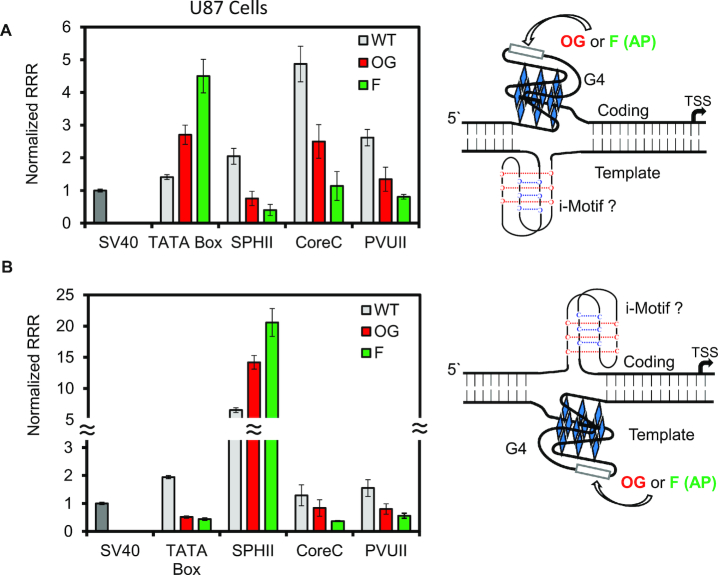
Each plasmid was then synthesized with the oxidatively modified G nucleotide OG installed at a hotspot for oxidation in the G-rich VEGF sequence following a previously established method (11,20). Additionally, knowledge that release of OG from the genome by OGG1 may yield an abasic site as the likely first intermediate in the repair process (18), and that the abasic site is important for gene regulation (Figure 1B) (11,13), led us to synthesize a tetrahydrofuran abasic site analog (i.e. F) at each position studied. The F analog was selected for its chemical stability during handling of the DNA, and this analog is processed nearly identically to authentic abasic sites by the endonuclease APE1 (64). Each site-specifically modified plasmid was transfected in the human cell lines U87 or HepG2; the data for the U87 cell line are provided in Figure 4. Because the trends for incorporation of the modified bases were the same in the HepG2 cell line, these data are provided in Supplementary Figure S5. When OG was located in the VEGF context at the site of the TATA box in the coding strand, expression increased by ∼3-fold relative to the all G-containing wild-type sequence in the cell lines studied (Figure 4A and Supplementary Figure S5); on the other hand, having the same sequence in the template strand with OG present led to a ∼4-fold decrease in Rluc expression relative to the wild-type state, again in all the cell lines studied (Figure 4B and Supplementary Figure S5). Having an AP site located at the same sites in the same context yielded an ∼4-fold increase in the expression on the coding strand and ∼4-fold decrease in expression on the template strand (Figure 4 and Supplementary Figure S5). The observations in these cells with the two modifications are consistent with our previous findings when the PQS replaced the TATA box of the SV40 promoter (11,25).
Figure 4.
The impact of modifying the VEGF PQS with OG or an abasic site analog F at various locations in the SV40 promoter. Data obtained with the G-rich sequence with and without modifications were placed in the (A) coding or (B) template strand of the promoter. The VEGF PQS installed, the site of the modifications, and their chemical structures are provided in Figure 1A.
In an additional permutation, we left the TATA box in place while adding the VEGF PQS before or after this key element. The new plasmids were studied with the G-rich sequence on the coding strand to better understand whether a PQS can function in conjunction with the TATA box. The VEGF PQS was installed either just upstream (5′) or downstream (3′) of the TATA box (Supplementary Figure S6). First, the VEGF PQS without a chemical modification produced similar Rluc expression levels as the wild-type SV40 promoter. Introduction of OG into the PQS 3′ to the TATA box led to ∼2-fold increase in Rluc expression, while introduction of the PQS with OG 5′ to the TATA box yielded ∼2.5-fold increase in Rluc expression. These observations confirm that oxidative modification of a promoter PQS to alter gene expression can still function when a TATA box is present in the promoter.
When the PQS and modifications were incorporated into the other more distant positions within the promoter, each location was found to yield a unique gene expression profile. In contrast to the modified VEGF PQSs at the TATA box position in the coding strand, placement of the same sequences in the same strand at the SPHII element found the modifications turned gene expression off (OG was 2.7-fold and F was 5.1-fold); however, on the template strand, introduction of modifications in the PQS context at the SPHII element led to an increase in gene expression (OG was 2.2-fold and F was 3.2-fold; Figure 4A, B, and Supplementary Figure S5). To reiterate, this result is opposite the strand bias observed when this PQS and modifications were studied closer to the TSS, replacing the TATA box. Next, the trends observed for the PQS with OG or F modifications at either the CoreC or PVUII enhancer elements found, regardless of the strand, the modifications OG or F in the VEGF PQS context attenuated gene expression (Figure 4A, B, and Supplementary Figure S5). In a final analysis, the PQS with OG or F were placed outside the context of the SV40 promoter to find the same expression levels as the native promoter, suggesting that expression level changes only occur when the modified sequence is in the promoter region (Supplementary Figure S4). These observations paint a very complex picture for determination of when and where oxidative modification of a promoter PQS will activate or deactivate transcription of a gene of interest.
Lastly, to determine whether promoter dependency impacted the observations, examination of the VEGF PQS with OG or F replacing the TATA box in the HSV-TK promoter of the luc gene on the same plasmid was studied. These studies used the unmodified Rluc gene as an internal standard. The findings for the TATA box VEGF PQS replacement identified similar values as observed for the same sequence, strand dependency, and modification status within the context of the SV40 promoter (Supplementary Figure S3). Next, the VEGF PQS in the WT or modified states were studied at positions -200 or -400 bp relative to the TSS in the HSV-TK promoter in the coding strand (Supplementary Figure S3). At the –200 bp position, OG or F led to activation of transcription that is in contrast to the results for a similar placement in the SV40 promoter (i.e. SPHII Figure 4A). At the –400 bp position the VEGF PQS with OG or F led to no significant change in expression similar to the SV40 promoter (Supplementary Figures S3 and S4). Additional studies to move the PQS through the HSV-TK promoter similar to the SV40 promoter were not pursued. Nonetheless, the findings in this second promoter identify that the VEGF PQS when modified impacts gene expression; however, there are differences between the two promoters studied suggesting long range interactions are also important in the type of response that will occur from oxidation of a PQS in a gene promoter. These observations further point to caution when drawing generalizations from the present data and all data regarding regulatory promoter PQSs.
Cell-based studies have built a case for folding of PQSs to G4s within the human genome and particularly gene promoters for regulating transcription leading to the claim G4s are epigenetic hallmarks in genomes (51). Our laboratory seeks to discover when oxidative modification of PQSs, as a result of oxidative stress, unmasks these epigenetic folds for gene regulation, thus ascribing the G modification OG as an epigenetic-like modification to facilitate the G4 folding process (6). The present observation is fascinating that OG or its initial repair product AP (modeled as F) turn transcription on when located in the VEGF PQS context positioned within an ∼200-bp window of the TSS in the SV40 promoter with dependency on location in this window. When the modified PQS was at –235 and further upstream, chemical modification of the sequence led to downregulation of transcription, regardless of the strand. These data in the model SV40 promoter define spatial constraints in which oxidative modification can unmask a G4 fold to upregulate transcription. We recognize these distances are most likely promoter dependent.
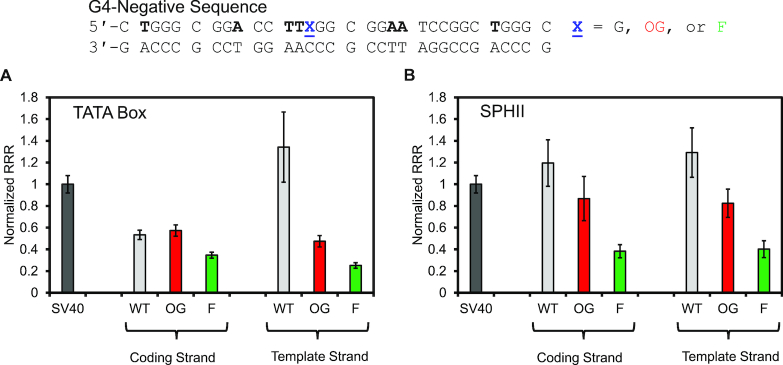
To address whether the PQS context was important for the gene expression changes observed, a set of control studies were conducted in a sequence not capable of adopting a G4 fold on the basis of a prior study (11). The sequence studied was judiciously selected to maintain the three SP1 transcription factor binding sites in the VEGF PQS while being incapable of G4 folding (44). The structure-switching negative sequence was studied at the TATA box and the SPHII sites in the SV40 promoter in both strands in the WT, OG- or F-modified states (Figure 5). Studies in the coding strand at the TATA box site found OG gave similar expression as the WT sequence and F was attenuated (Figure 5A); in contrast, the same sequence in the template strand found the WT state gave high expression and the OG- or F-modified plasmids produced >3-fold reduction in expression. This observation suggests the RNA pol II complex that forms near the TSS most likely interacts more strongly with the template strand when the duplex is denatured to initiate transcription, consistent with the current structural data (59). Next, studies at the SPHII enhancer site with the G4-negative sequence found regardless of the strand, OG slightly decreased expression and F resulted in a strong attenuation of the signal (Figure 5B). Previous studies found the presence of OG in an SP1 consensus sequence does decrease the binding affinity of the factor to the DNA (65), which would result in decreased expression as observed. The abasic site would be more disruptive to the protein-DNA interactions and consistent with this claim, the F was further detrimental to expression (Figure 5B).
Figure 5.
Gene expression measured in U87 cells for a G4-negative sequence in the WT, OG- or F-modified states in plasmids at the (A) TATA box or (B) SPHII sites in the SV40 promoter.
The studies in G4 negative sequences consistently show no change or decreased gene expression with a modification present (Figure 5). In contrast, gene expression was only found to increase when the modifications were in a sequence capable of B→G4 switching. We do recognize these studies are not definitive support for a G4 fold in the activation process, but they provide one more piece of evidence supporting this conclusion. Future studies are needed to add more support or refute G4 folds and their chemically modified state as gene regulatory units. As an additional note, the complementary strand to a PQS is a potential i-motif forming sequence that may also fold to aid in the gene expression changes observed. The present data cannot support or rule out the role of the i-motif in these processes. Future studies are needed to understand the interplay between G4s and i-motifs on complementary strands to impact gene expression when oxidatively modified.
Oxidative modification of a promoter PQS to drive a change in gene expression is dependent on the location from the TSS and coding versus template strand. We have found that close to the TSS, oxidation of the PQS increases gene expression in the coding strand and decreases gene expression on the template strand. Contrasting to this finding, movement of the PQS and oxidation status upstream found gene activation or deactivation was strand and promoter dependent (Figure 4 and Supplementary Figure S3). The ability of an oxidized PQS to activate transcription beyond ∼200 bp upstream of the TSS was not observed. This suggests to us that gene modulation via oxidatively modified PQSs may be most profound within ∼200 bp from the TSS. Beyond this approximate distance, G oxidation to OG decreases gene expression that most likely results from minimizing transcription factor binding to the DNA sequence. Previous studies have found OG in transcription factor consensus motifs decreases the stability of protein-DNA binding interactions (65–67), consistent with this claim.
Bioinformatic analysis of published results for possible redox-active PQSs
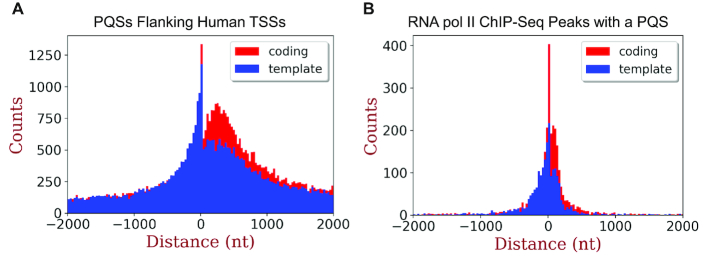
Where in the human genome is it possible for this proposed mechanism to function? To properly address this question, we are in the process of refining our ability to sequence the human genome for OG (68); in the meantime, we can use computational methods and previous studies to shed some light on this question. Bioinformatic inspection of the human genome to locate PQSs has found them to be enriched around TSSs (31–35). Further, enrichment of PQSs is similar on both the coding and template strand; but more on point is that these G-rich sequences are most enriched in the promoter within ∼200 bp upstream of the TSS (Figure 6A). On the basis of the present data, the human genome possesses many PQSs close enough to the TSS to be unmasked for gene regulation by oxidative stress. Thus, the number of folded G4s found by Balasubramanian and co-workers to regulate transcription under non-stressed conditions (51) is anticipated to increase in count under conditions of oxidative stress. To estimate the subset of PQSs in gene promoters poised for activation, we took RNA pol II ChIP-Seq data from U87 cells, inspected the peaks of polymerase enrichment for PQSs, and plotted them with respect to their location relative to the TSS (Figure 6B). This inspection found enrichment of poised RNA pol II binding sites with PQSs to be greatest within a few hundred nucleotides of the TSS on both coding (n = 3826) and template strands (n = 3010). Restriction of the population to those within 200 nucleotides of the TSS identified 746 poised genes with a PQS on the coding strand and 754 poised genes with a PQS on the template strand (Supplementary Table S2).
Figure 6.
Distribution of (A) PQSs and (B) RNA pol II ChIP-Seq enriched peaks with PQSs flanking human TSSs on the coding or template strand. Genome-wide the coding strand has 99,585 PQSs and the template strand has 94,588 PQSs flanking TSSs. Inspection of RNA pol II ChIP-Seq peaks of enrichment from U87 cells found 3,826 PQSs in the coding strand and 3,010 PQSs in the template strand.
Closer inspection of the gene ontology terms for genes with a PQS in the critical ∼200-bp window of sequence space upstream of the TSS in the RNA pol II ChIP-Seq data identified gene classes that could be modulated by the proposed mechanism (Figure 1B). There exist many gene ontology terms enriched in the list (Supplementary Table S3). Genes in the list that fit the hypothesis that G oxidation in a promoter PQS can modulate gene expression include those that respond to ionizing radiation, H2O2, ROS, or regulate DNA repair. There exist many other terms with positive enrichment in the analysis, as well as negative enrichment.
This finding could be fortuitous as PQSs and RNA pol II localization occur around TSSs; alternatively, the overlap does support the possibility for PQS-bearing promoters to be poised for oxidative modification to unmask G4 folds for gene regulation during oxidative stress. Analysis of these genome-wide results suggests similar counts of PQSs exist on either strand within a window of sequence space from the TSS for oxidative modification of a G nucleotide to impact gene expression.
In the final inspection of literature data for PQSs in the critical 200-bp window upstream of the TSS, we looked into the published G4-ChIP-Seq results (51). In this region 847 genes with PQSs were found nearly equally distributed across the coding and template strands (Supplementary Table S4). Inspection of gene ontology terms for these PQSs found enrichment in genes for the response to radiation, cellular response to stress, DNA repair, and cellular response to DNA damage stimulus (Supplementary Table S5). Changes in expression of these genes would be important during oxidative stress. This final inspection of the literature provides more support for a hypothesis of PQSs in gene promoters functioning as redox switches for regulation of genes that are involved in the stress response. Complete transcriptome profiling before and after oxidative stress to quantify properly how each gene changes for experimental validation to support these lists of genes will be undertaken in future studies. Boldogh, Ba and coworkers have found proinflammatory genes have a strong potential for regulation via G oxidation in a gene promoter (69), consistent with the hypothesis derived from the present work.
CONCLUSIONS
Experimental evidence is building for oxidative modification of a G nucleotide within a gene promoter to function as a chemical modification with regulatory potential (5–10). Many more studies are needed to better understand when, where, and how this occurs. Our laboratory and others have focused on oxidation of G-rich sequences with the potential for G4 formation within gene promoters for regulating mRNA levels (11,16,25). The present work took us a step forward by addressing possible spatial constraints within a promoter that define when oxidative modification activates transcription and when it diminishes transcription. We have found when the oxidatively modified PQS is close to the TSS, transcription is turned on when the chemistry occurs in the coding strand, while transcription is turned off by modification of the PQS in the template strand. So far, this rule has held with a variety of PQSs studied in our laboratory (11,25,27). In contrast, moving further upstream in the promoter found the effect of an oxidatively modified G to be location, strand, and promoter dependent with respect to the on or off switching potential. In the Xodo laboratory, they found oxidative modification G to OG in the KRAS PQS in the template strand positioned from –148 to –116 relative to the TSS turned transcription on (16,54), consistent with our studies in the SV40 promoter (Figure 4B). A few differences exist in our proposed mechanism for activation when the PQS is oxidized near the TSS relative to the proposal of Xodo and co-workers, and these could be best explained by the different PQSs studied and/or the concept that the promoter location of the modified PQS alters the critical protein players. Further work is needed to better understand the protein choreography for these pathways at each location. Established oncogene promoters with PQSs in the ∼200-bp window upstream of the TSS include KRAS (16), c-MYC (70), hTERT (71), PDGF-A (72), HIF-1α (73) and c-KIT (74). When the PQS was located further upstream from the TSS to positions >235 bps, oxidative modification of the DNA diminished gene expression. We have found a window of ∼200 bp within the TSS in which G oxidation to OG can impact gene expression in the model SV40 and HSV-TK promoters, although this window is likely different for other promoters, particularly those in humans. These findings are instructive for future efforts to better define when G oxidation to OG, and possibly other products, has epigenetic-like potential (6,9,12).
Supplementary Material
ACKNOWLEDGEMENTS
The HepG2 cells were a kind gift from Prof. Ryan Looper's laboratory at the University of Utah.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Cancer Institute for financial support of the project [R01 CA090689]; Oligonucleotide synthesis and Sanger sequencing were provided by the University of Utah Health Sciences Core facilities that are supported in part by a National Cancer Institute Cancer Center Support grant [P30 CA042014]. Funding for open access charge: National Institutes of Health.
Conflict of interest statement. None declared.
REFERENCES
- 1. Sohal R.S., Weindruch R.. Oxidative stress, caloric restriction, and aging. Science. 1996; 273:59–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lonkar P., Dedon P.C.. Reactive species and DNA damage in chronic inflammation: reconciling chemical mechanisms and biological fates. Int. J. Cancer. 2011; 128:1999–2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cadet J., Wagner J.R., Shafirovich V., Geacintov N.E.. One-electron oxidation reactions of purine and pyrimidine bases in cellular DNA. Int. J. Radiat. Biol. 2014; 90:423–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Berlett B.S., Stadtman E.R.. Protein oxidation in aging, disease, and oxidative stress. J. Biol. Chem. 1997; 272:20313–20316. [DOI] [PubMed] [Google Scholar]
- 5. Ba X., Boldogh I.. 8-Oxoguanine DNA glycosylase 1: beyond repair of the oxidatively modified base lesions. Redox Biol. 2018; 14:669–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Fleming A.M., Burrows C.J.. 8-Oxo-7,8-dihydroguanine, friend and foe: Epigenetic-like regulator versus initiator of mutagenesis. DNA Repair (Amst). 2017; 56:75–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Antoniali G., Malfatti M.C., Tell G.. Unveiling the non-repair face of the base excision repair pathway in RNA processing: A missing link between DNA repair and gene expression. DNA Repair (Amst). 2017; 56:65–74. [DOI] [PubMed] [Google Scholar]
- 8. Dickinson B.C., Chang C.J.. Chemistry and biology of reactive oxygen species in signaling or stress responses. Nat. Chem. Biol. 2011; 7:504–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Seifermann M., Epe B.. Oxidatively generated base modifications in DNA: Not only carcinogenic risk factor but also regulatory mark. Free Radic. Biol. Med. 2017; 107:258–265. [DOI] [PubMed] [Google Scholar]
- 10. Mittler R. ROS are good. Trends Plant Sci. 2016; 22:11–19. [DOI] [PubMed] [Google Scholar]
- 11. Fleming A.M., Ding Y., Burrows C.J.. Oxidative DNA damage is epigenetic by regulating gene transcription via base excision repair. Proc. Natl. Acad. Sci. U.S.A. 2017; 114:2604–2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Pan L., Zhu B., Hao W., Zeng X., Vlahopoulos S.A., Hazra T.K., Hegde M.L., Radak Z., Bacsi A., Brasier A.R. et al.. Oxidized guanine base lesions function in 8-oxoguanine DNA glycosylase1-mediated epigenetic regulation of nuclear factor kappaB-driven gene expression. J. Biol. Chem. 2016; 291:25553–25566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Antoniali G., Lirussi L., D’Ambrosio C., Dal Piaz F., Vascotto C., Casarano E., Marasco D., Scaloni A., Fogolari F., Tell G.. SIRT1 gene expression upon genotoxic damage is regulated by APE1 through nCaRE-promoter elements. Mol. Biol. Cell. 2014; 25:532–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Perillo B., Ombra M.N., Bertoni A., Cuozzo C., Sacchetti S., Sasso A., Chiariotti L., Malorni A., Abbondanza C., Avvedimento E.V.. DNA oxidation as triggered by H3K9me2 demethylation drives estrogen-induced gene expression. Science. 2008; 319:202–206. [DOI] [PubMed] [Google Scholar]
- 15. Pastukh V., Roberts J.T., Clark D.W., Bardwell G.C., Patel M., Al-Mehdi A.B., Borchert G.M., Gillespie M.N.. An oxidative DNA “damage” and repair mechanism localized in the VEGF promoter is important for hypoxia-induced VEGF mRNA expression. Am. J. Physiol. Lung Cell Mol. Physiol. 2015; 309:L1367–L1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cogoi S., Ferino A., Miglietta G., Pedersen E.B., Xodo L.E.. The regulatory G4 motif of the Kirsten ras (KRAS) gene is sensitive to guanine oxidation: implications on transcription. Nucleic Acids Res. 2018; 46:661–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hao W., Qi T., Pan L., Wang R., Zhu B., Aguilera-Aguirre L., Radak Z., Hazra T.K., Vlahopoulos S.A., Bacsi A. et al.. Effects of the stimuli-dependent enrichment of 8-oxoguanine DNA glycosylase1 on chromatinized DNA. Redox Biol. 2018; 18:43–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Allgayer J., Kitsera N., Bartelt S., Epe B., Khobta A.. Widespread transcriptional gene inactivation initiated by a repair intermediate of 8-oxoguanine. Nucleic Acids Res. 2016; 44:7267–7280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Tornaletti S., Maeda L.S., Kolodner R.D., Hanawalt P.C.. Effect of 8-oxoguanine on transcription elongation by T7 RNA polymerase and mammalian RNA polymerase II. DNA Repair (Amst). 2004; 3:483–494. [DOI] [PubMed] [Google Scholar]
- 20. Fleming A.M., Zhou J., Wallace S.S., Burrows C.J.. A role for the fifth G-track in G-quadruplex forming oncogene promoter sequences during oxidative stress: Do these “spare tires” have an evolved function. ACS Cent. Sci. 2015; 1:226–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Fleming A.M., Burrows C.J.. Formation and processing of DNA damage substrates for the hNEIL enzymes. Free Radic. Biol. Med. 2017; 107:35–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mangerich A., Knutson C.G., Parry N.M., Muthupalani S., Ye W., Prestwich E., Cui L., McFaline J.L., Mobley M., Ge Z. et al.. Infection-induced colitis in mice causes dynamic and tissue-specific changes in stress response and DNA damage leading to colon cancer. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:E1820–E1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Broxson C., Hayner J.N., Beckett J., Bloom L.B., Tornaletti S.. Human AP endonuclease inefficiently removes abasic sites within G4 structures compared to duplex DNA. Nucleic Acids Res. 2014; 42:7708–7719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bhakat K.K., Mantha A.K., Mitra S.. Transcriptional regulatory functions of mammalian AP-endonuclease (APE1/Ref-1), an essential multifunctional protein. Antioxid. Redox Signal. 2009; 11:621–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Fleming A.M., Zhu J., Ding Y., Burrows C.J.. 8-Oxo-7,8-dihydroguanine in the context of a promoter G-quadruplex is an on-off switch for transcription. ACS Chem. Biol. 2017; 12:2417–2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Redstone S.C.J., Fleming A.M., Burrows C.J.. Oxidative modification of the potential G-quadruplex sequence in the PCNA gene promoter can turn on transcription. Chem. Res. Toxicol. 2019; 32:437–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhu J., Fleming A.M., Burrows C.J.. The RAD17 promoter sequence contains a potential tail-dependent G-quadruplex that downregulates gene expression upon oxidative modification. ACS Chem. Biol. 2018; 13:2577–2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kang H.J., Kendrick S., Hecht S.M., Hurley L.H.. The transcriptional complex between the BCL2 i-motif and hnRNP LL is a molecular switch for control of gene expression that can be modulated by small molecules. J. Am. Chem. Soc. 2014; 136:4172–4185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. You C., Dai X., Yuan B., Wang J., Brooks P.J., Niedernhofer L.J., Wang Y.. A quantitative assay for assessing the effects of DNA lesions on transcription. Nat. Chem. Biol. 2012; 8:817–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Riedl J., Fleming A.M., Burrows C.J.. Sequencing of DNA lesions facilitated by site-specific excision via base excision repair DNA glycosylases yielding ligatable gaps. J. Am. Chem. Soc. 2015; 138:491–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Fleming A.M., Zhu J., Ding Y., Visser J.A., Burrows C.J.. Human DNA repair genes possess potential G-quadruplex sequences in their promoters and 5′-untranslated regions. Biochemistry. 2018; 57:991–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Huppert J.L., Balasubramanian S.. Prevalence of quadruplexes in the human genome. Nucleic Acids Res. 2005; 33:2908–2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Todd A.K., Johnston M., Neidle S.. Highly prevalent putative quadruplex sequence motifs in human DNA. Nucleic Acids Res. 2005; 33:2901–2907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Du Z., Zhao Y., Li N.. Genome-wide analysis reveals regulatory role of G4 DNA in gene transcription. Genome Res. 2008; 18:233–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Eddy J., Maizels N.. Gene function correlates with potential for G4 DNA formation in the human genome. Nucleic Acids Res. 2006; 34:3887–3896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Mi H., Muruganujan A., Casagrande J.T., Thomas P.D.. Large-scale gene function analysis with the PANTHER classification system. Nat. Protoc. 2013; 8:1551–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ernoult-Lange M., Omilli F., O’Reilly D.R., May E.. Characterization of the simian virus 40 late promoter: relative importance of sequences within the 72-base-pair repeats differs before and after viral DNA replication. J. Virol. 1987; 61:167–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Casaz P., Sundseth R., Hansen U.. Trans activation of the simian virus 40 late promoter by large T antigen requires binding sites for the cellular transcription factor TEF-1. J. Virol. 1991; 65:6535–6543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Schirm S., Jiricny J., Schaffner W.. The SV40 enhancer can be dissected into multiple segments, each with a different cell type specificity. Genes Dev. 1987; 1:65–74. [DOI] [PubMed] [Google Scholar]
- 40. Eadara J.K., Lutter L.C.. Determination of occupancies of the SPH and GT-IIC transcription factor binding motifs in SV40: Evidence for two forms of transcription elongation complex. Virology. 1996; 223:120–131. [DOI] [PubMed] [Google Scholar]
- 41. Moreau P., Hen R., Wasylyk B., Everett R., Gaub M.P., Chambon P.. The SV40 72 base repair repeat has a striking effect on gene expression both in SV40 and other chimeric recombinants. Nucleic Acids Res. 1981; 9:6047–6068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Everett R.D., Baty D., Chambon P.. The repeated GC-rich motifs upstream from the TATA box are important elements of the SV40 early promoter. Nucleic Acids Res. 1983; 11:2447–2464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Byrne B.J., Davis M.S., Yamaguchi J., Bergsma D.J., Subramanian K.N.. Definition of the simian virus 40 early promoter region and demonstration of a host range bias in the enhancement effect of the simian virus 40 72-base-pair repeat. Proc. Natl. Acad. Sci. U.S.A. 1983; 80:721–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Schafer G., Cramer T., Suske G., Kemmner W., Wiedenmann B., Hocker M.. Oxidative stress regulates vascular endothelial growth factor-A gene transcription through Sp1- and Sp3-dependent activation of two proximal GC-rich promoter elements. J. Biol. Chem. 2003; 278:8190–8198. [DOI] [PubMed] [Google Scholar]
- 45. Agrawal P., Hatzakis E., Guo K., Carver M., Yang D.. Solution structure of the major G-quadruplex formed in the human VEGF promoter in K+: insights into loop interactions of the parallel G-quadruplexes. Nucleic Acids Res. 2013; 41:10584–10592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Bochman M.L., Paeschke K., Zakian V.A.. DNA secondary structures: stability and function of G-quadruplex structures. Nat. Rev. Genet. 2012; 13:770–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kumar N., Patowary A., Sivasubbu S., Petersen M., Maiti S.. Silencing c-MYC expression by targeting quadruplex in P1 promoter using locked nucleic acid trap. Biochemistry. 2008; 47:13179–13188. [DOI] [PubMed] [Google Scholar]
- 48. Smestad J.A., Maher L.J. 3rd. Relationships between putative G-quadruplex-forming sequences, RecQ helicases, and transcription. BMC Med. Genet. 2015; 16:91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Nguyen G.H., Tang W., Robles A.I., Beyer R.P., Gray L.T., Welsh J.A., Schetter A.J., Kumamoto K., Wang X.W., Hickson I.D. et al.. Regulation of gene expression by the BLM helicase correlates with the presence of G-quadruplex DNA motifs. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:9905–9910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. David A.P., Margarit E., Domizi P., Banchio C., Armas P., Calcaterra N.B.. G-quadruplexes as novel cis-elements controlling transcription during embryonic development. Nucleic Acids Res. 2016; 44:4163–4173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hansel-Hertsch R., Beraldi D., Lensing S.V., Marsico G., Zyner K., Parry A., Di Antonio M., Pike J., Kimura H., Narita M. et al.. G-quadruplex structures mark human regulatory chromatin. Nat. Genet. 2016; 48:1267–1272. [DOI] [PubMed] [Google Scholar]
- 52. Fernando H., Sewitz S., Darot J., Tavare S., Huppert J.L., Balasubramanian S.. Genome-wide analysis of a G-quadruplex-specific single-chain antibody that regulates gene expression. Nucleic Acids Res. 2009; 37:6716–6722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Kendrick S., Hurley L.H.. The role of G-quadruplex/i-motif secondary structures as cis-acting regulatory elements. Pure Appl. Chem. 2010; 82:1609–1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Cogoi S., Xodo L.E.. G-quadruplex formation within the promoter of the KRAS proto-oncogene and its effect on transcription. Nucleic Acids Res. 2006; 34:2536–2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Verma A., Halder K., Halder R., Yadav V.K., Rawal P., Thakur R.K., Mohd F., Sharma A., Chowdhury S.. Genome-wide computational and expression analyses reveal G-quadruplex DNA motifs as conserved cis-regulatory elements in human and related species. J. Med. Chem. 2008; 51:5641–5649. [DOI] [PubMed] [Google Scholar]
- 56. Hegyi H. Enhancer-promoter interaction facilitated by transiently forming G-quadruplexes. Sci. Rep. 2015; 5:9165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Qin J.Y., Zhang L., Clift K.L., Hulur I., Xiang A.P., Ren B.Z., Lahn B.T.. Systematic comparison of constitutive promoters and the doxycycline-inducible promoter. PLoS ONE. 2010; 5:e10611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Zarrin A.A., Malkin L., Fong I., Luk K.D., Ghose A., Berinstein N.L.. Comparison of CMV, RSV, SV40 viral and Vlambda1 cellular promoters in B and T lymphoid and non-lymphoid cell lines. Biochim. Biophys. Acta. 1999; 1446:135–139. [DOI] [PubMed] [Google Scholar]
- 59. Sainsbury S., Bernecky C., Cramer P.. Structural basis of transcription initiation by RNA polymerase II. Nat. Rev. Mol. Cell Biol. 2015; 16:129–143. [DOI] [PubMed] [Google Scholar]
- 60. Yang C., Bolotin E., Jiang T., Sladek F.M., Martinez E.. Prevalence of the Initiator over the TATA box in human and yeast genes and identification of DNA motifs enriched in human TATA-less core promoters. Gene. 2007; 389:52–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Grabherr M.G., Pontiller J., Mauceli E., Ernst W., Baumann M., Biagi T., Swofford R., Russell P., Zody M.C., Di Palma F. et al.. Exploiting nucleotide composition to engineer promoters. PLoS ONE. 2011; 6:e20136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Holder I.T., Hartig J.S.. A matter of location: influence of G-quadruplexes on Escherichia coli gene expression. Chem. Biol. 2014; 21:1511–1521. [DOI] [PubMed] [Google Scholar]
- 63. Marusic M., Plavec J.. The effect of DNA sequence directionality on G-quadruplex folding. Angew. Chem. Int. Ed. Engl. 2015; 54:11716–11719. [DOI] [PubMed] [Google Scholar]
- 64. Schermerhorn K.M., Delaney S.. Transient-state kinetics of apurinic/apyrimidinic (AP) endonuclease 1 acting on an authentic AP site and commonly used substrate analogs: the effect of diverse metal ions and base mismatches. Biochemistry. 2013; 52:7669–7677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Ramon O., Sauvaigo S., Gasparutto D., Faure P., Favier A., Cadet J.. Effects of 8-oxo-7,8-dihydro-2′-deoxyguanosine on the binding of the transcription factor Sp1 to its cognate target DNA sequence (GC box). Free Rad. Res. 1999; 31:217–229. [DOI] [PubMed] [Google Scholar]
- 66. Hailer-Morrison M.K., Kotler J.M., Martin B.D., Sugden K.D.. Oxidized guanine lesions as modulators of gene transcription. Altered p50 binding affinity and repair shielding by 7,8-dihydro-8-oxo-2′-deoxyguanosine lesions in the NF-kappaB promoter element. Biochemistry. 2003; 42:9761–9770. [DOI] [PubMed] [Google Scholar]
- 67. Moore S.P., Toomire K.J., Strauss P.R.. DNA modifications repaired by base excision repair are epigenetic. DNA Repair (Amst). 2013; 12:1152–1158. [DOI] [PubMed] [Google Scholar]
- 68. Ding Y., Fleming A.M., Burrows C.J.. Sequencing the mouse genome for the oxidatively modified base 8-oxo-7,8-dihydroguanine by OG-Seq. J. Am. Chem. Soc. 2017; 139:2569–2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Visnes T., Cazares-Korner A., Hao W., Wallner O., Masuyer G., Loseva O., Mortusewicz O., Wiita E., Sarno A., Manoilov A. et al.. Small-molecule inhibitor of OGG1 suppresses proinflammatory gene expression and inflammation. Science. 2018; 362:834–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Brooks T.A., Hurley L.H.. The role of supercoiling in transcriptional control of MYC and its importance in molecular therapeutics. Nat. Rev. Cancer. 2009; 9:849–861. [DOI] [PubMed] [Google Scholar]
- 71. Chaires J.B., Trent J.O., Gray R.D., Dean W.L., Buscaglia R., Thomas S.D., Miller D.M.. An improved model for the hTERT promoter quadruplex. PLoS One. 2014; 9:e115580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Qin Y., Rezler E.M., Gokhale V., Sun D., Hurley L.H.. Characterization of the G-quadruplexes in the duplex nuclease hypersensitive element of the PDGF-A promoter and modulation of PDGF-A promoter activity by TMPyP4. Nucleic Acids Res. 2007; 35:7698–7713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. De Armond R., Wood S., Sun D., Hurley L.H., Ebbinghaus S.W.. Evidence for the presence of a guanine quadruplex forming region within a polypurine tract of the hypoxia inducible factor 1alpha promoter. Biochemistry. 2005; 44:16341–16350. [DOI] [PubMed] [Google Scholar]
- 74. Wei D., Husby J., Neidle S.. Flexibility and structural conservation in a c-KIT G-quadruplex. Nucleic Acids Res. 2015; 43:629–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.