Abstract
RecA is essential to recombinational DNA repair in which RecA filaments mediate the homologous DNA pairing and strand exchange. Both RecA filament assembly and the subsequent DNA strand exchange are directional. Here, we demonstrate that the polarity of DNA strand exchange is embedded within RecA filaments even in the absence of ATP hydrolysis, at least over short DNA segments. Using single-molecule tethered particle motion, we show that successful strand exchange in the presence of ATP proceeds with a 5′-to-3′ polarity, as demonstrated previously. RecA filaments prepared with ATPγS also exhibit a 5′-to-3′ progress of strand exchange, suggesting that the polarity is not determined by RecA disassembly and/or ATP hydrolysis. RecAΔC17 mutants, lacking a C-terminal autoregulatory flap, also promote strand exchange in a 5′-to-3′ polarity in ATPγS, a polarity that is largely lost with this RecA variant when ATP is hydrolyzed. We propose that there is an inherent strand exchange polarity mediated by the structure of the RecA filament groove, associated by conformation changes propagated in a polar manner as DNA is progressively exchanged. ATP hydrolysis is coupled to polar strand exchange over longer distances, and its contribution to the polarity requires an intact RecA C-terminus.
INTRODUCTION
Homologous recombination (HR) is the exchange of genetic information between identical or nearly identical DNA sequences. In bacteria, the major function of homologous recombination is the recombinational DNA repair of stalled or collapsed replication forks (1–4). Secondary functions include recombination during transformation and conjugation (5). In eukaryotes, recombinational DNA repair systems have also been re-purposed by evolution to promote crossover formation during meiosis I (6–8). The fundamental steps of the homologous recombination process include the alignment of homologous DNA sequences, invasion of a single-strand nucleoprotein filament into a homologous duplex and strand exchange. All these steps are catalyzed by recombinases. The bacterial RecA recombinase catalyzes these steps with significant efficiency in the absence of other accessory proteins.
The RecA nucleoprotein filament assembles, disassembles and promotes strand exchange with a predominant 5′-to-3′ polarity (9–20). However, the molecular basis of that polarity remains unclear. The observed polarity of RecA-mediated DNA strand exchange and filament disassembly from DNA relies on ATP hydrolysis to a large extent. In standard strand exchange reactions between a circular single-stranded DNA (ssDNA) and a linear duplex DNA, efficient and extensive DNA strand exchange coupled to ATP hydrolysis is reliably polar and proceeds 5′-to-3′ relative to the ssDNA to which the RecA protein filament is initially bound (11,14,17,19–24). In the absence of ATP hydrolysis, no polarity in strand exchange has been detected in numerous studies (11,17,21,22,25–28). This overall conclusion that polar strand exchange is linked to ATP hydrolysis has been contradicted by only one experiment reported in one study (29). The evidence for a requisite coupling between ATP hydrolysis and the phenomenon of polarity in strand exchange are thus substantial and are generally viewed as established (1,30–39).
Single-molecule approaches allow the examination of such issues with higher resolution. One of the recent efforts to examine RecA-mediated strand exchange with single molecule approaches noted that short strand exchange reactions in the presence of ATPγS proceeded with kinetics inconsistent with a bipolar reaction, but the direction of the suggested polarity was not determined (40).
RecA recombinases have to first assemble on single-stranded DNA to form RecA recombinase nucleoprotein filaments, the active component involved in the HR process. Previous work showed that this assembly process includes a slow nucleation step followed by rapid growth (41,42). Growth occurs not exclusively but predominantly at the 3′-proximal end (9,43,44). RecA also dissociates from the 5′-proximal end of the filament in a process coupled with ATP hydrolysis, leading to a net 5′-to-3′ growth (10,13,43,45,46). In E. coli, RecBCD initiates homologous recombinational repair of double-stranded breaks by recognizing, processing and generating a chi-containing, 3′-terminating single-strand DNA end for RecA assembly onto nucleoprotein filaments. Also, previous studies suggested that 3′-proximal end invasion by the RecA nucleoprotein filament is preferred to form a stable joint molecule that can subsequently be extended by a DNA polymerase (45,46). However, in the extended strand exchange phase, the RecA-mediated reaction proceeds with a 5′-to-3′ polarity (11,17,19,23,47,48) that would tend to eliminate any D-loop that formed. How can these observed polarities of RecA-mediated steps be reconciled to offer a consistent picture of the HR process? In this work, we used single-molecule experiments to visualize DNA strand exchange in real time. With relatively short RecA filaments (∼70–80 subunits), we demonstrate that successful DNA strand exchange events—those that result in the formation of stable products—are preferentially initiated at the 5′-proximal end of the bound ssDNA whether or not ATP is hydrolyzed. This observed 5′-proximal end preference for a successful reaction is not a recognition of that DNA end, but rather reflects the subsequent 5′-to-3′ polarity of strand exchange. Strand exchange polarity is thus an intrinsic property of the filament. There are thus two mechanistic components of a RecA-mediated DNA strand exchange reaction, which exhibit a pronounced polarity, one intrinsic to the filament that does not rely on ATP hydrolysis, and another (previously documented) that is layered onto the first that is dictated by ATP hydrolysis and mediates extensive strand exchange.
MATERIALS AND METHODS
Proteins and DNA substrates
Escherichia coli wild-type RecA protein and RecAΔC17 were purified as described (49). Hybrid DNA substrates were generated using the protocol described earlier (50,51), with all the primers listed in Supplementary Table SI. The short homologous hybrid DNA substrate used here is a 229/149 nt DNA, with a 40 nt overhang at both its 5′- and 3′-proximal ends. The long homologous hybrid DNA substrate is a 427/352 nt DNA, with a 40 and 35 nt gaps at its 5′- and 3′-proximal ends. Two other longer hybrid DNA substrates used in Supplementary Figure S2 included the 427/392 with only 5′-overhang (40 nt), and the 427/387 with only 3′-overhang (35 nt), in order to study the effect of single-stranded overhang of the duplex DNA on the invading process. ssDNA substrates were prepared by polymerase chain reaction (PCR) containing a phosphate-labeled primer followed by a lambda exonuclease digestion to remove the 5′-phosphate-labeled strand. In controlled experiments where surface-attachment was made through the 3′-digoxigenin end of DNA, the homologous hybrid DNA substrate is a 231/167 nt DNA, with a 30 nt and a 34 nt overhang at its 5′- and 3′-proximal ends. To generate the 3′-biotinylated-labeled 231 nt ssDNA, we used a 36 nt oligo A to anneal with 3′-biotin-labeled 45 nt primer and 186 nt ssDNA together. After the ligation, another complementary 36 nt oligo B (10-fold excess) was added to compete with free oligo A. ssDNA gap substrates, containing 264 nt secondary structure-free, AC repeat ssDNA, used in the filament assembly experiments were prepared as previously described (52,53).
Single-molecule TPM measurement and data analysis
Microscope imaging setup, coverglass reaction chambers and streptavidin-coated beads were prepared as previously described (53,54). Videos of experiments were acquired by a Newvicon camera (DAGE-MTI) using a custom software written in LabVIEW.
RecA nucleoprotein filaments were pre-assembled at 37°C for 10 min with specified cofactors before flowing into the coverglass chamber to initiate the reaction. Synaptic events were identified once stably tethered beads were observed when individually bead-attached, RecA-coated ssDNA nucleoprotein filaments attached to the surface-bound hybrid DNA molecules. To study the initial invading process, only transient tethers with a lifetime longer than 0.13 s were analyzed (50). The reaction buffer (pH 7.5) contains 25 mM Tris–HCl, 3 mM potassium glutamate, 10 mM magnesium acetate, 5 mM DTT, 5% glycerol as well as specified nucleotide cofactors in 2 mM. The initial Brownian motion (BM) value was obtained by calculating the standard deviation (SD) of tethered bead’s x-centroid positions using the first few frames up to ∼1.3 s. Only beads with symmetrical BM in x- and y-direction were analyzed.
‘Sum of the two-dice’ model
The observed initial BM distribution for successful and unsuccessful events can be described by a simple ‘sum of the two-dice’ model (see Supplemental Data for more details). Due to the differences in length and mechanical properties of a RecA-bound nucleoprotein filament, the filament makes a contribution to overall Brownian motion that is about ∼2.5 times greater than the unbound DNA. In the model, we arbitrarily divide our surface-bound DNA substrate into four segments (y1 to y4) that can contribute to BM and the RecA nucleoprotein filament into 10 segments (x1 to x10) that can make a similar segmental contribution to BM. The bead is attached to the incoming nucleoprotein filament. When tethered, the initial Brownian motion of the bead will reflect the point at which the filament first contacts the surface-linked DNA duplex, and will be the sum of the DNA segments between the contact point and the surface plus the nucleoprotein segments from the contact point to the end with the bead attached. In the case of ‘random collision’, a collision between RecA nucleoprotein filament and surface-bound DNA can occur anywhere, like rolling two independent dice (one with four faces and one with ten). The simulated initial BM distribution of this random process leads to a flat, wide shape (black square labeled curve, also see Supplementary Figure S1B for details). When there is a preference for the 5′- or 3′-proximal end segment of the RecA nucleoprotein filament, the simulated distribution of initial Brownian motion values is concentrated on small values (where the RecA nucleoprotein filament is contributing relatively little, Supplementary Figure S1C) or high values (where the RecA nucleoprotein filament contributes much more, Supplementary Figure S1D), respectively. These are indicated by red triangles and blue circles, respectively.
RESULTS
Synapse dwell time reflects synapse stability
RecA-mediated homologous recombination is initiated with a RecA nucleoprotein filament-mediated search for a homologous sequence to form a synapse intermediate (55–58). Synapse stability has been investigated previously (59–61). However, most approaches to date have been unable to separately define on rates and off rates. Here, we used the dwell time of transient tethered beads in the invading strand TPM experiments to directly reveal the rate constants defining synapse stability.
A variant of single-molecule tethered particle motion (TPM), utilized here, allows the visualization of RecA-mediated DNA strand exchange over short distances in real time (50,62). In this invading strand experiment, a hybrid DNA substrate containing ssDNA overhangs (40 nt) on both ends is prepared by annealing a long (229 nt) and a short (149 nt) single-stranded DNA and is anchored on an anti-digoxigenin-coated coverglass (Figure 1A). The inclusion of the single-stranded region in the hybrid DNA efficiently increases invading events of the initiation of strand exchange (50,51). A comparable single-stranded region is present at both ends of the duplex so as not to bias initiation at one end or the other. A pre-assembled RecA-coated ssDNA (229 nt), complementary to the long, tethered strand of the duplex, is labeled with a streptavidin-coated polystyrene bead using its 5′-biotin-proximal end for direct visualization of individual synaptic events.
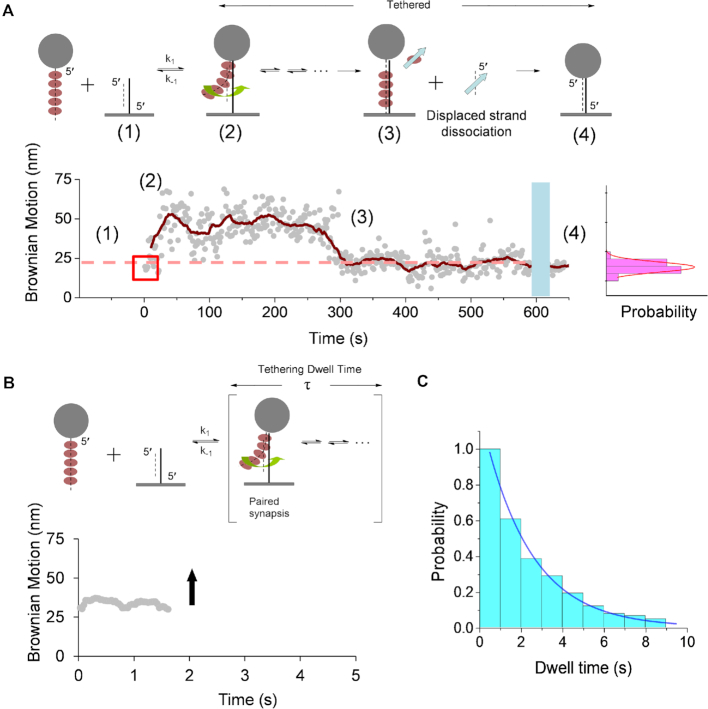
Figure 1.
The kinetic scheme and exemplary Brownian motion (BM) time-course for the RecA-mediated strand exchange reaction. (A) The kinetic scheme and experimental setup for synapse formation in successful strand exchange reactions. Bead-labeled RecA-ssDNA (229 nt) filaments were flowed into the reaction chamber containing surface-bound 229/149 hybrid DNA (stage 1). Tethered beads were observed only when a stable synaptic state was achieved (stage 2). The BM is calculated from the standard deviation of bead centroid position every 40 frames. The initial BM value (as highlighted by square) indicates the contact point between RecA filament and DNA. Strand exchange proceeds as BM increases (stage 2). The BM decrease between 250 and 320 s reflects RecA dissociation when strand exchange is completed (stage 3). A successful strand exchange reaction will lead to a final product of fully duplex DNA (229 bp). This is confirmed by extensive buffer wash to remove any excess RecA (shaded area), returning a BM ∼23 nm (stage 4), consistent with the BM of 229 bp dsDNA (histogram shown on the right). (B) An unsuccessful strand exchange reaction. Only transient tethers were observed, with bead disappearance (indicated by the upward arrow). Due to short dwell time of transient tethers (<2 s), the BM is shown by moving averages of 40 frames. (C) A histogram showing unsuccessful synapse lifetimes for RecA nucleoprotein filament with a sequence fully complementary to the surface-bound hybrid DNA in the presence of 2 mM ATP. Analysis of the synaptic state dwell time permits an evaluation of synapse stability. The data are fitted to a single exponential with 2.32 ± 0.08 s (N = 166).
As bead-attached RecA nucleoprotein filaments pair with the surface-bound hybrid DNA, a TPM bead signal appears. Tethered beads represent the invasion of a RecA nucleoprotein filament into the surface-bound duplex DNA to form the synapse. Nearly 20% of the tethered beads persisted for over several hundred seconds even after extensive buffer wash. This observation indicates that these synapses were stable and proceeded into the strand exchange step. The DNA-bead tethers undergo Brownian motion (BM), which is defined as the standard deviation of the bead centroid positions in the x-axis, as shown in Supplementary Figure S1. In TPM experiments, bead BM values are linearly correlated with DNA tether length. If strand exchange is completed, the bead will become stably tethered to the surface forming a 229 bp duplex DNA. Therefore, we can score the successful events by confirming that their final BM values are consistent with the fully 229 bp duplex DNA (histogram of Figure 1A) (50).
In addition to those successful tethered beads, the majority of the tethered beads (∼80%) were transient and not successful in completing strand exchange, suggesting that these transient synapses did not result in the completion of the strand exchange reaction. The transient RecA filament tethers generally existed for a few seconds or less. The dwell time of the synapse defines the lower limit of the off rate. The observation of the formation and detachment of these transient bead signals and the dwell time of individual synapses allow us to directly determine the factors affecting these unstable synaptic complexes.
Based on a simplified kinetic model, the synapse formation (stage 1, Figure 1A) is a rapid second-order, reversible process initiated by the collision between duplex DNA molecules and RecA nucleoprotein filaments (63–65). If successful, it is then followed by a slow multi-step, reversible, strand exchange process. The strand exchange is accompanied by an increase in Brownian motion, and then later by a decrease as RecA protein dissociates (stages 2 and 3 in Figure 1A). The tether remains stable after RecA dissociation as the bead remains attached to the dsDNA product (stage 4, Figure 1A; Supplementary Figure S2).
If unsuccessful, tethers appeared transiently and then disappeared (Figure 1B and Supplementary Figure S2). Limited by the frame interval of 33 ms, a threshold of 0.13 s (≥ four frames) (50) was chosen for analysis and counted as transient synaptic events. Hundreds of transient synaptic events were analyzed and their synapse dwell times are shown in histograms for homologous DNA with wild-type RecA unsuccessful in completing strand exchange (N = 166, 2 mM ATP; Figure 1C). These dwell times can be fitted to a single-exponential decay to obtain the synapse dwell times, 2.32 ± 0.08 s for wild-type RecA. In a control experiment containing homologous single-stranded DNA but without RecA, very few transient tethers (<10) were observed within 15 min, compared with over hundreds of events observed in reactions containing RecA. This helps to establish that registered events are RecA-mediated synaptic processes. In line with this, in experiments containing wild-type RecA-coated ssDNA with sequence heterologous to the surface-bound hybrid DNA, most of the transient tethers lasted shorter than 0.13 s (229/149 substrate, 2 mM ATP). The dwell time difference seen with the transient tethers obtained in experiments with homologous versus heterologous ssDNA coated with RecA suggests that the dwell time might also include unproductive events resulting from either initiation at not-preferred locations or reversal of otherwise successful initiation events (66).
Synapse stability and its dependencies
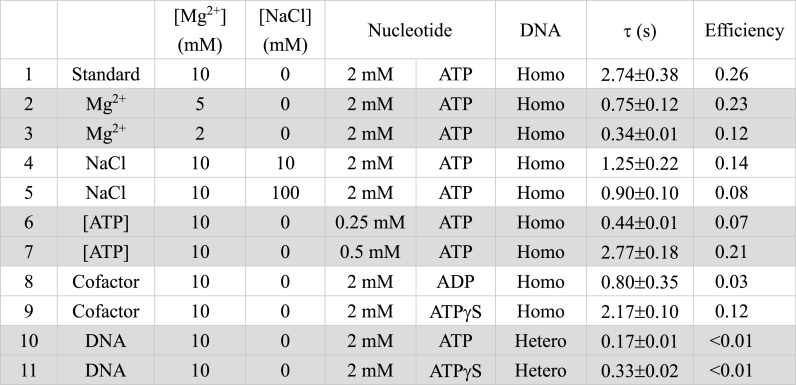
The stability of RecA nucleoprotein filaments directly affects the stability of synaptic states. When using a slightly longer 427/352 substrate, the dwell time is found to be 2.74 s (Table 1 and Supplementary Figure S3), similar to the one observed for the 229/149 substrate (∼2.32 s, Figure 1C, wild-type RecA, 2 mM ATP). These dwell times are sensitive to the concentration of divalent ions, salt concentration and nucleotide cofactor, as well as the sequence homology, as evident in the data summarized in Table 1. Our analysis below showed that these factors alter the synapse dwell time, revealing differences in synaptic stability, and in turn lead to observed differences in the efficiency of generating successful strand exchange products.
Table 1.
Lifetime of unsuccessful synapses and the strand exchange efficiency of 427/352 nt hybrid with 427 nt RecA-coated single-stranded DNA at pH 7.5
|
Length effects
A minimum length of DNA homology is thought to be necessary for the synapse formation in order to initiate the strand exchange process (67), while our previous work suggested that there is a wide range of synaptic filament size (50). The 229/149 DNA substrates with 229 nt RecA-coated single-stranded DNA have the transient dwell time of 2.32 ± 0.08 s (Figure 1C). Experiments using longer 427/352 DNA substrates with 427 nt RecA-ssDNA return a similar dwell time of 2.74 ± 0.38 s (Table 1). Using only 100 nt homologous ssDNA on the same 427/352 substrates, transient tethers also return a similar 2.43 ± 0.05 s dwell time. The lack of any dwell time dependence on homology lengths longer than 100 nt suggests that likely the minimum homology length is <100 nt (<33 RecA molecules), as expected based on studies establishing a minimum homology of ∼8 bp (55–57,67–69).
Magnesium ions and salt effects
Salt alters electrostatic screening and affects the stability of the nucleoprotein filaments (70). In addition, seven out of 25 amino acid residues in the C-terminal tail of E. coli RecA are negatively charged (71). Deletion of the C-terminal tail containing these negatively charged amino acid residues in RecA results in the enhancement in binding to duplex DNA, likely due to the reduction in electrostatic repulsion (49,72,73). Magnesium ions have been proposed to compensate the negative charges in the C-terminal tail of RecA to stabilize the RecA nucleoprotein filaments (71). Evidently, longer dwell times seen at higher magnesium concentrations (2.74 s for 10 mM Mg2+; 0.75 s for 5 mM Mg2+; 0.34 s for 2 mM Mg2+, all at 2 mM ATP and no NaCl, Table 1) are consistent with the idea that excess magnesium ions are capable of maintaining the stable synaptic state. This goes with the previous observation that the contour length of RecA nucleoprotein filament is extended 116–120% relative to duplex DNA in 1 mM magnesium ions, while at 10 mM magnesium ions, the extension is >150% at 1 mM ATPγS concentration (74,75). The negative charges in the C-terminal tail of RecA can form a network of surface salt bridges, which likely restrict access to DNA and to form a stable synaptic intermediate. Excess magnesium ions can disrupt the network of salt bridges and reduce the electrostatic repulsion between the negative charges in the RecA C-terminal tail and the DNA backbones to promote efficient DNA pairing and strand exchange (71). The decrease of synapse dwell time at increasing NaCl concentrations (over 0, 10 and 100 mM) is also consistent with the electrostatic nature of the RecA nucleoprotein filaments.
Nucleotide cofactor effects
Most ATPases, including those proteins possessing a RecA fold, exist in several nucleotide-dependent states: ATP-bound, ADP-bound or nucleotide-free states (76–82). The nucleotide states also dictate the substrate-binding affinity. RecA affinity for DNA declines with ADP or no nucleotide and increases with ATP or ATPγS (83). The nucleotide state bound to RecA also results in different structures of nucleoprotein filaments (compacted or extended form), when ATP is not being actively hydrolyzed (84,85). Additional structural alterations are produced by nucleic acids bound within RecA nucleoprotein filaments. Four states, O, Ac, Ao and P, have been proposed depending on the number of DNA strands bound to RecA and the status of a strand exchange reaction (35,71). The synapse dwell times were determined to 0.80, 2.17 and 2.74 s for ADP, ATPγS and ATP, respectively (Table 1). The short synapse dwell time of the ADP state is consistent with the model that the compact form of RecA nucleoprotein filament with ADP cannot form synapses efficiently (strand exchange efficiency ∼3%) (84). On the other hand, the ATP-like states (ATP and ATPγS) of RecA nucleoprotein filaments are active in forming synapses with longer dwell times. Our results did not show a significant difference for ATP or ATPγS states, suggesting that ATP hydrolysis is not essential for synapse formation. Since ATP binding is required for the extended RecA nucleoprotein filaments, the amount of ATP available determines the size of the continuous filament as well as its stability. We also determined the synapse dwell times at ATP concentrations below its Kd value (250 μM), around Kd (500 μM) and above (2 mM), and returned with 0.44, 2.77 and 2.74 s. At limited ATP concentration, the synapse dwell time is short, just a bit longer than the heterologous control at 2 mM ATP (0.17 s). In addition, the ATP-dependent dwell time is correlated with the RecA-mediated strand exchange efficiency (Table 1), suggesting that the ATP not only regulates the size and stability of RecA nucleoprotein filaments, but also affects the strand exchange efficiency decisively. To identify the successful events and calculate the strand exchange efficiency in ATPγS experiments, additional SDS washes were used to remove bound RecA from the products. These washes could potentially also disrupt surface–DNA or DNA–bead linkages, so the strand exchange efficiency can be underestimated for ATPγS experiments.
The correlation of synapse dwell time and the strand exchange efficiency observed here implies that the ability to stabilize the synaptic complex can potentially lead to an overall enhancement of strand exchange efficiency. On the other hand, the short synaptic dwell time of those complexes unlikely to be successful in progression into final products provides efficient turnover of RecA filaments and allows re-initiation of synaptic events at new locations.
The initial Brownian motion of RecA nucleoprotein filaments: strand exchange in the presence of ATP
RecA-mediated homologous recombination is initiated with a RecA nucleoprotein filament-mediated search for a homologous sequence (55–57). The subsequent strand exchange proceeds in the known 5′-to-3′ direction (polarity is with reference to the ssDNA bound within the RecA–ssDNA nucleoprotein filaments by convention) (11,19,20,86). The fundamental unit of homology recognition during the homology search is an 8 bp microhomology (55–57). In principle and by experimental observation (11,25,27,45,46,55), this initial pairing can occur at any location within the filament. However, subsequent steps often have a discrete polarity, and many pairing events are not productive. The bound DNA is organized into triplets (87). Once homology is located, subsequent strand exchange occurs in 3-nt increments, assuming the presence of contiguous homologous sequences (55). Our work set out to explore the polarity of these processes in more detail.
In our invading strand TPM experiment (Figure 1), the appearance of a tethered bead not only signals the formation of the synapse, but also offers information on the initial contact position of RecA nucleoprotein filaments on the duplex DNA. In TPM experiments, the apparent bead BM value is a function of DNA tether length, from surface attachment to the bead. As DNA tether length increases, the bead freedom of motion increases in a manner that translates directly to an increase in the observed Brownian motion. It has been shown that RecA binding to DNA stretches the DNA, and RecA nucleoprotein filaments have different mechanical properties than the unbound DNA alone (88). Formation of a fully coated and contiguous RecA filament on DNA increases the observed BM by a factor of ∼2.5 ((53,62) and this work). In our experiments, the initial observed Brownian motion depends upon the point at which the incoming RecA nucleoprotein filament (with bead attached at the 5′-proximal end) forms a synapse with the tethered duplex DNA, and is the sum of (i) the RecA nucleoprotein filament from the point of the synapse to the bead-attached end (x1 to xi, top, RecA-coated, Figure 2A) and (ii) the surface-bound duplex DNA from the point of synapse to the surface (y1 to yj, bottom, bare DNA). Synapses that occur closer to the 5′-end of the RecA-bound ssDNA will have lower initial BM due to a shorter RecA nucleoprotein filament contribution to the overall tether length (Figure 2D). Thus, the initial BM value (as defined in Figure 1A and Supplementary Figure S1A) offers valuable information on where the initial contact point of RecA nucleoprotein filaments occurs along the duplex DNA. We defined the initial BM value as the standard deviation of bead centroid position up to the first 1.3 s (Supplementary Figure S1A).
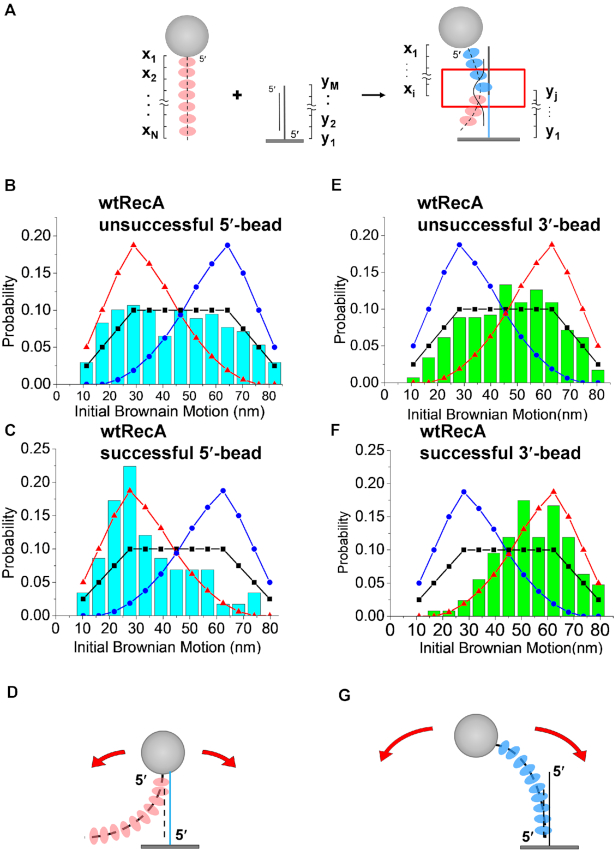
Figure 2.
Wild-type RecA nucleoprotein filaments have a 5′-proximal end preference for successful strand exchange. There is no end preference for unsuccessful reactions in the presence of ATP. (A) The initial Brownian Motion (BM) value reflects the initial contact position of the RecA nucleoprotein filaments and surface-immobilized homologous hybrid DNA. The initial BM includes the contribution of the RecA nucleoprotein filament (blue, xi) and duplex DNA (blue, yj). The 5′-DNA end is labeled with an asterisk for illustration purposes. (B–D) Histograms of initial BM values with beads attached to the 5′-proximal end of the RecA filaments (using 229 nt single-stranded DNA and 229/149 nt hybrid DNA). Simulations based on the sum of the two-dice model (see text and SuppIementary Data) are shown by the lines. The black line with squares refers to no preference for any site along the length of the RecA nucleoprotein filaments. The blue line with circles refers to the 3′-proximal end preference in the model, while the red line with triangles simulates an expected preference for the 5′-proximal end. (B) Initial BM histogram for the unsuccessful strand exchange (N = 169). The experimental data are consistent with no end preference. (C) Histograms of initial BM for the successful strand exchange (N = 58, blue bar for beads attached to 5′-proximal end). The experimental data are consistent with the red line, implying a 5′-proximal end preference. (D) An initiation point near the 5′-proximal end of the bead-attached nucleoprotein filament results in lower initial BM. (E–G) Histograms with beads attached to the 3′-end of the ssDNA bound by RecA filaments (using 231 nt single-stranded DNA and 231/167 nt hybrid DNA). (E) Histogram detailing initial BM values for the unsuccessful strand exchange events (N = 293), suggesting no site preference. (F) Histogram detailing initial BM values for successful strand exchange (N = 126, green bar for beads attached to 3′-proximal end), consistent with a 5′-proximal end preference. Due to the bead attached at the 3′-DNA end, the 5′-proximal end preference leads to a larger initial BM value. (G) Initiation near the 5′-proximal end of nucleoprotein filament results in a higher initial BM value when beads are attached to the 3′-end of the DNA bound within the filament.
Focusing on reactions with ATP, we compared the initial BM values for unsuccessful strand invasions to those that resulted in successful strand exchange. The initial BM values obtained from transient, unsuccessful synaptic events for wild-type RecA are compiled into the histogram shown in Figure 2B (N = 169, ATP, 229/149 substrate, data shown in bars). The histogram shows a rather wide, flat, continuous distribution, ranging from small BM values (∼10 nm) up to high BM values (∼90 nm). About 20% of the tethering events were successful in strand exchange, as verified by their stable presence after extensive buffer wash and by their final BM value of the expected product tether length (229 bp duplex DNA, Figure 1A). The initial BM value distribution of these successful tethers are shown in Figure 2C (N = 58). Surprisingly, the initial BM value of these successful tethers showed a narrow distribution with a peak around the relatively low value of 25 nm, significantly different from the flat distribution seen for the unsuccessful tethers. Analysis showed that the histograms of the unsuccessful (Figure 2B) and the successful (Figure 2C) tethers were statistically different (P-value = 0.5). Experiments using ATP in the presence of ATP regeneration system (4 units/ml pyruvate kinase and 1 mM phosphoenolpyruvate) returned with similar BM distributions (Supplementary Figure S4).
As mentioned, there are two segments contributing to the measured BM: the bead-attached RecA-coated ssDNA segment (x1 to xi) and the surface-bound segment (y1 to yj). The peak at the smaller initial BM value in the successful histogram (Figure 2B) indicated a smaller contribution of the RecA nucleoprotein segment in the observed BM. Considering that beads are attached on the 5′-proximal end of RecA nucleoprotein filaments, this inferred that the initial contact point for the successful tethers was at or near the 5′-proximal end segment of the RecA filament for these successful events (Figure 2D). We used a simple ‘sum of the two-dice model’ to simulate the initial BM distribution (see ‘Materials and methods’ section, Supplementary Figure S1B–D). In short, we could consider that the collision position between the RecA nucleoprotein filament and surface-bound DNA is determined by rolling two independent dice (one representing the RecA filament with 10 faces, and the other one is surface-bound DNA with 4 faces), with the faces of the dice dictating the location of filament and DNA. Thus, the observed BM value is the sum of the two-dice (Figure 2A). In the case of ‘random collision’ where the dice give random values, the model leads to a flat, wide distribution of initial BM values. However, if the dice are not random, where probabilities of each face are not equal, the initial BM distribution is peaked at either high or low BM values (Supplementary Figure S1B–D).
The initial BM distribution corresponded strikingly to the simulated 5′-proximal end preference distribution (red triangle labeled in Figure 2C, and Supplementary Figure S1C), with a high coefficient of correlation identified (see Supplementary Table SII). The simulated initial BM distribution of the random collision process (black square labeled curve and Supplementary Figure S1B) described the observed histogram of unsuccessful events pretty well (Figure 2B and Supplementary Table SII). This set of data (Figure 2B and C) clearly indicates that (i) initial interaction point of the RecA nucleoprotein filament (presumably the point of DNA pairing) could be anywhere along the length of the duplex DNA, and (ii) most successful strand exchange events resulted from the 5′-proximal end invasion of the RecA filament. This inferred that strand exchange completed with a 5′-to-3′ strand exchange polarity. Strand invasions could also occur at other locations but did not lead to measurable product formation.
Is it possible that the observed 5′-proximal end preference of RecA nucleoprotein filaments for successful strand exchange resulted from the steric hindrance caused by large 220-nm-sized beads? To exclude this possibility, we placed the surface-bound DNA upside down and had beads attached to the 3′-proximal end of the RecA filaments (see ‘Materials and methods’ section) to test if the end preference still existed. The histograms of the initial BM value for all of the unsuccessful tethers were similar whether beads were attached to the 3′-proximal end (Figure 2E) or the 5′-proximal end (Figure 2B), both showing a wide, flat distribution. However, the initial BM histogram for the successful events with beads attached at the 3′-proximal end of RecA filaments now shifted to a higher BM value (Figure 2F), significantly different from those events with beads attached at the 5′-proximal end (Figure 2C). The higher initial BM suggests larger contributions from the 3′-proximal end-bead-attached RecA segment, which indicates again that the contact point is closer to the 5′-proximal end segment of the RecA filaments (Figure 2G). Both bead attachment orientations return with the same conclusion: successful invading and strand exchange events are generally initiated at the 5′-proximal end of the bound ssDNA, and thus proceed in a 5′-to-3′ direction.
Experiments using the same design but a longer DNA length (427 nt RecA-coated ssDNA and 427/352 hybrid DNA) were carried out (Supplementary Figure S5). Different values of initial Brownian motion were observed, but the BM distribution trends remained the same: flat for unsuccessful (Supplementary Figure S5A) and a low BM value peak for successful (Supplementary Figure S5B, ATP) strand exchange reactions. This confirms the same 5′-proximal end preference for successful reactions. In summary, the polarity of strand exchange observed in the presence of ATP corresponds closely to the polarity reported broadly in the literature (11,14,17,19–24).
We wished to confirm that the single-stranded gaps at either end of the tethered duplex DNA were not affecting the outcome of the experiment. We, therefore, constructed two additional duplexes, placing the single-stranded gap uniquely at either the 3′-proximal end or 5′-proximal end of the surface-bound hybrid DNA (427/392 and 427/387 nt hybrid) with the same RecA-coated 427 nt complementary ssDNA (see Supplementary Figure S5C and D). Both of these DNAs generated the same result (in the presence of ATP), with successful DNA strand exchange initiating at the 5′-proximal end of the invading ssDNA. As was the case for the other experiments, the rapid initial pairing of RecA nucleoprotein filaments displayed no polarity preference on surface hybrid DNA for unsuccessful events.
The initial Brownian motion of RecA nucleoprotein filaments: strand exchange in the presence of ATPγS
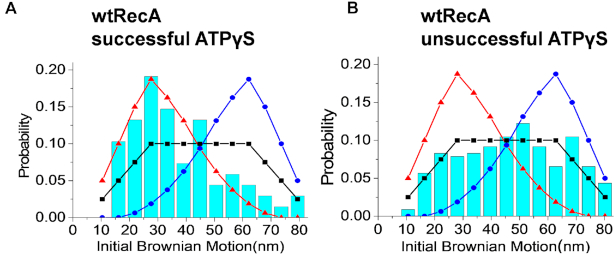
RecA nucleoprotein filaments display assembly and disassembly dynamics in the presence of ATP, as ATP hydrolysis triggers RecA dissociation from the 5′-proximal end of filaments (10,13,44,89). There is some indication that the polarity of strand exchange is enforced by a coupling to that dissociation, at least with certain RecA mutant proteins (90). A coupling of ATP hydrolysis to extensive unidirectional strand exchange (thousands of base pairs) has been demonstrated in multiple studies as described in ‘Introduction’ section. Could filament dynamics place a constraint on the observed 5′-to-3′ polarity observed for successful reactions? To address this, we conducted similar DNA strand exchange experiments using ATPγS, an ATP analog that is not appreciably hydrolyzed by RecA on the timescale of these experiments. RecA proteins were pre-incubated with ATPγS to form nucleoprotein filaments on the ssDNA with attached beads and then flowed into the reaction chamber to react with homologous hybrid DNA as before. Again we focused on the initial Brownian motion of observed tethers in the entire field, and also separately documented those that resulted in successful strand exchange reactions. The segments undergoing strand exchange in these experiments are sufficiently short so that complete strand exchange can be accomplished with ATPγS (17). Approximately 12% of the observed tethers underwent successful strand exchange in these experiments, measured as already described. Somewhat surprisingly, the initial BM of successful events again revealed a strong and unambiguous preference for the initiation of strand exchange at the 5′-proximal end of the invading ssDNA (Figure 3A), while unsuccessful events again showed no end preference (Figure 3B). With these relatively short DNA substrates, this is the same result as observed in the presence of ATP. Since there is very little RecA assembly/disassembly dynamics in the ATPγS reactions, the observed strong 5′-proximal end preference in successful strand exchange reactions cannot be attributed to a RecA filament disassembly process. The results infer an inherent 5′-to-3′ polarity of strand exchange embedded in the filament structure. Experiments using another non-hydrolyzable analog, AMP-PNP, are relatively inefficient in strand exchange. However, the limited data generated provided initial BM distributions (Supplementary Figure S6), very similar to the case of ATPγS. This result strongly counters the literature consensus (11,17,21,22,25–28) that strand exchange polarity is always tightly coupled to ATP hydrolysis.
Figure 3.
In the presence of ATPγS, wild-type RecA nucleoprotein filaments retain the 5′-proximal end preference for successful strand exchange, and exhibit no end preference for unsuccessful reactions. (A and B) This set of experiments used 5′-labeled 229 nt single-stranded DNA and 229/149 nt surface-bound hybrid DNA. (A) Histogram for initial BM values for successful strand exchange in the case of wild-type RecA filaments with beads attached at the 5′-end of the bound DNA in the presence of non-hydrolyzable ATPγS (N = 68). The experimental data are consistent with the 5′-proximal end preference (red line). (B) Histogram detailing initial BM values for the unsuccessful strand exchange with wild-type RecA in ATPγS (N = 229), suggesting no end preference.
The initial Brownian motion of RecAΔC17 nucleoprotein filaments
More than half of the last 25 amino acid residues in the C-terminal domain of RecA protein are either negatively charged (7 of the last 17 are Glu or Asp residues) or contain hydroxyl groups (six Ser or Thr residues), with an absence of positively charged residues (91). In addition, the last 24 residues are disordered in most previously determined crystal structures, leaving the precise orientation of this C-terminal tail unspecified (92,93). Expression of a RecAΔC17 mutant, where the last 17 amino acids of C-terminal are truncated, does not affect the UV resistance, the SOS response induction, or measured levels of recombination (94). In fact, RecAΔC17 has a higher binding affinity to dsDNA than the wild-type RecA and exhibits significant differences in pH- and magnesium ion-dependence for DNA strand exchange (49,90). The C-terminal tail has been proposed to act as a regulatory flap region to modulate the interaction between RecA monomers and other RecA functions (49,71,95).
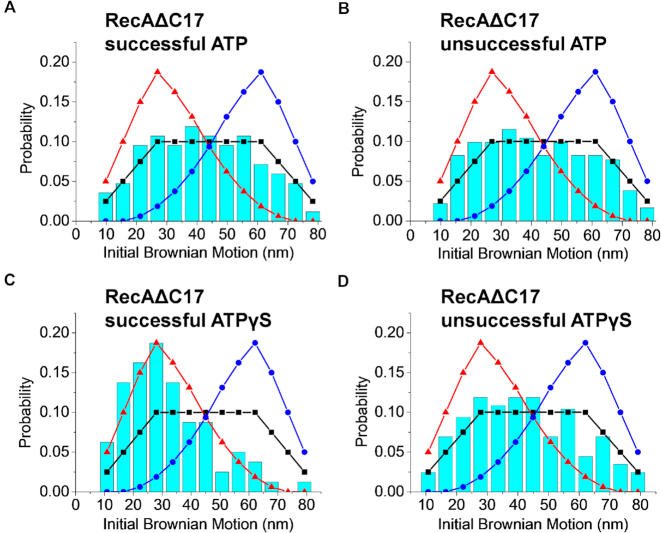
To determine if the RecA C-terminus affects the observed polarity of DNA strand exchange in this system, we carried out the same strand exchange experiments using RecAΔC17. We carried out the experiments in the presence of 2 mM ATP and 10 mM magnesium ions at pH 7.5. Approximately 2.6 ± 0.26% (N = 6) of the observed tethers led to successful DNA strand exchange in this experiment. Although the success rate of strand exchange declined markedly with this RecA variant under these conditions, control experiments containing no RecA in the reaction showed no successful tethers (0, N = 5, see Supplementary Figure S7). Controls with heterologous DNAs also showed no successful tethers (1.0 ± 0.20%) (50,62) so that the success rate, while low, is still significant. Over 80 successful events of RecAΔC17 mutants were analyzed. In contrast to what was observed with wtRecA, the initial BM value of successful and unsuccessful tethers of RecAΔC17 filaments both showed no particular initiation end preference with ATP, indicating that the polarity of the subsequent strand exchange was abolished in the absence of the C-terminal tail (Figure 4A and B). Both initial BM distributions can be well described by the random collision of the two-dice model (black squared curve).
Figure 4.
RecAΔC17 nucleoprotein filaments have no end preference for successful strand exchange in the presence of ATP, but retain the 5′-end preference in the presence of ATPγS. (A–D) This two sets of experiments used 5′-labeled 229 nt single-stranded DNA and 229/149 nt surface-bound hybrid DNA. (A) Histogram detailing initial BM values for successful RecAΔC17 nucleoprotein filaments with beads attached to the 5′-end of the bound DNA in the presence of ATP (N = 84). The experimental data indicate no end preference (black line). (B) Histogram detailing initial BM values for the unsuccessful RecAΔC17 nucleoprotein filaments (N = 182) in the presence of ATP shows no end preference. (C) Histogram detailing initial BM values for successful strand exchange by RecAΔC17 nucleoprotein filaments in the presence of ATPγS (N = 80) is most consistent with the 5′-proximal end preference model (red line). (D) Histogram detailing initial BM values for unsuccessful strand exchange by RecAΔC17 nucleoprotein filaments in the presence of ATPγS (N = 202) is most consistent with the no end preference model (black line).
We then carried out DNA strand exchange experiments with RecAΔC17 nucleoprotein filaments in the presence of 2 mM ATPγS. The reaction efficiency improved with this ATP analog, with approximately 8.9 ± 3.3% (N = 6) of the tethers leading to successful DNA strand exchange. The initial BM value of the RecAΔC17 successful tethers shows a clear peak at a small value (∼30 nm, Figure 4C), again strongly indicating initiation at the 5′-proximal end, similar to the wild-type RecA. Unsuccessful, transient tethers exhibited initial Brownian motion resembling the random process as seen in the wild-type RecA (Figure 4D).
Since the C-terminal tail acts as a regulatory flap region to modulate the interaction among RecA monomers, the apparent disappearance of polarity in the ATP reaction suggests that the C-terminal tail plays a modulating role in the observed 5′-to-3′ polarity of strand exchange reaction when ATP is hydrolyzed. However, the greatly reduced efficiency of this reaction could be due to other factors, which we examined next.
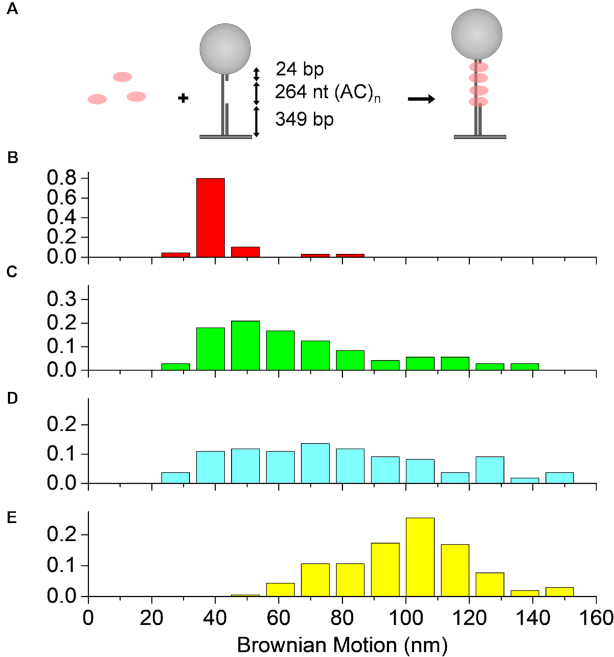
RecAΔC17 forms shorter nucleoprotein filaments
In the RecAΔC17 with ATP reactions, only ∼2.6% of overall tethers led to successful strand exchange products, significantly lower than that of the wtRecA (21%) under the same conditions. Even though RecAΔC17 mutants have a different optimal pH and Mg2+ range due to the truncation of negative charges in the C-terminus (71), the significantly reduced success efficiency led us to further examine the effect of the C-terminal truncation on the filament formation of RecAΔC17 mutants. We used the previously developed TPM assay (53,62,96) to determine the filament length of RecAΔC17 mutants under our reaction conditions in the presence of ATP or ATPγS. We prepared DNA substrates with a 264 nt gap containing only A and C residues to keep the strand free of secondary structure, flanked with duplex DNA (52), as shown in Figure 5A. We then monitored the filament assembly using the strand exchange reaction condition (pH 7.5). The BM levels of RecA filaments were determined 15 min after the introduction of wtRecA or RecAΔC17 mutants with the specified nucleotide co-factors, and the histograms are shown in Figure 5. The mean BM values of DNA substrates alone, RecAΔC17 mutant with ATP, and wtRecA with ATP exhibited a mean of 39.5, 54.2 and 101.5 nm, respectively. This indicated that RecAΔC17 mutants form relatively short nucleoprotein filaments in ATP, while wtRecA forms longer filaments. The DNA concentration in the current experiment is at picomolar levels, several million-fold lower than the DNA concentration used in previous work (49,97). The rates of DNA-dependent ATP hydrolysis by the mutant protein are not inherently greater than those of the wild-type protein (49,97). Thus, the apparent loss of polarity in the low-efficiency reactions with ATP may be a by-product of the action of relatively short and unusually dynamic filaments. Interestingly, when experiments were done using ATPγS, RecAΔC17 mutants form nucleoprotein filaments returning a broad BM distribution, with a mean value ∼80.6 nm (Figure 5D), increased from the 54.2 nm observed in the presence of ATP. The shift in the BM distribution shows that longer RecAΔC17 nucleoprotein filament lengths are formed in the presence of ATPγS. This longer filament length in ATPγS correlates well with the observed increase in successful strand exchange efficiency for RecAΔC17 mutants (8.9% for ATPγS and 2.6% for ATP). In the presence of ATP hydrolysis, the RecAΔC17 mutant protein likely forms short, discontinuous filaments. Strand exchange initiation events that would normally be eliminated might be occasionally stabilized by the formation of very transient short filaments on either side of the point of initiation. These would be quickly recycled by ATP hydrolysis. In the absence of ATP hydrolysis, a more continuous and static filament is formed, and the 5′-proximal end preference is restored.
Figure 5.
RecAΔC17 forms shorter nucleoprotein filaments in the presence of ATP. (A) Filament assembly experiments using ss/ds DNA substrates containing a 264 nt secondary-structure-free ssDNA gap at pH 7.5. (B) Histogram detailing initial BM values for DNA substrates in the absence of RecA exhibits a mean BM at ∼39.5 nm, N = 69. (C) Histogram detailing initial BM values for RecAΔC17 nucleoprotein filaments in the presence of ATP, with a mean of ∼54.2 nm (N = 72). (D) Histogram detailing initial BM values for RecAΔC17 nucleoprotein filaments in the presence of ATPγS, with a mean of ∼80.6 nm (N = 110). (E) Histogram detailing initial BM values for wild-type RecA nucleoprotein filaments, exhibiting a mean of ∼101.5 nm (N = 208). ATP and ATPγS concentrations are 2 mM for all experiments. All BM were measured 15 min after the introduction of 300 nM RecA (wild-type and mutant).
DISCUSSION
RecA promotes an extended unidirectional strand exchange, proceeding 5′-to-3′ relative to the ssDNA bound in a nucleoprotein filament, which is coupled to ATP hydrolysis (11,12,17,18). The present study documents a second and previously unobserved layer of polarity inherent within the filament. In the absence of ATP hydrolysis, the filament still has a pronounced tendency to promote strand exchange in the same 5′-to-3′ direction. The inability to detect this property in earlier studies may simply be due to the much longer length of DNA substrates used in the earlier work. We interpret the polarity arising without ATP hydrolysis as a conformation change, propagated unidirectionally from RecA subunit to RecA subunit in the filament as duplex DNA, is drawn into the filament and exchanged. The thermodynamics of the conformation change must limit reversal and structural factors must limit the reaction in the reverse direction. An ATPase-independent polarity in the strand exchange reaction has previously been seen in one experimental design (29), but its absence in numerous others (11,17,21,22,25–28) has led to the consensus that links polarity to ATP hydrolysis, a consensus that we now challenge. The present single molecule design with relatively short DNA substrates provides a new level of resolution and makes the ATPase-independent process more transparent.
The general previous failure to see strand exchange polarity in the absence of ATP hydrolysis (11,17,21,22,25–28) likely reflects a long-range effect of ATP hydrolysis in a long RecA filament. With much longer DNA substrates than those utilized here, the effect of ATP hydrolysis on polarity has been reproduced numerous times (11,17,21,22,25–28). We propose that with DNA substrates that are thousands of bp in length, DNA pairing at either end is readily detected and documented, as DNA pairing events are simply stabilized by longer regions of paired sequences. The intrinsic polarity is simply swamped out in these ensemble experiments and only revealed with single molecule approaches using short DNAs. With those same ensemble experiments with longer DNAs, ATP hydrolysis is necessary for extensive strand exchange (17), and the intrinsic polarity is strongly reinforced so that it can be observed (11,17,21,22,25–28).
Existing literature points to different end preferences of RecA filament during strand exchange. Some previous work using homology at either the 3′- or 5′-proximal ends of RecA-coated single-stranded DNA filaments actually documented that homology at the 3′-proximal end of RecA filament led to more stable joint molecule formation (64,65). As a result, it was previously proposed that the 3′-proximal end of the RecA filament is preferentially used for the successful formation of stable D-loops (45,98,99). Although 3′-DNA ends can be extended by DNA polymerase and a preference for that end might be expected, some early results could be explained by a tendency for RecA to disassemble from linear ssDNA at the 5′-proximal end, providing fewer opportunities for pairing. A similar phenomenon may limit successful pairing in the current experiments. More recent experiments using short-tailed substrates showed that 5′-proximal end substrates could be as reactive as 3′-proximal ending substrates (71), and 5′-proximal ending substrates formed more stable D-loops than the 3′-proximal ending ones. In addition, studies using DNA substrates with either short oligos or plasmid-sized ones both showed that the efficiency of RecA-mediated strand invasion is independent of the polarity of single-strand tails as long as RecA filaments contain more than 50 RecA molecules (27). The ATPase-independent polarity of strand exchange observed here over short DNA lengths provides insight that may reconcile at least some of these observations. There is clearly potential for RecA to mediate DNA pairing at any location where sufficient homology is present. The fate of those pairing events will be affected by DNA substrate structure, experimental conditions and additional properties of RecA as revealed here.
The lack of coupling between ATP hydrolysis and the polarity of DNA strand exchange, at least over short distances, may elucidate one part of the ATP hydrolytic cycle within the RecA filament. Propagation of strand exchange in one direction must have a thermodynamic explanation. The RecA-ssDNA filament, with subunits bound to ATP prior to DNA pairing, can take up a homologous duplex and pair it as previously shown (55–57). When 8 contiguous bp of homology are incorporated into the filament, a local transition occurs to a metastable strand exchange conformation. Strand exchange is then propagated in 3-nt (one RecA subunit) steps. Our work indicates that these steps/conformation changes are propagated unidirectionally along the filament, in the 5′-to-3′ direction relative to the initially bound ssDNA, in a process that does not require ATP hydrolysis. A conformation change in one RecA subunit presumably makes a similar conformation change more likely in the adjacent subunit, producing a cooperative and largely unidirectional wave traveling down the length of the filament (Figure 6). Our data cannot exclude the model that conformational change of DNA strands and their interaction with RecA’s bulky regions inside the filament may play a role in the transmission of the propagation, as the DNA strands are differently organized and positioned in the filament on each side of the strand exchange branch point.
Figure 6.
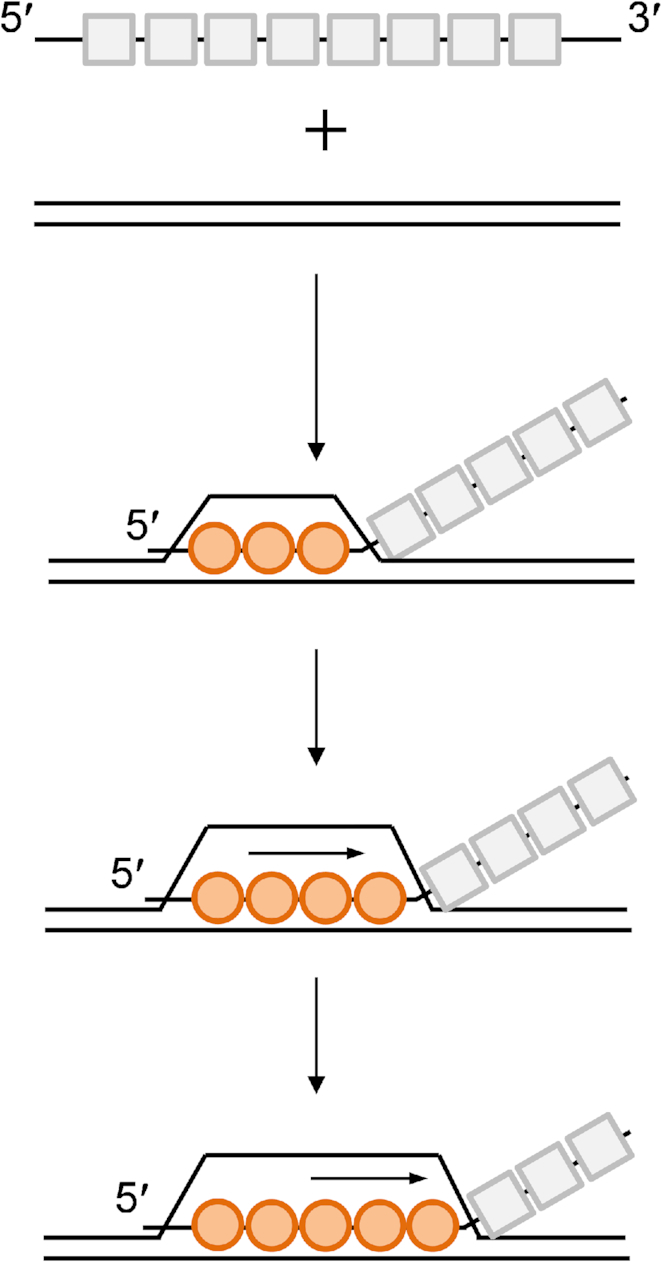
RecA strand exchange model with the polarity intrinsic to the RecA-ssDNA filaments. Structure of the RecA filament groove mediates an intrinsic strand exchange polarity. Successful strand exchange is initiated near the 5′-proximal end of RecA-ssDNA filaments, with an initial paired region of 8–9 bp (three RecA subunits). Strand exchange is propagated in 3 nt (one RecA subunit) steps. These steps are propagated unidirectionally along the filament, in the 5′-to-3′ direction relative to the initially bound ssDNA, in a process that does not require ATP hydrolysis. A conformation change in one RecA subunit makes a similar conformation change more likely in the adjacent subunit, producing a cooperative and largely unidirectional wave traveling down the length of the filament.
We also note that the promotion of a polar strand exchange reaction in the absence of ATP hydrolysis, at least over short distances, indicates that this reaction is not coupled to RecA subunit dissociation at the branch point. For the wild-type RecA protein, this argues strongly against some prominent models for RecA-mediated DNA strand exchange (42,90,100). One RecA variant that combines the ΔC17 deletion with the E38K mutation does exhibit an apparently tight coordination between subunit disassembly at the branch point and strand exchange (85), but we must now attribute that observation to a functional alteration associated with the ΔC17/E38K combination.
Summary
RecA protein promotes a strand exchange reaction with a defined polarity. A 5′-to-3′ polarity is intrinsic to the RecA–ssDNA nucleoprotein filament, and is independent of the RecA disassembly and ATP hydrolysis. The ATPase activity of RecA makes a contribution to extensive DNA strand exchange. ATP hydrolysis is coupled to nucleoprotein filament conformation changes that can promote DNA strand exchange over longer distances. Its contribution may require an intact RecA C-terminus.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Elizabeth Wood for purifying wild-type and mutant RecA proteins, and members in Li’s lab for discussion.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Ministry of Science and Technology of Taiwan [MOST 107–2113-M-002-010 to H.-W.L., MOST 107–2113-M-010–001 to H.-F.F.]; National Taiwan Univ career development grant to H.W. Li; United States National Institutes of Health [GM032335 to M.M.C.]. Funding for open access charge: MoST 107-2113-M-002-010.
Conflict of interest statement. None declared.
REFERENCES
- 1. Cox M.M. Recombinational DNA repair in bacteria and the RecA protein. Prog. Nucleic Acid Res. Mol. Biol. 1999; 63:311–366. [DOI] [PubMed] [Google Scholar]
- 2. Cox M.M., Goodman M.F., Kreuzer K.N., Sherratt D.J., Sandler S.J., Marians K.J.. The importance of repairing stalled replication forks. Nature. 2000; 404:37–41. [DOI] [PubMed] [Google Scholar]
- 3. Kowalczykowski S.C. Initiation of genetic recombination and recombination-dependent replication. Trends Biochem. Sci. 2000; 25:156–165. [DOI] [PubMed] [Google Scholar]
- 4. Kuzminov A. DNA replication meets genetic exchange: chromosomal damage and its repair by homologous recombination. Proc. Natl. Acad. Sci. U.S.A. 2001; 98:8461–8468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Babic A., Lindner A.B., Vulic M., Stewart E.J., Radman M.. Direct visualization of horizontal gene transfer. Science. 2008; 319:1533–1536. [DOI] [PubMed] [Google Scholar]
- 6. Campbell M.J., Davis R.W.. Toxic mutations in the recA gene of E. coli prevent proper chromosome segregation. J. Mol. Biol. 1999; 286:417–435. [DOI] [PubMed] [Google Scholar]
- 7. Li W., Ma H.. Double-stranded DNA breaks and gene functions in recombination and meiosis. Cell Res. 2006; 16:402–412. [DOI] [PubMed] [Google Scholar]
- 8. Shinohara A., Shinohara M.. Roles of RecA homologues Rad51 and Dmc1 during meiotic recombination. Cytogenet. Genome Res. 2004; 107:201–207. [DOI] [PubMed] [Google Scholar]
- 9. Bell J.C., Plank J.L., Dombrowski C.C., Kowalczykowski S.C.. Direct imaging of RecA nucleation and growth on single molecules of SSB-coated ssDNA. Nature. 2012; 491:274–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bork J.M., Cox M.M., Inman R.B.. RecA protein filaments disassemble in the 5′ to 3′ direction on single-stranded DNA. J. Biol. Chem. 2001; 276:45740–45743. [DOI] [PubMed] [Google Scholar]
- 11. Cox M.M., Lehman I.. Directionality and polarity in recA protein-promoted branch migration. Proc. Natl. Acad. Sci. U.S.A. 1981; 78:6018–6022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bedale W.A., Cox M.M.. Evidence for the coupling of ATP hydrolysis to the final (extension) phase of RecA protein-mediated DNA strand exchange. J. Biol. Chem. 1996; 271:5725–5732. [DOI] [PubMed] [Google Scholar]
- 13. Arenson T.A., Tsodikov O.V., Cox M.M.. Quantitative analysis of the kinetics of end-dependent disassembly of RecA filaments from ssDNA. J. Mol. Biol. 1999; 288:391–401. [DOI] [PubMed] [Google Scholar]
- 14. Shan Q., Cox M.M.. On the mechanism of RecA-mediated repair of double-strand breaks: no role for four-strand DNA pairing intermediates. Mol. Cell. 1998; 1:309–317. [DOI] [PubMed] [Google Scholar]
- 15. Shan Q., Bork J.M., Webb B.L., Inman R.B., Cox M.M.. RecA protein filaments: end-dependent dissociation from ssDNA and stabilization by RecO and RecR proteins. J. Mol. Biol. 1997; 265:519–540. [DOI] [PubMed] [Google Scholar]
- 16. Register J.C. 3rd, Griffith J.. The direction of RecA protein assembly onto single strand DNA is the same as the direction of strand assimilation during strand exchange. J. Biol. Chem. 1985; 260:12308–12312. [PubMed] [Google Scholar]
- 17. Jain S.K., Cox M.M., Inman R.B.. On the role of ATP hydrolysis in RecA protein-mediated DNA strand exchange. III. Unidirectional branch migration and extensive hybrid DNA formation. J. Biol. Chem. 1994; 269:20653–20661. [PubMed] [Google Scholar]
- 18. Cox J.M., Tsodikov O.V., Cox M.M.. Organized unidirectional waves of ATP hydrolysis within a RecA filament. PLoS Biol. 2005; 3:e52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kahn R., Cunningham R.P., DasGupta C., Radding C.M.. Polarity of heteroduplex formation promoted by Escherichia coli recA protein. Proc. Natl. Acad. Sci. U.S.A. 1981; 78:4786–4790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. West S.C., Cassuto E., Howard-Flanders P.. Heteroduplex formation by recA protein: polarity of strand exchanges. Proc. Natl. Acad. Sci. U.S.A. 1981; 78:6149–6153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Malkov V.A., Camerini-Otero R.D.. Dissociation kinetics of RecA protein-three-stranded DNA complexes reveals a low fidelity of RecA-assisted recognition of homology. J. Mol. Biol. 1998; 278:317–330. [DOI] [PubMed] [Google Scholar]
- 22. Shan Q., Cox M.M., Inman R.B.. DNA strand exchange promoted by RecA K72R. Two reaction phases with different Mg2+ requirements. J. Biol. Chem. 1996; 271:5712–5724. [DOI] [PubMed] [Google Scholar]
- 23. Gupta R.C., Golub E.I., Wold M.S., Radding C.M.. Polarity of DNA strand exchange promoted by recombination proteins of the RecA family. Proc. Natl. Acad. Sci. U.S.A. 1998; 95:9843–9848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Robu M.E., Inman R.B., Cox M.M.. RecA protein promotes the regression of stalled replication forks in vitro. Proc. Natl. Acad. Sci. U.S.A. 2001; 98:8211–8218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Dutreix M., Rao B.J., Radding C.M.. The effects on strand exchange of 5′ versus 3′ ends of single-stranded DNA in RecA nucleoprotein filaments. J. Mol. Biol. 1991; 219:645–654. [DOI] [PubMed] [Google Scholar]
- 26. Konforti B.B., Davis R.W.. ATP hydrolysis and the displaced strand are two factors that determine the polarity of RecA-promoted DNA strand exchange. J. Mol. Biol. 1992; 227:38–53. [DOI] [PubMed] [Google Scholar]
- 27. McIlwraith M.J., West S.C.. The efficiency of strand invasion by Escherichia coli RecA is dependent upon the length and polarity of ssDNA tails. J. Mol. Biol. 2001; 305:23–31. [DOI] [PubMed] [Google Scholar]
- 28. Reddy G., Jwang B., Rao B.J., Radding C.M.. Joints made by RecA protein in the interior of linear duplex DNA: effects of single-stranded ends, length of homology, and dynamic state. Biochemistry. 1994; 33:11486–11492. [DOI] [PubMed] [Google Scholar]
- 29. Rosselli W., Stasiak A.. Energetics of RecA-mediated recombination reactions. Without ATP hydrolysis RecA can mediate polar strand exchange but is unable to recycle. J. Mol. Biol. 1990; 216:335–352. [DOI] [PubMed] [Google Scholar]
- 30. Bianco P.R., Tracy R.B., Kowalczykowski S.C.. DNA strand exchange proteins: a biochemical and physical comparison. Front. Biosci. 1998; 3:D570–D603. [DOI] [PubMed] [Google Scholar]
- 31. Kowalczykowski S.C., Eggleston A.K.. Homologous pairing and DNA strand-exchange proteins. Annu. Rev. Biochem. 1994; 63:991–1043. [DOI] [PubMed] [Google Scholar]
- 32. Kowalczykowski S.C., Dixon D.A., Eggleston A.K., Lauder S.D., Rehrauer W.M.. Biochemistry of homologous recombination in Escherichia coli. Microbiol. Rev. 1994; 58:401–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Cox M.M., Lehman I.R.. Enzymes of general recombination. Annu. Rev. Biochem. 1987; 56:229–262. [DOI] [PubMed] [Google Scholar]
- 34. Cox M.M. Why does RecA protein hydrolyse ATP. Trends Biochem. Sci. 1994; 19:217–222. [DOI] [PubMed] [Google Scholar]
- 35. Cox M.M. The bacterial RecA protein as a motor protein. Annu. Rev. Microbiol. 2003; 57:551–577. [DOI] [PubMed] [Google Scholar]
- 36. Cox M.M. The RecA protein. Bacterial Chromosome. 2004; 369–388. [Google Scholar]
- 37. Lusetti S.L., Cox M.M.. The bacterial RecA protein and the recombinational DNA repair of stalled replication forks. Annu. Rev. Biochem. 2002; 71:71–100. [DOI] [PubMed] [Google Scholar]
- 38. Roca A.I., Cox M.M.. RecA protein: structure, function, and role in recombinational DNA repair. Prog. Nucleic Acid Res. Mol. Biol. 1997; 56:129–223. [DOI] [PubMed] [Google Scholar]
- 39. Ghosal D., Lowe J.. Collaborative protein filaments. EMBO J. 2015; 34:2312–2320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. van der Heijden T., Modesti M., Hage S., Kanaar R., Wyman C., Dekker C.. Homologous recombination in real time: DNA strand exchange by RecA. Mol. Cell. 2008; 30:530–538. [DOI] [PubMed] [Google Scholar]
- 41. Pugh B.F., Cox M.M.. General mechanism for RecA protein binding to duplex DNA. J. Mol. Biol. 1988; 203:479–493. [DOI] [PubMed] [Google Scholar]
- 42. Cox M.M. Motoring along with the bacterial RecA protein. Nat. Rev. Mol. Cell Biol. 2007; 8:127–138. [DOI] [PubMed] [Google Scholar]
- 43. Shan Q., Cox M.M.. RecA filament dynamics during DNA strand exchange reactions. J. Biol. Chem. 1997; 272:11063–11073. [DOI] [PubMed] [Google Scholar]
- 44. Joo C., McKinney S.A., Nakamura M., Rasnik I., Myong S., Ha T.. Real-time observation of RecA filament dynamics with single monomer resolution. Cell. 2006; 126:515–527. [DOI] [PubMed] [Google Scholar]
- 45. Konforti B.B., Davis R.W.. 3′ homologous free ends are required for stable joint molecule formation by the RecA and single-stranded binding proteins of Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 1987; 84:690–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Konforti B.B., Davis R.W.. The preference for a 3′ homologous end is intrinsic to RecA-promoted strand exchange. J. Biol. Chem. 1990; 265:6916–6920. [PubMed] [Google Scholar]
- 47. Chow S., Honigberg S., Bainton R., Radding C.. Patterns of nuclease protection during strand exchange. recA protein forms heteroduplex DNA by binding to strands of the same polarity. J. Biol. Chem. 1986; 261:6961–6971. [PubMed] [Google Scholar]
- 48. Stasiak A., Egelman E.H., Howard-Flanders P.. Structure of helical RecA-DNA complexes: III. The structural polarity of RecA filaments and functional polarity in the RecA-mediated strand exchange reaction. J. Mol. Biol. 1988; 202:659–662. [DOI] [PubMed] [Google Scholar]
- 49. Lusetti S.L., Wood E.A., Fleming C.D., Modica M.J., Korth J., Abbott L., Dwyer D.W., Roca A.I., Inman R.B., Cox M.M.. C-terminal deletions of the Escherichia coli RecA protein. Characterization of in vivo and in vitro effects. J. Biol. Chem. 2003; 278:16372–16380. [DOI] [PubMed] [Google Scholar]
- 50. Fan H.F., Cox M.M., Li H.W.. Developing single-molecule TPM experiments for direct observation of successful RecA-mediated strand exchange reaction. PLoS One. 2011; 6:e21359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Lu C.-H., Li H.-W.. DNA with different local torsional states affects RecA-mediated recombination progression. ChemPhysChem. 2017; 18:584–590. [DOI] [PubMed] [Google Scholar]
- 52. Chung C., Li H.W.. Direct observation of RecBCD helicase as single-stranded DNA translocases. J. Am. Chem. Soc. 2013; 135:8920–8925. [DOI] [PubMed] [Google Scholar]
- 53. Wu H.Y., Lu C.H., Li H.W.. RecA-SSB interaction modulates RecA nucleoprotein filament formation on SSB-wrapped DNA. Sci. Rep. 2017; 7:11876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Fan H.F., Li H.W.. Studying RecBCD helicase translocation along Chi-DNA using tethered particle motion with a stretching force. Biophys. J. 2009; 96:1875–1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Qi Z., Redding S., Lee J.Y., Gibb B., Kwon Y., Niu H., Gaines W.A., Sung P., Greene E.C.. DNA sequence alignment by microhomology sampling during homologous recombination. Cell. 2015; 160:856–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Yang D., Boyer B., Prevost C., Danilowicz C., Prentiss M.. Integrating multi-scale data on homologous recombination into a new recognition mechanism based on simulations of the RecA-ssDNA/dsDNA structure. Nucleic Acids Res. 2015; 43:10251–10263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Prentiss M., Prevost C., Danilowicz C.. Structure/function relationships in RecA protein-mediated homology recognition and strand exchange. Crit. Rev. Biochem. Mol. Biol. 2015; 50:453–476. [DOI] [PubMed] [Google Scholar]
- 58. Lee J.Y., Terakawa T., Qi Z., Steinfeld J.B., Redding S., Kwon Y., Gaines W.A., Zhao W., Sung P., Greene E.C.. Base triplet stepping by the Rad51/RecA family of recombinases. Science. 2015; 349:977–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Yancey-Wrona J.E., Camerini-Otero R.D.. The search for DNA homology does not limit stable homologous pairing promoted by RecA protein. Curr. Biol. 1995; 5:1149–1158. [DOI] [PubMed] [Google Scholar]
- 60. Bazemore L.R., Takahashi M., Radding C.M.. Kinetic analysis of pairing and strand exchange catalyzed by RecA. Detection by fluorescence energy transfer. J. Biol. Chem. 1997; 272:14672–14682. [DOI] [PubMed] [Google Scholar]
- 61. Mani A., Braslavsky I., Arbel-Goren R., Stavans J.. Caught in the act: the lifetime of synaptic intermediates during the search for homology on DNA. Nucleic Acids Res. 2010; 38:2036–2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Hsu H.F., Ngo K.V., Chitteni-Pattu S., Cox M.M., Li H.W.. Investigating Deinococcus radiodurans RecA protein filament formation on double-stranded DNA by a real-time single-molecule approach. Biochemistry. 2011; 50:8270–8280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Register J.C. 3rd, Christiansen G., Griffith J.. Electron microscopic visualization of the RecA protein-mediated pairing and branch migration phases of DNA strand exchange. J. Biol. Chem. 1987; 262:12812–12820. [PubMed] [Google Scholar]
- 64. Riddles P.W., Lehman I.R.. The formation of paranemic and plectonemic joints between DNA molecules by the recA and single-stranded DNA-binding proteins of Escherichia coli. J. Biol. Chem. 1985; 260:165–169. [PubMed] [Google Scholar]
- 65. Riddles P.W., Lehman I.R.. The formation of plectonemic joints by the recA protein of Escherichia coli. Requirement for ATP hydrolysis. J. Biol. Chem. 1985; 260:170–173. [PubMed] [Google Scholar]
- 66. Danilowicz C., Yang D., Kelley C., Prevost C., Prentiss M.. The poor homology stringency in the heteroduplex allows strand exchange to incorporate desirable mismatches without sacrificing recognition in vivo. Nucleic Acids Res. 2015; 43:6473–6485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Shen P., Huang H.V.. Homologous recombination in Escherichia coli: dependence on substrate length and homology. Genetics. 1986; 112:441–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. De Vlaminck I., van Loenhout M.T., Zweifel L., den Blanken J., Hooning K., Hage S., Kerssemakers J., Dekker C.. Mechanism of homology recognition in DNA recombination from dual-molecule experiments. Mol. Cell. 2012; 46:616–624. [DOI] [PubMed] [Google Scholar]
- 69. Hsieh P., Camerini-Otero C.S., Camerini-Otero R.D.. The synapsis event in the homologous pairing of DNAs: RecA recognizes and pairs less than one helical repeat of DNA. Proc. Natl. Acad. Sci. U.S.A. 1992; 89:6492–6496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Ragunathan K., Liu C., Ha T.. RecA filament sliding on DNA facilitates homology search. eLife. 2012; 1:e00067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Lusetti S.L., Shaw J.J., Cox M.M.. Magnesium ion-dependent activation of the RecA protein involves the C terminus. J. Biol. Chem. 2003; 278:16381–16388. [DOI] [PubMed] [Google Scholar]
- 72. Benedict R.C., Kowalczykowski S.C.. Increase of the DNA strand assimilation activity of recA protein by removal of the C terminus and structure-function studies of the resulting protein fragment. J. Biol. Chem. 1988; 263:15513–15520. [PubMed] [Google Scholar]
- 73. Tateishi S., Horii T., Ogawa T., Ogawa H.. C-terminal truncated Escherichia coli RecA protein RecA5327 has enhanced binding affinities to single- and double-stranded DNAs. J. Mol. Biol. 1992; 223:115–129. [DOI] [PubMed] [Google Scholar]
- 74. Egelman E.H., Stasiak A.. Structure of helical RecA-DNA complexes. Complexes formed in the presence of ATP-gamma-S or ATP. J. Mol. Biol. 1986; 191:677–697. [DOI] [PubMed] [Google Scholar]
- 75. Honigberg S.M., Gonda D.K., Flory J., Radding C.M.. The pairing activity of stable nucleoprotein filaments made from recA protein, single-stranded DNA, and adenosine 5′-(gamma-thio)triphosphate. J. Biol. Chem. 1985; 260:11845–11851. [PubMed] [Google Scholar]
- 76. Romberg L., Vale R.D.. Chemomechanical cycle of kinesin differs from that of myosin. Nature. 1993; 361:168–170. [DOI] [PubMed] [Google Scholar]
- 77. Taylor E.W. Cell motility. Variations on the theme of movement. Nature. 1993; 361:115–116. [DOI] [PubMed] [Google Scholar]
- 78. Berchtold H., Reshetnikova L., Reiser C.O., Schirmer N.K., Sprinzl M., Hilgenfeld R.. Crystal structure of active elongation factor Tu reveals major domain rearrangements. Nature. 1993; 365:126–132. [DOI] [PubMed] [Google Scholar]
- 79. Pai E.F., Kabsch W., Krengel U., Holmes K.C., John J., Wittinghofer A.. Structure of the guanine-nucleotide-binding domain of the Ha-ras oncogene product p21 in the triphosphate conformation. Nature. 1989; 341:209–214. [DOI] [PubMed] [Google Scholar]
- 80. Schlichting I., Almo S.C., Rapp G., Wilson K., Petratos K., Lentfer A., Wittinghofer A., Kabsch W., Pai E.F., Petsko G.A. et al.. Time-resolved X-ray crystallographic study of the conformational change in Ha-Ras p21 protein on GTP hydrolysis. Nature. 1990; 345:309–315. [DOI] [PubMed] [Google Scholar]
- 81. Milburn M.V., Tong L., deVos A.M., Brunger A., Yamaizumi Z., Nishimura S., Kim S.H.. Molecular switch for signal transduction: structural differences between active and inactive forms of protooncogenic ras proteins. Science. 1990; 247:939–945. [DOI] [PubMed] [Google Scholar]
- 82. Wang J. Nucleotide-dependent domain motions within rings of the RecA/AAA(+) superfamily. J. Struct. Biol. 2004; 148:259–267. [DOI] [PubMed] [Google Scholar]
- 83. Kowalczykowski S.C., Krupp R.A.. DNA-strand exchange promoted by RecA protein in the absence of ATP: implications for the mechanism of energy transduction in protein-promoted nucleic acid transactions. Proc. Natl. Acad. Sci. U.S.A. 1995; 92:3478–3482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Yu X., Egelman E.H.. Structural data suggest that the active and inactive forms of the RecA filament are not simply interconvertible. J. Mol. Biol. 1992; 227:334–346. [DOI] [PubMed] [Google Scholar]
- 85. Yu X., Egelman E.H.. Direct visualization of dynamics and co-operative conformational changes within RecA filaments that appear to be associated with the hydrolysis of adenosine 5′-O-(3-thiotriphosphate). J. Mol. Biol. 1992; 225:193–216. [DOI] [PubMed] [Google Scholar]
- 86. Menetski J.P., Bear D.G., Kowalczykowski S.C.. Stable DNA heteroduplex formation catalyzed by the Escherichia coli RecA protein in the absence of ATP hydrolysis. Proc. Natl. Acad. Sci. U.S.A. 1990; 87:21–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Chen Z., Yang H., Pavletich N.P.. Mechanism of homologous recombination from the RecA–ssDNA/dsDNA structures. Nature. 2008; 453:489–494. [DOI] [PubMed] [Google Scholar]
- 88. Hegner M., Smith S.B., Bustamante C.. Polymerization and mechanical properties of single RecA-DNA filaments. Proc. Natl. Acad. Sci. U.S.A. 1999; 96:10109–10114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Galletto R., Amitani I., Baskin R.J., Kowalczykowski S.C.. Direct observation of individual RecA filaments assembling on single DNA molecules. Nature. 2006; 443:875–878. [DOI] [PubMed] [Google Scholar]
- 90. Britt R.L., Haruta N., Lusetti S.L., Chitteni-Pattu S., Inman R.B., Cox M.M.. Disassembly of Escherichia coli RecA E38K/ΔC17 nucleoprotein filaments is required to complete DNA strand exchange. J. Biol. Chem. 2010; 285:3211–3226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Sancar A., Stachelek C., Konigsberg W., Rupp W.D.. Sequences of the recA gene and protein. Proc. Natl. Acad. Sci. U.S.A. 1980; 77:2611–2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Story R.M., Weber I.T., Steitz T.A.. The structure of the E. coli recA protein monomer and polymer. Nature. 1992; 355:318–325. [DOI] [PubMed] [Google Scholar]
- 93. Story R.M., Steitz T.A.. Structure of the recA protein-ADP complex. Nature. 1992; 355:374–376. [DOI] [PubMed] [Google Scholar]
- 94. Larminat F., Defais M.. Modulation of the SOS response by truncated RecA proteins. Mol. Gen. Genet. 1989; 216:106–112. [DOI] [PubMed] [Google Scholar]
- 95. Eggler A.L., Lusetti S.L., Cox M.M.. The C terminus of the Escherichia coli RecA protein modulates the DNA binding competition with single-stranded DNA-binding protein. J. Biol. Chem. 2003; 278:16389–16396. [DOI] [PubMed] [Google Scholar]
- 96. Lu C.-H., Yeh H.-Y., Su G.-C., Ito K., Kurokawa Y., Iwasaki H., Chi P., Li H.-W.. Swi5–Sfr1 stimulates Rad51 recombinase filament assembly by modulating Rad51 dissociation. Proc. Natl. Acad. Sci. U.S.A. 2018; 115:E10059–E10068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Eggler A.L., Lusetti S.L., Cox M.M.. The C terminus of the Escherichia coli RecA protein modulates the DNA binding competition with single-stranded DNA-binding protein. J. Biol. Chem. 2003; 278:16389–16396. [DOI] [PubMed] [Google Scholar]
- 98. Konforti B.B., Davis R.W.. DNA substrate requirements for stable joint molecule formation by the RecA and single-stranded DNA-binding proteins of Escherichia coli. J. Biol. Chem. 1991; 266:10112–10121. [PubMed] [Google Scholar]
- 99. Radding C.M. Helical interactions in homologous pairing and strand exchange driven by RecA protein. J. Biol. Chem. 1991; 266:5355–5358. [PubMed] [Google Scholar]
- 100. Howard-Flanders P., West S.C., Stasiak A.. Role of RecA protein spiral filaments in genetic recombination. Nature. 1984; 309:215–219. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.