Abstract
Stem cell identity and plasticity are controlled by master regulatory genes and complex circuits also involving non-coding RNAs. Circular RNAs (circRNAs) are a class of RNAs generated from protein-coding genes by backsplicing, resulting in stable RNA structures devoid of free 5’ and 3’ ends. Little is known of the mechanisms of action of circRNAs, let alone in stem cell biology. In this study, for the first time, we determined that a circRNA controls mesenchymal stem cell (MSC) identity and differentiation. High-throughput MSC expression profiling from different tissues revealed a large number of expressed circRNAs. Among those, circFOXP1 was enriched in MSCs compared to differentiated mesodermal derivatives. Silencing of circFOXP1 dramatically impaired MSC differentiation in culture and in vivo. Furthermore, we demonstrated a direct interaction between circFOXP1 and miR-17–3p/miR-127–5p, which results in the modulation of non-canonical Wnt and EGFR pathways. Finally, we addressed the interplay between canonical and non-canonical Wnt pathways. Reprogramming to pluripotency of MSCs reduced circFOXP1 and non-canonical Wnt, whereas canonical Wnt was boosted. The opposing effect was observed during generation of MSCs from human pluripotent stem cells. Our results provide unprecedented evidence for a regulatory role for circFOXP1 as a gatekeeper of pivotal stem cell molecular networks.
INTRODUCTION
Human mesenchymal stem cells (MSCs) are multipotent cells that can be isolated from various adult or perinatal tissues and possess the ability to self-renew in culture and differentiate into mesodermal derivatives, including osteocytes, chondrocytes and adipocytes (1–3). Due to this differentiation potential and other properties to regenerate injured tissues indirectly via growth factor secretion and immunomodulation, MSCs hold promise for regenerative medicine. In particular, MSCs are used in bone reconstruction therapies and are frequently associated with biomaterials or subjected to ex vivo stimuli to induce proper maturation into fully functional osteocytes and chondrocytes (4). The therapeutic efficacy of MSCs hinges upon a fine control of MSC lineage specification. Thus, it is crucial to gain a deeper understanding of the molecular mechanisms that regulate their differentiation (5–7). Factors involved in fate decision and intermediate or final stages of MSC differentiation, such as Wnt and transforming growth factor-β, have attracted increasing attention from the scientific community (8–10). On the other hand, little is known about the molecular mechanisms that regulate the maintenance of human MSC identity and their uncommitted state. Even though roles for epidermal growth factor receptor (EGFR) and non-canonical Wnt signaling have been documented (11–13), the integration of these signaling pathways with epigenetic regulators, such as non-coding RNAs (ncRNAs), is not understood. Therefore, the aim of this study was to uncover molecular networks that sustain the undifferentiated state and self-renewal of MSCs in an epigenetic perspective.
Mammalian cells contain thousands of RNA molecules that do not code for proteins, but play key roles in the regulation of physiological processes (14–16). Recent research has indicated that microRNAs (miRNAs) and long non-coding RNAs (lncRNAs) regulate the differentiation and cell fate decisions of MSCs (17–19). Circular RNAs (circRNAs) are a recently discovered class of ncRNAs. Although the existence of circRNAs was proposed more than 20 years ago, for many years they were thought to be functionless byproducts of mRNA splicing (20–22). Nevertheless, recent studies have identified a large number of endogenous circRNAs in various tissues, at different developmental stages, and in many organisms under diverse conditions of growth and stress, thus hinting at a relevant functional role of circRNAs in cellular biology and pathophysiology (23–25).
CircRNAs may be generated from exons, introns or intron-containing exons by a back-splicing reaction that covalently links an upstream 3′-splice site to a downstream 5′-splice site, leading to a closed loop structure (26). This particular conformation is reported to increase the stability of circRNA, compared to its linear counterpart. Even though the mechanisms underlying these events are not fully understood, recent studies have demonstrated that the presence of specific mammalian genomic features, such as reverse complementary sequences in the flanking introns and the activity of specific RNA-binding proteins, enhance circRNA biogenesis (27–30).
Concerning the biological function of circRNAs, their mechanisms of action have been largely unexplored. It has been proposed that some circRNAs play an important role in gene regulation by acting as competing endogenous RNAs (ceRNAs). For instance, miRNA ‘sponging’ is a mechanism of action of ceRNA, as shown for SRY and CDR1 as in neuronal tissues (24,31) and for HIPK3 in various cancers (32). In addition, circRNAs can promote the expression of their parental genes by regulating the RNA Pol II transcription complex in the nucleus (33).
Other mechanisms for circRNAs have been proposed, e.g. as hubs for protein interaction, as shown for circ-Mbl, which interacts with the Mbl protein to compete for the splicing of its linear counterpart (29). Furthermore, circ-Foxo3 has been demonstrated to regulate cell cycle progression by forming ternary complexes with CDK2 and p21 (34). Moreover, it has recently been shown that circRNAs can be translated efficiently into small truncated peptides, even though the molecular activity of this type of circRNA-derived protein is not yet understood (35). Finally, high conservation of circRNAs across species and their tissue- and developmental stage-specific expression suggest their role in cell identity and fate determination during development (23,24,36–39).
In the current study, we report the unique role of a circRNA originating from the FOXP1 gene (circFOXP1) in the maintenance of MSC identity and regulation of differentiation. CircFOXP1 acts as a miRNA sponge targeting miR-17–3p and miR-127–5p, and promotes proliferation and differentiation of MSCs, supporting the hypothesis of circRNAs as major players in stem cell fate decision-making processes.
MATERIALS AND METHODS
Cell line, cell culture and treatments
MSCs and HSFs were cultured in α-minimum essential medium supplemented with 20% fetal bovine serum (FBS; Life Technologies, cat. no. 10270–106) at 37°C and 5% CO2. MSCs were isolated from both cord blood and bone marrow, as described previously (40–43). The MSC identity was confirmed by the immunophenotype profile (1): cells were positive for the MSC surface antigens CD73, CD90 and CD105, and were negative for the hematopoietic markers CD45 and CD34 (data not shown). The human material was obtained after informed consent from healthy donors, and the study was approved by the internal Ethics Committee. All experiments were performed according to the amended Declaration of Helsinki.
Adipo-, osteo- and chondrogenic differentiation of MSCs was induced using hMSC Osteogenic Bullet Kit (Lonza, cat. no. PT-3002), hMSC Chondro Bullet Kit (Lonza, cat. no. PT-3003) and hMSC Adipogenic Bullet Kit (Lonza, cat. no. PT-3004) according to the manufacturer’s instructions. Differentiation was confirmed by staining with Oil Red O (Sigma Aldrich, cat. no. O1391) for adipogenesis, Alizarin Red (Sigma Aldrich, cat. no. 130–22-3) for osteogenesis and Alcian Blue (Sigma Aldrich, cat. no. A5268) for chondrogenesis.
HEK-293T cells were obtained from the American Type Culture Collection (ATCC) and were cultured in Dulbecco’s modified Eagle medium supplemented with 10% FBS. All cells were routinely tested for mycoplasma using the MycoAlert Mycoplasma Detection kit (Lonza, cat. no. LT07).
Human induced pluripotent stem cells (hiPSCs) derived from MSCs were generated and characterized by uSTEM company (uSTEM, Padova, Italy). uSTEM also generated BJ fibroblasts-hiPSCs used to obtain hiPSC-derived MSCs, following the protocol described elsewhere (44).
Transcription was blocked by adding 1 μg/ml actinomycin D (Sigma Aldrich, cat. no. A9415) or dimethyl sulphoxide (Sigma Aldrich, cat. no 472301) as a control to the cell culture medium.
Microarray analysis
Sample preparation and microarray hybridization were performed according to the manufacturer’s protocol (Arraystar). Briefly, circRNAs were treated with Rnase R to remove linear RNAs. Then, each sample was amplified and transcribed into fluorescent cRNA using the random priming method with a Super RNA Labeling Kit (Arraystar). The labeled cRNA was hybridized onto an Arraystar Human Circular RNA Microarray (Arraystar V1.0). The array was scanned with the Agilent Scanner G2505C, and raw data were extracted by Agilent Feature Extraction software (version 11.0.1.1). Identification of circRNAs followed circBASE database nomenclature (45).
RNA isolation, reverse transcription and qRT-PCR analysis
The total RNA from cell lines was isolated using TRIzol reagent (Ambion, cat. no. 15596–026). For RNA isolation from adipo, osteo and cartilage healthy tissues, samples were disrupted using Tissue Tearor (Biospec Products). RNA concentration and quality were verified using a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies).
For the quantitative real time PCR (qRT-PCR) assay, cDNA was synthesized from 1 μg of total RNA with SuperScript IV VILO (Invitrogen, cat. no. 11756500). The cDNA was diluted 10-fold and 1 μl used as template for qRT-PCR analysis using SYBR Select Master Mix for CFX (Life Technologies, cat. no. 4472937) on a CFX96 thermal cycler (BioRad) following the manufacturer’s instruction. According to the MIQE guidelines (46), amplification efficiency, correlation coefficient and slope of standard curves generated using serial dilutions for each primer pair were evaluated (Supplementary Table S1). The relative expression levels of the selected targets were determined using the ΔΔCt method and normalized, where not differentially specified, to ACTB mRNA levels. In the case of miRNA, the miScript II RT Kit (Qiagen, cat. no. 218161) and miScript SYBR Green PCR Kit (Qiagen, cat. no. 218073) were used, according to the manufacturer’s protocols.
qRT-PCR products were resolved on 2% agarose gel. The DNA products were gel purified using the Wizard SV Clean-Up system (Promega, cat. no. A9281), according to the manufacturer’s protocol, and submitted to Sanger sequencing for sequence validation. All primers used in this study, listed in Supplementary Table S2, were designed using Primer 3 software.
DNA construct and mutagenesis
To obtain the expression of circFOXP1, the genomic region with the Alu sequence was amplified using Q5 High-Fidelity DNA Polymerase (NEB, cat. no. M0491). The polymerase chain reaction (PCR) products were inserted into the pEYFP-C1 vector. The luciferase reporter was constructed by subcloning the circFOXP1 fragment directly downstream of the Renilla luciferase cassette into the psiCHECK2 vector. Mutation of each miRNA-binding site was performed using the Q5 Site-Directed Mutagenesis Kit (NEB, cat. no. E0554S). For the CRISPR/Cas9 assay, gRNA cloning vectors were constructed using pUC-gRNA cloning vector. All constructs were verified by sequencing. All primers used are listed in Supplementary Table S2.
RNase R treatment of total RNA
A 2 μg sample of total RNA was treated with 3 U/μg of Rnase R (Epicentre Biotechnologies, cat. no. RNR07250) or water as a control (Mock) for 20 min at 37°C. Digested RNA was subsequently purified using an RNeasy MinElute Cleanup Kit (Qiagen, cat. no. 74204).
Nuclear and cytoplasmic fractionation
Nuclear/cytoplasmic fractionation was performed as follows. Cells were washed with cold phosphate-buffered saline (PBS), harvested by scraping and centrifuged for 5 min at 500 × g. Cell pellets were lysed in 5× (v/v) ice-cold Buffer A (10 mM Tris–HCl, pH 7.4, 10 mM NaCl, 3 mM MgCl2, 10% glycerol, 1 mM ethylenediaminetetraacetic acid (EDTA), 0.5 mM dithiothreitol (DDT) and protease inhibitor cocktail) for 10 min at 4°C. Lysates were cleared by centrifugation for 5 min at 1200 × g and 4°C, and the supernatant was collected and saved as the cytoplasmic fraction on ice. The crude nuclear pellet was resuspended in 250 μl of Buffer B (10 mM Tris–HCl, pH 7.5, 15 mM KCl, 30 mM NaCl, 10% glycerol, 1 mM EDTA, 0.3% sodium deoxycholate, 0.5% IGEPAL, 0.5 mM DTT, and 1× protease inhibitor cocktail) and centrifuged for 5 min at 1200 × g and 4°C. The pellet was resuspended in 5 × (v/v) ice-cold Buffer F (10 mM Tris–HCl, pH 7.0, 100 mM NaCl, 30 mM Na Pyrophosphate (Na4P2O7), 50 mM NaF, 5 mM ZnCl2, 1% Triton X-100 and 1× protease inhibitor cocktail), incubated for 10 min on ice and then centrifuged for 5 min at 12 000 × g and 4°C. The nuclear supernatant was collected on ice.
RNA FISH
In situ hybridization was performed using biotinylated DNA oligonucleotides (probes) specific for the circFOXP1 sequence or negative control (Supplementary Table S2). MSCs at 80–90% confluence were fixed in 4% paraformaldehyde for 20 min. After prehybridization (PBS/0.5% Triton X-100), cells were incubated with probes in hybridization buffer (40% formamide, 10% dextran sulfate, 4× saline-sodium citrate buffer, 10 mM DDT and 1 mg/ml yeast transfer RNA) at 37°C overnight. Signals were detected using a tyramide-conjugated Alexa 568 fluorochrome tyramide signal amplification kit (Invitrogen, cat. no. T20934). Nuclei were counterstained with 4′,6-diamidino-2-phenylindole (Invitrogen, cat. no. D1306). Images were acquired using a Leica TCS SP2 confocal microscope, with an HCX PL APO IBD.BL 63 × /1.4 objective.
Oligonucleotide transfection
A predesigned siRNA targeting human circ_FOXP1 (si-circ_FOXP1) and negative control (scrambled siRNA, siSCRL) were purchased from Integrated DNA Technologies. The sequences used are listed in Supplementary Table S2. MSCs were transfected with siRNA at a final concentration of 10 nM using Lipofectamine RNAiMAX (Invitrogen, cat. no. 13778030), according to the manufacturer’s protocol. Cells were grown for 48 h post-transfection and harvested.
Cell proliferation assay
Transfected MSCs were seeded onto 24-well plates in culture medium containing serum and maintained for 2 days at 37°C. The cells were harvested, and the cell number was determined using a Coulter counter, as described previously (47).
Luciferase reporter assay
At 24 h prior to transfection, HEK-293T cells were seeded in 24-well plates at a density of 1 × 105. The cells were cotransfected with 0.5 μg of reporter vectors and 15 pmol of miRNA mimic. After 48 h, the luciferase activity was measured with the dual-luciferase reporter assay system (Promega, cat. no. E1910). For each assay, one internal control (Firefly Luciferase reporter, FLuc) and one negative control (miRNA negative control) were used. To take into account the transfection efficiency variability, the RLuc activity was first normalized to the FLuc activity. The fold change was determined by comparing the activity of each miRNA mimic with that of the miRNA negative control.
RNA pull-down assay
The RNA pull-down experiment was performed in at least three independent biological samples, as described previously (34,48), with minor modifications. Briefly, 1 × 107 HEK-293T cells were cross-linked with ultraviolet light at 4000 mJ/cm2 in ice-cold PBS and then pelleted at 1300 × g for 5 min. The pellet was resuspended in ice-cold Lysis Buffer (50 mM Tris–HCl, pH 7.5, 100 mM NaCl, 1% IGEPAL, 1 mM DDT, 0.1 U/μl RNasin (Promega, cat. no. N2111) and protease inhibitor cocktail). The cell lysates were incubated overnight at 37°C with 3 μg of biotinylated DNA oligonucleotides (probes), specific for the circFOXP1 sequence or negative control in two volumes of Hybridization Buffer (50 mM Tris–HCl, pH 7.5, 700 mM NaCl, 6 mM EDTA, 1% IGEPAL, 15% formamide, 1 mM DDT, 0.1 U/μl RNasin and protease inhibitor cocktail). A total of 50 μl of washed Streptavidin C1 magnetic beads (Invitrogen, cat. no. 65001) were blocked for 1 h at room temperature with 0.5 mg/ml yeast total RNA and 1 mg/ml bovine serum albumin. Subsequently, blocked beads were added to each binding reaction and incubated for 2 h at 37°C. The beads were sequentially washed five times with Wash Buffer (2× saline-sodium citrate buffer, 0.5% IGEPAL, 1 mM phenylmethylsulphonyl fluoride and 0.1 mM DDT). After treating with DNase, RNAs were extracted by an miRNeasy Mini Kit (Qiagen, cat. no. 217004) and analyzed by quantitative real-time PCR.
Surgical, cell injection and follow-up procedures
Wistar albino male rats (Rattus norvegicus) were supplied by Envigo® and allowed to acclimatize for at least 7 days before the start of surgical procedures, which were all approved by the UK Home Office and Local Research Ethics Committee. All procedures were performed under sterile conditions. Isoflurane inhalation was used for induction (5%) and maintenance (2.5–3%) of anesthesia. To avoid hypothermia during procedure, a heat pad was placed under the surgical surface, and subcutaneous pre-medications were given as follows: Buprenorphine 0.05 mg/kg, Ibuprofen 4 mg/kg, Synulox® antibiotic 0.5 ml and 5cc warm normal saline. Skin was prepared using chlorhexidine and the leg was draped. A longitudinal skin incision over the antero-medial side of the leg was made, followed by dissection of subcutaneous tissue and exposure of the tibia. Surrounding muscles and soft tissue were carefully detached from around the bone. Under 0.9% saline irrigation, a fracture/osteotomy was created in the mid-shaft using a rotatory burr and a 1-mm gap created and maintained with a stainless steel spacer. Stripping of the periosteum and endosteum was performed to a distance equal to one bone diameter of the diaphysis proximally and distally using a periosteal stripper externally and the bent end of a needle (23G) internally. A needle was introduced retrogradely from the fracture site toward the tibial tuberosity. This needle was used as guide for second needle introduced in an antegrade direction. Once the latter reached the fracture site, the spacer was inserted and the needle was then passed into the distal medullary canal and fixed into the distal fragment. The skin was sutured using an absorbable monofilament Monocryle 4/0. Another dose of Buprenorphine (0.05 mg/kg) was given after 24 h to control post-operative pain.
Three weeks after this procedure to create the fracture/osteotomy, cultured cells were trypsinized, processed and resuspended in 150 μl PBS, a 26G needle and 1 ml syringe were used to deliver the cells in vivo. Cells were injected into the fracture site using the novel Z-track technique to prevent backflow leakage and to avoid cell loss after injection. Rats were randomized into two groups and injected as follows: 4 × 105 circFOXP1-KD BM-MSCs were injected into the experimental group (n = 6) and similar number of scramble knockdown BM-MSCs was injected into the control group (n = 6).
Animals were closely monitored and no local or systemic side effects were noticed post-injection. Animals were X-rayed weekly for 8 weeks to follow the progression of the healing process.
Mechanical analysis was performed in 4-point bending using a mechanical testing machine (Zwick/Roell). A load (12.5N) was applied, producing a bending moment at the fracture site of 0.05 N m, which was considered to represent the bending moment that would occur on the tibia during normal walking. Intact legs were used as controls for each subject.
Computational and statistical analysis
Raw data of profiled circRNAs were normalized by the quantile method, as implemented by GeneSpring GX v11.5.1 software (Agilent Technologies). Differential expression determination of circRNA binding with multiple miRNAs, the details of the miRNA response elements and the initial miRNA target prediction were performed with proprietary algorithms at Arraystar, then further processed as detailed below.
The batch effect on raw data was taken into account and smoothened using the ComBat method (49). Sample distances were assessed on normalized data with principal component analysis calculated by the ‘prcomp’ function in base-R. Selection of differentially expressed features (circRNAs) was performed using fold change and unpaired t-tests with cutoffs on unadjusted P-values.
Selected miRNAs were used as inputs to build the interaction network to find genes for the pathway analysis with the CyTargetLinker plugin (using the available miRTarbase, TargetScan and microCosm modules) in Cytoscape version 3.1.0. Pathway enrichment analyses of the resulting gene lists were performed in Cytoscape using the ClueGO plugin screening GO terms, Reactome and KEGG databases; exported results were further processed and displayed with R.
All statistical tests were performed using Prism 7 (GraphPad). Statistical parameters are reported in figure legends and include: number of replicates analyzed (n), dispersion and precision measures (mean ± standard deviations or standard error of the mean) and statistical significance (P-value). All data were normally distributed according to the Shapiro–Wilk normality test and then statistically assessed by two-tailed Student’s t-test or one-way ANOVA test followed by Dunnett’s multiple comparisons post-hoc test as indicated in figure legends. In figures, asterisks mean *P < 0.05, **P < 0.01, ***P < 0.001, ns = not significant. P values <0.05 and lower were considered significant. All experiments were performed in triplicate biological replicates.
RESULTS
MSCs possess a unique circRNA expression profile compared to differentiated fibroblasts
To determine which circRNAs may play a role in MSC definition and fate decision, a high-throughput screening assay was first performed. Primary MSCs isolated from both human bone marrow (adult stem cell source; BM-MSCs, n = 3) and umbilical cord blood (perinatal/fetal stem cell source; CB-MSCs, n = 3) were used to exclude tissue source-related molecular biases. Moreover, human skin fibroblasts (differentiated stromal cell counterpart; HSFs, n = 3) were also analyzed to rule out circRNAs associated with generic stromal identity and the differentiated state (Supplementary Table S3).
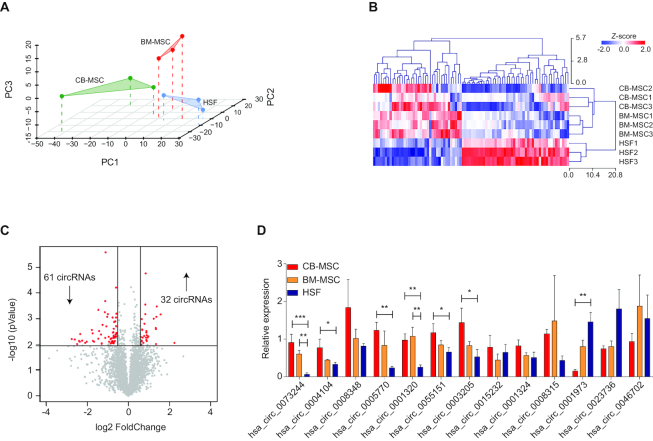
First, circRNAs were identified by microarray analysis of total RNA. Interestingly, an integrated principal component analysis (PCA) showed that the three cell populations cluster independently (Figure 1A), suggesting homogeneous and well-defined, tissue-specific circRNA expression profiles. Considering that BM-MSCs represent the gold standard in the MSC research field and that HSFs represent the differentiated stromal population, differentially expressed circRNAs between BM-MSCs and HSFs were selected for a clustering analysis along with CB-MSCs, applying a fold-change filter based on P-values ≤ 0.05. Intriguingly, unsupervised hierarchical clustering showed that CB-MSCs cluster together with BM-MSCs, while both of them were distant from HSFs (Figure 1B). Among the selected circRNAs, 93 showed differential expression, with a fold change ≥ 1.5. In detail, 32 were upregulated, whereas 61 were downregulated in MSCs compared to HSFs (Figure 1C).
Figure 1.
Mesenchymal stem cells (MSCs) possess a unique circular RNA (circRNA) expression profile compared to differentiated fibroblasts. (A) Principal component analysis of the circRNA expression profiles of bone marrow MSCs (BM-MSCs), cord blood MSCs (CB-MSCs) and human skin fibroblasts (HSFs). Each cell type was assayed in triplicate. (B) Heat map of the most differentially expressed circRNAs (cut-off was set to -1.5 > fold change >1.5) between MSCs and HSFs, with columns representing circRNAs and rows representing samples. The circRNAs were classified according to the Pearson correlation (Z-score). (C) A volcano plot was used to visualize differential circRNA expression between MSCs and HSFs. The red dots represent the differentially expressed circRNAs with statistical significance (P < 0.01); downward arrows indicate downregulated circRNAs and upward arrows indicate upregulated circRNAs. (D) The relative transcriptional level of the top 10 differentially expressed circRNAs was confirmed by qRT-PCR analysis of RNAs extracted from independent biological replicates of BM-MSCs, CB-MSCs and HSFs. Data in panel (D) are shown as means ± standard deviation (n = 3), statistical analysis was performed by one-way analysis of variance followed by the Dunnett correction; *P < 0.05, **P < 0.01, ***P < 0.001.
Subsequently, a subset of top differentially expressed circRNAs (n = 10 upregulated and n = 3 downregulated) were selected for further validation with independent biological replicates. CircRNA expression levels were quantified by qRT-PCR with divergent primers to distinguish circRNAs from linear counterparts. Those that were consistent with the microarray data are shown in Figure 1D. Finally, the presence of head-to-tail junction sequences was confirmed by Sanger sequencing for differentially expressed circRNAs (Supplementary Figure S1).
CircFOXP1 is a marker of undifferentiated MSCs
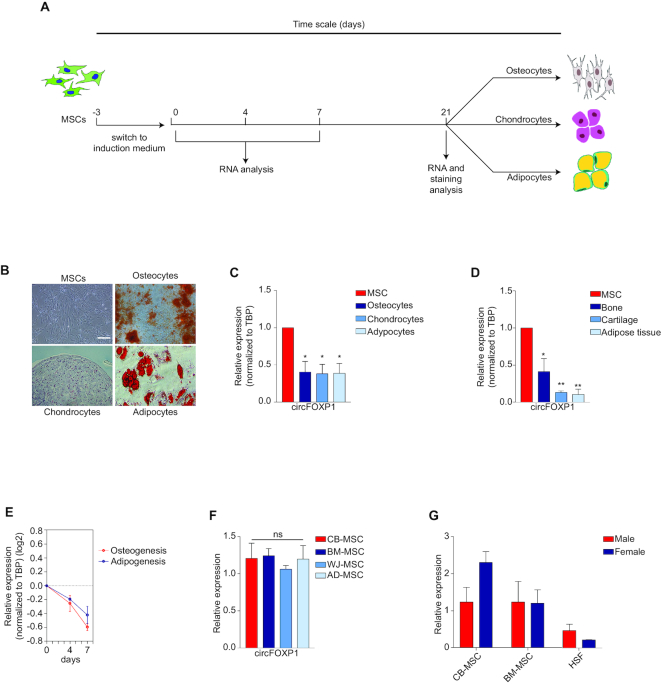
To address whether differentially expressed circRNAs actively participate in the MSC regulatory molecular network, expression changes of validated top-upregulated MSC circRNAs were examined during lineage specification. Well-established cell culture models for in vitro differentiation into mesodermal derivatives (osteo-, chondro- and adipocytes) were implemented, involving specific staining protocols and molecular analyses (Figure 2A and B; Supplementary Figure S2A). qRT-PCR analysis showed that hsa_circ_0001320, a circRNA originating from the FOXP1 gene (termed as circFOXP1), was the strongest regulated circRNA among the validated top-upregulated circRNAs in all differentiated mesodermal cell types (Figure 2C and Supplementary Figure S2B). Importantly, these data were confirmed in primary human tissues (Figure 2D). In addition, time-course analysis revealed that circFOXP1 was downregulated shortly following the first days of differentiation (Figure 2E), suggesting that its expression is strictly related to the undifferentiated MSC state. Intriguingly, circFOXP1 showed the same trend of its linear counterpart (Supplementary Figure S2C), which may hint at a correlation between circFOXP1 and FOXP1 mRNA transcriptional levels, as already reported for other circRNAs and their linear counterparts (50,51). To corroborate these data, we also analyzed circFOXP1 expression in MSCs isolated from other sources (Wharton’s jelly and adipose tissue, as additional perinatal/fetal and adult sources, respectively), and the same expression levels were observed within each group (Figure 2F). Finally, CB-MSCs and BM-MSCs, along with HSFs, derived from male and female donors showed that circFOXP1 expression is gender independent (Figure 2G). Altogether, these data indicate that circFOXP1 is a specific molecular marker for undifferentiated MSCs.
Figure 2.
CircFOXP1 is a specific mesenchymal stem cell (MSC) marker. (A) Schematic representation of the experimental outline. MSCs were grown for 3 days in the presence of serum. Starting from day 0, MSCs were induced to differentiate into osteocytes, chondrocytes and adipocytes. At different time points, the MSCs were analyzed for Alizarin Red S, Alcian Blue and Oil Red O staining positivity and the gene expression profile, as indicated. (B) Representative images taken after 21 days of differentiation. An image of MSCs at the start of differentiation is included; scale bar, 200 μm. (C) qRT-PCR for circFOXP1 in MSCs, osteocytes, chondrocytes and adipocytes at the end of MSC differentiation. (D) qRT-PCR for circFOXP1 in MSCs as well as bone, cartilage and adipose tissue. (E) The relative transcriptional level of circFOXP1 as MSCs (day 0) differentiated into osteocytes (day 4) and adipocytes (day 7) was measured by qRT-PCR. Transcriptional levels were normalized to those of MSCs and were represented in log2 scale. (F) qRT-PCR for circFOXP1 in MSCs originating from other sources, including Wharton’s jelly (WJ-MSCs) and adipose tissue (AD-MSCs). (G) The transcriptional level of circFOXP1 in bone marrow MSCs (BM-MSCs), cord blood MSCs (CB-MSCs) and human skin fibroblasts (HSFs) isolated from male and female individuals was measured by qRT-PCR. The relative expression levels in panels (C-E) were normalized to TBP. Data in panel (G) are shown as means ± standard deviation (n = 3); statistical analysis was performed by the Wilcoxon–Mann–Whitney non-parametric test. Data in panels (C), (D) and (F) are shown as means ± standard error of the mean (n = 3); statistical analysis was performed by one-way analysis of variance followed by the Dunnett correction; ns, not significant; *P < 0.05, **P < 0.01.
CircFOXP1 is an exon-based RNase-resistant RNA that localizes to the cytoplasm
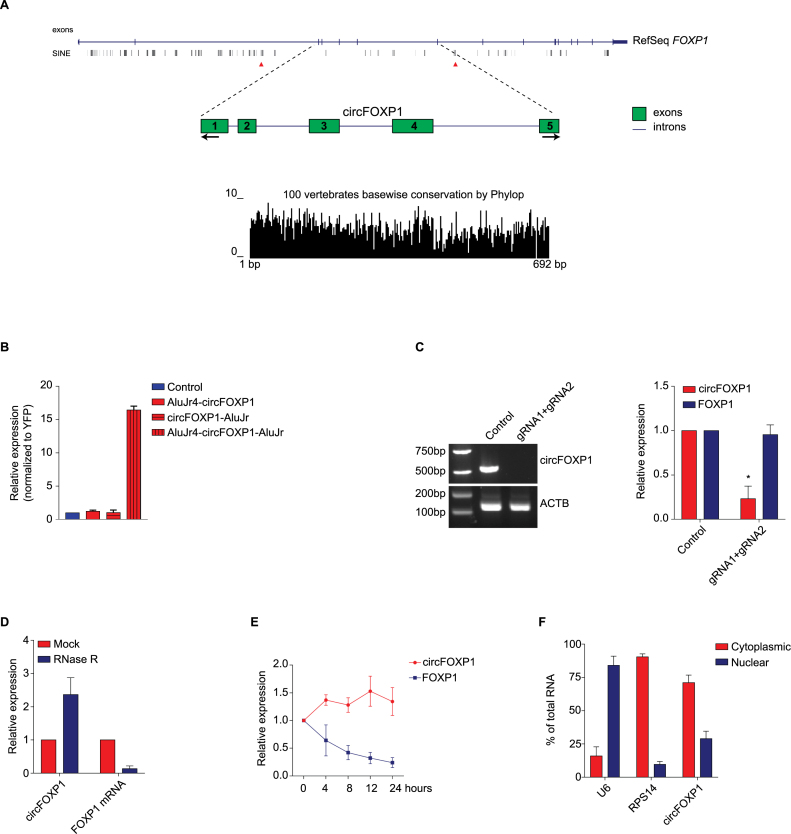
The genomic structure of circFOXP1 was obtained from the UCSC genome database (http://genome.ucsc.edu/). Its full genomic length is 38 225 bp, while its spliced length is 692 bp, comprised of five exons with high conservation across 100 species of vertebrates (Figure 3A). Subsequently, the mechanism of circFOXP1 biogenesis was investigated. It has been proposed that complementary Alu sequences in an inverted orientation across long flanking introns can facilitate the formation of circular RNAs (27,52,53). Along this hypothesis, the introns flanking circFOXP1 were analyzed. Interestingly, two inverted short-interspersed elements, AluJr4 (18 267 bp downstream the 3’-end of circFOXP1) and AluJr (5492 bp upstream the 5’-end of circFOXP1), showing a highly reverse complementarity (71% identity over 239 nt; Supplementary Figure S3A), were detected. Thus, two 500-nt regions containing these Alu sequences were cloned into the pEYFP expression vector in different combinations (Supplementary Figure S3B). Modulation of the circRNA transcriptional level was detected by qRT-PCR analysis after transfection of human embryonic kidney (HEK)-293T cells with the generated expression vectors. According to previous analyses, the presence of two inverted complementary elements in the expression vector was essential for the formation of circFOXP1, while circularization was not increased when one or both Alu sequences were missing (Figure 3B). To validate the role of Alu sequences in the circularization in a more physiological and refined setting, we deleted the repeated sequence using CRISPR/Cas9 technology. Two gRNAs were designed to target the external ends of the AluJr4 region (Supplementary Figure S3C), and they were co-transfected in HEK-293T cells stably expressing Cas9. We confirmed the deletion of AluJr4 sequence by PCR. As previously demonstrated, the absence of Alu region inhibited the circularization of circFOXP1 (Figure 3C).
Figure 3.
CircFOXP1 is an exon-based RNase-resistant RNA that localizes to the cytoplasm. (A) Schematic representation of the genomic regions of the FOXP1 gene from which circFOXP1 is originated, showing flanking Alu repeats and other short interspersed nuclear elements. Red arrows indicate the complementary Alu sequences (AluJr4 and AluJr). Black arrows represent divergent primers binding to the transcribed genomic region of circFOXP1. Conservation across 100 vertebrate species of the circFOXP1 sequence is illustrated. (B) HEK-293T cells were transfected with the expression plasmids (pEYFP-: control, AluJr4-circFOXP1, circ-FOXP1-AluJr and AluJr4-circFOXP1-AluJr), and at 48 h post-transfection, the amount of circFOXP1 was measured by qRT-PCR and normalized to yellow fluorescent protein (YFP). (C) Normal PCR data for CRISPR/Cas9-mediated deletions of AluJr4 region and qRT-PCR for the reduction of circFOXP1 expression. (D) qRT-PCR was used to measure the abundance of circFOXP1 and FOXP1 mRNA after RNase R treatment. The amounts of circFOXP1 and FOXP1 mRNA were normalized to the amounts of the mock-treated samples. (E) qRT-PCR was used to determine the abundance of circFOXP1 and FOXP1 mRNA in MSCs treated with actinomycin D at the indicated time points. (F) Cytoplasmic and nuclear fractions of cellular RNA were analyzed for circFOXP1 expression by qRT-PCR and expressed as a percentage of the input. U6 and RPS14 mRNA were used as reference RNAs for nuclear and cytoplasmic fractions, respectively. Data in panel (C) are shown as the means ± standard error of the mean (n = 3), statistical analysis was performed by two-tailed Student’s t-test; *P < 0.05, ***P < 0.001.
Subsequently, total RNA was treated with RNase R to analyze the stability of circFOXP1. Indeed, RNase R is able to degrade linear RNAs, but not circRNAs, thanks to its exonuclease activity. As expected, circFOXP1 showed higher resistance to RNase R digestion, compared to the linear mRNA control (Figure 3D). Moreover, the circFOXP1 transcript exhibited a half-life >24 h after treatment with actinomycin D, while its linear counterpart showed a half-life <4 h (Figure 3E). These analyses provided additional support that circFOXP1 has a bona fide circRNA structure.
Finally, since recent reports have proposed that the function of circRNAs is associated with their subcellular compartmentalization; nuclear/cytoplasmic fractionation was performed on MSC samples to determine circFOXP1 localization. CircFOXP1 was enriched in the cytoplasmic fraction, as shown by qRT-PCR analysis (Figure 3F). CircFOXP1 localization was further addressed by fluorescence in situ hybridization (FISH), which directly revealed and confirmed a clear cytoplasmic localization (Supplementary Figure 3D). Altogether, these results demonstrate that circFOXP1 is an abundant and highly stable circRNA present in the cytoplasm of MSCs.
CircFOXP1 maintains MSC differentiation capacity in vitro
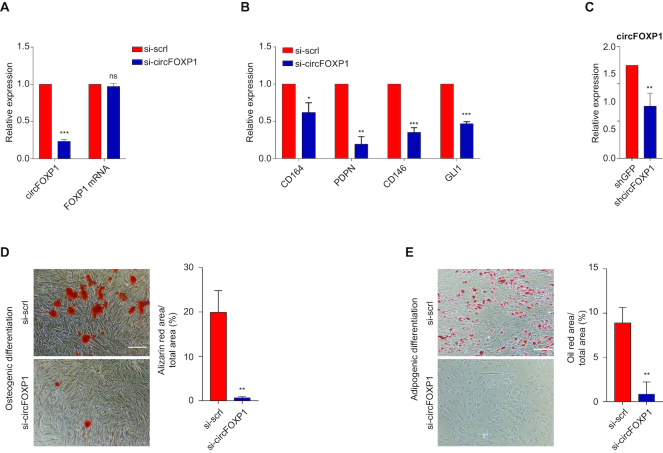
To investigate the biological role of circFOXP1 in MSCs, a knockdown experiment was performed. A small interfering RNA (siRNA) targeting the back-splice junction sequence of circFOXP1 and a scrambled negative control siRNA sequence were designed (Supplementary Figure S4A). These siRNAs were transfected into MSCs, and the effect of siRNA-mediated knockdown of circFOXP1 was assessed after 48 h. As expected, the treatment effectively silenced the circular transcripts, whereas it did not affect the expression of linear mRNAs (Figure 4A). A subsequent cell proliferation assay showed that the downregulation of circFOXP1 markedly reduced MSC growth (Supplementary Figure S4B). In addition, the MTT assay also revealed that the cell proliferation of silenced MSCs was reduced by 20% after 48 h (Supplementary Figure S4C).
Figure 4.
The biological role of circFOXP1 is to maintain mesenchymal stem cells (MSCs) in their undifferentiated state. (A) The knockdown efficiency of circFOXP1 and FOXP1 mRNA for si-circFOXP1 was measured by qRT-PCR. Both the circRNA and linear mRNA levels in control scrambled siRNA (si-control) were set at 1. The RNA levels in si-circFOXP1 cells are shown as a fold change with respect to the control. (B) The relative transcriptional levels of the indicated genes were measured by qRT-PCR in MSCs transfected with si-control or si-circFOXP1. (C) The knockdown efficiency of circFOXP1 for shcircFOXP1 was measured by qRT-PCR. The circRNA level in control shRNA (shGFP) was set at 1. (D) Left, representative images of Alizarin Red S staining of MSCs grown for 21 days in osteogenic induction medium after circFOXP1 knockdown. Right, quantification of Alizarin Red S staining via dissolving the dye and subsequent absorbance measurement at 405 nm; scale bar, 200 μm. (E) Left, representative images of Oil Red O staining of MSCs grown for 14 days in adipogenic induction and maintenance medium after circFOXP1 knockdown. Right, quantification of Oil Red O staining via dissolving the dye and subsequent absorbance measurement at 405 nm; scale bar, 100 μm. Data in panels (A), (B) and (C) are shown as means ± standard error of the mean (n = 3), statistical analysis was performed by two-tailed Student’s t-test; *P < 0.05, **P < 0.01, ***P<0.001.
Next, mRNA levels of MSC cell surface markers were measured to investigate whether circFOXP1 knockdown had consequences on the molecular profile of undifferentiated MSCs (54). qRT-PCR analysis revealed that MSC marker CD164, PDPN, CD146 and GLI1 (44,54–56) expression levels were reduced after 7 days (Figure 4B). In agreement with these findings, flow cytometry analysis of CD73, CD90, CD105 and CD146 showed that the CD90+/CD146+ and CD73+/CD105+ double-positive populations were less abundant in si-circFOXP1 MSCs than si-scrl MSCs (Supplementary Figure S4D).
To establish whether circFOXP1 is essential for the maintenance of undifferentiated MSCs in long-term culture, we transduced MSCs with a lentivirus expressing small hairpin RNA (shRNA) for circFOXP1 to generate a stable knockdown. As previously observed after siRNA treatment, shRNA effectively silenced circFOXP1 transcripts and did not affect the expression of FOXP1 mRNA (Figure 4C and Supplementary Figure S4E). Then, control and circFOXP1 knockdown (circFOXP1-KD) MSCs were induced to undergo osteogenic and adipogenic differentiation and analyzed after 21 days. After osteogenic induction, circFOXP1-KD MSCs showed a significantly lower matrix mineralization, as evidenced by Alizarin Red S staining compared with the negative control (Figure 4D). A similar analysis was conducted to measure the capacity of MSCs to differentiate into the adipocytic lineage after circFOXP1-KD. Oil Red O-stained circFOXP1-KD MSCs showed low accumulation of intracellular lipid droplets, compared to the negative control (Figure 4E). Taken together, the aforementioned results indicate that circFOXP1 acts as a regulator of the transcriptional program sustaining mesenchymal stem cell identity.
CircFOXP1 regulates bone repair by MSCs
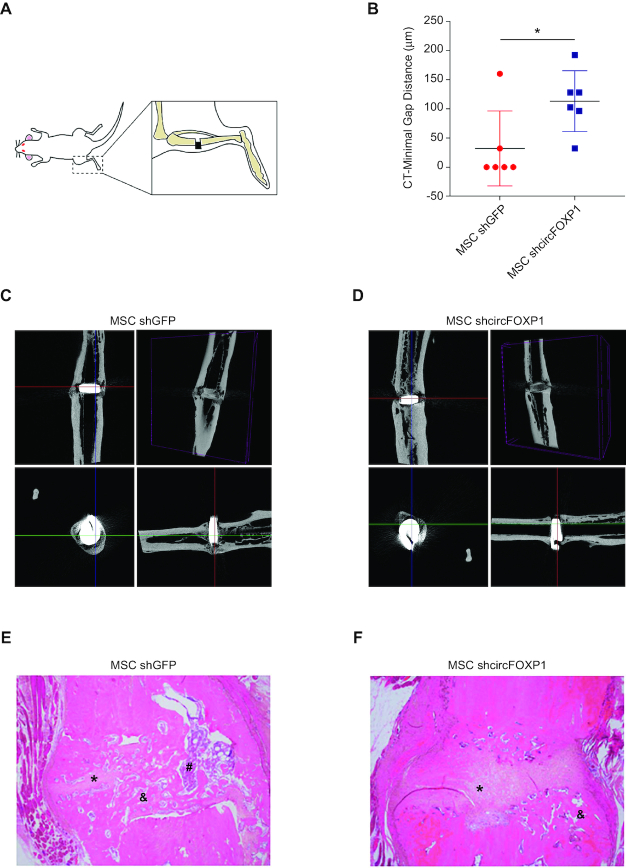
To establish whether circFOXP1-KD MSCs lose mesenchymal identity and properties in vivo, we established an atrophic non-union model in the rat femur (Figure 5A). Three weeks after osteotomy, MSCs transduced with sh-circFOXP1 or a control shRNA were injected into the gap. Rats were monitored after cell injection every week for 10 weeks by X-ray to assess progression of the healing process. In the group treated with circFOXP1-KD MSCs only two out of six rats developed bone union, one of which was only partial. By contrast, in the control group, full union was achieved in five out of six animals (P-value > 0.05, Fisher’s exact test) (Figure 5B).
Figure 5.
CircFOXP1 knockdown reduce bone repair properties of MSCs. (A) Schematic representation of experiment. (B) Box-and-whisker plot indicated measurements of bone fracture after MSC treatment. (C andD) Three sectional images and 3D reconstruction of micro-CT analysis after 8 week MSCs injection. (E andF) Hematoxylin/Eosin staining showed fibrotic tissue (*), osteocyte cells (&) and osteoblast cells (#). Data in panel (B) are shown as means ± standard deviation (n = 6); statistical analysis was performed by two-tailed Student’s t-test; *P < 0.05.
Postmortem clinical examination of the tibiae that developed normal union in the control group showed normal rigidity and were able to withstand a simulated weight bearing test. In contrast, non-union bones in the circFOXP1-KD group revealed obvious motion at the osteotomy site. When the leg was held from one end, deflection of the shaft due to gravity was observed. Therefore, these were not eligible for mechanical testing.
3D images from micro-CT visualization were reconstructed to give a detailed view at the osteotomy site. The control group showed good callus formation and bone bridges filling the gap (Figure 5C and Supplementary Figure S5A), whereas in the circFOXP1-KD group no bone bridge was found at the osteotomy site and fracture line was clearly detectable, with a minimal gap distance in excess of 100 μm (Figure 5D and Supplementary Figure S5B). In a particular case, one of the samples in the circFOXP1-KD group was initially considered to be a union, but only marginal callus formation and bone bridges were later observed. Consequently, healing was considered to be suboptimal, also considering the gap width of 96.091 μm, whereas successful unions all displayed gaps of >50 μm (Supplementary Figure S5C and Supplementary Table S4). Reconstruction of micro-CT data into 3D images gave detailed information about callus shape at the osteotomy site and showed the presence or absence of bone bridges between fracture ends, discriminating unions and non-union (Figure 5C and D).
Finally, histological examination showed normal union with bone bridges connecting fracture ends in the control groups, while the circFOXP1-KD group showed non-union filled with fibrous tissue and no bone bridges (Figure 5E and F).
CircFOXP1 acts as a competing endogenous RNA interacting with multiple miRNAs
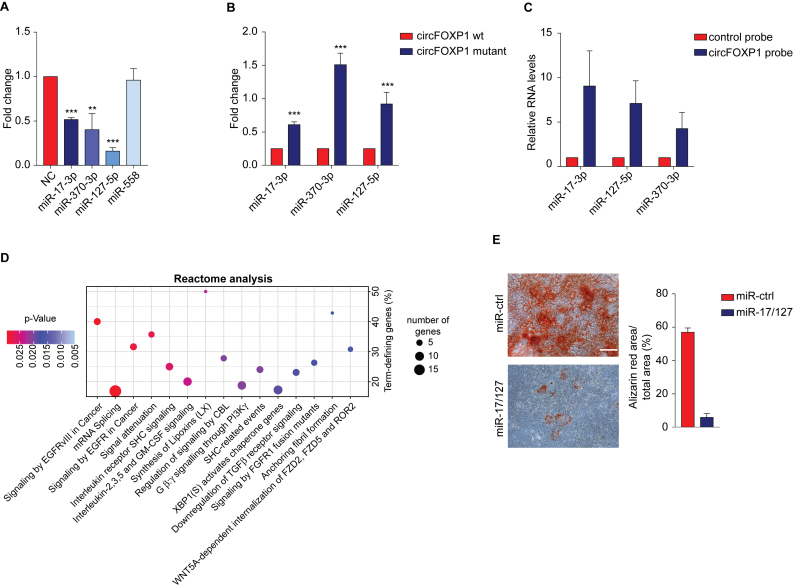
Previous research has proposed that circFOXP1 may act as a ceRNA by sequestering miRNAs present in the cytoplasm to inhibit translation of their target mRNAs (27,32,57). Considering the cytoplasmic localization of circFOXP1, it is conceivable that it plays a role in MSCs as a ceRNA. To screen miRNAs ‘sponged’ by circFOXP1, Arraystar proprietary algorithms based on the public databases TargetScan and miRanda were used and the highest ranking candidate miRNAs were selected. To validate whether the miRNAs selected could interact with circFOXP1, a luciferase reporter assay was performed. The sequence of circFOXP1 was inserted immediately downstream of the Renilla luciferase reporter gene (RLuc+circFOXP1), and then each candidate interacting miRNA was co-transfected with the luciferase reporter into HEK-293T cells. MiR-17-3p, miR-127-5p and miR-370-3p were able to reduce the Renilla luciferase reporter activity, compared to the negative control, while miR-558 was not able to interact with the circFOXP1 sequence (Figure 6A). Subsequently, at least one miRNA-binding site defined by the Arraystar proprietary algorithms was confirmed using the miRDB and CircInteractome databases (58,59) (Supplementary Figure S6A and Supplementary Table S5). Then, each confirmed miRNA target site in the circFOXP1 sequence was mutated, and the Renilla luciferase assay was repeated. No significant effect on luciferase levels was observed, demonstrating the specificity of the miRNA–circFOXP1 target site interaction (Figure 6B). Next, using a pull-down assay with biotin-labeled oligonucleotides complementary to the back-splice junction of circFOXP1, the direct interaction between miR-127-5p/miR-17-3p/miR-370-3p and circFOXP1 in its native circular structure was validated. HEK-293T cells were transfected with circFOXP1 and each miRNA, and the cell lysates were subjected to the pull-down assay. Equal amounts of circFOXP1 were observed in the cell lysates mixed with the specific or control probes (Supplementary Figure S6B). Consistent with the Renilla luciferase reporter assay, the circRNA probe pulled down more than 5-fold circFOXP1, compared to the control probe (Supplementary Figure S6C). Noticeably, pull-down with the circFOXP1-specific probe, but not the control probe, yielded not only circFOXP1 but also miR-17–3p, miR-127–5p and miR-370–3p (Figure 6C). The presence of the head-to-tail junction sequence of circFOXP1 in the pull-down assay was confirmed by Sanger sequencing after resolving on an agarose gel (Supplementary Figure S6D).
Figure 6.
CircFOXP1 acts as a competing endogenous RNA for multiple miRNAs. (A) Luciferase reporter assay for Renilla luciferase activity of RLuc-circFOXP1 in HEK-293T cells co-transfected with miRNA mimics. (B) Luciferase reporter assay for Renilla luciferase activity of RLuc-circFOXP1 or RLuc-circFOXP1 mutant in HEK-293T cells co-transfected with miRNA mimics. (C) Lysates prepared from HEK-293T cells co-transfected with circFOXP1 and miRNA mimics were subjected to the RNA pull-down assay. The interaction of miR-17–3p, miR-127–5p and miR-370–3p with circFOXP1 was measured by qRT-PCR. (D) Reactome analysis of miRNA targets. The top 15 ranked Reactome pathways are shown. The y- and x-axis indicate the enrichment factor (% of genes involved relative to the total number of pathway genes) and pathway name, respectively. The size of the dots indicates the number of genes involved in the respective annotation category, while the color denotes the statistical significance of the enrichment. (E) Mineralized nodule formation as assessed by Alizarin Red staining; scale bar, 200 μm. Data in panels (A) and (B) are shown as means ± standard deviation (n = 3); statistical analysis was performed by two-tailed Student’s t-test; **P < 0.01, ***P < 0.001.
To explore the molecular mechanisms whereby circFOXP1 supports MSC identity, a circRNA–miRNA–mRNA regulatory molecular axis was delineated using the Cytoscape tool. Among the miRNAs found to be sponged by circFOXP1, only miR-17–3p and miR-127–5p demonstrated a potentially relevant role in the regulation of different molecular pathways (Supplementary Figure S6E). Reactome and Gene Ontology (GO) database analyses performed on the target genes of miR-17–3p and miR-127–5p indicated a significant correlation with EGFR and non-canonical Wnt signaling (Figure 6D and Supplementary Figure S6F), which are associated with MSC proliferation and self-renewal (11–13). To validate the bioinformatics analysis experimentally, we addressed the expression levels of the direct target mRNAs of miR-17–3p and miR-127–5p. qRT-PCR analysis showed that WNT5A, ROR2, PIK3CA and NRAS were downregulated upon circFOXP1 knockdown (Supplementary Figure S6G).
We finally investigated whether the capacity of circFOXP1 to maintain MSCs depends on miR-17–3p/miR-127–5p inhibition. Synthetic mimics of these miRNAs were transfected into MSCs. Next, miR-17–3p/miR-127–5p overexpressing MSCs were induced to differentiate toward the osteogenic lineage. Overexpression of miR-17–3p and miR-127–5p inhibited osteogenic differentiation as indicated by lower levels of Alizarin Red staining (Figure 6E). Taken together, these results indicate that circFOXP1 acts as a regulator of MSC identity through inhibition of miR-17-3p/miR-127-5p activity.
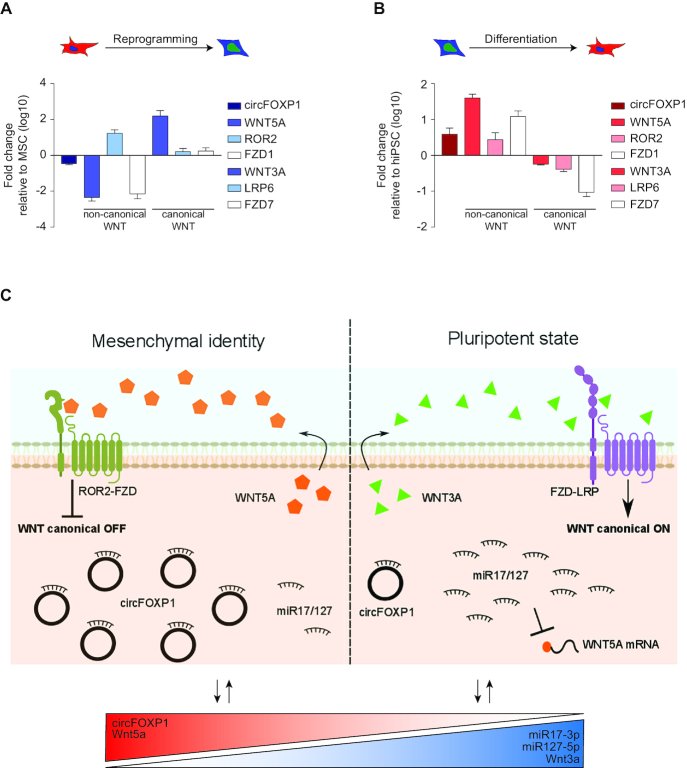
To address a broader role for circFOXP1 at the interplay between non-canonical and canonical Wnt pathways, MSCs were reprogrammed to human-induced pluripotent stem cells (hiPSCs), stably changing their stem cell identity. To maintain pluripotency, hiPSCs activate the canonical and disengage the non-canonical Wnt pathways, as reported by the literature (60,61). Upon reprogramming (Supplementary Figure S7A), circFOXP1 was downregulated (Figure 7A). At the same time non-canonical Wnt5a ligand and Fzd1 co-receptor gene expression was silenced, whereas canonical Wnt3a ligand, Lrp6 and Fzd7 receptors mRNAs were upregulated (Figure 7A). Furthermore, consistent with circFOXP1 deregulation, miR17-3p and miR127-5p were upregulated (Supplementary Figure S7C). To confirm these data, we also generated MSCs from hiPSCs (Supplementary Figure S7B) to address the opposite process. In accordance with the previous results, circFOXP1 was upregulated in the newly generated hiPSC-derived MSCs (Figure 7B). In detail, downregulation of canonical Wnt3a, Lrp6 and Fzd7 was detected, whereas non-canonical Wnt5a, Ror2 and Fzd1 were strongly upregulated (Figure 7B).
Figure 7.
circFOXP1 reinforce non-canonical Wnt pathway and contrast canonical Wnt signaling. (A and B) The relative transcriptional levels of circFOXP1 and Wnt molecular players were quantified by qRT-PCR analysis in MSC-derived hiPSCs and in hiPSC-derived MSCs, respectively. (C) Graphical visualization of the circFOXP1-centered molecular circuit that sustains MSC identity. Data in panels (A) and (B) are shown as means ± standard error of the mean (n = 3).
DISCUSSION
In the past couple of decades, the concept of a self-reinforcing transcriptional regulatory network has driven most of the pluripotent stem cell research (60,62). In particular, the stimuli necessary to exit this regulatory network and enter differentiation have been investigated intensively (63,64). Indeed, the current paradigm considers stem cells in their undifferentiated state as poised to differentiate in response to transient signaling cascades, while still ensuring the maintenance of cell identity through multiple rounds of cell division. Controlling the equilibrium between stem cell self-renewal and cell fate specification is indispensable for maintaining tissue homeostasis. The deregulation of these processes leads to loss of cell identity (65,66), an essential characteristic to be controlled for consistent regenerative medicine applications. Notwithstanding their importance, self-reinforcing regulatory networks and the molecular mechanism governing the transition of MSCs from the undifferentiated state to mesodermal commitment are still undefined, in particular with respect to ncRNAs.
In the past few years, ncRNAs have attracted much attention as fundamental players in the regulation of molecular networks related to differentiation pathways (67,68). Among ncRNAs, circRNAs have emerged as novel regulators of physiological cell functions (23,24,31,32). In this work, for the first time, we determined the MSC circRNAome defined as the complete set of circRNAs detectable in a given cell type. Compared with HSF, representative of a terminally differentiated state, we found more downregulated than upregulated circRNAs in MSCs. This was in accordance with previous works which showed that circRNAs were upregulated during differentiation (36,69), while a reduced number of circRNAs were found in proliferating compared to non-proliferating cells (70). These observations could be explained by circRNAs get passively diluted by cell proliferation or vice versa accumulated in non-proliferating cells (terminally differentiated or quiescent) due to their high stability. In our work, we well characterized and functionally defined the key role played by one highly expressed circRNA in MSCs, hsa_circ_0001320, derived from the FOXP1 gene (termed as circFOXP1). Although FOXP1 mRNA can produce more than 30 different circRNAs, as previously detected in other cell lines (23,36,71), only circFOXP1 had consistently higher expression in MSCs. CircFOXP1 is composed of five exons and is flanked on both sides by long introns containing many Alu repeats. Previous studies have indicated that the presence of inverted repeats of Alu sequences (IRAlu) may facilitate RNA transcript circularization (23,24,27,36). Consistent with this concept, our results also support the role of RNA pairing by IRAlu in the enhanced generation of circRNAs.
Our results demonstrate that circFOXP1 exerts a pivotal role in MSCs on the support of their multipotent identity, and that it is also involved in the regulation of MSC differentiation. Indeed, circFOXP1 was consistently downregulated across all in vitro generated mesodermal derivatives studied, compared to undifferentiated MSCs. This was confirmed in human biopsies of bone, cartilage and adipose tissue from healthy individuals.
Furthermore, our data show that solely circFOXP1, but not FOXP1 mRNA, regulates MSC identity both in vitro and following transplantation: circFOXP1 knockdown rapidly reduced cell surface proteins of MSCs correlated with high colony formation and dramatically decreased MSC differentiation in culture. These results were confirmed in vivo, where circFOXP1-KD MSCs lose their ability to regenerate bone in a created non-union defect in rats.
Defining the signaling network activated in adult stem cells is an essential step toward understanding their capabilities to differentiate and regenerate tissues. For instance, MSCs express a set of transcripts for genes encoding signaling receptors linked to stem cell survival and growth (AXL, PDGFR and EGFR) and self-renewal (EGFR, EPHR and FGFR) (72–74).
Consistent with the observed abundant and stable cytoplasmic localization of circFOXP1, we hypothesized and confirmed that it acts as a ‘sponge’ for multiple miRNAs, reinforcing the idea that a single circRNA may target simultaneously different miRNAs. Strikingly, bioinformatics showed that the combined action of miR-17–3p and miR-127–5p may regulate growth, survival and balance between undifferentiated and differentiated MSCs, via, for instance, EGFR and non-canonical Wnt signaling. Indeed, EGFR and non-canonical Wnt signaling must be correctly activated in MSCs to maintain multipotency and confer effective regenerative capacity (13,75–79). A deeper analysis of non-canonical and canonical Wnt pathways during reprogramming to pluripotency of MSCs or during the opposite process hinted at a role for circFOXP1 modulating this interplay. In MSCs, elevated levels of circFOXP1 sustain non-canonical and consequently inhibit canonical Wnt pathway. In hiPSCs, the low abundance of circFOXP1 allow for miR17-3p/miR127-5p-mediated suppression of non-canonical Wnt pathway, reinforcing endogenous canonical Wnt signaling (Figure 7C).
In summary our work describes, for the first time, the role of a circRNA in the control of the developmental potential of human adult and fetal MSCs. In our model, an abundant circRNA derived from the FOXP1 gene was able to preserve the MSC multipotent state by sponging multiple miRNAs. This functional interaction is fundamental to inhibit miRNA activity and avoid interference of signaling cascades associated with stemness and differentiation. Although it is likely that other mechanisms contribute to this biological control, our data describe a circRNA-centered molecular circuit exerting a regulatory function in MSCs. circFOXP1 should be regarded as an essential gatekeeper of MSC identity.
DATA AVAILABILITY
Raw and normalized data files for the microarray analysis have been deposited in the NCBI Gene Expression Omnibus under accession number GSE122178.
Supplementary Material
ACKNOWLEDGEMENTS
Authors thank Marta Dossena and Ilaria Panzeri for important discussions and technical support. We are grateful to all the members of the Cell Factory lab for insightful conversations.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
‘Ricerca Corrente Fondazione IRCCS Ca’ Granda Ospedale Maggiore Policlinico Milano’ 2018. Funding for open access charge: Ricerca corrente 2018, Ministero della Salute. HORIZON 2020 - ORTHOUNION (ORTHOpedic randomized clinical trial with expanded bone-marrow mesenchymal stem cells and bioceramics versus autograft in long bone non-UNIONs), project number 733288.
Conflict of interest statement. None declared.
REFERENCES
- 1. Dominici M., Le Blanc K., Mueller I., Slaper-Cortenbach I., Marini F., Krause D., Deans R., Keating A., Prockop D., Horwitz E.. Minimal criteria for defining multipotent mesenchymal stromal cells. The International Society for Cellular Therapy position statement. Cytotherapy. 2006; 8:315–317. [DOI] [PubMed] [Google Scholar]
- 2. Bianco P., Robey P.G., Simmons P.J.. Mesenchymal stem cells: revisiting history, concepts, and assays. Cell Stem Cell. 2008; 2:313–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kilian K.A., Bugarija B., Lahn B.T., Mrksich M.. Geometric cues for directing the differentiation of mesenchymal stem cells. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:4872–4877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Undale A.H., Westendorf J.J., Yaszemski M.J., Khosla S.. Mesenchymal stem cells for bone repair and metabolic bone diseases. Mayo Clin. Proc. 2009; 84:893–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sacchetti B., Funari A., Michienzi S., Di Cesare S., Piersanti S., Saggio I., Ferrari S., Robey P.G., Riminucci M. et al.. Self-renewing osteoprogenitors in bone marrow sinusoids can organize a hematopoietic microenvironment. Cell. 2007; 131:324–336. [DOI] [PubMed] [Google Scholar]
- 6. Medici D., Shore E.M., Lounev V.Y., Kaplan F.S., Kalluri R., Olsen B.R.. Conversion of vascular endothelial cells into multipotent stem-like cells. Nat. Med. 2010; 16:1400–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Xiao L., Sobue T., Esliger A., Kronenberg M.S., Coffin J.D., Doetschman T., Hurley M.M.. Disruption of the Fgf2 gene activates the adipogenic and suppresses the osteogenic program in mesenchymal marrow stromal stem cells. Bone. 2010; 47:360–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mendez-Ferrer S., Michurina T.V., Ferraro F., Mazloom A.R., Macarthur B.D., Lira S.A., Scadden D.T., Ma’ayan A., Enikolopov G.N., Frenette P.S.. Mesenchymal and haematopoietic stem cells form a unique bone marrow niche. Nature. 2010; 466:829–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Takada I., Kouzmenko A.P., Kato S.. Wnt and PPARgamma signaling in osteoblastogenesis and adipogenesis. Nat. Rev. Rheumatol. 2009; 5:442–447. [DOI] [PubMed] [Google Scholar]
- 10. Wei X., Mao Z., Hou Y., Lin L., Xue T., Chen L., Wang H., Yu C.. Local administration of TGFbeta-1/VEGF165 gene-transduced bone mesenchymal stem cells for Achilles allograft replacement of the anterior cruciate ligament in rabbits. Biochem. Biophys. Res. Commun. 2011; 406:204–210. [DOI] [PubMed] [Google Scholar]
- 11. Yu S., Geng Q., Ma J., Sun F., Yu Y., Pan Q., Hong A.. Heparin-binding EGF-like growth factor and miR-1192 exert opposite effect on Runx2-induced osteogenic differentiation. Cell Death Dis. 2013; 4:e868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhu J., Shimizu E., Zhang X., Partridge N.C., Qin L.. EGFR signaling suppresses osteoblast differentiation and inhibits expression of master osteoblastic transcription factors Runx2 and Osterix. J. Cell Biochem. 2011; 112:1749–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dickinson S.C., Sutton C.A., Brady K., Salerno A., Katopodi T., Williams R.L., West C.C., Evseenko D., Wu L., Pang S. et al.. The Wnt5a receptor, receptor tyrosine Kinase-Like orphan receptor 2, is a predictive cell surface marker of human mesenchymal stem cells with an enhanced capacity for chondrogenic differentiation. Stem Cells. 2017; 35:2280–2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Huarte M. The emerging role of lncRNAs in cancer. Nat. Med. 2015; 21:1253–1261. [DOI] [PubMed] [Google Scholar]
- 15. O’Connell R.M., Rao D.S., Chaudhuri A.A., Baltimore D.. Physiological and pathological roles for microRNAs in the immune system. Nat. Rev. Immunol. 2010; 10:111–122. [DOI] [PubMed] [Google Scholar]
- 16. Sayed D., Abdellatif M.. MicroRNAs in development and disease. Physiol. Rev. 2011; 91:827–887. [DOI] [PubMed] [Google Scholar]
- 17. Ivey K.N., Srivastava D.. MicroRNAs as regulators of differentiation and cell fate decisions. Cell Stem Cell. 2010; 7:36–41. [DOI] [PubMed] [Google Scholar]
- 18. Flynn R.A., Chang H.Y.. Long noncoding RNAs in cell-fate programming and reprogramming. Cell Stem Cell. 2014; 14:752–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sacchetti B., Fatica A., Sorci M., Sorrentino A., Signore M., Cerio A., Felicetti F., Feo A., Pelosi E., Care A. et al.. Effect of miR-204&211 and RUNX2 control on the fate of human mesenchymal stromal cells. Regen. Med. Res. 2017; 5:2. [DOI] [PubMed] [Google Scholar]
- 20. Sanger H.L., Klotz G., Riesner D., Gross H.J., Kleinschmidt A.K.. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc. Natl. Acad. Sci. U.S.A. 1976; 73:3852–3856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Nigro J.M., Cho K.R., Fearon E.R., Kern S.E., Ruppert J.M., Oliner J.D., Kinzler K.W., Vogelstein B.. Scrambled exons. Cell. 1991; 64:607–613. [DOI] [PubMed] [Google Scholar]
- 22. Capel B., Swain A., Nicolis S., Hacker A., Walter M., Koopman P., Goodfellow P., Lovell-Badge R.. Circular transcripts of the testis-determining gene Sry in adult mouse testis. Cell. 1993; 73:1019–1030. [DOI] [PubMed] [Google Scholar]
- 23. Salzman J., Gawad C., Wang P.L., Lacayo N., Brown P.O.. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One. 2012; 7:e30733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Memczak S., Jens M., Elefsinioti A., Torti F., Krueger J., Rybak A., Maier L., Mackowiak S.D., Gregersen L.H., Munschauer M. et al.. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013; 495:333–338. [DOI] [PubMed] [Google Scholar]
- 25. Zhu P., Zhu X., Wu J., He L., Lu T., Wang Y., Liu B., Ye B., Sun L., Fan D. et al.. IL-13 secreted by ILC2s promotes the self-renewal of intestinal stem cells through circular RNA circPan3. Nat. Immunol. 2019; 20:183–194. [DOI] [PubMed] [Google Scholar]
- 26. Starke S., Jost I., Rossbach O., Schneider T., Schreiner S., Hung L.H., Bindereif A.. Exon circularization requires canonical splice signals. Cell. Rep. 2015; 10:103–111. [DOI] [PubMed] [Google Scholar]
- 27. Jeck W.R., Sorrentino J.A., Wang K., Slevin M.K., Burd C.E., Liu J., Marzluff W.F., Sharpless N.E.. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013; 19:141–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhang X.O., Wang H.B., Zhang Y., Lu X., Chen L.L., Yang L.. Complementary sequence-mediated exon circularization. Cell. 2014; 159:134–147. [DOI] [PubMed] [Google Scholar]
- 29. Ashwal-Fluss R., Meyer M., Pamudurti N.R., Ivanov A., Bartok O., Hanan M., Evantal N., Memczak S., Rajewsky N., Kadener S.. circRNA biogenesis competes with pre-mRNA splicing. Mol. Cell. 2014; 56:55–66. [DOI] [PubMed] [Google Scholar]
- 30. Conn S.J., Pillman K.A., Toubia J., Conn V.M., Salmanidis M., Phillips C.A., Roslan S., Schreiber A.W., Gregory P.A., Goodall G.J.. The RNA binding protein quaking regulates formation of circRNAs. Cell. 2015; 160:1125–1134. [DOI] [PubMed] [Google Scholar]
- 31. Hansen T.B., Jensen T.I., Clausen B.H., Bramsen J.B., Finsen B., Damgaard C.K., Kjems J.. Natural RNA circles function as efficient microRNA sponges. Nature. 2013; 495:384–388. [DOI] [PubMed] [Google Scholar]
- 32. Zheng Q., Bao C., Guo W., Li S., Chen J., Chen B., Luo Y., Lyu D., Li Y., Shi G. et al.. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat. Commun. 2016; 7:11215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Li Z., Huang C., Bao C., Chen L., Lin M., Wang X., Zhong G., Yu B., Hu W., Dai L. et al.. Exon-intron circular RNAs regulate transcription in the nucleus. Nat. Struct. Mol. Biol. 2015; 22:256–264. [DOI] [PubMed] [Google Scholar]
- 34. Du W.W., Yang W., Liu E., Yang Z., Dhaliwal P., Yang B.B.. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res. 2016; 44:2846–2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Legnini I., Di Timoteo G., Rossi F., Morlando M., Briganti F., Sthandier O., Fatica A., Santini T., Andronache A., Wade M. et al.. Circ-ZNF609 is a circular RNA that can be translated and functions in myogenesis. Mol. Cell. 2017; 66:22–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Rybak-Wolf A., Stottmeister C., Glazar P., Jens M., Pino N., Giusti S., Hanan M., Behm M., Bartok O., Ashwal-Fluss R. et al.. Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol. Cell. 2015; 58:870–885. [DOI] [PubMed] [Google Scholar]
- 37. Shen T., Han M., Wei G., Ni T.. An intriguing RNA species–perspectives of circularized RNA. Protein Cell. 2015; 6:871–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. You X., Vlatkovic I., Babic A., Will T., Epstein I., Tushev G., Akbalik G., Wang M., Glock C., Quedenau C. et al.. Neural circular RNAs are derived from synaptic genes and regulated by development and plasticity. Nat. Neurosci. 2015; 18:603–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wang P.L., Bao Y., Yee M.C., Barrett S.P., Hogan G.J., Olsen M.N., Dinneny J.R., Brown P.O., Salzman J.. Circular RNA is expressed across the eukaryotic tree of life. PLoS One. 2014; 9:e90859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Barilani M., Lavazza C., Vigano M., Montemurro T., Boldrin V., Parazzi V., Montelatici E., Crosti M., Moro M., Giordano R. et al.. Dissection of the cord blood stromal component reveals predictive parameters for culture outcome. Stem Cells Dev. 2015; 24:104–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Barilani M., Lavazza C., Boldrin V., Ragni E., Parazzi V., Crosti M., Montelatici E., Giordano R., Lazzari L.. A chemically defined Medium-Based strategy to efficiently generate clinically relevant cord blood mesenchymal stromal colonies. Cell Transplant. 2016; 25:1501–1514. [DOI] [PubMed] [Google Scholar]
- 42. Ragni E., Montemurro T., Montelatici E., Lavazza C., Vigano M., Rebulla P., Giordano R., Lazzari L.. Differential microRNA signature of human mesenchymal stem cells from different sources reveals an “environmental-niche memory” for bone marrow stem cells. Exp. Cell Res. 2013; 319:1562–1574. [DOI] [PubMed] [Google Scholar]
- 43. Ragni E., Vigano M., Rebulla P., Giordano R., Lazzari L.. What is beyond a qRT-PCR study on mesenchymal stem cell differentiation properties: how to choose the most reliable housekeeping genes. J. Cell. Mol. Med. 2013; 17:168–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Barilani M., Banfi F., Sironi S., Ragni E., Guillaumin S., Polveraccio F., Rosso L., Moro M., Astori G., Pozzobon M. et al.. Low-affinity nerve growth factor receptor (CD271) heterogeneous expression in adult and fetal mesenchymal stromal cells. Sci. Rep. 2018; 8:9321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Glazar P., Papavasileiou P., Rajewsky N.. circBase: a database for circular RNAs. RNA. 2014; 20:1666–1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Bustin S.A., Benes V., Garson J.A., Hellemans J., Huggett J., Kubista M., Mueller R., Nolan T., Pfaffl M.W., Shipley G.L. et al.. The MIQE guidelines: minimuminformation for publication of quantitative real-time PCR experiments. Clin. Chem. 2009; 55:611–622. [DOI] [PubMed] [Google Scholar]
- 47. Jeyapalan Z., Deng Z., Shatseva T., Fang L., He C., Yang B.B.. Expression of CD44 3′-untranslated region regulates endogenous microRNA functions in tumorigenesis and angiogenesis. Nucleic Acids Res. 2011; 39:3026–3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ranzani V., Rossetti G., Panzeri I., Arrigoni A., Bonnal R.J., Curti S., Gruarin P., Provasi E., Sugliano E., Marconi M. et al.. The long intergenic noncoding RNA landscape of human lymphocytes highlights the regulation of T cell differentiation by linc-MAF-4. Nat. Immunol. 2015; 16:318–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Johnson W.E., Li C., Rabinovic A.. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007; 8:118–127. [DOI] [PubMed] [Google Scholar]
- 50. Enuka Y., Lauriola M., Feldman M.E., Sas-Chen A., Ulitsky I., Yarden Y.. Circular RNAs are long-lived and display only minimal early alterations in response to a growth factor. Nucleic Acids Res. 2016; 44:1370–1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Yu C.Y., Li T.C., Wu Y.Y., Yeh C.H., Chiang W., Chuang C.Y., Kuo H.C.. The circular RNA circBIRC6 participates in the molecular circuitry controlling human pluripotency. Nat. Commun. 2017; 8:1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Zhang Y., Xue W., Li X., Zhang J., Chen S., Zhang J.L., Yang L., Chen L.L.. The biogenesis of nascent circular RNAs. Cell. Rep. 2016; 15:611–624. [DOI] [PubMed] [Google Scholar]
- 53. Kramer M.C., Liang D., Tatomer D.C., Gold B., March Z.M., Cherry S., Wilusz J.E.. Combinatorial control of Drosophila circular RNA expression by intronic repeats, hnRNPs, and SR proteins. Genes Dev. 2015; 29:2168–2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Chan C.K.F., Gulati G.S., Sinha R., Tompkins J.V., Lopez M., Carter A.C., Ransom R.C., Reinisch A., Wearda T., Murphy M. et al.. Identification of the human skeletal stem cell. Cell. 2018; 175:43–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Shi Y., He G., Lee W.C., McKenzie J.A., Silva M.J., Long F.. Gli1 identifies osteogenic progenitors for bone formation and fracture repair. Nat. Commun. 2017; 8:2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Crisan M., Yap S., Casteilla L., Chen C.W., Corselli M., Park T.S., Andriolo G., Sun B., Zheng B., Zhang L. et al.. A perivascular origin for mesenchymal stem cells in multiple human organs. Cell Stem Cell. 2008; 3:301–313. [DOI] [PubMed] [Google Scholar]
- 57. Chen L.L. The biogenesis and emerging roles of circular RNAs. Nat. Rev. Mol. Cell Biol. 2016; 17:205–211. [DOI] [PubMed] [Google Scholar]
- 58. Dweep H., Sticht C., Pandey P., Gretz N.. miRWalk–database: prediction of possible miRNA binding sites by “walking” the genes of three genomes. J. Biomed. Inform. 2011; 44:839–847. [DOI] [PubMed] [Google Scholar]
- 59. Dudekula D.B., Panda A.C., Grammatikakis I., De S., Abdelmohsen K., Gorospe M.. CircInteractome: A web tool for exploring circular RNAs and their interacting proteins and microRNAs. RNA Biol. 2016; 13:34–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Fagnocchi L., Cherubini A., Hatsuda H., Fasciani A., Mazzoleni S., Poli V., Berno V., Rossi R.L., Reinbold R., Endele M. et al.. A Myc-driven self-reinforcing regulatory network maintains mouse embryonic stem cell identity. Nat. Commun. 2016; 7:11903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Nusse R. Wnt signaling and stem cell control. Cell Res. 2008; 18:523–527. [DOI] [PubMed] [Google Scholar]
- 62. Boiani M., Scholer H.R.. Regulatory networks in embryo-derived pluripotent stem cells. Nat. Rev. Mol. Cell. Biol. 2005; 6:872–884. [DOI] [PubMed] [Google Scholar]
- 63. Kunath T., Saba-El-Leil M.K., Almousailleakh M., Wray J., Meloche S., Smith A.. FGF stimulation of the Erk1/2 signalling cascade triggers transition of pluripotent embryonic stem cells from self-renewal to lineage commitment. Development. 2007; 134:2895–2902. [DOI] [PubMed] [Google Scholar]
- 64. Gattazzo F., Urciuolo A., Bonaldo P.. Extracellular matrix: a dynamic microenvironment for stem cell niche. Biochim. Biophys. Acta. 2014; 1840:2506–2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Petersson M., Reuter K., Brylka H., Kraus A., Schettina P., Niemann C.. Interfering with stem cell-specific gatekeeper functions controls tumour initiation and malignant progression of skin tumours. Nat. Commun. 2015; 6:5874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Vermeulen L., Morrissey E., van der Heijden M., Nicholson A.M., Sottoriva A., Buczacki S., Kemp R., Tavare S., Winton D.J.. Defining stem cell dynamics in models of intestinal tumor initiation. Science. 2013; 342:995–998. [DOI] [PubMed] [Google Scholar]
- 67. Zhang W., Dong R., Diao S., Du J., Fan Z., Wang F.. Differential long noncoding RNA/mRNA expression profiling and functional network analysis during osteogenic differentiation of human bone marrow mesenchymal stem cells. Stem Cell Res. Ther. 2017; 8:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Dong R., Du J., Wang L., Wang J., Ding G., Wang S., Fan Z.. Comparison of long noncoding RNA and mRNA expression profiles in mesenchymal stem cells derived from human periodontal ligament and bone marrow. Biomed. Res. Int. 2014; 2014:317853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Kristensen L.S., Okholm T.L.H., Veno M.T., Kjems J.. Circular RNAs are abundantly expressed and upregulated during human epidermal stem cell differentiation. RNA Biol. 2018; 15:280–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Bachmayr-Heyda A., Reiner A.T., Auer K., Sukhbaatar N., Aust S., Bachleitner-Hofmann T., Mesteri I., Grunt T.W., Zeillinger R., Pils D.. Correlation of circular RNA abundance with proliferation–exemplified with colorectal and ovarian cancer, idiopathic lung fibrosis, and normal human tissues. Sci. Rep. 2015; 5:8057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Dahl M., Daugaard I., Andersen M.S., Hansen T.B., Gronbaek K., Kjems J., Kristensen L.S.. Enzyme-free digital counting of endogenous circular RNA molecules in B-cell malignancies. Lab. Invest. 2018; 98:1657–1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Krampera M., Pasini A., Rigo A., Scupoli M.T., Tecchio C., Malpeli G., Scarpa A., Dazzi F., Pizzolo G., Vinante F.. HB-EGF/HER-1 signaling in bone marrow mesenchymal stem cells: inducing cell expansion and reversibly preventing multilineage differentiation. Blood. 2005; 106:59–66. [DOI] [PubMed] [Google Scholar]
- 73. Neuss S., Becher E., Woltje M., Tietze L., Jahnen-Dechent W.. Functional expression of HGF and HGF receptor/c-met in adult human mesenchymal stem cells suggests a role in cell mobilization, tissue repair, and wound healing. Stem Cells. 2004; 22:405–414. [DOI] [PubMed] [Google Scholar]
- 74. Ozaki Y., Nishimura M., Sekiya K., Suehiro F., Kanawa M., Nikawa H., Hamada T., Kato Y.. Comprehensive analysis of chemotactic factors for bone marrow mesenchymal stem cells. Stem Cells Dev. 2007; 16:119–129. [DOI] [PubMed] [Google Scholar]
- 75. Liu X., Qin J., Luo Q., Bi Y., Zhu G., Jiang W., Kim S.H., Li M., Su Y., Nan G. et al.. Cross-talk between EGF and BMP9 signalling pathways regulates the osteogenic differentiation of mesenchymal stem cells. J. Cell. Mol. Med. 2013; 17:1160–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Platt M.O., Roman A.J., Wells A., Lauffenburger D.A., Griffith L.G.. Sustained epidermal growth factor receptor levels and activation by tethered ligand binding enhances osteogenic differentiation of multi-potent marrow stromal cells. J. Cell. Physiol. 2009; 221:306–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Chandra A., Lan S., Zhu J., Siclari V.A., Qin L.. Epidermal growth factor receptor (EGFR) signaling promotes proliferation and survival in osteoprogenitors by increasing early growth response 2 (EGR2) expression. J. Biol. Chem. 2013; 288:20488–20498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Cai S.X., Liu A.R., He H.L., Chen Q.H., Yang Y., Guo F.M., Huang Y.Z., Liu L., Qiu H.B.. Stable genetic alterations of beta-catenin and ROR2 regulate the Wnt pathway, affect the fate of MSCs. J. Cell. Physiol. 2014; 229:791–800. [DOI] [PubMed] [Google Scholar]
- 79. Bilkovski R., Schulte D.M., Oberhauser F., Gomolka M., Udelhoven M., Hettich M.M., Roth B., Heidenreich A., Gutschow C., Krone W. et al.. Role of WNT-5a in the determination of human mesenchymal stem cells into preadipocytes. J. Biol. Chem. 2010; 285:6170–6178. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw and normalized data files for the microarray analysis have been deposited in the NCBI Gene Expression Omnibus under accession number GSE122178.