Abstract
Background
Elymus L. is the largest genus in the tribe Triticeae Dumort., encompassing approximately 150 polyploid perennial species widely distributed in the temperate regions of the world. It is considered to be an important gene pool for improving cereal crops. However, a shortage of molecular marker limits the efficiency and accuracy of genetic breeding for Elymus species. High-throughput transcriptome sequencing data is essential for gene discovery and molecular marker development.
Results
We obtained the transcriptome dataset of E. sibiricus, the type species of the genus Elymus, and identified a total of 8871 putative EST-SSRs from 6685 unigenes. Trinucleotides were the dominant repeat motif (4760, 53.66%), followed by dinucleotides (1993, 22.47%) and mononucleotides (1876, 21.15%). The most dominant trinucleotide repeat motif was CCG/CGG (1119, 23.5%). Sequencing of PCR products showed that the sequenced alleles from different Elymus species were homologous to the original SSR locus from which the primer was designed. Different types of tri-repeats as abundant SSR motifs were observed in repeat regions. Two hundred EST-SSR primer pairs were designed and selected to amplify ten DNA samples of Elymus species. Eighty-seven pairs of primer (43.5%) generated clear and reproducible bands with expected size, and showed good transferability across different Elymus species. Finally, thirty primer pairs successfully amplified ninety-five accessions of seventeen Elymus species, and detected significant amounts of polymorphism. In general, hexaploid Elymus species with genomes StStHHYY had a relatively higher level of genetic diversity (H = 0.219, I = 0.330, %P = 63.7), while tetraploid Elymus species with genomes StStYY had low level of genetic diversity (H = 0.182, I = 0.272, %P = 50.4) in the study. The cluster analysis showed that all ninety-five accessions were clustered into three major clusters. The accessions were grouped mainly according to their genomic components and origins.
Conclusions
This study demonstrated that transcriptome sequencing is a fast and cost-effective approach to molecular marker development. These EST-SSR markers developed in this study are valuable tools for genetic diversity, evolutionary, and molecular breeding in E. sibiricus, and other Elymus species.
Electronic supplementary material
The online version of this article (10.1186/s12870-019-1825-8) contains supplementary material, which is available to authorized users.
Keywords: Elymus genus, E. sibiricus, Transcriptome sequencing, EST-SSRs development, Transferability, Genetic relationship
Background
The tribe Triticeae Dumort., an economically important tribe in the grass family (Poaceae), contains three important cereal crops closely related to human life and civilization, namely, wheat (Triticum aestivum L.), barley (Hordeum vulgare L.) and rye (Secale cereale L.), as well as many economically valuable forage grasses, such as crested wheatgrass (Agropyron cristatum L.), bottlebrush grass (Hystrix patula), sheepgrass [Leymus chinensis (Trin.) Tzvel.], and Siberian wildrye (Elymus sibiricus L.) [1]. Elymus L. is the largest genus in the tribe Triticeae Dumort., encompassing approximately 150 polyploid perennial species widely distributed in the temperate regions of the world [2, 3]. The diverse origins and ecological habitats may cause genetic variation among and within different Elymus species and populations that could be captured and used in plant breeding and improvement programs [4, 5].
As one of the main mode of speciation, polyploidy (autopolyploidy and allopolyploidy) is a natural hybridization process [6]. It is well known that Elymus arose through hybridization between representatives of different genera [2, 7]. Previous cytological studies suggested that five basic genomes (St, H, Y, P and W) donated by different diploid species constitute Elymus species [8]. Different genomic constitution and the long-term differentiation among different species have made Elymus form a pattern of reticulate evolution and possess rich genetic basis than their diploid parents [9, 10]. The Elymus species thus are ideal materials for studying the formation mechanism and evolution pattern of polyploidy species. Furthermore, Asia is the main origin and genetic differentiation center of Elymus [11, 12]. Many Elymus species like E. sibiricus and E. nutans occur naturally in Eastern Mongolia, the Himalayas, and Western and Northern China [13]. Previous studies mainly focused on the classification between Elymus and their relatives, the origin determination of the ancestor species including St, H and Y genome, and the evolution relationships between basic genomes based on cytogenetics and molecular sequences [12, 14–16]. Information on molecular phylogeny and genetic structure among different Elymus species is limited, but necessary for germplasm collection, conservation and utilization. The phylogenetic relationships and genetic components among different Elymus species are largely unknown because of the limited detection ability of specific or unique nuclear gene sequences [9, 10, 12, 17–19]. Genetic diversity analysis among phylogenetically related species is based on the development of transferable and orthologous molecular markers. Expressed sequence tag-derived simple sequence repeat markers (EST-SSRs) are the markers of choice, because they are abundant, co-dominant, highly polymorphic, and are easily transferable among phylogenetically related species [13]. In previous studies, three expressed sequence tag (EST) libraries were developed and annotated for Pseudoroegneria spicata, a mixture of both Elymus wawawaiensis and E. lanceolatus, and a Leymus cinereus × L. triticoides interspecific hybrid [20]. EST-SSR primers developed from three perennial diploid Triticeae species were used to produce amplicons in these three species, and EST-SSR primers derived from Thinopyrum bessarabicum and Th. elongatum had greater transferability to each other than those derived from the St-genome Pseudoroegneria spicata due to close relationship between Jb and Je genomes [21]. Moreover, EST-SSR markers are critical for genetic relationship analysis, genetic mapping, and DNA fingerprinting for many crops and forage grasses [13, 22–27]. Mott et al. developed simple sequence repeat markers based on Elymus expressed sequence tag sequence, and used a subset of the 23 most polymorphic SSRs to analyze genetic diversity of seven North American Elymus, Pseudoroegneria and Pascopyrum species [28].
Recently, the advent of next-generation sequencing (NGS), especially de novo transcriptome sequencing, had provided a cost-effective approach to identify microsatellite loci [23, 29, 30]. You et al. identified 5278 SSRs in taro (Colocasia esculenta) transcriptome data, and finally used 62 polymorphic markers for taro genetic diversity study [31]. Our previous transcriptome sequencing in E. sibiricus have generated and identified 185,523 unigenes and more than 30,000 differentially expressed transcripts (DETs), which provided important genetic resources and sequence information for developing EST-SSR markers in this study [32].
The objectives of the study were to (i) develop EST-SSR markers from E. sibiricus transcriptome sequencing and verify the transferability among different Elymus species; (ii) to evaluate the genetic diversity and genetic relationships among 17 polyploidy Elymus species based on the developed EST-SSR markers; (iii) to elucidate the phylogenetic relationships and genetic differentiation between StH, StY and StHY genome in Elymus species. Characterization of the genetic components among and within Elymus genomes will contribute to the understanding of the origin and evolution mechanism in polyploidy species.
Results
Frequency and distribution of EST-SSR markers
A total of 4,598,845 contigs were obtained after de novo assembly. The total number of unigenes with paired-end reads was 135,433. The total length of the unigenes was 97,059,505, with an average length of 716.66 bp and N50 value of 1269. Among the 135,433 unigenes, the length of 109,476 unigenes (80.83%) ranged from 200 to 1000 bp, the length of 21,630 (15.97%) ranged from 1000 to 3000 bp, and 4327 unigenes (3.20%) were more than 3000 bp. The length distribution of the unigenes was shown in Additional file 1: Figure S1. Of the 135,433 unigenes, 57,756 (42.65%) unigenes were successfully annotated in at least one database (Table 1). According to Nr annotation, most BLAST hits were detected from Aegilops tauschii (22.38%), followed by Hordeum vulgare (18.01%) and Triticum urartu (12.88%) (Additional file 2: Figure S2).
Table 1.
Function annotation of the E. sibiricus transcriptom
| Anno Database | Annotated Number | Percentage (%) |
|---|---|---|
| COG Annotation | 13,813 | 10.20% |
| GO Annotation | 34,044 | 25.14% |
| KEGG Annotation | 16,352 | 12.07% |
| KOG Annotation | 25,954 | 19.16% |
| Pfam Annotation | 31,603 | 23.33% |
| Swissprot Annotation | 24,992 | 18.45% |
| Nr Annotation | 55,738 | 41.16% |
| All Annotated | 57,756 | 42.65% |
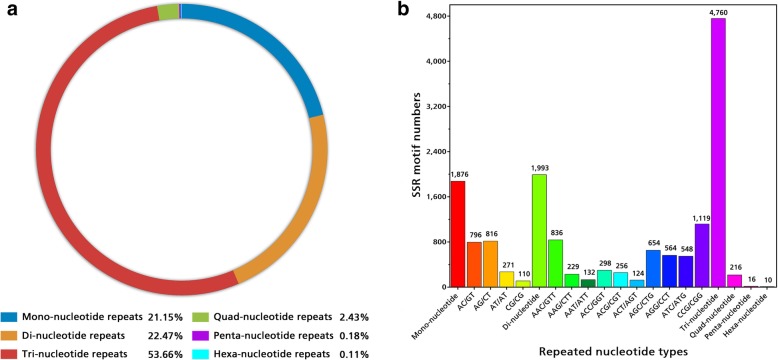
A total of 8871 putative EST-SSRs were identified from 6685 unigenes (Table 2). An average of one EST-SSR was found every 6.2 kb. Among these potential SSRs, six types of motifs were identified, of which trinucleotides (4760, 53.66%), dinucleotides (1993, 22.47%) and mononucleotides (1876, 21.15%) were the most abundant SSRs (Fig. 1a). EST-SSRs with five tandem repeats (40.71%) were the most common type, followed by six (23.48%), ten (12.95%), seven (7.49%), eleven (5.16%), eight (3.29%), nine (2.25%) and twelve (2.04%) tandem repeats. The remaining tandem repeats were less than 1%. Among the SSRs identified, the most dominant trinucleotide repeat motif was CCG/CGG (1119, 23.5%) and AAC/GTT (836, 17.6%), while motifs of AG/CT (816, 40.9%) and AC/GT (796, 39.9%) were represented in the dinucleotide repeats (Fig. 1b).
Table 2.
The distribution of the EST-SSRs based on the number of repeat units
| Repeats | Mono- | Di- | Tri- | Quad- | Penta- | Hexa- | Total | Percentage (%) |
|---|---|---|---|---|---|---|---|---|
| 5 | 0 | 0 | 3400 | 195 | 12 | 4 | 3611 | 40.71 |
| 6 | 0 | 944 | 1115 | 21 | 1 | 2 | 2083 | 23.48 |
| 7 | 0 | 426 | 235 | 0 | 2 | 1 | 664 | 7.49 |
| 8 | 0 | 282 | 10 | 0 | 0 | 0 | 292 | 3.29 |
| 9 | 0 | 199 | 0 | 0 | 1 | 0 | 200 | 2.25 |
| 10 | 1041 | 107 | 0 | 0 | 0 | 1 | 1149 | 12.95 |
| 11 | 425 | 33 | 0 | 0 | 0 | 0 | 458 | 5.16 |
| 12 | 178 | 2 | 0 | 0 | 0 | 1 | 181 | 2.04 |
| 13 | 88 | 0 | 0 | 0 | 0 | 0 | 88 | 0.99 |
| 14 | 60 | 0 | 0 | 0 | 0 | 0 | 60 | 0.68 |
| 15 | 42 | 0 | 0 | 0 | 0 | 1 | 43 | 0.48 |
| 16 | 25 | 0 | 0 | 0 | 0 | 0 | 25 | 0.28 |
| 17 | 8 | 0 | 0 | 0 | 0 | 0 | 8 | 0.09 |
| 18 | 3 | 0 | 0 | 0 | 0 | 0 | 3 | 0.03 |
| 19 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0.01 |
| 20 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.00 |
| 21 | 2 | 0 | 0 | 0 | 0 | 0 | 2 | 0.02 |
| 22 | 3 | 0 | 0 | 0 | 0 | 0 | 3 | 0.03 |
| Total | 1876 | 1993 | 4760 | 216 | 16 | 10 | 8871 | |
| Percentage (%) | 21.15 | 22.47 | 53.66 | 2.43 | 0.18 | 0.11 |
Fig. 1.
Characteristics of identified SSR. Six types of motif and their percentage (a), different types of tandem repeats and their percentage (b)
Development and transferability of novel EST-SSRs
Two hundred EST-SSR primer pairs were designed and selected to amplify ten selected DNA samples. Eighty-seven pairs of primer (43.5%) generated clear and reproducible bands with expected size, and showed good transferability across different Elymus species. Forty-four primers (22%) generated non-polymorphic bands. Eighteen primers (9%) produced bands with unexpected size, and the remaining fifty-one (25.5%) primers didn’t produce any bands. Finally, a total of thirty polymorphic EST-SSRs were used for the genetic diversity analysis of 480 Elymus individual plants.
Verification of repeat motif types across different species
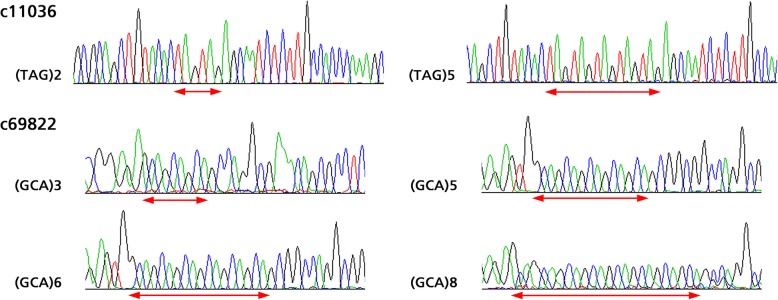
To determine the authenticity of EST-SSR primers, amplicons from 17 different Elymus species for two EST-SSRs were sequenced. In all of the cases, the sequenced alleles from different Elymus species were homologous to the original SSR locus from which the primer was designed. Marker polymorphisms among the 17 Elymus species were found due to variation in number of repeats of SSR motifs. According to the sequencing results of expected bands generated from primer c11036, E. sibiricus had five TAG repeats, the remaining sixteen species had two TAG repeats. For primer c69822, E. antiquus had eight GCA repeats, E. nutans, E. ciliaris and E. panormitanus had three GCA repeats, E. sibiricus had five GCA repeats, and the remaining twelve Elymus species had six GCA repeats (Figs. 2 and 3).
Fig. 2.
Comparative electropherogram analysis of two EST-SSR loci (c11036 and c69822) among different species of Elymus
Fig. 3.
Alignment of sequences obtained from selected PCR bands amplified by two primers (a, c11036; b, c69822) in seventeen Elymus species. The expected repeat motif types were marked in bold letters
Genetic diversity analysis of StH, StHY, and StY genome combinations
Thirty EST-SSR primers generated 572 bands. The number of amplified bands ranged from 5 (c11036) to 25 (c66150, c67290 and c68713), with an average of 19.1 (Table 3). The percentage of polymorphic bands among Elymus species was 100%. The expected heterozygosity (He) ranged from 0.73 to 0.95, with an average of 0.92. The observed heterozygosity (Ho) ranged from and 0.89 to 1.00, with an average of 0.98. The polymorphism information content (PIC) value ranged from 0.28 (c61134 and c69822) to 0.43 (c68713), with an average of 0.36.
Table 3.
Allelic diversity of thirty used EST-SSR markers in ninety-five accessions of Elymus
| Primer | SSRs | Forward primer | Reverse primer | Tm (°C) | T | M | TP | PP | He | Ho | PIC |
|---|---|---|---|---|---|---|---|---|---|---|---|
| c11036 | (TAG)5 | CACTTGGGTGTGCAAAAGAA | TTCCCCTAGCAGGCAGATTA | 58 | 5 | 0 | 5 | 100.0% | 0.73 | 0.90 | 0.32 |
| c24738 | (AGC)5 | ATTTTGGCTATTGAGCGTGG | GAAGATGGGCGACAAATTCT | 58 | 22 | 0 | 22 | 100.0% | 0.94 | 0.92 | 0.36 |
| c40721 | (GAG)5 | GAGGGTAATGACCGTCTGGA | TCTGGCCATGTTGTTGTTGT | 58 | 21 | 0 | 21 | 100.0% | 0.94 | 0.97 | 0.39 |
| c40810 | (ACA)6 | ATCTTCTATCACGGCCAACG | TTGATGGCACATTGAGATCC | 58 | 15 | 0 | 15 | 100.0% | 0.91 | 0.92 | 0.35 |
| c45646 | (TTG)5 | CTGATGATGATGGCCAGTTG | GAGAGGTCATTCCGCATGTT | 60 | 19 | 0 | 19 | 100.0% | 0.94 | 0.96 | 0.42 |
| c47765 | (GCT)5 | GAGAGAGACACACGGAAGGC | GACAGAGAGCTCGCAGGAG | 62 | 23 | 0 | 23 | 100.0% | 0.94 | 1.00 | 0.36 |
| c56077 | (TGA)5 | GGCCAATCAAATTGGAACAT | AAGGCCTCCGAGCTAAAAAG | 56 | 13 | 0 | 13 | 100.0% | 0.91 | 0.89 | 0.37 |
| c57159 | (TGT)5 | CGCCGCTTATCACTTTTGTT | ACTAATGATGTCGCCGATCC | 58 | 21 | 0 | 21 | 100.0% | 0.93 | 1.00 | 0.35 |
| c60959 | (ACT)5 | CATTGTTGGCGTTCAATCAC | CCGTTAGCTCATCCAAGCTC | 58 | 15 | 0 | 15 | 100.0% | 0.91 | 0.96 | 0.38 |
| c61134 | (TCC)6 | CCTCCCAGGTGACACGTACT | GGTAGGGGGCGTAAGAAGAG | 64 | 11 | 0 | 11 | 100.0% | 0.85 | 1.00 | 0.28 |
| c62352 | (CGG)6 | TTTTAGAGCAGCAGCAGCAA | GACCCGGAGAAGATCAACAA | 58 | 22 | 0 | 22 | 100.0% | 0.93 | 1.00 | 0.32 |
| c64785 | (CTT)5 | TCCCTTTGTCTCCCTCTCCT | ACAATCACGGCGATAAGGTC | 60 | 18 | 0 | 18 | 100.0% | 0.92 | 0.94 | 0.33 |
| c65566 | (CGC)5 | TCCTCGTCTCCTCCCTCC | CTCGTCCTCGTAGTGCTCCT | 60 | 23 | 0 | 23 | 100.0% | 0.95 | 1.00 | 0.41 |
| c66150 | (CGG)5 | CTCAGATCGTCCTCCGTCAC | CTCCTCCTGCTGCTCGTC | 60 | 25 | 0 | 25 | 100.0% | 0.95 | 1.00 | 0.34 |
| c66252 | (CTT)6 | GGAGGAGGAGAACTTCCTGG | TCACCGGAAAAACACATTCA | 56 | 21 | 0 | 21 | 100.0% | 0.94 | 1.00 | 0.33 |
| c66352 | (CTG)5 | GACTACTGGAGGCGGACTTG | GAGAGCAGGCAGAGAGCCTA | 64 | 19 | 0 | 19 | 100.0% | 0.93 | 1.00 | 0.31 |
| c66859 | (AAG)5 | AGCGACCTGGACATGAACTC | GAATCCACCCTGAAGGATCA | 60 | 20 | 0 | 20 | 100.0% | 0.94 | 0.96 | 0.41 |
| c67170 | (CCG)5 | GGACCTCCCACAAAGTGAAA | AGTAGCTCCCCACCACGAG | 60 | 22 | 0 | 22 | 100.0% | 0.94 | 0.99 | 0.38 |
| c67290 | (CGG)5 | AGACGAGGACGTCGGTGTT | AGACACGCGAGAGGGTAGAC | 60 | 25 | 0 | 25 | 100.0% | 0.95 | 1.00 | 0.41 |
| c68346 | (AGC)5 | CGAGTTGTGTCACCCAGAGA | ACAAGCAAGGCATACCCAAG | 60 | 16 | 0 | 16 | 100.0% | 0.92 | 1.00 | 0.37 |
| c68713 | (CGG)6 | GTTCGTCGTCTCCGCATC | AAGAAGCACAGGCCAAGC | 56 | 25 | 0 | 25 | 100.0% | 0.95 | 1.00 | 0.43 |
| c69557 | (GCG)5 | ACCAGGCGATTTATGGACAG | GTGACCGCAACAATGACAAC | 60 | 19 | 0 | 19 | 100.0% | 0.94 | 0.95 | 0.38 |
| c69822 | (GCA)5 | CAGCCGAAAATTTCCGTAGA | GCCATTTGAGGAGGACAAAA | 58 | 20 | 0 | 20 | 100.0% | 0.93 | 1.00 | 0.28 |
| c70441 | (CGG)5 | TGGAGGAGACGGTAGAGGTG | CAAGAATAAACGGGAGCGAA | 58 | 21 | 0 | 21 | 100.0% | 0.93 | 1.00 | 0.33 |
| c70704 | (AAG)5 | ACCTCAACCGTGCTCTCAAG | TCCTTGGCCATCAACTTCTC | 60 | 19 | 0 | 19 | 100.0% | 0.94 | 1.00 | 0.40 |
| c70817 | (GAC)5 | AGCCATGCTCAGGACCATAC | TACATTCTGCTGCTTGGCAC | 60 | 17 | 0 | 17 | 100.0% | 0.93 | 0.99 | 0.41 |
| c71552 | (GCC)5 | CACTTCTCTCTCTCCCTCGC | CCGTGGATGACAACGGTTAC | 62 | 19 | 0 | 19 | 100.0% | 0.93 | 1.00 | 0.36 |
| c75219 | (GGA)5 | CACTCGCAACCGAGGAAG | CGGAGTCCAACGTCGTCTAC | 58 | 16 | 0 | 16 | 100.0% | 0.92 | 1.00 | 0.38 |
| c79438 | (TAA)5 | TGAATGATTCTCGCGAAGTG | GAGGAGGGCAAACAACAAAA | 58 | 20 | 0 | 20 | 100.0% | 0.93 | 0.99 | 0.36 |
| c82957 | (GCC)6 | GAGAACAACGGGCAAGAAGA | GGGACGATTTGGAACAGCTA | 60 | 20 | 0 | 20 | 100.0% | 0.94 | 0.98 | 0.40 |
| Mean | 19.1 | 0 | 19.1 | 100.0% | 0.92 | 0.98 | 0.36 |
T total number of amplified bands, M number of monomorphic bands, TP total number of polymorphic bands, PP percentage of polymorphism, He expected heterozygosity, Ho observed heterozygosity, PIC polymorphic information content
In general, hexaploid Elymus species with genomes StStHHYY had a relatively higher level of genetic diversity (Na = 1.637, Ne = 1.369, H = 0.219, I = 0.330, %P = 63.7), while tetraploid Elymus species with genomes StStYY genomes had relatively lower level of genetic diversity (Na = 1.504, Ne = 1.309, H = 0.182, I = 0.272, %P = 50.4) in the study (Table 4). At the species level, the highest level of genetic diversity was found in E. cylindricus (Na = 1.734, Ne = 1.398, H = 0.239, I = 0.363, %P = 73.4), followed by E. nutans (Na = 1.633, Ne = 1.368, H = 0.218, I = 0.329, %P = 63.3) and E. sibiricus (Na = 1.573, Ne = 1.331, H = 0.197, I = 0.297, %P = 57.3). The lowest level of genetic diversity was found in E. antiquus with only two individuals (Na = 1.309, Ne = 1.219, H = 0.128, I = 0.187, %P = 30.9). Further, we compared the genetic diversity among different geographical groups in E. nutans and E. sibiricus. For E. sibiricus, the high level of genetic diversity was found in Western Tien-shan group (Geo-7, H = 0.230, %P = 66.7%), the lowest level of genetic diversity was found in Mongolian Plateau group (Geo-2, H = 0.183, %P = 52.4%). For E. nutans the highest level of genetic diversity was found in Xinjiang Tianshan group (Geo-12, H = 0.234, %P = 68.9%), and the lowest level of genetic diversity was found in Southeastern Qinghai-Tibetan Plateau group (Geo-9, H = 0.208, %P = 59.8%).
Table 4.
Genomic classification, geographic information and genetic diversity analysis of 95 accessions among three Elymus genomes
| Gen | Species | Accession | SS | Geographic information | Genetic analysis | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Origins | La(N) | Lo(E) | Al(m) | Na | Ne | H | I | %P | ||||
| StH | Elymus sibiricus | PI504462 | 5 | Xining, Qinghai, China (Geo-1) | 36.7° | 101.8° | 2300 | 1.605 | 1.358 | 0.211 | 0.317 | 60.5 |
| PI531669 | 6 | Xining, Qinghai, China (Geo-1) | 36.6° | 101.8° | 2400 | 1.603 | 1.351 | 0.208 | 0.313 | 60.3 | ||
| PI499456 | 6 | Shandan, Gansu, China (Geo-1) | 38.8° | 101.1° | 1750 | 1.652 | 1.368 | 0.221 | 0.334 | 65.2 | ||
| PI639859 | 2 | Aba, Sichuan, China (Geo-1) | 33.1° | 102.6° | 3320 | 1.268 | 1.189 | 0.111 | 0.162 | 26.8 | ||
| PI655199 | 6 | Longriba, Sichuan, China (Geo-1) | 32.1° | 102.6° | 3280 | 1.610 | 1.322 | 0.196 | 0.300 | 61.0 | ||
| PI499453 | 6 | Inner Mongolia, China (Geo-2) | 43.9° | 116.1° | 1000 | 1.580 | 1.309 | 0.188 | 0.287 | 58.0 | ||
| PI665507 | 6 | Ulaanbaatar, Mongolia (Geo-2) | 48.1° | 108.7° | 1524 | 1.600 | 1.321 | 0.195 | 0.297 | 60.0 | ||
| PI634231 | 3 | Mongolia (Geo-2) | 47.6° | 112.4° | 747 | 1.470 | 1.306 | 0.179 | 0.265 | 47.0 | ||
| PI610886 | 6 | Dzuunburen, Mongolia (Geo-2) | 50.0° | 105.7° | 920 | 1.659 | 1.384 | 0.226 | 0.341 | 65.9 | ||
| W621576 | 2 | Hatgal, Mongolia (Geo-2) | 50.3° | 99.8° | 2042 | 1.309 | 1.219 | 0.128 | 0.187 | 30.9 | ||
| PI598782 | 6 | Buryat, Russia (Geo-3) | 52.2° | 109.3° | 900 | 1.596 | 1.318 | 0.193 | 0.295 | 59.6 | ||
| PI598789 | 6 | Chita, Russia (Geo-3) | 52.1° | 113.5° | 700 | 1.635 | 1.364 | 0.216 | 0.327 | 63.5 | ||
| PI610991 | 3 | Buryat, Russia (Geo-3) | 51.9° | 104.8° | 630 | 1.400 | 1.259 | 0.151 | 0.225 | 40.0 | ||
| PI655095 | 6 | Bayshint, Mongolia (Geo-4) | 49.7° | 90.3° | 1509 | 1.685 | 1.398 | 0.235 | 0.354 | 68.5 | ||
| PI611014 | 6 | Gorno Altay, Russia (Geo-4) | 50.3° | 87.7° | 950 | 1.607 | 1.325 | 0.196 | 0.299 | 60.7 | ||
| PI634228 | 6 | Gorno Altay, Russia (Geo-4) | 51.2° | 85.6° | 1250 | 1.573 | 1.314 | 0.188 | 0.287 | 57.3 | ||
| PI655084 | 6 | Gorno Altay, Russia (Geo-4) | 51.3° | 85.6° | 1090 | 1.619 | 1.341 | 0.204 | 0.310 | 61.9 | ||
| PI639757 | 6 | Gorno Altay, Russia (Geo-4) | 51.7° | 85.8° | 340 | 1.591 | 1.306 | 0.187 | 0.287 | 59.1 | ||
| PI655140 | 6 | Urumqi, Xinjiang, China (Geo-5) | 43.5° | 87.0° | 1650 | 1.640 | 1.383 | 0.225 | 0.337 | 64.0 | ||
| PI595182 | 6 | Urumqi, Xinjiang, China (Geo-5) | 43.9° | 87.9° | 1250 | 1.677 | 1.392 | 0.233 | 0.351 | 67.7 | ||
| PI595180 | 4 | Urumqi, Xinjiang, China (Geo-5) | 43.8° | 87.9° | 1600 | 1.509 | 1.313 | 0.184 | 0.275 | 50.9 | ||
| PI619577 | 5 | Urumqi, Xinjiang, China (Geo-5) | 43.1° | 87.6° | 2360 | 1.575 | 1.351 | 0.205 | 0.307 | 57.5 | ||
| PI499468 | 6 | Urumqi, Xinjiang, China (Geo-5) | 43.7° | 87.9° | 1770 | 1.626 | 1.373 | 0.220 | 0.331 | 62.6 | ||
| PI499462 | 2 | Urumqi, Xinjiang, China (Geo-5) | 43.6° | 87.9° | 1050 | 1.268 | 1.189 | 0.111 | 0.162 | 26.8 | ||
| PI598780 | 6 | Vladivostok, Russia (Geo-6) | 43.2° | 132.0° | 200 | 1.552 | 1.288 | 0.176 | 0.270 | 55.2 | ||
| PI628699 | 5 | Sovetskaya Gavan, Russia (Geo-6) | 49.0° | 140.3° | 50 | 1.631 | 1.338 | 0.205 | 0.314 | 63.1 | ||
| PI628706 | 6 | Sovetskaya Gavan, Russia (Geo-6) | 48.6° | 139.9° | 40 | 1.650 | 1.385 | 0.227 | 0.342 | 65.0 | ||
| PI598778 | 6 | Kazakhstan (Geo-7) | – | – | – | 1.692 | 1.393 | 0.233 | 0.353 | 69.2 | ||
| PI659930 | 5 | Kochkorka, Kyrgyzstan (Geo-7) | 41.7° | 75.7° | 2438 | 1.622 | 1.376 | 0.222 | 0.332 | 62.2 | ||
| PI659942 | 6 | Kochkorka, Kyrgyzstan (Geo-7) | 41.7° | 75.5° | 2627 | 1.687 | 1.401 | 0.236 | 0.356 | 68.7 | ||
| 5.2 | 1.573 | 1.331 | 0.197 | 0.297 | 57.3 | |||||||
| StHY | Elymus cylindricus | WEcy3 | 9 | Yongdeng, Gansu, China | 36.7° | 103.3° | 2300 | 1.734 | 1.398 | 0.239 | 0.363 | 73.4 |
| StHY | Elymus dahuricus | WEda5 | 8 | Shandan, Gansu, China | 38.8° | 101.1° | 1750 | 1.694 | 1.374 | 0.225 | 0.344 | 69.4 |
| StHY | Elymus excelsus | WEex1 | 6 | Yongdeng, Gansu, China | 36.7° | 103.3° | 2300 | 1.654 | 1.364 | 0.218 | 0.331 | 65.4 |
| StHY | Elymus nutans | PI499612 | 6 | Xining, Qinghai, China (Geo-8) | 36.7° | 101.3° | 2450 | 1.652 | 1.391 | 0.229 | 0.344 | 65.2 |
| PI619586 | 6 | Dulan, Qinghai, China (Geo-8) | 35.8° | 97.6° | 2950 | 1.659 | 1.363 | 0.218 | 0.331 | 65.9 | ||
| PI531645 | 6 | Qinghai, China (Geo-8) | – | – | – | 1.720 | 1.438 | 0.255 | 0.382 | 72.0 | ||
| Xiahe15 | 4 | Xiahe, Gansu, China (Geo-8) | 35.0° | 102° | 3100 | 1.532 | 1.304 | 0.184 | 0.278 | 53.2 | ||
| PI639852 | 5 | Xiahe, Gansu, China (Geo-8) | 35.1° | 102.4° | 2960 | 1.608 | 1.358 | 0.212 | 0.320 | 60.8 | ||
| Xiahe48 | 5 | Xiahe, Gansu, China (Geo-8) | 34.6° | 102.1° | 3100 | 1.650 | 1.371 | 0.221 | 0.335 | 65.0 | ||
| PI655186 | 5 | Xiahe, Gansu, China (Geo-8) | 35.2° | 102.5° | 2830 | 1.617 | 1.368 | 0.217 | 0.326 | 61.7 | ||
| PI655193 | 4 | Maqu, Gansu, China (Geo-8) | 34.0° | 102.1° | 3280 | 1.561 | 1.339 | 0.200 | 0.300 | 56.1 | ||
| PI655195 | 5 | Maqu, Gansu, China (Geo-8) | 34.0° | 102.8° | 3350 | 1.587 | 1.336 | 0.201 | 0.304 | 58.7 | ||
| PI655192 | 5 | Maqu, Gansu, China (Geo-8) | 34.1° | 102.2° | 3660 | 1.656 | 1.403 | 0.236 | 0.353 | 65.6 | ||
| Luqu28 | 5 | Luqu, Gansu, China (Geo-8) | 34.5° | 102.7° | 3100 | 1.593 | 1.338 | 0.201 | 0.304 | 59.3 | ||
| PI639855 | 5 | Luqu, Gansu, China (Geo-8) | 34.7° | 102.5° | 3060 | 1.657 | 1.406 | 0.238 | 0.355 | 65.7 | ||
| Luqu27 | 5 | Luqu, Gansu, China (Geo-8) | 34.4° | 102.5° | 3100 | 1.698 | 1.406 | 0.241 | 0.363 | 69.8 | ||
| PI628698 | 6 | Tianzhu, Gansu, China (Geo-8) | 37.2° | 102.8° | 2860 | 1.694 | 1.398 | 0.237 | 0.357 | 69.4 | ||
| PI499451 | 6 | Lanzhou, Gansu, China (Geo-8) | 36.1° | 103.8° | 1520 | 1.691 | 1.404 | 0.238 | 0.359 | 69.1 | ||
| PI499611 | 4 | Lanzhou, Gansu, China (Geo-8) | 37.3° | 102.8° | 2300 | 1.586 | 1.363 | 0.213 | 0.319 | 58.6 | ||
| PI639858 | 6 | Aba, Sichuan, China (Geo-9) | 33.1° | 102.7° | 3460 | 1.649 | 1.367 | 0.218 | 0.329 | 64.9 | ||
| PI619521 | 6 | Luhuo, Sichuan, China (Geo-9) | 31.8° | 100.5° | 3740 | 1.647 | 1.375 | 0.222 | 0.335 | 64.7 | ||
| W622112 | 4 | Luhuo, Sichuan, China (Geo-9) | 31.5° | 100.5° | 3190 | 1.589 | 1.375 | 0.218 | 0.324 | 58.9 | ||
| PI619569 | 6 | Dege, Sichuan, China (Geo-9) | 31.9° | 99.0° | 4110 | 1.661 | 1.368 | 0.221 | 0.336 | 66.1 | ||
| PI619520 | 6 | Barkam, Sichuan, China (Geo-9) | 32.0° | 102.7° | 3300 | 1.642 | 1.353 | 0.213 | 0.323 | 64.2 | ||
| W622107 | 6 | Kangding, Sichuan, China (Geo-9) | 30.0° | 101.8° | 3800 | 1.673 | 1.392 | 0.232 | 0.350 | 67.3 | ||
| PI639862 | 4 | Kangding, Sichuan, China (Geo-9) | 30.0° | 101.9° | 3110 | 1.516 | 1.300 | 0.180 | 0.272 | 51.6 | ||
| W623602 | 5 | Litang, Sichuan, China (Geo-9) | 29.9° | 100.3° | 3780 | 1.603 | 1.361 | 0.212 | 0.318 | 60.3 | ||
| PI619516 | 3 | Sichuan, China (Geo-9) | – | – | – | 1.406 | 1.264 | 0.154 | 0.228 | 40.6 | ||
| PI619527 | 6 | Lhasa, Tibet, China (Geo-10) | 29.7° | 91.1° | 4460 | 1.694 | 1.401 | 0.238 | 0.359 | 69.4 | ||
| PI619530 | 6 | Lhasa, Tibet, China (Geo-10) | 30.0° | 90.6° | 4020 | 1.645 | 1.355 | 0.212 | 0.323 | 64.5 | ||
| PI619526 | 6 | Gongbogyamda, Tibet, China (Geo-10) | 30.0° | 93.1° | 3600 | 1.673 | 1.380 | 0.228 | 0.345 | 67.3 | ||
| PI619589 | 6 | Nedong, Tibet, China (Geo-10) | 29.3° | 91.8° | 3800 | 1.656 | 1.365 | 0.219 | 0.332 | 65.6 | ||
| PI619533 | 6 | Nedong, Tibet, China (Geo-10) | 29.2° | 91.8° | 3470 | 1.638 | 1.347 | 0.209 | 0.319 | 63.8 | ||
| PI619590 | 6 | Changdu, Tibet, China (Geo-10) | 30.7° | 97.3° | 3740 | 1.642 | 1.363 | 0.218 | 0.329 | 64.2 | ||
| PI619592 | 4 | Changdu, Tibet, China (Geo-10) | 31.1° | 97.2° | 4200 | 1.511 | 1.312 | 0.183 | 0.275 | 51.1 | ||
| PI619525 | 6 | Tibet, China (Geo-10) | 31.3° | 98.0° | 3100 | 1.647 | 1.365 | 0.217 | 0.329 | 64.7 | ||
| PI619522 | 4 | Gongjue, Tibet, China (Geo-10) | 30.9° | 98.3° | 4100 | 1.551 | 1.347 | 0.202 | 0.301 | 55.1 | ||
| PI619532 | 5 | Yangbajain, Tibet, China (Geo-10) | 30.1° | 90.5° | 4150 | 1.607 | 1.359 | 0.212 | 0.319 | 60.7 | ||
| W623617 | 5 | Tibet, China (Geo-10) | 30.2° | 99.9° | 4000 | 1.624 | 1.352 | 0.210 | 0.319 | 62.4 | ||
| PI547394 | 6 | Inner Mongolia, China (Geo-11) | – | – | – | 1.645 | 1.349 | 0.211 | 0.322 | 64.5 | ||
| PI499450 | 5 | Inner Mongolia, China (Geo-11) | – | – | – | 1.668 | 1.407 | 0.237 | 0.355 | 66.8 | ||
| PI628675 | 6 | Hotan, Xinjiang, China (Geo-12) | 36.3° | 80.0° | 3300 | 1.691 | 1.398 | 0.236 | 0.356 | 69.1 | ||
| PI619519 | 6 | Burqin, Xinjiang, China (Geo-12) | 47.7° | 86.9° | 450 | 1.715 | 1.392 | 0.236 | 0.359 | 71.5 | ||
| PI619575 | 5 | Habahe, Xinjiang, China (Geo-12) | 47.9° | 86.2° | 1200 | 1.663 | 1.392 | 0.232 | 0.348 | 66.3 | ||
| PI564956 | 6 | Pakistan (Geo-13) | – | – | – | 1.684 | 1.389 | 0.232 | 0.350 | 68.4 | ||
| W610220 | 6 | Gilgit, Pakistan (Geo-13) | 35.0° | 74.0° | 2736 | 1.666 | 1.384 | 0.228 | 0.344 | 66.6 | ||
| PI406466 | 6 | Russia (Geo-14) | – | – | – | 1.671 | 1.383 | 0.228 | 0.344 | 67.1 | ||
| StHY | Elymus tangutorum | WEta2 | 5 | Minle, Gansu, China | 38.4° | 100.8° | 2310 | 1.654 | 1.394 | 0.231 | 0.347 | 65.4 |
| 5.5 | 1.637 | 1.369 | 0.219 | 0.330 | 63.7 | |||||||
| StY | Elymus abolinii | PI499585 | 5 | Urumqi, Xinjiang, China | 43.8° | 88.1° | 1900 | 1.617 | 1.378 | 0.220 | 0.329 | 61.7 |
| StY | Elymus antiquus | PI564957 | 2 | Nedong, Tibet, China | 29.2° | 91.8° | 3550 | 1.309 | 1.219 | 0.128 | 0.187 | 30.9 |
| StY | Elymus burchan-buddae | PI636649 | 4 | Xiahe, Gansu, China | 35.2° | 102.5° | 3000 | 1.600 | 1.363 | 0.214 | 0.322 | 60.0 |
| PI655143 | 7 | Barkam, Sichuan, China | 32.0° | 102.6° | 3300 | 1.720 | 1.401 | 0.239 | 0.363 | 72.0 | ||
| PI655210 | 1 | Litang, Sichuan, China | 30.2° | 99.9° | 4000 | – | – | – | – | – | ||
| StY | Elymus ciliaris | PI531575 | 7 | Beijing, China | 39.9° | 116.3° | 60 | 1.685 | 1.387 | 0.230 | 0.348 | 68.5 |
| StY | Elymus gmelinii | PI639761 | 3 | Gorno Altay, Russia | 50.6° | 86.5° | 950 | 1.460 | 1.299 | 0.174 | 0.259 | 46.0 |
| PI655100 | 5 | Zhaosu, Xinjiang, China | 43.2° | 81.2° | 2370 | 1.551 | 1.320 | 0.189 | 0.286 | 55.1 | ||
| W621543 | 2 | Erdenebulgan, Miongolia | 50.2° | 101.1° | 1676 | 1.280 | 1.198 | 0.116 | 0.169 | 28.0 | ||
| StY | Elymus longearistatus | PI401282 | 1 | Tehran, Iran | 36.1° | 51.4° | 2850 | – | – | – | – | – |
| StY | Elymus macrochaetus | PI564944 | 4 | Alma Ata, Kazakhstan | 43.1° | 76.9° | 2000 | 1.509 | 1.321 | 0.186 | 0.278 | 50.9 |
| StY | Elymus nevskii | PI564925 | 1 | Novosibirsk, Russia | 54.8° | 83.1° | 150 | – | – | – | – | – |
| PI632570 | 4 | Bayan-Olgii, Mongolia | 49.0° | 89.8° | 1848 | 1.488 | 1.299 | 0.175 | 0.262 | 48.8 | ||
| StY | Elymus panormitanus | PI254866 | 2 | Sirsank, Mosul, Iraq | 37.6° | 44.7° | 200 | 1.315 | 1.223 | 0.130 | 0.190 | 31.5 |
| StY | Elymus semicostatus | PI564964 | 3 | Pakistan | – | – | – | 1.420 | 1.260 | 0.155 | 0.232 | 42.0 |
| StY | Elymus tibeticus | PI639828 | 3 | Lhasa, Tibet, China | 29.3° | 91.0° | 3500 | 1.414 | 1.260 | 0.154 | 0.230 | 41.4 |
| WEti4 | 8 | Tianzhu, Gansu, China | 37.0° | 103.1° | 2400 | 1.696 | 1.406 | 0.239 | 0.360 | 69.6 | ||
| 3.6 | 1.504 | 1.309 | 0.182 | 0.272 | 50.4 | |||||||
Gen, Genome; SS, sample size; La, Latitude; Lo, Longitude; Al, Altitude; Na, observed number of alleles; Ne, effective number of alleles; H, Nei’s genetic diversity; I, Shannon’s Information index; %P, the percentage of polymorphic loci; Geo-1 to Geo-7, different populations of Elymus sibiricus with different geographic origins; Geo-8 to Geo-14, different populations of Elymus nutans with different geographic origins
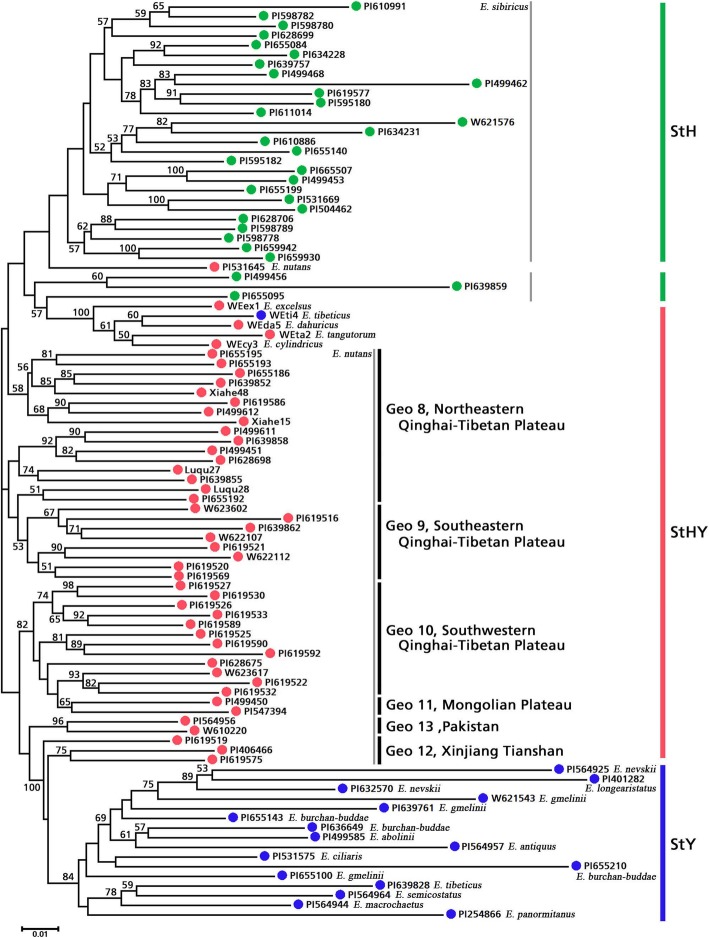
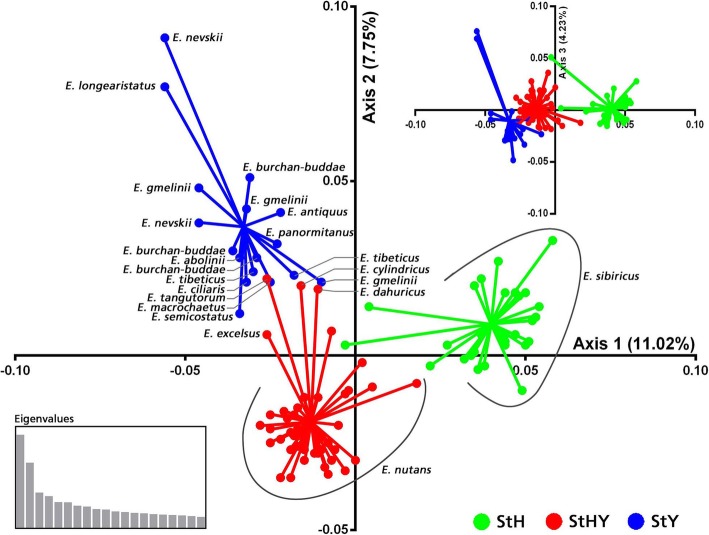
The cluster analysis showed that all ninety-five accessions were clustered into three major clusters (Fig. 4). Cluster 1 contained 30 StH, 13 StHY and 1 StY genome accessions. Cluster 2 contained 16 StHY genome accessions. Cluster 3 contained 16 StY and 19 StHY genome accessions. The results observed in the principal coordinate analysis (PCoA) were in agreement with the cluster analysis. The first three principal components explained 23% of the total variation. A moderate, but clear separation between different genomes was released (Fig. 5). According to PCoA results, two species, E. nevskii and E. longearistatus had a close genetic relationship. Four StHY genome species (E. cylindricus, E. dahuricus, E. excelsus and E. tangutorum) formed one group and had a closer genetic relationship with some StY genome species (Figs. 4 and 5).
Fig. 4.
A neighbor-joining (NJ) dendrogram tree showing the genetic relationship among Elymus accessions based on EST-SSRs. Only bootstrap values higher than 50% are presented. Three types of Elymus genome were represented by different colors, green (StH), red (StHY) and blue (StY). Besides, different geographic groups of E. nutans were annotated. The corresponding detailed information for the 95 Elymus accessions is shown in Table 4
Fig. 5.
Principal coordinate analysis (PCoA) for the first three axes generated from 95 Elymus accessions based on EST-SSR markers
In addition, analysis of molecular variance (AMOVA) was used to evaluate variance components among and within different accessions, species and genomes. The results revealed that 10% of variation occurred among species, whereas 90% of genetic variation existed within species (90%) (Table 5). Despite different genomes, more than 80% of total variance existed within Elymus accessions (86% for StH, 83% for StHY and 81% for StY, respectively), while less than 20% of genetic existed among accessions.
Table 5.
Analysis of molecular variance (AMOVA) for different Elymus genomes
| Source | Degrees of freedom (df) | Sum of squares (SS) | Mean square (MS) | Variance components | Total variance (%) | P-value |
|---|---|---|---|---|---|---|
| StH, StHY and StY genomes | ||||||
| Among genomes | 2 | 2272.668 | 1136.334 | 0.000 | 0% | |
| Among species within genomes | 14 | 2577.613 | 184.115 | 11.059 | 10% | 0.001 |
| Within species | 466 | 45,687.781 | 98.042 | 98.042 | 90% | 0.001 |
| Total | 482 | 50,538.062 | 109.102 | 100% | ||
| StH genome | ||||||
| Among accessions | 29 | 4453.518 | 153.570 | 13.463 | 14% | 0.001 |
| Within accessions | 126 | 10,549.117 | 83.723 | 83.723 | 86% | |
| Total | 155 | 15,002.635 | 97.187 | 100% | ||
| StHY genome | ||||||
| Among species | 4 | 984.661 | 246.165 | 4.710 | 4% | 0.001 |
| Among accessions within species | 43 | 6974.159 | 162.190 | 13.767 | 13% | 0.001 |
| Within accesions | 214 | 19,048.986 | 89.014 | 89.014 | 83% | 0.001 |
| Total | 261 | 27,007.805 | 107.491 | 100% | ||
| StY genome | ||||||
| Among species | 10 | 1592.952 | 159.295 | 2.890 | 3% | 0.004 |
| Among accessions within species | 6 | 815.011 | 135.835 | 15.970 | 16% | 0.001 |
| Within accesions | 48 | 3846.990 | 80.146 | 80.146 | 81% | 0.001 |
| Total | 64 | 6254.954 | 99.006 | 100% | ||
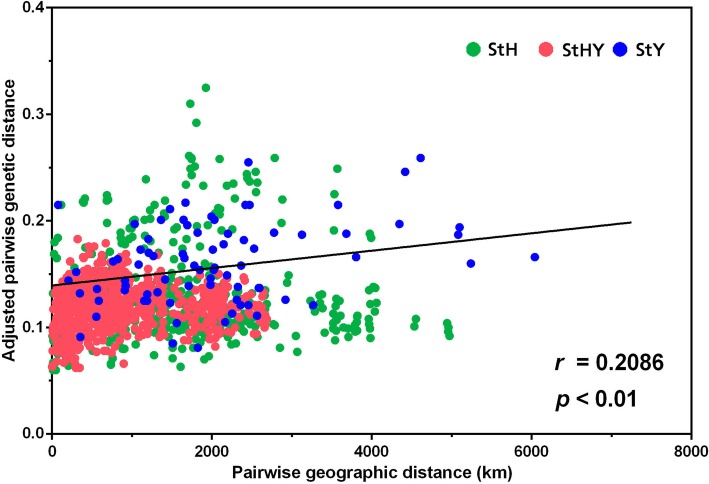
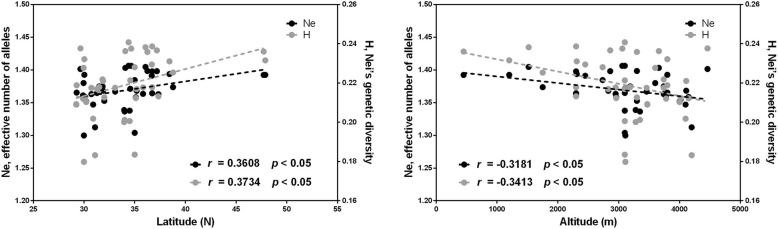
The Mantel test was used to investigate the correlations between genetic information and environmental factors, including geographic distance, latitude and altitude. The regression analysis with 9999 permutations showed a strong positive correlation between Nei’s genetic distance and geographic distance (r = 0.2086, p < 0.01) (Fig. 6). There were no significant correlations between genetic diversity and latitude and altitude at the species level. A positive correlations were found between effective number of alleles (Ne), Nei’s genetic diversity (H) and latitude (r = 0.3608, p < 0.05 and r = 0.3734, p < 0.05, respectively) for StHY genomes accessions. But Ne and H had a negatives correlation with altitude (r = − 0.3181, p < 0.05 and r = − 0.3413, p < 0.05, respectively) (Fig. 7).
Fig. 6.
Regression analysis between pairwise geographic distance and adjusted pairwise genetic distance of 95 Elymus accessions
Fig. 7.
Regression analysis between the effective number of alleles, Nei’s genetic diversity (H) and environmental factors (latitude and altitude) for StHY genome accessions
Genetic structure and genetic differentiation analysis
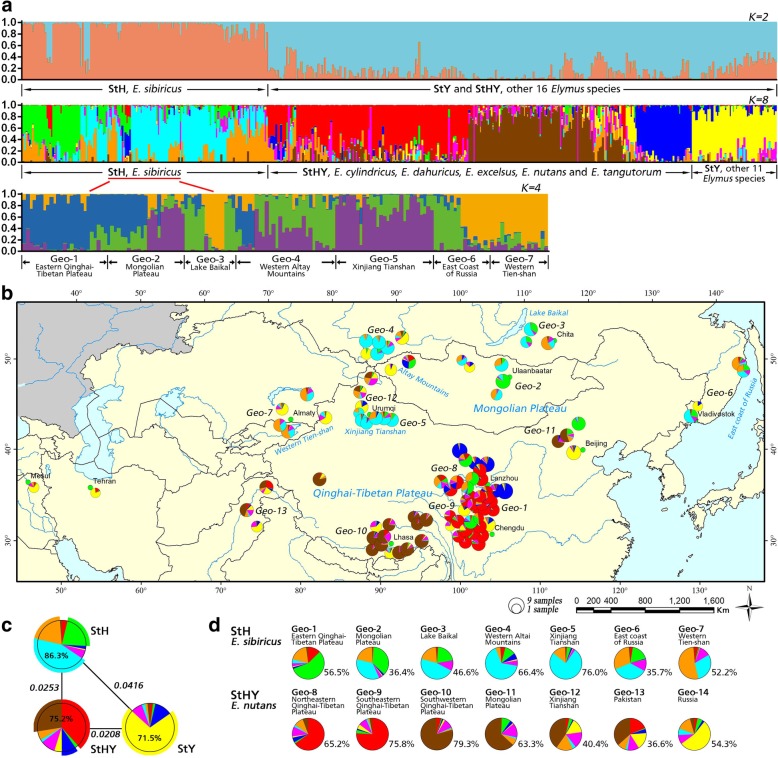
The genetic structure of 480 individuals from 95 Elymus accessions was analyzed using STRUCTURE software. Based on maximum likelihood and K (ΔK) values, the optimal number of groups was two.
As shown in Fig. 8a, E. sibiricus (StH) accessions could be easily separated from other sixteen Elymus species with StY and StHY genomes. We further investigated the internal genetic structure of these Elymus species. E. sibiricus accessions were assigned to four subgroups. Eleven StY genomes species were assigned to the same group. The remaining accessions with StHY genomes were assigned to other groups. There was no obvious relationship between geographic origin and genetic structure in Elymus accessions. For example, twenty-three individuals of five accessions from Geo-2 geographical groups were clustered into two groups.
Fig. 8.
The structure analysis of 95 Elymus accessions based on Bayesian inferred from STRUCTURE program with 30 developed EST-SSRs. a STRUCTURE output with K = 2 and K = 8 showing the population structure among 480 Elymus individuals. Different vertical lines represent an individual genotype and different colors represent genetic stock. Besides, the structure analysis among 30 E. sibiricus accessions was performed based on K = 4; (b) The geographic distribution of the 95 Elymus accessions inferred with Structure across K = 8. The pie charts in the map represent the proportion of each accession and the size of each pie is proportional to sample size from 1 to 9 (Table 4); (c) The genetic distance among the StH, StHY and StY genomes. At K = 8, the proportion of each genome was described by using the pies, of which the protruding sectors belonged to the genome itself; (d) The mean ancestry in each of the eight clusters among 14 geographic groups of E. sibiricus and E. nutans. The percentage of the largest proportion was showed in the graph
Based on results from STRUCTURE analysis, genetic components of each accession was presented using pie graph (Fig. 8b). Based on the different genetic components within different genomes, the highest genetic distance was found between StH and StY genome (0.0416) (Fig. 8c). While StHY genome had a similar genetic distance with StH (0.0253) and StY (0.0208). Besides, we further investigated the probable ancestor origin among and within seven E. sibiricus and seven E. nutans groups (Fig. 8d). Geographical groups: Geo-1, Geo-4, Geo-5 and Geo-7 had more than 50% of same genetic components each. Particularly, E. sibiricus accessions from Xingjiang Tianshan (Geo-5) group shared 76% of genetic components. For E. nutans, some geographic groups had similar ancestor origin, for example, Geo-8 and Geo-9 originating from Eastern Qinghai-Tibetan Plateau shared more than 65% of genetic components (65.2% for Geo-8 and 75.8% for Geo-9). Accessions from Russia (Geo-14) had different ancestor origin compared with other geographical groups.
Discussion
The development of EST-SSRs based on E. sibiricus transcriptome database
In E. sibiricus, marker-assisted selection (MAS) and molecular breeding lag behind other forage species due to the lack of effective molecular markers systems. Simple sequence repeat (SSR) markers are considered to be one of the most important marker systems for plant genetic and breeding studies due to their high polymorphism, high abundance, co-dominance, and genome-wide distribution. Compared with genomic SSRs, expressed sequence tag-derived simple repeat markers (EST-SSRs) are easily transferred among related species owing to the regions being more evolutionary conserved than non-coding sequences. Some previous studies reported the development of expressed sequence tags (ESTs) and simple sequence repeat (SSR) markers for several grasses in the Triticeae. A library of 10,368 ESTs, including 442 SSRs, was developed using hybrids of Elymus lanceolatus and E. wawawaiensis [20]. Until now, only 45,580 EST sequences for Elymus species were included in the National Center for Biotechnology Information (NCBI) database. Recently, the advent of next-generation sequencing (NGS) technologies, such as transcriptome sequencing (RNA-Seq), provided a fast, reliable and cost-effective tool for identifying and developing large-scale EST-SSR markers [33]. These EST-SSR markers derived from RNA-Seq can be used for genetic diversity analysis, genetic linkage map construction, and marker-assisted selection breeding, etc. [25, 26]. There have been few reports on transcriptome analysis about E. sibiricus, the type species of Elymus genus. In this study, among 135,433 assembled unigene sequences, 8871 potential EST-SSRs were identified. The EST-SSR frequency was 4.94%, which was higher than sheepgrass (4.38%) and rice (3.57%) [30, 34]. The distribution density was one SSR per 6.20 kb, which was higher than previous reports in Leymus chinensis (1/10.78 kb) and Medicago sativa (1/12.06 kb), but lower than Paeonia suffruticosa (1/9.24 kb), and Melilotus albus (1/3.99 kb) [30, 33, 35, 36]. Some possible explanations for the difference of the SSRs frequency in expressed sequence tags could be the different genetic basis of various plant species, SSR search criteria, as well as the mining tools used. In many organisms, the extensive distribution of trinucleotide repeats in coding sequences is a sign of the effects of selection, indicating that these SSRs were selected against possible frameshift mutations [30]. In this study, nucleotide repeat from mono- to hexa-nucleotide were detected in 8871 potential SSRs from 135,433 unigenes. Tri-nucleotide repeats were the most abundant SSR motifs. Particularly, CCG/CGG was predominant trinucleotide repeats, followed by AAC/GTT, which was similar with previous reports in rice, barley, wheat, and sheepgrass [30, 37]. Kantety et al. reported that the di-nucleotide repeat motifs existed in similar frequencies in ESTs from various cereal species such as GA/CT and GC/CG [38]. However, in our study, AG/CT (40.9%) was the most abundant di-nucleotide repeat motif in E. sibiricus, which was the same as in annual ryegrass [39]. Our results suggested that most frequent motif repeats might vary between forage grasses and other cereal species. In addition, these ETS-SSR markers developed from E. sibiricus transcriptome data showed good transferability across different Elymus species, suggesting these EST-SSR markers are useful tools for further genetic diversity analysis and molecular breeding for Elymus species.
Phylogenetic relationship of StH, StHY and StY genome combinations
Elymus is a diverse, geographically widespread allopolyploid genus which includes multiple distinct genomic combinations. Cytological studies suggest that five basic genomes, namely, the St, Y, H, P and W in various combinations constitute Elymus species [11]. Of the five basic genomes, the St genome derived from Pseudoroegneria is a fundamental genome that exists in all Elymus species [40]. The H, P and W genomes are derived from the genera Hordeum, Agropyron and Australopyrum of Triticeae, respectively [12]. However, the accurate origin of the Y genome has not yet been identified, although this genome is present in the majority of the Asiatic Elymus species. Previous studies indicated that the StH genome Elymus species is allotetraploid that combines the genomes of Pseudoroegneria (St) and Hordeum (H) [2, 10]. The heterologous hexaploid species, StHY, may undergo two hybridization events, the combination of the St and Y genomes formed tetraploid StY genome, the second hybridization event involved in the combination of the StY and H genome [12, 18]. Genetic diversity analysis in different genomic combinations will facilitate the understanding of the evolution process and genetic differentiation among different species. The phylogenetic relationships of the StH, StY and StHY genome Elymus species have been reported by using molecular sequences [9, 10, 12, 17–19]. In this study, we used newly developed EST-SSR markers to analyze the phylogenetic relationships of 17 StH, StY and StHY genome Elymus species collected from diverse geographical regions. Based on our results, hexaploid StHY species had higher level of genetic diversity (H = 0.219, %P = 63.7) than the other two tetraploid species (StH, H = 0.197, %P = 57.3 and StY, H = 0.182, %P = 50.4). Two hybridization events of the StHY genome species could have contributed to the higher genetic diversity. Liu et al. reported that the hexaploid had two genomic types of ITS sequences, while all the tetraploid Elymus species had only one genomic type of ITS sequences, suggesting that hexaploid Elymus species had higher level of genetic diversity [12]. In addition, our results showed significant heterogeneity among the three genomes Elymus species based on EST-SSRs. The cluster analysis showed all accessions were grouped into three major clusters. In general, the vast majority of accessions could easily be distinguished according to their different genome constitutions. However, the phylogenetic tree based on EST-SSR markers also indicated a multiple origins of polyploids in the evolutionary process of same Elymus species. For example, different accessions of the StHY genome species were clustered in different clusters, which may suggest different maternal lineages of the polyploid genus. Phylogeny relationship of the St genome in Elymus L. sensu lato based on one nuclear DNA and two chloroplast genes showed Pseudoroegneria and Hordeum served as the St and H genome diploid ancestors, and Pseudoroegneria served as maternal donor of the St genome for Elymus s. l. [40, 41]. Meanwhile, differences in the levels and patterns of nucleotide diversity of the rbcL gene implied that the St genome lineages in the species of Elymus s. l. have differently evolutionary potentials [41]. In this study, all 95 Elymus accessions showed a certain degree of genetic similarity due to a shared St genome from Pseudoroegneria. All E. sibiricus accessions and thirteen E. nutans accessions were grouped into Cluster 1, suggesting that the two species had closer genetic relationship. E. sibiricus and E. nutans shared St and H genome. In addition, previous phylogenetic relationships in Elymus based on ITS and chloroplast trnL-F sequences also suggested the St and Y genomes may have the same origin [12]. In this study, almost all StY genome species from Asian were grouped together. A previous study showed that 9 StStYY genome species including E. abolinii, E. ciliaris, E. gmelinii, E. longearistatus, E. nevskii, E. semicostatus were grouped together with 100 bootstrap value, and suggested the Asian StStYY tetraploids probably represent a single evolutionary lineage with subsequent introgression [42]. Hybridization and polyploidization are the major driving force in the diversity and evolution of the genus Elymus. Hybridizations between different ancestral diploid genera had formed the novel allopolyploid species [7, 17]. As a major mechanism of evolution and speciation, polyploidy Elymus species form diverse genotypes and phenotypes to adapt to the different ecological niches (especially in high altitude and high latitude regions) by inducing genomic replication, gene expression, and increasing the complexity of regulatory networks [43, 44]. Although these Elymus species have the different genome combinations, gene flow (Nm) existed between different species (16.07 for StH and StHY, 6.65 for StH and StY, and 11.72 for StHY and StY), suggesting that no strict reproductive barriers exist among the three genomes.
Conservation implications
Phenotypically and genetically diverse germplasm is a potentially valuable source for the improvement of the desired agronomic trait [13]. Wild germplasm could provide advantageous alleles like improved stress tolerance, forage quality, and higher yield for modifying currently used cultivars by hybridization and introgression. The collection and preservation of rich and specific germplasm resources of wild relative species will benefit the utilization of excellent traits and special resistance genes [1, 4, 5]. According to AMOVA analysis, larger genetic variation was found within Elymus species. This result was in agreement with previous genetic studies of Elymus species which found that the majority of variation was apportioned within populations or geographic regions [45]. Hence, a considerable amount of overall genetic variation of Elymus species could be captured when sampling a larger number of plants from Elymus population. Meanwhile, Mantel test indicated that a strong positive correlation between Nei’s genetic distance and geographic distance (r = 0.2086, p < 0.01) was found for Elymus accessions, which suggested more genetic diversity and variation could be captured in the wide range of geographical regions. Based on our data, accessions from Qinghai-Tibetan Plateau and Tianshan mountain had high level of genetic diversity. Therefore, these wild accessions with rich genetic diversity could be used as important genetic resources for future Elymus breeding programs.
Conclusions
In this study, we developed 87 polymorphic EST-SSR markers that showed good transferability across different Elymus species. Secondly, 30 EST-SSR markers were used to analyze the genetic diversity of 95 accessions of 17 Elymus species. Our results showed that hexaploid Elymus species with genomes StStHHYY had a relatively higher level of genetic diversity, while tetraploid Elymus species with genomes StStYY had low level of genetic diversity. The cluster analysis showed that all 95 accessions were clustered into three major clusters. The accessions were grouped mainly according to their genomic components and origins. In general, this study demonstrated that transcriptome sequencing is a fast and cost-effective approach to molecular marker development. These EST-SSR markers developed in this study are valuable tools for genetic diversity, evolutionary, and molecular breeding in E. sibiricus, and other Elymus species.
Methods
Development of EST-SSR markers derived from transcriptome of E. sibiricus
Our previous study had constructed cDNA libraries and sequenced abscission zone tissue samples of E. sibiricus based on next-generation sequencing (NGS) [32]. The obtained raw reads (NCBI SRA: SRX2617497) were preprocessed to filter adaptor sequences, low-quality sequences, and reads with quality less than Q30 using the FASTX toolkit. The Trinity program was employed to assemble the de novo transcriptome clean reads [46]. The assembled unigene sequences were directly identified in simple sequence repeat identification tool program (MicroSatellite), of which the parameters were set for mono-, di-, tri-, tetra-, penta-, and hexa- nucleotide motifs as a minimum repeat number of 12, 6, 5, 5, 4 and 4, respectively. The EST-SSRs were designed using Primer 3 (http://primer3.sourceforge.net) based on the MISA result.
Plant materials for genetic diversity analysis
A total of ninety-five accessions of seventeen Elymus species were collected, including thirty accessions with genomes StH (2n = 4× = 28), forty-eight accessions with genomes StHY (2n = 6× = 42) and seventeen accessions with genomes StY (2n = 4× = 28). A total of 480 individual plants were included. Materials were obtained from the U.S. Department of Agriculture Germplasm Resources Information Network (GRIN) and Lanzhou University. These accessions originated from their primary distribution areas in Asia with a broad latitudinal (29.2° to 54.8° N) and elevational (40 to 4460 m) range. Particularly, a total of 156 E. sibiricus (StH genome) individual plants were grouped into seven geographical groups with different geographic origin, including Eastern Qinghai-Tibetan Plateau (Geo-1), Mongolian Plateau (Geo-2), Lake Baikal (Geo-3), Western Altai Mountains (Geo-4), Xinjiang Tianshan (Geo-5), East coast of Russia (Geo-6) and Western Tien-shan (Geo-7) (Table 4 and Fig. 8). A total of 234 E. nutans (StHY genome) individual plants were grouped into seven geographical groups, including Northeastern Qinghai-Tibetan Plateau (Geo-8), Southeastern Qinghai-Tibetan Plateau (Geo-9), Southwestern Qinghai-Tibetan Plateau (Geo-10), Mongolian Plateau (Geo-11), Xinjiang Tianshan (Geo-12), Pakistan (Geo-13) and Russia (Geo-14).
Plant materials for verification of repeat motif types across different species
Seventeen individual plants which represented the three genomes (StH, StY and StHY) were used to validate the EST-SSR markers. These accessions included PI639859, PI619589, PI499585, PI564957, PI 636649, PI531575, PI655100, PI 410282, PI 564944, PI 632570, PI254866, PI564964, WEti4, WEcy3, WEda5, WEex1 and WEta2.
DNA extraction and genotyping
Seeds of all accessions were germinated in a greenhouse (25/15°Cday/night temperature) until 8 weeks old. Young leaf tissues of 480 individuals were collected for genomic DNA extraction (sodium dodecyl sulfate, SDS methods) [13]. Each accession was represented by 1 to 9 individuals, with an average of 5.1 (details in Table 4). The quantity and quality of DNA samples were determined using the NanoDrop ND1000 spectrophotometer (Thermo Scientific, Waltham, MA, USA) and agarose gel electrophoresis, then diluted to 25 ng/μL and stored at − 20 °C.
Two hundred EST-SSR primer pairs were randomly selected from 8871 potential SSRs and were synthesized by Shanghai Sangon Biological Engineering Technology (Shanghai, China). A total of 20 individual plants from 17 Elymus species were selected for primer screening. Each primer was amplified twice to check whether it produce clear and reproducible bands. Finally, EST-SSRs with high transferability, polymorphism and repeatability were used to genotype 480 individual plants. The PCR amplification and EST-SSR genotyping as well as eletrophoresis were carried out as described by Xie et al. [47].
Data analysis
The amplified bands were considered as present (1) and absent (0), and only clear and reproducible bands were considered. The expected heterozygosity (He), observed heterozygosity (Ho) and polymorphism information content (PIC) value were calculated as the previous methods [13, 48]. The expected heterozygosity formula is as follows: He = 1- ∑pi 2, where pi is frequency of the ith allele. The number of heterozygotes is determined by direct count method. PIC was calculated for each primer according to the formula: PIC = 1 – p2 – q2, where p is frequency of present band and q is frequency of absent band. Genetic diversity parameters including observed number of alleles (Na), effective number of alleles (Ne), Nei’s genetic diversity (H), Shannon’s information index (I) and the percentage of polymorphic loci (% P) were calculated by using POPGENE v 1.31 program (Edmonton, AB, Canada) [49]. A neighbor-joining (NJ) tree was displayed by using of MEGA v 5 software based on the operations supported in PowerMarker v 3.25, of which the probabilities for each node was assessed by bootstrap analysis using 1000 replicates [50, 51]. A principal coordinates analysis (PCoA) was constructed in GenAlEx v 6.5 [52]. Correlations between pairwise genetic distance and adjusted pairwise geographic distance were calculated by GenAlEx v 6.5 based on the mantel test with 9999 permutations. Person relation analysis was used to test the correlations between genetic parameter (Ne, effective number of alleles and H, Nei’s genetic diversity) and environmental factors (latitude and altitude). The analysis of molecular variance (AMOVA) was used to investigate the total genetic variation among genomes, within species and within populations using GenAlEx v 6.5. The program STRUCTURE v 2.3.4 was used to analyze the genetic structure of four hundred and eighty individuals using a Markov chain Monte Carlo (MCMC) algorithm. Assuming an admixture model sample and correlated allele frequencies, 20 independent runs were performed for all K values (ranged from 1 to 11), each with 10,000 MCMC interactions and 10,000 replications. The delta K method was employed to determine the optimal K value for all the data set [53].
Additional files
Figure S1. Length distribution of all unigenes. The x-axis represents the size of all unigenes, and the y-axis represents the number of all unigenes with a certain length. (JPG 594 kb)
Figure S2. Characteristics of the homology search of the unigene library of E. sibiricus against the Nr database, species distribution of top ten BLAST hits for each unigene with a cut-off of 1E-5. (JPG 147 kb)
Acknowledgements
This work was supported by Chinese National Basic Research Program (2014CB138704), the program for Changjiang Scholars and Innovative Research Team in University (IRT13019), Chinese National Natural Science Foundation (No. 31302023) and 111 program (B12002).
Funding
This study was financially supported by grants from the Chinese National Basic Research Program (973 Program) (No. 2014CB138704), the Chinese National Natural Science Foundation (No. 31302023).
Availability of data and materials
Raw sequencing data used for EST-SSR marker development are available in NCBI SRA: SRX2617497 (https://www.ncbi.nlm.nih.gov/biosample/6545378). Other datasets supporting the conclusions of this article are included within the article and its additional files.
Abbreviations
- AMOVA
Analysis of molecular variance
- DET
Differentially expressed transcript
- EST-SSR
Expressed sequence tag-derived simple sequence repeat markers
- H
Nei’s genetic diversity
- He
The expected heterozygosity
- Ho
observed heterozygosity
- I
Shannon’s information index
- Na
number of alleles
- Ne
effective number of alleles
- NGS
next-generation sequencing
- PCoA
Principal coordinates analysis
- PIC
polymorphism information content
Authors’ contributions
WX and YW conceived and designed the research, and WX also wrote the manuscript. YZ performed the research, analyzed the data, and wrote this manuscript. YZ, JZ, NW and JY analyzed the data. FN also wrote this manuscript. All authors read and approved the manuscript.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Zongyu Zhang, Email: zhangzy17@lzu.edu.cn.
Wengang Xie, Phone: +86-931-891-3014, Email: xiewg@lzu.edu.cn.
Yongqiang Zhao, Email: zhaoyq16@lzu.edu.cn.
Junchao Zhang, Email: zhangjch16@lzu.edu.cn.
Na Wang, Email: wangn17@lzu.edu.cn.
Fabrice Ntakirutimana, Email: olifabu@gmail.com.
Jiajun Yan, Email: yanjj510@yahoo.com.cn.
Yanrong Wang, Phone: +86-931-891-891-2496, Email: yrwang@lzu.edu.cn.
References
- 1.Lu B. Diversity and conservation of the Triticeae genetic resources. Chinese Biodiversity. 1995;3:63–68. [Google Scholar]
- 2.Dewey DR. The genomic system of classification as a guide to intergeneric hybridization with the perennial Triticeae. In: Gustafson JP, editor. Gene manipulation in plant improvement. New York: Columbia University Press; 1984. [Google Scholar]
- 3.Löve À. Conspectus of the Triticeae. Feddes Repert. 1984;95:425–521. [Google Scholar]
- 4.Zeng J, Cao W, Hucl P, Yang Y, Xue A, Chi D, Fedak G. Molecular cytogenetic analysis of wheat - Elymus repens introgression lines with resistance to Fusarium head blight. Genome. 2013;56:75–82. doi: 10.1139/gen-2012-0130. [DOI] [PubMed] [Google Scholar]
- 5.Cainong JC, Bockus WW, Feng Y, Chen P, Qi L, Sehgal SK, Danilova TV, Koo D-H, Friebe B, Gill BS. Chromosome engineering, mapping, and transferring of resistance to Fusarium head blight disease from Elymus tsukushiensis into wheat. Theor Appl Genet. 2015;128:1019–1027. doi: 10.1007/s00122-015-2485-1. [DOI] [PubMed] [Google Scholar]
- 6.Buerkle CA, Morris RJ, Asmussen MA, Rieseberg LH. The likelihood of homoploid hybrid speciation. Heredity. 2000;84:441–451. doi: 10.1046/j.1365-2540.2000.00680.x. [DOI] [PubMed] [Google Scholar]
- 7.Lu BR, Salomon B. Differentiation of the StY genomes in Elymus species as referred by meiotic paring in interspecific hybrids and its evolutionary significance. Biodivers Sci. 2004;12:213–226. [Google Scholar]
- 8.Wang RRC, Jensen KB. Chapter 3: Wheatgrasses and Wildryes. In: Ram J. Singh (ed.) Genetic Resources, Chromosome Engineering, and Crop Improvement. Vol. 5 Forage Crops, CRC Press, Boca Raton, FL. 2009;Pp. 41–79. Available from: https://core.ac.uk/download/pdf/12107989.pdf
- 9.Mason-Gamer RJ, Burns MM, Naum M. Reticulate evolutionary history of a complex group of grasses: phylogeny of Elymus StStHH allotetraploids based on three nuclear genes. PLoS One. 2010;5:e10989. doi: 10.1371/journal.pone.0010989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fan X, Sha LN, Wang XL, Zhang HQ, Kang HY, Wang Y, Zhou YH. Phylogeny and molecular evolution of the Acc1 gene within the StH genome species in Triticeae (Poaceae) Gene. 2013;529:57–64. doi: 10.1016/j.gene.2013.07.017. [DOI] [PubMed] [Google Scholar]
- 11.Lu BR. The genus Elymus L. in Asia. Taxonomy and biosystematics with special reference to genomic relationships. In: Wang RRC, Jensen KB, Jaussi C, eds. Proceedings of the 2nd International Triticeae Symposium, Logan, UT, USA. Logan, UT, USA: The Utah State University Press; 1994. Available from: https://www.researchgate.net/publication/305386590_The_genus_Elymus_in_Asia_Taxonomy_and_biosystematics_with_special_reference_to_genomic_relationships
- 12.Liu Q, Ge S, Tang H, Zhang X, Zhu G, Lu B. Phylogenetic relationships in Elymus (Poaceae: Triticeae) based on the nuclear ribosomal internal transcribed spacer and chloroplast trnL-F sequences. New Phytol. 2006;170:411–420. doi: 10.1111/j.1469-8137.2006.01665.x. [DOI] [PubMed] [Google Scholar]
- 13.Zhang Z, Xie W, Zhang J, Zhao X, Zhao Y, Wang Y. Phenotype- and SSR-based estimates of genetic variation between and within two important Elymus species in western and northern China. Genes. 2018;9:147. doi: 10.3390/genes9030147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang RRC. Genome relationships in the perennial Triticeae based on diploid hybrids and beyond. Hereditas. 1992;116:133–136. doi: 10.1111/j.1601-5223.1992.tb00812.x. [DOI] [Google Scholar]
- 15.Sun G. Genetic diversity of rbcL gene in Elymus trachycaulus complex and their phylogenetic relationships to several Triticeae species. Genet Resour Crop Evol. 2007;54:1737–1746. doi: 10.1007/s10722-006-9183-4. [DOI] [Google Scholar]
- 16.Yang CR, Baum BR, Chen WH, Zhang HQ, Liu XY, Fan X, Sha LN, Kang HY, Wang Y, Zhou YH. Genomic constitution and taxonomy of the Chinese hexaploids Elymus cylindricus and E. breviaristatus (Poaceae: Triticeae) Bot J Linn Soc. 2016;182:650–657. doi: 10.1111/boj.12469. [DOI] [Google Scholar]
- 17.Sun D, Sun G. Untangling nucleotide diversity and evolution of the H genome in polyploid Hordeum and Elymus species based on the single copy of nuclear gene DMC1. PLoS One. 2012;7:e50369. doi: 10.1371/journal.pone.0050369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fan X, Sha LN, Dong ZZ, Zhang HQ, Kang HY, Wang Y, Wang XL, Zhang L, Ding CB, Yang RW, Zheng YL, Zhou YH. Phylogenetic relationships and Y genome origin in Elymus L. sensu lato (Triticeae; Poaceae) based on single-copy nuclear Acc1 and Pgk1 gene sequences. Mol Phylogenet Evol. 2013;69:919–928. doi: 10.1016/j.ympev.2013.06.012. [DOI] [PubMed] [Google Scholar]
- 19.Gao G, Deng JB, Gou XM, Wang Q, Ding CB, Zhang L, Zhou YH, Yang RW. Phylogenetic relationships among Elymus and related diploid genera (Triticeae: Poaceae) based on nuclear rDNA ITS sequences. Biologia. 2015;70:183–189. Available from: https://www.degruyter.com/view/j/biolog.2015.70.issue-2/biolog-2015-0019/biolog-2015-0019.xml?rskey=tASnWI&result=10
- 20.Bushman BS, Larson SR, Mott IW, Cliften PF, Wang RRC, Chatterton NJ, Hernandez AG, Ali S, Kim RW, Thimmapuram J, Gong G, Liu L, Mikel MA. Development and annotation of perennial Triticeae ESTs and SSR markers. Genome. 2008;51:779–788. doi: 10.1139/G08-062. [DOI] [PubMed] [Google Scholar]
- 21.Wang RRC, Larson SR, Jensen KB. Differential transferability of EST-SSR primers developed from diploid species Pseudoroegneria spicata, Thinopyrum bessarabicum, and Th. elongatum. Genome. 2017;60(6):530–536. doi: 10.1139/gen-2016-0157. [DOI] [PubMed] [Google Scholar]
- 22.Rouf Mian MA, Saha MC, Hopkins AA, Wang ZY. Use of tall fescue EST-SSR markers in phylogenetic analysis of cool-season forage grasses. Genome. 2005;48:637–647. doi: 10.1139/g05-029. [DOI] [PubMed] [Google Scholar]
- 23.Karan M, Evans DS, Reilly D, Schulte K, Wright C, Innes D, Holton TA, Nikles DG, Dickinson GR. Rapid microsatellite marker development for African mahogany (Khaya senegalensis, Meliaceae) using next-generation sequencing and assessment of its intra-specific genetic diversity. Mol Ecol Resour. 2012;12:344–353. doi: 10.1111/j.1755-0998.2011.03080.x. [DOI] [PubMed] [Google Scholar]
- 24.Huang L, Huang X, Yan H, Yin G, Zhang X, Tian Y, Zhang Y, Jiang X, Yan Y, Ma X, Peng Y, Zhou J, Nie G. Constructing DNA fingerprinting of Hemarthria cultivars using EST-SSR and SCoT markers. Genet Resour Crop Evol. 2014;61:1047–1055. doi: 10.1007/s10722-014-0107-4. [DOI] [Google Scholar]
- 25.Huang LK, Yan HD, Zhao XX, Zhang XQ, Wang J, Frazier T, Yin G, Huang X, Yan DF, Zang WJ, Ma X, Peng Y, Yan YH, Liu W. Identifying differentially expressed genes under heat stress and developing molecular markers in orchardgrass (Dactylis glomerata L.) through transcriptome analysis. Mol Ecol Resour. 2015;15:1497–1509. doi: 10.1111/1755-0998.12418. [DOI] [PubMed] [Google Scholar]
- 26.Zhao X, Huang L, Zhang X, Wang J, Yan D, Li J, Tang L, Li X, Shi T. Construction of high-density genetic linkage map and identification of flowering-time QTLs in orchardgrass using SSRs and SLAF-seq. Sci Rep. 2016;6:29345. doi: 10.1038/srep29345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mott IW, Larson SR, Jones TA, Robins JG, Jensen KB, Peel MD. A molecular genetic linkage map identifying the St and H subgenomes of Elymus (Poaceae: Triticeae) wheatgrass. Genome. 2011;54:819–828. doi: 10.1139/g11-045. [DOI] [PubMed] [Google Scholar]
- 28.Mott IW, Larson SR, Bushman BS. Simple sequence repeat (SSR) markers for Elymus, Pseudoroegneria and Pascopyrum species (Triticeae: Gramineae) Plant Genet Resour - C. 2011;9(4):489–494. doi: 10.1017/S1479262111000694. [DOI] [Google Scholar]
- 29.Davey JW, Hohenlohe PA, Etter PD, Boone JQ, Catchen JM, Blaxter ML. Genome-wide genetic marker discovery and genotyping using next-generation sequencing. Nat Rev Genet. 2011;12:499–510. doi: 10.1038/nrg3012. [DOI] [PubMed] [Google Scholar]
- 30.Chen S, Huang X, Yan X, Liang Y, Wang Y, Li X, Peng X, Ma X, Zhang L, Cai Y, Ma T, Cheng L, Qi D, Zheng H, Yang X, Li X, Liu G. Transcriptome analysis in sheepgrass (Leymus chinensis): a dominant perennial grass of the Eurasian steppe. PLoS One. 2013;8:e67974. doi: 10.1371/journal.pone.0067974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.You Y, Liu D, Liu H, Zheng X, Diao Y, Huang X, Hu Z. Development and characterisation of EST-SSR markers by transcriptome sequencing in taro (Colocasia esculenta (L.) Schoot) Mol Breeding. 2015;35:134. doi: 10.1007/s11032-015-0307-4. [DOI] [Google Scholar]
- 32.Xie W, Zhang J, Zhao X, Zhang Z, Wang Y. Transcriptome profiling of Elymus sibiricus, an important forage grass in Qinghai-Tibet plateau, reveals novel insights into candidate genes that potentially connected to seed shattering. BMC Plant Biol. 2017;17:78. doi: 10.1186/s12870-017-1026-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu J, Cai C, Cheng F, Cui H, Zhou H. Characterisation and development of EST-SSR markers in tree peony using transcriptome sequences. Mol Breeding. 2014;34:1853–1866. doi: 10.1007/s11032-014-0144-x. [DOI] [Google Scholar]
- 34.Victoria FC, da Maia LC, de Oliveira AC. In silico comparative analysis of SSR markers in plants. BMC Plant Biol. 2011;11:15. doi: 10.1186/1471-2229-11-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu Z, Chen T, Ma L, Zhao Z, Zhao PX, Nan Z, Wang Y. Global transcriptome sequencing using the Illumina platform and the development of EST-SSR markers in autotetraploid alfalfa. PLoS One. 2013;8:e83549. doi: 10.1371/journal.pone.0083549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yan Z, Wu F, Luo K, Zhao Y, Yan Q, Zhang Y, Wang Y, Zhang J. Cross-species transferability of EST-SSR markers developed from the transcriptome of Melilotus and their application to population genetics research. Sci Rep. 2017;7:17959. doi: 10.1038/s41598-017-18049-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.La Rota M, Kantety RV, Yu JK, Sorrells ME. Nonrandom distribution and frequencies of genomic and EST-derived microsatellite markers in rice, wheat, and barley. BMC Genomics. 2005;6:23. doi: 10.1186/1471-2164-6-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kantety RV, La Rota M, Matthews DE, Sorrells ME. Data mining for simple sequence repeats in expressed sequence tags from barley, maize, rice, sorghum and wheat. Plant Mol Biol. 2002;48:501–510. doi: 10.1023/A:1014875206165. [DOI] [PubMed] [Google Scholar]
- 39.Pan L, Huang T, Yang Z, Tang L, Cheng Y, Wang J, Ma X, Zhang X. EST-SSR marker characterization based on RNA-sequencing of Lolium multiflorum and cross transferability to related species. Mol Breeding. 2018;38:80. doi: 10.1007/s11032-018-0775-4. [DOI] [Google Scholar]
- 40.Dong ZZ, Fan X, Sha LN, Wang Y, Zeng J, Kang HY, Zhang HQ, Wang XL, Zhang L, Ding CB, Yang RW, Zhou YH. Phylogeny and differentiation of the St genome in Elymus L. sensu lato (Triticeae; Poaceae) based on one nuclear DNA and two chloroplast genes. BMC Plant Biol. 2015;15:179. doi: 10.1186/s12870-015-0517-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dong Zhen-Zhen, Fan Xing, Sha Li-Na, Zeng Jian, Wang Yi, Chen Qian, Kang Hou-Yang, Zhang Hai-Qin, Zhou Yong-Hong. Phylogeny and molecular evolution of the rbcL gene of St genome in Elymus sensu lato (Poaceae: Triticeae) Biochemical Systematics and Ecology. 2013;50:322–330. doi: 10.1016/j.bse.2013.05.005. [DOI] [Google Scholar]
- 42.Mason-Gamer RJ. Phylogeny of a genomically diverse group of Elymus (Poaceae) allopolyploids reveals multiple levels of reticulation. PLoS One. 2013;8:e78449. doi: 10.1371/journal.pone.0078449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Otto SP. The evolutionary consequences of polyploidy. Cell. 2007;131:452–462. doi: 10.1016/j.cell.2007.10.022. [DOI] [PubMed] [Google Scholar]
- 44.Blanc G, Wolfe KH. Functional divergence of duplicated genes formed by polyploidy during Arabidopsis evolution. Plant Cell. 2004;16:1679–1691. doi: 10.1105/tpc.021410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ma X, Zhang XQ, Zhou YH, Bai SQ, Liu W. Assessing genetic diversity of Elymus sibiricus (Poaceae: Triticeae) population from Qinghai-Tibet plateau by ISSR markers. Biochem Syst Ecol. 2008;36:514–522. doi: 10.1016/j.bse.2008.03.003. [DOI] [Google Scholar]
- 46.Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, Chen Z, Mauceli E, Hacohen N, Gnirke A, Rhind N, di Palma F, Birren BW, Nusbaum C, Lindblad-Toh K, Friedman N, Regev A. Full-length transcriptome assembly from RNA-seq data without a reference genome. Nat Biotechnol. 2011;29:644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xie W, Zhao X, Zhang J, Wang Y, Liu W. Assessment of genetic diversity of Siberian wild rye (Elymus sibiricus L.) germplasms with variation of seed shattering and implication for future genetic improvement. Biochem Syst Ecol. 2015;58:211–218. doi: 10.1016/j.bse.2014.12.006. [DOI] [Google Scholar]
- 48.Weir BS. Genetic data analysis. Methods for discrete population genetic data. Massachusetts: Sinauer Associates, Inc. Sunderland; 1996. [Google Scholar]
- 49.Yeh FC, Boyle TJB. Population genetic analysis of co-dominant and dominant markers and quantitative traits. Belg J Bot. 1997;129:157. [Google Scholar]
- 50.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu K, Muse SV. PowerMarker: an integrated analysis environment for genetic marker analysis. Bioinf Appl Notes. 2005;21:2128–2129. doi: 10.1093/bioinformatics/bti282. [DOI] [PubMed] [Google Scholar]
- 52.Peakall R, Smouse PE. GenAlEx 6.5: genetic analysis in excel. Population genetic software for teaching and research - an update. Bioinformatics. 2012;28:2537–2539. doi: 10.1093/bioinformatics/bts460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Evanno G, Regnaut S, Goudet J. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol. 2005;14:2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Length distribution of all unigenes. The x-axis represents the size of all unigenes, and the y-axis represents the number of all unigenes with a certain length. (JPG 594 kb)
Figure S2. Characteristics of the homology search of the unigene library of E. sibiricus against the Nr database, species distribution of top ten BLAST hits for each unigene with a cut-off of 1E-5. (JPG 147 kb)
Data Availability Statement
Raw sequencing data used for EST-SSR marker development are available in NCBI SRA: SRX2617497 (https://www.ncbi.nlm.nih.gov/biosample/6545378). Other datasets supporting the conclusions of this article are included within the article and its additional files.