Abstract
Background
Previous study demonstrated that extracellular ATP could promote cell migration and invasion in multiple human cancers. Till now, the pro-invasive mechanisms of ATP and P2RX6, a preferred receptor for ATP, are still poorly studied in RCC.
Methods
Bioinformatics analysis was performed to identify the differentially expressed genes during RCC different stages. Tissue microarray, IHC staining and survival analysis was respectively used to evaluate potential clinical function. In vitro and in vivo assays were performed to explore the P2RX6 biological effects in RCC progression.
Results
We found that ATP might increase RCC cells migration and invasion through P2RX6. Mechanism dissection revealed that ATP-P2RX6 might modulate the Ca2+-mediated p-ERK1/2/MMP9 signaling to increase the RCC cells migration and invasion. Furthermore, METTL14 implicated m6A modification in RCC and down-regulated P2RX6 protein translation. In addition, human clinical survey also indicated the positive correlation of this newly identified signaling in RCC progression and prognosis.
Conclusions
Our findings revealed that the newly identified ATP-P2RX6-Ca2+-p-ERK1/2-MMP9 signaling facilitates RCC cell invasion and metastasis. Targeting this novel signaling pathway with small molecules might help us to develop a new approach to better suppress RCC progression.
Electronic supplementary material
The online version of this article (10.1186/s13046-019-1223-y) contains supplementary material, which is available to authorized users.
Keywords: Renal cell carcinoma, P2RX6, m6A, Ca2+, p-ERK1/2/MMP9
Background
Renal cell carcinoma (RCC) is the most lethal of all urological malignancies, accounting for about 3.8% of all new malignancies, with about 65,340 new cases and 14,970 deaths estimated for 2018 in the United States alone [1, 2] . Situation in other countries is also roughly the same [3]. However, RCC is basically radiotherapy and chemotherapy resistant and best treated by resection [4–6]. Although tyrosine kinase inhibitor (TKI)-based antiangiogenic therapy is the standard treatment for metastatic RCC (mRCC) and has improved patients’ prognosis, but the effect is still limited due to drug resistance [7, 8]. Thus, it is urgent to elucidate the underlying mechanisms of RCC earlier metastasis and find out more clinical indicator and therapeutic target for RCC metastasis.
Adenosine 5′-triphosphate (ATP) is the main form of intracellular energy for all types of cell and played different roles in multiple cancers development [9, 10]. It is also released extracellular, both in physiological and pathological conditions, where it mediates various biological functions via activation of P2 receptors [11–13]. The P2 receptors have two subfamilies in mammalian cells, P2X and P2Y. P2X family of ligand-gated ion channel receptors have seven members, P2RX1–7, which all constitute non-selective cation channels proposed to be functional under a homo- or heterotrimeric association [11, 14]. P2Y family of G protein-coupled receptors, including P2RY1, 2, 4, 6, 11, 12, 13, 14 [15, 16], which can modulate the adenylate cyclase-cyclic adenosine 3′, 5′-monophosphate pathway or stimulate the phospholipase C (PLC)-Ca2+ signaling pathway [17].
Previous reports revealed that prinergic receptor subtypes were involved in the development of multiple tumors, such as prostate, bladder, breast et al. [9, 10, 18]. It is demonstrated that P2RY1 and P2RY2 receptors were expressed and involved in prostate cancer cell proliferation; P2RX5 receptors were involved in cell differentiation and P2RX7 receptors were involved in cell death. In human squamous cell carcinoma, P2RY2, P2RX5 and P2RX7 seem to correlate with cell proliferation, differentiation and cell death, respectively [19]. Here, we found that P2RX6, a preferred receptor for ATP, contributed to the invasion and metastasis of RCC cells. However, the potential signaling pathway, especially in RCC metastasis, is still not clear.
N6-methyladenosine (m6A), a predominant internal modification of RNA in higher eukaryotes, aroused people’s attention. This mechanism involves two important catalytic proteins, demethylase and methyltransferase [20]. Recently, numbers of studied showed that m6A modification played a crucial role in multiple tumor development [21–23], such as nuclear RNA export, transcription splicing, cell fate determination and protein translation control [24–26]. Previous study revealed that methyltransferase-like 14 (METTL14) could suppress the metastatic potential of hepatocellular carcinoma by modulating m6A dependent primary microRNA processing [22]. In addition, hypoxia could induce the breast cancer stem cell phenotype by HIF-dependent and ALKBH5-mediated m6A of NANOG mRNA [23]. However, the status of m6A modification and the underlying regulatory mechanisms in RCC remained incompletely understood.
Materials and methods
Bioinformatics analysis
We obtained the KIRC patient data from the TCGA database collection (https://xenabrowser.net/datapages/). The database is generated by the TCGA Research Network: http://cancergenome.nih.gov/. GO pathway analysis was obtained from http://geneontology.org/. Gene expression and survival data were got from http://ualcan.path.uab.edu/. The demographic information such as gender, age, BMI and tumor clinical characteristics such as gene expression, OS and TNM status were collected. Clinical TNM staging were based on the UICC 8th edition [27] and we excluded patients without TNM stage information from the analysis.
Cell lines and chemicals
The OS-RC-2, 786-O, and HEK-293 cells were obtained from Cell Bank of the Chinese Academy of Sciences (Shanghai, China). SN12-PM6 cell line was kindly provided by Dr. Qingbo Huang from the Department of Urology, Chinese PLA General Hospital, Beijing, China. SW839 cell line was kindly provided by Dr. Chawnshang Chang from George Whipple Lab for Cancer Research, University of Rochester Medical Center, Rochester, NY, 14646 USA. All cell lines were expanded to passage 3, stored in aliquots in liquid nitrogen, and were used for fewer than 4 months after receipt or resuscitation from cryopreservation. Dulbecco’s Modified Eagle’s Medium (DMEM, Gibco, USA) was used to culture cells above. All cells described above were cultured at 37 °C in 5% CO2 and tested for mycoplasma every 6 months using the Universal Mycoplasma Detection Kit (ATCC, USA).
Lentivirus packaging
The standard calcium chloride transfection method was performed. The pMD2G envelope plasmid and psAX2 packaging plasmid, with interest gene knockdown/ overexpression plasmid were transfected into HEK293T cells for 48 h to generate the lentivirus supernatant, oligo sequences were listed in Additional file 1: Table S1. The lentivirus supernatants were harvest through a 0.45 μm nitrocellulose filter and used immediately or frozen in − 80 °C for later use.
Wound healing migration assay
Cells were seeded into six-well plates at a density that after 24 h of growth, they reached approximately 60% confluence. Gently and slowly scrape the single layer with a new pipette tip across the center of the well. Then, washed wells twice with PBS to remove the detached cells and replenished with fresh medium. The cells were grown for an additional 24 h. Images were captured of the monolayer using a microscope. The gap distance was quantitatively evaluated using ImageJ v1.51 (Wayne Rasband, USA). The relative migration was determined by setting the number of average migration distance in control group as one. The experiments were tested in triplicate.
Transwell invasion assay
Cell invasion capacity was assessed by a matrigel invasion assay using matrigel coating 8.0 μm filter membranes. Cells (5 × 104) in 150 μL of serum-free medium were plated onto each filter, with 600 μL of 10% serum-containing medium placed in the lower chamber and then incubated for 12 h in cell culture incubator at 37 °C in a 5% CO2 atmosphere. After 12 h, removed the cells on the upper surface of the filters with cotton swabs. Filters were fixed with 4% paraformaldehyde in both sides and washed with PBS, and stained with crystal violet. Cells that had invaded to the lower surface of the filter were counted with Image-pro Plus v6.0 (Media Cybernetics, USA) in 5 randomly selected fields. The relative invasion was determined by setting the number of invading cells in control group as one. The experiments were tested in triplicate.
RNA extraction and quantitative real-time PCR analysis (qRT-PCR)
Trizol reagent (Invitrogen, USA) was used to extract the total RNA from cells or tissues according to the manufacturer’s protocol. PrimeScript RT reagent Kit (Takara, Japan) was used to synthesize cDNAs. RT-qPCR was performed with KAKA SYBR FAST qPCR Kit (Kapa Biosystems, USA) using a 7900HT Fast Real-Time PCR System (Applied Biosystems, Japan). All primers were listed in Additional file 1: Table S1. The expression level was normalized to endogenous small nuclear RNA U6 or GAPDH. The 2−ΔΔCt method was used to analyze the expression level relative to the endogenous control.
Western immunoblotting analysis (WB)
Cells were lysed on ice using RIPA buffer plus phosphatase inhibitors and protease inhibitors. For western blot analysis, 25 μg of protein extracts were loaded to 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis gels and transferred to nitrocellulose membranes. Primary antibody was used to incubate the membranes at 4 °C overnight and secondary antibody for 1 h with room temperature. GAPDH or Actin expression were used as loading control. The antibodies used were listed in Additional file 1: Table S1.
Immunofluorescence assay (IF)
Cells were grown on coverslips and fixed in 4% paraformaldehyde at room temperature for 10 min. After PBS washing, the cells were blocked with 3% BSA at 37 °C for 30 min and incubated at 4 °C with anti-p-ERK1/2 or MMP9 overnight, and then probed with a tetramethylrhodamine isothiocyanate (TRITC)-conjugated secondary antibody (Sigma, USA) at 37 °C for 2 h. Subsequently, cells were stained with DAPI and observed under a fluorescence microscope.
Measurement of intracellular calcium ion concentration
We measured the calcium ion ((Ca2+)i) concentration according to the method in the published papers [28, 29]. Briefly, cells were stained with 5 μM Fluo-3 AM and washed with physiological solution (125 mM NaCl, 5 mM KCl, 1 mM MgCl2, 10 mM HEPES, 5 mM glucose, and 1 mM CaCl2). Then, cells were treated with ATP and fluorescent images were scanned every 5 s using a confocal microscope (IX70 Fluoview, Olympus, Japan; excitation wavelength 488 nm, emission wavelength 530 nm). The changes in (Ca2+)i were calculated as follows: Change in (Ca2+)i = (Fmax-F0) / F0. F, the fluorescence intensity; F0, the basal fluorescence intensity before treatment; Fmax, the maximum level of fluorescence intensity.
m6A quantification assay
The m6A content in the total RNAs were measured with the m6A RNA methylation quantification kit (ab185912, Abcam, UK). Briefly, 200 ng RNAs were coated on assay wells. According to the manufacturer’s instructions, detection antibody solution and capture antibody solution were then added to assay wells separately in a suitable diluted concentration. Reading the absorbance at 450 nm and quantifying the m6A levels. Statistical analysis were performed based on the standard curve.
Mouse tail vein injection xenograft models
For the in vivo metastasis assays, luciferase labeled OS-RC-2 cells stably expressing OE-P2RX6 or pWPI-vector were injected into the tail vein of 5 weeks old BALB/c nude mice (Sipper-BK laboratory animal Company, Shanghai, China). All mice were maintained in an SPF level environment and were manipulated according to protocols approved by the Institutional Animal Care and Use Committee of the Shanghai Tenth People’s hospital. Each tumor cell sub-line was injected into ten mice (2 X 106 per mouse). IVIS Lumina imaging system (Calipers, Hopkinton, USA) was used to observe tumor metastasis. After 8 weeks, sacrificed the mice, harvested the tissues and fixed in 10% neutral PB-buffered formalin (pH 7.4). The fixed samples were then embedded in paraffin. The sections were stained with H&E and antibody for analyzing the presence of metastases.
Immunohistochemical staining (IHC)
IHC was performed on the samples from the human RCC tissues and mouse xenografted tumors, as described previously [30]. Briefly, the samples were fixed in 4% neutral buffered paraformaldehyde and embedded in paraffin and cut into 5 μm slices. After deparaffnization, hydration, and antigen retrieval, these sections were incubated with corresponding primary antibodies, incubated with biotinylated secondary antibodies (Vector Laboratories, Burlingame, CA, USA) and then visualized by VECTASTAIN ABC peroxidase system and 3, 3′-diaminobenzidine (DAB) kit (Vector Laboratories). The slides were scored by two experienced pathologist without the knowledge of patient outcome. The expression was assessed semi-quantitatively as follows: negative (−) < 5%, 5–25% (+, weak positive), 25–50% (++, moderate positive) and > 50% (+++, strong positive). Negative and weak positive expressions were defined as low expression, while positive and strong positive expressions were defined as high expression. Image-pro Plus v6.0 (Media Cybernetics, USA) was used to quantify the integral optical density (ISO) in 5 randomly selected fields.
Clinical samples
A total of 17 RCC samples and paired para-tumor tissues were obtained for patients who underwent partial or radical nephrectomy in Department of Urology, Shanghai Tenth People’s Hospital, Tongji University (Shanghai, China). The 238 specimens which were included in tissue microarray, Kaplan-Meier survival analysis and Cox regression analysis were obtained from Department of Urology database, these samples had corresponding at least 5 years follow-up information. The fresh tissues were frozen in liquid nitrogen to protect the protein and RNA away from degradation. The use of human tissues was approved by the ethics committee of Shanghai Tenth People’s Hospital.
Statistical analysis
All data were analyzed using the Graphpad 7.0 (GraphPad Software, USA) and SPSS 23 (IBM, USA). Results are expressed as Means ± S.D. from at least 3 independent experiments. Student’s T Test was used for comparing two groups data and one-way ANOVA followed by individual comparisons with Dunnett’s test was use for comparing more than two groups data. All data was considered significantly when P < 0.05.
Results
P2RX6 is highly expressed and associated with poor prognosis of RCC through TCGA database
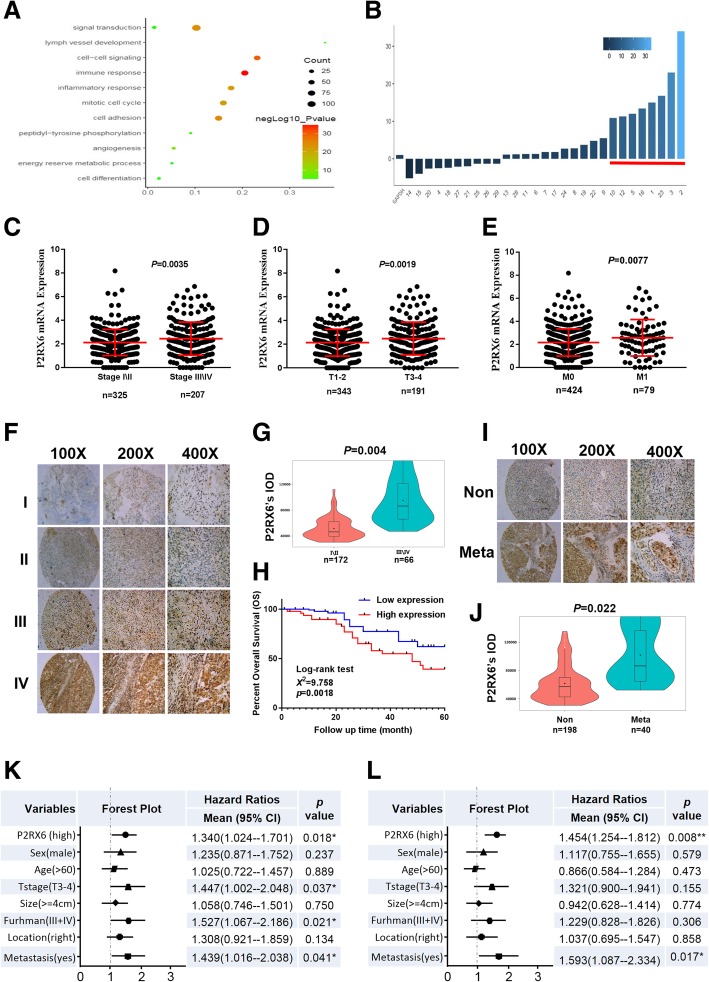
The candidates selecting process was described as Additional file 8: Figure S1 A. We downloaded 534 samples clinical information from TCGA database (Additional file 2: Table S2) and got 628 differentially expressed genes (DEGs) with the screening criteria G4/G1 > 3 and P < 0.001. Validations of 44 DEGs were got by intersected with GO pathway analysis (Fig. 1a) top six signaling pathway. Meanwhile, 29 DEGs were consistent with their survival outcome. Furthermore, we used qRT-PCR in 10 paired clinical renal tumor and para-tumors samples and revealed that 8 DEGs increased in renal tumor samples significantly (Fig. 1b). Specific information in the screening process and GO analysis result can be seen in Additional file 3: Table S3. Among the remaining 8 DEGs (ADRA2A, P2RX6, XCL2, XCL1, CCL7, TRIM54, TNFRSF18 and SPOCK1), the expression of ADRA2A, the top one, in KIRC based on individual cancer is lower than the normal tissues (Additional file 8: Figure S1 B), but patients with lower expressed ADRA2A have a better overall survival (OS) (Additional file 8: Fig. S1 C). On the other hand, P2RX6 had the dramatically differential expression between normal and tumor tissues in our clinical samples (Fig. 1b). Meanwhile, TCGA clinicopathologic correlation analysis suggested that P2RX6 expression associated with RCC pathological stage, pathological grade, metastasis (**P = 0.0035, 0.0019, 0.0077, respectively) (Fig. 1c-e). In addition, P2RX6 mRNA expression correlated with RCC patients OS, indicating higher expression of P2RX6 associated with RCC poor prognosis (**P = 0.00175) (Addtional file 8: Figure S1 D). Meanwhile, subsequent loss-of-functional experiments also confirmed that P2RX6 might be the key candidate what we were looking for.
Fig. 1.
P2RX6 is highly expressed and associated with RCC poor prognosis. a 628 DEGs’ Go pathway enrichment analysis. b qRT-PCR assay validation of 29 DEGs mRNA expression in 10 paired clinical tumor compared with para-tumor samples. c TCGA database analysis revealed P2RX6 expression is higher in stage III and stage IV (n = 207) compared with stage I and stage II (n = 325) (**P = 0.0035). d TCGA database analysis revealed P2RX6 expression is higher in T3 and T4 (n = 191) compared with T1 and T2 (n = 343) (**P = 0.0019). e TCGA database analysis revealed P2RX6 expression is higher in M1 (n = 79) compared with M0 (n = 424) (**P = 0.0077). (F-L) Human clinical studies for P2RX6 expression in RCC patients. f Human clinical samples analysis revealed P2RX6 expression is higher in higher RCC pathological stage samples. g Violin plot quantitative analysis for Fig. 1f. h Kaplan-Meier analysis for different P2RX6 protein expression in RCC patients (**P = 0.0018). i Representative IHC image of P2RX6 expression in Non and Meta group tissue samples. j Violin plot quantitative analysis for Fig. 1i. k Univariate Cox regression analysis and forest map. l Multivariate cox regression analyses and forest map. Non means non-metastatic group, and Meta means metastatic group. * P < 0.05, ** P < 0.01
Together, results from Fig. 1a-e and Additional file 8: Figure S1 A-D suggested that P2RX6 might be involved in RCC progression and function as a potential biomarker in prognostic value.
Human clinical studies for P2RX6 expression in RCC patients
Next, to link the analysis result from TCGA RCC database, we analyzed IHC staining results from 238 human RCC samples which revealed the median number of P2RX6 expression was IOD 61725.5. Clinicopathologic correlation analysis proved that P2RX6 expression associated with RCC pathological stage (**P = 0.004) (Fig. 1f-g, Additional file 4: Table S4). In addition, P2RX6 protein expression correlated with RCC patients OS, indicating higher expression of P2RX6 associated with RCC poor prognosis (**P = 0.0018) (Fig. 1h). Besides, P2RX6 was also associated with RCC metastasis (*P = 0.022) (Fig. 1i-j, Additional file 4: Table S4).
In addition, we explored the correlation between P2RX6 and RCC clinical pathological characteristics. Correlation regression analysis of 238 samples demonstrated that high expression of P2RX6 was clearly associated with T stage (**P = 0.004), Fuhrman grade (*P = 0.039) and metastasis (*P = 0.022) (Additional file 5: Table S5). Meanwhile, we implemented univariate and multivariate cox regression models to analyze the correlation of the multiple variables with OS of 238 RCC patients. Univariate analysis demonstrated that a higher level of P2RX6 (hazard ratio, HR = 1.340; 95% confidence interval, CI = 1.024--1.701; *P = 0.018), a higher T stage (HR = 1.447; 95% CI = 1.002--2.048; *P = 0.037), higher Furhman grade (HR = 1.527; 95% CI = 1.067--2.186; *P = 0.021) and more metastasis (HR = 1.439; 95% CI = 1.016--2.038; *P = 0.041) were potently correlated with OS (Fig. 1k). Multivariate analysis substantiated that a higher P2RX6 expression level (HR = 1.454; 95% CI = 1.254--1.812; **P = 0.008) and more metastasis (HR = 1.593; 95% CI = 1.087--2.334; *P = 0.017) were markedly associated with OS (Fig. 1l).
Taken together, these results indicated that RCC patients’ samples are in agreement with online database suggesting P2RX6 was higher expressed in RCC tissues than normal renal tissues and correlated with poor prognosis.
P2RX6 promotes RCC cell migration and invasion in vitro
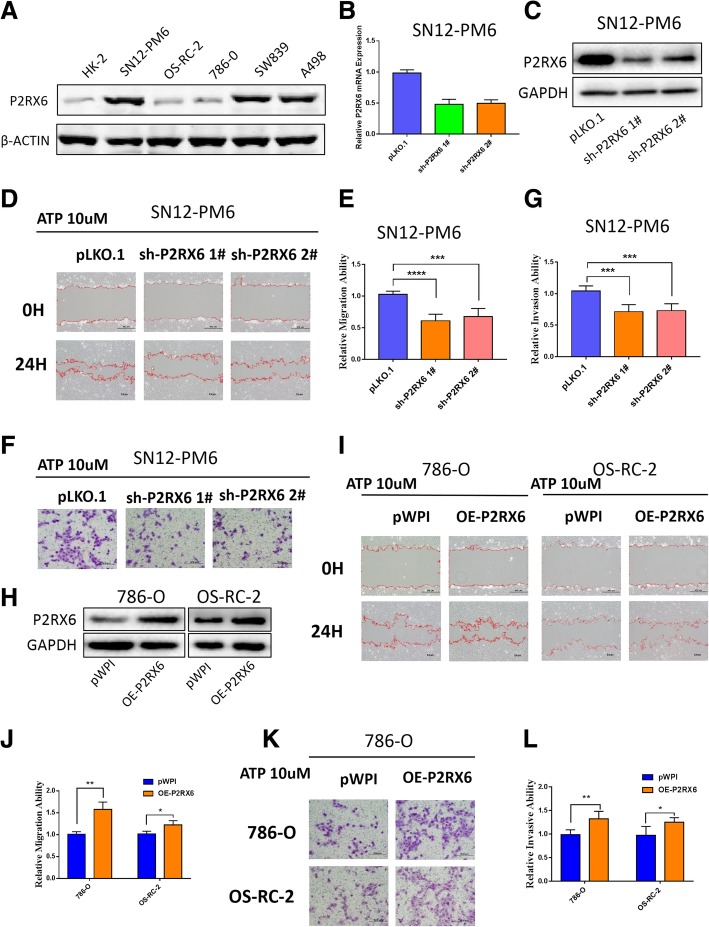
We first tested P2RX6 protein expression in multiple RCC cell lines (SN12-PM6, OS-RC-2, 786-O, SW839, A498) vs. normal renal proximal tubule epithelial cell line HK-2, and found its expression was elevated in most RCC cell lines (Fig. 2a). Previous study demonstrated that ATP induced breast cancer cells migration and invasion. To investigate ATP effect on RCC, we treated OS-RC-2 cells with different concentration of ATP. Results from “wound-healing” migration assay revealed that 10uM ATP increased a substantial migration of the cells into the scratched area (Additional file 9: Figure S2 A). Similar results were also obtained when we performed transwell invasion assay (Additional file 9: Figure S2 B-C).
Fig. 2.
P2RX6 mediated the ATP-enhanced RCC migration/invasion. a WB validation of P2RX6 protein expression was elevated in multiple RCC (SN12-PM6, OS-RC-2, 786-O, SW839, A498) vs. normal renal proximal tubule epithelial cell line HK-2. b qRT-PCR assay validation of P2RX6 mRNA level knocking-down efficiency and c WB validation of P2RX6 protein level knocking-down efficiency when knocking down using sh-P2RX6#1, sh-P2RX6#2 in SN12-PM6 cells. d Migration assay after using 2 specific P2RX6 shRNAs in SN12-PM6 cells treated with ATP, PLKO.1-vector as control. e Quantitative analysis for Fig. 2D. f Transwell assay were performed after using 2 specific P2RX6 shRNAs in SN12-PM6 cells treated with ATP, PLKO.1-vector as control. g Quantitative analysis for Fig. 2f. h WB validated P2RX6 was overexpressed when using P2RX6-cDNA in 786-O and OS-RC-2 cells. i Migration assay after P2RX6 overexpression in 786-O and OS-RC-2 cells treated with ATP, pWPI-vector as control. j Quantitative analysis for Fig. 2i. k Transwell assay after P2RX6 overexpression in 786-O and OS-RC-2cells treated with ATP, PWPI-vector as control. l Quantitative analysis for Fig. 2k. * P < 0.05, ** P < 0.01, *** P < 0.001, **** P < 0.0001
Then, we therefore focused on whether P2RX6 played an impressive role in this energy metabolism. Then, we applied the interruption approaches using two specific shRNAs (sh-P2RX6 1# and 2#) to block P2RX6 expression (see their suppressing effects on mRNA level in Fig. 2b and protein level in Fig. 2c), and results revealed that suppressed P2RX6 expression led to block the ATP-increased cell migration (Fig. 2d-e) and invasion (Fig. 2f-g) in SN12-PM6 cells. We also performed the rescue assay with another cell line, SW839, and we found the result was coincident with SN12-PM6’s phenotype (Additional file 9: Figure S2 D-I). Next, we used the opposite approach via adding P2RX6-cDNA (Fig. 2h) into 786-O and OS-RC-2 cell, and results revealed that increased P2RX6 expression conversely led to increase ATP effects on up-regulating cell migration (Fig. 2i-j) and invasion (Fig. 2k-l) in both cells.
Together, results from Fig. 2a-l suggested ATP might function via this newly identified P2RX6 to increase RCC cell migration and invasion.
Mechanism dissection of how P2RX6 elevates the RCC migration and invasion: via altering Ca2+ influx ion channel signaling
Previous studies indicated that parts of P2RX receptors family involved in membrane trafficking, ion permeation (including calcium permeability) and ATP binding [31, 32]. Also, ATP elicited increases in intracellular Ca2+ concentration could be repressed by the specific antagonist [33].
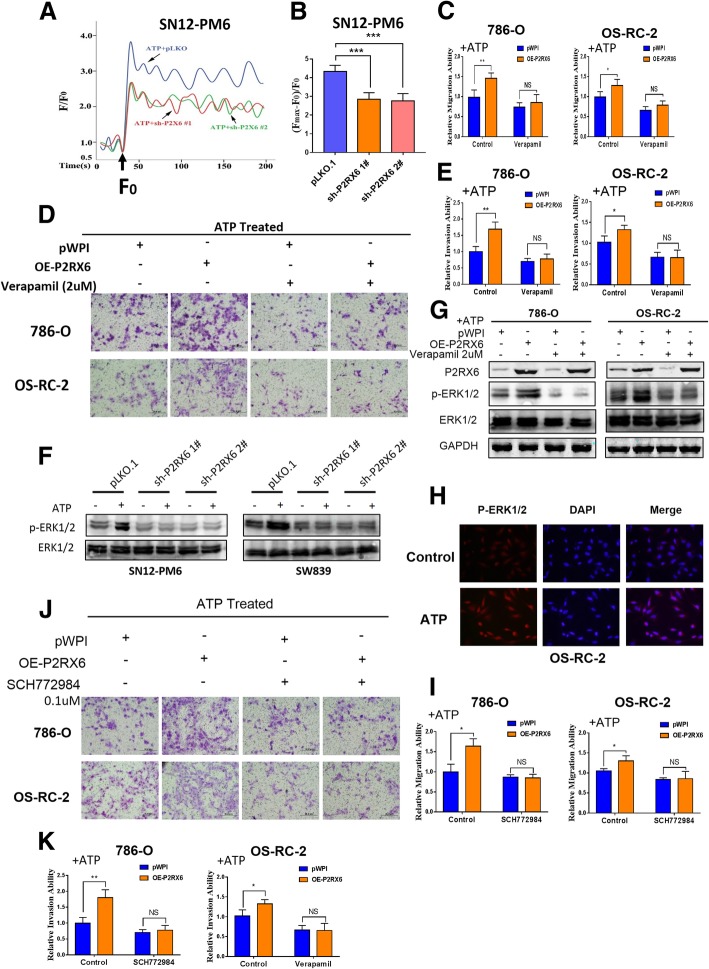
To further explore P2RX6’s function in RCC, the intracellular Ca2+ level (Ca2+)i was be measured in response to agonist ATP. 10 μM ATP caused the immediate and rapid increase in (Ca2+)i in SN12-PM6, which was significantly decreased in P2RX6-knocked-down group compared to control group (Fig. 3a-b), suggesting P2RX6 participated in the process ATP-induced Ca2+ influx activity.
Fig. 3.
The ATP-P2RX6-Ca2+ signaling mediated the MAPK family ERK1/2 activation to promote RCC migration/invasion. a Intracellular Ca2+ levels were determined in SN12-PM6 to evaluate the P2RX6 effect. Arrows indicate the points at which ATP (10 μM) was added. b The net change in Ca2+ levels was normalized to (Fmax-F0)/F0. c Migration assay showed Ca2+ inhibitor verapamil (2uM) could block ATP-P2RX6 induced cell migration in 786-O and OS-RC-2 cells. d Transwell assay showed Ca2+ inhibitor verapamil (2uM) could block ATP-P2RX6 induced cell migration in 786-O and OS-RC-2 cells. e Quantitative analysis for Fig. 3d. f Western Immunoblotting for MAPK family ERK1/2 and p-ERK1/2 expression using sh-P2RX6 1# and 2# treat with/without DHT, GAPDH as an internal control. g WB for MAPK family ERK1/2 and p-ERK1/2 expression using verapamil treat with/without over-express P2RX6, GAPDH as an internal control. h Immunofluorescence showed that ATP could stimulate the expression of p-ERK1/2 in both the cellular cytoplasm and nucleus. i Migration assay showed ERK1/2 inhibitor SCH772984 (0.1uM) could block over-expression P2RX6 induced cell migration in 786-O and OS-RC-2 cells. j Transwell assay showed ERK1/2 inhibitor SCH772984 (0.1uM) could block over-expression P2RX6 induced cell invasion in 786-O and OS-RC-2 cells. k Quantitative analysis for Fig. 3j. N.S. = not significant. * P < 0.05, ** P < 0.01, *** P < 0.001
To identify which specific mechanism the ATP-P2RX6 might promote RCC cell migration and invasion, we then applied the interruption approaches with Ca2+ specific inhibitor, verapamil, to examine their interruption effects on the ATP-P2RX6-increased cell migration and invasion. The results revealed that adding the 2uM verapamil selectively block Ca2+ influx could lead to block ATP-increased cell migration and invasion (Fig. 3c-e) in 786-O and OS-RC-2 cells.
Together, results from Fig. 3a-e using rescue assay concluded that ATP-P2RX6 might function via altering Ca2+ influx to increase the RCC cell migration and invasion.
Mechanism dissection of how ATP-P2RX6 axis can facilitate RCC migration and invasion: via increasing the MAPK family ERK1/2 phosphorylation signaling
Next, in order to explore the intra-cellular downstream signals to mediate the ATP-P2RX6-increased the RCC cell migration and invasion, we focused on the MAPK family ERK1/2 signals since our team recently contributed to this signaling pathway in recent years [34–36]. Meanwhile, early studies indicated the Ca2+ influx could effect on the MARK signaling to influence various human cancer cells [37–39].
We first demonstrated that adding 10uM ATP led to activate MARK family ERK1/2 via increasing their phosphorylation in RCC cell lines. We then added P2RX6-shRNA or Ca2+ inhibitor verapamil, and results revealed that adding these shRNAs or inhibitor could result in decreasing the phosphorylation of MARK family ERK1/2 (Fig. 3f-g). IF assay demonstrated that ATP treatment up-regulated p-ERK1/2 expression in both cytoplasm and nucleus, indicating that ATP not only stimulates p-ERK1/2 expression but also accelerates its translocation from cytoplasm to nucleus in RCC (Fig. 3h). Meanwhile, adding inhibitor SCH772984 0.1uM specific to ERK1/2 could interrupt the ATP-enhanced RCC migration (Fig. 3i) and invasion (Fig. 3j-k) in both 786-O and OS-RC-2 cells.
Together, results from Fig. 3f-k suggested that ATP-P2RX6-Ca2+ axis may function via increasing the MARK family ERK1/2 phosphorylation to enhance RCC cell migration and invasion.
Mechanism dissection of how ATP-P2RX6-Ca2+ −p-ERK1/2 axis augments RCC cell migration and invasion: via promoting MMP9 expression
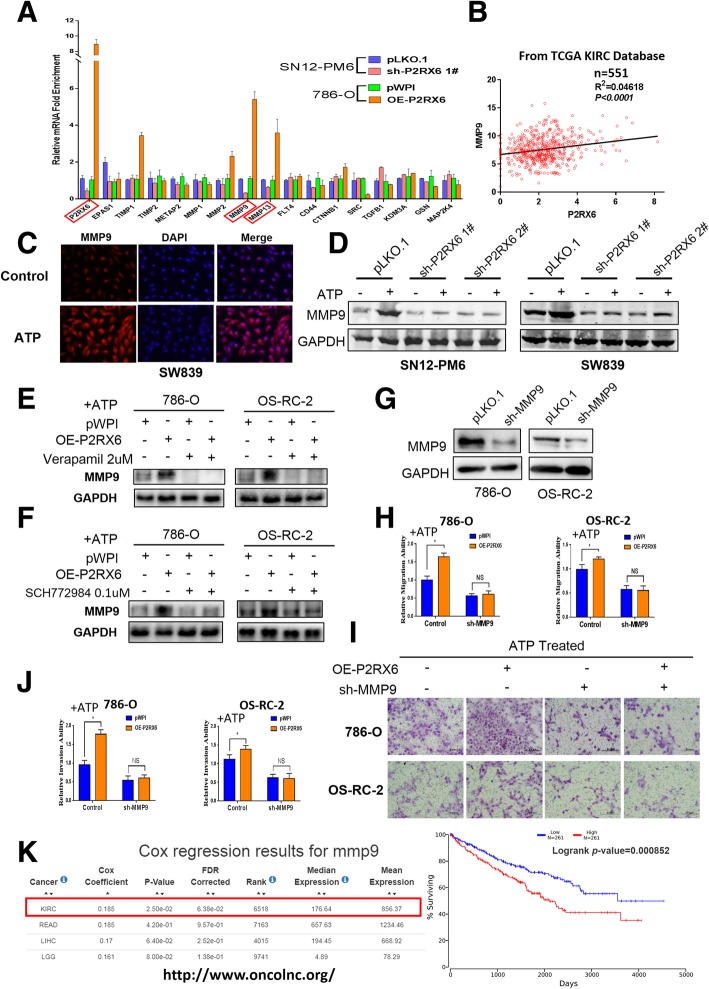
To search the downstream potential genes that can mediate the ATP-P2RX6-Ca2+ −p-ERK1/2-increased RCC cell migration and invasion, we then screen several metastasis related genes that expressed in both SN12-PM6 and 786-O cells, and results revealed that manipulating P2RX6 could produce consistent changes in MMP9 and MMP13 mRNA expression (Fig. 4a). TCGA database indicated that P2RX6 has positive correlation with MMP9 and MMP13 mRNA expression (Fig. 4b, Additional file 10: Figure S3 A). Besides, IF assay previously demonstrated that ATP treatment up-regulated MMP9 expression in RCC (Fig. 4c). WB analyses further confirmed MMP9 protein level was induced after adding 10uM ATP and knocking down P2RX6 could abrogate this effect (Fig. 4d). However, MMP13 failed to obtain this result (Additional file 10: Figure S3 B).
Fig. 4.
The ATP-P2RX6-Ca2+ signaling mediated the MAPK family ERK1/2 activation to promote RCC migration/invasion. a qRT-PCR assay for screening some metastasis genes after manipulating P2RX6 in SN12-PM6 and 786-O cells. b Correlation analysis for P2RX6 and MMP9 from TCGA database. c Immunofluorescence showed that ATP could stimulate the expression of MMP9. d WB of MMP9 using P2RX6-shRNA. e WB of MMP9 using Ca2+ inhibitor verapamil (2uM). f WB of MMP9 using ERK1/2 inhibitor SCH772984 (0.1uM). WB to show MMP9 Knockdown efficiency in J82 and 5637 cells. g WB of MMP9 using MMP9-shRNA. h Migration assay using sh-MMP9 in 786-O and OS-RC-2 cells treated with ATP. i Transwell assay after sh-MMP9 in 786-O and OS-RC-2 cells treated with ATP. Scale bar, 200 μm. j Quantitative analysis for Fig. 4i. k MMP9 gene information on http://www.oncolnc.org/ and Kaplan-Meier analysis for MMP9 mRNA expression in RCC patients (***P = 0.000852). N.S. = not significant. * P < 0.05, ** P < 0.01, *** P < 0.001
Then we focused on the MMP9 as both its mRNA and protein expressions were all elevated after adding ATP. The interruption approaches via adding the P2RX6-shRNA, Ca2+ inhibitor verapamil or ERK1/2 inhibitor SCH772984 all resulted in attenuating MMP9 expression at protein level (Fig. 4d-f). Importantly, adding MMP9-shRNA effectively suppressed MMP9 expression (Fig. 4g), which then resulted in blocking the P2RX6-induced RCC cell migration and invasion in OS-RC-2 and 786-O cells (Fig. 4h-j). In addition, MMP9 mRNA expression correlated with RCC patients OS, indicating higher expression of MMP9 associated with RCC poor prognosis (***P = 0.000852) (Fig. 4k, figure obtained from OncoLnc website http://www.oncolnc.org/).
Together, results from Fig. 4a-k suggested that ATP-P2RX6-Ca2+ −p-ERK1/2 axis may function via maximizing the metastasis gene MMP9 expression to facilitate RCC migration and invasion.
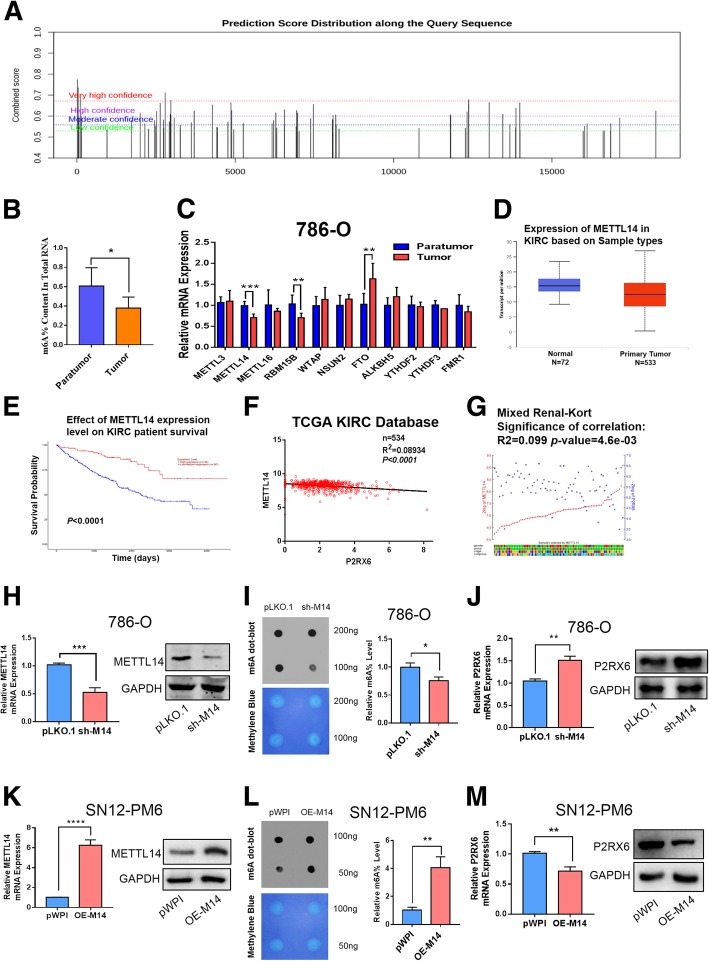
The m6A modification is decreased in RCC and P2RX6 can be regulated by METTL14
Recent studies have shown that RNA methylation contributes to RCC development [20], so we sought to find out whether the high expression of P2RX6 in RCC was regulated by m6A methylation. To explore the potential mechanism of m6A modification in P2RX6, firstly, we predicted P2RX6 potential m6A sites with m6A Finder (http://m6a.renlab.org/) and found that P2RX6 has numbers of very high confidence m6A sites distribution (Fig. 5a, Additional file 6: Table S6). Then, with the colorimetric m6A quantification assay, we measured the levels of m6A in the total RNAs of 7 paired renal tumor tissues and adjacent tissues. We found that m6A levels were decreased in renal tumor tissues compared with their adjacent tissues (Fig. 5b). Recently, numbers of m6A modification related proteins have been identified, including NSUN2, FTO, METTL3, METTL14, ALKBH5 and YTHDF2 et al. [40]. We measured the expression levels of eleven m6A modification related genes in seven paired renal tumor tissues. We found that METTL14 and RBM15B mRNA expression significantly decreased, while FTO significantly increased in tumor tissues (Fig. 5c). Then, analyzing TCGA KIRC database, we found METTL14 mRNA was decreased in tumor tissues compared to normal tissues (Fig. 5d). Meanwhile, Kaplan-Meier analysis demonstrated lower METTL14 expression has worse OS result (***P < 0.0001) (Fig. 5e) (Fig. 5d-e obtained from UALCAN website http://ualcan.path.uab.edu/) [41]. Interesting, METTL14 has a negative correlation with P2RX6 gene in TCGA database (R2 = 0.089, ***P < 0.0001) (Fig. 5f) and website tool R2 (https://hgserver1.amc.nl/) (R2 = 0.099, ***P < 0.0046) (Fig. 5g, Additional file 7: Table S7). However, RBM15B has lower expression in tumor tissues and lower expression could result in worse OS, also RBM15B has a positive correlation with P2RX6 expression in TCGA database (Additional file 11: Figure S4 A-C). Meanwhile, FTO has higher expression in tumor tissues and a negative correlation with P2RX6 expression, but survival analysis has no significantly difference between two groups (Additional file 11: Figure S4 D-F). Combined with the above results, METTL14, an important m6A methyltransferase that has a positive correlation with m6A levels, was then considered as the candidate molecule for aberrant m6A modification in RCC.
Fig. 5.
The m6A modification is decreased in RCC and P2RX6 can be regulated by METTL14. a P2RX6 potential m6A sites distribution. b The m6A contents of total RNAs in tumor tissues paired with para-tumor tissues (n = 7). c The mRNA levels of m6A modification associated genes in 7 pairs of RCC tumor and para-tumor tissues. d Expression of METTL14 in TCGA KIRC. e Kaplan-Meier analysis for METTL14 expression level on KIRC patients’ OS. f Correlation between METTL14 and P2RX6 from TCGA database. g Correlation between METTL14 and P2RX6 from website R2. h Validation of METTL14 mRNA and protein level knocking-down efficiency in 786-O cells. i The m6A contents of total RNAs in 786-O. Dot blot assay analyzed the Poly(A)+ RNAs isolated from pLKO and sh-M14 group cells. RNAs were equally loaded by two fold serial dilution. Methylene blue was used to detect equal mRNA loading in two groups. j P2RX6 mRNA and protein expression level after knocking-down METTL14. k Validation of METTL14 mRNA and protein level knocking-down efficiency in OS-RC-2 cells. l The m6A contents of total RNAs in SN12-PM6 cell line. m P2RX6 mRNA and protein expression level after over-expression METTL14. M14 = METTL14, N.S. = not significant. * P < 0.05, ** P < 0.01, *** P < 0.001
Next, we implemented the interruption approaches using specific shRNA (sh-M14) to block METTL14 expression (Fig. 5h) and then examined m6A levels in the RCC cells. Consistently, knocking down METTL14 led to decrease m6A levels in 786-O cell (Fig. 5i) and increase P2RX6 mRNA and protein level (Fig. 5j). Furthermore, after over-expressing METTL14 (OE-M14) in SN12-PM6 cell line, the results are consistent with the knocking-down data (Fig. 5k-m).
Together, results from Fig. 5a-m suggested METTL14 might abrogate P2RX6 protein level via m6A methylated modification.
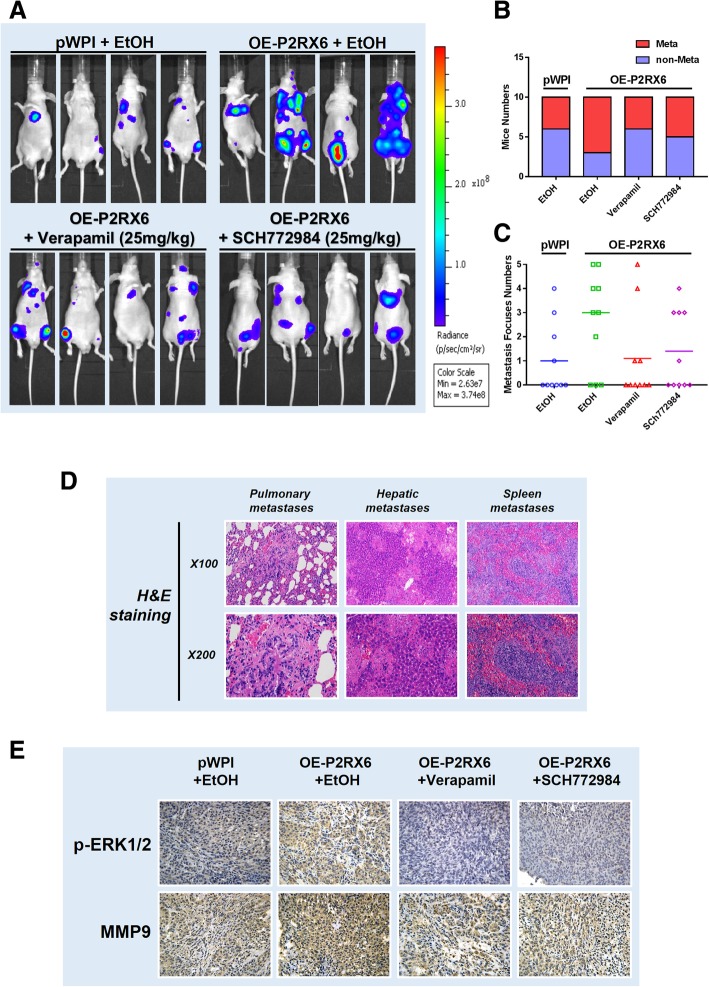
Preclinical study using in vivo mouse model confirms that the ATP-P2RX6-Ca2+ −p-ERK1/2-MMP9 axis increases RCC metastasis
To confirm above in vitro cell lines data in the in vivo mouse model, we injected xenografted RCC OS-RC-2 cells expressing firefly luciferase into BALB/c nude mice tail vein [42]. After 8 weeks of implantation, the mice were sacrificed, the metastatic sites were further examined. The results indicated that mice received OE-P2RX6 injection saliently developed more metastatic tumors than the vehicle group (Fig. 6a-b). Importantly, using small molecules of Ca2+ influx (verapamil) or p-ERK1/2(SCH772984) to suppress the ATP-P2RX6-Ca2+ −p-ERK1/2 signaling all led to suppress the RCC progression and metastasis (Fig. 6c). In addition, anatomic studies were carried out and the histological staining were performed to confirm the tumor type (Fig. 6d).
Fig. 6.
Preclinical study using mouse model to confirm ATP increased RCC metastasis via P2RX6-Ca2+ −p-ERK1/2-MMP9 axis. a The experimental scheme. The tumor metastases in nude mice implanted with OS-RC-2 cells. The nude mice were divided into 4 groups: pWPI-vector + EtOH (Mock), OE-P2RX6 + EtOH, OE-P2RX6 + Verapamil, OE-P2RX6 + SCH772984. Mice were sacrificed after 8 weeks were assessed for metastasis. The IVIS image for monitoring tumor and metastasis. b Quantitative analysis for Fig. 6a. c Number of metastasis foci in each groups. d Hematoxylin and eosin (H&E) staining were performed to confirm the tumor type. e Representative IHC images and quantification of p-ERK1/2 and MMP9 expression on mice metastasis foci
IHC staining also testified that the expression of p-ERK1/2, MMP9, were higher in OE-P2RX6 group mice compared to the vehicle control, and using small molecules of Ca2+ influx or p-ERK1/2 to suppress the P2RX6-Ca2+ −p-ERK1/2 signaling all led to suppress those OE-P2RX6-increased p-ERK1/2-MMP9 signaling (Fig. 6e).
Together, preclinical study results from in vivo RCC mouse model (Fig. 6a-e) were in agreement with in vitro cell lines data illustrating ATP-OE-P2RX6 could enhance RCC metastasis via altering the ATP-P2RX6-Ca2+ p-ERK1/2-MMP9 signaling.
As summarized in Fig. 7a-b, the m6A-suppressed P2RX6 activation promotes renal cancer cells migration and invasion through ATP-induced Ca2+ influx modulating ERK1/2 phosphorylation and MMP9 signalling pathway.
Fig. 7.
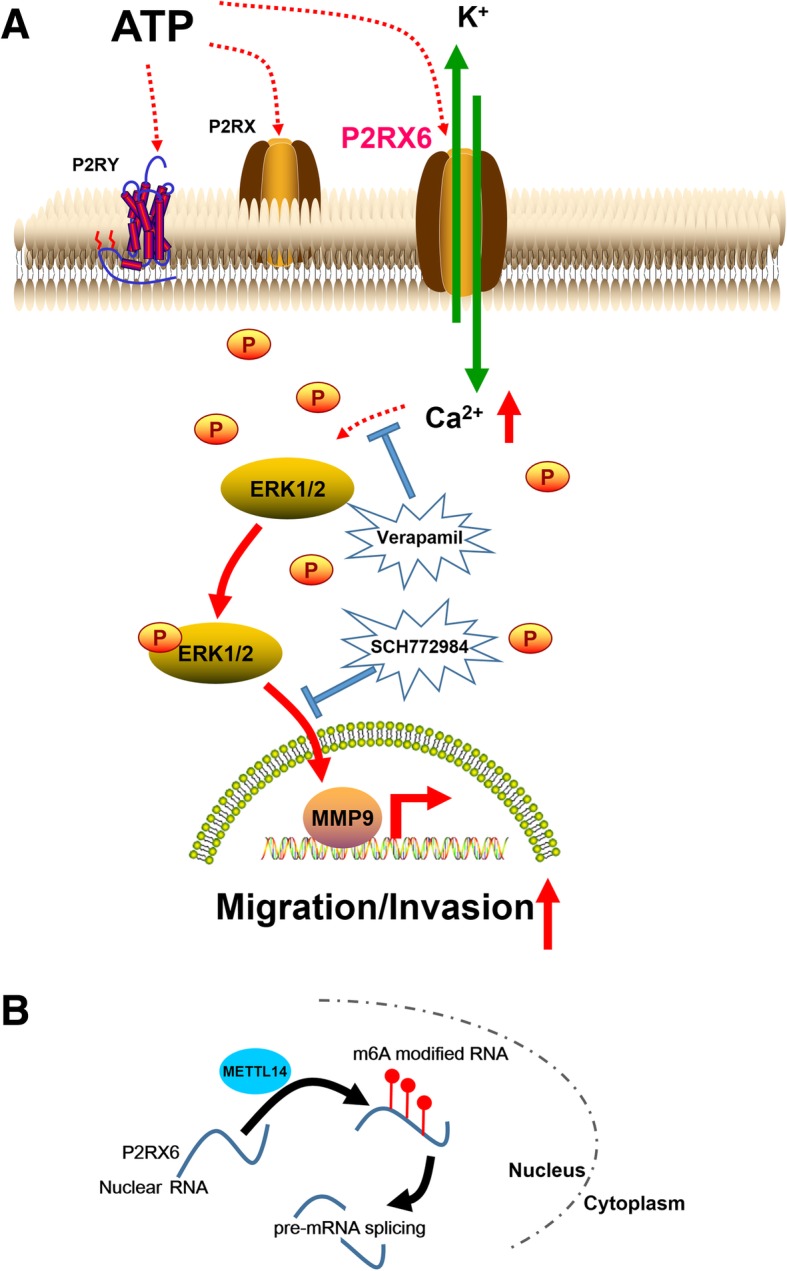
The cartoon model of METTL14-P2RX6-Ca2+ −p-ERK1/2-MMP9 axis signal on RCC cell migration and invasion. a P2RX6’s specific mechanism on metastasis. b METTL14’s specific mechanism on regulation P2RX6 mRNA m6A methylation
Discussion
A number of studies have found that tumor microenvironment extracellular ATP might play a detrimental role in tumor progression [9, 10, 43] and initiate signaling pathways through activating membrane receptors. Previous reports demonstrated that extracellular ATP activates P2RY2 and promotes prostate cancer cells invasion and metastasis [44]. Li et al. revealed that ATP or UTP could activate EGFR and ERK1/2 and P2RY2 could suppress ATP-induced phosphorylation of EGFR and ERK1/2. Besides, among P2RX receptors, P2RX7 draw many attentions. P2RX7 activation releases important pro-inflammatory cytokines such as interleukin-1β and fever-inducing prostaglandin E2, which is crucial for initiation of the inflammatory signaling cascade [31, 45–47]. As such, studies have recently shown that P2RX7 participated in tumors metastasis, and it is up-regulated compared with normal tissues, including chronic lymphocyte leukemia, melanoma, neuroblastoma, prostate, breast and thyroid cancers [13, 33, 48, 49].
In recent years, P2RX6 function has been researched widely [50, 51]. In transiently transfected HEK293 cell, it has been reported that the recombinant homomeric P2RX6 complexes reach the plasma membrane. There are also researchers proved that P2RX6 can only play a role at the cell surface in the synergy of P2X4 or P2X2 [52, 53]. Our preliminary data showed that ATP could promote RCC migration and invasion and P2RX6 might play a crucial role according to bioinformatics analysis. Till now, there is no report about P2RX6 specific mechanism in RCC metastasis.
In this study, we found membrane protein P2RX6, whose higher expression could be detected in several RCC cells compared with HK2 normal cell. Clinical survey and TCGA database analysis performed that P2RX6 expression was obviously correlated with RCC pathological stage, pathological grade and organ metastasis. We therefore focused on P2RX6 for further analysis. Preclinical studies using in vitro multiple RCC cells and in vivo mouse model all demonstrated P2RX6 might function via ATP-P2RX6-Ca2+ −p-ERK1/2-MMP9 axis to drive RCC cells invasion and metastasis.
As one of the most important second messengers in intracellular signaling network, Ca2+ flux and Ca2+-dependent signaling also played a pivotal role in cancer progression [54, 55]. Upon initiation of the Ca2+ ion channel and phosphorylation of its substrate, an intracellular calcium signaling cascade is activated that mediates a variety of biological processes by coordinating with other abnormal signals and modulating the activity of subsequent transcription factors [56–58]. This study illustrated that verapamil could block the Ca2+ influx, which is caused by P2RX6. Elucidation of this mechanism facilitates the identification of new RCC biomarkers and helps in the development of new targeted therapies.
Previous studies reported that ERK can be activated by Ca2+ and then phosphorylates and activates various transcription factors and other molecular effectors [59]. Here, we found that Ca2+ signal as elicited by ATP via P2RX6 caused ERK1/2 phosphorylation. Indeed, application of 0.1uM SCH772984, a concentration that had no effect on ERK1/2 phosphorylation, could then blunt the ATP-enhanced RCC cell migration and invasion. This result demonstrated that the P2RX6 signal and the Ca2+ signal synergistically induced ERK1/2 phosphorylation.
So far, studies have shown that a large number of m6A-related enzymes play a key role in tumor development. These enzymes are mainly divided into two categories, one is methyltransferase (METTL3/14/16, WTAP, KIAA1429 and RBM15/15B et al) and the other is demethylase (FTO and ALKBH5 et al) [20]. Interestingly, with screening the potential m6A candidates, we identified that METTL14 could suppress P2RX6 mRNA and protein level. That is to say, METTL14 may increase P2RX6 pre-mRNA splicing by mediating P2RX6 mRNA level m6A methylation. The specific mechanism is not clear, but it provides a direction for subsequent research.
Conclusions
In conclusion, our preclinical studies using multiple in vitro cell lines and in vivo mouse models as well as human clinical studies all suggest that ATP-P2RX6-Ca2+ −p-ERK1/2-MMP9 axis facilitate RCC migration and invasion. The involvement of purine receptors in cancer cell biology is a promising area of research that can increase understanding of the processes that lead to cancer metastasis. This may lead to the development of new drug approaches in RCC treatment.
Additional files
Table S1. Oligonucleotide Sequences, Antibody and Inhibitor used in this study. (PPTX 62 kb) (PPTX 61 kb)
Table S2. TCGA Clinical data. (XLSX 59 kb) (XLSX 58 kb)
Table S3. Candidates selecting process & GO analysis result. (XLSX 49 kb)
Table S4. P2RX6 IHC analysis of tissues microarray. (XLSX 10 kb) (XLSX 9 kb)
Table S5. Clinical characteristics of 238 RCC patients according to P2RX6 expression levels. (PPTX 43 kb) (PPTX 42 kb)
Table S6. P2RX6 m6A in plain text from. (XLSX 15 kb) (XLSX 14 kb)
Table S7. Correlation between METTL14 and P2RX6. (XLSX 12 kb) (XLSX 11 kb)
Figure S1. Candidates selecting and P2RX6 gene bioinformatics characteristic. A P2RX6 gene selecting process flowchart. B ADRA2A gene expression in different stages. C Kaplan-Meier analysis for ADRA2A mRNA expression in RCC patients. D P2RX6 gene information on http://www.oncolnc.org/ and Kaplan-Meier analysis for P2RX6 mRNA expression in RCC patients (**P = 0.00175). (TIF 1943 kb)
Figure S2. ATP effects on migration/invasion of OS-RC-2 cells and validating P2RX6 gene function in SW839 cell line. A In vitro wound-healing motility assay with 3, 10, 30uM ATP treatment in OS-RC-2 cells. B Transwell invasion assays were performed with 3, 10, 30uM ATP treatment in OS-RC-2 cells. C Quantitative analysis for Fig. S1B. D qRT-PCR assay validation of P2RX6 mRNA level knocking-down efficiency and E WB validation of P2RX6 protein level knocking-down efficiency when knocking down using sh-P2RX6#1 in SW839 cells. F Migration assay after using sh-P2RX6#1 in SW839 cells treated with ATP, PLKO.1-vector as control. G Quantitative analysis for Fig. S2 F.H Transwell assay were performed after using sh-P2RX6#1 in SW839 cells treated with ATP, PLKO.1-vector as control. I Quantitative analysis for Fig. S2 H. * indicated P < 0.05. And ** indicated P < 0.01, *** indicated P < 0.001. (TIF 2967 kb)
Figure S3. Excluding MMP13 as downstream target gene. A Correlation analysis for P2RX6 and MMP13 from TCGA database. B WB validation of MMP13 protein level after treating with ATP. (TIF 786 kb)
Figure S4. Excluding RBM15B and FTO as target methylated gene. A RBM15B has lower expression in tumor tissues and B lower expression could result in a worse OS, and C has a positive correlation with P2RX6 expression in TCGA database. D FTO has higher expression in tumor tissues and E survival analysis has no significantly difference between two groups. F FTO has a negative correlation with P2RX6 expression (TIF 939 kb)
Acknowledgements
Not applicable.
Funding
This work was supported by the National Natural Science Foundation of China (31570775, 81602216 and 81772705), Natural Science Foundation of Shanghai (16ZR1426500), Three Year Plan of Action Program from Shen Kang (16CR3062B), Cultivation Program of Renji Hospital (PYZY16–006).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- RCC
Renal cell carcinoma
- m6A
N6-methyladenosine
- ATP
Adenosine 5′-triphosphate
- qRT-PCR
quantitative real-time PCR analysis
- WB
Western immunoblotting
- IF
Immunofluorescence
- IHC
Immunohistochemical
Authors’ contributions
D-KG, WZ and J-HZ conceived the study; D-KG, J-HZ and J-JM performed most of the experiments; WZ and WX performed the bioinformatic analyses; Y-HC, Y-FX and G-HH helped with various experiments and statistical analyses; J-HZ, WZ and WX supervised the study; DK G wrote the manuscript. All authors approved final version of the manuscript.
Ethics approval
All human tissue samples were obtained with patient consent and the approval of the Institutional Clinical Ethics Review Board at Shanghai Tenth People’s Hospital. All animal studies were performed with an approved protocol by the Institutional Animal Care and Use Committee of Shanghai Tenth People’s Hospital.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing financial interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Dongkui Gong, Email: gongdk2007@whu.edu.cn.
Jin Zhang, Email: med-zhangjin@vip.sina.com.
Yonghui Chen, Email: cyh1488@163.com.
Yunfei Xu, Email: xuyunfeibb@sina.com.
Junjie Ma, Email: 13776188630@163.com.
Guanghui Hu, Email: 15hu_guanghui@tongji.edu.cn.
Yiran Huang, Email: huangyrrenji@163.com.
Junhua Zheng, Email: zhengjh0471@sina.com.cn.
Wei Zhai, Email: jacky_zw2002@hotmail.com.
Wei Xue, Email: xuewei@renji.com.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 2.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018. [DOI] [PubMed]
- 3.Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 4.Morris MR, Latif F. The epigenetic landscape of renal cancer. Nat Rev Nephrol. 2017;13:47–60. doi: 10.1038/nrneph.2016.168. [DOI] [PubMed] [Google Scholar]
- 5.Cavaliere C, D'Aniello C, Pepa CD, Pisconti S, Berretta M, Facchini G. Current and emerging treatments for metastatic renal cell carcinoma. Curr Cancer Drug Targets. 2018;18:468–479. doi: 10.2174/1568009617666170209094030. [DOI] [PubMed] [Google Scholar]
- 6.Miao D, Margolis CA, Gao W, Voss MH, Li W, Martini DJ, Norton C, Bosse D, Wankowicz SM, Cullen D, et al. Genomic correlates of response to immune checkpoint therapies in clear cell renal cell carcinoma. Science. 2018;359:801–806. doi: 10.1126/science.aan5951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van der Mijn JC, Mier JW, Broxterman HJ, Verheul HM. Predictive biomarkers in renal cell cancer: insights in drug resistance mechanisms. Drug Resist Updat. 2014;17:77–88. doi: 10.1016/j.drup.2014.10.003. [DOI] [PubMed] [Google Scholar]
- 8.Qu L, Ding J, Chen C, Wu ZJ, Liu B, Gao Y, Chen W, Liu F, Sun W, Li XF, et al. Exosome-transmitted lncARSR promotes Sunitinib resistance in renal Cancer by acting as a competing endogenous RNA. Cancer Cell. 2016;29:653–668. doi: 10.1016/j.ccell.2016.03.004. [DOI] [PubMed] [Google Scholar]
- 9.Janssens R, Boeynaems JM. Effects of extracellular nucleotides and nucleosides on prostate carcinoma cells. Br J Pharmacol. 2001;132:536–546. doi: 10.1038/sj.bjp.0703833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shabbir M, Ryten M, Thompson C, Mikhailidis D, Burnstock G. Purinergic receptor-mediated effects of ATP in high-grade bladder cancer. BJU Int. 2008;101:106–112. doi: 10.1111/j.1464-410X.2007.07293.x. [DOI] [PubMed] [Google Scholar]
- 11.Chadet S, Jelassi B, Wannous R, Angoulvant D, Chevalier S, Besson P, Roger S. The activation of P2Y2 receptors increases MCF-7 breast cancer cells migration through the MEK-ERK1/2 signalling pathway. Carcinogenesis. 2014;35:1238–1247. doi: 10.1093/carcin/bgt493. [DOI] [PubMed] [Google Scholar]
- 12.Burnstock G, Fredholm BB, North RA, Verkhratsky A. The birth and postnatal development of purinergic signalling. Acta Physiol (Oxf) 2010;199:93–147. doi: 10.1111/j.1748-1716.2010.02114.x. [DOI] [PubMed] [Google Scholar]
- 13.Burnstock G, Verkhratsky A. Long-term (trophic) purinergic signalling: purinoceptors control cell proliferation, differentiation and death. Cell Death Dis. 2010;1:e9. doi: 10.1038/cddis.2009.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.North RA. Molecular physiology of P2X receptors. Physiol Rev. 2002;82:1013–1067. doi: 10.1152/physrev.00015.2002. [DOI] [PubMed] [Google Scholar]
- 15.Burnstock G. Therapeutic potential of purinergic signalling for diseases of the urinary tract. BJU Int. 2011;107:192–204. doi: 10.1111/j.1464-410X.2010.09926.x. [DOI] [PubMed] [Google Scholar]
- 16.Li WH, Qiu Y, Zhang HQ, Tian XX, Fang WG. P2Y2 receptor and EGFR cooperate to promote prostate Cancer cell invasion via ERK1/2 pathway. PLoS One. 2015;10:e0133165. doi: 10.1371/journal.pone.0133165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Burnstock G. Purine and pyrimidine receptors. Cell Mol Life Sci. 2007;64:1471–1483. doi: 10.1007/s00018-007-6497-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gow IF, Thomson J, Davidson J, Shennan DB. The effect of a hyposmotic shock and purinergic agonists on K+(Rb+) efflux from cultured human breast cancer cells. Biochim Biophys Acta. 2005;1712:52–61. doi: 10.1016/j.bbamem.2005.04.002. [DOI] [PubMed] [Google Scholar]
- 19.Greig AV, Linge C, Healy V, Lim P, Clayton E, Rustin MH, McGrouther DA, Burnstock G. Expression of purinergic receptors in non-melanoma skin cancers and their functional roles in A431 cells. J Invest Dermatol. 2003;121:315–327. doi: 10.1046/j.1523-1747.2003.12379.x. [DOI] [PubMed] [Google Scholar]
- 20.Dai D, Wang H, Zhu L, Jin H, Wang X. N6-methyladenosine links RNA metabolism to cancer progression. Cell Death Dis. 2018;9:124. doi: 10.1038/s41419-017-0129-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin S, Choe J, Du P, Triboulet R, Gregory RI. The m(6)a methyltransferase METTL3 promotes translation in human Cancer cells. Mol Cell. 2016;62:335–345. doi: 10.1016/j.molcel.2016.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ma JZ, Yang F, Zhou CC, Liu F, Yuan JH, Wang F, Wang TT, Xu QG, Zhou WP, Sun SH. METTL14 suppresses the metastatic potential of hepatocellular carcinoma by modulating N(6) -methyladenosine-dependent primary MicroRNA processing. Hepatology. 2017;65:529–543. doi: 10.1002/hep.28885. [DOI] [PubMed] [Google Scholar]
- 23.Zhang C, Samanta D, Lu H, Bullen JW, Zhang H, Chen I, He X, Semenza GL. Hypoxia induces the breast cancer stem cell phenotype by HIF-dependent and ALKBH5-mediated m(6)A-demethylation of NANOG mRNA. Proc Natl Acad Sci U S A. 2016;113:E2047–E2056. doi: 10.1073/pnas.1602883113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han D, Fu Y, Parisien M, Dai Q, Jia G, et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature. 2014;505:117–120. doi: 10.1038/nature12730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang X, Zhao BS, Roundtree IA, Lu Z, Han D, Ma H, Weng X, Chen K, Shi H, He C. N(6)-methyladenosine modulates messenger RNA translation efficiency. Cell. 2015;161:1388–1399. doi: 10.1016/j.cell.2015.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Geula S, Moshitch-Moshkovitz S, Dominissini D, Mansour AA, Kol N, Salmon-Divon M, Hershkovitz V, Peer E, Mor N, Manor YS, et al. Stem cells. m6A mRNA methylation facilitates resolution of naive pluripotency toward differentiation. Science. 2015;347:1002–1006. doi: 10.1126/science.1261417. [DOI] [PubMed] [Google Scholar]
- 27.Brierley JD, Gospodarowicz MK, Wittekind C. TNM classification of malignant Tumours. UICC International Union against Cancer. 8th edn: Wiley-Blackwell; 2017. p. 199.
- 28.Jin H, Ham SA, Kim MY, Woo IS, Kang ES, Hwang JS, Lee K-W, Kim HJ, Roh GS, Lim D-S, et al. Activation of peroxisome proliferator-activated receptor-δ attenuates glutamate-induced neurotoxicity in HT22 mouse hippocampal cells. J Neurosci Res. 2012;90:1646–1653. doi: 10.1002/jnr.23053. [DOI] [PubMed] [Google Scholar]
- 29.Jin H, Eun SY, Lee JS, Park SW, Lee JH, Chang KC, Kim HJ. P2Y2receptor activation by nucleotides released from highly metastatic breast cancer cells increases tumor growth and invasion via crosstalk with endothelial cells. Breast Cancer Res. 2014;16:R77. doi: 10.1186/bcr3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhai W, Sun Y, Jiang M, Wang M, Gasiewicz TA, Zheng J, Chang C. Differential regulation of LncRNA-SARCC suppresses VHL-mutant RCC cell proliferation yet promotes VHL-normal RCC cell proliferation via modulating androgen receptor/HIF-2alpha/C-MYC axis under hypoxia. Oncogene. 2016;35:4866–4880. doi: 10.1038/onc.2016.19. [DOI] [PubMed] [Google Scholar]
- 31.Surprenant A, North RA. Signaling at purinergic P2X receptors. Annu Rev Physiol. 2009;71:333–359. doi: 10.1146/annurev.physiol.70.113006.100630. [DOI] [PubMed] [Google Scholar]
- 32.North R. Alan. P2X receptors. Philosophical Transactions of the Royal Society B: Biological Sciences. 2016;371(1700):20150427. doi: 10.1098/rstb.2015.0427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jelassi B, Anchelin M, Chamouton J, Cayuela ML, Clarysse L, Li J, Gore J, Jiang LH, Roger S. Anthraquinone emodin inhibits human cancer cell invasiveness by antagonizing P2X7 receptors. Carcinogenesis. 2013;34:1487–1496. doi: 10.1093/carcin/bgt099. [DOI] [PubMed] [Google Scholar]
- 34.Zhai W, Sun Y, Guo C, Hu G, Wang M, Zheng J, Lin W, Huang Q, Li G, Zheng J, Chang C. LncRNA-SARCC suppresses renal cell carcinoma (RCC) progression via altering the androgen receptor(AR)/miRNA-143-3p signals. Cell Death Differ. 2017;24:1502–1517. doi: 10.1038/cdd.2017.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen C, Xue S, Zhang J, Chen W, Gong D, Zheng J, Ma J, Xue W, Chen Y, Zhai W, Zheng J. DNA-methylation-mediated repression of miR-766-3p promotes cell proliferation via targeting SF2 expression in renal cell carcinoma. Int J Cancer. 2017;141:1867–1878. doi: 10.1002/ijc.30853. [DOI] [PubMed] [Google Scholar]
- 36.Zhai W, Ma J, Zhu R, Xu C, Zhang J, Chen Y, Chen Z, Gong D, Zheng J, Chen C, et al. MiR-532-5p suppresses renal cancer cell proliferation by disrupting the ETS1-mediated positive feedback loop with the KRAS-NAP1L1/P-ERK axis. Br J Cancer. 2018;119:591–604. doi: 10.1038/s41416-018-0196-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hsu SS, Huang CJ, Cheng HH, Chou CT, Lee HY, Wang JL, Chen IS, Liu SI, Lu YC, Chang HT, et al. Anandamide-induced Ca2+ elevation leading to p38 MAPK phosphorylation and subsequent cell death via apoptosis in human osteosarcoma cells. Toxicology. 2007;231:21–29. doi: 10.1016/j.tox.2006.11.005. [DOI] [PubMed] [Google Scholar]
- 38.Sindreu CB, Scheiner ZS, Storm DR. Ca2+ −stimulated adenylyl cyclases regulate ERK-dependent activation of MSK1 during fear conditioning. Neuron. 2007;53:79–89. doi: 10.1016/j.neuron.2006.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moreno-Ortega AJ, Buendia I, Mouhid L, Egea J, Lucea S, Ruiz-Nuno A, Lopez MG, Cano-Abad MF. CALHM1 and its polymorphism P86L differentially control ca(2)(+)homeostasis, mitogen-activated protein kinase signaling, and cell vulnerability upon exposure to amyloid beta. Aging Cell. 2015;14:1094–1102. doi: 10.1111/acel.12403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fu Ye, Dominissini Dan, Rechavi Gideon, He Chuan. Gene expression regulation mediated through reversible m6A RNA methylation. Nature Reviews Genetics. 2014;15(5):293–306. doi: 10.1038/nrg3724. [DOI] [PubMed] [Google Scholar]
- 41.Chandrashekar DS, Bashel B, Balasubramanya SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi BVSK, Varambally S. UALCAN: a portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia. 2017;19:649–658. doi: 10.1016/j.neo.2017.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brown R, L. Richardson K, C. Kalinowski F, Epis M, Horsham J, Kabir T, H. De Pinho M, Beveridge D, M. Stuart L, Wintle L, Leedman P: Evaluation of MicroRNA delivery In Vivo 2018. [DOI] [PubMed]
- 43.Yokdang N, Tellez JD, Tian H, Norvell J, Barsky SH, Valencik M, Buxton IL. A role for nucleotides in support of breast cancer angiogenesis: heterologous receptor signalling. Br J Cancer. 2011;104:1628–1640. doi: 10.1038/bjc.2011.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li WH, Qiu Y, Zhang HQ, Liu Y, You JF, Tian XX, Fang WG. P2Y2 receptor promotes cell invasion and metastasis in prostate cancer cells. Br J Cancer. 2013;109:1666–1675. doi: 10.1038/bjc.2013.484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Di Virgilio F. Liaisons dangereuses: P2X(7) and the inflammasome. Trends Pharmacol Sci. 2007;28:465–472. doi: 10.1016/j.tips.2007.07.002. [DOI] [PubMed] [Google Scholar]
- 46.Barbera-Cremades M, Baroja-Mazo A, Gomez AI, Machado F, Di Virgilio F, Pelegrin P. P2X7 receptor-stimulation causes fever via PGE2 and IL-1beta release. FASEB J. 2012;26:2951–2962. doi: 10.1096/fj.12-205765. [DOI] [PubMed] [Google Scholar]
- 47.Pelegrin P: targeting interleukin-1 signaling in chronic inflammation: focus on P2X(7) receptor and Pannexin-1. Drug news Perspect 2008, 21:424–433. [DOI] [PubMed]
- 48.Solini A, Cuccato S, Ferrari D, Santini E, Gulinelli S, Callegari MG, Dardano A, Faviana P, Madec S, Di Virgilio F, Monzani F. Increased P2X7 receptor expression and function in thyroid papillary cancer: a new potential marker of the disease? Endocrinology. 2008;149:389–396. doi: 10.1210/en.2007-1223. [DOI] [PubMed] [Google Scholar]
- 49.Deli T, Varga N, Adam A, Kenessey I, Raso E, Puskas LG, Tovari J, Fodor J, Feher M, Szigeti GP, et al. Functional genomics of calcium channels in human melanoma cells. Int J Cancer. 2007;121:55–65. doi: 10.1002/ijc.22621. [DOI] [PubMed] [Google Scholar]
- 50.Ormond SJ, Barrera NP, Qureshi OS, Henderson RM, Edwardson JM, Murrell-Lagnado RD. An uncharged region within the N terminus of the P2X6 receptor inhibits its assembly and exit from the endoplasmic reticulum. Mol Pharmacol. 2006;69:1692–1700. doi: 10.1124/mol.105.020404. [DOI] [PubMed] [Google Scholar]
- 51.Barrera NP, Ormond SJ, Henderson RM, Murrell-Lagnado RD, Edwardson JM. Atomic force microscopy imaging demonstrates that P2X2 receptors are trimers but that P2X6 receptor subunits do not oligomerize. J Biol Chem. 2005;280:10759–10765. doi: 10.1074/jbc.M412265200. [DOI] [PubMed] [Google Scholar]
- 52.Jones CA, Vial C, Sellers LA, Humphrey PP, Evans RJ, Chessell IP. Functional regulation of P2X6 receptors by N-linked glycosylation: identification of a novel alpha beta-methylene ATP-sensitive phenotype. Mol Pharmacol. 2004;65:979–985. doi: 10.1124/mol.65.4.979. [DOI] [PubMed] [Google Scholar]
- 53.Collo G, North RA, Kawashima E, Merlo-Pich E, Neidhart S, Surprenant A, Buell G. Cloning OF P2X5 and P2X6 receptors and the distribution and properties of an extended family of ATP-gated ion channels. J Neurosci. 1996;16:2495–2507. doi: 10.1523/JNEUROSCI.16-08-02495.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kuchay S, Giorgi C, Simoneschi D, Pagan J, Missiroli S, Saraf A, Florens L, Washburn MP, Collazo-Lorduy A, Castillo-Martin M, et al. PTEN counteracts FBXL2 to promote IP3R3- and ca 2+ −mediated apoptosis limiting tumour growth. Nature. 2017;546:554–558. doi: 10.1038/nature22965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bononi A, Giorgi C, Patergnani S, Larson D, Verbruggen K, Tanji M, Pellegrini L, Signorato V, Olivetto F, Pastorino S, et al. BAP1 regulates IP3R3-mediated ca 2+ flux to mitochondria suppressing cell transformation. Nature. 2017;546:549–553. doi: 10.1038/nature22798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bakowski D, Parekh AB. Regulation of store-operated calcium channels by the intermediary metabolite pyruvic acid. Curr Biol. 2007;17:1076–1081. doi: 10.1016/j.cub.2007.05.041. [DOI] [PubMed] [Google Scholar]
- 57.Jing J, He L, Sun A, Quintana A, Ding Y, Ma G, Tan P, Liang X, Zheng X, Chen L, et al. Proteomic mapping of ER-PM junctions identifies STIMATE as a regulator of ca 2+ influx. Nat Cell Biol. 2015;17:1339–1347. doi: 10.1038/ncb3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.L-j S, H-q J, G-p L, Tian T, Ma G-l, Y-x L, Z-x L, R-l P, Li R-h, H-l P, et al. LncRNA CamK-A regulates Ca2+−signaling-mediated tumor microenvironment remodeling. Mol Cell. 2018;72:71–83. doi: 10.1016/j.molcel.2018.08.014. [DOI] [PubMed] [Google Scholar]
- 59.Ashwell JD. The many paths to p38 mitogen-activated protein kinase activation in the immune system. Nat Rev Immunol. 2006;6:532–540. doi: 10.1038/nri1865. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Oligonucleotide Sequences, Antibody and Inhibitor used in this study. (PPTX 62 kb) (PPTX 61 kb)
Table S2. TCGA Clinical data. (XLSX 59 kb) (XLSX 58 kb)
Table S3. Candidates selecting process & GO analysis result. (XLSX 49 kb)
Table S4. P2RX6 IHC analysis of tissues microarray. (XLSX 10 kb) (XLSX 9 kb)
Table S5. Clinical characteristics of 238 RCC patients according to P2RX6 expression levels. (PPTX 43 kb) (PPTX 42 kb)
Table S6. P2RX6 m6A in plain text from. (XLSX 15 kb) (XLSX 14 kb)
Table S7. Correlation between METTL14 and P2RX6. (XLSX 12 kb) (XLSX 11 kb)
Figure S1. Candidates selecting and P2RX6 gene bioinformatics characteristic. A P2RX6 gene selecting process flowchart. B ADRA2A gene expression in different stages. C Kaplan-Meier analysis for ADRA2A mRNA expression in RCC patients. D P2RX6 gene information on http://www.oncolnc.org/ and Kaplan-Meier analysis for P2RX6 mRNA expression in RCC patients (**P = 0.00175). (TIF 1943 kb)
Figure S2. ATP effects on migration/invasion of OS-RC-2 cells and validating P2RX6 gene function in SW839 cell line. A In vitro wound-healing motility assay with 3, 10, 30uM ATP treatment in OS-RC-2 cells. B Transwell invasion assays were performed with 3, 10, 30uM ATP treatment in OS-RC-2 cells. C Quantitative analysis for Fig. S1B. D qRT-PCR assay validation of P2RX6 mRNA level knocking-down efficiency and E WB validation of P2RX6 protein level knocking-down efficiency when knocking down using sh-P2RX6#1 in SW839 cells. F Migration assay after using sh-P2RX6#1 in SW839 cells treated with ATP, PLKO.1-vector as control. G Quantitative analysis for Fig. S2 F.H Transwell assay were performed after using sh-P2RX6#1 in SW839 cells treated with ATP, PLKO.1-vector as control. I Quantitative analysis for Fig. S2 H. * indicated P < 0.05. And ** indicated P < 0.01, *** indicated P < 0.001. (TIF 2967 kb)
Figure S3. Excluding MMP13 as downstream target gene. A Correlation analysis for P2RX6 and MMP13 from TCGA database. B WB validation of MMP13 protein level after treating with ATP. (TIF 786 kb)
Figure S4. Excluding RBM15B and FTO as target methylated gene. A RBM15B has lower expression in tumor tissues and B lower expression could result in a worse OS, and C has a positive correlation with P2RX6 expression in TCGA database. D FTO has higher expression in tumor tissues and E survival analysis has no significantly difference between two groups. F FTO has a negative correlation with P2RX6 expression (TIF 939 kb)
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.