Abstract
Background
Rhodotorula toruloides is a promising platform organism for production of lipids from lignocellulosic substrates. Little is known about the metabolic aspects of lipid production from the lignocellolosic sugar xylose by oleaginous yeasts in general and R. toruloides in particular. This study presents the first proteome analysis of the metabolism of R. toruloides during conversion of xylose to lipids.
Results
Rhodotorula toruloides cultivated on either glucose or xylose was subjected to comparative analysis of its growth dynamics, lipid composition, fatty acid profiles and proteome. The maximum growth and sugar uptake rate of glucose-grown R. toruloides cells were almost twice that of xylose-grown cells. Cultivation on xylose medium resulted in a lower final biomass yield although final cellular lipid content was similar between glucose- and xylose-grown cells. Analysis of lipid classes revealed the presence of monoacylglycerol in the early exponential growth phase as well as a high proportion of free fatty acids. Carbon source-specific changes in lipid profiles were only observed at early exponential growth phase, where C18 fatty acids were more saturated in xylose-grown cells. Proteins involved in sugar transport, initial steps of xylose assimilation and NADPH regeneration were among the proteins whose levels increased the most in xylose-grown cells across all time points. The levels of enzymes involved in the mevalonate pathway, phospholipid biosynthesis and amino acids biosynthesis differed in response to carbon source. In addition, xylose-grown cells contained higher levels of enzymes involved in peroxisomal beta-oxidation and oxidative stress response compared to cells cultivated on glucose.
Conclusions
The results obtained in the present study suggest that sugar import is the limiting step during xylose conversion by R. toruloides into lipids. NADPH appeared to be regenerated primarily through pentose phosphate pathway although it may also involve malic enzyme as well as alcohol and aldehyde dehydrogenases. Increases in enzyme levels of both fatty acid biosynthesis and beta-oxidation in xylose-grown cells was predicted to result in a futile cycle. The results presented here are valuable for the development of lipid production processes employing R. toruloides on xylose-containing substrates.
Electronic supplementary material
The online version of this article (10.1186/s13068-019-1478-8) contains supplementary material, which is available to authorized users.
Keywords: Rhodotorula toruloides, Proteome, Xylose, Lipid production
Background
Successful replacement of fossil diesel fuels with biological oils currently faces a number of challenges such as competing technologies, including electric vehicles, as well as unsustainable biological oil production practices such as clearing rainforests for palm oil cultivation [1, 2]. There is also an increasing competition for lipids from the food sector due to a growing population, increasing standards of human nutrition, and sustainability concerns. Lipids are required in animal feeds, for instance oils from wild pelagic fish are used to feed fish in aquaculture. So-called “single cell oils” (SCOs) are lipids derived from microorganisms, which represent a potentially more sustainable source of lipids to partially replace fish-derived oils [3]. Some microorganisms can produce lipids from lignocellulose-based substrates. The five-carbon (C5) monosaccharide xylose is the second-most abundant sugar in plant biomass and its utilization by microorganisms is regarded as a challenge to reach a sustainable valorization of plant biomass and commercialization of lignocellulosic fuels and chemicals [4]. Some oleaginous yeasts can convert xylose to lipids up to more than 20% of its dry cell weight: 22% Rhodotorula glutinis [5], 58% Trichosporon fermentans [6], 37% Cryptococcus curvatus [7], 33% Cryptococcus albidus [8], 42% Rhodotorula toruloides [9–11]. Cultivation on xylose in combination with glucose was also investigated using Lipomyces starkeyi with 61% lipids of dry cell weight (DCW) produced [12], while engineering of Yarrowia lipolytica strain enabled it to assimilate xylose [13, 14]. Microbial production of lipids from xylose was furthermore investigated in filamentous fungi: Cunninghamella echinulata (lipids constituted 58% DCW), Mortierella isabellina (lipids constituted 66% DCW) [15].
Establishment of sustainable lucrative microbial lipid production in industrial scale includes: (i) use of complex substrate (generated from plant biomass and containing a mixture of C6 and C5- sugars and lignocellulose-derived inhibitors); (ii) energy-consuming aerated cultivation; (iii) production of oil with high value compositional fatty acid (FA) profile including n − 3 long-chain polyunsaturated FA; (iv) production of value-added by-products. The basidiomycete yeast R. toruloides is relatively fast growing, can assimilate pentoses [11], is resistant to lignocellulosic inhibitors [16], has a beneficial FA composition [9] including n − 3 linolenic acid, and produces carotenoids [17]. R. toruloides is one of the most promising species for lipid production with reported lipid yields of up to 65.5% DCW under high cell density cultivation with low-nitrogen feeding [18].
Rhodotorula toruloides was originally applied in 1980 as a production organism for industrial-scale production of oil substitutes to replace cocoa butter [19]. The mono-unsaturated 18-carbon FA oleic acid (C18:1 n − 9) is the major FA and is accumulated to ~ 55% of total FA in R. toruloides Y4. Less than half of this amount, ~ 20% composes palmitic acid (C16:0, saturated). Stearic (C18:0) and linoleic (C18:2 n − 6) acid can each reach up to 10% of total FA [11, 18, 20]. A number of R. toruloides strains have been subjected to genome sequencing [21–25], genetic engineering protocols are established [26–30] and the yeast is recognized as a novel platform organism for production of oleochemicals [31, 32]. A number of cultivation conditions of R. toruloides have therefore been examined, e.g. growth on mixed substrates [33] or different cultivation modes on lignocellulosic substrates [34, 35] and lipid production at sulfate [36] or phosphate-limitation [37]. Improved genetically modified strains of R. toruloides for conversion of biomass have been generated [38].
The molecular physiology of R. toruloides during lipid production from different sugar substrates have been investigated using both proteomics [39, 40] and transcriptomics [25, 41, 42] approaches. Meanwhile, the molecular physiology during conversion of xylose into lipids in R. toruloides and in oleaginous yeasts in general has been poorly investigated. Xylose assimilation involves the enzymes NAD(P)H-dependent xylose reductase (XR) and NADH-dependent xylitol dehydrogenase (XDH). XR reduces xylose to xylitol, which is then oxidized by XDH to xylulose. Xylulose is subsequently phosphorylated by xylulose kinase before entering the pentose phosphate pathway (PPP). R. toruloides also possesses the enzyme phosphoketolase, which cleaves xylulose-5-phosphate to acetyl phosphate and glyceraldehyde-3-phosphate [43]. Unlike glucose assimilation, xylose assimilation requires NADPH due to the initial reduction of xylose into xylitol by XR. The cellular NADPH pool can be replenished by a number of pathways including PPP, the malic enzyme (ME) transhydrogenase cycle or the cytoplasmatic NADP+-dependent isocitrate dehydrogenase. While xylose utilization by oleaginous yeasts is poorly understood, a number of studies have investigated xylose metabolism in ethanol-producing yeasts: e.g. in engineered Saccharomyces cerevisiae [44], Schefferomyces stipitis [45], and Ogataea polymorpha [46]. These studies reported that the carbon source affected levels of proteins involved in e.g. glucose repression, sugar transport, gluconeogenesis and amino acid catabolism. In the current study, comparative whole-proteome analysis was employed to study lipid formation in R. toruloides cultivated on either glucose or xylose as sole carbon source under conditions of nitrogen limitation.
Results
Batch cultivation of Rhodotorula toruloides
Growth, sugar consumption and lipid production of R. toruloides in medium containing either glucose or xylose as a carbon source were monitored in batch cultivations (Fig. 1). The C/N ratio was set to 75 in order to provide sufficient carbon surplus while averting inhibitory effect of nitrogen limitation. Sugar concentration was set at 40 g/l, similar to levels obtained in lignocellulose hydrolysate [47]. Fermentations were performed at 25 °C to minimize energy input, which is beneficial for industrial processes [48]. Cell growth was monitored by optical density (OD) and DCW measurements every 8 h. Samples for lipid and protein analysis were taken at early exponential phase (8 or 16 h), late exponential (16 or 40 h) and lipid production phase (64 or 96 h), with the later sampling-points referring to xylose, as indicated in Fig. 1.
Fig. 1.
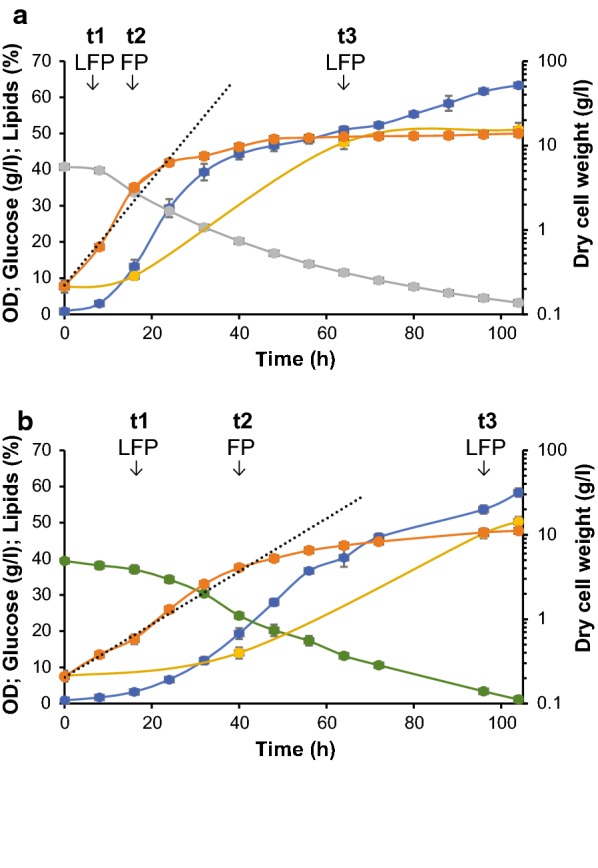
Batch cultivation of R. toruloides in chemically defined medium containing either glucose (a) or xylose (b). Cultivations of R. toruloides in a total volume of 0.5 l medium were performed in triplicate at pH 5.5 and 25 °C in 0.7–l bioreactors. Figure shows OD (blue), DCW (orange), glucose (gray), xylose (green), lipids (yellow), DCW trendline (interrupted gray line). Sample points for fatty acids profile (F), lipid classes (L) and proteome analysis (P) at early exponential (t1), late exponential (t2) and lipid production (t3) phase are all indicated. Mean values of three independent cultivation experiments are shown with error bars indicating one standard deviation
The maximum growth rate of R. toruloides on glucose was almost twice that of xylose, which is also reflected in an uptake rate of glucose during exponential phase that is roughly double the xylose uptake rate (Table 1). The exponential growth phase on glucose lasted approximately 24 h compared to 40 h for xylose, after which the observed increase in DCW and OD slowed down (Fig. 1). The xylose level was lower than the glucose level after 72 h, even though the growth rate on xylose was lower. Possibly, nitrogen was depleted in xylose-based medium later than in the medium containing glucose [49–51]. Growth on xylose resulted in lower final biomass yield and concentration as compared to glucose (Table 1, Fig. 1). This could indicate a higher ATP maintenance requirement of xylose-grown cells during the longer growth phase as compared to growth on glucose.
Table 1.
Physiological parameters for batch cultivation of R. toruloides CBS 14 in minimal medium containing either glucose or xylose
| Glucose | Xylose | |
|---|---|---|
| Lag phase (h) | 8 | 16 |
| Expontential phase (h) | 54 | 80 |
| μmax (h−1) | 0.14 | 0.07 |
| Final DCW (g/l) | 13.8 ± 0.13 | 11.1 ± 0.1 |
| Starting sugar (g/l) | 40.8 ± 0.3 | 39.4 ± 0.25 |
| Residual sugar (g/l) | 3.19 ± 0.81 | 1.1 ± 1 |
| Final YX/S (gDW/g glucose−1) | 0.36 ± 0.004 | 0.28 ± 0.003 |
| Lipid content at t0 (%)a | 7.59 ± 1.54 | |
| Lipid content at t2 (%)a | 10.58 ± 1.00 | 13.99 ± 1.5 |
| Lipid content at t3 (%)a | 47.51 ± 1.81 | 47.05 ± 1.4 |
| Final YL/S (g/g) | 0.19 ± 0.01 | 0.15 ± 0.009 |
at0 represents the beginning of cultivation, t2—late exponential growth phase, t3—lipid accumulation phase
Significant lipid accumulation was observed at the end of the exponential growth phase, which was attributed to nitrogen depletion from the culture medium as commonly observed for cultivations of oleaginous yeasts [19]. Although sugar levels decreased further after the late exponential phase, particularly in glucose (Fig. 1, cf. glucose at 64 and 96 h), this only resulted in a slight increase in lipid content. The final lipid content under both growth conditions was approximately 51%, while final lipid yields were slightly higher on glucose, following the same trend as the biomass yield (Table 1). Interestingly, the higher lipid yield on glucose is comparable to yields reported previously [11], although the previous study reported similar biomass yields between glucose and xylose, which was not observed in the present study. In addition, the previous study reported lower lipid content of xylose-grown cells compared to glucose-grown cells. This disparity between studies could be explained by differences in the C/N ratio used for cultivation [cf. 65 [11] and 75 (our study)] or elemental medium composition [52]. Meanwhile, R. toruloides CCT 0783 showed similar effects of glucose and xylose on the biomass yield, while lipid yields remained unaffected [34, 53].
Lipid classes
Samples were taken for lipid class profiling to investigate whether the distribution of lipid classes changed during the cultivation (Fig. 1, Table 2). Monoacylglycerols (MAGs) were detected in the early exponential phase. MAGs are not commonly reported in yeast lipidomes [20, 54, 55], although it has been observed in cultivations of R. toruloides NP11 [56] and Y. lipolytica [57]. The presence of MAGs in R. toruloides could be connected with diacylglycerol lipase activity, which cleaves one FA chain from diacylglycerols (DAGs) [58], as Rhodotorula spp. are used in the food industry as sources of lipases [59]. Furthermore, a high proportion of free FA was observed in R. toruloides compared to other species where free FA are commonly below 5% of total lipids [60, 61]. MAGs were only detected during early exponential phase of the R. toruloides cultivations, while the percentage of DAGs and free FA increased towards late exponential phase. This indicates that the absence of MAGs in late stage is not necessarily due to absence of lipase activity in R. toruloides.
Table 2.
Lipid class composition of R. toruloides presented as percentage of total identified lipids
| t1 | t3 | |||||||
|---|---|---|---|---|---|---|---|---|
| Glucose | Xylose | Glucose | Xylose | |||||
| % of lipid classes | mg/gDCW | % of lipid classes | mg/gDCW | % of lipid classes | mg/gDCW | % of lipid classes | mg/gDCW | |
| Phospholipids | 15.09 ± 0.34 | 13.71 | 16.27 ± 1.5 | 17.56 | 4.11 ± 0.34 | 19.53 | 3.9 ± 0.21 | 18.37 |
| Monoacylglycerols | 1.02 ± 0.33 | 0.93 | 1.35 ± 0.5 | 1.46 | nd | nd | nd | nd |
| Diglycerols | 8.35 ± 0.7 | 7.59 | 7.88 ± 0.26 | 8.50 | 4.9 ± 0.3 | 23.28 | 4.02 ± 0.36 | 18.93 |
| Sterols | 14.11 ± 0.34 | 12.82 | 14.61 ± 0.79 | 15.76 | 6.49 ± 0.3 | 30.83 | 5.67 ± 0.15 | 26.71 |
| Free fatty acids | 38.15 ± 0.9 | 34.66 | 40.77 ± 3.1 | 43.99 | 11.9 ± 0.3 | 56.54 | 10.80 ± 0.25 | 50.87 |
| Triacylglycerols | 23.82 ± 1.7 | 21.64 | 19.39 ± 4.5 | 20.92 | 72.64 ± 0.76 | 345.11 | 75.61 ± 0.63 | 356.12 |
| TAG/PL | 1.58 | 1.19 | 17.66 | 19.37 | ||||
| Total lipid content | 9.085 ± 1 | 10.79 ± 1 | 47.51 ± 1.8 | 47.1 ± 1.4 | ||||
Samples were collected during early exponential phase (t1) and lipid production phase (t3). Total lipids were not determined at t1, but instead the values reported here are the mean total lipid contents between the first two total lipid measurements (Fig. 1), assuming a linear increase over time. Absolute levels of lipid classes were determined from the mean percentages of lipid classes and mean value of total lipid content
DCW dry cell weight, TAG triacylglycerols, PL phospholipids
Lipid accumulation in R. toruloides is mainly in the form of neutral lipids as triacylglycerols (TAGs), irrespective of the carbon source (Table 2). The percentage of sterols was also approximately twofold higher by the time the cells entered stationary phase compared to early exponential phase. In contrast, phospholipid proportions remained essentially unchanged, which was reflected by a pronounced increase in TAGs/phospholipids ratio compared to S. cerevisiae [54] or Y. lipolytica [60]. The high TAGs/phospholipid ratio in R. toruloides suggested that the accumulated neutral lipids are stored in giant lipid droplets rather than the cell membrane. The R. toruloides lipid droplet proteome was shown to drive a formation of giant lipid droplets in this organism [56]. Even at the start of cultivation, the observed TAGs/phospholipid ratio in R. toruloides was higher than that what has previously been reported for S. cerevisiae [54, 62] and Y. lipolytica [60, 63, 64].
No major differences in the proportions of lipid classes in R. toruloides were observed between cultivations containing either xylose and glucose. The high similarity in final relative content of TAGs in cells cultivated in medium containing glucose and xylose fits well with absence of differences in final total lipid content in examined cells.
Fatty acid profiles
Samples were taken at different R. toruloides growth phases for lipid analysis to determine whether any changes in lipid classes and content correlates with changes in the FA profile (Fig. 1, Table 3). The dominant FA under all conditions was oleic acid. The final FA compositions agreed somewhat with previous reports [11, 18]. The content of linoleic acid and stearic acid has previously been reported to be roughly 13% for each of the total FA content in R. toruloides strains CBS 14 and Y4 [11, 18]. However, in the present study we found that the amount of linoleic acid was almost 1.7 times lower than stearic acid. This difference can be due to differences in media composition and cultivation conditions: R. toruloides cultivations were carried out at 25 °C in the present study compared to 30 °C in previous reports [11, 18]. Higher proportions of polyunsaturated FA have previously been reported to accumulate in R. glutinis when this yeast was cultivated at 25 °C as compared to 30 °C [65]. Small amounts of heptadecenoic (C17:1) and heptadecanoic (C17:0) acid could also be detected in some samples (Table 3). Production of heptadecenoic acid has previously been reported in the two closely related species Rhodotorula babjevae and Rhodotorula glutinis [66, 67].
Table 3.
Fatty acid profiles of R. toruloides during early exponential phase and lipid accumulation phase following batch cultivation in medium containing either glucose or xylose (presented as percentage of total identified, average ± standard deviation)
| t0 | t1 | t2 | t3 | ||||
|---|---|---|---|---|---|---|---|
| Glc = Xyl | Xylose | Glucose | Xylose | Glucose | Xylose | Glucose | |
| C14:0 | 0.98 ± 0.25 | 1.03 ± 0.26 | 0.91 ± 0.03 | 1.23 ± 0.09 | 1.25 ± 0.05 | 1.28 ± 0.04 | 1.1 ± 0.07 |
| C16:0 | 14.35 ± 1.9 | 17.91 ± 2.5 | 17.16 ± 0.01 | 26.63 ± 0.22 | 25.76 ± 0.24 | 26.73 ± 0.26 | 25.55 ± 0.3 |
| C16:1 | 1.39 ± 0.28 | 0.96 ± 0.1 | 0.51 ± 0.02 | 0.98 ± 0.05 | 0.53 ± 0.01 | 1.00 ± 0.04 | 0.57 ± 0.05 |
| C17:0 | n.d. | 0.76 ± 0.13 | n.d. | n.d. | n.d. | n.d. | n.d. |
| C17:1 | 0.75 ± 0.18 | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. |
| C18:0 | 3.84 ± 0.44 | 5.55 ± 0.57 | 7.63 ± 0.5 | 8.11 ± 0.28 | 9.31 ± 0.19 | 7.73 ± 0.22 | 7.83 ± 0.56 |
| C18:1 (n − 9) | 66.66 ± 2.28 | 46.27 ± 3.05 | 36.99 ± 1.3 | 45.26 ± 0.5 | 47.23 ± 0.61 | 45.07 ± 0.7 | 47.25 ± 1.06 |
| C18:2 (n − 6) | 6.62 ± 0.33 | 20.36 ± 4.16 | 23.56 ± 0.71 | 13.51 ± 0.69 | 11.58 ± 0.5 | 13.83 ± 0.57 | 13.06 ± 0.68 |
| C18:3 (n − 3) | 3.38 ± 0.86 | 5.74 ± 1.9 | 11.25 ± 0.98 | 2.78 ± 0.15 | 2.94 ± 0.22 | 2.99 ± 0.19 | 3.01 ± 0.25 |
| C24:0 | 0.9 ± 0.01 | 1.15 ± 0.13 | 0.66 ± 0.05 | 0.73 ± 0.05 | 0.76 ± 0.02 | 0.67 ± 0.05 | 0.91 ± 0.13 |
| Avg length | 17.4891 | 17.5942 | 17.4104 | 17.3038 | 17.3546 | 17.3084 | 17.3586 |
| Avg desaturation | 0.9218 | 1.0517 | 1.1837 | 0.816 | 0.7974 | 0.827 | 0.8297 |
Samples were collected at the beginning of cultivation (t0) as well as during early exponential phase (t1), late exponential phase (t2) and lipid production phase (t3). Polyunsaturated fatty acids were counted for the number of unsaturations according to [18]
n.d. not detected
The second major FA detected was either palmitic or linoleic acid, and this was growth phase dependent even while the average chain length remained relatively unchanged. Some differences in the FA chain lengths were observed compared to previous reports of the Y4 strain of R. toruloides [18], although the dominance of oleic acid, the average chain length and saturation levels were in agreement with published data.
Carbon source-specific changes in lipid profiles of R. toruloides were observed only at early exponential phase, where C18 FA were more saturated in xylose compared to glucose. Wiebe et al. observed a higher desaturation of C18 FA and an increase in palmitic acid proportion in response to xylose in R. toruloides CBS 14 [11], which was not observed in this study. Combined, it shows that medium composition and cultivation conditions (especially temperature) more significantly affect FA composition than carbon source and growth phase.
Effect of growth phases on proteome composition of R. toruloides
As glucose and xylose assimilation proceed through distinct catabolic pathways, we anticipated that alternative carbon sources result in adjustments of the R. toruloides proteome. To investigate this, we examined relative proteome differences at different growth phases (Fig. 1). A total of 2995 proteins (corresponding to 36.7% of the genome) could consistently be identified in at least two replicates of each tested condition (Additional file 1). Early and late exponential and stationary (lipid production) phases were compared separately for glucose and xylose cultures, to evaluate the effect of growth phase.
Of the 2995 identified proteins, 795 and 902 were differentially expressed when comparing early exponential and with stationary phase for xylose and glucose cultures, respectively (Additional files 2: Fig. S1, 3, 4). A core of 251 upregulated and 168 downregulated proteins were found when comparing lipid accumulation to early exponential phase in cells grown on both glucose and xylose, whereas a large part of the proteins were regulated specifically by carbon substrate (Additional file 2: Fig. S2).
Gene set analysis (GSA) performed to identify systemic changes revealed that genes involved in ribosome biogenesis and function (Additional file 2: Fig. S1) were downregulated during the cultivation on both sugars. These observed changes are more likely associated with the growth phase rather than lipogenesis, as transcriptional downregulation of ribosomes is connected to inactivation of the target of rapamycin (TOR) signaling pathway upon nitrogen depletion [68]. A similar response has been reported for the R. toruloides strain NP11 as well as for the oleaginous yeast Y. lipolytica [69].
Proteins showing the largest differential expression during lipid accumulation phase on both glucose and xylose were identified. Proteins increasing five to eightfold in abundance included urea transporter, NCS2 allantoate transporter, major facilitator superfamily (MFS) transporter of unknown specificity, siderochrome-iron transporter, amine oxidase, perilipin-like protein and aspartic proteinase (Additional files 2: Fig. S1, 3, 4). The observed upregulation of proteins involved in the assimilation of nitrogen agreed well with the expected cessation of the nitrogen catabolite repression (NCR) under conditions of nitrogen limitation [25]. Regarding genes that are downregulated at the later growth phase, little consensus could be found between the two carbon sources (Additional files 2: Fig. S1, 3, 4).
Carbon source-dependent proteome changes
Comparison between carbon source for each time point resulted in the identification of 457, 230 and 181 proteins for early exponential, late exponential and lipid accumulation stage, respectively (Additional files 2: Fig. S3, 5, 6, 7). It was observed that 43 upregulated and 6 downregulated proteins were common between all time points in xylose as compared to glucose-grown cells (Additional file 2: Fig. S4). Additionally, 104 proteins were regulated similarly in response to xylose at early and late exponential phase, 5 proteins at late exponential and lipid accumulation phase and 7 proteins at early exponential and lipid accumulation phase. A majority of differentially expressed proteins were growth phase-specific between xylose and glucose.
Only limited conclusions could be drawn from gene set analysis of these results (Fig. 2): proteins involved in ribosomal function were among the top upregulated proteins in early exponential phase on glucose, which is consistent with the observed higher growth rate on this carbon sources (Fig. 2, Table 1).
Fig. 2.
GO term enrichment analysis of differentially expressed proteins in R. toruloides grown on glucose or xylose during early exponential growth phase (a), late exponential growth phase (b), lipid accumulation phase (c). For each GO term showing significant change (rank score of ≥ 5), the direction and significance of the relative changes in protein levels are shown, together with the total number of proteins within each GO term
Proteins involved in transport (MFS transporters), metabolism of C5 sugar (xylitol dehydrogenase, l-iditol 2-dehydrogenase) and NADPH generation (NADPH-dependent alcohol dehydrogenase) were among the top upregulated proteins on xylose at all time points (3- to 11-fold increase), while there were little similarities among the top downregulated proteins. Mitochondrial ME was among the top downregulated proteins in xylose-grown yeast at early stationary phase.
As the untargeted GSA did not elucidate systemic transcriptional responses, we proceeded with targeted analysis of the largest observed transcriptional changes complemented with metabolic pathways and genes suspected to be affected by different carbon sources.
Sugar transport
The expression levels of various sugar transporters show a carbon source-specific regulation. Thirteen putative sugar transporters were identified in the proteome dataset, of which eight (Rhto_01630, Rhto_03448, Rhto_07444, Rhto_06801, Rhto_06080, Rhto_01923, Rhto_00228, Rhto_07706) had significantly elevated levels (5.6- to 338-fold) for xylose cultivations as compared to glucose, suggesting their involvement in xylose transport (Fig. 3 and Additional file 8). Conversely, three transporters (Rhto_04266, Rhto_00984, Rhto_06016) with 16-fold elevated levels in cells fermenting glucose are likely involved in glucose transport. The relatively high level of the transporter Rhto_06080 irrespective of carbon source during exponential phase and Rhto_06016 and Rhto_01923 at stationary phase might indicate that these proteins function as unspecific sugar transporters. The transporter Rhto_01923 might have a high affinity to both sugars, while Rhto_06016 to xylose only. Lower levels of sugar transporters induced by glucose rather than xylose suggest high efficiency of expressed transporters.
Fig. 3.

Heatmap showing relative abundance levels (log-transformed value of label-free quantification) of proteins involved in sugar transport during batch cultivation of R. toruloides in minimal medium containing either glucose or xylose
Xylose assimilation
Proteins predicted to be involved in xylose assimilation were highly upregulated in the presence of xylose, in particular xylitol dehydrogenase (Fig. 4 and Additional file 8). Four NADPH-dependent XRs where strongly upregulated (6.8- to 68.5-fold) at all three time points during xylose cultivations. Besides xylose reductase (Rhto_03963), also aldo/keto reductases (Rhto_00641, Rhto_06555, Rhto_00602) are therefore likely involved in xylose assimilation.
Fig. 4.
The figure shows a map of the central carbon metabolism of R. toruloides augmented with log2-transformed fold change of protein levels relative to the level at glucose early exponential phase. For most proteins, colour scale ranges from − 4 (blue) to + 4 (red). For proteins with very large changes (*), the scale ranges from − 4 (blue) to + 10 (red). More gene and protein names, annotations and expression abundance data are included in Additional file 8
Apart from induction of proteins involved in xylose assimilation, the absence of glucose from the xylose-containing medium also resulted in a strong upregulation (64- to 1000-fold) of enzymes involved in assimilation of alternative carbon sources (beta-glucosidase Rhto_05582; mandelate racemase/muconate lactonizing enzyme Rhto_05818) (Additional files 5, 6, 7 and Fig. 4).
Glycolytic proteins and TCA cycle
Differences in expression levels of glycolytic and tricarboxylic acid (TCA) cycle proteins were less pronounced than those of sugar transporters, although this might be expected due to dominance of post-translational regulation of glycolytic enzymes in yeast [70, 71]. Apart from upregulation of phosphoglycerate mutase during growth on xylose, no significant differences were observed in the levels of enzymes involved in glycolysis, PPP and phosphoketolase metabolic routes (Additional file 8, Fig. 4). From these data it was therefore not possible to infer whether xylulose-5-phosphate is metabolized via phosphoketolase or the non-oxidative steps of PPP and further glycolysis.
Oleaginous yeasts under nitrogen limitation are known to transfer citrate from the mitochondrion into the cytoplasm thereby reducing the flow of metabolites through the TCA cycle. In contrast to previous work [25] where higher C/N ratios were used, only aconitate hydrotase and succinate dehydrogenase showed moderate decreased levels during lipid accumulation in the present study.
NADPH generation
Each addition of a two-carbon unit during fatty acid elongation requires one molar equivalent of NADPH. The observed upregulation of PPP enzymes during lipid accumulation on both sugar substrates would suggest that NADPH generation in R. toruloides occurs mainly through PPP (Fig. 4 and Additional file 8). Such a scenario was further supported by the lack of significant differences in the levels of NAD(P)-dependent isocitrate dehydrogenase or cytosolic ME. Meanwhile, the upregulation of two other enzymes of the transhydrogenation cycle, i.e. pyruvate carboxylase and l-malate dehydrogenase (2.1-fold), suggests that this path has some contribution into regeneration of NADPH during lipogenesis.
In addition, we identified five aldehyde dehydrogenases (Rhto_05838, Rhto_04425, Rhto_04310, Rhto_06724, Rhto_04543) that might use NAD(P) as cofactor. A number of these dehydrogenases increased their expression during lipid accumulation (Additional file 8), while also some response to carbon source could be observed. Protein Rhto_01922 (potentially mitochondrial), annotated as alcohol dehydrogenase (NADPH), was among the top upregulated proteins (832-fold) on xylose at all time points.
Oleaginicity
The levels of ATP-citrate lyase (Rhto_03915), acetyl-CoA carboxylase (Rhto_02004) as well as both subunits of FA synthase (FAS; Rhto_02139 and Rhto_02032) were upregulated in R. toruloides during conversion of both xylose and glucose into lipids (Additional file 8). Apart from cytoplasmic ME, our results agree with earlier studies showing upregulation of key enzymes of lipid production machinery during lipogenesis [25, 39].
Levels of pyruvate decarboxylase decreased twofold during lipid accumulation on both carbon sources while levels of alcohol dehydrogenase (ADH, Rhto_03062) decreased sharply by 588- and 1024-fold on glucose and xylose, respectively. This suggests rechannelling of carbon towards more energy-efficient carbon utilization, the pyruvate dehydrogenase—citrate synthase—ATP-citrate lyase path rather than to pyruvate decarboxylase—acetaldehyde dehydrogenase—acetyl-CoA synthetase path.
Beta-oxidation
The R. toruloides proteomic data suggested that peroxisomal beta-oxidation triggered by nitrogen starvation was more pronounced in cells cultivated on xylose-based substrate than on glucose. The levels of enzymes involved in peroxisomal oxidation (Rhto_03890, Rhto_03776, Rhto_05407, Rhto_07118, Rhto_00300, Rhto_06581) was 8- to 16-fold higher in xylose than in glucose-grown cells already during the exponential growth phase (Additional file 8). The lipid production phase coincided with a decrease in levels of some enzymes of peroxisomal oxidation (Rhto_00300, Rhto_04298) in xylose-grown cells while levels of others (Rhto_02517, Rhto_05520) increased in glucose-grown cells. These differences might indicate that TOR signaling is affected by carbon source in line with [72].
Similar trends were observed for mitochondrial beta-oxidation: 8–128 times upregulation of the enzymes (Rhto_04971, Rhto_06738, Rhto_01625, Rhto_00397) happened in xylose-grown cells already in the beginning of cultivation, however, in glucose-grown cells 2.3–8 times upregulation (Rhto_04971, Rhto_05797, Rhto_00397) occurred only at lipid accumulation phase.
Simultaneous upregulation of the two inverse pathways beta-oxidation and FA biosynthesis in cells grown on xylose during induction of lipogenesis is expected to result in a futile cycle, which may partially explain the lower final biomass yields on xylose as compared to glucose.
TAG synthesis
The majority of enzymes involved in TAG synthesis did not change in protein levels during R. toruloides cultivation with the exception of a 4.1-fold increase in the level of glycerol-3-phosphate dehydrogenase (Rhto_02273) on both carbon sources as well as a twofold increase in the level of the phosphatidic acid biosynthetic 1-acyl-sn-glycerol-3-phosphate acyltransferase (Rhto_06718) in glucose-grown cells (Additional file 8). An increase in TAG lipase (Rhto_00361) level was observed during the lipid accumulation phase on xylose, that is probably counteracted by increase of perilipins, which protect lipids in droplets from degradation. The observed increase of TAG coincided with elevated absolute levels of DAGs and free FAs during stationary phase. However, MAGs were only detected during early exponential phase.
Phospholipids
Observed differences in levels of phospholipid biosynthetic enzymes suggest that phospholipid biosynthesis is affected by the choice of carbon source. Xylose-grown cells displayed a 2.5-fold increase in phosphatidate cytidylyltransferase (Rhto_01718) levels (converting phosphatidic acid to CDP-DAG), twofold upregulation of phosphatidyl-N-methylethanolamine N-methyltransferase (Rhto_03783, forming phosphatidyl-N-dimethylethanolamine) and 2.3-fold downregulation of CDP-diacylglycerol-inositol 3-phosphatidyltransferase (Rhto_02741, synthesizing phosphatidyl-1D-myo-inositol) during lipid production phase (Additional file 8). In glucose-treated cells the level of phosphatidylserine decarboxylase (Rhto_03399, forming phosphatidylethanolamine) was upregulated 16 times. Meanwhile, analysis of lipid classes did not reveal any difference in proportion of phospholipids in R. toruloides cells in response to carbon source.
Mevalonate pathway
The mevalonate pathway is involved in the synthesis of dolichol, carotenoids and sterols from for acetyl-CoA and ATP and is, therefore, in competition with FA biosynthesis. Some enzymes of this pathway were regulated differentially in R. toruloides in response to carbon source. For instance, during the lipid production phase in glucose-treated cells, the level of acetyl-CoA C-acetyltransferase (Rhto_02048) decreased 3.5-fold. Xylose-grown cells displayed a twofold increase in the enzyme geranylgeranyl diphosphate synthase (Rhto_02504, Additional file 8). Phytoene dehydrogenase (Rhto_04602) was upregulated in stationary phase cells irrespective of carbon source.
Amino acid biosynthesis
While we did not anticipate that carbon source would affect other metabolic pathways, we did observe changes in e.g. amino acid biosynthesis. A 2.5-fold decrease in the levels of leucine biosynthetic enzymes was observed in both xylose- and glycose-grown cells upon the initiation of the lipid production phase (Additional file 8), while the downregulation of enzymes involved in the biosynthesis of proline (proline synthetase associated protein Rhto_04810) and leucine (branched-chain-amino acid aminotransferase Rhto_05760; Rhto_08045; acetolactate synthase Rhto_03298; acetolactate synthase small subunit Rhto_03988) was detected in xylose- as compared to glucose-grown cells at the exponential phase. This might be related to the decrease of protein biosynthesis in xylose-treated cells.
The downregulation of enzymes involved in the synthesis of leucine (3-isopropylmalate dehydratase Rhto_07040; branched-chain-amino acid aminotransferase Rhto_05760; acetolactate synthase Rhto_03298; ketol-acid reductoisomerase Rhto_04566) and the interconversion of methionine and cysteine (cystathionine beta-lyase Rhto_02122) was observed during lipid production in glucose-grown R. toruloides cells compared to earlier growth phase. Lipid accumulation in Y. lipolytica has previously been shown to be accompanied by downregulation of leucine biosynthesis [69].
Oxidative stress response
Levels of proteins involved in glutathione metabolism increased in R. toruloides cells cultivated on xylose but also during lipid production, possibly as a response to oxidative stress induced under these conditions. Levels of lactoylglutathione lyase (Rhto_06289), glutathione peroxidase (Rhto_00225), glutathione S-transferases (Rhto_03923, Rhto_00450) and NAD(P)H-dependent S-(hydroxymethyl) glutathione dehydrogenase (Rhto_03559) increased during exponential phase in xylose-grown cells compared to glucose-grown cells (Additional file 8). Levels of lactoylglutathione lyase (Rhto_06289), glutathione S-transferases (Rhto_00450, Rhto_03923) and glutathione peroxidase (Rhto_00225) also increased during the lipid production phase in xylose-grown cells relative to exponential growth phase. The levels of glutathione peroxidase (Rhto_00225) and glutathione S-transferase (Rhto_03923) also increased during the lipid accumulation phase in glucose-grown R. toruloides cells. Formate dehydrogenase (Rhto_06042) was detected among top upregulated enzymes during cultivation in xylose-based medium. Formate dehydrogenase is involved in the oxidation of formic acid to carbon dioxide as the final step of glutathione-dependent formaldehyde detoxification [73].
Activation of enzymes involved in carotenoid biosynthesis during lipid accumulation might be connected with oxidative stress response. Phytoene dehydrogenase (Rhto_04602) was upregulated during lipid accumulation as compared to exponential growth phase in R. toruloides cells grown on both substrates (Additional file 8). Additionally, we detected increased levels of geranylgeranyl diphosphate synthase (Rhto_02504) in glucose-grown cells.
Nitrogen starvation response
A number of parallel changes were observed in both glucose- and xylose-grown R. toruloides cells in response to nitrogen starvation. The quicker depletion of nitrogen from glucose-containing medium meant that differences in levels of proteins involved in the nitrogen starvation response were first observed in glucose-grown cells during exponential phase. Therefore, the levels of amino acid transmembrane transporters (Rhto_01344, Rhto_00398, Rhto_07341) and proteins involved in amino acid degradation (branched-chain amino acid aminotransferase Rhto_05760; tyrosine aminotransferase Rhto_01136; aromatic amino acid aminotransferase I Rhto_07034; l-asparaginase Rhto_07901) appeared lower in xylose-grown R. toruloides cells compared to glucose-grown cells during early exponential phase as the xylose-grown cells had not yet fully exhausted their nitrogen source and hence not activated their nitrogen starvation response (Additional file 8). Further depletion of nitrogen from the medium during the cultivation in xylose eventually triggered a nitrogen starvation response that was consistent between the two carbon sources, such as increased levels of glutamate dehydrogenase, allantoicase, urea and amino acid transporters and autophagy-related proteins. Nitrogen starvation induces autophagic processes, liberating intracellular space for growth of lipid droplet [25, 53]. We observed upregulation of several autophagic proteins (Rhto_05541, Rhto_01208, Rhto_07813) in R. toruloides cells at lipid accumulation as compared to exponential phase on both substrates (Additional file 8). Additionally, at this stage in xylose-grown cells, the levels of autophagy-related protein Rhto_06526, Rhto_02366 and Rhto_07138 were upregulated. Interestingly, several proteins involved in autophagy (Rhto_06526, Rhto_01636, Rhto_05541) were upregulated in cells cultivated in xylose as compared to glucose at early exponential phase. This might indicate stress response to the absence of glucose.
Discussion
A number of previous proteomic and transcriptomic studies have investigated metabolism of xylose in ethanol-producing yeasts [44–46], while this study of R. toruloides represents the first proteomic investigation of an oleaginous yeast during conversion of xylose to lipids. As the major constituent of hemicellulose, xylose is the second-most abundant monosaccharide in nature after glucose [4].
Many notable differences were observed between the proteomes of cells cultivated in medium containing either glucose or xylose. Glucose- and xylose-grown cells had similar lipid content and profiles, but xylose-grown cells displayed lower biomass yields, in agreement with previous observations [34]. The growth rate of xylose-grown R. toruloides cells in exponential phase was roughly half of the growth rate on glucose. The observed decrease in the levels of ribosomal proteins and other proteins associated with translation of mRNA in xylose-grown cells was the most prominent change in proteome between the two sugar substrates and is likely rather connected with differences in the growth rates.
The lower biomass yield and growth rate are possibly an effect of higher ATP demands by R. toruloides when grown on xylose. In the presence of excess sugar, and in particular during lipid accumulation, beta-oxidation is a futile cycle, and as proteins involved in beta-oxidation were upregulated during growth on xylose it is feasible that this futile cycle is underlying the higher ATP requirements. This is potentially an effect of absence of glucose, which would otherwise down-regulate beta-oxidation. Abolition of this pathway is, therefore, expected to have a significant beneficial impact on not only lipid formation but also on growth in excess of sugar with more pronounced effect on xylose as compared to glucose. Disruption of peroxisome biogenesis in Y. lipolytica has previously been reported to impair β-oxidation, which resulted in increased lipid accumulation from glucose [74]. However, another study showed that disruption of peroxisome biogenesis in R. toruloides had the opposite effect, namely a decreased lipid production in glucose medium [75].
There were indications of that sugar transport is the underlying reason for the observed differences in physiology between glucose- and xylose-grown R. toruloides cells. A number of candidate glucose- and xylose-specific transporters were identified from the R. toruloides proteome datasets. As assimilation on xylose required strongly increased levels of some transport proteins, while only a reduced growth rate could be obtained, it is suggested that the xylose transporters are less efficient than those responsible for glucose transport. It is possible that the applied protein extraction methods have a bias against membrane proteins; however, improving xylose transport could be a valuable strategy for further optimization of R. toruloides for use of lignocellulosic biomass.
We have detected expression of S. cerevisiae homologs of members of glucose catabolite repression pathway in R. toruloides. However, apart from the monosaccharide transporter, Rhto_06016 (homolog of SNF3), we did not find any homologs to S. cerevisiae proteins-members of glucose induction pathway in genome of R. toruloides, which in the absence of glucose represses proteins required for glucose assimilation, such as glucose transporters [76]. Our results showed that expression of some of glucose transporters was lower in xylose-grown cells. It remains unclear which mechanisms in R. toruloides provide function of glucose induction pathway. Possibly, release from strong inhibition of glucose-induced proteins can provide high fitness of microorganism in nutrient poor environment, such as soil and phyllosphere—natural habitats of R. toruloides [77].
Analysis of metabolic routes that can be used by R. toruloides to assimilate sugars did not elucidate a dominant pathway. As the main catabolic flux of glucose is funneled through glycolysis, this results in a lower flux through xylulose-5-P compared to catabolism of xylose. From this reduced availability of xylulose-5-P one might expect a downregulation of phosphoketolase during growth on glucose, however, its protein level showed no significant change as response to the carbon source. One possibility is that once cells pre-cultured on glucose were transferred to xylose medium, the xylose flux into cells was low enough that PPP enzymes levels did not have to be increased further to ensure efficient xylose catabolism.
The phosphoketolase reaction normally enables higher efficiency of carbon metabolism since bypasses the wasteful decarboxylation pyruvate to acetyl-CoA, which causes a loss of one third of the carbon substrate, while phosphoketolase can produce an acetyl residue directly from a C5-substrate. However, under conditions of excess of carbon and simultaneous nitrogen limitation, the carbon saving phosphoketolase reaction may become unnecessary, which could result in xylulose-5-P being redirected towards PPP. Such a scenario would also explain the decrease in phosphoketolase levels during the lipid production phase on both substrates since the NADPH-regenerating reactions of PPP occur upstream of xylulose-5-P and therefore phosphoketolase is not expected to be regulated by NADPH demand. It was speculated in a previous study that assimilation of xylose via phosphoketolase would enable slightly higher lipid yields when grown on xylose (0.34 g/g sugar) as compared to growth on glucose (0.32 g/g) [78]. It should be noted that partial assimilation of xylose via PPP would still be required for synthesis of essential nucleic acid precursors as well as regeneration of NADPH for lipogenesis.
NADPH generation for lipid synthesis was assumed to occur through PPP although the observed increase of the levels of transhydrogenase pathway enzymes indicates that ME could contribute to NADP+ reduction during stationary phase. Previous studies showed that in other oleaginous yeasts, Y. lipolytica and L. starkeyi PPP provides major contribution to NADPH pool during lipogenesis [79, 80]. It has been previously demonstrated, that overexpression of genes encoding native ME slightly increased lipid accumulation in R. toruloides [75]. Interestingly, an increase in levels of a putative NADP+-dependent alcohol dehydrogenase was observed, which suggests that oxidation of one or more alcohols into the corresponding aldehydes could contribute to supply of NADPH pool. Aldehyde dehydrogenase could also potentially generate NADPH, but the substrate specificity of this enzyme must first be determined. Lipid synthesis is an NADPH demanding process, but the absence of significant changes in the levels of NADPH-regenerating enzymes in the present study would suggest that NADPH availability is not limiting lipid accumulation in xylose-grown R. toruloides cells.
Additional metabolic processes displaying significant changes in protein levels between glucose- and xylose-grown R. toruloides cells included biosynthesis of amino acids and phospholipids as well as oxidative stress response. Similar effect of xylose on ethanol-producing yeasts has been reported previously [44–46]. Some of these effects, such as downregulation of leucine biosynthesis and upregulation of xylose transporters during growth on xylose were reminiscent of proteomic changes observed in ethanol-producing yeasts during cultivation on either xylose or glucose. The increased levels of two of the three enzymes involved in the glutathione-dependent formaldehyde detoxification pathway during cultivation in xylose-based medium was curious, as there was no obvious source of formaldehyde formation equivalent to what occurs during assimilation of methylated substrates such as methanol or methylated amines.
As the difference in R. toruloides growth rate were likely underlying many of the detected protein levels, it would be beneficial to perform continuous cultivations on both carbon sources to provide additional insight on the response on protein levels. Future studies should characterize proteome of R. toruloides cells with higher lipid content generated from xylose-based substrate. In addition, multicomponent lignocellulose-based medium could provide valuable information for further optimization of hemicellulose fermentation by R. toruloides. While proteomics measurements are instrumental to inform how transcriptional regulation affects metabolism, further augmenting with transcriptomics analysis could provide a higher depth of analysis due to its larger coverage of the total genome.
Conclusions
This study is the first investigation of proteome of oleaginous yeast R. toruloides during conversion of xylose into lipids. Our study showed two times lower maximum growth rate and sugar uptake rate of R. toruloides cells in xylose as compared to glucose-based substrate. Proteome analysis revealed significantly lower levels of ribosomal proteins and translation associated factors in xylose-grown cells as compared to glucose-grown cells. In addition, xylose-grown cells contained higher levels of enzymes involved in peroxisomal beta-oxidation and oxidative stress response and lower levels of enzymes involved in leucine biosynthesis compared to cells cultivated on glucose. The levels of enzymes involved in sugar transport and phospholipid biosynthesis differed in response to carbon source. In addition, the proteomic data reported here identifies several potential target sugar transporters (Rhto_01630, Rhto_03448, Rhto_07444, Rhto_06801, Rhto_06080, Rhto_01923) and enzymes involved in peroxisomal beta-oxidation (Rhto_03890, Rhto_03776, Rhto_05407, Rhto_07118, Rhto_00300, Rhto_06581) for genetic engineering to optimize xylose metabolism.
Methods
Strain and media
Rhodotorula toruloides CBS 14 was obtained from the Westerdijk Fungal Biodiversity Institute (Utrecht, the Netherlands) and stored at − 80 °C in 50% v/v glycerol. For cultivation experiments, frozen R. toruloides stocks were revived on solid YPD medium (20 g/l glucose, 20 g/l peptone and 10 g/l yeast extract, 16 g/l agar) and incubated for 3 days at 25 °C. R. toruloides was precultured in 200 ml YPD liquid medium in 500 ml Erlenmeyer flasks and incubated for 3 days at 25 °C on an orbital shaker at 150 rpm. The chemically defined cultivation medium used for growth experiments contained: 1.173 g/l (NH4)2SO4; 1.7 g/l yeast nitrogen base without amino acids or ammonium sulfate; 3 g/l KH2PO4; 0.5 g/l MgSO4–7H2O; 40 g/l carbon source (either xylose or glucose). The carbon/nitrogen (C/N) ratio of the medium was set to 75. 0.07% (v/v) polypropylene glycol was used to prevent adhesion and foaming of yeast cells to glass surfaces within the bioreactors.
Yeast cultivation
Batch cultivations were performed in 0.7–l bioreactors (Multifors 2, Infors-HTBottmingen, Switzerland). Cultivation experiments for each carbon source (glucose and xylose) for analysis of lipid accumulation were performed in triplicate. R. toruloides precultures were washed twice with a saline solution (9 g/l NaCl) and then used to inoculate the bioreactors to a final optical density at 600 nm (OD600) of approx. 1.0 in a total volume of 0.5 l chemically defined cultivation medium containing either glucose or xylose as carbon source. Batch cultivations were performed at pH 5.5 and 25 °C. The dissolved oxygen tension (DOT) was kept above 20 by regulating a stirring speed between 200 and 600 rpm at an aeration rate of 1 l/min. Samples for high-performance liquid chromatography (HPLC) analysis as well as OD600 and DCW measurements were taken every 8 h. Samples for lipid analysis were taken at early exponential phase, late exponential phase and early stationary phase for each cultivation experiment.
Analytical methods
Cell OD600 was measured using an Ultrospec 1100 pro spectrophotometer (GE Healthcare, Chicago IL). Yeast cell biomass was determined by cell dry weight measurement from 2 ml of culture samples. The samples were centrifuged at 17,000×g, washed twice with deionized water, incubated in an oven at 105 °C for 72 h and then weighed. Each DCW measurement was done in triplicate. HPLC measurements of ethanol, acetate and glycerol were performed as described previously [81]. Yeast lipids were extracted using the method described by Folch et al. [82] with some modifications [83]. Briefly, 100 mg of freeze-dried yeast cells were suspended in 1 ml 1 M HCl and soaked for 15 min followed by 1 h incubation at 75 °C. 5 ml of chloroform:methanol (2:1, v/v) was added to the sample tube, which was then vortexed for 15 min at maximum speed. The sample tube was centrifuged at 5000×g for 3 min at room temperature followed by the transfer of the lower lipid layer to a tube of known weight. The extraction was repeated with remaining upper layer of the sample. The lipid phases from the first and second extractions were pooled followed by evaporation of the solvent under N2 gas flow. The total amount of lipids was determined gravimetrically. Total lipid content was determined at the start of fermentation instead of at early exponential phase due to a lack of material.
Dried lipid samples were dissolved in 0.5 ml hexane and methylated with BF3 according to Appelqvist [84]. Methyl esters were analyzed by gas chromatography (GC) as described previously [83, 85]. The standard mixture 68A (Nu-Check, Elysian, MN) and retention times were used to identify FA. The double binding or unsaturation index (UI) was calculated as
Lipid class composition was determined according to Olsen and Henderson [86] with slight modifications. Samples were diluted to a final concentration of 1 g/l in hexane, and 5 µl of each sample was then applied with a CAMAG thin-layer chromatography (TLC) Sampler ATS4 (Camag, Switzerland) 2 cm from the base edge of the TLC plates (pre-coated with silica gel TLC plates (20 × 10 cm; Silicagel 60; 0.20 mm layer, Merck, Darmstadt, Germany) in 2 mm bands with an application speed of 250 nl/s. N2 was used as spray gas. All samples were applied in duplicate, and the distance between tracks was 9.8 mm. Separation of the lipid classes was executed with a CAMAG Automatic Developing Chamber 2 (ADC 2) (Camag, Switzerland). Lipid classes were separated using a hexane:diethyl ether:acetic acid (85:15:2; v/v/v) mobile phase. Afterwards, separation procedure plates were submerged in a solution of 3% (w/v) cupric acetate in 8% (v/v) phosphoric acid and then charred for 20 min at 140 °C. Quantitative analysis of the separated lipid classes was done by scanning the plates with a CAMAG TLC Scanner 3 (Camag, Switzerland). The scanning was performed at a speed of 20 mm/s and a data resolution of 100 μm/step, with a slit dimension of 6.00 × 0.45 mm at a wavelength of 350 nm. Lipid classes were identified by comparison to external standards (TLC 18-4A, Nu-Chek Prep, Elysian, USA; Ergosterol, PHR1512, Sigma-Aldrich, Sweden). The mode Savitsky-Golay 7 was used for data filtering. Manual baseline and peak correction were used when necessary.
Preparation of samples for proteome analysis
Samples for proteome analysis were taken at early exponential phase (8 or 16 h), late exponential (16 or 40 h) and lipid production phase (64 or 96 h), with the later sampling-points referring to xylose. Cell pellets were washed and frozen. Cell pellets were thawed and resuspended in 500 μl of a solution of 4% (w/v) SDS, 10 mM DTT and 100 mM Tris–HCl (pH 8). Resuspended cell pellets were then transferred to 2 ml FastPrep tubes containing ca 500 μl of a 3:1 (w/w) glass bead mixture of bead sizes 106 μm (Sigma-Aldrich, product no. G4649) and 425–600 μm (Sigma-Aldrich, product no. G8772), respectively. Cells were disrupted using a FastPrep homogenizer (MP Biomedicals, CA) with three 45-s cycles at full speed interspersed with 5 min cooling on ice between runs. The tubes were then centrifuged for 15 min at 20,000×g, and the supernatant was transferred to fresh tubes. An upper red layer of the supernatant could be observed at this point and was carefully avoided. The bottom layer was used to generate tryptic peptides according to the Filter-Aided Sample Prep (FASP) protocol [87]. Briefly, 50 μl of each FastPrep extract was incubated for 10 min at 95 °C, and thereafter processed according to the FASP protocol (in Microcon YM-30). Tryptic peptides were desalted using STAGE spin tips, and redissolved in 30 μl loading solution consisting of 2% (v/v) acetonitrile and 0.05% (w/v) trifluoroacetic acid in water. Approximate peptide concentrations were determined using a NanoDrop 2000 instrument (ThermoFisher Scientific, MA) and the A205 protocol [88].
LC–MS/MS analysis
Approximately, 1 μg of tryptic peptides was loaded onto a trap column (Acclaim PepMap100, C18, 5 µm, 100 Å, 300 µm i.d. × 5 mm) and then backflushed onto a 50 cm × 75 μm analytical column (Acclaim PepMap RSLC C18, 2 μm, 100 Å, 75 μm i.d. × 50 cm, nanoViper) for liquid chromatography-mass spectrometry (LC–MS/MS) analysis. A 120 min gradient from 4 to 40% solution B (80% [v/v] acetonitrile, 0.1% [v/v] formic acid) was used for separation of the peptides, at a flow rate of 300 nl/min. The Q-Exactive mass spectrometer was set up as follows (Top12 method): a full scan (300–1600 m/z) at R = 70.000 was followed by (up to) 12 MS2 scans at R = 35,000, using an NCE setting of 28 eV. Singly charged precursors and precursors with charge > 5 were excluded for MS/MS. Dynamic exclusion was set to 20 s.
Bioinformatics
The raw MS data were analyzed using MaxQuant [89] version 1.5.3.40 and proteins were identified and quantified using the MaxLFQ algorithm [90]. The data were searched against the UniProt R. toruloides proteome (UP000016926; 8138 sequences) supplemented with common contaminants such as human keratin and bovine serum albumin. In addition, reversed sequences of all protein entries were concatenated to the database to allow for estimation of false discovery rates. The tolerance levels used for matching to the database were 4.5 ppm for MS and 20 ppm for MS/MS. Trypsin/P was used as digestion enzyme and 2 miscleavages were allowed. Carbamidomethylation of cysteine was set as fixed modification and protein N-terminal acetylation, oxidation of methionines and deamidation of asparagines and glutamines were allowed as variable modifications. All identifications were filtered to achieve a protein false discovery rate (FDR) of 1%. Perseus version 1.5.2.6 [91] were used for data analysis, and the quantitative values were log2-transformed, and grouped according to carbon source. Proteins were only considered detected if they were present in at least two replicates on one of the two carbon sources. Missing values were imputed by drawing numbers from a normal distribution centred around the level of detection of the mass spectrometer. Student’s t tests including permutation-based FDR (p < 0.05) were used to detect significant differences between groups and volcano plots were constructed by plotting the negative logarithm of the p values against the fold difference between two groups in a scatter plot, as shown in Additional file 2: Fig. S1. Differential expression between groups was assigned if proteins had a significant t test and showed ≥ 2-fold difference. Due to increased variation in growth dynamics at early stationary phase, we used the p values (p ≤ 0.05) instead of permutation-based FDR for t-test truncation to detect differences in cells cultivated with glucose or xylose as sole carbon source at the late stage of cultivation. Individual proteins were referenced by their UniProt accession numbers as described for the R. toruloides NP11 genome annotation [25]. Gene set analysis was performed as described earlier [60] using available functional genome annotation [25].
Additional files
Additional file 1. Complete list of proteins detected during batch cultivation of R. toruloides in minimal medium containing either glucose or xylose and their relative expression values.
Additional file 2: Fig. S1. GO term enrichment analysis and volcano plots of differentially expressed proteins in R. toruloides during either exponential growth phase or lipid production phase in medium containing either glucose (A) or xylose (B). For each GO term showing significant change (rank score of ≥ 5), the direction and significance of the relative changes in protein levels are shown, together with the total number of proteins within each GO term. Fig. S2. Venn diagram of differentially expressed proteins in R. toruloidesduring either exponential growth phase or lipid production phase in medium containing either glucose (yellow circle) or xylose (blue circle). Number in red or blue colour indicates up or downregulated proteins, respectively, in cells during lipid accumulation as compared to exponential phase. Fig. S3. Volcano plots of differentially expressed proteins in R. toruloides cultivated on either glucose or xylose during early exponential growth phase (A), late exponential growth phase (B), or lipid accumulation phase (C). Fig. S4. Venn diagram of differentially expressed proteins in R. toruloidescultivated on either glucose or xylose during early exponential growth phase (t1, yellow circle), late exponential growth phase (t2, brown circle), or lipid accumulation phase (t3, green cycle). Number in red or blue colour indicates up or downregulated proteins, respectively, in cells grown on xylose as compared to cells grown on glucose.
Additional file 3. Relative levels of differentially expressed proteins in glucose-grown R. toruloides during either exponential cultivated phase or lipid production phase.
Additional file 4. Relative levels of differentially expressed proteins in xylose-grown R. toruloides during either exponential cultivated phase or lipid production phase.
Additional file 5. Relative levels of differentially expressed proteins in R. toruloides cultivated on either glucose or xylose during early exponential growth phase.
Additional file 6. Relative levels of differentially expressed proteins in R. toruloides cultivated on either glucose or xylose during late exponential growth phase.
Additional file 7. Relative levels of differentially expressed proteins in R. toruloides grown on either glucose or xylose during lipid accumulation phase.
Additional file 8. Relative levels of proteins involved in central carbon metabolism of R. toruloides.
Acknowledgements
We thank Dr. Tomas Linder for proofreading the manuscript.
Abbreviations
- PPP
pentose phosphate pathway
- SCOs
single cell oils
- DCW
dry cell weight
- FA
fatty acid
- XR
xylose reductase
- XDH
xylitol dehydrogenase
- ME
malic enzyme
- OD
optical density
- MAGs
monoacylglycerols
- DAGs
diacylglycerols
- TAGs
triacylglycerol
- GSA
gene set analysis
- TOR
target of rapamycin
- FAS
fatty acid synthase
- ADH
alcohol dehydrogenase
- MFS
major facilitator superfamily
- NCR
nitrogen catabolite repression
- TCA
tricarboxylic acid
- HPLC
high-performance liquid chromatography
- GC
gas chromatography
- TLC
thin-layer chromatography
- LC–MS
liquid chromatography–mass spectrometry
Authors’ contributions
IAT provided major contributions to study design and performed a major part of laboratory work, data analysis and writing the manuscript. JBr, JB and SS performed chemical analysis, data evaluation and was involved in manuscript writing. NM performed parts of the laboratory work and data analysis. MSk and MOA performed major parts of laboratory work. JN provided a major contribution for study design, data analysis and final manuscript writing. MS and EK coordinated the project and provided major contributions to study design, data analysis and final manuscript writing. All authors read and approved the final manuscript.
Funding
Financial support was provided by a strategic grant of The Swedish Research Council for Environment, Agricultural Sciences and Spatial Planning (Formas), Grant Number 213-2013-80 and Formas Mobility Grant 2016-00767.
Availability of data and materials
All the data analyzed in this study are included in this manuscript and its additional files. The proteomics data has been deposited to the ProteomeXchange consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository [92] with the dataset identifier PXD012332.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Ievgeniia A. Tiukova, Email: tiukova@chalmers.se
Jule Brandenburg, Email: Jule.Brandenburg@slu.se.
Johanna Blomqvist, Email: johanna.blomqvist@nmbu.no.
Sabine Sampels, Email: sabine.sampels@slu.se.
Nils Mikkelsen, Email: nils.mikkelsen@slu.se.
Morten Skaugen, Email: morten.skaugen@nmbu.no.
Magnus Ø. Arntzen, Email: magnus.arntzen@nmbu.no
Jens Nielsen, Email: nielsenj@chalmers.se.
Mats Sandgren, Email: mats.sandgren@slu.se.
Eduard J. Kerkhoven, Email: eduardk@chalmers.se
References
- 1.McDonald BC, de Gouw JA, Gilman JB, Jathar SH, Akherati A, Cappa CD, et al. Volatile chemical products emerging as largest petrochemical source of urban organic emissions. Science. 2018;359(6377):760–764. doi: 10.1126/science.aaq0524. [DOI] [PubMed] [Google Scholar]
- 2.Wallington TJ, Anderson JE, Kurtz EM, Tennison PJ. Biofuels, vehicle emissions, and urban air quality. Faraday Discuss. 2016;189:121–136. doi: 10.1039/C5FD00205B. [DOI] [PubMed] [Google Scholar]
- 3.Blomqvist J, Pickova J, Tilami SK, Sampels S, Mikkelsen N, Brandenburg J, et al. Oleaginous yeast as a component in fish feed. Sci Rep. 2018;8(1):15945. doi: 10.1038/s41598-018-34232-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sanchez Nogue V, Karhumaa K. Xylose fermentation as a challenge for commercialization of lignocellulosic fuels and chemicals. Biotechnol Lett. 2015;37(4):761–772. doi: 10.1007/s10529-014-1756-2. [DOI] [PubMed] [Google Scholar]
- 5.Easterling ER, French WT, Hernandez R, Licha M. The effect of glycerol as a sole and secondary substrate on the growth and fatty acid composition of Rhodotorula glutinis. Bioresour Technol. 2009;100(1):356–361. doi: 10.1016/j.biortech.2008.05.030. [DOI] [PubMed] [Google Scholar]
- 6.Zhu LY, Zong MH, Wu H. Efficient lipid production with Trichosporon fermentans and its use for biodiesel preparation. Bioresour Technol. 2008;99(16):7881–7885. doi: 10.1016/j.biortech.2008.02.033. [DOI] [PubMed] [Google Scholar]
- 7.Evans CT, Scragg AH, Ratledge C. Regulation of citrate efflux from mitochondria of oleaginous and non-oleaginous yeasts by long-chain fatty acyl-CoA esters. Eur J Biochem. 1983;132(3):617–622. doi: 10.1111/j.1432-1033.1983.tb07408.x. [DOI] [PubMed] [Google Scholar]
- 8.Hansson L, Dostálek M. Influence of cultivation conditions on lipid production by Cryptococcus albidus. Appl Microbiol Biotechnol. 1986;24(1):12–18. doi: 10.1007/BF00266278. [DOI] [Google Scholar]
- 9.Jagtap SS, Rao CV. Production of D-arabitol from D-xylose by the oleaginous yeast Rhodosporidium toruloides IFO0880. Appl Microbiol Biotechnol. 2018;102(1):143–151. doi: 10.1007/s00253-017-8581-1. [DOI] [PubMed] [Google Scholar]
- 10.Moreton RS. Physiology of lipid accumulation yeast. In: Moreton RS, editor. Single cell oil. London: Longman; 1988. pp. 1–32. [Google Scholar]
- 11.Wiebe MG, Koivuranta K, Penttila M, Ruohonen L. Lipid production in batch and fed-batch cultures of Rhodosporidium toruloides from 5 and 6 carbon carbohydrates. BMC Biotechnol. 2012;12:26. doi: 10.1186/1472-6750-12-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhao X, Kong X, Hua Y, Feng B, Zhao Z. Medium optimization for lipid production through co-fermentation of glucose and xylose by the oleaginous yeast Lipomyces starkeyi. Eur J Lipid Sci Technol. 2008;110(5):405–412. doi: 10.1002/ejlt.200700224. [DOI] [Google Scholar]
- 13.Ledesma-Amaro R, Nicaud JM. Metabolic engineering for expanding the substrate range of Yarrowia lipolytica. Trends Biotechnol. 2016;34(10):798–809. doi: 10.1016/j.tibtech.2016.04.010. [DOI] [PubMed] [Google Scholar]
- 14.Li H, Alper HS. Enabling xylose utilization in Yarrowia lipolytica for lipid production. Biotechnol J. 2016;11(9):1230–1240. doi: 10.1002/biot.201600210. [DOI] [PubMed] [Google Scholar]
- 15.Fakas S, Papanikolaou S, Batsos A, Galiotou-Panayotou M, Mallouchos A, Aggelis G. Evaluating renewable carbon sources as substrates for single cell oil production by Cunninghamella echinulata and Mortierella isabellina. Biomass Bioenerg. 2009;33(4):573–580. doi: 10.1016/j.biombioe.2008.09.006. [DOI] [Google Scholar]
- 16.Hu C, Zhao X, Zhao J, Wu S, Zhao ZK. Effects of biomass hydrolysis by-products on oleaginous yeast Rhodosporidium toruloides. Bioresour Technol. 2009;100(20):4843–4847. doi: 10.1016/j.biortech.2009.04.041. [DOI] [PubMed] [Google Scholar]
- 17.Lee JJ, Chen L, Shi J, Trzcinski A, Chen WN. Metabolomic profiling of Rhodosporidium toruloides grown on glycerol for carotenoid production during different growth phases. J Agric Food Chem. 2014;62(41):10203–10209. doi: 10.1021/jf502987q. [DOI] [PubMed] [Google Scholar]
- 18.Li Y, Zhao Z, Bai F. High-density cultivation of oleaginous yeast Rhodosporidium toruloides Y4 in fed-batch culture. Enzyme Microbial Technol. 2007;41(3):312–317. doi: 10.1016/j.enzmictec.2007.02.008. [DOI] [Google Scholar]
- 19.Ratledge C, Wynn J. The biochemistry and molecular biology of lipid accumulation in oleaginous microorganisms. Adv Appl Microbiol. 2002;51:1–52. doi: 10.1016/S0065-2164(02)51000-5. [DOI] [PubMed] [Google Scholar]
- 20.Wei Y, Siewers V, Nielsen J. Cocoa butter-like lipid production ability of non-oleaginous and oleaginous yeasts under nitrogen-limited culture conditions. Appl Microbiol Biotechnol. 2017;101(9):3577–3585. doi: 10.1007/s00253-017-8126-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hu J, Ji L. Draft genome sequences of Rhodosporidium toruloides strains ATCC 10788 and ATCC 10657 with compatible mating types. Genome Announc. 2016;4(2):e00098-16. doi: 10.1128/genomeA.00098-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kumar S, Kushwaha H, Bachhawat AK, Raghava GP, Ganesan K. Genome sequence of the oleaginous red yeast Rhodosporidium toruloides MTCC 457. Eukaryot Cell. 2012;11(8):1083–1084. doi: 10.1128/EC.00156-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morin N, Calcas X, Devillers H, Durrens P, Sherman DJ, Nicaud JM, et al. Draft genome sequence of Rhodosporidium toruloides CECT1137, an oleaginous yeast of biotechnological interest. Genome Announc. 2014;2(4):e00641-14. doi: 10.1128/genomeA.00641-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sambles C, Middelhaufe S, Soanes D, Kolak D, Lux T, Moore K, et al. Genome sequence of the oleaginous yeast Rhodotorula toruloides strain CGMCC 2.1609. Genom Data. 2017;13:1–2. doi: 10.1016/j.gdata.2017.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhu Z, Zhang S, Liu H, Shen H, Lin X, Yang F, et al. A multi-omic map of the lipid-producing yeast Rhodosporidium toruloides. Nat Commun. 2012;3:1112. doi: 10.1038/ncomms2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Coradetti ST, Pinel D, Geiselman GM, Ito M, Mondo SJ, Reilly MC, et al. Functional genomics of lipid metabolism in the oleaginous yeast Rhodosporidium toruloides. Elife. 2018;7:e32110. doi: 10.7554/eLife.32110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koh CM, Liu Y, Du M, Ji L. Molecular characterization of KU70 and KU80 homologues and exploitation of a KU70-deficient mutant for improving gene deletion frequency in Rhodosporidium toruloides. BMC Microbiol. 2014;14:50. doi: 10.1186/1471-2180-14-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lin X, Wang Y, Zhang S, Zhu Z, Zhou YJ, Yang F, et al. Functional integration of multiple genes into the genome of the oleaginous yeast Rhodosporidium toruloides. FEMS Yeast Res. 2014;14(4):547–555. doi: 10.1111/1567-1364.12140. [DOI] [PubMed] [Google Scholar]
- 29.Liu Y, Koh CM, Sun L, Hlaing MM, Du M, Peng N, et al. Characterization of glyceraldehyde-3-phosphate dehydrogenase gene RtGPD1 and development of genetic transformation method by dominant selection in oleaginous yeast Rhodosporidium toruloides. Appl Microbiol Biotechnol. 2013;97(2):719–729. doi: 10.1007/s00253-012-4223-9. [DOI] [PubMed] [Google Scholar]
- 30.Zhang S, Skerker JM, Rutter CD, Maurer MJ, Arkin AP, Rao CV. Engineering Rhodosporidium toruloides for increased lipid production. Biotechnol Bioeng. 2016;113(5):1056–1066. doi: 10.1002/bit.25864. [DOI] [PubMed] [Google Scholar]
- 31.Park YK, Nicaud JM, Ledesma-Amaro R. The engineering potential of Rhodosporidium toruloides as a workhorse for biotechnological applications. Trends Biotechnol. 2018;36(3):304–317. doi: 10.1016/j.tibtech.2017.10.013. [DOI] [PubMed] [Google Scholar]
- 32.Yaegashi J, Kirby J, Ito M, Sun J, Dutta T, Mirsiaghi M, et al. Rhodosporidium toruloides: a new platform organism for conversion of lignocellulose into terpene biofuels and bioproducts. Biotechnol Biofuels. 2017;10:241. doi: 10.1186/s13068-017-0927-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bommareddy RR, Sabra W, Maheshwari G, Zeng AP. Metabolic network analysis and experimental study of lipid production in Rhodosporidium toruloides grown on single and mixed substrates. Microb Cell Fact. 2015;14:36. doi: 10.1186/s12934-015-0217-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bonturi N, Matsakas L, Nilsson R, Christakopoulos P, Miranda E, Berglund K, et al. Single cell oil producing yeasts Lipomyces starkeyi and Rhodosporidium toruloides: selection of extraction strategies and biodiesel property prediction. Energies. 2015;8:5040–5052. doi: 10.3390/en8065040. [DOI] [Google Scholar]
- 35.Fei Q, O’Brien M, Nelson R, Chen X, Lowell A, Dowe N. Enhanced lipid production by Rhodosporidium toruloides using different fed-batch feeding strategies with lignocellulosic hydrolysate as the sole carbon source. Biotechnol Biofuels. 2016;9:130. doi: 10.1186/s13068-016-0542-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu S, Zhao X, Shen H, Wang Q, Zhao ZK. Microbial lipid production by Rhodosporidium toruloides under sulfate-limited conditions. Bioresour Technol. 2011;102(2):1803–1807. doi: 10.1016/j.biortech.2010.09.033. [DOI] [PubMed] [Google Scholar]
- 37.Wu S, Hu C, Jin G, Zhao X, Zhao ZK. Phosphate-limitation mediated lipid production by Rhodosporidium toruloides. Bioresour Technol. 2010;101(15):6124–6129. doi: 10.1016/j.biortech.2010.02.111. [DOI] [PubMed] [Google Scholar]
- 38.Diaz T, Fillet S, Campoy S, Vazquez R, Vina J, Murillo J, et al. Combining evolutionary and metabolic engineering in Rhodosporidium toruloides for lipid production with non-detoxified wheat straw hydrolysates. Appl Microbiol Biotechnol. 2018;102(7):3287–3300. doi: 10.1007/s00253-018-8810-2. [DOI] [PubMed] [Google Scholar]
- 39.Liu H, Zhao X, Wang F, Li Y, Jiang X, Ye M, et al. Comparative proteomic analysis of Rhodosporidium toruloides during lipid accumulation. Yeast. 2009;26(10):553–566. doi: 10.1002/yea.1706. [DOI] [PubMed] [Google Scholar]
- 40.Shi J, Feng H, Lee J, Ning Chen W. Comparative proteomics profile of lipid-cumulating oleaginous yeast: an iTRAQ-coupled 2-D LC-MS/MS analysis. PLoS ONE. 2013;8(12):e85532. doi: 10.1371/journal.pone.0085532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Qi F, Zhao X, Kitahara Y, Li T, Ou X, Du W, et al. Integrative transcriptomic and proteomic analysis of the mutant lignocellulosic hydrolyzate-tolerant Rhodosporidium toruloides. Eng Life Sci. 2017;17:249–261. doi: 10.1002/elsc.201500143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang Y, Zhang S, Zhu Z, Shen H, Lin X, Jin X, et al. Systems analysis of phosphate-limitation-induced lipid accumulation by the oleaginous yeast Rhodosporidium toruloides. Biotechnol Biofuels. 2018;11:148. doi: 10.1186/s13068-018-1134-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Evans CT, Ratledge C. Induction of xylulose-5-phosphate phosphoketolase in a variety of yeasts grown ond-xylose: the key to efficient xylose metabolism. C Arch Microbiol. 1984;139:48–52. doi: 10.1007/BF00692711. [DOI] [Google Scholar]
- 44.Salusjarvi L, Kankainen M, Soliymani R, Pitkanen JP, Penttila M, Ruohonen L. Regulation of xylose metabolism in recombinant Saccharomyces cerevisiae. Microb Cell Fact. 2008;7:18. doi: 10.1186/1475-2859-7-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yuan T, Ren Y, Meng K, Feng Y, Yang P, Wang S, et al. RNA-Seq of the xylose-fermenting yeast Scheffersomyces stipitis cultivated in glucose or xylose. Appl Microbiol Biotechnol. 2011;92(6):1237–1249. doi: 10.1007/s00253-011-3607-6. [DOI] [PubMed] [Google Scholar]
- 46.Kim OC, Suwannarangsee S, Oh D, Kim S, Seo J, Kim C, et al. Transcriptome analysis of xylose metabolism in the thermotolerant methylotrophic yeast Hansenula polymorpha. Bioprocess Biosyst Eng. 2013;36:1509. doi: 10.1007/s00449-013-0909-3. [DOI] [PubMed] [Google Scholar]
- 47.Tiukova IA, de Barros Pita W, Sundell D, Haddad Momeni M, Horn SJ, Stahlberg J, et al. Adaptation of Dekkera bruxellensis to lignocellulose-based substrate. Biotechnol Appl Biochem. 2014;61(1):51–57. doi: 10.1002/bab.1145. [DOI] [PubMed] [Google Scholar]
- 48.Karlsson H, Ahlgren S, Sandgren M, Passoth V, Wallberg O, Hansson PA. A systems analysis of biodiesel production from wheat straw using oleaginous yeast: process design, mass and energy balances. Biotechnol Biofuels. 2016;9:229. doi: 10.1186/s13068-016-0640-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Evans CT, Ratledge C. Influence of nitrogen metabolism on lipid accumulation by Rhodosporidium toruloides CBS 14. J Gen Microbiol. 1984;130:1705–1710. [Google Scholar]
- 50.Shen H, Jin G, Hu C, Gong Z, Bai F, Zhao ZK. Effects of dilution rate and carbon-to-nitrogen ratio on lipid accumulation by Rhodosporidium toruloides under chemostat conditions. Sheng Wu Gong Cheng Xue Bao. 2012;28(1):56–64. [PubMed] [Google Scholar]
- 51.Turcotte G, Kosaric N. The effect of C/N ratio on lipid production by Rhodosporidium toruloides ATCC 10788. Biotechnol Lett. 1989;11(9):637. doi: 10.1007/BF01025273. [DOI] [Google Scholar]
- 52.Ismail KS, Sakamoto T, Hasunuma T, Zhao XQ, Kondo A. Zinc, magnesium, and calcium ion supplementation confers tolerance to acetic acid stress in industrial Saccharomyces cerevisiae utilizing xylose. Biotechnol J. 2014;9(12):1519–1525. doi: 10.1002/biot.201300553. [DOI] [PubMed] [Google Scholar]
- 53.Bonturi N, Crucello A, Viana AJC, Miranda EA. Microbial oil production in sugarcane bagasse hemicellulosic hydrolysate without nutrient supplementation by a Rhodosporidium toruloides adapted strain. Process Biochem. 2017;57:16–25. doi: 10.1016/j.procbio.2017.03.007. [DOI] [Google Scholar]
- 54.Ejsing CS, Sampaio JL, Surendranath V, Duchoslav E, Ekroos K, Klemm RW, et al. Global analysis of the yeast lipidome by quantitative shotgun mass spectrometry. Proc Natl Acad Sci USA. 2009;106(7):2136–2141. doi: 10.1073/pnas.0811700106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Klose C, Surma MA, Gerl MJ, Meyenhofer F, Shevchenko A, Simons K. Flexibility of a eukaryotic lipidome—insights from yeast lipidomics. PLoS ONE. 2012;7(4):e35063. doi: 10.1371/journal.pone.0035063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhu Z, Ding Y, Gong Z, Yang L, Zhang S, Zhang C, et al. Dynamics of the lipid droplet proteome of the Oleaginous yeast Rhodosporidium toruloides. Eukaryot Cell. 2015;14(3):252–264. doi: 10.1128/EC.00141-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Beopoulos A, Verbeke J, Bordes F, Guicherd M, Bressy M, Marty A, et al. Metabolic engineering for ricinoleic acid production in the oleaginous yeast Yarrowia lipolytica. Appl Microbiol Biotechnol. 2014;98(1):251–262. doi: 10.1007/s00253-013-5295-x. [DOI] [PubMed] [Google Scholar]
- 58.Van Heusden GP, Nebohacova M, Overbeeke TL, Steensma HY. The Saccharomyces cerevisiae TGL2 gene encodes a protein with lipolytic activity and can complement an Escherichia coli diacylglycerol kinase disruptant. Yeast. 1998;14(3):225–232. doi: 10.1002/(SICI)1097-0061(199802)14:3<225::AID-YEA215>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 59.Strehaiano P, Ramon-Portugal F, Taillandier P. Yeasts as biocatalysts. In: Querol A, Fleet G, editors. Yeasts in food and beverages. Berlin: Springer; 2006. [Google Scholar]
- 60.Kerkhoven EJ, Pomraning KR, Baker SE, Nielsen J. Regulation of amino-acid metabolism controls flux to lipid accumulation in Yarrowia lipolytica. NPJ Syst Biol Appl. 2016;2:16005. doi: 10.1038/npjsba.2016.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rattray JB, Schibeci A, Kidby DK. Lipids of yeasts. Bacteriol Rev. 1975;39(3):197–231. doi: 10.1128/br.39.3.197-231.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Casanovas A, Sprenger RR, Tarasov K, Ruckerbauer DE, Hannibal-Bach HK, Zanghellini J, et al. Quantitative analysis of proteome and lipidome dynamics reveals functional regulation of global lipid metabolism. Chem Biol. 2015;22(3):412–425. doi: 10.1016/j.chembiol.2015.02.007. [DOI] [PubMed] [Google Scholar]
- 63.Beopoulos A, Mrozova Z, Thevenieau F, Le Dall MT, Hapala I, Papanikolaou S, et al. Control of lipid accumulation in the yeast Yarrowia lipolytica. Appl Environ Microbiol. 2008;74(24):7779–7789. doi: 10.1128/AEM.01412-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hein EM, Hayen H. Comparative lipidomic profiling of Saccharomyces cerevisiae and four other hemiascomycetous yeasts. Metabolites. 2012;2(1):254–267. doi: 10.3390/metabo2010254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Granger LM, Perlot P, Goma G, Pareilleux A. Kinetics of growth and fatty acid production of Rhodotorula glutinis. Appl Microbiol Biotechnol. 1992;37(1):13–17. doi: 10.1007/BF00174195. [DOI] [Google Scholar]
- 66.Brandenburg J, Poppele I, Blomqvist J, Puke M, Pickova J, Sandgren M, et al. Bioethanol and lipid production from the enzymatic hydrolysate of wheat straw after furfural extraction. Appl Microbiol Biotechnol. 2018;102(14):6269–6277. doi: 10.1007/s00253-018-9081-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rezanka T, Kolouchova I, Sigler K. Precursor directed biosynthesis of odd-numbered fatty acids by different yeasts. Folia Microbiol (Praha). 2015;60(5):457–464. doi: 10.1007/s12223-015-0388-9. [DOI] [PubMed] [Google Scholar]
- 68.Hardwick JS, Kuruvilla FG, Tong JK, Shamji AF, Schreiber SL. Rapamycin-modulated transcription defines the subset of nutrient-sensitive signaling pathways directly controlled by the Tor proteins. Proc Natl Acad Sci USA. 1999;96(26):14866–14870. doi: 10.1073/pnas.96.26.14866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kerkhoven EJ, Kim YM, Wei S, Nicora CD, Fillmore TL, Purvine SO, et al. Leucine Biosynthesis Is Involved in Regulating High Lipid Accumulation in Yarrowia lipolytica. Biol. 2017;8(3):e00857-17. doi: 10.1128/mBio.00857-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Daran-Lapujade P, Rossell S, van Gulik WM, Luttik MA, de Groot MJ, Slijper M, et al. The fluxes through glycolytic enzymes in Saccharomyces cerevisiae are predominantly regulated at posttranscriptional levels. Proc Natl Acad Sci USA. 2007;104(40):15753–15758. doi: 10.1073/pnas.0707476104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.van den Brink J, Canelas AB, van Gulik WM, Pronk JT, Heijnen JJ, de Winde JH, et al. Dynamics of glycolytic regulation during adaptation of Saccharomyces cerevisiae to fermentative metabolism. Appl Environ Microbiol. 2008;74(18):5710–5723. doi: 10.1128/AEM.01121-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhang J, Vaga S, Chumnanpuen P, Kumar R, Vemuri GN, Aebersold R, et al. Mapping the interaction of Snf1 with TORC1 in Saccharomyces cerevisiae. Mol Syst Biol. 2011;7:545. doi: 10.1038/msb.2011.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Popov VO, Lamzin VS. NAD(+)-dependent formate dehydrogenase. Biochem J. 1994;301(Pt 3):625–643. doi: 10.1042/bj3010625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Xue Z, Sharpe PL, Hong SP, Yadav NS, Xie D, Short DR, et al. Production of omega-3 eicosapentaenoic acid by metabolic engineering of Yarrowia lipolytica. Nat Biotechnol. 2013;31(8):734–740. doi: 10.1038/nbt.2622. [DOI] [PubMed] [Google Scholar]
- 75.Zhang S, Ito M, Skerker JM, Arkin AP, Rao CV. Metabolic engineering of the oleaginous yeast Rhodosporidium toruloides IFO0880 for lipid overproduction during high-density fermentation. Appl Microbiol Biotechnol. 2016;100(21):9393–9405. doi: 10.1007/s00253-016-7815-y. [DOI] [PubMed] [Google Scholar]
- 76.Kaniak A, Xue Z, Macool D, Kim JH, Johnston M. Regulatory network connecting two glucose signal transduction pathways in Saccharomyces cerevisiae. Eukaryot Cell. 2004;3(1):221–231. doi: 10.1128/EC.3.1.221-231.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yurkov AM. Yeasts of the soil—obscure but precious. Yeast. 2018;35(5):369–378. doi: 10.1002/yea.3310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Papanikolaou S, Aggelis G. Lipids of oleaginous yeasts. Part I; Biochemistry of single cell oil production. Eur J Lipid Sci Technol. 2011;113:1031–1051. doi: 10.1002/ejlt.201100014. [DOI] [Google Scholar]
- 79.Beopoulos A, Nicaud JM, Gaillardin C. An overview of lipid metabolism in yeasts and its impact on biotechnological processes. Appl Microbiol Biotechnol. 2011;90(4):1193–1206. doi: 10.1007/s00253-011-3212-8. [DOI] [PubMed] [Google Scholar]
- 80.Vorapreeda T, Thammarongtham C, Cheevadhanarak S, Laoteng K. Repertoire of malic enzymes in yeast and fungi: insight into their evolutionary functional and structural significance. Microbiology. 2013;159(Pt 12):2548–2557. doi: 10.1099/mic.0.065342-0. [DOI] [PubMed] [Google Scholar]
- 81.Fredlund E, Blank LM, Schnurer J, Sauer U, Passoth V. Oxygen- and glucose-dependent regulation of central carbon metabolism in Pichia anomala. Appl Environ Microbiol. 2004;70(10):5905–5911. doi: 10.1128/AEM.70.10.5905-5911.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Folch J, Lees M, Sloane Stanley GH. A simple method for the isolation and purification of total lipides from animal tissues. J Biol Chem. 1957;226(1):497–509. [PubMed] [Google Scholar]
- 83.Brandenburg J, Blomqvist J, Pickova J, Bonturi N, Sandgren M, Passoth V. Lipid production from hemicellulose with Lipomyces starkeyi in a pH regulated fed-batch cultivation. Yeast. 2016;33(8):451–462. doi: 10.1002/yea.3160. [DOI] [PubMed] [Google Scholar]
- 84.Appelqvist L. Rapid methods of lipid extraction and fatty acid methyl ester preparation for seed and leaf tissue with special remarks on preventing accumulation of lipid contaminants. Chem Sci. 1968;28:551–570. [Google Scholar]
- 85.Olstorpe M, Pickova J, Kiessling A, Passoth V. Strain- and temperature-dependent changes of fatty acid composition in Wickerhamomyces anomalus and Blastobotrys adeninivorans. Biotechnol Appl Biochem. 2014;61(1):45–50. doi: 10.1002/bab.1130. [DOI] [PubMed] [Google Scholar]
- 86.Olsen RE, Henderson RJ. The rapid analysis of neutral and polar marine lipids using double-development HPTLC and scanning densitometry. J Exp Mar Biol Ecol. 1989;129(2):189–197. doi: 10.1016/0022-0981(89)90056-7. [DOI] [Google Scholar]
- 87.Wisniewski JR. Filter-Aided Sample Preparation: the Versatile and Efficient Method for Proteomic Analysis. Methods Enzymol. 2017;585:15–27. doi: 10.1016/bs.mie.2016.09.013. [DOI] [PubMed] [Google Scholar]
- 88.Simonian MH. Spectrophotometric determination of protein concentration. Curr Protoc Toxicol. 2004;Appendix 3:A 3G 1-7. [DOI] [PubMed]
- 89.Tyanova S, Temu T, Cox J. The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat Protoc. 2016;11(12):2301–2319. doi: 10.1038/nprot.2016.136. [DOI] [PubMed] [Google Scholar]
- 90.Cox J, Hein MY, Luber CA, Paron I, Nagaraj N, Mann M. Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol Cell Proteom. 2014;13(9):2513–2526. doi: 10.1074/mcp.M113.031591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Tyanova S, Temu T, Sinitcyn P, Carlson A, Hein MY, Geiger T, et al. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat Methods. 2016;13(9):731–740. doi: 10.1038/nmeth.3901. [DOI] [PubMed] [Google Scholar]
- 92.Vizcaino JA, Cote RG, Csordas A, Dianes JA, Fabregat A, Foster JM, et al. The PRoteomics IDEntifications (PRIDE) database and associated tools: status in 2013. Nucleic Acids Res. 2013;41(Database issue):D1063–D1069. doi: 10.1093/nar/gks1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. Complete list of proteins detected during batch cultivation of R. toruloides in minimal medium containing either glucose or xylose and their relative expression values.
Additional file 2: Fig. S1. GO term enrichment analysis and volcano plots of differentially expressed proteins in R. toruloides during either exponential growth phase or lipid production phase in medium containing either glucose (A) or xylose (B). For each GO term showing significant change (rank score of ≥ 5), the direction and significance of the relative changes in protein levels are shown, together with the total number of proteins within each GO term. Fig. S2. Venn diagram of differentially expressed proteins in R. toruloidesduring either exponential growth phase or lipid production phase in medium containing either glucose (yellow circle) or xylose (blue circle). Number in red or blue colour indicates up or downregulated proteins, respectively, in cells during lipid accumulation as compared to exponential phase. Fig. S3. Volcano plots of differentially expressed proteins in R. toruloides cultivated on either glucose or xylose during early exponential growth phase (A), late exponential growth phase (B), or lipid accumulation phase (C). Fig. S4. Venn diagram of differentially expressed proteins in R. toruloidescultivated on either glucose or xylose during early exponential growth phase (t1, yellow circle), late exponential growth phase (t2, brown circle), or lipid accumulation phase (t3, green cycle). Number in red or blue colour indicates up or downregulated proteins, respectively, in cells grown on xylose as compared to cells grown on glucose.
Additional file 3. Relative levels of differentially expressed proteins in glucose-grown R. toruloides during either exponential cultivated phase or lipid production phase.
Additional file 4. Relative levels of differentially expressed proteins in xylose-grown R. toruloides during either exponential cultivated phase or lipid production phase.
Additional file 5. Relative levels of differentially expressed proteins in R. toruloides cultivated on either glucose or xylose during early exponential growth phase.
Additional file 6. Relative levels of differentially expressed proteins in R. toruloides cultivated on either glucose or xylose during late exponential growth phase.
Additional file 7. Relative levels of differentially expressed proteins in R. toruloides grown on either glucose or xylose during lipid accumulation phase.
Additional file 8. Relative levels of proteins involved in central carbon metabolism of R. toruloides.
Data Availability Statement
All the data analyzed in this study are included in this manuscript and its additional files. The proteomics data has been deposited to the ProteomeXchange consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository [92] with the dataset identifier PXD012332.




