Abstract
Background
Natural and artificial selection leads to changes in certain regions of the genome resulting in selection signatures that can reveal genes associated with the selected traits. Selection signatures may be identified using different methodologies, of which some are based on detecting contiguous sequences of homozygous identical-by-descent haplotypes, called runs of homozygosity (ROH), or estimating fixation index (FST) of genomic windows that indicates genetic differentiation. This study aimed to identify selection signatures in a paternal broiler TT line at generations 7th and 16th of selection and to investigate the genes annotated in these regions as well as the biological pathways involved. For such purpose, ROH and FST-based analysis were performed using whole genome sequence of twenty-eight chickens from two different generations.
Results
ROH analysis identified homozygous regions of short and moderate size. Analysis of ROH patterns revealed regions commonly shared among animals and changes in ROH abundance and size between the two generations. Results also suggest that whole genome sequencing (WGS) outperforms SNPchip data avoiding overestimation of ROH size and underestimation of ROH number; however, sequencing costs can limited the number of animals analyzed. FST-based analysis revealed genetic differentiation in several genomic windows. Annotation of the consensus regions of ROH and FST windows revealed new and previously identified genes associated with traits of economic interest, such as APOB, IGF1, IGFBP2, POMC, PPARG, and ZNF423. Over-representation analysis of the genes resulted in biological terms of skeletal muscle, matrilin proteins, adipose tissue, hyperglycemia, diabetes, Salmonella infections and tyrosine.
Conclusions
Identification of ROH and FST-based analyses revealed selection signatures in TT line and genes that have important role in traits of economic interest. Changes in the genome of the chickens were observed between the 7th and 16th generations showing that ancient and recent selection in TT line may have acted over genomic regions affecting diseases and performance traits.
Electronic supplementary material
The online version of this article (10.1186/s12864-019-5811-1) contains supplementary material, which is available to authorized users.
Keywords: Runs of homozygosity, Fixation index, FST, Artificial selection, Gallus gallus
Background
Artificial selection of animals lead to changes on particular genomic regions that affect traits of economic interest, as well as traits involved in adaptation to climatic and stress conditions, immune response, and disease resistance [1]. Thus, selection signature regions are printed along the genome as a result of selection pressure. Detecting selection signatures is important for a better understanding of population history and genetic mechanisms affecting phenotypic differentiation in humans, livestock and wild animals [2]. Understanding how selection acts on livestock populations may also benefit breeding programs in order to improve traits of economic interest in these animals, such as chicken breeds which have been intensively selected for fast growth and muscle development [3]. Detection approaches rely on scanning the genome for regions of homozygosity, as well as on estimating allele or haplotype frequency differences between populations or generations within a population. There are several statistical methods for these analyses, such as extended haplotype homozygosity (EHH) [4], integrated haplotype score (iHS) [5], runs of homozygosity (ROH) [2], and FST statistics [6].
Runs of homozygosity are regions in the genome containing contiguous homozygous genotypes identical by descent (IBD), i.e. regions where the pairs of alleles are most likely inherited from a common ancestor [2]. Recent studies used this approach to better understand human diseases [7–9], human ancestry [10], and population structure and traits of interest in livestock species, such as cattle [11–14], swine [15], poultry [16, 17], and sheep [18]. The fixation index (FST), first defined by Wright [19], is a measure that exploits differences in allele frequencies to infer the genetic differentiation between populations or generations [20]. A certain locus under selection pressure changes its frequency over the generations. Thus, high values of FST indicate candidate selection signatures due to differences in locus frequency among populations or across generations. Previous studies have reported important selection signatures in Virginia [21] and Brazilian broiler and layer chicken lines [22] using this method.
Embrapa Swine and Poultry, a Brazilian National Research Center, has been raising experimental chicken populations under selection since the 1970’s. One of these lines is the paternal broiler line called TT, which has been under multi-trait selection since 1992 [23]. Identification of selection signatures in chicken lines can help understand which regions underwent selection pressure over time and how their biological mechanisms act to express the traits of interest, such as muscle growth and fat deposition. In this sense, we aimed to investigate selection signatures in TT broiler line by detecting ROH in the 7th and 16th generations, raised in the years of 1998 and 2007, respectively, and estimating FST statistic between these two generations. The identification of those regions will provide better understanding of artificial selection effects on broiler lines, and may point out candidate genes and biological mechanisms underlying performance traits.
Methods
Ethics statement
This study followed experimental protocols pertinent to animal experimentation with the approval of the Embrapa Swine and Poultry Ethics Committee on Animal Utilization (CEUA) in Concordia, Santa Catarina State, Brazil, on resolution number 011/2010. It followed the rules of National Council of Animal Experimentation Control (CONCEA) in accordance with international guidelines to guarantee animal welfare.
Chicken population
Chickens used in this study were from a broiler line developed by the Embrapa Swine and Poultry National Research Center. This line, called TT, was originated from Cornish and White Plymouth Rock breeds, that has been under a multi-trait selection process focused on body weight, feed conversion, cut yields, breast weight, abdominal fat, and other traits, since 1992 [17, 23, 24]. The chickens were raised in open sided poultry houses, receiving commercial broiler diet and water ad libitum [17, 24]. Chickens were euthanized by cervical dislocation at 42 days of age. Two groups of animals from this line were analyzed, 14 chickens (half male and half female) from the seventh-generation (7th) raised in the year of 1998 and 14 male chickens from the sixteenth-generation (16th) raised in 2007.
The performance of birds from the 7th generation, as hatched average live weight at 35 days of age was 2272 g; the breast area in the live bird was 96,1 cm2 and the individual feed conversion rate (FCR) for males, from 36 to 43 days of age was 2268 g. The performance of birds from 16th, as hatched average live weight at 42 days of age was 2457 g; the breast area was 112 cm2 and the individual FCR from 43 to 49 days of age was 2798 g [17]. Note that the age of selection for BW and BA has changed from 35 to 42 days of age in the described period, as well as the FCR evaluation period, which has changed from 36 to 43 days in 1998 to 43–49 days of age in 2007.
Sequencing and quality control
Whole genome sequencing (WGS) data of 28 chickens were used in this study. Animals were individually sequenced to a minimum coverage of 11.4x using the HiSeq2500 (Illumina) platform, and the alignment of reads was done against the chicken genome assembly (Gallus_gallus-5.0, UCSC) chicken reference genome using Bowtie2 [25]. Detailed information about library preparation, sequencing, quality control of reads, alignment and SNP and INDEL identification are fully described in Boschiero et al. [22] and Moreira et al. [26]. Variants identified in sexual, mitochondrial, random or unplaced chromosomes were removed from our analysis.
Principal component analysis
Genetic relationship between the 28 animals was assessed with a principal component analysis (PCA) using the SNP dataset (n = 9,914,904). The analysis was performed using the SNPRelate package of Bioconductor by means of an in-house script in R.
Identification of runs of homozygosity
The identification of ROH was chosen to obtain information about selection signatures and how they are shared between animals in both generations. Analyses were performed using PLINK v1.9 software [27, 28], which uses a sliding window approach: a window, with a minimum size, slides across the genome, calling a segment if it is in accordance with the parameters established and the threshold of calculated proportion of homozygous windows overlapping each SNP in that segment. The parameters used in the analysis were set based on Ceballos et al. [29] and they are listed in Table 1.
Table 1.
PLINK parameters for run of homozygosity (ROH) analysis
| Parameter | Value | Definition |
|---|---|---|
| -homozyg-snp | 50 | Minimum number of SNP required to consider a ROH; |
| -homozyg-kb | 300 | Size (Kb) of the sliding window; |
| -homozyg-density | 50 | Minimum density required to consider a ROH; |
| -homozyg-gap | 1000 | Maximum size (Kb) between two SNP to be considered in the same ROH; |
| -homozyg-window-snp | 50 | Number of SNP present in the sliding window; |
| -homozyg-window-het | 3 | Number of heterozygous SNP allowed in a ROH; |
| -homozyg-window-missing | 5 | Number of missing calls allowed in a ROH; |
| -homozyg-window-threshold | 0.05 | Proportion of overlapping windows that must be called homozygous to define a given SNP as in a homozygous segment. |
Dataset of the 28 animals comprised 9,914,904 SNP, and all INDEL were excluded. The parameter -homozyg-group was also used to obtain information of the overlapping ROH (pools), i.e., ROH that appeared in at least two animals in the same region of the genome. The output plink.hom.overlap shows each ROH of each animal overlapping and their respective union (uROH) and consensus sequences (cROH), besides their genome position, size and number of SNP. The consensus ROH (cROH, i.e. a consensus segment of ROH that appeared in a common region in at least two animals) of the pools were used for annotation and enrichment analysis, to avoid randomly assigned ROH and to represent what changed and what is conserved between the animals [30, 31]. In addition, we used an in-house script in R to check the overlap between the regions of all cROH and the ROH previously identified in the TT Reference population (originated from TT broiler line) by Marchesi et al. [17], given the positional coordinates (chromosome, start and end) of these regions and considering at least one overlapped base pair.
Genomic inbreeding coefficients
Individual genomic inbreeding coefficients were calculated based on ROH data (FROH), as defined by McQuillan et al. [32], to know if there was a difference of inbreeding between the 7th and 16th generations. FROH was calculated as:
where LROH is the total size of ROH in the genome and Laut is the total size of autosomal genome covered by SNP of an individual (933.071 Mb, Gallus_gallus-5.0 chicken reference genome - UCSC).
FST analysis
This method was applied to compare the two generations, i.e. to identify selection signatures by estimating the differences in allele frequency between the 7th and 16th generations. The fixation index was calculated according to Weir and Cockerham’s pairwise estimator method [33] using VCFtools v.1.16 software [34], in which SNP and INDEL analyses were run separately, comprising datasets of 9,914,904 SNP and 793,603 INDEL. The same parameters used recently in chickens by Boschiero et al. [22] were applied: FST values were calculated using overlapping windows of 20 Kb size sliding by steps of 10 Kb size. Windows with less than 10 SNP or 5 INDEL were removed, and all negative values were set to zero. FST values of the remaining windows were ranked, and those equal or above 0.3 were considered as candidate selection signatures. The software BEDTools [35] was used to check if there were equivalent regions identified in both datasets.
Functional analysis
Functional analysis was performed to identify genes annotated within the candidate selection signature regions identified and, consequently, the biological mechanisms that may be involved with traits of adaptation and performance. Such information was obtained assessing the position (start and end coordinates) of the candidate selection signatures (cROH and FST windows ≥0.3) in the chicken genome available at BioMart Ensembl genome browser platform (Ensembl Genes release 94, Gallus_gallus-5.0 assembly) [36].
We also assessed the genes annotated in the candidate selection signatures under different perspectives in order to understand the effects of selection on TT line in different periods. First, we investigated changes that occurred between the 7th and 16th generations: (i) cROH of regions shared exclusively among animals of the 7th; (ii) cROH of regions shared exclusively among animals of the 16th; (iii) cROH of regions that were shared among at least four animals of the 16th more than animals of 7th; (iv) FST SNP windows (≥ 0.3); and (v) FST INDEL windows (≥ 0.3). In addition, we looked for genes annotated in (vi) cROH of regions shared with 12 or more animals (among the 28), to identify regions probably related to chicken domestication or even, specialization into broilers.
Functional enrichment was performed using MeSH Enrichment and Semantic Analysis, Bioconductor’s package [37, 38], in R software [39] to investigate if there was overrepresentation of any biological processes and components. For such purpose, datasets of genes annotated in the specific candidate selection signatures previously mentioned (i-vi) were analyzed separately. The p-value was adjusted using the Benjamin-Hochberg false discovery rate (FDR) method [40].
Overlapping selection signatures with QTL
In order to confirm the role of the selection signatures detected herein in the regulation of important phenotypes in chickens, we investigated the overlap with QTL associated with traits of economic interest. The analysis was performed using an in-house script in R to overlap the regions of all cROH, FST SNP and INDEL windows against the QTLs available at the Chicken QTL database [41], given the positions (chromosome, start and end) of these regions and considering at least one overlapped base pair. Particularly, we also analyzed if there was overlap of candidate selection signatures with QTL associated with fat deposition previously identified by Moreira et al. [42] in the TT Reference population, originated from an expansion of TT line in 2007 for genomic studies purpose [17].
Results
Principal component analysis
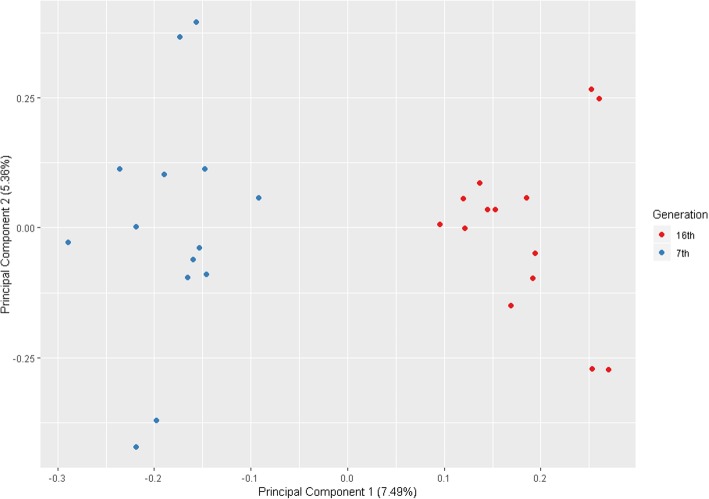
Principal component analysis using genomic data revealed a cluster separation between animals of the 7th and the 16th generations (Fig. 1). The distinct clustering demonstrated that genome data successfully separate these animals accordingly to their generation.
Fig. 1.
Principal component analysis using genomic data (SNP = 9,914,904) for 7th and 16th generations
Runs of homozygosity
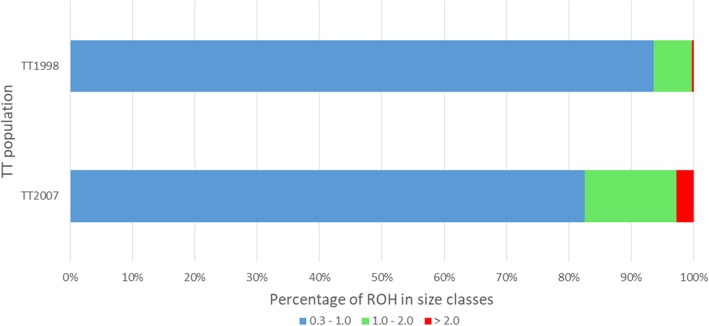
Analysis of the whole genome sequence data of the 28 animals (14 of the 7th and 14 of the 16th generation) with PLINK’s sliding window approach identified 5721 ROH (1944 in the 7th and 3777 in the 16th generation animals) (Additional file 1). The average number of segments per animal was lower in the 7th (138.9 ROH/animal) than in the 16th generation (269.8 ROH/animal) (Table 2). The ROH presented small and moderate sizes, ranging between 300 Kb and 4.9 Mb, and most of them had sizes smaller than 1.0 Mb in both generations (1821 ROH in the 7th and 3120 in the 16th generation animals). A change in the distribution of ROH sizes was also observed between both generations (Fig. 2). The proportion of ROH smaller than 1.0 Mb decreased (93.7% in the 7th to 82.6% in 16th generation) while the proportion of ROH with sizes between 1.0 and 2.0 Mb increased (6.1% in the 7th to 14.7% in the 16thgeneration) as well as ROH bigger than 2.0 Mb (0.3% in the 7th to 2.7% in 16th generation). The average total size of ROH per animal was 73.2 Mb in the 7th and 188.6 Mb in the 16th generation (Table 2).
Table 2.
ROH features for each animal of 7th and 16th generations
| IID | Number of ROHa | Total sizeb (Kb) | Mean size of segments (Kb) | FROH |
|---|---|---|---|---|
| 7th_01 | 130 | 60,492.6 | 465.328 | 0.0648 |
| 7th_02 | 212 | 125,062.0 | 589.913 | 0.1340 |
| 7th_03 | 131 | 63,732.1 | 486.504 | 0.0683 |
| 7th_04 | 61 | 25,416.8 | 416.668 | 0.0272 |
| 7th_05 | 65 | 30,014.6 | 461.763 | 0.0322 |
| 7th_06 | 85 | 38,303.9 | 450.634 | 0.0411 |
| 7th_07 | 183 | 103,302.0 | 564.491 | 0.1107 |
| 7th_08 | 148 | 71,381.7 | 482.309 | 0.0765 |
| 7th_09 | 209 | 138,608.0 | 663.195 | 0.1486 |
| 7th_10 | 219 | 128,120.0 | 585.024 | 0.1373 |
| 7th_11 | 94 | 42,055.7 | 447.401 | 0.0451 |
| 7th_12 | 175 | 82,420.5 | 470.974 | 0.0883 |
| 7th_13 | 188 | 95,302.7 | 506.929 | 0.1021 |
| 7th_14 | 44 | 20,107.3 | 456.985 | 0.0215 |
| Means (CV) of the 7th generation | 139 (43.8%) | 73,165.7 (54.8%) | 503,437 (13.9%) | 0,0784 (54.8%) |
| 16th_01 | 245 | 164,355.0 | 670.839 | 0.1761 |
| 16th_02 | 241 | 183,336.0 | 760.729 | 0.1965 |
| 16th_03 | 254 | 174,906.0 | 688.608 | 0.1875 |
| 16th_04 | 280 | 200,919.0 | 717.567 | 0.2153 |
| 16th_05 | 268 | 188,023.0 | 701.579 | 0.2015 |
| 16th_06 | 318 | 200,111.0 | 629.281 | 0.2145 |
| 16th_07 | 256 | 181,422.0 | 708.681 | 0.1944 |
| 16th_08 | 290 | 196,574.0 | 677.843 | 0.2107 |
| 16th_09 | 289 | 206,456.0 | 714.381 | 0.2213 |
| 16th_10 | 283 | 215,635.0 | 761.963 | 0.2311 |
| 16th_11 | 275 | 187,931.0 | 683.386 | 0.2014 |
| 16th_12 | 254 | 181,634.0 | 715.095 | 0.1947 |
| 16th_13 | 242 | 167,875.0 | 693.697 | 0.1799 |
| 16th_14 | 282 | 191,333.0 | 678.484 | 0.2051 |
| Means (CV) of the 16th generation | 270 (8.3%) | 188,607.9 (7.7%) | 700.152 (4.9%) | 0.2021 (7.7%) |
IID individual identification, CV coefficient of variation %
aTotal number of ROH identified in each animal
bTotal size of autosomal genome covered by ROH
Fig. 2.
Percentage of ROH in 7th and 16th generations distributed in size classes. A decrease in the proportion of ROH smaller than 1.0 Mb in 16th is observed, meanwhile the proportions of ROH with sizes above 1.0 MB increased
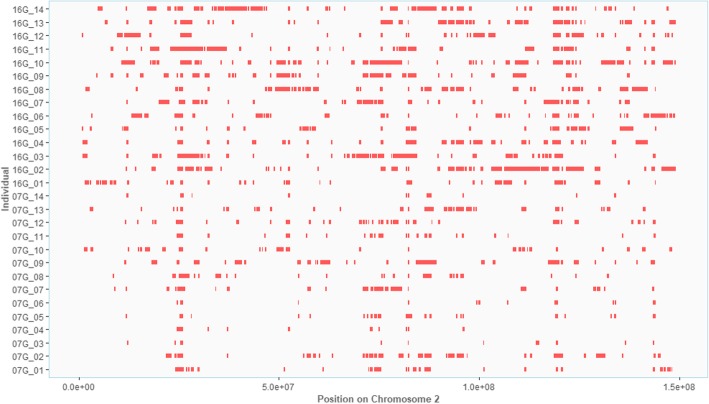
ROH were identified in all chromosomes, except on GGA16 and GGA30–32 (Table 3). Figure 3 represents all ROH, with their proportional sizes, distributed across GGA2 for the 28 animals. Regions where ROH is presented in more than one animal formed a sharing pattern. Figures of other chromosomes are provided as Additional file 2. The four longest ROH (> 4 Mb) were located on different macro chromosomes (GGA2: 116,060,874 – 120,088,450; GGA5: 39,097,092 – 43,183,508; GGA3: 25,504,098 – 29,639,462; GGA4: 69,071,022 – 73,960,022). Overlaps of ROH from at least two animals established 1941 pools (Additional file 3). There was one pool of ROH shared among the 28 animals, and it was located in the GGA2 with a consensus sequence of 300.2 Kb (82,146,603 – 82,446,837). Furthermore, most of the pools consisted in regions shared among two to seven animals (74.4%). There were 87 regions with ROH commonly shared with at least 12 animals, and most of them identified on GGA1.
Table 3.
Summary of runs of homozygosity (ROH), pools of ROH, FST SNP windows, and FST INDEL windows by chromosome in TT population in all animals from 7th and 16th generations
| GGA | Size (Mb) | Number of ROH | Number of pools | Number of FST SNP windows (≥ 0.3) | Number of FST INDEL windows (≥ 0.3) |
|---|---|---|---|---|---|
| 1 | 196.20 | 1230 | 415 | 53 | 65 |
| 2 | 149.56 | 1087 | 338 | 28 | 28 |
| 3 | 111.30 | 609 | 228 | 28 | 23 |
| 4 | 91.28 | 582 | 212 | 5 | 4 |
| 5 | 59.83 | 392 | 137 | 6 | 9 |
| 6 | 35.47 | 211 | 77 | 4 | 5 |
| 7 | 36.95 | 267 | 85 | 17 | 21 |
| 8 | 29.96 | 213 | 66 | 1 | 2 |
| 9 | 24.09 | 106 | 39 | 6 | 10 |
| 10 | 20.44 | 106 | 39 | – | 2 |
| 11 | 20.22 | 114 | 36 | 1 | 1 |
| 12 | 19.95 | 90 | 41 | – | – |
| 13 | 18.41 | 107 | 35 | 1 | 2 |
| 14 | 15.60 | 101 | 32 | – | 1 |
| 15 | 12.76 | 81 | 32 | – | – |
| 17 | 10.96 | 72 | 22 | – | – |
| 18 | 11.05 | 87 | 28 | 1 | – |
| 19 | 9.98 | 61 | 16 | – | 2 |
| 20 | 14.11 | 54 | 19 | – | – |
| 21 | 6.86 | 26 | 7 | – | – |
| 22 | 4.73 | 13 | 4 | 2 | 3 |
| 23 | 5.79 | 21 | 7 | 1 | – |
| 24 | 6.28 | 30 | 12 | – | – |
| 25 | 2.91 | 7 | 1 | – | – |
| 26 | 5.31 | 9 | 1 | – | – |
| 27 | 5.66 | 16 | 5 | – | – |
| 28 | 4.97 | 25 | 6 | – | – |
| 33 | 1.65 | 4 | 1 | – | – |
Fig. 3.
Genome wide distribution of runs of homozygosity (ROH) in TT population. Size and location of ROH in chromosome 2 for each animal are represented in parallel. Patterns of shared ROH can be observed in some regions of the chromosome. In addition, a higher frequency of ROH in animals of 16G was observed
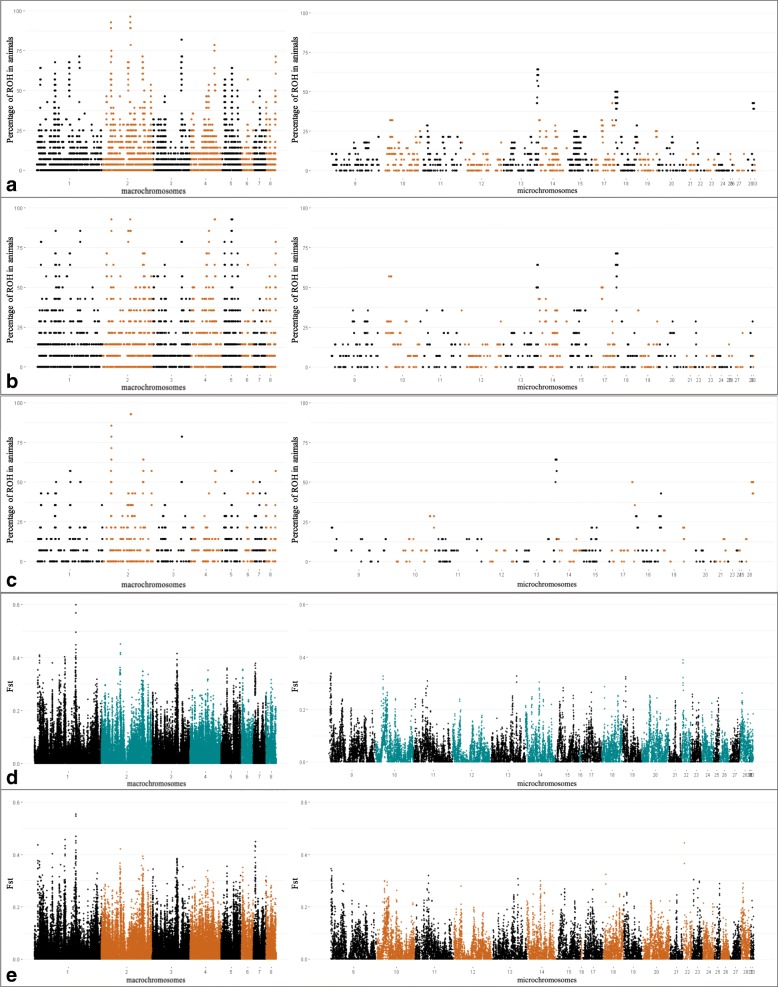
Regions commonly shared among the animals become more frequent in the 16th generation. We observed a greater number of ROH pools shared among animals from 16th generation than animals from 7th generation (Fig. 4-a, b and c).
Fig. 4.
Manhattan plot of genomic regions and percentage of animals that shares the candidate selection signatures as well, SNPs and INDELs Fst windows. a: ROH pools detected in all the 28 animals; b: ROH pools detected in the 14 birds from 16th generation; c: ROH pools detected in the 14 birds from 7th generation; d: SNPs Fst windows; e: INDELS Fst windows. For a, b and c, the X-axis represents the chromosomes, and Y-axis shows the proportion of animals that shares the ROH pools. For d and e, the X-axis represents the chromosomes, and Y-axis shows the Fst values
Genomic inbreeding coefficients
Individual genomic inbreeding coefficients based on ROH (FROH) were calculated for both generations (Table 2). Mean, maximum and minimum individual FROH for animals of the 7th generation were 0.0784, 0.1340, and 0.0215, respectively, with a coefficient of variation (CV) of 52.8%. For animals of the 16th, the mean, maximum and minimum individual FROH were 0.2021, 0.2213, and 0.1761 (CV = 7.4%).
FST windows
FST analysis identified 91,638 and 86,404 windows for SNP and INDEL datasets, respectively, after removing windows with less than 10 SNP and five INDEL. The number of markers per window ranged from 10 to 1562 SNP (average of 216.3 SNP/window) and from five to 72 INDEL (average of 18.2 INDEL/window). Mean FST values for SNP and INDEL datasets were 0.040 and 0.038, respectively, while the highest FST values were 0.598 and 0.555.
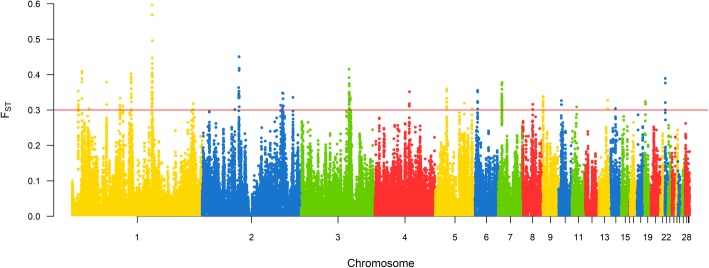
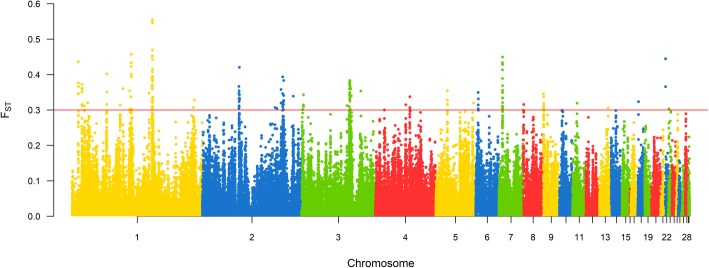
Windows with FST values equal or higher than 0.3 were considered candidate selection signatures. There were 178 windows using SNP dataset (Fig. 5) and 154 windows using INDEL dataset (Fig. 6) above this threshold value (FST ≥ 0.3). More information about these windows are available in the Additional files 5 and 6. Most of these windows were in the macrochromosomes (Table 3) and approximately 87% of the INDEL windows overlapped with SNP windows (Fig. 4 – c and d).
Fig. 5.
Manhattan plot of genome wide distribution of FST windows for SNP dataset. Red line represents threshold of 0.3, windows above this value were considered candidate selection signature
Fig. 6.
Manhattan plot of genome wide distribution of FST windows for INDEL dataset. Red line represents threshold of 0.3, windows above this value were considered candidate selection signatures
Genes in consensus selection signature regions
Annotation analysis using the Ensembl genome browser revealed 5681 genes annotated in the 1941 cROH pools shared among at least two of the 28 animals (Additional file 7). Annotation of FST windows (Fst ≥0.3) identified 56 and 60 genes for SNP and INDEL datasets, respectively (Additional files 8 and 9). Since a great part of SNP windows overlapped with INDEL windows, 37 of these genes were common for both datasets (Table 4). In addition, about 46.1% of SNP FST windows and 37.7% of INDEL FST windows overlapped with cROH. Thus, we found 34 genes annotated in cROH in common with genes annotated in FST (SNP and/or INDEL) windows (Table 4).
Table 4.
Genes annotated commonly between selection signatures of two or more datasets
| Gene ID | Gene name | Description |
|---|---|---|
| F ST SNP and INDEL windows (≥0.3) datasets | ||
| ENSGALG00000000242 | EBF2 | early B cell factor 2 |
| ENSGALG00000002370 | SH2D4B | SH2 domain containing 4B |
| ENSGALG00000002414 | TSPAN14 | tetraspanin 14 |
| ENSGALG00000004045 | AGAP1 | ArfGAP with GTPase domain, ankyrin repeat and PH domain 1 |
| ENSGALG00000004116 | TRPM8 | transient receptor potential cation channel subfamily M member 8 |
| ENSGALG00000004129 | SPP2 | secreted phosphoprotein 2 |
| ENSGALG00000007555 | CCND1 | cyclin D1 |
| ENSGALG00000007556 | LTO1 | LTO1, ABCE1 maturation factor |
| ENSGALG00000012542 | RASD2 | RASD family member 2 |
| ENSGALG00000015402 | C3orf38 | chromosome 3 open reading frame 38 |
| ENSGALG00000015403 | EPHA3 | EPH receptor A3 |
| ENSGALG00000015570 | GPR63 | G protein-coupled receptor 63 |
| ENSGALG00000015573 | FHL5 | four and a half LIM domains 5 |
| ENSGALG00000016518 | PHKA2 | phosphorylase kinase regulatory subunit alpha 2 |
| ENSGALG00000016522 | PPEF1 | protein phosphatase with EF-hand domain 1 |
| ENSGALG00000016529 | CDKL5 | cyclin dependent kinase like 5 |
| ENSGALG00000016541 | novel gene | BEN domain containing 2 |
| ENSGALG00000016543 | NHS | NHS actin remodeling regulator |
| ENSGALG00000022866 | ZNF654 | zinc finger protein 654 |
| ENSGALG00000026372 | novel gene | -- |
| ENSGALG00000028376 | FGF19 | fibroblast growth factor 19 |
| ENSGALG00000032974 | ADAMTS2 | ADAM metallopeptidase with thrombospondin type 1 motif 2 |
| ENSGALG00000033076 | novel gene | -- |
| ENSGALG00000035116 | STAG1 | stromal antigen 1 |
| ENSGALG00000035393 | LRRC14B | leucine rich repeat containing 14B |
| ENSGALG00000035906 | YTHDC1 | YTH domain containing 1 |
| ENSGALG00000036204 | novel gene | -- |
| ENSGALG00000036327 | NGEF | neuronal guanine nucleotide exchange |
| ENSGALG00000036730 | MRPS35 | mitochondrial ribosomal protein S35 |
| ENSGALG00000036938 | RALYL | RALY RNA binding protein like |
| ENSGALG00000038154 | YAP1 | Yes associated protein 1 |
| ENSGALG00000038730 | GIGYF2 | GRB10 interacting GYF protein 2 |
| ENSGALG00000039139 | TNS3 | tensin 3 |
| ENSGALG00000039690 | STMN2 | stathmin 2 |
| ENSGALG00000039738 | SLC9A3 | solute carrier family 9 member A3 |
| ENSGALG00000040264 | C9H2ORF82 | chromosome 9 open reading frame, human C2orf82 |
| ENSGALG00000042764 | COG5 | component of oligomeric golgi complex 5 |
| cROH and F ST SNP windows (≥0.3) | ||
| ENSGALG00000000242 | EBF2 | early B cell factor 2 |
| ENSGALG00000001153 | AUTS2 | AUTS2, activator of transcription and developmental regulator |
| ENSGALG00000003705 | VPS13C | vacuolar protein sorting 13 homolog C |
| ENSGALG00000004045 | AGAP1 | ArfGAP with GTPase domain, ankyrin repeat and PH domain 1 |
| ENSGALG00000004116 | TRPM8 | transient receptor potential cation channel subfamily M member 8 |
| ENSGALG00000006237 | PKN2 | protein kinase N2 |
| ENSGALG00000007555 | CCND1 | cyclin D1 |
| ENSGALG00000007556 | LTO1 | LTO1, ABCE1 maturation factor |
| ENSGALG00000015402 | C3orf38 | chromosome 3 open reading frame 38 |
| ENSGALG00000015403 | EPHA3 | EPH receptor A3 |
| ENSGALG00000016518 | PHKA2 | phosphorylase kinase regulatory subunit alpha |
| ENSGALG00000016522 | PPEF1 | protein phosphatase with EF-hand domain 1 |
| ENSGALG00000016529 | CDKL5 | cyclin dependent kinase like 5 |
| ENSGALG00000022866 | ZNF654 | zinc finger protein 654 |
| ENSGALG00000025253 | gga-mir-1694 | gga-mir-1694 |
| ENSGALG00000028376 | FGF19 | fibroblast growth factor 19 |
| ENSGALG00000030580 | RPS6KA5 | ribosomal protein S6 kinase A5 |
| ENSGALG00000032958 | AMPH | amphiphysin |
| ENSGALG00000034119 | novel gene | collagen type XV alpha 1 chain |
| ENSGALG00000035116 | STAG1 | stromal antigen 1 |
| ENSGALG00000035906 | YTHDC1 | YTH domain containing 1 |
| ENSGALG00000036938 | RALYL | RALY RNA binding protein like |
| ENSGALG00000038730 | GIGYF2 | GRB10 interacting GYF protein 2 |
| ENSGALG00000040167 | TPD52 | tumor protein D52 |
| cROH and F ST INDEL windows (≥0.3) datasets | ||
| ENSGALG00000000242 | EBF2 | early B cell factor 2 |
| ENSGALG00000000667 | EDN2 | endothelin 2 |
| ENSGALG00000004045 | AGAP1 | ArfGAP with GTPase domain, ankyrin repeat and PH domain 1 |
| ENSGALG00000004116 | TRPM8 | transient receptor potential cation channel subfamily M member 8 |
| ENSGALG00000007555 | CCND1 | cyclin D1 |
| ENSGALG00000007556 | LTO1 | LTO1, ABCE1 maturation factor |
| ENSGALG00000008163 | PSME4 | proteasome activator subunit 4 |
| ENSGALG00000015402 | C3orf38 | chromosome 3 open reading frame 38 |
| ENSGALG00000015403 | EPHA3 | EPH receptor A3 |
| ENSGALG00000016518 | PHKA2 | phosphorylase kinase regulatory subunit alpha |
| ENSGALG00000016522 | PPEF1 | protein phosphatase with EF-hand domain 1 |
| ENSGALG00000016529 | CDKL5 | cyclin dependent kinase like 5 |
| ENSGALG00000022866 | ZNF654 | zinc finger protein 654 |
| ENSGALG00000025789 | gga-mir-6614 | gga-mir-6614 |
| ENSGALG00000027632 | ACYP2 | acylphosphatase 2 |
| ENSGALG00000027960 | GRPR | gastrin releasing peptide receptor |
| ENSGALG00000028376 | FGF19 | fibroblast growth factor 19 |
| ENSGALG00000034516 | SHISA6 | shisa family member 6 |
| ENSGALG00000035116 | STAG1 | stromal antigen 1 |
| ENSGALG00000035906 | YTHDC1 | YTH domain containing 1 |
| ENSGALG00000036810 | novel gene | -- |
| ENSGALG00000036938 | RALYL | RALY RNA binding protein like |
| ENSGALG00000038730 | GIGYF2 | GRB10 interacting GYF protein 2 |
| ENSGALG00000039102 | TOX | thymocyte selection associated high mobility |
| ENSGALG00000040322 | novel gene | -- |
| cROH and F ST SNP and INDEL windows (≥0.3) datasets | ||
| ENSGALG00000000242 | EBF2 | early B cell factor 2 |
| ENSGALG00000004045 | AGAP1 | ArfGAP with GTPase domain, ankyrin repeat and PH domain 1 |
| ENSGALG00000004116 | TRPM8 | transient receptor potential cation channel subfamily M member 8 |
| ENSGALG00000007555 | CCND1 | cyclin D1 |
| ENSGALG00000007556 | LTO1 | LTO1, ABCE1 maturation factor |
| ENSGALG00000015402 | C3orf38 | chromosome 3 open reading frame 38 |
| ENSGALG00000015403 | EPHA3 | EPH receptor A3 |
| ENSGALG00000016518 | PHKA2 | phosphorylase kinase regulatory subunit alpha |
| ENSGALG00000016522 | PPEF1 | protein phosphatase with EF-hand domain 1 |
| ENSGALG00000016529 | CDKL5 | cyclin dependent kinase like 5 |
| ENSGALG00000022866 | ZNF654 | zinc finger protein 654 |
| ENSGALG00000028376 | FGF19 | fibroblast growth factor 19 |
| ENSGALG00000035116 | STAG1 | stromal antigen 1 |
| ENSGALG00000035906 | YTHDC1 | YTH domain containing 1 |
| ENSGALG00000036938 | RALYL | RALY RNA binding protein like |
| ENSGALG00000038730 | GIGYF2 | GRB10 interacting GYF protein 2 |
Based on Biomart Ensembl database, some of the genes commonly annotated in Fst and cROH regions, plays a role in biological processes involved in traits of economic interest in chicken or in other model animals; all the biological processes related to the genes mentioned in Table 4, are available at Additional file 10. There were genes involved in lipid metabolic processes, glucose metabolism and homeostasis and adipose tissue development. Other genes were described to be involved in muscle cell differentiation, muscle tissue development, and constituents of skeletal muscle. Moreover, there were a group of genes related to different types of behavior, such as grooming, locomotion, fear response, feeding behavior, aggressiveness, and social, exploration and maternal behaviors. Genes involved in the immune humoral system, differentiation, proliferation, homeostasis and chemotaxis of B cells, and regulation of cytokines production were also annotated in regions of the candidate selection signatures.
In order to complement the Fst analysis we identified genes in cROH regions that were either exclusive between the 7th and 16th generations. There were 71 genes annotated in the regions that were in homozygosity only in the 7th generation and 1881 genes annotated in regions of cROH shared only among animals of the 16th generation. We also identified genes in cROH regions that had changed between the 7th and 16th generations. For that, we considered regions that either increased or decreased by at least four animals in cROH regions between generations. There were 1318 genes annotated on these regions. For example, the gene IGF-I (GGA1 55,335,204 – 55,383,631) was annotated in a cROH region (GGA1 55,149,208 – 55,359,089) shared between seven animals of the 7th and 13 animals of the 16th. Additional file 11 presents genes annotated in these regions and that were previously associated with traits of economic interest in chickens..
These different gene lists were used to perform MeSH overrepresentation analysis, with the purpose of having an integrated knowledge of biological processes may be involved in the selection of TT line. MeSH analysis indicates if there is an overrepresentation of a particular group of genes in a biological category, such as anatomy, diseases or phenomena and processes. In this sense, overrepresentation analysis resulted in eight different biological terms: ‘matrilin proteins’, ‘skeletal muscle’, ‘Salmonella infections in animals’, ‘adipose tissue’, ‘cystatins’, ‘tyrosine’, ‘pregnancy in diabetics’, and ‘hyperglycemia’. Table 5 presents each one of these terms and their respective gene counts and p-values.
Table 5.
MeSH enrichment analysis of genes annotated in candidate selection signatures
| MeSH term (MeSH ID) | Gene count | P-value | BHa | Datasetb |
|---|---|---|---|---|
| Matrilin Proteins (D064235) | 3 | 0.00008523 | 0.01605961 | a |
| Muscle, Skeletal (D018482) | 9 | 0.00037904 | 0.04927546 | a |
| Salmonella Infections, Animal (D012481) | 2 | 0.02170325 | 0.02893767 | b |
| Adipose Tissue (D000273) | 9 | 0.00246916 | 0.0157409 | c |
| Cystatins (D015891) | 2 | 0.00018075 | 0.01590614 | d |
| 2 | 0.00019528 | 0.02089512 | e | |
| Tyrosine (D014443) | 3 | 0.00085140 | 0.03746136 | d |
| Pregnancy in Diabetics (D011254) | 1 | 0.00703107 | 0.03515533 | d |
| 1 | 0.00730629 | 0.03653145 | e | |
| Hyperglycemia (D006943) | 1 | 0.01748688 | 0.04066230 | d |
| 1 | 0.01816762 | 0.04223941 | e |
aBenjamini & Hochberg procedure for controlling false discovery rate (FDR) [40]
bAnalysis considering dataset of: (a) 232 genes annotated in cROH of pools shared between at least 12 animals of both generations; (b) 71 genes annotated in cROH of pools shared exclusively between 7th animals; (c) 1318 genes annotated in cROH of pools shared between at least four 16th animals more than 7th’s; (d) 56 genes annotated in FST windows of SNP data (FST ≥ 0.3); and (e) 60 genes annotated in FST windows of INDEL data (FST ≥ 0.3)
Selection signatures in overlap with QTL in the TT line
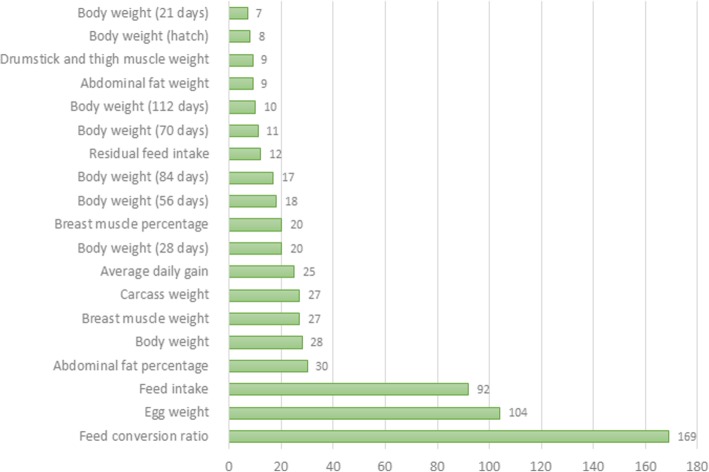
To identify selection signatures associated with quantitative traits, we investigated the overlap between the regions of cROH, FST SNP and INDEL windows (≥ 0.3) with QTL regions previously associated with traits of economic interest in chickens available at the Chicken QTL database (release 37). About 72.8% of the 1941 cROH overlapped with QTL regions (n = 2617). There were also overlap of 60.1% of the 178 FST SNP windows and 68.2% of the 154 FST INDEL windows with QTL regions (n = 107 and 105, respectively). These QTL regions were associated with 143 different traits (Additional file 12) of which some are very important for broilers’ breeding program goals such as feed conversion rate, feed intake, average daily gain, body weight, breast muscle weight, and others (Figs. 7, 8 and 9).
Fig. 7.
Manhattan plot of genome wide distribution of FST windows for INDEL dataset. Red line represents threshold of 0.3, windows above this value were considered candidate selection signatures
Fig. 8.
Manhattan plot of genome wide distribution of FST windows for INDEL dataset. Red line represents threshold of 0.3, windows above this value were considered candidate selection signatures
Fig. 9.
Manhattan plot of genome wide distribution of FST windows for INDEL dataset. Red line represents threshold of 0.3, windows above this value were considered candidate selection signature
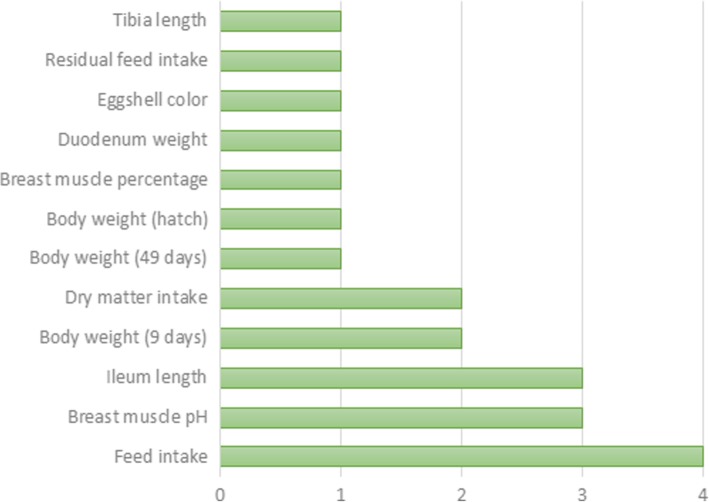
We also identified cROH regions that overlapped with 10 QTL previously mapped for fatness traits on GGA5, GGA9, GGA10, GGA13, GGA15, and GGA27 in the same population utilized herein (TT Reference Population) (Table 6) [42]. There was no overlap between the FST windows (≥ 0.3) and these QTLs mapped for fatness.
Table 6.
QTLs associated with fat traits in TT Reference Population overlapping with consensus runs of homozygosity (cROH)
| Trait (QTL ID)a | Chr | QTL position (start-end)b | cROH position (start - end) |
|---|---|---|---|
| ABFW (160520) | 5 | 38,000,437 – 38,996–916 | 38,015,470 – 38,234,917 |
| 38,425,606 – 38,473,340 | |||
| ABFW (160521) | 10 | 7,000,336 – 7,998,549 | 6,978,426 – 7,049,244 |
| 7,474,909 – 7,543,996 | |||
| 7,931,784 – 7,932,642 | |||
| ABFW (160522) | 13 | 3,002,617 – 3,998,616 | 3,572,237 – 3,641,314 |
| ABFP (160525) | 5 | 38,000,437 – 38,996–916 | 38,015,470 – 38,234,917 |
| 38,425,606 – 38,473,340 | |||
| ABFP (160526) | 10 | 7,000,336 – 7,998,549 | 6,978,426 – 7,049,244 |
| 7,474,909 – 7,543,996 | |||
| 7,931,784 – 7,932,642 | |||
| ABFP (160527) | 13 | 3,002,617 – 3,998,616 | 3,572,237 – 3,641,314 |
| SKINW (160529) | 15 | 6,000,311 – 6,999,944 | 6,175,697 – 6,362,475 |
| 6,473,966 – 6,475,580 | |||
| SKINW (160531) | 24 | 5,000,105 – 5,999,010 | 4,720,727 – 5,060,139 |
| 5,187,457 – 5,646,905 | |||
| 5,899,715 – 5,962,715 | |||
| SKINP (160534) | 9 | 4,000,836 – 4,999,336 | 4,664,760 – 4,860,555 |
| SKINP (160530) | 15 | 6,000,311 – 6,999,944 | 6,175,697 – 6,362,475 |
| 6,473,966 – 6,475,580 |
ABFW abdominal fat weight, ABFP abdominal fat percentage, SKINW skin weight, SKINP skin percentage
aQTLID from QTL chicken database
bPositions in the Gallus_gallus-5.0 version of the chicken genome
Discussion
Runs of homozygosity
ROH studies have addressed human evolution and diseases, conservation and evolution of wild species, and genomic features of livestock animals [2, 7, 13, 17]. The investigation of ROH as selection signatures using different generations of the TT line provided two new insights. First, the identification of important selection signatures that may refer to periods preceding the generations under study. Second, it allowed the comparison of how these selection signatures were shared among the individuals and how they have changed over the generations. Since shared ROH is an indication of regions under selection [18], the consensus regions of ROH (cROH) were used in this study to understand the possible biological consequences of selection in this broiler line.
The ROH identified in animals of TT line presented small to moderate sizes, ranging from 300 Kb to 4.9 Mb. Short ROH are most probable to be IBD genomic regions inherited from ancient ancestors indicating long term selection [43]. Over the generations, IBD segments tends to break down due to recombination events by repeated meiosis. Thus, ROH size is associated with the degree of shared parental ancestry and for how long it was passed across generations [32] and, as expected, short ROH regions shared between animals of the 7th and 16th generations encompassed genes associated with traits of interest (Additional file 10). It is important to mention that not all short ROH are IBD and a proportion of them may be identical-by-state (IBS) due to genetic drift, as well as population bottlenecks, and therefore some authors recommend caution in attributing these regions as candidate selection signatures [18, 44, 45]. The minimum size of 300 Kb was set in our analysis to detect ROH, considering that the use of WGS data calls ROH with smaller sizes compared to SNPchip data. A limitation in our study is that we were not able to determine which proportion of ROH is attributed to genetic drift and may lead to false positives. However, strategies were adopted to improve the chances to detect true selection signatures associated with the selection program. They were (i) using ROH regions in common with at least two animals, (ii) overlapping these regions with the Chicken QTL database, and (iii) performing MeSH overrepresentation analysis. These combined strategies reduce the chance to detect candidate selection signatures due to genetic drift.
In a study with offspring from animals of the 16th generation, Marchesi et al. [17] identified ROH in 1279 chickens using a high-density 600 K genotyping array data. They adopted the same parameters used in the present study, except for the minimum size of ROH and number of heterozygous SNP allowed. As expected, regions of ROH were commonly shared between both populations. More than 98% of the cROH identified herein in animals of the 16th generations, overlapped with ROH identified in the study of Marchesi et al. [17], corroborating our findings (Additional file 4). However, a higher number of ROH per animal and ROH with smaller sizes were identified in the 28 animals compared to those of Marchesi et al. [17]. We suggest that the reasons for identifying shorter ROH relies on the higher resolution of WGS data in comparison with SNPchip data. This difference in ROH calling was also observed in another study with feline that used both types of dataset for the same individuals [46].
It is also important to highlight that low coverage WGS data may present higher error rate of variant calling in comparison with SNPchip data, and this may lead to inaccuracy of ROH calling [29]. Thus, we followed parameters based on Ceballos et al. [29], which demonstrated equivalent results to SNPchip data’s results, when dealing with low coverage WGS. Moreover, in order to extend the chances of detecting accurate ROH, we opted to investigate consensus regions of ROH, i.e. regions of ROH in common with at least two animals, that might indicate regions under selection [18, 30]. All these observations corroborate with our suggestion that the smaller size and higher number of ROH possibly relies on the better resolution of WGS.
In our study, an increase in ROH abundance was observed, i.e. between the 7th and 16th generations there was an increase on the average number of ROH segments per animal and in the average size of segment per animal (Fig. 2 and Table 2). Investigation of the history of the breeding program indicate that these differences may have occurred due to a bottleneck effect. Marchesi et al. [17] estimated the Ne in TT line backing to 200 generations ago and reported a decay in Ne, especially in the last five generations, ranging from 157 to 113 chickens (Ne of TT Reference Population). Thus, when a population size is reduced, the average of heterozygosity in a certain locus is expected to decline, depending on the Ne [47, 48]. The occurrence of a bottleneck effect is supported by the observed increase in the mean genomic inbreeding coefficient from the 7th to the 16th generation (7th FROH = 0.078 and 16th FROH = 0.202). It is worth to mention that, even if mating between close related individuals is avoided some level of inbreeding is unavoidable, because TT line is a closed population [17].
Furthermore, the percentage of animals sharing a ROH region increased from the 7th to the 16th generation (Fig. 4). Mastrangelo et al. [18] reported similar observations of an increase in the abundance of ROH in a sheep breed it and suggested that a decrease in the effective population size (Ne) had occurred resulting in recent and historical autozygosity events. Thus, we suggest that the increase in homozygous regions across generations in TT line is consequence of selection pressure over genomic regions that are important to the breeding program’s goals jointly to reduction on the Ne and inbreeding. In fact, genes associated with traits of economic interest, such as the APOB, POMC, PPARG and other genes (Additional file 10), were annotated in regions shared with more animals of the 16th than with 7th generation, supporting that the regions containing these genes were under selection pressure in the respective period.
FST windows
An alternative approach applied in this study for identification of selection signatures was the genetic differentiation method based on allele frequency differences called FST statistics. Previous studies have used this method for detecting selection signature in livestock species, such as broilers [14, 21, 22, 49, 50]. Here we compared two groups of animals of TT broiler line, 10 generations distant from each other. The parameters were the same used by Boschiero et al. [22], in the comparison of the TT line against a layer line. The authors states that windows of 20 Kb allow a finer resolution of the regions in addition to windows with sufficient number of markers, considering that the amount of variants in a window is essential for increasing the power of the analysis [22, 51]. This intent was achieved in our results since we obtained an average of 216 SNP/window and 18 INDEL/window, which were similar to the results obtained by Boschiero et al. [22] with averages of 268 SNP/window and 26 INDEL/window. Furthermore, there was a considerable number of windows in common between SNP and INDEL datasets with 87% of overlapping, a fact also observed by Boschiero et al. [22].
Estimates of FST range from zero, meaning no genetic difference between the subpopulations, up to 1.0, meaning complete genetic differentiation [52]. Although there is not a determined threshold to capture regions that indicate genetic differentiation as a candidate selection signature, some authors use a threshold for the top 0.1% values of FST [21, 22]. Here we established a threshold value of 0.3 in order to obtain regions that might be in a differentiation process in the TT line. Only 0.002% of the windows had FST values above this threshold, for SNP and INDEL datasets, and the highest estimated values were 0.598 and 0.555 for SNP and INDEL datasets, respectively. In addition, as discussed by Boschiero et al. [22], combining strategies to detect selection signatures minimizes the occurrence of false positives.
Selection signatures of broiler performance and adaptation
Besides identifying regions in the chicken genome under selection pressure, knowing the genes annotated in these regions and how they biologically act is essential for understanding how the selection signatures contributed to the current phenotype of the evaluated animals. Since TT broiler line is under multi-trait selection since 1992 aiming to improve body weight, feed conversion, cut yields, breast weight, viability, fertility, and hatchability and to reduce abdominal fat [17, 23], it is expected that genes influencing the performance of these traits are under selection pressure.
Therefore, investigating which genes were annotated in the candidate selection signatures regions identified in both FST and ROH analysis helps to understand the biological mechanisms that affected the construction and evolution of the phenotype of TT line. In this sense, genes involved with traits of economic interest were identified in these regions. The genes IGFB2, TGFB2, HOXD9, HOXD10, POMC SPP1, SPP2, and IGF1 were some of the genes annotated in the candidate selection signatures of TT line and that were previously found in other selection signatures and associated with traits such as growth, body weight and composition, abdominal fat, organogenesis and feed intake and consumption [22, 50, 53–66]. Furthermore, we identified a group of genes annotated in the selection signatures that are involved with structural constituents, cell differentiation, and development of muscle tissue: ACTC1, AKAP6, ATP2A2, KCNMA1, MYO1B, MYO1C, MYO1E, MYO1F, MYO6, MYO7A, MYO10, MYO16, TPM4, VCL, and V1PR1 [22, 67]. Selection signatures identified in our analysis also indicate regions involved in lipid metabolism and adipose tissue development, encompassing the ADCY2, AKAP6, APOB, ATPR2, IGFBP2, PLA2R1, PPARG, SCARB1 and ZNF423 genes [22, 68–77].
Chickens raised in production systems are under several stressful conditions that can affect performance and the immune system of these animals [78, 79]. Stress challenged animals respond by changing their response behavior, metabolic rates, and functioning of cardiovascular and immune systems [78]. Thus, a selective pressure over genomic regions controlling responses to stressor conditions may occur, and the selection signatures identified with the ROH analysis shows a class of genes involved in these aspects: ACE, BAG1, CACNA1C, ELP2, HSPA8, MOCOS, MRTO4, MYH9, NSUN2, PAX5, PQLC2 and TRPM8 [16, 17, 80, 81].
Changes in TT line across the generations
Enrichment analysis using MeSH was performed in order to provide a better integrated view of the changes that occurred. [82]. Adipose tissue was in overrepresentation among the genes of these regions, what is expected since selection for growth in broilers, could lead to elevated fat deposition [83]. In addition, overlaps of cROH regions with QTL associated with fat deposition in TT Reference population support that regions affecting these traits were indirectly selected across generations in TT line. Hyperglycemia was also overrepresented and it is possibly a consequence of fat deposition in these animals. The excess of adipose tissue in chickens may lead to a condition similar to the early stage of type 2 diabetes in humans, manifesting hyperglycemia and exogenous insulin resistance [84, 85]. Another overrepresentation was ‘pregnancy in diabetics’, and, as it is known, pregnancy is not a biological mechanism of birds. However, Nadaf et al. [83] discuss that some QTL associated with chicken fatness have genes playing a role in obesity and diabetes in humans, and since MeSH is a tool that comprises animals in general, this association may have be done due to genes with similar functions.
Cystatins was among genes annotated in regions that underwent allele frequency changes during the 7th and 16th generations. Cystatin is a superfamily of reversible competitive inhibitors of cysteine proteases such as calpains, cathepsins, and ficins, and the cystatin system have important roles in protein turnover, antigen presentation and disease immunity [86, 87]. As well, tyrosine was overrepresented among the genes of FST SNP windows, which is considered a nonessential amino acid in animals [88].
The ROH analysis also provided information about regions that may indicate selection in a period that precedes the studied generations. In these regions there were overrepresentation of genes involved in skeletal muscle and the matrilin proteins, both important for the growth of chickens. Matrilin is a four-member family of proteins composing extracellular matrix of some tissues as cartilage, a connective tissue. They bind to collagen-containing fiber and other matrix constituents and can form oligomers [89].
Furthermore, the abundant amount of overlaps between cROH and FST windows and QTL regions associated with traits of economic interest in broilers, such as feed conversion, feed intake, growth, and abdominal fat, enforces the results of candidate selection signatures involved in performance traits. This fact together corroborates that TT line have been selected for growth and muscle deposition for a long period with a possible consequence of increased fat deposition, and for some traits such as proteic turnover and metabolism of tyrosine.
Conclusion
Regions under selection pressure in a paternal broiler line were investigated in this study. Using ROH analysis, we were able to identify regions that were inherited backing to common ancestors since the beginning of the broiler line origin, how these regions were shared between the animals of both generations, and what has changed in the genetic make-up of the TT line by selection between the 7th and 16th generations. FST-based analysis revealed regions that changed between generations. Annotation and enrichment analysis revealed the selection program affected genes and biological processes involved in skeletal muscle, cartilage and adipose tissues development. The investigation of selection signatures provided valuable insights about genes and biological processes involved in performance, adaptation and disease traits.
Additional files
A text file with information of the 5721 ROH identified in the analysis with the 28 chickens. Information comprises individual identification (IID), chromosome (CHR), start (POS1) and end (POS2) positions, size (KB), number of SNP in each ROH (NSNP), and density of SNP (DENSITY). (TXT 256 kb)
A compressed file of images (TIFF format) of genome wide distribution of runs of homozygosity (ROH) in TT population for each chromosome. (RAR 144 kb)
A text file with information about pools of overlapping ROH among the 28 chickens. Information comprises pool identification (POOL), family identification (FID), individual identification (IID), chromosome (CHR), start (BP1) and end (BP2) positions, size (KB), and number of SNP in each ROH of in the pool. Each pool has n + 2 lines: the n lines are respective to the n individuals in overlap and the last two represents the union and consensus regions of the overlap. (TXT 776 kb)
A text file with information of FST windows using SNP dataset. Information comprises chromosome (CHR), start (START) and end (END) positions, number of variants (NVAR), and weighted FST values (FST) for each window. (XLSX 19935 kb)
A text file with information of FST windows using INDEL dataset. Information comprises chromosome (CHR), start (START) and end (END) positions, number of variants (NVAR), and weighted FST values (FST) for each window. (TXT 2830 kb)
List of the 5681 genes annotated in the 1941 consensus regions of runs of homozygosity (cROH). (TXT 2578 kb)
List of the 56 genes annotated in the 178 FST windows (≥0.3) using SNP dataset. (XLS 5840 kb)
List of the 60 genes annotated in the 154 FST windows (≥0.3) using INDEL dataset. (XLS 79 kb)
Table S1: Genes annotated commonly between selection signatures of two or more datasets. The datasets were the 1941 consensus regions of ROH (cROH), the FST SNP windows (> 0.3), and the FST INDEL windows (> 0.3). (XLS 83 kb)
Table S2: Genes previously associated with traits of interest located in candidate selection signatures. Those located in consensus regions of ROH presents the number of animals sharing the common region, and those located in FST windows present the respective weighted FST values. (XLSX 394 kb)
positional information of overlaps between QTL from the QTL database with candidate selection signatures of ROH and FST windows (SNP and INDEL datasets). (DOCX 20 kb)
positional information of overlaps between cROH of animals of the 7th and 16th generations with ROH of animals of the TT Reference Population. (XLSX 1198 kb)
Acknowledgments
Not applicable.
Abbreviations
- ABFP
abdominal fat percentage
- ABFW
abdominal fat weight
- CEUA
Ethics Committee on Animal Utilization
- CONCEA
National Council of Animal Experimentation Control
- cROH
consensus region of runs of homozygosity
- EHH
extended haplotype homozygosity
- FDR
false discovery rate
- FROH
inbreeding coefficient based on runs of homozygosity
- FST
fixation index
- IBD
identical-by-descent
- IBS
identical-by-state
- iHS
integrated haplotype score
- INDEL
insertions and deletions
- Ne
effective population size
- QTL
quantitative trait loci
- ROH
runs of homozygosity
- SKINP
skin percentage
- SKINW
skin weight
- SNP
single nucleotide polymorphism
- UCSC
University of California, Santa Cruz
- uROH
union region of runs of homozygosity
- WGS
whole genome sequencing
Authors’ contributions
OACA, GCMM, FMR, CB and LLC conceived the idea of this research and participated in the experimental design. OACA and GCMM performed data analyses. OACA drafted the manuscript. OACA, GCMM, FMR, CB, LLC, MCL, AMGI, JOP, and FJN collaborated with interpretation, discussion and writing of the manuscript. AMGI, JOP, MCL, and LLC provided the experimental environment and data analysis support. All authors have read and approved the final manuscript.
Funding
This study was financed by the Brazilian Agricultural Research Corporation – Embrapa (project number 01.11.07.002.04.02), by the thematic project (2014/08704–0) from São Paulo Research Foundation (FAPESP) and by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior – Brasil (CAPES) – Finance Code 001. The National Council of Scientific and Technological Development (CNPq) grant number 481755/2007–1 from the Brazilian Government funded the TT Reference Population development. Almeida received a fellowship from CAPES, Finance Code 001. Moreira received fellowships from FAPESP, grants 14/21380–9 (in cooperation agreement with CAPES), and 16/00569–1 as well, fellowship from CNPq, − Brazil (153113/2018–9). Coutinho and Ledur are recipients of productivity fellowship from CNPq. The authors thank CAPES, CNPq, and FAPESP for the financial support. The foment agencies provided financial support to generate the data, however, they do not participate on the design of the study, sample collection, analysis, data interpretation and in writing the manuscript.
Availability of data and materials
All data generated from the analyses of this work are public and included in this article in the main manuscript or as additional files. All the SNPs utilized were submitted to European Variation Archive (EVA) – EMBL-EBI, accession PRJEB25004 and to dbSNP (NCBI) with the submitter handle “LBA_ESALQ”. Additional datasets and scripts used in the analyses are available from the corresponding author under reasonable request.
Ethics approval and consent to participate
This study followed experimental protocols pertinent to animal experimentation with the approval of the Embrapa Swine and Poultry Ethics Committee on Animal Utilization (CEUA) in Concordia, Santa Catarina State, Brazil, on resolution number 011/2010. It followed the rules of National Council of Animal Experimentation Control (CONCEA) in accordance with international guidelines to guarantee animal welfare.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Octávio Augusto Costa Almeida, Email: octaviocosta@usp.br.
Gabriel Costa Monteiro Moreira, Email: gcmmoreira@usp.br.
Fernanda Marcondes Rezende, Email: frezende@ufu.br.
Clarissa Boschiero, Email: clarissaboschi@yahoo.com.
Jane de Oliveira Peixoto, Email: jane.peixoto@embrapa.br.
Adriana Mercia Guaratini Ibelli, Email: adriana.ibelli@embrapa.br.
Mônica Corrêa Ledur, Email: monica.ledur@embrapa.br.
Francisco José de Novais, Email: francisco.novais@usp.br.
Luiz Lehmann Coutinho, Phone: +55-19-3429-4434, Email: llcoutinho@usp.br.
References
- 1.Keller SR, Taylor DR. History, chance and adaptation during biological invasion: separating stochastic phenotypic evolution from response to selection. Ecol Lett. 2008;11:852–866. doi: 10.1111/j.1461-0248.2008.01188.x. [DOI] [PubMed] [Google Scholar]
- 2.Ceballos FC, Joshi PK, Clark DW, Ramsay M, Wilson JF. Runs of homozygosity: windows into population history and trait architecture. Nat Rev Genet Nature Publishing Group. 2018;19:220–234. doi: 10.1038/nrg.2017.109. [DOI] [PubMed] [Google Scholar]
- 3.Al-Nasser A, Al-Khalaifa H, Al-Saffar A, Khalil F, Al-Bahouh M, Ragheb G, et al. Overview of chicken taxonomy and domestication. Worlds Poult Sci J. 2007;63:285–300. doi: 10.1017/S004393390700147X. [DOI] [Google Scholar]
- 4.Sabeti PCC, Reich DEE, Higgins JMM, Levine HZPZP, Richter DJJ, Schaffner SFF, et al. Detecting recent positive selection in the human genome from haplotype structure. Nature. 2002;419:832–837. doi: 10.1038/nature01140. [DOI] [PubMed] [Google Scholar]
- 5.Voight BF, Kudaravalli S, Wen X, Pritchard JK. A map of recent positive selection in the human genome. PLoS Biol. 2006;4:0446–0458. doi: 10.1371/journal.pbio.0040446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lewontin RC, Krakauer J. Distribution of gene frequency as a test of the theory of the selective neutrality of polymorphisms. Genetics. 1973;74:175–195. doi: 10.1093/genetics/74.1.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Christofidou P, Nelson CP, Nikpay M, Qu L, Li M, Loley C, et al. Runs of homozygosity: association with coronary artery disease and gene expression in monocytes and macrophages. Am J Hum Genet. 2015;97:228–237. doi: 10.1016/j.ajhg.2015.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lencz T, Lambert C, DeRosse P, Burdick KE, Morgan TV, Kane JM, et al. Runs of homozygosity reveal highly penetrant recessive loci in schizophrenia. Proc Natl Acad Sci. 2007;104:19942–19947. doi: 10.1073/pnas.0710021104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Orloff Mohammed S., Zhang Li, Bebek Gurkan, Eng Charis. Integrative Genomic Analysis Reveals Extended Germline Homozygosity with Lung Cancer Risk in the PLCO Cohort. PLoS ONE. 2012;7(2):e31975. doi: 10.1371/journal.pone.0031975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pemberton TJ, Absher D, Feldman MW, Myers RM, Rosenberg NA, Li JZ. Genomic patterns of homozygosity in worldwide human populations. Am J Hum Genet. 2012;91:275–292. doi: 10.1016/j.ajhg.2012.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lashmar SF, Visser C, van Marle-Köster E, Muchadeyi FC. Genomic diversity and autozygosity within the SA Drakensberger beef cattle breed. Livest Sci Elsevier BV. 2018;212:111–119. doi: 10.1016/j.livsci.2018.04.006. [DOI] [Google Scholar]
- 12.Purfield Deirdre C, Berry Donagh P, McParland Sinead, Bradley Daniel G. Runs of homozygosity and population history in cattle. BMC Genetics. 2012;13(1):70. doi: 10.1186/1471-2156-13-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Szmatoła T, Gurgul A, Ropka-Molik K, Jasielczuk I, Zabek T, Bugno-Poniewierska M. Characteristics of runs of homozygosity in selected cattle breeds maintained in Poland. Livest Sci. 2016;188:72–80. doi: 10.1016/j.livsci.2016.04.006. [DOI] [Google Scholar]
- 14.Zhao F, McParland S, Kearney F, Du L, Berry DP. Detection of selection signatures in dairy and beef cattle using high-density genomic information. Genet Sel Evol Genetics Selection Evolution. 2015;47:1–12. doi: 10.1186/s12711-014-0081-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bosse Mirte, Megens Hendrik-Jan, Madsen Ole, Paudel Yogesh, Frantz Laurent A. F., Schook Lawrence B., Crooijmans Richard P. M. A., Groenen Martien A. M. Regions of Homozygosity in the Porcine Genome: Consequence of Demography and the Recombination Landscape. PLoS Genetics. 2012;8(11):e1003100. doi: 10.1371/journal.pgen.1003100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fleming DS, Koltes JE, Markey AD, Schmidt CJ, Ashwell CM, Rothschild MF, et al. Genomic analysis of Ugandan and Rwandan chicken ecotypes using a 600 k genotyping array. BMC Genomics BMC Genomics. 2016;17:1–16. doi: 10.1186/s12864-015-2294-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marchesi JAP, Buzanskas ME, Cantão ME, Ibelli AMG, Peixoto JO, Joaquim LB, et al. Relationship of runs of homozygosity with adaptive and production traits in a paternal broiler line. Animal [internet]. 2017 [cited 2018 mar 24];1–9. Available from: http://www.ncbi.nlm.nih.gov/pubmed/29065939 [DOI] [PubMed]
- 18.Mastrangelo S, Tolone M, Sardina MT, Sottile G, Sutera AM, Di Gerlando R, et al. Genome-wide scan for runs of homozygosity identifies potential candidate genes associated with local adaptation in Valle del Belice sheep. Genet Sel Evol BioMed Central. 2017;49:1–10. doi: 10.1186/s12711-016-0283-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wright S. The genetical structure of populations. Ann Eugenics. 1951;15:323–354. doi: 10.1111/j.1469-1809.1949.tb02451.x. [DOI] [PubMed] [Google Scholar]
- 20.Weir BS. Estimating F-statistics : a historical view. 2015. p. 79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lillie M, Sheng ZY, Honaker CF, Andersson L, Siegel PB, Carlborg O. Genomic signatures of 60 years of bidirectional selection for 8-week body weight in chickens. Poult Sci. 2018;97:781–790. doi: 10.3382/ps/pex383. [DOI] [PubMed] [Google Scholar]
- 22.Boschiero C, Moreira GCM, Gheyas AA, Godoy TF, Gasparin G, Mariani PDSC, et al. Genome-wide characterization of genetic variants and putative regions under selection in meat and egg-type chicken lines. BMC Genomics [Internet]. 2018; [cited 2018 Mar 19];19:83. Available from: http://www.ncbi.nlm.nih.gov/pubmed/29370772. [DOI] [PMC free article] [PubMed]
- 23.Venturini GC, Stafuzza NB, Cardoso DF, Baldi F, Ledur MC, Peixoto JO, et al. Association between ACTA1 candidate gene and performance, organs and carcass traits in broilers. Poult Sci. 2015;94:2863–2869. doi: 10.3382/ps/pev285. [DOI] [PubMed] [Google Scholar]
- 24.Nunes BDN, Ramos SB, Savegnago RP, Ledur MC, Nones K, Klein CH, et al. Genetic parameters for body weight, carcass chemical composition and yield in a broiler-layer cross developed for QTL mapping. Genet Mol Biol [Internet] 2011;34:429–34. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3168183&tool=pmcentrez&rendertype=abstract [DOI] [PMC free article] [PubMed]
- 25.Langmead B, Salzberg SL. Fast gapped-read alignment with bowtie 2. Nat Methods [Internet] 2012 [cited 2013 Sep 17];9:357–9. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3322381&tool=pmcentrez&rendertype=abstract [DOI] [PMC free article] [PubMed]
- 26.Moreira G. C. M., Godoy T. F., Boschiero C., Gheyas A., Gasparin G., Andrade S. C. S., Paduan M., Montenegro H., Burt D. W., Ledur M. C., Coutinho L. L. Variant discovery in a QTL region on chromosome 3 associated with fatness in chickens. Animal Genetics. 2015;46(2):141–147. doi: 10.1111/age.12263. [DOI] [PubMed] [Google Scholar]
- 27.Purcell Shaun, Neale Benjamin, Todd-Brown Kathe, Thomas Lori, Ferreira Manuel A.R., Bender David, Maller Julian, Sklar Pamela, de Bakker Paul I.W., Daly Mark J., Sham Pak C. PLINK: A Tool Set for Whole-Genome Association and Population-Based Linkage Analyses. The American Journal of Human Genetics. 2007;81(3):559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Purcell S. PLINK 1.9. p. https://www.cog-genomics.org/plink2.
- 29.Ceballos FC, Hazelhurst S, Ramsay M. Assessing runs of homozygosity: a comparison of SNP Array and whole genome sequence low coverage data. BMC Genomics BMC Genomics. 2018;19:1–12. doi: 10.1186/s12864-017-4368-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang Q, Guldbrandtsen B, Bosse M, Lund MS, Sahana G. Runs of homozygosity and distribution of functional variants in the cattle genome. BMC Genomics BMC Genomics. 2015;16:1–16. doi: 10.1186/1471-2164-16-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Peripolli E, Stafuzza NB, Munari DP, Lima ALF, Irgang R, Machado MA, et al. Assessment of runs of homozygosity islands and estimates of genomic inbreeding in Gyr (Bos indicus) dairy cattle. BMC Genomics BMC Genomics. 2018;19:1–13. doi: 10.1186/s12864-017-4368-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McQuillan R, Leutenegger AL, Abdel-Rahman R, Franklin CS, Pericic M, Barac-Lauc L, et al. Runs of homozygosity in European populations. Am J Hum Genet. 2008;83:359–372. doi: 10.1016/j.ajhg.2008.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weir BS, Cockerham CC. Estimating F-Statistics for the Analysis of Population Structure. Evolution (N. Y). [Internet]. Society for the Study of Evolution; 1984 [cited 2017 Nov 7]; 38:1358. Available from: http://www.jstor.org/stable/2408641?origin=crossref [DOI] [PubMed]
- 34.Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA, et al. The variant call format and VCFtools. Bioinformatics. 2011;27:2156–2158. doi: 10.1093/bioinformatics/btr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zerbino DR, Achuthan P, Akanni W, Amode MR, Barrell D, Bhai J, et al. Ensembl 2018. Nucleic Acids Res. 2018;46:D754–D761. doi: 10.1093/nar/gkx1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yu G. Using meshes for MeSH term enrichment and semantic analyses. Bioinformatics. 2018;34:3766–3767. doi: 10.1093/bioinformatics/bty410. [DOI] [PubMed] [Google Scholar]
- 38.Morota G. R package for MeSH enrichment analysis - GitHub. 2014. [Google Scholar]
- 39.Team RDC. R: a language and environment for statistical computing. 2008. p. https://www.R-project.org.
- 40.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing [internet]. J. R. Stat. Soc. Ser. B. WileyRoyal statistical society; 1995 [cited 2018 may 10]. p. 289–300. Available from: https://www.jstor.org/stable/2346101.
- 41.Chicken QTL database (release 37). 2018.
- 42.Moreira GCM, Boschiero C, Cesar ASM, Reecy JM, Godoy TF, Trevisoli PA, et al. A genome-wide association study reveals novel genomic regions and positional candidate genes for fat deposition in broiler chickens. BMC Genomics [Internet]. BioMed Central; 2018 [cited 2018 Jun 7];19:374. Available from: http://www.ncbi.nlm.nih.gov/pubmed/29783939 [DOI] [PMC free article] [PubMed]
- 43.Howrigan DP, Simonson MA, Keller MC. Detecting autozygosity through runs of homozygosity: a comparison of three autozygosity detection algorithms. BMC Genomics. 2011;12. [DOI] [PMC free article] [PubMed]
- 44.Purfield DC, Mcparland S, Wall E, Berry DP. The distribution of runs of homozygosity and selection signatures in six commercial meat sheep breeds. PLoS One. 2017;12:1–23. doi: 10.1371/journal.pone.0176780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kardos M, Luikart G, Bunch R, Dewey S, Edwards W, Mcwilliam S, et al. Whole-genome resequencing uncovers molecular signatures of natural and sexual selection in wild bighorn sheep. Mol Ecol. 2015;24:5616–5632. doi: 10.1111/mec.13415. [DOI] [PubMed] [Google Scholar]
- 46.Farias FHG, Tomlinson C, Labuda J, Perez-Camargo G, Middleton R, Warren WC. The practical use of genome sequencing data in the management of a feline colony pedigree. BMC Vet Res BMC Veterinary Research. 2017;13:1–9. doi: 10.1186/s12917-017-1144-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wright S. Evolution in Mendelian Populations. Genetics. 1931;16:97–159. doi: 10.1093/genetics/16.2.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nei M, Maruyama T, Chakraborty R. The bottleneck effect and genetic variability in populations. Evolution (N Y) 1975;29:1–10. doi: 10.1111/j.1558-5646.1975.tb00807.x. [DOI] [PubMed] [Google Scholar]
- 49.Ma Y, Wei J, Zhang Q, Chen L, Wang J, Liu J, et al. A genome scan for selection signatures in pigs. PLoS One. 2015;10:1–18. doi: 10.1371/journal.pone.0116850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gholami Mahmood, Erbe Malena, Gärke Christian, Preisinger Rudolf, Weigend Annett, Weigend Steffen, Simianer Henner. Population Genomic Analyses Based on 1 Million SNPs in Commercial Egg Layers. PLoS ONE. 2014;9(4):e94509. doi: 10.1371/journal.pone.0094509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Willing EM, Dreyer C, van Oosterhout C. Estimates of genetic differentiation measured by fst do not necessarily require large sample sizes when using many snp markers. PLoS One. 2012;7:1–7. doi: 10.1371/journal.pone.0042649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Holsinger KE, Weir BS. Genetics in geographically structured populations: defining, estimating and interpreting FST. Nat Rev Genet. 2009;10:639–650. doi: 10.1038/nrg2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bai Y, Sun G, Kang X, Han R, Tian Y, Li H, et al. Polymorphisms of the pro-opiomelanocortin and agouti-related protein genes and their association with chicken production traits. Mol Biol Rep. 2012;39:7533–7539. doi: 10.1007/s11033-012-1587-y. [DOI] [PubMed] [Google Scholar]
- 54.Bennett AK, Hester PY, Spurlock DEM. Polymorphisms in vitamin D receptor, osteopontin, insulin-like growth factor 1 and insulin, and their associations with bone, egg and growth traits in a layer - broiler cross in chickens. Anim Genet. 2006;37:283–286. doi: 10.1111/j.1365-2052.2006.01439.x. [DOI] [PubMed] [Google Scholar]
- 55.Bhattacharya TK, Chatterjee RN, Dushyanth K, Paswan C, Shukla R, Shanmugam M. Polymorphism and expression of insulin-like growth factor 1 (IGF1) gene and its association with growth traits in chicken. Br Poult Sci. 2015;56:398–407. doi: 10.1080/00071668.2015.1041098. [DOI] [PubMed] [Google Scholar]
- 56.Bian LH, Wang SZ, Wang QG, Zhang S, Wang YX, Li H. Variation at the insulin-like growth factor 1 gene and its association with body weight traits in the chicken. J Anim Breed Genet. 2008;125:265–270. doi: 10.1111/j.1439-0388.2008.00739.x. [DOI] [PubMed] [Google Scholar]
- 57.Zhou H, Mitchell AD, McMurtry JP, Ashwell CM, Lamont SJ. Insulin-like growth factor-I gene polymorphism associations with growth, body composition, skeleton integrity, and metabolic traits in chickens. Poult Sci. 2005;84:212–219. doi: 10.1093/ps/84.2.212. [DOI] [PubMed] [Google Scholar]
- 58.Agarwal SK, Cogburn LA, Burnside J. Comparison of gene expression in normal and growth hormone receptor-deficient dwarf chickens reveals a novel growth hormone regulated gene. Biochem Biophys Res Commun. 1995;206:153–160. doi: 10.1006/bbrc.1995.1022. [DOI] [PubMed] [Google Scholar]
- 59.Li ZH, Li H, Zhang H, Wang SZ, Wang QG, Wang YX. Identification of a single nucleotide polymorphism of the insulin-like growth factor binding protein 2 gene and its association with growth and body composition traits in the chicken. J Anim Sci. 2006;84:2902–2906. doi: 10.2527/jas.2006-144. [DOI] [PubMed] [Google Scholar]
- 60.Leng L, Wang S, Li Z, Wang Q, Li H. A polymorphism in the 3′-flanking region of insulin-like growth factor binding protein 2 gene associated with abdominal fat in chickens. Poult Sci. 2009;88:938–942. doi: 10.3382/ps.2008-00453. [DOI] [PubMed] [Google Scholar]
- 61.Yamagishi T, Ando K, Nakamura H, Nakajima Y. Expression of the Tgfβ2 gene during Chick embryogenesis. Anat Rec. 2012;295:257–267. doi: 10.1002/ar.22400. [DOI] [PubMed] [Google Scholar]
- 62.Darzi Niarami M, Masoudi AA, Vaez Torshizi R. Association of Single Nucleotide Polymorphism of GHSR and TGFB2 genes with growth and body composition traits in sire and dam lines of a broiler chicken. Anim Biotechnol. 2014;25:13–22. doi: 10.1080/10495398.2013.803478. [DOI] [PubMed] [Google Scholar]
- 63.Tang S, Ou J, Sun D, Zhang Y, Xu G, Zhang Y. A novel 62-bp indel mutation in the promoter region of transforming growth factor-beta 2 (TGFB2) gene is associated with body weight in chickens. Anim Genet. 2011;42:108–112. doi: 10.1111/j.1365-2052.2010.02060.x. [DOI] [PubMed] [Google Scholar]
- 64.Tickle C. The contribution of chicken embryology to the understanding of vertebrate limb development. Mech Dev. 2004;121:1019–1029. doi: 10.1016/j.mod.2004.05.015. [DOI] [PubMed] [Google Scholar]
- 65.Izpisúa-Belmonte JC, Tickle C, Dollé P, Wolpert L, Duboule D. Expression of the homeobox Hox-4 genes and the specification of position in chick wing development. Nature. 1991;350:585–589. doi: 10.1038/350585a0. [DOI] [PubMed] [Google Scholar]
- 66.Wardlaw SL. Obesity as a neuroendocrine disease: lessons to be learned from proopiomelanocortin and melanocortin receptor mutations in mice and men. J Clin Endocrinol Metab. 2001;86:1442–1446. doi: 10.1210/jcem.86.4.7388. [DOI] [PubMed] [Google Scholar]
- 67.Sellers James R. Myosins: a diverse superfamily. Biochimica et Biophysica Acta (BBA) - Molecular Cell Research. 2000;1496(1):3–22. doi: 10.1016/S0167-4889(00)00005-7. [DOI] [PubMed] [Google Scholar]
- 68.Sato K, Abe H, Kono T, Yamazaki M, Nakashima K, Kamada T, et al. Changes in peroxisome proliferator-activated receptor gamma gene expression of chicken abdominal adipose tissue with different age, sex and genotype. Anim Sci J. 2009;80:322–327. doi: 10.1111/j.1740-0929.2009.00639.x. [DOI] [PubMed] [Google Scholar]
- 69.Ji Bo, Ernest Ben, Gooding Jessica R, Das Suchita, Saxton Arnold M, Simon Jean, Dupont Joelle, Métayer-Coustard Sonia, Campagna Shawn R, Voy Brynn H. Transcriptomic and metabolomic profiling of chicken adipose tissue in response to insulin neutralization and fasting. BMC Genomics. 2012;13(1):441. doi: 10.1186/1471-2164-13-441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Larkina TA, Sazanova AL, Fomichev KA, Barkova OY, Malewski T, Jaszczak K, et al. HMG1A and PPARG are differently expressed in the liver of fat and lean broilers. J Appl Genet. 2011;52:225–228. doi: 10.1007/s13353-010-0023-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sun YN, Gao Y, Qiao SP, Wang SZ, Duan K, Wang YX, et al. Epigenetic DNA methylation in the promoters of peroxisome proliferator-activated receptor γ in chicken lines divergently selected for fatness. J Anim Sci. 2014;92:48–53. doi: 10.2527/jas.2013-6962. [DOI] [PubMed] [Google Scholar]
- 72.Sato K, Matsushita K, Matsubara Y, Kamada T, Akiba Y. Adipose tissue fat accumulation is reduced by a single intraperitoneal injection of peroxisome proliferator-activated receptor gamma agonist when given to newly hatched chicks. Poult Sci. 2008;87:2281–2286. doi: 10.3382/ps.2008-00086. [DOI] [PubMed] [Google Scholar]
- 73.Matsubara Y, Aoki M, Endo T, Sato K. Characterization of the expression profiles of adipogenesis-related factors, ZNF423, KLFs and FGF10, during preadipocyte differentiation and abdominal adipose tissue development in chickens. Comp Biochem Physiol - B Biochem Mol Biol Elsevier Inc. 2013;165:189–195. doi: 10.1016/j.cbpb.2013.04.002. [DOI] [PubMed] [Google Scholar]
- 74.Hermier D, Forgez P, Chapman MJ. A density gradient study of the lipoprotein and apolipoprotein distribution in the chicken, Gallus domesticus. Biochim Biophys Acta (BBA)/Lipids Lipid Metab. 1985;836:105–118. doi: 10.1016/0005-2760(85)90226-7. [DOI] [PubMed] [Google Scholar]
- 75.Schumaker VN, Phillips ML, Chatterton JE. Apolipoprotein B and low-density lipoprotein structure: implications for biosynthesis of triglyceride-rich lipoproteins. Adv Protein Chem. 1994;45:205–248. doi: 10.1016/S0065-3233(08)60641-5. [DOI] [PubMed] [Google Scholar]
- 76.Zhang S, Shi H, Li H. Cloning and tissue expression characterization of the chicken APOB gene. Anim Biotechnol. 2007;18:243–250. doi: 10.1080/10495390701574887. [DOI] [PubMed] [Google Scholar]
- 77.Zhang S., Li H., Shi H. Single Marker and Haplotype Analysis of the Chicken Apolipoprotein B Gene T123G and D9500D9- Polymorphism Reveals Association with Body Growth and Obesity. Poultry Science. 2006;85(2):178–184. doi: 10.1093/ps/85.2.178. [DOI] [PubMed] [Google Scholar]
- 78.Renaudeau D, Collin A, Yahav S, De Basilio V, Gourdine JL, Collier RJ. Adaptation to hot climate and strategies to alleviate heat stress in livestock production. Animal. 2012;6:707–728. doi: 10.1017/S1751731111002448. [DOI] [PubMed] [Google Scholar]
- 79.Lara LJ, Rostagno MH. Impact of heat stress on poultry production. Animals. 2013;3:356–369. doi: 10.3390/ani3020356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sun L, Lamont SJ, Cooksey AM, McCarthy F, Tudor CO, Vijay-Shanker K, et al. Transcriptome response to heat stress in a chicken hepatocellular carcinoma cell line. Cell Stress Chaperones. 2015;20:939–950. doi: 10.1007/s12192-015-0621-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chuang HH, Neuhausser WM, Julius D. The super-cooling agent icilin reveals a mechanism of coincidence detection by a temperature-sensitive TRP channel. Neuron. 2004;43:859–869. doi: 10.1016/j.neuron.2004.08.038. [DOI] [PubMed] [Google Scholar]
- 82.Morota G., Peñagaricano F., Petersen J. L., Ciobanu D. C., Tsuyuzaki K., Nikaido I. An application of MeSH enrichment analysis in livestock. Animal Genetics. 2015;46(4):381–387. doi: 10.1111/age.12307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Nadaf Javad, Pitel Frédérique, Gilbert Hélène, Duclos Michel J., Vignoles Florence, Beaumont Catherine, Vignal Alain, Porter Tom E., Cogburn Larry A., Aggrey Samuel E., Simon Jean, Le Bihan-Duval Elisabeth. QTL for several metabolic traits map to loci controlling growth and body composition in an F2 intercross between high- and low-growth chicken lines. Physiological Genomics. 2009;38(3):241–249. doi: 10.1152/physiolgenomics.90384.2008. [DOI] [PubMed] [Google Scholar]
- 84.Akiba Y, Chida Y, Takahashi T, Ohtomo Y, Sato K, Takahashi K. Persistent hypoglycemia induced by continuous insulin infusion in broiler chickens. Br Poult Sci. 1999;40:701–705. doi: 10.1080/00071669987124. [DOI] [PubMed] [Google Scholar]
- 85.Renema RA, Rustad ME, Robinson FE. Implications of changes to commercial broiler and broiler breeder body weight targets over the past 30 years. Worlds Poult Sci J. 2007;63:457–472. doi: 10.1017/S0043933907001572. [DOI] [Google Scholar]
- 86.Ochieng J, Chaudhuri G. Cystatin superfamily. J Health Care Poor Underserved. 2010;21:51–70. doi: 10.1353/hpu.0.0257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Turk ">Vito. Cystatins: Biochemical and structural properties, and medical relevance. Frontiers in Bioscience. 2008;Volume(13):5406. doi: 10.2741/3089. [DOI] [PubMed] [Google Scholar]
- 88.Wu G. Dietary requirements of synthesizable amino acids by animals: a paradigm shift in protein nutrition. J Anim Sci Biotechnol. 2014;5:1–12. doi: 10.1186/2049-1891-5-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wagener R, Ehlen HWA, Ko YP, Kobbe B, Mann HH, Sengle G, et al. The matrilins - adaptor proteins in the extracellular matrix. FEBS Lett. 2005;579:3323–3329. doi: 10.1016/j.febslet.2005.03.018. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A text file with information of the 5721 ROH identified in the analysis with the 28 chickens. Information comprises individual identification (IID), chromosome (CHR), start (POS1) and end (POS2) positions, size (KB), number of SNP in each ROH (NSNP), and density of SNP (DENSITY). (TXT 256 kb)
A compressed file of images (TIFF format) of genome wide distribution of runs of homozygosity (ROH) in TT population for each chromosome. (RAR 144 kb)
A text file with information about pools of overlapping ROH among the 28 chickens. Information comprises pool identification (POOL), family identification (FID), individual identification (IID), chromosome (CHR), start (BP1) and end (BP2) positions, size (KB), and number of SNP in each ROH of in the pool. Each pool has n + 2 lines: the n lines are respective to the n individuals in overlap and the last two represents the union and consensus regions of the overlap. (TXT 776 kb)
A text file with information of FST windows using SNP dataset. Information comprises chromosome (CHR), start (START) and end (END) positions, number of variants (NVAR), and weighted FST values (FST) for each window. (XLSX 19935 kb)
A text file with information of FST windows using INDEL dataset. Information comprises chromosome (CHR), start (START) and end (END) positions, number of variants (NVAR), and weighted FST values (FST) for each window. (TXT 2830 kb)
List of the 5681 genes annotated in the 1941 consensus regions of runs of homozygosity (cROH). (TXT 2578 kb)
List of the 56 genes annotated in the 178 FST windows (≥0.3) using SNP dataset. (XLS 5840 kb)
List of the 60 genes annotated in the 154 FST windows (≥0.3) using INDEL dataset. (XLS 79 kb)
Table S1: Genes annotated commonly between selection signatures of two or more datasets. The datasets were the 1941 consensus regions of ROH (cROH), the FST SNP windows (> 0.3), and the FST INDEL windows (> 0.3). (XLS 83 kb)
Table S2: Genes previously associated with traits of interest located in candidate selection signatures. Those located in consensus regions of ROH presents the number of animals sharing the common region, and those located in FST windows present the respective weighted FST values. (XLSX 394 kb)
positional information of overlaps between QTL from the QTL database with candidate selection signatures of ROH and FST windows (SNP and INDEL datasets). (DOCX 20 kb)
positional information of overlaps between cROH of animals of the 7th and 16th generations with ROH of animals of the TT Reference Population. (XLSX 1198 kb)
Data Availability Statement
All data generated from the analyses of this work are public and included in this article in the main manuscript or as additional files. All the SNPs utilized were submitted to European Variation Archive (EVA) – EMBL-EBI, accession PRJEB25004 and to dbSNP (NCBI) with the submitter handle “LBA_ESALQ”. Additional datasets and scripts used in the analyses are available from the corresponding author under reasonable request.