Abstract
Trebouxiophyceae (Chlorophyta) is a species-rich class of green algae with a remarkable morphological and ecological diversity. Currently, there are a few completely sequenced mitochondrial genomes (mtDNA) from diverse Trebouxiophyceae but none from lichen symbionts. Here, we report the mitochondrial genome sequence of Trebouxia sp. TR9 as the first complete mtDNA sequence available for a lichen-symbiont microalga. A comparative study of the mitochondrial genome of Trebouxia sp. TR9 with other chlorophytes showed important organizational changes, even between closely related taxa. The most remarkable change is the enlargement of the genome in certain Trebouxiophyceae, which is principally due to larger intergenic spacers and seems to be related to a high number of large tandem repeats. Another noticeable change is the presence of a relatively large number of group II introns interrupting a variety of tRNA genes in a single group of Trebouxiophyceae, which includes Trebouxiales and Prasiolales. In addition, a fairly well-resolved phylogeny of Trebouxiophyceae, along with other Chlorophyta lineages, was obtained based on a set of seven well-conserved mitochondrial genes.
Subject terms: Evolution, Plant sciences
Introduction
The use of organelle genomic information has become a common practice for comparative studies and phylogenetic analyses of entire genomes (phylogenomics). The sequencing of organelle genomes provides valuable information about the evolution of both the organelles and the organisms that carry them. Green photosynthetic eukaryotic organisms include both Chlorophyta and Streptophyta phyla. Chlorophyta comprises unicellular and multicellular green algae, whereas Streptophyta contains both green algae and embryophytes1. Initially, morphology and ultrastructural data allowed for distinguishing four classes within Chlorophyta: Chlorophyceae, Prasinophyceae, Trebouxiophyceae and Ulvophyceae, in alphabetical order2. Later, molecular data contributed to elucidating the evolution of chlorophytes, corroborating the initial hypothesis of the antiquity of Prasinophyceae, which gave rise to the remaining Chlorophyta classes3,4. Regarding this issue, the phylogenetic relationships among chlorophytes remain controversial, especially at higher taxonomic levels (order, class).
Chloroplast genomes are especially attractive for evolutionary studies of photosynthetic eukaryotes. Currently, the number of complete sequences of organellar genomes in the NCBI databases is more than fifteen-fold greater in streptophytes than in chlorophytes (2,492 and 161 are available in the NCBI databases, respectively); among the 161 genomes from chlorophytes, only 56 correspond to mitogenomes. This imbalance is more remarkable among Trebouxiophyceae, since the availability of chloroplast genomes has increased in the few last years (approximately 30 chloroplast genomes are available in the NCBI databases, most of them are published)1,5,6. In contrast, almost a dozen mitogenomes are currently available, and some of them were recently published, including those of Chlorella heliozoae, Micractinium conductrix7 and Botryococcus braunii8.
Trebouxiophyceae have a wide range of lifestyles, including free-living species, endosymbionts of heliozoa (‘Chlorella’-like green algae)9, plants (e.g., Coccomyxa)10, mutualistic or parasitic associations with invertebrates11, non-photosynthetic microalgae (e.g., Helicosporidium and Prototheca)12 and symbionts of fungi (e.g., Trebouxia, Asterochloris, Symbiochloris, Myrmecia and others)13–15.
Within the Trebouxiophyceae, 22 genera are known to be involved in symbiosis with lichen thalli15. However, there is no completely sequenced mitochondrial genome from any lichen microalga, since only four regions of the mtDNA of Trebouxia aggregata are available in GenBank (accessions EU123944, EU123947, EU123948 and EU123949). The genus Trebouxia is one of the most species-rich microalgal genera, comprising non-motile coccoid green algae that are present in approximately one half of all lichens15. The initial number of Trebouxia species formally described on the basis of phenotypic characters was approximately 3016. This number has increased in recent years after the application of phylogenetic species concepts (e.g.17–19). However, the lack of closed complete genomes that can be used as references has hindered the systematic study of the molecular evolution of the members of the Trebouxia genus. Trebouxia sp. TR9 is a phycobiont of the lichen Ramalina farinacea (L.) Ach., which has been extensively studied in recent years in relation to many ecological and physiological traits20–26. However, molecular analyses of this Trebouxia species have been restricted to a few molecular markers from both nuclear and chloroplast genomes (e.g.26–30) without consideration of the mitochondrial genome. In this study, we report the complete sequence of the mitochondrial genome of Trebouxia sp. TR9, determined from high-throughput Roche 454 pyrosequencing. We compare its structure, organization and gene content with other mitochondrial genomes reported for Trebouxiophyceae and other Chlorophyta microalgae and provide a phylogenetic reconstruction on the basis of seven selected mitochondrial genes (cob, cox1, nad1, nad2, nad4, nad5 and nad6).
Results
Structural features of the completely assembled mtDNA of Trebouxia sp. TR9
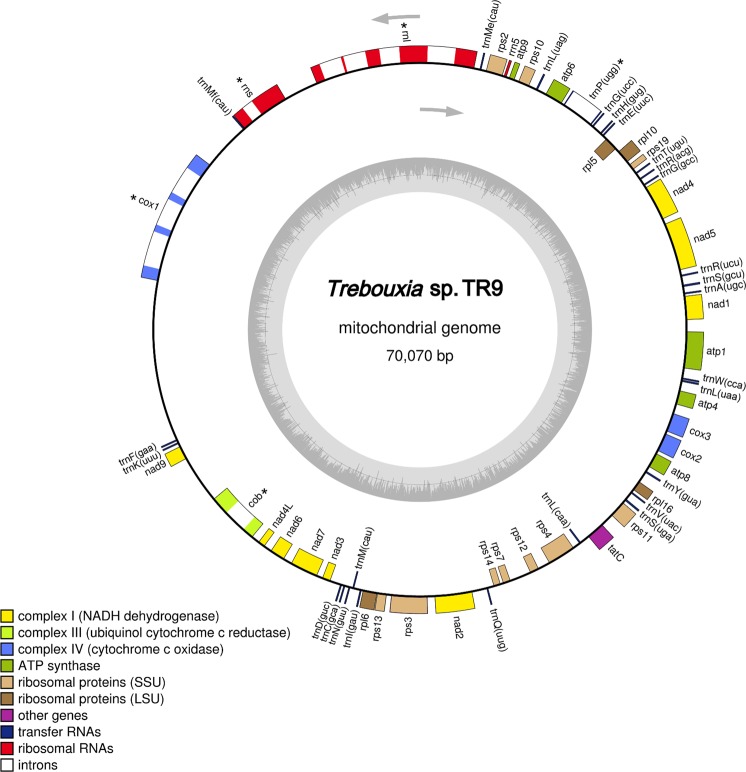
The mitochondrial genome (mtDNA) of Trebouxia sp. TR9 (Fig. 1) is a circular molecule of 70,070 bp with a GC content of 32.7% and a total of 67 genes. Thirty-three genes encoded conserved proteins, including nine subunits of the electron transport complex I (nad1, nad2, nad3, nad4, nad4L, nad5, nad6, nad7 and nad9), one subunit of complex III (cob), three subunits of complex IV (cox1, cox2 and cox3), five F0 subunits of the ATP-synthase complex (atp1, atp4, atp6, atp8, and atp9), fourteen ribosomal proteins: ten for the small ribosomal subunit (rps2, rps3, rps4, rps7, rps10, rps11, rps12, rps13, rps14 and rps19) and four for the large ribosomal subunit (rpl5, rpl6, rpl10 and rpl16), the TatC membrane protein, and four genes encoding putative LAGLIDADG homing endonucleases (LHEs). In addition, 27 tRNA genes and three genes for ribosomal RNAs (rrnl, rrns and rrn5) were identified in the Trebouxia sp. TR9 mtDNA. Regarding the tRNAs, a total of 26 tRNA genes were identified with RNAweasel and tRNAscan-SE, whereas with ARAGORN, we found 27 tRNAs, including an additional trnP (ugg). Three tRNA genes with different sequences and the same anticodon (cau) were identified for tRNA-Met. Three tRNA genes with different sequences and anticodons were found for tRNA-Leu, and two tRNA genes with different sequences and anticodons were found for tRNA-Gly, tRNA-Ile, tRNA-Arg and tRNA-Ser. For the remaining 13 tRNAs, only one gene each was found. The additional tRNA gene found with ARAGORN corresponded to tRNA-Pro (ugg) spanning from positions 9,751 to 11,174, with a group II intron of 1,350 bp predicted with RNAWEASEL. Regarding intron content, a total of ten introns were identified within the mtDNA of Trebouxia sp. TR9 with RNAweasel. Nine of them were group I introns, and only one belonged to group II (Fig. 1). Group I introns were located within the genes rrnL, rrnS, cob and cox1. Most of them belonged to group IB (within the genes cox1 and rrnL), followed by group IA (within genes rrnS and rrnL) and a single intron of group ID (within the gene cob). Intron sizes ranged from 500 to 1,443 bp within the genes rrnS and cox1 (third intron), respectively. Only introns within the genes cob and cox1 included open reading frames (ORFs) encoding homing endonucleases (HEs), with either a single or two LAGLIDADG motifs in each of them. As stated above, a group II intron was found in the gene coding tRNA-Pro (ugg).
Figure 1.
Gene map of the complete mitochondrial genome of the microalga Trebouxia sp. TR9. Genes shown inside the circle are transcribed clockwise, and genes outside are transcribed counter clockwise. Asterisks indicate genes with introns.
Phylogenetic analyses of Trebouxiophyceae and other Chlorophyta lineages based on seven mitochondrial genes
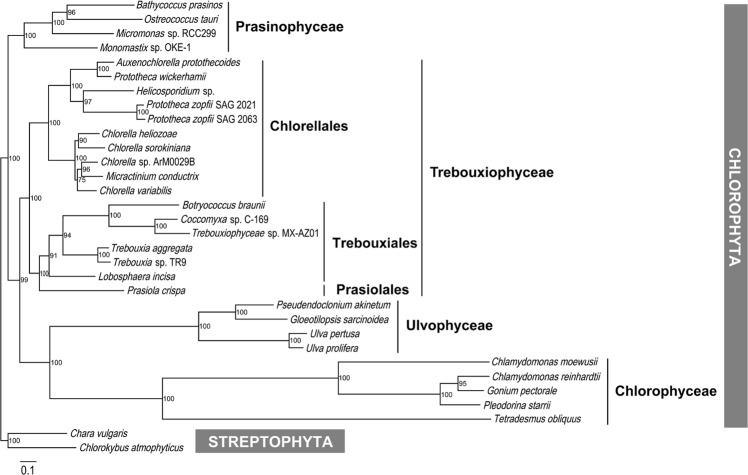
Here, we present a phylogenetic reconstruction of chlorophytes (Fig. 2), including species from different divisions and two streptophytes as outgroups, Chlorokybus atmophyticus and Chara vulgaris (see Table S1 for accessions). All analyses were based on a nucleotide sequence alignment of 9,032 bp, including the sequences without introns of seven mitochondrial genes (cob, cox1, nad1, nad2, nad4, nad5 and nad6), which are conserved among all the studied chlorophytes. The phylogram in Fig. 2 shows four major clades: the first clade included the Prasinophyceae, the second clade included the Trebouxiophyceae, the third clade included the Ulvophyceae, and the fourth clade included the Chlorophyceae. Our phylogenetic reconstruction shows that Trebouxia sp. TR9 is closely related to Trebouxia aggregata, another lichen phycobiont. The two lichen microalgae were included within a sub-clade, along with Botryococcus braunii, Coccomyxa spp., Lobosphaera incisa, Micractinium conductrix and Prasiola crispa. This sub-clade II, including Trebouxiales and Prasiolales, is a sister of another sub-clade I that includes Chlorellales.
Figure 2.
Phylogram based on the sequence analysis of seven mitochondrial genes from 32 green algal species (Table S1). Bootstrap values are indicated in the nodes.
Gene content of the mtDNAs from Trebouxiophyceae and other chlorophytes
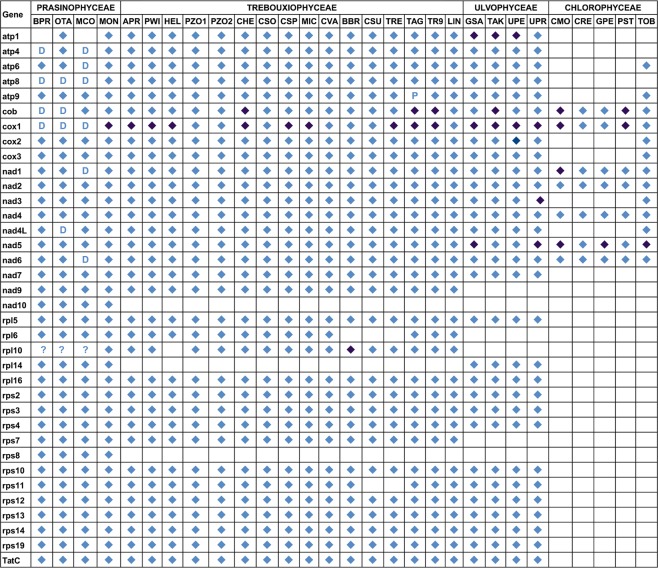
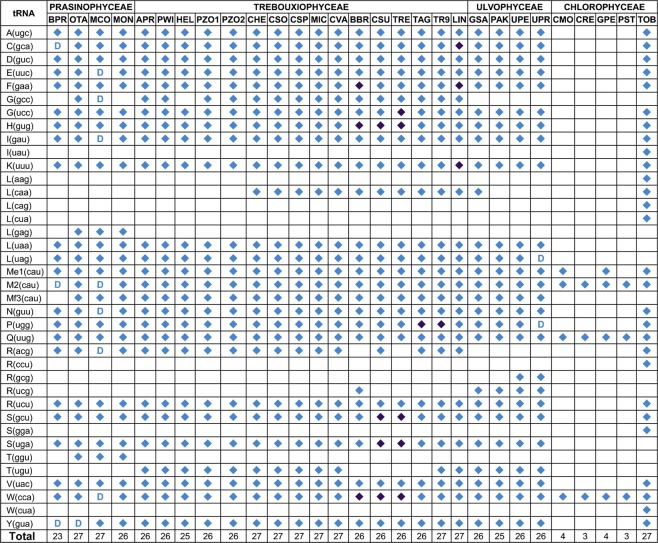
Figures 3 and 4 show the repertoire of genes coding conserved proteins and tRNAs, respectively, in a number of Chlorophyta algae belonging to different classes (Prasinophyceae, Trebouxiophyceae, Ulvophyceae and Chlorophyceae). At least 26 genes coding for proteins were shared by the studied species belonging to Prasinophyceae, Trebouxiophyceae and Ulvophyceae. Conversely, the studied Chlorophyceae, except Tetradesmus obliquus, showed extensive gene loss (most of them coding for ribosomal proteins and tRNAs). Several genes seemed to be lost in certain species within an algal class. For instance, the rpl6 and rps11 genes are present in all the studied Trebouxiophyceae except Coccomyxa sp. C169 and Trebouxiophyceae sp. MX-AZ01. Other genes seemed to be retained in specific algal classes, as is the case of the rpl10 gene.
Figure 3.
Gene repertoires of the mtDNAs from the green algal mtDNAs examined in this study. Diamonds indicate the presence of a standard gene. A “D” denotes gene duplications. Light and dark blue diamonds indicate the presence or absence of introns, respectively. “P” and “?” indicate a partial sequence and uncertainty, respectively.
Figure 4.
Transfer RNA repertoires of the mtDNAs from the green algal mtDNAs examined in this study. A diamond or a “D” indicates the presence of a standard gene if it was duplicated. The presence or absence of introns is indicated by light and dark blue diamonds, respectively.
In this study, we identified the rpl10 gene in the mitochondrial genome of all the studied Trebouxiophyceae, Prasinophyceae, Micromonas sp., Monomastix sp. and Ostreococcus tauri (Table S2). The hypothetical mitochondrial ribosomal L10 proteins would have variable sizes ranging from 510 to 867 aa. Additionally, the rpl10 gene was located downstream of rps19 in all the studied Trebouxiophyceae except for Auxenochlorella protothecoides and Prototheca wickerhamii, in which rpl10 mapped downstream of rps10. The genes downstream of rpl10 were more variable, including genes encoding proteins, rRNAs and tRNAs. Botryococcus braunii had a group II intron of 2,598 bp in the rpl10 gene (positions 1,288 to 3,885 in the nucleotide sequence with accession number NC_027722), which was predicted with the program RNAweasel. This intron was the only group II intron within a protein-coding gene found in the Trebouxiophycean algae analysed in this study.
Comparative analysis of the structure of the mtDNAs from Trebouxiophyceae and other chlorophytes
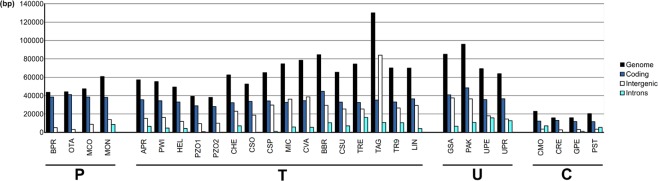
A comparison of the structure of the mtDNAs from different chlorophytes (Fig. 5) showed strikingly variable sizes among Trebouxiophyceae, ranging from 38,164 bp in Prototheca zopfii SAG 2063 to more than 130,000 bp that results from the sum of the partial sequences of Trebouxia aggregata available in GenBank. The complete mitochondrial genome of Trebouxia sp. TR9 had identical repertoires of genes coding for proteins and tRNAs as the lichen-symbiont alga Trebouxia aggregata, whose mitogenome has not been completely sequenced (except trnT (ugu) and a partial sequence of atp9, probably due to the incompleteness of the available sequences) (Figs 3 and 4). Such repertoires were approximately the same as that of free-living Trebouxiophyceae. Thus, the symbiotic association with the mycobiont does not seem to have any impact on the gene content of the mtDNA in the two studied Trebouxia species.
Figure 5.
Total lengths (bp) of coding, intronic, and intergenic sequences in the chlorophyte mtDNAs examined in this study. Species names are abbreviated as in Table S1. The systematic classification is indicated at the bottom (P: Prasinophyceae, T: Trebouxiophyceae, U: Ulvophyceae, C: Chlorophyceae). Data for Trebouxia aggregata were obtained from partial sequences (accessions EU123944, EU123947, EU123948 and EU123949).
The remarkable enlargement of the mtDNAs in Trebouxiophyceae and Ulvophyceae with respect to other chlorophytes was due to the presence of more introns and larger intergenic spacers (Fig. 5). The most extreme difference in the mitogenome size (of at least 59,986 bp) was found between the two closely related Trebouxia microalgae. This difference is mostly due to non-coding regions, which included 37,029 bp and 94,900 bp in Trebouxia sp. TR9 and T. aggregata, respectively, while the coding regions were quite similar: 33,041 bp and 35,156 bp in Trebouxia sp. TR9 and T. aggregata, respectively. A more detailed picture of these differences can be observed in Fig. S1, which shows the genetic maps of four regions of the mtDNA of T. aggregata (accessions EU123944, EU123947, EU123948 and EU123949) and their counterparts in Trebouxia sp. TR9. This figure depicts important differences in the total lengths due to longer intergenic regions in T. aggregata, which contrasts with the more compact structure observed in Trebouxia sp. TR9. Furthermore, this difference was not due to any large insertion/deletion at a specific part of the genome but to small increases or decreases in every intergenic region. Figure S1 also shows high synteny between the mtDNAs from these two Trebouxia species. The whole-genome alignment of the Trebouxia sp. TR9 mtDNA along with other chlorophytes (Fig. S2) showed high conservation of many coding regions, along with remarkable rearrangements, using the steptophyte Mesostigma viride as a reference (accession NC_008240). The most compact and smallest mitochondrial genomes belonged to the Chlorophyceae, which showed intensive gene loss (Fig. 5).
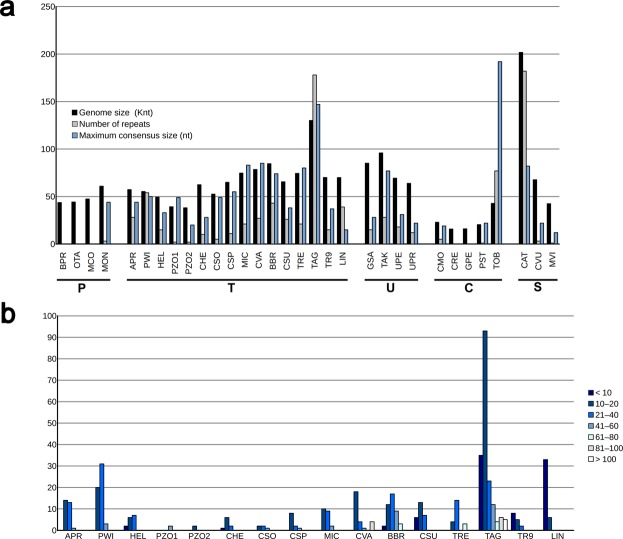
To determine if the expanding/contracting regions were related to the presence of repeats, we searched for tandem repeats (trs) using the program “Tandem repeats finder”. Tandem repeats are mainly characterized by the number of copies and their consensus size (the length of the repeat unit). Our results (Fig. 6a) indicated that Trebouxia sp. TR9 had one of the lowest numbers of trs of the studied Trebouxiophyceae, along with Chlorella heliozoae, Chlorella sorokiniana, Chlorella sp., Helicosporidium sp. and Prototheca zopfii, all them with less than 15 trs. The remaining studied Trebouxiophyceae had a number of trs between 20 and 60, except for Trebouxia aggregata, which displayed the largest number of trs of 178. These trs were mostly located within intergenic spacers (169 out of 178). Figure 6b shows the distribution of the consensus size for all trs found in the mtDNAs from Trebouxiophyceae. The consensus size of the trs was highly heterogeneous among the studied Trebouxiophyceae (Fig. 6b). Generally, the trs with larger consensus sizes were found in mtDNAs with a high number of trs. In our analysis, we found large trs of more than 140 bp of period size in only two chlorophytes belonging to different classes: Trebouxia aggregata and Tetradesmus obliquus, with maximum consensus sizes of 147 and 192, respectively (Fig. 6a). Notably, within each algal class, the species with the largest mtDNA also has the highest number and consensus size of trs (e.g., Monomastix sp. within the Prasinophyceae, Trebouxia aggregata among the Trebouxiophyceae, Tetradesmus obliquus among the Chlorophyceae and Chlorokybus atmophyticus among the Streptophyta). The loosely packed mitochondrial genome of T. aggregata and the more compact mitochondrial genome of Trebouxia sp. TR9 may be a consequence of either an expansion or contraction of intergenic regions, respectively. Such expansion/contraction of intergenic regions may have occurred by gain/loss of tandem repeat units, i.e., by varying the number of copies rather than by insertion/deletion of fragments of different sizes.
Figure 6.
Tandem repeats in the green algal mtDNAs examined in this study. (a) Genome sizes, number of repeats found and maximum consensus size. (b) Frequency of tandem repeats by length in Trebouxiophyceae. The systematic classification is indicated at the bottom (P: Prasinophyceae, T: Trebouxiophyceae, U: Ulvophyceae, C: Chlorophyceae, S: Streptophyta).
Diversity of intron content of the Chlorophyta algae mitochondrial genomes
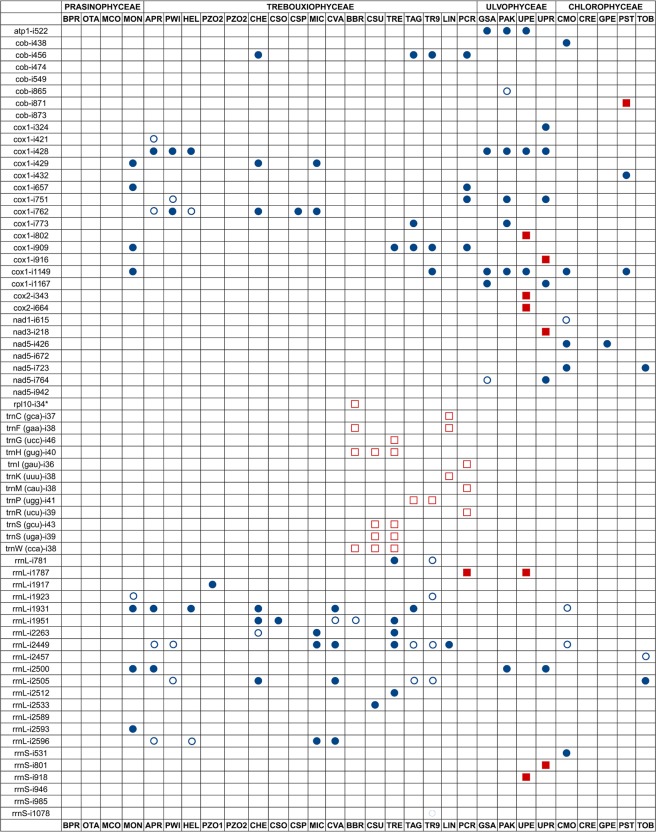
The coding regions of many genes are interrupted by introns in a variety of genetic systems and organisms. To date, four main types of introns have been distinguished based on their splicing mechanism: spliceosome introns, nuclear and archaeal tRNA introns, group I introns and group II introns. As far as we know, our study provides the first compilation of data about the variety of introns, including both group I and group II introns, in the mitochondrial genomes from a number of chlorophytes (Fig. 7). A total of 91 introns were found in the mtDNA of the studied Trebouxiophyceae: 66 group I and 25 group II introns. Most group I introns were found either in certain protein-coding genes (cob and cox1) or the gene encoding the LSU rDNA. Group II introns were mostly found within genes coding certain tRNAs, except two introns inserted within the rpl10 and rrnL genes of the microalgae B. braunii (Trebouxiales) and P. crispa (Prasiolales), respectively. In other chlorophytes analysed in this study belonging to other classes different from Trebouxiophyceae, a total of 77 introns was observed: 42 group I and 35 group II introns. A number of ORFs were coded within introns and corresponded to putative LAGLIDADG homing endonucleases in the case of group I introns and maturases or reverse transcriptases in the case of group II introns.
Figure 7.
Distribution of introns among the green algal mtDNAs examined in this study. Blue circles and red squares indicate the presence of either a group I or group II intron, respectively. Filled symbols and empty symbols denote introns containing or lacking ORFs, respectively. The insertion positions are provided after the gene names and are preceded by an “i”. Positions are relative to Mesostigma viride mtDNA (for protein-coding and tRNA genes) and Escherichia coli (for rRNA genes).
As stated above, we identified the rpl10 gene of B. braunii as the first protein-coding gene bearing a group II intron in the mitochondrial genome of a Trebouxiophyceae (Table S2). The gene for the ribosomal protein L10 (rpl10) is present in a wide diversity of land plants and mitochondrial genomes of algal streptophytes. However, this gene has remained unidentified and largely unannotated in the records of sequenced mitochondrial genomes from green algae. In most cases, this gene actually corresponded to conserved ORFs of unknown function. In other cases, this gene has remained unannotated (Table S2).
One of the most striking features of the mtDNAs analysed in our study was the relatively high number of group II introns disrupting a variety of genes coding tRNAs in a specific group of Trebouxiophyceae (Fig. 7). Indeed, a total of 12 genes coding tRNAs had a group II intron: trnC (gca), trnF (gaa), trnG (ucc), trnH (gug), trnI (gau), trnK (uuu), trnM (cat), trnP (ugg), trnR (ucu), trnS (gcu), trnS (uga) and trnW (cca). Some of these introns were unannotated in their respective genomic sequences (Table S2). All the Trebouxiophyceae with introns in genes coding tRNAs belonged to clade II (Fig. 2), including Trebouxiales and Prasiolales. tRNAs are fundamental components of the translation machinery and in the regulation of gene expression and several other biological processes.
Only two Trebouxia algae analysed in this study had a group II intron within the gene coding tRNA-Pro (ugg). In T. aggregata, this intron has not been annotated (position 6,937 to 8,915 in the sequence, GenBank accession number EU123949). Another Trebouxia phycobiont associated with the lichen Rhizocarpon geographicum (Trebouxia sp. RG in this study) showed the partial sequence of an intron within the trnP (ugg) gene (position 1 to 156 in the sequence, accession number JN847694). In the three Trebouxia algae, the trnP gene with a group II intron had a conserved position upstream of the atp6 gene. Comparison of the intergenic spacer between the trnP and atp6 genes in the three Trebouxia algae showed different lengths, including 115, 326 and 1,152 bp in Trebouxia sp. TR9, Trebouxia sp. RG and T. aggregata, respectively. Interestingly, a total of six trs were found in this spacer in T. aggregata, one of them with a period size of 95, whereas no trs were found in the other two Trebouxia algae with shorter intergenic spacers. This finding reinforces the notion of an enlargement of the mitochondrial genome of T. aggregata due to the duplication of sequences within intergenic spacers rendering the observed high number of trs. The trnP (ugg) gene without any introns was also present in all the studied chlorophytes except for those belonging to Chlamydomonadales, which lack this gene (Fig. 4).
Discussion
The phylum Chlorophyta comprises morphologically and ecologically diverse green algae, which have traditionally been included within three major clades: Ulvophyceae, Trebouxiophyceae, and Chlorophyceae (UTC clade). Moreover, the replacement of “UTC clade” with the term “core Chlorophyta” has been proposed to indicate the previous UTC taxa plus additional classes1,4,31. The class Chlorophyceae is considered to be a monophyletic group32–34. However, the monophyly of Ulvophyceae and Trebouxiophyceae is not strongly supported by several molecular studies, and their polyphyly has been suggested in several publications29,31,34. The monophyly/polyphyly of Trebouxiophyceae depends on the selected sequences and taxon sampling. In some studies, Chlorellales is placed in a clade independent of other Trebouxiophyceae (e.g., Trebouxiales)5,6,31,34,35, whereas other studies support the monophyly of Trebouxiophyceae36,37. The topology of the phylogram obtained here (Fig. 2) is consistent with the placement of Chlorellales with Trebouxiales in a single clade, as proposed by other phylogenetic reconstructions based on a higher number of mitochondrial genes7 and plastid genes7,35,36,38; however, it is noteworthy that this reconstruction is based on a rather limited variety of algal groups. Some authors state that sampling across different algal groups is a prerequisite for deriving a reliable phylogenetic classification of the core Chlorophyta31. Moreover, in several phylogenetic studies based on chloroplast sequences, the inferred topologies were dependent upon the data set and the method of analysis, differing mainly with respect to the relative positions of the major lineages in the core Chlorophyta39.
As previously stated, the symbiotic association does not seem to influence the gene content of the mtDNA in the two studied Trebouxia species. This observation contrasts with the reduction in genome content observed in other symbiotic relationships, such as bacterial endosymbionts of insects40. However, our findings are consistent with the absence of organellar genomic reduction observed in some green algae involved in other types of symbiotic relationships, which tend to have larger mtDNAs7, and suggest that symbiosis may promote larger mtDNAs. In addition, the gene content of the mtDNA from Trebouxiophyceae was similar to that of several streptophytes38, indicating their conservation during the evolution from a common ancestor.
The hypothetical expansion or contraction of intergenic regions in the mitogenomes of the studied Trebouxia algae by gain/loss of tandem repeat units may have occurred in other algal groups, such as Streptophyta algae. Within this algal group, Chlorokybus atmophyticus has a large mtDNA (201,763 nt) with very extended intergenic spacers41 and 182 trs, whereas Mesostigma viride has a more compact mtDNA (42,424 bp)42 and a single predicted tr. In this line, we observed a certain parallelism with previous studies of obligate intracellular livestock pathogens43. In the referred studies, the bacterium Ehrlichia ruminantium displayed a lower coding ratio due to unusually long intergenic regions related to an active process of genome expansion/contraction. This process was targeted at trs in non-coding regions, based on the addition or removal of 150-bp tandem units and seemed to be specific to E. ruminantium. This finding agrees with previously proposed mechanisms of tr deletion or amplification through DNA slippage44. Moreover, E. ruminantium seemed to be capable of rapidly undergoing genomic rearrangements upon exposure to novel environmental conditions43. It has been proposed that mitochondrial genomic architecture is shaped by two types of mtDNA repair45: (i) within genes, in which gene conversion would maintain low mutation rates, and (ii) within non-coding regions, in which expansion(s) and rearrangements may be explained by break-induced replication (BIR). Both processes can explain the low mutation rates in coding sequences and the striking expansions of non-coding sequences. The same argument has been proposed for the mitochondrial genome from several Dunaliella species46, which have undergone massive levels of mitochondrial genomic expansion. Moreover, BIR within organelle systems is known to be inaccurate and cause rearrangements and expansions in Arabidopsis thaliana47. It is plausible that the intergenic regions in the Trebouxia mitochondrial genomes would also be shaped via BIR. This mechanism may explain the expansion and disarray observed in the intergenic regions of the mitogenomes from T. aggregata in relation to those from Trebouxia sp. TR9.
Several studies indicated that plants are the only group of eukaryotes other than Reclinomonas (Excavata) that still retain the gene rpl10 in their mitochondrial genomes38,48,49. In this study, we found retention of the rpl10 gene in the mitochondrial genome of representatives of certain Chlorophyta classes (e.g., Prasinophyceae and Trebouxiophyceae) and its absence in other classes (e.g., Chlorophyceae and Ulvophyceae). These observations, along with its possible pseudogenization in some lineages, are consistent with the model of evolution of the rpl10 gene in plants proposed by Kubo and Arimura49. According to this model, this gene was originally in the mitochondrial genomes. Then, it was lost from most eukaryotic lineages except plants. However, certain plant lineages lack rpl10 in their mitochondrial genomes and have a nuclear-encoded rpl10 because of the duplication of the rpl10 gene transferred from the chloroplast to the nucleus. This copy of rpl10 seems to functionally compensate for the lack of the mitochondrial rpl10 gene in any subcellular compartment. This model remains to be demonstrated in chlorophytes.
As far as we know, our study provides the first compilation of data about the variety of introns, which includes group II introns disrupting tRNA genes, in a specific algal group within Trebouxiophyceae (Fig. 7). Introns disrupting tRNA genes were found in a variety of forms and different genetic systems in all the three kingdoms of life, being particularly abundant in archaeal and eukaryotic genomes50. Currently, there is a certain controversy on the origin of tRNA introns. An “intron-first” hypothesis suggests that a large part of introns present in all primordial tRNA genes have been lost during evolution. After intron loss, the two halves were joined in the genome, rendering an intron-less tRNA gene51. Alternatively, the “intron-late” hypothesis suggests the insertion of introns after the establishment of primordial tRNA genes52. The existence of split tRNAs is consistent with the second hypothesis51. In our study, several pieces of evidence for the gain of a modern intron by tRNA genes can be observed. First, the presence of introns within tRNA genes is restricted to a single clade among Trebouxiophyceae (clade II in Fig. 2). Moreover, in the case of the trnP (ugg) gene, which was exclusively found in Trebouxia microalgae, mature parts of tRNA-Pro (ugg) are highly homologous to those of the same tRNA from other related Trebouxiophyceae. Thus, the trnP (ugg) gene from the common ancestor of Trebouxia algae likely acquired its introns over the course of evolution. A parallel picture can be observed in other intron-bearing tRNA genes. For instance, the intron of the trnH (gug) and trnW (cca) genes was probably acquired by the common ancestor of these three Trebouxiophyceae: B. braunii, Coccomyxa sp. C-169 and Trebouxiophyceae sp. MX-AZ01. Similarly, the intron of the trnS (gcu) and trnS (uga) genes was probably acquired by the common ancestor of Coccomyxa sp. C-169 and Trebouxiophyceae sp. MX-AZ01. In this scenario, the relatively recent intron gain by tRNA genes, which was found in a single algal species in this study, might be the result of a modern acquisition rather than a loss during evolution [e.g., trnC (gca) and trnK (uuu) genes in L. incisa; trnG (ucc) gene in Trebouxiophyceae sp. MX-AZ01; trnI (gau), trnM (cau) and trnR (ucu) genes in B. braunii].
The structural analyses of the mitochondrial genome of Trebouxiophyceae and other Chlorophyta algae reported in this study contribute considerably to understanding the evolution of the mitochondrial genomes in the most ancestral photosynthetic eukaryotes. Our investigation stresses the importance of providing new sequences of mitochondrial genomes of green algae to find new features that may be crucial to establishing evolutionary patterns in different algal lineages and to more precisely delineate such lineages based on phylogenetic analyses.
Methods
Phycobiont isolation and culture conditions
Trebouxia sp. TR9 was isolated from the lichen Ramalina farinacea (L.) Ach.53 and cultured in Bold 3N medium54 in a growth chamber at 15 °C under a 14-h/10-h light/dark cycle (lighting conditions: 25 μmol m−2 s−1).
DNA isolation and sequencing and genome assembly and annotation
DNA extraction and purification were performed according to the protocol used by Ausubel et al.55. The purified DNA was sequenced using 454 GS FLX Titanium technology (454 Life Sciences, Roche, Basel, Switzerland) at Lifesequencing facilities (Parc Cièntific, Universitat de València, Spain). The 454 pyrosequencing reads were assembled using Mira assembly software56. Contigs corresponding to the mitochondrial genomes were selected using BLASTn, BLASTx and tBLASTx57 against a local database of mitochondrial genomes from Viridiplantae built from the NCBI nucleotide databases. To connect the different contigs and corroborate the genome circularity, a number of primers were designed (Table 1). PCR was performed in a 96-well LabCycler (SensoQuest Biomedizicnische Elektronik) using EmeraldAmp GT PCR Master Mix (Takara Bio Inc., Shiga, Japan). PCR products were purified using Illustra GFX PCR DNA (GE Healthcare Life Science, Buckinghamshire, England) and sequenced with an ABI 3100 Genetic Analyzer using an ABI BigDyeTM Terminator Cycle Sequencing Ready Reaction Kit (Applied Biosystems, Foster City, California). Most of the genes and open reading frames (ORFs) were identified using the MFannot organelle genome annotator (http://megasun.bch.umontreal.ca/cgi-bin/mfannot/mfannotInterface.pl). The unannotated rpl10 genes were identified using BLAST tools. Motifs for both RPL10 proteins and homing endonucleases were found with BLAST and MotifSearch available at https://www.genome.jp/tools/motif/. tRNA genes were localized using RNAweasel58, tRNAscan-SE59 and ARAGORN60.
Table 1.
List of primers used for DNA amplification and sequencing.
| Name | Sequence | Position | Length | Direction |
|---|---|---|---|---|
| MT_61K_769_F | AGTTTACGGAATTATAACAGCG | 776–797 | 22 | forward |
| MT_61K_1838_R | TACGTTGATTTAGCAAACCAATG | 1823–1845 | 23 | reverse |
| MT_61K_23618_F | AGTAGAGACACAACATCATTAAC | 23192–23214 | 23 | forward |
| MT_61K_24958_R | GAGCTGACGACAGCCATG | 24521–24538 | 18 | reverse |
| MT_61K_60869_R | GAAAGTGGCTCTTCCAGCA | 58242–58260 | 19 | reverse |
| MT_61K_59310_F | TGTGTTTACCTATTTCACCAAG | 59759–59780 | 22 | forward |
| MT_10K_431_F | ACACCTAGTTGGTATTGCTTTG | 60470–60491 | 22 | forward |
| MT_10K_654_R | GGTGTTTGAAAGATAGACTGCA | 60690–60711 | 22 | reverse |
| MT_10K_9453_F | GCATATCGTCAAATGTCATTG | 69290–69310 | 21 | forward |
| MT_10K_9858_R | CAAGTATTGAGTAGCGGCGT | 69693–69712 | 20 | reverse |
Phylogenetic analyses
Phylogenetic reconstructions were performed based on seven mitochondrial genes from 32 algal species (see Table S1 for accessions). Alignments were performed with Muscle61 with Geneious R1062 and trimmed with GBLOCKs63, with options for less stringent selection. The less stringent selection of blocks allowed for smaller final blocks, gap positions within the final blocks and less strict flanking positions.
For the maximum-likelihood (ML) analyses, the concatenated nucleotide matrix of 32 taxa and 9,032 bp were analysed with the GTR + G + I model of nucleotide substitution, which was selected according to the automatic model selection of PhyML64. Data sets were subjected to ML with PhyML64. Bootstrap probabilities65 were calculated to estimate the robustness of the clades from 100 replicates in the data. The consensus tree was drawn with FigTree66.
Additional analyses
Whole-genome alignments were performed with MultiPipMaker67. Gene maps were constructed with Geneious R1062. Tandem repeats were found using the program Tandem repeats finder68.
Accession codes
The complete mitochondrial genome sequence generated in this study has been deposited under the GenBank accession number MH917293.
Supplementary information
Acknowledgements
This study was funded by grants CGL2016-79158-P and CGL2016-80259-P from the Ministry of Economy and Competitiveness (MINECO, Spain) and FEDER and PROMETEO/2017/039 Excellence in Research (Generalitat Valenciana, Spain). The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author Contributions
E.B., E.D. and L.C. conceived the study. F.M. generated DNA sequence data and performed the genome assemblies. E.C. and F.M. performed the annotations. A.M., E.D., F.G. and F.M. performed the genomic and phylogenetic analyses. E.B., E.D. and L.C. wrote the manuscript. E.D. and F.M. generated the figures. All authors reviewed the manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-44700-7.
References
- 1.Ruhfel BR, Gitzendanner MA, Soltis PS, Soltis DE, Burleigh JG. From algae to angiosperms-inferring the phylogeny of green plants (Viridiplantae) from 360 plastid genomes. BMC Evol. Biol. 2014;14:23-2148-14-23. doi: 10.1186/1471-2148-14-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pröschold, T. & Leliaert, F. In Unravelling the Algae (eds Brodie, J. & Lewis, J.) 123–153 (CRC Press, Taylor & Francis Group, New York, 2007).
- 3.Friedl, T. & Rybalka, N. In Progress in Botany 73 (eds Lüttge, U., Beyschlag, W., Büdel, B. & Francis, D.) 259–280 (Springer, Berlin, Heidelberg, 2012).
- 4.Leliaert F, et al. Phylogeny and Molecular Evolution of the Green Algae. Crit. Rev. Plant Sci. 2012;31:1–46. doi: 10.1080/07352689.2011.615705. [DOI] [Google Scholar]
- 5.Lemieux C, Otis C, Turmel M. Chloroplast phylogenomic analysis resolves deep-level relationships within the green algal class Trebouxiophyceae. BMC Evol. Biol. 2014;14:211-014-0211-2. doi: 10.1186/s12862-014-0211-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Turmel M, Otis C, Lemieux C. Dynamic Evolution of the Chloroplast Genome in the Green Algal Classes Pedinophyceae and Trebouxiophyceae. Genome Biol. Evol. 2015;7:2062–2082. doi: 10.1093/gbe/evv130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fan W, Guo W, Van Etten JL, Mower JP. Multiple origins of endosymbionts in Chlorellaceae with no reductive effects on the plastid or mitochondrial genomes. Sci. Rep. 2017;7:10101-017-10388-w. doi: 10.1038/s41598-017-10388-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blifernez-Klassen, O. et al. Complete Chloroplast and Mitochondrial Genome Sequences of the Hydrocarbon Oil-Producing Green Microalga Botryococcus braunii Race B (Showa). Genome Announc4, 10.1128/genomeA.00524-16 (2016). [DOI] [PMC free article] [PubMed]
- 9.Pröschold T, Darienko T, Silva PC, Reisser W, Krienitz L. The systematics of Zoochlorella revisited employing an integrative approach. Environ. Microbiol. 2011;13:350–364. doi: 10.1111/j.1462-2920.2010.02333.x. [DOI] [PubMed] [Google Scholar]
- 10.Trémouillaux-Guiller J, Huss VA. A cryptic intracellular green alga in Ginkgo biloba: ribosomal DNA markers reveal worldwide distribution. Planta. 2007;226:553–557. doi: 10.1007/s00425-007-0526-y. [DOI] [PubMed] [Google Scholar]
- 11.Gustavs, L., Schiefelbein, U., Darienko, T. & Pröschold, T. In Algal and Cyanobacteria Symbioses 169–208 (World Scientific, Europe, 2015).
- 12.Figueroa-Martínez F, Nedelcu AM, Smith DR, Adrian RP. When the lights go out: the evolutionary fate of free-living colorless green algae. New Phytol. 2015;206:972–982. doi: 10.1111/nph.13279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Škaloud P, Steinova J, Ridka T, Vancurova L, Peksa O. Assembling the challenging puzzle of algal biodiversity: species delimitation within the genus Asterochloris (Trebouxiophyceae, Chlorophyta) J. Phycol. 2015;51:507–527. doi: 10.1111/jpy.12295. [DOI] [PubMed] [Google Scholar]
- 14.Moya P, et al. Myrmecia israeliensis as the primary symbiotic microalga in squamulose lichens growing in European and Canary Island terricolous communities. Fottea. 2018;18:72–85. doi: 10.5507/fot.2017.022. [DOI] [Google Scholar]
- 15.Muggia, L., Leavitt, S. & Barreno, E. The hidden diversity of lichenised Trebouxiophyceae (Chlorophyta). Phycologia, 503–524 (2018).
- 16.Friedl T. Comparative ultrastructure of pyrenoids in Trebouxia (Microthamniales, Chlorophyta) Pl Syst Evol. 1989;164:145–159. doi: 10.1007/BF00940435. [DOI] [Google Scholar]
- 17.Sadowska-Deś AD, et al. Integrating coalescent and phylogenetic approaches to delimit species in the lichen photobiont Trebouxia. Mol. Phylogenet. Evol. 2014;76:202–210. doi: 10.1016/j.ympev.2014.03.020. [DOI] [PubMed] [Google Scholar]
- 18.Leavitt SD, et al. Fungal specificity and selectivity for algae play a major role in determining lichen partnerships across diverse ecogeographic regions in the lichen-forming family Parmeliaceae (Ascomycota) Mol. Ecol. 2015;24:3779–3797. doi: 10.1111/mec.13271. [DOI] [PubMed] [Google Scholar]
- 19.Molins A, Moya P, García-Breijo FJ, Reig-Armiñana J, Barreno E. A multi-tool approach to assess microalgal diversity in lichens: isolation, Sanger sequencing, HTS and ultrastructural correlations. The Lichenologist. 2018;50:123–138. doi: 10.1017/S0024282917000664. [DOI] [Google Scholar]
- 20.Casano LM, et al. Two Trebouxia algae with different physiological performances are ever-present in lichen thalli of Ramalina farinacea. Coexistence versus Competition? Environ. Microbiol. 2011;13:806–818. doi: 10.1111/j.1462-2920.2010.02386.x. [DOI] [PubMed] [Google Scholar]
- 21.del Hoyo A, et al. Oxidative stress induces distinct physiological responses in the two Trebouxia phycobionts of the lichen Ramalina farinacea. Ann. Bot. 2011;107:109–118. doi: 10.1093/aob/mcq206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Álvarez R, et al. Different strategies to achieve Pb-tolerance by the two Trebouxia algae coexisting in the lichen Ramalina farinacea. J. Plant Physiol. 2012;169:1797–1806. doi: 10.1016/j.jplph.2012.07.005. [DOI] [PubMed] [Google Scholar]
- 23.del Campo EM, et al. The genetic structure of the cosmopolitan three-partner lichen Ramalina farinacea evidences the concerted diversification of symbionts. FEMS Microbiol. Ecol. 2013;83:310–323. doi: 10.1111/j.1574-6941.2012.01474.x. [DOI] [PubMed] [Google Scholar]
- 24.Casano, L. M., Braga, M. R., Álvarez, R., del Campo, E. M. & Barreno, E. Differences in the cell walls and extracellular polymers of the two Trebouxia microalgae coexisting in the lichen Ramalina farinacea are consistent with their distinct capacity of immobilizing extracellular Pb. Plant Science (2015). [DOI] [PubMed]
- 25.Centeno DC, Hell AF, Braga MR, Del Campo EM, Casano LM. Contrasting strategies used by lichen microalgae to cope with desiccation-rehydration stress revealed by metabolite profiling and cell wall analysis. Environ. Microbiol. 2016;18:1546–1560. doi: 10.1111/1462-2920.13249. [DOI] [PubMed] [Google Scholar]
- 26.Moya P, Molins A, Martínez-Alberola F, Muggia L, Barreno E. Unexpected associated microalgal diversity in the lichen Ramalina farinacea is uncovered by pyrosequencing analyses. PLoS One. 2017;12:e0175091. doi: 10.1371/journal.pone.0175091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.del Campo EM, Casano LM, Gasulla F, Barreno E. Suitability of chloroplast LSU rDNA and its diverse group I introns for species recognition and phylogenetic analyses of lichen-forming Trebouxia algae. Mol. Phylogenet. Evol. 2010;54:437–444. doi: 10.1016/j.ympev.2009.10.024. [DOI] [PubMed] [Google Scholar]
- 28.del Campo EM, Casano LM, Barreno E. Evolutionary implications of intron-exon distribution and the properties and sequences of the RPL10A gene in eukaryotes. Mol. Phylogenet. Evol. 2013;66:857–867. doi: 10.1016/j.ympev.2012.11.013. [DOI] [PubMed] [Google Scholar]
- 29.Martínez-Alberola, F. Caracterización genómica del microalga Trebouxia sp. TR9 aislada del liquen Ramalina farinacea (L.) Ach. mediante secuenciación masiva. PhD Dissertation. Universitat de València, http://roderic.uv.es/handle/10550/48824 (2015).
- 30.Hinojosa-Vidal Ernesto, Marco Francisco, Martínez-Alberola Fernando, Escaray Francisco J., García-Breijo Francisco J., Reig-Armiñana José, Carrasco Pedro, Barreno Eva. Characterization of the responses to saline stress in the symbiotic green microalga Trebouxia sp. TR9. Planta. 2018;248(6):1473–1486. doi: 10.1007/s00425-018-2993-8. [DOI] [PubMed] [Google Scholar]
- 31.Fučíková K, et al. New phylogenetic hypotheses for the core Chlorophyta based on chloroplast sequence data. Frontiers in Ecology and Evolution. 2014;2:63. [Google Scholar]
- 32.Turmel M, Brouard JS, Gagnon C, Otis C, Lemieux C. Deep division in the Chlorophyceae (Chlorophyta) revealed by chloroplast phylogenomic analyses. J. Phycol. 2008;44:739–750. doi: 10.1111/j.1529-8817.2008.00510.x. [DOI] [PubMed] [Google Scholar]
- 33.Tippery NP, Fučíková K, Lewis PO, Lewis LA. Probing the monophyly of the Sphaeropleales (Chlorophyceae) using data from five genes. J. Phycol. 2012;48:1482–1493. doi: 10.1111/jpy.12003. [DOI] [PubMed] [Google Scholar]
- 34.Sun L, et al. Chloroplast Phylogenomic Inference of green algae relationships. Scientific Reports. 2016;6:20528. doi: 10.1038/srep20528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lemieux C, Vincent AT, Labarre A, Otis C, Turmel M. Chloroplast phylogenomic analysis of chlorophyte green algae identifies a novel lineage sister to the Sphaeropleales (Chlorophyceae) BMC Evol. Biol. 2015;15:264-015-0544-5. doi: 10.1186/s12862-015-0544-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fang L, et al. Improving phylogenetic inference of core Chlorophyta using chloroplast sequences with strong phylogenetic signals and heterogeneous models. Mol. Phylogenet. Evol. 2018;127:248–255. doi: 10.1016/j.ympev.2018.06.006. [DOI] [PubMed] [Google Scholar]
- 37.Leliaert F, et al. Chloroplast phylogenomic analyses reveal the deepest-branching lineage of the Chlorophyta, Palmophyllophyceae class. nov. Sci. Rep. 2016;6:25367. doi: 10.1038/srep25367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Turmel M, Otis C, Lemieux C. Tracing the evolution of streptophyte algae and their mitochondrial genome. Genome Biol. Evol. 2013;5:1817–1835. doi: 10.1093/gbe/evt135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Turmel M, Otis C, Lemieux C. Divergent copies of the large inverted repeat in the chloroplast genomes of ulvophycean green algae. Sci. Rep. 2017;7:994-017-01144-1. doi: 10.1038/s41598-017-01144-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wernegreen JJ. Ancient bacterial endosymbionts of insects: Genomes as sources of insight and springboards for inquiry. Exp. Cell Res. 2017;358:427–432. doi: 10.1016/j.yexcr.2017.04.028. [DOI] [PubMed] [Google Scholar]
- 41.Turmel M, Otis C, Lemieux C. An unexpectedly large and loosely packed mitochondrial genome in the charophycean green alga Chlorokybus atmophyticus. BMC Genomics. 2007;8:137. doi: 10.1186/1471-2164-8-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Turmel M, Otis C, Lemieux C. The complete mitochondrial DNA sequence of Mesostigma viride identifies this green alga as the earliest green plant divergence and predicts a highly compact mitochondrial genome in the ancestor of all green plants. Mol. Biol. Evol. 2002;19:24–38. doi: 10.1093/oxfordjournals.molbev.a003979. [DOI] [PubMed] [Google Scholar]
- 43.Frutos R, et al. Comparative genomic analysis of three strains of Ehrlichia ruminantium reveals an active process of genome size plasticity. J. Bacteriol. 2006;188:2533–2542. doi: 10.1128/JB.188.7.2533-2542.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lovett ST. Encoded errors: mutations and rearrangements mediated by misalignment at repetitive DNA sequences. Mol. Microbiol. 2004;52:1243–1253. doi: 10.1111/j.1365-2958.2004.04076.x. [DOI] [PubMed] [Google Scholar]
- 45.Christensen AC. Plant mitochondrial genome evolution can be explained by DNA repair mechanisms. Genome Biol. Evol. 2013;5:1079–1086. doi: 10.1093/gbe/evt069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Del Vasto M, et al. Massive and widespread organelle genomic expansion in the green algal genus Dunaliella. Genome Biol. Evol. 2015;7:656–663. doi: 10.1093/gbe/evv027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Davila JI, et al. Double-strand break repair processes drive evolution of the mitochondrial genome in Arabidopsis. BMC Biol. 2011;9:64-7007-9-64. doi: 10.1186/1741-7007-9-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mower JP, Bonen L. Ribosomal protein L10 is encoded in the mitochondrial genome of many land plants and green algae. BMC Evol. Biol. 2009;9:265-2148-9-265. doi: 10.1186/1471-2148-9-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kubo N, Arimura S. Discovery of the rpl10 gene in diverse plant mitochondrial genomes and its probable replacement by the nuclear gene for chloroplast RPL10 in two lineages of angiosperms. DNA Res. 2010;17:1–9. doi: 10.1093/dnares/dsp024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yoshihisa T. Handling tRNA introns, archaeal way and eukaryotic way. Front. Genet. 2014;5:213. doi: 10.3389/fgene.2014.00213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Di Giulio M. The non-monophyletic origin of the tRNA molecule and the origin of genes only after the evolutionary stage of the last universal common ancestor (LUCA) J. Theor. Biol. 2006;240:343–352. doi: 10.1016/j.jtbi.2005.09.023. [DOI] [PubMed] [Google Scholar]
- 52.Cavalier-Smith T. Intron phylogeny: a new hypothesis. Trends Genet. 1991;7:145–148. doi: 10.1016/0168-9525(91)90102-V. [DOI] [PubMed] [Google Scholar]
- 53.Gasulla F, Guera BE. A rapid and effective method for isolating lichen phycobionts into axenic culture. Symbiosis. 2010;51:175–179. doi: 10.1007/s13199-010-0064-4. [DOI] [Google Scholar]
- 54.Bold HC, Parker BC. Some supplementary attributes in the classification of Chlorococcum species. Arch. Mikrobiol. 1962;42:267–288. doi: 10.1007/BF00422045. [DOI] [PubMed] [Google Scholar]
- 55.Ausubel, F. M. et al. In Current protocols in molecular biology (John Wiley & Sons, Inc., New York, 2003).
- 56.Chevreux B, et al. Using the miraEST assembler for reliable and automated mRNA transcript assembly and SNP detection in sequenced ESTs. Genome Res. 2004;14:1147–1159. doi: 10.1101/gr.1917404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Altschul SF, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lang BF, Laforest MJ, Burger G. Mitochondrial introns: a critical view. Trends Genet. 2007;23:119–125. doi: 10.1016/j.tig.2007.01.006. [DOI] [PubMed] [Google Scholar]
- 59.Lowe TM, Chan PP. tRNAscan-SE On-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 2016;44:W54–7. doi: 10.1093/nar/gkw413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Laslett D, Canback B. ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucleic Acids Res. 2004;32:11–16. doi: 10.1093/nar/gkh152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kearse M, et al. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 2000;17:540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- 64.Guindon S, et al. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 65.Felsenstein J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 66.Rambaut, A. FigTree: Tree figure drawing tool. Institute of Evolutionary Biology, University of Edinburgh1.3.1 (2008).
- 67.Schwartz S, et al. MultiPipMaker and supporting tools: Alignments and analysis of multiple genomic DNA sequences. Nucleic Acids Res. 2003;31:3518–3524. doi: 10.1093/nar/gkg579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Benson G. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 1999;27:573–580. doi: 10.1093/nar/27.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.