Abstract
In microorganisms, a number of metalloproteins including PerR are found to regulate gene expression in response to environmental reactive oxygen species (ROS) changes. However, discovery of similar regulatory mechanisms remains elusive within mammalian cells. As an antioxidant metalloenzyme that maintains intracellular ROS homeostasis, copper zinc superoxide dismutase (SOD1) has high affinity for DNA in solution and in cells. Here, we explored the regulatory roles of SOD1 in the expression of genes in response to ROS changes within mammalian cells. SOD1-occupied DNA sites with distinct sequence preference were identified. Changing ROS levels both were found to impact DNA–SOD1 interactions in solution and within HeLa cells. GGA was one of the base triplets that had direct contact with SOD1. DNA–SOD1 interactions were observed to regulate the ROS-responsive expression of functional genes including oncogenes and amyotrophic lateral sclerosis-linked genes in transcriptional phases. Our results confirm another function of SOD1, acting as a H2O2-responsive regulatory protein in the expression of numerous mammalian genes.
INTRODUCTION
A few of microbial metalloproteins including PerR were found to regulate expression of many genes in response to externally environmental reactive oxygen species (ROS) changes (1). However, similar regulatory proteins remain elusive in the expression of mammalian genes responsive to intracellular ROS changes so far. The primary antioxidant enzyme copper zinc superoxide dismutase (SOD1) catalyzes the rapid conversion of superoxide anion into hydrogen peroxide (H2O2), maintaining homeostasis of intracellular ROS (2). On the one hand, increasing evidence indicates that SOD1 plays a regulatory role in diverse cellular processes (3–5) including redox signaling (3), respiration (4) and nutrient sensing (5) via controlling cellular ROS levels (2). On the other hand, the observations performed in solution (6–8) and cells (9), as well as bioinformatic analysis (10), suggest that SOD1 might be a DNA binding protein linked to regulation of gene expression, which is supported by both the higher content of wild-type (WT) SOD1 and its mutants in the nucleus than that in other compartments (11,12). Furthermore, high activity of amyotrophic lateral sclerosis (ALS)-linked SOD1 mutants also suggests that SOD1 might have another noncanonical function not enzymatic activity contributing to ALS (13). We previously indicated the added divalent metal ion-dependent DNA cleavage activity of WT SOD1 and DNA as an accelerating template in the aggregation of WT SOD1 and its mutants (6–8), also implying affinity of SOD1 for DNA. However, it remains unclear in mammalian cells whether SOD1 can regulate gene expression responsive to intracellular ROS changes as a DNA binding protein.
Here, we employed multiple methods to explore the regulatory roles of SOD1 in the expression of functional genes in response to intracellular ROS changes via binding to functional human DNA elements. SOD1 was found to show a ROS dependent distribution within a mammalian cell. SOD1-occupied DNA sequences with different preference were identified in the vicinity of transcriptional start sites. GGA was one of the base triplets that directly contact SOD1. Inactivation- and demetallation-mediated conformational changes altered both sequence preference and affinity of SOD1 interactions with DNA. Moreover, SOD1 binding was observed to regulate the ROS-responsive expression of numerous genes, especially oncogenes and ALS-linked genes, in transcriptional phases. A comparison of regulation in the expression of oncogenes and ALS-linked genes suggested that SOD1 deficiency-mediated reduction of SOD1 binding to DNA activates both the genes to promote onset and development of cancer and those to delay ALS development. Our results confirm that SOD1 functions as a H2O2-responsive regulatory protein in the expression of numerous mammalian genes in addition to acting as an antioxidant enzyme, and indicate that, similar to that within microbial cells, there is a metalloprotein-based regulatory mechanism of the gene expression in response to ROS changes within mammalian cells.
MATERIALS AND METHODS
Cell culture
HeLa cells were obtained from China Center for Type Culture Collection (CCTCC) and maintained at 37°C in a saturating humidity atmosphere containing 5% CO2. HeLa cells were cultured in DMEM medium (Gibco; C11995500BT) supplemented with 1% penicillin/streptomycin (Gibco; 15140122), and 10% fetal bovine serum (FBS) (Gibco; 26140079).
Immunofluorescence
Immunofluorescence was observed to examine the intracellular distribution of SOD1. HeLa cells were seeded in a 6-well plate at a density of 2 × 105 cells/well in the DMEM culture medium. After washing with PBS, cells were incubated first for 4 h in the medium mixed respectively with 0.4 mM H2O2, 2 mM GSH, 50 μM LD100, 1 mM paraquat (Sigma; 36541), 5 μg/ml 4-nitroquinoline-N-oxide (4NQO) (Sigma; N8141), and 10 mM 3-amino-1,2,4-triazole (AT) (Sigma; A8056). 0.4 mM H2O2 was usually used to provide an intracellular oxidative environment (9). Unless otherwise specified, the use of H2O2 in subsequent cell experiments was 0.4 mM. Following removal of the culture medium, the cells were rinsed twice with PBS, fixed with ice-cold 4% paraformaldehyde for 15 min in the DMEM culture medium and permeated 3 × 5 min with 0.1% Triton X-100 in PBS. Then, the cells were blocked with 5% goat serum, incubated overnight at 4°C with Alexa Fluor 488 conjugated anti-human SOD1 primary antibody (Abcam; ab199733) (1:100 dilution) in PBS supplemented with 4% FBS, and washed for 4 × 10 min with PBS. Finally, the cells were counterstained for 10 min with 0.6 μM DAPI (Life Technologies; D21490) in PBS, washed for 4 × 10 min with PBS and mounted in glycerol + 10% 10 × PBS. All fluorescent images were captured with a Leica confocal microscopy. The fluorescence intensity of SOD1 in the nucleus and cytoplasm were quantified using ImageJ.
SOD1 knockdown within HeLa cells
To verify that SOD1 binding to DNA can regulate gene expression, SOD1 knockdown was performed. The validated siRNA of SOD1 was purchased from commercial supplier (Ambion; AM51331). Briefly, after seeding HeLa cells at 40–50% confluent, they were transfected with 90 pmol siRNA in the presence of 20 μl Lipofectamine RNAiMAX (Invitrogen; 13778075) in 4 ml completed DMEM with 600 μl Opti-MEM (Gibco; 31985070) at 37°C for 24 h. In the next day, cells were transfected again under the same condition to obtain higher transfection effect. The performance of SOD1 knockdown in HeLa cells had been systematically evaluated as previous reports (14) under the tested conditions.
EMSA
EMSA was used to study the binding of SOD1 to DNA in solution. The 12 bp biotin labelled probes were generated by annealing the sense and anti-sense strands of synthetic oligonucleotides and incubated first with SOD1 in the DNA-binding buffer (10 mM Tris–HCl, pH 7.5, 50 mM KCl and 1 mM DTT) with 1% formaldehyde (Sigma; F8775) at room temperature for 30 min. Then, 0.125 M glycine was added to stop the binding reactions. The binding mixture was subjected to electrophoresis at room temperature on a 6.5% polyacrylamide gel at 100 V for 40 min in 0.5× TBE buffer (Sigma; T3913). Following electrophoresis, the DNAs were transferred to positively charged nylon membrane (Roche; 11209299001) at 360 mA for 45 min in 0.5× TBE, and crosslinked to the membrane at 120 mJ/cm2 using a commercial UV-light crosslinking instrument. After detected the biotin-labeled DNAs by Chemiluminescent Nucleic Acid Detection Module (Thermo Scientific; 89880), the membranes were placed in a film cassette, and exposed to X-ray film (Sigma; Z380156) for 2–5 min.
DNase I footprinting assay with capillary electrophoresis
DNase I footprinting assay was performed to probe DNA bases with which SOD1 directly contacts. The 6-FAM labelled probes were generated by annealing the sense and anti-sense strands of synthetic oligonucleotides. All SOD1 binding reactions in the presence of formaldehyde were carried out as described in EMSA. The binding reactions in the absence of formaldehyde were performed in DNA-binding buffer (10 mM Tris–HCl, pH 7.5, 50 mM KCl and 1 mM DTT) at room temperature for 30 min. The additional incubation was performed for 2 min in the pH 8.0 40 mM Tris–HCl containing 10 mM MgSO4, 1 mM CaCl2 and RQ1 RNase-Free DNase (Promega; M6101) at 37°C. The cleavage reaction was stopped by adding 2 μl Stop Buffer (20 mM EGTA, pH 8.0) at 65°C for 1 min, followed by DNA extraction and precipitation. Samples were analyzed by ABI 3730XL DNA analyzer (Life Technologies).
Small-angle X-ray scattering (SAXS)
To obtain the structural insight into SOD1 binding to DNA, SAXS experiments were performed. SAXS experiments were carried out at Shanghai Synchrotron Radiation Facility beamline BL16B1. Following incubation of 150 μM SOD1 with or without 60 mM EDTA or LD100 at 37°C for 12 h, data were collected for 100 μl samples. For the complex of S1-SOD1, the mixture containing 100 μM SOD1 and 100 μM of S1 was incubated for 24 h in pH 7.4 20 mM Tris–HCl at 37°C. Before data collection, the sample was centrifuged for 10 min at 10 000 rpm at 4°C to remove the potential aggregates. The X-ray wavelength was 1.25 Å with a sample-to-detector distance of 1850 mm. The scattering vector q was 4πsin θ/λ, and 2θ was the scattering angle. X-ray exposures between 200 and 500 s were taken for all samples and matching the buffer blank, and the scattering of buffer was subtracted. Datasets were merged with PRIMUS. The real-space P(r) distribution, radius of gyration (Rg) and maximum distance Dmax were calculated with GNOM. Twenty independent ab initio modeling jobs were performed using DAMMIN, and averaged by DAMAVER.
Circular dichroism
To observe effects of the specific inhibitor LD100 on the SOD1 structure, circular dichroism (CD) experiments were performed at 25°C on a Chariscan CD photomultiplier (Applied Photophsics Limited) equipped with a Quantum Northwest TC125 temperature controller, and the path length of the quartz cuvette was 1.0 cm. The scanning speed was 100 nm/min, and the response time was 0.2 s. All spectra were collected between 200 and 260 nm, and the baseline was corrected using signals of the buffer.
Assay of SOD1 activity
SOD1 activity was determined by measuring the inhibition of tetrazolium salt reduction using HT Superoxide Dismutase Assay Kit (Trevigen; 7501-500-K). Briefly, after suspending the cells in 5–10 volumes of cold 1× Cell Extraction Buffer and incubating the cell suspensions on ice with periodic vortexing for 30 min, the cleared cell lysate was obtained by centrifugation at 10 000 × g for 10 min at 4°C. Mn- and Fe-SODs were inactivated by adding 800 μl of ice-cold chloroform/ethanol (37.5/62.5 (v/v)) to 500 μl of the cell lysate, shaking for 30 s, and then centrifuging at 2500 × g for 10 min. After determining the protein concentration of the cell lysate by BCA Protein Assay Kit (Thermo Scientific; 23225), serial dilutions of cell extracts were obtained by adding 1× SOD buffer. 25 μl diluted cell extracts were added into 150 μl Master Mix. The reactions of SOD1 activity assay were initiated by adding 25 μl 1× Xanthine solution. Three parallel experiments were performed in each experiment, and the activity was normalized by the control group.
HADDOCK docking
To obtain the structural insight into SOD1 binding to DNA, the HADDOCK runs were performed using the Easy interface for HADDCOK 2.2 (15). The structure of SOD1 (PDB: 1SPD) was downloaded from the PDB database. 3D-DART (16) was used to generate all the nucleic acid structure models. As experimental input parameters, all residues on the surface of SOD1 were marked as active, and the entire DNA strand was selected as active. HADDOCK clustered 81 structures in nine clusters, which represents 40.5% of the water-refined models HADDOCK generated. The statistics of the top 1 cluster (HADDOCK: −195.5 ± 23.3) was used to construct the structural model of DNA–SOD1.
Statistical analysis
All statistical tests were done using Microsoft Excel, and the statistical analysis of data was indicated in each figure. For comparison between each sample group and its control group, unpaired Student's t test was used. Parallel experiments were also indicated in each figure. Data were presented as means ± SD (indicated within each figure). P values less than 0.05 (*P < 0.05, **P < 0.01, ***P < 0.001) were considered statistically significant.
RESULTS AND DISCUSSION
H2O2 promotes SOD1 nuclear translocation
SOD1 is mainly localized in cytosol, but changes in the intracellular levels of H2O2 might alter its subcellular localization. To examine the distribution of SOD1 within a mammalian cell under varied redox conditions, immunofluorescence was used to localize SOD1 within HeLa cells respectively exposed to H2O2, GSH and the efficient and specific SOD1 inhibitor LD100 that we designed based on the structure and catalytic mechanism of SOD1 (Supplementary Figure S1A) (9). The immunofluorescence imaging showed that the oxidant H2O2 translocated SOD1 to the nucleus, and the reductant GSH elevated its content in cytosol (Figure 1). The SOD1 content was elevated in cytosol of the cells exposed to LD100 (Figure 1), because the LD100 inhibition of SOD1 resulted in reduced levels of H2O2 (9). AT, a catalase inhibitor, elevated endogenous H2O2 in the cell and translocated SOD1 to the nucleus (Supplementary Figure S1B and C), indicating that endogenous H2O2 also regulates SOD1 nuclear localization. Within the cells treated with O2•−-generating agent (4NQO) and paraquat (Supplementary Figure S1D), both the level of H2O2 and the content of SOD1 in the nucleus had comparable increase (Supplementary Figure S1B and C), because SOD1 is a high efficient catalyst of the dismutation of O2•− to H2O2. These results indicated that SOD1 displays a redox-dependent distribution in a mammalian cell.
Figure 1.
H2O2 promotes SOD1 nuclear translocation. Immunofluorescence using the antibody against SOD1 (green) and DAPI (blue, a nucleus-staining dye) indicated a redox-dependent distribution within HeLa cells. After HeLa cells were treated respectively with or without 0.4 mM H2O2, 2 mM GSH and 50 μM LD100 for 4 h, immunofluorescence staining was performed. Scale bars, 25 μm. Finally, we quantified the fluorescence intensity of SOD1 in the nucleus and cytoplasm using ImageJ, and then obtained the percentage of SOD1 in the nucleus (n > 20). Data are mean ± SD.
SOD1 occupies DNA sites with distinct sequence preference
To elucidate the potential interactions between SOD1 and DNA in a mammalian cell under varied redox conditions, we performed chromatin immunoprecipitation-sequencing (ChIP-Seq) tests with the immunoprecipitation (IP) grade anti-SOD1 antibody directed toward SOD1 in a HeLa cell treated respectively with and without (control) H2O2, GSH and LD100. The ChIP samples were digested with endo-exonuclease (micrococcal nuclease, ChIP grade) to address theoretical limitations in ChIP-Seq and to enhance resolution. We used the anti-RNA polymerase II antibody and normal rabbit IgG to construct positive and negative control IPs, respectively. The significantly high levels of GAPDH promoter signals relative to inputs in positive control IPs indicated the successful construction of ChIP (Supplementary Figure S2A). In the GSH treated cells, DNA samples obtained by ChIP were too little to perform sequencing, because the GSH-mediated translocation of SOD1 toward cytosol decreased the content SOD1 in the nucleus (Figure 1).
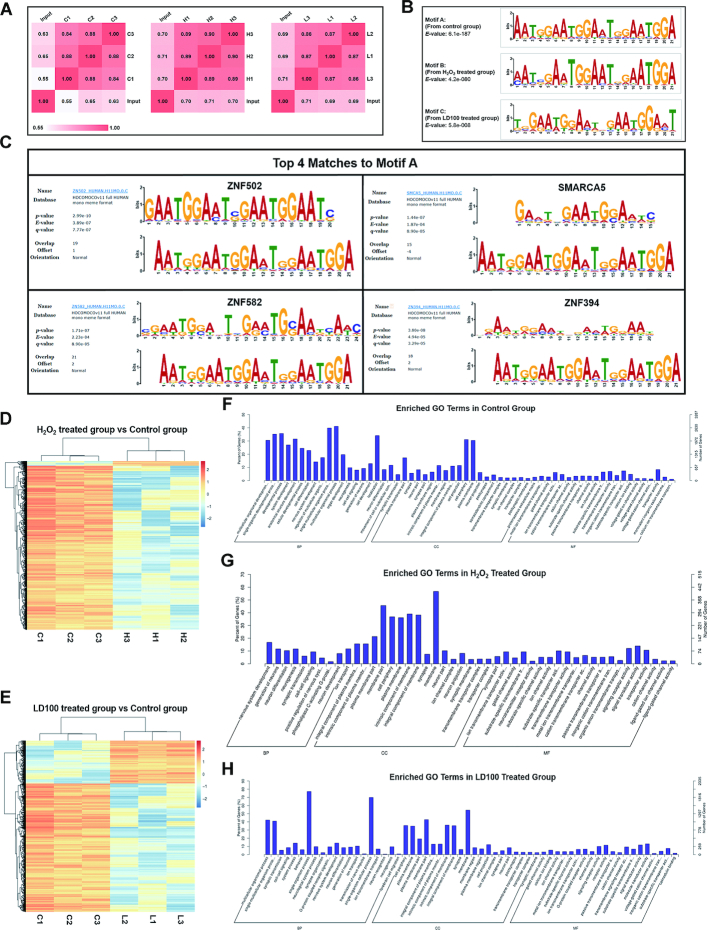
Sequencing of the ChIP samples produced clean reads respectively from control, H2O2- and LD100-treated cell groups through trimming the raw reads. Pearson correlations showed that a satisfactory correlation occurred between parallel samples (Figure 2A), and the high-throughput sequencing data are reliable. After these clean reads were aligned against homo sapiens genomes, we obtained uniquely mapping ChIP-Seq reads to homo sapiens genomes for control, H2O2- and LD100-treated groups, respectively. Following mapping reads to the genomes, peak calling approaches were used to estimate SOD1-occupied DNA sites within a cell genome. 45 139, 4697 and 21 293 peaks were obtained respectively in control, H2O2 and LD100 treated groups (Supplementary Figure S2B). The treatment with H2O2 and LD100 not only drastically reduced the number but also slightly reduced the enrichment of peaks (Supplementary Figure S2C). Then, the significant DNA motifs were obtained from our ChIP-Seq data (Supplementary Figure S2D). Annotation of the motifs in the control group provided the SOD1-bound DNA motifs with high statistical significance, indicating that DNA–SOD1 interactions occur through significant sequence preference (Figure 2B), not in a random pattern. Thus, the ChIP-Seq experiments confirmed the previously observed affinity of SOD1 for DNA (6,7,9,10). The high-affinity sites were found to be strongly enriched near the summits of the ChIP-Seq peaks, and a substantial fraction of high-affinity sites within accessible genomic regions are occupied.
Figure 2.
SOD1 binds to DNA with different sequence preferences under varied redox conditions. (A) Person correlations between parallel ChIP-Seq samples in control (designed as C1, C2 and C3), H2O2 (H1, H2 and H3), and LD100 (L1, L2 and L3) treated groups. Unless otherwise specified, the labels for all ChIP-Seq samples were the same as here. All the ChIP-Seq data were representative of three independent experiments. (B) Typical SOD1 binding motifs under varied redox conditions. (C) Motif logo matching between motif A (bottom) and four consensus motifs (top), determined by TOMTOM (MEME). (D and E) Hierarchical clustering analyses of fold enrichment values in control, H2O2 and LD100 treated groups. (F–H) Enriched GO terms of all ChIP-Seq samples in control (F), H2O2 (G), and LD100 treated groups (H).
Under normal conditions, motif A was the sequence with the highest statistical significance for SOD1 binding in the genome. Comparing these motifs with databases showed that the motif A in the control ChIP-Seq group matched perfectly with the transcription factors ZNF394-, ZNF502-, ZNF582- and SMARCA5-bound motifs (Figure 2C), implying that some regulatory roles of SOD1 may be similar to those of these transcription factors in gene expression. Moreover, both processing of ChIP-Seq-derived peak regions and annotation of genomic loci provided distributions of all peaks in homo sapiens genomes (Supplementary Figure S3A), indicating that a plenty of SOD1 binding events (15.78%) occurs in the vicinity (within 3000 bp) of transcriptional start site (TSS) (Supplementary Figure S3B). Compared with the control group, both H2O2 and LD100 treated groups had fewer peaks near TSS (Supplementary Figure S3B). In addition, the treatment with H2O2 and LD100 also slightly reduced the enrichment of peaks in the vicinity of TSS (Supplementary Figure S2C). This vicinity of binding sites to a TSS is important in determining whether a SOD1 binding event affects transcription. Therefore, these results suggest that SOD1 binding to DNA could regulate expression of the target genes in transcriptional phases.
The analogous sequences of the motif A in H2O2- and LD100-treated groups were motifs B and C, respectively. However, motifs B and C showed less statistical significance than the motif A (Figure 2B), suggesting that the treatment with either H2O2 or LD100 weaken interactions between SOD1 and these motifs in homo sapiens genomes. The sequences in the motif B with high statistical significance were similar to those in the motif A (Supplementary Figure S2D), indicating that oxidative environment resulted from exogenous H2O2 hardly impacts the DNA sequence preference of SOD1 occupancy, but impairs the intensity of SOD1 binding to DNA. However, the treatment with LD100 resulted in the motifs with high statistical significance which are remarkably distinct from the motifs A and B (Supplementary Figure S2D), indicating that SOD1 inhibition has noticeable influence on both DNA sequence preference and intensity of SOD1 binding. These results are expected because both inhibition-mediated SOD1 conformational changes (Supplementary Figure S4A-C) and low H2O2 content (17) could alter both DNA sequence preference and affinity of the SOD1 occupancy.
To find differences in SOD1 binding sites and in their fold enrichment in the genomes following treatment with either H2O2 or LD100 in comparison with the genome in control, we performed hierarchical clustering using the fold enrichment values in all samples. On the one hand, the treatment with H2O2 resulted in reduced fold enrichment values of most peaks comparing with the untreated samples, but significant differences were not found in the enrichment pattern (Figure 2D), an indication that H2O2 reduces interactions of SOD1 with DNA, but does not have significant impact on the pattern of SOD1 interactions. On the other hand, fold enrichment values were found to decrease for three-quarters of peaks, but to increase for one quarter of peaks in the case of LD100 treatment (Figure 2E), suggesting that the intracellular SOD1 inhibition not only reduces the affinity of SOD1 for DNA, but also alters the pattern of SOD1 interactions with DNA owing to the inhibition-mediated conformational changes of SOD1. Together, these results indicated that both H2O2 exposure and LD100 inhibition all can reduce the affinity of SOD1 for DNA, but the latter even alter the pattern of SOD1 interactions with DNA, as indicated in the motif analysis.
Gene ontology (GO) classification of the genes was performed in the range of 2000 bp around the peak summits to understand functions of the SOD1-occupied DNA sites. The enriched GO terms in control displayed a large body of genes involved in nervous system development, transmembrane transportation, and neurogenesis, generation and differentiation of neurons, suggesting that SOD1 binding may regulate many key genes in the nervous system (Figure 2F). The treatment with either H2O2 or LD100 was found to decrease the numbers of genes regulated by SOD1 binding (Figure 2G and H). Therefore, both H2O2 and LD100-mediated redox dyshomeostasis could weaken the SOD1 binding to DNA likely altering SOD1 structures, which may result in a decrease in the number of nervous systems and neurons linked-genes regulated by the SOD1 binding.
DNA–SOD1 interactions are further verified in solution
To probe the sequence-specificity of SOD1 binding to DNA, seven dsDNA sequences (12 bp, S1–S7, Figure 3A) were selected according to the above-described DNA sequence preference of SOD1 occupancy (Supplementary Figure S2D) and their 5′-ends were labelled by biotin. The binding of SOD1 to these DNA fragments with varied sequence preference was first examined in pH 7.4 buffer under varied redox conditions by electrophoretic mobility shift assay (EMSA). The EMSA tests performed with S1 of the highest preference indicated that all S1 molecules were completely saturated by SOD1 at ≥1:2 of their molar ratios (Figure 3B), revealing that the optimal stoichiometry of SOD1 binding to this DNA molecule is 1:2, and at least parts of the specific SOD1 binding bases or base pairs are contained in this DNA sequence. Changing buffer pH did not impact the formation of S1-SOD1 complexes (Figure 3C). Moreover, the binding of SOD1 to all of the designed DNA sequences was observed by EMSA under the tested conditions. The simultaneous appearance of DNA–SOD1 complexes and free DNAs (Figure 3D) indicated that interactions between SOD1 and S4, S5 or S6 were weak and SOD1 binding did not occur for S2, S3 and S7. These observations were supported by the quantitative fluorescence anisotropy tests that demonstrated the high affinity (Kd = 8.93 nM) of SOD1 for S1 and no affinity for the sequence S7 under the tested conditions (Supplementary Figure S5A and B). These results suggested that the DNA–SOD1 interactions identified by EMSA and affinity tests were consistent with the DNA sequence preference of SOD1 occupancy (Supplementary Figure S2D). To further confirm the dsDNA sequence-specificity of SOD1 interactions, the competition of two DNA sequences for SOD1 was tested. The results showed that the labelled S1 was replaced by the unlabelled S1 of increased concentrations from the labelled S1-SOD1 complexes (Figure 3E), but not by the unlabeled S2 (Figure 3F), revealing that the formation of DNA–SOD1 complexes was dependent on the base pair sequence in S1. Moreover, the tests with the single-stranded S1 (ssS1) showed that SOD1 completely saturated ssS1 at 1:2 like the double-stranded form of S1, and the affinity of SOD1 for the complementary strand of ssS1 was much weaker than dsS1 (Supplementary Figure S6), as indicated by the tests performed using the mixture of ssS1 and its complementary strand under tested conditions (Supplementary Figure S6). Comparing the tested dsDNA sequences indicated that the quadruplet GGAA in S1 is essential for SOD1 binding to DNA. In addition, the antioxidant activity of SOD1 is not significantly affected by the presence of dsS1 or dsS7 (Supplementary Figure S7A and B).
Figure 3.
Verification of DNA–SOD1 interactions in vitro. (A) The selected DNA sequences (S1–S7, 12 bp) with different SOD1 binding ability. (B and C) EMSA showed that the formation of S1-SOD1 complexes was dependent on concentrations of SOD1 in 10 mM pH 7.4 PBS, and pH did not impact the binding of SOD1 to DNA in the tested range of pH. (D) EMSA indicated that SOD1 binds to the selected DNA sequences with different preference under tested conditions. (E and F) The competition performed with excess of the unlabelled S1 and S2 further confirms the formation of S1-SOD1 complexes.
To determine which DNA bases are directly involved in the binding of SOD1, DNase I footprinting assay was carried out with a 40 bp dsDNA (SL, Figure 4A) designed based on the above-mentioned sequence preference and EMSA results of DNA–SOD1 interactions. Formaldehyde was used to fix potential interactions between DNA and SOD1 prior to footprinting. This cross-linking led to the formation of a linear 200 bp pentamer of SL (Figure 4B). Capillary electrophoresis analysis of SOD1 occupied sites in its range of 161–200 bp showed that SOD1 associated with DNA through the direct contact with the base sequence GGA or GAA (Figure 4A and B), which is slightly different from the results from protein array tests (10), because SOD1 can be cross-linked to the 3′ or 5′-end of DNA (18). To exclude influence of the cross-linking on the sequence specificity of SOD1 binding, footprinting assays were also carried out with the SL-SOD1 complex treated without formaldehyde, providing a similar result, but the difference being one base (T) because of low resolution (Figure 4C and D). The combination of both lines of evidence not only confirmed that the ChIP-Seq data-derived DNA sequence preference of SOD1 binding is reliable, but also revealed that the triplet GGA is one of the base sequences directly involved in DNA–SOD1 interactions. In addition, the molecular docking simulation of DNA–SOD1 interactions suggested that the α-helix on the surface of a SOD1 monomer sticks into the major groove of the triplet GGA-containing dsS1 stretch to form a stable complex (Figure 4E), which is not only in line with the results from DNase I footprinting assay, but also supported by the solution small-angle X-ray scattering (SAXS) structure that proved the direct contact of the SOD1 α-helix to DNA in the major groove of DNA (Figure 4F).
Figure 4.
Binding model of DNA–SOD1 complexes. (A) The SL bases directly involved in the DNA–SOD1 interactions fixed by 1% formaldehyde cross-linking were determined by capillary electropherograms following DNase I digestion of 5′-FAM-labelled pentameric SL. Stars represent the sites protected by SOD1. (C) Capillary electropherograms were generated from the SL-SOD1 complexes treated without 1% formaldehyde following DNase I digestion of 5′-FAM-labelled SL. (B and D) Cartoon representation of SL highlights the SOD1 binding DNA sites resulted from DNase I footprinting tests in (A) and (C). (E) The structural docking model for S1-DNA complex. The purple regions in DNA represent the potential SOD1 binding sites in S1 motif. α-helix, red; β-sheet, yellow; random coil and DNA, green; SOD1 binding sites, purple. (F) A final concentration of 100 μM SOD1 and 100 μM of S1 was incubated in 20 mM Tris–HCl (pH 7.4) at 37°C for 24 h. Before data collection, the sample was centrifuged at 10,000 rpm at 4°C for 10 min to remove the potential aggregates. Based on SAXS results, space-filling models of the S1-SOD1 complexes were constructed. Space-filling models of the S1-SOD1 complexes derived from SAXS data are depicted in grey, with HADDOCK model of this complexes docked into mesh envelope.
DNA–SOD1 binding is impacted by H2O2 and Demetallization
The formation of dsS1-SOD1 complexes was examined respectively under varied redox conditions and under the conditions containing the specific chelating agent of zinc or inhibitor of SOD1. The solution redox conditions were altered by respective addition of H2O2 and GSH. SOD1 was found to be inactivated by adding H2O2 in a H2O2 concentration-dependent manner, but SOD1 retained 85% activity at 0.4 mM H2O2 (Figure 5A). The formation of S1-SOD1 complexes was dramatically inhibited when H2O2 ≥1 mM (Figure 5B), which was in agreement with the results obtained by ChIP-Seq (Figure 2B), and also with the SOD1 structural changes resulted from H2O2 oxidation of SOD1 (19). Oxidation of the histidine residues coordinated to the metal ions causes partial metal ion release form SOD1, resulting in considerable exposure of hydrophobic surfaces (20). However, the presence of GSH slightly decreased SOD1 activity (Figure 5C), and elevated the formation of S1-SOD1 complexes in a GSH concentration-dependent manner (Figure 5D). These results indicated that the binding of SOD1 to DNA is reduced in oxidative environments, but intensified in reductive environments.
Figure 5.
SOD1 binding to DNA depends on redox environments and its conformation. (A and B) The treatment with H2O2 for 4 h reduced not only the activity of SOD1 (A) but also the formation of S1-SOD1 complexes (B). (C and D) The treatment with GSH for 4 h slightly reduced the activity of SOD1 (C) but significantly elevated the formation of S1-SOD1 complexes (D). (E and H) The specific inhibitor of SOD1 (LD100, E) and the chelator of Zn2+ (TPEN, H) both prevented SOD1 from interactions with DNA under tested conditions. (F and G) The formation of S1-A4V (F) and S1-H46R/H48Q (G) complexes were dependent on concentrations of SOD1 in 10 mM pH 7.4 PBS. Data are mean of triplicate samples ± SD (*P < 0.05, **P < 0.01, ***P < 0.001; unpaired Student's t test), and all error bars are SD.
S1-SOD1 complexes were observed to rapidly disappear upon addition of the specific SOD1 inhibitor LD100 (Figure 5E), indicating that the inactivation-mediated SOD1 conformational relaxation (Supplementary Figure S4A-C), similar to the structural changes caused by mutation (21), can prevent SOD1 from binding to DNA. In support of this, DNA binding of two ALS-linked SOD1 mutants A4V and H46R/H48Q was examined under the tested conditions. The mutation A4V does not impact the activity and structure of SOD1, but H46R/H48Q eliminates the activity of and significantly alters the structure of SOD1 owing to the removal of copper (22). The results indicated that the binding of A4V to dsS1 was similar to that of wild-type SOD1 (Figure 5F), but the binding of H46R/H48Q was dramatically reduced (Figure 5G), supported by their affinity constants for dsS1 of Kd = 5.03 (A4V) and 33.7 nM (H46R/H48Q) (Supplementary Figure S5C and D). These revealed that DNA binding can be regulated by structural alterations of SOD1.
Similarly, removal of Zn2+ from SOD1 by the specific Zn2+ chelator TPEN resulted in the rapid disappearance of S1-SOD1 complexes (Figure 5H), because this demetallation can also alter structures of a metalloenzyme (23), and Zn2+ plays a structural role in SOD1 (2). In brief, SOD1 is a H2O2-responsive DNA binding protein both in solution and within a cell, and the formation of DNA–SOD1 complexes is regulated by its conformational changes.
SOD1 regulates gene expression through binding to DNA
To explore the regulatory roles of SOD1 binding in gene expression under varied redox conditions, we carried out global mRNA-sequencing (mRNA-Seq) on SOD1 knockdown cells and cells treated with LD100. Joint analysis of mRNA-Seq and ChIP-Seq data not only produced overlapped genes of differentially expressed genes in SOD1 knockdown cells and genes closest to TSS in ChIP-Seq samples under normal conditions, but also displayed that the expression of 1486 genes would be regulated in transcriptional phases by SOD1 binding, but 413 genes among them were also regulated by changing SOD1 activity (Figure 6A, and Supplementary Figure S8A). Moreover, 637 and 849 genes of 1486 genes were upregulated and downregulated respectively upon SOD1 knockdown (Figure 6B). In addition, we also found that other 732 genes were only regulated by changing SOD1 activity (Supplementary Figure S8A). These results indicate that SOD1 can play a regulatory role in the expression of these genes by either binding to DNA (Figure 6B) or by changing intracellular ROS levels. To understand the regulatory significance of SOD1 binding, more examinations were carried out on ALS- and cancer-linked ChIP-Seq and mRNA-Seq data of the genes, because SOD1 has become a key target in drug design of ALS and cancer treatment (24,25). The results indicated that the 209 regulated genes were linked to nervous system development (Supplementary Figure S9 and Supplementary Table S1), and the expression of their half was controlled by SOD1 binding rather than the enzymatic activity of SOD1 because the specific inactivation of SOD1 with LD100 did not impact their expression (Supplementary Figure S9). It is worthy that four ALS-linked genes UNC13A, NRG1, INA and PTK6 (26–29) were found to be modulated via SOD1 binding but not via SOD1 enzymatic activity (Figure 6C). In addition, the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment of 1486 genes showed that SOD1 binding impacted onset and development of multiple kinds of cancer via regulating the expression of oncogenes (Figure 6D).
Figure 6.
SOD1 regulates expression of numerous genes in transcriptional phases. (A) A hierarchical clustering analysis of the differentially expressed genes (DEGs) caused by DNA binding of SOD1. After overlapping the genes in control group of ChIP-Seq and the DEGs in SOD1 knockdown group of RNA-Seq, the heat map was preparation by normalized log10(FPKM+1) of the DEGs. (B) A volcano plot of log2(fold change) vs. the -log10(padj) for the DEGs in (A). (C, E and F) Relative FPKM values of some key genes in SOD1 knockdown and LD100 treated HeLa cells (INA, UNC13A, PTK6 and NRG1 in (C); PBX2 and FGFR4 in (E); TUSC2 and CRTC3 in (F). (D) An enriched KEGG pathway scatterplot of the DEGs in (A) shows the most affected 20 signaling pathways with high statistical significance. (G) A heat map of the genes regulates by SOD1 at transcriptional levels, drawn using normalized log10(FPKM+1) of the genes. (H and K) Genome browser views of the SOD1 binding sites near TSS of all genes in (G) and in (J) (red for the ChIP-Seq under normal conditions; blue for the Input). (I and J) RT-pPCR tests for the SOD1-regulated genes in knockdown HeLa cells (TUSC2, CRTC3, KCNAB2, SPEG, QRFP, MAF1, DNM2, NFIC, PTK6, TNFRSF25 and FGFR4 in (I); INA, UNC13A and NRG1 in (J)). Data are mean of triplicate samples ± SD (*P < 0.05, **P < 0.01, ***P < 0.001; unpaired Student's t test), and all error bars are SD.
To establish the regulatory role of SOD1 binding in the expression of oncogenes, in-depth analysis was performed on mRNA-Seq data. Because high expression of PBX2 and FGFR4 promotes cancer cell growth through both suppression of apoptosis (30) and regulation of cellular pathways to enhance drug resistance (31), the elevated expression of PBX2 and FGFR4 in SOD1 knockdown cells (Figure 6E) indicated that this SOD1 deficiency can enable the survival of cancer cells from both apoptosis and anti-cancer treatment through DNA–SOD1 interactions. However, SOD1 inhibition was found to reduce the expression of FGFR4. Moreover, SOD1 knockdown resulted in elevated PTK6 and reduced TUSC2 and CRTC3 (Figure 6C and F), indicating that this downregulation of SOD1 promotes tumorigenesis by PTK6 (32), regulates the early pathogenesis of lung tumors by TUSC2 (33), and reduces the transcription of human T-Cell leukemia virus type 1 by CRTC3 (34). In brief, these results revealed that the SOD1 deficiency activates the genes to promote onset and development of cancer through reducing its binding to DNA. Then, SOD1 binding to DNA around TSS can control the expression of multiple oncogenes in transcriptional phases.
The SOD1 binding sites were further analyzed in the range of 3000 bp from TSS, because 15.78% of SOD1 binding sites in homo sapiens genomes are close to TSS (Supplementary Figure S3B). Analysis revealed that SOD1 may regulate 442 genes during the transcription phase (Supplementary Figure S8B), among which 22 genes, including oncogenes (e.g., PTK6, PBX2, FGFR4, TUSC2, CRTC3), voltage-gated ion channel-associated genes (e.g. KCNAB2, CBARP), muscle- and nerve-linked genes (e.g., QRFP, PTK6, SPEG, OLFM1), transcription-related genes (e.g. NFIC, BEND3, MAF1, SNRNP70), were found to be most likely regulated by SOD1 binding in transcriptional phases (Figure 6G and H). RT-qPCR determinations of mRNA showed that the expression of a subset (CRTC3, SPEG, QRFP, MAF1, DNM2, TNFRSF25 and FGFR4) selected from these regulated genes was significantly altered in SOD1 knockdown cells (Figure 6I). The observation for regulation of the oncogenes is in line with the conclusions from the in-deep analysis of mRNA-Seq data (Figure 6C, E and F). Our reported results showed that the expression level of SOD1 in the normal cell RWPE-1 was <0.3-fold than that of the cancer cell DU145 (14). Compared with DU145 cells, the relative expression changes in the subset of genes in RWPE-1 cells were consistent with the results in SOD1 knockdown HeLa cells (Supplementary Figure S10A). Furthermore, the expressions of CRTC3 and FGFR4 in HeLa cells were also respectively downregulated and upregulated with increasing concentrations of H2O2 (Supplementary Figure S10B), which indicated that the regulation of CRTC3 and FGFR4 by SOD1 binding was dependent on H2O2 concentrations. Overall, these results further verified the regulatory role of SOD1 in transcriptional phases of expression of the genes involved in cancer, transcription, voltage-gated ion channels and nervous systems.
SOD1 knockdown was also found to regulate the expression of ALS-linked genes including INA, UNC13A, PTK6, NRG1, SPEG and QRFP (26–29,35,36). This SOD1 deficiency elevated the expression of UNC13A, PTK6 and NRG1, and reduced the expression of INA (Figure 6C). The expressions of UNC13A, INA, and NRG1 were further verified by RT-qPCR (Figure 6J). In addition, SOD1 binding sites were also observed to appear around the TSS of UNC13A, PTK6, NRG1 and INA (Figure 6H and K). Accordingly, DNA–SOD1 interactions might prolong the lifespan of sporadic ALS patients by UNC13A (26), negatively modulate the acetylcholine receptor by PTK6 (27), regulate the axoglial and neuromuscular development in the pathogenesis and progression of ALS by NRG1 (28), and control intracellular transport to axons and dendrites by INA (29). Moreover, the genes encoding QRFP (neural polypeptide) and SPEG (regulating myocyte cytoskeletal development) were downregulated by SOD1 knockdown (Figure 6I). Their downregulation through transcription causes neurological or muscular abnormalities (35,36), which might also have contribution to ALS. Overall, the regulation of expression of these ALS-linked genes by affinity interactions of SOD1 with DNA might be in favor of delaying ALS progression.
CONCLUSIONS
In summary, the nuclear translocation and DNA binding of SOD1 take place in turn upon elevation of the intracellular H2O2 content. SOD1 binds to DNA mainly in the vicinity of TSS in a sequence-specific manner, and readily forms complexes with GGA-containing DNA stretches. Both sequence preference and affinity of SOD1 interactions with DNA depends on SOD1 conformation. Changing intracellular ROS content also impacts the affinity of SOD1 for DNA. SOD1 binding can regulate the ROS-responsive expression of cancer- and ALS-linked genes in transcriptional phases. A comparison of regulation in expression of the oncogenes and ALS-linked genes suggests that SOD1 deficiency-mediated reduction of SOD1 interactions with DNA activates both the genes to promote onset and development of cancer and those likely to delay ALS progression, implying there is a correlation between cancer progression and ALS repression. Accordingly, targeting both SOD1 and DNA–SOD1 interactions should be a promising avenue to design anti-cancer and -ALS strategies.
The evidence presented here not only confirms, for the first time to our knowledge, a new function of SOD1: acting as a H2O2-responsive regulatory protein in expression of numerous mammalian functional genes, but also indicates that there is a metalloprotein-based regulatory mechanism of the gene expression in response to ROS changes within mammalian cells, similar to that within microbial cells.
DATA AVAILABILITY
All Chip-Seq datasets are available in GEO, accession number: GSE124082. Sequence tracks of genes regulated by SOD1 binding to DNA were provided as Supplementary Data.
Supplementary Material
ACKNOWLEDGEMENTS
We thank staff of beamline BL16B1 at Shanghai Synchrotron Radiation Facility, China, for their assistance in data collection. We also thank Novogene for their assitance in ChIP-Seq.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Natural Science Foundation of China [21271079, 21771073]; Fundamental Research Funds for the Central Universities [2018CXZZ027]; self-determined research fund of CCNU from the colleges’ basic research and operation of MOE; The support from 111 Program [B17019]. Funding for open access charge: National Natural Science Foundation of China.
Conflict of interest statement. None declared.
REFERENCES
- 1. Imlay J.A. Transcription factors that defend bacteria against reactive oxygen species. Annu. Rev. Microbiol. 2015; 69:93–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Sheng Y., Abreu I.A., Cabelli D.E., Maroney M.J., Miller A.F., Teixeira M., Valentine J.S.. Superoxide dismutases and superoxide reductases. Chem. Rev. 2014; 114:3854–3918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Reczek C.R., Chandel N.S.. ROS-dependent signal transduction. Curr. Opin. Cell Biol. 2015; 33:8–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Reddi R., Culotta V.C.. SOD1 integrates signals from oxygen and glucose to repress respiration. Cell. 2013; 152:224–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Tsang K., Chen M., Cheng X., Qi Y., Chen Y., Das I., Li X.X., Vallat B., Fu L.W., Qian C.N. et al.. SOD1 phosphorylation by mTORC1 couples nutrient sensing and redox regulation. Mol. Cell. 2018; 70:502–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Jiang W., Han Y., Pan Q., Shen T., Liu C.. Roles of exogenous divalent metals in the nucleolytic activity of Cu,Zn superoxide dismutase. J. Biol. Inorg. Chem. 2006; 11:835–848. [DOI] [PubMed] [Google Scholar]
- 7. Jiang W., Han Y., Zhou R., Zhang L., Liu C.. DNA is a template for accelerating the aggregation of copper, zinc superoxide dismutase. Biochemistry. 2007; 46:5911–5923. [DOI] [PubMed] [Google Scholar]
- 8. Zhao D., Zhang S., Meng Y., Dong X., Zhang D., Liang Y., Wang L., Liu C.. Polyanion binding accelerates the formation of stable and low-toxic aggregates of ALS linked SOD1 mutant A4V. Proteins. 2014; 82:3356–3372. [DOI] [PubMed] [Google Scholar]
- 9. Tsang K., Liu Y., Thomas J., Zhang Y.J., Zheng X.F.S.. Superoxide dismutase 1 acts as a nuclear transcription factor to regulate oxidative stress resistance. Nat. Commun. 2014; 5:3446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hu S., Xie Z., Onishi A., Yu X., Jiang L., Lin J., Rho H., Woodard C., Wang H., Jeong J.S. et al.. Profiling the human protein-DNA interactome reveals ERK2 as a transcriptional repressor of interferon signaling. Cell. 2009; 139:610–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Barbosa L.F., Cerqueira F.M., Macedo A.F.A., Garcia C.C.M., Angeli J.P.F., Schumacher R.I., Sogayar M.C., Augusto O., Carrì M.T., Mascio P.D. et al.. Increased SOD1 association with chromatin, DNA damage, p53 activation, and apoptosis in a cellular model of SOD1-linked ALS. BBA-Mol. Basis Dis. 2010; 1802:462–471. [DOI] [PubMed] [Google Scholar]
- 12. Cereda C., Leoni E., Milani P., Pansarasa O., Mazzini G., Guareschi S., Alvisi E., Ghiroldi A., Diamanti L., Bernuzzi S. et al.. Altered intracellular localization of SOD1 in leukocytes from patients with sporadic amyotrophic lateral sclerosis. PLoS One. 2013; 8:e75916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Borchelt R., Lee M.K., Slunt H.S., Guarnieri M., Xu Z.S., Wong P.C., Brown R.H. Jr, Price D.L., Sisodia S.S., Cleveland D.W.. Superoxide dismutase 1 with mutations linked to familial amyotrophic lateral sclerosis possesses significant activity. Proc. Natl. Acad. Sci. U.S.A. 1994; 91:8292–8296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Li X., Chen Y., Zhao J., Shi J., Wang M., Qiu S., Hu Y., Xu Y., Cui Y., Liu C. et al.. The specific inhibition of SOD1 selectively promotes apoptosis of cancer cells via regulation of the ROS signaling network. Oxid. Med. Cell Longev. 2019; 2019:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. van Zundert G.C.P., Rodrigues J.P.G.L.M., Trellet M., Schmitz C., Kastritis P.L., Karaca E., Melquiond A.S.J., van Dijk M., de Vries S.J., Bonvin A.M.J.J.. The HADDOCK2.2 webserver: User-friendly integrative modeling of bimolecular complexes. J. Mol. Biol. 2015; 428:720–725. [DOI] [PubMed] [Google Scholar]
- 16. Van Dijk M., Bonvin A.M.J.J.. 3D-DART: a DNA structure modelling server. Nucleic Acids Res. 2009; 37:W235–W239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Dong X., Zhang Z., Zhao J., Lei J., Chen Y., Li X., Chen H., Tian J., Zhang D., Liu C. et al.. The rational design of specific SOD1 inhibitors via copper coordination and their application in ROS signaling research. Chem. Sci. 2016; 7:6251–6262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Conaway C.C., Whysner J., Verna L.K., Williams G.M.. Formaldehyde mechanistic data and risk assessment: endogenous protection from dna adduct formation. Pharmacol. Ther. 1996; 71:29–55. [DOI] [PubMed] [Google Scholar]
- 19. Bosco D.A., Morfini G., Karabacak N.M., Song Y., Gros-Louis F., Pasinelli P., Goolsby H., Fontaine B.A., Lemay N., McKenna-Yasek D. et al.. Wild-type and mutant SOD1 share an aberrant conformation and a common pathogenic pathway in ALS. Nat. Neurosci. 2010; 13:1396–1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rakhit R., Cunningham P., Furtos-Matei A., Dahan S., Qi X.F., Crow J.P., Cashman N.R., Kondejewski L.H., Chakrabartty A.. Oxidation-induced misfolding and aggregation of superoxide dismutase and its implications for amyotrophic lateral sclerosis. J. Biol. Chem. 2002; 277:47551–47556. [DOI] [PubMed] [Google Scholar]
- 21. McCaffrey P. SOD1 mutant protein gets loose in ALS. Lancet Neurol. 2006; 5:119. [DOI] [PubMed] [Google Scholar]
- 22. Winkler D.D., Schuermann J.P., Cao X., Holloway S.P., Borchelt D.R., Carroll M.C., Proescher J.B., Culotta V.C., Hart J.P.. Structural and biophysical properties of the pathogenic SOD1 variant H46R/H48Q. Biochemistry. 2009; 48:3436–3447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sigel H., Martin R.B.. Coordinating properties of the amide bond. Stability and structure of metal ion complexes of peptides and related ligands. Chem. Rev. 1982; 82:385–426. [Google Scholar]
- 24. Ralph S., Radcliffe P.A., Day D.M., Carthy J.M., Leroux M.A., Lee D.C.P., Wong L., Bilsland L.G., Greensmith L., Kingsman S.M. et al.. Silencing mutant SOD1 using RNAi protects against neurodegeneration and extends survival in an ALS model. Nat. Med. 2005; 11:429–433. [DOI] [PubMed] [Google Scholar]
- 25. Huang P., Feng L., Oldham E.A., Keating M.J., Plunkett W.. Superoxide dismutase as a target for the selective killing of cancer cells. Nature. 2000; 407:390–395. [DOI] [PubMed] [Google Scholar]
- 26. Diekstra P., Vught P.W.J., Rheenen W., Koppers M., Pasterkamp R.J., Es M.A., Schelhaas H.J., Visser M., Robberecht W., Damme P.V. et al.. UNC13A is a modifier of survival in amyotrophic lateral sclerosis. Neurobiol. Aging. 2012; 33:630. [DOI] [PubMed] [Google Scholar]
- 27. Ferraiuolo L., Bono J.P.D., Heath P.R., Holden H., Kasher P., Channon K.M., Kirby J., Shaw P.J.. Transcriptional response of the neuromuscular system to exercise training and potential implications for ALS. J. Neurochem. 2009; 109:1714–1724. [DOI] [PubMed] [Google Scholar]
- 28. Song F., Chiang P., Wang J., Ravits J., Loeb J.A.. Aberrant neuregulin1 signaling in amyotrophic lateral sclerosis. J. Neuropathol. Exp. Neurol. 2012; 71:104–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Rouleau A., Clark A.W., Rooke K., Pramatarova A., Krizus A., Suchowersky O., Julien J., Figlewicz D.. SOD1 mutation is assosiated with accumulation of neurofilaments in amyotrophic lateral scelaries. Ann. Neurol. 1996; 39:128–131. [DOI] [PubMed] [Google Scholar]
- 30. Qiu Y., Song B., Zhao G., Deng B., Makino T., Tomita Y., Wang J., Luo W., Doki Y., Aozasa K. et al.. Expression level of Pre B cell leukemia homeobox 2 correlates with poor prognosis of gastric adenocarcinoma and esophageal squamous cell carcinoma. Int. J. Oncol. 2010; 36:651–663. [DOI] [PubMed] [Google Scholar]
- 31. Turkington R.C., Longley D.B., Allen W.L., Stevenson L., McLaughlin K., Dunne P.D., Blayney J.K., Salto-Tellez M., Van Schaeybroeck S., Johnston P.G.. Fibroblast growth factor receptor 4 (FGFR4): a targetable regulator of drug resistance in colorectal cancer. Cell Death Dis. 2014; 5:e1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Peng M., Ball-Kell S.M., Franks R.R., Xie H., Tyner A.L.. Protein tyrosine kinase 6 regulates mammary gland tumorigenesis in mouse models. Oncog. 2013; 2:e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Prudkin L., Behrens C., Liu D.D., Zhou X., Ozburn N.C., Bekele B.N., Minna J.D., Moran C., Roth J.A., Ji L.. Loss and reduction of Fus1 protein expression is a frequent phenomenon in the pathogenesis of lung cancer. Clin. Cancer Res. 2008; 14:41–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Koga H., Ohshima T., Shimotohno K.. Enhanced activation of Tax-dependent transcription of human T-cell leukemia virus type I (HTLV-I) long terminal repeat by TORC3. J. Biol. Chem. 2004; 279:52978–52983. [DOI] [PubMed] [Google Scholar]
- 35. do Rego C., Leprince J., Chartrel N., Vaudry H., Costentin J.. Behavioral effects of 26RFamide and related peptides. Peptides. 2006; 27:2715–2721. [DOI] [PubMed] [Google Scholar]
- 36. Hsieh C., Yoshizumi M., Endege W.O., Kho C., Jain M.K., Kashiki S., Santos R., Lee W., Perrella M.A., Lee M.. APEG-1, a novel gene preferentially expressed in aortic smooth muscle cells, is down-regulated by vascular injury. J. Biol. Chem. 1996; 271:17354–17359. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All Chip-Seq datasets are available in GEO, accession number: GSE124082. Sequence tracks of genes regulated by SOD1 binding to DNA were provided as Supplementary Data.