Abstract
Importance:
Risk score models predicting the progression of age-related macular degeneration (AMD) to its advanced forms may be useful for targeting high-risk individuals for lifestyle changes that reduce risk for AMD progression, helping with differential diagnosis of AMD and its subtypes, identifying high-risk subjects for participation in clinical trials, and selecting appropriate therapies.
Objective:
To develop and validate a predictive model for progression to advanced stages of AMD in 2 independent cohorts.
Design:
Participants in a validation cohort and an independent derivation population were classified into 5 stages of AMD based on ocular examination and fundus photographs at baseline. Progression was defined as either eye progressing from stage 1, 2, or 3 to either stage 4 or stage 5 at any follow-up visit to the end of the study. Cox proportional hazards models were used for progression analyses. Covariates included demographic and environmental factors, 6 variants in 5 genes, and baseline AMD grades in both eyes. The algorithm developed with the derivation sample was assessed for calibration and discrimination in the validation data set.
Setting:
Clinic populations and referrals.
Participants:
The derivation population comprised 2914 subjects with 809 progressors. The independent validation cohort comprised 980 individuals with no, early, or intermediate AMD in at least one eye at baseline, of whom 294 progressed to advanced stages of geographic atrophy or neovascular disease.
Main Outcome Measure:
Progression to advanced AMD.
Results:
For the model with all nongenetic and genetic factors, the respective C statistics for progression to advanced AMD in the derivation and validation samples were 0.858 and 0.750 at 5 years and 0.884 and 0.809 at 10 years, and models also discriminated risk for progression to geographic atrophy and neovascular disease separately. For unilateral or bilateral intermediate AMD, 5-year cumulative incidence rates of progression to advanced AMD were 10% with the low-risk score and 50% with the high-risk score; for unilateral advanced disease, the progression rates were 22% and 80% for the fellow eye.
Conclusions and Relevance:
The risk prediction model was validated in an independent study of progression from no, early, or intermediate stages to advanced subtypes of AMD and will be useful for research, clinical trials, and personalized medicine.
WE HAVE PREVIOUSLY developed risk prediction models based on demographic, environmental, and genetic factors that can predict the occurrence of age-related macular degeneration (AMD) and its progression from early and intermediate stages to the advanced forms of geographic atrophy (GA) and neovascular disease (NV).1-7 In our most recent and expanded model,6 we considered progression in both eyes, incorporated macular drusen size in both eyes and presence of unilateral advanced AMD at baseline, accounted for time-varying rates of progression by using AMD grades at all follow-up visits, extended the follow-up period to 12 years, and included a larger number of participants. We also calculated absolute risks for individuals with a specific set of demographic, ocular, and genetic risk factors, adjusting for competing risks (death from other causes) according to age and sex of the subject. We validated our model by splitting the total sample in half, deriving the model from 1 subsample, and testing it in the other subsample. The areas under the receiver operating characteristic curve (AUCs) for progression at 10 years in the model with genetic factors, macular drusen size, and environmental covariates were 0.915 and 0.908 in the 2 subsamples. We also demonstrated that the sample sizes needed for clinical trials that test the effectiveness of new therapies would be lower if genetic susceptibilities were considered in the design.6
For this study, we developed a modified risk prediction model and evaluated the validity of the model in an independent cohort of subjects being followed up in an observational study of risk factors for progression to advanced forms of AMD. Risk score models may be useful for targeting high-risk individuals for lifestyle changes that reduce risk for AMD progression, helping with differential diagnosis of AMD and its subtypes, identifying high-risk subjects for participation in clinical trials, and selecting appropriate therapies.
METHODS
PHENOTYPE AND PROGRESSION DATA
The derivation study cohort is based on subjects in the Age-Related Eye Disease Study.6,8 Follow-up time was 0.5 to 13.0 years (mean, 8.8 years). The independent validation cohort consisted of white patients (excluding first-degree relatives) who were enrolled in our ongoing studies to identify genetic and environmental factors for onset and progression of macular degeneration.2,9-13 Subjects were derived from clinic populations and referrals and were prospectively followed up for 0.10 to 17.9 years (mean, 6.2 years). Participants were classified using the Clinical Age-Related Maculopathy Staging System,14 based on ocular examination and grading of fundus photographs at baseline, into 5 stages: normal or stage 1 in both eyes (essentially free of age-related macular abnormalities or having only a few small drusen), early AMD or stage 2 in the worse eye (mild changes including multiple small drusen, nonextensive intermediate drusen, and/or pigment abnormalities), intermediate AMD or stage 3 in the worse eye (drusen with a diameter ≥125 μm, extensive intermediate drusen), stage 4 in one eye (advanced dry AMD with central or noncentral GA), and stage 5 with advanced NV AMD in one eye at baseline. Both cohorts were classified using this system. Because category 3 in the original Age-Related Eye Disease Study classification included noncentral GA and category 4 included both advanced forms of AMD as well as vision loss regardless of phenotype,8 we reclassified these groups independent of visual acuity level into Clinical Age-Related Maculopathy Staging System grades 4 (GA) and 5 (NV) as described herein.
Progression was defined as either eye progressing from stage 1, 2, or 3 to either stage 4 or stage 5 at any follow-up visit to the end of the study within each individual. In a subgroup analysis, we classified progressors to each advanced stage of AMD separately as progression to GA and progression to NV in the fellow eye. Time to progression was recorded for the first eye to progress if both eyes were at risk and for the fellow eye if one eye was at risk. Individuals were considered progressors if there was no advanced AMD in either eye at baseline and they developed advanced AMD in at least one eye during follow-up or if they had advanced AMD in one eye at baseline and progressed to advanced AMD in the fellow eye during follow-up. For subjects in the former group, we controlled for baseline grade in each eye, evaluated the time to progression in each eye, and used the earlier of the 2 progression times if both eyes progressed at different times. For subjects in the latter group, we controlled for AMD category in the affected eye at baseline (ie, Clinical Age-Related Maculopathy Staging System grades 4 and 5) and AMD grade in the nonadvanced eye at baseline and evaluated the time to progression in the fellow eye. Detailed drusen phenotype information was not included because it was not available for the validation cohort.
Demographic and risk factor data, including education, smoking history, and body mass index (BMI; calculated as weight in kilograms divided by height in meters squared), were obtained at the baseline visit from questionnaires and height and weight measurements. Antioxidant supplement use was not incorporated into this model. This variable was a weak predictor in our previous models4,6 and was not available for all subjects in the validation cohort. The research protocol was approved by institutional review boards and all participants signed informed consent statements. Research adhered to the tenets of the Declaration of Helsinki.
GENOTYPE DATA
The following 6 common single-nucleotide polymorphisms associated with AMD were evaluated in both cohorts: (1) complement factor H (CFH) Y402H (rs1061170)15-18;(2) CFH rs1410996, an independently associated single-nucleotide polymorphism variant within intron 14 of CFH2; (3) ARMS2/HTRA1 (rs10490924) on chromosome 1019-23; (4) complement component 2 (C2) E318D (rs9332739)2,24; (5) complement factor B (CFB) R32Q (rs641153)2,24; and (6) complement component 3 (C3) R102G (rs2230199).9,25 Genotyping was performed using primer mass extension and matrix-assisted laser desorption/ionization-time-of-flight mass spectrometry analysis (MassEXTEND method of Sequenom) at the Broad Institute Center for Genotyping and Analysis, Cambridge, Massachusetts.
STATISTICAL ANALYSIS
Analyses were performed using the Cox proportional hazards model to evaluate relationships between progression of AMD and the following variables: genotypes, age (<65, 65-74, and ≥75 years), sex, education (high school or less, more than high school), cigarette smoking (never, past, current), and BMI (<25, 2529.9, and ≥30). Hazard ratios (HRs) and 95% CIs were calculated for demographic, behavioral, ocular, and genetic factors. The method for calculation of the AMD progression risk score based on regression coefficients of all demographic, environmental, genetic, and ocular factors has been reported previously.4,6
Survival analysis was used to determine 5-year and 10-year cumulative incidence rates of AMD for individual subjects with various risk factor levels at baseline, adjusting for competing mortality risks according to age and sex as previously described.6 The total risk score was used to estimate HRs for specific subjects relative to a subject with no risk factors and thereby to estimate the AMD survival curve for individual subjects based on specific levels of risk factors using the baseline option of PROC PHREG in SAS statistical software version 9.2 (SAS Institute, Inc). The probability of AMD progression was estimated over different periods for individual subjects.
The AUCs were obtained for progression within 5 and 10 years. In addition, an age-adjusted C statistic based on the curve was calculated to assess the probability that the risk score based on the group of risk factors in that model from a random progressor was higher than the corresponding risk score from a random nonprogressor within the same 10-year age group.26 Confidence limits were obtained and C statistics were compared between competing models.27 To assess discrimination, the model obtained from the derivation sample was applied to the validation sample and the AUC was determined for both samples. To assess the calibration of the model, the subjects in the validation sample were subdivided into age-specific deciles of predicted 5-year AMD risk as obtained from the model including genes, environment, and baseline AMD status based on the derivation sample. The observed number of subjects who progressed over 5 years was compared with the expected number of subjects who progressed to advanced AMD over 5 years, estimated from Kaplan-Meier curves obtained for individual subjects and adjusted for competing mortality risks (as obtained from 2006 life tables). This comparison was performed for age-specific risk deciles as determined from the person-specific risk scores. A Hosmer-Lemeshow test statistic was then computed to compare the observed and predicted counts by risk decile summed over all age groups. In addition, 5-year cumulative incidence estimates were obtained for individual subjects based on specific demographic, environmental, genetic, and ocular risk factors and were adjusted for competing mortality risks. These 5-year cumulative incidence estimates were then stratified by age-specific quartiles and were calculated separately for subjects with and without advanced AMD in one eye at baseline.
RESULTS
The validation sample comprised 980 individuals, of whom 294 progressed to either GA or NV in at least one eye and 686 did not progress during the course of the study. There were 144 progressors to GA (stage 4) and 150 progressors to NV (stage 5) in at least one eye. Corresponding numbers for the derivation sample were 809 progressors among a total of 2914 subjects, with 355 progressors to GA and 454 progressors to NV. The mean (SD) ages of progressors and nonprogressors at baseline were 74.1 (7.5) and 73.2 (7.8) years, respectively, in the validation sample, compared with 70.2 (5.2) and 68.1 (4.7) years for the corresponding numbers in the derivation sample.
Univariate associations between baseline demographic, environmental, and genetic factors and progression to advanced AMD in the derivation and validation samples are shown in Table 1. In both samples, progressors were older, had less education (statistically significant in the derivation sample but not in the validation sample), and were more likely to be current smokers. Higher BMI was associated with progression in the derivation sample. Women were more likely to progress in the validation sample. Regarding the genetic factors, both cohorts had a higher percentage of progressors with the CC (risk) genotype for CFH rs1061170 and the CC (risk) genotype for CFH rs1410996. There were more progressors with the TT (risk) genotype for ARMS2/HTRA1 rs10490924 and the GG (risk) genotype for C3 rs2230199. For the derivation sample, progressors had a lower frequency for the C allele (protective) of C2 rs9332739 and the T allele (protective) of CFB rs641153. For the validation cohort, results were in the same direction for C2 but CFB was not related. In both cohorts, ocular findings indicate a higher percentage of progressors in the fellow eye for subjects with advanced AMD in one eye at baseline compared with the percentage among those not having advanced AMD in either eye. Furthermore, progression was associated with a higher baseline grade (ie, 3 vs 2) in the nonadvanced eye regardless of whether the fellow eye had advanced AMD.
Table 1.
Univariate Associations Between Baseline Demographic, Environmental, Macular, and Genetic Variables and Incident Advanced Age-Related Macular Degeneration
| Variable | Derivation Sample |
Validation Sample |
||||
|---|---|---|---|---|---|---|
| Progressors, No. (%) (n = 809) |
Nonprogressors, No. (%) (n = 2105) |
P Valuea |
Progressors, No. (%) (n = 294) |
Nonprogressors, No. (%) (n = 686) |
P Valuea |
|
| Age, y | <.001 | .05 | ||||
| <65 | 110 (13) | 455 (21) | 27 (9) | 93 (14) | ||
| 65-74 | 496 (61) | 1403 (67) | 114 (39) | 269 (39) | ||
| ≥75 | 203 (25) | 247 (12) | 153 (52) | 323 (47) | ||
| Sex | .87 | .006 | ||||
| Female | 455 (56) | 1193 (57) | 184 (63) | 362 (53) | ||
| Male | 354 (44) | 912 (43) | 110 (37) | 323 (47) | ||
| Education | <.001 | .35 | ||||
| ≤High school | 315 (39) | 652 (31) | 120 (41) | 256 (37) | ||
| >High school | 494 (61) | 1453 (69) | 174 (59) | 429 (63) | ||
| Smoking | <.001 | .04 | ||||
| Never | 317 (39) | 1060 (50) | 117 (40) | 301 (44) | ||
| Past | 422 (52) | 939 (45) | 146 (50) | 146 (50) | ||
| Current | 70 (9) | 106 (5) | 31 (11) | 39 (6) | ||
| BMI | .003 | .91 | ||||
| <25 | 243 (30) | 714 (34) | 99 (34) | 239 (35) | ||
| 25-29 | 331 (41) | 894 (42) | 129 (44) | 288 (42) | ||
| ≥30 | 235 (29) | 497 (24) | 66 (22) | 158 (23) | ||
| Genotype | ||||||
| CFH rs1061170 (Y402H) | <.001 | <.001 | ||||
| TT | 129 (16) | 762 (36) | 46 (16) | 191 (28) | ||
| CT | 357 (44) | 974 (46) | 138 (47) | 332 (48) | ||
| CC | 323 (40) | 369 (18) | 110 (37) | 162 (24) | ||
| CFH rs1410996 | <.001 | <.001 | ||||
| TT | 30 (4) | 348 (17) | 13 (4) | 78 (11) | ||
| CT | 246 (30) | 966 (46) | 85 (29) | 290 (42) | ||
| CC | 533 (66) | 791 (38) | 196 (67) | 317 (46) | ||
| ARMS2/HTRA1 rs10490924 (A69S) | <.001 | <.001 | ||||
| GG | 251 (31) | 1228 (58) | 98 (33) | 351 (51) | ||
| GT | 392 (48) | 745 (35) | 133 (45) | 279 (41) | ||
| TT | 166 (21) | 132 (6) | 63 (21) | 55 (8) | ||
| C2 rs9332739 (E318D) | <.001 | .11 | ||||
| GG | 782 (97) | 1937 (92) | 277 (94) | 625 (91) | ||
| CG/CC | 27 (3) | 168 (8) | 17 (6) | 60 (9) | ||
| CFB rs641153 (R32Q) | <.001 | .81 | ||||
| CC | 755 (93) | 1760 (84) | 257 (87) | 595 (87) | ||
| CT/TT | 54 (7) | 345 (16) | 37 (13) | 90 (13) | ||
| C3 rs2230199 (R102G) | <.001 | .001 | ||||
| CC | 384 (47) | 1289 (61) | 141 (48) | 399 (58) | ||
| CG | 353 (44) | 729 (35) | 124 (42) | 244 (36) | ||
| GG | 72 (9) | 87 (4) | 29 (10) | 42 (6) | ||
| Advanced AMD in one eye at baselineb | <.001 | <.001 | ||||
| Neither eye | 543 (67) | 1955 (93) | 181 (62) | 584 (85) | ||
| Grade 4 | 52 (6) | 8 (0) | 29 (10) | 11 (2) | ||
| Grade 5 | 214 (26) | 142 (7) | 84 (29) | 90 (13) | ||
| Grade in nonadvanced eye at baseline (advanced AMD in fellow eye)b | <.001 | .06 | ||||
| 1 | 4 (2) | 39 (26) | 5 (4) | 10 (9) | ||
| 2 | 31 (12) | 56 (37) | 28 (25) | 30 (30) | ||
| 3 | 231 (87) | 55 (37) | 80 (71) | 61 (60) | ||
| Grade in both eyes at baseline (advanced AMD in neither eye)b | <.001 | <.001 | ||||
| 1/1 or 1/2 | 32 (6) | 1160 (59) | 2 (1) | 290 (50) | ||
| 2/2 | 25 (5) | 243 (12) | 41 (23) | 129 (22) | ||
| 1/3 or 2/3 | 89 (16) | 335 (17) | 15 (8) | 48 (8) | ||
| 3/3 | 397 (73) | 217 (11) | 123 (68) | 117 (20) | ||
Abbreviations: AMD, age-related macular degeneration; BMI, body mass index (calculated as weight in kilograms divided by height in meters squared).
Based on Pearson χ2 for variables with 2 categories and χ2 test for trend for variables with more than 2 categories.
Grades are based on Clinical Age-Related Maculopathy Staging System score14:1 indicates no AMD; 2, early AMD; and 3, intermediate AMD.
Incidence rates of AMD according to baseline AMD grade compared with individuals with intermediate AMD (stage 3) in 1 or both eyes are displayed in Table 2. This group was selected as the reference group because it had the largest number of progressors. Incidence rates were comparable in the 2 samples and indicate dramatically higher rates in eyes with stage 3 vs 2 in both samples. Furthermore, the risk of incident advanced AMD is higher if the fellow eye had advanced AMD, particularly if the fellow eye had stage 4 AMD (GA).
Table 2.
Incidence Rate of Advanced Age-Related Macular Degeneration in the Derivation and Validation Samples Over 12 Years According to Baseline Age-Related Macular Degeneration Grade
| AMD Grades in Both Eyesa |
Derivation Sample |
Validation Sample |
||||||
|---|---|---|---|---|---|---|---|---|
| Progressors, No. |
Person- years |
Incidence Rate/ 1000 Person-years |
IRR (95% CI) | Progressors, No. |
Person- years |
Incidence Rate/ 1000 Person-years |
IRR (95% CI) | |
| 1/1, 1/2, or 2/2 | 57 | 14 881.0 | 3.8 | 0.06 (0.05-0.08) | 43 | 2476.4 | 17.4 | 0.21 (0.15-0.29) |
| 1/3, 2/3, or 3/3 | 486 | 8140.0 | 59.7 | 1 [Reference] | 138 | 1633.4 | 84.5 | 1 [Reference] |
| 1/4, 2/4, or 3/4 | 52 | 322.0 | 161.5 | 2.70 (2.03-3.60) | 29 | 179.4 | 161.7 | 1.91 (1.28-2.86) |
| 1/5, 2/5, or 3/5 | 214 | 2363.5 | 90.5 | 1.52 (1.29-1.78) | 84 | 849.3 | 98.9 | 1.17 (0.89-1.54) |
Abbreviations: AMD, age-related macular degeneration; IRR, incidence rate ratio.
Grades are based on Clinical Age-Related Maculopathy Staging System score14:1 indicates no AMD; 2, early AMD; 3, intermediate AMD; 4, geographic atrophy; and 5, neovascular disease.
Multivariate associations between risk factors and progression to advanced AMD for both samples are presented in Table 3. There were significant effects of age, smoking, BMI, ARMS2/HTRA1, CFH rs1410996, C2, CFB, and C3 on risk of progression in the derivation sample. Older age and smoking were significantly related to progression in the validation sample (individuals aged ≥75 vs <65 years: HR = 2.6; 95% CI, 1.7-4.1; P < .001; and current smoking vs never: HR = 2.2; 95% CI, 1.4-3.3; P < .001), while the effect of BMI was not seen. The directions of the effects for 5 of the 6 single-nucleotide polymorphisms were similar in the validation sample compared with the derivation sample, while CFB was not related in the validation sample. Consistent with the results shown in Table 2, having one eye with advanced AMD at baseline increased risk of progression in both samples. To evaluate this more specifically, subgroup analyses were performed separately for progression to GA and NV (eTable 1 and eTable 2, http://www.jamaophth.com). The ARMS2/HTRA1 TT genotype was more strongly related to NV than GA in the validation sample (HR = 3.2 for NV and HR = 1.9 for GA). Having GA in one eye was a strong risk factor for progression to GA in the fellow eye (HR = 3.8 in the derivation sample; HR =2.6 in the validation sample) but not to NV. Having NV at baseline in one eye was a risk factor for progression to NV in the fellow eye (HR = 1.6 in the derivation sample; HR = 1.3 in the validation sample) but not to GA.
Table 3.
Multivariate Association Between Demographic, Environmental, Genetic, and Advanced Age-Related Macular Degeneration Grade and Incident Advanced Age-Related Macular Degeneration
| Variable | Derivation Sample |
Validation Sample |
||
|---|---|---|---|---|
| HR (95% CI)a | P Value | HR (95% CI)a | P Value | |
| Progressors/nonprogressors, No. | 809/2105 | 294/685 | ||
| Age, y | ||||
| <65 | 1 [Reference] | 1 [Reference] | ||
| 65-74 | 1.4 (1.1-1.7) | .002 | 1.5 (1.0-2.3) | .08 |
| ≥75 | 2.0 (1.6-2.5) | <.001 | 2.6 (1.7-4.1) | <.001 |
| Sex | ||||
| Female | 1 [Reference] | 1 [Reference] | ||
| Male | 1.0 (0.8-1.1) | .95 | 1.0 (0.8-1.2) | .81 |
| Education | ||||
| ≤High school | 1 [Reference] | 1 [Reference] | ||
| >High school | 0.9 (0.8-1.0) | .08 | 0.8 (0.6-1.0) | .11 |
| Smoking | ||||
| Never | 1 [Reference] | 1 [Reference] | ||
| Past | 1.2 (1.1-1.4) | .007 | 1.0 (0.8-1.4) | .71 |
| Current | 1.6 (1.3-2.1) | <.001 | 2.2 (1.4-3.3) | <.001 |
| BMI | ||||
| <25 | 1 [Reference] | 1 [Reference] | ||
| 25-29 | 1.1 (0.9-1.3) | .18 | 1.2 (0.9-1.5) | .32 |
| ≥30 | 1.3 (1.1-1.6) | .006 | 1.1 (0.8-1.5) | .71 |
| Genotype | ||||
| CFH rs1061170 (Y402H) | ||||
| TT | 1 [Reference] | 1 [Reference] | ||
| CT | 1.1 (0.9-1.1) | .50 | 1.2 (0.8-1.7) | .49 |
| CC | 1.3 (1.0-1.6) | .08 | 1.2 (0.7-1.9) | .50 |
| ARMS2/HTRA1 rs10490924 (A69S) | ||||
| GG | 1 [Reference] | 1 [Reference] | ||
| GT | 1.6 (1.3-1.8) | <.001 | 1.4 (1.1-1.8) | .01 |
| TT | 2.3 (1.8-2.8) | <.001 | 2.5 (1.8-3.5) | <.001 |
| CFH rs1410996 | ||||
| TT | 1 [Reference] | 1 [Reference] | ||
| CT | 2.0 (1.3-2.9) | <.001 | 1.1 (0.5-2.0) | .92 |
| CC | 2.7 (1.8-4.2) | <.001 | 1.7 (0.8-3.4) | .14 |
| C2 rs9332739 (E318D) | ||||
| GG | 1 [Reference] | 1 [Reference] | ||
| CG/CC | 0.5 (0.4-0.8) | .002 | 0.7 (0.4-1.1) | .11 |
| CFB rs641153 (R32Q) | ||||
| CC | 1 [Reference] | 1 [Reference] | ||
| CT/TT | 0.6 (0.4-0.8) | <.001 | 1.0 (0.7-1.4) | .97 |
| C3 rs2230199 (R102G) | ||||
| CC | 1 [Reference] | 1 [Reference] | ||
| CG | 1.2 (1.1-1.4) | .004 | 1.3 (1.0-1.6) | .047 |
| GG | 1.6 (1.2-2.0) | .001 | 1.6 (1.0-2.3) | .04 |
| Grade in each eye for individuals without advanced AMD at baselineb | ||||
| 1/1, 1/2, or 2/2 | 0.09 (0.07-0.1) | <.001 | 0.3 (0.1-0.4) | <.001 |
| 1/3, 2/3, or 3/3 | 1 [Reference] | 1 [Reference] | ||
| 1/4, 2/4, or 3/4 | 2.2 (1.6-2.9) | <.001 | 1.4 (0.9-2.1) | .15 |
| 1/5, 2/5, or 3/5 | 1.2 (1.0-1.4) | .06 | 1.0 (0.8-1.3) | .94 |
Abbreviations: AMD, age-related macular degeneration; BMI, body mass index (calculated as weight in kilograms divided by height in meters squared); HR, hazard ratio.
Multivariate models control for all variables in the table simultaneously.
Grades are based on Clinical Age-Related Maculopathy Staging System score14:1 indicates no AMD; 2, early AMD; 3, intermediate AMD; 4, geographic atrophy; and 5, neovascular disease.
The C statistics for progression to advanced AMD at 5 and 10 years after baseline for the derivation and validation samples are shown in Table 4. For the derivation and validation models in the overall sample, the C statistics (SE) were 0.858 (0.009) and 0.750 (0.022) for progression to advanced AMD at 5 years and 0.884 (0.007) and 0.809 (0.025) for progression at 10 years, indicating very good discriminative ability. For progression to GA and NV separately, the C statistics (SE) were 0.860 (0.013) and 0.831 (0.013), respectively, at 5 years for the derivation sample and 0.737 (0.029) and 0.704 (0.029) at 5 years for the validation sample.
Table 4.
Area Under the Curve Statistics for Progression to Advanced Age-Related Macular Degeneration, Geographic Atrophy, and Neovascular Disease at 5 and 10 Years After Baseline
| Follow-up | AUC (SE) |
|
|---|---|---|
| Derivation Samplea |
Validation Sampleb |
|
| 5 y | ||
| All advanced AMD | 0.858 (0.009) | 0.750 (0.022) |
| Geographic atrophy | 0.860 (0.013) | 0.737 (0.029) |
| Neovascular disease | 0.831 (0.013) | 0.704 (0.029) |
| 10y | ||
| All advanced AMD | 0.884 (0.007) | 0.809 (0.025) |
| Geographic atrophy | 0.882 (0.010) | 0.719 (0.032) |
| Neovascular disease | 0.881 (0.009) | 0.753 (0.028) |
Abbreviations: AMD, age-related macular degeneration; AUC, area under the curve.
Number of progressors for derivation sample: all advanced AMD, n = 809; geographic atrophy, n = 355; and neovascular disease, n = 454.
Number of progressors for validation sample: all advanced AMD, n = 294; geographic atrophy, n = 144; and neovascular disease, n = 150.
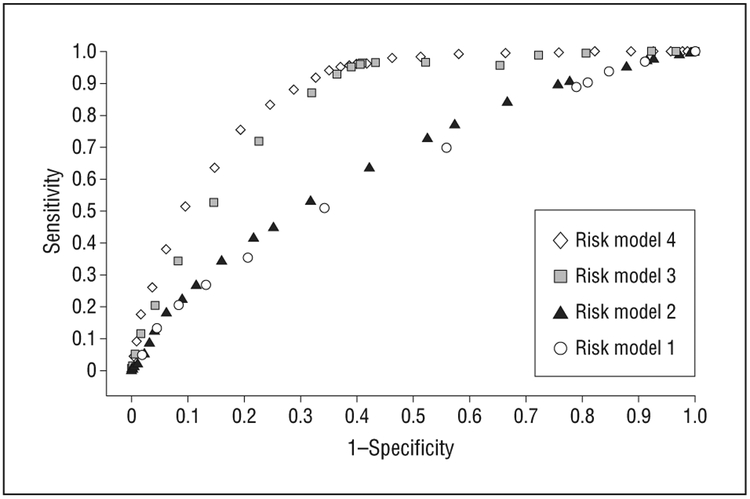
Receiver operating characteristic curves for 4 models are displayed in Figure 1, including the following variables: age, sex, education (model 1), addition of smoking and BMI (model 2), addition of macular phenotypes (model 3), and all of these variables plus the genetic variants (model 4, used in Table 3). There was a substantial increase in AUC between risk model 2 (AUC [SE], 0.61 [0.014]) and risk model 3 (AUC [SE], 0.82 [0.010]) (P < .001) with the addition of baseline AMD status and an additional increase in AUC between model 3 and model 4 (AUC [SE], 0.86 [0.009]) (P < .001) with the addition of genetic variables.
Figure 1.
Receiver operating characteristic curves for 5-year progression to advanced age-related macular degeneration according to risk model, including model 1 (age, sex, education), model 2 (model 1 plus smoking and body mass index), model 3 (model 2 plus baseline age-related macular degeneration grade), and model 4 (all of these variables plus the genetic variants), for the derivation sample.
We calculated positive and negative predictive values (Table 5) using 81% for sensitivity and 77% for specificity, which corresponded to a risk score cutoff of 1.5 in the full model. For the derivation sample, the positive predictive value was 42% (929 individuals with a risk score ≥1.5, of whom 391 progressed) and the negative predictive value was 95% (1976 individuals with a risk score <1.5, of whom 1885 did not progress). The corresponding values for the validation sample were a positive predictive value of 52% (255 individuals, of whom 132 progressed) and a negative predictive value of 84% (346 individuals, of whom 292 did not progress). There was also a striking increase in risk with increasing category of the risk score in both samples, with up to a 70% to 75% progression rate at 5 years for the highest score.
Table 5.
Relationship Between Risk Score and Progression to Advanced Age-Related Macular Degeneration at 5 Years in the Derivation and Validation Samples
| Risk Score or Predictive Value | Derivation Sample |
Validation Sample |
|---|---|---|
| Risk score, No. of subjects (%)a | ||
| All categories | ||
| <0.5 | 1529 (2) | 243 (12) |
| 0.5-0.9 | 141 (12) | 34 (24) |
| 1.0-1.4 | 306 (17) | 69 (26) |
| 1.5-1.9 | 448 (32) | 94 (43) |
| 2.0-2.4 | 309 (46) | 103 (53) |
| 2.5-2.9 | 142 (60) | 46 (61) |
| 3.0-4.0 | 30 (70) | 12 (75) |
| 2 Categories | ||
| <1.5 | 1976 (5) | 346 (16) |
| ≥1.5 | 929 (42) | 255 (52) |
| Predictive value, % | ||
| Positiveb | 42 | 52 |
| Negativec | 95 | 84 |
Numbers indicate subjects in each risk score category who either progressed within 5 years or had at least 5 years’ follow-up; percentages indicate the percentages of progressors in each risk category. Risk score is based on the full model including age, sex, baseline age-related macular degeneration grade in both eyes, education, body mass index, and 6 single-nucleotide polymorphisms in 5 genes.
Indicates the probability of progression within 5 years given that the risk score is 1.5 or higher.
Indicates the probability of no progression within 5 years given that the risk score is less than 1.5.
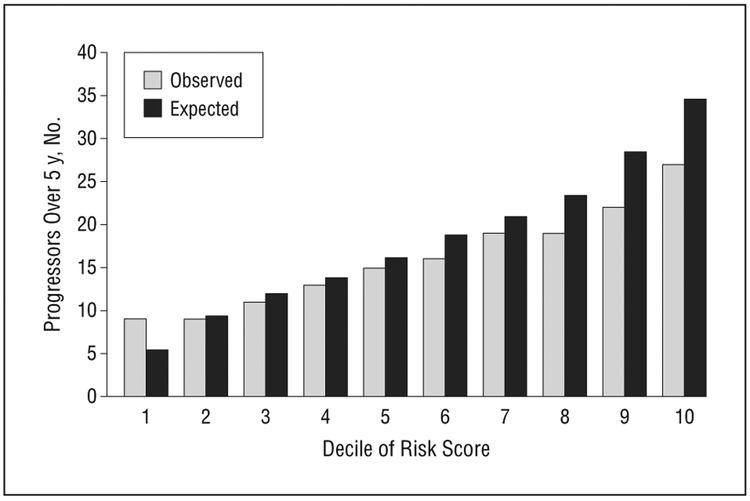
Calibration of the risk prediction model was assessed in the validation sample based on the model with all risk factors fit to the derivation sample, for the baseline intermediate and unilateral advanced groups (Figure 2). The observed and expected counts within age-specific risk deciles in the test sample were not significantly different (Hosmer-Lemeshow χ2 = 5.1, P = .54 for no advanced AMD in either eye and χ2 = 4.8, P = .58 for advanced AMD in one eye at baseline), indicating adequate calibration of the model in an independent sample of subjects in a study of progression to advanced AMD. Results based on discrimination and calibration in the validation sample indicated that the risk model would likely perform well in other white populations of individuals at risk for progression to advanced AMD.
Figure 2.
Calibration of the risk model for progression to advanced age-related macular degeneration comparing the observed and expected number of progressors in the validation data set according to deciles of the risk score from the derivation sample.
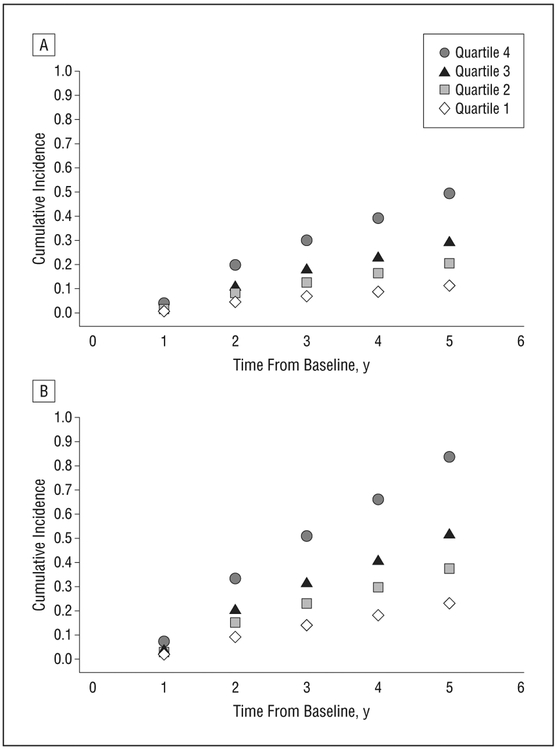
The cumulative incidence of progression to advanced AMD by age-specific quartile of risk score adjusted for competing mortality risks was calculated separately for individuals with intermediate AMD in 1 or both eyes (Figure 3A) and with unilateral advanced AMD in one eye and intermediate AMD in the fellow eye (Figure 3B) at baseline. Among individuals with the same macular phenotypes at baseline, the cumulative incidence of advanced AMD varied substantially according to the risk score. For the unilateral or bilateral intermediate AMD group, the lowest risk quartile group had a 10% rate of progression at 5 years, whereas the highest risk quartile group had a 40% rate of progression. Among individuals with unilateral advanced AMD at baseline, the rate of progression in the fellow eye was about 22% for the lowest risk quartile and increased to greater than 80% for the highest risk quartile, indicating substantial separation of risk of progression when applying the risk prediction score among subjects with the same macular phenotype at baseline.
Figure 3.
Cumulative incidence of progression to advanced age-related macular degeneration by age-specific quartile of risk score for individuals with intermediate age-related macular degeneration in one or both eyes at baseline (A) and with unilateral advanced age-related macular degeneration and intermediate age-related macular degeneration in the fellow eye at baseline (B) in the derivation sample.
COMMENT
We developed and validated our risk prediction model for AMD progression in 2 independent cohorts. The performance of the risk score was evaluated by applying prediction rules from the derivation sample to the validation sample for all progressors and for progression to GA and NV separately. In the validation data set, the AUCs showed very good discrimination, indicating that risk scores for cases in general were higher than for controls.28 Calibration was also good as assessed by agreement between the predicted risk and observed proportion developing advanced AMD, indicating that the predicted probability of progression as derived from the derivation data set was in reasonable agreement with the observed probability of progression in the validation data set, within risk deciles. We defined a positive score as a risk score of at least 1.5, corresponding to a sensitivity and specificity of approximately 80%. The positive and negative predictive values for the validation sample were approximately 52% and 84%, respectively, indicating that about 50% of subjects with a positive score developed advanced AMD over 5 years compared with only 16% of those with a negative score.
In the evaluation of progression to advanced subtypes separately, the ARMS2/HTRA1 TT genotype had a stronger association with progression to NV than GA in the validation sample. This observation supports our previous genome-wide association studies showing that the ARMS2/HTRA1 risk genotype confers greater risk of NV.29,30 We also found that the type of advanced AMD in one eye at baseline was a risk factor for progression to that same subtype in the other eye in both cohorts. Results from our previous sibling correlation analyses also suggested that the subtype of advanced AMD in one eye predicted development of the same subtype in the fellow eye.30
We have derived several prediction models for AMD.1-7 Notably, as shown in this study and previously, the models can distinguish rate of progression over time among individuals with the same macular phenotype at baseline, which can guide selection of subjects for clinical trials. In another report, we also assessed transitions to different stages of macular disease in a model that included baseline drusen status, the genes we assessed in this article, and genes in the high-density lipoprotein pathway (LIPC, CETP, ABCA1) as well as other pathways including COL8A1 in the collagen–extracellular matrix pathway.7,31-34 That study resulted in 5- and 10-year AUCs of 0.883 and 0.895, respectively.7 We did not include these recently reported genes in this article because they were not included in the derivation sample.
Two other recent articles35,36 described risk models for progression to advanced AMD using methods reported previously.4 One report included 2 of the 6 genes used in our models and mentioned a calibration statistic in a small cohort of patients with intermediate AMD previously treated with laser and missing 1 of the model components. The AUCs were 0.73 to 0.87.35 Ying et al36 evaluated data on 63 progressors to GA using a model without genes but including a night vision score and hypertension, with AUCs of 0.68 to 0.76.
Advantages of the current study include the evaluation of the predictive power of various combinations of demographic, environmental, genetic, and baseline macular phenotypes based on large, well-characterized populations of white patients in 2 independent cohorts. Additional strengths include the standardized collection of risk factor information, direct measurements of height and weight, and classification of maculopathy by standardized ophthalmic examinations and grading of fundus photographs.
To our knowledge, this is the first report of a risk prediction model for AMD progression incorporating non-genetic and genetic risk factors that was externally validated in a large, independent, prospective cohort comprising patients with and without AMD and with all stages of maculopathy. There is increasing interest in personalized medicine and individual risk prediction for AMD and its progression. Our models can be used to compute genetic load as well as comprehensive risk scores that incorporate demographic, macular, and environmental factors.1-7 These risk algorithms will be useful for research, clinical trials, and personalized medicine.
Supplementary Material
Acknowledgments
Funding/Support: This work was supported by grant R01-EY11309 from the National Institutes of Health, the Massachusetts Lions Eye Research Fund Inc, unrestricted grants from Research to Prevent Blindness Inc, the Macula Vision Research Foundation, and the Age-Related Macular Degeneration Research Fund, Ophthalmic Epidemiology and Genetics Service, Tufts Medical Center, Tufts University School of Medicine, Boston, Massachusetts.
Footnotes
Conflict of Interest Disclosures: Tufts Medical Center has filed patent applications for materials related to this work.
Previous Presentation: This paper was presented at the 2012 Annual Meeting of the Association for Research in Vision and Ophthalmology; May 8, 2012; Fort Lauderdale, Florida.
Contributor Information
Johanna M. Seddon, Department of Ophthalmology, New England Eye Center, Tufts Medical Center, Boston, Massachusetts..
Robyn Reynolds, Ophthalmic Epidemiology and Genetics Service, New England Eye Center, Tufts Medical Center, Boston, Massachusetts..
Yi Yu, Ophthalmic Epidemiology and Genetics Service, New England Eye Center, Tufts Medical Center, Boston, Massachusetts..
Bernard Rosner, Tufts University School of Medicine, and Channing Laboratory, Harvard Medical School, Boston, Massachusetts..
REFERENCES
- 1.Seddon JM, George S, Rosner B, Klein ML. CFH gene variant, Y402H,and smoking, body mass index, environmental associations with advanced age-related macular degeneration. Hum Hered. 2006;61 (3):157–165. [DOI] [PubMed] [Google Scholar]
- 2.Maller J, George S, Purcell S, et al. Common variation in three genes, including a noncoding variant in CFH, strongly influences risk of age-related macular degeneration. Nat Genet. 2006;38(9):1055–1059. [DOI] [PubMed] [Google Scholar]
- 3.Seddon JM, Francis PJ, George S, Schultz DW, Rosner B, Klein ML. Association of CFH Y402H and LOC387715 A69S with progression of age-related macular degeneration. JAMA. 2007;297(16):1793–1800. [DOI] [PubMed] [Google Scholar]
- 4.Seddon JM, Reynolds R, Maller J, Fagerness JA, Daly MJ, Rosner B. Prediction model for prevalence and incidence of advanced age-related macular degeneration based on genetic, demographic, and environmental variables. Invest Ophthalmol Vis Sci. 2009;50(5):2044–2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reynolds R, Hartnett ME, Atkinson JP, Giclas PC, Rosner B, Seddon JM. Plasma complement components and activation fragments: associations with age-related macular degeneration genotypes and phenotypes. Invest Ophthalmol Vis Sci. 2009;50(12):5818–5827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Seddon JM, Reynolds R, Yu Y, Daly MJ, Rosner B. Risk models for progression to advanced age-related macular degeneration using demographic, environmental, genetic, and ocular factors. Ophthalmology. 2011;118(11):2203–2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yu Y, Reynolds R, Rosner B, Daly MJ, Seddon JM. Prospective assessment of genetic effects on progression to different stages of age-related macular degeneration using multistate Markov models. Invest Ophthalmol Vis Sci. 2012;53 (3):1548–1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Age-Related Eye Disease Study Research Group. A randomized, placebo-controlled, clinical trial of high-dose supplementation with vitamins C and E, beta carotene, and zinc for age-related macular degeneration and vision loss: AREDS report No. 8. Arch Ophthalmol. 2001;119(10):1417–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maller JB, Fagerness JA, Reynolds RC, Neale BM, Daly MJ, Seddon JM. Variation in complement factor 3 is associated with risk of age-related macular degeneration. Nat Genet. 2007;39(10):1200–1201. [DOI] [PubMed] [Google Scholar]
- 10.Fagerness JA, Maller JB, Neale BM, Reynolds RC, Daly MJ, Seddon JM. Variation near complement factor I is associated with risk of advanced AMD. Eur J Hum Genet. 2009;17(1):100–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Seddon JM, Cote J, Davis N, Rosner B. Progression of age-related macular degeneration: association with body mass index, waist circumference, and waisthip ratio. Arch Ophthalmol. 2003;121(6):785–792. [DOI] [PubMed] [Google Scholar]
- 12.Seddon JM, Santangelo SL, Book K, Chong S, Cote J. A genomewide scan for age-related macular degeneration provides evidence for linkage to several chromosomal regions. Am J Hum Genet. 2003;73(4):780–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Seddon JM, Cote J, Page WF, Aggen SH, Neale MC. The US twin study of age-related macular degeneration: relative roles of genetic and environmental influences. Arch Ophthalmol. 2005;123(3):321–327. [DOI] [PubMed] [Google Scholar]
- 14.Seddon JM, Sharma S, Adelman RA. Evaluation of the clinical age-related maculopathy staging system. Ophthalmology. 2006;113(2):260–266. [DOI] [PubMed] [Google Scholar]
- 15.Klein RJ, Zeiss C, Chew EY, et al. Complement factor H polymorphism in age-related macular degeneration. Science. 2005;308(5720):385–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Edwards AO, Ritter R III, Abel KJ, Manning A, Panhuysen C, Farrer LA. Complement factor H polymorphism and age-related macular degeneration. Science. 2005; 308(5720):421–424. [DOI] [PubMed] [Google Scholar]
- 17.Haines JL, Hauser MA, Schmidt S, et al. Complement factor H variant increases the risk of age-related macular degeneration. Science. 2005;308(5720):419–421. [DOI] [PubMed] [Google Scholar]
- 18.Hageman GS, Anderson DH, Johnson LV, et al. A common haplotype in the complement regulatory gene factor H (HF1/CFH) predisposes individuals to age-related macular degeneration. Proc Natl Acad Sci U S A. 2005;102(20):7227–7232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jakobsdottir J, Conley YP, Weeks DE, Mah TS, Ferrell RE, Gorin MB. Susceptibility genes for age-related maculopathy on chromosome 10q26. Am J Hum Genet. 2005;77(3):389–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rivera A, Fisher SA, Fritsche LG, et al. Hypothetical LOC387715 is a second major susceptibility gene for age-related macular degeneration, contributing independently of complement factor H to disease risk. Hum Mol Genet. 2005;14 (21):3227–3236. [DOI] [PubMed] [Google Scholar]
- 21.Dewan A, Liu M, Hartman S, et al. HTRA1 promoter polymorphism in wet age-related macular degeneration. Science. 2006;314(5801):989–992. [DOI] [PubMed] [Google Scholar]
- 22.Yang Z, Camp NJ, Sun H, et al. A variant of the HTRA1 gene increases susceptibility to age-related macular degeneration. Science. 2006;314(5801):992–993. [DOI] [PubMed] [Google Scholar]
- 23.Kanda A, Chen W, Othman M, et al. A variant of mitochondrial protein LOC387715/ARMS2, not HTRA1, is strongly associated with age-related macular degeneration. Proc Natl Acad Sci U S A. 2007;104(41):16227–16232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gold B, Merriam JE, Zernant J, et al. ; AMD Genetics Clinical Study Group. Variation in factor B (Bf) and complement component 2 (C2) genes is associated with age-related macular degeneration. Nat Genet. 2006;38(4):458–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yates JR, Sepp T, Matharu BK, et al. ; Genetic Factors in AMD Study Group. Complement C3 variant and the risk of age-related macular degeneration. N Engl J Med. 2007;357(6):553–561. [DOI] [PubMed] [Google Scholar]
- 26.Hanley JA, McNeil BJ. The meaning and use of the area under a receiver operating characteristic (ROC) curve. Radiology. 1982;143(1):29–36. [DOI] [PubMed] [Google Scholar]
- 27.Rosner B, Glynn RJ. Power and sample size estimation for the Wilcoxon rank sum test with application to comparisons of C statistics from alternative prediction models. Biometrics. 2009;65(1):188–197. [DOI] [PubMed] [Google Scholar]
- 28.Choi BC. Slopes of a receiver operating characteristic curve and likelihood ratios for a diagnostic test. Am J Epidemiol. 1998;148(11):1127–1132. [DOI] [PubMed] [Google Scholar]
- 29.Sobrin L, Reynolds R, Yu Y, et al. ARMS2/HTRA1 locus can confer differential susceptibility to the advanced subtypes of age-related macular degeneration. Am J Ophthalmol. 2011;151(2):345–352, e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sobrin L, Ripke S, Yu Y, et al. Heritability and genome-wide association study to assess genetic differences between advanced age-related macular degeneration subtypes. Ophthalmology. 2012;119(9):1874–1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Neale BM, Fagerness J, Reynolds R, et al. Genome-wide association study of advanced age-related macular degeneration identifies a role of the hepatic lipase gene (LIPC). Proc Natl Acad Sci U S A. 2010;107(16):7395–7400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reynolds R, Rosner B, Seddon JM. Serum lipid biomarkers and hepatic lipase gene associations with age-related macular degeneration. Ophthalmology. 2010; 117(10):1989–1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Seddon JM, Reynolds R, Rosner B. Associations of smoking, body mass index, dietary lutein, and the LIPC gene variant rs10468017 with advanced age-related macular degeneration. Mol Vis. 2010;16:2412–2424. [PMC free article] [PubMed] [Google Scholar]
- 34.Yu Y, Bhangale TR, Fagerness J, et al. Common variants near FRK/COL10A1 and VEGFA are associated with advanced age-related macular degeneration. Hum Mol Genet. 2011;20(18):3699–3709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Klein ML, Francis PJ, Ferris FL III, Hamon SC, Clemons TE. Risk assessment model for development of advanced age-related macular degeneration. Arch Ophthalmol. 2011;129(12):1543–1550. [DOI] [PubMed] [Google Scholar]
- 36.Ying GS, Maguire MG; Complications of Age-Related Macular Degeneration Prevention Trial Research Group. Development of a risk score for geographic atrophy in Complications of Age-Related Macular Degeneration Prevention Trial. Ophthalmology. 2011;118(2):332–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.