Summary
In the presence of DNA damage, cells delay the entry into mitosis, presumably to allow time for repair. Methods to detect the delay of mitosis in a multicellular model organism, Drosophila melanogaster, are described here. These include the collection of embryos and larvae, irradiation with x-rays to damage DNA, and fixing and staining of tissues with an antibody to phosphorylated histone H3 to measure the mitotic index. These methods should be useful in identifying potential mutants that are unable to regulate mitosis following DNA damage.
Keywords: Drosophila, embryo, larvae, cell cycle, mitosis, DNA damage, checkpoint
1. Introduction
Cell proliferation is essential for growth and maintenance of all organisms. Equally important is the need to inhibit cell proliferation in response to extra- cellular and intracellular conditions such as the lack of nutrients or damaged and incompletely replicated DNA. Inability to regulate cell division in the presence of damaged DNA can compromise the genetic integrity of daughter cells. As such, mutational loss of checkpoint mechanisms that sense the presence of DNA defects and regulate the cell-division cycle as needed can increase genome instability and predisposition to cancer (1). This chapter describes methods to assay for regulation of mitosis in the presence of damaged DNA in Drosophila melanogaster. Ionizing radiation (x-rays) is used to induce DNA damage, but the methods described here could also be used in combination with other DNA damaging radiation such as γ -rays and ultraviolet (UV). A commercially available antibody to a phosphorylated serine on histone H3 (PH3 [2,3]), a mitosis-specific antigen, is used to identify mitotic cells. These methods can be used to test potential checkpoint mutants for their ability to regulate mitosis. Methods for detection of mitotic regulation in two different developmental stages, embryo and larvae, are described. Thus, maternal effect lethal or embryonic lethal mutants (i.e., when mutations prevent development beyond embryonic stages) can be assayed as embryos, while larval lethal mutants (i.e., when mutations permit development to larval stages) can be assayed at either stage. In summary, this chapter describes methods for collection of embryos and larvae, irradiation to induce DNA damage, and fixing and staining to detect changes in mitotic index.
2. Materials
- Containers for fly culture:
- Collection cages—large plexiglass containers sealed at one end with insect screen. These hold flies during egg collection.
- Egg collection baskets—plexiglass cylinders sealed at one end with nylon netting.
A soft paint brush.
Small (35×10 mm) plastic petri dish.
Large (100×15 mm) plastic petri dish lid.
Two pair fine forceps, e.g., no. 5 Watchmaker.
Dissecting microscope.
Nutator or other rocking device to mix samples during incubation.
Phosphate buffered saline (PBS): 140 mM NaCl, 2.6 mM KCl, 10 mM Na2HPO4,1.8 mM KH2PO4, pH 7.4.
Fix solution for larval tissues: 1X PBS, 5% formaldehyde, 0.3% Triton-X-100 (made fresh).
Fix solution for embryos: 1X PBS, 10% formaldehyde (made fresh).
PBTx: 1X PBS, 0.3% Triton-X-100.
Block solution; PBTx plus 10% Normal Goat Serum.
Rabbit anti-PH3 antibody (Upstate Biotech), diluted 1:1000 in block just prior to use.
Anti-rabbit secondary antibodies conjugated to FITC or rhodamine.
Flouromount-G (Southern Biotechnology Associates, Inc.).
Hoechst 33258 for staining DNA.
Microscope slides and coverslips.
Compound fluorescence microscope.
Heptane.
50% bleach (made fresh in water).
Methanol.
X-ray source.
3. Methods
Subheadings 3.1.–3.4. apply to embryos and Subheadings 3.5.–3.8. apply to larvae. Methods outline (1) embryo collection and irradiation; (2) fixation;(3) staining to visualize mitotic cells; (4) data collection and interpretation; (5) collection and aging of embryos to reach appropriate larval stages; (6) irradiation; (7) dissection to obtain imaginal discs; and (8) fixation, staining, and interpretation of data.
3.1. Embryo Collection and Irradiation
A working knowledge of Drosophila culture is assumed but may be found in (4,6). Flies and embryos are kept in a humidified incubator at 25°C throughout the procedure except for the brief interval needed for irradiation. Time intervals are adjusted for embryo development at 25°C and should be adhered to faithfully.
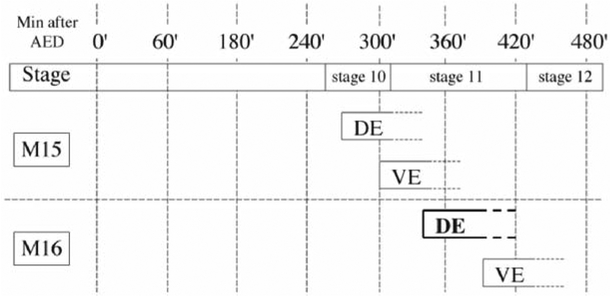
Collect embryos on a grape-agar plate for 60 min and discard (see Note 1). This precollection removes embryos that are abnormally older because adult females hold their eggs in the absence of fresh food. Next, use a fresh grape-agar plate to collect embryos for 10 min. These are the embryos you will use. Let embryos age for 325 min; this will allow cells of the dorsal epidermis to reach interphase of embryonic cell-division cycle 16 ((7); Fig. 1).
Cut the agar slab (with embryos) in half. Expose one half to 570 rads of x-rays, which is the LD50 for this stage in embryogenesis; keep the other half, which serves as the unirradiated control, in the incubator (see Note 2).
Return irradiated embryos to the incubator and let experimental and control samples incubate for 10–20 min. The duration of this step depends on how fast the following steps can be performed. The key is to place embryos in the fix at exactly 20 min after irradiation.
Fig. 1.
Timetable for mitosis in embryonic cell cycle 15 and 16 (M15 and M16, respectively). The approximate time of mitotic entry for the dorsal and ventral epidermis (de and ve, respectively) is diagrammed. The time after egg deposition (AED) is shown for 25°C.
3.2. Fixation
This step is carried out at room temperature (RT).
Squirt dH2O onto embryos and loosen them from agar using a soft paint brush. Pour water with embryos into the egg collection basket and rinse with dH2O to remove yeast. Blot with tissue paper to remove excess dH2O.
Dechorionate embryos (remove the chorion) by placing basket in 50% bleach for 2 min. Swirl embryos occasionally. Remove basket and rinse embryos extensively with dH2O to remove all bleach. Blot to remove excess dH2O.
Transfer basket to a petri dish containing heptane. Prewet 1 mL plastic pipet tip by pipeting heptane up and down a few times; this prevents embryo from sticking to the side of pipet tips. Trim the end of the pipet tip to enlarge the opening; this prevents tissue damage. Use a Pipetman® to transfer embryos to 20-mL glass vial containing 3 mL of fix solution. Add 3 mL heptane to glass vial and rock the vial for 20 min at RT.
Remove most of the lower phase (aqueous fix layer). Add 10 mL MeOH to glass vial and shake vigorously for 30 s to remove the vitelline membrane that encloses the embryo. Let embryos settle (see Note 3).
MeOH and heptane will separate into two phases; embryos trapped at the interface did not lose the vitelline membrane and are not useful for antibody staining. Collect embryos only from the bottom of glass vial and transfer to a new microfuge tube. Wash embryos 3 times with MeOH. These can be stored at –20°C in MeOH for several months or processed for antibody staining directly.
3.3. Antibody Staining
All steps are performed at RT. Starting with step 6, samples should be kept in the dark by wrapping in foil.
Using a Pipetman with a cut off tip, transfer embryos to a new 1.5-mL microfuge tube. Remove excess MeOH.
Rehydrate embryos with three rinses of PBTx. Rock embryos in the last rinse for 5 min.
Replace the last PBTx rinse with block solution and incubate for at least 1 h while rocking.
Replace block solution with the primary antibody (1:1000 rabbit anti-PH3 in block) and rock for 2 h.
Wash three times for 15 min with block solution.
Add secondary antibody solution, diluted 1:500 in block solution, and rock for 2 h (see Note 4).
Wash three times for 15 min with PBTx.
Replace the last wash with 1 mL Hoechst solution (10 μg/mL in PBTx) and rock for 4 min.
Wash three times for 15 min with PBTx.
Mount embryos in Fluoromount-G and analyze.
3.4. Data Collection and Interpretation
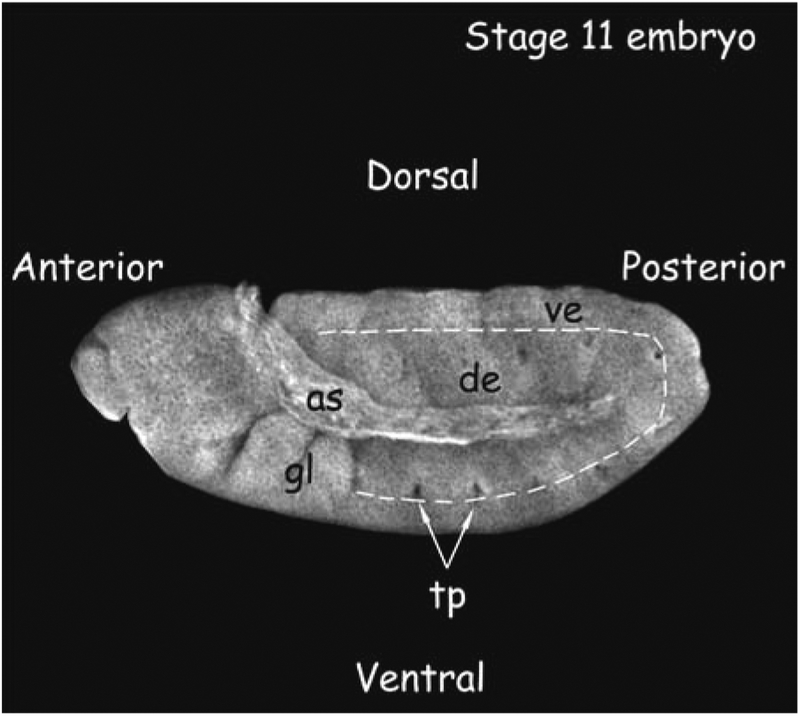
Use a fluorescent microscope with a ×10 or 20 objective to identify embryos of the correct orientation and correct age. It is easiest to use Hoechst staining to identify morphological markers (Fig. 2).
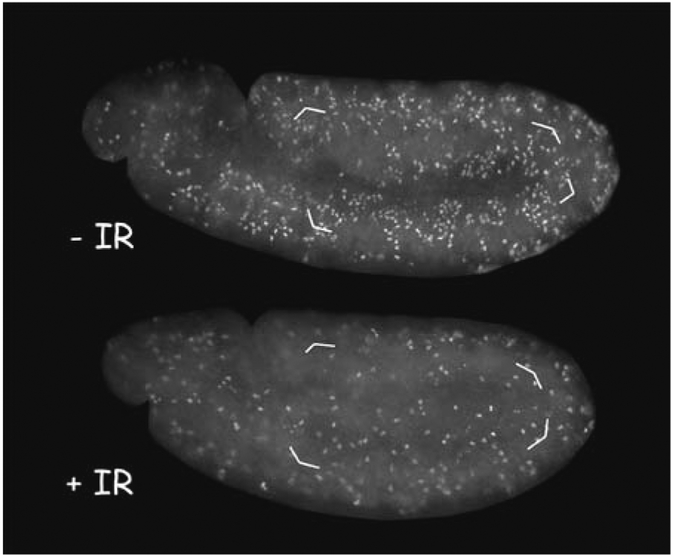
Using a higher power objective (´×40–100), quantify the total number of PH3- positive cells within the entire dorsal epidermis (de) of the embryo. It is easiest to begin counting mitotic cells just below the last gnathal lobe (gl) and continue around the amnioserosa to include the entire dorsal epidermis (Figs. 2 and 3).
Count at least 10 embryos each from unirradiated and irradiated samples. Wild- type embryos typically show about 85% reduction in mitotic index at 20 min after irradiation (reduction from111 ±14 to 17 ±11 mitotic cells in Fig. 3).
Fig. 2.
Stage 11 embryo. This embryo was stained with Texas Red–conjugated Wheat Germ Agglutinin (Molecular Probes) to visualize major morphological markers. The dorsal and ventral epidermis (de and ve, respectively) are marked. The gnathal lobes (gl) and trachael pits (tp) are characteristic of this stage embryo. as = amnioserosa. Embryo staging is as in (13).
Fig. 3.
. Stage 11 embryos were irradiated with 0 (–IR) or 570 rad (+IR) of x-rays before fixing and staining with an anti-PH3 antibody to visualize mitotic cells. Quantification of PH3-stained cells in the dorsal epidermis (shown enclosed by brackets) from similar embryos show a reduction in mitoses in irradiated embryos.
3.5. Collecting and Aging to Obtain Third Instar Larvae
A working knowledge of fly culture is assumed but may be found in (4,6).
Place Drosophila adults in a molasses agar bottle seeded with yeast and allow egg deposition for 2–4 h; adjust collection time to avoid a high density of embryos (see Note 5).
Remove adults from bottle.
Age embryos for 4 d at 25°C to reach late third instar stage of larval development, during which larvae crawl out of the food and up the bottle wall (see Note 6).
3.6. Irradiation
In wild-type larvae, inhibition of mitosis is apparent at 1 h after irradiation in both wing and eye-antennal imaginal discs (8). This is because cells in mitosis at the time of irradiation have exited mitosis, and further entry into mitosis is prevented by irradiation. Mitoses resume by 6 h after irradiation, indicating that the activation of the DNA damage checkpoint is transient.
Use a soft paintbrush wetted with water to transfer larvae into a petri dish containing water (see Note 7).
Irradiate larvae in petri dish with 4000 rads of x-rays, which is the LD50 for this stage in development (see Note 8).
Allow larvae to recover at 25°C. The recovery time will depend on how fast the following dissection steps can be performed. The key is to be able to incubate tissues in fix solution by 1 h after irradiation.
3.7. Dissection
This step is performed at room temperature. During fixing and staining, larval tissues remain in microfuge tubes. To prevent tissue loss, allow adequate time for tissues to settle to bottom of tubes and use a Pipetman instead of vacuum aspiration to remove all solutions.
Place larvae in a drop of PBS on the underside of a petri plate lid.
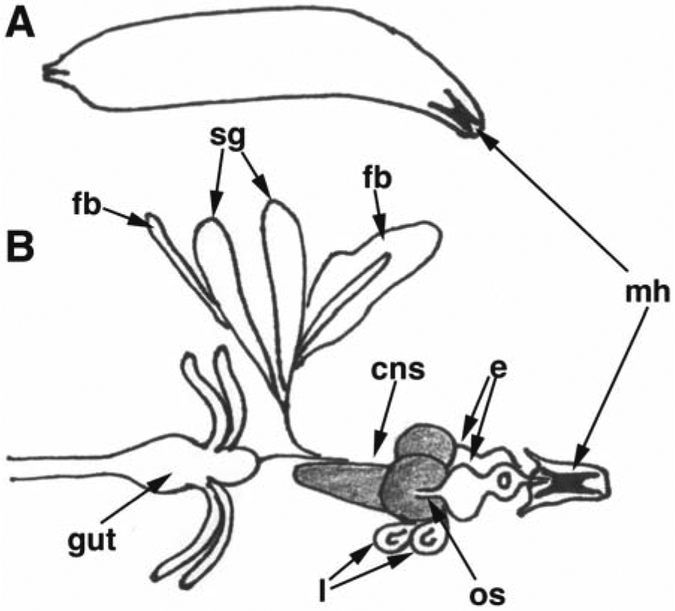
Extricate imaginal discs. This is best done by first grabbing the protruding mouth hooks with one pair of forceps and holding the larva approximately two-thirds down the body length with the other pair of forceps. Once forceps are in place, pull the mouthhooks away from the larval body. In addition to the translucent imaginal discs, salivary glands, optic lobes, fat, and the gut will come out attached to the mouthooks (Fig. 4[9]). Use forceps to remove salivary glands, fat, and gut tissue. Eye-antennal imaginal discs sit atop the optic lobes and are attached via the optic nerve stalk; leave all attached to mouthooks. Use forceps to grab mouthooks and transfer tissues to a fresh drop of PBS.
Using this method of dissection, it is difficult to consistently obtain wing discs. Another method of dissection more conducive to obtaining wing discs involves pinching the larvae in half with forceps, then turning the anterior portion of the larvae inside out by using forceps to push the anterior mouth hooks through the new opening.
Repeat to obtain imaginal discs from 5 to 10 larvae.
Fig. 4.
. Schematic representation of a third instar larva (A) and larval tissues (B): mh = mouthhooks; sg = salivary glands; fb = fat body; cns = central nervous system (optic lobes and ventral nerve cord, shaded); os= optic nerve stalk; e = eye-antennae imaginal disc; l = leg imaginal disc. Only two representative masses of fat body are shown.
3.8. Fixation and Staining
Transfer all dissected tissues to a 1.5-mL microfuge tube containing 1 mL fixing solution. Gently rock for 20 min.
Allow tissues to settle to the bottom and use a pipet to remove fix solution (see Note 9). Wash for 10 min with 1 mL of PBTx while gently rocking. Repeat the washing step twice (see Note 10).
Replace wash solution with the primary antibody (rabbit anti-PH3 diluted 1:1000 in block solution; at least 300 μL/tube); incubate at 4°C for 12 or more h with rocking.
Replace the primary antibody with 1 mL PBTx and wash for 10 min with rocking. Repeat wash twice.
Dilute secondary antibody 1:500 in block solution (see Note 4). Add at least 300 μL/ tube and incubate for at least 2 h at RT. Remove and discard secondary antibody. This and all subsequent incubation steps are carried out in the dark by wrapping samples in foil.
Replace secondary antibody solution with 1 mL PBTx and wash for 10 min with rocking. Repeat wash twice.
Add 1 mL of PBTx with 10 μg/mL of Hoescht 33528 and incubate for 2 min with rocking.
Replace Hoechst solution with 1 mL PBTx and wash for 5 min with rocking. Repeat twice.
To mount tissues onto microscope slide, use a wide-mouthed 200-μL pipet tip to withdraw a mouth hook with attached discs and place on microscope slide.
Remove most of PBTx on slide using a pipet.
Drop 50 μL of Flourmount-G onto tissues. Use forceps or tungsten needles to pull eye-antennal and wing discs away from the other tissues. To free eye-antennal discs, sever the optic stalk, which attaches each eye-antennal disc to each optic lobe of the brain. This step needs to be performed quickly because Flourmount-G will thicken fast.
Cover with coverslip and seal with clear fingernail polish.
Image discs using a compound fluorescence microscope. The eye-antennal and wing discs are sac-like epithelial structures that are comprised of two cell layers: an apical peripodial layer composed of large squamous cells, and a basal columnar layer (10,11) The columnar cells contribute to adult structures, but peripodial cells do not contribute to the adult. Both cell types show inhibition of mitosis after DNA damage. Thus, the number of mitotic cells showing PH3 stain can be quantified for the whole disc and compared with irradiated and non-irradiated larvae (see Notes 11 and 12).
4. Notes
Several drops of yeast paste in water are placed onto grape-agar plates to induce egg laying. Yeast paste should be blotted dry with a paper towel in order to prevent flies from sticking to it.
If the x-ray generator has not been properly calibrated, Lethal Dose 50 (LD50) should be empirically determined for an individual x-ray machine as “the dose that yields 50% mortality.” LD50 for various developmental stages of Drosophila can be found in (4).
When adding MeOH at the end of fixation, make sure there is still a heptane layer; this helps to trap embryos that did not lose their vitelline membrane at the interface. If there is not a discrete heptane layer, add 1–2 mL of heptane and shake for 30 s; this should restore the heptane layer. It is possible to lose up to 50% of embryos at the interface between MeOH and heptane during the fixation step.
Secondary antibodies are preabsorbed to remove nonspecifically binding antibodies. This is done by diluting the secondary antibody in block solution at 1:10 and incubating with an equal volume of fixed embryos for at least 2 h. The antibody solution is then removed and stored in a separate tube for up to 6 mo. It should be diluted 50-fold just before use to give a working dilution of 1:500.
Sparse embryo collections might result from adults either too young or old. Conversely, competition for resources will slow Drosophila development such that few larvae will be in the wandering stage on d 4. To avoid overcrowded conditions, adjust embryo collection time based on female fecundity, or use a spatula to transfer a small section of agar along with embryos to a new bottle.
At 25°C, the wandering third instar larval stage lasts approx 24 h and is followed by pupariation, where larvae become immobile. Third instar larvae undergoing pupariation will move slowly and should be avoided if dissecting eye-antennal discs to assay for the mitotic checkpoint. Eye-antennal discs from older animals begin folding and are difficult to image.
Oxygen deprivation (hypoxia) can halt cell cycle proliferation; take care not to submerge larvae in water during and after irradiation. Easy to recognize, hypoxic larvae move sluggishly and die if unable to move from water. Conversely, crawling third instar larvae move rapidly, and care should be taken to prevent escape, which might ensue if there is too little water in the petri dish.
LD50 for various developmental stages of Drosophila can be found in (4). Always irradiate wild-type larvae along with mutant larvae to control for a functional x-ray source.
Problems with antibody staining (i.e., little or no signal) can often be traced back to over-fixing. Remove fix promptly.
Imaginal tissues are incredibly fragile. After fixation, tissues can be left at 4°C for up to 24 h if necessary; incubating longer can lead to excessive tissue degradation. If at all possible, antibody staining should begin immediately after fixation.
Detailed description of imaginal discs can be found in (4). Both the eye-antennal disc and the wing disc are large, easy to identify, and useful in assaying for the mitotic checkpoint. These discs contribute to the Drosophila adult by forming the eye-antennae and the wing, respectively, during metamorphosis. During third instar larval development, eye-antennal discs have a well defined region of mitotic cells posterior to the morphogenetic furrow (Fig. 5; (12)), as well as asynchronous mitoses. Wing disc mitoses occur randomly during the larval third instar. The wing imaginal disc is loosely attached to the cluster of imaginal discs and brain that leave the body with the mouthhooks, and is easily lost during dissection and subsequent incubation steps. Allowing ample time for tissues to settle to the bottom of the tube helps prevent the loss of wing discs. The eye-antennal disc, on the other hand, which is attached to the brain and the mouthooks, is difficult to lose.
If the background fluorescence is too high, try incubating larval tissues in block solution for 1 h before adding primary antibody, or reduce the amount of time the tissues are in secondary antibody. If the signal is too low, try incubating tissues in primary antibody overnight at room temperature instead of 4°C.
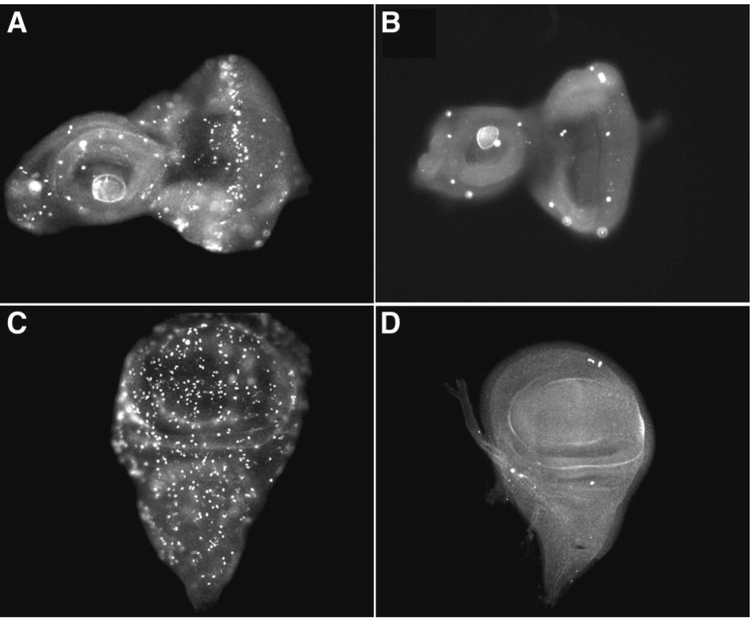
Fig. 5.
. Mitoses in eye-antennal and wing discs with and without irradiation. All imaginal discs have been incubated with an antibody against PH3 to detect cells in mitosis. (A,B) Eye-antennal and (C,D) wing imaginal discs from larvae that were not irradiated (A,C), or irradiated with 4000 rads of x-rays (B,D) and allowed to recover for 1 h.
Acknowledgments
We thank Maria Pagratis and Mark Robida for critical reading of the manuscript, and Anita Wichmann for technical assistance with Fig. 5. Work in the Su lab is supported by grants from the American Cancer Society (RPG-99– 166–01-CCG) and the National Institutes of Health (RO1-GM66441). A. P. and B. R. J. are supported by a NIH pre-doctoral training grant.
References
- 1.Zhou BB and Elledge SJ (2000) The DNA damage response: putting check- points in perpective. Nature 408, 433–439. [DOI] [PubMed] [Google Scholar]
- 2.Hendzel MJ, Wei Y, Mancini MA, et al. (1997) Mitosis-specific phosphorylation of histone H3 initiates primarily within pericentromeric heterochromatin during G2 and spreads in an ordered fashion coincident with mitotic chromosome condensation. Chromosoma 106, 348–360. [DOI] [PubMed] [Google Scholar]
- 3.Su TT, Sprenger F, DiGregorio PJ, Campbell SD, and O’Farrell PH (1998) Exit from miosis in Drosophila syncytial embryos requires proteolysis and cyclin degradation, and is associated with localized dephosphorylation. Genes Dev. 12, 1495–1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ashburner M (1989) Drosophila: A Laboratory Handbook. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. [Google Scholar]
- 5.Sullivan W, Ashburner M, and Hawley RS, eds. (2000) Appendix 3. In: Drosophila Protocols. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY: pp. 655–659. [Google Scholar]
- 6.Sisson JC (2000) Culturing large populations of Drosophila for protein biochemistry. In: Protocols Drosophila (Sullivan W, Ashburner M, and Hawley RS, eds.). Cold Spring Harbor Laboratory, Cold Spring Harbor, NY: pp. 541–551. [Google Scholar]
- 7.Foe VE, Odell GM, and Edgar BA (1993) Mitosis and morphogenesis in the Drosophila embryo In: The Development of Drosophila melanogaster (Bate M and Martinez Arias A, eds.). Cold Spring Harbor Laboratory, Cold Spring Harbor, NY: pp. 149–300. [Google Scholar]
- 8.Brodsky MH, Sekelsky JJ, Tsang G, Hawley RS, and Rubin GM (2000) mus304 encodes a novel DNA damage checkpoint protein required during Drosophila development. Genes Dev. 14, 666–678. [PMC free article] [PubMed] [Google Scholar]
- 9.Wolff T (2000) Histological techniques for the Drosophila eye. Part I: Larva and pupa. In: Protocols Drosophila (Sullivan W, Ashburner M, and Hawley RS, eds). Cold Spring Harbor Laboratory, Cold Spring Harbor, NY: pp. 201–227. [Google Scholar]
- 10.Fristrom D and Fristrom JW (1993) The metamorphic development of the adult epidermis In: The Development of Drosophila melanogaster (Bate M and Martinez Arias A, eds). Cold Spring Harbor Laboratory, Cold Spring Harbor, NY: pp. 843–897. [Google Scholar]
- 11.Wolff T and Ready DF (1993) Pattern formation in the Drosophila retina In: The Development of Drosophila melanogaster (Bate M and Martinez Arias A, eds). Cold Spring Harbor Laboratory, Cold Spring Harbor, NY: pp. 1277–1326. [Google Scholar]
- 12.Thomas BJ, Gunning DA, Cho J, and Zipursky L (1994) Cell cycle progression in the developing Drosophila eye: roughex encodes a novel protein required for the establishment of G1. Cell 77, 1003–1014. [DOI] [PubMed] [Google Scholar]
- 13.Campos-Ortega JA and Hartenstein V (1985) The Embryonic Development of Drosophila melanogaster. Springer-Verlag, Berlin. [Google Scholar]