Abstract
The evolution of genetic mechanisms for host immune evasion and anti-malarial resistance has enabled the Plasmodium falciparum malaria parasite to inflict high morbidity and mortality on human populations. Most studies of P. falciparum genetic diversity have focused on single-nucleotide polymorphisms (SNPs), assisting the identification of drug resistance-associated loci such as the chloroquine related crt and sulfadoxine-pyrimethamine related dhfr. Whilst larger structural variants are known to impact adaptation, for example, mdr1 duplications with anti-malarial resistance, no large-scale, genome-wide study on clinical isolates has been undertaken using whole genome sequencing data. By applying a structural variant detection pipeline across whole genome sequence data from 2,855 clinical isolates in 21 malaria-endemic countries, we identified >70,000 specific deletions and >600 duplications. Most structural variants are rare (48.5% of deletions and 94.7% of duplications are found in single isolates) with 2.4% of deletions and 0.2% of duplications found in >5% of global isolates. A subset of variants was present at high frequency in drug-resistance related genes including mdr1, the gch1 promoter region, and a putative novel duplication of crt. Regional-specific variants were identified, and a companion visualisation tool has been developed to assist web-based investigation of these polymorphisms by the wider scientific community.
Subject terms: Genome informatics, Malaria
Introduction
Plasmodium falciparum malaria imposes a heavy morbidity and mortality burden, with an estimated 216 million new cases and 446,000 deaths in 2016 alone, with ~90% of the burden in sub-Saharan Africa1. An understanding of the genomic diversity of P. falciparum parasites could provide insights into novel phenotypes that impact responses to antimalarials and other control measures, as well as host-pathogen interactions. Single nucleotide polymorphism (SNP) based analyses have revealed insights into drug resistance, molecular barcodes for continental origin2, transmission dynamics3, multiplicity of infection4 and regions under selective pressure related to immunological and anti-malarial treatment pressure5. In comparison, investigations of structural variants (SVs), such as insertions, deletions and duplications, have been sparse. This is despite SVs making an important contribution to genomic diversity and comprising many nucleotides of heterogeneity. In particular, copy number variants (CNVs; large indels and duplications) are thought to be widespread in the P. falciparum genome6.
Malaria parasites are exposed to strong selection from the human immune response and treatment with antimalarial drugs. Subsequently, CNVs have often been found in association with specific P. falciparum phenotypes, such as drug resistance. Duplications of mdr1 have been shown to underlie a multi-drug resistance phenotype, with these variants now present at high population frequencies in Southeast Asia7, with copy number altering the parasite response to multiple anti-malarial drugs8. Recently, we identified a novel promoter duplication for gch1 at near-fixation in a Malawi population5, which is distinct from the whole gene duplication observed in Southeast Asia known to contribute to sulfadoxine-pyrimethamine (SP) resistance. In general, such regional genetic variation may arise from differences in drug regimens, mosquito vectors, and host immunity, but is poorly understood.
Given their importance, a genome-wide structural variant map for P. falciparum with country and regional resolution should provide insights with which to better understand the impacts of treatment regimes, assess changes in parasite diversity, and ultimately inform the roll-out of anti-malarial drugs and other control initiatives. The advent of microarray technologies, such as genomic hybridisation arrays (CGH), has improved methods for detecting and confirming known SVs9,10. However, studies of this type have typically featured modest sample sizes and focused on the exome of lab-adapted isolates. The largest array-based study in P. falciparum clinical isolates (n = 122) identified 134 high-confidence CNVs across the parasite exome, established they were more common in South American than African or Southeast Asian populations, and identified several loci including mdr1, rh2b, and histidine-rich proteins II and III to be under positive selection10. Recently whole genome sequencing platforms, which produce a greater depth of short or long reads, have been used to detect SVs in P. falciparum strains11,12, and have potentially finer resolution than array-based approaches. Coupled with bioinformatic advances in detection algorithms, there is now capacity to accurately characterise a broader range of SV types. For example, extremes in coverage can identify duplications and deletions, split sequences and alternative de novo assembly-based approaches can detect a number of other types, including inversions and large insertions and deletions13.
By analysing whole genome sequencing data from 2,855 clinical isolates and focusing on robust genomic regions (82.6%) of the AT-rich P. falciparum genome, we present the first comprehensive genomic map of SVs within the global P. falciparum population, with a focus on CNVs. We identify a total of 70,257 high quality deletions (mean 440.56 per isolate, median size 26 bp) and 601 high quality duplications (mean 0.43 per isolate, median size 1,478 bp), contrasting with an average of 24,495 SNPs and 33,479 small indels (<15 bp) per isolate. Several variants were found to be geographically specific and highlight novel structural variants with roles in antigenic variation, drug resistance, and host-pathogen interactions. We confirm specific variants using several alternative approaches, including the analysis of PacBio long-read data of P. falciparum laboratory strains.
Results
Distribution of variant type, size and location
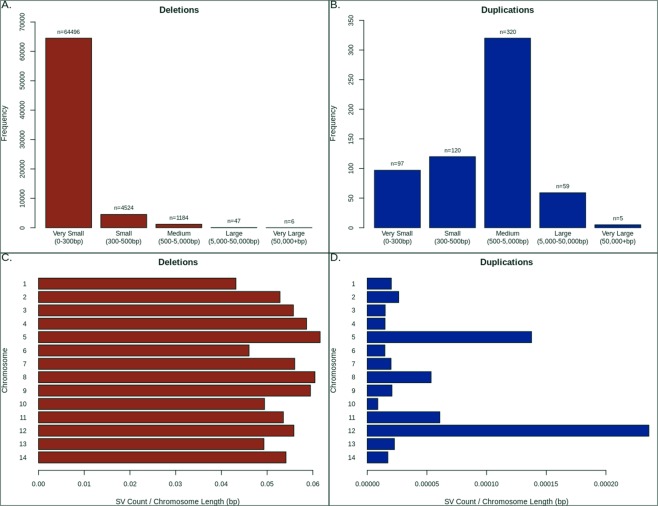
The 2,855 isolates represented 21 countries across Africa (Central, East, West), Asia (South, Southeast) and South America (Supplementary Table 1), and all displayed minimal evidence of multiplicity of infection (based on genome-wide SNPs) and non-anomalous coverage (see Methods). Using a structural variant (SV) discovery pipeline based primarily upon DELLY13, we identified more than 1 million putative variants deletions and duplications relative to the 3D7 reference genome, across robust regions (~83%; excluding regions that were highly variable or within 100 kbp of a chromosome end) of the P. falciparum genome. SVs of length greater than 300 bp were present in 4,941 genes, including in known drug resistance candidates (e.g. crt, mdr1, gch1) and indel/duplication hotspots, such as within the liver stage antigen LSA114, the gametocyte specific Pf11-114 and invasion-related Rh2b15 and EBA175 (PF3D7_0731500)16 genes (see Supplementary Table 2 for the 117 genes with >1% SV frequency). This raw set was filtered using a population-based SV analysis pipeline (see Methods), and the resulting dataset of 70,858 ‘high-quality’ variants included 70,257 deletions (mean 440.56 per isolate, median size 26 bp; 91.8% very small or micro <300 bp) and 601 duplications (mean 0.43 per isolate, median 1,478 bp; 16.1% very small or micro <300 bp). Most duplications (94.7%) and half of deletions (34,065 deletions; 48.6%) were unique to single isolates (total: 34,634; 34,065 deletions and 569 duplications) (Fig. 1). Both deletions and duplications tend to occur within intergenic regions (intergenic/genic ratio: deletions 1.42, duplications 2.15), and there is disproportional increase in the density of duplications in chromosomes 5 and 12, due to known drug resistance loci (mdr1, gch1) (Fig. 1). The most frequent genes with high-quality SVs (>300 bp) reveals loci that include both deletions and duplications (e.g. LSA3), and clusters of duplications (e.g. in chromosome 11: FP3, ApiAP2, CYP19B, FP2A, TREP) (Supplementary Table 3). Therefore, a (1 kbp) window-based analysis was used to identify regions with overlapping but distinct SVs, thereby assisting with refining their breakpoints. For deletions, 24,947 (of 27,388) windows (78.1% genic) were represented, compared to 2,441 windows (80.4% genic) for duplications.
Figure 1.
High quality variants by position, length and per-chromosome. (A) Distribution of deletions by size categories. (B) Distribution of duplications by size categories. (C) Distribution of distinct form of deletion across each chromosome. (D) Distribution of distinct forms of duplication across each chromosome.
Frequently occurring specific variants
Previous work has shown that SVs present in a relatively high proportion of the global population are consistent with evidence of phenotypic selection10, we therefore prioritised common variants, that is, present in at least 1% of our isolates (29 or more). In total we identify 7,618 common variants of which only two are duplications, one being the previously identified 436 bp gch1 promoter region duplication5 and the other being a 252 bp intergenic duplication (11:1029648–1029899; between PF3D7_1126300 and PF3D7_1126400) and exclusive to Southeast Asia. Of all common variants, 2,780 (36.5%) are genic and the median length is 25 bp (see Supplementary Table 4 for the subset with global frequency >35%). At a minimum frequency of 5%, there were 1,676 variants with median length 27 bp, including 723 (43.1%) genic and 1 duplication (gch1). We focused primarily on SVs in excess of 300 bp in length, as DELLY calling is considered more reliable at this threshold13. Only 36 loci or regions with common variants greater than 300 bp in length (Supplementary Table 5) were identified, including four intergenic deletions (size range: 605 to 1,023 bp), and one 553 bp deletion in LSA3 (PF3D7_0220000). Interestingly the 553 bp deletion in LSA3 is present primarily in Southeast Asia, particularly Thailand (14.6%), Laos (11.5%), and Myanmar (11.2%) (Global 5.5%; Africa 0.1%, America 0.0%, Asia 10.2%), and may represent region-specific host-directed selection. Two of the intergenic variants show evidence of continental differences (Allele frequency difference: FST score >0.2), including a 1,015 bp deletion in chromosome 9 upstream of gexp22 (PF3D7_0935500) (FST: 0.227; Africa 13.2%, America 70.8%, Asia 41.7%) and a 605 bp deletion in chromosome 12 upstream of ap2mu (PF3D7_1218300) (FST 0.249; 0.2% Africa, 0.0% America, 33.6% Asia).
Exploration of structural variation in anti-malarial resistance candidates
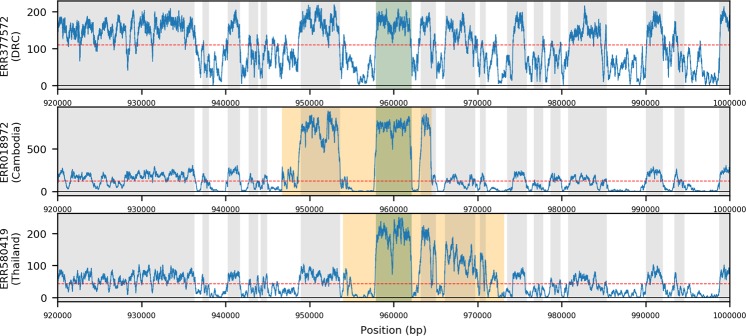
Previous analyses have revealed evidence of structural variation in loci associated drug resistance (e.g. gch1). Given that SVs can have a significant impact on gene expression and anti-malarial resistance, we focused an analysis on the identification of novel structural variants in candidate genes (dhfr, dhps, kelch13, mdr1, gch1, crt). Despite removing mixed infections determined using genome-wide SNPs, it is possible for heterogeneous duplications (i.e. differences in genetic copies) to display as mixed genotype calls, therefore we extended our ‘high-quality’ dataset to include those duplications previously excluded for high rates of the heterozygous genotype. The resulting ‘high-quality relaxed’ dataset included 91,936 putative variants (70,257 deletions: mean 440.56 per isolate, median size 26 bp; 21,679 duplications: mean 10.92 per isolate, median 1,354 bp). To minimise the number of false positives, we manually verified the alignments for all candidate regions specifically mentioned here. Overall, no high-quality SVs were identified in SP resistance associated dhfr (PF3D7_0417200) or dhps (PF3D7_0810800) genes, or artemisinin resistance associated kelch13 (PF3D7_1343700). We identify 115 specific duplication types containing mdr1 in 189 isolates, primarily in Southeast Asia (Southeast Asia 12.9%; Cambodia 9.5%, Myanmar 11.9%, Papua New Guinea 3.8%, Thailand 29.0%, Vietnam 5.9%), and near absent in Africa (Africa 0.10%; Ghana 0.2%) (Fig. 2), consistent with previous reports17. Similarly, tandem duplications are also present in KE01 (Kenya) and KH01 (Cambodia) within our complementary PacBio-based dataset (n = 13).
Figure 2.
Coverage plots showing examples of isolates with three types of mdr1 duplications. (A) No duplication in a Democratic Republic of Congo isolate. (B) Duplication in a Cambodian isolate. (C) Duplication in a Thai isolate; Blue traces represents the per base coverage for each isolate. Orange region indicates the predicted structural variant; Green region indicates the gene of interest, Grey indicates neighbouring genes; horizontal line is the median coverage for the isolate.
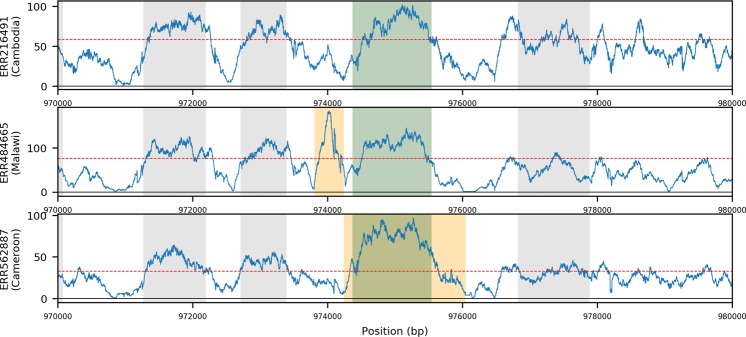
The whole gene duplication of gch1 (PF3D7_1224000) and a recently identified 436 bp gch1 promoter duplication may be linked to SP resistance5. We identify 307 isolates with 135 distinct forms of whole gene duplication across gch1 (9.9% of the total dataset) (Fig. 3). Similar whole gene tandem duplications were present in PacBio isolates for 7G8 and KH02, as a triplication in GB4, and as a triplication with an inverted middle copy in DD2; this being consistent with the existing literature12. In contrast, 491 high quality isolates are positive for the previously identified 436 bp specific ‘promoter region’ duplication (14.0% of total). We confirm this duplication being present at near-fixation in Malawi (89.5%), frequent in the rest of East Africa (Tanzania 78.5%, Kenya 31.6%), maintained in West Africa (Gambia 6.1%, Ghana 4.3%, Guinea 22.2%) and Central Africa (Democratic Republic of Congo 26.3%), but absent from all Asian and American isolates (Regional FST 0.554). No such duplication was found in any of the validation isolates with PacBio sequencing data (n = 13), though none of these were isolated from Malawi. These data therefore support the gch1 promoter duplication being present at notable frequency across Africa, and the need for further functional characterisation of any potential role in SP resistance.
Figure 3.
Coverage plots showing examples of isolates with three types of gch1 duplications. (A) No duplication in a Cambodian isolate. (B) Promoter duplication in a Malawian isolate. (C) Gene Duplication in a Cameroon isolate; Blue traces represent the per base coverage for each isolate. Orange region indicates the predicted structural variant; Green region indicates the gene of interest, Grey indicates neighbouring genes; horizontal line is the median coverage for the isolate.
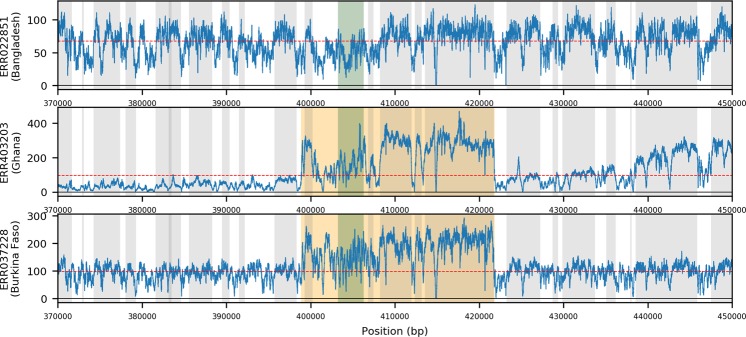
Finally, we uncover evidence of duplications of crt (PF3D7_0709000) across 35 West African and 2 Cambodian isolates. A 22.9 kbp duplication of crt is present and consistent across 32 isolates sourced in West Africa (4.3%), specifically sub-populations isolated in Burkina Faso (n = 14, 29.2%), Ghana (n = 15, 3.4%), Guinea (n = 1, 0.85%) and Mali (n = 1, 1.8%). Those 32 isolates display 26 specific variants, the consensus of which suggests that the duplication is most likely approximately 22,893 bp in length, and therefore includes several genes (PF3D7_0708900 (sco1), PF3D7_0709000 (crt), PF3D7_0709050 (small nucleolar RNA), PF3D7_0709100 (cg1), PF3D7_0709200 (glp3) and PF3D7_0709300 (cg2)) (Fig. 4). A further three isolates (2 from Ghana, 1 from Burkina Faso) display a similar 28.7 kbp duplication, which also includes PF3D7_0708800 (heat shock protein 110) and may be under different selective pressure. Beyond Africa, two Cambodian isolates feature smaller 702 bp and 11,844 bp duplications. No crt duplication was present in the validation (PacBio) dataset (n = 13). To explore the specific variability of crt in West Africa, we calculated the abundance of resistance-associated haplotypes (alleles) directly from raw reads, finding that only both CVMNK (chloroquine susceptible) and CVIET (chloroquine resistant) haplotypes were present in all 35 duplication-positive isolates; this compared to 20.3% (133/656) of isolates with evidence for mixed haplotypes but no evidence of duplication (Supplementary Table 6). Increasing the stringency on the calling of genotypes led to the retention of a disproportional number of mixed haplotypes in those samples with duplications (at least 2 (10) reads required to call haplotypes: duplications 33/35 (24/35) vs. no duplications 99/133 (44/133)). For those isolates with mixed haplotype calls (n = 168), the levels of crt gene to chromosome 7-wide coverage were greater in the duplication group (ratio median: duplication group (n = 35) 1.10 vs non-duplication (n = 133) 0.74; Wilcoxon P = 1.7 × 10−10). There was no difference in the coverage ratio within the non-duplication group (ratio median: mixed haplotypes (n = 133) 0.74 vs. single haplotype (n = 523) 0.76; Wilcoxon P = 0.09). These observations lend support to the robustness of duplications in crt. The proportion of CVIET haplotype reads in the duplication-positive group (median 0.53; IQR: 0.31–0.57; Supplementary Fig. 1) suggested a degree of parity of the carriage of chloroquine susceptible and resistant forms, and contrasted with other West African isolates without duplications (Wilcoxon P = 0.02; Supplementary Fig. 1). It is unclear, without additional transcriptional analysis, whether these forms are expressed independently though we hypothesise that the presence of both forms may allow individual parasites to benefit from the resistance form whilst reducing associated fitness costs. If so, a heterogeneous duplication of this sort may represent a more evolutionarily resilient form of crt-associated resistance.
Figure 4.
Coverage plots showing examples of isolates with three types of crt duplications. (A) No duplication in a Bangladesh isolate. (B) Gene duplication in a Ghanaian isolate. (C) Gene duplication in a Burkina Faso isolate; Blue traces represents the per base coverage for each isolate. Orange region indicates the predicted structural variant; Green region indicates the gene of interest, Grey indicates neighbouring genes; horizontal line is the median coverage for the isolate.
Population-Specific Variants
Regional differences (across West Africa, Central Africa, East Africa, South Asia, Southeast Asia, South America) in SV frequencies were quantified with FST analysis for all high-quality variants (median (range): deletions 0.002 (0–0.613); duplications 0.001 (0–0.554)). A total of 153 high quality variants (152 deletions and one duplication) have strong regional differences (FST > 0.2), including: (i) an Asia-specific 59 bp deletion within the hypothetical protein PF3D7_0312900 (FST 0.613, 69.8% South Asia, 70.9% Southeast Asia, 0.0% Rest of the World), (ii) a 40 bp South America-specific deletion in the putative histone deacetylase PF3D7_1472200 (FST 0.497, 54.2% South America, 0.0% Rest of the World), and (iii) the 436 bp gch1 promoter region duplication (FST 0.554, 78.0% East Africa, 17.2% Central Africa, 6.8% West Africa, 0.0% Rest of the World) (Supplementary Table 7).
Extending our analysis to the full list of SVs detected by the DELLY pipeline that were confirmed using alternative coverage-based approaches (CNVNator or Control-FREEC software), we identity several SVs with biogeographical specificity. Non-drug resistance candidates include a 169 bp deletion within the rhoptry-associated membrane antigen PF3D7_0707300 (FST 0.354; 46.0% Africa, 0.6% Asia, 0.0% America) and a 370 bp deletion in the ring-infected erythrocyte surface antigen PF3D7_0102200 (FST 0.213; Africa 40.3%, Asia 4.2%, America 4.2%) with elevation in West and Central Africa (FST 0.321; West Africa 66.1%, Central Africa 36.1%, East Africa 4.1%, South Asia 50.9%, Southeast Asia 2.5%, South America 4.2%). We also identify a near Africa-specific 586 bp deletion within the C-terminal of reticulocyte binding protein 2 homologue b (rh2b, PF3D7_1335300) (FST: 0.334; 58.4% Africa, 12.5% America, 1.3% Asia) and a 29 bp deletion in rhopH2 (PF3D7_0929400) (FST 0.288; 73.7% Africa, 100% America, 40.8% Asia), knockdown of which has been shown to inhibit parasite growth within host erythrocytes18. These findings were supported by manual inspection of coverage depth and split read support.
Discussion
This large and geographically comprehensive study of SVs in P. falciparum characterises both known and novel variants, the latter occurring in loci associated with antimalarial resistance, host-pathogen interactions, and disease severity. Deletions represent the bulk of SVs (>99%) identified, primarily due to an abundance of shorter forms (median 26 bp) in comparison to duplications (median 713 bp). We find that 48.6% of high quality deletions and 65.0% of high quality duplications were found in single isolates, in line with previous work with smaller sample sizes including a recent study which found that approximately half of structural variants were only present in one of 16 isolates9. Previous large-scale studies have often overlooked the role of smaller structural variants (15 to 500 bp), defining and applying a minimum size of 500 bp. Our results demonstrate that a significant number (97.7%) of high quality variants are present in the 15 to 500 bp size range, indicating that previous studies may have under-estimated the full range of genomic variants within the P. falciparum genome. This finding that most SVs are under 500 bp in size is consistent with previous studies in various species9,19.
Population-specific SVs suggest evidence of localised selective pressure10. These include the drug resistance associated mdr1 and gch1 genes, and a striking novel 22.9 kbp duplication of the chloroquine resistance associated gene crt, for which isolates are positive for both the CVMNK (chloroquine susceptible) and CVIET (chloroquine resistant) forms of the gene across multiple independent West African sub-populations. Whilst, the putative crt duplications need to be confirmed, the detection of those variants in isolates with a balanced number of CVMNK and CVIET haplotypes was robust to increasing the stringency on the number of supporting sequencing reads. It is unclear whether dual-carriage of these variants would allow expression of both or either forms of the crt transporter, though it is likely that this could allow individual parasites to benefit from chloroquine resistance with a reduced fitness cost. This finding is similar to previously identified alternatively spliced forms of crt in eastern Sudan which were hypothesised to facilitate ‘switching’ between chloroquine resistant and susceptible isoforms20. Further short ~29 bp and ~430 bp deletions identified here at low frequency in crt may reflect the specific deletions identified in that same study. Follow up studies, particularly with culture-adapted clinical isolates in which this duplication is present, are required to properly characterise in vitro phenotypes. We also present further characterisation of the promoter region duplication for the SP resistance associated gene gch1, previously identified in Malawi5. This additional analysis confirms the duplication is at near-fixation in Malawi, and highlights its presence across Central and East Africa, including notably high frequencies in Tanzania, Kenya, Guinea and the DRC. Further this genetic region has been shown to be under positive selection in Malawi using SNP-based metrics5.
Our results demonstrate that application of our pipeline can enhance the speed and capacity of high throughput structural variant discovery. However, this is not without limitations, especially as we rely upon validity of the underlying mapping, for which some regions (such as those which are highly variable or repetitive) are known to be difficult to characterise. To resolve this issue, we excluded known highly variable regions from our analysis, such as var, rifin and stevor genes and subtelomeric regions. However, in doing so we prevent discovery of true variants within these regions, including duplications in AT-rich loci12. In addition, all variants found were identified relative to the 3D7 reference strain, consistent with the approach taken in other studies9,10. Given that 3D7 is most likely an African strain, SVs within African isolates may be artificially under-represented due to those variants also being present within 3D7. Further, the discovery stage of our pipeline inherits the limitations of those tools, such as an inability to infer high quality inversions. This risk was limited by prioritising those variants that were identified by DELLY with support from an alternative discovery software (CNVNator or Control-FREEC).
The approach taken in this study, as with standard SNP discovery, requires single-genotype samples, preventing investigation of more complex isolates. By pre-screening for non-complex infections and also filtering on rates of predicted genotype, we were able to reduce false positive calls but also removed several highly likely variants that presented with a high prevalence of predicted heterozygous calls and potentially underestimate the total number of duplications. Notable candidate variants excluded from our high-quality dataset but supported by manual inspection of coverage depth or similar variants within the existing scientific literature include 102 putative deletions within the glycophorin A binding, invasion-critical gene EBA175 (PF3D7_0731500)16, the most prominent being a 424 bp deletion in 1,492 isolates (30.5% of isolates). We also identify a 586 bp deletion elevated in Africa (58.4% Africa, 12.5% America, 1.3% Asia) within Rh2b, a gene that plays a key role in erythrocyte invasion15. Similar deletions have previously been identified (and validated here) in the T996 P. falciparum line21 and in isolates from Senegal, where it is possibly associated with the utilisation of neuraminidase-sensitive invasion pathways22. Another variant is a 29 bp deletion in rhopH2 (PF3D7_0929400), which has a reduced prevalence in Asian populations, and knockdowns of which have been shown to inhibit parasite growth within host erythrocytes16.
Our final count of 70,858 high quality specific variants assumes that each SV is distinct by their specific base-pair location. This means that we identify variants which arose from similar evolutionary events but may place insufficient emphasis on variants with a shared phenotypic impact. Previous studies collapsed analysis to a locus level, but risk overlooking complex structural variation within the same gene. This challenge was partially resolved via our windows-based approach, whereby variants are grouped due to their presence within a 1 kbp window.
Overall, our work presents a set of high-quality SVs, some population specific, which are likely to have functional consequences for drug resistance and erythrocyte invasion. An extended list of further structural variation requires both technological advances, such as low cost long read platforms with low error rates, as well as computational and algorithmic advances that assemble genomes to high accuracy and require less hands-on filtering. To facilitate further exploration of our full set of global structural variation by the wider scientific community we have developed a visualisation and analysis tool. This resource will assist much-needed genomic investigations into P. falciparum, potentially leading to biological insights for the development of disease control measures.
Methods
Sequence data
Illumina raw sequence data from more than 3,500 isolates in the Pf3k project were downloaded from the European Nucleotide Archive (see the project website, https://www.malariagen.net/projects/pf3k). The raw sequences were aligned to the P. falciparum 3D7 genome using bwa-mem software (settings: –c 100 –T 50)23, resulting in a mean coverage of 70.7-fold (Genic: 91.1-fold, Intergenic: 41.8-fold). SNPs and small indels (<15 bp) were called using SAMtools/BCFtools24 (default settings, version 1.6.1) and GATK software25 (settings: “-T UnifiedGenotyper -ploidy 1 -glm BOTH -allowPotentiallyMisencodedQuals 2”, version 3.4–46). Isolates bearing abnormal coverage less than 20-fold or greater than 300-fold coverage were excluded to reduce false positive rates. Similarly, isolate samples with complex infections were excluded by accessing multiplicity of infection (MOI) as rates of missing and heterozygous SNP calls (>20%) and using estMOI software4 (>20% genome with MOI > 1; established using the standard point of inflection approach5). Specific SVs were further filtered by their phasing, as predicted by DELLY, as a secondary control against cryptic mixed infections (see below for the stringent heterozygous genotype filtering). After quality control, our dataset included 2,855 isolates representing West Africa (n = 691), Central Africa (n = 344), East Africa (n = 464), South Asia (n = 43), Southeast Asia (n = 1,291), and South America (n = 22). The Pf3k Illumina data was supplemented by PacBio sequences (ERP009847) from 13 laboratory strains12,26,27, including 7G8 (Brazil), IT (Brazil), HB3 (Honduras), GA01 (Gabon), GN01 (Guinea), GB4 (Ghana), SN01 (Senegal), CD01 (Congo), KE01 (Kenya), SD01 (Sudan), DD2 (Indochina), KH01 (Cambodia), and KH02 (Cambodia).
Structural variant discovery
Structural variants were predicted from short read alignments against the latest 3D7 reference assembly using DELLY (v0.7.3), which has been found to be robust across a range of organisms13. Smaller SVs, between 15 and 300 bp, were also identified with DELLY using the -i argument that utilises only soft-clipped read support. DELLY calling is considered more reliable for variants greater than 300 bp in length13, and our discussion of results is therefore predominantly of this group but recognises the presence of shorter variants of high frequency having strong support. The DELLY genotyping model (which assumes diploidy) was employed to call heterozygous SVs. Variants longer than 100,000 base pairs were excluded as a conservative filter for erroneous calls. Variants identified within 100 kbp of a chromosome end and in var, rifin and stevor genes were removed due to established difficulties in accurately mapping these regions28.
Population-based SV Filtering and analysis
The SV-Pop (version 1.0) pipeline was utilised for post-discovery, population-based filtering of SVs and is publicly available from https://github.com/mattravenhall/SV-Pop. Population-wide filters were applied to exclude those variants with mean DELLY quality scores below 0.9, missingness >10%, an absence of paired read support, or homozygous reference calls frequency >10%. Variants were also removed if they displayed a heterozygous genotype frequency greater than 30%, as these suggest cryptic mixed infections not identified at the SNP calling stage. This filter was not applied for the candidate gene analysis. Regional hotspots were identified using sums of isolates with variants in 1 kbp sliding windows with a 500 bp step size. Ultimately our approach produced three lists of candidate SVs: (i) over 1 million putative variants identified by DELLY alone (raw dataset), (ii) our primary “high quality” set of 70,858 SVs following filtering by SV-Pop (high-quality dataset), and (iii) 92,313 “high quality relaxed” candidate SVs following relaxation of the SV-Pop parameters to include heterozygous duplications (high-quality relaxed dataset).
Validation
The whole genomes from the 13 laboratory strains were used to validate any putative deletions and duplications detected by applying our discovery pipeline to the 2,855 isolates. Manual verification was performed for all SVs found in regions specifically mentioned in this manuscript (e.g. in drug resistance genes), and involved examination of per-base coverage plots and read pair alignments with the 2,855 Illumina samples. To further confirm high quality SVs, deletions and duplications were also identified using CNVNator (v0.3.2; bin size of 400 bp)29 and by Control-FREEC (v11.0; window size 100 bp, window step 50 bp, ploidy of 1)11,30 software. Concordance statistics between the variants detected on the DELLY pipeline and these alternative approaches were derived on a per variant basis in 1 kbp windows. Using either CNVNator and/or Control-FREEC we confirmed 97.4% for all high-quality variants detected using our DELLY pipeline, and there was 95.3% concordance for the subset of variants greater than 100 bp.
Visualisation of variants
All SVs can be viewed using an online tool (http://genomics.lshtm.ac.uk/PfGlobalSV/) (developed using SV-pop platform software31), where the full list of isolates (n = 2,855) with ENA codes are presented. This tool and our analysis compare variant frequencies between populations. Specifically, multi-population FST statistics were calculated between continent (Africa, Asia, South America) and region-based sub-populations (West Africa, Central Africa, East Africa, South Asia, Southeast Asia, South America) for both windows and variants using Nei’s method32.
Calculation of crt haplotype abundance
To determine the variability of crt in duplication positive isolates, we conducted strict match read counts with high quality pre-alignment reads for five specific haplotypes. Haplotype sequences were 25 base pairs long, and included CVMVK (TGTATGTGTAATGAATAAAATTTTT), CVIET (TGTATGTGTAATTGAAACAATTTTT), CVIDT (TGTATGTGTAATTGATACAATTTTT), CVMET (TGTATGTGTAATGGAAACAATTTTT), and CVMNT (TGTATGTGTAATGAATACAATTTTT). Only CVIET and CVMNK haplotypes were observed in West Africa, and the proportion of CVIET reads for each isolate was calculated. These analyses were performed on the high-quality relaxed dataset.
Supplementary information
Acknowledgements
The Medical Research Council UK funded eMedLab computing resource was used for data analysis. M.R. is funded by the Biotechnology and Biological Sciences Research Council (Grant Number BB/J014567/1). T.G.C. received funding from the MRC UK (Grant Nos MR/K000551/1, MR/M01360X/1, MR/N010469/1, MR/R020973/1) and BBSRC UK (BB/R013063/1). S.C. received funding from the Medical Research Council UK grants (MR/R020973/1) and the BBSRC UK (BB/R013063/1).
Author Contributions
M.R. conducted the data analysis, E.B.D. produced the assemblies. C.J.S. and D.A.B. provided data. M.R., S.C. and T.G.C. designed the study, and wrote the first draft of the manuscript. All authors read and approved the final manuscript.
Data Availability
For the primary short read data set, public accession numbers for the raw sequence data analysed are contained in SRA studies ERP000190 and ERP000199, as well as being accessible from the Pf3k project website (https://www.malariagen.net/projects/pf3k). Raw PacBio sequence data is available from the European Nucleotide Archive (ERP009847).
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Susana Campino and Taane G. Clark jointly supervised this work.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-44599-0.
References
- 1.World Health Organization. World Malaria Report 2016. WHO Press (WHO Press, 2016).
- 2.Preston, M. D. et al. A barcode of organellar genome polymorphisms identifies the geographic origin of Plasmodium falciparum strains. Nat. Commun. 5 (2014). [DOI] [PMC free article] [PubMed]
- 3.Daniels RF, et al. Modeling malaria genomics reveals transmission decline and rebound in Senegal. Proc Natl Acad Sci USA. 2015;112:7067–72. doi: 10.1073/pnas.1505691112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Assefa SA, et al. estMOI: estimating multiplicity of infection using parasite deep sequencing data. Bioinformatics. 2014;30:1292–1294. doi: 10.1093/bioinformatics/btu005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ravenhall, M. et al. Characterizing the impact of sustained sulfadoxine/pyrimethamine use upon the Plasmodium falciparum population in Malawi. Malar. J. 15 (2016). [DOI] [PMC free article] [PubMed]
- 6.Ribacke U, et al. Genome wide gene amplifications and deletions in Plasmodium falciparum. Mol. Biochem. Parasitol. 2007;155:33–44. doi: 10.1016/j.molbiopara.2007.05.005. [DOI] [PubMed] [Google Scholar]
- 7.Price RN, et al. Mefloquine resistance in Plasmodium falciparum and increased pfmdr1 gene copy number. Lancet. 2004;364:438–447. doi: 10.1016/S0140-6736(04)16767-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sidhu ABS, Valderramos SG, Fidock D. A. pfmdr1 mutations contribute to quinine resistance and enhance mefloquine and artemisinin sensitivity in Plasmodium falciparum. Mol. Microbiol. 2005;57:913–926. doi: 10.1111/j.1365-2958.2005.04729.x. [DOI] [PubMed] [Google Scholar]
- 9.Cheeseman IH, et al. Gene copy number variation throughout the Plasmodium falciparum genome. BMC Genomics. 2009;10:353. doi: 10.1186/1471-2164-10-353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheeseman IH, et al. Population Structure Shapes Copy Number Variation in Malaria Parasites. Mol. Biol. Evol. 2015;33:msv282-. doi: 10.1093/molbev/msv282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sepúlveda N, et al. A Poisson hierarchical modelling approach to detecting copy number variation in sequence coverage data. BMC Genomics. 2013;14:128. doi: 10.1186/1471-2164-14-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miles A, et al. Indels, structural variation, and recombination drive genomic diversity in Plasmodium falciparum. Genome Res. 2016;26:1288–99. doi: 10.1101/gr.203711.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rausch T, et al. DELLY: structural variant discovery by integrated paired-end and split-read analysis. Bioinformatics. 2012;28:i333–i339. doi: 10.1093/bioinformatics/bts378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Claessens A, et al. RecQ helicases in the malaria parasite Plasmodium falciparum affect genome stability, gene expression patterns and DNA replication dynamics. PLoS Genet. 2018;14(7):e1007490. doi: 10.1371/journal.pgen.1007490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Campino S, et al. A forward genetic screen reveals a primary role for Plasmodium falciparum Reticulocyte Binding Protein Homologue 2a and 2b in determining alternative erythrocyte invasion pathways. PLoS Pathog. 2018;14(11):e1007436. doi: 10.1371/journal.ppat.1007436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tolia NH, Enemark EJ, Sim BKL, Joshua-Tor L. Structural basis for the EBA-175 erythrocyte invasion pathway of the malaria parasite Plasmodium falciparum. Cell. 2005;122:183–93. doi: 10.1016/j.cell.2005.05.033. [DOI] [PubMed] [Google Scholar]
- 17.Amato R, et al. Genetic markers associated with dihydroartemisinin-piperaquine failure in Plasmodium falciparum malaria in Cambodia: a genotype-phenotype association study. Lancet. Infect. Dis. 2017;17:164–173. doi: 10.1016/S1473-3099(16)30409-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Counihan, N. A. et al. Plasmodium falciparum parasites deploy RhopH2 into the host erythrocyte to obtain nutrients, grow and replicate. Elife6 (2017). [DOI] [PMC free article] [PubMed]
- 19.Conrad DF, Andrews TD, Carter NP, Hurles ME, Pritchard JK. A high-resolution survey of deletion polymorphism in the human genome. Nat. Genet. 2006;38:75–81. doi: 10.1038/ng1697. [DOI] [PubMed] [Google Scholar]
- 20.Gadalla NB, et al. Alternatively spliced transcripts and novel pseudogenes of the Plasmodium falciparum resistance-associated locus pfcrt detected in East African malaria patients. J. Antimicrob. Chemother. 2015;70:116–23. doi: 10.1093/jac/dku358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Taylor HM, Grainger M, Holder AA. Variation in the expression of a Plasmodium falciparum protein family implicated in erythrocyte invasion. Infect. Immun. 2002;70:5779–89. doi: 10.1128/IAI.70.10.5779-5789.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jennings CV, et al. Molecular analysis of erythrocyte invasion in Plasmodium falciparum isolates from Senegal. Infect. Immun. 2007;75:3531–8. doi: 10.1128/IAI.00122-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li H. Toward better understanding of artifacts in variant calling from high-coverage samples. Bioinformatics. 2014;30:2843–2851. doi: 10.1093/bioinformatics/btu356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McKenna A, et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Otto TD, et al. Long read assemblies of geographically dispersed Plasmodium falciparum isolates reveal highly structured subtelomeres. Wellcome Open Res. 2018;3:52. doi: 10.12688/wellcomeopenres.14571.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Benavente ED, et al. Global genetic diversity of var2csa in Plasmodium falciparum with implications for malaria in pregnancy and vaccine development. Sci Rep. 2018;8(1):15429. doi: 10.1038/s41598-018-33767-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Samad H, et al. Imputation-based population genetics analysis of Plasmodium falciparum malaria parasites. PLoS Genet. 2015;11(4):e1005131. doi: 10.1371/journal.pgen.1005131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Abyzov A, Urban AE, Snyder M, Gerstein M. CNVnator: an approach to discover, genotype, and characterize typical and atypical CNVs from family and population genome sequencing. Genome Res. 2011;21:974–84. doi: 10.1101/gr.114876.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Boeva V, et al. Control-FREEC: a tool for assessing copy number and allelic content using next-generation sequencing data. Bioinformatics. 2012;28:423–425. doi: 10.1093/bioinformatics/btr670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ravenhall M, et al. SV-Pop: Population-based structural variant analysis and visualization. BMC Bioinformatics. 2019;20:136. doi: 10.1186/s12859-019-2718-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nei M. Analysis of gene diversity in subdivided populations. Proc. Natl. Acad. Sci. USA. 1973;70:3321–3. doi: 10.1073/pnas.70.12.3321. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
For the primary short read data set, public accession numbers for the raw sequence data analysed are contained in SRA studies ERP000190 and ERP000199, as well as being accessible from the Pf3k project website (https://www.malariagen.net/projects/pf3k). Raw PacBio sequence data is available from the European Nucleotide Archive (ERP009847).