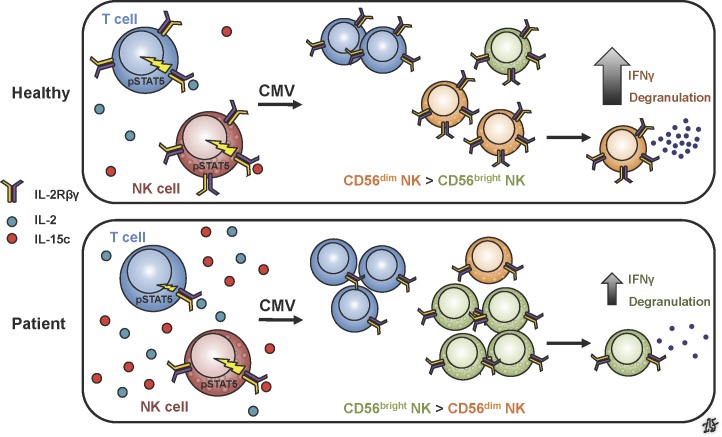
A novel homozygous mutation in human IL2RB results in decreased IL-2Rβ protein expression and dysregulated IL-2/15 signaling. This hypomorphic mutation leads to decreased regulatory T cell frequency and an abnormal NK cell compartment, with clinical manifestations of autoimmunity and susceptibility to CMV.
Abstract
The pleiotropic actions of interleukin-2 (IL-2) are essential for regulation of immune responses and maintenance of immune tolerance. The IL-2 receptor (IL-2R) is composed of IL-2Rα, IL-2Rβ, and IL-2Rγ subunits, with defects in IL-2Rα and IL-2Rγ and their downstream signaling effectors resulting in known primary immunodeficiency disorders. Here, we report the first human defect in IL-2Rβ, occurring in two infant siblings with a homozygous IL2RB mutation in the WSXWS motif, manifesting as multisystem autoimmunity and susceptibility to CMV infection. The hypomorphic mutation results in diminished IL-2Rβ surface expression and dysregulated IL-2/15 signaling, with an anticipated reduction in regulatory T cells. However, in contrast to the IL-2Rβ−/− animal model, which lacks NK cells, these siblings demonstrate an expansion of NK cells, particularly the CD56bright subset, and a lack of terminally differentiated NK cells. Thus, the early-onset autoimmunity and immunodeficiency are linked to functional deficits arising from altered IL-2Rβ expression and signaling in T and NK cells.
Graphical Abstract
Introduction
Onset of autoimmunity in infants is an increasingly recognized herald of primary immunodeficiency disorders (PIDs). Serving as the prototype for monogenic early-onset autoimmunity, immune dysregulation polyendocrinopathy enteropathy X-linked (IPEX) syndrome caused by deleterious mutations in FOXP3 established a clear mechanism by which loss of immune tolerance resulted from impaired regulatory T cell (T reg) development or function (Powell et al., 1982; Bennett et al., 2001). Owing to enhanced sensing of and dependence on IL-2 by T regs, perturbations in the IL-2R and its downstream signaling pathways result in IPEX-like syndromes (Alroqi and Chatila, 2016).
IL-2 is an essential immunoregulatory cytokine produced primarily by T cells after antigen stimulation, exerting its effects via the trimeric IL-2R expressed on T cells and natural killer (NK) cells (Waldmann, 2006; Zhang et al., 2013). This receptor consists of α (IL-2Rα), β (IL-2Rβ), and γ (IL-2Rγ) chains (Wang et al., 2005), the latter of which is shared among several cytokine receptors (for IL-2, -4, -7, -9, -15, and -21). IL-2Rβ is shared with the IL-15R, which, unlike IL-2R, involves the transpresentation of IL-15 with IL-15Rα (IL-15 complex; IL-15c) by antigen-presenting cells to IL-2Rβ/IL-2Rγ–expressing cells. Signal transduction for IL-2R and IL-15R is mediated by IL-2Rβ and IL-2Rγ, with common downstream signaling proteins Janus kinase 3 (JAK3) and signal transducer and activator of transcription 5 (STAT5; Waldmann, 2006).
The intertwined actions of IL-2 and IL-15 are evident in disease states resulting from defects within the IL-2/15 axis. Deficiency of IL-2Rγ or JAK3 results in SCID with absent T cells and NK cells, while IL-2Rα deficiency is characterized as an IPEX-like syndrome with impaired T cells but relatively unaffected NK cells (Noguchi et al., 1993; Macchi et al., 1995; Russell et al., 1995; Roifman, 2000; Caudy et al., 2007). Further downstream of IL-2R/15R, deficiency of STAT5b results in an IPEX-like syndrome with more devastating effects on NK cells (Kofoed et al., 2003; Bernasconi et al., 2006). Although Gilmour et al. (2001) previously described a patient with a T−B+NK− SCID phenotype and diminished IL-2Rβ expression, a causative variant IL2RB was not found. Rather than absence of T cells, IL-2Rβ−/− mouse models demonstrate extensive T cell proliferation and reduction in T reg and NK cells, with autoimmunity proving fatal by 12 wk of age (Suzuki et al., 1995), affirming the critical roles of IL-2 and IL-15 in development and function.
Here, we report the clinical and immunological phenotypes of two siblings with the first described human IL-2Rβ defect. This hypomorphic mutation results in dysregulated IL-2/15 signaling, elevated plasma IL-2/15 levels, reduced T reg frequency, and an abnormal NK compartment, which, in concert, lead to autoimmunity and CMV susceptibility.
Results and discussion
Clinical and immunological phenotypes of siblings with the homozygous IL2RB mutation
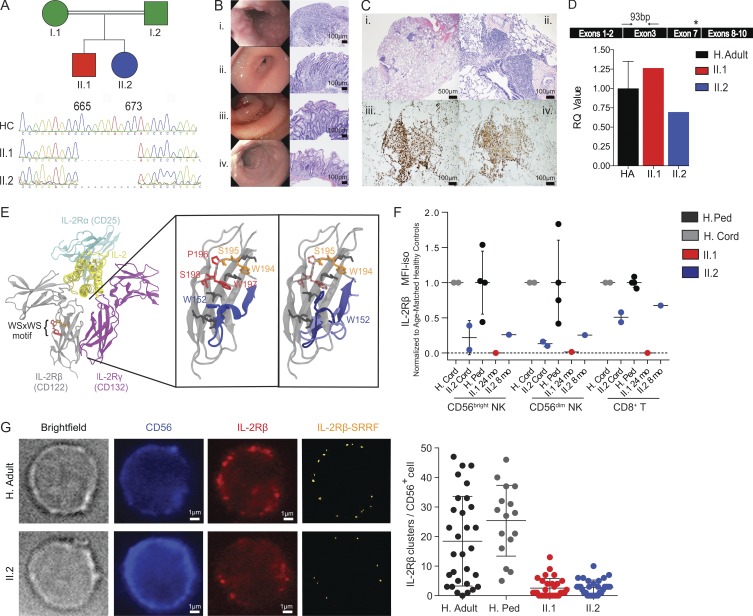
We investigated a 2-yr-old boy (II.1) born to consanguineous parents (I.1 and I.2) from Tajikistan (Fig. 1 A). At 1 mo old, the patient had failure to thrive and diarrhea, which worsened after 2-mo-old immunizations. Biopsies retrieved by endoscopy revealed marked submucosal lymphoid hyperplasia of the esophagus, accompanied by gastritis, duodenitis, and colitis (Fig. 1 B). CMV was also found in the bronchoalveolar lavage, intestinal specimens, and blood (1,717 copies/ml). Subsequent evaluations found eczematous dermatitis, food allergy, lymphocytic interstitial pneumonitis (Fig. 1 C), EBV viremia (7,201 copies/ml), and a positive direct Coombs (without anemia).
Figure 1.
Identification of an IL2RB homozygous mutation in patients with lymphoproliferation and autoimmunity. (A) Pedigree of the consanguineous parents with two affected children and chromatogram showing the 9-nucleotide deletion in the IL2RB gene. (B) Patient II.1 endoscopy of the (i) esophagus with severe nodularity with the submucosal lymphoid hyperplasia; (ii) stomach with edema of the antrum with chronic active gastritis; (iii) duodenum with erythema, edema, and loss of villous profile; and (iv) colon with small superficial aphthous ulcerations with chronic active colitis. Bars, 100 µm. (C) Patient II.2 bronchoscopy shows (i) lymphocytic interstitial pneumonia pattern; (ii) patchy peri-airway and intraalveolar macrophage accumulation, with a predominant CD8+ and CD4+ T lymphocyte infiltrate (iii and iv). Bars, 500 or 100 µm. (D) IL2RB gene expression quantification relative to actin (HA in black, n = 3, 1 ± 0.35; one experiment). Cartoon shows primer position (black arrows) relative to deletion (asterisk). RQ, relative quantification. (E) 3D modeling of the IL-2Rβ protein structure, with disruption of the WSXWS motif in the patients (from left to right): the IL-2R complex is shown in ribbons. The WSXWS motif is shown in red and orange. In the expanded view of the IL-2Rβ protein D2 domain with the WSXWS motif, the three residues that are deleted in the patient are shown in red (translucent in patient). Adjacent residues involved in the π cation-stacking ladder are shown in dark gray. (F) Normalized surface IL-2Rβ protein expression mean fluorescence intensity (MFI) in CD56bright NK, CD56dim NK, and CD8+ T cells. The MFI is calculated as IL-2Rβ MFI − isotype control MFI normalized to the mean for each cell type. Healthy cord (H. cord) and pediatric controls (H. Ped; age 12–30 mo) were normalized independently. For cord comparisons, two experiments with different healthy cord donors are shown. For pediatric comparisons, one experiment with four healthy pediatric controls is shown. Ped, pediatric. (G) Representative bright-field and immunofluorescence images of CD56 and IL-2Rβ staining are shown for healthy adult (H. Adult; top) and patient II.2 (bottom). Number of IL-2Rβ clusters per cell in one healthy adult (18.43 ± 15.16, cells = 30), three pediatric controls (25.38 ± 12, cells = 16), patient II.1 (2.567 ± 3.256, cells = 30), and patient II.2 (2.8 ± 2.497, cells = 30) is shown as a dot graph where each dot represents one cell. Bars, 1 µm. Two experiments were performed. Mean ± SD is shown.
Immunophenotyping of peripheral blood mononuclear cells (PBMCs) from patient II.1 revealed a predominance of CD8+ T cells and NK cells (Table 1). Further T cell phenotyping showed increased memory (CD45RO+) T cells, with a distribution of central memory and effector memory subsets similar to those of healthy adults (Table 1 and Fig. S1 A). Additionally, the proportion of CD4+CD25+FOXP3+ T regs was decreased in the patient. Analysis of B cell subsets demonstrated increased memory B cells, with a predominance of isotype-switched B cells (Fig. S1 B), and associated elevated serum immunoglobulin levels. The patient also exhibited high NK cell counts, particularly CD56bright NK cells (Fig. S1 C). Equivalent frequencies of myeloid populations (dendritic cells and monocyte subsets) compared with healthy controls were observed (Fig. S1 D).
Table 1. Immunological laboratory characteristics of siblings with homozygous IL2RB mutation.
| Variable | Proband (II.1) | Sibling (II.2) | Reference range |
|---|---|---|---|
| White blood cell differential | |||
| Absolute neutrophil count (cells/µl) | 3,560 | 1,210 | 1,500–6,000 |
| Absolute monocyte count (cells/µl) | 1,980 | 670 | 280–1,380 |
| Absolute eosinophil count (cells/µl) | 250 | 400 | 80–420 |
| Absolute basophil count (cells/µl) | 70 | 10 | 10–70 |
| Absolute lymphocyte count (cells/µl) | 4,352 | 3,372 | 1,800–9,000 |
| Lymphocyte subsets | |||
| CD3+ T (cells/µl) | 2,313 | 1,648 | 2,500–5,600 |
| CD4+ T (cells/µl) | 738 | 1,144 | 1,800–4,000 |
| CD8+ T (cells/µl) | 1,395 | 422 | 590–1,600 |
| CD4+:CD8+ ratio | 0.53 | 2.71 | 1.2–6.2 |
| CD4+CD45RA+ naive T (% CD4) | 45 | 61.1 | 77–94 |
| CD4+CD62L+CD27+ naive T (% CD4) | 44 | 52.13 | |
| CD4+CD62L+CD27+CD45RO+ central memory T (% CD4) | 36 | 31.12a | |
| CD4+CD62L−CD27−CD45RO+ effector memory T (% CD4) | 4.3 | 14.8a | |
| CD4+CD45RO+ memory T (% CD4) | 55 | 38.3 | 3–16 |
| CD4+CD25++FOXP3++ regulatory T (% CD4) | 0.3 | 0.33a | 1.8–8.3 |
| CD8+CD45RA+ naive T (% CD8) | 62 | 82.62a | |
| CD8+CD62L+CD27+ naive T (% CD8) | 4 | 64.4a | |
| CD8+CD45RO+ memory T (% CD8) | 39 | 8.04a | |
| CD8+CD62L+CD27+CD45RO+ central memory T (% CD8) | 1 | 3.06a | |
| CD8+CD62L−CD27−CD45RO+ effector memory T (% CD8) | 30 | 9.59a | |
| CD19+ B (cells/µl) | 662 | 749 | 430–3,000 |
| CD3−CD16+/56+ NK (cells/µl) | 1,235 | 932 | 170–830 |
| CD3−CD16+/56+ NK (% lymphocytes) | 28 | 28 | 3–14 |
| CD3−CD56dim NK (% lymphocytes) | 6.61a | 5.49a | |
| CD3−CD56bright NK (% lymphocytes) | 5.79a | 12a | |
| IL-2R studies | |||
| CD3+CD132[IL-2Rγ]+ T (%) | 100 | NA | 99–100 |
| Soluble IL-2Rα (pg/ml) | 1,147 | 1421 | ≤1,033 |
| TCR excision circles (copies/µl) | 115.2 | 86.4 | >40 |
| Serum immunoglobulin levels | |||
| IgG (mg/dl) | 1,753 | 793 | 140–533 |
| IgA (mg/dl) | 168 | 100 | 0–40 |
| IgM (mg/dl) | 186 | 53 | 0–84 |
| IgE (IU/ml) | 781 | NA | 0–29 |
| NK cell studies | |||
| NK cell function (lytic units) | 6.6 | NA | ≥2.6 |
| CD107a mobilization (%) | 23 | NA | 11–35 |
College of American Pathologists/Clinical Laboratory Improvement Amendments (CAP/CLIA) representative laboratory data are shown for each patient at age 4 mo. Numbers in boldface indicate values above the normal range for age-matched controls in the laboratory in which the study was performed. CAP/CLIA laboratory studies were performed at Children’s Hospital Colorado Clinical Laboratory, Advanced Diagnostic Laboratories National Jewish Health, Mayo Medical Laboratories, and Cincinnati Children’s Diagnostic Immunology Laboratory. Age-appropriate reference ranges are provided where available. NA, not applicable.
Laboratory studies performed under research study protocol, at age 6 mo for sibling, and age 2 yr for the proband. Complete immunophenotype data are provided in Fig. S1.
A chromosomal microarray revealed multiple large regions of homozygosity, the most notable of which involved 22q12.1-13.1, encompassing IL2RB. Whole-exome sequencing in the kindred identified multiple variants of unknown significance (Table S1), of which IL2RB showed the lowest minor allele frequency score and greatest likelihood of pathogenicity. This IL2RB mutation is a homozygous 9-nucleotide in-frame deletion that results in a loss of three amino acids (NM_000878.3; p.Pro222_Gln225del). Sanger sequencing confirmed homozygosity for the mutation in the proband and his then in utero sister (Fig. 1 A), and heterozygosity for the parents.
Immunophenotyping of patient II.2 at birth (cord blood) and 4 mo demonstrated similar abnormalities to the proband (Table 1 and Fig. S1). Clinically, she presented with CMV viremia (28,294 copies/ml) and severe autoimmune hemolytic anemia at 2 mo and later lymphocytic interstitial pneumonitis at 4 mo. For management of their autoimmune manifestations, both patients were initially treated with methylprednisolone (2 and 3 mo, respectively), after which they transitioned to sirolimus (19 and 6 mo, respectively), until their eventual hematopoietic stem cell transplantations (HSCTs). If not maintained on ganciclovir/valganciclovir, the CMV viremia for both patients would escalate, with peak viral loads of 154,386 and 362,908 copies/ml, respectively. Despite antiviral therapy, neither patient cleared CMV before HSCT. The peak EBV load for patient II.1 was 20,703 copies/ml, while patient II.2 never contracted EBV. In contrast to patient II.1, patient II.2 was diagnosed in utero; therefore, her pathology was examined from birth before any infection or immunosuppression, best capturing the immunopathogenesis of this IL2RB mutation.
Homozygous IL2RB mutation results in altered protein structure and decreased protein expression
While the homozygous deletion in IL2RB does not diminish transcript levels (Fig. 1 D), its location is predicted to disrupt the highly conserved TrpSerXaaTrpSer (WSXWS) extracellular motif (Hilton et al., 1996), a motif common to type I cytokine receptors influencing receptor folding, trafficking, binding, and signaling (Yoshimura et al., 1992; Baumgartner et al., 1994; Dagil et al., 2012). In silico three-dimensional (3D) protein modeling simulations of the deletion showed that residue W152 and the associated segment are ejected from the interaction region involved in the formation of a π cation-stacking ladder that provides structural stability and facilitates signal transduction in the prolactin receptor (Dagil et al., 2012; Fig. 1 E). To assess the downstream effects of this in silico prediction, IL-2Rβ protein expression was evaluated by flow cytometry. Both patients showed markedly reduced surface IL-2Rβ expression in CD56bright NK and CD56dim NK cells, which express the highest levels of IL-2Rβ. IL-2Rβ expression was also decreased in CD8+ and CD4+ T cells (Figs. 1 F and S2 A). Given the dramatic reduction of surface IL-2Rβ expression observed in patient II.1, decreased IL-2Rβ surface expression on CD56+ NK cells was confirmed in both patients by immunofluorescence and stream real-time superresolution (SRRF) microscopy (Fig. 1 G).
Homozygous IL2RB mutation results in dysregulated IL-2 and IL-15 signaling
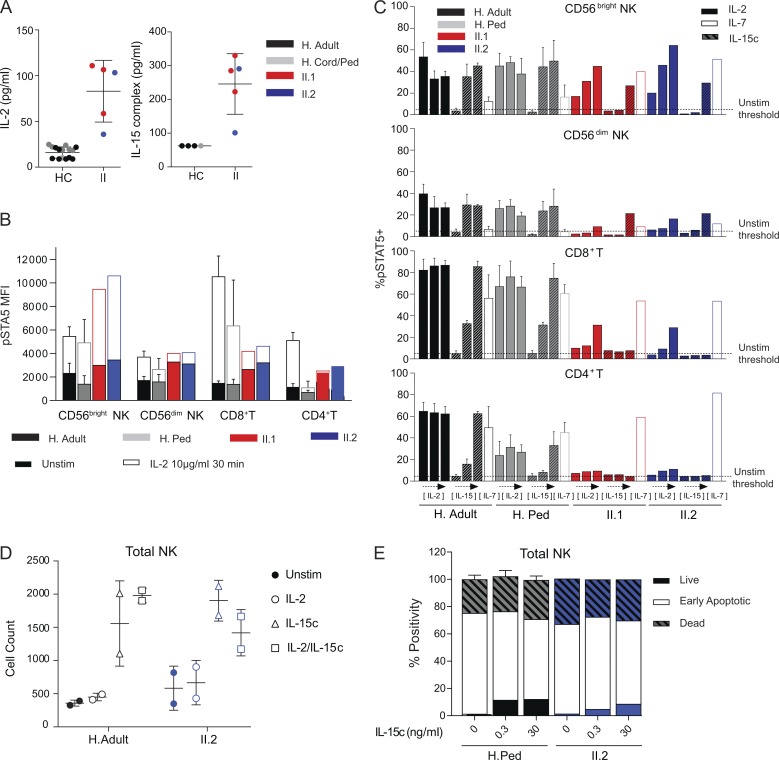
Given the reduced IL-2Rβ surface expression, IL-2 and IL-15 plasma levels and their signaling pathway were evaluated. Plasma levels of IL-2 and IL-15c (IL-15/15Rα) in both patients, measured at multiple time points, were significantly higher than that of their age-matched controls (Fig. 2 A). Plasma elevations in other proinflammatory cytokines were not observed (Fig. S2 C). These data are in sharp contrast to those of IL-2Rα deficiency, where numerous cytokines are elevated (Goudy et al., 2013). The selective elevation of IL-2 and IL-15 in both patients may be due to the inability of a hypomorphic receptor to act as a “sink” for these cytokines. To determine if the excess cytokine level translated into constant basal elevated signaling in vivo, STAT5 phosphorylation (pSTAT5) in the absence of any exogenous stimuli (basal level) was evaluated (Fig. 2 B, filled box). Indeed, both patients displayed elevated basal pSTAT5 levels in CD56bright NK, CD56dim NK, CD8+, and CD4+ T cells, consistent with continuous in vivo IL-2Rβ stimulation.
Figure 2.
IL2RB homozygous patients have increased IL-2 and IL-15c plasma levels, dysregulated STAT5 phosphorylation, and IL-15c–induced NK cell survival. (A) Plasma levels of IL-2 and IL-15c in patients (IL-2 = 83.08 ± 15.1, IL-15c = 246 ± 40.1, n = 3 for II.1 and n = 2 for II.2), and healthy controls (HC; IL-2 = 16.72 ± 5.8, Il-15c = 62.5 ± 0). For IL-2 ELISA measurements, there were n = 5 adult, n = 3 pediatric (age 12–30 mo), n = 7 cord controls, and four experiments; for IL-15c, ELISA, there were n = 3 adult and n = 2 pediatric (age 12–30 mo) controls, and two experiments. Ped, pediatric. (B) Basal pSTAT5 MFI (no stimulation [unstim], filled) and maximal response (IL-2 at 10 µg/ml for 30 min, unfilled) in CD56bright and CD56dim NK, CD8+, and CD4+ T cells. (C) Percentage of CD56bright and CD56dim NK, CD8+, and CD4+ T cells with pSTAT5 signal above unstimulated level in response to 30-min stimulation with IL-2 (0.1, 1.0, or 10 µg/ml, left to right, filled bar), IL-15c (0.1, 10, or 1,000 ng/ml, left to right, hashed bar), or IL-7 (1 µg/ml, unfilled bar). Unstimulated %pSTAT5+ level is set with 95% of cells below threshold (dashed line). For B and C, one experiment with II.1 age 24 mo and II.2 age 8 mo compared with healthy adults (H. adult, n = 3) and pediatric controls (age 12–30 mo, H. ped, n = 3) is shown. (D) Total number of live NK cells cultured with no cytokines, IL-2, IL-15c or IL-2, and IL-15c for 100 h from healthy adults (n = 2) and patient II.2 (n = 2, two experiments). (E) Percentage of Annexin V-7−AAD− (live), Annexin V+7-AAD− (early apoptotic), and Annexin V+7-AAD+ (dead) NK cells cultured with no cytokine or with IL-15c (0.3 or 30 ng/ml) for 110 h. One experiment with healthy adults (n = 2) and pediatric controls (age 12–30 mo, n = 3) shown. Mean ± SD is shown.
In vitro signaling responses to increasing concentrations of IL-2 and IL-15c for 30 min (Fig. 2 C) or 15 min (Fig. S2 D) in CD8+ and CD4+ T cells were dramatically reduced in both patients. Additionally, for both patients, CD4+ T cells did not respond to any of the cytokine concentrations or stimulation time points tested, while CD8+ T cells responded only to the highest concentration of IL-2 but not IL-15c. However, the pSTAT5 response to IL-7 in these subsets was comparable to that of healthy controls, negating an intrinsic inability to phosphorylate STAT5 (Fig. 2 C). In contrast, CD56bright NK cells were capable of responding to IL-2 and IL-15c in a time- and dose-dependent manner (Fig. 2, B and C; and Fig. S2 D). Remarkably, despite the higher pSTAT5 basal level in both patients’ NK cells, the pSTAT5 maximal level in the CD56bright NK population was much higher than that of adult or pediatric controls. This finding is in contrast to the CD56dim NK population, where the patients’ cells also showed a higher pSTAT5 basal level, but the pSTAT5 maximal response was similar to that of healthy controls (Fig. 2, B and C). This differential IL-2Rβ expression and signaling pattern is likely to account for the selective expansion of CD56bright NK cells. Heterozygosity for the IL2RB deletion, as observed in the parents, resulted in moderately decreased IL-2Rβ expression, but did not impair STAT5 phosphorylation induced by IL-2 or IL-15c (Fig. S2, B and E). Other cytokine signaling pathways such as STAT1 and STAT3 remained intact for the patients (Fig. S2 F).
Next, we assessed the functional consequences of dysregulated IL-2/15 signaling by survival and proliferation studies. In patient II.2 and healthy controls, the absolute cell numbers of live NK cells increased when incubated in vitro, with IL-15c concentrations equivalent to that of the patient’s plasma levels (Fig. 2 D). We also assessed apoptosis using Annexin V and aminoactinomycin D (7-AAD) as a measure of early and late apoptosis at different IL-15c concentrations. Patient II.2’s NK compartment demonstrated an increase in live cell frequency proportional to increasing IL-15c concentrations, with no apparent changes in the early apoptotic cell frequency (Fig. 2 E). This increase in absolute number and frequency did not correlate with increased proliferation in II.2 (data not shown). Interestingly, no apparent increase in survival or proliferation was noted in CD8+ T cells when exposed to the same concentrations of IL-15c that NK cells responded to in the survival studies (data not shown). This finding suggests that for the patient’s NK cells, even with decreased IL-2Rβ surface expression, the signaling networks downstream of IL-2/15R remain functional. Thus, the continuous in vivo IL-2Rβ stimulation by IL-12/15c likely accounts for the NK cell survival. In regard to proliferative responses, CD4+ and CD8+ T cell proliferation in response to anti-CD3 stimulation was induced in a dose-dependent fashion and modestly reduced in both patients compared with pediatric and adult controls (Fig. S2 G for patient II.2; data not shown for patient II.1). In contrast to IL-2Rβ−/− mice, which demonstrate a severe defect in proliferation, T cells from a hypomorphic IL2RB patient can still proliferate in response to TCR stimulus. Collectively, the immunophenotypic and functional data support a hypomorphic nature for this IL2RB mutation, resulting in decreased, but not absent, protein expression and dysregulated signaling capability.
Hypomorphic IL2RB mutation results in dysregulated NK cell compartment
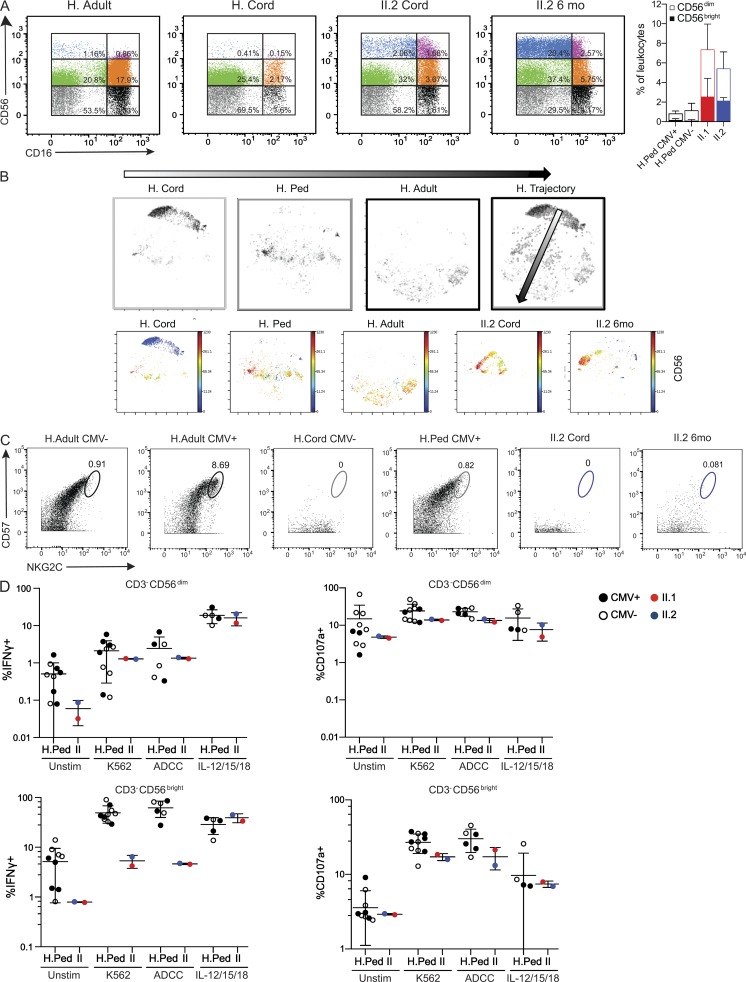
Given the abnormal expansion of CD56bright NK cells, we compared the NK cell compartment of both patients to healthy controls of different ages. In healthy donors, peripheral blood NK cells comprise two main subsets: CD56dimCD16+ NK cells and CD56brightCD16− NK cells, the latter of which are believed to be a developmental precursor to the former (De Maria et al., 2011). Compared with age-matched controls, both patients demonstrated a marked increase of CD56brightCD16− and CD56brightCD16+ NK cells (Fig. 3 A). This NK skewing is likely secondary to their underlying genetic defect, and not their infectious history, given that both CMV− and CMV+ pediatric controls did not demonstrate this CD56bright expansion (Fig. 3 A). Interestingly, this expansion became more pronounced in patient II.2 as she aged (Figs. 3 A and S1 C). These data suggest an abnormal developmental progression of NK cells toward the CD56dimCD16+ endpoint.
Figure 3.
IL2RB patients possess a dysregulated NK cell compartment. (A) Representative dot plots show NK cell subsets dissected based on CD56 versus CD16 from the CD3−CD19−CD14−CD33− gate. Bar graph shows the frequency of CD3−CD56dim (unfilled) and CD3−CD56bright (filled) NK cells from CMV+ (n = 5) or CMV− (n = 5) healthy pediatric (H. Ped) donors and patients. Two experiments shown. (B) viSNE maps represent each cell from the CD3−CD19−CD14−CD33−HLADR−LILRB1lo population as a dot. The location of the cell clusters for each healthy control in the 2D viSNE map represent the NK cell compartment in infancy (H. cord, cord blood, n = 6, 3 CMV+ and 3 CMV−), childhood (H. Ped, age 12 mo, n = 10, 5 CMV+ and 5 CMV−), and adulthood (H. Adult, n = 6, 3 CMV+ and 3 CMV−). The merge of these three viSNE maps illustrates a healthy “NK trajectory” (H. Trajectory). Below are each of the visNE maps described above colored by CD56 signal intensity. Two experiments with data merged and normalized together for analysis. (C) Representative dot plots for CD57 and NKG2C from the CD3−CD19−CD14−CD33−HLADR−LILRB1lo parent gate (two experiments). Circles indicate CD57+NKG2Chi cells. (D) Functional assay of CD3−CD56dim and CD3−CD56bright NK cells from CMV+ (closed circles) or CMV− (open circles) healthy pediatric donors (age 12–30 mo) and patients II.1 (24 mo) and II.2 (10 mo). Intracellular expression of IFNγ and surface expression of CD107a were assessed after PBMC stimulation with K562 cells (n = 10), anti-CD20–coated Raji cells (n = 6), and IL-12/15 (10 ng/ml) and IL-18 (100 ng/ml; n = 4). Stimulated %CD107a+ and %IFNγ+ frequencies were based on a 5% positivity threshold set on the unstimulated samples. One experiment was performed. Mean ± SD is shown. Unstim, no stimulation.
To further dissect the NK cell compartment, PBMCs from patient II.2 and healthy controls of different ages and CMV status were analyzed via mass cytometry, assessing 35 different NK cell specific parameters, including multiple activating and inhibitory NK cell receptors and maturation markers. The t-distributed stochastic neighbor-embedding algorithm (viSNE) was applied to visualize different human NK cell populations on a 2D map at single-cell resolution (Amir et al., 2013). viSNE clustering of healthy cord blood samples, healthy pediatric and adult controls, and patient II.2 at cord and 6 mo of age revealed an abnormal NK cell compartment in the patient (Fig. 3 B). As a healthy individual ages, the NK cell compartment moves from the top right region of the viSNE map to the bottom left (Fig. 3, B and H, trajectory). In contrast, the NK cell compartment of patient II.2 at cord and 6 mo occupies a completely different region of the visNE map, indicating strikingly different phenotypic characteristics based on the 35-parameter analysis (Fig. 3 B). Despite the unusual NK phenotype, levels of perforin and granzyme B contained in CD56bright and CD56dim NK cells in II.2 were similar to those of the healthy control (data not shown). By clinical assay standards, patient II.1 demonstrated intact K562 killing (chromium release; Table 1). However, clinical studies were performed at 4 mo of age when CMV PCR demonstrated 1,700 copies/ml, while Fig. 3 studies were performed at 24 mo when CMV viremia reached >100,000 copies/ml.
It has been shown that during CMV infection there is preferential expansion of mature NK cells (CD56dimCD16+) expressing the activating CD94-NKG2C receptor complex, at which time these cells also express CD57, a well-established maturation marker (Gumá et al., 2004; Lopez-Vergès et al., 2011; Muccio et al., 2016). Given the chronic CMV infection history in the patients, we further investigated the CD57+NKG2Chi CMV-associated NK cell subset. Patient II.2 lacked CD57+NKG2Chi NK cells, even when comparing her status from birth (CMV−) to 6 mo when she was significantly viremic (Fig. 3 C). Particularly, CD57 expression in the CD56dim NK cells was almost absent, suggesting an abnormality in NK cell maturation. To functionally assess NK cells, cytotoxicity against tumor cells (K562) and antibody-dependent cellular cytotoxicity (ADCC; Raji cells with anti-CD20) studies were performed. The patients demonstrated markedly decreased IFNγ production and CD107a mobilization in response to both K562 and ADCC conditions in the CD56bright subpopulation, which is surprising given the significant expansion of this population in the patients. In the CD56dim subpopulation, the patients demonstrate a mild reduction in IFNγ production and CD107a mobilization compared with the healthy controls in K562 and ADCC conditions (Fig. 3 D). Thus, the accumulation of immature CD56bright NK cells, abnormal “NK trajectory” based on 35 NK phenotypic and maturation markers, markedly decreased CD57 expression, and decreased IFNγ production and CD107a mobilization suggest a developmental functional defect in the IL2RB hypomorphic patients, which may contribute to their CMV susceptibility.
Hypomorphic IL2RB mutation and persistent CMV infection result in T cell oligoclonality in patient II.1
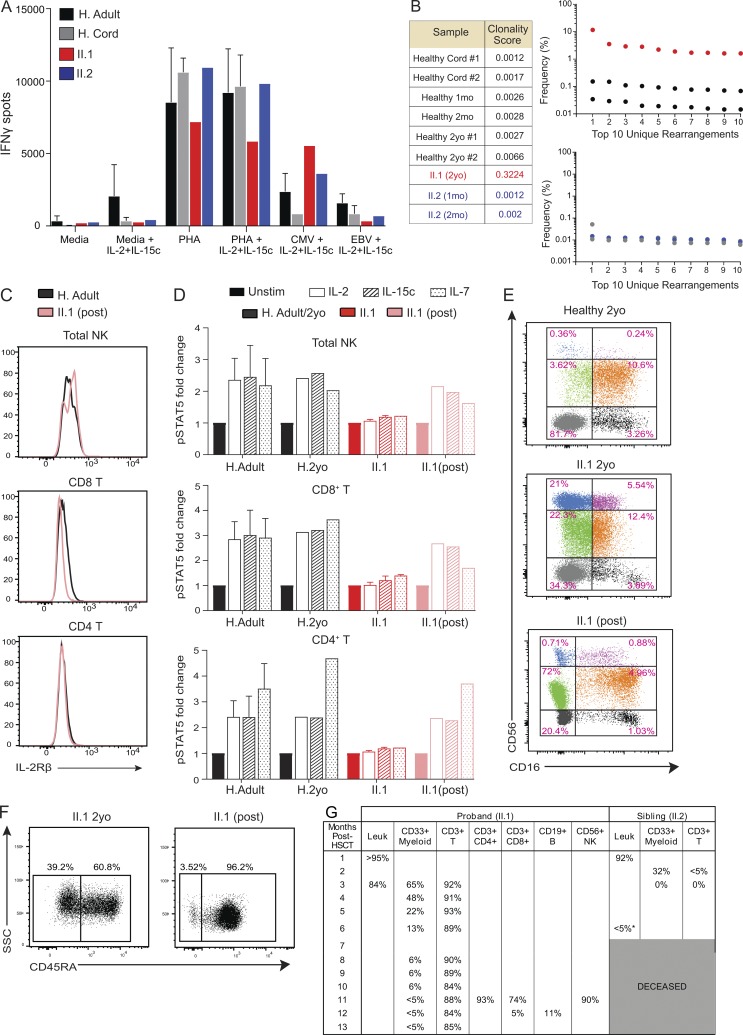
Since a defect in controlling chronic CMV infection by NK cells was observed, we also evaluated T cell responses against CMV. In the presence of ongoing viremia, patient II.1 demonstrated robust IFNγ production in response to CMV peptides compared with II.2 and healthy controls (Fig. 4 A). These data highlight a striking contrast to IL-2Rα–deficient patients, in whom severely diminished CMV-specific T cell responses are observed (Goudy et al., 2013).
Figure 4.
T cell immunophenotypic and functional characterization in patient II.1. (A) Production of IFNγ in response to PHA, EBV, and CMV ± IL-2/15c was measured by ELISPOT. Healthy adults (H. Adult; n = 7) and cord blood samples (H. Cord; n = 3) were used with patient samples (n = 1 for each) in one experiment. (B) TCR sequencing was performed in patients and healthy controls, and clonality scores are shown. Graphs show the frequency of the top 10 TCR rearrangements unique to patients and their respective age-matched healthy controls. Each numbered TCR rearrangement is different and unique for each patient and control. (C) Histograms of IL-2Rβ surface protein expression in patient II.1 post-HSCT and healthy control. (D) Normalized pSTAT5 responses in cell types as indicated. PBMCs were stimulated with IL-2 (1 µg/ml) or IL-15c (1 µg/ml) for 30 min. Fold change in MFI is calculated as pSTAT5 MFI for each condition normalized to the unstimulated (unstim) condition for each individual. Data merged from two experiments with II.1 pre- and post-HSCT compared with healthy adults (H. Adult, n = 7) and healthy 2 yr old (n = 1). (E) Representative dot plots show NK cell subsets dissected based on CD56 versus CD16 from the CD3−CD19−CD14−CD33− gate from one experiment. (F) Representative dot plots show SSC versus CD45RA+ from the CD3+ gate in patient II.1 pre- and post-HSCT. Two experiments shown. (G) Percentage of lymphoid and myeloid chimerism in patients II.1 and II.2 post-HSCT. mo, months old; yo, years old. Mean ± SD is shown.
TCR repertoire analysis also demonstrated significant oligoclonality in patient II.1 (Fig. 4 B). The frequencies of unique TCR rearrangements in this patient were also markedly increased, with a resultant clonality score of 0.3224. In contrast, before CMV infection, patient II.2 demonstrated a TCR rearrangement frequency distribution similar to that of age-matched healthy controls, with a clonality score of 0.0012–0.002 (Fig. 4 B). This suggests that the oligoclonal expansion observed in patient II.1 is likely related to the uncontrolled CMV infection.
Clinical outcomes of siblings with hypomorphic IL2RB mutation
Due to the severe and recalcitrant nature of their disease manifestations, both patients underwent myeloablative HSCT. Following HSCT, patient II.1 had complete resolution of his chronic diarrhea, bloody stools, severe eczema, pulmonary infiltrates, and CMV viremia. Consistent with this clinical improvement, patient II.1 regained normal IL-2Rβ surface protein expression in CD4+ T and NK cells (Fig. 4 C). In vitro stimulation studies also showed complete recovery of pSTAT5 levels in response to IL-2 and IL-15c (Fig. 4 D). Moreover, CD56bright and CD56bright NK cell subset frequencies and T cell memory phenotype skewing normalized after transplant (Fig. 4, E and F), validating HSCT as a likely curative treatment for this disease. Chimerism evaluation demonstrated robust lymphoid engraftment at the time of these studies, with poor myeloid reconstitution (Fig. 4 G). After failed HSCT at 8 mo of age, patient II.2 succumbed to complications of thrombotic microangiopathy and eventual multiorgan failure.
In summary, we have identified a novel primary immunodeficiency disease caused by the first described human IL2RB mutation, revealing previously unappreciated insights into IL-2/15 signaling and NK cell biology. The IL2RB mutation leads to a deletion in the WSXWS motif, resulting in reduced IL-2Rβ expression and dysregulated downstream signaling. As might be expected for a defect in IL-2 signaling, this mutation clinically mirrors an IPEX-like syndrome. Additionally, as with IL-2Rα deficiency, these IL-2Rβ–deficient patients demonstrated decreased T reg frequency, skewing toward memory T cells, and lymphocytic infiltration into multiple tissues. Clinically, both IL-2Rα and IL-2Rβ defects fail to control CMV infection, yet the distinct cytokine signaling consequences underlying these defects suggest a critical role for mature NK cells in mediating this control (Caudy et al., 2007; Goudy et al., 2013). Like IL-2Rα–deficient patients and those treated with anti–IL-2Rα therapy, the altered IL-2/15 signaling in IL-2Rβ–deficient patients leads to a selective expansion of the immature CD56bright NK cell subset, and consequently a lower frequency of terminally differentiated CD57+ NK cells.
The excess plasma IL-2/15 cytokines in IL-2Rβ–deficient patients may explain the counterintuitive overrepresentation of both CD8+ T cells (II.1) and NK cells (II.1 and II.2). In a setting in which receptor signaling is compromised but ligand availability is high, cell types with the highest receptor levels (memory CD8+ T and NK) would be able to achieve the necessary signaling threshold to expand and survive. The fact that both the plasma IL-2/15 cytokine levels and the distribution of NK cell populations normalized in the patient after successful HSCT is consistent with this prediction.
We describe the first known human IL-2Rβ defect as the molecular basis of a novel immune dysregulation syndrome. With this study, we not only broaden the spectrum of monogenic PIDs, but also expand our understanding of IL-2/15 receptor biology. While there are ∼50 PIDs that have some impact on NK cells (five of which affect primarily NK cells), none result in an expansion of CD56bright NK cells with resultant autoimmunity and immunodeficiency (Mace and Orange, 2016). Our findings highlight the critical role of IL-2/15 signaling in T and NK cell development and antiviral immunity and provide insight into the mechanisms that regulate the intricate interface between self-tolerance and host immunity.
Materials and methods
Study approval
Human samples were obtained from the Allergy and Immunology Clinic at Children’s Hospital Colorado. Age-appropriate consent and assent was obtained. All human donors were enrolled under study protocol 16-0918, approved by the Institutional Review Board of the Research Compliance Office at the University of Colorado.
Whole-exome sequencing
Whole-exome sequencing was performed in patient II.1 and his parents using the Agilent SureSelect Clinical Research Exome kit. Sequence reads were aligned to the reference human genome (GRCh37) using Novoalign (V3.03.01; http://www.novocraft.com). Variants were called with HaplotypeCaller using GATK’s best practices. Single nucleotide variants and insertion/deletions were functionally annotated with SnpEff (http://snpeff.sourceforge.net) and filtered to retain only moderate- and high-effect variants. A total of 343 nonsynonymous nonsense and frameshift variants were identified with a minor allele frequency <0.1%. A novel homozygous recessive disruptive in-frame deletion in IL2RB (NM_000878.3; p.Pro222_Ser224del) was identified in patient II.1, resulting in the deletion of three amino acids. Both parents were heterozygous for the variant. Sanger sequencing confirmed the variant in patients II.1, II.2, and their parents. A region flanking the predicted mutation was sequenced by Eton Bioscience using the forward primer 5′-GTGTTCCTGCAGTTGATCAGC-3′ and the reverse primer 5′-CTCCCTCCAAGTTGTCCACG-3′. Sequence alignment image was generated using Geneious software (Biomatters).
IL2RB qPCR
RNA was isolated from PAXgene Blood RNA tubes (PreAnalytix), and cDNA was created with GoScript Reverse Transcription System (Promega) according to the manufacturer’s recommendations. The following probes were designed and ordered from Integrated DNA Technologies: CD122FWD, 5′-TCCCAGTTCACATGCTTCTAC-3′, PBR/56-FAM, 5′-AACATCTCCTGTGTCTGGAGCCAA-3′, and REV 5′-CACTTCCTGCCAAGTCCAT-3′. For Actin beta gene, Hs.PT.39a.22214847 primers and probe were used. Whole-exome sequencing data have been desposited in the European Genome-Phenome Archive with accession ID EGAS00001003599.
IL-2Rβ 3D modeling
IL-2Rβ was modeled from the crystal structure of heterotrimeric IL-2R in complex with IL-2 (PDB: 2ERJ; Stauber et al., 2006). Residues 6–209 from chain β of the crystal structure were used to model the extracellular domain. The transmembrane residues 215–239 were modeled as an α-helix using the Molefacture plugin in Visual Molecular Dynamics (Humphrey et al., 1996). The extracellular domain crystal structure, together with the modeled α-helix, were then used as templates to model residues 6–244 with MODELLER (Eswar et al., 2006). The resulting model was embedded in a mammalian membrane with a composition based on rat liver plasma membrane (Keenan and Morré, 1970). The Solvate and Autoionize plugins in VMD were used to solvate the system and add NaCl to a concentration of 150 mM. The total system size is 101,100 atoms. To model the deletion of residues 196–198, two additional systems were constructed by shifting the residues immediately above or below in sequence into the positions vacated by 196–198. Each of the three systems was simulated for 100 ns using Nanoscale Molecular Dynamics (Phillips et al., 2005).
Flow cytometry analysis
PBMCs were stained with anti–CD3-BV510 (BioLegend; 300435), anti–CD8-APC (BD; 555369), anti–CD4-PacBlue (BD; 558116), anti–CD19-APC-Cy7 (BD; 561743), and anti–CD56-PE (BioLegend; 318306). Anti–IL-2Rβ-FITC (clone TU27; Invitrogen; 11-1228-42), anti–IL-2Rβ-BV515 (clone Mik-Bta3; BD; 564688), or mouse IgG1 isotype controls (BD; 554679 or 564416) were applied either before (surface) or after (total) permeabilization. Permeabilization was performed with BD Cytofix/Cytoperm and Perm/Wash buffer (BD; 55471), and Foxp3 staining was performed after treatment with Transcription Factor Staining Buffer Set (eBioscience; 00-5523-00). For pSTAT5 analysis, fresh PMBCs were stimulated with IL-2 (Peprotech; 200-02, 0.1, 1.0, or 10 µg/ml), IL-15c (IL-15 (Peprotech; 200-15, 0.1, 10, or 1,000 ng/ml) complexed with IL-15Rα (R&D Systems; 147-IR); or IL-7 (Peprotech; 200-07, 1 µg/ml), for 15 or 30 min at room temperature, and fixed with 1.6% paraformaldehyde (PFA; Thermo Fisher Scientific; 28908). Cells were surface stained with anti–CD4-PacBlue (BD; 558116), anti–CD3-BV570 (BioLegend; 300435), anti–CD16-BV711 (BD; 563127), anti–CD56-PE (BioLegend; 318306), and anti-CD45Ra-AF700 (BD; 560673). Following permeabilization with ice-cold MeOH for 8 min, cells were stained with anti–pSTAT5-AF647 (BD; 612599) or mouse IgG1-AF647 isotype control (Invitrogen; MA5-181868), anti–pSTAT1-AF488 (BD; 612596), and anti–pSTAT6-PECy7 (BioLegend; 686013). For survival analysis, PBMCs (II.1 at age 18 mo and 2 yr, II.2 at 4 and 6 mo) were treated with IL-2 (100 pg/ml) and/or IL-15 (300 pg/ml) complexed with IL-15Rα (300 pg/ml) for 110 h. Ghost Dye Red 780 (Tonbo Biosciences; 13-0865-T100), Annexin V FITC (BD; 556419), and 7-AAD (Thermo Fisher Scientific; 00-6993-50) were added to determine live/dead. The total live number of cells was calculated based on Ghost Dye staining. To assess the percentage of live, early apoptotic, and dead NK cells, Annexin V−7-AAD−, Annexin V+7-AAD−, and Annexin V+7-AAD+ were assessed respectively. Proliferation was induced using different concentrations of anti-CD3 (BD; 555336) and anti-CD28 (2.5 µg/ml; BioLegend; 302914). Proliferation analysis was performed with CellTrace Violet (Thermo Fisher Scientific; C34557) according to the manufacturer’s protocol. Data were acquired in a BD LSR X20 flow cytometer (BD Biosciences) except the proliferation assay, which was acquired in a CytoFLEX flow cytometer (Beckman Coulter). All data were analyzed with FlowJo software (TreeStar).
Immunofluorescence and SRRF microscopy
PMBCs were fixed using 4% PFA and then blocked using 5% goat serum in PBS and stained with anti–CD56-PE (BioLegend; 318306) and anti–IL-2Rβ-APC (clone TU27; BioLegend; 339007). Cells were seeded onto poly-l-lysine coverslips (neuVitro GG-22-1.5-pll) and allowed to adhere for 1 d. Coverslips were mounted using ProLong Gold antifade reagent (Thermo Fisher Scientific; P36930), and slides were cured for 24 h before imaging. Samples were imaged using the HILO illumination mode in a Zeiss Elyra P.1 inverted microscope system using a 63× TIRF objective and additional 1.6× magnification. For SRRF imaging, 100 frames were acquired with a 22-ms exposure time per frame and a 640-nm laser operating at 2% laser power. Images were imported to Fiji, and the SRRF algorithm was applied using the “NanoJ-SRRF” Fiji plugin as previously described (Gustafsson et al., 2016). The number of IL-2Rβ clusters was detected using the Analyze Particles function in Fiji, with an area of 25 pixels squared considered permissive. Data from 30 cells per sample were plotted with GraphPad Prism.
Serological assessments
Levels of IL-2 and IL-15 complex (IL-15 complexed with IL-15Rα) were analyzed using commercially available IL-2 ELISA (Invitrogen; 88-7025-22) and IL-15 complex ELISA (R&D Systems; DY6924), according to the manufacturer’s protocol. Cytokine plasma concentrations were quantified by electrochemiluminescence immunoassay using the U-Plex human proinflammatory Combo 1 panel MSD (Meso Scale Discovery) plates according to each manufacturer’s instructions. For ELISPOT immunoprecipitation, sterile clear plates (Millipore; MAIPS4510) were coated with anti-IFNγ monoclonal antibody (Thermo Fisher Scientific; M700A, 1:1,000 in PBS). PBMCs were plated at 100 µl/well (200,000 cells/well). Peptides were diluted in complete medium for final concentration of 2 µg/ml per well (JPT Innovative Peptide Solutions; PepMix HCMVA PM-PP65-1, and PepMix EBV PM-EBV-BARF1). In some wells, IL-15/IL-15Rα complex (IL-15 from Peprotech; 200-15, complexed with IL-15Rα from R&D Systems; 147-IR, 300 pg/ml) and IL-2 (Peprotech; 200-02, 100 pg/ml) were added and biotinylated anti-human IFNγ polyclonal antibody (Thermo Fisher Scientific; M701B, 1:1,000 in PBS/4% BSA) was added. Plates were scanned using an ELISPOT reader (Cellular Technology Limited; S6 Universal Analyzer) and analyzed using Cellular Technology Limited 5.2.12 Professional Analyzer software.
NK cell degranulation and cytokine production
Thawed PBMCs were plated at a final concentration of 5 × 105 cells/ml in 96-well deep-well-bottom plates. These cells were mixed with K562 target cells at an effector-to-target ratio of 1:1 in the presence of protein transport inhibitor cocktail (Thermo Fisher Scientific; 00498003) and anti–CD107a-APC-CY7 (BioLegend; 328630). IL-12 (10 ng/ml), IL-15 (10 ng/ml), and IL-18 (100 ng/ml) were added as a positive control for IFNγ production. Cells were incubated for 4 h at 37°C. For ADCC assay, Raji cells were first coated with anti-CD20 mAb (nonfucosylated Rituximab; InvivoGen) for 15 min at room temperature. PBMCs were then mixed with Raji target cells at an effector-to-target ratio of 1:1. Cells were then incubated for 4 h at 37°C in the presence of protein transport inhibitor cocktail (Thermo Fisher Scientific; 00498003) and anti–CD107a-APC-CY7 (BioLegend; 328630). After incubation, cells were washed and stained for extracellular receptors, permeabilized (fixation/permeabilization buffer; eBioscience), and stained for anti–CD56-PE (BioLegend; 318306), anti–NKG2D-PE Dazzle 594 (BioLegend; 320828), anti–CD3-BV711 (BioLegend; 325218), anti–DNAM-PE-CY7 (BioLegend; 338316), anti–CD16-BV605 (BioLegend; 302039), anti–CD57-PERCP-CY5.5 (BioLegend; 359622), anti-PLZF (BD; 563490), anti–FcεRI-FITC (Millipore; FCABS400), anti–IFNγ-BV421 (BioLegend; 502520), and Live/Dead Fixable aqua dead cell staining (Thermo Fisher Scientific; L34965).
Mass cytometry (cytometry time-of-flight) analysis
Whole blood was collected into heparinized vacutainers for separation of plasma and PBMCs using Ficoll Paque Plus (GE Healthcare; 17-1440-03). For pSTAT1 and pSTAT3 responses, cells were resuspended in RPMI and stimulated with 40 KU/ml IFN-α (PBL; 11101-02). Cells were fixed with 1.6% PFA (Thermo Fisher Scientific; 28908) before staining. Fixed cells were stored at −80°C and thawed on the day of barcoding and/or staining. NK analysis was performed with the following antibodies:CD57-Y/89 (HD57); HLADR-Cd/112 (Tu36); CD3-In/153 (UCHT); CD38-Pr/141 (HIT2); CD69-Nd/142 (FN50); CD33-Nd/143 (WM53); CD14-Nd/143 (M5E5); CD2 Nd/144 (RPA-2.10); LILRB1-Nd/145 (GHI/75); CD19-Nd/146 (HIB19); CD8-Sm/147 (SK1); CD4-Sm/149 (SK3); CD62L-Eu/151 (DREG-56); KIR2DS4-Eu/153 (179315); KIR2DS2-Sm/152 (polyclonal); NKp46-G/155 (9E2); NKG2D-Gd/156 (1D11); TIGIT-Gd/157 (741182); 2B4-Gd/158 (C1.7); DNAM-1-Tb/159 (DX11); FAS-L-Gd/160 (NOK-1); Nkp30-Dy/161 (P30-15); Siglec-7-Dy/162 (S7.7); NKG2C-Dy163 (134522); NKp44-Dy164 (P44-8); CD96-Ho/165 (NK92.39); KIR2DL1-Er/166 (143211); CD94-Er/167 (DX22); CXCR6-Er168 (K041E5); PD1-Tm/169 (EH12.2H7); KIR2DL5-Er/170 (UP-R1); NKG2A-Yb/171 (131411); NTB-A-Tb/172 (NT-7); KIR3DL1-Yb/173 (DX-9); CD56-Tb/174 (NCAM16.2); KIR2DL3-Lu/175 (180701); CD16-Bi/209 (3G8); FCRγ-Sm/152 (polyclonal); Syk-Nd/150 (4D10.2); Ki67-Yb/176 (Ki-67); and Perforin-Yb/176 (B-D48). Immunophenotyping was performed with the following antibodies: CD45-Y/89 (HI30; BioLegend); CD3-Nd/144 (UCHT1; BD); CD45Ra-Nd/153 (HI100; BioLegend); CCR7-Sm/152 (G043H7; BD), CD14-Sm/154 (M5E2; BioLegend); CD16-Eu/153 (B73.1; eBioscience); CD4-Gd/155 (SK3; BioLegend), CD8-Nd/142 (SK1; BioLegend); CD1c-Dy/161 (L161; BioLegend); CD27-Yb/162 (L128; BD); CD19-Dy/163 (SJ25C1; Santa Cruz); CD11b-Sm/209 (B-ly6; BioLegend); HLADR-Er/170 (L243; BioLegend); pSTAT1-Yb/173 (58D6; CST); and pSTAT3-Gd/158 (4/pSTAT-3; Fluidigm). Barcoding methodology was adapted from Zunder et al. (2015). Data were acquired on a Helios instrument (Fluidigm). To make all samples maximally comparable, data were acquired using internal metal isotope bead standards and normalized as previously described (Finck et al., 2013). Files were debarcoded using the Matlab Debarcoder Tool (Zunder et al., 2015). Data were analyzed with FlowJo and Cytobank. For viSNE map generation, equal cell numbers from every control and patient sample were analyzed together to allow for between-map comparisons (Fig. 2 B).
TCR sequencing
DNA was isolated from whole-blood samples with a DNeasy Blood & Tissue Kit (Qiagen; 69504). DNA was sent to Adaptive Biotechnologies for deep-resolution sequencing of the β-chain using the ImmunoSEQ platform.
Statistics
Statistical analysis was performed with Prism (GraphPad Software). The Shapiro–Wilk test was used to determine normal distribution of the samples. Healthy control subjects and patients were systematically analyzed as two groups by using Mann–Whitney U tests unless otherwise stated.
Online supplemental material
Fig. S1 presents additional immunophenotypic data for patients II.1 and II.2. Additional characterization of protein expression, serum cytokine levels, signaling, and cell survival for this IL2RB mutation is provided in Fig. S2. The list of variants of unknown significance from the whole-exome sequencing report for patient II.1 is shown in Table S1.
Supplementary Material
Acknowledgments
We thank the patients and their family. We are also grateful to Jared Klarquist for his intellectual input and helpful comments. We also thank Kimberly Jordan and Jennifer McWilliams from the Human Immune Monitoring Shared Resource for their assistance with ELISPOTs.
E. Vendrame is supported by National Institutes of Health (NIH) grants T32AI007502 and IL1TR001084. C. Galambos is supported by the Jayden DeLuca Foundation, The Linda Crnic Institute for Down Syndrome, and NIH grant 2R56HL068702-13. J.C. Gumbart is supported by NIH grant R01GM123169 and Georgia Tech through a Cullen-Peck Fellowship. Simulations were run on the Extreme Science and Engineering Discovery Environment, supported by National Science Foundation grant ACI-1548562. M.R. Verneris is supported by National Institute of Allergy and Infectious Diseases grant R01AI100879. J.R. Kelsen is supported by National Institutes of Diabetes and Digestive and Kidney Diseases grant K23DK100461. K.E. Sullivan is supported by the Wallace Chair of Pediatrics. S. Ogolla and R. Rochford are supported by NIH grant R01 CA102667. C.A. Blish is a John and Tashia Morgridge Faculty Scholar in Pediatric Translational Medicine from the Stanford Maternal Child Health Institute. C.A. Blish is also funded by the Burroughs Wellcome Fund Investigators in the Pathogenesis of Infectious Diseases and NIH grant U01AI13130201. R.M. Kedl and I.Z. Fernandez are supported by National Institute of Allergy and Infectious Diseases grants AI101205, AI126899, and T32AI007405. R.M. Baxter, J.E. Garcia-Perez, and E.W.Y. Hsieh are supported by the National Institute of Arthritis and Musculoskeletal and Skin Diseases grant K23AR070897, the Boettcher Foundation Webb-Waring Biomedical research grant, and the Jeffrey Model Translational Research award.
The authors declare no competing financial interests.
Author contributions: I.Z. Fernandez, R.M. Baxter, R.M. Kedl, C.M. Dutmer, and E.W.Y. Hsieh enrolled the patients on study, conceived the study, developed the reagents, performed experiments, analyzed data, and wrote the manuscript. D.S. Kong and T. Ranganath performed experiments. J.E. Garcia-Perez performed experiments and assisted in data analysis. E. Vendrame and T. Ranganath performed mass cytometry experiments. S. Ogolla, R. Quinones, H. Eissa, M.R. Verneris, and R. Rochford assisted with acquisition of pediatric age samples. H. Eissa, M.R. Verneris, and R. Quinones supervised the HSCT for both patients. E.F. de Zoeten identified the index patient, enrolled him into the Children’s Hospital of Philadelphia Very Early Onset Inflammatory Bowel Disease study for whole-exome sequencing, and provided clinical care and insights into the clinical and immunological phenotypes. J. Black and C. Galambos provided histopathology studies and images. K. Lundquist and J.C. Gumbart performed 3D in silico protein prediction models. N. Dawany, J.R. Kelsen, and K.E. Sullivan performed and analyzed whole-exome sequencing data. C.A. Blish provided intellectual guidance on this project and contributed to manuscript writing. All authors read and approved the manuscript prior to submission.
References
- Alroqi F.J., and Chatila T.A.. 2016. T Regulatory Cell Biology in Health and Disease. Curr. Allergy Asthma Rep. 16:27 10.1007/s11882-016-0606-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amir A.D., Davis K.L., Tadmor M.D., Simonds E.F., Levine J.H., Bendall S.C., Shenfeld D.K., Krishnaswamy S., Nolan G.P., and Pe’er D.. 2013. viSNE enables visualization of high dimensional single-cell data and reveals phenotypic heterogeneity of leukemia. Nat. Biotechnol. 31:545–552. 10.1038/nbt.2594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumgartner J.W., Wells C.A., Chen C.M., and Waters M.J.. 1994. The role of the WSXWS equivalent motif in growth hormone receptor function. J. Biol. Chem. 269:29094–29101. [PubMed] [Google Scholar]
- Bennett C.L., Christie J., Ramsdell F., Brunkow M.E., Ferguson P.J., Whitesell L., Kelly T.E., Saulsbury F.T., Chance P.F., and Ochs H.D.. 2001. The immune dysregulation, polyendocrinopathy, enteropathy, X-linked syndrome (IPEX) is caused by mutations of FOXP3. Nat. Genet. 27:20–21. 10.1038/83713 [DOI] [PubMed] [Google Scholar]
- Bernasconi A., Marino R., Ribas A., Rossi J., Ciaccio M., Oleastro M., Ornani A., Paz R., Rivarola M.A., Zelazko M., and Belgorosky A.. 2006. Characterization of immunodeficiency in a patient with growth hormone insensitivity secondary to a novel STAT5b gene mutation. Pediatrics. 118:e1584–e1592. 10.1542/peds.2005-2882 [DOI] [PubMed] [Google Scholar]
- Caudy A.A., Reddy S.T., Chatila T., Atkinson J.P., and Verbsky J.W.. 2007. CD25 deficiency causes an immune dysregulation, polyendocrinopathy, enteropathy, X-linked-like syndrome, and defective IL-10 expression from CD4 lymphocytes. J. Allergy Clin. Immunol. 119:482–487. 10.1016/j.jaci.2006.10.007 [DOI] [PubMed] [Google Scholar]
- Dagil R., Knudsen M.J., Olsen J.G., O’Shea C., Franzmann M., Goffin V., Teilum K., Breinholt J., and Kragelund B.B.. 2012. The WSXWS motif in cytokine receptors is a molecular switch involved in receptor activation: insight from structures of the prolactin receptor. Structure. 20:270–282. 10.1016/j.str.2011.12.010 [DOI] [PubMed] [Google Scholar]
- De Maria A., Bozzano F., Cantoni C., and Moretta L.. 2011. Revisiting human natural killer cell subset function revealed cytolytic CD56(dim)CD16+ NK cells as rapid producers of abundant IFN-gamma on activation. Proc. Natl. Acad. Sci. USA. 108:728–732. 10.1073/pnas.1012356108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eswar N., Webb B., Marti-Renom M.A., Madhusudhan M.S., Eramian D., Shen M.Y., Pieper U., and Sali A.. 2006. Comparative protein structure modeling using Modeller. Curr. Protoc. Bioinformatics. Chapter 5:Unit-5.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finck R., Simonds E.F., Jager A., Krishnaswamy S., Sachs K., Fantl W., Pe’er D., Nolan G.P., and Bendall S.C.. 2013. Normalization of mass cytometry data with bead standards. Cytometry A. 83:483–494. 10.1002/cyto.a.22271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmour K.C., Fujii H., Cranston T., Davies E.G., Kinnon C., and Gaspar H.B.. 2001. Defective expression of the interleukin-2/interleukin-15 receptor beta subunit leads to a natural killer cell-deficient form of severe combined immunodeficiency. Blood. 98:877–879. 10.1182/blood.V98.3.877 [DOI] [PubMed] [Google Scholar]
- Goudy K., Aydin D., Barzaghi F., Gambineri E., Vignoli M., Ciullini Mannurita S., Doglioni C., Ponzoni M., Cicalese M.P., Assanelli A., et al. . 2013. Human IL2RA null mutation mediates immunodeficiency with lymphoproliferation and autoimmunity. Clin. Immunol. 146:248–261. 10.1016/j.clim.2013.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gumá M., Angulo A., Vilches C., Gómez-Lozano N., Malats N., and López-Botet M.. 2004. Imprint of human cytomegalovirus infection on the NK cell receptor repertoire. Blood. 104:3664–3671. 10.1182/blood-2004-05-2058 [DOI] [PubMed] [Google Scholar]
- Gustafsson N., Culley S., Ashdown G., Owen D.M., Pereira P.M., and Henriques R.. 2016. Fast live-cell conventional fluorophore nanoscopy with ImageJ through super-resolution radial fluctuations. Nat. Commun. 7:12471 10.1038/ncomms12471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilton D.J., Watowich S.S., Katz L., and Lodish H.F.. 1996. Saturation mutagenesis of the WSXWS motif of the erythropoietin receptor. J. Biol. Chem. 271:4699–4708. 10.1074/jbc.271.9.4699 [DOI] [PubMed] [Google Scholar]
- Humphrey W., Dalke A., and Schulten K.. 1996. VMD: visual molecular dynamics. J. Mol. Graph. 14:33–38: 27–28. 10.1016/0263-7855(96)00018-5 [DOI] [PubMed] [Google Scholar]
- Keenan T.W., and Morré D.J.. 1970. Phospholipid class and fatty acid composition of golgi apparatus isolated from rat liver and comparison with other cell fractions. Biochemistry. 9:19–25. 10.1021/bi00803a003 [DOI] [PubMed] [Google Scholar]
- Kofoed E.M., Hwa V., Little B., Woods K.A., Buckway C.K., Tsubaki J., Pratt K.L., Bezrodnik L., Jasper H., Tepper A., et al. . 2003. Growth hormone insensitivity associated with a STAT5b mutation. N. Engl. J. Med. 349:1139–1147. 10.1056/NEJMoa022926 [DOI] [PubMed] [Google Scholar]
- Lopez-Vergès S., Milush J.M., Schwartz B.S., Pando M.J., Jarjoura J., York V.A., Houchins J.P., Miller S., Kang S.M., Norris P.J., et al. . 2011. Expansion of a unique CD57+NKG2Chi natural killer cell subset during acute human cytomegalovirus infection. Proc. Natl. Acad. Sci. USA. 108:14725–14732. 10.1073/pnas.1110900108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macchi P., Villa A., Giliani S., Sacco M.G., Frattini A., Porta F., Ugazio A.G., Johnston J.A., Candotti F., O’Shea J.J., et al. . 1995. Mutations of Jak-3 gene in patients with autosomal severe combined immune deficiency (SCID). Nature. 377:65–68. 10.1038/377065a0 [DOI] [PubMed] [Google Scholar]
- Mace E.M., and Orange J.S.. 2016. Genetic Causes of Human NK Cell Deficiency and Their Effect on NK Cell Subsets. Front. Immunol. 7:545 10.3389/fimmu.2016.00545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muccio L., Bertaina A., Falco M., Pende D., Meazza R., Lopez-Botet M., Moretta L., Locatelli F., Moretta A., and Della Chiesa M.. 2016. Analysis of memory-like natural killer cells in human cytomegalovirus-infected children undergoing αβ+T and B cell-depleted hematopoietic stem cell transplantation for hematological malignancies. Haematologica. 101:371–381. 10.3324/haematol.2015.134155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguchi M., Yi H., Rosenblatt H.M., Filipovich A.H., Adelstein S., Modi W.S., McBride O.W., and Leonard W.J.. 1993. Interleukin-2 receptor gamma chain mutation results in X-linked severe combined immunodeficiency in humans. Cell. 73:147–157. 10.1016/0092-8674(93)90167-O [DOI] [PubMed] [Google Scholar]
- Phillips J.C., Braun R., Wang W., Gumbart J., Tajkhorshid E., Villa E., Chipot C., Skeel R.D., Kalé L., and Schulten K.. 2005. Scalable molecular dynamics with NAMD. J. Comput. Chem. 26:1781–1802. 10.1002/jcc.20289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell B.R., Buist N.R., and Stenzel P.. 1982. An X-linked syndrome of diarrhea, polyendocrinopathy, and fatal infection in infancy. J. Pediatr. 100:731–737. 10.1016/S0022-3476(82)80573-8 [DOI] [PubMed] [Google Scholar]
- Roifman C.M. 2000. Human IL-2 receptor alpha chain deficiency. Pediatr. Res. 48:6–11. 10.1203/00006450-200007000-00004 [DOI] [PubMed] [Google Scholar]
- Russell S.M., Tayebi N., Nakajima H., Riedy M.C., Roberts J.L., Aman M.J., Migone T.S., Noguchi M., Markert M.L., Buckley R.H., et al. . 1995. Mutation of Jak3 in a patient with SCID: essential role of Jak3 in lymphoid development. Science. 270:797–800. 10.1126/science.270.5237.797 [DOI] [PubMed] [Google Scholar]
- Stauber D.J., Debler E.W., Horton P.A., Smith K.A., and Wilson I.A.. 2006. Crystal structure of the IL-2 signaling complex: paradigm for a heterotrimeric cytokine receptor. Proc. Natl. Acad. Sci. USA. 103:2788–2793. 10.1073/pnas.0511161103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki H., Kündig T.M., Furlonger C., Wakeham A., Timms E., Matsuyama T., Schmits R., Simard J.J., Ohashi P.S., Griesser H., et al. . 1995. Deregulated T cell activation and autoimmunity in mice lacking interleukin-2 receptor beta. Science. 268:1472–1476. 10.1126/science.7770771 [DOI] [PubMed] [Google Scholar]
- Waldmann T.A. 2006. The biology of interleukin-2 and interleukin-15: implications for cancer therapy and vaccine design. Nat. Rev. Immunol. 6:595–601. 10.1038/nri1901 [DOI] [PubMed] [Google Scholar]
- Wang X., Rickert M., and Garcia K.C.. 2005. Structure of the quaternary complex of interleukin-2 with its alpha, beta, and gammac receptors. Science. 310:1159–1163. 10.1126/science.1117893 [DOI] [PubMed] [Google Scholar]
- Yoshimura A., Zimmers T., Neumann D., Longmore G., Yoshimura Y., and Lodish H.F.. 1992. Mutations in the Trp-Ser-X-Trp-Ser motif of the erythropoietin receptor abolish processing, ligand binding, and activation of the receptor. J. Biol. Chem. 267:11619–11625. [PubMed] [Google Scholar]
- Zhang T., Scott J.M., Hwang I., and Kim S.. 2013. Cutting edge: antibody-dependent memory-like NK cells distinguished by FcRγ deficiency. J. Immunol. 190:1402–1406. 10.4049/jimmunol.1203034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zunder E.R., Finck R., Behbehani G.K., Amir A.D., Krishnaswamy S., Gonzalez V.D., Lorang C.G., Bjornson Z., Spitzer M.H., Bodenmiller B., et al. . 2015. Palladium-based mass tag cell barcoding with a doublet-filtering scheme and single-cell deconvolution algorithm. Nat. Protoc. 10:316–333. 10.1038/nprot.2015.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.