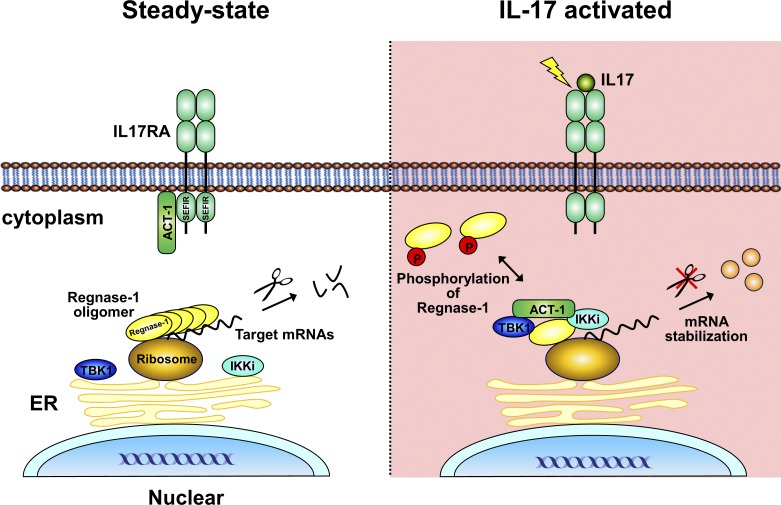
The endoribonuclease Regnase-1 suppresses inflammation through RNA degradation. Here, we show that Regnase-1 is phosphorylated and inactivated by the Act1-TBK1-IKKi axis during IL-17 stimulation. Moreover, this phosphorylation substantially contributes to the mRNA stabilization needed for amplification of TH17-cell–mediated inflammation.
Abstract
Regnase-1 (also known as Zc3h12a or MCPIP-1) is an endoribonuclease involved in mRNA degradation of inflammation-associated genes. Regnase-1 is inactivated in response to external stimuli through post-translational modifications including phosphorylation, yet the precise role of phosphorylation remains unknown. Here, we demonstrate that interleukin (IL)-17 induces phosphorylation of Regnase-1 in an Act1-TBK1/IKKi–dependent manner, especially in nonhematopoietic cells. Phosphorylated Regnase-1 is released from the endoplasmic reticulum (ER) into the cytosol, thereby losing its mRNA degradation function, which leads to expression of IL-17 target genes. By using CRISPR/Cas-9 technology, we generated Regnase-1 mutant mice, in which IL-17–induced Regnase-1 phosphorylation is completely blocked. Mutant mice (Regnase-1AA/AA and Regnase-1ΔCTD/ΔCTD) were resistant to the IL-17–mediated inflammation caused by T helper 17 (Th17) cells in vivo. Thus, Regnase-1 plays a critical role in the development of IL-17–mediated inflammatory diseases via the Act1-TBK1-IKKi axis, and blockade of Regnase-1 phosphorylation sites may be promising for treatment of Th17-associated diseases.
Graphical Abstract
Introduction
The immune system combats microorganisms by inducing expression of a variety of cytokines and cell surface molecules, as well as a myriad of antimicrobial proteins and substances (Beutler, 2009; Takeuchi and Akira, 2010; Iwasaki and Medzhitov, 2015). In response to various external stimuli, multiple inflammation-related genes are induced; these are tightly controlled in a rapid, coordinated, and transient fashion. It is becoming clear that mRNA stability/instability contributes to rapid regulation of such gene products.
The LPS-inducible protein Regnase-1, also known as Zc3h12a and MCPIP-1 (Liang et al., 2008), comprises a protein family together with Regnase-2, -3, and -4. These proteins contain a single Cys-Cys-Cys-His (CCCH)-type zinc finger domain and a Pilt-N-terminus (PIN)-like domain, both of which may be involved in RNA recognition and binding (Jura et al., 2012; Akira, 2013). Regnase-1 (MCPIP-1 or ZC3H12a)–deficient mice develop spontaneous autoimmune diseases, accompanied by splenomegaly and lymphadenopathy (Matsushita et al., 2009; Liang et al., 2010). We previously showed that Regnase-1 is an endoribonuclease involved in the destabilization of interleukin (IL)-6 and IL-12 mRNA, as well as Regnase-1 mRNA itself, via the stem-loop structure present in the 3′-untranslated region (UTR) of these genes (Matsushita et al., 2009). Regnase-1 is present in a variety of cells (Mizgalska et al., 2009; Iwasaki et al., 2011), and its target mRNAs have been revealed in a variety of cell types; they include cytokines, chemokines, transcription factors, inflammation-associated genes, microRNAs, antiapoptotic genes, T cell surface markers, DNA demethylation genes, and several viral RNAs (Suzuki et al., 2011; Uehata et al., 2013; Garg et al., 2015; Mino et al., 2015; Kochan et al., 2016). In response to external stimuli that use MyD88 signaling, Regnase-1 protein is phosphorylated by IκB kinases (IKKs), followed by ubiquitin-dependent degradation (Iwasaki et al., 2011). Concomitantly, IKK-dependent activation of NF-κB induces expression of a variety of inflammation-associated genes, including Regnase-1 mRNA (Liang et al., 2008; Iwasaki et al., 2011). Thus, Regnase-1 suppresses unnecessary inflammatory reactions under unstimulated conditions, whereas it acts as a negative feedback mechanism during an inflammatory response, suppressing an excessively protracted inflammatory reaction. We previously reported that the initial phosphorylation of Regnase-1 by IL-1 receptor–associated kinase (IRAK) 1 (IRAK1) is required for its subsequent IKK–mediated phosphorylation and proteasome–mediated degradation (Iwasaki et al., 2011). Recently, Regnase-1 and Roquin-1/2 were shown to be regulated by mucosa-associated lymphoid tissue lymphoma translocation protein 1 in response to TCR stimulation (Uehata et al., 2013; Jeltsch et al., 2014).
In this study, we generated two types of knock-in mice, in which IKK phosphorylation sites in Regnase-1 were blocked (Regnase-1 S435A/S439A [AA] mutant) and the C-terminal portion of Regnase-1 was deleted, thus resulting in a lack of phosphorylation (Regnase-1 ΔCTD), respectively. Using these mice, we assessed the role of phosphorylation sites within Regnase-1 in vivo. Both mutant mice showed resistance to experimental autoimmune encephalomyelitis (EAE), with higher resistance in Regnase-1 ΔCTD mice. Interestingly, IL-17 stimulation–induced phosphorylation of the Regnase-1 AA mutant but not of the Regnase-1 ΔCTD mutant. This phosphorylation was mediated by Act1-dependent TANK-binding kinase 1 (TBK1)/IKKi (or IKKi/IKK epsilon) activation and induced release of Regnase-1 from the ER. Phosphorylation of Regnase-1 by TBK1/IKKi and its release from the ER was sufficient to suppress the ability of Regnase-1 to destabilize mRNA. Our findings reveal the mechanism by which Regnase-1 controls the mRNA stability of inflammatory genes during IL-17 stimulation and provide clear evidence for the dominant role of Regnase-1 in IL-17–mediated inflammation.
Results
Regnase-1 S435A/S439A (AA) mutation prevents IKK–mediated phosphorylation and attenuates EAE disease severity
To determine how IKK–mediated phosphorylation and degradation of Regnase-1 control cytokine expression in the immune response, we generated knock-in mice harboring two amino acid substitutions of Ser435 and Ser439 with Ala in Regnase-1 protein (Fig. S1, A and B).
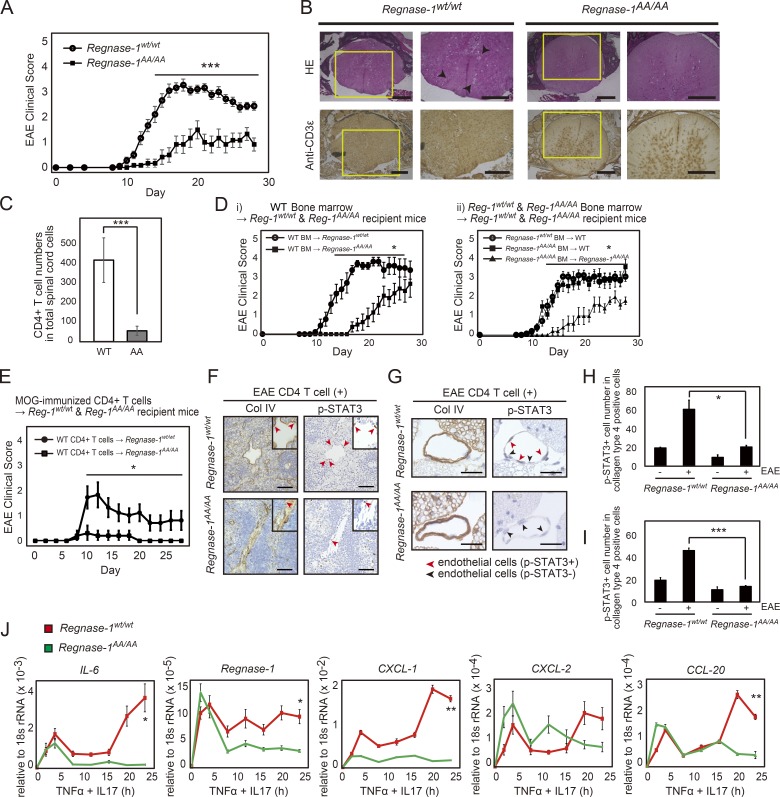
As expected, LPS-stimulated Regnase-1AA/AA macrophages did not demonstrate Regnase-1 protein degradation (Fig. S1 C). Mutant protein level was significantly elevated over time in Regnase-1AA/AA macrophages, compared with WT macrophages. Phosphorylation of Regnase-1 by IRAKs, detected as a lagging band in electrophoresis (Fig. S1 C), was regularly observed in Regnase-1AA/AA macrophages. Regnase-1AA/AA mice demonstrated delayed onset and slow progression of EAE compared with control mice (Fig. 1 A). Histological analysis of the spinal cord revealed a significant decrease in inflammation, demyelination, axon degeneration, and T cell infiltration, particularly CD4+ T cells into neuronal tissue in Regnase-1AA/AA mice (Fig. 1, B and C). The in vitro differentiation ability of naive CD4+ T cells into T helper (Th) 1 (Th1), Th17, or T regulatory (T reg) cells was not impaired in Regnase-1AA/AA mice (Fig. S2 A). Bone marrow chimeras suggested that the improvement in the EAE disease in Regnase-1AA/AA mice was caused by nonhematopoietic cells rather than immune cells (Fig. 1 D).
Figure 1.
Regnase-1 AA mutation attenuates EAE disease severity. (A) EAE clinical scores of WT (n = 15) and Regnase-1AA/AA (n = 12) mice. ***, P < 0.005. (B) Histological analysis of frozen sections stained with hematoxylin–eosin (HE; top row) and anti-CD3ε (bottom row). Arrows indicate inflammatory cellular infiltrates. Scale bars, 200 µm. (C) CD4+ T cell numbers (n = 3) in spinal cord cells (1.0 × 105 cells) at 15 d after immunization were analyzed by flow cytometry. ***, P < 0.005. (D) EAE clinical scores of chimeric mice generated by (i) transfer of WT bone marrow (BM) cells into WT (Reg-1wt/wt) or Regnase-1AA/AA (Reg-1AA/AA) mice (each n = 8) or (ii) transfer of WT or Regnase-1AA/AA bone marrow cells into WT mice (each n = 12) or Regnase-1AA/AA mice (each n = 6). *, P < 0.05. (E) EAE clinical scores of WT or Regnase-1AA/AA mice after intravenous transfer of pathogenic CD4+ T cells (each n = 5). *, P < 0.05. (F and G) Histological analysis of endothelial cell inflammation in the spleen (F) and the fifth lumbar vertebra of the spinal cord (G) at 12 h after transfer. Sections were stained with anti–type-IV collagen (Col IV) and anti-phospho-STAT3 (p-STAT3) antibodies. Red and black arrows indicate phospho-STAT3–positive and –negative cells in blood vessel endothelial cells, respectively. Scale bars, 50 µm. (H and I) The relative number of anti-phospho-STAT3–positive cells in F and G, respectively. Measurement was performed at three times. *, P < 0.05; ***, P < 0.005. (J) Quantitative PCR analysis of IL-6, Regnase-1, CXCL-1, CXCL2, and CCL-20 mRNA in WT and Regnase-1AA/AA MEFs. Cells were stimulated with TNF-α and IL-17A for 0–24 h. Data were collected from four independent experiments. *, P < 0.05; **, P < 0.01. rRNA, ribosomal RNA.
To further confirm this putative mechanism, we used a transfer EAE model, in which myelin oligodendrocyte glycoprotein (MOG)–specific autoreactive CD4+ T cells, containing a highly enriched and activated population of Th17 cells, were intravenously transferred into WT and Regnase-1AA/AA mice (Ogura et al., 2008; Arima et al., 2012). Disease progression was completely suppressed in Regnase-1AA/AA mice (Fig. 1 E). In this model, activated Th17 cells can induce signal transducer and activator of transcription 3 (STAT3) activation and inflammation in endothelial cells of the dorsal blood vessel in the fifth lumbar vertebra of the spinal cord in a manner dependent on IL-6 and IL-17, which are primarily secreted from Th17 cells. Immunohistological analysis of phosphorylated STAT3 in spleen and fifth lumbar vertebra endothelial cells (type IV collagen-positive) revealed a reduced number of phospho-STAT3 cells in Regnase-1AA/AA mice (Fig. 1, F–I). These findings reveal a critical role for IKK–mediated degradation of Regnase-1 in controlling the Th17 cell–associated inflammatory response.
IL-17 activates both NF-κB and MAPK signaling pathways but is a weak activator of NF-κB (Gaffen, 2009). In nonhematopoietic cells, cooperative stimulation of IL-6 and IL-17 leads to IL-6 overexpression in a feedback amplification loop of NF-κB and STAT3 activation (Ogura et al., 2008; Lee et al., 2012). We measured cytokine and chemokine production after stimulation with a combination of IL-17A and TNF-α or IL-6. In MEFs and liver sinusoidal endothelial cells (LSECs) from Regnase-1AA/AA mice, expression levels of IL-6, Regnase-1, CXCL-1, CXCL-2, and CCL-20 mRNA were reduced, particularly in later stages of stimulation, compared with expression levels in cells from WT mice (Fig. 1 J and Fig. S2 B). Protein levels of IL-6, CXCL-1, and CXCL-2 were also lower in LSECs from Regnase-1AA/AA mice (Fig. S2 C). Thus, IKK–mediated phosphorylation and degradation of Regnase-1 occur via NF-κB activation upon IL-17A stimulation in nonhematopoietic cells, resulting in the expression of inflammatory cytokine mRNA.
Regnase-1 is phosphorylated by TBK1 and IKKi in the IL-17 receptor (IL-17R) signaling pathway
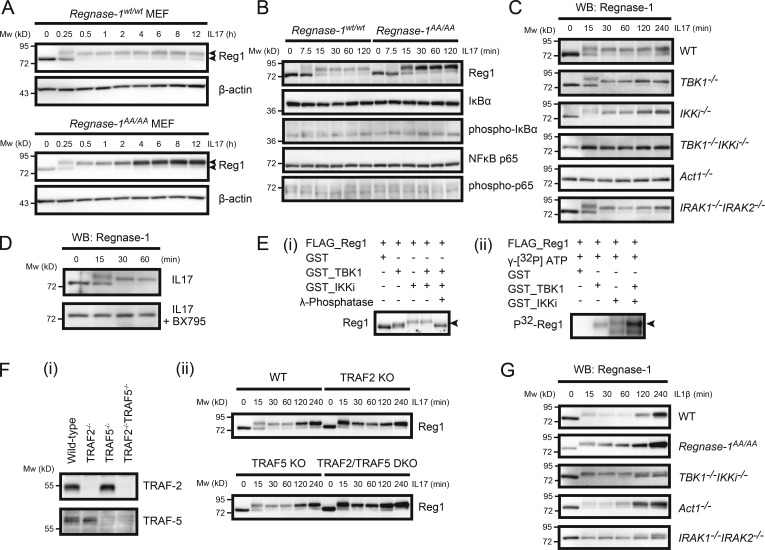
IL-17A treatment induced Regnase-1 phosphorylation, detected as a mobility-shifted band in electrophoresis (Fig. 2 A). In WT MEFs, the Regnase-1 phosphorylation pattern was similar to that observed in IRAK1- and IRAK2–mediated phosphorylation upon LPS or IL-1 stimulation (Iwasaki et al., 2011), although a dramatic disappearance of Regnase-1 did not occur during IL-17A stimulation (Fig. 2 A). IL-17A stimulation resulted in weak activation of NF-κB (Fig. 2 B), indicating that weak IKK activation in the IL-17A signaling pathway caused weak degradation of Regnase-1 during IL-17A stimulation. Indeed, the phosphorylated form of Regnase-1 AA mutant protein gradually accumulated in Regnase-1AA/AA MEFs during IL-17A stimulation.
Figure 2.
Regnase-1 phosphorylation by TBK1 and IKKi upon IL-17 stimulation. (A–D) Immunoblotting analysis of Regnase-1 (Reg1) and β-actin (A) and Regnase-1, IκB, phospho-IκB, NF-κB p65, and phospho-NF-κB p65 (B) in WT and Regnase-1AA/AA MEFs; (C) Regnase-1 in WT, TBK1-deficient, IKKi-deficient, TBK1 and IKKi double-deficient, Act1-deficient, and IRAK-1 and IRAK-2 double-deficient MEFs; (D) Regnase-1 in WT MEFs treated without (top) or with (bottom) BX795 (50 µM). MEFs were stimulated with IL-17A (50 ng/ml) for the indicated time. Two arrows in A indicate phosphorylated (top) and unphosphorylated (bottom) forms of Regnase-1. (E) In vitro phosphorylation of Regnase-1 by TBK1 and IKKi. Purified Regnase-1, obtained from Regnase-1–deficient MEFs expressing FLAG-tagged Regnase-1 AA mutant, was incubated with recombinant TBK1 and/or IKKi in the presence or absence of γ-phosphatase for 3 h. Regnase-1 phosphorylation was analyzed by immunoblotting (i) and [32P]-autoradiography (ii). The arrows indicate phosphorylated Regnase-1. (F) IL-17–induced Regnase-1 phosphorylation in WT, TRAF2-deficient, TRAF5-deficient, and TRAF2/TRAF5 double-deficient MEF cell lines. Immunoblotting analysis of TRAF-2 and TRAF-5 (i) and Regnase-1 (ii) in these MEF cell lines. MEFs were stimulated with IL-17A (50 ng/ml) for the indicated times. (G) Immunoblotting analysis of Regnase-1 expression in WT, Regnase-1AA/AA, TBK1/IKKi double-deficient, Act1-deficient, and IRAK1/IRAK2 double-deficient MEFs. Cells were stimulated with IL-1β for 0–240 min. Mw, molecular weight. WB, Western blotting.
To identify kinases responsible for the phosphorylation of Regnase-1 in the IL-17A signaling pathway, we examined a series of MEFs lacking individual kinases. We found that Regnase-1 phosphorylation did not occur in MEFs lacking both TBK1 and IKKi, or Act1, an adaptor protein indispensable for the IL-17R signaling pathway (Fig. 2 C). MEFs lacking either TBK1 or IKKi displayed Regnase-1 phosphorylation in response to IL-17A, indicating that TBK1 and IKKi have Regnase-1 kinase activity, independent of each other (Fig. 2 C). Although the phosphorylation pattern of Regnase-1 induced by IRAKs appeared to be similar to that caused by TBK1 and IKKi, Regnase-1 phosphorylation occurred in IRAK1/IRAK2 double-deficient MEFs (Fig. 2 C), indicating that IRAK1 and IRAK2 are dispensable for IL-17–mediated Regnase-1 phosphorylation. Treatment of MEFs with BX795, an inhibitor of both TBK1 and IKKi, inhibited Regnase-1 phosphorylation upon IL-17A stimulation (Fig. 2 D). Phosphorylation of recombinant Regnase-1 by recombinant TBK1 and/or IKKi in vitro was detected as a mobility-shifted pattern by immunoblotting and 32P autoradiography (Fig. 2 E). Thus, TBK1 and IKKi are both responsible for Regnase-1 phosphorylation, in response to IL-17A. However, TRAF2 and TRAF5, known as important downstream mediators of the IL-17R signaling pathway (Sun et al., 2011), are dispensable for Regnase-1 phosphorylation during IL-17A stimulation (Fig. 2 F). IL-1β–induced Regnase-1 phosphorylation also occurred in TBK1/IKKi double-deficient MEFs (Fig. 2 G). Therefore, TBK1 and IKKi may phosphorylate Regnase-1 in an IRAK-independent manner, and IRAKs may phosphorylate Regnase-1 independently of TBK1 and IKKi.
Act1 contributes to TBK1/IKKi–mediated Regnase-1 phosphorylation by interacting with Regnase-1
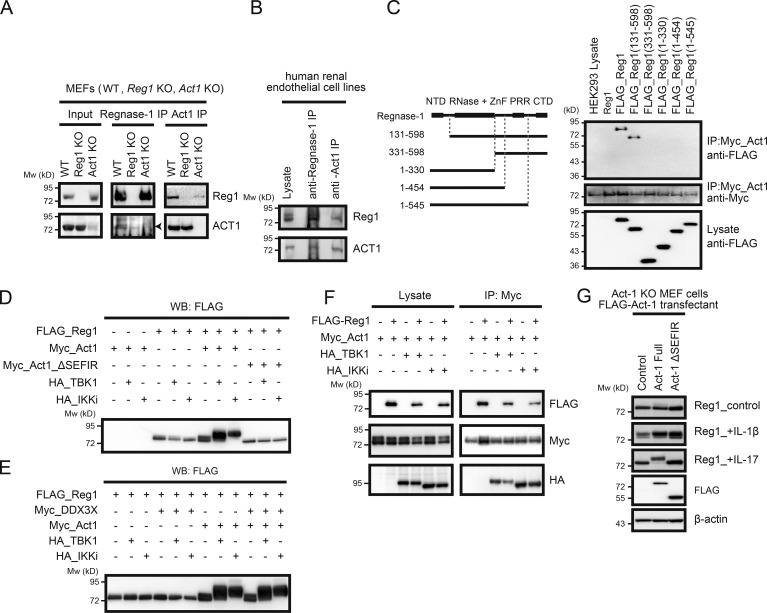
Coimmunoprecipitation using both anti-Regnase-1 and anti-Act1 antibodies demonstrated that Regnase-1 binds to Act1 (Fig. 3, A and B). Coimmunoprecipitation also revealed that Regnase-1 bound to Act1 via its C-terminal domain (Fig. 3 C). We next coexpressed Regnase-1 with Act1, TBK1, IKKi, or DDX3X in HEK293 cells. The phosphorylated, mobility-shifted band of Regnase-1 appeared when coexpressed with Act1 but not with TBK1 or IKKi; a more intense band of phosphorylated Regnase-1 was detected when it was coexpressed with Act1 plus TBK1 or Act1 plus IKKi (Fig. 3 D). In contrast, the level of phosphorylated Regnase-1 decreased when it was coexpressed with the Act1 mutant lacking the C-terminal SEFIR domain (Act1 ΔSEFIR) plus TBK1 or Act1 ΔSEFIR plus IKKi (Fig. 3 D). DDX3X, which has been reported to interact with both Act1 and TBK1 in the IL17R signaling pathway (Soulat et al., 2008; Somma et al., 2015), did not affect Regnase-1 phosphorylation driven by an Act1/TBK1/IKKi axis (Fig. 3 E). These results suggest that the interaction of Regnase-1 with Act1 and TBK1 or IKKi is indispensable for its phosphorylation. Coimmunoprecipitation analysis showed that Act1 interacts with TBK1, IKKi, and Regnase-1 (Fig. 3 F). Act1-deficient MEFs stably expressing FLAG (DYKDDDK)-Act1 ΔSEFIR failed to show Regnase-1 phosphorylation in response to IL-17A stimulation (Fig. 3 G). These results indicate that the binding of Act1 to Regnase-1 through their C-terminal domains results in increased accessibility of Regnase-1 to TBK1 and IKKi and enables simultaneous phosphorylation of Regnase-1 and Act1 by TBK1 or IKKi.
Figure 3.
Act1 plays a pivotal role in Regnase-1 phosphorylation induced by IL-17. (A and B) Coimmunoprecipitation of Regnase-1 (Reg1) and Act1 from primary MEFs. Cell lysates from WT, Regnase-1–deficient, and Act1–deficient MEFs (A) or a human renal glomerular endothelial cell line (B) were coimmunoprecipitated by either anti–Regnase-1 or anti-Act1 antibody. Eluted proteins were analyzed with anti–Regnase-1 and anti-Act1 antibodies. (C) Coimmunoprecipitates of Act1 with full-length or N- or C-terminally truncated forms of Regnase-1. All constructs were independently expressed using HEK293 cells. A control (untagged Regnase-1) and FLAG-tagged Regnase-1 variants (left) were coimmunoprecipitated with Myc-tagged Act1. Eluted proteins were subjected to immunoblotting analysis (right) with anti-FLAG and anti-Myc antibodies. (D and E) Immunoblotting analysis of FLAG-tagged Regnase-1 in HEK293 cells transfected with FLAG–Regnase-1, Myc-tagged Act1 (D), Myc-tagged Act1 mutant (Myc-Act1 ΔSEFIR; D), Myc-tagged DDX3X (E), HA-tagged TBK1 and HA-tagged IKKi (D and E). (F) Immunoblotting analysis of Act1-deficient MEFs stably expressing FLAG-tagged Act1 (full-length and ΔSEFIR mutant). Cells were stimulated with IL-1β and IL-17 for 1 h, lysed, and analyzed by immunoblotting of Regnase-1, FLAG-Act1, and β-actin. (G) Coimmunoprecipitation of FLAG-tagged Regnase-1, Myc-tagged Act1, HA-tagged TBK1, and HA-tagged IKKi. Cell lysates from HEK293 transfectants were mixed and coimmunoprecipitated by anti–Myc-coated beads. Eluted proteins were subjected to immunoblotting analysis with anti-FLAG, anti-Myc, and anti-HA antibodies. Mw, molecular weight.
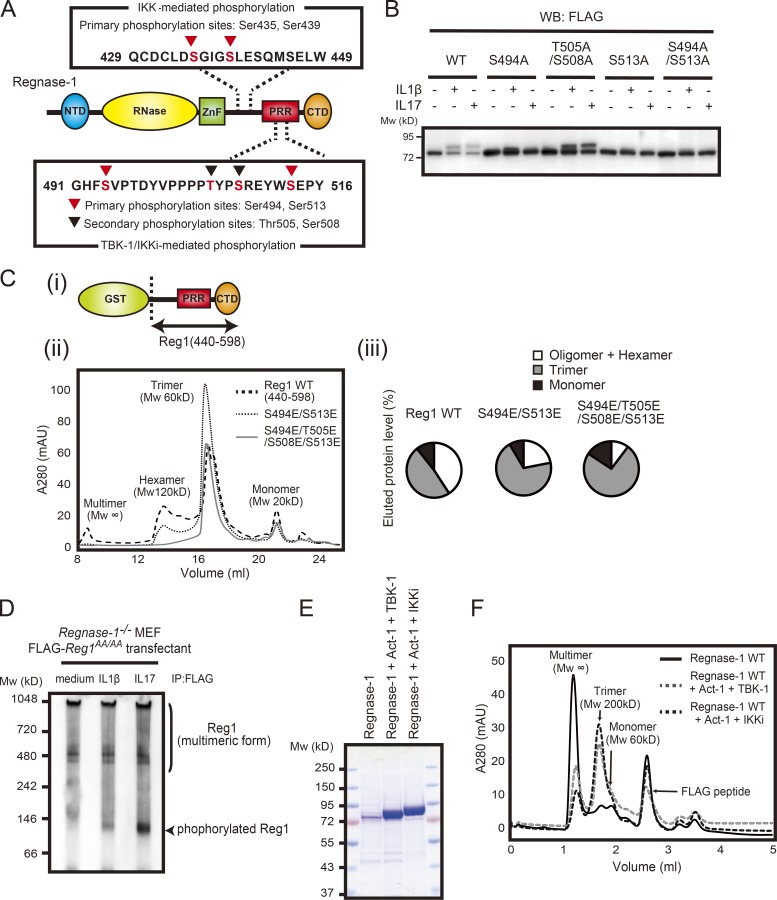
Phosphorylation of residues in the proline-rich region dissociates oligomerized Regnase-1
We attempted to identify Regnase-1 amino acid residues phosphorylated by TBK1 and IKKi. Five Regnase-1 residues were identified as critical phosphorylation sites (Fig. 4 A and Table S1). One of the five residues (Ser439) corresponded to the phosphorylation target of IKKs. The other four residues (Ser494, Thr505, Ser508, and Ser513) were located in the proline-rich region of Regnase-1 and did not contain any consensus sequences for phosphorylation. Substitution of Ser494 and Ser513 with alanine resulted in the disappearance of phosphorylated Regnase-1 when stimulated by IL-1β or IL-17A in HeLa cells (Fig. 4 B), indicating that these are the central residues involved in the electrophoretic mobility changes in phosphorylated Regnase-1 and that they are phosphorylation sites shared by IRAKs and TBK1/IKKi.
Figure 4.
Phosphorylation of Regnase-1 results in dissociation of oligomerized Regnase-1. (A) Domain schematic of Regnase-1 and mapping of the phosphorylation sites by IKKs (IKKα and IKKβ) and TBK1/IKKi. NTD, N-terminal domain; ZnF, zinc-finger domain; PRR, proline-rich region; CTD, C-terminal domain. (B) Immunoblotting analysis of Regnase-1 in HeLa cells transfected with Regnase-1 mutants (WT, S494A, T505A/S508A, S513A, and S494A/S513A). Cells were stimulated with IL-1β (10 ng/ml) and IL-17A (50 ng/ml) for 1 h. WB, Western blotting. (C) i: Diagram of the GST-fused Regnase-1 (Reg1) construct (440–598). ii: Gel filtration of WT and mutated (S494E/S513E or S494E/T505E/S508E/S513E) Regnase-1 (440–598). The molecular weight of each elution peak was estimated by using molecular weight standard markers and defined as multimer (Mw: ∞), hexamer (Mw: 120 kD), trimer (Mw: 60 kD), or monomer (Mw: 20 kD). iii: The eluted fractions (multimer + hexamer, trimer, and monomer) were quantified as a percentage of total eluted proteins. mAU, milli-absorbance unit. (D) Native-PAGE and immunoblotting analysis of Regnase-1. Regnase-1 was obtained from Regnase-1–deficient MEFs expressing FLAG-tagged Regnase-1 AA mutant stimulated with IL-1β (10 ng/ml) and IL-17A (50 mg/ml) for 1 h. (E) SDS-PAGE of purified Regnase-1 proteins from Expi-293F (Thermo Fisher Scientific) cells transiently expressing FLAG-tagged Regnase-1 with Myc-tagged Act1 and HA-tagged TBK1 or HA-tagged IKKi. Samples were stained with Coomassie brilliant blue. (F) Gel filtration analysis of purified Regnase-1 in E. The molecular weight of each elution peak was estimated by using molecular weight standard markers and defined as multimer (Mw: ∞), trimer (Mw: 200 kD), or monomer (Mw: 60 kD). Mw, molecular weight.
Previous studies revealed that Regnase-1 can form homo-oligomers (Yokogawa et al., 2016; Wilamowski et al., 2018). The Regnase-1 proline-rich region is reportedly involved in Regnase-1 oligomerization, and the expression of a Regnase-1 truncated mutant lacking this region leads to increased levels of its targeted mRNAs (Suzuki et al., 2011; Lin et al., 2013). To mimic phosphorylation, we introduced glutamic acid substitution at four phosphorylation sites in the proline-rich region. Oligomerization of Regnase-1 fragments (WT, S494E/S513E, and S494E/T505E/S508E/S513E) was analyzed by using gel filtration chromatography. The WT fragment formed several oligomers with high molecular weight, but oligomerization was inhibited in the mutants (Fig. 4 C); this indicated that introduction of phosphoserine and phosphothreonine residues into the proline-rich region promoted the dissociation of oligomerized Regnase-1. To examine the molecular assembly of phosphorylated Regnase-1, we used native PAGE analysis of Regnase-1 purified from MEFs stably expressing an N-terminal FLAG-tagged Regnase-1; IL-1β or IL-17A stimulation induced the appearance of a low-molecular-weight form of Regnase-1 (Fig. 4 D). Finally, we performed gel filtration analysis of full-length Regnase-1 protein. Regnase-1 protein was purified from Expi293F cells that transiently expressed Regnase-1, Act1, TBK1, or IKKi (Fig. 4 E). Unphosphorylated Regnase-1 formed primarily oligomers of high molecular weight, whereas phosphorylated Regnase-1 was eluted as a mixture of oligomers and trimers (Fig. 4 F). These results suggest that phosphorylation within the proline-rich region of Regnase-1 affects interactions required for self-assembly of Regnase-1 and promotes transformation from oligomerized to trimeric forms.
Regnase-1 phosphorylation changes its subcellular localization from the ER to cytosol
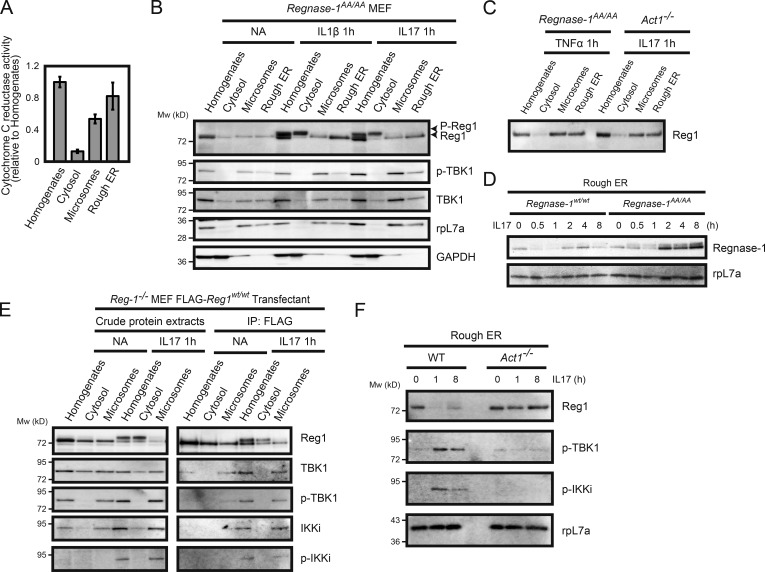
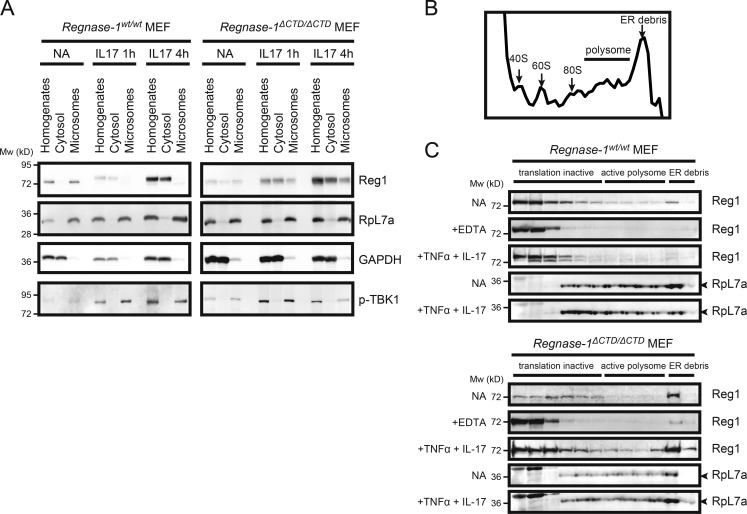
Regnase-1 proteins are found in rough ER membrane fractions (Mino et al., 2015). We isolated cellular compartments such as the ER membrane, microsomes, and soluble cytoplasmic fractions from cell homogenates of MEFs stimulated by IL-1β or IL-17A and analyzed their protein distribution with immunoblotting. We confirmed that nicotinamide adenine dinucleotide phosphate–cytochrome c reductase, a known marker of ER membrane protein, was more abundant in the isolated membrane fraction (Fig. 5 A). All phosphorylated Regnase-1 protein was present in the cytoplasm, whereas unphosphorylated Regnase-1 remained localized to ribosome-containing organelles (Fig. 5 B). To facilitate precise characterization of the subcellular localization of phosphorylated Regnase-1, we used MEFs from Regnase-1 AA mutant mice, in which Regnase-1 was resistant to IKK–mediated degradation. This cytosolic distribution of Regnase-1 was not observed in TNF-α–stimulated MEFs or in IL-17A–stimulated Act1-deficient MEFs, in which Regnase-1 was not phosphorylated (Fig. 5 C); this indicated that Regnase-1 phosphorylation and subsequent dissociation of Regnase-1 oligomers caused translocation of Regnase-1 from ER to the cytoplasm. Interestingly, the quantity of unphosphorylated Regnase-1 in the ER fraction was greater in Regnase-1AA/AA MEFs than in WT cells with increased time after IL-17A stimulation (Fig. 5 D). This finding suggests that, during IL-17A stimulation, the regulation of Regnase-1–targeted mRNA levels is mediated by unphosphorylated Regnase-1 protein attached to the ER.
Figure 5.
Regnase-1 phosphorylation and its translocation from the ER membrane. (A) Isolation of ER membrane–containing subcellular organelles from cell homogenates. The purity of each fraction was evaluated through the measurement of activity of cytochrome c reduction by NADP reduced (NADPH)–cytochrome c reductase (n = 4). (B and C) Immunoblotting analysis of subcellular organelle fractions. Regnase-1 (Reg1), ribosomal protein L7a (rpL7a; ER marker), and GAPDH (cytoplasmic marker) in the cell homogenates, soluble cytoplasmic fraction, microsomes, and rough ER membranes. Fractions were prepared from Regnase-1AA/AA MEFs stimulated with IL-1β (10 ng/ml) and IL-17A (50 ng/ml) for 1 h (B) and from Regnase-1AA/AA and Act1-deficient MEFs stimulated with TNF-α (20 ng/ml) and IL-17A (50 ng/ml) for 1 h (C). Arrows indicate phosphorylated (top) and unphosphorylated (bottom) forms of Regnase-1. (D) Immunoblotting analysis of Regnase-1 and rpL7a in rough ER membranes prepared from WT and Regnase-1AA/AA MEFs stimulated with IL-17A (50 ng/ml) for 0–8 h. (E) FLAG-tagged Regnase-1 was immunoprecipitated from cell organelle fractions of Regnase1-deficient MEFs expressing FLAG-tagged Regnase-1 AA mutant stimulated with or without IL-17A (50 ng/ml) for 1 h. Immunoprecipitates were subjected to immunoblotting analysis of Regnase-1, TBK1, phospho-TBK1, IKKi, and phospho-IKKi. (F) Immunoblotting analysis of Regnase-1, phospho-TBK1, phospho-IKKi, and rpL7a in the ER membrane fractions isolated from WT and Act1-deficient MEFs. Cells were stimulated with IL-17A (50 ng/ml) for 0, 1, and 8 h.
We found that Regnase-1 interacted with phosphorylated TBK1 and phosphorylated IKKi at the microsomes, but not in the soluble cytoplasmic fraction (Fig. 5 E). Act1 was responsible for phosphorylation of both TBK1 and IKKi at the ER in response to IL-17A, and Act1 promoted kinase–mediated phosphorylation of Regnase-1 (Fig. 5 F). These results suggest that Act1 acts as a signal transducer to promote phosphorylation of TBK1, IKKi, and Regnase-1 on the ER membrane after IL-17A stimulation.
IL-17–mediated Regnase-1 phosphorylation results in attenuated degradation of its target mRNAs
We measured the levels of IL-6 mRNA in MEFs that were pretreated with TNF and then stimulated with IL-17A alone. IL-6 mRNA induction was strongly enhanced during IL-17A stimulation, which caused Regnase-1 phosphorylation in WT MEFs (Fig. 6 A). IL-6 mRNA induction was suppressed in Regnase-1 AA mutant and TBK1/IKKi double-deficient cells during IL-17A stimulation; notably, it was significantly inhibited in the double-deficient MEFs (Fig. 6 B). TBK1/IKKi double-deficient cells did not show Regnase-1 phosphorylation and maintained Regnase-1 subcellular localization at ribosome-containing organelles in response to IL-17A (data not shown).
Figure 6.
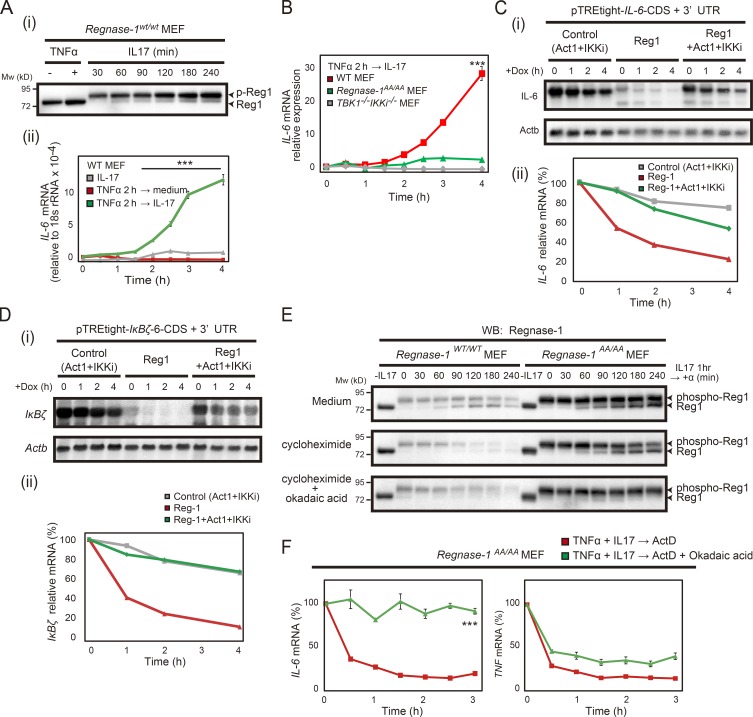
Regnase-1 phosphorylation results in attenuation of its degradation of IL-6 mRNA. (A) Immunoblotting analysis of Regnase-1 (Reg1; i) and quantitative PCR (qPCR) analysis of IL-6 mRNA (ii). WT MEFs sequentially stimulated with TNF-α for 2 h, followed by IL-17A for 0–4 h. qPCR data were collected from three independent experiments. ***, P < 0.005. (B) IL-6 mRNA expression in WT and Regnase-1AA/AA and TBK1/IKKi double-deficient MEFs stimulated TNF-α for 2 h followed by IL-17A for 0–4 h. ***, P < 0.005. (C and D) i: Autoradiography of IL-6 (top) and Actb (bottom; C) or IκBζ (top) and Actb (bottom; D) mRNA levels in Tet-off HEK293 cells cotransfected with either a pTRE-tight-IL6-CDS+3′-UTR (C) or a pTRE-tight-IκBζ-CDS+3′-UTR vector (D) together with expression plasmids of control (Act1+IKKi), Regnase-1, or Regnase-1+Act1+IKKi. Total mRNA was prepared from cells after treatment with doxycycline (Dox) for 0–4 h, then subjected to Northern blotting. ii: Relative IL-6 mRNA (C) and IκBζ mRNA (D) levels in Tet-off HEK293 cells during the doxycycline treatment. (E) Immunoblotting analysis of Regnase-1 in Regnase-1AA/AA MEFs. Cells were treated with IL-17A (50 ng/ml) for 1 h and then incubated in medium containing nothing (control), cycloheximide (100 µM), or both cycloheximide and okadaic acid (0.5 µM) for 0–240 min. WB, western blotting. (F) qPCR analysis of IL-6 and TNF mRNA levels in Regnase-1AA/AA MEFs treated with TNF-α (20 ng/ml) and IL-17A (50 ng/ml) for 2 h (n = 4) and then incubated in medium containing ActD (5 µg/ml) or both ActD and okadaic acid (0.5 µM) for 0–180 min. ***, P < 0.005. Mw, molecular weight.
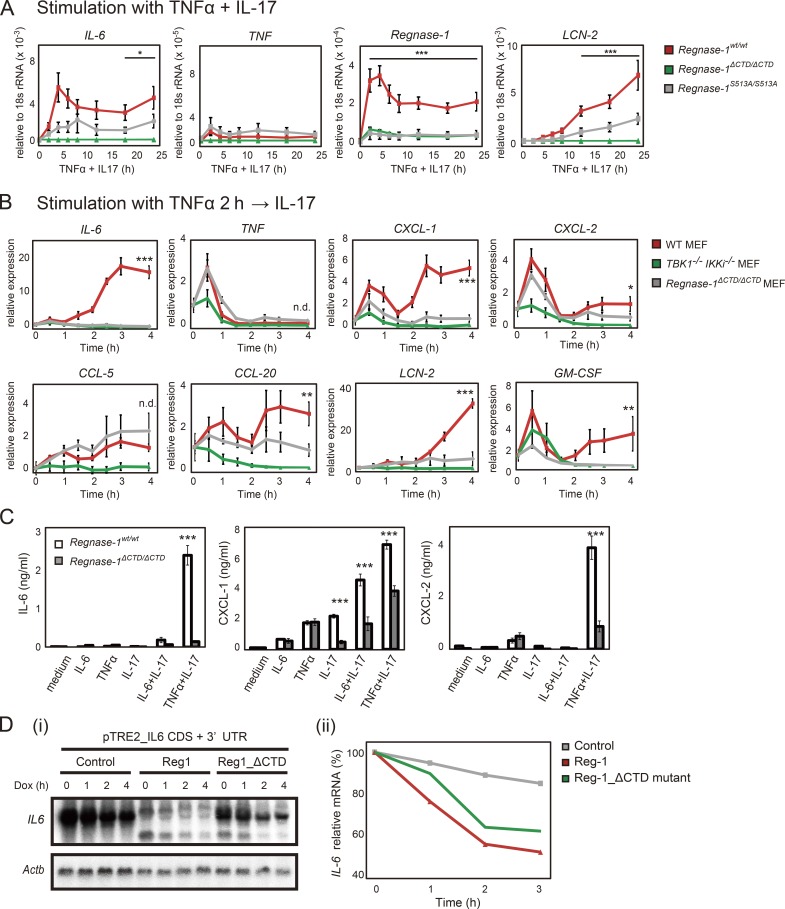
We evaluated the effect of Regnase-1 phosphorylation on the mRNA decay ability using the Tet-off inducible system. Regnase-1 was readily phosphorylated when coexpressed with Act1 and IKKi. IL-6 mRNA degradation by Regnase-1 was blocked by coexpression with Act1 and IKKi (Fig. 6 C). The mRNA of transcription factor IκBζ is a known target for Regnase-1 (Garg et al., 2015) and is translated in the cytosol, whereas target IL-6 mRNA is translated in the ER. Degradation of IκBζ mRNA was also blocked by coexpression of Regnase-1 with Act-1 and IKKi (Fig. 6 D). These findings strongly indicate that phosphorylated Regnase-1 lacks the ability to degrade its target mRNAs.
To investigate the mechanism of target mRNA suppression in Regnase-1AA/AA cells, we used immunoblotting analysis of phosphorylated and unphosphorylated Regnase-1 during the recovery phase after discontinuation of IL-17A stimulation. Unphosphorylated Regnase-1 protein gradually emerged during the recovery phase in WT MEFs and was observed in the presence of a protein synthesis inhibitor (Fig. 6 E). The increase in unphosphorylated Regnase-1 protein levels was markedly enhanced in Regnase-1AA/AA MEFs compared with WT cells. In contrast, unphosphorylated Regnase-1 did not emerge in the presence of okadaic acid, an inhibitor of protein phosphatase-1 and -2A (Fig. 6 E), indicating that the appearance of unphosphorylated Regnase-1 is mediated by a phosphatase. IL-6 mRNA stability was also enhanced in the presence of okadaic acid during the recovery phase after TNF-α and IL-17A treatment of Regnase-1AA/AA MEFs (Fig. 6 F). Thus, conversion of phosphorylated to unphosphorylated Regnase-1 may be involved in the suppression of Regnase-1 target mRNAs observed in Regnase-1AA/AA cells during IL-17A stimulation.
A C-terminally truncated mutation and an S513A mutation of Regnase-1 (Regnase-1 ΔCTD and Regnase-1 S513A) inhibit IL-17–mediated phosphorylation and result in enhanced protein stability
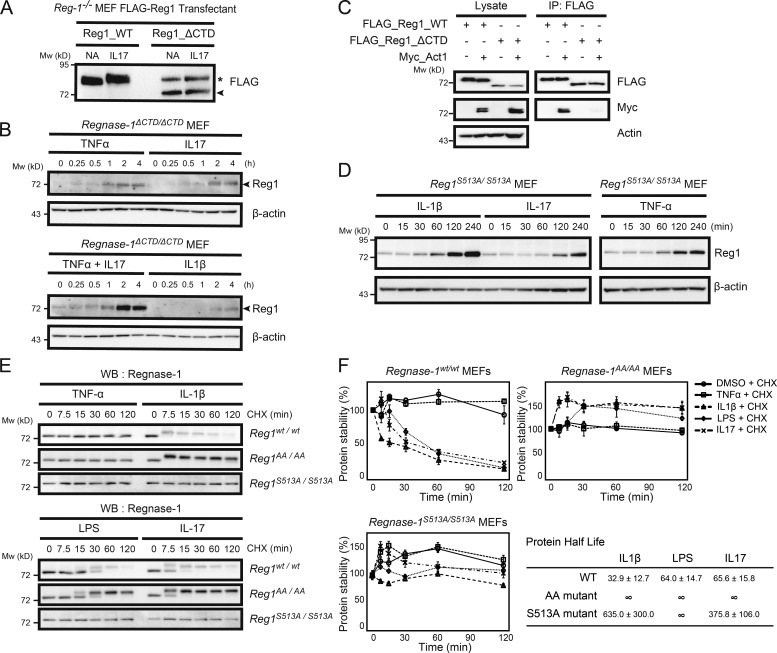
By exploring Regnase-1 mutants resistant to IL-17–mediated phosphorylation, we found that removal of the C-terminal domain (essential for interaction with Act1) blocked phosphorylation upon IL-17A stimulation (Fig. 7 A). We successfully obtained mutant mice with a 1-bp deletion in codon 517, causing a frameshift mutation (Regnase-1 ΔCTD, Fig. S3, A and B). When MEFs from Regnase-1ΔCTD/ΔCTD mutant mice were stimulated with TNF-α, IL-17A, and IL-1β, the amount of Regnase-1 ΔCTD protein was increased, without the mobility-shifted band representing its phosphorylation (Fig. 7 B). Furthermore, Regnase-1 ΔCTD protein showed significantly reduced phosphorylation during IL-17A stimulation compared with WT (Fig. 7 C), indicating that Regnase-1 ΔCTD lacks Act1-binding ability and remains unphosphorylated. We generated mutant mice containing an alanine substitution at Ser513 (Fig. S3 C). TNF-α, IL-17A, and IL-1β stimulation of MEFs obtained from Regnase-1S513A/S513A mice induced up-regulation of unphosphorylated Regnase-1 mutant protein in the same manner as that of the Regnase-1 ΔCTD mutation (Fig. 7 D). We investigated the protein stability of Regnase-1 ΔCTD and S513A mutant proteins. MEFs from Regnase-1AA/AA Regnase-1S513A/S513A mutant mice were stimulated with TNF-α, IL-1β, LPS, and IL-17A in the presence of the translation inhibitor cycloheximide. Levels of Regnase-1 protein also did not exhibit significant reduction during IL-1β, LPS, and IL-17A stimulation of Regnase-1S513A/S513A mutant cells (Fig. 7, E and F). Thus, Regnase-1 S513A mutation resulted in enhanced protein stability based on resistance to phosphorylation and degradation induced by IL-17A stimulation.
Figure 7.
Regnase-1 frameshift mutation and alanine mutation at the TBK1/IKKi phosphorylation site provide resistance to Regnase-1 degradation induced by proinflammatory cytokine stimulation. (A) Immunoblotting of lysates of Regnase-1-deficient (Reg-1–/–) MEFs stably expressing FLAG-tagged Regnase-1 (Reg1; WT and ΔCTD mutant). Cells were stimulated with or without IL-17A for 1 h and probed with anti-FLAG antibody. Regnase-1ΔCTD mutant and nonspecific protein bands are indicated by the arrowhead and asterisk, respectively. NA, no addition. (B) Immunoblotting of Regnase-1 and β-actin in Regnase-1ΔCTD/ΔCTD MEFs stimulated as indicated for 0–4 h. Arrowheads indicate Regnase-1. (C) Coimmunoprecipitation of FLAG-tagged WT Regnase-1 or the ΔCTD mutant with Act1, TBK1, and IKKi in HEK293 cells. Immunoprecipitated Regnase-1 was subjected to immunoblotting as indicated. IP, immunoprecipitation. (D) Immunoblotting analysis of Regnase-1 and β-actin in Regnase-1S51A/S513A MEFs stimulated with IL-1β, IL-17, and TNF-α for 0–4 h. (E and F) Comparison of protein half-life of Regnase-1 variants. Regnase-1wt/wt, Regnase-1AA/AA, and Regnase-1S513A/S513A MEFs were exposed to cycloheximide (CHX) and stimulated with TNF-α, IL-1β, LPS, and IL-17 for 0–120 min. (E) Immunoblotting analysis of Regnase-1 after stimulation as indicated. WB, western blotting. (F) Regnase-1 protein measured by immunoblotting and normalized to β-actin control. Protein half-lives are shown. Data were collected from three independent experiments. Mw, molecular weight.
ΔCTD mutation protects binding of Regnase-1 to the ER membrane during IL-17A stimulation
We next investigated the binding pattern of Regnase-1 ΔCTD in ribosome-containing organelles. Regnase-1 ΔCTD remained bound to microsomes after IL-17A stimulation. Regnase-1 ΔCTD protein levels were increased in microsomes (Fig. 8 A). Regnase-1 has been shown to bind to translationally active ribosomes assembled on polysomes (Mino et al., 2015). We confirmed Regnase-1 binding to translationally active polysomes in WT and Regnase-1ΔCTD/ΔCTD MEFs stimulated with TNF-α and IL-17A (Fig. 8 B). Localization of Regnase-1 in translationally active polysomes was increased in mutant MEFs stimulated with these cytokines compared with unstimulated MEFs; in contrast, Regnase-1 localization on polysomes was reduced in stimulated WT MEFs (Fig. 8 C). Regnase-1 localization on translationally active polysomes was dissolved in the presence of EDTA, indicating that Regnase-1 binds to polysomes through its interaction with ribosomes (Fig. 8 C).
Figure 8.
Regnase-1 ΔCTD mutation inhibits changes in its subcellular localization. (A) Immunoblotting of indicated proteins in subcellular organelles isolated from WT and Regnase-1ΔCTD/ΔCTD MEFs after IL-17A stimulation for indicated times. (B and C) Analysis of polysome fractions isolated from WT and Regnase-1ΔCTD/ΔCTD cell lysates stimulated with or without TNF-α and IL-17A for 4 h. (B) UV absorbance profile of sucrose-gradient fractions (10–50% sucrose) after ultracentrifugation of cell lysates. (C) Immunoblotting of Regnase-1 (Reg1) and RpL7a in sucrose-gradient fractions. Mw, molecular weight. NA, no addition.
Regnase-1 ΔCTD and S513A mutations abolish IL-17–mediated inflammatory responses
The conflicting pattern of protein localization between WT and mutant Regnase-1 implicates that Regnase-1 ΔCTD mutation degrades its target mRNA expression, even during inflammatory stimulation. Both costimulation and sequential stimulation with TNF-α plus IL-17A induced a time-dependent increase in Regnase-1 target mRNAs in WT MEFs but not in mutant MEFs (Fig. 9, A and B). In addition, protein levels of IL-6, CXCL-1, and CXCL-2 after 24-h stimulation with IL-17A plus either IL-6 or TNF-α were significantly lower in Regnase-1ΔCTD/ΔCTD MEFs compared with WT MEFs (Fig. 9 C). To exclude the possibility that a C-terminal truncation of Regnase-1 affects its RNase activity, we investigated the mRNA decay of IL-6 mRNA in Tet-off HEK293 cells expressing Regnase-1 ΔCTD mutant. Importantly, Regnase-1 ΔCTD mutant exhibited a level of IL-6 mRNA degradation comparable to that of WT Regnase-1 (Fig. 9 D). Thus, Regnase-1 ΔCTD protein maintains its subcellular localization at ribosome-containing organelles after IL-17A stimulation and acts as a negative regulator, inhibiting enhancement of mRNA stability and subsequent IL-17A–mediated inflammatory cytokine production.
Figure 9.
Regnase-1 ΔCTD and S513A mutations reduce mRNA stability of Regnase-1-targeted genes during IL-17 stimulation. (A) Quantitative PCR (qPCR) of indicated mRNAs in WT, Regnase-1ΔCTD/ΔCTD, and Regnase-1S513A/S513A MEFs (n = 4). Cells were stimulated as indicated for 0–24 h. *, P < 0.05; ***, P < 0.005. (B) qPCR analysis of IL-6, TNF, CXCL-1, CXCL-2, CCL-5, CCL20, LCN-2, and GM-CSF expression in WT, Regnase-1 ΔCTD mutant, and TBK1/IKKi double-deficient MEFs. Cells were stimulated with TNF-α (20 ng/ml) for 2 h, followed by IL-17A (50 ng/ml) for 0–4 h. *, P < 0.05; **, P < 0.01; ***, P < 0.005. n.d., no difference. (C) Production of IL-6, CXCL-1, and CXCL-2 by WT and Regnase-1ΔCTD/ΔCTD MEFs in response to exposure to IL-6 (20 ng/ml), TNF-α (20 ng/ml), IL-17A (50 ng/ml), IL-6+IL-17A, or TNF-α+IL-17A for 24 h. Protein levels in cell supernatants were assessed by ELISA (n = 3). ***, P < 0.005. (D) i: Autoradiography of IL-6 (top) and Actb (bottom) mRNA levels in Tet-off HEK293 cells cotransfected with a pTRE-tight-IL6-CDS+3′-UTR vector, together with expression plasmids of control (empty vector), Regnase-1, or ΔCTD mutant. Total mRNA was prepared from cells after treatment with doxycycline (Dox) for 0–4 h, then subjected to Northern blotting. ii: Relative IL-6 mRNA levels in Tet-off HEK293 cells during doxycycline treatment. GM-CSF, granulocyte macrophage CSF; Mw, molecular weight.
Regnase-1 ΔCTD mutation markedly reduces EAE disease severity
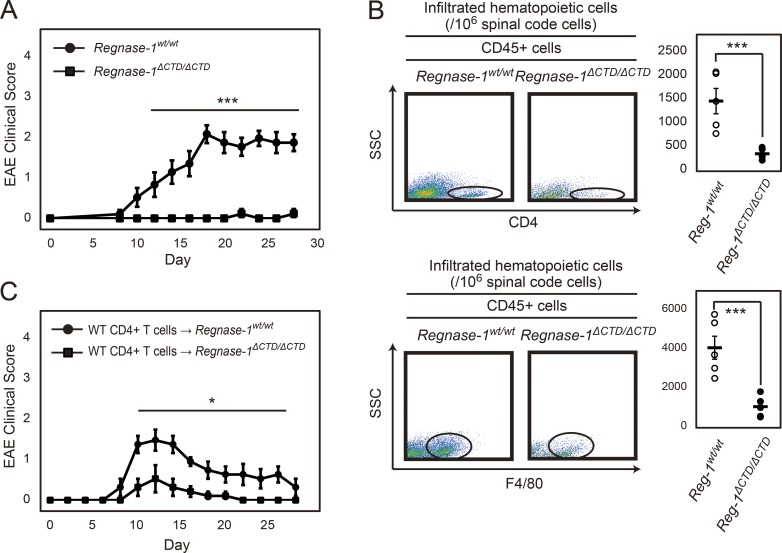
Finally, we induced EAE in Regnase-1ΔCTD/ΔCTD mice. EAE severity was strongly attenuated in these mice compared with WT mice (Fig. 10 A). Flow cytometric analysis of mice at 28 d after immunization revealed significantly reduced infiltration of CD4+ T cells and macrophages into neuronal tissue in Regnase-1ΔCTD/ΔCTD mice (Fig. 10 B); Th1 and Th17 cell numbers in spleen and lymph nodes were similar between WT and mutant mice, except for Th1 cells from spleen, which were increased in mutant mice (Fig. S4). After transfer of EAE-pathogenic CD4+ T cells, the EAE clinical score was significantly reduced in both Regnase-1ΔCTD/ΔCTD mice and Regnase-1AA/AA mice (Fig. 10 C). Thus, Regnase-1 ΔCTD mutation may suppress EAE pathogenesis via continuous inhibition of STAT3 activation in endothelial cells, preventing Th17–mediated inflammation required to infiltrate these cells into nerve organs.
Figure 10.
Regnase-1 ΔCTD mutation enhances anti-inflammatory response and blocks EAE pathogenesis. (A) EAE clinical scores of WT (n = 8) and Regnase-1ΔCTD/ΔCTD mice (n = 8) for 28 d. ***, P < 0.005. (B) Flow cytometry of spinal cord cells at 28 d after immunization. Representative plots and statistics (each n = 5) of CD4+ T cells (top) and F4/80+ macrophages (bottom) among CD45+ cells in spinal cord cells (SCC; 106 cells) from WT and Regnase-1ΔCTD/ΔCTD mice. ***, P < 0.005. (C) EAE clinical scores of mice after intravenous transfer of WT EAE-pathogenic CD4+ T cells into WT or Regnase-1CTD/CTD mice (each n = 5) for 28 d. *, P < 0.05.
Discussion
We generated a Regnase-1 AA mutant, in which two serine residues phosphorylated by IKKs were mutated to alanine residues. IL-17A stimulation induced rapid phosphorylation of Regnase-1 in both WT and Regnase-1 AA mutant MEFs. This phosphorylation was IKK independent and contributed to reduced degradation of Regnase-1 targeted mRNAs through its dissociation from ribosome-containing organelles. In our experiments, phosphorylation was sustained beyond the duration necessary for degradation, which ensures moderate expression of target mRNAs. We observed the appearance of unphosphorylated Regnase-1 in the late phase of IL-17A stimulation, which may suppress a set of mRNAs regulated by Regnase-1 protein and lead to reduced EAE severity in Regnase-1 AA mutant mice. The accumulation of unphosphorylated Regnase-1 protein in Regnase-1 AA mutant cells occurred as a result of NF-κB activation to induce Regnase-1 expression, as well as through dephosphorylation of phosphorylated Regnase-1 by endogenous phosphatases. Regnase-1 dephosphorylation resulted in recovery of mRNA decay activity through attachment to the ER. Thus, Regnase-1 may inhibit expression of its target mRNAs in response to weak stimuli via reversible dephosphorylation and rapid relocation to the ER.
We also generated Regnase-1 ΔCTD mutant mice, which expressed Regnase-1 protein lacking the C-terminal domain required for interaction with Act1. These mice also showed reduced EAE severity and Th17–mediated inflammation; thus, the phenotype was similar to that of Regnase-1AA/AA mice, although the mechanism differed between these mice. Regnase-1 AA mutant protein is more abundant than WT protein because of its resistance to IKK–mediated phosphorylation; this appears to be beneficial in EAE. Regnase-1 ΔCTD mutant protein showed a lower level of expression in steady state and increased expression upon stimulation with some inflammatory cytokines, promoting accumulation of unphosphorylated Regnase-1. Another mutation, Regnase-1 S513A, also resulted in down-regulation of the mutant protein level and an increase in unphosphorylated Regnase-1 during cytokine stimulation in a manner similar to that of the Regnase-1 ΔCTD mutant. The low level of Regnase-1 ΔCTD and S513A proteins in steady-state conditions may result from autoregulation of its own mRNA, as Regnase-1 mRNA contains a self-binding site in its 3′-UTR (Iwasaki et al., 2011). Our finding that IL-17–mediated phosphorylation of Regnase-1 causes self-regulation of its mRNA is consistent with a previous report that Regnase-1 mRNA was stabilized during IL-17A stimulation (Somma et al., 2015). The importance of protein phosphorylation for RNA degradation is similar to that observed in a mitogen-induced RNA-binding protein, tristetraprolin (TTP); notably, a phosphorylation-deficient mutant form of the protein demonstrates a low expression level but has a strong effect on target mRNA destabilization when stimulated with LPS (Ross et al., 2015). The TTP mutant also undergoes activation-dependent changes in subcellular localization (Brook et al., 2006). These studies provide an important insight into the regulatory role of phosphorylation of Regnase-1 and TTP, not only in signaling that leads to protein degradation but also in attenuating their mRNA destabilization activities to avoid subcellular accumulation of unphosphorylated/active proteins.
In the IL-17R signaling pathway, IL-17R and Act1 form a complex with a homotypical interaction between their conserved SEFIR domains in the cytoplasm; the complex is increased in an IL-17–dependent manner (Qian et al., 2007). Interaction of Act1 with IKKi promotes Act1 phosphorylation upon IL-17 stimulation (Bulek et al., 2011). TBK1 can also phosphorylate Act1 and cause enhanced NF-κB activation in the IL-17R signaling pathway (Qu et al., 2012). IL-17 stimulation induces formation of an Act1–RNA protein complex in both nucleus and cytoplasm, which stabilized mRNAs of inflammatory genes (Herjan et al., 2018), indicating that Act1 is a major signal transducer for IL-17R signaling. Activated Act1 encounters Regnase-1 on the ER when stimulated with IL-17. This engagement promotes Regnase-1 phosphorylation by TBK1 and IKKi, thereby reinforcing Regnase-1 translocation from the ER to the cytoplasm and inhibiting its mRNA degradation ability.
Previous studies have shown that Regnase-1 can act as a negative regulator for IL-17R signaling by destabilizing several mRNAs involved in the IL-17 response, and that it contributes to the suppression of Th17 cell–associated autoimmune diseases (Jeltsch et al., 2014; Garg et al., 2015; Monin et al., 2017). Indeed, Regnase-1 is highly expressed in injured tissues of psoriatic patients (Monin et al., 2017). We have revealed a new dimension to the Regnase-1–mediated mRNA regulation, which is controlled by its phosphorylation. This mechanism may explain why Regnase-1 cannot suppress autoimmune inflammation, regardless of excess expression. Our model suggests that Regnase-1 is constitutively phosphorylated and inactivated in injured tissues of autoimmune patients, which are exposed to proinflammatory cytokines (e.g., members of the IL-1 family, as well as IL-17).
There are several reports of RNA-binding proteins that stabilize specific mRNAs in the IL-17 response (Anderson, 2008; Sun et al., 2011; Herjan et al., 2013, 2018). Regnase-1, an RNA-binding RNase, destabilizes specific mRNAs at steady state and is rapidly inactivated during IL-17 stimulation. Costimulation with TNF-α and IL-17A dramatically reduced target mRNA production in Regnase-1 ΔCTD mutant cells, as well as in TBK1/IKKi double-deficient cells. These results strongly suggest that interactions among Regnase-1, Act1, and TBK1/IKKi govern expression of these inflammatory genes induced by IL-17A stimulation and that Regnase-1 plays a pivotal role in regulating the inflammatory response in response to IL-17. Blockade of Regnase-1 phosphorylation may provide novel strategies for the treatment of Th17-associated diseases.
Materials and methods
Generation of Regnase-1 S435A/S439A (AA) knock-in mice
Genomic DNA containing the Regnase-1 gene was isolated from ES cells (GSI-1). An ∼12-kbp genomic fragment encompassing exon 5 and exon 6 and downstream of the Regnase-1 gene termination was subcloned into a pCR-TOPO vector (Thermo Fisher Scientific). A targeting vector was designed to replace Ser435 and Ser439 residues at exon 6 with Ala by site-directed mutagenesis. A neomycin-resistant gene flanked by two loxP sites was inserted into the intron between exons 5 and 6. The linearized vector was introduced into GSI-1 ES cells by electroporation. The targeted ES cells were screened and identified by genomic PCR and Southern blotting. These cells were microinjected into blastocysts from C57BL/6 mice. Chimeric male mice were bred with C57BL/6 female mice to produce F1 heterozygous mice. F1 mice were further bred with CAG-Cre transgenic mice to remove the neomycin gene cassette. After removal of the CAG-Cre allele by breeding with C57BL/6 mice, Regnase-1 AA heterozygous (Regnase-1AA/+) mice were backcrossed with C57BL/6 mice for at least ten generations, then intercrossed to obtain Regnase-1AA/AA homozygotes. All mouse experiments were approved by the Animal Research Committee of the Research Institute for Microbial Diseases, Osaka University.
Generation of Regnase-1 frameshift mutant and S513A/S513A mutant alleles
Regnase-1 frameshift mutant mice containing a frameshift mutation in the C-terminus and Regnase-1 S513A/S513A mutant mice were designed and constructed by nonprofit organization Biotechnology Research and Development (Drs. M. Ikawa and M. Okabe, Osaka University, Osaka, Japan) with the clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9 genome-editing technology. Guide RNA sequences used in this study were as follows: 5′-GTGGGTGGGGGTAATGGGTA-3′ and 5′-CCTACCCATCCAGAGTAC-3′. Each gRNA sequence was cloned in-frame into CRISPR/Cas9 vector pSpCas9(BB)-2A-Puro PX459 (Addgene; no. 62988; Ran et al., 2013). The single-strand oligodeoxynucleotide sequence (103 bases, 5′-CCACCGACTATGTGCCCCCGCCACCCACCTACCCATCCAGAGAGTATTGGGCTGAGCCGTATCCATTACCCCCACCCACTCCTGTCCTTCAGGAGCCCCAGAG-3′) was synthesized for the S513A mutation. The PX459 vectors and single-strand oligodeoxynucleotide were introduced together into fertilized eggs from C57BL/6 × C57BL/6 mice by using previously described methods (Mashiko et al., 2014). These mouse embryos were transferred into the oviduct of pseudopregnant Crl:CD1 (ICR) females. Gene mutations in the Regnase-1 allele were identified by DNA sequencing. Mutant mice that contained a frameshift mutation, resulting in the expression of the C-terminal truncated Regnase-1 protein (Regnase-1 ΔCTD), were intercrossed to obtain Regnase-1 ΔCTD homozygotes.
Plasmids
Regnase-1 expression vectors, including pFLAG-CMV2 (Sigma) and pcDNA3.1-Myc, and viral expression vectors, such as pMRX-FLAG-Regnase-1-ires-puro, have previously been described (Matsushita et al., 2009; Iwasaki et al., 2011). Truncated versions of Regnase-1 lacking the N or C terminus were constructed by PCR amplification of Regnase-1 cDNA, which was inserted into the pFLAG-CMV2 vector. Point mutations of Regnase-1 expression constructs were prepared by using the Quickchange II Site-Directed Mutagenesis Kit (Agilent Technologies). For overexpression in Escherichia coli, the portion of Regnase-1 containing the proline-rich domain and C-terminal domain (441–598) was amplified from Regnase-1 cDNA and inserted into the pGEX-6P vector (GE Healthcare). Myc-Act1, human influenza hemagglutinin epitope (HA)-TBK1, and HA-IKKi were PCR amplified from each cDNA and inserted in-frame into a pcDNA3.1 vector. The pTRE-tight-IL-6-CDS+3′-UTR vector has previously been described (Matsushita et al., 2009). The IκBζ-CDS + 3′UTR (315–3815) was PCR amplified from its cDNA and inserted in-frame into a pTRE-tight vector (Clontech).
Reagents and cells
Recombinant mouse IL-17A was purchased from R&D Systems. Recombinant mouse IL-1β, mouse TNF-α, mouse IL-6, and human IL-17A were purchased from Biolegend. A recombinant anti–Regnase-1 rabbit monoclonal antibody was generated with previously described methods by Chugai Pharmaceutical Co. Ltd., Japan (Suzuki et al., 2016). Four New Zealand White rabbits (Kitayama Labs) were immunized four times with purified Regnase-1 protein. All rabbit experiments, including immunization, humane sacrifice, and collection of whole bloods and spleens, were approved by the Institutional Animal Care and Use Committee at Chugai. Anti-Act1 (H-300), anti-IκB-α (C-21), and anti-NF-κB p65 (C-20) antibodies were purchased from Santa Cruz Biotechnology. Anti-RPL7A (15340–1-AP) was from Proteintech. Anti-FLAG M2, anti-Myc (9E10), and anti-HA (12CA5) antibodies; FLAG M2 affinity gel; and FLAG×3 peptide were purchased from Sigma. Anti–phospho-IκBα (Ser32/36), anti–phospho-NF-κB p65 (Ser468), anti–phospho-STAT3 (Tyr705), anti–phospho-TBK1 (Ser172), and anti–phospho-IKKε (Ser172) antibodies were purchased from Cell Signaling Technology. Anti-CD3ε and anti–type IV collagen antibodies were purchased from Abcam. LPS (Salmonella minnesota R595) and BX795 were purchased from InvivoGen. Okadaic acid and cycloheximide were purchased from Wako Pure Chemicals. Bone marrow–derived macrophages were generated by culturing bone marrow cells with Roswell Park Memorial Institute 1640 medium containing 20 ng/ml macrophage CSF (Peprotech). HeLa cells were purchased from the American Type Culture Collection. WT, Regnase-1AA/AA, Regnase-1ΔCTD/ΔCTD, and Regnase-1S513A/S513A MEFs were prepared from mouse embryos at 13.5 d of gestation. Regnase-1–deficient MEFs were prepared as previously described (Iwasaki et al., 2011). Act1-deficient MEFs were generated from Traf3ip2ADJM mice, a kind gift from Dr. Y. Matsushima (Matsushima et al., 2010). To maintain the properties of primary cultured cells, the above MEFs were used for all experiments before they had undergone five passages. Regnase-1−/−, TBK1−/−, IKKi−/−, and TBK1−/−/IKKi−/− MEFs were prepared as described previously (Hemmi et al., 2004; Kawagoe et al., 2008; Matsushita et al., 2009). IKKα−/−/IKKβ−/− MEFs were a gift from Dr. I. Verma (Li et al., 2000). An endothelial cell line was prepared from human renal glomerular endothelial cells (ScienCell Research Laboratories). The cells were immortalized by lentiviral infection with the SV40 T antigen (Cosmo Bio). Effector cluster of differentiation (CD)-4+ T cells induced by in vitro differentiation were generated in accordance with previously reported methods (Zhu et al., 2010). For induction of Th1, Th17, or induced regulatory T (iTreg) cells, naive CD4+ T cells (1.0 × 106 cells) were plated on an anti–CD3ε-coated (BD Bioscience; 10 µg/ml) 96-well plate, activated by anti-CD3ε (1 µg/ml) and anti-CD28 antibodies (1 µg/ml), and cultured under Th1, Th17, or iT reg differentiation conditions for 3 d: 10 µg/ml anti–IL-4 (BD Bioscience) and 10 ng/ml IL-12 (Peprotech) for Th1; 10 µg/ml anti–IL-4, 10 µg/ml anti–IFN-γ (BD Bioscience), 10 ng/ml TGF-β (Peprotech), 30 ng/ml IL-6, and 50 ng/ml IL-23 (Peprotech) for Th17; 10 µg/ml anti-IL-4, 10 µg/ml anti-IFN-γ, and 10 ng/ml TGF-β for iT reg. LSECs were prepared as previously described (Smedsrød and Pertoft, 1985). Cell lines expressing Regnase-1 and Act1 variants (full-length and ΔSEFIR) with N-terminal FLAG epitope tags were constructed by retroviral infection of Regnase-1−/− immortalized MEFs and Act1−/− immortalized MEFs with virus-containing culture supernatant, which was harvested from Plat-E retroviral packaging cells that were transfected with pMRX-FLAG-Regnase-1-ires-puro and pMRX-FLAG-Act1-ires-puro (Morita et al., 2000), respectively. Cells were cultured and maintained in DMEM containing 2 µg/ml puromycin.
EAE model
Conventional EAE was induced by immunization with MOG (35–55) peptide (AnaSpec). An emulsion of MOG (35–55) peptide (300 ng/mouse) and complete Freund’s adjuvant (CFA; InvivoGen) was mixed at a 1:1 ratio and injected subcutaneously into mice. After intraperitoneal injection of pertussis toxin on days 0 and 2, mice were monitored daily and assessed by clinical scoring between 7 and 28 d after immunization. Clinical score was measured by using a previously defined scale (Huseby et al., 2001). For construction of bone marrow chimera mice, bone marrow cells (3.0–5.0 × 107 cells/ml) were intravenously injected into gamma-ray–irradiated 4–5-wk-old mice (10 Gy). At least 4 wk later, chimeric mice were challenged by induction of EAE. EAE induction, induced by passive transfer of pathogenic CD4+ T cells, was performed according to previously described methods (Arima et al., 2012). Briefly, WT mice were sacrificed after 10 d of injection of MOG (35–55) peptide/CFA and pertussis toxin. CD4+ T cells were isolated from splenocytes (4 × 106 cells) and cocultured with MOG-peptide–pulsed irradiated splenocytes. CD4+ T cells, isolated from cocultured cells by using CD4 (L3T4) microbeads and an autoMACS separator (Miltenyi), were intravenously injected into WT, Regnase-1AA/AA, and Regnase-1ΔCTD/ΔCTD mice (1.5 × 107 cells/mouse). Frozen sections were prepared from the spinal cord, lymph nodes, and spleen by using a Leica CM 1850 cryostat (Leica), then immunostained with the indicated antibodies, VECTASTAIN Elite ABC rabbit IgG kit (Vector Science), and DAB Peroxidase Substrate kit (Vector Science). The sections were then stained with hematoxylin–eosin; and images of the sections were acquired by using a BZ-9000 microscope (Keyence). Acquired images were processed by using HS ALL software in the BZ-II analyzer (Keyence). To prepare spinal cord sections, we used previously described methods (Kawamoto, 2003).
Flow cytometry
The following antibodies were prepared for flow cytometry: PerCP-Cy5.5–conjugated anti-mouse CD4, PE-conjugated anti-mouse IL-17A, FITC-conjugated anti-mouse IFN-γ, PE-conjugated anti-mouse CD25 (BD Bioscience), and Alexa-647–conjugated anti-mouse Foxp3 (Biolegend). Spinal cord cells were stained by anti-CD4 and F4/80 antibodies (Biolegend). CD4+ T cells were cultured with 100 nM PMA (Sigma), 1 µM inonomycin (Sigma), and GolgiPlug (BD Bioscience) for 2 h at 37°C. Intracellular staining (IFN-γ, IL-17A, and Foxp3) was performed after permeation and fixation of cells by using either the Cytofix/Cytoperm intracellular staining kit (BD Bioscience; for IFN-γ and IL-17A) or the Foxp3/Transcription factor staining buffer set (Affymetrix; for Foxp3).
Immunoprecipitation
Primary MEFs (WT, Regnase-1–deficient, and Act1 deficient) and HEK293 cells transiently expressing FLAG- or Myc-tagged proteins were used for immunoprecipitation. Cells were disrupted by ultrasonication. After removal of cell debris by centrifugation at 20,000 ×g for 5 min, the cell lysate was incubated with either anti–Regnase-1, anti-Act1 (D-11; Santa Cruz), anti-FLAG M2, or anti-Myc (Sigma) antibodies bound to rProtein A Sepharose Fast Flow, Protein G Sepharose 4 Fast Flow (both GE Healthcare), or Dynabeads Protein G (Thermo Fisher Scientific) for 1 h at 4°C. Beads were then washed twice with Tris buffer. Immunoprecipitated proteins were eluted with 3× SDS sample buffer and subjected to 10% SDS-PAGE (PAGE).
Gel filtration analysis
The C-terminal segment of Regnase-1 (441–598) was produced as a glutathione S-transferase (GST)–fusion protein in Escherichia coli Rosetta2 (DE3) cells (Merck Millipore). The cells were lysed in Bugbuster Protein extraction reagent (Merck Millipore) supplemented with protease inhibitors. The fusion protein was purified from the cell lysate by affinity chromatography using Glutathione Sepharose 4B (GE Healthcare) and cleaved in Tris buffer with PreScission Protease (GE Healthcare) to release the Regnase-1 segment. After the addition of 1% 3-(3-cholamidopropyl)diethylammonio-1 propanesulfonate (CHAPS) to increase protein solubility, the GST protein was removed with a Glutathione Sepharose 4B column. Phosphorylated Regnase-1 was purified from Expi293F cells (Thermo Fisher Scientific) transiently expressing FLAG–Regnase-1, Myc-Act1, and HA-TBK1 or HA-IKKi. After transfection using the ExpiFectamine 293 transfection kit (Thermo Fisher Scientific) and cultured for 3 d, cells were suspended in 20 mM Tris-HCl, pH 7.4, and 150 mM NaCl supplemented with Complete mini protease inhibitor and PhosStop phosphatase inhibitor cocktails (Roche), then ruptured by using a Dounce homogenizer. FLAG–Regnase-1 was purified from cell homogenates by incubation with anti-FLAG M2 affinity gel for 1 h at 4°C. Bound proteins were eluted by 0.15 mg/ml FLAG × 3 peptide (Sigma) in Tris buffer and dialyzed for 12 h to remove peptide. Purified Regnase-1 protein was loaded onto a Superdex 200 gel filtration column (GE Healthcare). The apparent molecular weight of the eluted peaks was estimated from the elution patterns of the molecular weight markers for gel filtration chromatography (Sigma).
In vitro kinase assay
Regnase-1 protein was obtained from MEFs stably expressing FLAG–Regnase-1. Cells were disrupted using a water bath ultrasonicator (Bioruptor Plus; Diagenode). Regnase-1 was purified with FLAG M2 affinity gel (Sigma). The following recombinant proteins were prepared for in vitro kinase assay: GST protein (Sigma), GST-tagged TBK1 (Sigma), GST-tagged IKKi (Thermo Fisher Scientific), and Lambda protein phosphatase (New England BioLabs). FLAG–Regnase-1 (50 µg/ml) was incubated in 0.2 M Hepes, pH 7.0, 20 mM MgCl2, 2 mM ATP (or 50 µCi [γ-32P] ATP), and 1.0 M mannitol for 4 h at 30°C. Samples were mixed with 3× SDS sample buffer (Iwasaki et al., 2011) and separated by 10% SDS-PAGE. Regnase-1 was detected by immunoblotting. Phosphorylated proteins in the presence of [γ-32P]ATP were visualized by autoradiography.
Isolation of subcellular fractions
Separation and isolation of the ER membrane fraction were performed in accordance with previously described methods (Lerner et al., 2003). MEF cells (5 × 107 cells) were suspended in a homogenization buffer (10 mM Hepes-KOH, pH 7.5, 10 mM KoAc, 1.5 mM Mg(OAc)2, 2 mM dithiothreitol, 1 mM PMSF, and 200 U/ml RNaseOUT ribonuclease inhibitor [Thermo Fisher Scientific]), then ruptured by a Dounce homogenizer. To separate microsomal and cytosolic fractions, homogenates were subjected to low-speed centrifugation (1,500 ×g) for 5 min. The supernatant was further subjected to high-speed centrifugation at 65,000 ×g (Beckman TLA 45 rotor) for 20 min at 4°C. The microsomal pellet was resuspended in Tris buffer. To isolate the ER membrane fraction, homogenates were mixed with 2.5 M sucrose in HEPES-potassium-magnesium (HKM) buffer [50 mM Hepes-KOH, 150 mM KoAc, and 5 mM Mg(OAc)2] in a 1:4 ratio. One-half milliliter of 0.25 M sucrose in HKM buffer and 0.75 ml of 1.3 M sucrose in HKM buffer were layered over the 2-ml mixture. After centrifugation at 500,000 ×g (Beckmann TLA 100.3 rotor) for 45 min at 4°C, ER membranes in the 1.3 M/2.0 M sucrose interface were extracted and diluted in HKM buffer. After centrifugation at 500,000 ×g for 20 min at 4°C, membranes were resuspended in Tris buffer. Separation of microsomes and ER membranes from homogenates was detected by cytochrome c reductase (nicotinamide adenine dinucleotide phosphate) assay kit (Sigma).
Protein stability assay
WT, Regnase-1AA/AA, and Regnase-S513A/S513A MEFs were incubated for 0–120 min with either 20 ng/ml TNF-α, 10 ng/ml IL-1β, 100 ng/ml LPS, or 50 ng/ml IL-17A in the presence of 5 µg/ml cycloheximide. Cells were lysed by using Tris buffer containing 1% NP-40 (Nacalai Tesque) and Complete Mini (Roche), then centrifuged for 5 min at 20,000 ×g. Cell supernatants were mixed with 3× SDS sample buffer and subjected to 10% SDS-PAGE. The intensity of protein bands was quantified by using National Institutes of Health ImageJ software.
Polysome profiling
WT and Regnase-1 ΔCTD MEF cells (1–2 × 108 cells) were pretreated with 5 µg/ml cycloheximide for 10 min before cell collection. Collected cells were suspended in 1 ml homogenization buffer supplemented with 100 µg/ml cycloheximide and hypotonically lysed by several strokes of a Dounce homogenizer. To solubilize membrane-containing cell fractions, 10% digitonin was added to a final concentration of 2%. After a low-speed centrifugation (1,500 ×g) for 5 min, the lysate supernatant was loaded on top of a linear gradient of 10–60% sucrose polysome buffer (50 mM Hepes-KOH, pH 7.5, 150 mM KCl, 10 mM MgSO4, 2 mM dithiothreitol, 1 mM PMSF, 100 µg/ml cycloheximide, and 100 U/ml RNaseOUT ribonuclease inhibitor). After ultracentrifugation at 160,000 ×g for 2 h at 4°C in a Beckman SW41 Ti rotor, fractions were collected from the top of a sucrose gradient using a piston gradient fractionator (Biocomp Instruments), transferred into UV-Star 96-well plate (Greiner Bio-one), and monitored by UV absorbance at 260 nm.
Quantitative PCR analysis
Total RNA was purified with the High Pure RNA isolation kit (Roche) and reverse transcribed with the ReverTra Ace kit (Toyobo). For quantitative PCR, PCR amplification for cDNA was performed with Thunderbird Probe qPCR Mix (Toyobo). TaqMan probes for mouse IL-6, TNF, LCN-2, granulocyte macrophage (GM)–CSF, CXCL-1, CXCL-2, CCL-5, and CCL-20 were purchased from Applied Biosystems. Fluorescence was detected by a Viia7 Real-time PCR system (Applied Biosystems). mRNA expression levels were normalized by comparison with 18S ribosomal RNA expression.
The Tet-off system
HEK293 Tet-off cells (3.0 × 106 cells) were cotransfected with either pTRE-tight-IL6-CDS + 3′-UTR or pTRE-tight-IκBζ-CDS + 3′-UTR, as described previously (Matsushita et al., 2009), together with a series of expression vectors encoding Regnase-1, Regnase-1ΔCTD, Act1, and IKKi. After 3 h, cells were subdivided into three 60-mm culture dishes and cultured overnight. Suppression of IL6-CDS + 3′-UTR or pTRE-tight-IκBζ-CDS + 3′-UTR expression was induced by culturing cells in the presence of 1 µg/ml doxycycline.
Statistical analysis
Statistical analysis of differences between two groups was performed with Student’s t test (two-tailed). P < 0.05 was considered statistically significant. Error bars represent the mean ± SEM.
Data availability
The data are available from the corresponding author on reasonable request.
Online supplemental material
Fig. S1 shows the scheme for the construction of Regnase-1 S435A/S439A double knock-in mice and shows that cell lysates from wild-type and these mutant macrophages stimulated with LPS have been analyzed by immunoblotting with the indicated antibodies. Fig. S2 shows the flow cytometric pattern of CD4+ T cell subsets after in vitro differentiation from naive CD4+ T cells and the comparison of mRNA and protein levels in WT and Regnase-1AA/AA LSECs stimulated with several proinflammatory cytokines. Fig. S3 shows the scheme for the construction of Regnase-1 frameshift mutant and Regnase-1 S513A knock-in mice using the CRISPR/Cas9 system. Fig. S4 shows the flow cytometric pattern of Th1 and Th17 cells in lymph node cells and splenocytes from WT and Regnase-1ΔCTD/ΔCTD mice at 28 d after EAE immunization and the numbers of Th1 and Th17 cells. Table S1 indicates the amino acids of Regnase-1 that are phosphorylated when coexpressed with Act1 and TBK1 or IKKi in HEK293 cells.
Supplementary Material
Acknowledgments
We thank our laboratory members: E. Kamada for secretarial assistance and R. Kawaguchi, C. Funamoto, and N. Kitagaki for technical assistance. We thank Y. Takada and Dr. K. Saito for measurements and analysis of mass spectrometry, Dr. Y. Matsushima for providing Act1-deficient MEFs, and Dr. M. Ikawa and Dr. M. Okabe for the construction of Regnase-1 ΔCTD mutant mice. We also thank Ryan Chastain-Gross from Edanz Group for editing a draft of this manuscript.
This work was supported by the Japanese Ministry of Education, Culture, Sports, Science, and Technology (MEXT)’s Specially Promoted Research (15H05704); the Japan Society for the Promotion of Science through the Funding Program for World-Leading Innovative R&D on Science and Technology (FIRST Program); and the National Institutes of Health (P01 A1070167).
S. Akira has research support from Chugai Pharmaceutical Co. Ltd. The terms of this arrangement have been reviewed and approved by Osaka University in accordance with its policy on objectivity in research. The authors declare no additional competing financial interests.
Author contributions: H. Tanaka performed most of the experiments. Y. Arima, D. Kamimura, and Y. Tanaka performed cell transplantation experiments. T. Uehata generated Regnase-1AA/AA mice. N. Takahashi generated antibodies. H. Tanaka, Y. Arima, D. Kamimura, T. Satoh, K. Maeda, M. Murakami, and S. Akira analyzed the data. H. Tanaka and S. Akira designed the study and prepared the manuscript.
References
- Akira S. 2013. Regnase-1, a ribonuclease involved in the regulation of immune responses. Cold Spring Harb. Symp. Quant. Biol. 78:51–60. 10.1101/sqb.2013.78.019877 [DOI] [PubMed] [Google Scholar]
- Anderson P. 2008. Post-transcriptional control of cytokine production. Nat. Immunol. 9:353–359. 10.1038/ni1584 [DOI] [PubMed] [Google Scholar]
- Arima Y., Harada M., Kamimura D., Park J.H., Kawano F., Yull F.E., Kawamoto T., Iwakura Y., Betz U.A., Márquez G., et al. . 2012. Regional neural activation defines a gateway for autoreactive T cells to cross the blood-brain barrier. Cell. 148:447–457. 10.1016/j.cell.2012.01.022 [DOI] [PubMed] [Google Scholar]
- Beutler B. 2009. Microbe sensing, positive feedback loops, and the pathogenesis of inflammatory diseases. Immunol. Rev. 227:248–263. 10.1111/j.1600-065X.2008.00733.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brook M., Tchen C.R., Santalucia T., McIlrath J., Arthur J.S., Saklatvala J., and Clark A.R.. 2006. Posttranslational regulation of tristetraprolin subcellular localization and protein stability by p38 mitogen-activated protein kinase and extracellular signal-regulated kinase pathways. Mol. Cell. Biol. 26:2408–2418. 10.1128/MCB.26.6.2408-2418.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulek K., Liu C., Swaidani S., Wang L., Page R.C., Gulen M.F., Herjan T., Abbadi A., Qian W., Sun D., et al. . 2011. The inducible kinase IKKi is required for IL-17-dependent signaling associated with neutrophilia and pulmonary inflammation. Nat. Immunol. 12:844–852. 10.1038/ni.2080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaffen S.L. 2009. Structure and signalling in the IL-17 receptor family. Nat. Rev. Immunol. 9:556–567. 10.1038/nri2586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg A.V., Amatya N., Chen K., Cruz J.A., Grover P., Whibley N., Conti H.R., Hernandez Mir G., Sirakova T., Childs E.C., et al. . 2015. MCPIP1 endoribonuclease activity negatively regulates interleukin-17-mediated signaling and inflammation. Immunity. 43:475–487. 10.1016/j.immuni.2015.07.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemmi H., Takeuchi O., Sato S., Yamamoto M., Kaisho T., Sanjo H., Kawai T., Hoshino K., Takeda K., and Akira S.. 2004. The roles of two IkappaB kinase-related kinases in lipopolysaccharide and double stranded RNA signaling and viral infection. J. Exp. Med. 199:1641–1650. 10.1084/jem.20040520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herjan T., Yao P., Qian W., Li X., Liu C., Bulek K., Sun D., Yang W.P., Zhu J., He A., et al. . 2013. HuR is required for IL-17-induced Act1-mediated CXCL1 and CXCL5 mRNA stabilization. J. Immunol. 191:640–649. 10.4049/jimmunol.1203315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herjan T., Hong L., Bubenik J., Bulek K., Qian W., Liu C., Li X., Chen X., Yang H., Ouyang S., et al. . 2018. IL-17-receptor-associated adaptor Act1 directly stabilizes mRNAs to mediate IL-17 inflammatory signaling. Nat. Immunol. 19:354–365. 10.1038/s41590-018-0071-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huseby E.S., Sather B., Huseby P.G., and Goverman J.. 2001. Age-dependent T cell tolerance and autoimmunity to myelin basic protein. Immunity. 14:471–481. 10.1016/S1074-7613(01)00127-3 [DOI] [PubMed] [Google Scholar]
- Iwasaki A., and Medzhitov R.. 2015. Control of adaptive immunity by the innate immune system. Nat. Immunol. 16:343–353. 10.1038/ni.3123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki H., Takeuchi O., Teraguchi S., Matsushita K., Uehata T., Kuniyoshi K., Satoh T., Saitoh T., Matsushita M., Standley D.M., and Akira S.. 2011. The IκB kinase complex regulates the stability of cytokine-encoding mRNA induced by TLR-IL-1R by controlling degradation of regnase-1. Nat. Immunol. 12:1167–1175. 10.1038/ni.2137 [DOI] [PubMed] [Google Scholar]
- Jeltsch K.M., Hu D., Brenner S., Zöller J., Heinz G.A., Nagel D., Vogel K.U., Rehage N., Warth S.C., Edelmann S.L., et al. . 2014. Cleavage of roquin and regnase-1 by the paracaspase MALT1 releases their cooperatively repressed targets to promote T(H)17 differentiation. Nat. Immunol. 15:1079–1089. 10.1038/ni.3008 [DOI] [PubMed] [Google Scholar]
- Jura J., Skalniak L., and Koj A.. 2012. Monocyte chemotactic protein-1-induced protein-1 (MCPIP1) is a novel multifunctional modulator of inflammatory reactions. Biochim. Biophys. Acta. 1823:1905–1913. 10.1016/j.bbamcr.2012.06.029 [DOI] [PubMed] [Google Scholar]
- Kawagoe T., Sato S., Matsushita K., Kato H., Matsui K., Kumagai Y., Saitoh T., Kawai T., Takeuchi O., and Akira S.. 2008. Sequential control of Toll-like receptor-dependent responses by IRAK1 and IRAK2. Nat. Immunol. 9:684–691. 10.1038/ni.1606 [DOI] [PubMed] [Google Scholar]
- Kawamoto T. 2003. Use of a new adhesive film for the preparation of multi-purpose fresh-frozen sections from hard tissues, whole-animals, insects and plants. Arch. Histol. Cytol. 66:123–143. 10.1679/aohc.66.123 [DOI] [PubMed] [Google Scholar]
- Kochan J., Wawro M., and Kasza A.. 2016. IF-combined smRNA FISH reveals interaction of MCPIP1 protein with IER3 mRNA. Biol. Open. 5:889–898. 10.1242/bio.018010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J., Nakagiri T., Oto T., Harada M., Morii E., Shintani Y., Inoue M., Iwakura Y., Miyoshi S., Okumura M., et al. . 2012. IL-6 amplifier, NF-κB-triggered positive feedback for IL-6 signaling, in grafts is involved in allogeneic rejection responses. J. Immunol. 189:1928–1936. 10.4049/jimmunol.1103613 [DOI] [PubMed] [Google Scholar]
- Lerner R.S., Seiser R.M., Zheng T., Lager P.J., Reedy M.C., Keene J.D., and Nicchitta C.V.. 2003. Partitioning and translation of mRNAs encoding soluble proteins on membrane-bound ribosomes. RNA. 9:1123–1137. 10.1261/rna.5610403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Estepa G., Memet S., Israel A., and Verma I.M.. 2000. Complete lack of NF-kappaB activity in IKK1 and IKK2 double-deficient mice: Additional defect in neurulation. Genes Dev. 14:1729–1733. [PMC free article] [PubMed] [Google Scholar]
- Liang J., Wang J., Azfer A., Song W., Tromp G., Kolattukudy P.E., and Fu M.. 2008. A novel CCCH-zinc finger protein family regulates proinflammatory activation of macrophages. J. Biol. Chem. 283:6337–6346. 10.1074/jbc.M707861200 [DOI] [PubMed] [Google Scholar]
- Liang J., Saad Y., Lei T., Wang J., Qi D., Yang Q., Kolattukudy P.E., and Fu M.. 2010. MCP-induced protein 1 deubiquitinates TRAF proteins and negatively regulates JNK and NF-kappaB signaling. J. Exp. Med. 207:2959–2973. 10.1084/jem.20092641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin R.J., Chien H.L., Lin S.Y., Chang B.L., Yu H.P., Tang W.C., and Lin Y.L.. 2013. MCPIP1 ribonuclease exhibits broad-spectrum antiviral effects through viral RNA binding and degradation. Nucleic Acids Res. 41:3314–3326. 10.1093/nar/gkt019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashiko D., Young S.A., Muto M., Kato H., Nozawa K., Ogawa M., Noda T., Kim Y.J., Satouh Y., Fujihara Y., and Ikawa M.. 2014. Feasibility for a large scale mouse mutagenesis by injecting CRISPR/Cas plasmid into zygotes. Dev. Growth Differ. 56:122–129. 10.1111/dgd.12113 [DOI] [PubMed] [Google Scholar]
- Matsushima Y., Kikkawa Y., Takada T., Matsuoka K., Seki Y., Yoshida H., Minegishi Y., Karasuyama H., and Yonekawa H.. 2010. An atopic dermatitis-like skin disease with hyper-IgE-emia develops in mice carrying a spontaneous recessive point mutation in the Traf3ip2 (Act1/CIKS) gene. J. Immunol. 185:2340–2349. 10.4049/jimmunol.0900694 [DOI] [PubMed] [Google Scholar]
- Matsushita K., Takeuchi O., Standley D.M., Kumagai Y., Kawagoe T., Miyake T., Satoh T., Kato H., Tsujimura T., Nakamura H., and Akira S.. 2009. Zc3h12a is an RNase essential for controlling immune responses by regulating mRNA decay. Nature. 458:1185–1190. 10.1038/nature07924 [DOI] [PubMed] [Google Scholar]
- Mino T., Murakawa Y., Fukao A., Vandenbon A., Wessels H.H., Ori D., Uehata T., Tartey S., Akira S., Suzuki Y., et al. . 2015. Regnase-1 and Roquin regulate a common element in inflammatory mRNAs by spatiotemporally distinct mechanisms. Cell. 161:1058–1073. 10.1016/j.cell.2015.04.029 [DOI] [PubMed] [Google Scholar]
- Mizgalska D., Wegrzyn P., Murzyn K., Kasza A., Koj A., Jura J., Jarzab B., and Jura J.. 2009. Interleukin-1-inducible MCPIP protein has structural and functional properties of RNase and participates in degradation of IL-1beta mRNA. FEBS J. 276:7386–7399. 10.1111/j.1742-4658.2009.07452.x [DOI] [PubMed] [Google Scholar]
- Monin L., Gudjonsson J.E., Childs E.E., Amatya N., Xing X., Verma A.H., Coleman B.M., Garg A.V., Killeen M., Mathers A., et al. . 2017. MCPIP1/Regnase-1 restricts IL-17A- and IL-17C-dependent skin inflammation. J. Immunol. 198:767–775. 10.4049/jimmunol.1601551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita S., Kojima T., and Kitamura T.. 2000. Plat-E: An efficient and stable system for transient packaging of retroviruses. Gene Ther. 7:1063–1066. 10.1038/sj.gt.3301206 [DOI] [PubMed] [Google Scholar]
- Ogura H., Murakami M., Okuyama Y., Tsuruoka M., Kitabayashi C., Kanamoto M., Nishihara M., Iwakura Y., and Hirano T.. 2008. Interleukin-17 promotes autoimmunity by triggering a positive-feedback loop via interleukin-6 induction. Immunity. 29:628–636. 10.1016/j.immuni.2008.07.018 [DOI] [PubMed] [Google Scholar]
- Qian Y., Liu C., Hartupee J., Altuntas C.Z., Gulen M.F., Jane-Wit D., Xiao J., Lu Y., Giltiay N., Liu J., et al. . 2007. The adaptor Act1 is required for interleukin 17-dependent signaling associated with autoimmune and inflammatory disease. Nat. Immunol. 8:247–256. 10.1038/ni1439 [DOI] [PubMed] [Google Scholar]
- Qu F., Gao H., Zhu S., Shi P., Zhang Y., Liu Y., Jallal B., Yao Y., Shi Y., and Qian Y.. 2012. TRAF6-dependent Act1 phosphorylation by the IκB kinase-related kinases suppresses interleukin-17-induced NF-κB activation. Mol. Cell. Biol. 32:3925–3937. 10.1128/MCB.00268-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran F.A., Hsu P.D., Wright J., Agarwala V., Scott D.A., and Zhang F.. 2013. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 8:2281–2308. 10.1038/nprot.2013.143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross E.A., Smallie T., Ding Q., O’Neil J.D., Cunliffe H.E., Tang T., Rosner D.R., Klevernic I., Morrice N.A., Monaco C., et al. . 2015. Dominant suppression of inflammation via targeted mutation of the mRNA destabilizing protein tristetraprolin. J. Immunol. 195:265–276. 10.4049/jimmunol.1402826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smedsrød B., and Pertoft H.. 1985. Preparation of pure hepatocytes and reticuloendothelial cells in high yield from a single rat liver by means of Percoll centrifugation and selective adherence. J. Leukoc. Biol. 38:213–230. 10.1002/jlb.38.2.213 [DOI] [PubMed] [Google Scholar]
- Somma D., Mastrovito P., Grieco M., Lavorgna A., Pignalosa A., Formisano L., Salzano A.M., Scaloni A., Pacifico F., Siebenlist U., and Leonardi A.. 2015. CIKS/DDX3X interaction controls the stability of the Zc3h12a mRNA induced by IL-17. J. Immunol. 194:3286–3294. 10.4049/jimmunol.1401589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soulat D., Bürckstümmer T., Westermayer S., Goncalves A., Bauch A., Stefanovic A., Hantschel O., Bennett K.L., Decker T., and Superti-Furga G.. 2008. The DEAD-box helicase DDX3X is a critical component of the TANK-binding kinase 1-dependent innate immune response. EMBO J. 27:2135–2146. 10.1038/emboj.2008.126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun D., Novotny M., Bulek K., Liu C., Li X., and Hamilton T.. 2011. Treatment with IL-17 prolongs the half-life of chemokine CXCL1 mRNA via the adaptor TRAF5 and the splicing-regulatory factor SF2 (ASF). Nat. Immunol. 12:853–860. 10.1038/ni.2081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki H.I., Arase M., Matsuyama H., Choi Y.L., Ueno T., Mano H., Sugimoto K., and Miyazono K.. 2011. MCPIP1 ribonuclease antagonizes dicer and terminates microRNA biogenesis through precursor microRNA degradation. Mol. Cell. 44:424–436. 10.1016/j.molcel.2011.09.012 [DOI] [PubMed] [Google Scholar]
- Suzuki K., Tsunoda H., Omiya R., Matoba K., Baba T., Suzuki S., Segawa H., Kumanogoh A., Iwasaki K., Hattori K., and Takagi J.. 2016. Structure of the Plexin Ectodomain Bound by Semaphorin-Mimicking Antibodies. PLoS One. 11:e0156719. 10.1371/journal.pone.0156719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi O., and Akira S.. 2010. Pattern recognition receptors and inflammation. Cell. 140:805–820. 10.1016/j.cell.2010.01.022 [DOI] [PubMed] [Google Scholar]
- Uehata T., Iwasaki H., Vandenbon A., Matsushita K., Hernandez-Cuellar E., Kuniyoshi K., Satoh T., Mino T., Suzuki Y., Standley D.M., et al. . 2013. Malt1-induced cleavage of regnase-1 in CD4(+) helper T cells regulates immune activation. Cell. 153:1036–1049. 10.1016/j.cell.2013.04.034 [DOI] [PubMed] [Google Scholar]
- Wilamowski M., Gorecki A., Dziedzicka-Wasylewska M., and Jura J.. 2018. Substrate specificity of human MCPIP1 endoribonuclease. Sci. Rep. 8:7381 10.1038/s41598-018-25765-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokogawa M., Tsushima T., Noda N.N., Kumeta H., Enokizono Y., Yamashita K., Standley D.M., Takeuchi O., Akira S., and Inagaki F.. 2016. Structural basis for the regulation of enzymatic activity of Regnase-1 by domain-domain interactions. Sci. Rep. 6:22324 10.1038/srep22324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J., Yamane H., and Paul W.E.. 2010. Differentiation of effector CD4 T cell populations. Annu. Rev. Immunol. 28:445–489. 10.1146/annurev-immunol-030409-101212 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data are available from the corresponding author on reasonable request.